ABSTRACT
A single inner centromere protein (INCENP) found throughout eukaryotes modulates Aurora B kinase activity and chromosomal passenger complex (CPC) localization, which is essential for timely mitotic progression. It has been proposed that INCENP might act as a rheostat to regulate Aurora B activity through mitosis, with successively higher activity threshold levels for chromosome alignment, the spindle checkpoint, anaphase spindle transfer and finally spindle elongation and cytokinesis. It remains mechanistically unclear how this would be achieved. Here, we reveal that the urochordate, Oikopleura dioica, possesses two INCENP paralogs, which display distinct localizations and subfunctionalization in order to complete M-phase. INCENPa was localized on chromosome arms and centromeres by prometaphase, and modulated Aurora B activity to mediate H3S10/S28 phosphorylation, chromosome condensation, spindle assembly and transfer of the CPC to the central spindle. Polo-like kinase (Plk1) recruitment to CDK1 phosphorylated INCENPa was crucial for INCENPa-Aurora B enrichment on centromeres. The second paralog, INCENPb was enriched on centromeres from prometaphase, and relocated to the central spindle at anaphase onset. In the absence of INCENPa, meiotic spindles failed to form, and homologous chromosomes did not segregate. INCENPb was not required for early to mid M-phase events but became essential for the activity and localization of Aurora B on the central spindle and midbody during cytokinesis in order to allow abscission to occur. Together, our results demonstrate that INCENP paralog switching on centromeres modulates Aurora B kinase localization, thus chronologically regulating CPC functions during fast embryonic divisions in the urochordate O. dioica.
Abbreviations: CCAN: constitutive centromere-associated network; CENPs: centromere proteins; cmRNA: capped messenger RNA; CPC: chromosomal passenger complex; INCENP: inner centromere protein; Plk1: polo-like kinase 1; PP1: protein phosphatase 1; PP2A: protein phosphatase 2A; SAC: spindle assembly checkpoint; SAH: single α-helix domain
KEYWORDS: Aurora B kinase, Inner Centromere Protein (INCENP), tunicate, kinetochore, cytokinesis, abscission
Introduction
The correct segregation of chromosomes is critical to the division of somatic cells and the production of viable gametes in the germline. In eukaryotic cells this relies on proper regulation of contacts between spindle microtubules and chromosomes at macromolecular kinetochores built on centromeric chromatin. Although the presence of the kinetochore can be dated back to the last eukaryotic common ancestor, kinetochore composition across species has diverged through gene losses, duplications, and inventions [1]. Functionally comparative analyses of kinetochore modules in different evolutionary lineages can be informative in assessing the extent to which the various regulatory pathways are conserved or adapted to specific constraints. The chromosomal passenger complex (CPC) is a central actor in multiple mechanisms at centromeres and kinetochores, ensuring faithful segregation of chromosomes to daughter nuclei during mitosis.
The multifunctionality of the CPC is reflected in its dynamic subcellular localizations [2]. From late S phase to early prophase, it is found on chromosome arms before being progressively confined to the inner centromere by prometaphase. As anaphase begins, the CPC moves from centromeres to the central spindle and the equatorial cortex before finally becoming concentrated at the midbody during telophase and cytokinesis. In early mitosis, the CPC regulates chromosome structure, removal of cohesins from chromosome arms, spindle formation, kinetochore assembly and the correction of erroneous kinetochore-microtubule attachments towards satisfaction of the Spindle Assembly Checkpoint (SAC). Once amphitelic attachments are established, the CPC coordinates chromosome segregation with central spindle stabilization after anaphase onset, and it is further required for contractile ring constriction and abscission during cytokinesis [3].
The CPC is composed of the catalytic module consisting of Aurora B kinase and the C-terminal (IN box) of the Inner Centromere protein (INCENP), and the localization module consisting of Survivin, Borealin and the N terminal (CEN) of INCENP, which are linked together by the central scaffolding region of INCENP. These three proteins in the localization module form a triple helical bundle closely juxtaposed to the BIR (baculovirus inhibitor of apoptosis repeat) domain of Survivin [4], which recognizes the phosphorylated histone H3 Thr3 (H3pT3) epitope created by the mitotic kinase Haspin at the inner centromere [5–7]. This inner centromeric localization module of the CPC maintains sister chromatid cohesion and mediates assembly of the inner kinetochore for detection of tension loss, while the centromeric kinase module is required for tension-dependent error correction in kinetochore-microtubule attachment and coordination of SAC silencing upon chromosome bi-orientation [8,9]. In the kinase module, the IN box crowns the small lobe of Aurora B and first generates a partially active conformation of Aurora B through an allosteric mechanism. Subsequently, phosphorylation (in trans) in a TSS motif in the C-terminal of INCENP and Thr232 in the T-loop of the kinase domain, by Aurora B itself, achieves full activation [10]. The central region of INCENP contains a single α-helix (SAH) domain flanked by phospho-regulatory domains, and the N-terminal of the SAH domain binds microtubules. It has been proposed that this domain may act as a flexible “dog leash”, which allows Aurora B residing at the end of the complex to reach dynamic substrates at the outer kinetochore while anchoring the localization module to the centromere [11,12].
Aurora B phosphorylates numerous targets to exert regulatory effects during mitosis [13]. An important determinant of substrate phosphorylation is proximity to Aurora B kinase, which is opposed by PP1 and PP2A phosphatase activities. PP1 dephosphorylates H3pS10 and pS28 to coordinate chromosome condensation with mitotic progression [14], and through interaction with Repo-Man [15], counteracts Haspin-mediated spreading of H3pT3 to chromosome arms, permitting centromeric concentration of Aurora B until (pro)metaphase. PP1/PP2A counteracts a midzone Aurora B gradient and promotes chromosome decondensation and nuclear envelope reassembly during anaphase [16]. PP2A-B56 further opposes Aurora B phosphorylation of MKLP2 to promote abscission [17]. In addition to opposing phosphatases, there is interplay between the CPC and other mitotic kinases. Plk1 precedes the CPC to the centromere in Drosophila cells. Upon centromeric CPC recruitment, Plk1 binds INCENP and is phosphorylated to its active state by Aurora B, creating a basis for the kinetochore-microtubule attachment/detachment cycle [18]. In contrast, Plk1 is recruited to centromeres by CDK1-phosphorylated INCENP in human cells [19].
In assessing transitions in CPC activity, one model proposes that an increasing gradient of Aurora B kinase activity is required for progression through mitosis [20], with successively higher threshold levels for chromosome alignment, the spindle checkpoint, anaphase spindle transfer and finally spindle elongation and cytokinesis. Through employing a series of INCENP mutants, assaying subsequent Aurora B kinase activity, and correlating this with cell cycle progression, it was suggested that INCENP might act as a rheostat in regulating changing levels of Aurora B activity through mitosis. However, given that a normal cell does not employ a graded series of such “mutant” INCENP paralogs, it remains unclear as to how such a rheostat would function in wild type cells. Many species do have some CPC regulatory component paralogs (Borealin/Australin in Drosophila, Survivin-A/B in nonmammalian vertebrates, Dasra-A/B in some vertebrates, or Aurora B/C in mammals) though it is thought that some such paralogs might function principally during meiosis [21] or in certain developmental contexts [22–24]. Thus far the only known INCENP paralog that has been described is found in chicken, where it is expressed at very low levels and is unable to substitute for canonical INCENP [20].
Here we show that the marine planktonic chordate, O. dioica, a member of the closest extant group to vertebrates [25] possesses two INCENP paralogs. O. dioica has an exceptionally short chordate life cycle (6 days at 15 ℃) in which a very fast embryonic developmental mitotic phase [26], is followed by rapid post-metamorphic growth through extensive recourse to endoreduplicative cell cycle variants [27–30]. This chordate genome has undergone strong secondary compaction to 70 Mb [31,32], and is distributed over a diploid chromosome number of 2N = 6. Accompanying these features in this rapidly evolving lineage are a number of innovations in the cell cycle regulatory complement [33]. Among these, we demonstrate in this study that unlike the chicken, both O. dioica INCENP paralogs are expressed at significant levels and have distinct temporal and spatial roles during mitotic progression. We propose an INCENP paralog switching model that drives rapid and irreversible transitions between CPC states during fast embryonic cleavages.
Results
O. dioica lacks most components of the CCAN
O. dioica has a small chromosome set, very rapid embryonic divisions and an unusually amplified core cell cycle regulatory complement compared to most invertebrates [33]. The kinetochore is an important regulatory nexus in ensuring faithful chromosome segregation that also exhibits plasticity in its composition across different evolutionary lineages [1]. To gain insight into possible genetic adaptations central to the division machinery, we assessed the kinetochore network composition of O. dioica by in-depth homology searches using orthologous sequences from a variety of metazoans. We focused on structural determinants of kinetochore assembly, as well as proteins that have established roles in kinetochore signaling. We found reduced subsets of most essential components, including CenH3/CENP-A at the inner centromere, Knl1, Mis12 and Ndc80 (KMN network) at the outer kinetochore, all SAC components, as well as Rod and Spindly in the RZZ complex (Figure 1(a)). However, we could not identify any components of the constitutive centromere-associated network (CCAN) at the inner kinetochore. It is hypothesized that the CCAN was present in the last eukaryotic common ancestor, and has been subsequently lost multiple times in many eukaryotic lineages [1]. We can not rule out the possibility that homology detection of CCAN components is compromised by rapid protein sequence evolution in O. dioica. Nonetheless, it would appear that the CCAN has undergone very substantial modifications in O. dioica, with possible corresponding impacts on mechanisms of centromere and kinetochore regulation compared to other current models.
