Abstract
Pseudomonas aeruginosa is an opportunistic bacterial pathogen that causes various acute and chronic infections. It is intrinsically resistant to a variety of antibiotics. However, production of pyocins during SOS response sensitizes P. aeruginosa to quinolone antibiotics by inducing cell lysis. The polynucleotide phosphorylase (PNPase) is a conserved phosphate-dependent 3′–5′ exonuclease that plays an important role in bacterial response to environmental stresses and pathogenesis by influencing mRNA and small RNA stabilities. Previously, we demonstrated that PNPase controls the type III and type VI secretion systems in P. aeruginosa. In this study, we found that mutation of the PNPase coding gene (pnp) increases the bacterial resistance to ciprofloxacin. Gene expression analyses revealed that the expression of pyocin biosynthesis genes is decreased in the pnp mutant. PrtR, a negative regulator of pyocin biosynthesis genes, is upregulated in the pnp mutant. We further demonstrated that PNPase represses the expression of PrtR on the post-transcriptional level. A fragment containing 43 nucleotides of the 5′ untranslated region was found to be involved in the PNPase mediated regulation of PrtR. Overall, our results reveled a novel layer of regulation on the pyocin biosynthesis by the PNPase in P. aeruginosa.
Keywords: Pseudomonas aeruginosa, polynucleotide phosphorylase, ciprofloxacin resistance, PrtR, pyocins
Introduction
Pseudomonas aeruginosa causes acute and chronic infections in immunocompromised patients (Balasubramanian et al., 2013). Emergence of drug-resistant P. aeruginosa strains greatly increases the difficulty of clinical treatment. Fluoroquinolone antibiotics have been used to treat P. aeruginosa infections (Andriole, 2005; Klodzinska et al., 2016). P. aeruginosa encodes multiple resistant determinants against fluoroquinolone antibiotics, such as multidrug efflux systems and pyocyanin (Subedi et al., 2018; Fan et al., 2019). However, chromosomally encoded pyocin biosynthesis genes increase the bacterial susceptibility to fluoroquinolone antibiotics (Brazas and Hancock, 2005; Sun et al., 2014; Chen et al., 2017). Ninety percent of P. aeruginosa strains produce pyocins, and each P. aeruginosa strain usually produces multiple types of the pyocins (Michel-Briand and Baysse, 2002; Ghequire and De Mot, 2014). Expression of the pyocin biosynthesis genes is activated by PrtN, while a λ CI homologous protein PrtR directly represses the transcription of prtN (Matsui et al., 1993). Genotoxic agents, including fluoroquinolone antibiotics and mitomycin-C, cause DNA damages, leading to the activation of RecA and subsequent SOS response. The activated RecA induces cleavage of PrtR, resulting in derepression of PrtN and production and release of pyocins, which are accompanied by lysis of the producer cells (Penterman et al., 2014).
Polynucleotide phosphorylase (PNPase) is a highly conserved exonuclease that degrades both RNA and ssDNA. In the presence of Mg2+ and inorganic phosphate (Pi), PNPase displays a 3′–5′ exoribonuclease activity. Meanwhile, PNPase can polymerize rNDP into RNA independent of a template. Thus, PNPase plays an important role in RNA metabolism in both prokaryotic and eukaryotic organisms (Cardenas et al., 2009, 2011; Cameron et al., 2018). In addition, in the presence of either Fe3+ or Mn2+ PNPase can polymerize dNDPs into ssDNA without a template. It also possesses a 3′–5′ exodeoxyribonuclease activity (Chou and Singer, 1971; Gillam and Smith, 1974; Beljanski, 1996). PNPase contains two PH domains at the N-terminus, forming a catalytic core and C-terminal RNA binding KH and S1 domains (Bermudez-Cruz et al., 2005; Briani et al., 2007; Fernandez-Ramirez et al., 2010). In addition, PNPase interacts with ribonuclease E, RNA helicase RhlB and enolase in certain species of Gram-negative bacteria, forming a RNA degradosome that plays an important role in mRNA decay (Carpousis, 2007; Nurmohamed et al., 2009). PNPase has been shown to be involved in bacterial responses to environmental stresses (Leszczyniecka et al., 2004; Cameron et al., 2018). In Yersinia and Campylobacter jejuni, PNPase is crucial for the growth at low temperatures (Haddad et al., 2009; Henry et al., 2012). In Escherichia coli and Bacillus subtilis, PNPase protects the bacterium against oxidative stresses mainly by promoting repair of oxidatively damaged DNA (Hayakawa et al., 2001; Cardenas et al., 2009, 2011; Wu et al., 2009) and contributes to bacterial survival upon UV radiation (Cardenas et al., 2009, 2011; Rath et al., 2012). PNPase has also been shown to be involved in the virulence of bacterial pathogens, including Yersinia, Salmonellae, and Helicobacter pylori (Rosenzweig et al., 2007; Hu and Zhu, 2015; Chen et al., 2016; Engman et al., 2016).
Previously, we demonstrated that PNPase is an essential gene in P. aeruginosa. Deletion of the KH and S1 domains results in downregulation of the type III secretion system and upregulation of the type VI secretion system (Chen et al., 2016). However, the role of PNPase in P. aeruginosa response to environmental stresses, such as antibiotics remains unknown. Here in this study, we found that mutation of the pnp increases the bacterial tolerance to fluoroquinolone antibiotics due to downregulation of the pyocin biosynthesis genes. We further demonstrated that the 5′-untranslated region (5′-UTR) of the prtR mRNA is involved in the PNPase mediated translational repression. Therefore, our results revealed a novel regulatory mechanism of pyocin production and the related bacterial resistance against ciprofloxacin.
Materials and Methods
Bacterial Strains, Growth Conditions, Plasmids and Primers
The bacterial strains, plasmids and primers used in this study were listed in Table 1 (Furste et al., 1986; Hoang et al., 1998; Choi and Schweizer, 2006; Sun et al., 2014; Chen et al., 2016, 2017). All bacterial strains were cultured in Luria–Bertani (LB) broth (5 g/L Nacl, 5 g/L yeast extract and 10 g/L tryptone, pH 7.4) at 37∘C with agitation at 200 rpm. All chromosomal gene mutations were generated as described previously (Hoang et al., 1998).
TABLE 1.
