Phosphorylation of substrate proteins by kinases regulates signaling pathways and cellular mechanisms. Aberrant signaling is a hallmark of cancer. We investigated quantitative associations between kinase-substrate pairs of more than 30 thousand phosphorylation sites in breast tumors and xenografts, finding auto-phosphorylating kinases and kinase-substrate pairs associated with specific cancer subtypes, druggable targets, and ones that show clinically-related or immune signatures. Our study connects driver kinases to their signaling effects toward informing rational targeted treatments of individual breast tumors.
Keywords: Breast cancer, Phosphorylation, Signaling Molecules*, Kinases*, Drug targets*, Mass Spectrometry, Bioinformatics
Graphical Abstract
Highlights
Our search identifies 2,134 kinase-substrate phosphosite pairs in breast cancer.
CDKs and MAPKs are dominant regulators of trans substrate-phosphorylation.
Druggability, outcomes, and immune signatures related to kinase-substrates.
Experimentally validated activated phosphosites of ERBB2, EIF4EBP1, and EGFR.
Abstract
Aberrant phospho-signaling is a hallmark of cancer. We investigated kinase-substrate regulation of 33,239 phosphorylation sites (phosphosites) in 77 breast tumors and 24 breast cancer xenografts. Our search discovered 2134 quantitatively correlated kinase-phosphosite pairs, enriching for and extending experimental or binding-motif predictions. Among the 91 kinases with auto-phosphorylation, elevated EGFR, ERBB2, PRKG1, and WNK1 phosphosignaling were enriched in basal, HER2-E, Luminal A, and Luminal B breast cancers, respectively, revealing subtype-specific regulation. CDKs, MAPKs, and ataxia-telangiectasia proteins were dominant, master regulators of substrate-phosphorylation, whose activities are not captured by genomic evidence. We unveiled phospho-signaling and druggable targets from 113 kinase-substrate pairs and cascades downstream of kinases, including AKT1, BRAF and EGFR. We further identified kinase-substrate-pairs associated with clinical or immune signatures and experimentally validated activated phosphosites of ERBB2, EIF4EBP1, and EGFR. Overall, kinase-substrate regulation revealed by the largest unbiased global phosphorylation data to date connects driver events to their signaling effects.
Mutations and alterations in cancer dysregulate kinases and signaling cascades. Large-scale studies of breast cancer have discovered drivers, with genomic and expression changes, in kinases of the PI3K/Akt signaling and TP53/RB signaling/cell-cycle checkpoint pathways (1, 2). However, genomic findings provide only indirect inference of phosphorylation activity. Further, large-scale proteomic studies using reverse phase protein array (RPPA)1 are limited to coverages of ∼200 proteins and phosphoproteins with available antibodies (3–5). The impact of candidate driver events on direct signaling has therefore been seldomly explored in the corresponding tumors. Although functional experiments in in vivo systems or model organisms have enabled controlled assessment of downstream effects, they must be complemented by in vivo observations that account for the molecular complexity of each tumor.
Mass spectrometry (MS) is evolving rapidly and has cataloged tens of thousands of phosphorylation sites (phosphosites). Recent studies by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) using liquid chromatography (LC) MS/MS have generated proteomic/phosphoproteomic data sets that have more deeply profiled the cancer proteome (2, 6, 7), providing an opportunity to evaluate regulation of phospho-signaling in cancer. Other MS-based studies focusing on the kinome have highlighted kinases that are disrupted in cancer (8) and identified complementary genomic and proteomic alterations in a handful of signaling pathways (9). However, characterization of kinase-substrate interaction at a single-residue level has been largely limited to in vitro and in silico predictions using sequence-based analysis of kinase binding motifs (10–14). Direct observation of kinase-substrate associations in patient tumors can complement these studies to help reveal their regulation mechanisms.
In this study, we characterized phospho-signaling in cancer by analyzing the phosphoproteomic profiles of 77 human breast tumors characterized in TCGA and CPTAC projects (1, 2) and validating predictions using an independent cohort of 24 breast cancer patient-derived xenografts (PDXs). Approximately 60% (387/630) of phosphosites are significantly associated with expression of their respective proteins across 91 unique kinases in EGFR, PKC, STE20, and other families. MAP kinases and CDKs are involved in wide-spread trans-correlations with downstream substrates. Only selected substrate phosphosites (∼5%, 1747/38,710) are associated by kinases in this breast cancer cohort, suggesting cancer-specific usage of signaling modules. Notably, our analysis revealed 806 potentially associated phosphosites by protein kinases not captured by current experimental or motif-based predictions, including ERBB2, EGFR, MAPKs, and CDKs. Landscapes of associations revealed adjacent phosphosites showing similar or distinct regulations. Finally, druggable kinase-substrate pair analysis validated singleton driver events of AKT1, ERBB2, and RAF1, while exposing potentially druggable pairs and cascades downstream of several MAP kinases not apparent at the genomic level. Overall, our findings emphasize that evaluating signaling effects in patients' tumor through a systematic, high-throughput manner could ultimately prove clinically useful.
EXPERIMENTAL PROCEDURES
Sample Description
Samples of human breast cancer have been described in CPTAC marker papers (2, 6). The corpus is comprised of 77 breast cancer samples showing unimodal distribution in proteomes, their 3 technical replicates, and 3 normal breast samples. Samples of the 24 PDX breast cancer were as described previously (15).
Data Generation
TCGA Genomics Data
The TCGA somatic mutation data, level-3 segment-based copy number data, and level-3 normalized RNA expression data were downloaded from firehose (archive date October 17, 2014). We then converted the segment-based copy number data to the gene-based copy number data by using the RefSeq database (version 20130727). The CNV ploidy number is divided by 2 and then log2-transformed to obtain the final CNV levels for analysis. We also log2-transformed the RSEM values of RNA expression data.
TCGA RPPA Data
Normalized RPPA data of TCGA tumors were downloaded from The Cancer Protein Atlas (TCPA, archive date 2015–10−30). The RPPA data were normalized across batches using replicates-based normalization (RBN) as previously described (16).
Global Proteomics Data
Global proteomics data for the human samples were downloaded from the Mertins et al. breast cancer study (2). Global proteomics data for the PDX samples were downloaded from the Huang et al. breast cancer PDX study (15). As previously described, 2-component Gaussian mixture model-based normalization algorithm was used to normalize the data and accomandate both consistently and differentially-expressed proteins and phosphosites within each sample. Further, proteins and phosphosites used in this study were required to have observed (nonmissing) iTRAQ ratios in at least 30 samples and an overall standard deviation larger than 0.5 (across all samples where they were observed).
To obtain phosphoprotein level used in our kinase-substrate analysis, we used the WGCNA package (17) to collapse phosphosite expression level within each protein. For proteins with three or more phosphosites, we used connectivity based collapsing using the phosphosite with the highest connectivity according to a signed weighted correlation network adjacency matrix. For proteins with two phosphosites, we select the phosphosites with the highest mean value.
Protein and Phosphorylation Databases
UniProt
We applied HotSpot3D (v1.1.1) which accesses crystal structures from RCSB Protein Data Bank (PDB) and calculates residue distances using the average distance-measure option in preprocessing (structures processed January 2017) (18). We used a custom Perl script to retrieve phosphosites, active sites, and binding sites (i.e. features matching MOD_RES, ACT_SITE, BINDING, or SITE) from UniProt (date accessed 1/30/2017) (19).
PhosphositePlus (20, 21)
For phosphosite comparison, we downloaded the data from file Phosphorylation_site_dataset.gz (Downloaded: 2/13/2019) to extract phosphosite mapping to human proteins and reverse-translated to their unique genomic positions using transvar (22). For kinase-substrate pair analysis, we downloaded the file Kinase_Substrate_Dataset.gz (Downloaded: 2/11/2016) and extracted the kinase-substrate proteins pairs. To identify phosphosites known in cancer, we filtered the disease-associated sites database for cancer terms (ex. cancer, *oma, and leukemia) using the file Disease-associated_sites.gz (Downloaded: 2/11/2016). The sites that were not matched to a valid genomic coordinate by TransVar (22) were excluded and the remainder sites were further reviewed. We ultimately retained 261 sites, where 84 unique sites were quantified in our dataset.
Phospho.ELM
We downloaded the phospho.ELM database from PhosphositePlus (2/11/2016). We then extracted the phosphosites mapping to human proteins and reverse-translated to their unique genomic positions using transvar (22).
PhosphoNetwork
We downloaded the Supplementary Tables from Newman et al. (10) and derived the predicted kinase-substrate pairs from the file comKSI.csv, this file combined refined kinase-substrate pairs to 719 kinase-substrate pairs as described. We then further filtered out the pairs already observed in PhosphositePlus and combined the remaining pairs with pairs from PhosphositePlus for analysis.
NetworKIN
We downloaded the NetworKIN output (networkin_human_predictions_3.1.tsv.gz, download date: 3/20/2018) from KinomeXplorer (12) (http://networkin.info/). We then standardized the kinase names to HUGO symbols using mapping from the human kinome (23) and custom scripts. The processing resulted in 423,574 predicted kinase-substrate phosphosite pairs with mapped gene names, of which 412,696 pairs were within the kinase/substrate genes investigated in our quantitative analyses and were thus used for further analyses.
Bioinformatics Analyses
Cross Data Type and Database Integration
All gene names were converted to HUGO Gene Nomenclature Committee's approved gene names for comparison across levels and datasets. To match the exact phosphosite (ex. PIK3CA:NP_006209.2:s312) across databases, all phosphosites were reverse-mapped to their respective genomic positions (ex. chr3:g.178921452_178921454) using transvar (22).
Kinase-Substrate Pairs Regression Analysis
We obtained 3245 unique kinase-substrate pairs in the PhosphositePlus database (20, 21) and an additional 1752 kinase-substrate pairs from the PhosphoNetwork database (24). We then screened all kinase-substrate-phosphosite pairs within these known kinase-substrate protein pairs. For each kinase-substrate-phosphosite pair to be tested, we required both kinase protein/phosphoprotein expression and phosphosite phosphorylation to be observed in at least 10 samples in the respective datasets and the overlapped dataset. We then applied the linear regression model as implemented in glm function in R to test for the relation between kinase and substrate phosphosite. The tests are independently conducted for cis and trans interactions in the cohorts of 77 human and 24 PDX breast cancer samples using linear regression models. Linear regression modeling is generally based on estimating coefficients of independent variables that give the best fit to a dependent variable within the context of a random error. For the ith trial for kinase phosphosite abundance in the cis associations, kinase phosphosite abundance, Ai, depends on kinase protein expression, Si, and error, Ei,
For the ith trial for kinase phosphosite abundance in the trans associations, substrate phosphosite abundance, Ai, depends on kinase phosphoprotein expression, Ki, substrate protein expression, Si, and error, Ei,
where the beta coefficients are determined by least-squares calculation. The resulting p values were adjusted using the Benjamini-Hochberg procedure to FDR.
We determined kinase-phosphosite pairs as validated if they showed p value under 0.05 and positive regression coefficients in the PDX cohort. For the kinase-phosphosite pairs showing top significant associations in the regression analysis, we calculated the average of phosphorylation level for each of the substrate phosphosite and protein expression of the kinase within each of 4 breast cancer subtypes (Basal, Her2, LumA and LumB) for display in Fig. 2 and 3.
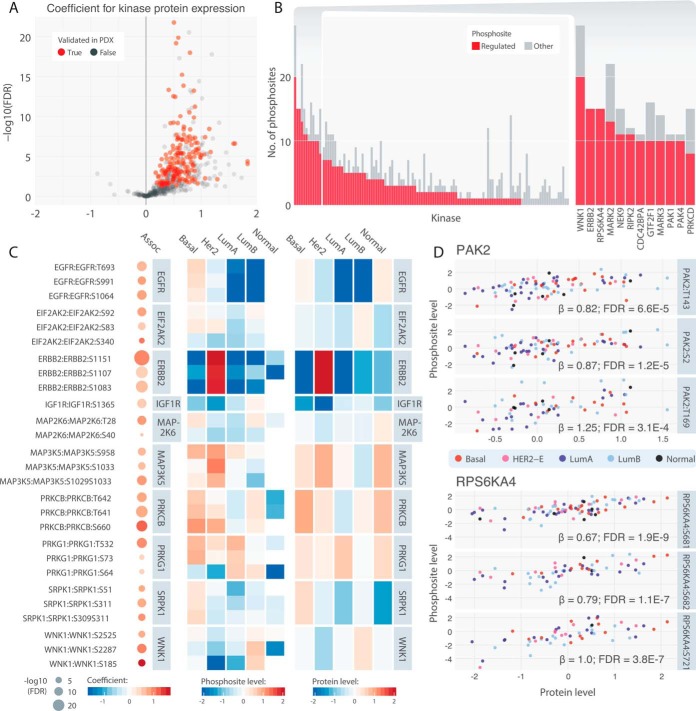
Fig. 2.
