Significance
Gene fusions are tumor-specific genomic aberrations and are among the most powerful biomarkers and drug targets in translational cancer biology. The advent of RNA-sequencing technologies over the last decade has provided a unique opportunity for detecting novel fusions via deploying computational algorithms on public sequencing databases. However, precise fusion detection algorithms are still out of reach. We develop Data-Enriched Efficient PrEcise STatistical fusion detection (DEEPEST), a highly specific and efficient statistical pipeline specially designed for mining massive sequencing databases and apply it to all 33 tumor types and 10,500 samples in The Cancer Genome Atlas database. We systematically profile the landscape of detected fusions via classic statistical models and identify several signatures of selection for fusions in tumors.
Keywords: gene fusion, cancer genomics, bioinformatics, pan-cancer analysis, TCGA
Abstract
The extent to which gene fusions function as drivers of cancer remains a critical open question. Current algorithms do not sufficiently identify false-positive fusions arising during library preparation, sequencing, and alignment. Here, we introduce Data-Enriched Efficient PrEcise STatistical fusion detection (DEEPEST), an algorithm that uses statistical modeling to minimize false-positives while increasing the sensitivity of fusion detection. In 9,946 tumor RNA-sequencing datasets from The Cancer Genome Atlas (TCGA) across 33 tumor types, DEEPEST identifies 31,007 fusions, 30% more than identified by other methods, while calling 10-fold fewer false-positive fusions in nontransformed human tissues. We leverage the increased precision of DEEPEST to discover fundamental cancer biology. Namely, 888 candidate oncogenes are identified based on overrepresentation in DEEPEST calls, and 1,078 previously unreported fusions involving long intergenic noncoding RNAs, demonstrating a previously unappreciated prevalence and potential for function. DEEPEST also reveals a high enrichment for fusions involving oncogenes in cancers, including ovarian cancer, which has had minimal treatment advances in recent decades, finding that more than 50% of tumors harbor gene fusions predicted to be oncogenic. Specific protein domains are enriched in DEEPEST calls, indicating a global selection for fusion functionality: kinase domains are nearly 2-fold more enriched in DEEPEST calls than expected by chance, as are domains involved in (anaerobic) metabolism and DNA binding. The statistical algorithms, population-level analytic framework, and the biological conclusions of DEEPEST call for increased attention to gene fusions as drivers of cancer and for future research into using fusions for targeted therapy.
Gene fusions are known to drive some cancers and can be highly specific and personalized therapeutic targets; some of the most famous fusions are the BCR–ABL1 fusion in chronic myelogenous leukemia (CML), the EML4–ALK fusion in non-small lung cell carcinoma, TMPRSS2–ERG in prostate cancer, and FGFR3–TACC3 in a variety of cancers including glioblastoma multiforme (1–4). Since fusions are generally absent in healthy tissues, they are among the most clinically relevant events in cancer to direct targeted therapy and to be used as effective diagnostic tools in early detection strategies using RNA or proteins; moreover, as they are truly specific to cancer, they have promising potential as neo-antigens (5–7).
Because of this, clinicians and large sequencing consortia have made major efforts to identify fusions expressed in tumors via screening massive cancer sequencing datasets (8–12). However, these attempts are limited by critical roadblocks: current algorithms suffer from high false-positive (FP) rates and unknown false-negative (FN) rates. Thus, ad hoc choices have been made in calling and analyzing fusions including taking the consensus of multiple algorithms and filtering lists of fusions using manual approaches (13–15). These approaches lead to what third-party reviews agree is imprecise fusion discovery and bias against discovering novel oncogenes (15–17). This suboptimal performance becomes more problematic when fusion detection is deployed on large cancer sequencing datasets that contain thousands or tens of thousands of samples. In such scenarios, precise fusion detection must overcome the problem of multiple hypothesis testing: each algorithm is testing for fusions thousands of times, a regime known to introduce FPs. To overcome these problems, the field has turned to consensus-based approaches, where multiple algorithms are run in parallel (10), and a metacaller allows “voting” to produce the final list of fusions. This is also unsatisfactory, as it introduces FNs.
Both shortcomings in the ascertainment of fusions by existing algorithms and using recurrence alone to assess fusions’ function have limited the use of fusions to discover new cancer biology. As one of many examples, a recent study of more than 400 pancreatic cancers found no recurrent gene fusions, raising the question of whether this is due to high FN rates or whether this means that fusions are not drivers in the disease (18). Recurrence of fusions is currently one of the only standards in the field used to assess the functionality of fusions, but the most frequently expressed fusions may not be the most carcinogenic (19); on the other hand, there may still be many undiscovered gene fusions that drive cancer.
Thus, the critical question “Are gene fusions underappreciated drivers of cancer?” is still unanswered. In this paper, we first provide an algorithm that has significant advance in precision for unbiased fusion detection at exon boundaries in massive genomics datasets. The algorithm, Data-Enriched Efficient PrEcise STatistical fusion detection (DEEPEST), is a second-generation fusion algorithm with significant computational and algorithmic advance over our previously developed MACHETE (Mismatched Alignment Chimera Tracking Engine) algorithm (20). A key innovation in DEEPEST is its statistical test of fusion prevalence across populations, which can identify FPs in a global unbiased manner.
The precision and efficient implementation of DEEPEST allowed us to conduct an unbiased screen for expressed fusions occurring at annotated exon boundaries (based on GRCh38) in a cohort of 10,521 RNA-sequencing datasets, including 9,946 tumor samples and 575 normal (tumor adjacent) samples, across the entire 33 tumor types of The Cancer Genome Atlas (TCGA). Beyond recovery of known fusions, DEEPEST discovers fusions with potentially important implications in cancer biology that had not been detected by previous studies.
While frequent recurrence of gene fusions has been considered a hallmark of a selective event during tumor initiation, and this recurrence has historically been the only evidence available to support that a fusion drives a cancer, private or very rare gene fusions are beginning to be considered potential functional drivers (21). However, the high FP rates in published algorithms prevent a statistical analysis of whether reported private or rare gene fusions exhibit a signature of selection across massive tumor transcriptome databases, such as TCGA. We have formulated statistical tests for nonneutral selection of fusion expression by calculating the expected rates of rarely recurrent gene fusions and partner genes, enrichment of gene families such as kinase genes or those curated in Catalog Of Somatic Mutations In Cancer (COSMIC) (22), and enrichment for protein domains or pairs of protein domains present exclusively in fusions. These analyses reveal a significant signal for selection of gene fusions. The statistical tests provide a basis for identifying candidate oncogenes and driver and druggable fusions.
To illustrate one of our findings, a large fraction of ovarian serous cystadenocarcinoma tumors has until now lacked explanatory drivers beyond nearly universal TP53 mutations and defects in homologous recombination pathways. Because TP53 mutations create genome instability, a testable hypothesis is that TP53 mutations permit the development of rare or private driver fusions in ovarian cancers, and the fusions have been missed due to biases in currently available algorithms. We apply DEEPEST to RNA-sequencing (RNA-Seq) data from bulk tumors and find that 94.6% of the ovarian tumors we screened have detectable fusions, half of the ovarian cancer tumors express gene fusions involving a known COSMIC gene, and 36% have fusions involving genes in a kinase pathway.
In summary, DEEPEST is an advance in accuracy for fusion detection in massive RNA-Seq datasets. The algorithm is reproducible, publicly available, and can be easily run in a dockerized container (Materials and Methods). Its results have important biological implications: DEEPEST, applied in conjunction with statistical analysis to the entire TCGA database, reveals a signature of fusion expression consistent with the existence of under-appreciated drivers of human cancer, including selection for rare or private gene fusions with implications from basic biology to the clinic.
Results
DEEPEST Is a Statistical Algorithm for Gene Fusion Discovery in Massive Public Databases.