Bacterial strains, plasmids and primers used in this study.
| Strain/Plasmid/Primer | Description | Source/Purpose |
| P. aeruginosa | ||
| PAK | Wild type strain of Pseudomonas aeruginosa | David Bradley |
| ΔKH-S1 | PAK with pnp (KH and S1) deletion | Chen et al., 2016 |
| ΔKH-S1 /Tn7T-pnp | PAKΔKH-S1 with pnp inserted on chromosome with mini-Tn7T insertion; | Chen et al., 2016 |
| PAKΔPA0614 | PAK deleted of PA0614 | This study |
| PAKΔPA0629 | PAK deleted of PA0629 | This study |
| PAKΔprtN | PAK deleted of prtN | This study |
| ΔKH-S1ΔPA0614 | PAKΔKH-S1 deleted of PA0614 | This study |
| ΔKH-S1ΔPA0629 | PAKΔKH-S1 deleted of PA0629 | This study |
| ΔKH-S1ΔprtN | PAKΔKH-S1 deleted of prtN | This study |
| PAK/pMMB67EH | PAK containing plasmid pMMB67EH | This study |
| ΔKH-S1/pMMB67EH | PAKΔKH-S1 containing plasmid pMMB67EH | This study |
| PAK/pMMB67EH-prtR | PAK containing plasmid pMMB67EH-prtR | Chen et al., 2017 |
| Plasmid | ||
| pEX18Tc | Gene replacement vector; Tcr, oriT+, sacB+ | Hoang et al., 1998 |
| pUC18T-mini-Tn7T-Tc | mini-Tn7 base vector from insertion into chromosome attTn7 site; Tcr | Choi and Schweizer, 2006 |
| pUC18T-mini-Tn7T-PprtR-lacZ | prtR promoter of PAK fused to promoterless lacZ on pUC18T-mini-Tn7T | Sun et al., 2014 |
| pMMB67EH | Expression vector with tac promoter;Apr | Furste et al., 1986 |
| pUCP20 (no promoter) | Escherichia-Pseudomonas shuttle vector; no promoter; Ampr | This study |
| pUCP20(no promoter) -pRkaraRed(43)-PrtR-His | 6 × His-tagged PrtR driven by the PBAD promoter with 43 bp of the 5′-UTR sequence on pUCP20(no promoter) | This study |
| pUCP20(no promoter) -pRkaraRed(15)-PrtR-His | 6 × His-tagged PrtR driven by the PBAD promoter with 15 bp of the 5′-UTR sequence on pUCP20(no promoter) | This study |
| pUCP20(no promoter) -pRkaraRed(43)-GFP | GFP driven by the PBAD promoter with 43 bp of the 5′-UTR sequence on pUCP20(no promoter) | This study |
| pUCP20(no promoter) -pRkaraRed(15)-GFP | GFP driven by the PBAD promoter with 15 bp of the 5′-UTR sequence on pUCP20(no promoter) | This study |
| Primer | Sequence (5′→3′) | Function |
| PA0636-RT-S | TGGAAGACCCGGCAGAAG | RT-PCR |
| PA0636-RT-AS | CGTTGAGCTTGGACAGATCCT | RT-PCR |
| PA0614-RT-S | CGCTGCCTGCCAAGGA | RT-PCR |
| PA0614-RT-AS | ATCAGTACCCAGAGCGGCATT | RT-PCR |
| PA0629-RT-S | GTGGAGAACCTCAATTACAG | RT-PCR |
| PA0629-RT-AS | TAGGTGTTGTCGGCAATC | RT-PCR |
| prtR-RT-S | GATGCGCAACCTGAAGCA | RT-PCR |
| prtR-RT-AS | TGAATGGTGTTCTGCGAAACC | RT-PCR |
| prtN-RT-S | CGACGATAGCCACAAG | RT-PCR |
| prtN-RT-AS | GGATGCGATGCTGTC | RT-PCR |
| lexA-RT-S | AATCCCGCCTTCTTCAAT | RT-PCR |
| lexA-RT-AS | AATGCCGATGTCCTTCAT | RT-PCR |
| recA-RT-S | ATATCAAGAACGCCAACT | RT-PCR |
| recA-RT-AS | TAGAACTTCAGTGCGTTA | RT-PCR |
| BamHI-PprtR-lacZ-S# | CGCGGATCC GAGCCAGGACCAGTTCGTTGGC | Transcriptional fusion |
| HindIII-lacZ-AS | ATTATAAAGCTT TTATTTTTGACACCAGACCAACTGG | Transcriptional fusion |
| SacI-PBAD-S | CCAAGAGCTC TTATGACAACTTGACGGC | Translational fusion |
| HindIII-prtR-AS | ATTATAAAGCTT TCAGTGGTGGTGGTGGTGGTGACCTCCCC GCACCAGGGACGGGCCGC | Translational fusion |
| XhoI- prtR(43)-GFP S | CCGCTCGAG TAGGCTCTTTACAGAAAATCCATCGGTCTGTAGA TTGCCGAGCATGAGTAAAGGAGAAGAACTTTTCACTG | Translational fusion |
| XhoI- prtR(15)-GFP S | CCGCTCGAG TGTAGATTGCCGAGCATGAGTAAAGGAGAAGAA CTTTTCACTG | Translational fusion |
| HindIII –GFP-AS | CCCAAGCTT TTATTTGTATAGTTCATCCATGCCATG | Translational fusion |
#The endonuclease cutting sites are underlined.
Minimum Inhibitory Concentration and Survival Assay
Minimum inhibitory concentrations were determined by the twofold serial dilution method as described previously (Fan et al., 2019). Overnight bacterial cultures were diluted 1:50–1:100 in LB and cultured at 37∘C until the OD600 reached 0.8–1.0. The bacterial concentration was adjusted to 1 × 105 CFU/ml and 200 μl of the bacteria was added to each well of a 96-well plate (Corning). The plate was incubated for 24 h at 37∘C without agitation. The Minimum inhibitory concentration was recorded as the lowest concentration of antibiotic that inhibited visible growth. For the survival assay, bacteria were grown to an OD600 of 1.0 at 37∘C. Then the bacteria were treated with ciprofloxacin at indicated concentrations at 37∘C with agitation at 200 rpm. The numbers of live cells before and after antibiotic treatment were determined by serial dilution and plating assay.