Figure 1.
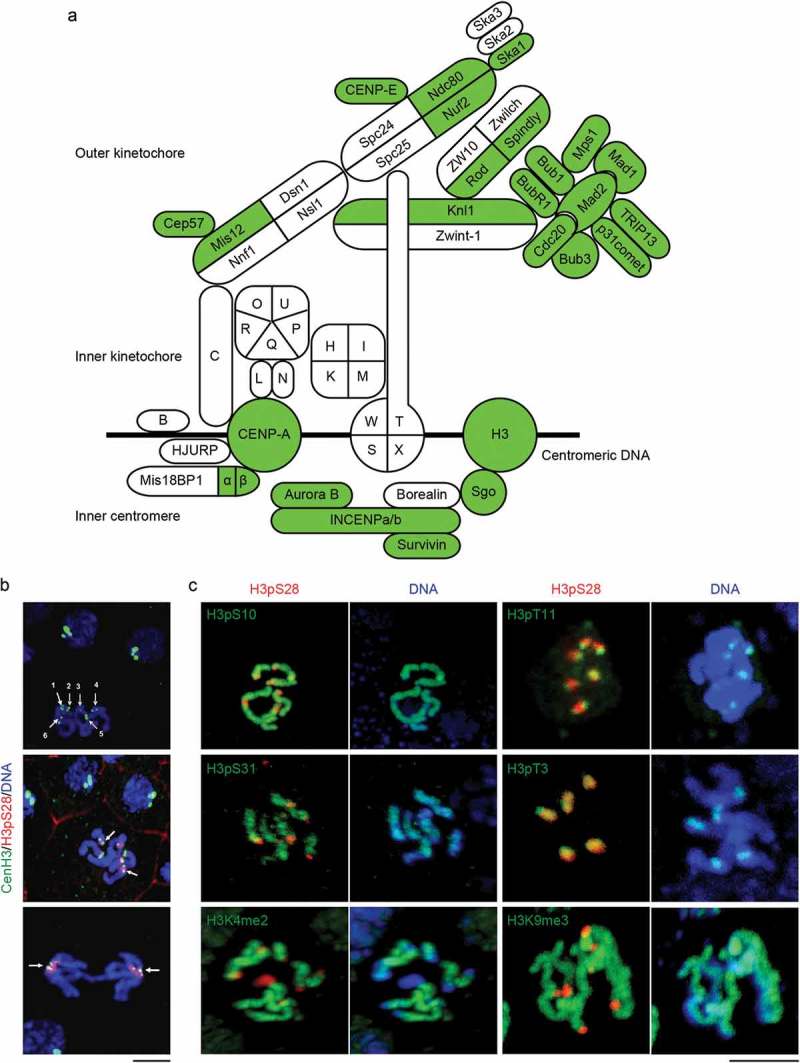
O. dioica kinetochore and inner centromere complements and epigenetic marks. (a) Orthologs that were detected (green) in the O. dioica genome are projected onto the human complement (modified from [1]). Centromere proteins (CENPs) of the CCAN are represented by single letters. (b) Identification of O. dioica centromeres using CenH3. Top left: Six monocentric chromosomes in mitotic phase were indicated by numbers. Middle: CenH3-GFP (arrow) flanked H3pS28 signal on the centromeres. Bottom: CenH3-GFP localized at the centrosome proximal side of separating sister chromatids at anaphase. (c) The distribution of major histone H3 phosphorylation and methylation modifications on O. dioica chromosomes. H3pT3 and H3pS28 localized on the inner centromeres, and H3pT11 localized on the outer centromeres. In striking contrast, H3pS10, H3pS31, H3K4me2 and H3K9me3 localized on the chromosome arms. Bars, 3 µm.
O. dioica centromeres are enriched for CenH3, H3pT3, H3pT11 and H3pS28 but lack several conserved marks
To initiate assessment of centromere regulation during O. dioica cell division, we exploited the O. dioica centromeric histone variant CenH3/CENP-A. To determine the deposition and the localization of odCenH3 within chromatin, fertilized O. dioica oocytes were injected with cmRNA encoding an odCenH3-eGFP fusion protein and allowed to develop until hatched (4–5 h post fertilization). Whereas interphase cells showed odCenH3-eGFP clustered at the nuclear periphery, odCenH3 was found flanking a single region within each of the six monocentric chromosomes in mitotic cells (Figure 1(b)). Furthermore, odCenH3-eGFP positive spots focused at the centrosome proximal side of the separating sister chromatids during anaphase, as expected from centromeres subjected to spindle pulling forces.
In addition to deposition of CenH3 as a key epigenetic marker defining centromeres, several additional epigenetic marks have been reported to decorate pericentromeres and centromeres of mitotic chromosomes in different species. We investigated the spatial and temporal distribution of histone modifications, focusing on histone H3 phosphorylations. O. dioica mitotic chromosomes exhibited strong enrichment of H3pT3, H3pT11 and H3pS28 at centromeric regions, whereas H3pS10 and H3pS31 were distributed uniformly along chromosome arms (Figure 1(c)). H3pS10 and H3pT11 could readily be detected from early prophase onwards, with H3pT11 peaking in prophase and disappearing as chromosomes aligned at the metaphase plate. In contrast, the centromeric marks H3pS28 and H3pT3 were observed only from prometaphase onwards, with H3pT3 removed at the onset of anaphase (Fig S1). Whereas H3pT3 and H3pS28 decorated the inner centromere during mitosis, centromeric H3pT11 staining resolved into two outer centromere spots, reflecting the localization pattern for CenH3 (Figure 1(b)).
Trimethylation of H3K9 is a hallmark of peri-centromeric heterochromatin while H3K4 dimethylation marks inner centromeres in several animals. In O. dioica, however, neither modification was enriched at centromeres. Instead H3K4me2 started to spread along chromosome arms at the onset of prometaphase but was excluded from centromeric regions throughout mitosis (Figure 1(c)). Taken together, O. dioica mitotic centromeres are decorated with the H3 phospho marks H3pT3, H3pT11 and H3pS28, but lack the H3 methylation marks characteristic of centromeres in other species.
Aurora B is the sole kinase responsible for H3S10 and H3S28 phosphorylation in O. dioica somatic cells
In contrast to H3pT3 and H3pT11, H3pS28 has not been reported to be enriched at the inner centromeres in other animals. To understand its regulation, we turned to Aurora B, a critical mitotic kinase reported to phosphorylate H3S28 in addition to H3S10. O. dioica genome queries and subsequent phylogenetic analysis revealed a single Aurora B homolog, in addition to an Aurora A homolog. Expression profiling of Aurora B showed its expression to be highest during developmental stages predominated by somatic mitotic proliferation (Figure 2(a)), consistent with its function during mitosis.
Figure 2.
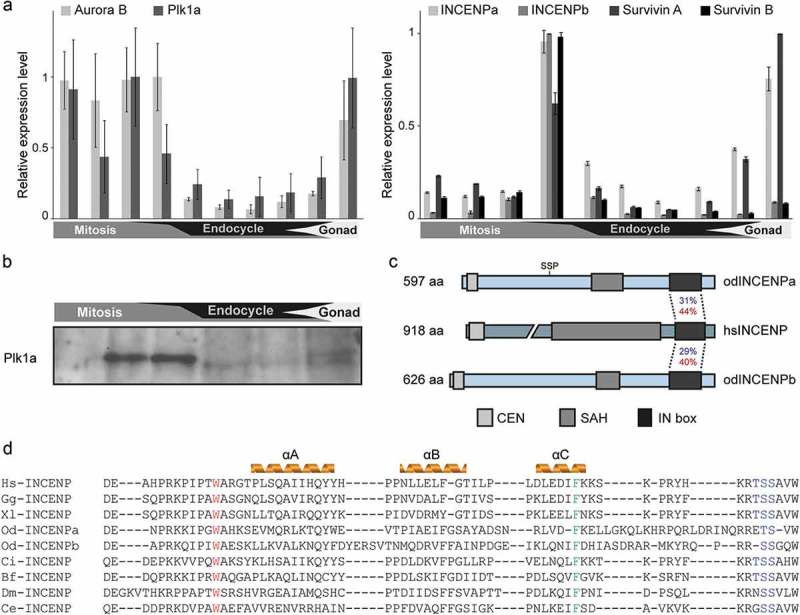
Temporal expression pattern of the CPC components and Plk1a during O. dioica development. (a) Developmental expression analysis of core CPC components and Plk1a. Aurora B and Plk1a (left), and two coexisting INCENP and Survivin paralogs (right), were highly expressed during mitotic proliferation and gonad development. Paired histogram bars correspond to the following developmental stages from left to right: oocytes, 2–8 cells, 1 hpf, hatching, metamorphosis, Day 2, Day 3, Day 4, Day 5 and Day 6. Mean RT-qPCR values normalized to EF1β transcripts (n = 3) are shown with SE. (b) Plk1a protein was enriched during mitotic development as determined by western blot using odPlk1a antibody. Equal amounts of whole animal lysates from various stages were loaded. (c) Domain comparison of Homo sapiens and O. dioica INCENP homologs. N terminal CEN, single α-helix (SAH) and C terminal IN box domains are indicated. Identities (blue) and similarities (red) between O. dioica INCENP paralog IN boxes and the single human ortholog IN box are shown. The CDK1 consensus phosphorylation site (SS209P) is positioned on INCENPa and is absent in INCENPb. (d) Sequence alignment of the INCENP IN box with predicted secondary structure in several model species. W801 (Xenopus numbering), involved in Aurora B interaction, is marked in red and F837, involved in Aurora B activation, is marked in green. Aurora B phosphorylation sites “TSS” are marked in blue. Hs, Homo sapiens; Gg, Gallus gallus; Xl, Xenopus laevis; Od, Oikopleura dioica; Ci, Ciona intestinalis; Bf, Branchiostoma floridae; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans.