Cis kinase-phosphosite pairs. A, Volcano plots highlighting positively cis-associated pairs. Associations that were validated in the 24 PDX models (p < 0.05) are colored in red. B, Counts of cis-associated phosphosites among all corresponding phosphosites for each kinase. Kinases with more than 7 cis-associated phosphosites are shown individually in a close-up barplot (right). C, Top associated cis pairs and their average kinase protein and phosphosite levels in each of the breast cancer subtypes. On the left panel, each dot represents a cis association identified by the regression analysis, where its size represents the significance and color represents the regression coefficient. The middle and right panels illustrate the average cis-associated phosphosite expression and kinase protein expression, respectively, in each of the breast cancer subtypes. D, Correlations of kinase protein level of PAK2 and PRS6KA4 and their respective top 3 cis-associated phosphosites. Each dot represents one tumor sample colored according to its subtype.
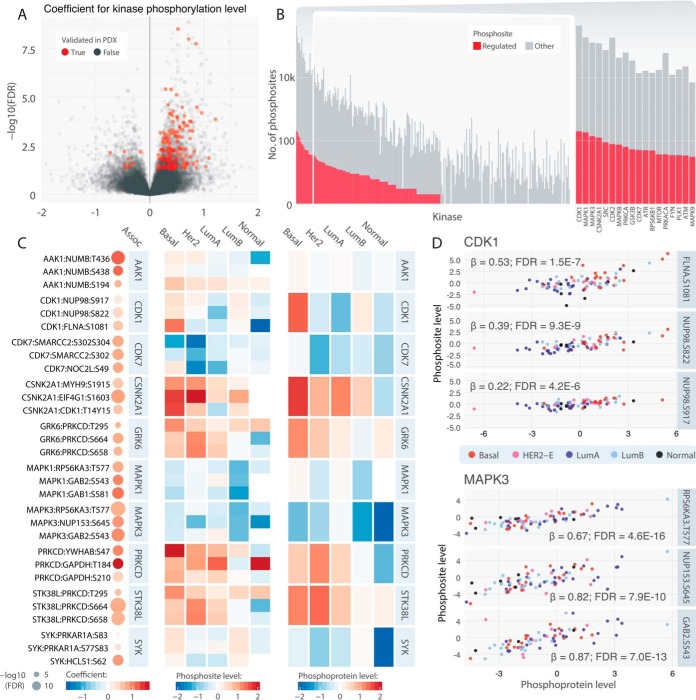
Fig. 3.
Trans kinase-substrate phosphosite pairs. A, Volcano plots highlighting positively associated pairs. Associations that were validated in PDX models (p < 0.05) are colored in red. B, Counts of trans-associated phosphosites among all corresponding phosphosites for each kinase. Kinases with more than 30 trans-associated phosphosites are shown individually in a close-up barplot (right). C, Top associated trans pairs and their average kinase phosphoprotein and substrate phosphosite levels in each of the breast cancer subtypes. On the left panel, each dot represents a trans association identified by the regression analysis, where its size represents the significance and color represents the regression coefficient. The middle and right panels illustrate the average trans-associated substrate phosphosite expression and kinase protein expression, respectively, in each of the breast cancer subtypes. D, Correlations of kinase phosphoprotein level of CDK1 and MAPK3 and their respective top 3 trans-associated phosphosites. Each dot represents one tumor sample colored according to its subtype.
Finally, we compared our results of trans association to 10 simulated kinase-substrate datasets by distributing the same kinases in even proportions to all incidences of current substrate phosphosites and conducted the kinase-substrate association testing in 574,356 randomized pairs with sufficient quantifications. Only 2.82% (16, 194) showed positive association using the same criteria (regression coefficient β > 0 and FDR < 0.05). We then determined the significance of our observed proportion to the simulated results using the Fisher's exact test.
Kinase-Substrate Pair Validation Using Thresholds Defined in PDX Data
We used sensitivity/specificity analyses to determine the thresholds derived from the PDX cohort used for validating kinase-substrate pairs found in the human cohort. In this analysis, the correlated significance (with regression coefficient β > 0 and FDR < 0.05) of tested kinase-substrate pairs with sufficient PDX data (cis n = 396; trans n = 27,585) is considered as the response variable and the kinase regression coefficients are considered as the predictor variables (supplemental Fig. S2). For cis analysis, the area under curve (AUC) for the regression coefficient of the kinase protein in the PDX cohort is moderate at 0.5464; but at our threshold coefficient of 0.1, we could achieve sensitivity of 0.98 and specificity of 0.09. For trans analysis, the AUC for the regression coefficient of the kinase phosphoprotein in the PDX cohort is better at 0.612; and at our threshold coefficient of 0.1, we could achieve sensitivity of 0.46 and specificity of 0.71.
Kinase Group and Family Enrichment Analysis
To test whether kinases showing significant cis or trans correlations are enriched in kinase groups and families, we applied the one tailed Fisher's exact test under a null hypothesis that the odds ratio of associated kinases in the family are not greater. The universe of kinases for each of the cis and trans tests was defined as the total tested kinase, and the 2-by-2 table was constructed by (1) whether the kinase belonged to the kinase group/family, and (2) whether the kinase had any significant correlations.
Structural and Cophosphorylation Analysis
We used HotSpot3D (18) to generate pairwise linear and 3D distances between residues within 1,288 proteins having available PDB structures. The active sites were mapped based on data from the RSCB PDB, as of January 2017 (http://www.rcsb.org/pdb/home/home.do). Pearson's correlation coefficients and adjusted p values were then calculated for each pair of two phosphosites within these proteins. We limited this correlation analysis to pairs of phosphosites jointly observed in at least 5 samples in the breast cancer cohort. For linear examinations of association landscapes, the lolliplots were generated and modified using the PCGP protein painter (http://explore.pediatriccancergenomeproject.org/proteinPainter).
Druggable Kinase-Substrate Pairs and Cascades Analysis
We compiled a list 76 druggable genes along with their respective drugs, from established public databases as previously described (15). We then limited the analysis to the 68 genes with per-sample average RSEM value greater than 100. We searched for significantly associated kinase-substrate pairs where either the kinase or substrate belonged to the set of druggable genes. The score of each pair was calculated as the sum of standardized kinase and substrate phosphosite levels:
where μ is the mean and σ is the standard deviation.
To identify druggable outliers in the conventional method, we scanned for events at the somatic mutation, CNV, RNA, and protein expression levels for each gene. CNV, RNA, and protein expression outliers were identified as the ones greater than 2 interquartile ranges (IQR) above median, as previously described (2, 15). We then complemented this conventional single-event analysis by identifying outliers using the kinase-substrate score.
To define the druggable cascade of each of the druggable kinase, we extracted all significantly associated cis phosphosites and their first-degree correlated trans substrate phosphosites. We then expanded one level beyond, extracting additional trans phosphosites associated with the phosphoprotein level of the first-degree substrates. The resulting cascades were visualized using heatmaps showing levels of each protein/phosphoprotein/phosphosite and a network diagram using Cytoscape (25).
Clinical and Immune Correlation Analysis
For each kinase-substrate pair, We conducted association analysis between the kinase-substrate score (defined above) of this pair for each patient and pathological stage, survival, radiation therapy, and immune signatures adjusted for their PAM50 subtypes. For continuous variables, including pathological stage and immune scores, we used a Gaussian linear regression. For whether the sample has gone through radition therapy, we used a logistic regression model. We used the Cox proportional Hazards model for survival analysis. The resulting p values were adjusted to FDR using the Benjamini-Hochberg procedure.
Western Blotting Validation
Frozen PDX tumor samples from the Washington University Human in Mouse (WHIM) collection were extracted for soluble proteins using PULSE Tubes (FT500-ND, Pressure BioSciences) in RIPA lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1% NP-40, 0.1% SDS, 0.25% sodium deoxycholate, protease inhibitors (EDTA-free Complete Mini, Roche), 1 mm EDTA, 1 mm PMSF and phosphatase inhibitors (1 mm NaPPi, 1 mm Na3VO4, 10 mm NaF)). Tumor lysates were clarified by centrifugation for 5 mins at 13,000 rpm, measured for protein concentration by Bradford assay, and boiled in SDS sample buffer for immunoblotting. Proteins (20 μg/lane) were separated on Any kDa Mini-PROTEAN TGX Precast Gels (Bio-Rad), transferred to Immobilon PVDF membrane (Millipore), blocked with 5% skim milk in TBS/0.1% Tween-20 (TBST) for 30 mins at room temperature and incubated overnight at 4 °C with primary antibodies. The following primary antibodies were used: mouse monoclonal HER2 antibody (MS730-P0, Thermo, 1:1500), rabbit polyclonal phospho-HER2 (S1151) antibody (AP3781c, Abgent, 1:1000), rabbit monoclonal 4E-BP1 antibody (#9644, Cell Signaling Technology, 1:2000), rabbit polyclonal phospho-EIF4EBP1 (S65) antibody (AP0032, Abclonal, 1:1000) and rabbit polyclonal actin (A2066, Sigma, 1:5000). After washing with TBST, membranes were incubated with appropriate HRP-conjugated secondary antibodies for 1 h at room temperature, followed by washing, and developed using a chemiluminescence substrate (SuperSignal West Dura, Thermo) and visualization on a ChemiDoc Imaging System (Bio-Rad). Image Lab software version 3.0 (Bio-Rad) was used for image acquisition and analysis of the blots. The software identifies lanes, detects bands, and generates an analysis table quantifying the volume intensity of each band. Protein levels were quantitated by densitometry.
Statistical testing of the results was conducted using linear regression models, where the quantity measured through Western blotting is the dependent variable and the quantification obtained through high throughput proteomics is the independent variable, with the date/replicates of the blotting experiment as the covariate.
SRM Validation
Stable Isotope-Labeled Peptides
Crude stable isotope-labeled peptides (SI peptides) were synthesized with 13C/15N on C-terminal lysine or arginine (New England Peptide, Gardner, MA). The SI peptides were dissolved individually in 15% acetonitrile (ACN) and 0.1% formic acid (FA) at a concentration of 1.5 mm (according to the nominal peptide quantity provided by the vendor) and stored at −80 °C. A mixture of these SI peptides was made with a final concentration of 10 pmol/μl for each peptide.
SRM Assay Development
To evaluate the peptide quality and select the best responsive transitions for each peptide, 500 fmol/μl of heavy peptide mixtures were subjected to high-resolution mass spectrometry (MS) analysis using the Orbitrap Fusion Lumos instrument (Thermo Fisher Scientific, San Jose, CA) running in the HCD mode (26). The resulting RAW files were processed with DTARefinery (27) and MS-GF+ (28, 29) to match against the RefSeq human protein sequence database, released on April 18, 2017 (46,174 proteins) for generating a list of MS/MS fragment ions derived from SI peptides. The 6 most intensive fragment ions for each peptide were initially selected based on corresponding MS/MS spectra. The collision energies for individual transitions were obtained by using empirical equations from the Skyline software (30). Second, LC-SRM was used to further evaluate all heavy peptides for the LC performance (e.g. the stability of peptide retention time), MS response (e.g. reliable heavy peptides identification and S/N>3), transition interferences, and endogenous peptide detectability by spiking them into water and the pooled wild type samples. In the end, 3 or more transitions per peptide were selected for configuration of the final panel of assays for reproducible targeted quantification. The transitions for each precursor were provided in supplemental Table S23.
The intended use of these assays is the Tier 2 measurements, with the ability to measure repeatedly sets of analytes of interest within and across samples by spiked in internal standards for each analyte, for confident detection and precise quantification (31). Response curves were generated for the peptides using triplicate measurements of varying concentrations of heavy peptides and a constant concentration of light peptides spiked into a tryptic digest of a previously characterized human breast cancer xenograft (32) (supplemental Fig. S13). For peptide IPLENLQIIR, the concentrations of heavy peptide were 0, 20, 50, 100, 200, 800, and 1600 amol/μl and the concentration of light peptide was 125 amol/μl; 5 μl of the sample at each data point was injected for analysis by LC-SRM. Its limit of detection (LOD) and lower limit of quantification (LLOQ) were both determined at 20 amol/μl level. For the two phosphopeptides NGLQSCPIKED(pS)FLQR and ELVEPL(pT)PSGEAPNQALLR, the concentrations of heavy peptide were 0, 0.4, 0.8, 1.6, 3.125, 6.25, 12.5, 25, 50, 100, and 200 fmol/μl and the concentration of light peptide was 200 fmol/μl; they were spiked into the same breast tumor xenograft sample (100 μg peptide), followed by phosphopeptide enrichement (see below) and LC-SRM analysis. The LOD and LLOQ for both phosphopeptides were 0.4 fmol/μl and 1.6 fmol/μl, respectively. The LOD was determined by S/N>3. The LLOQ was determined by both S/N>10 and CV<20%. The inter-day CVs were also determined (supplemental Fig. S14).