RNA Extraction, Reverse Transcription, and Quantitative RT-PCR
Overnight bacterial cultures were diluted 1:50–1:100 into fresh LB with and without 0.016 μg/ml ciprofloxacin and grown to an OD600 of 0.8–1.0. Total RNA was isolated with an RNeasy Mini kit (Tiangen Biotech, Beijing, China) and cDNA was synthesized with a PrimeScript Reverse Transcriptase (TaKaRa, Dalian, China). 1 μg RNA was used for reverse transcription. In the quantitative RT-PCR experiment, the cDNA was mixed with specific forward and reverse primers and the SYBR Premix Ex TaqTM II (TaKaRa). The CFX Connect Real-Time system (Bio-Rad, United States) was used to perform the quantitative RT-PCR. rpsL, which encodes the 30S ribosomal protein S12 was used as an internal control.
Western Blotting
Samples from the same number of bacterial cells were loaded onto 10 or 12% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel. Then the proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane and probed with a GFP antibody or a mouse monoclonal antibody against the 6 × His tag (1:2000; Cell Signaling Technology, United States) at room temperature for 1–2 h or overnight at 4∘C. Then the PVDF membrane was washed with 1 × phosphate-buffered saline (1 × PBS, 5.4 mM KCl, 20 mM Na2HPO4, 274 mM NaCl, 4 mM KH2PO4, pH 7.4) containing 2% 24 times. Next, the PVDF membrane was incubated with an anti-rabbit IgG (1: 2,000; Promega, United States) at room temperature for 1.5 h. Signals were detected by an ECL Plus kit (Millipore). The signals were visualized by a Bio-Rad molecular imager (ChemiDocXRS). The RNA polymerase α subunit RpoA was used as a loading control (with an antibody from Biolegend).
Promoter Activity Assay
The promoter region of the prtR gene was amplified by PCR with the primers shown in Table 1. The PCR product was fused with the coding sequence of lacZ. The PprtR-lacZ fusion was inserted into the chromosome of P. aeruginosa strains via a miniTn7 vector (Choi and Schweizer, 2006). To measure the expression level of LacZ, the bacteria were grown to an OD600 of 0.5, and then treated with ciprofloxacin at indicated concentrations for 3 h. The β-galactosidase activities were measured as described previously (Weng et al., 2016). Briefly, each sample (0.5 ml bacteria) was collected by centrifugation and resuspended in 1.5 ml Z buffer (60 mM NaH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 10 mM KCl and 50 mM β-mercaptoethanol, pH 7.0). 0.5 ml of the suspension was mixed with 10 μl 0.1% SDS (BBI Life Sciences, Shanghai, China) and 10 μl chloroform (BBI Life Sciences, Shanghai, China), and then vortexed for 10–15 s. The remaining 1 ml was used for OD600 measurement. 100 μl ONPG (40 mg/ml; Sigma, United States) was added to each sample, followed by incubation at 37∘C. When the color turned into light yellow, 0.5 ml 1 M Na2CO3 was added to the mixture to stop the reaction. OD420 was measured, and the time was recorded. The β-galactosidase activity (Miller units) was calculated as (1000 × OD420)/(T × V × OD600). T, reaction time (minute); V, bacteria volume (ml).
Results
PNPase Influences the Bacterial Resistance to Ciprofloxacin
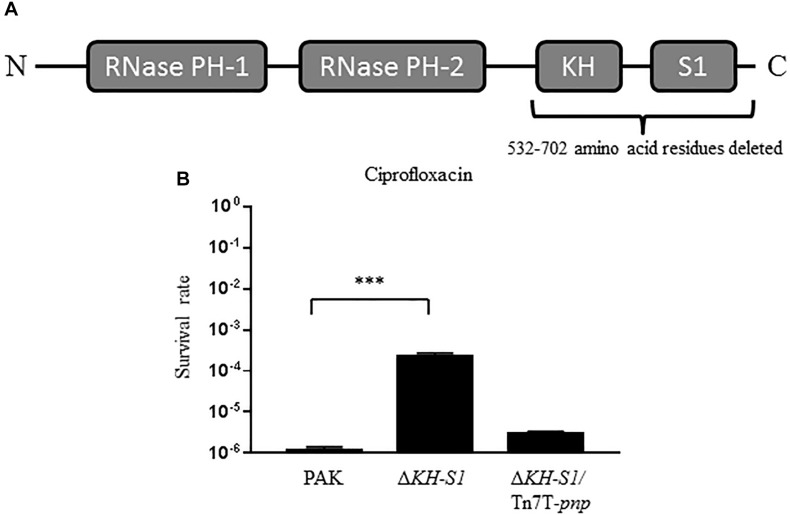
To test the role of PNPase in antibiotic resistance of P. aeruginosa, we determined the MICs of various antibiotics against wild type PAK and an isogenic mutant with the deletion of the KH-S1 domains of PNPase (ΔKH-S1) (Figure 1A; Chen et al., 2016). The two strains displayed similar levels of resistance (MICs) to most of the tested antibiotics, including erythromycin, carbenicillin and gentamicin. However, the MICs of ciprofloxacin and ofloxacin were increased four and two fold in the ΔKH-S1 mutant, respectively (Table 2). Complementation with a pnp gene restored the bacterial susceptibility (Table 2). Consistent with the MIC test results, in the presence of 0.16 μg/ml (1 × MIC) ciprofloxacin, deletion of the KH-S1 domains increased the bacterial survival rate by approximately 100-fold, which was restored by complementation with a pnp gene (Figure 1B).
FIGURE 1.
Bacterial survival rates under the treatment of ciprofloxacin. (A) Domains of the PNPase of Pseudomonas aeruginosa. (B) PAK, ΔKH-S1 mutant and the complemented strain (ΔKH-S1/Tn7T-pnp) were grown to an OD600 of 1.0 at 37∘C and treated with 0.16 μg/ml ciprofloxacin for 6 h. At indicated time points, the bacterial survival rates were determined by serial dilution and plating assay. ∗∗∗p < 0.001 by Student’s t-test.
TABLE 2.
Bacterial susceptibilities to antibiotics.
|
MIC (μg/ml) |
|||||
| Strain | Ciprofloxacin | Ofloxacin | Carbenicillin | Erythromycin | Gentamicin |
| PAK | 0.16 | 1.5 | 150 | 125 | 0.625 |
| ΔKH-S1 | 0.64 | 3 | 150 | 125 | 0.625 |
| ΔKH-S1/Tn7T-pnp | 0.16 | 1.5 | 150 | 125 | – |
“–” Indicates that the complemented strain is resistant to gentamicin due to the miniTn7T insertion.