In hatched animals, active Aurora B was enriched at centromeric regions coinciding with the increase of the H3pS28 signal in metaphase chromosomes, followed by relocation to the central spindle upon anaphase onset (Figure 3(a)). Interestingly, localization of active Aurora B to chromosome arms preceded H3pS28 but coincided with onset of H3S10 phosphorylation (Fig S1). These data are consistent with Aurora B being the kinase responsible for both H3S10 and H3S28 phosphorylation in O. dioica. To further evaluate this, hatched animals were pre-incubated in the proteasome inhibitor MG132 for one hour, arresting cells in (pro)metaphase as judged by condensed chromatin and the presence of high levels of H3pS10 and H3pS28. Subsequent inhibition of Aurora B with AZD1152 for more than one hour completely abrogated S10 and S28 phosphorylation throughout the animal trunk, while maintaining mitotic arrest with condensed DNA (Figure 3(b)). These data suggested Aurora B as the sole kinase responsible for H3S10 and H3S28 phosphorylation during O. dioica somatic development.
Figure 3.
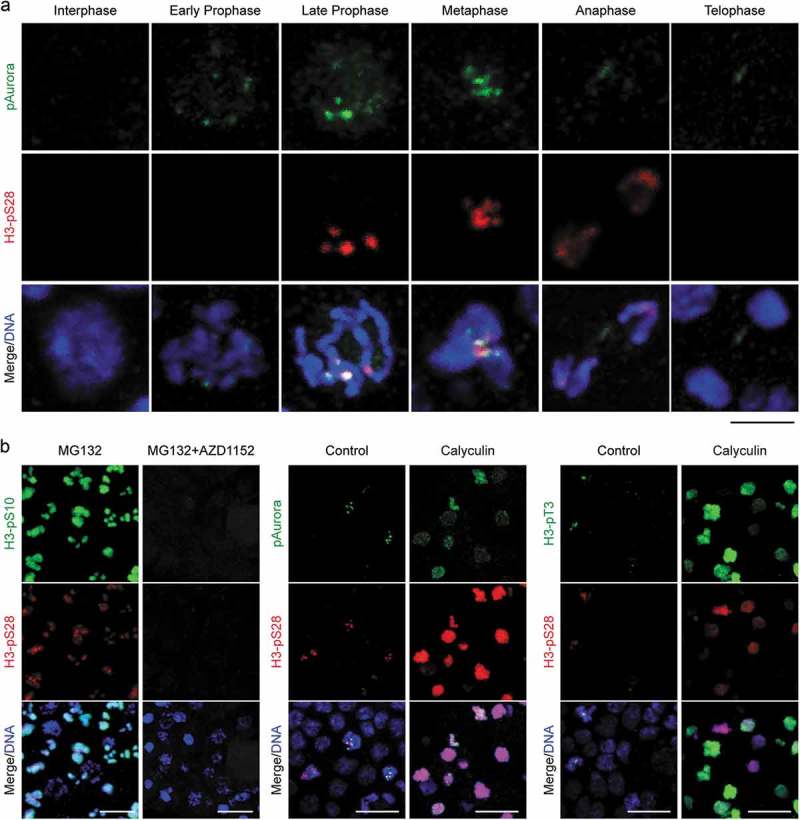
The counteracting effects of protein phosphatases on Aurora B kinase activity in somatic cells. (a) Temporal distribution of Aurora B in mitosis. Aurora B localized on chromosome arms at early prophase, enriched on centromeres from late prophase until metaphase coinciding with the onset of H3pS28 on centromeres, relocated to the central spindle at anaphase and midbody at telophase. Bar, 3 µm. (b) Mitotic H3pS28 on centromeres was governed by Aurora B to antagonize protein phosphatases. Left: Inhibition of Aurora B (AZD1152) after metaphase arrest abolished H3pS10 and H3pS28 in mitotic cells. Middle: Inhibition of protein phosphatases by Calyculin induced Aurora B and H3pS28 redistribution along chromosome arms. Right: Inhibition of protein phosphatases also induced H3pT3 redistribution along chromosome arms. Bars, 10 µm.
Given that H3pS10 was present throughout chromatin from prophase onwards, whereas H3pS28 initiated at centromeres during prometaphase and remained restricted to inner centromeric chromatin, we questioned how a single kinase could generate two H3 phosphorylation marks with such different spatial and temporal dynamics. Inhibition of protein phosphatase 1 (PP1) has been shown to induce premature prophase phosphorylation of H3S28 and H3pS28 is reportedly more sensitive than H3pS10 to PP1 activity [14], suggesting that this could underlie the apparent differences between these two phospho-marks. Indeed, inhibition of protein phosphatase activity using Calyculin in developing O. dioica embryos induced Aurora B and H3pT3 redistribution along chromosome arms, and accordingly, H3pS28 spread throughout chromatin (Figure 3(b)). The difference between H3pS10 and H3pS28 could therefore be reconciled by the balancing phosphatase activity of PP1 isoforms, with Aurora B activity accumulating at centromeres as mitosis progresses to overcome counteracting protein phosphatase activity.
INCENP paralog deployments correlate with temporal and spatial activity of Aurora B
Aurora B has been shown to be the catalytic core of the CPC, with ancillary subunits such as INCENP providing a scaffold to modulate activity and localization. Interestingly, the O. dioica genome contained two INCENP paralogs, INCENPa and INCENPb, both of which possessed clear Aurora B binding domains (IN box) and Aurora B phosphorylation consensus sites (Figure 2(c and d)). As for Aurora B, both INCENP and Survivin paralogs were highly expressed during developmental stages dominated by mitotic proliferation (Figure 2(a)).
Reasoning that the scaffolding and activating, INCENPa and INCENPb paralogs might impose differential localization and activity on Aurora B, fertilized oocytes injected with cmRNA encoding eGFP-fusions of INCENPa or INCENPb were allowed to develop until hatched and analyzed for localization of each paralog. Interestingly, INCENPa was found distributed throughout the nucleus during prophase, then showed brief enrichment at centromeres coinciding with onset of H3S28 phosphorylation, followed by loss of this enrichment at metaphase (Figure 4(a)). Since this same pattern was observed in the presence of the proteasome inhibitor MG132 (Fig S2), this appears to arise through delocalization of INCENPa as opposed to its degradation. Inhibition of Aurora B by AZD1152, resulted in loss of late prophase enrichment of INCENPa at centromeres, suggesting that Aurora B activity is required for this pulsed INCENPa enrichment.
Figure 4.
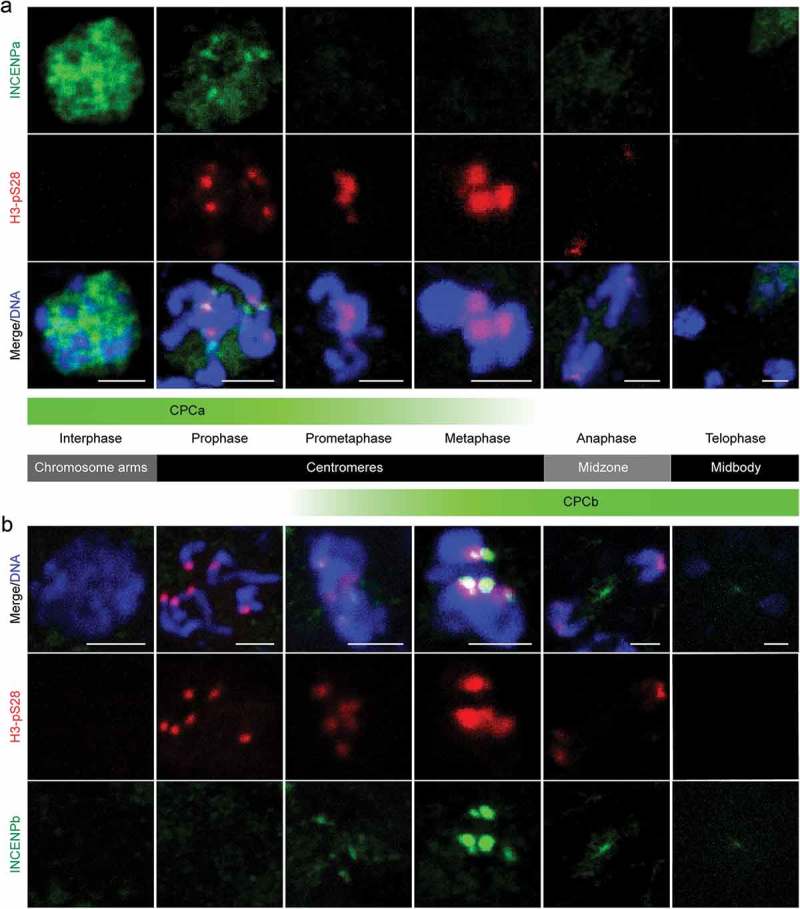
INCENP paralog deployments correlate with temporal and spatial activity of Aurora B in mitosis. (a) INCENPa localized throughout the nucleus during interphase, focused on centromeres during early prophase, coinciding with the onset of centromeric H3pS28, and disappeared afterwards. Bars, 2 µm. (b) INCENPb was undetectable during interphase and prophase, was first detected on centromeres during prometaphase and peaked during metaphase, coinciding with maximum levels of H3pS28. INCENPb relocated to the central spindle during anaphase, and concentrated in the midbody during telophase. Bars, 2 µm. The diagram in the middle illustrated the localization of CPCa and CPCb, representing INCENPa and INCENPb respectively, during each mitotic phase.