Phosphopeptides Enrichment by IMAC
Two hundred micrograms of tryptic peptides from each sample spiked with 1000 fmol/μl of each of the crude SIL peptides were subjected to phosphopeptides enrichment via immobilized metal affinity chromatography (IMAC) (33, 34). The in-house-made IMAC tip was capped in a tip-end with a 20-μm polypropylene frits disk followed by packing with Ni-NTA silica resin (Qiagen, Hilden, Germany). First, Ni2+ ions were removed by adding 50 mm EDTA in 1 m NaCl. The tip was then activated with 100 mm FeCl3 and equilibrated with 6% (v/v) acetic acid at pH 3.0 before sample loading. Tryptic peptides were dissolved in 6% (v/v) acetic acid and loaded onto the IMAC tip. Followed by 1% (v/v) trifluoroacetic acid, 80% ACN, and 6% (v/v) acetic acid washing steps, the bound phosphopeptides were eluted by 200 mm NH4H2PO4 and then desalted by SDB-XC StageTips (35) and dried under vacuum.
LC-SRM
The enriched phosphopeptides or the unmodified peptide samples with spiked SIL peptides were dissolved in 2% ACN/0.1% FA and analyzed using a TSQ Vantage triple quadruple mass spectrometer (Thermo Fisher Scientific) equipped with a nanoACQUITY UPLC system (Waters, Milford, MA). Peptide samples were loaded onto an ACQUITY UPLC BEH 1.7-μm C18 column (100 μm i.d. × 10 cm). The mobile phases were (A) 0.1% FA in water and (B) 0.1% FA in ACN. Two microliter of the unmodified peptide sample (0.3 μg) and 5 fmol/μl SIL peptides were loaded onto the column and separated at a flow rate of 400 nL/min using a 100-min gradient profile as follows (min:%B): 11:0.5, 13.5:0.5, 17:8, 25:13, 55:20, 80:38.5, 85:95, 89:50, 90:95, 91:0.5. For enriched phosphopeptide samples, all of the eluent from IMAC was dissolved in 10 μl and 4.5 μl were loaded onto the column and separated with the same LC gradient. The LC column is operated at a temperature of 44 °C. The parameters of the triple quadruple instrument were set as follows: 0.7 fwhm Q1 and Q3 resolution, and 1 s cycle time. Data were acquired in time-scheduled SRM mode (retention time window: 10 min) without blinding the samples.
Data Analysis
SRM data were analyzed using Skyline software (version 4.2). The total peak area ratios of endogenous light peptides and their heavy isotope-labeled internal standards (i.e. L/H peak area ratios) were calculated for quantification. Peak detection and integration were carried out based on two criteria: (1) same retention time and (2) similar relative SRM peak intensity ratios across multiple transitions between light (endogenous) peptides and the heavy SIL peptide standards. All data were manually inspected to ensure correct peak detection and accurate integration.
RESULTS
Detect and Characterize Phosphosites in Breast Cancer
We analyzed genomics and proteomics data from 77 breast cancer and 3 normal breast samples (supplemental Table S1) characterized by TCGA(1) and CPTAC(2) (Fig. 1, Experimental Procedures). Genomic analysis of TCGA DNA-Seq, SNP-array, and RNA-Seq data provided comprehensive assessment of somatic mutations, copy number variations (CNV), and mRNA expression, respectively. The CPTAC breast cancer project quantified global protein and phosphosite expression levels using the Isobaric Tag for Relative and Absolute Quantitation (iTRAQ) technology. We further used proteogenomic data from 24 independent breast cancer patient-derived xenografts (PDXs, supplemental Table S1) (15) generated using the same technologies and bioinformatic pipeline for validation.
Fig. 1.
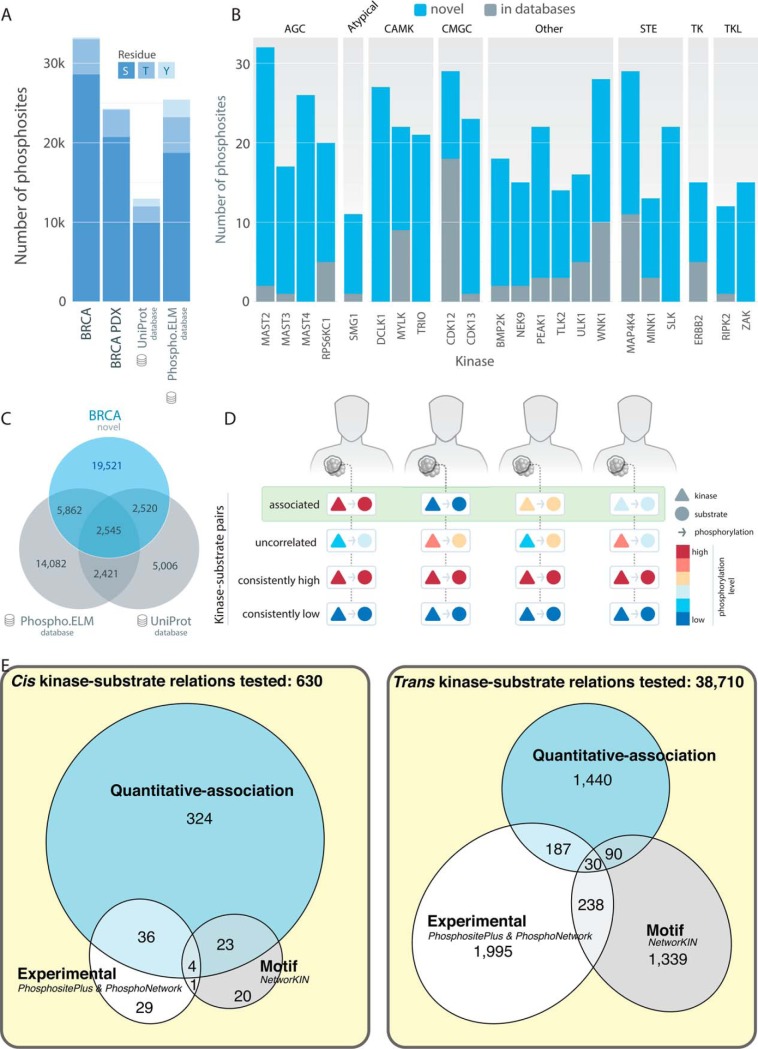
Landscape of the 33,239 quantified phosphosites in breast cancer. A, Distribution of phosphosites and counts of serine (S), threonine (T), and tyrosine (Y) residues in the 77 breast cancer (BRCA) and 24 PDX cohorts compared with the UniProt and Phospho.ELM databases. B, Number of novel and known phosphosites from UniProt and Phospho.ELM detected in kinases of major kinase groups. Only kinases having more than 10 novel sites are shown (supplemental Table S2). C, Venn diagram showing quantified phosphosites in human breast tumors compared with phosphosites in the UniProt and Phospho.ELM databases. D, Diagram of correlated, uncorrelated, consistently high and consistently low kinase-substrate pairs in breast cancer samples. E, Venn diagrams showing the numbers of cis or trans-associated phospho-pairs identified through quantitative association in this study out of the total numbers of tested pairs, compared with experimental validated relations in PhosphositePlus and PhosphoNetwork and kinase-motif predicted kinase-phosphosites in NetworKIN.
As previously described (2), 33,239 out of the 62,679 confidently identified phosphosites passed additional missing value and standard deviation filters and were used for downstream analyses (Methods). This catalog of breast cancer phosphosites added 19,521 phosphosites not currently found in the 12,492 and 24,910 genomically mapped human phosphosites from the UniProt (37) and the Phospho.ELM databases (38), respectively (Fig. 1A, 1C; 13,883 sites not found using protein coordinate matching in the PhosphositePlus database (20, 21)). These phosphosites covered 315 of the 523 previously described human kinases (23, 39). Kinases with a significant number of novel phosphosites may have underappreciated regulatory roles in breast cancer, including MAST2 (30 novel phosphosites) and MAST4 (26) from the AGC kinase group, CDK13 (22) and CDK12 (11) from the CMGC kinase group, SLK (22) and MAP4K4 (18) from the STE kinase group, and ERBB2 (10) from the TK group (Fig. 1B, supplemental Table S2). Compared with other quantified sites, the 84 detected, known cancer-associated phosphosites (Experimental Procedures, supplemental Table S3), including AKT1 pT308, GSK3B pS9, and MTOR pS2448, exhibited higher standard deviation in both breast cancer and PDX samples (Wilcoxon rank sum test, p = 2.6e-03 and 2.6e-04, respectively, supplemental Fig. S1), implicating their differential regulation across tumors.
In this study, we aim to identify quantitatively correlated kinase-substrate pairs that are concordant in their relative abundance across samples (Fig. 1D). We hypothesize these phosphosites are regulated in a patient-specific manner, contributing to cross-sample variation in wiring of signaling networks. We examined two classes of kinase-substrate relations: (1) cis associations whereby a kinase protein expression is correlated with its own phosphosites and (2) trans associations whereby a kinase phosphoprotein expression is correlated with a substrate phosphosite level (Experimental Procedures). We specifically analyzed sites showing substantial variations (Observed across 30 samples and standard deviation > 0.5) across breast tumors (Experimental Procedures), noting that there may be other kinase-substrate pairs that remain in steady states across breast tumors (i.e. consistently high or low) not addressed by our current approach and dataset (Fig. 1D).
To achieve this goal, we curated and screened 4997 pairs of human kinase and substrate proteins based on the PhosphositePlus and PhosphoNetwork databases (20, 24). We then compared the quantitatively correlated pairs we identified at the phosphosite level with experimentally observed kinase-substrate phosphosite pairs in PhosphositePlus (phosphosites observed in vivo or in vitro) and PhosphoNetwork (CEASAR: kinase-substrate map generated through in vivo sites and bioinformatics algorithm), as well as pairs based on kinase binding-motif prediction using NetworKIN3.0 (11, 12). Our correlation approach overlapped previously identified pairs and added new observations at the phosphosite level (Fig. 1E). For cis associations, 40 experimentally observed cis-pairs showed significant correlations. For trans associations, we observed a notable enrichment of both experimentally observed pairs (Fisher's exact test, 2.2-fold enrichment, p = 5.5e-22) and kinase binding-motif predicted pairs (Fisher's exact test, 1.7-fold enrichment, p = 8.3e-07), suggesting that our quantitative-correlation approach affords an independent and complementary method that can identify potential kinase-substrate phosphosite pairs. Notably, our approach identified a substantial amount of 1440 trans associated pairs in addition to those found by in vitro experiments or binding-motif analysis.
Phosphosites in Auto-phosphorylated Kinases Across Breast Cancer Subtypes
Theoretically, a phosphosite's abundance will correlate with its protein level as determined by stoichiometry. However, among the wide-spread molecular changes in tumors, many are presumed to be “passengers” without clear functional effects and validated functional events can often be prioritized by those affecting downstream changes, such as genomic truncation/CNV/methylation associated with mRNA gene expression (40–42) or mRNA changes associated with protein expression (15, 43). Thus, we hypothesize cis-association will enable us to select for meaningful protein level changes and their functional phosphosites. Further, those phosphosites with increased abundance above expectations might suggest autophosphorylation, a hallmark functionality of many kinases involved in oncogenic signaling.
For cis analysis, protein expression of 120 kinases known to exhibit auto-phosphorylation were evaluated with relation to peptide abundance of their 630 phosphosites using a linear regression model (Experimental Procedures). Protein abundance measures do not always guarantee the activation of the phosphosites of the same protein: 61.4% (387/630) of the tested kinase-substrate relations showed significant positive associations (regression coefficient β > 0 and FDR < 0.05) in breast cancer (Fig. 2A, supplemental Table S4). Of the identified cis-associated pairs, 98.4% with sufficient PDX data (179/182) showed positive correlation (β > 0.1, corresponding to sensitivity of 0.98 and specificity of 0.09 [Methods], supplemental Fig. S2) in the cohort of 24 breast cancer PDXs (15) and considered validated (supplemental Table S5).
At the kinase group or family level, enrichments of cis-associated kinase genes within the group or family imply their diverse regulation and possible functional quantity changes across breast tumors. Using the background of all tested kinases, the AGC kinase group was most significantly enriched with cis associations (Fisher's Exact Test, p = 0.024, Experimental Procedures), with all 17 tested genes showing significant cis correlations with their respective phosphosites, followed by the STE group (14/14 genes, Fisher's Exact Test, p = 0.049, supplemental Table S6). Notably, at the kinase family level, we observed significant cis-association in all 8 of the STE20 kinases in the analysis, including PAK1/2/4, STK3/4/24, and MAP4K1/3 (supplemental Table S7).