Downregulation of Pyocin Biosynthesis Genes Contributes to the Increased Resistance to Ciprofloxacin in the ΔKH-S1 Mutant
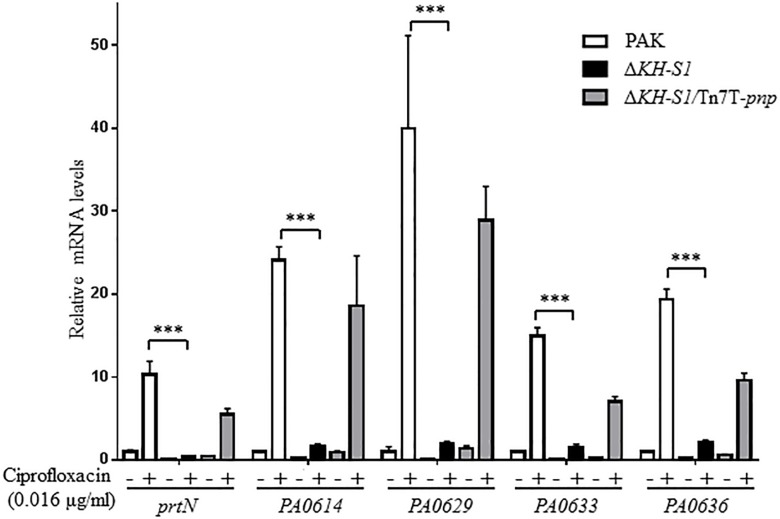
In our previous transcriptome analysis of the ΔKH-S1 mutant, no alternation was observed on the expression of the multidrug efflux system genes, whereas the pyocin biosynthesis genes were downregulated (Chen et al., 2016). Due to the role of pyocins in the bacterial susceptibility to ciprofloxacin (Brazas and Hancock, 2005; Sun et al., 2014; Chen et al., 2017), we verified the expression levels of the R-type (PA0614) and F-type pyocins (PA0629, PA0633, and PA0636) genes by real time PCR (Nakayama et al., 2000; Michel-Briand and Baysse, 2002). Due to the difference in the MICs of ciprofloxacin to wild type PAK and the ΔKH-S1 mutant, we treated both strains with 0.016 μg/ml ciprofloxacin (1/10 MIC to PAK), which did not affect the growth of both strains. In the presence or absence of ciprofloxacin, the mRNA levels of the pyocin biosynthesis genes were lower in the ΔKH-S1 mutant than those in wild type PAK. Complementation with a pnp gene restored the mRNA levels in the ΔKH-S1 mutant (Figure 2). In PAK, the resistance to ciprofloxacin was increased upon deletion of prtN, PA0614, and PA0629, which encode the transcriptional activator for the pyocin biosynthesis genes, a holin- and a lysozyme-like protein, respectively (Table 3). However, deletion of those genes in the ΔKH-S1 mutant did not further increase the resistant level (Table 3), indicating that the repression of pyocin biosynthesis genes might result in the increased resistance to ciprofloxacin.
FIGURE 2.
Expression levels of pyocin biosynthesis genes in the ΔKH-S1 mutant. PAK, ΔKH-S1 and the complemented strain were grown to an OD600 of 0.8–1.0 at 37∘C with or without 0.016 μg/ml ciprofloxacin, followed by RNA extraction. The mRNA levels of prtN, PA0614, PA0629, PA0633, and PA0636 were determined by real-time PCR with rpsL as the internal control. ∗∗∗p < 0.001 by Student’s t-test.
TABLE 3.
Bacterial susceptibilities to ciprofloxacin.
| Strain | MIC (μg/ml) |
| PAK | 0.16 |
| ΔPA0614 | 0.32 |
| ΔPA0629 | 0.32 |
| ΔprtN | 0.32 |
| ΔKH-S1 | 0.64 |
| ΔKH-S1ΔPA0614 | 0.64 |
| ΔKH-S1ΔPA0629 | 0.64 |
| ΔKH-S1ΔprtN | 0.64 |
| PAK/pMMB67EH | 0.16 |
| PAK/pMMB67EH-prtR-His | 0.64 |
| ΔKH-S1/pMMB67EH | 0.64 |
The PrtR Protein Level Is Increased in the ΔKH-S1 Mutant
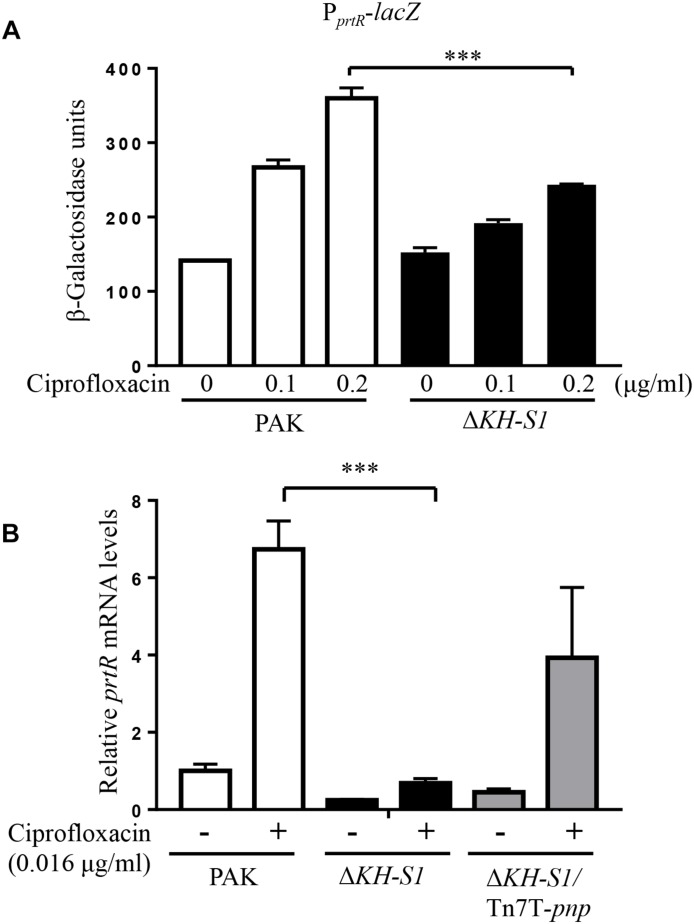
PrtR directly represses the transcription of prtN that encodes the transcriptional activator of the pyocin biosynthesis genes (Matsui et al., 1993). Since the mRNA level of prtN was lower in the ΔKH-S1 mutant (Figure 2), we suspected that the PrtR protein level might be higher in the ΔKH-S1 mutant. To test the protein level of PrtR, we utilized a C-terminal 6 × His-tagged prtR driven by its native promoter (designated as PprtR-prtR-His) (Figure 3A; Sun et al., 2014). Indeed, the PrtR-His level was higher in the ΔKH-S1 mutant than that in PAK in the presence or absence of ciprofloxacin (Figure 3B). In addition, overexpression of prtR in PAK increased the MIC of ciprofloxacin by fourfold and enhanced the survival rate in the presence of ciprofloxacin to the similar level as that of the ΔKH-S1 mutant (Figure 3C and Table 3). These results suggest that the increased resistance to ciprofloxacin is likely due to the higher protein level of PrtR in the ΔKH-S1 mutant.
