Lipid droplets (LDs) and peroxisomes form a functional alliance to maintain lipid and energy homeostasis. Chang et al. provide mechanistic insights by describing a tethering complex that comprises LD-localized M1 Spastin and peroxisomal ABCD1 at LD–peroxisome contact sites and supports fatty acid trafficking.
Abstract
Lipid droplets (LDs) are neutral lipid storage organelles that transfer lipids to various organelles including peroxisomes. Here, we show that the hereditary spastic paraplegia protein M1 Spastin, a membrane-bound AAA ATPase found on LDs, coordinates fatty acid (FA) trafficking from LDs to peroxisomes through two interrelated mechanisms. First, M1 Spastin forms a tethering complex with peroxisomal ABCD1 to promote LD–peroxisome contact formation. Second, M1 Spastin recruits the membrane-shaping ESCRT-III proteins IST1 and CHMP1B to LDs via its MIT domain to facilitate LD-to-peroxisome FA trafficking, possibly through IST1- and CHMP1B-dependent modifications in LD membrane morphology. Furthermore, LD-to-peroxisome FA trafficking mediated by M1 Spastin is required to relieve LDs of lipid peroxidation. M1 Spastin’s dual roles in tethering LDs to peroxisomes and in recruiting ESCRT-III components to LD–peroxisome contact sites for FA trafficking may underlie the pathogenesis of diseases associated with defective FA metabolism in LDs and peroxisomes.
Introduction
Nonvesicular trafficking pathways exploit the dense cytoplasmic packing of organelles to enable functional alliances through interorganelle associations. Such associations are mediated by tethering proteins, which create contact sites for direct channeling of metabolites, lipids, and ions between organelles (Wong et al., 2019). A major challenge has been to identify the diverse array of proteins involved in generating organelle contact sites and to understand how they mediate nonvesicular transport.
Among the organelles that engage in nonvesicular transport are lipid droplets (LDs; Schuldiner and Bohnert, 2017; Henne et al., 2018). LDs stockpile fatty acids (FAs) as neutral lipids and release FAs as building materials for lipid synthesis and protein modification (Pol et al., 2014; Hashemi and Goodman, 2015; Walther et al., 2017). When nutrient availability is low, LDs transfer FAs to mitochondria and peroxisomes for β-oxidation, a crucial process that generates precursors for mitochondrial oxidative phosphorylation supporting energy production (Finn and Dice, 2006; Poirier et al., 2006). In addition, LDs transfer FAs to peroxisomes for other steps in FA metabolism, including β-oxidation of very long chain FAs, α-oxidation of branched chain FAs, bile acid and ether phospholipid synthesis, and docosahexaenoic acid generation (Wanders, 2013; Lodhi and Semenkovich, 2014; Islinger et al., 2018). Aberrant FA metabolism in LDs is associated with severe physiological consequences, including neurological diseases and lipodystrophy (Welte, 2015; Kory et al., 2016). Defects in peroxisomal FA metabolism and biogenesis also lead to accumulation of LDs, as demonstrated by patients with adrenoleukodystrophy (ALD) and Zellweger syndrome (Schaumburg et al., 1972; Baes et al., 1997; Engelen et al., 2012), suggesting a functional alliance between LDs and peroxisomes.
LD–mitochondria and LD–peroxisome contact sites are thought to facilitate direct channeling of FAs across these organelles’ boundaries and to prevent toxicity from free cytosolic FAs (Unger et al., 2010; Nguyen et al., 2017; Schuldiner and Bohnert, 2017; Henne et al., 2018). While much attention has focused on characterizing contact sites between LDs and mitochondria, the machinery that creates LD–peroxisome contact sites and their functional significance has remained a mystery.
Recently, possible protein candidates involved in LD–peroxisome contact sites and/or associated FA metabolism have emerged from studies of the pathogenesis of hereditary spastic paraplegias (HSPs), a group of inherited neurological disorders with a prominent clinical feature of lower-extremity spasticity (Welte, 2015; Blackstone, 2018). Mutations in the gene encoding Spastin are the most common cause of HSP and impact both LD and peroxisomal function. Depletion of Spastin in Drosophila melanogaster and Caenorhabditis elegans leads to aberrant FA metabolism in LDs (Papadopoulos et al., 2015). In addition, HSP patient–derived cells with mutations in Spastin showed impaired peroxisome movement and distribution (Wali et al., 2016), coinciding with increased lipid peroxidation and reduced energy production possibly caused by defective FA trafficking to peroxisomes. Intriguingly, some ALD patients with mutations in a peroxisomal membrane FA transporter, ATP binding cassette subfamily D member 1 (ABCD1), also manifest neurological symptoms such as spasticity (Maris et al., 1995; Shaw-Smith et al., 2004; Zhan et al., 2013; Lodhi and Semenkovich, 2014; Koutsis et al., 2015; Balicza et al., 2016). Together, these findings raise the possibility that Spastin and/or ABCD1 could function at LD–peroxisome contact sites to facilitate interorganelle FA trafficking.