Top proteins with high percentage of cis-associated phosphosites (Fig. 2B) included well-known and novel breast cancer proteins, such as ERBB2 (15 significant cis-regulated phosphosites), RPS6KA4 (15), NEK9 (11), RIPK2 (11), CDC42BPA (10), and GTF2F1 (10). Further, at least 7 cis-associated phosphosites in each of ERBB2, PAK4, NEK9, and RIPK2 were validated in the PDX cohort (β > 0.1, supplemental Table S5). Various cis-regulations exhibited molecular distinctions across breast cancer subtypes (Fig. 2C), exemplified by elevated ERBB2 protein and phosphorylation in HER2-E breast cancers. Other subtype-specific regulated pairs identified were WNK1 up-regulated in luminal A, and EGFR and SRPK1 up-regulated in basal breast cancers. Closer examination revealed clear correlations between levels of PAK2 protein and its pS2 and pT143 phosphosites, as well as PRS6KA4 protein and its pS681 and pS682 phosphosites (Fig. 2D).
We hypothesized that the observed cis associations may indicate kinase autophosphorylation if each unit change in kinase protein resulted in an even greater change in phosphosite level (β > 1). Using this approach, we identified 41 cis-regulated sites that showed the steep increase, including CDK9 pT186, RAF1 pS43, BRAF pS151/S729, and PRS6KA4 pS737 (supplemental Fig. S3), and thus potentially were regulated in part by the autophosphorylation. Among the 41 pairs, only CDK9 pT186, PRKCB pS660, and PRKCD pS645 were validated as auto-phosphorylated in PhosphositePlus (20, 21), warranting further validation to phosphosites discovered using the quantitative-correlation approach.
Correlated Kinase-Substrate Pairs in Breast Cancer
For trans analysis, we surveyed 7404 kinase-substrate protein pairs and a total of 38,710 kinase-substrate phosphosite relations with sufficient observations in our data set (Experimental Procedures). We applied a linear model using phosphosite abundance of the substrate as the dependent variable, phosphoprotein expression of the kinase as the independent variable. To avoid associations because of co-expression, we correct for the protein expression of the substrate using it as a covariate. Although only 4.51% (1,747/38,710) of the tested relations showed significant positive associations (Fig. 3A, supplemental Table S8), this correlated proportion is still significantly higher than that found using simulated pairs (Experimental Procedures, Fisher's Exact Test, p < 2.2e-16). Further, Only 143 and 81 of the 1,747 trans pairs were identified when we modeled the kinase-substrate relations using protein and mRNA levels of the kinase, respectively, suggesting that a limited fraction of the identified kinase-substrate pairs are likely because of co-expression of the kinase and substrate (supplemental Fig. S4). Phosphoprotein levels of the kinase are required to identify 1576 of the trans associations, consistent with a mathematical model of phosphosignaling reactions (44) and suggesting the identification of signaling pairs.
Of the identified trans-regulatory pairs, 45.5% with sufficient PDX data (407/894) were confirmed (β > 0.1, corresponding to sensitivity of 0.46 and specificity of 0.71 (Experimental Procedures), supplemental Fig. S2) in breast cancer PDXs (15) (supplemental Table S9). The moderate validation rate may be because of increased sensitivity to micro-environment in trans pairs and larger sample sizes potentially required to firmly establish trans associations (supplemental Fig. S3). To reveal global patterns of trans associations less affected by potential noise and lack of power, we sought to identify kinase families enriching for trans-associated kinase genes among the background of all tested kinases. The identified trans-regulations are congregated in 165 kinases, which showed the most significant enrichment in MAPK (Fisher's Exact Test, p = 0.00045), CDK (Fisher's Exact Test, p = 0.0026), and PKC (Fisher's Exact Test, p = 0.0061) families (supplemental Table S7).
Multiple kinases with the highest number of trans associations have been previously implicated in breast cancer. Particularly, ATM (33 trans-associated substrate phosphosites) and ATR (44), proteins known to orchestrate the DNA damage response (DDR) pathway, were significantly associated with phosphosites of chromatin-associated SMARCC2, MCM2, histone-lysine methyltransferase KMT2A, and the DNA damage checkpoint protein TP53BP1 (supplemental Fig. S5). In the PI3K pathway, kinases associated with trans-associations of more than 20 phosphosites included GSK3B (35), RPS6KB1 (39), MTOR (29), and RPS6KA1 (21, supplemental Fig. S5). We also observed specific interactions, such as the inhibitory phosphorylation of GSK3A by AKT1 (AKT1:GSK3A pS278, linear regression, FDR = 0.034) (45).
CDK1 (178 trans-associated phosphosites), CDK2 (72), and CDK7 (44) showed a wide-spread effect of trans-associations on substrates, including NUP98 and FLNA, supporting their central roles in cell cycle signaling. MAPK1 (148 trans-associated phosphosites), MAPK3 (111), MAPK8 (50), MAPK14 (18), and MAPK9 (15) were also associated with up-regulation of phosphosites of multiple downstream substrates, including RPS6KA3/5, GAB1/2, MAP2K1, and CIC (Fig. 3C, 3D, Supplementary Information).
We further investigated the kinases with highest fraction of trans-correlated phosphosites among all tested phosphosites. Twenty-six kinases satisfied the criteria of having at least 2 significant trans correlated phosphosites and fraction of correlated/tested sites greater than 10%, including some of the kinases with the highest numbers of correlated phosphosites: CDK7 (44 correlated/323 tested), MAPK1 (148/1201), ATR (44/378), RPS6KB1 (39/338), and MAPK3 (111/1063). Of note, this analysis also prioritized additional kinases with high fractions of correlated substrate phosphosites, including MAP2K5 (3/3), AAK1 (5/12), RIPK1 (3/13), MAP2K6 (9/40), and STK36 (4/20) whose phosphoprotein level changes likely affect their downstream phosphosignaling.
Sequence and Structural Patterns Correlated with Phosphosite Expression
Our cis and trans analyses identified pairs of phosphosites on the same protein showing concordant regulation patterns, including those in ERBB2, PRKCB, and WNK1. We hypothesized that phosphosites in spatial proximity would be affected by the same regulators and would show similar phosphorylation patterns. For each pair of phosphosites, we calculated the correlation coefficient of their levels and compared that to their proximity on PDB structures, as determined by HotSpot3D (18, 45) (supplemental Table S10, Methods). We found a significant correlation between phosphosite levels with both 3D distances (measured in ångströms) of the phosphosite pairs (Spearman correlation, p = 3.5e-08, rho = −0.40) and linear distances (measured in amino acid residue counts, Spearman correlation, p = 7.9e-04, rho = −0.25, supplemental Fig. S6), confirming co-regulation yet potentially divergent abundance in adjacent phosphosites.
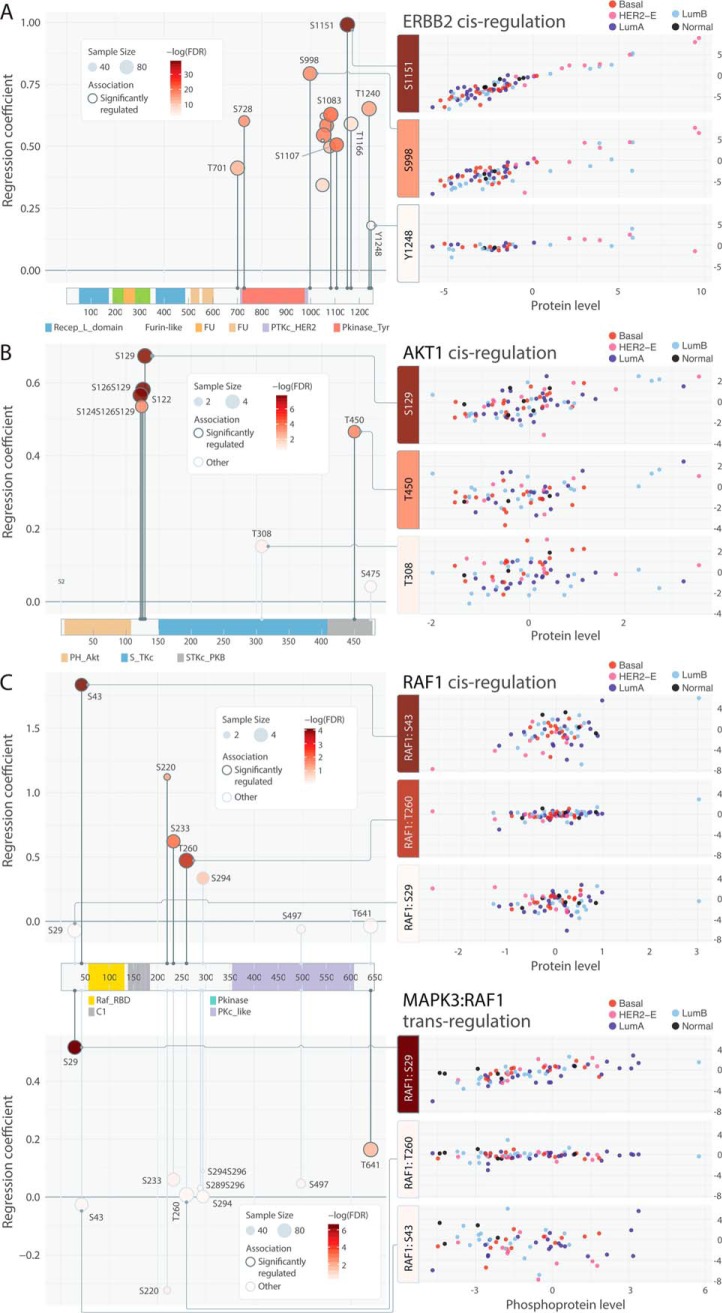
Phosphosites with strong cis-associations may help detect activating events correlated with high kinase expression. We identified ERBB2 pS1151/S998/T1240 as showing the most significant cis-associations with particularly high levels in HER2-E tumors (linear regression, β > 0.65; FDR < 6.2e-16) and may serve as complementary HER2 biomarkers to the tyrosine residues targeted by available antibodies, such as pY1221 and pY1248 (Fig. 4A). Notably, pT1240 has been shown to drastically reduce in phosphorylation level upon either trastuzumab and Bis-Fab 1235 (antagonists of HER2) treatments whereas pY1248 shows an increase (46), demonstrating their differential regulation upon inhibition. On the other hand, poorly correlated phosphosites in kinases with strong cis-effects may suggest additional post-translational modification mechanisms. For example, we observed strong association between AKT1 protein and phosphorylation levels for sites pS122/S126/S129 (linear regression, FDR < 8.2E-08, notation such as S126129/S124S126S129 in Figs., Supplementary Tables, and texts indicates dual/triple phosphorylation of the phosphosites in the same detected peptide). However, the associations for pT308/S475 (linear regression, FDR > 0.38) are considerably weaker, even with similar observed sample sizes (Fig. 4B). Other discordant sites (supplemental Fig. S7) include the strongly associated ABL1 pS637/S737/T800/T871 versus the nonassociated pS16/S588/S828; the strongly associated PTK2 pS390/S570/S708/S910 versus the nonassociated pY576/S840; and the associated RIPK1 pS320/S610 versus the nonassociated pS330/S416.
Fig. 4.
Patterns of associated phosphosites on primary sequences and 3D structures. A, Consistent cis-associations of phosphosites identified in ERBB2. B, Discordant cis-associations of phosphosites identified in AKT1. (C) Cis and trans-associations by MAPK3 of RAF1 phosphosites. D, Cis-associated phosphosites p.T202 and p.Y204 in spatial proximity adjacent to the active site p.D166 of MAPK3 as in PDB structure 4QTB (67). E, Trans-associated phosphosites p.S201 and p.S207 (by MAP3K5) are found in spatial proximity to the active sites, including p.D179 and p.T211, of MAP2K6, which is co-crystalized with an ATP analog as in PDB structure 3VN9(68).
In RAF1, we observed strong cis-associations at pS43/T260/S233/S220 (FDR < 0.0358), but not pS29 (linear regression, FDR = 0.84, Fig. 4C). This association pattern is complemented by the trans-regulation of RAF1 by MAPK3; RAF1 pS29/T241 are tightly correlated with MAPK3 phosphoprotein expression (linear regression, FDR < 0.032) when controlled for RAF1 protein expression, indicating MAPK3 may specifically regulate RAF1 phosphorylation at these sites.