FIGURE 3.
Expression of PrtR in the ΔKH-S1 mutant. (A) Fragments of the prtR promoter region fused with the prtR-His or a promoterless lacZ gene. (B) Protein levels of PrtR-His in PAK and the ΔKH-S1 mutant carrying the PprtR-prtR-His on the bacterial chromosome. The bacterial cells were grown to an OD600 of 1.0, and then incubated with or without 0.06 μg/ml ciprofloxacin for 1 h. The PrtR-His levels were determined by Western blotting. RpoA was used as the loading control. (C) PAK containing an empty vector or the prtR overexpression plasmid was grown to an OD600 of 1.0 and treated with 0.16 μg/ml ciprofloxacin for 6 h. At indicated time points, the bacterial survival rate was determined by serial dilution and plating. ∗∗∗p < 0.001 by Student’s t-test.
PNPase Affects the Expression of PrtR at the Post-transcription Level Through Its 5′-UTR
To understand the mechanism of the increased PrtR level, we examined the promoter activity by utilizing a transcriptional fusion of lacZ reporter gene with the promoter of prtR (PprtR-lacZ). The presence of ciprofloxacin induced the lacZ expression in wild type PAK, however, the lacZ expression levels in the ΔKH-S1 mutant were lower than those in PAK in the presence of the same concentrations of ciprofloxacin (Figure 4A). Consistent with the above results, the mRNA level of prtR was lower in the ΔKH-S1 mutant (Figure 4B), which might be due to an auto-repression of PrtR (Sun et al., 2014). Nevertheless, this result indicates that the upregulation of PrtR in the ΔKH-S1 mutant might be mediated through a post-transcriptional mechanism.
FIGURE 4.
The promoter activity and mRNA level of prtR in the ΔKH-S1 mutant. (A) Expression of PprtR-lacZ in PAK and the ΔKH-S1 mutant. The bacteria were grown to an OD600 of 0.5, and then treated with ciprofloxacin at indicated concentrations for 3 h, followed by the β–galactosidase assay. ∗∗∗p < 0.001 by Student’s t-test. (B) PAK, ΔKH-S1 and the complemented strain were grown to an OD600 of 0.8–1.0 at 37∘C with or without 0.016 μg/ml ciprofloxacin. The mRNA levels of prtR were determined by real-time PCR with rpsL as the internal control. ∗∗∗p < 0.001 by Student’s t-test.
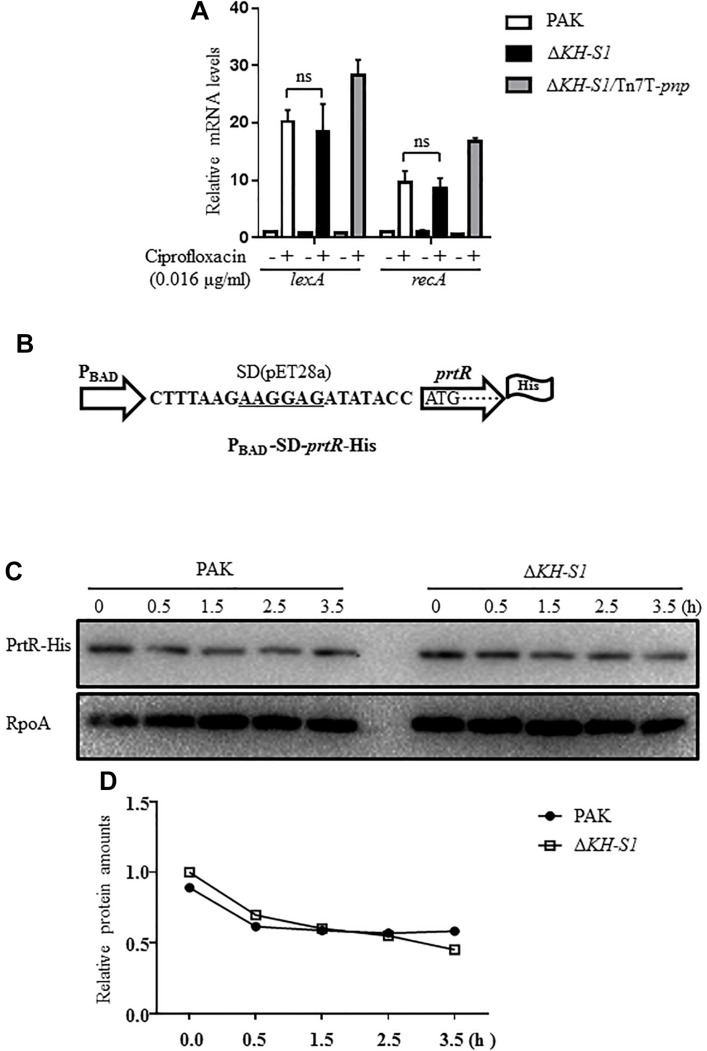
Previous studies demonstrated that the stability of PrtR is regulated by RecA in response to genotoxic agents (Sun et al., 2014). Treatment with ciprofloxacin induced similar expression levels of recA and lexA in the ΔKH-S1 mutant and PAK, indicating a similar level of SOS response (Figure 5A). To examine the PrtR protein stability, we constructed a C-terminal 6 × His-tagged PrtR driven by an inducible PBAD promoter with an exogenous ribosome binding site from the vector pET28a, resulting in PBAD-SD-prtR-His (Figure 5B). In the absence of ciprofloxacin, the levels of the PrtR-His were similar in the ΔKH-S1 mutant and PAK. Treatment with ciprofloxacin resulted in a similar degradation rate of the PrtR-His in both strains (Figures 5C,D).