We engineered a statistical algorithm, DEEPEST, to discover and estimate the prevalence of gene fusions in massive numbers of datasets. Here, we have applied DEEPEST to 10,000 datasets, but in principle, DEEPEST can be applied to 100,000, 1 million, or more samples. DEEPEST includes key innovations such as controlling FPs arising from analysis of massive RNA-Seq datasets for fusion discovery, a problem conceptually analogous to multiple hypothesis testing via P values, which cannot be solved by direct application of common false-discovery rate (FDR)-controlling procedures, which rely on the assumption of a uniform distribution of P values under the null hypothesis.
The DEEPEST pipeline contains 2 main computational steps: 1) junction nomination component which is run on a subset of all samples to be analyzed, called “the discovery set”; and 2) statistical testing of nominated junctions on all analyzed samples, “the test set.” In this paper, we have used all samples as the discovery set, but this set could be a fraction of RNA-Seq data if desired.
Step 1 includes running KNIFE (known and novel isoform explorer) method to detect chimeric junctions (23), defined as a splicing event between 2 distinct genes, whose exons are on the same chromosome and within the distance of 1 MB, and a method based on the MACHETE algorithm (20) to detect chimeric junctions with partner exons being farther than 1 MB from each other or on different chromosomes/strands (Fig. 1). Putative fusions are nominated from the initial database by using a null statistical model of read-alignment profiles that models the effect of junction sequence composition and gene abundance in generating FP fusions (SI Appendix and Materials and Methods). This step relies on extensive computational engineering, which restructures the MACHETE pipeline into an efficient reproducible publicly available workflow based on dockerized containers, using the Common Workflow Language (CWL). Another advance in DEEPEST over MACHETE is further improvement of sensitivity by including gold standard cancer fusions in the junction nomination step of MACHETE, which makes DEEPEST easily portable to clinical settings where clinicians desire precise identification of a set of known fusions. For this purpose, we used fusions curated in ChimerDB 3.0 (24).
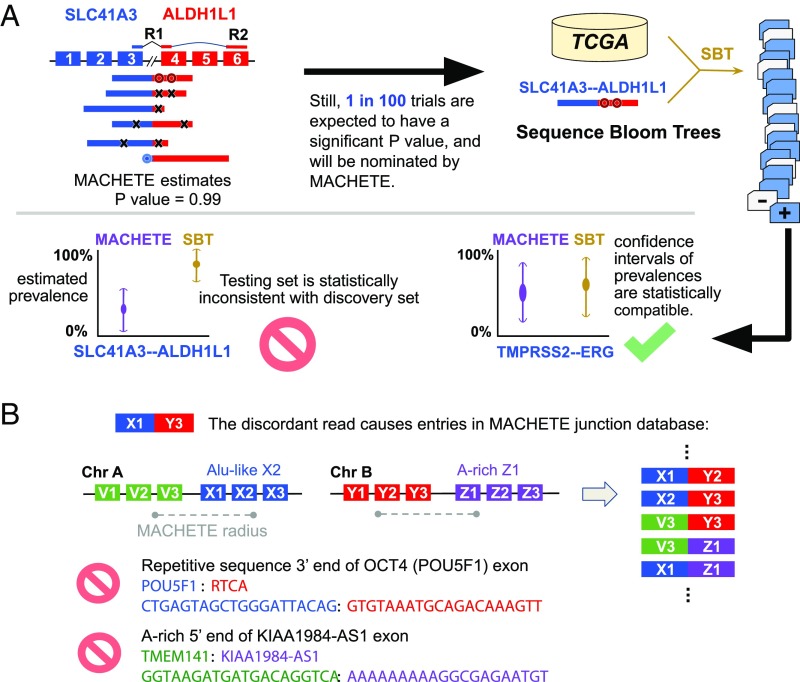
Fig. 1.
Origin and identification of FP from running DEEPEST on thousands of samples. (A) DEEPEST uses all reads, including those censored by other algorithms, to generate an empirical P value for each candidate fusion. SBTs, together with further statistical modeling, are used to identify FP arising from testing on multiple samples, some of which are reported by other algorithms (SI Appendix, Fig. S4A). The first black arrow shows the motivation for designing the SBT step. (B) cDNA or mapping artifacts result in the inclusion of exon–exon junctions from all combinations of exons within a fixed genomic radius of X1 with all exons in the radius of Y3. Some such exon junctions will include degenerate sequences that cannot be mapped uniquely, and thus DEEPEST blinds itself to detection of fusions containing such highly degenerate sequences (for example, due to Alu exonization) or with polyA stretches at the 5′ end.
In Step 2, the statistical refinement step, DEEPEST uses rigorous statistical approaches based on orthogonal sequence level queries via the sequence bloom tree (SBT) (25), a method that indexes the sequence composition of genomic datasets and can rapidly query whether specific k-mers appear in the corpus. This step is modular and can in principle be applied to any fusion discovery algorithm to identify FPs resulting from multiple testing, a major challenge brought on by running discovery algorithms on massive datasets. Fusions nominated by the junction nomination component are subjected to a secondary statistical test: they are efficiently tested in the discovery set along with an arbitrarily large number of added samples in the test set, here tens of thousands of samples, by rapid queries using SBT. This step further decreases the FP identification of fusions beyond MACHETE, which has been already shown to have better specificity than any other published algorithm (20). Intuitively, this step checks whether the prevalence of fusions found by running MACHETE (or KNIFE) is statistically consistent with the estimated prevalence using a string-query based approach (such as SBT). Since the SBT has perfect sensitivity by searching merely by looking at fusion-junctional sequences, samples could be positive for a fusion by SBT yet negative by MACHETE, which requires discordant reads to nominate fusions (20). For a fusion to be called by DEEPEST, it should have an SBT detection frequency that is statistically consistent with the estimated prevalence by the junction nomination component and additionally pass statistical filtering such as a test for repetitive sequences near exon boundaries (Fig. 1 and SI Appendix).
DEEPEST does not require human guidance, is fully automated, and can be applied to any paired-end RNA-Seq database by leveraging the massive computational power of cloud platforms. A web-based user-friendly version of the pipeline has been implemented on the Seven Bridges Cancer Genomics Cloud (CGC) (26), which allows a user to run the workflow either by uploading RNA-Seq data or using RNA databases already available on CGC. (Currently, the average cost of running the workflow for a single TCGA sample on the cloud is roughly $3.) Moreover, most parts are portable as they are dockerized and can be easily exported to many platforms using a description given by the CWL (27).
DEEPEST Improves Sensitivity and Specificity of Fusion Detection.
We first evaluated DEEPEST FP and FN rates on fusion positive benchmarking datasets used by third parties to assess the performance of 14 state-of-the-art algorithms (28). On each dataset, DEEPEST has 100% positive-predictive value (PPV), the ratio of the number of true positive calls to the total number of calls, higher than all 14 other state-of-the-art algorithms (SI Appendix, Fig. S1), DEEPEST has a comparable, although numerically higher, PPV than PRADA (Pipeline for RNA-Sequencing Data Analysis) (29), which is the next best algorithm. For this analysis, we only applied the first component of DEEPEST, which is based on MACHETE, as the SBT refinement step utilizes the statistical power across a large cohort of samples, which is not the case for simulated datasets.
Because simulations can only model errors with known sources, it is common for algorithms to perform differently on real and simulated data; for example, simulated data do not model reverse transcriptase template switching or chimeras arising from ligation or PCR artifacts. Thus, in addition to evaluating the performance of DEEPEST on simulated data, we performed a thorough computational study of DEEPEST performance on real data. To evaluate the FP rate of DEEPEST on real data, we applied it to several hundred normal datasets, including Genotype-Tissue Expression (GTEx) (30) and TCGA normal samples. Notably, DEEPEST calls fewer fusions in GTEx samples than does STAR-Fusion (28) (SI Appendix, Fig. S2A), an algorithm used in a recent pan-cancer TCGA analysis (10). In addition, DEEPEST reports fewer fusions (509 fusions) on TCGA normal samples compared with the 3,128 calls in the same samples by TumorFusions (8) (SI Appendix, Fig. S2B), which is a TCGA fusion list based on PRADA (29). This provides evidence that, unlike other algorithms, DEEPEST retains the specificity seen in simulations in real tissue samples.
