Xia et al. show that constricted migration causes DNA damage and a cell cycle block that is rescued by antioxidant plus myosin II inhibition or overexpressed repair factors. Cell cycle progression exhibits a threshold in the mechanically induced DNA damage.
Abstract
Migration through 3D constrictions can cause nuclear rupture and mislocalization of nuclear proteins, but damage to DNA remains uncertain, as does any effect on cell cycle. Here, myosin II inhibition rescues rupture and partially rescues the DNA damage marker γH2AX, but an apparent block in cell cycle appears unaffected. Co-overexpression of multiple DNA repair factors or antioxidant inhibition of break formation also exert partial effects, independently of rupture. Combined treatments completely rescue cell cycle suppression by DNA damage, revealing a sigmoidal dependence of cell cycle on excess DNA damage. Migration through custom-etched pores yields the same damage threshold, with ∼4-µm pores causing intermediate levels of both damage and cell cycle suppression. High curvature imposed rapidly by pores or probes or else by small micronuclei consistently associates nuclear rupture with dilution of stiff lamin-B filaments, loss of repair factors, and entry from cytoplasm of chromatin-binding cGAS (cyclic GMP-AMP synthase). The cell cycle block caused by constricted migration is nonetheless reversible, with a potential for DNA misrepair and genome variation.
Introduction
“Go-or-grow” posits that cell migration and cell cycle are mutually exclusive in space and time (Giese et al., 1996; Garay et al., 2013). Some go-or-grow mechanisms in 3D are now being modeled with Transwell pores (Beadle et al., 2008; Wolf et al., 2013; Harada et al., 2014), and for large pores, migration from contact-inhibited monolayers on the top into sparse microenvironments on the bottom promotes cell cycle reentry and growth, whereas small constricting pores seem overall disruptive (Fig. 1 A). Constricted migration causes nuclear lamina breaks (Harada et al., 2014), nuclear rupture (Denais et al., 2016; Raab et al., 2016; Irianto et al., 2017), and excess DNA damage based on immunostained foci of phospho-histone-2AX (γH2AX; Irianto et al., 2017; Pfeifer et al., 2018). However, at least one DNA damage marker (53BP1) shows no increase when immunostained (Irianto et al., 2017; Pfeifer et al., 2018), suggesting that puncta of overexpressed GFP-53BP1 in live-cell imaging (Denais et al., 2016; Raab et al., 2016) are not indicative of damage (Belin et al., 2015) and instead reflect segregation of mobile nuclear proteins into chromatin-poor pockets (Irianto et al., 2016). Accurate imaging of DNA damage sites is indeed nontrivial (Britton et al., 2013), and γH2AX foci counts after constricted migration appear to increase only ∼50% across cell cycle stages, even when blocking cell cycle (Pfeifer et al., 2018). On the other hand, cell cycle checkpoints for DNA damage (Houtgraaf et al., 2006) could in principle be activated reversibly by constricted migration and thereby shed light on 3D mechanisms of go-or-grow.
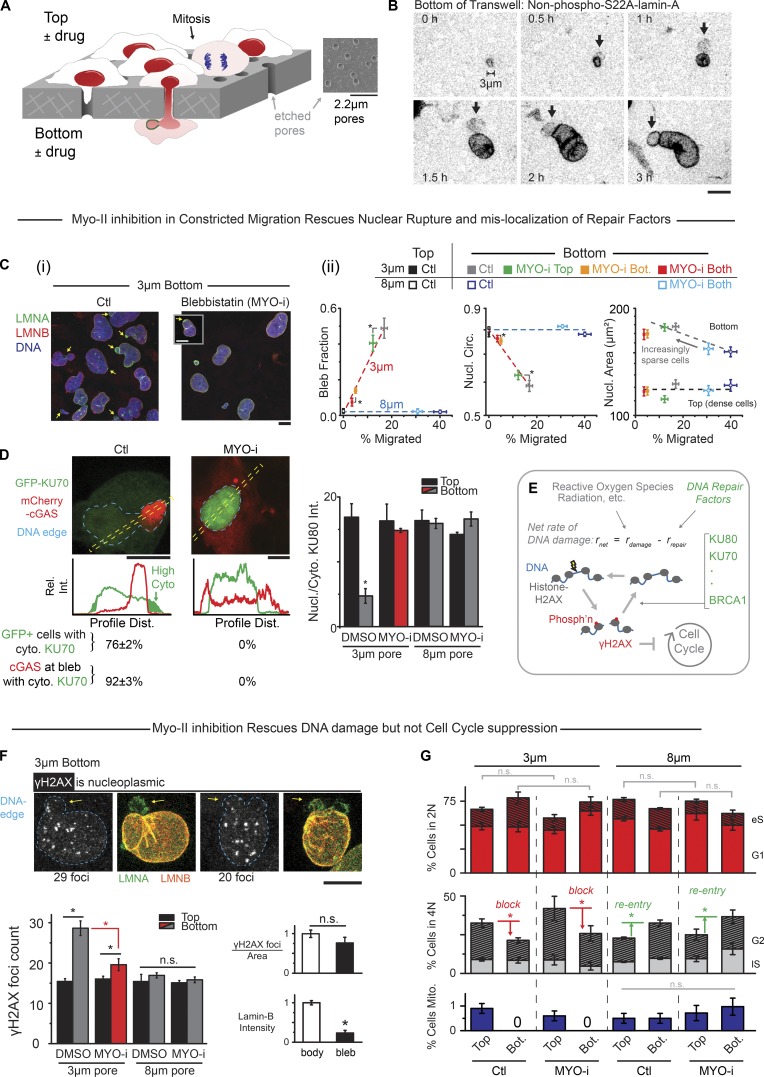
Figure 1.
MYO-i on bottom rescues nuclear rupture and DNA damage but not cell cycle suppression. (A) Nuclei rupture in constricted migration through Transwells of customized pore size that also allow asymmetric exposure to drugs. (B) Time-lapse images of A549 cell expressing GFP-lamin-A as nonphosphorylatable S22A and emerging from a 3-µm pore, with a bleb (arrows) forming at the leading tip of the nucleus. GFP-lamin-A accumulates in the bleb independently of S22 phosphorylation (see also Fig. S1 B). (Ci) In constricted migration of U2OS cells, nuclear blebs form with lamina disruptions (arrows) except when blebbistatin (MYO-i) is on bottom (Bot.). Inset: Rupture occurs occasionally (arrow) with MYO-i. (Cii) Addition of MYO-i to the 3-µm bottom or both sides of a Transwell greatly reduces migration and nuclear (Nucl.) blebs but increases circularity (Circ.). The 8-µm Transwell is used as a control (ctl; >100 cells per condition, n ≥ 3 experiments *, Pxy < 0.05; Pxy is the joint probability obtained by multiplying px and py, which assumes independence; Wang, 2013). (D) U2OS cells expressing DNA repair protein GFP-KU70 and cytoplasmic DNA-binding protein mCherry-cGAS migrated through 3-µm pores; most cells showed GFP-KU70 mislocalized to cytoplasm, and mCherry-cGAS accumulated in the nuclear bleb in control (Ctl). MYO-i prevents such nucleocytoplasmic exchange (n = 3 experiments). Bar graph: Endogenous DNA repair factor KU80 also mislocalizes to cytoplasm (cyto.), except with MYO-i or with larger pores that eliminate blebs (50∼300 cells, n > 3 experiments, *, P < 0.05). Dist., distance; Rel. Int., relative intensity. (E) DNA breaks constantly form and are repaired, but if net DNA damage is high, then damage checkpoints block cell cycle progression. Phosph’n denotes phosphorylation. (F) Foci of γH2AX (white in image) are not enriched in nuclear blebs (arrows) after 3-µm pore migration. Bar graphs: γH2AX foci measured in confocal projections are in excess on bottom except with MYO-i or with larger pores. Compared with the nuclear body, blebs are low in lamin-B as expected but equal in foci density (>100 cells, n = 5 experiments; *, P < 0.05). n.s., not significant. (G) Using EdU spike-in to label replicating DNA during Transwell migration, DNA stain intensity and EdU were used to identify a cell as 2N (nonreplicated genome) or 4N (fully replicated genome) and as G1, early S (eS), late S (lS), or G2 (see Fig. S1 G). When contact-inhibited cells migrate through large (8-µm) pores into sparse microenvironments, cells reenter cell cycle. Constricting (3-µm) pores block cell cycle and suppress mitosis (Mito.), regardless of MYO-i. No significant difference is seen between 2N/4N populations on bottom or top after treatment with blebbistatin for both 3- and 8-µm pores (n.s.; >400 cells per condition, n = 3 experiments; *, P < 0.05). All scale bars: 10 µm.
Migration through commercially available constricting pores (3-µm diameter) but not large pores (8-µm) is driven by myosin II, with glioblastoma cells using up-regulated myosin II to push or pull their nuclei through brain slices and small pores (Beadle et al., 2008; Ivkovic et al., 2012). Myosin II is used in other cell types to pull the nucleus as a “piston” through porous extracellular matrix pores (Petrie et al., 2014). Inhibition of myosin II slows invasion through small circular pores or rectangular channels (Harada et al., 2014; Thomas et al., 2015; Thiam et al., 2016), but activation can also impede invasion (Surcel et al., 2015). Effects of myosin II on nuclear rupture and DNA integrity remain equally unclear, but actomyosin tension on the front of the nucleus might explain rupture and bleb formation at the high-curvature leading tip of a squeezed nucleus. Transwells allow drugs such as a myosin II inhibitor to be added to one side or the other and thereby address mechanisms of nuclear pushing or pulling, and Transwells can also be etched to fine-tune the fit of the nucleus in the pore (Fig. 1 A) to determine curvature-related mechanisms of nuclear rupture and its effects. Such mechanisms in myosin II–driven migration of cells can in principle also be compared with results from aspiration of a nucleus into a micropipette with the same circular cross-section as a Transwell pore, but with the advantage that the nucleus responds in aspiration to a well-defined stress while the cytoskeleton is depolymerized.
Nuclear rupture and excess γH2AX have been reported for hypercontractile cancer cells in standard culture conditions (Takaki et al., 2017) and for cardiac cells in intact embryonic hearts subjected to a sudden spike in contractility (Cho et al., 2019). Sources of damage remain unclear and potentially include membrane-permeable reactive oxygen species, but cytoplasmic nucleases have also been speculated to enter ruptured nuclei (Denais et al., 2016; Raab et al., 2016). Blebs of chromatin that form at rupture sites accumulate cyclic GMP-AMP synthase (cGAS), which enters from cytoplasm and that could in principle bind γH2AX to inhibit repair in the bleb (Liu et al., 2018a). However, such blebs show no excess γH2AX, which tentatively argues against nuclease entry (Irianto et al., 2017), but we nonetheless examine here one cytoplasmic nuclease, TREX1, that has been seen to accumulate in disrupted micronuclei (Maciejowski et al., 2015). DNA repair factors clearly mislocalize to cytoplasm for hours, based on immunostaining as well as live-imaging of GFP constructs (Xia et al., 2018), but it remains uncertain whether any DNA damage relates to nuclear depletion of such factors if not nuclear entry of nucleases or other factors. Similar questions apply to micronuclei that form in mitosis with missegregated chromosomes (independently of actomyosin) and that somehow exhibit DNA damage as well as depletion of lamin-B and DNA repair factors (Hatch et al., 2013; Liu et al., 2018b). Because lamin filaments are stiff (Turgay et al., 2017), the frequent observation of lamin-B loss from curved regions of nuclear membrane might seem physically sensible, but effects of deformation rate, chromatin stress, and nuclear envelope tension are equally conceivable. We ultimately relate rupture and dysregulation of the DNA damage response to curvature and rate rather than tension, and we begin with molecular rescue approaches to constricted migration as a 3D form of go-or-grow.
Results
Myosin II inhibition rescues nuclear envelope rupture and some DNA damage
As a cell forces its nucleus through a highly constricting pore (3-µm diameter, ∼10-µm length), distortions last for hours, and a nuclear bleb forms early near the highly curved leading edge (Fig. 1 B). Such blebs are consistent with observations of nuclear lamina rupture in constricted migration that is either driven by a serum gradient (Harada et al., 2014; Denais et al., 2016; Raab et al., 2016; Irianto et al., 2017) or proceeds, as here, in the absence of any serum gradient (Harada et al., 2014; Denais et al., 2016; Raab et al., 2016; Irianto et al., 2017). Blebs are detectable in a large fraction of nuclei (∼50%) even after the 24 h of migration, and blebs remain low in lamin-B (Fig. 1 Ci) but accumulate in lamin-A regardless of lamin-A’s phosphorylation state (Fig. 1 B, real-time imaging; or Fig. S1, A and B, immunostained protein). Importantly, inhibition of myosin II with blebbistatin eliminates nuclear blebs while decreasing migration rate (Fig. 1 Cii). Blebbistatin also causes nuclei to be more rounded and slightly less spread, consistent with its effects on many cell types adhering to rigid 2D substrates (Khatau et al., 2009).