M1 Spastin is an isoform generated from the first translation initiation codon of Spastin (Claudiani et al., 2005). It contains an N-terminal hydrophobic hairpin (Hp) motif that helps distribute it to LDs and other membrane compartments (Reid et al., 2005; Connell et al., 2009; Park et al., 2010; Papadopoulos et al., 2015). The Hp motif is followed by a microtubule interacting and trafficking (MIT) domain that selectively binds two ESCRT-III (endosomal sorting complex required for transport III) proteins: increased sodium tolerance 1 (IST1) and charged multivesicular body protein 1B (CHMP1B; Reid et al., 2005; Agromayor et al., 2009; Renvoisé et al., 2010). Unlike most ESCRT-III components, IST1 and CHMP1B form external coats on positively curved membranes (McCullough et al., 2015). M1 Spastin also harbors a microtubule binding domain and an ATPase-associated with diverse cellular activities (AAA) ATPase domain implicated in remodeling microtubule networks (White et al., 2007; Roll-Mecak and Vale, 2008). Thus, M1 Spastin is a multimodular ATPase combining functions of LD association and ESCRT-III interaction. Nonetheless, a unifying mechanism that explains how different domains of Spastin operate together, specifically in FA metabolism at LDs and peroxisomes, is lacking.
Here, we provide evidence that M1 Spastin and ABCD1 form a tethering complex that draws LDs and peroxisomes together to facilitate LD-to-peroxisome FA trafficking. M1 Spastin promotes LD–peroxisome contact formation by inserting its Hp motif into the LD lipid monolayer and using a peroxisome-interacting (PXI) region to form a complex with ABCD1 on peroxisomes. Furthermore, M1 Spastin controls LD-to-peroxisome FA trafficking by recruiting IST1 and CHMP1B to LDs. This indicates a novel role for ESCRT-III proteins in FA trafficking, possibly through modifying LD membrane morphology. Finally, we show that Spastin-mediated FA trafficking is necessary for reducing peroxidated lipids in LDs. Together, these findings reveal a novel FA trafficking pathway orchestrated by M1 Spastin in conjunction with ESCRT-III and ABCD1 at LD–peroxisome contact sites, with relevance for understanding diseases involving mutations affecting Spastin and ABCD1.
Results
M1 Spastin promotes LD–peroxisome contact formation
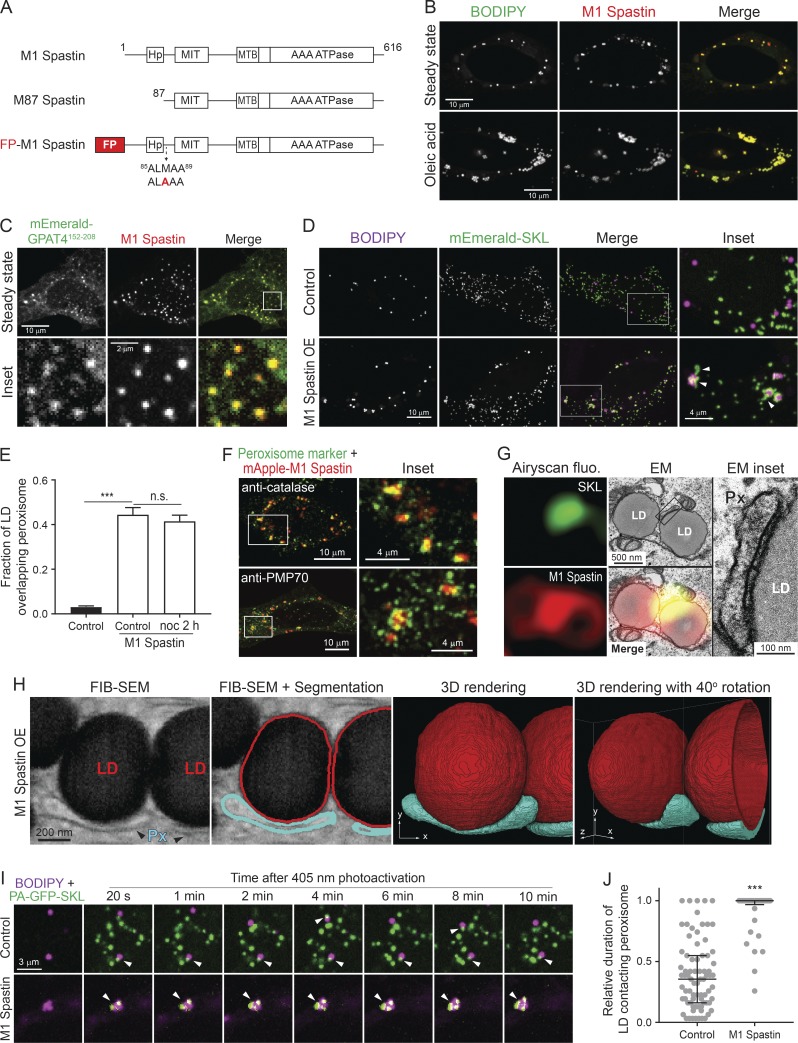