We ran DEEPEST on the entire TCGA corpus: 9,946 tumor samples across all 33 tumor types. DEEPEST detects 31,007 fusions across TCGA. Consistent with what is known about tumor type-specific gene fusion expression, DEEPEST reports the highest abundance of fusions in sarcoma (SARC), uterine corpus endometrial carcinoma (UCEC), and esophageal carcinoma (ESCA) tumor types and the fewest number of detected fusions in thyroid carcinoma (THCA), testicular germ cell tumors (TGCT), and uveal melanoma (UVM). We provide the description of tumor types in SI Appendix, Fig. S5.
While calling significantly fewer fusions in normal samples, DEEPEST identifies significantly more fusions in TCGA tumor samples compared with 2 most recent surveys of the same samples (8, 10), the latter is based on STAR-Fusion that is more sensitive in simulated data. While some fusion algorithms might exhibit better sensitivity (at the cost of higher FP rates) on simulated datasets, DEEPEST is more sensitive in real cancer datasets (SI Appendix, Fig. S2 C and D). When samples shared between 3 studies are considered, DEEPEST detects much more fusions (29,820 fusions, compared with 23,624 fusions in ref. 10 and 19,846 fusions by TumorFusions) and substantially fewer calls in real normal datasets (SI Appendix, Fig. S2 A and B), suggesting that the modeling used by DEEPEST is a better fit for real data. DEEPEST-only fusions are enriched in cancers known to have high genomic instability (ESCA, ovarian carcinoma [OV], stomach adenocarcinoma, and SARC) compared with fusions found only by TumorFusions and ref. 10 (SI Appendix, Fig. S2D). Together, this implies that DEEPEST is more specific on simulated and real data and identifies more high confidence fusions on real data.
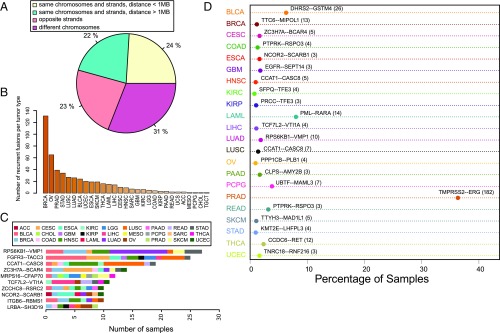
Because fusions between exons that are closer to each other than 1 MB in the reference genome and transcribed on the same strand could be due to local DNA variation or transcriptional or posttranscriptional splicing, for example, into circular RNA (circRNA) (31), we define an “extreme fusion” to be a fusion that joins exons that are farther than 1 MB apart, are on opposite strands, or are on different chromosomes and profile the distribution of DEEPEST-called fusions as a function of extreme characteristics. Around 24% of fusions have both partner genes transcribed from the same chromosome and strand and within 1 MB, 22% are on the same chromosome and strand but separated by at least 1 MB, 23% are strand crosses with genes being on opposite strands, and 31% are interchromosomal fusions (Fig. 2A).
Fig. 2.
The landscape of detected fusions. (A) The relative position of the partner exons in the detected fusions. (B) The number of recurrent fusions for each tumor type. (C) The most recurrent fusion for each tumor type. (D) Fusions with the most diverse tumor types.
DEEPEST finds 1,486 recurrent fusions (512 distinct recurrent fusions), called in at least 2 tumors within a tumor type (Fig. 2B). Many gene fusions are detected in diverse cancers, for example, MRPS16–CFAP70 and FGFR3–TACC3 (10 cancer types) (Fig. 2C). Restricted to a single tumor type, most fusions have low levels of recurrent gene fusions with exceptions of the well-known TMPRSS2–ERG in prostate adenocarcinoma (PRAD) (182 samples, 36.3% of tumor samples), PML–RARA in acute myeloid leukemia (LAML) (14 samples, 8% of tumor samples), and DHRS2–GSTM4 in bladder urothelial carcinoma (BLCA) (Fig. 2D).
Around 41% of DEEPEST’s 31,007 fusions (12,196 fusions) had not been detected by previous fusion studies on TCGA (SI Appendix, Fig. S2C). Far fewer fusions are found only by one of the other algorithms (4,402 fusions in TumorFusions and 5,860 fusions in ref. 10) (SI Appendix, Fig. S2C). We further investigated DEEPEST-only fusions and queried them through FusionHub portal (https://fusionhub.persistent.co.in/) to see if they are present in any other fusion database and found that 9,272 distinct fusions (i.e., gene pairs) were not present in any other fusion database (Dataset S1). Included in this list are 157 previously unreported recurrent fusions (Dataset S1 and SI Appendix, Fig. S3), including a recurrent fusion for PRAD involving SCHLAP1, a long noncoding RNA (LncRNA) known to have driving oncogenic activities in the prostate cancer (32).
Statistical Analysis of Fusion Prevalence in Large Data Queries Improves Precision of Calls.
The SBT refinement step is a critical innovation of the DEEPEST pipeline and substantially improved its specificity by removing fusion calls likely to be FPs. Such fusions passed the first component of DEEPEST due to multiple hypothesis testing but were flagged by the statistical SBT refinement step (SI Appendix, Fig. S4A).
The main power of the refinement step is it is agnostic to gene name or ontology (such as pseudogenes, paralogs, synonymous genes, and duplicated genes) used by conventional filters, and which can lead to true positives being removed from lists (e.g., RUNX1–RUNX1T1). Furthermore, most fusions (89%) removed during the SBT refinement step would have passed such filters (SI Appendix, Fig. S4B).
To further evaluate the performance of the SBT-based refinement component, we extracted 2 groups of fusions from the fusions called by the junction nomination component: 1) likely FPs (LFPs) that are fusions with SBT hits in the GTEx samples and 2) likely true positives (LTPs) that contain fusions shared between the DEEPEST first component, ref. 10, and TumorFusions. Note that LFP fusions are defined on the basis of GTEx data, whereas the SBT refinement step does not “touch” GTEx data, meaning that the LFP fusion set can be used as a test set for the specificity of the SBT refinement step. A fusion is removed by the SBT refinement step if its 2-sided binomial P value is less than 0.05, an arbitrary and conventional statistical threshold. To evaluate the precision of the SBT step, we treated each 2-sided P value as a continuous measure and stratified the P values for LFP and LTP sets (SI Appendix, Fig. S4C). The P values of LFP fusions (with GTEx SBT hits) are significantly smaller than those of LTP fusions (Mann–Whitney U test, P 2.2e−16), implying that the majority of them are filtered out by the SBT statistical refinement step. On the other hand, LTPs possess higher 2-sided P values, which indicates that they would pass the refinement step. In summary, while the statistical refinement step improves DEEPEST calls, the approach outlined is also a general methodology for increasing the precision of RNA variant calls on massive datasets.
DEEPEST Identifies LncRNAs Are Prevalent in Fusions.
A major category of fusions identified by DEEPEST are fusions that involve LncRNAs, which have been overlooked by other methods due to bioinformatic or heuristic filters. Fusions involving well-studied (and thus named) LncRNAs have been appreciated to have oncogenic potential (21, 33, 34). Due to their functions in regulating cellular homeostasis, fusions that involve LncRNA have the potential to contribute or drive oncogenic phenotypes (34, 35). Genome-wide pan-cancer analyses have previously not profiled those with the LINC annotation. We found that fusions involving LncRNAs (as annotated by the Ensembl database release 89) are abundant in tumors (10% of fusion calls involve LncRNAs and 20% of tumors are found to have at least one fusion involving a LncRNA) (Dataset S4). A large fraction of LncRNA fusions ( or 994 fusions) involves LINC RNAs, which have been overlooked by previous methods due to their biases and heuristic filters for discarding “uncharacterized” genes.
Prevalence of Fusions Found by DEEPEST Is Correlated with Genome Instability.