On rigid substrates in 2D, the microtubule organizing center (MTOC) is generally in front of a migrating nucleus and sets the direction for migration (Gomes et al., 2005; Raab and Discher, 2017). However, constricting pores set the direction of 3D migration, and the MTOC is seen to often lag the nucleus (Fig. S1 C). In 3D migration, actin-rich protrusions lead the way and are followed by myosin II assemblies (Mogilner and Odde, 2011). Consistent with this, inhibition of myosin II only on the bottom of a Transwell proves sufficient to suppress migration rate as well as nuclear rupture (Figs. 1 Cii and S1 Di). Little effect is seen with drug only on top, consistent with minimal diffusion of drug through pores (Fig. S1, Di and Dii). The results thus suggest that myosin II pulls a nucleus through a small pore and ruptures the front of a nucleus that spreads as the emerging cell does (Fig. 1 Cii).
Myosin II inhibition eliminates mislocalization of nuclear factors into the cytoplasm, such as DNA repair factors KU70 and KU80 (as a GFP construct or endogenous protein, respectively; Figs. 1 D and S1 E). Mislocalization coincides with entry into nuclear blebs of cytoplasmic DNA-binding protein mCherry-cGAS (Fig. 1 D), and of course blebbistatin blocks such cGAS entry. Large pores (8-µm diameter) still require large distortions of nuclei but do not cause mislocalization of nuclear factors (Fig. 1 D, bar graph). DNA repair factors help maintain DNA damage at low steady-state levels, which is important because elevated levels of damage can block cell cycle (Fig. 1 E; Sancar et al., 2004).
Foci of γH2AX in fixed and immunostained nuclei accumulate throughout the nucleoplasm after constricted migration, but myosin II inhibition largely suppresses excess foci (Fig. 1 F, bar graph; and Fig. S1 F). Nuclear blebs, which are low in lamin-B, rarely exhibit excess foci, which seems to spatially decouple mechanisms of nuclear lamina rupture from mechanisms of excess DNA damage. Furthermore, large pores do not affect the basal level of γH2AX foci, just as they had no effect on rupture (Fig. 1 D). Despite the clear effects of blebbistatin, a persistent excess of DNA damage even within a nonruptured, myosin II–inhibited nucleus is consistent with segregation of nuclear factors away from chromatin in constricted migration (Irianto et al., 2016; Bennett et al., 2017). Nonetheless, functional evidence of DNA damage and its rescue would certainly add confidence to such conclusions from image analysis.
Cell cycle progression, measured as a percentage of cells with duplicated genomes (i.e., 4N rather than 2N; Fig. S1 G), increases after cells migrate through 8-µm pores from the Transwell top where the crowded cells are contact inhibited (Fig. 1 G). In contrast, and regardless of myosin II inhibition, a decrease in percentage of 4N is observed after cells migrate through 3-µm pores, with no mitotic cells detected in fixed samples (Fig. 1 G). Live-cell imaging further showed no obvious detachment (or death) of any cells, and dividing cells were seen only on 8-µm bottom (Fig. S1 H). Importantly, blebbistatin does not affect the percentage of 2N cells on top of a Transwell or on bottom (for both 3- and 8-µm pores; not significant [n.s.] in Fig. 1 G), and it has no effect on percentage of 2N cells in 2D culture (Fig. S1 G, bottom). Blebbistatin’s partial rescue of DNA damage therefore does not affect cell cycle suppression caused by constricted migration.
Overexpressed DNA repair factors or added antioxidant rescue some DNA damage
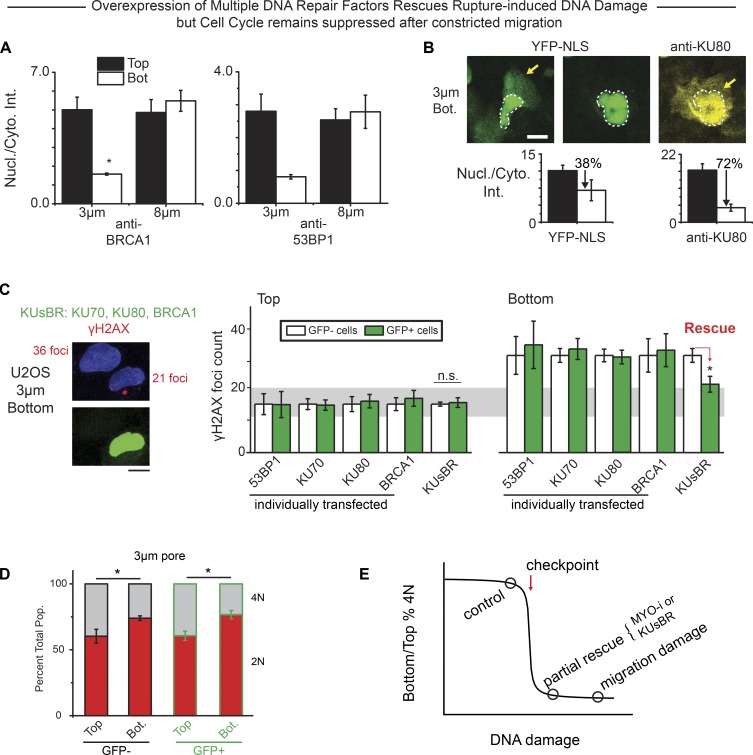
A steady-state level of DNA damage (D) results from sources (S), such as oxidizing species, in balance with DNA repair factors (R), with the simplest calculation of the steady state indicating D ∼ S/R (Cho et al., 2019). Mislocalization of R to cytoplasm is sustained for hours after nuclear envelope rupture based on imaging of GFP constructs in live cells on rigid substrates (Xia et al., 2018), which would predict increased D. Intensity ratios for nuclear/cytoplasm of multiple repair factors in cells fixed and immunostained also remain low after migration (24-h assay) through constricting pores relative to large pores and also relative to cells that have not migrated (Fig. 2, A and B). A much smaller YFP–nuclear localization signal (YFP-NLS) construct shows a nuclear/cytoplasm intensity that is only slightly low (Fig. 2 B), consistent with comparatively rapid nuclear reentry of such constructs (Raab et al., 2016). We hypothesized that some migration-induced DNA damage could be rescued by overexpression of key DNA repair factors, because partial knockdown of some DNA repair factors (KU80, BRCA1, etc.) but not others (53BP1) increases DNA damage, and such damage seems additive (Irianto et al., 2017). Indeed, simultaneous cooverexpression of KU70, KU80, and BRCA1 (designated KUsBR, as GFP constructs; Fig. S2 A) partially rescued the migration-induced DNA damage, whereas overexpression of the individual factors as well as GFP-53BP1 showed no significant effects (Fig. 2 C). Nuclear blebs were still apparent, suggesting that the elevated nuclear levels of multiple key factors facilitated the repair. However, a cell cycle defect in terms of 4N suppression is still seen after 3-µm pore migration (Fig. 2 D).
Figure 2.
Overexpression of multiple DNA repair factors rescues DNA damage independently of rupture in constricted migration, but cell cycle suppression persists. (A) Constricted migration causes DNA repair proteins BRCA1 and 53BP1 to mislocalize from the nucleus (Nucl.) into the cytoplasm (Cyto.; more than five fields of view each; n > 3 experiments; *, P < 0.05). Bot., bottom; Int., intensity; anti-, antibody-stained. (B) As a nucleus squeezes out of a constriction, both endogenous KU80 (yellow) and overexpressed YFP-NLS (green) are mislocalized to cytoplasm, but in nuclei that have fully exited, KU80 remains cytoplasmic while YFP-NLS is mostly nuclear. Bar graphs: Overall nuclear/cytoplasm (Nucl./Cyto.) ratio is thus lower for KU80 than for YFP-NLS after 3-µm pore migration (more than five fields of view each; n > 3 experiments per condition; *, P < 0.05). (C) Co-overexpression of three DNA repair factors (KUsBR) partially rescues excess DNA damage after constricted migration despite nuclear rupture, whereas rescue is not achieved by overexpression of any single repair protein (200–500 cells per condition, n > 3 experiments per condition; *, P < 0.05.). n.s., not significant. (D) The 4N population (Pop.) of both control and KUsBR cells remain suppressed despite the partial rescue of migration-induced DNA damage (200–500 cells per condition, n > 3 experiments per condition; *, P < 0.05). All scale bars: 10 µm. (E) A critical amount of DNA damage is hypothesized to block cell cycle progression.
Neither myosin II inhibition nor repair factor overexpression rescued the cell cycle block, and so we hypothesized that the source of DNA damage could be key to revealing a sigmoidal relationship between cell cycle and DNA damage (Fig. 2 E). Free radicals and other oxidants can permeate membranes and cause DNA damage (Fig. 3 A; Mello Filho et al., 1984; Imlay et al., 1988; Kryston et al., 2011; Mahaseth and Kuzminov, 2016), and even a 0.5-h exposure to H2O2 increases γH2AX foci counts and DNA damage in single-cell electrophoretic comet assays (exhibiting similar approximately three- to fourfold increases in Fig. S2, Bi and Bii). Such results complement evidence of DNA oxidation (Fig. S2 C). Excess repair factors can also reduce H2O2-induced DNA damage in 2D culture (Fig. S2 D). After constricted migration, nuclear oxidative stress is ∼30% higher based on labeling by a redox dye (Fig. 3 A). We therefore added a membrane-permeable antioxidant, glutathione, as a reduced monoethyl-ester (GSH-MEE, denoted GSH in the figures) that reduces H2O2-induced DNA damage in 2D cultures (Figs. 3 A and S2 B, right); GSH-MEE has likewise been shown to reduce oxidative stress and genomic instability in pluripotent stem cells in 2D cultures (Skamagki et al., 2017). Partial rescue of DNA damage by GSH-MEE does not affect migrated cell numbers or nuclear blebs (Fig. 3, B and C). The cell cycle block persists with antioxidant (Fig. 3 D), which is again consistent with only a partial rescue of DNA damage.
Figure 3.
Complete rescue of DNA damage and cell cycle defect requires antioxidant and MYO-i, revealing a bimodal dependence of cell cycle on DNA damage. (A) Reactive oxygen species (ROS) damage DNA but can be neutralized by reduced glutathione, including a membrane-permeable ethyl ester (GSH-MEE, denoted GSH in the figure). Constricted migration increases ROS based on intensity of MitoSOX dye (>50 cells per condition, n = 3 experiments; *, P < 0.05). Bot., bottom. (B) GSH-MEE partially rescues the excess DNA damage after constricted migration. Complete rescue of DNA damage requires both MYO-i or KUsBR and GSH-MEE on both sides of the Transwell membrane, but drugs on bottom suffice (>80 cells per condition, n = 3 experiments; *, P < 0.05). All scale bars: 10 µm. Ctl, control. (C) GSH-MEE alone does not affect constricted migration or nuclear rupture (more than three fields of view, n = 3 experiments; *, P < 0.05). n.s., not significant. (D) Both control and GSH-MEE show 4N suppression after constricted migration, but the cell cycle block is rescued by GSH-MEE+MYO-i or KUsBR even with MYO-i only on bottom (>80 cells per condition, n = 3 experiments; *, P < 0.05). Pop., population. (E) For a pore size–dependent test of cell cycle suppression, we enlarged commercial pores by NaOH etching to novel diameters of 2.20, 2.30, 3.85, 3.95, and 5.0 µm. The plot shows that the percentage of 4N remains suppressed on bottom unless DNA damage is fully rescued. The Hill equation, y = A + Bm/[(x − 1)m + Cm], has fit parameters (m = 15, A = 0.67, B = 0.22, C = 0.23, R2 = 0.78) and goes through error bars for 13 of 15 data points. Data from pore migration rescue experiments (expts.; Figs. 1, 2, and 3) are also fitted. Representative error bars (8 µm and control) for pore and pipette experiments are shown (>50 cells per condition, n = 3 experiments).