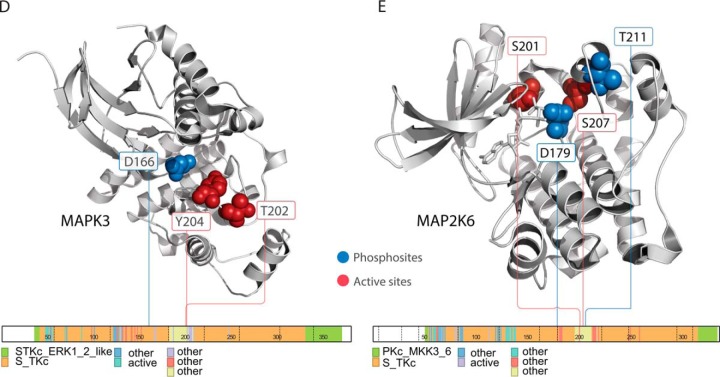
Intriguingly, several regulated phosphosites reside in structural proximity to active sites of kinases (supplemental Table S11, S12). MAPK3 (ERK1) pT202/Y204, which are known to be phosphorylated by MAP2K1 and MAP2K2 (MEK1 and MEK2) to trigger its activation (47, 48), are adjacent to its active site pD166 (Fig. 4D). These two sites showed significant cis-regulation by MAPK3 protein, albeit not trans-association by MAP2K1/2 phosphoprotein. In MAP2K6, MAP3K5-regulated pS207 and an adjacent site pS201 are in its catalytic domain near active sites pD179 and pT211 (Fig. 4E). These phosphosites may alter the biochemical properties of the active site and affect the activity level of the corresponding kinase.
Kinase-Substrate Pairs Associated with Treatment Options
To inform targeted treatment, druggability analysis has traditionally focused on detecting single activating events, for example EGFR mutation, PIK3CA mutation, and ERBB2 amplification (49, 50). Although these events can predict treatment response, we reason that co-occurrence of elevated kinase-substrate phosphorylation can confirm aberrant activation and reveal new treatment options. We conducted an in silico druggability analysis for the significant cis and trans kinase-substrate pairs by screening 68 druggable genes with potential inhibitors (15) that are abundantly expressed in our breast cancer samples (Experimental Procedures). We used abundance levels of both the kinase and substrate phosphosite to construct a kinase-substrate pair score and identified samples showing 2 standard deviations above cohort medians in their scores (Experimental Procedures).
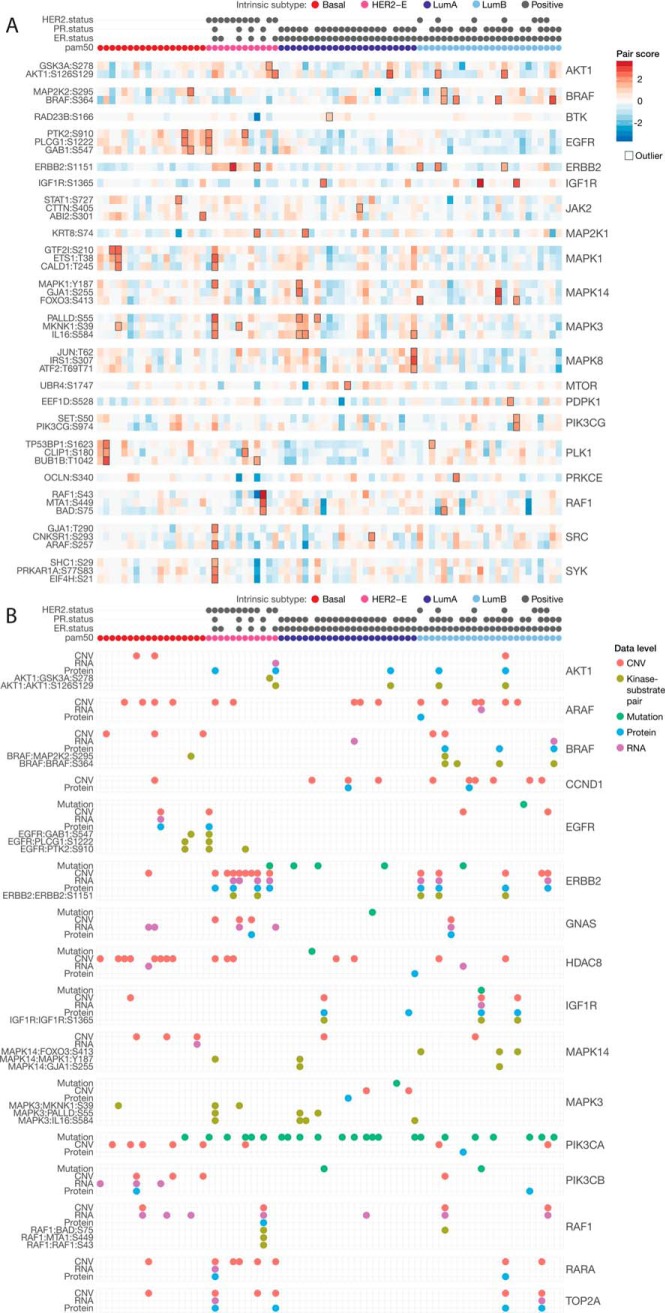
Among the 286 associated kinase-substrate pairs, we identified 164 overexpressed events among 113 unique pairs (Fig. 5A, supplemental Table S13). Overexpression of the cis pair ERBB2:ERBB2 pS1151 were found in 5 HER2-positive samples. The cis BRAF:BRAF pS364 pair and the trans MAPK14:FOXO3 pS413 pairs were found exclusively in 4 and 3 luminal B breast cancers, respectively. SRC and SYK-regulated trans pairs also showed higher levels in luminal B breast cancers. Pairs associated with MAP kinases, such as MAPK3:PALLD pS55, MAPK3:IL16 pS584 and MAPK8:JUN pT62, showed higher levels and several pair overexpression in luminal A breast cancers. In samples without prominent ERBB2 signaling, we observed other alternative overexpression pairs triggered by other receptor tyrosine kinases, including EGFR regulated trans pairs and IGF1R-regulated cis pair IGF1R: IGF1R pS1365, which is a conserved phosphosite found in 10 species (51). In the 24 breast cancer PDXs, we also discovered overexpression of many regulated pairs, including ERBB2:ERBB2 pS1151, BRAF:BRAF pS364, and EGFR:PLCG1 pS1222 (supplemental Table S14, supplemental Fig. S8), underscoring their prevalence in breast cancer.
Fig. 5.
Druggability analysis of single and paired events in 77 breast cancer samples. A, Heatmap of kinase-substrate pairs where the kinase or the substrate is a potential druggable target. The sample-pair showing outlier pair event is outlined. Only the top 3 associated pairs were shown for each kinase when there were more than 3 pairs showing kinase-substrate outliers. B, Druggable events identified in the mutation, CNV, RNA, protein, and phospho-pair level for breast cancer samples in the same subtype order.
We compared the candidate targets identified through paired druggability analysis and conventional single driver analysis (Fig. 5B, supplemental Table S15), where we compiled mutations, CNV, RNA, and protein levels of the same 68 expressed, potentially druggable genes (Experimental Procedures). We observed concordant single driver events with overexpression pair events for kinases, including AKT1, BRAF, ERBB2, IGF1R, and RAF1. Samples with ERBB2, IGF1R and RAF1 copy number amplified outliers often show high expression of the respective kinases-substrate pairs (Fig. 5B). However, single driver events do not guarantee activated signaling of their associated kinase-substrate pairs. For example, some samples with PIK3CA mutation, ERBB2 mutation, or RAF1 copy number amplification did not show overexpressed kinase-substrate pairs associated with the respective kinases (Fig. 5B). Observing concurrent activation of downstream targets using phosphoproteomics data is needed to further confirm these effects and treatment options for each patient.
On the other hand, active signaling events may occur in samples without mutations or expression aberrations of the kinase. This is particularly evident in both human and PDX samples with outlier MAPK3 and MAPK14 trans pairs (Fig. 5B, supplemental Fig. S9, supplemental Table S16). Only 2 out of 7 samples with outlier MAPK3 trans pairs showed MAP3K1 mutations and none of the 5 samples with outlier MAPK14 trans pairs carried MAP3K1 or MAP2K4 mutations (supplemental Fig. S10), suggesting MAP kinases may be activated by other upstream signaling mechanisms rather than directly being altered at the sequence or expression levels. We also observed some of these overexpression pair events in absence of single driver events for AKT1, BRAF, and EGFR (Fig. 5B) that require further investigation.
Activated Phosphosignaling Cascades Derived from Connected Pairs
We further extended the analysis from pairs to two-level signaling cascades that include the first and second degree substrates of the druggable kinase (Methods). We hypothesize in these signaling cascades, the activated kinases could trigger phosphorylation of multiple downstream targets at multiple steps, thereby presenting heightened opportunities for targeted inhibition treatment.
Out of 28 kinases associated with at least one cis or trans-associated phosphosite, 16 also had second-degree substrates (Supplemental Table 17). AKT1, BRAF, EGFR, JAK2, PRKCE, and PLK1 showed cis and trans interactions that may help confirm activity levels (Fig. 6, supplemental Fig. S11). In addition to its cis associations, AKT1 also associated with phosphosites of ILF3, ETS1, and GSK3A. In turn, GSK3A is associated with 3 phosphosites of NDRG1, 2 sites of RICTOR, and 1 site each in LARP1 and ANKS1A (Fig. 6A). BRAF also has multiple cis-associated phosphosites and is associated with phospho-MAP2K1/2 (MEK1/2), which are associated with downstream phosphosites at MTA1, KRT8, and PRKCD (Fig. 6B). Finally, high levels of EGFR phosphorylation were mostly observed in basal subtype breast cancers and these tumors also exhibited high phosphosite levels in GAB1, PLCG1, FAM129B, and PTK2; PTK2 phosphoprotein is further associated with phosphosites of PPP1R13L and PXN (Fig. 6C). The association of the primary druggable kinase with its signaling cascade could strengthen the rationale for targeted inhibition in tumors showing co-activation.
Fig. 6.
Druggable kinase-substrate cascades originating from (A) AKT1 (B) BRAF and (C) EGFR in the 77 breast cancer samples. The samples in the heatmap were ordered by the phosphoprotein level of each of the kinases (derived using phosphosites with the highest connectivity, Methods). For each node in each network diagram, the color represents the relative level of basal compared with luminal A/B breast cancers, where blue indicates higher level in luminal and red indicates higher level in basal tumors. For the edges, the darkness of the color is scaled by the correlation coefficient and the width is scaled by -log(FDR) of the association.
Kinase-Substrate Pairs Correlated with Clinical and Immune Features
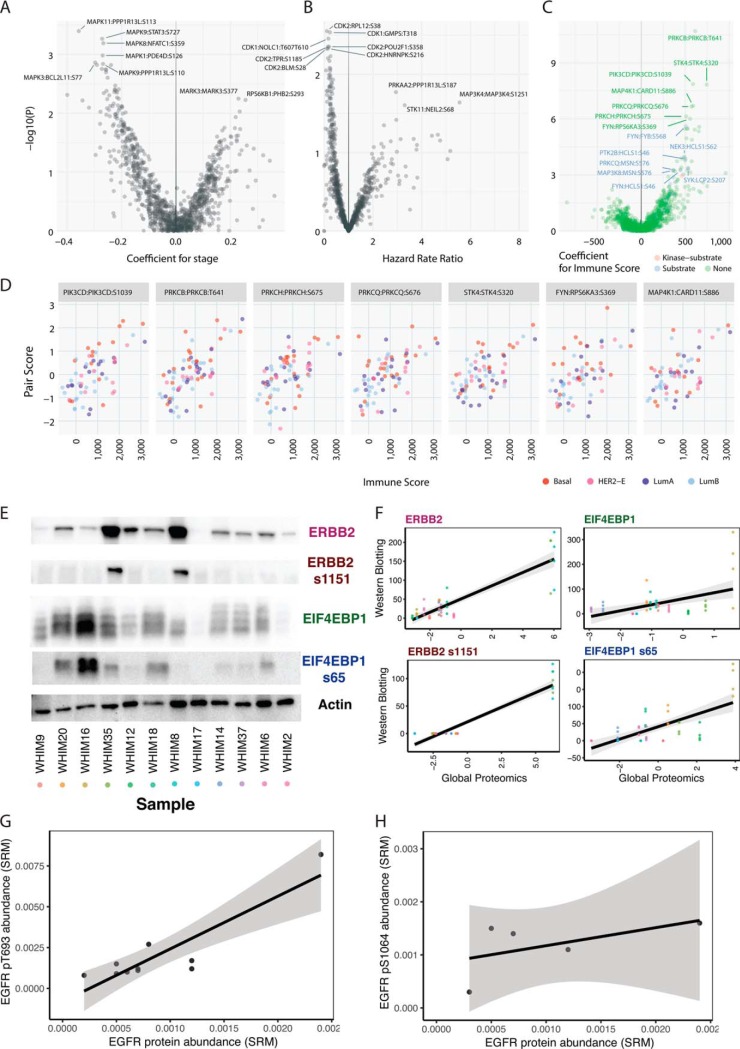
Last, we sought to identify kinase-substrate pairs associated with clinical characteristics of breast cancer. For each kinase-substrate pair, we conducted a regression analysis using the aforementioned kinase-substrate score as the independent variable versus pathological stage or immune scores as the dependent variable, adjusting for their PAM50 subtypes as a covariate (Experimental Procedures). One hundred seventy-five pairs showed potential association (p < 0.05) with pathogical stage (Fig. 7A, supplemental Table S18). Kinase-substrate pairs stemmed from MAP kinases, including MAPK11:PPP1R13L pS113, MAPK9:STAT3 pS727, and MAPK8:NFATC1 pS359 showed top correlations with earlier clinical stages whereas MARK3:MARK3 pS377 and RPS6KB1:PHB2 pS293 are associated with later stages. We also conducted a survival analysis using the kinase-substrate score as the predictor and the PAM50 subtypes as a covariate in a Cox proportional hazards model, finding 83 pairs are potentially associated with survival (Fig. 7B, supplemental Table S19). Notably, these pairs are dominated by 32 CDK1 and 29 CDK2 trans-regulated pairs correlated with longer overall survival (Hazard Rate Ratio < 1) in the breast cancer cohort. Signaling pairs associated with poor survival include MAP3K11:MAP3K11 pS789S793, PRKAA2:PPP1R13L pS187, and NEK9:NEK9 pS868, supporting previous results showing the functionality of NEK9 in MAPK/MEK signaling and resistance to PI3K inhibitor treatment (52).