In contrast, INCENPb was undetectable during prophase, but became enriched at centromeres from prometaphase onwards and peaked during metaphase, which coincided with maximum levels of H3pS28. Afterwards, INCENPb localized to the central spindle during anaphase, and concentrated in the midbody at telophase (Figure 4(b)). Thus, an intriguing composite picture for localization of INCENP paralogs emerges, mirroring the localization pattern of active Aurora B (Figure 3(a)), with INCENPa and Aurora B (CPCa) colocalizing during early mitosis controlling the induction of chromosome-wide H3pS10 during early prophase and centromeric H3pS28 during late prophase. Complementary INCENPb dynamics reflected the dominant pool of centromeric CPC (CPCb) from metaphase onwards, which relocalized to the central spindle after anaphase onset.
Mutation of a putative Plk1 docking site on INCENPa abrogates centromeric enrichment of CPCa during late prophase
Considering that CPCa and CPCb sequentially localized on centromeres, we asked how this dynamic localization pattern was achieved. In other species, H3pT3 and H2ApT120 on the inner centromeres and kinetochores are required to recruit the CPC [34], which might be also the case in O. dioica, since H3pT3 appears on centromeres coinciding with H3pS28 (Fig S1). In addition, the activity of several prominent mitotic kinases, such as CDK1 and Plk1 also affect CPC localization in other model organisms [12,19]. Searching the O. dioica genome revealed the presence of two Plk1 orthologs, Plk1a and Plk1b. In contrast to Plk1b, the developmental expression profile, overall transcript level and the mitotic degradation signal of Plk1a indicated it represented the principal ortholog regulating mitosis (Figure 2(a)). Western blots of total animal lysates from several developmental time points, covering proliferative stages and endocycling stages (lacking mitosis) using an antibody targeting Plk1a further confirmed its mitotic expression (Figure 2(b)). Using an antibody recognizing active Plk1 (Plk1-pT210), Plk1 activity was found to accumulate predominantly at the centrosomes throughout mitosis and to the midbody following chromosome segregation, which was supported by co-staining with α-tubulin (Fig S3A). Importantly, whereas nuclear active Plk1 appeared generally diffuse, a brief pulse of enrichment at centromeres was apparent during late prophase, coinciding with onset of centromeric Aurora B recruitment and concomitant H3pS28 enrichment (Figure 5(a and b)). Consistent with observations in other models, Plk1 colocalized with Aurora B at the central spindle following anaphase onset (Figure 5(b)).
Figure 5.
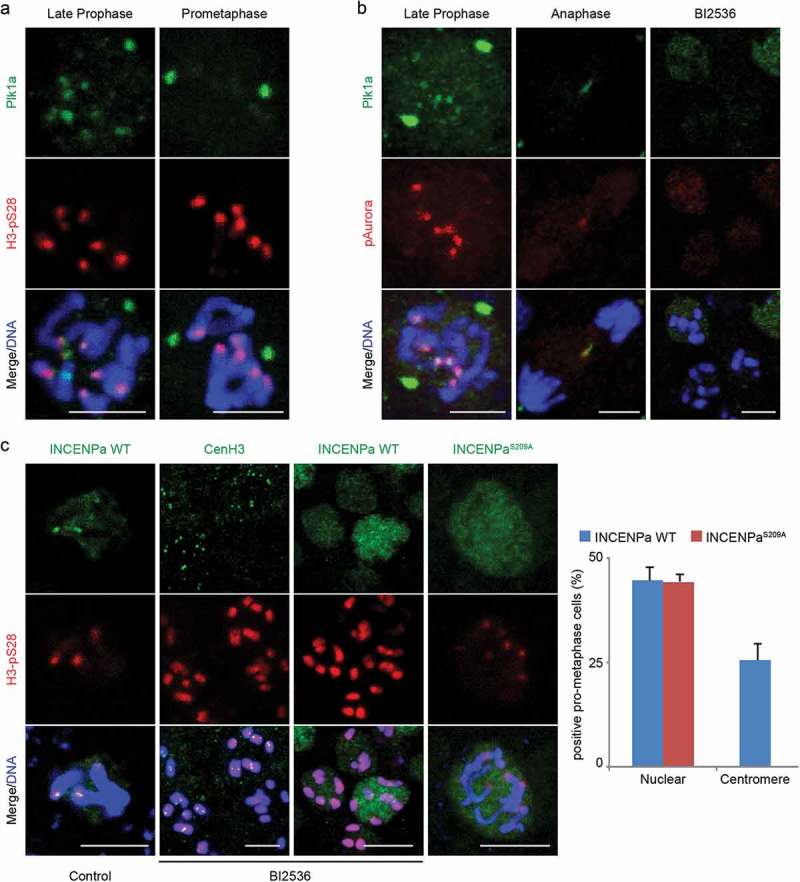
Plk1 docking on INCENPa is a prerequisite for centromeric enrichment of CPCa during late prophase. (a) Plk1 localized on centrosomes and briefly enriched on centromeres during late prophase concomitant with the onset of centromeric H3pS28. (b) Plk1 colocalized with Aurora B on centromeres during late prophase and on the central spindle during anaphase. Inhibition of Plk1 (BI2536) abolished Plk1 staining on centromeres and centrosomes, and Aurora B spread along chromosome arms. (c) Plk1 inhibition resulted in centromeric H3pS28 and INCENPa spreading throughout chromosomes, without affecting CenH3 on centromeres. The INCENPaS209A mutant failed to localize to centromeres. The graph on the right shows the percentage of pro-metaphase cells that showed nuclear or centromeric INCENP localization for INCENPa WT or the S209A mutant. Mean values (n = 3) are shown with SE. Bars, 3 µm.
To assess the roles for Plk1a in mitotic progression, embryos at various developmental stages were incubated with the Plk1 inhibitor BI2536. Plk1 activity was essential for embryonic development as inhibition induced rapid developmental arrest, with penetrant phenotypes apparent within 30 minutes of BI2536 treatment (Fig S3B). Animals accumulated mitotic cells with individually condensed chromosomes that failed to congress, similar to monopolar spindle defects in other animals [35]. Cells that did manage to congress their chromosomes displayed pronounced defects in nuclear membrane disassembly, as judged from aberrant Lamin and nuclear pore complex (NPC) accumulations (Fig S3C).
Interestingly, Plk1 inhibition rapidly removed active Plk1 from centrosomes and centromeres. Concurrently, active Aurora B dispersed along chromosome arms (Figure 5(b)). Accordingly, centromeric H3pS28 and INCENPa spread throughout chromosomes following treatment with BI2536 (Figure 5(c)). The observation that Plk1 inhibition did not affect the centromeric localization of CenH3 (Figure 5(c)) argued against a general effect on centromere identity. Rather, it suggested that Plk1 could be involved in regulating the relocalization of CPCa from chromosome arms to centromeres. The C-terminus of Plk1 contains polo box domains that specifically associate with phospho-epitopes, with S-pS/T-P providing the optimal binding consensus site [36]. In human cell lines, priming phosphorylation of INCENP by CDK1 resulted in the subsequent recruitment of Plk1 to this phospho-epitope [19]. Interestingly, INCENPa, but not INCENPb, contained an S-S-P sequence that could constitute an analogous interaction site for Plk1 (Figure 2(c)). To evaluate whether this sequence could function to recruit Plk1 and induce centromeric relocalization of CPCa, an INCENPa cmRNA variant lacking this recruitment site (INCENPaS209A) was injected in fertilized oocytes. Scoring cells in prophase to metaphase, both the WT and S209A variant were comparably able to localize to the nucleus during prophase. However, exogenously added INCENPaS209A failed to localize to centromeres, in sharp contrast to injected INCENPa WT (Figure 5(c)). Together these data suggest association of Plk1a with INCENPa as a prerequisite for centromeric enrichment of CPCa.