Combinations rescue both DNA damage and cell cycle
Complete rescue of both the cell cycle block after constricted migration (Fig. 3 D) and the DNA damage (Fig. 3 B) is achieved by combining antioxidant with myosin II inhibition. Single-cell electrophoretic comet assays (in both alkaline and neutral conditions) confirm that DNA damage is higher after constricted migration (Fig. S2 E), and the approximately twofold increases in apparent mobility approximate the increases in γH2AX foci counts measured after constricted migration (from ∼13 on top to ∼25 foci on bottom for control in Fig. 3 B) and are also consistent with past measurements (Irianto et al. [2017] showed an increase from ∼14 to 25 AU under alkaline conditions). Importantly, only the excess damage on bottom is rescued by GSH-MEE+MYO-i, while a basal level of apparent damage remains.
Myosin II inhibition suppresses migration and nuclear bleb formation regardless of antioxidant (compare Fig. 3 C to Fig. 1 Cii). Adding the combination of blebbistatin and antioxidant only to the bottom of the Transwell is sufficient to suppress all of the cell cycle defect in addition to most of the excess γH2AX foci (blue bars in Fig. 3, B and D). The results suggest antioxidant prevents the DNA damage that otherwise persists after myosin II inhibition (Fig. 1, F and G). Similarly, the combination of antioxidant and KUsBR repair factors also rescued the excess DNA damage and cell cycle (green bars in Fig. 3, B and D), suggesting that the excess oxidative DNA damage can be repaired unless DNA repair factors are partially lost from the nucleus. The results collectively indicate that cell cycle progression exhibits a threshold in the excess DNA damage caused by constricted migration (Fig. 3 E).
Cell cycle threshold versus DNA damage: dependence on pore size
To assess the possible generality of a strongly sigmoidal relationship between cell cycle and DNA damage in migration through pores, we needed to make Transwell pores with a wide range of diameters. Unlike small pores (3 µm) studied above, migration through large pores (8 µm) promotes cell cycle reentry and has no effect on basal γH2AX foci counts (Fig. 1, F and G; Pfeifer et al., 2018). Transwells with 2.2-µm-diameter pores were made from commercially available 1-µm pores by adding NaOH to etch the polyester (similarly for 4- and 5-µm pores from 3-µm pores; Fig. S2 F). After extensive washing, cells were added to the top of the custom-made Transwells as in all studies above, and after 24 h, γH2AX foci were counted and cell cycle phases were measured. A pore diameter of ∼4 µm is the critical pore size: relative to cells on top and relative to large pores, migrated cells show excess DNA damage and a slight suppression in cell cycle, whereas smaller pores (2.2 to 3 µm) cause an increasing excess in DNA damage and a deep but constant defect in cell cycle progression (Fig. 3 E). The combined data are steplike, based on simple statistical tests (Fig. S2 Gi), and are fitted better by a strongly sigmoidal function compared with a line (Fig. S2 Gii). A high cooperativity exponent, m >> 1, typically indicates many interactions upstream (such as in supramolecular γH2AX foci) that amplify signals to downstream regulators.
High curvature (at high rates) drives nuclear envelope rupture
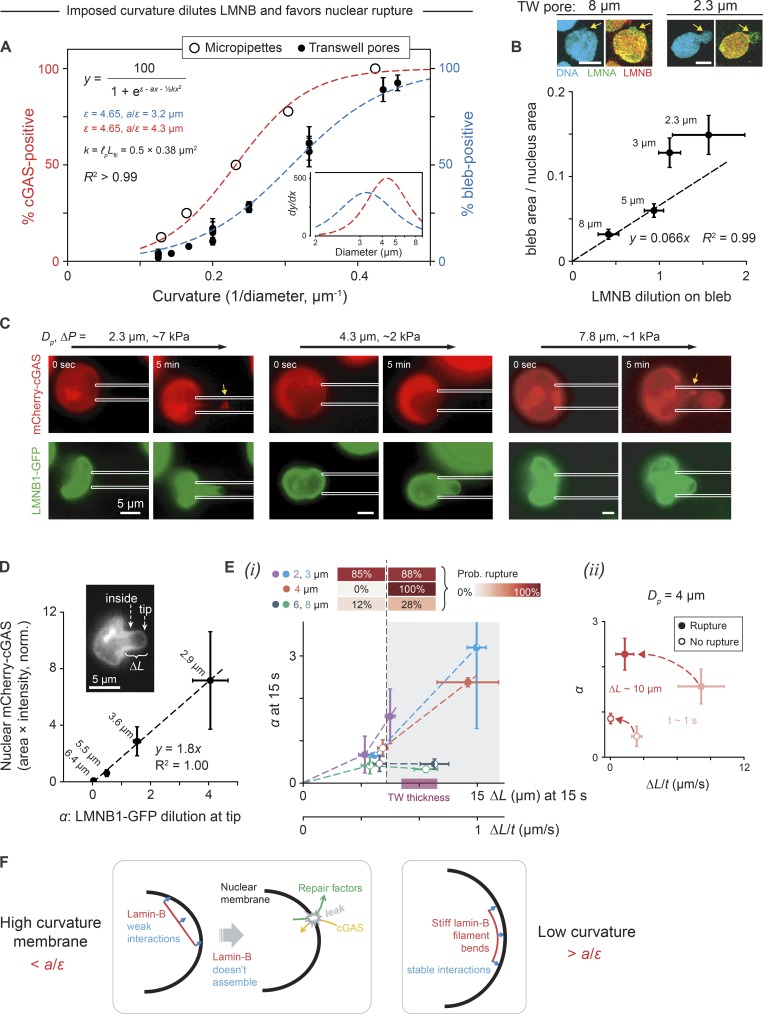
Given the dependence of DNA damage, and thus cell cycle suppression, on Transwell pore size, we next quantified the pore size dependence of nuclear envelope rupture (Fig. 1, C and D; and Fig. 2 A) in Transwell migration and then in micropipette aspiration. We hypothesized that controlled rates of aspiration into micropipettes could ultimately explain the linear dependence observed between bleb formation (nuclear rupture) and migration rate achieved with blebbistatin (Fig. 1 Cii). Past studies with micropipette aspiration of larunculin-treated, detached cells have shown that partial knockdown of Lamin-A in A549 gives nuclei that aspirate several-fold more quickly at physiologically relevant low pressures, and these results correlated with migration through constricting pores that was also several-fold faster and with tumor expansion in vivo that was again several-fold faster (Harada et al., 2014).
U2OS cell migration through custom-etched Transwells quantified the frequency of nuclear blebs at the end of a 24-h migration period (which tends to be cGAS+, per Fig. 1 D) as a sigmoidal function of pore curvature calculated as the inverse of pore diameter (Fig. 4 A, blue curve). Smaller pores impose greater curvature on migrating cells and somehow cause higher rates of nuclear envelope rupture. To assess on time scales of minutes whether such rupture reflects the intrinsic limits of nuclear strength or stability, micropipette aspiration was applied to detached, latrunculin-treated U2OS cells using micropipettes of varying diameter (Dp) and a wide range of pressures (Fig. 4 A, red curve). Pressures of kiloPascal or less are relevant to migration stresses, with nuclear aspiration rate increasing with aspiration pressure (Harada et al., 2014). Monitoring of nuclear cGAS entry as an indicator of nuclear envelope rupture reveals again a sigmoidal relationship between rupture frequency and curvature (Fig. 4 A, red curve). Surprisingly, some of the highest calculated tensions (which is calculated as aspiration pressure × radius, and which assumes pressure is fully sustained by the nuclear envelope rather than chromatin) do not consistently rupture nuclei; instead, rupture frequency correlates best with curvature and with pressure (Fig. S3, A–D). Importantly, the smallest curvatures cause nuclear rupture even at the lowest pressures and rates. The curvature dependence fits a statistical physics model of a single filament that is either attached to or else detached from a curved surface (see Materials and methods); the results also conform to a continuum model of lamina bending with a curvature4 dependence for lamina dilution (Pfeifer et al., 2018).
Figure 4.
High-curvature pores favor nuclear envelope rupture and lamin-B dilution, especially for rapid distension. (A) Latrunculin-treated U2OS cells transfected with mCherry-cGAS and GFP-lamin-B1 detached from the coverslip and were pulled under controlled pressure into micropipettes of varying diameter, Dp. The probability of nuclear rupture, as indicated by nuclear entry of mCherry-cGAS, increases with 1/Dp (we ignore a factor of 2 in the conventional definition of curvature). Rupture probability shows the same functional dependence on curvature for active migration through Transwell pores, where nuclear blebs demark rupture. Fit function and parameters are per the single filament model (see Materials and methods); also, y = Axm/(B + xm) fits equally well with m ≈ 4, consistent with a continuum model (Pfeifer et al., 2018; six or more cells per Dp; ≥50 cells per Transwell pore diameter; Mean ± SEM). Inset: Derivatives of fits indicate a critical pipette/pore diameter of ∼4 µm, consistent with Fig. 3 E. (B) Smaller pores increase nuclear bleb size and lamin-B dilution (i.e., nucleus-to-bleb lamin-B intensity ratio minus 1). Dashed line is fitted to first two points, and the origin (≥11 cells per Transwell pore (TW) diameter; SEM). Images: Nuclear blebs indicated by arrows. Scale bars: 10 µm. (C) Accumulation of mCherry-cGAS at the aspirated tip of the nucleus (arrow) indicates nuclear rupture, which coincides with GFP-LMNB1 dilution. sec, second. (D) Tip dilution of lamin-B1 (α) is relative to inside intensity (inset), and mCherry-cGAS accumulation is normalized (norm.) to both cytoplasmic intensity and pipette cross section (as a maximum area). Binned points show that higher α correlates with more cGAS inside the nucleus at t ≈ 5 min. Each point is labeled by mean pipette diameter for cells within that bin (36 cells total, ≥5 cells per bin; SEM). (Ei) For every aspirated cell at t ≈ 15 s, nuclear extension ΔL into the pipette shows that low or slow ΔL associates with low α (unshaded region), and small diameter (2–4 µm) is needed for high α (shaded region). Filled portions of data points indicate the fraction of nuclei within that bin that rupture by t ≈ 5 min, with upper heatmap indicating rupture probabilities (Prob.) for low or high ΔL (36 cells total, three or more cells per bin; SEM). (Eii) Data for all cells aspirated with Dp = 4 µm at t ≈ 1 s (light red) and at ΔL = 10 µm (i.e., Transwell filter thickness). Fast nuclear entry and rupture occurs at ΔP = 4.6 ± 1.2 kPa, whereas slow entry and no rupture occurs at ΔP = 3.2 ± 1.0 kPa (six cells total, three cells per bin; SEM). (F) Lamin-B filaments are too stiff to bend along high-curvature nuclear membranes, and are thus depleted, but can stably interact with low-curvature membranes.
Lamin-B dilution on blebs after Transwell migration indeed increases with pore curvature (Fig. 4 B); the size of the major bleb increases and most dramatically versus dilution at a pore size of 3–5 µm (ignoring second blebs and blebless scars of disrupted lamin-B). The 3–5-µm size range corresponds well with the curvature-dependent peak for rupture (Fig. 4 A, inset). Dilution at the tip of the aspirated nucleus was similarly quantified with GFP-lamin-B1 (Fig. 4, C and D) and with the nuclear pore factor GFP-Nup153 (Fig. S3 E), and both tend to dilute more at higher curvature. Lamin-B1 dilution clearly correlates spatiotemporally with loss of nuclear envelope integrity, based on accumulation of mCherry-cGAS at the aspirated tip. High curvature thus favors lamin-B1 disruption and nuclear rupture in both Transwell pores and micropipettes.
For every aspirated cell, the extension of the nucleus into the pipette was measured at an early time point (t = 15 s of a total aspiration time of ∼5 min). Across all pipette diameters, low extension leads to minimal lamin-B1 dilution (Fig. 4, E–I; and Fig. S3 F). In small pipettes (2–4 µm), lamin-B1 dilution increases sharply with extension and extension rate, whereas in large pipettes (6- or 8-µm diameter), lamin-B1 dilution remains low (Fig. 4 Ei) except at very high extension rates (Fig. S3 Fii). To assess whether nuclear extension or nuclear extension rate determine nuclear rupture, data for the critical pipette size Dp = 4 µm was plotted separately for rupture (high pressure) or no rupture (low pressure) at an initial time point and at 10-µm extension, which corresponds to the thickness of a Transwell (Fig. 4 Eii). Rupture clearly associates with larger lamin-B dilution and higher extension rates. Importantly, the lack of rupture at low rates of aspiration can explain why myosin II inhibition in constricted migration through similar-size pores, which dramatically reduces migration rate, also effectively eliminates nuclear envelope rupture (Figs. 1 D and S1 E).