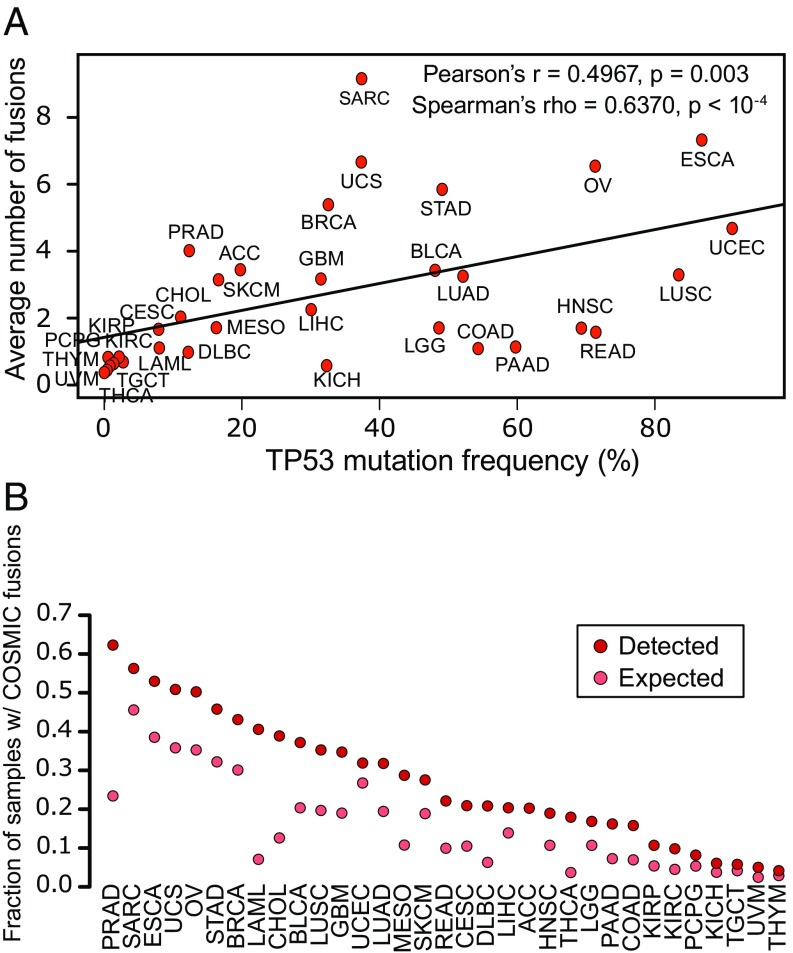
As another computational test of whether DEEPEST maintains high precision in a variety of solid tumors that have more complex cytogenetics than LAML, we tested whether the abundance of detected fusions per cancer is correlated with the mutation rate of TP53, which is an orthogonal measure of a tumor’s genome instability (36). The average number of fusions per sample identified by DEEPEST has a higher (and significant) correlation with TP53 mutation rate across tumor types compared with ref. 10 and TumorFusions (Pearson correlation: 0.497, 0.38, and 0.31, respectively; Spearman’s correlation: 0.637, 0.596, and 0.54, respectively; Fig. 3A). We found that there is a significant correlation between fusion abundance and TP53 mutation frequency as 2 orthogonal measures of genomic instability. DEEPEST calls more fusions in tumors with high TP53 mutation rates in less cytogenetically complex tumors, while retaining tight control of FPs in other samples.
Fig. 3.
Association of fusions with genomic instability. (A) DEEPEST fusions are significantly correlated with the TP53 mutation frequency. (B) Detected fusions are highly enriched in genes cataloged by COSMIC. For all tumor types, except for uterine carcinosarcoma (UCS), UCEC, pheochromocytoma and paraganglioma (PCPG), UVM, TGCT, kidney chromophobe (KICH), thymoma (THYM), and adrenocortical carcinoma (ACC), the observed fraction significantly exceeds the expected fraction based on the null hypothesis of random pairing (Bonferroni-corrected FDR 0.05).
DEEPEST Identifies a Positive Selection on Fusions Containing Known Oncogenes.
Gene fusions could arise from random reassortment of DNA sequences due to deficiencies in the structural integrity of DNA in tumors with no functional impact, or they can be driving events, such as for BCR–ABL1 in chronic myelogenous leukemia (1). In our first global computational tests of whether fusions are passengers or drivers, we tested whether DEEPEST-called fusions are enriched in genes known to play oncogenic roles. Since functional gene ontologies are not used by DEEPEST, this analysis provides an independent test of whether fusions are enriched for genes in known cancer pathways. For each tumor type, we tested for enrichment of the 719 genes present in the COSMIC Cancer Gene Census database (22).
Under the null hypothesis of random pairing of genes in fusions, we compared the observed to expected fraction of samples containing at least one fusion with a COSMIC gene by conditioning on the total number of fusions detected for a tumor type, where tumor types with more detected fusions are expected to have a higher ratio of samples with COSMIC fusions (Materials and Methods). The largest enrichment for COSMIC genes is in PRAD (3-fold change vs expected fraction: P 1e−6), THCA (4.9-fold change vs expected fraction; P 1e−6), and LAML (5.6-fold change vs expected fraction; P 1e−6) (Fig. 3B and Dataset S3). This is expected because the most frequent gene fusions in PRAD involve the ETS family of transcription factors (which are cataloged as COSMIC genes), THCA tumors are highly enriched for kinase fusions, and LAML is a disease where fusions, including known drivers, have been intensively studied, and therefore their partners are annotated as COSMIC genes. Most tumor types lack prevalent recurrent gene fusions, and thus there is no a priori bias that fusions will be enriched for COSMIC genes in other tumor types.
In PRAD, SARC, ESCA, UCS, and OV, the fraction of samples with fusions containing a COSMIC partner exceeds 50%, a rate much greater than expected by chance, the null fraction of samples with COSMIC fusions is 45% for SARC and less than 40% for other tumor types (Fig. 3B and Dataset S3). In more than 90.7% (Bonferroni-corrected FDR 0.05) of the tumor samples we studied, COSMIC genes are statistically enriched above the background rate. Together this is strong evidence for a positive selection pressure on gene fusions in various tumor types, including cancers such as OV, where fusions are currently not considered to play a driving role.
Statistical Analysis of Rare Fusions Shows a Selection in More Than 11% of TCGA Tumors.
Fusion recurrence is considered to be evidence that a fusion plays a driving role. This argument grew out of work focused on point mutations in cancer genomes (37). However, the total number of possible gene fusions (the sample space) greatly exceeds the sample space of point mutations. The number of potential gene fusions scales quadratically with the number of genes in the genome (in the samples we analyzed, genes were expressed). This means that there are up to 625 million potential gene fusions, more than an order of magnitude greater than the number of possible point mutations that is bounded by the number of protein-coding bases in the transcribed genome (). Therefore, fusions could be strongly selected for in tumors even without observing high levels of recurrence. If a moderate fraction of human genes could function as oncogenes when participating in fusions, rare fusion expression is expected in a population-level survey, even one as large as the TCGA cohort.
To account for this effect, we formalized a statistical test for whether the prevalence of rare recurrent fusions fits a model of neutral selection by a null distribution where fusion expression arises by chance, the theory of which was worked out in ref. 38 (Materials and Methods). We mapped the probability of observing recurrent gene fusions to a familiar problem in statistics: if balls (corresponding to the number of observed fusions) are thrown into boxes (corresponding to the total number of possible gene pairs), how many boxes are expected to have or more balls? In other words, given the number of detected fusions, how many of them are expected to be called for at least samples?