CPCb is delocalized from centromeres upon Plk1 and Aurora B inhibition
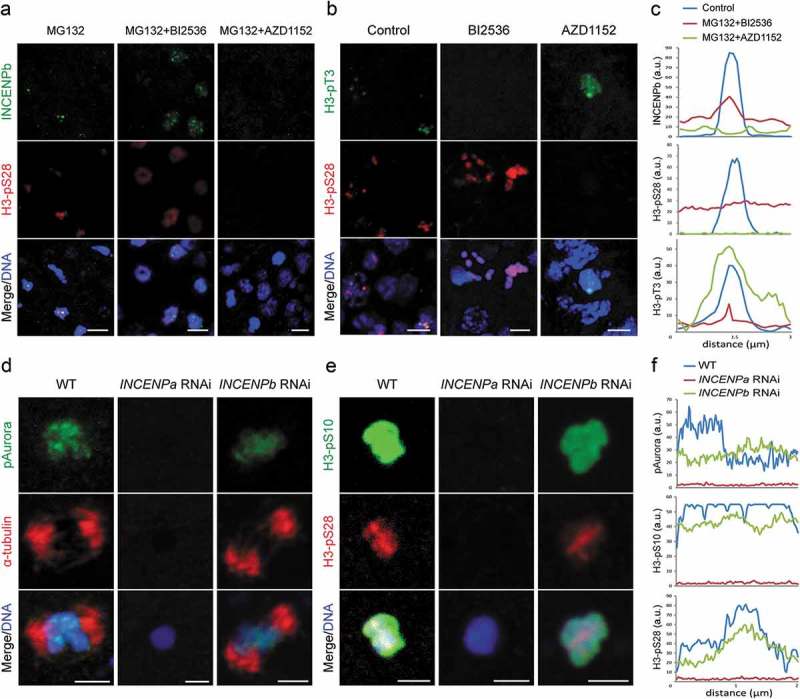
When analyzing the Plk1 inhibition phenotype in more detail, we noticed that nuclei arrested in a typical monopolar prometaphase displayed much lower overall levels of H3pS28 than cells that arrested later during metaphase with congressed chromosomes. Considering the timing of the metaphase arrest, this suggested that CPCb is also deregulated upon Plk1 inhibition. To circumvent the dominant prometaphase arrest upon Plk1 inhibition, animals injected with cmRNA encoding INCENPb-eGFP were allowed to hatch, arrested in metaphase by brief incubation with MG132 (1 h) followed by Plk1 inhibition. Whereas INCENPb localization was restricted to H3pS28 positive centromeres in MG132 arrested cells, inhibition of Plk1 led to a pronounced delocalization from centromeres and induced chromosome-wide diffusion of INCENPb (Figure 6(a and c)). As INCENPb neither contains an obvious Plk1 binding epitope nor colocalizes with Plk1a, the effects of Plk1 inhibition on CPCb localization are likely indirect.
Figure 6.
Effects of Plk1 or Aurora B inhibition on CPCb localization and phenotypes arising from INCENP paralog knockdowns. (a) INCENPb was enriched on centromeres and colocalized with H3pS28 in metaphase arrested cells (MG132 treatment). Inhibition of Plk1 (BI2536) resulted in a significant delocalization of INCENPb and H3pS28 from centromeres to chromosome arms. Inhibition of Aurora B (AZD1152) abolished INCENPb and H3pS28 staining on chromosomes. (b) Inhibition of Plk1 (BI2536) abolished H3pT3 on centromeres, whereas inhibition of Aurora B (AZD1152) resulted in some H3pT3 delocalization from centromeres to chromosome arms. Bars, 3 µm. (c) Line graphs show fluorescence intensity (a.u.: arbitrary units) of INCENPb, H3pS28 and H3pT3 under respective treatment conditions in (a) and (b). The x axis represents distance along chromosomes. (d) Aurora B was absent on chromosomes and the meiotic spindle was not formed at meiotic metaphase I in INCENPa RNAi oocytes. In INCENPb RNAi oocytes, Aurora B localized to chromosomes and meiotic spindles assembled. (e) Neither H3pS10 nor H3pS28 were present on chromosomes and chromosomes were not condensed at meiotic metaphase I in INCENPa RNAi oocytes. In INCENPb RNAi oocytes H3pS10 localized on bivalents and H3pS28 localized to centromeres. Bars, 2 µm. (f) Line graphs show fluorescence intensity (a.u.: arbitrary units) of pAurora, H3pS10 and H3pS28 under respective treatment conditions in (d) and (e). The x axis represents distance along chromosomes.
Interestingly, inhibition of Aurora B resulted in exclusion of INCENPb from the chromosomes in metaphase arrest cells, implying Aurora B activity is even more critical for centromeric localization of CPCb at metaphase (Figure 6(a and c)). We further assessed H3pT3, the centromeric adaptor of CPC, under the circumstances of Plk1 and Aurora B inhibition. Strikingly, inhibition of Plk1 totally abolished H3pT3 from chromosomes, while inhibition of Aurora B caused a general delocalization of H3pT3 from centromeres with spreading along chromosome arms (Figure 6(b and c)). Taken together, these data suggest the existence of a conserved spatial and temporal feedback loop where Plk1a and Aurora B activities are a prerequisite for Haspin mediated H3pT3 deposition and CPCa recruitment on centromeres, and also provide a rationale for CPCb spreading upon initial CPCa delocalization.
CPCa is required for meiotic spindle assembly and phosphorylation of H3S10 and H3S28
Given the intriguing distribution patterns of INCENPa and INCENPb, their roles in Aurora B activity were studied by RNAi. Due to high levels of maternal transcript stocks in oocytes, INCENP paralogs were targeted during oogenesis to deplete their mRNAs in oocytes. The oocytes laid by the injected females were then fertilized by wild type sperm to observe phenotypic effects. Knockdown efficiencies of target genes were confirmed by RT-qPCR, with no significant off-target effects on the paralogous gene (Fig S4). Depletion of the respective INCENP paralogs gave rise to distinct phenotypes. INCENPa RNAi resulted in defective completion of meiosis (see below). In contrast, INCENPb RNAi oocytes extruded two polar bodies as normal after fertilization. During mitotic exit of the first cell division, cleavage furrows formed and ingressed normally. However, the ingressing furrow arrested before completing abscission and regressed. As a result, binucleate one-celled embryos entered the next cell cycle round and cleavage furrow regression occurred again. Finally, embryos arrested with multiple nuclei in a single cell (Video S1).
Our previous work had shown that Aurora B promoted meiotic spindle assembly during prometaphase I under the control of CDK1d/Cyclin Ba [37]. Thus, we investigated how Aurora B activity and meiotic spindle assembly were affected in these knockdown oocytes. Aurora B was absent from chromosomes and the meiotic spindle was not established in INCENPa RNAi oocytes. In distinct contrast, in INCENPb RNAi oocytes, Aurora B still located on centromeres and meiotic spindles formed with three bivalents aligned on the equatorial plate (Figure 6(d and f)). We further examined H3pS10 and H3pS28 under these conditions. In INCENPa RNAi oocytes, neither H3pS10 nor H3pS28 were present. In addition, chromosomes were not condensed, and three pairs of homologous chromosomes could not be distinguished. In INCENPb RNAi oocytes, H3pS10 was present on whole chromosomes and H3pS28 was enriched on centromeres, similar to wild type oocytes arrested at meiotic metaphase I (Figure 6(e and f)). Neither H3pS10 nor H3pS28 distributions were affected during mitosis of INCENPb RNAi embryos (Fig S5). Taken together, these results indicate that CPCa is involved in phosphorylation of H3S10 and H3S28, chromosome condensation and meiotic spindle assembly, whereas CPCb is not required for these meiotic and early mitotic events.
INCENPb is critical for the localization and activity of aurora B on the central spindle and midbody during mitotic exit
Aurora B-dependent pathways regulating contractile ring constriction, cleavage furrow ingression and abscission have been well established. Since the abscission defective phenotype of INCENPb RNAi embryos was obvious during the first several embryonic cell divisions, we examined pAurora staining during this period (Figure 7). Aurora B located on chromosome arms in interphase, on centromeres during late prophase and metaphase, similar to wild type control embryos (Figure 7(a-c)). During anaphase, Aurora B staining on the central spindle was weaker than wild type. This signal became very weak on the central spindle during telophase when the separated chromosomes have decondensed and reformed daughter nuclei, and dropped to undetectable levels on the midbody during cytokinesis. This contrasted wild type, where Aurora B signal remained strong on the central spindle and midbody during late mitosis. These observations suggested that INCENPb was critical for maintaining the localization and activity of Aurora B on the central spindle during anaphase and telophase, and on the midbody during cytokinesis.
Figure 7.