Lamin-B filaments have high affinity for nuclear envelope (because of farnesylation and lamin-B receptor) but are stiff with a persistence length of ∼0.5 µm or larger measured for lamins in mouse cells (Turgay et al., 2017). Such filaments will tend to dissociate from a nuclear envelope of high Gaussian curvature. We hypothesize that lamin-B filaments are therefore depleted from high-curvature regions but stably interact with low-curvature regions (Fig. 4 F). This physical picture applies most clearly to the leading tip of the nucleus pulled passively into micropipettes or during cell migration through Transwell pores of varying diameter. This physical picture is also modeled in the fit to both types of data (Fig. 4 A). Although the noted rate effects at intermediate curvature are possibly attributable to lamin-A (see Discussion), the model could generally apply to curved membranes, including micronuclei.
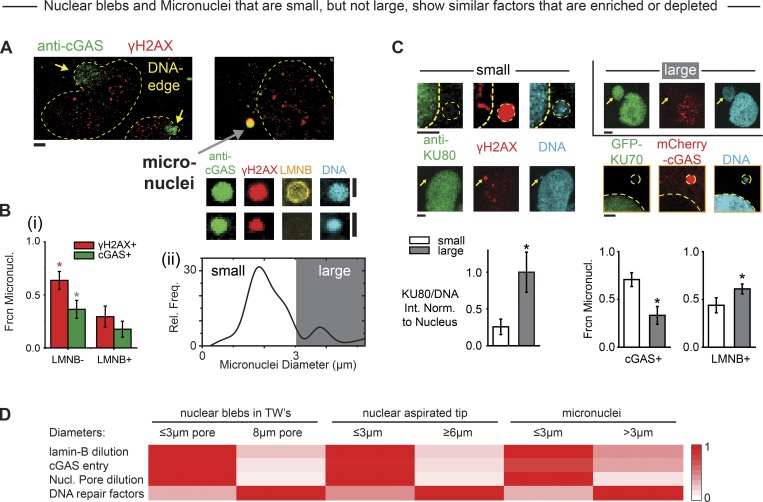
Micronuclei of high curvature also show high cGAS, low lamin-B, and low KU80
Nuclear blebs generated in constricted migration of interphase cells through various pore sizes (Fig. 4 B) resemble micronuclei generated in cell division (Fig. 5 A), and we noticed that micronuclei generated in cell division also vary in size and curvature. We hypothesized that such a variation could provide an alternative approach to studies of nuclear envelope curvature effects. Micronuclei in 2D culture are in particular known to be γH2AX+ and to lack lamin-B, independent of cytoskeleton (Hatch et al., 2013; Harding et al., 2017; Liu et al., 2018b), and the DNA-stained micronuclei (Fig. 5 A) that are most visible within sparsely spread cells on a Transwell bottom tend to be cGAS+ and γH2AX+ when lacking in lamin-B (Fig. 5 Bi). Micronuclei also tend to be small (<3 µm) but are sometimes large (>3 µm; Fig. 5 Bii). Small, high-curvature micronuclei further lack KU70/KU80 repair factors in addition to being cGAS+ and lamin-B deficient (Fig. 5 C).
Figure 5.
Small micronuclei and ruptured nuclei exhibit many similarities. (A) Endogenous cGAS binds DNA in both nuclear blebs (yellow arrows) and many micronuclei (gray arrow). Many micronuclei are γH2AX+, and some lack lamin-B (LMNB). (B) Composition of micronuclei assessed after constricted migration shows the following. (Bi) More of the low LMNB micronuclei are γH2AX+ and cGAS+, expressed as a fraction of the micronuclei counted (Frcn Micronucl.; for 30–40 micronuclei per condition from three experiments; *, P < 0.05). (Bii) A broad distribution of small and large micronuclei (n = 68 from three experiments; *, P < 0.05). Rel. Freq., relative frequency. (C) Small micronuclei (<3 µm) tend to have low KU80 (left two images), low GFP-KU70 (bottom right image), and low LMNB but high cGAS (bar graphs; 30–40 micronuclei per condition from three experiments; *, P < 0.05). For KU80/DNA, intensity normalization (Int. Norm.) is done by normalizing to the main nucleus. All scale bars: 2 µm. (D) Heatmap summary of similarities between nuclear blebs, nuclear aspiration, and micronuclei from various experiments in terms of lamin-B dilution, cGAS entry from cytoplasm, nuclear (Nucl.) pore dilution, and levels of DNA repair factors (as explained in Materials and methods, Heatmap comparisons of curvature-associated nuclear envelope rupture properties). TW, Transwell.
Importantly, micronuclei can clearly be double-positive as (cGAS+, γH2AX+; Fig. 5 A), and so our observations that nuclear blebs are not double-positive (Fig. 5 A) are unlikely to relate to inhibition of repair by cGAS (Liu et al., 2018a). Another notable difference is that antioxidant fails to affect DNA damage in micronuclei (Fig. S4 Ai). Nonetheless, many structural similarities are evident between small micronuclei, micropipette-induced rupture, and nuclear blebs after constricted migration (Fig. 5 D), including depletion of nuclear pores (Fig. S4 Aii; Hatch et al., 2013; Liu et al., 2018b). The fact that lamin-B filaments are stiff (with a persistence length of approximately ≥0.5 µm [Turgay et al., 2017]) and have high affinity for nuclear envelope suggests once again that high-curvature membranes oppose stable assembly of lamin-B (Fig. 4 F), even though defects in other processes including nuclear import/export are also likely important.
Alternatives: TREX1 nuclease overexpression does not affect DNA damage or cell cycle
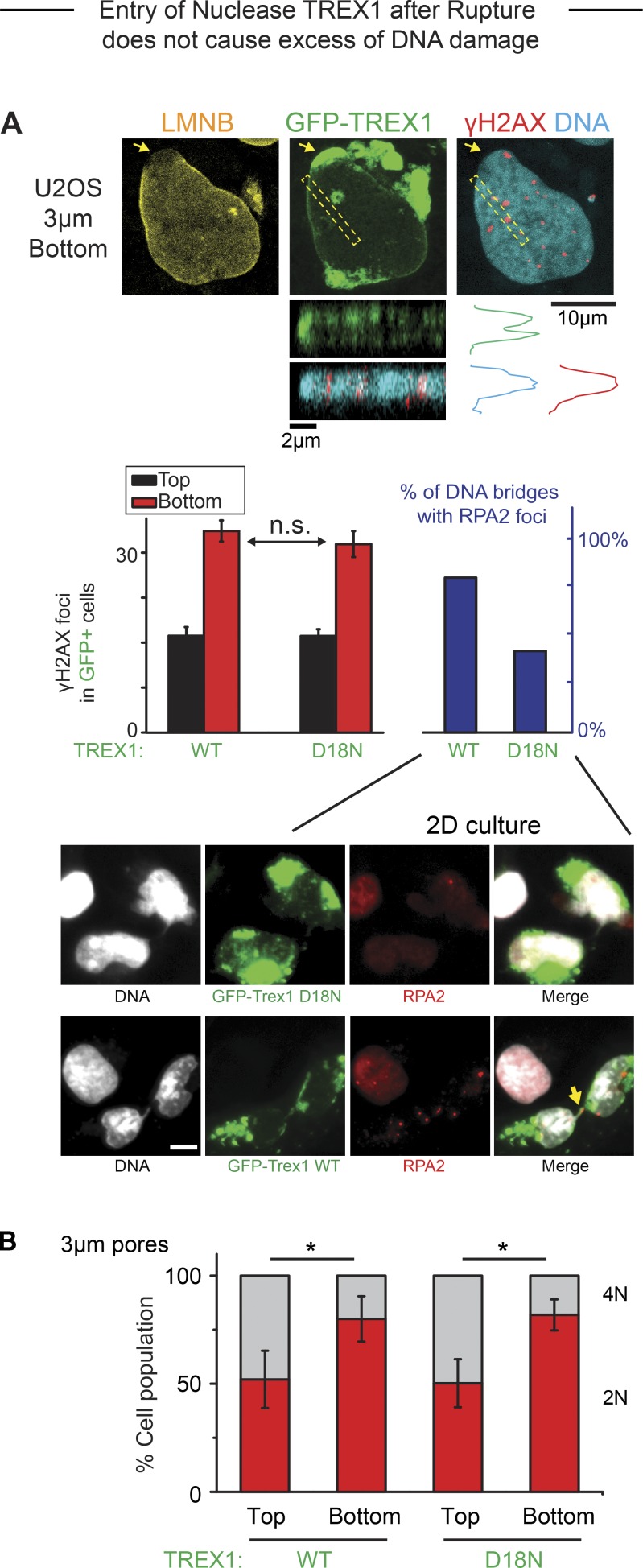
Other pathways and factors could of course affect DNA damage as well as cell cycle. Entry of cytoplasmic factors into a ruptured nucleus has been speculated to include nucleases (Denais et al., 2016; Raab et al., 2016) and certainly includes cGAS as cited. TREX1 exonuclease and the cytoplasmic DNA-binding protein cGAS (MB21D1 gene) are both expressed in U2OS cells (Fig. S4 B). TREX1 is a single-exon, transmembrane protein normally in the endoplasmic reticulum with a cytoplasmic-facing exonuclease domain; knockout mice do not exhibit excessive DNA damage, increased mutations, or cancer but do exhibit autoimmune inflammation (especially in heart) that relates to cGAS activation (Morita et al., 2004; Maciejowski et al., 2015; Rice et al., 2015; Yan, 2017). Disrupted micronuclei accumulate TREX1, and overexpression of TREX1 in WT cells can show a significant net increase in nuclease activity (Maciejowski et al., 2015), which prompted a similar approach here. Overexpressed GFP-TREX1 or an inactive mutant (D18N-TREX1) show some accumulation on nuclear blebs and show expected differential effects on chromatin bridges but show no obvious nucleoplasmic localization nor any differences in DNA damage on the bleb or in the nucleus before and after constricted migration(Fig. 6 A). Cell cycle defects are also unaffected (Fig. 6 B). Endogenous cGAS likewise accumulates in the bleb (Fig. S4 C; compare also to mCherry-cGAS, Fig. 1 D) and shows no obvious colocalization with γH2AX (Fig. S4 C) despite recent suggestions from laser-induced nuclear damage (Liu et al., 2018a). The bleb also immunostains for acetyl-histone-H3 (Fig. S4 D), which is a likely marker of euchromatin (Bannister and Kouzarides, 2011) and could relate to the restricted binding of cGAS to the bleb. Nuclear loss of the transcriptional coactivator YAP1 (Fig. S4 E) is also consistent with epigenetic changes and could directly affect cell cycle (Dong et al., 2007) as well as the DNA damage response (Strano et al., 2005).
Figure 6.
Entry of TREX1 after rupture does not cause elevation of DNA damage. (A) Overexpressed nuclease GFP-TREX1 localizes to site of nuclear rupture after constricted migration (arrow), but γH2AX foci are not enriched. A representative z-stack image confirms that TREX1 is a transmembrane protein. Bar graphs (left): Both WT and inactive (D18N) GFP-TREX1 show the same excess DNA damage after constricted migration. Functionality of WT TREX1 was confirmed by an increase in RPA2 foci on mitotic bridges of DNA (right; >150 cells per condition, n = 3 experiments; *, P < 0.05). Representative images: Overexpression of WT TREX1 (GFP-TREX1-WT) causes formation of RPA2 foci on DNA bridges (yellow arrow) after mitosis, confirming nuclease activity. With overexpression of inactive mutant TREX1 (GFP-TREX1-D18N), fewer cells exhibit such RPA2 foci. n.s., not significant. Scale bar: 10 μm. (B) Suppression of 4N is the same for WT and mutant GFP-TREX1 after constricted migration (>150 cells per condition, n = 3 experiments; *, P < 0.05).