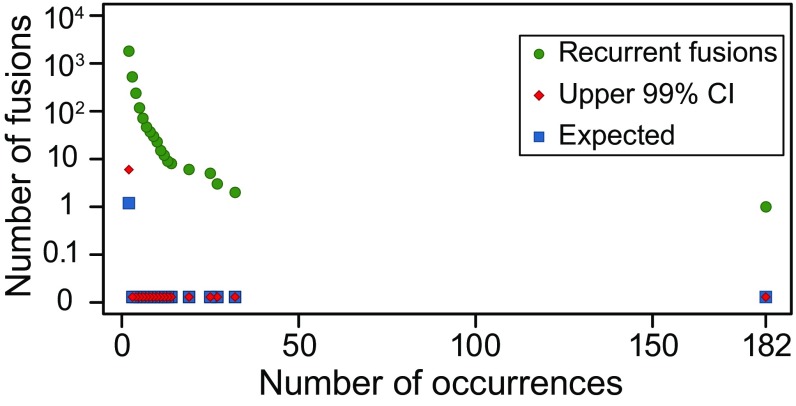
The most prevalent fusions expected under neutral selection would be observed only 2 times, and we would expect to observe only 5 such fusions (Fig. 4), making this and thousands of other fusions highly unlikely to be observed under the null hypothesis. Controlling for multiple hypothesis testing, this analysis recovers several known recurrent fusions including TMPRSS2–ERG, PML–RARA, FGFR3–TACC3, and DHRS2–GSTM4 (4, 39, 40).
Fig. 4.
Statistical analysis of recurrent fusions. Observed number of recurrent fusions that occur more than x times is significantly higher than the expectation and the upper 99% CI expected in the null (Benjamini–Yekutieli FDR control at level 0.01).
This analysis reveals evidence that recurrent fusions are selected for in diverse tumors; RPS6KB1–VMP1, a fusion between the ribosomal protein kinase (41) and a vacuolar protein (VMP1) present in 8 tumor types, is the most prevalent detected gene fusion, after TMPRSS2–ERG, across the entire TCGA cohort and supports findings by previous studies that these fusions have a driving role (42, 43) (Fig. 2C). Globally, 14% of the fusions (1,486) found by DEEPEST are observed at higher rates than expected by chance (P 1e−6); more than 11.9% of tumors (1,181) have recurrent fusions (Fig. 4 and Dataset S1).
Recurrently Fused Genes Distinguish Tumors from Nonneoplastic Tissue and Are Fused in More Than 30% of TCGA Tumors.
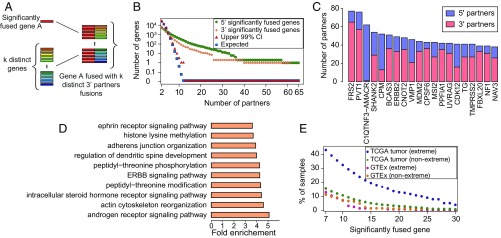
If many genes could serve as oncogenic fusion partners, fusions under selection could be private, yet partners could be much more prevalent than would be expected by chance. To test whether 3′ or 5′ partner genes are overrepresented in fusions found by DEEPEST in the TCGA cohort, we used the “balls in boxes” null distribution above, where boxes correspond to all possible 3′ (respectively 5′) partners (expressed genes) and balls correspond to the total number of fusion pairs (i.e., 31,007 fusions) detected across all samples. We map the coincidence of balls in one box to distinct 5′ (resp. 3′) partner genes being paired with one 3′ (resp. 5′) partner and call genes with statistically significant numbers of 5′ and 3′ partners “significantly fused” (Fig. 5 A and B).
Fig. 5.
(A) Significantly fused genes: those paired with multiple partners (counted by counting distinct fusion pairs). (B) Significantly fused genes are observed at rates higher than expected by chance; genes with k 10 partners would be unlikely to occur by random assortment of genes into fusions (FDR-corrected P 0.01). (C) Genes with the highest number of distinct gene partners. (D) GO enrichment analysis of significantly fused genes shows enrichment in pathways known to regulate cancer growth. (E) Tumors are highly enriched for extreme fusions involving significantly fused genes.
The number of significantly fused 5′ and 3′ partners is large: DEEPEST reports 864 recurrent 5′ partners and 378 recurrent 3′ partners, both having P values of 1e−5 (Fig. 5B), when only 110 genes with more than 6 partners would be expected by chance (Dataset S2); 190 and 48 genes are found in fusions as significantly fused 5′ and 3′ partner genes with more than 12 partners, respectively, when no such genes would be expected by chance. The most significant 5′ partner gene is FRS2, a docking protein that is critical in FGF receptor signaling (44); FRS2 fusions are detected in 52 tumors or in 0.5% of TCGA cases. Other highly significant recurrent partners include PVT1, ERBB2 (HER2), known oncogenes, and tumor suppressors such as MDM2, which negatively regulates TP53 (45) and UVRAG (46) (Fig. 5C). The most promiscuous 3′ partner genes are CPM, a gene regulating innate immune development (39 partners), and the gene C1QTNF3–AMACR (61 partners) (Fig. 5C). Other genes with the highest numbers of distinct 5′ partners include CDK12, a cyclin-dependent kinase emerging as a target in cancer therapy (47), and well-known tumor suppressors such as RAD51B (48). We also found 31 noncoding RNAs as significantly fused genes. PVT1, noncoding RNAs of unknown function: AC134511.1, AC025165.3, and LINC00511 have the most 3′ partners; and BCAR4, PVT1, and noncoding RNAs of unknown function: AP005135.1 and AC020637.1 have the most 5′ partners (Dataset S2). While some of these noncoding RNAs such as PVT1 (49), LINC00511 (50), and BCAR4 (51) have been shown to act as oncogenes, our findings call for further investigation into the potential driving roles of other significantly fused noncoding RNAs.
While the list of significantly fused genes includes well-known cancer genes as described above, only 15.5% of them (193 genes) are currently annotated as COSMIC genes, i.e., 888 significantly fused genes are previously unreported candidate oncogenes, calling for further functional investigation of these genes. This is an enrichment for COSMIC annotation but is not exhaustive: significantly fused genes are a large class of potential oncogenes and tumor suppressors that function through gene rearrangement rather than gain or loss of function through point mutation. To functionally annotate significantly fused partner genes, we carried out gene ontology (GO) enrichment analysis and found the highest enrichment (Binomial test, Bonferroni-corrected FDR 0.05) in cancer pathways such as androgen signaling pathway, ERBB signaling pathway, and ephrin receptor signaling pathway (Fig. 5D and Dataset S2).
To further support the role of significantly fused genes in cancer, we evaluated the rate that such genes are detected in TCGA tumor and GTEx samples as a function of 1) the number of partners and 2) the nature of the rearrangement underlying the fusion: “extreme” events that bring together 2 exons that are farther than 1 MB apart, on opposite strands, or on different chromosomes in the reference, and all other events are “nonextreme” (Fig. 5E). Nonextreme events could arise through small scale genomic duplication or transcriptional readthrough coupled to “back-splicing” to generate circRNA (20, 23). Globally, DEEPEST detects fusions including significantly fused genes (10 partners) at a much higher rate in TCGA tumors (7,050 fusions in of samples) than in GTEx controls (29 such fusions in of samples), despite GTEx samples being sequenced at an average depth of 50 million reads, roughly similar depth to tumor samples.
The deviation between the fraction of such fusions in TCGA versus GTEx increases with the number of partners of the significantly fused gene such that among those with at least 23 partners, only 2 fusions are detected in GTEx (0.7% of samples), while 1,845 such fusions are detected in 1,202 TCGA tumors (12.1% of samples). Notably, the 2 fusions detected in GTEx are PVT1–MYC and FRS2–CPSF6, both fusions are “nonextreme,” splicing detected between 2 genes transcribed in the same orientation with promoters 200 kB from each other, events which could arise from somatic or germline variation or transcriptional readthrough.
Fusions involving significantly fused genes in TCGA and GTEx samples have distinguishing structural features. The large majority of fusions in tumors arise from extreme rearrangements (SI Appendix, Fig. S6) regardless of the number of partners a significantly fused gene contains. More than 90% of fusions in TCGA that involve significantly fused genes with at least 23 partners are extreme, whereas no such GTEx fusions are extreme (SI Appendix, Fig. S6). This again implies a tumor-specific selection for extreme fusions, which increase the complexity of partners available to significantly fused genes. Together, analysis of rare recurrent gene fusions and recurrent 3′ and 5′ partners identify hundreds of candidate oncogenes, which constitute a significant fraction of gene fusions.