Fig. 7.
Clinical association of kinase-substrate pairs. A, Volcano plot showing association of kinase-substrate pairs with pathological stage. Positive coefficient denotes higher kinase-substrate scores associating with more advanced pathological stage. B, Volcano plot showing association of kinase-substrate pairs with survival. Hazard rate ratios greater than 1 denote higher kinase-substrate scores associating with poor survival. C, Volcano plot showing association of kinase-substrate pairs with transcriptome-based immune signature score, as calculated by the ESTIMATE algorithm. Positive coefficients denote higher kinase-substrate scores associating with higher immune scores. The color of each pair indicates whether its kinase or substrate belongs to the immune gene list used by ESTIMATE. D, Top kinase-substrate pairs (p < 1e-6) associated with immune scores where each dot indicates one breast cancer sample. E, Western blotting of ERBB2 (HER2), ERBB2 s1151 (p.HER2 S1151), EIF4EBP1 (4E-BP1), and EIF4EBP1 s65 (pEIF4E-BP1 s65) and actin control in 12 breast cancer PDX samples. F, Correlation between protein/phosphoprotein expression measured through Western blotting versus high-throughput proteomics. Each dot indicates one sample in one experimental replicate. The dots are colored by the samples listed in (E). Four replicates were quantified through Western blotting for statistical analyses. G, Correlation between EGFR protein levels and p.T693 phosphosite level as measured through selected reaction monitoring (SRM). H, Correlation between EGFR protein levels and p.S1064 phosphosite level as measured through SRM. For (G) and (H), each data point represent samples with quantifications in a cohort of 12 independent human breast cancer samples.
We identified 160 pairs potentially associated (linear regression, p < 0.05, 40 pairs with FDR < 0.05) with transcriptomically derived immune score calculated using the ESTIMATE algorithm (53) (Experimental Procedures, Fig. 7C, 7D, supplemental Table 20). We then specifically asked whether the associated pairs overlapped directly with the immune cell signature genes (53). Our analysis showed that 15 pairs positively associated with the immune scores have their respective substrates as immune signature genes with significant enrichment (15/160 versus 11/918, Fisher's Exact Test, p = 3.6e-07). Notably, the top pairs among the other associated 145 pairs also involve kinases known to be of critical importance in immune systems, including PRKCQ (54), MAP4K1 (55), FYN (56), SYK (57), and JAK2 (58) (Fig. 7C, top 6 pairs shown in Fig. 7D).
Experimental Validation of Regulated Pairs from Global Proteomics
We applied two independent technologies, Western blotting and selected reaction monitoring (SRM), to validate multiple kinase-substrate pairs discovered in our global proteomics dataset (Methods). We first conducted Western blotting to validate our observations of up-regulated proteins and phosphosites found in the high throughput proteomics approach using PDX tumors from the Washington University Human in Mouse (WHIM) collection (15, 59). Specifically, we measured expression of ERBB2, ERBB2 pS1151, EIF4EBP1, and EIF4EBP1 pS65 in 12 PDX breast tumors where these markers showed expressed variability in mass spectrometry data (Fig. 7E). For all 4 markers, we found significant associations between expression quantified through Western blotting and mass spectrometry (Fig. 7F, linear regression, p < 0.0005).
For ERBB2, the HER2-positive WHIM8 and WHIM35 showed notably high ERBB2 and ERBB2 pS1151 in both quantifications (Fig. 7E, 7F). For EIF4EBP1, Western blotting showed substantial increase of EIF4EBP1 and EIF4EBP1 pS65 for WHIM16, the top EIF4EBP1-expresser in MS data. However, the expression levels in other tumors were lower, not fully recapitulating the dynamic range in MS quantification (Fig. 7E, 7F). Of note, the samples were ordered from left to right (WHIM9 to WHIM2) by their pair scores of MAPK3: EIF4EBP1 pS65, an experimentally validated regulated pairs (60) also identified in our study. However, although their phosphosite levels do distinctly segregate into the highly expressed and poorly expressed groups, our results also demonstrate that EIF4EBP1 pS65 is likely regulated by other contextual factors in vivo.
We then applied a targeted proteomics approach SRM to validate cis-regulated pairs found in EGFR in 12 independent primary human breast tumor samples (Methods, Fig. 7G, 7H). For both phosphosites EGFR pT693 and pS1064, we observed positive associations. EGFR pT693 is significantly associated with EGFR proteins (p = 5.74E-04); EGFR pS1064 was not associated but we only obtained EGFR pS1064 quantifications in 5 samples. EGFR pT693 maps to the juxtamembrane region of the protein (61) and was found to be upregulated in mutant PIK3CA knockin cells (62). Overall, the validation conducted in separate sample sets using different technologies validate the association we identified for ERBB2, EIF4EBP1, and EGFR.
DISCUSSION
We present a systematic discovery of quantitatively correlated kinase-substrate pairs in breast cancer (Fig. 1D). The high-throughput dataset generated by LC-MS/MS enabled global assessment of 33,239 phosphosites, 19,521 of which were not observed in two of the most comprehensive phosphosite databases, UniProt and Phospho.ELM (Fig. 1A, 1B). Our analysis allowed us to identify 2134 (387 cis and 1,747 trans) kinase/substrate correlation relationships; using the same bioinformatics pipeline, analysis based on RPPA-detected phosphosites only found 4 (supplemental Table S21) and seldom interrogated more than one phosphosite on a protein, further stressing the need of evaluating interactions through global phosphoproteomics in patient samples. This result clearly suggests that more regulated phosphosites in cancer will emerge with additional, even larger mass spectrometry-based proteome studies. Although our analyses have enlarged the catalogue of phospho-regulations, larger sample sizes will be required to fully establish findings and to discover weaker associations. Specifically, we identified many of cis/trans correlations in larger sets above 70 samples with concurrently observed kinase-substrate data (supplemental Fig. S2). Further, our current datasets have limited observations in multiple kinases and substrates and even more sensitive approaches will be required to assess their phosphorylation states. For example, this serine-rich dataset may also be complemented by observations using other techniques enriching for tyrosine residues (63–65), which may enhance findings of auto-phosphorylated tyrosines in receptor tyrosine kinases, including EGFR and HER2. Increased sample sizes will also enable more complex modeling of the multi-to-multi kinase/substrate relationships not fully accounted for in our current work.
We identified 61.4% (387/630) phosphosites of kinases showing significant cis-association, many of which were concentrated in known or in nominated breast cancer proteins, such as ERBB2, RRPS6KA4, NEK9, RIPK2, and PAK1 (Fig. 2). In contrast, only 4.51% (1747/38,710) trans kinase-substrate pairs showed significant association (Fig. 3). For example, PRKACA has only 23 out of 1,652 corresponding substrate sites being regulated and PRKCZ only has 1 out of 542. It is possible that trans substrate usage may be highly tissue-specific and that some of the previously curated pairs do not interact in breast cancer. Another possibility is that some kinase-substrate pairs established through in vitro evidence are not relevant in physiological environments, although the validation rate for in vivo and in vitro pairs do not differ significantly (supplemental Table S22). Future investigation using data across tissue and cancer types would be pivotal in addressing whether we observe tissue-specific usage of kinase-substrate pairs. Such studies could also reveal the consistently high/low pairs (Fig. 1D) in each cancer type and highlight cancer-specific signaling. In addition to analyses of kinase-substrate pairs activated in specific breast cancer subtypes (Fig. 2, 3,6), expanded cohorts should also enable discovery of pairs associated with specific drivers or histological features.
Our approach validates and complements experimentally or binding motif-predicted phosphosites regulated by kinase (Fig. 1E). Although the regulated sites we discovered are based on observation using primary tumors, their casual relationships can be further validated by experimental perturbation or time-series analyses. Methodologies unifying diverse predictions and observations will be required to understand how each phosphosite is regulated.
The kinase-substrate pairs we discovered also complement previous studies focusing on singleton drivers of cancer. Conventionally, pathways were mostly constructed by linking single candidate driver genes (such as significantly mutated genes or focally amplified genes) through known interactions. Our approach detects the association of the signaling pairs in vivo, and thus directly validates the signaling impact of driver events. This approach also enabled us to build relevant subcascades stemming from potentially druggable kinases AKT1, BRAF, and EGFR (Fig. 6). To compare with other network-generating studies, we also constructed a network of all observed regulations (supplemental Fig. S12). However, such approaches may obfuscate activated subnetworks, as downstream phosphorylation targets could be mediated by multiple kinases.
This first large-scale examination of over 33,000 phosphosites in breast cancers creates a foundation for druggable analysis of kinase-substrate pairs beyond singleton druggable events (Fig. 5). Predictive value of response to targeted treatment has been limited in samples for some clearly-defined driver events in cancer. For example, PIK3CA mutation status cannot conclusively predict treatment response to PI3K inhibitor in clinical studies (66). Co-occurrence of downstream activating events, as we have observed for AKT1, BRAF, ERBB2, IGF1R, and RAF1 (Fig. 5B), may further support targeted inhibition. In both breast cancer PDXs showing ERBB2:ERBB2 pS1151 cis outlier pairs identified using both global proteomics and Western blotting (supplemental Fig. S9), lapatinib treatment significantly reduced tumor growth (15). Further, we previously observed varied response to PI3K inhibition therapy in 3 of the breast cancer PDX models, where both WHIM18 and WHIM20 responded to PI3K/mTOR inhibitors and WHIM16 was resistant (15). WHIM18 and WHIM20 also showed notably high pair scores of the cis pair AKT1:AKT1 pS126S129 than WHIM16 (supplemental Fig. S9). Larger data sets using tumors undergoing treatments, such as the ongoing PDXNet projects establishing and investigating thousands of PDXs, will be instrumental for identifying treatment responses and formally testing the hypothesis that activated signaling pairs can accurately predict response to inhibition.
Further, we identified outlier kinase-substrate pairs in samples without singleton events for kinases including EGFR, MAPK3, and MAPK14 (Fig. 5B). Inhibition of the MAP2K1/2 (MEK1/2) upstream of MAP kinases surpressed the MAPK signaling pathway and its combinatory treatment with RTK inhibitors have resulted in tumor regression of triple-negative breast tumors (8). Our discovery of MAPK mediated pairs reveals therapeutic opportunities. Finally, resistance mechanisms often consist of rewiring of signaling pathways and could be further explored through high-throughput proteomics and approaches developed in this study.
Signaling networks are crucially important in cancer. However, large-scale omic studies to date have mainly focused on singling-out individual driver events and rarely investigated their signaling impact. Studying kinase-substrate relations in vivo, and most particularly in tumor samples from patients undergoing therapy, will uncover the wiring of signaling networks in each tumor and likely improve treatment design.
DATA AVAILABILITY
All SRM results including the assay characterization data are organized as Skyline files on the Panorama server (36) and can be accessed via the link https://panoramaweb.org/kinasesubstrate.url.
Supplementary Material
Acknowledgments
We thank The Cancer Genome Atlas (cancergenome.nih.gov) and CPTAC as the source of primary data and members of the CPTAC Research Network for helpful discussions.
Footnotes
* This work was supported by the National Cancer Institute grants U24CA160035 to R.R.T. and M.J.E., U24CA210972 to D.F., L.D., and S.H.P., U24CA211006 to L.D. and R.G., and National Human Genome Research Institute grant U01HG006517 to L.D. The authors declare no competing financial interests.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- RPPA
- reverse-phase protein array
- LC
- liquid chromatography
- PDX
- patient-derived xenograft.