Cell cycle defect after constricted migration: reversible and possibly mutagenic
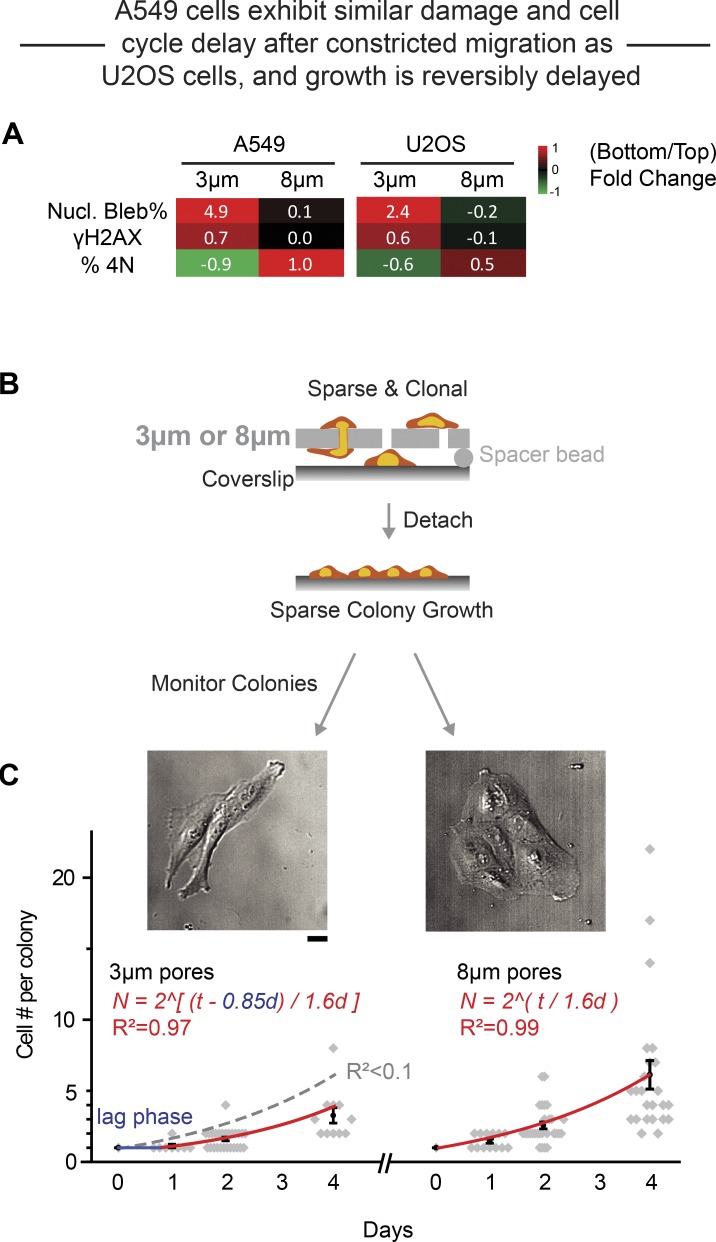
A cell cycle block caused by DNA damage could be irreversible, leading to cell senescence or death. Conversely, an irreversible excess of γH2AX is also evident in apoptosis (Lu et al., 2006), and constricted migration can cause apoptosis (Harada et al., 2014). On the other hand, a cell cycle block could be reversible, with either proper repair of the damage or else misrepair. We observe that the total number of cells (top plus bottom) are nearly constant after 24 h of constricted migration, even though the initial wave of cells requires only a few hours to cross a Transwell. Also, we do not observe floating cells that commonly indicate cell death. To determine therefore whether the defect in cell cycle is reversible and also generalizable after constricted migration, we studied A549 lung carcinoma cells, which are quantitatively similar to U2OS cells in terms of nuclear envelope rupture, DNA damage, and cell cycle suppression (Fig. 7 A). The epithelial-like A549 cells were chosen because they migrate relatively slowly in 2D and thus grow in trackable colonies. We clonally expanded A549 cells and placed a collagen-coated coverslip under the Transwells with a spacer of ∼2 µm, and the coverslips with cells attached sparsely were then transferred to standard cultures to monitor proliferation (Fig. 7 B). After migration through 8-µm pores, the population-average doubling time in colonies after transfer to a coverslip was similar to standard 2D growth (∼1.6 d). However, after migration through 3-µm pores, a 0.85-d delay in growth was evident before the same 1.6-d doubling time (Fig. 7 C). Expansion after constricted migration is thus consistent with repair of DNA damage and reversibility of the cell cycle delay (Figs. 1, 2, and 3).
Figure 7.
Cell expansion after constricted migration reveals a growth delay. (A) Heatmaps summarize the bottom-to-top fold-change in percentage nuclear (Nucl.) blebs, DNA damage, and percentage 4N for 3- and 8-µm pore migration of both A549 and U2OS cells (log2Ratio). (B) Schematics: Clonal A549 cells migrated through Transwells to a collagen coverslip placed under the Transwells with a ∼2-µm spacer, and the coverslips with sparsely attached cells were then transferred to standard cultures to monitor proliferation. (C) After migration through 8-µm pores, the population-average doubling time in colonies after transfer to a coverslip is similar to standard 2D growth (∼1.6 d, right curve, exponential fit, n > 4 experiments). However, after migration through 3-µm pores, a 0.85-d delay in growth is evident using the same 1.6-d doubling time. If the delay is set to zero, then the exponential fit with 1.6-d doubling time fits poorly (gray dashed fit, R2 < 0.1). Scale bar: 20 µm.
For cells that proliferate after a spike in DNA damage, misrepair of the damage could in principle lead to chromosomal changes, although selection for preexisting chromosomal differences within a starting population might also contribute to any detected genome variation. Genomic changes have been reported from sequencing of micronuclei (Liu et al., 2018b) and from metaphase spreads of hypercontractile cancer cells (Takaki et al., 2017). Genome variation was therefore studied using single nucleotide polymorphism (SNP) arrays (with necessary control studies; Fig. S5 A) after migration through small or large pores compared with 2D cultures. We again started with clonally expanded A549 cells because U2OS cells exhibit high genomic instability and many micronuclei (Zhang et al., 2015). Cells were migrated three times, including detachment and expansion, with a final expansion of randomly chosen single cells to ∼106 cells for genomic analyses (Fig. S5 B). Although all 10 randomly chosen clones obtained after migration through 8-µm pores or from nonmigrated 2D control cultures were statistically the same, 3 of 5 clones obtained after migration through 3-µm pores differed significantly from the others and each other (Fig. S5 C). Moreover, these three clones from 3-µm pores were also significantly different from each other. For comparison, A549 tumors in stiff subcutaneous sites of immunodeficient mice (NSG) exhibit genomic variation (Fig. S5 D) that is much higher than control cultures that proliferated for similar duration but is similar in magnitude to that achievable in the more controlled in vitro studies of constricted migration.
Discussion
Nuclear rupture has been clearly demonstrated during cell migration in vivo with YFP-NLS constructs (Denais et al., 2016; Raab et al., 2016; Irianto et al., 2017), and findings here with rigid Transwells suggest that similar studies of GFP-repair factors should provide further insight (Fig. 2 A), especially in combination with cell cycle markers. However, in considering the broader applicability of the present results to such tissue processes, a number of factors need to be considered, including tissue compliance and permeability as well as pore length and shape (noting that the pores in Transwells and micropipettes are circular in cross-section rather than square or rectangular per the various channel devices intended for similar studies). Transwell rigidity certainly requires the cell and nucleus to deform strongly, and so deformable channels (Pathak and Kumar, 2012) would likely reveal a smaller equivalent diameter than the critical diameter reported here (Fig. 3 E). Transwell impermeability to liquid flow implies likely roles for aquaporins based on migration through similarly impermeable channels (Stroka et al., 2014), but the effects on transport of other solutes, especially oxidants (Fig. 3 A), are as unknown as the in vivo effects.
Pro-migration and pro-damage effects of high myosin II activity in constricted migration (Fig. 1) seem consistent with results for hypercontractile cells in 2D rigid cultures (Takaki et al., 2017), which have also suggested DNA damage and a potential for genome variation. However, myosin II knockout in yeast has the opposite effect in that it increases aneuploidy (Rancati et al., 2008), and so perhaps an optimum myosin II activity is needed for maintaining genome integrity. High-curvature regions of interphase nuclei in 2D culture are prone to rupture and excess DNA damage, which are also favored by high contractility and rigid substrates independent of migration (Xia et al., 2018). Because lamin-B filaments are stiff, with a persistence length of ∼0.5 µm (Turgay et al., 2017), they will not bend and bind a nuclear envelope with high Gaussian curvature. This explanation seems consistent with curvature effects of constricting pores, micropipette aspiration, and micronuclei. However, high rates of imposing curvature that result from high pressures (Fig. 4 Eii) or high myosin II (Fig. 1) are also more damaging than low rates of imposing curvature (at low pressures), particularly for intermediate curvatures of ∼3–4-µm diameter. Because lamin-A behaves as a viscous polymer layer (Harada et al., 2014), low rates of extension and curvature generation (unlike high rates) should allow more lamin-A to flow and stabilize lamin-B’s attachment to a highly curved nuclear envelope (Fig. 4 F). Studies of lamin-A knockdown cells at various migration and passive extension rates should lend deeper physical insight.
Myosin II inhibition slows migration through pores but only partially rescues the excess DNA damage after constricted migration, which is consistent with a failure to rescue cell cycle. Squeezing dozens of mobile proteins, especially DNA repair factors, away from the dense chromatin in the pore could explain some of the excess DNA damage (Irianto et al., 2017; Pfeifer et al., 2018). Such squeezing into low-density regions of chromatin likely explains the large intranuclear accumulations of GFP-53BP1 during migration through narrow channels (Denais et al., 2016; Raab et al., 2016; Irianto et al., 2017) independent of DNA damage (Irianto et al., 2017; Pfeifer et al., 2018). KU70 and KU80 are likely more important than 53BP1 and even BRCA1 in the KUsBR combination, because BRCA1 foci correlated with cell cycle effect after constricted migration (Pfeifer et al., 2018) and because KU70 and KU80 are known to bind to DNA ends in a stoichiometric complex. DNA damage measurements based on γH2AX are nonetheless consistent with results from both alkaline and neutral comet assays on cells isolated from Transwells, including at least one rescue condition (Fig. S2 E). Although constricting pores cause mislocalization of the MTOC (Fig. S1 C), which could delay cell cycle, the mislocalization lasts only the few hours that are required for a cell to migrate through the pore (Fig. 1 B). DNA damage and a cell cycle checkpoint are ultimately consistent with simple orthogonal measures of a sigmoidal delay, illustrating mechanical control of cell cycle as one mechanism of go-or-grow.
Possibilities of misrepair
Misrepair of DNA damage during the cell cycle delay seems conceivable, but multiple models might explain the genomic variation detectable after several rounds of constricted migration and expansion (Fig. S5). One model is that the constricting 3-µm pores simply select genomically distinct cells within the clonally expanded starting population. Since 12 randomly generated clones were all statistically the same, the distinct cells in a selection-based model would have to exist at low probability of P < 0.08 (= 1/12) within the starting population. A second model is that migration and expansion introduce unique genomic changes in the pure starting population at an overall mutation rate () of = 20∼25% among the many conditions, which reflects three clones being different among the 15 measured clones. The clones from 3-µm pores thus exhibit a mutation rate of 60% (three of five) that is threefold greater than . Conversely, the probability of showing no mutations is 75–80% (100% − ), so the probability that no mutations are measured in all 10 clones from both the 8-µm pores and 2D is p = (75–80%)10 = 0.05–0.10. No significant copy number variation differences between all clones from 8-µm and 2D conditions is thus unlikely to be by chance in this second model. More cloning and genomic analysis might add further statistical significance to migration-induced variation relative to the selection model, but genomic analyses are terminal and will always be limited to such statistical analyses of probabilities. To more definitively distinguish such models or perhaps identify other mechanisms, monitoring the same living cell for the emergence of genome changes is a method that needs to be developed.
Physical effectors of genome variation in vivo seem equally important to pursue, because meta-analyses of complex human tumors indicate that tumor cells in stiff tissues tend to harbor more mutations than tumors in soft tissues such as leukemias and other hematopoietic cancers (Pfeifer et al., 2017). Stiffer tissues have more collagen (Pfeifer et al., 2017) and should have lower porosity. Effects of immune cells and inflammation could be equally important, although the mice used here (Fig. S5) lack an acquired immune system that can limit growth of solid tumors with high genomic variation (Rizvi et al., 2015). Even if mutagenesis is proven not to occur after constricted migration in vivo, the primary evidence here from rescuable DNA damage reveals what seems a sensible and possibly general mechano-regulated threshold for cell cycle progression.
Materials and methods
Cell culture
U2OS and C2C12 cells were cultured in DMEM high-glucose medium (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin (Sigma). A549 cells were cultured in Ham’s F12 medium (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were incubated at 37°C and 5% CO2. A549 cells stably expressing GFP-S22-lamin-A were first treated with shLMNA (Sigma; TRCN0000061833: 5′-CGACTGGTGGAGATTGACAAT-3′) to achieve a stable knockdown background per previous studies (Buxboim et al., 2014).