FIGURE 5.
PrtR protein stabilities in PAK and the ΔKH-S1 mutant. (A) PAK, ΔKH-S1 and the complemented strain were grown to an OD600 of 0.8–1.0 at 37∘C with or without 0.016 μg/ml ciprofloxacin. The mRNA levels of lexA and recA were determined by real-time PCR with rpsL as the internal control. ns, not significant by Student’s t-test. (B) The C-terminal 6 × His-tagged prtR is driven by an inducible PBAD promoter with an exogenous ribosome binding site (designated as PBAD-SD-prtR-His). The ribosome binding sequence was underlined. (C,D) Strains carrying the PBAD-SD-prtR-His were grown to an OD600 of 0.6–0.8 at 37∘C, followed by induction with 0.2% arabinose for 1.5 h. Then, 500 μg/ml chloramphenicol and 0.016 μg/ml ciprofloxacin were added to the medium. At the indicated time points, bacterial cells of each strain were collected and the levels of PrtR-His were determined by Western blotting. RpoA was used as the loading control. The relative intensity of each band was quantified by ImageJ.
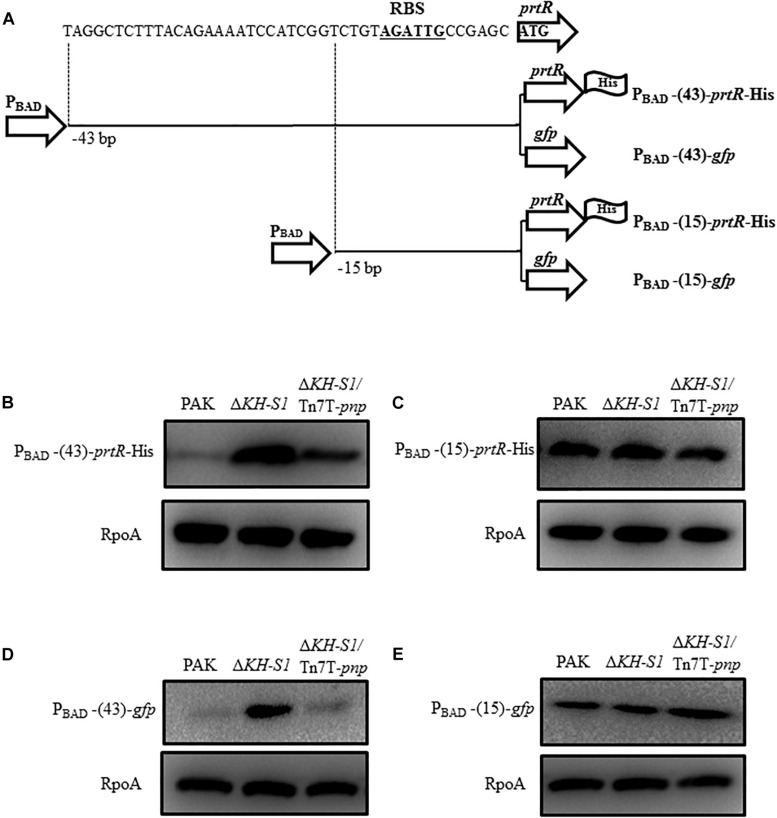
We then examined whether the translation of the prtR mRNA is affected in the ΔKH-S1 mutant. Since the 5′ untranslated region (5′-UTR) of a mRNA is usually involved in the translational regulation, we constructed a 6 × His-tagged prtR driven by an exogenous PBAD promoter with 43 bp of the prtR 5′-UTR sequence (Figure 6A). The translation of the PrtR was higher in the ΔKH-S1 mutant (Figure 6B). To identify the region involved in the post-transcriptional regulation, we reduced the 5′-UTR sequence to 15 bp, resulting in PBAD-15-prtR-His (Figure 6A). From this construct, similar levels of PrtR-His were observed in the ΔKH-S1 mutant and wild type PAK (Figure 6C). As the coding region might be involved in the translational regulation, we replaced the prtR coding sequence with a gfp gene, resulted in PBAD-43-gfp and PBAD-15-gfp, respectively (Figure 6A). Fusion with the 43 bp 5′-UTR of prtR resulted in higher GFP level in the ΔKH-S1 mutant, which was restored by complementation with a pnp gene (Figure 6D). However, reduction of the 5′-UTR to 15 bp resulted in similar levels of GFP (Figure 6E). These results suggest that the 5′-UTR of the prtR mRNA might be involved in the PNPase mediated post-transcriptional regulation of PrtR.
FIGURE 6.
Translational regulation of prtR by the PNPase. (A) Structures of the 6 × His-tagged prtR fusions. PBAD-43-prtR-His and PBAD-15-prtR-His represent 6 × His-tagged prtR driven by the PBAD promoter with 43 and 15 bp of the 5′-UTR sequence of the prtR gene, respectively. The prtR open reading frame was replaced by a gfp gene, resulting in PBAD-43-gfp and PBAD-15-gfp. The potential ribosome binding site (RBS) was shown in bold underlined letters. Strains containing the prtR-His or gfp expression plasmid were grown to an OD600 of 1.0 and then induced with 0.2% arabinose for 1.5 h. Protein levels of PrtR (B,C) and GFP (D,E) were determined by Western blotting. RpoA was used as the loading control.
Discussion
In this study, we found that deletion of the KH-S1 domains of the PNPase increased the bacterial resistance to fluoroquinolone antibiotics. We further demonstrated that the PrtR level is increased in the ΔKH-S1 mutant, which reduces the PrtN expression, resulting in downregulation of the pyocin biosynthesis genes in the presence of ciprofloxacin.
The PNPase is a conserved exoribonuclease that degrades single stranded RNA. It contains two N-terminal PH domains that possess the ribonuclease activity, and C-terminal KH and S1 domains that are involved in the binding of RNAs. The PNPase plays an important role in the maturation of rRNAs and tRNAs. Besides, the PNPase has been shown to control gene expression through sRNAs. In Salmonella typhimurium, Hfq independent sRNAs CsrB, CsrC, and CopA are initially cleaved by RNase E, followed by degradation by PNPase (Viegas et al., 2007). In E. coli, PNPase degrades the sRNAs SgrS, GlmY, MicA, and RyhB when they are not bond to Hfq (Andrade et al., 2012). Meanwhile, PNPase also increases the stability of certain Hfq-bond sRNAs (Bandyra et al., 2016). For instance, deletion of pnp in E. coli resulted in reduced level of ArcZ, a negative regulator of mutS. Consequently, upregulation of mutS in the pnp mutant decreases bacterial spontaneous mutation rate (Chen and Gottesman, 2017).