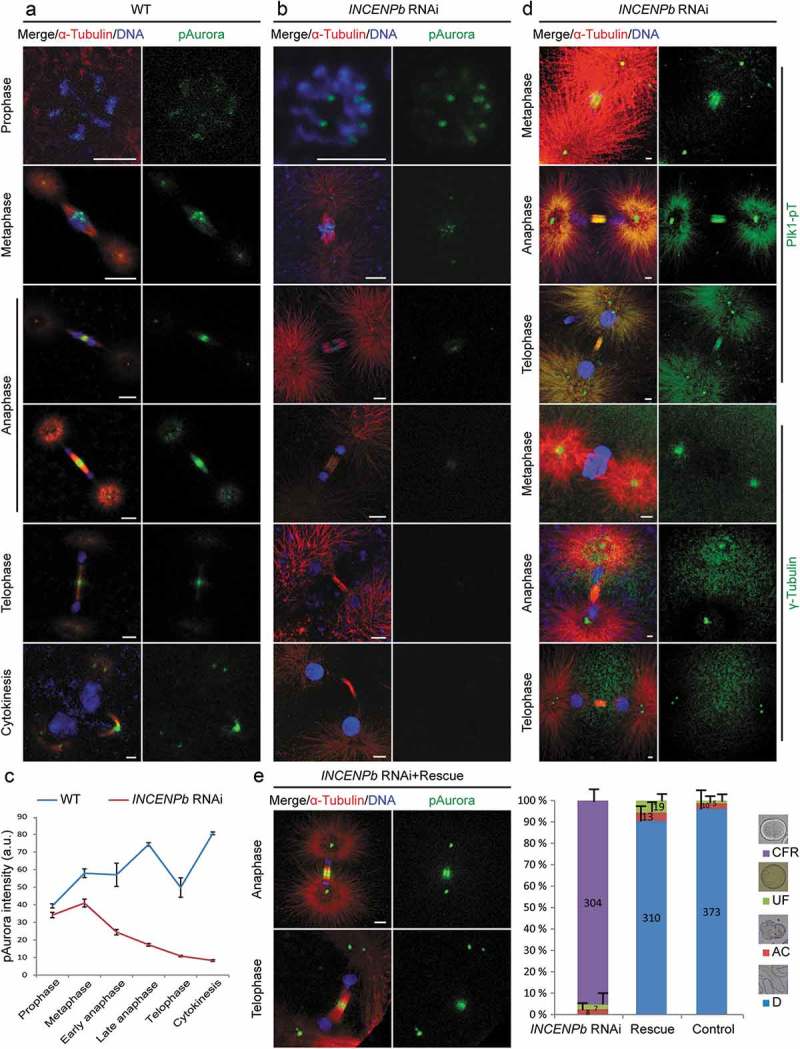
INCENPb is required for the localization and activity of Aurora B on the central spindle and midbody during late mitosis. (a) In wild type early embryonic division, Aurora B localized on chromosome arms at prophase, on centromeres at metaphase, relocated to the central spindle at early anaphase, and was enriched on the midbody during telophase and cytokinesis. (b) In INCENPb RNAi embryos, Aurora B localized on chromosome arms at prophase, and on centromeres at metaphase. Aurora B signal on the central spindle was weak at early anaphase, decreased further at late anaphase and dropped below detectable levels on the midbody during telophase and cytokinesis. (c) Fluorescence intensity (arbitrary units) of pAurora throughout M-phase in wild-type versus INCENPb RNAi embryos. SE bars are shown for n = 3 replicates. (d) INCENPb knockdown did not affect the localization of Plk1 or centrosome integrity. Active Plk1 localized normally on centromeres at metaphase, the central spindle at anaphase, the midbody at telophase, and on centrosomes during all phases. γ-tubulin staining showed normal integrity of centrosomes and clear centriole disengagement at telophase. (e) Exogenous RNAi-resistant INCENPb cmRNA fully rescued the division defects caused by endogenous INCENPb knockdown. Right panel: in INCENPb RNAi group, more than 95% of embryos experienced cleavage furrow regression defects and arrested as aneuploid; in the rescue group, ~90% of INCENPb RNAi embryos progressed normally through embryogenesis, similar to the control group. Mean values of 3 independent experiments are shown with SE, and the number of embryos evaluated in the INCENPb knockdown, rescue and control groups indicated directly on the histogram bars. CFR, cleavage furrow regression; UF, unfertilized; AC, abnormal cleavage; D, developed normally. Left panel: Aurora B signal was restored on the central spindle during anaphase and on the midbody during telophase. Bars, 2 µm.
We next assessed the integrity of the central spindle and centrosomes in INCENPb RNAi embryos, using active Plk1 and γ-tubulin as indicators of intact central spindle and centrosomes. Plk1 showed normal localization (Figure 7(d)): on centromeres at metaphase, on the central spindle at anaphase, on the midbody at telophase and consistently on centrosomes, just as in wild type (Fig S3A). The observation that γ-tubulin staining showed one spot on each centrosome at metaphase and anaphase, and two spots at telophase indicated that centrosome integrity and centriole disengagement were not affected.
To further support that the defects of decreased Aurora B activity on the central spindle and midbody in INCENPb RNAi embryos were caused specifically by the depletion of INCENPb, exogenous RNAi-resistant INCENPb cmRNA was supplied together with INCENPb RNAi in gonad injections. Successful knockdown of endogenous INCENPb in oocytes was confirmed by qPCR (Fig S4). After fertilization with wild type sperm, these INCENPb RNAi rescue embryos divided normally. Normal localization of Aurora B was observed in rescue embryos, with robust Aurora B signal restored on the central spindle at anaphase and on the midbody during telophase and cytokinesis (Figure 7(e)). Thus, INCENPb is required for sustained localization of the CPC at the central spindle and midbody during mitotic exit, and its absence results in an inability to complete abscission. The INCENPa paralog cannot compensate for these late CPC functions.
Discussion
Sequential activities of mitotic kinases enhance the sensitivity of signaling cascades, reinforcing catalytic nonlinearity to achieve fast, switch-like, bistable and irreversible responses [38]. Bistable oscillations are evolutionarily convergent mechanisms that are particularly employed in the cell cycle. Examples are the Rb-E2F switch at G1/S [39], Wee1/Myt1-Cdc25-CDK1/Cyclin B regulation during early embryonic divisions [40], and the three-tiered Raf-MEK-Erk, MAP kinase cascade [38]. Here, we report that functional specialization of INCENP paralogs provides a switch in CPC kinase activities to reinforce transitions during late mitotic events during rapid embryonic cell cycles. Mathematical models and experimental validations propose that scaffold proteins in signaling cascades might specify, accelerate or insulate catalytic reactions in distinct contexts [41]. Sensitivity of kinase activation increases as the scaffold membership increases [42]. Importantly, scaffold proteins also have the potential to redirect evolutionarily conserved signaling modules [43]. Through modulation of scaffold concentrations at different subcellular compartments, localized signaling sources are widely employed in gradient sensing. A concentration gradient of Aurora B activity emanating from the centromere is proposed to act on outer kinetochore components to promote disassembly of kinetochore-microtubule attachments during error correction [13] though this model is debated [44]. Ultrasensitivity in CPC functions generated by switching of paralogs of the INCENP scaffolding protein could sharpen thresholds for catalytic activities in fast mitotic cycles.
Our main findings are summarized in the INCENP paralog switching model (Figure 8). INCENPa modulates Aurora B activity early in mitosis, which is critical for H3S10/S28 phosphorylation, chromosome condensation, Aurora B centromeric localization, spindle assembly, and Aurora B transfer from centromeres to the central spindle. CDK1 phosphorylates INCENPa to prime Plk1a docking, important for INCENPa-Aurora B enrichment on centromeres, potentially through a Haspin-H3pT3-CPC positive feedback loop. INCENP switching occurs on centromeres during prometaphase, with INCENPb-Aurora B subsequently translocating to the central spindle at anaphase onset. Though INCENPb is not required for CPC functions on centromeres during (pro)metaphase and at the central spindle in early anaphase, INCENPb becomes essential for sustained localization and activity of Aurora B on the central spindle at late anaphase and on the midbody during telophase and cytokinesis in order for abscission to occur.
Figure 8.
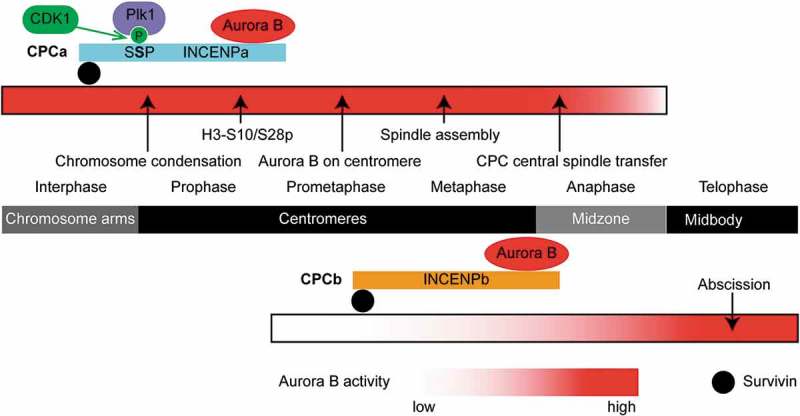
INCENP paralog switching model. Localization of the CPC at each mitotic phase is indicated in the dark middle bar. Major mitotic events are indicated by arrows. Levels of Aurora B activity are represented by degree of red shading for CPCa (top) and CPCb (bottom). CDK1 phosphorylation of (SS209P) in INCENPa and subsequent recruitment of Plk1 are shown. INCENPa modulates Aurora B activity during early mitosis, which is critical for H3S10/S28 phosphorylation, chromosome condensation, Aurora B centromeric localization, spindle assembly, and Aurora B transfer from centromeres to the central spindle. INCENPb appears on centromeres during prometaphase, then INCENPb-Aurora B translocates to the central spindle at anaphase onset. INCENPb becomes essential for localization and activity of Aurora B on the central spindle at late anaphase and on the midbody during telophase, cytokinesis and completion of abscission.
INCENP can integrate multiple kinase signaling cascades on the centromere [45]. We tested the influence of kinase activities from Plk1 and Aurora B on centromeric localizations of two INCENP paralogs. Inhibition assays showed that Plk1 was crucial for Haspin-mediated H3pT3 and CPCa recruitment to centromeres, but less so for CPCb. The abolishment of centromeric H3pT3 upon Plk1 inhibition was more severe than upon Aurora B inhibition (partial diffusion along chromatin), which differs from comparable H3pT3 delocalization effects in mammalian cells under these respective inhibitions [46,47]. Although INCENPb normally concentrated on centromeres and the central spindle, its depletion did not cause obvious defects from metaphase to early anaphase. One explanation is that INCENPa compensates the loss of INCENPb on centromeres and the central spindle to maintain sufficient Aurora B activity during this time period, as exemplified in functional overlaps between Aurora B and C in early mammalian development [22], or in the functional equivalence of Australin and Borealin in Drosophila S2 cells [21]. In O. dioica, the switching of INCENP paralogs in active CPC complexes is initiated at the centromere in order to drive late mitotic events.