Immunostaining
Cells were fixed in 4% formaldehyde (Sigma) for 15 min, followed by 15-min permeabilization by 0.5% Triton X-100 (Sigma), 30-min blocking by 5% BSA (Sigma), and overnight incubation in primary antibodies at 4°C. The antibodies used include lamin-A/C (1:500, sc-7292, mouse; Santa Cruz), lamin-B (1:500, goat, sc-6217; Santa Cruz; 1:500, rabbit, ab16048; Abcam), γH2AX (1:500, mouse, 05-636-I; Sigma), 53BP1 (1:300, rabbit, NB100-304; Novus), KU80 (1:500, rabbit, C48E7; Cell Signaling), BRCA1 (1:500, mouse, sc-6954; Santa Cruz), cGas (1:500, rabbit, D1D3G; Cell Signaling), Pericentrin (1:500, rabbit, ab4448; Abcam), acetyl-H3 (1:500, rabbit, ab47915; Abcam), anti-nuclear pore complex proteins antibody (1:500, mouse, ab24609; Abcam), and replication protein A2 (RPA2; 1:500, mouse, ab2175; Abcam). Finally, after 90-min incubation in secondary antibodies (1:500, donkey anti-mouse, -goat, or -rabbit, A21202, A31571, A11056, A21206, and A31573; Thermo Fisher Scientific), the cells’ nuclei were stained with 8 µM Hoechst 33342 (Thermo Fisher Scientific) for 15 min. When used, 1 µg/ml phalloidin-tetramethylrhodamine isothiocyanate (Sigma) was added to cells for 45 min just before Hoechst staining. For avidin, 15 µg/ml FITC-conjugated avidin was added after BSA blocking and incubated for 1 h at 37°C, as described in Radisky et al. (2005). MitoSOX (M36008; Thermo Fisher Scientific) was applied to live samples for 10–15 min at 3.8 × 10−3 µg/µl before fixation. Carboxyfluorescein diacetate (CFDA; Vybrant CFDA SE Cell Tracer Kit, V12883; Thermo Fisher Scientific) was applied at 2 µM in live samples as instructed.
EdU labeling and staining
As regularly done in other cell cycle studies (e.g., Lee et al., 2013), cell cycle phase was determined using a combination of EdU and DNA intensity, even though such intensities are continuously distributed. EdU (10 µM; Abcam) was added to Transwell membrane 1 h before fixation and permeabilization. After permeabilization, samples were stained with 100 mM Tris (pH 8.5; Sigma), 1 mM CuSO4 (Sigma), 100 µM Cy5 azide dye (Cyandye), and 100 mM ascorbic acid (Sigma) for 30 min at room temperature. Samples were thoroughly washed and then underwent immunostaining as described above.
Transwell migration
Migration assays were performed using 24-well inserts with 3- and 8-µm-pore filters (Corning) with 2 × 106 and 1 × 105 pores/cm2, respectively. 1.5 × 105 cells were seeded on top of the pore filters. The medium, supplemented with 10% FBS and 1% penicillin-streptomycin, was added to both the top and bottom of each 24-well insert such that there was no nutrient gradient across the pore filter. After incubating for ∼24 h at 37°C and 5% CO2, the entire filter was fixed and stained with desired antibodies following standard immunofluorescence protocol. If drug treatment was required, drugs including 20 µM of blebbistatin and 1 mg/ml GSH-MEE (G-275-500; GoldBio) were added at top, bottom, or both sides of the membrane 2 h after seeding. In this case, cells attach and block the pore first so that drugs will not diffuse through the membrane, as confirmed by CFDA staining (Fig. S1 Dii).
Transwell migration to glass
Approximately 105 and 103 A549 cells obtained from a clone were seeded over the 3- and 8-µm-pore filters, respectively. Each insert was placed over a 15-mm-diameter microscopic slide coverslip in a Petri dish; before use, the coverslip was coated with 100 µg/ml collagen I solution for 1 h. Beads with 2–3 µm were added between coverslip and Transwell membrane to create space allowing cells to migrate to bottom. Medium, supplemented with 10% FBS and 1% penicillin-streptomycin was added to the top of each insert. Medium was also added to the Petri dish such that there was no nutrient gradient across the pore filter. After incubating for ∼24 h at 37°C and 5% CO2, the coverslips had sparse single cells on them, and the entire coverslip was transferred to a standard 24-well plate. Medium was added, and the samples were ready for live monitoring.
Transwell pore etching
Porous membrane cell culture inserts are commercially available from Corning; they come in a limited assortment of pore diameters, including 1, 3, 5, and 8 µm. To generate intermediate pore sizes, such as 4 or 6 µm, the polycarbonate membranes were etched with 2 M NaOH in a 60°C incubator. 3-µm-pore membranes were incubated for 72 min and 120 min to generate 4- and 5-µm-diameter pores, respectively. The same conditions were used to etch 5-µm-pore membranes to generate 6- and 7-µm-diameter pores. To achieve pores of <3-µm diameter, 1-µm-pore membranes were irradiated with 365 nm ultraviolet light (Spectroline XX-15A, 0.7 A) for 30 min per side, and then etched with 9 M NaOH at room temperature (22°C). Etching for 4.5 and 5 h yielded 2.2- and 2.3-µm-diameter pores, respectively. In every case, after etching, membranes were thoroughly washed with MilliQ water and dried under vacuum. Etched membranes were sterilized with UV irradiation before being used in migration assays. Etching conditions were adapted from Cornelius et al. (2007). The sizes of etched pores imaged by confocal microscopy were measured/confirmed by ImageJ (National Institutes of Health; Fig. S2 F). Expected etched pore sizes matched well with measured pore sizes. Additionally, etched 4- and 5-µm pores, for instance, have the same distributions as the commercially available 3-µm ones and are sufficiently tight that the 4-µm pore is not a combination of 3- and 5-µm pores.
Optical microscopy
Conventional epifluorescence images were taken using an Olympus IX71 microscope with a 40×/0.6-NA objective and a digital EMCCD camera (Cascade 512B; Photometrics), and confocal imaging was done on a Leica TCS SP8 system with a 63×/1.4-NA oil-immersion objective. Fixed samples were either immersed in PBS for epifluorescence or mounted with antifade (Prolong Gold, P10144; Thermo Fisher Scientific) for confocal microscopy at room temperature. All fluorochromes (secondary antibodies) are listed in Immunostaining. Higher-NA lenses give higher-resolution images and should increase foci counts. Only Fig. S2 (B and D) in this study and images in Irianto et al. (2017); Fig. 3 Ci) were taken using the 40×/0.6 NA objective, and control cells consistently show ∼10 γH2AX foci. Other images of γH2AX here and in Irianto et al. (2017) (Fig. 1, C and D) were all taken with a 63×/1.4 NA objective, and control cells consistently show ∼15 γH2AX foci. Raw images were used directly for image quantification, and ImageJ was the only software used for analysis.
Scanning electron microscopy
Scanning electron microscopy of etched Transwell membranes was performed with an FEI Quanta 600 FEG Mark II ESEM, operated in wet environmental mode.
Live imaging
For live imaging in Fig. S1, we used an EVOS FL Auto Imaging System with 10× or 20× objective in normal culture conditions (37°C, 5% CO2, and complete culture medium as specified above).
Image analysis
All imaging analysis was done with Fiji ImageJ in blinded analyses with uniform adjustment of brightness and contrast. Blebs were counted manually, as they have very distinguishable features (such as lack of lamin-B in the bleb) when we overlay lamin-A and lamin-B together, as shown in Fig. S1 D. Foci counting was performed on z-stack projections automatically by the ImageJ built-in algorithm “Find Maxima,” which identifies in an image local maxima that exceed a user-defined threshold that is the same for all samples, experimental and control, within a given experiment. Sample images and counts have been added in Figs. 1 F, 2 C, and S2 B as references. We validated our automated foci counts with comet assay (Fig. S2 B). Two sets of results were linearly correlated. All foci counts were done on raw images instead of processed images so that adjusting contrast only affected visualization. Intensity quantification was also done automatically in ImageJ after the region of interest was selected. In the case of MitoSox intensity, cells were selected in ImageJ, followed by automated intensity quantification (integrated intensity). The MitoSox intensity after H2O2 treatment only increases 50%, although massive oxidative damage is present. Therefore, a weaker increase in MitoSox intensity at bottom is expected. For small differences, P values were calculated in Excel 2013. The fact that intensity measurement was automated improved confidence in this result.
Alkaline comet assay
Alkaline comet assays were performed according to the manufacturer’s protocol (OxiSelect Comet Assay Kit, STA-350; Cell Biolabs). Briefly, cells were trypsinized, mixed with liquefied agarose at 37°C, placed dropwise onto the supplied glass slide (75 µl per glass well), and incubated for 15 min at 4°C for the agarose to gel. Lysis buffer from the kit was then added to the solidified gel and incubated for 45 min, followed by an additional 30-min incubation with alkaline solution. Electrophoresis was performed at 300 mA for 30 min in alkaline solution followed by a deionized water wash and a 70% ethanol wash, before samples were air dried overnight. Finally, DNA dye in the kit was added to each sample for 15 min, followed by epifluorescence imaging as described above.
Immunoblotting
Cells were trypsinized and pelleted (∼1.5 million cells, counted each time). Pellets were resuspended in 1× lithium dodecyl sulfate lysis buffer supplemented with 1% protease and 1% phosphatase inhibitors and sonicated on ice (3 × 15 × 1-s pulses, intermediate power setting). After resting for 30 min on ice, samples were denatured at 80°C with 0.5% β-mercaptoethanol (vol/vol) for 10 min. Samples were loaded onto bis-Tris 4–12% gradient gels (equal volume, usually 4 µl per 15-well gel) for electrophoresis (100 V × 10 min; 160 V × 55 min) and then transferred (iBlot; Life Technologies; settings P3, 7 min) to blotting membrane. Band intensities were quantified using Fiji/ImageJ, relative to local background levels flanking the specific bands. Antibodies were used at 1:100 dilution as mentioned above. β-Actin (mouse, sc-47778; Santa Cruz) was used as normalizer.
Transfection
GFP-TRX1 WT and D18N are from Addgene (27219, 27220). GFP-BRCA1 (Addgene plasmid 71116) was a gift from Dr. Daniel Durocher of the Lunenfeld-Tanenbaum Research Institute, Toronto, Canada; GFP-KU70 and GFP-KU80 were gifts from Dr. Stuart L. Rulten of the University of Sussex, Brighton, UK (Grundy et al., 2013); GFP-Nup153 is from Addgene (64268; Duheron et al., 2014); lamin-B1-GFP was a gift from Dr. Harald Herrmann-Lerdon of Universitätsklinikum Erlangen, Erlangen, Germany, and Tatjana Wedig of the German Cancer Research Center, Heidelberg, Germany; and mCherry-cGAS was a gift from R.A. Greenberg (Harding et al., 2017). Cells were passaged 24 h before transfection. siLMNA and siControl were purchased from Dharmacon (siLMNA: L-004978-00, 5′-GAAGGAGGGUGACCUGAUA-3′, 5′-UCACAGCACGCACGCACUA-3′, 5′-UGAAAGCGCGCAAUACCAA-3′, and 5′-CGUGUGCGCUCGCUGGAAA-3′; nontargeting siRNA: D-001810-10, 5′-UGGUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAUGUUGUGUGA-3′, 5′-UGGUUUACAUGUUUUCUGA-3′, and 5′-UGGUUUACAUGUUUUCCUA-3′). A complex of GFPs (0.2–0.5 ng/ml) and 1 µg/ml Lipofectamine 2000 (Invitrogen) was prepared according to manufacturer’s instructions and then added for 24 h (GFPs) to cells in corresponding medium supplemented with 10% FBS. KUsBR consists of GFP-KU70, GFP-KU80, and GFP-BRCA1 (0.2–0.5 ng/ml each).
Neutral comet assay
Neutral comet assays were performed following the manufacturer’s protocol (CometAssay HT, 4252-040-K; Trevigen). Briefly, melted agarose cooled to 37°C was mixed with the trypsinized cells. The mixture was placed dropwise onto the glass slide from the kit (30 µl per glass well), and incubated for 10 min at 4°C in the dark. The slide was then immersed in the supplied lysis solution for 1 h at 4°C, followed by a 30-min incubation with neutral electrophoresis buffer. Electrophoresis was performed at 15 V (1 V/cm between electrodes) for 45 min. The slide was then washed using DNA precipitation solution for 30 min, followed by a 30-min wash with 70% ethanol. After being air dried in the dark overnight, the samples were stained using DNA dye (SYBR Gold Nucleic Acid Gel Stain, S11494; Invitrogen) for 30 min and viewed using an epifluorescence microscopy.