Previously, we demonstrated that PNPase regulates type VI secretion system through degradation of the sRNAs RsmY and RsmZ (Chen et al., 2016). In this study, we found that a 43-nucleotide 5′-UTR of the prtR mRNA is required for the PNPase mediated translational repression. Reduction of the 5′-UTR to 15-nucleotide resulted in the similar levels of the PrtR protein in the ΔKH-S1 mutant and wild type strain. The 5′-UTR might control gene expression through several mechanisms. For example, formation of a hairpin structure might block the ribosome binding site. PNPase might affect the secondary structure by recruiting an endonuclease. Another possibility is that a sRNA might anneal to the 5′-UTR, which alters the secondary structure or directly blocks the ribosome binding site. In addition, PNPase might directly bind to an mRNA though its KH-S1 domains, which affects the translation. To examine whether PNPase can directly bind to the 5′-UTR of the prtR mRNA, we performed an RNA electrophoretic mobility shift assay. However, no interaction was observed (data not show). It might be possible that another protein is required to facilitate the interaction. Further studies are needed to elucidate the regulatory mechanism.
Pyocins are chromosomally encoded bacteriocins produced by most of P. aeruginosa strains. Production and release of pyocins under environmental stresses such as the presence of genotoxic agents might provide an advantage in the competition against other bacteria (Michel-Briand and Baysse, 2002). A recent study revealed that R-type pyocins play an important role in the competition among various P. aeruginosa strains during the infection of cystic fibrosis patients (Oluyombo et al., 2019). In addition, when pyocins are released through cell lysis, the liberated chromosomal DNA and other components function as the matrix for biofilm formation (Turnbull et al., 2016). However, for the individual pyocins producer cells, the release of pyocins leads to cell death. Therefore, the production of pyocins should be under a tight control. Our study here revealed a novel post-transcriptional regulation on the key regulator PrtR. Further studies are needed to elucidate the molecular details of the regulatory mechanism and the signaling pathway.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
ZF, WW, and SJ conceived and designed the experiments. ZF, HC, ML, XP, WF, HR, and RC performed the experiments. YJ, WW, FB, ZC, and SJ analyzed the data. ZF, WW, and SJ wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (41831287, 31670130, 81670766, 31870130, and 31600110), Science and Technology Program of Sichuan Province (2018JZ0069), Science and Technology Committee of Tianjin (17JCQNJC09200), the “Fundamental Research Funds for the Central Universities,” Nankai University (63191521), and the Ph.D. Candidate Research Innovation Fund of Nankai University.
References
- Andrade J. M., Pobre V., Matos A. M., Arraiano C. M. (2012). The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18 844–855. 10.1261/rna.029413.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriole V. T. (2005). The quinolones: past, present, and future. Clin. Infect. Dis. 41(Suppl. 2), S113–S119. 10.1086/428051 [DOI] [PubMed] [Google Scholar]
- Balasubramanian D., Schneper L., Kumari H., Mathee K. (2013). A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 41 1–20. 10.1093/nar/gks1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyra K. J., Sinha D., Syrjanen J., Luisi B. F., De Lay N. R. (2016). The ribonuclease polynucleotide phosphorylase can interact with small regulatory RNAs in both protective and degradative modes. RNA 22 360–372. 10.1261/rna.052886.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beljanski M. (1996). De Novo synthesis of DNA-like molecules by polynucleotide phosphorylase in vitro. J. Mol. Evol. 42 493–499. [DOI] [PubMed] [Google Scholar]
- Bermudez-Cruz R. M., Fernandez-Ramirez F., Kameyama-Kawabe L., Montanez C. (2005). Conserved domains in polynucleotide phosphorylase among eubacteria. Biochimie 87 737–745. 10.1016/j.biochi.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Brazas M. D., Hancock R. E. (2005). Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49 3222–3227. 10.1128/aac.49.8.3222-3227.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briani F., Del Favero M., Capizzuto R., Consonni C., Zangrossi S., Greco C., et al. (2007). Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie 89 145–157. 10.1016/j.biochi.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Cameron T. A., Matz L. M., De Lay N. R. (2018). Polynucleotide phosphorylase: not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet. 14:e1007654. 10.1371/journal.pgen.1007654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas P. P., Carrasco B., Sanchez H., Deikus G., Bechhofer D. H., Alonso J. C. (2009). Bacillus subtilis polynucleotide phosphorylase 3′-to-5′ DNase activity is involved in DNA repair. Nucleic Acids Res. 37 4157–4169. 10.1093/nar/gkp314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas P. P., Carzaniga T., Zangrossi S., Briani F., Garcia-Tirado E., Deho G., et al. (2011). Polynucleotide phosphorylase exonuclease and polymerase activities on single-stranded DNA ends are modulated by RecN, SsbA and RecA proteins. Nucleic Acids Res. 39 9250–9261. 10.1093/nar/gkr635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A. J. (2007). The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61 71–87. 10.1146/annurev.micro.61.080706.093440 [DOI] [PubMed] [Google Scholar]
- Chen F., Chen G., Liu Y., Jin Y., Cheng Z., Liu Y., et al. (2017). Pseudomonas aeruginosa oligoribonuclease contributes to tolerance to ciprofloxacin by regulating pyocin biosynthesis. Antimicrob. Agents Chemother. 61:e2256-16. 10.1128/aac.02256-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Gottesman S. (2017). Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev. 31 1382–1395. 10.1101/gad.302547.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Weng Y., Zhu F., Jin Y., Liu C., Pan X., et al. (2016). Polynucleotide phosphorylase regulates multiple virulence factors and the stabilities of small RNAs RsmY/Z in Pseudomonas aeruginosa. Front. Microbiol. 7:247. 10.3389/fmicb.2016.00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H., Schweizer H. P. (2006). mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1 153–161. 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