An Aurora B activity gradient emanating from the central spindle coordinates chromosome decondensation with nuclear envelope reformation (NER) [16]. This gradient provides spatial information to position the cleavage furrow [48]. INCENP interacts with actin [49], or MKLP2 in a Myosin II-dependent manner [50], to target the CPC to the equatorial cortex and initiate cleavage furrow ingression. In INCENPb knockdown embryos, cleavage furrows formed and progressed as normal, and NER occurred during telophase, even though Aurora B localization to the central spindle was substantially reduced at early anaphase and became undetectable thereafter (Figure 7). One interpretation is that initial localization of the CPC to the central spindle is sufficient to coordinate post-metaphase mitotic events and that maintenance of CPC in the midzone observed in wildtype embryos is not strictly required for these events. Alternatively, despite its inability to maintain CPC localization at the spindle midzone, INCENPa may be able to compensate for the absence of INCENPb for CPC function in these processes. Finally, we cannot exclude that a minor contribution from residual INCENPb levels in these knockdown assays is sufficient for these events. We can, however, conclude that critical levels of INCENPb to sustain Aurora B kinase activity are required for completion of cytokinesis.
Translocation of Aurora B from chromosomes to the central spindle and the increase of Aurora B activity on the midbody are essential for orderly mitotic exit [51]. INCENPb becomes chronologically critical for the localization and activity of Aurora B on the central spindle and midbody. A comparative proteomics analysis has shown that Aurora B dramatically switches its binding partners in the mitosis to cytokinesis transition, with many involved in microtubule binding and cytoskeleton organization [52,53]. Intermediate filaments such as vimentin, GRAP (glial fibrillary acidic protein) and desmin are components of intercellular bridges, and phosphorylation by Aurora B can lead to their disassembly [54]. Aurora B inactivates RhoA at end of cleavage ingression by phosphorylating MgcRacGAP and inducing its RhoGAP activity towards RhoA [55], and it has been proposed that the GAP activity of MgcRacGAP inactivates Rac to suppress branched F-actin production and drive contractile ring constriction [56]. Aurora B phosphorylates the C-terminal of the kinesin-6 family motor protein MKLP1 on the central spindle from anaphase onset. Phosphorylated MKLP1 is enriched on the midbody to maintain furrow ingression during telophase and to accumulate abscission machinery for cytokinesis [57,58]. High Aurora B activity regulates ordered distribution of the ESCRT-III subunit CHMP4C on the midbody arms before its incorporation to the Flemming body to promote abscission [59]. In O. dioica, it appears that the INCENPb paralog may be subfunctionally specialized and required for such regulatory roles of the CPC during mitotic exit.
In summary, this study reveals how two INCENP paralogs modulate CPC activity and localization to complete a chordate mitotic cycle. This provides evidence of a link between gene duplication in core cell cycle regulatory machineries and functional specialization in regulating orderly mitotic events. This also poses numerous intriguing questions regarding the plasticity of essential mitotic regulators as an adaptation to evolutionary constraints in diverse eukaryotes in executing fundamental cell division processes.
Materials and methods
Ortholog identification
We employed two methods to find orthologs for O. dioica kinetochore and inner centromere complements. tBLASTn was first employed to search the O. dioica genome using orthologous sequences from human or Ciona intestinalis as queries. If BLAST failed to detect orthologs, iterated sensitive homology search using the HMMER web server [60] with a permissive E value was used to identify divergent homologs. In some cases, initial hits from other species were aligned and used as queries. The resulting list of O. dioica orthologs was then further assessed by blast in NCBI and conserved motif predictions based on experimental evidence in the literature.
Animal culture and in vitro fertilization
O. dioica were cultured as described [61]. For in vitro fertilization, mature female animals were transferred to watch glasses with 5 ml artificial seawater (Red Sea Salt 30 g/l, final salinity 30 PSU) and left to spawn. Sperm solutions were prepared from 3–4 male animals in 10 ml artificial seawater kept on ice. Solutions were checked for viability using a Nikon Eclipse E400 microscope and 50 µl aliquots were used to fertilize the oocytes spawned from one female. After 2.5 minutes, fertilized eggs were washed with artificial seawater. Embryos were left to develop at 18℃ to the desired stage.
Immunostaining
Samples were processed for immunofluorescence and analyzed by confocal microscopy as described [33]. Prior to tubulin staining, animals were allowed to hatch and transferred to watch glasses with seawater containing 500 nM Paclitaxel (Sigma). Animals were incubated for 15 minutes and fixed in 4% paraformaldehyde/0.1 M MOPS pH 7.5/0.5 M NaCl/0.1% Triton X-100. Commercial antibodies are shown in Table S1 and custom made antibodies, rabbit polyclonal affinity-purified anti-Lamin 1 (acetyl-QSPISLPPLSGST-amide) and anti-Plk1a (acetyl-AVDNLPENEEQGYARP-amide and acetyl-LKQDGSTQTFLARKLNT-amide) were obtained from 21st Century Biochemicals, Marlboro, MA. Primary antibodies were diluted 1:100 (1:50 for Plk1-pT210) and incubated for 2 weeks. Alexa Fluor secondary antibodies, anti-rabbit and anti-mouse Alexa488, anti-rat and anti-mouse Alexa568 (Molecular Probes), were used at 1:500 dilutions and incubated for 1–2 weeks.
Confocal fluorescence microscopy and image processing
Immunofluorescence images were acquired on a Leica SP5 confocal microscope with an APO 40.0x/1.25 oil objective. All images within the same figure were collected with identical acquisition settings. To obtain the entire spindle, some images were acquired as z stacks with 1-µm optical sectioning and processed with a maximum intensity projection method using Leica Application Suite X software. Background was reduced by applying a 3-pixel radius filter in Image J. Fluorescence intensity was also quantified in Image J. For signals surrounding centromeres, lines of equal length were drawn along the chromosomes with centromeres positioned in the middle. Intensity values were acquired using plot profile analysis and plotted as line graphs. For quantitative comparison of pAurora signals, three Regions of Interest (ROI) were assessed: at centromeres in prophase and metaphase, and at central spindle and midbody in anaphase, telophase and cytokinesis. Intensities were measured using histogram analysis. Mean values and standard error were calculated from the measurements.
Time-lapse imaging
Fertilized oocytes were put in chamber slides and filmed at 15℃. Time-lapse imaging was performed under a Nikon Eclipse TE2000-S microscope equipped with a 20×/NA0.45 objective and a monochrome camera (IMAGINGSOURCE, DMK 23UP1300). Images were captured at 10-second intervals for 2 h using IC Capture 2.4. The movie was created in QuickTime at a playing speed of 6 frames per second.
Chemical treatments
Embryos were transferred to 6-well plates coated with 2% agarose and chemicals were diluted in 5 ml artificial seawater at stated concentrations: BI2536, 10 μM (Axon), Calyculin A, 250 nM (LC Laboratories), Paclitaxel, 500 nM (Sigma), AZD1152, 20 μM (Selleckchem), and MG-132, 20 μM (Calbiochem). For Calyculin A treatment, animals were not transferred to the chemical before 7 h post fertilization. After incubation with respective inhibitors for 2 h, animals were collected for immunostaining. Control animals were incubated in the corresponding concentrations of the DMSO solvent.
Microinjection of capped mRNA and dsRNA
eGFP fusion construction, capped mRNA and dsRNA synthesis, and microinjections in embryos or gonads were performed as described previously [62]. Briefly, 1-cell embryos were injected with 400 ng/µl capped mRNA encoding eGFP-fusion INCENP paralogs and collected shortly after hatching for immunostaining using anti-GFP antibody in combination with anti-H3pS28 to detect the localizations of INCENP proteins. To knockdown INCENP paralogs, anesthetized day 4 females were injected with 400 ng/µl dsRNA targeting a 400 ~ 550 bp fragment within the coding region and raised until spawning. Aliquots of spawned oocytes were collected for RT-qPCR to calculate knockdown efficiencies, and remaining oocytes were fertilized with fresh sperm solution to observe phenotypes. For rescue assays, a dsRNA resistant INCENPb-eGFP fusion construct was synthesized by creating numerous silent third-codon point mutations within the dsRNA targeting region (GenScript).
RNA was isolated from 50 oocytes following the manufacturer’s protocol using the RNAqueous-Micro Kit (Ambion). Total RNA (200 ng) was reverse transcribed using M-MLV reverse transcriptase (Life Technologies) with oligo-dT primers. RT-qPCR was performed in 20 µl reactions using cDNA equivalent to 2 ng total RNA, 500 nM gene specific forward and reverse primers and iQTM SYBR_Green Supermix (Bio-Rad). Samples were run on a CFX96 (BioRad). Elongation factor 1-beta expression levels were used as normalization controls. Statistical significance of knockdown efficiencies was assessed by the Student’s T-test.
Funding Statement
This work was supported by a PhD fellowship from the Dept. of Biological Sciences, University of Bergen https://www.uib.no (H.F.) and Norwegian Research Council grants [183690/S10 NFR-FUGE and 133335/V40] https://www.forskningsradet.no/no/Forsiden/1173185591033 to (E.M.T.); Norges Forskningsråd [183690/S10 NFR-FUGE]; Norges Forskningsråd [133335/V40]; Universitetet i Bergen.
Acknowledgments
We thank A. Aasjord and K. N. Nøkling for providing animals from the Oikopleura culture facility. We also thank Ines Drinnenberg, Institut Curie, Paris, for helpful discussions regarding the CCAN.