Establishment of A549 tumors in vivo
For each injection, ∼106 cells were suspended in 100 µl ice-cold PBS and 25% Matrigel (BD) and injected subcutaneously into the flank of nonobese diabetic/severe combined immunodeficient mice with null expression of IL-2R gamma chain (NSG mice). Mice were obtained from the University of Pennsylvania Stem Cell and Xenograft Core. All animal experiments were planned and performed according to institutional animal care and use committee protocols. The tumors were grown for 4 wk, and tumor cells from mice were then isolated for SNP array.
Micropipette aspiration
U2OS cells were transfected overnight with mCherry-cGAS and lamin-B1-GFP or with GFP-Nup153. Before aspiration, cells were detached using 0.05% trypsin-EDTA (Life Technologies) and then incubated in 0.2 µg/ml latrunculin-A (Sigma) and 8 µM Hoechst 33342 (Thermo Fisher Scientific) for 30 min at 37°C, as described previously (Pajerowski et al., 2007). During aspiration, cells were suspended in PBS with 1% BSA and 0.2 µg/ml latrunculin-A. Imaging was done on a Nikon TE300 epifluorescence microscope with a 60×/1.25 NA oil-immersion objective and a digital EMCCD camera (Cascade, Photometrics). Aspiration pressures (approximate kiloPascals) were varied over a wide range (see Fig. S3), and aspiration was monitored over ∼5 min.
Calculating lamin-B dilution
To quantify lamin-B dilution on a nuclear bleb, we measured the average (background-subtracted) lamin-B intensities of the entire on-bleb and off-bleb regions of a blebbed nucleus. Lamin-B dilution is the ratio of the off-bleb intensity to the on-bleb intensity minus 1, so that dilution = 0 if the bleb exhibits zero lamin-B deficiency. Lamin-B dilution at the leading tip of an aspirated nucleus is calculated in the same way, except on-bleb and off-bleb intensities are replaced by tip and inside intensities (Fig. 4 D, inset), which are measured within boxes of area approximating the pipette diameter squared. Dilution of nuclear pore complex proteins (GFP-Nup153) was quantified in the same way (Figs. 5 D and S3 E).
Single-filament model
We consider a single lamin-B filament that is stiff with a measurable length Lfil and persistence length ℓp (Turgay et al., 2017), and the filament is either attached to the nuclear membrane or else detached. The molecular partition function is thus
| (1) |
We choose the detached state as the reference state with Edetached = 0, but the energy of the attached state results from a competition between two terms:
| (2) |
where Ebinding and Ebending, respectively, are a negative binding energy that favors filament attachment and a positive energy due to bending disruption of the interaction by the curved nuclear envelope. Expanding Ebending in curvature (1/D) yields
| (3) |
where a/D captures the contact area between the nuclear membrane and the lamin-B filament (), and the last term is simply a bending energy with filament bending modulus (). These expansion terms increase (i.e., become more positive) for higher membrane curvature, which opposes filament adsorption. The probability of the detached state is then
| (4) |
In the high-curvature limit, a/D and the bending term become large such that
and , consistent with high probability of lamin-B loss and nuclear envelope rupture in high-curvature Transwell pores and micropipettes (Figs. 4 and S4). In the low-curvature limit (1/D), , assuming E >> kBT, which is consistent with strong attachment.
When fitting the curvature dependence of nuclear envelope rupture in Transwell pores and micropipettes (Fig. 4 A), we use the values ℓp = 0.5 µm and Lfil = 0.38 µm from Turgay et al. (2017) to set the bending modulus k in the quadratic term: . The ratio between the interaction energy a and the binding energy E provides a critical length scale that corresponds to the inflection point of the rupture-versus-curvature plot (Fig. 4 A, inset). Pore/pipette diameters smaller than a/E are in the high-curvature regimen and favor lamin-B desorption; diameters larger than a/E are in the low-curvature regimen and favor lamin-B attachment.
Nuclear indentation by atomic force microscopy (AFM) during fluorescent imaging
U2OS cells transfected with mCherry-cGAS and siLMNA were replated on coverslips at a density of 60,000 cells/cm2 and cultured overnight. Coverslips were mounted in the fluid cell of a hybrid AFM system (MFP-3D-BIO, software: MFP-3D+Igor Pro 6.05; Asylum Research, Oxford Instruments), which has a base plate and X-Y scanner that rest on an inverted optical fluorescence microscope (Olympus IX81 with 40×/0.60-NA objective). Experiments were performed in closed liquid cell at temperature of ∼29°C in high-glucose DMEM with 10% serum and buffered at pH 7.4 with 25 mM Hepes to prevent cell death in the absence of CO2 (Frigault et al., 2009). Cells were indented using MSCT-AUHW cantilevers (Bruker) with nominal spring constants 0.03–0.1 N/m, nominal tip radius 10–40 nm, and nominal tip height 2.5–8 µm. Before each experiment, cantilever spring constants were calibrated via the thermal fluctuations method to determine indentation forces. Then, a fluorescent cell was chosen for probing by AFM based on expression pattern; for example, expression of mCherry in the cytoplasm. The cantilever was positioned on the top of the nucleus, and the nucleus was locally compressed with forces of ∼10–40 nN. When the cantilever deflection reached the predefined set point, the tip would dwell on the spot for a predefined time of 100 s before the cantilever was retracted and detached from the cell. Simultaneous fluorescent images were captured every 10 or 15 s for the entire probing cycle, including before force was applied and after the cantilever was retracted for several minutes. In multiple independent experiments that cumulated to hundreds of pokes on nuclei inside cells, mCherry-cGAS reliably ruptured in. In addition, poking over the nucleus rarely showed cytoplasmic rupture (∼1%), consistent with nucleus-specific rupture. The same protocol has been used in Xia et al. (2018). The pressure that the tip exerted on the cell surface during typical maximum indentation of ∼4 µm was estimated as P = F/(πa)2, where F is the force and (πa)2 is the contact area between the tip and the cell for given indentation. The contact radius was calculated from the averaged nominal values of the pyramid tip specified by the manufacturer and given with , (Rico et al., 2005), where δ is the indentation and 2θ is the vertex angle of the pyramid tip.
Genome (SNP array) analysis
DNA isolation was done with Blood and Cell Culture DNA Mini Kit (Qiagen) per the manufacturer’s instructions. DNA samples were genotyped at the Center for Applied Genomics Core in the Children’s Hospital of Philadelphia, PA, for SNP using array HumanOmniExpress-24 BeadChip Kit (Illumina). For this array, >700,000 probes have an average interprobe distance of ∼4 kb along the entire genome. For each sample, the Genomics Core provided the data as GenomeStudio files (Illumina). The data were then exported to Matlab to convert SNP array data to single nucleotide variation data. To compare different samples, probes with no call (because of either low read intensity or being located outside the call cluster) were removed from further analysis.
Hierarchical clustering of SNP array results was performed based on the level of difference for each clone compared with 8-µm clone 0. Clustering was done in Matlab by calculating the pairwise distance first (“psidt”), followed by optical leaf ordering (“linkage” and “optimalleaforder”) and generation of a dendrogram (“dendrogram”). The P values are calculated between each clone and the 8-µm clone 0, using Kolmogorov–Smirnov (López-Bigas and Ouzounis, 2004; Mavrakis et al., 2010), with asterisks indicating P < 0.05 and showing that only the bottommost three clones (after 3-µm migration) have genomes that differ significantly. These three clones also exhibit changes of >40 Mb. This is important because the uncertainty associated with reproducibility was determined by running the same DNA sample on three SNP arrays. We determined a variation of ∼40 Mb (from the 3,100-Mb array; Fig. S5 A), with differences likely reflecting imperfections in the array of probes or the hybridization process. Furthermore, SNP array linearity was determined by mixing DNA from two very different clones in different ratios, which showed clear linearity when the signal was >40 Mb.
Heatmap comparisons of curvature-associated nuclear envelope rupture properties
The numbers in the heatmap were obtained from Figs. 1, 4, 5, S3, and S4 and additional literature. (1) LaminB dilution: Numbers of the first four columns were obtained from Fig. 4 directly. For micronuclei, as there is no bleb on micronuclei, the numbers reflected the fraction of micronuclei that are lamin-B–positive or –negative (Fig. 5 C). (2) cGAS entry: Numbers of the first four columns were obtained from Figs. 4 and 5 C directly. (3) Nuclear pore dilution: The numbers of the first two columns were taken from Fig. S4 Aii. Aspirated tip data come from the passive pulling of detached, latrunculin-treated U2OS cells overexpressing GFP-Nup153 into small or large micropipettes (Fig. S3 E). For micronuclei, the numbers reflected the average coverage of the nucleoporins (NUPs) on different sizes of micronuclei in Liu et al. (2018b). (4) Fraction of DNA repair factors in the nucleus: Numbers of the first two columns were obtained from Fig. 1 directly. The aspiration experiments in the third and fourth columns were done previously in Irianto et al. (2016). For micronuclei, the numbers reflected KU80/DNA in Fig. 5 C, and the results are consistent with the RPA trend in Liu et al. (2018b). The row of cGAS is normalized to the highest value, as all experiments are comparable. The nuclear pore dilution in each case is normalized to the highest value within its own dataset. In other cases, the first four columns of each row were normalized to the highest value, as the experiments are comparable. Tithe micronuclei results (rows 1, 3, and 4) are normalized within their own dataset, as some criteria and data sources are slightly different.
Data sources are as follows. Nuclear blebs in Transwells: lamin-B dilution, Fig. 4 B; cGAS entry, Fig. 4 A; nuclear pore dilution: Fig. S4 Aii; DNA repair factors, Fig. 1 D. Nuclear aspirated tip: lamin-B dilution, Fig. 4 D; cGAS entry, Fig. 4 D; nuclear pore dilution, Fig. S3 E; DNA repair factors, Irianto et al. (2016). Micronuclei: lamin-B dilution, Fig. 5 C; cGAS entry, Fig. 5 C; nuclear pore dilution, Liu et al. (2018b); DNA repair factors, Fig. 5 C.
Statistical analysis
All statistical analyses were performed using Microsoft Excel 2013. Fig. 3 E fitting was done in Python 2.7 via Spyder 3.2.4 (curve_fit scipy). Unless otherwise noted, statistical comparisons were made by unpaired two-tailed Student’s t test and were considered significant if P < 0.05. Unless mentioned otherwise, all plots show mean ± SEM, and n indicates the number of samples, cells, or wells quantified in each experiment. Figure legends specify the exact meaning of n for each figure (Cumming et al., 2007).
Online supplemental material
Fig. S1 shows phospho-lamin-A, MTOC, and myosin II effects in and on constricted migration and/or rupture. Fig. S2 addresses repair factors, reactive oxygen species, and MYO-i effects on DNA damage and cell cycle, including use of etched pores for migration. Fig. S3 shows that nuclear envelope rupture frequency correlates more with curvature and pressure than with applied tension. Fig. S4 describes the composition of micronuclei, nuclear blebs, and ruptured nuclei. Fig. S5 provides genomic variation measurements after constricted migration.
Supplementary Material
Acknowledgments
The authors thank Dr. Stuart L Rulten from University of Sussex for various DNA damage–response protein plasmids used in this study and Dr. Qinnan Zhang from University of Notre Dame for schematic drawing.
The authors in this study were supported by the National Institutes of Health National Cancer Institute under Physical Sciences Oncology Center Award U54 CA193417; National Heart, Lung, and Blood Institute Award R21 HL128187; the United States–Israel Binational Science Foundation; Charles Kaufman Foundation Award KA2015-79197; and National Science Foundation Materials Research Science and Engineering Center at Penn as well as grant agreement CMMI 15-48571. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
The authors declare no competing financial interests.
Author contributions: Y. Xia, C.R. Pfeifer, and D.E. Discher designed experiments. R.A. Greenberg contributed key materials, methods, and discussion. Y. Xia, C.R. Pfeifer, K. Zhu, J. Irianto, D. Liu, K. Pannell, E.J. Chen, L.J. Dooling, M.P. Tobin, M. Wang, I.L. Ivanovska, and L.R. Smith performed and analyzed experiments. Y. Xia, C.R. Pfeifer, and D.E. Discher wrote the manuscript.