- Chou J. Y., Singer M. F. (1971). Deoxyadenosine diphosphate as a substrate and inhibitor of polynucleotide phosphorylase of Micrococcus luteus. II. Inhibition of the initiation of adenosine diphosphate polymerization by deoxyadenosine diphosphate. J. Biol. Chem. 246 7497–7504. [PubMed] [Google Scholar]
- Engman J., Negrea A., Sigurlasdottir S., Georg M., Eriksson J., Eriksson O. S., et al. (2016). Neisseria meningitidis polynucleotide phosphorylase affects aggregation, adhesion, and virulence. Infect. Immun. 84 1501–1513. 10.1128/iai.01463-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Xu C., Pan X., Dong Y., Ren H., Jin Y., et al. (2019). Mechanisms of RsaL mediated tolerance to ciprofloxacin and carbenicillin in Pseudomonas aeruginosa. Curr. Genet. 65 213–222. 10.1007/s00294-018-0863-3 [DOI] [PubMed] [Google Scholar]
- Fernandez-Ramirez F., Bermudez-Cruz R. M., Montanez C. (2010). Nucleic acid and protein factors involved in Escherichia coli polynucleotide phosphorylase function on RNA. Biochimie 92 445–454. 10.1016/j.biochi.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Furste J. P., Pansegrau W., Frank R., Blocker H., Scholz P., Bagdasarian M., et al. (1986). Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48 119–131. 10.1016/0378-1119(86)90358-6 [DOI] [PubMed] [Google Scholar]
- Ghequire M. G., De Mot R. (2014). Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol. Rev. 38 523–568. 10.1111/1574-6976.12079 [DOI] [PubMed] [Google Scholar]
- Gillam S., Smith M. (1974). Enzymatic synthesis of deoxyribo-oligonucleotides of defined sequence. Properties of the enzyme. Nucleic Acids Res. 1 1631–1647. 10.1093/nar/1.12.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad N., Burns C. M., Bolla J. M., Prevost H., Federighi M., Drider D., et al. (2009). Long-term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity. Appl. Environ. Microbiol. 75 7310–7318. 10.1128/aem.01366-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa H., Kuwano M., Sekiguchi M. (2001). Specific binding of 8-oxoguanine-containing RNA to polynucleotide phosphorylase protein. Biochemistry 40 9977–9982. 10.1021/bi010595q [DOI] [PubMed] [Google Scholar]
- Henry A., Shanks J., van Hoof A., Rosenzweig J. A. (2012). The Yersinia pseudotuberculosis degradosome is required for oxidative stress, while its PNPase subunit plays a degradosome-independent role in cold growth. FEMS Microbiol. Lett. 336 139–147. 10.1111/j.1574-6968.12000.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212 77–86. 10.1016/s0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Hu J., Zhu M. J. (2015). Defects in polynucleotide phosphorylase impairs virulence in Escherichia coli O157:H7. Front. Microbiol. 6:806. 10.3389/fmicb.2015.00806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodzinska S. N., Priemel P. A., Rades T., Morck Nielsen H. (2016). Inhalable antimicrobials for treatment of bacterial biofilm-associated sinusitis in cystic fibrosis patients: challenges and drug delivery approaches. Int. J. Mol. Sci. 17:E1688. 10.3390/ijms17101688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M., DeSalle R., Kang D. C., Fisher P. B. (2004). The origin of polynucleotide phosphorylase domains. Mol. Phylogenet. Evol. 31 123–130. 10.1016/j.ympev.2003.07.012 [DOI] [PubMed] [Google Scholar]
- Matsui H., Sano Y., Ishihara H., Shinomiya T. (1993). Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J. Bacteriol. 175 1257–1263. 10.1128/jb.175.5.1257-1263.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel-Briand Y., Baysse C. (2002). The pyocins of Pseudomonas aeruginosa. Biochimie 84 499–510. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Takashima K., Ishihara H., Shinomiya T., Kageyama M., Kanaya S., et al. (2000). The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38 213–231. [DOI] [PubMed] [Google Scholar]
- Nurmohamed S., Vaidialingam B., Callaghan A. J., Luisi B. F. (2009). Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J. Mol. Biol. 389 17–33. 10.1016/j.jmb.2009.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluyombo O., Penfold C. N., Diggle S. P. (2019). Competition in biofilms between cystic fibrosis isolates of Pseudomonas aeruginosa is shaped by R-Pyocins. mBio 10:e01828-18. 10.1128/mBio.01828-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J., Singh P. K., Walker G. C. (2014). Biological cost of pyocin production during the SOS response in Pseudomonas aeruginosa. J. Bacteriol. 196 3351–3359. 10.1128/jb.01889-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D., Mangoli S. H., Pagedar A. R., Jawali N. (2012). Involvement of pnp in survival of UV radiation in Escherichia coli K-12. Microbiology 158(Pt 5), 1196–1205. 10.1099/mic.0.056309-0 [DOI] [PubMed] [Google Scholar]
- Rosenzweig J. A., Chromy B., Echeverry A., Yang J., Adkins B., Plano G. V., et al. (2007). Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett. 270 255–264. 10.1111/j.1574-6968.2007.00689.x [DOI] [PubMed] [Google Scholar]
- Subedi D., Vijay A. K., Willcox M. (2018). Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: an ocular perspective. Clin. Exp. Optom. 101 162–171. 10.1111/cxo.12621 [DOI] [PubMed] [Google Scholar]
- Sun Z., Shi J., Liu C., Jin Y., Li K., Chen R., et al. (2014). PrtR homeostasis contributes to Pseudomonas aeruginosa pathogenesis and resistance against ciprofloxacin. Infect. Immun. 82 1638–1647. 10.1128/iai.01388-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull L., Toyofuku M., Hynen A. L., Kurosawa M., Pessi G., Petty N. K., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7:11220. 10.1038/ncomms11220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas S. C., Pfeiffer V., Sittka A., Silva I. J., Vogel J., Arraiano C. M. (2007). Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 35 7651–7664. 10.1093/nar/gkm916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Chen F., Liu Y., Zhao Q., Chen R., Pan X., et al. (2016). Pseudomonas aeruginosa enolase influences bacterial tolerance to oxidative stresses and virulence. Front. Microbiol. 7:1999. 10.3389/fmicb.2016.01999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Jiang Z., Liu M., Gong X., Wu S., Burns C. M., et al. (2009). Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry 48 2012–2020. 10.1021/bi801752p [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the manuscript and/or the supplementary files.