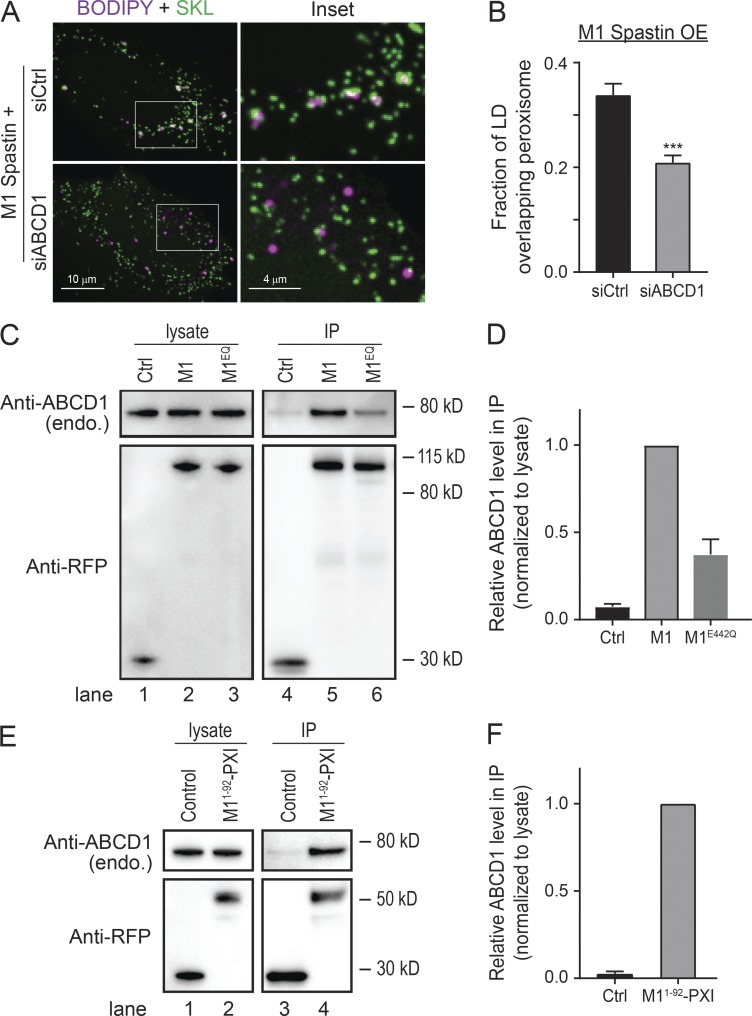
To test a role of M1 Spastin in regulating the association between LDs and peroxisomes, we appended the fluorescence protein (FP) mApple to the N-terminus of M1 Spastin after mutating its second start codon (M87A) to eliminate expression of M87 Spastin (Fig. 1 A). When overexpressed in HeLa cells, mApple-M1 Spastin colocalizes with boron-dipyrromethene (BODIPY)–labeled LDs, both under steady-state conditions and during LD biogenesis induced by oleic acid treatment (Fig. 1 B; Papadopoulos et al., 2015). We also observed colocalization of mApple-M1 Spastin with GPAT4152–208 (Fig. 1 C), a membrane marker for nascent LDs (Wilfling et al., 2013). Therefore, M1 Spastin appears to associate with LDs at different stages of LD biogenesis. Using mEmerald-SKL to label peroxisomes, we found that peroxisomes normally have intermittent contacts with LDs (Fig. 1 D, top panels). Upon overexpression of M1 Spastin, however, a significant increase in association between LDs and peroxisomes occurs (Fig. 1 D, bottom panels), with LD-associated peroxisomes frequently displaying an elongated morphology (Fig. 1 D, inset). Quantification of the fraction of LD signal overlapping peroxisome signal revealed a significant increase in LD–peroxisome colocalization by M1 Spastin overexpression (Fig. 1 E). This enhanced association of LDs and peroxisomes was not affected by nocodazole treatment (Fig. 1 E), suggesting that microtubule dynamics are not required for M1 Spastin to mediate LD–peroxisome association. Immunostaining of peroxisomal proteins, including luminal catalase and membrane-associated PMP70, showed peroxisomes in close association with LDs decorated with mApple-M1-Spastin (Fig. 1 F). The enhancement of LD–peroxisome association by M1 Spastin overexpression was also observed in U-2 OS osteosarcoma cells and MRC-5 fibroblasts (Fig. S1, A and B). In addition, overexpression of M87 Spastin, a Spastin isoform devoid of Hp motif residing in the cytoplasm (Fig. S1, C and D), did not enhance LD–peroxisome association (Fig. S1 E). These observations indicated that M1 Spastin, but not M87 Spastin, plays a major role in mediating LD–peroxisome association and strengthens the notion that affecting microtubule dynamics alone has no effect on LD–peroxisome association.