REFERENCES
- 1. TCGA. (2012) Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., Gatza M. L., Wilkerson M., Perou C. M., Yellapantula V., Huang K.-l., Lin C., McLellan M. D., Yan P., Davies S. R., Townsend R. R., Skates S. J., Wang J., Zhang B., Kinsinger C. R., Mesri M., Rodriguez H., Ding L., Paulovich A. G., Fenyö D., Ellis M. J., and Carr S. A. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 534, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gujral T. S., Karp R. L., Finski A., Chan M., Schwartz P. E., MacBeath G., and Sorger P. (2013) Profiling phospho-signaling networks in breast cancer using reverse-phase protein arrays. Oncogene. 32, 3470–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akbani R., Ng P. K. S., Werner H. M. J., Shahmoradgoli M., Zhang F., Ju Z., Liu W., Yang J.-Y., Yoshihara K., Li J., Ling S., Seviour E. G., Ram P. T., Minna J. D., Diao L., Tong P., Heymach J. V., Hill S. M., Dondelinger F., Städler N., Byers La Meric-Bernstam F., Weinstein J. N., Broom B. M., Verhaak R. G. W., Liang H., Mukherjee S., Lu Y., and Mills G. B. (2014) A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 5, 3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Al-Ejeh F., Miranda M., Shi W., Simpson P. T., Song S., Vargas A. C., Saunus J. M., Smart C. E., Mariasegaram M., Wiegmans A. P., Chenevix-Trench G., Lakhani S. R., and Khanna K. K. (2014) Kinome profiling reveals breast cancer heterogeneity and identifies targeted therapeutic opportunities for triple negative breast cancer. Oncotarget. 5, 3145–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H., Liu T., Zhang Z., Payne Samuel H., Zhang B., McDermott Jason E., Zhou J.-Y., Petyuk Vladislav A., Chen L., Ray D., Sun S., Yang F., Chen L., Wang J., Shah P., Cha Seong W., Aiyetan P., Woo S., Tian Y., Gritsenko Marina A., Clauss Therese R., Choi C., Monroe Matthew E., Thomas S., Nie S., Wu C., Moore Ronald J., Yu K.-H., Tabb David L., Fenyö D., Bafna V., Wang Y., Rodriguez H., Boja Emily S., Hiltke T., Rivers Robert C., Sokoll L., Zhu H., Shih I.-M., Cope L., Pandey A., Zhang B., Snyder Michael P., Levine Douglas A., Smith Richard D., Chan Daniel W., Rodland Karin D., Carr Steven A., Gillette Michael A., Klauser Karl R., Kuhn E., Mani D. R., Mertins P., Ketchum Karen A., Thangudu R., Cai S., Oberti M., Paulovich Amanda G., Whiteaker Jeffrey R., Edwards Nathan J., McGarvey Peter B., Madhavan S., Wang P., Chan Daniel W., Pandey A., Shih I.-M., Zhang H., Zhang Z., Zhu H., Cope L., Whiteley Gordon A., Skates Steven J., White Forest M., Levine Douglas A., Boja Emily S., Kinsinger Christopher R., Hiltke T., Mesri M., Rivers Robert C., Rodriguez H., Shaw Kenna M., Stein Stephen E., Fenyo D., Liu T., McDermott Jason E., Payne Samuel H., Rodland Karin D., Smith Richard D., Rudnick P., Snyder M., Zhao Y., Chen X., Ransohoff David F., Hoofnagle Andrew N., Liebler Daniel C., Sanders Melinda E., Shi Z., Slebos Robbert J. C., Tabb David L., Zhang B., Zimmerman Lisa J., Wang Y., Davies Sherri R., Ding L., Ellis Matthew J. C., Townsend R. R., Barrès V., Ouellet V., Lafontaine J., Tonin P. N., Provencher D. M., Mes-Masson A. M., Cai R. L., Yan-Neale Y., Cueto M. A., Xu H., Cohen D., Network CGAR, Network CGAR, Collins B. C., Gillet L. C., Rosenberger G., Röst H. L., Vichalkovski A., Gstaiger M., Aebersold R., Cope L., Wu R. C., Shih I., Wang T. L., Creighton C. J., Hernandez-Herrera A., Jacobsen A., Levine D. A., Mankoo P., Schultz N., Du Y., Zhang Y., Larsson E., Sheridan R., Network CGAR, al. e, Dizon D. S., Damstrup L., Finkler N. J., Lassen U., Celano P., Glasspool R., Crowley E., Lichenstein H. S., Knoblach P., Penson R. T., Fanjul-Fernández M., Quesada V., Cabanillas R., Cadiñanos J., Fontanil T., Obaya A., Ramsay A. J., Llorente J. L., Astudillo A., Cal S., López-Otín C., Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C. al. e, Gadducci A., Lanfredini N., Sergiampietri C., Gong F., Miller K. M., Hanahan D., Weinberg R. A., Jovanovic M., Rooney M. S., Mertins P., Przybylski D., Chevrier N., Satija R., Rodriguez E. H., Fields A. P., Schwartz S., Raychowdhury R. al. e, Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M. S., Gramolini A. O., Morris Q., Hallett M. T. al. e, Köbel M., Huntsman D., Gilks C. B., Komili S., Silver P. A., Kuo K. T., Guan B., Feng Y., Mao T. L., Chen X., Jinawath N., Wang Y., Kurman R. J., Shih I., Wang T. L., Langfelder P., Horvath S., Liu J., Matulonis U. A., Lu P., Vogel C., Wang R., Yao X., Marcotte E. M., Mackay H. J., Hirte H., Colgan T., Covens A., MacAlpine K., Grenci P., Wang L., Mason J., Pham P. A., Tsao M. S. al. e, Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bähler J., McEllin B., Camacho C. V., Mukherjee B., Hahm B., Tomimatsu N., Bachoo R. M., Burma S., Meng H., Tian L., Zhou J., Li Z., Jiao X., Li W. W., Plomann M., Xu Z., Lisanti M. P., Wang C., Pestell R. G., Mertins P., Yang F., Liu T., Mani D. R., Petyuk V. A., Gillette M. A., Clauser K. R., Qiao J. W., Gritsenko M. A., Moore R. J. al. e, Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., CPTAC N, al. e, Mielcarek M., Zielonka D., Carnemolla A., Marcinkowski J. T., Guidez F., Modesitt S. C., Sill M., Hoffman J. S., Bender D. P., Group G. O., Pradeep S., Kim S. W., Wu S. Y., Nishimura M., Chaluvally-Raghavan P., Miyake T., Pecot C. V., Kim S. J., Choi H. J., Bischoff F. Z. al. e, Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S. al. e, Ruggles K. V., Tang Z., Wang X., Grover H., Askenazi M., Teubl J., Cao S., McLellan M. D., Clauser K. R., Tabb D. L. al. e, Tang J., Cho N. W., Cui G., Manion E. M., Shanbhag N. M., Botuyan M. V., Mer G., Greenberg R. A., Vang R., Levine D. A., Soslow R. A., Zaloudek C., Shih I., Kurman R. J., Verhaak R. G., Tamayo P., Yang J. Y., Hubbard D., Zhang H., Creighton C. J., Fereday S., Lawrence M., Carter S. L., Mermel C. H. al. e, Vogel C., Marcotte E. M., Woo S., Cha S. W., Na S., Guest C., Liu T., Smith R. D., Rodland K. D., Payne S., Bafna V., Woodbine L., Gennery A. R., Jeggo P. A., Wu L., Candille S. I., Choi Y., Xie D., Jiang L., Li-Pook-Than J., Tang H., Snyder M., Yang L., Duff M. O., Graveley B. R., Carmichael G. G., Chen L. L., Yang J. Y., Yoshihara K., Tanaka K., Hatae M., Masuzaki H., Itamochi H., Takano M., Ushijima K., Tanyi J. L., Coukos G., Network CGATR, al. e, Yi B., Zhang Y., Zhu D., Zhang L., Song S., He S., Zhang B., Li D., Zhou J., Yu G., Zhang B., Bova G. S., Xu J., Shih I., Wang Y., Zhang B., Li H., Riggins R. B., Zhan M., Xuan J., Zhang Z., Hoffman E. P., Clarke R., Wang Y., Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., CPTAC N, et al. (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 166, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J. C., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J. C., and Liebler D. C. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature. 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duncan J. S., Whittle M. C., Nakamura K., Abell A. N., Midland A. A., Zawistowski J. S., Johnson N. L., Granger D. A., Jordan N. V., Darr D. B., Usary J., Kuan P. F., Smalley D. M., Major B., He X., Hoadley K. A., Zhou B., Sharpless N. E., Perou C. M., Kim W. Y., Gomez S. M., Chen X., Jin J., Frye S. V., Earp H. S., Graves L. M., and Johnson G. L. (2012) Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 149, 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fleuren E. D., Zhang L., Wu J., and Daly R. J. (2016) The kinome 'at large' in cancer. Nat. Rev. Cancer. 16, 83–98 [DOI] [PubMed] [Google Scholar]
- 10. Newman R. H., Hu J., Rho H. S., Xie Z., Woodard C., Neiswinger J., Cooper C., Shirley M., Clark H. M., Hu S., Hwang W., Jeong J. S., Wu G., Lin J., Gao X., Ni Q., Goel R., Xia S., Ji H., Dalby K. N., Birnbaum M. J., Cole P. A., Knapp S., Ryazanov A. G., Zack D. J., Blackshaw S., Pawson T., Gingras A. C., Desiderio S., Pandey A., Turk B. E., Zhang J., Zhu H., and Qian J. (2013) Construction of human activity-based phosphorylation networks. Mol. Syst. Biol. 9, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jorgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., and Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell. 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horn H., Schoof E. M., Kim J., Robin X., Miller M. L., Diella F., Palma A., Cesareni G., Jensen L. J., and Linding R. (2014) KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods. 11, 603–604 [DOI] [PubMed] [Google Scholar]
- 13. Ochoa D., Jonikas M., Lawrence R. T., El Debs B., Selkrig J., Typas A., Villen J., Santos S. D., and Beltrao P. (2016) An atlas of human kinase regulation. Mol. Syst. Biol. 12, 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Creixell P., Schoof E. M., Simpson C. D., Longden J., Miller C. J., Lou H. J., Perryman L., Cox T. R., Zivanovic N., Palmeri A., Wesolowska-Andersen A., Helmer-Citterich M., Ferkinghoff-Borg J., Itamochi H., Bodenmiller B., Erler J. T., Turk B. E., and Linding R. (2015) Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell. 163, 202–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang K. L., Li S., Mertins P., Cao S., Gunawardena H. P., Ruggles K. V., Mani D. R., Clauser K. R., Tanioka M., Usary J., Kavuri S. M., Xie L., Yoon C., Qiao J. W., Wrobel J., Wyczalkowski M. A., Erdmann-Gilmore P., Snider J. E., Hoog J., Singh P., Niu B., Guo Z., Sun S. Q., Sanati S., Kawaler E., Wang X., Scott A., Ye K., McLellan M. D., Wendl M. C., Malovannaya A., Held J. M., Gillette M. A., Fenyo D., Kinsinger C. R., Mesri M., Rodriguez H., Davies S. R., Perou C. M., Ma C., Reid Townsend R., Chen X., Carr S. A., Ellis M. J., and Ding L. (2017) Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat. Commun. 8, 14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J., Lu Y., Akbani R., Ju Z., Roebuck P. L., Liu W., Yang J. Y., Broom B. M., Verhaak R. G., Kane D. W., Wakefield C., Weinstein J. N., Mills G. B., and Liang H. (2013) TCPA: a resource for cancer functional proteomics data. Nat. Methods. 10, 1046–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langfelder P., and Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niu B., Scott A. D., Sengupta S., Bailey M. H., Batra P., Ning J., Wyczalkowski M. A., Liang W. W., Zhang Q., McLellan M. D., Sun S. Q., Tripathi P., Lou C., Ye K., Mashl R. J., Wallis J., Wendl M. C., Chen F., and Ding L. (2016) Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat. Genet. 48, 827–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2014) PhosphoSitePlus: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou W., Chen T., Chong Z., Rohrdanz M. A., Melott J. M., Wakefield C., Zeng J., Weinstein J. N., Meric-Bernstam F., Mills G. B., and Chen K. (2015) TransVar: a multilevel variant annotator for precision genomics. Nat. Methods. 12, 1002–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 24. Hu J., Rho H. S., Newman R. H., Zhang J., Zhu H., and Qian J. (2014) PhosphoNetworks: a database for human phosphorylation networks. Bioinformatics. 30, 141–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu C., Shi T., Brown J. N., He J., Gao Y., Fillmore T. L., Shukla A. K., Moore R. J., Camp D. G. 2nd, Rodland K. D., Qian W. J., Liu T., and Smith R. D. (2014) Expediting SRM assay development for large-scale targeted proteomics experiments. J Proteome Res. 13, 4479–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Welton J. L., Khanna S., Giles P. J., Brennan P., Brewis I. A., Staffurth J., Mason M. D., and Clayton A. (2010) Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteomics 9, 1324–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim S., Gupta N., and Pevzner P. A. (2008) Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 7, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S., and Pevzner P. A. (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., Grant R. P., Hoofnagle A. N., Huttenhain R., Koomen J. M., Liebler D. C., Liu T., MacLean B., Mani D. R., Mansfield E., Neubert H., Paulovich A. G., Reiter L., Vitek O., Aebersold R., Anderson L., Bethem R., Blonder J., Boja E., Botelho J., Boyne M., Bradshaw R. A., Burlingame A. L., Chan D., Keshishian H., Kuhn E., Kinsinger C., Lee J. S., Lee S. W., Moritz R., Oses-Prieto J., Rifai N., Ritchie J., Rodriguez H., Srinivas P. R., Townsend R. R., Van Eyk J., Whiteley G., Wiita A., and Weintraub S. (2014) Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tabb D. L., Wang X., Carr S. A., Clauser K. R., Mertins P., Chambers M. C., Holman J. D., Wang J., Zhang B., Zimmerman L. J., Chen X., Gunawardena H. P., Davies S. R., Ellis M. J., Li S., Townsend R. R., Boja E. S., Ketchum K. A., Kinsinger C. R., Mesri M., Rodriguez H., Liu T., Kim S., McDermott J. E., Payne S. H., Petyuk V. A., Rodland K. D., Smith R. D., Yang F., Chan D. W., Zhang B., Zhang H., Zhang Z., Zhou J. Y., and Liebler D. C. (2016) Reproducibility of differential proteomic technologies in CPTAC fractionated xenografts. J. Proteome Res. 15, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsai C. F., Wang Y. T., Chen Y. R., Lai C. Y., Lin P. Y., Pan K. T., Chen J. Y., Khoo K. H., and Chen Y. J. (2008) Immobilized metal affinity chromatography revisited: pH/acid control toward high selectivity in phosphoproteomics. J. Proteome Res. 7, 4058–4069 [DOI] [PubMed] [Google Scholar]