DEEPEST finds higher enrichment of significantly fused genes in more tumors compared with other TCGA fusions lists (SI Appendix, Fig. S7), calling significantly fused genes with 10 partners in ; and with 20 partners in more samples compared with recent studies (10 partners: DEEPEST: 3,705; ref. 10: 2,570; TumorFusions: 2,787 samples; 20 partners: DEEPEST: 1,479; ref. 10: 823; TumorFusions: 958 samples). While 7.6% of DEEPEST fusions have a gene with 20 partners, only 4.8% and 5.6% of fusions in ref. 10 and TumorFusions, respectively, have such genes. FRS2 is found to have the highest number of partners in all 3 lists; however, DEEPEST identifies 65 3′ partners, which is larger than other 2 lists: 41 and 52 partners by ref. 10 and TumorFusions, respectively.
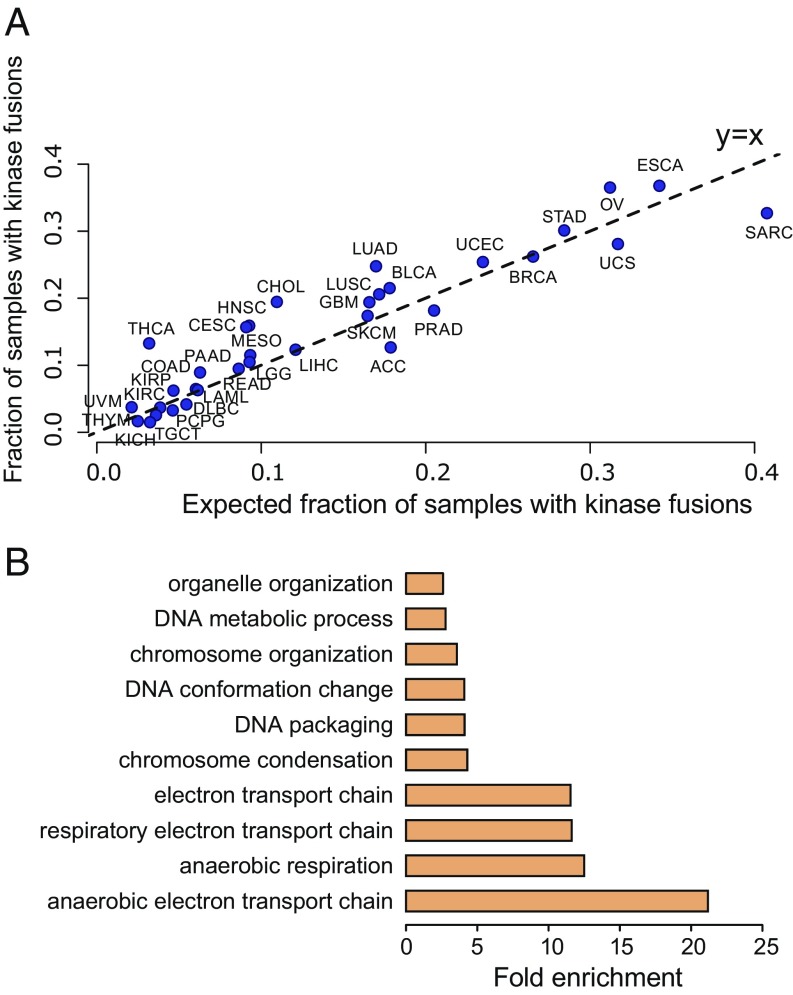
Lung Adenocarcinoma and Serous OV Have High Statistical Enrichment for Kinase Fusions.
The most common genetic lesions in OV and lung adenocarcinoma (LUAD) is TP53 mutation, present in 85.8% of OV and 52.12% of LUAD cases (cBioPortal; retrieved 2018 Nov 19) (52), although there is a debate in the literature that this prevalence is an underestimate. However, TP53 mutations are not sufficient to cause cancers (53). In OV, the explanatory driving events are as yet unknown (54). We tested the hypothesis that genome instability in OV could generate fusions responsible for driving some fraction of these cancers, which might have been missed because of shortcomings in fusion detection sensitivity. The rate of kinase fusions is statistically significantly higher than would be expected by chance, supporting a selection for and driving role of kinase fusions in these tumor types. DEEPEST predicts that 37% of ovarian tumors (Binomial test, P 1e−5) and 25% of lung adenocarcinoma tumors (Binomial test, P 1e−5) contain kinase fusions (Fig. 6A and Dataset S3), a rate higher than what would be expected based on the null assumption of random pairing of genes in fusions. Other cancers with high enrichment of kinase fusions include: THCA (13.3% of samples; P 1e−6), head and neck squamous cell carcinoma (HNSC) (16% of samples; P 1e−6), and cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (15.7% of samples; P = 7.7e−5) (Fig. 6A and Dataset S3).
Fig. 6.
Protein domain analysis. (A) Analysis of the fraction of samples containing kinase fusions reveals that THCA, CHOL, LUAD, OV, and many other tumor types have significant high enrichment of kinase fusions in addition of high overall rates. (B) GO analysis identifies enrichment of cellular metabolism and DNA organization in the protein domains enriched in all fusion transcripts.
Positive Selection for Fusions to Rewire the Cancer Proteome.
To test if there is selection on the protein domains included in fusions, we compared the rate at which each protein domain occurs in the reference proteome to its prevalence in the DEEPEST-called fusion proteome. This analysis identified a set of 120 domains that are statistically enriched in fusion proteins. The most highly enriched domains are AT_hook, a DNA binding motif found for example in the SWISNF complex, NUP84-NUP100, a domain present in some nucleoporins, and Per1 a domain involved in lipid remodeling, all present at 15 times higher frequency than the reference proteome (P 1e−10) (Dataset S4). Tyrosine kinase domains are 1.8-fold enriched in fusions compared with the reference proteome (P 1e−10). To functionally characterize the 120 domains enriched in fusions proteins, we performed GO enrichment analysis using the dcGOR R package (55) and identified overrepresented biological processes among these domains (Binomial test, Benjamini–Yekutieli-corrected FDR 0.05): the enriched domains were involved in (anaerobic) electron transport, chromosome condensation and organization, and DNA metabolism or organization (Fig. 6B and Dataset S4).
To find the set of domain pairs enriched in fusions, we compared the observed frequency of each domain pair against the null probability of random pairing between domains; 226 domain pairs are enriched above background (Bonferroni-corrected FDR 0.05), among the highest enriched domain pairs are NHR2–RUNT, RUNT–TAFH, and RUNT–zf-MYND in the in-frame fusion protein RUNX1–RUNX1T1 detected in LAML samples (Dataset S4).
Because enrichment of protein domain pairs could be sensitive to how we model the null distribution, we formulated a test for selection of fusion proteins containing 2 in-frame domains where the “most pessimistic” null distribution for our problem can be computed in closed form. This analysis considers only fusions whose 5′ and 3′ parent genes contain only one annotated domain. Out of 3,388 fusions with 1-domain parental genes, 681 fusions with 2 domains were observed, whereas only 282 were expected by chance under a closed-form, conservative null distribution (P 1e−5) (SI Appendix), strong evidence for selection of such fusions that couple intact domains in the fusion protein.
In addition to the above enrichment, 17% of all DEEPEST fusions result in proteins that have protein domain pairs that do not exist in the reference proteome. These pairs include well-known driving fusions such as the domain pairs Pkinase_Tyr–TACC and I-set–TACC in FGFR3–TACC3 but also include 9,500 other domain pairs not found in the reference proteome, which implies their potential for tumor-specific function (Dataset S4).
Discussion
Some of the first oncogenes were discovered with statistical modeling that linked inherited mutations and cancer risk (56). The advent of high-throughput sequencing has promised the discovery of novel oncogenes, which can inform basic biology and provide therapeutic targets or biomarkers (57, 58). However, unbiased methodologies for the discovery of novel oncogenic gene fusions have been only partially successful.