References
- Bannister A.J., and Kouzarides T.. 2011. Regulation of chromatin by histone modifications. Cell Res. 21:381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle C., Assanah M.C., Monzo P., Vallee R., Rosenfeld S.S., and Canoll P.. 2008. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell. 19:3357–3368. 10.1091/mbc.e08-03-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin B.J., Lee T., and Mullins R.D.. 2015. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-1/2 that promotes efficient DNA repair. [corrected]. eLife. 4:e07735 10.7554/eLife.07735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.R., Pfeifer C.R., Irianto J., Xia Y., Discher D.E., and Liu A.J.. 2017. Elastic-Fluid Model for DNA Damage and Mutation from Nuclear Fluid Segregation Due to Cell Migration. Biophys. J. 112:2271–2279. 10.1016/j.bpj.2017.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S., Coates J., and Jackson S.P.. 2013. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202:579–595. 10.1083/jcb.201303073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A., Swift J., Irianto J., Spinler K.R., Dingal P.C., Athirasala A., Kao Y.R., Cho S., Harada T., Shin J.W., and Discher D.E.. 2014. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr. Biol. 24:1909–1917. 10.1016/j.cub.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Vashisth M., Abbas A., Majkut S., Vogel K., Xia Y., Ivanovska I.L., Irianto J., Tewari M., Zhu K., et al. 2019. Mechanosensing by the Lamina Protects against Nuclear Rupture, DNA Damage, and Cell-Cycle Arrest. Dev. Cell.:S1534-5807(19)30317-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius T., Apel P.Y., Schiedt B., Trautmann C., Toimil-Molares M., Karim S., and Neumann R.. 2007. Investigation of nanopore evolution in ion track-etched polycarbonate membranes. Nucl. Instrum. Methods Phys. Res. B. 265:553–557. 10.1016/j.nimb.2007.10.004 [DOI] [Google Scholar]
- Cumming G., Fidler F., and Vaux D.L.. 2007. Error bars in experimental biology. J. Cell Biol. 177:7–11. 10.1083/jcb.200611141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., and Lammerding J.. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science. 352:353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., and Pan D.. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 130:1120–1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duheron V., Chatel G., Sauder U., Oliveri V., and Fahrenkrog B.. 2014. Structural characterization of altered nucleoporin Nup153 expression in human cells by thin-section electron microscopy. Nucleus. 5:601–612. 10.4161/19491034.2014.990853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigault M.M., Lacoste J., Swift J.L., and Brown C.M.. 2009. Live-cell microscopy - tips and tools. J. Cell Sci. 122:753–767. 10.1242/jcs.033837 [DOI] [PubMed] [Google Scholar]
- Garay T., Juhász É., Molnár E., Eisenbauer M., Czirók A., Dekan B., László V., Hoda M.A., Döme B., Tímár J., et al. 2013. Cell migration or cytokinesis and proliferation?--revisiting the “go or grow” hypothesis in cancer cells in vitro. Exp. Cell Res. 319:3094–3103. 10.1016/j.yexcr.2013.08.018 [DOI] [PubMed] [Google Scholar]
- Giese A., Loo M.A., Tran N., Haskett D., Coons S.W., and Berens M.E.. 1996. Dichotomy of astrocytoma migration and proliferation. Int. J. Cancer. 67:275–282. [DOI] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., and Gundersen G.G.. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463. 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Grundy G.J., Rulten S.L., Zeng Z., Arribas-Bosacoma R., Iles N., Manley K., Oliver A., and Caldecott K.W.. 2013. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 32:112–125. 10.1038/emboj.2012.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T., Swift J., Irianto J., Shin J.W., Spinler K.R., Athirasala A., Diegmiller R., Dingal P.C., Ivanovska I.L., and Discher D.E.. 2014. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 204:669–682. 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J., and Greenberg R.A.. 2017. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 548:466–470. 10.1038/nature23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E.M., Fischer A.H., Deerinck T.J., and Hetzer M.W.. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 154:47–60. 10.1016/j.cell.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtgraaf J.H., Versmissen J., and van der Giessen W.J.. 2006. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revasc. Med. 7:165–172. 10.1016/j.carrev.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Imlay J.A., Chin S.M., and Linn S.. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 240:640–642. 10.1126/science.2834821 [DOI] [PubMed] [Google Scholar]
- Irianto J., Pfeifer C.R., Bennett R.R., Xia Y., Ivanovska I.L., Liu A.J., Greenberg R.A., and Discher D.E.. 2016. Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol. Biol. Cell. 27:4011–4020. 10.1091/mbc.e16-06-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J., Xia Y., Pfeifer C.R., Athirasala A., Ji J., Alvey C., Tewari M., Bennett R.R., Harding S.M., Liu A.J., et al. 2017. DNA Damage Follows Repair Factor Depletion and Portends Genome Variation in Cancer Cells after Pore Migration. Curr. Biol. 27:210–223. 10.1016/j.cub.2016.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivkovic S., Beadle C., Noticewala S., Massey S.C., Swanson K.R., Toro L.N., Bresnick A.R., Canoll P., and Rosenfeld S.S.. 2012. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol. Biol. Cell. 23:533–542. 10.1091/mbc.e11-01-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau S.B., Hale C.M., Stewart-Hutchinson P.J., Patel M.S., Stewart C.L., Searson P.C., Hodzic D., and Wirtz D.. 2009. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 106:19017–19022. 10.1073/pnas.0908686106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryston T.B., Georgiev A.B., Pissis P., and Georgakilas A.G.. 2011. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat. Res. 711:193–201. 10.1016/j.mrfmmm.2010.12.016 [DOI] [PubMed] [Google Scholar]
- Lee K.Y., Fu H., Aladjem M.I., and Myung K.. 2013. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J. Cell Biol. 200:31–44. 10.1083/jcb.201206084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang H., Wu X., Ma D., Wu J., Wang L., Jiang Y., Fei Y., Zhu C., Tan R., et al. 2018a Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 563:131–136. 10.1038/s41586-018-0629-6 [DOI] [PubMed] [Google Scholar]
- Liu S., Kwon M., Mannino M., Yang N., Renda F., Khodjakov A., and Pellman D.. 2018b Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 561:551–555. 10.1038/s41586-018-0534-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bigas N., and Ouzounis C.A.. 2004. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 32:3108–3114. 10.1093/nar/gkh605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Zhu F., Cho Y.-Y., Tang F., Zykova T., Ma W.Y., Bode A.M., and Dong Z.. 2006. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol. Cell. 23:121–132. 10.1016/j.molcel.2006.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., Li Y., Bosco N., Campbell P.J., and de Lange T.. 2015. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 163:1641–1654. 10.1016/j.cell.2015.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaseth T., and Kuzminov A.. 2016. Prompt repair of hydrogen peroxide-induced DNA lesions prevents catastrophic chromosomal fragmentation. DNA Repair (Amst.). 41:42–53. 10.1016/j.dnarep.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis K.J., Wolfe A.L., Oricchio E., Palomero T., de Keersmaecker K., McJunkin K., Zuber J., James T., Khan A.A., Leslie C.S., et al. 2010. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 12:372–379. 10.1038/ncb2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello Filho A.C., Hoffmann M.E., and Meneghini R.. 1984. Cell killing and DNA damage by hydrogen peroxide are mediated by intracellular iron. Biochem. J. 218:273–275. 10.1042/bj2180273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A., and Odde D.. 2011. Modeling cellular processes in 3D. Trends Cell Biol. 21:692–700. 10.1016/j.tcb.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G., Daly G., Lindahl T., and Barnes D.E.. 2004. Gene-targeted mice lacking the Trex1 (DNase III) 3′-->5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell. Biol. 24:6719–6727. 10.1128/MCB.24.15.6719-6727.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski J.D., Dahl K.N., Zhong F.L., Sammak P.J., and Discher D.E.. 2007. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 104:15619–15624. 10.1073/pnas.0702576104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak A., and Kumar S.. 2012. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc. Natl. Acad. Sci. USA. 109:10334–10339. 10.1073/pnas.1118073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., Koo H., and Yamada K.M.. 2014. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 345:1062–1065. 10.1126/science.1256965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer C.R., Alvey C.M., Irianto J., and Discher D.E.. 2017. Genome variation across cancers scales with tissue stiffness - an invasion-mutation mechanism and implications for immune cell infiltration. Curr. Opin. Syst. Biol. 2:103–114. 10.1016/j.coisb.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer C.R., Xia Y., Zhu K., Liu D., Irianto J., Garcia V.M.M., Millan L.M.S., Niese B., Harding S., Deviri D., et al. 2018. Constricted migration increases DNA damage and independently represses cell cycle. Mol. Biol. Cell. 29:1948–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., and Discher D.E.. 2017. Matrix rigidity regulates microtubule network polarization in migration. Cytoskeleton (Hoboken). 74:114–124. 10.1002/cm.21349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.M., Manel N., and Piel M.. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 352:359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Radisky D.C., Levy D.D., Littlepage L.E., Liu H., Nelson C.M., Fata J.E., Leake D., Godden E.L., Albertson D.G., Nieto M.A., et al. 2005. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 436:123–127. 10.1038/nature03688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G., Pavelka N., Fleharty B., Noll A., Trimble R., Walton K., Perera A., Staehling-Hampton K., Seidel C.W., and Li R.. 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 135:879–893. 10.1016/j.cell.2008.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Rodero M.P., and Crow Y.J.. 2015. Human disease phenotypes associated with mutations in TREX1. J. Clin. Immunol. 35:235–243. 10.1007/s10875-015-0147-3 [DOI] [PubMed] [Google Scholar]
- Rico F., Roca-Cusachs P., Gavara N., Farré R., Rotger M., and Navajas D.. 2005. Probing mechanical properties of living cells by atomic force microscopy with blunted pyramidal cantilever tips. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 72:021914 10.1103/PhysRevE.72.021914 [DOI] [PubMed] [Google Scholar]
- Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. 2015. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 348:124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L.A., Unsal-Kaçmaz K., and Linn S.. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39–85. 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- Skamagki M., Correia C., Yeung P., Baslan T., Beck S., Zhang C., Ross C.A., Dang L., Liu Z., Giunta S., et al. 2017. ZSCAN10 expression corrects the genomic instability of iPSCs from aged donors. Nat. Cell Biol. 19:1037–1048. 10.1038/ncb3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S., Monti O., Pediconi N., Baccarini A., Fontemaggi G., Lapi E., Mantovani F., Damalas A., Citro G., Sacchi A., et al. 2005. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol. Cell. 18:447–459. 10.1016/j.molcel.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Stroka K.M., Jiang H., Chen S.H., Tong Z., Wirtz D., Sun S.X., and Konstantopoulos K.. 2014. Water permeation drives tumor cell migration in confined microenvironments. Cell. 157:611–623. 10.1016/j.cell.2014.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surcel A., Ng W.P., West-Foyle H., Zhu Q., Ren Y., Avery L.B., Krenc A.K., Meyers D.J., Rock R.S., Anders R.A., et al. 2015. Pharmacological activation of myosin II paralogs to correct cell mechanics defects. Proc. Natl. Acad. Sci. USA. 112:1428–1433. 10.1073/pnas.1412592112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki T., Montagner M., Serres M.P., Le Berre M., Russell M., Collinson L., Szuhai K., Howell M., Boulton S.J., Sahai E., and Petronczki M.. 2017. Actomyosin drives cancer cell nuclear dysmorphia and threatens genome stability. Nat. Commun. 8:16013 10.1038/ncomms16013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam H.R., Vargas P., Carpi N., Crespo C.L., Raab M., Terriac E., King M.C., Jacobelli J., Alberts A.S., Stradal T., et al. 2016. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 7:10997 10.1038/ncomms10997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.G., Yenepalli A., Denais C.M., Rape A., Beach J.R., Wang Y.L., Schiemann W.P., Baskaran H., Lammerding J., and Egelhoff T.T.. 2015. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J. Cell Biol. 210:583–594. 10.1083/jcb.201502039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y., Eibauer M., Goldman A.E., Shimi T., Khayat M., Ben-Harush K., Dubrovsky-Gaupp A., Sapra K.T., Goldman R.D., and Medalia O.. 2017. The molecular architecture of lamins in somatic cells. Nature. 543:261–264. 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. 2013. Joint probability distribution. In Dubitzky W., Wolkenhauer O., Cho K.H., Yokota H., eds. Encyclopedia of Systems Biology, Springer, New York, 1063 10.1007/978-1-4419-9863-7_426 [DOI] [Google Scholar]
- Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A.L., Hoffman R.M., Figdor C.G., Weiss S.J., and Friedl P.. 2013. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201:1069–1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Ivanovska I.L., Zhu K., Smith L., Irianto J., Pfeifer C.R., Alvey C.M., Ji J., Liu D., Cho S., et al. 2018. Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol. 217:3796–3808. 10.1083/jcb.201711161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N. 2017. Immune diseases associated with TREX1 and STING dysfunction. J. Interferon Cytokine Res. 37:198–206. 10.1089/jir.2016.0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., and Pellman D.. 2015. Chromothripsis from DNA damage in micronuclei. Nature. 522:179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.