Figure 1.
M1 Spastin promotes LD–peroxisome contact formation. (A) Diagrams of M1 Spastin, M87 Spastin, and FP-tagged M1 Spastin construct. Amino acid number and protein domains are indicated. The mutated residue is labeled in red. (B) mApple-M1 Spastin colocalizes with BODIPY-493/503–labeled LDs in HeLa cells in steady state (top) or following 300 µM oleic acid treatment for 16 h (bottom). Representative confocal images are shown. (C) mApple-M1 Spastin colocalizes with mEmerald-GPAT4152–208 in HeLa cells. Representative confocal images are shown. (D) Association between BODIPY-665/676–labeled LDs and mEmerald-SKL–labeled peroxisomes in control or mApple-M1 Spastin-overexpressing (OE) HeLa cells. Representative confocal MIP images are shown. White arrowheads indicate elongated peroxisomes. (E) Fraction of LD overlapping peroxisome as described in D. mApple-M1 Spastin-expressing cells were further treated with nocodazole (noc). Means ± SEM are shown (22–41 cells from three independent experiments). ***, P < 0.001; n.s., not significant. (F) Association between mApple-M1 Spastin and peroxisomes immunostained with catalase (top) and PMP70 (bottom) antibodies in HeLa cells. Representative confocal MIP images are shown. (G) CLEM images of an LD–peroxisome contact site in mApple-M1 Spastin and mEmerald-SKL expressing HeLa cells treated with 15 µM oleic acid for 16 h. Px, peroxisome. (H) A single 8-nm FIB-SEM slice (left) at LD–peroxisome contacts in mApple-M1 expressing HeLa cells treated with 15 µM oleic acid for 16 h. LDs and peroxisomes (Px) were segmented and reconstructed to reveal LD–peroxisome contacts in 3D. (I) Association between BODIPY-665/676–labeled LDs and peroxisomes labeled by PA-GFP-SKL in control and in mApple-M1 Spastin-overexpressing HeLa cells monitored by confocal microscopy. White arrowheads indicate LD–peroxisome contacts. (J) Relative duration of LD contacting peroxisome as described in I. Median ± interquartile ranges are shown (37–77 LDs from two to three independent experiments). ***, P < 0.001.
We next used correlative light-EM (CLEM) to examine the nature of LD–peroxisome association in M1 Spastin–overexpressing cells. In correlated Airyscan and transmission EM images, elongated peroxisomes were visualized tightly attached to the surface of LDs (Fig. 1 G), indicating LD–peroxisome contacts. Increased electron density was seen at LD–peroxisome contacts (Fig. 1 G, inset), prefiguring the existence of protein and/or membrane complexes that might serve to bridge these organelles. We applied focused ion beam-scanning EM (FIB-SEM) to examine LD–peroxisome contacts in 3D at 8-nm isotropic resolution in mApple-M1 Spastin–overexpressing cells. Similar to conventional transmission EM, we observed elongated peroxisomes tightly associated with LDs in single FIB-SEM slices (Fig. 1 H). With the volumetric information provided by FIB-SEM, the contact sites throughout the entirety of peroxisome and LD surfaces were visualized. Interestingly, the contour of the contacting peroxisomes mimicked that of the LDs. The association between the peroxisome and LD spanned most of the length of the peroxisome, suggesting tethering sites with a large surface area (Fig. 1 H and Video 1).