DEEPEST is a unified, reproducible statistical algorithm to detect gene fusions in large-scale RNA-Seq datasets without human-guided filtering. DEEPEST has significantly lower FP rates than other algorithms. The unguided DEEPEST filters have not sacrificed detection of known true positives. Further, DEEPEST assigns a statistical score that can be used to prioritize fusions on the basis of statistical support, rather than the absolute read counts supporting the fusion. Such a statistical score is unavailable in other algorithms but of potential scientific and clinical utility as the discovery rate and the tradeoff between sensitivity and specificity of DEEPEST can be tuned by modifying the threshold on scoring.
Although many likely driving and druggable gene fusions have been identified by high-throughput sequencing, studies reporting them have either a nontested or nontrivial FP rate even using heuristic or ontological filters, making those fusions unreliable for clinical use. Similar problems also limit the sensitivity in screens of massive datasets to discover fusions, novel oncogenes, or signatures of evolutionary advantage for rare or private gene fusions. To illustrate DEEPEST’s potential clinical contribution, we integrated DEEPEST-detected fusions with a recent curated precision oncology knowledge base (OncoKB) (59), with drugs stratified by evidence that they interact with specific proteins and found druggable fusions in 327 tumors (3.3%) (SI Appendix, Fig. S8). However, the current list of druggable fusions is biased toward genes that have classically been considered oncogenes, and mainly kinases. By increasing the precision of fusion calls, the analysis in this paper opens the door for further functional and clinical studies to better identify drug targets and repurpose already validated drugs that could target fusions.
The DEEPEST algorithm improves detection of gene fusions that have been missed by other algorithms’ lists of “high confidence” gene fusions. Analysis of these gene fusions uncovers fundamental cancer biology. First, we find evidence that gene fusions are more prevalent than previously thought in a variety of tumors including high grade serous ovarian cancers. Second, our computational analysis suggests the hypothesis that gene fusions involving kinases and perhaps other genes are a contributing driver of these cancers. Further, fusions in ovarian and most tumor types are under selection to include gene families that are known to drive cancers, such as kinases and genes annotated as COSMIC.
DEEPEST allows for rigorous and unbiased quantification of gene fusions at annotated exonic boundaries and for tests of whether partners in gene fusions that may be rare or private are present at greater frequencies than would be expected due to chance. The results in this paper establish statistical evidence that gene fusions and the partner genes involved in fusions are under a much greater selective pressure than previously appreciated: under a highly stringent definition of an enriched partner, more than 10 to 20% of all TCGA tumors profiled harbor a gene fusion including a gene under selection by tumors to be involved in gene fusions that require large scale genomic rearrangement. Future work profiling normal cohorts will distinguish whether fusions including these genes that found in GTEx arise due to variation at the DNA sequence, transcriptional, or posttranscriptional levels. Significantly, this discovery required comprehensive statistical analysis of rare gene fusions, using large numbers of samples to increase power to detect selection for gene fusion expression by tumors.
Together, the results in this paper lead to a model that fusions may be lesions like point mutations, present across tumors rather than tumor-defining, and suggests that by focusing on one tumor type to detect recurrence, and by relying on classical metrics for recurrence and selection, some important cancer biology is lost. Further, the computational evidence in this paper suggests rare fusions are drivers of a substantial fraction of tumors.
Materials and Methods
An Enhanced Statistical Fusion Detection Framework for Large-Scale Genomics.
We used the discovery set to generate a list of fusions passing MACHETE statistical bar (Fig. 1). We then queried all datasets for any fusions found in any discovery set (Fig. 1) and estimated the incidence of each fusion with SBTs (25). SBTs are data structures developed to quickly query many files of data of short-read sequences from RNA-Seq data (and other data) for a particular sequence. These structures build on the concept of Bloom filters. SI Appendix contains technical details about the methodology used. Next, we used standard binomial CIs to test for consistency of the rate that fusions were present in the samples used in MACHETE discovery step and the rate that they were found in the SBT. Fusion sequences that were more prevalent across the entire dataset that is statistically compatible with the predicted prevalence from the discovery set were excluded from the final list of fusions (Fig. 1).
For intuition on why this step is important, consider the scheme in Fig. 1: a candidate exon-exon junction sequence that could be generated from sequencing error results in a read that has been generated by a single gene being more similar in sequence to a read that spans a fusion between 2 homologous genes. There is a difference between MACHETE and SBT, which will lead to both FPs and FNs by the SBT step: SBTs will not consider the alignment profile of all reads aligning to a junction (as MACHETE does), including reads with errors or evidence of other artifacts, as such reads that would have mismatches with the query sequence and are consequently censored by the SBT (these reads would be FN by the SBT). The same censoring leads the SBT, like other algorithms, to have a high FP rate due to: 1) FP intrinsic to the Bloom filters used in the SBT; 2) even if the bloom filter itself has a null FP rate, SBT may falsely identify a putative fusion due to events such as described above (and depicted in Fig. 1). As another example of a FP from reason 2 above, if a single artifact (e.g., a ligation artifact between 2 highly expressed genes) in a single sample passes MACHETE statistical threshold in the discovery step, it will be included as a query sequence for the SBT step, and the SBT could detect it at a high frequency because the statistical models used by MACHETE are not used by the SBT (Fig. 1). Testing for the consistency of the rate of each sequence being detected in the discovery set with its prevalence estimated by SBT controls for the multiple testing bias leading to increased FPs from the SBT (Fig. 1).
We built SBT index files for all TCGA samples across various TCGA projects using SBT default parameters (k-mer index size 20 and a minimum count of 3 for adding a k-mer to a bloom filter). The 40mer flanking the fusion junction (20 nucleotides on the 5′ side and 20 nucleotides on the 3′ side) is retrieved for each fusion nominated by the junction nomination component and each TCGA tumor type is queried for the fasta file containing all 40mers for the fusions called by the first component. For the sensitivity threshold, which determines the required fraction of k-mers in the query sequence that should be found for a hit, we used a more stringent value of 0.9 (instead of default value 0.8) to improve the specificity of DEEPEST. After querying, the detection frequencies of each fusion junction by the first component and SBT are compared, and if they are statistically consistent, the fusion could pass the SBT refinement step; otherwise, it would be discarded. Technical details of the statistical framework, postprocessing of DEEPEST output files, and SBT query step are provided in SI Appendix.
Null Probability for Recurrent Fusions.
For genes, there are possible fusions. If fusions are detected by DEEPEST across all tumors, let be the number of recurrent fusions. In our analysis, there are different gene names in DEEPEST report files and we have reported fusions. The probability that no fusion is recurrent can be computed using the Poisson approximation for the birthday coincidences problem with , . As shown in Fig. 4, the expected value of the number of recurrent fusions (with a frequency of at least 2) is 5.
Calculations for the Expected Number of Recurrent 5′ and 3′ Partners.
As a test of the likelihood of observing our results, we use a statistical model of the probability of observing as many or more recurrent 5′ and 3′ partners under the assumption that the genes in each fusion pair are randomly chosen from all expressed genes. To do this, we use the generalized birthday model from ref. 38. First, we consider all expressed genes as boxes, which represent each potential 5′ partner gene. Next, we consider the distribution of the number of distinct 3′ partners for each gene if 3′ partner genes, which we take to be numbered balls, were thrown at random into boxes. When the first ball arrives in the box , this represents that the first observed fusion on our list has gene on its 5′ side. At the end of this process, we have thrown fusions (balls) into boxes. For a given , the number of balls occupying a single box, we can calculate the probability of having boxes with at least balls. The distribution of has been shown to be a Poisson distribution (ref. 38, theorem 2.2), where . We perform the following calculations to find significant recurrent 5′ gene partners. For each , we find the expected number and the 99% upper CI of the number of boxes (5′ genes) that have at least balls (distinct 3′ partners) according to the null distribution. For statistical analysis, a significance level of 0.01 was considered. Moreover, since we are testing multiple hypotheses in our analysis, we adopt the Benjamini–Hochberg–Yekutieli FDR control procedure (60) and correct the significance value for each . For each , we construct the CI at level (1 − corrected significance level) (Fig. 5B). Similarly, we can find the expectation and upper CI (after Benjamini–Hochberg–Yekutieli correction) for each number of recurrent 3′ genes that have at least 5′ gene partners (Fig. 5B). We provide a table of P values for each observation of the number of recurrent genes with at least partners in Fig. 5 by the formula 1 − F(#3′(5′) partners with at least 5′ (3′) partners, where F(⋅) is the cumulative distribution function of the Poisson distribution (Dataset S2).