- 34. Tsai C. F., Hsu C. C., Hung J. N., Wang Y. T., Choong W. K., Zeng M. Y., Lin P. Y., Hong R. W., Sung T. Y., and Chen Y. J. (2014) Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Anal. Chem. 86, 685–693 [DOI] [PubMed] [Google Scholar]
- 35. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 36. Sharma V., Eckels J., Schilling B., Ludwig C., Jaffe J. D., MacCoss M. J., and MacLean B. (2018) Panorama public: a public repository for quantitative data sets processed in Skyline. Mol. Cell. Proteomics 17, 1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. The UniProt Consortium (2018) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46, 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dinkel H., Chica C., Via A., Gould C. M., Jensen L. J., Gibson T. J., and Diella F. (2011) Phospho.ELM: a database of phosphorylation sites–update. Nucleic Acids Res. 39, D261–D267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chartier M., Chenard T., Barker J., and Najmanovich R. (2013) Kinome Render: a stand-alone and web-accessible tool to annotate the human protein kinome tree. PeerJ. 1, e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ciriello G., Miller M. L., Aksoy B. A., Senbabaoglu Y., Schultz N., and Sander C. (2013) Emerging landscape of oncogenic signatures across human cancers. Nat. Gen. 45, 1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bailey M. H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M. C., Kim J., Reardon B., Ng P. K., Jeong K. J., Cao S., Wang Z., Gao J., Gao Q., Wang F., Liu E. M., Mularoni L., Rubio-Perez C., Nagarajan N., Cortes-Ciriano I., Zhou D. C., Liang W. W., Hess J. M., Yellapantula V. D., Tamborero D., Gonzalez-Perez A., Suphavilai C., Ko J. Y., Khurana E., Park P. J., Van Allen E. M., Liang H., Group M. C. W., Cancer Genome Atlas Research, N., Lawrence M. S., Godzik A., Lopez-Bigas N., Stuart J., Wheeler D., Getz G., Chen K., Lazar A. J., Mills G. B., Karchin R., and Ding L. (2018) Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang K. L., Mashl R. J., Wu Y., Ritter D. I., Wang J., Oh C., Paczkowska M., Reynolds S., Wyczalkowski M. A., Oak N., Scott A. D., Krassowski M., Cherniack A. D., Houlahan K. E., Jayasinghe R., Wang L. B., Zhou D. C., Liu D., Cao S., Kim Y. W., Koire A., McMichael J. F., Hucthagowder V., Kim T. B., Hahn A., Wang C., McLellan M. D., Al-Mulla F., Johnson K. J., Cancer Genome Atlas Research, N., Lichtarge O., Boutros P. C., Raphael B., Lazar A. J., Zhang W., Wendl M. C., Govindan R., Jain S., Wheeler D., Kulkarni S., Dipersio J. F., Reimand J., Meric-Bernstam F., Chen K., Shmulevich I., Plon S. E., Chen F., and Ding L. (2018) Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355–370.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., Gatza M. L., Wilkerson M., Perou C. M., Yellapantula V., Huang K. L., Lin C., McLellan M. D., Yan P., Davies S. R., Townsend R. R., Skates S. J., Wang J., Zhang B., Kinsinger C. R., Mesri M., Rodriguez H., Ding L., Paulovich A. G., Fenyo D., Ellis M. J., Carr S. A., and NCI CPTAC (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heinrich R., Neel B. G., and Rapoport T. A. (2002) Mathematical models of protein kinase signal transduction. Mol. Cell. 9, 957–970 [DOI] [PubMed] [Google Scholar]
- 45. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., and Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 46. Scheer J. M., Sandoval W., Elliott J. M., Shao L., Luis E., Lewin-Koh S. C., Schaefer G., and Vandlen R. (2012) Reorienting the Fab domains of trastuzumab results in potent HER2 activators. PLoS ONE 7, e51817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rossomando A. J., Wu J., Michel H., Shabanowitz J., Hunt D. F., Weber M. J., and Sturgill T. W. (1992) Identification of Tyr-185 as the site of tyrosine autophosphorylation of recombinant mitogen-activated protein kinase p42mapk. Proc. Natl. Acad. Sci. U.S.A. 89, 5779–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seger R., and Krebs E. G. (1995) The MAPK signaling cascade. FASEB J. 9, 726–35 [PubMed] [Google Scholar]
- 49. Drebin J. A., Link V. C., Stern D. F., Weinberg R. A., and Greene M. I. (1985) Down-modulation of an oncogene protein product and reversion of the transformed phenotype by monoclonal antibodies. Cell 41, 697–706 [DOI] [PubMed] [Google Scholar]
- 50. Rubio-Perez C., Tamborero D., Schroeder M. P., Antolin A. A., Deu-Pons J., Perez-Llamas C., Mestres J., Gonzalez-Perez A., and Lopez-Bigas N. (2015) In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27, 382–396 [DOI] [PubMed] [Google Scholar]
- 51. Safaei J., Manuch J., Gupta A., Stacho L., and Pelech S. (2011) Prediction of 492 human protein kinase substrate specificities. Proteome Sci. 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mundt F., Rajput S., Li S., Ruggles K. V., Mooradian A. D., Mertins P., Gillette M. A., Krug K., Guo Z., Hoog J., Erdmann-Gilmore P., Primeau T., Huang S., Edwards D. P., Wang X., Wang X., Kawaler E., Mani D. R., Clauser K. R., Gao F., Luo J., Davies S. R., Johnson G. L., Huang K. L., Yoon C. J., Ding L., Fenyo D., Ellis M. J., Townsend R. R., Held J. M., Carr S. A., and Ma C. X. (2018) Mass spectrometry-based proteomics reveals potential roles of NEK9 and MAP2K4 in resistance to PI3K inhibition in triple-negative breast cancers. Cancer Res. 78, 2732–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P. W., Levine D. A., Carter S. L., Getz G., Stemke-Hale K., Mills G. B., and Verhaak R. G. W. (2013) Inferring tumor purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Altman A., and Villalba M. (2002) Protein kinase C theta (PKC theta): A key enzyme in T cell life and death. J. Biochem. 132, 841–846 [DOI] [PubMed] [Google Scholar]
- 55. Yin H. L., Shi Z. B., Jiao S., Chen C. C., Wang W. J., Greene M. I., and Zhou Z. C. (2012) Germinal center kinases in immune regulation. Cell Mol. Immunol. 9, 439–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ben Mkaddem S., Murua A., Flament H., Titeca-Beauport D., Bounaix C., Danelli L., Launay P., Benhamou M., Blank U., Daugas E., Charles N., and Monteiro R. C. (2017) Lyn and Fyn function as molecular switches that control immunoreceptors to direct homeostasis or inflammation. Nat. Commun. 8, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mocsai A., Ruland J., and Tybulewicz V. L. (2010) The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat. Rev. Immunol. 10, 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Villarino A. V., Kanno Y., and O'Shea J. J. (2017) Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol. 18, 374–384 [DOI] [PubMed] [Google Scholar]
- 59. Li S., Shen D., Shao J., Crowder R., Liu W., Prat A., He X., Liu S., Hoog J., Lu C., Ding L., Griffith O. L., Miller C., Larson D., Fulton R. S., Harrison M., Mooney T., McMichael J. F., Luo J., Tao Y., Goncalves R., Schlosberg C., Hiken J. F., Saied L., Sanchez C., Giuntoli T., Bumb C., Cooper C., Kitchens R. T., Lin A., Phommaly C., Davies S. R., Zhang J., Kavuri M. S., McEachern D., Dong Y. Y., Ma C., Pluard T., Naughton M., Bose R., Suresh R., McDowell R., Michel L., Aft R., Gillanders W., DeSchryver K., Wilson R. K., Wang S., Mills G. B., Gonzalez-Angulo A., Edwards J. R., Maher C., Perou C. M., Mardis E. R., and Ellis M. J. (2013) Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin T. A., Kong X., Haystead T. A., Pause A., Belsham G., Sonenberg N., and Lawrence J. C. Jr. (1994) PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 266, 653–656 [DOI] [PubMed] [Google Scholar]
- 61. Tong J. F., Taylor P., and Moran M. F. (2014) Proteomic Analysis of the Epidermal Growth Factor Receptor (EGFR) Interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Mol. Cell. Proteomics 13, 1644–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu X. Y., Renuse S., Sahasrabuddhe N. A., Zahari M. S., Chaerkady R., Kim M. S., Nirujogi R. S., Mohseni M., Kumar P., Raju R., Zhong J., Yang J., Neiswinger J., Jeong J. S., Newman R., Powers M. A., Somani B. L., Gabrielson E., Sukumar S., Stearns V., Qian J., Zhu H., Vogelstein B., Park B., and Pandey A. (2014) Activation of diverse signalling pathways by oncogenic PIK3CA mutations. Nat. Commun. 5, 4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sharma K., D'Souza R. C., Tyanova S., Schaab C., Wisniewski J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 64. Hochgrafe F., Zhang L., O'Toole S. A., Browne B. C., Pinese M., Porta Cubas A., Lehrbach G. M., Croucher D. R., Rickwood D., Boulghourjian A., Shearer R., Nair R., Swarbrick A., Faratian D., Mullen P., Harrison D. J., Biankin A. V., Sutherland R. L., Raftery M. J., and Daly R. J. (2010) Tyrosine phosphorylation profiling reveals the signaling network characteristics of Basal breast cancer cells. Cancer Res. 70, 9391–9401 [DOI] [PubMed] [Google Scholar]
- 65. Croucher D. R., Hochgrafe F., Zhang L., Liu L., Lyons R. J., Rickwood D., Tactacan C. M., Browne B. C., Ali N., Chan H., Shearer R., Gallego-Ortega D., Saunders D. N., Swarbrick A., and Daly R. J. (2013) Involvement of Lyn and the atypical kinase SgK269/PEAK1 in a basal breast cancer signaling pathway. Cancer Res. 73, 1969–1980 [DOI] [PubMed] [Google Scholar]
- 66. Gonzalez-Angulo A. M., and Blumenschein G. R. Jr. (2013) Defining biomarkers to predict sensitivity to PI3K/Akt/mTOR pathway inhibitors in breast cancer. Cancer Treat. Rev. 39, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chaikuad A., Tacconi E. M., Zimmer J., Liang Y., Gray N. S., Tarsounas M., and Knapp S. (2014) A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol. 10, 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Matsumoto T., Kinoshita T., Matsuzaka H., Nakai R., Kirii Y., Yokota K., and Tada T. (2012) Crystal structure of nonphosphorylated MAP2K6 in a putative auto-inhibition state. J. Biochem. 151, 541–549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SRM results including the assay characterization data are organized as Skyline files on the Panorama server (36) and can be accessed via the link https://panoramaweb.org/kinasesubstrate.url.