Author contributions
Conceptualization: H.F., C.C. and E.M.T. Funding acquisition: E.M.T. Supervision: C.C. and E.M.T. Investigation: H.F., M.R., A.M., C.C. and E.M.T. Writing: H.F., M.R., C.C. and E.M.T.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].van Hooff JJE, Tromer E, van Wijk LM, et al. Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 2017;18:1559–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ruchaud S, Carmena M, Earnshaw WC.. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. [DOI] [PubMed] [Google Scholar]
- [3].Carmena M, Wheelock M, Funabiki H, et al. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jeyaprakash AA, Klein UR, Lindner D, et al. Structure of a Survivin–borealin–INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. [DOI] [PubMed] [Google Scholar]
- [5].Kelly AE, Ghenoiu C, Xue JZ, et al. Survivin reads phosphorylated histone H3 Threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang F, Dai J, Daum JR, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yamagishi Y, Honda T, Tanno Y, et al. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. [DOI] [PubMed] [Google Scholar]
- [8].Haase J, Bonner MK, Halas H, et al. Distinct roles of the chromosomal passenger complex in the detection of and response to errors in kinetochore-microtubule attachment. Dev Cell. 2017;42:640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hengeveld RCC, Vromans MJM, Vleugel M, et al. Inner centromere localization of the CPC maintains centromere cohesion and allows mitotic checkpoint silencing. Nat Commun. 2017;8:15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sessa F, Mapelli M, Ciferri C, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–391. [DOI] [PubMed] [Google Scholar]
- [11].Samejima K, Platani M, Wolny M, et al. The inner centromere protein (INCENP) coil is a single α-helix (SAH) domain that binds directly to microtubules and is important for chromosome passenger complex (CPC) localization and function in mitosis. J Biol Chem. 2015;290:21460–21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wheelock MS, Wynne DJ, Tseng BS, et al. Dual recognition of chromatin and microtubules by INCENP is important for mitotic progression. J Cell Biol. 2017;216:925–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lampson MA, Cheeseman IM. Sensing centromere tension: aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goto H, Yasui Y, Nigg EA, et al. Aurora-B phosphorylates histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. [DOI] [PubMed] [Google Scholar]
- [15].Qian J, Lesage B, Beullens M, et al. PP1/Repo-Man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal Aurora B targeting. Curr Biol. 2011;21:766–773. [DOI] [PubMed] [Google Scholar]
- [16].Afonso O, Matos I, Pereira AJ, et al. Feedback control of chromosome separation by a midzone Aurora B gradient. Science. 2014;345:332–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fung SYS, Kitagawa M, Liao P-J, et al. Opposing activities of Aurora B kinase and B56-PP2A phosphatase on MKLP2 determine abscission timing. Curr Biol. 2017;27:78–86. [DOI] [PubMed] [Google Scholar]
- [18].Carmena M, Pinson X, Platani M, et al. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 2012;10:e1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Goto H, Kiyono T, Tomono Y, et al. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat Cell Biol. 2006;8:180–187. [DOI] [PubMed] [Google Scholar]
- [20].Xu Z, Ogawa H, Vagnarelli P, et al. INCENP-Aurora B interactions modulate kinase activity and chromosome passenger complex localization. J Cell Biol. 2009;187:637–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gao S, Giansanti MG, Buttrick GJ, et al. Australin: a chromosomal passenger protein required specifically for Drosophila melanogaster male meiosis. J Cell Biol. 2008;180:521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fernández-Miranda G, Trakala M, Martín J, et al. Genetic disruption of Aurora B uncovers an essential role for Aurora C during early mammalian development. Development. 2011;138:2661–2672. [DOI] [PubMed] [Google Scholar]
- [23].Niedzialkowska E, Wang F, Porebski PJ, et al. Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol Biol Cell. 2012;23:1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sampath SC, Ohi R, Leismann O, et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. [DOI] [PubMed] [Google Scholar]
- [25].Delsuc F, Tsagkogeorga G, Lartillot N, et al. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. [DOI] [PubMed] [Google Scholar]
- [26].Stach T, Winter J, Bouquet J-M, et al. Embryology of a planktonic tunicate reveals traces of sessility. Proc Natl Acad Sci USA. 2008;105:7229–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ganot P, Thompson EM. Patterning through differential endoreduplication in epithelial organogenesis of the chordate, Oikopleura dioica. Dev Biol. 2002;252:59–71. [DOI] [PubMed] [Google Scholar]
- [28].Ganot P, Kallesøe T, Thompson EM. The cytoskeleton organizes germ nuclei with divergent fates and asynchronous cycles in a common cytoplasm during oogenesis in the chordate Oikopleura. Dev Biol. 2007;302:577–590. [DOI] [PubMed] [Google Scholar]
- [29].Ganot P, Bouquet J-M, Kallesøe T, et al. The Oikopleura coenocyst, a unique chordate germ cell permitting rapid, extensive modulation of oocyte production. Dev Biol. 2007;302:591–600. [DOI] [PubMed] [Google Scholar]
- [30].Ganot P, Moosmann-Schulmeister A, Thompson EM. Oocyte selection is concurrent with meiosis resumption in the coenocystic oogenesis of Oikopleura. Dev Biol. 2008;324:266–276. [DOI] [PubMed] [Google Scholar]
- [31].Denoeud F, Henriet S, Mungpakdee S, et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Seo HC, Kube M, Edvardsen RB, et al. Miniature genome in the marine chordate Oikopleura dioica. Science. 2001;294:2506. [DOI] [PubMed] [Google Scholar]
- [33].Campsteijn C, Øvrebø JI, Karlsen BO, et al. Expansion of Cyclin D and CDK1 paralogs in Oikopleura dioica, a chordate employing diverse cell cycle variants. Mol Biol Evol. 2012;29:487–502. [DOI] [PubMed] [Google Scholar]
- [34].Krenn V, Musacchio A. The Aurora B kinase in chromosome bi-orientation and spindle checkpoint signaling. Front Oncol. 2015;5:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Glover DM, Leibowitz MH, McLean DA, et al. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. [DOI] [PubMed] [Google Scholar]
- [36].Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. [DOI] [PubMed] [Google Scholar]
- [37].Feng H, Thompson EM. Specialization of CDK1 and cyclin B paralog functions in a coenocystic mode of oogenic meiosis. Cell Cycle. 2018;17:1425–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].O’Shaughnessy EC, Palani S, Collins JJ, et al. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2010;144:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yao G, Lee TJ, Mori S, et al. A bistable Rb–E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. [DOI] [PubMed] [Google Scholar]
- [40].Pomerening JR, Sontag ED, Ferrell JE. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5:346–351. [DOI] [PubMed] [Google Scholar]
- [41].Greenwald EC, Redden JM, Dodge-Kafka KL, et al. Scaffold state switching amplifies, accelerates, and insulates protein kinase C signaling. J Biol Chem. 2014;289:2353–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci USA. 2000;97:5818–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Resnick TD, Satinover DL, MacIsaac F, et al. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev Cell. 2006;11:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ghenoiu C, Wheelock MS, Funabiki H. Autoinhibition and Polo-dependent multisite phosphorylation restrict activity of the histone H3 kinase Haspin to mitosis. Mol Cell. 2013;52:734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang F, Ulyanova NP, van der Waal MS, et al. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol. 2011;21:1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fuller BG, Lampson MA, Foley EA, et al. Midzone activation of Aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Landino J, Norris SR, Li M, et al. Two mechanisms coordinate the recruitment of the chromosomal passenger complex to the plane of cell division. Mol Biol Cell. 2017;28:3634–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kitagawa M, Fung SYS, Onishi N, et al. Targeting Aurora B to the equatorial cortex by MKlp2 is required for cytokinesis. PLoS One. 2013;8:e64826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kitagawa M, Lee SH. The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front Cell Dev Biol. 2015;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ozlü N, Monigatti F, Renard BY, et al. Binding partner switching on microtubules and Aurora-B in the mitosis to cytokinesis transition. Mol Cell Proteomics. 2010;9:336–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pike T, Brownlow N, Kjaer S, et al. PKCɛ switches Aurora B specificity to exit the abscission checkpoint. Nat Commun. 2016;7:13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kawajiri A, Yasui Y, Goto H, et al. Functional significance of the specific sites phosphorylated in Desmin at cleavage furrow: aurora-B may phosphorylate and regulate type III intermediate filaments during cytokinesis coordinatedly with Rho-kinase. Mol Biol Cell. 2003;14:1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Minoshima Y, Kawashima T, Hirose K, et al. Phosphorylation by Aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell. 2003;4:549–560. [DOI] [PubMed] [Google Scholar]
- [56].Canman JC, Lewellyn L, Laband K, et al. Inhibition of Rac by the GAP activity of Centralspindlin is essential for cytokinesis. Science. 2008;322:1543–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Neef R, Klein UR, Kopajtich R, et al. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr Biol. 2006;16:301–307. [DOI] [PubMed] [Google Scholar]
- [58].Steigemann P, Wurzenberger C, Schmitz MHA, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. [DOI] [PubMed] [Google Scholar]
- [59].Capalbo L, Mela I, Abad MA, et al. Coordinated regulation of the ESCRT-III component CHMP4C by the chromosomal passenger complex and centralspindlin during cytokinesis. Open Biol. 2016;6:160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bouquet J-M, Spriet E, Troedsson C, et al. Culture optimization for the emergent zooplanktonic model organism Oikopleura dioica. J. Plankton Res. 2009; 31: 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Øvrebø JI, Campsteijn C, Kourtesis I, et al. Functional specialization of chordate CDK1 paralogs during oogenic meiosis. Cell Cycle. 2015;14:880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