Software Availability.
DEEPEST workflow, in which all needed softwares are preinstalled, and all custom scripts used for analysis of fusions are available at ref. 27. Also, a publicly available online tool with web interface is available on the Cancer Genomics Cloud at ref. 26.
Supplementary Material
Acknowledgments
We thank Steven Artandi for useful discussions, and members of J.S.’s laboratory for feedback on the manuscript. J.S. is supported by National Institute of General Medical Sciences Grant R01 GM116847, NSF Faculty Early Career Development Program Award MCB-1552196, a McCormick–Gabilan Fellowship, and a Baxter Family Fellowship. J.S. is also an Alfred P. Sloan Fellow in Computational & Evolutionary Molecular Biology. R.D. is supported by Cancer Systems Biology Scholars Program Grant R25 CA180993. This research benefited from the use of credits from the NIH Cloud Credits Model Pilot, a component of the NIH Big Data to Knowledge Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.T. is a guest editor invited by the Editorial Board.
Data deposition: DEEPEST workflow, with all needed softwares preinstalled, have been deposited in GitHub, https://github.com/salzmanlab/DEEPEST-Fusion. Also, a publicly-available online tool with web interface is available for the DEEPEST algorithm on the Cancer Genomics Cloud platform, https://cgc.sbgenomics.com/public/apps#jordanski.milos/deepest-fusion/deepest-fusion/. All custom scripts used to generate the figures have been deposited in GitHub, https://github.com/salzmanlab/DEEPEST-Fusion/tree/master/custom_scripts.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900391116/-/DCSupplemental.
References
- 1.Hungerford D. A., A minute chromosome in human chronic granulocytic leukemia. Science 132, 1497–1499 (1960). [Google Scholar]
- 2.Soda M., et al. , Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Tomlins S. A., et al. , Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 10, 177–188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh D., et al. , Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337, 1231–1235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J., Mardis E. R., Maher C. A., INTEGRATE-neo: A pipeline for personalized gene fusion neoantigen discovery. Bioinformatics 33, 555–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ragonnaud E., Holst P., The rationale of vectored gene-fusion vaccines against cancer: Evolving strategies and latest evidence. Ther. Adv. Vaccin. 1, 33–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X. S., Mardis E. R., Applications of immunogenomics to cancer. Cell 168, 600–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X., et al. , TumorFusions: An integrative resource for cancer-associated transcript fusions. Nucleic Acids Res. 46, D1144–D1149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alaei-Mahabadi B., Bhadury J., Karlsson J. W., Nilsson J. A., Larsson E., Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc. Natl. Acad. Sci.U.S.A. 113, 13768–13773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Q., et al. , Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 23, 227–238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stransky N., Cerami E., Schalm S., Kim J. L., Lengauer C., The landscape of kinase fusions in cancer. Nat. Commun. 5, 4846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshihara K., et al. , The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 34, 4845–4854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Wu N., Liu J., Wu Z., Dong D., Fusioncancer: A database of cancer fusion genes derived from RNA-seq data. Diagn. Pathol. 10, 131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abate F., et al. , Bellerophontes: An RNA-Seq data analysis framework for chimeric transcripts discovery based on accurate fusion model. Bioinformatics 28, 2114–2121 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Liu S., et al. , Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 44, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrara M., et al. , State of art fusion-finder algorithms are suitable to detect transcription-induced chimeras in normal tissues? BMC Bioinform. 14, S2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S., Vo A. D., Qin F., Li H., Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci. Rep. 6, 21597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey P., et al. , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Saramäki O. R., et al. , TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin. Cancer Res. 14, 3395–3400 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Hsieh G., et al. , Statistical algorithms improve accuracy of gene fusion detection. Nucleic Acids Res. 45, e126–e126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latysheva N. S., Babu M. M., Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res. 44, 4487–4503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes S. A., et al. , Cosmic: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabo L., et al. , Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 16, 126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee M., et al. , Chimerdb 3.0: An enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res. 45, D784–D789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon B., Kingsford C., Fast search of thousands of short-read sequencing experiments. Nat. Biotechnol. 34, 300–302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordanski M., Dehghannasiri R., Salzman J., DEEPEST-Fusion App. Cancer Genomics Cloud. https://cgc.sbgenomics.com/public/apps#jordanski.milos/deepest-fusion/deepest-fusion/ Deposited 13 May 2019. [Google Scholar]
- 27.Dehghannasiri R., Jordanski M., Salzman J., DEEPEST-Fusion. GitHub. https://github.com/salzmanlab/DEEPEST-Fusion Deposited 8 January 2019. [Google Scholar]
- 28.Haas B., et al. , Star-fusion: Fast and accurate fusion transcript detection from RNA-seq. BioRxiv, page 120295 (24 March 2017).
- 29.Torres-García W., et al. , Prada: Pipeline for RNA sequencing data analysis. Bioinformatics 30, 2224–2226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lonsdale J., et al. , The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salzman J., Gawad C., Wang P. L., Lacayo N., Brown P. O., Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prensner J. R., et al. , The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45, 1392–1398(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin C., Yang L., Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol. 28, 287–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huarte M., The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Kopp F., Mendell J. T., Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forment J. V., Kaidi A., Jackson S. P., Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 12, 663–670 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Rowley J. D., Chromosome translocations: Dangerous liaisons revisited. Nat. Rev. Cancer 1, 245–250 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Henze N., A Poisson limit law for a generalized birthday problem. Stat. Probab. Lett. 39, 333–336 (1998). [Google Scholar]
- 39.Kakizuka A., et al. , Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses rar with a novel putative transcription factor, PML. Cell 66, 663–674 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Luo W., et al. , GSTM4 is a microsatellite-containing EWS/FLI target involved in Ewing’s sarcoma oncogenesis and therapeutic resistance. Oncogene 28, 4126–4132(2009). [DOI] [PubMed] [Google Scholar]
- 41.Cai C., et al. , miR-195 inhibits tumor progression by targeting RPS6KB1 in human prostate cancer. Clin. Cancer Res. 21, 4922–4934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inaki K., et al. , Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res. 21, 676–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blum A. E., et al. , Rna sequencing identifies transcriptionally viable gene fusions in esophageal adenocarcinomas. Cancer Res. 76, 5628–5633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadari Y. R., Gotoh N., Kouhara H., Lax I., Schlessinger J., Critical role for the docking-protein FRS2 in FGF receptor-mediated signal transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 98, 8578–8583 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubbutat M. H. G., Jones S. N., Vousden K. H., Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Liang C., et al. , Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 8, 688–698 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Lui G. Y. L., Grandori C., Kemp C. J., CDK12: An emerging therapeutic target for cancer. J. Clin. Pathol. 71, 957–962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thacker J., The RAD51 gene family, genetic instability and cancer. Cancer Lett. 219, 125–135 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Guan Y., et al. , Amplification of pvt1 contributes to the pathophysiology of ovarian and breast cancer. Clin. Cancer Res. 13, 5745–5755 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Sun C.-C., et al. , Long intergenic noncoding RNA 00511 acts as an oncogene in non–small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Therapy-Nucleic Acids 5, e385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing Z., et al. , lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data, Cancer Discov. 2, 401–404(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martincorena I., et al. , High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowtell D. D., et al. , Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 15, 668–679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang H., dcGOR: An R package for analysing ontologies and protein domain annotations. PLoS Comput. Biol. 10, e1003929 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knudson A. G., Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U.S.A. 68, 820–823 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cibulskis K., et al. , Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawrence M. S., et al. , Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakravarty D., et al. , OncoKB: A precision oncology knowledge base. JCO Precis. Oncol. 1, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y., Yekutieli D., The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.