We further examined the dynamics of LD–peroxisome interaction by labeling peroxisomes with photoactivatable (PA) GFP attached to SKL (PA-GFP-SKL). Applying photoactivation to a few small regions containing LDs, a subpopulation of peroxisomes was then tracked. In control cells, we observed both stable and transient LD–peroxisome contacts during 10 min of imaging (Fig. 1 I, top panels; and Video 2), suggesting the existence of endogenous tethers bridging these organelles. In M1 Spastin–overexpressing cells, most of the highlighted peroxisomes were stably tethered to LDs (Fig. 1 I, bottom panels; and Video 3). We further quantified the relative duration of LDs contacting peroxisomes during 10 min of imaging and found that M1 Spastin significantly extends the contact time (Fig. 1 J). These results suggested that M1 Spastin may function as part of a tether for LD–peroxisome contacts.
Endogenous Spastin is required for LD–peroxisome contact formation
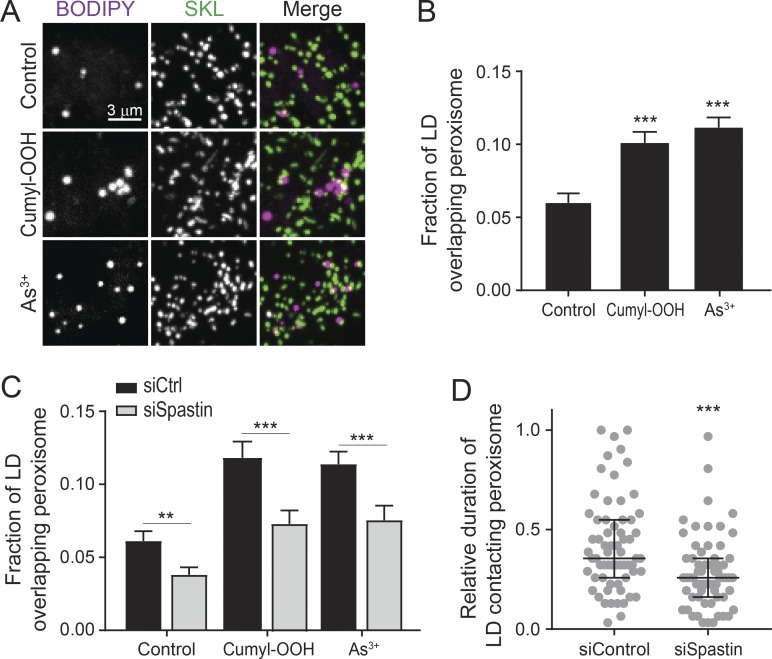
Since peroxisomes are essential for maintaining redox homeostasis (Wanders, 2013), we wondered if oxidative stress could regulate LD–peroxisome contacts. We found that LD–peroxisome contacts were enhanced in control cells following cumene hydroperoxide (cumyl-OOH) or NaAsO2 (As3+) treatment (Fig. 2, A and B), which induces lipid peroxidation (van der Kraaij et al., 1990; Ramos et al., 1995). These observations suggested that LD–peroxisome contacts may have a role in handling lipid peroxidation. Knockdown of Spastin by siRNA transfection, which reduces both M1 and M87 Spastin expression (Fig. S2 A), led to a decrease in LD–peroxisome contacts in both control and stressed cells (Fig. 2 C). We further applied photoactivation experiments to examine the dynamic LD–peroxisome association; knockdown of Spastin significantly reduced the duration of LDs contacting peroxisomes (Fig. 2 D). Since overexpression of M87 Spastin alone did not enhance LD–peroxisome contact formation (Fig. S1 E), these data suggest that endogenous M1 Spastin serves a major role in mediating LD–peroxisome contact formation.
Figure 2.
Endogenous Spastin is required for LD–peroxisome contact formation. (A) Association between LDs and peroxisomes in control HeLa cells or HeLa cells treated with cumyl-OOH or As3+ for 30 min. Representative confocal MIP images are shown. (B) Fraction of LD overlapping peroxisome as described in A. Means ± SEM are shown (31–40 cells from three to four independent experiments). (C) Fraction of LD overlapping peroxisome in siCtrl and siSpastin-transfected HeLa cells in control condition or under oxidative stress as described in A. Means ± SEM are shown (21–28 cells from three independent experiments). (D) Relative duration of LD contacting peroxisome (as described in Fig. 1, I and J) in siControl and siSpastin-transfected HeLa cells. Median ± interquartile ranges are shown (64–65 LDs from three independent experiments). **, P < 0.01; ***, P < 0.001.
M1 Spastin’s Hp motif directly inserts into the lipid monolayer of LDs
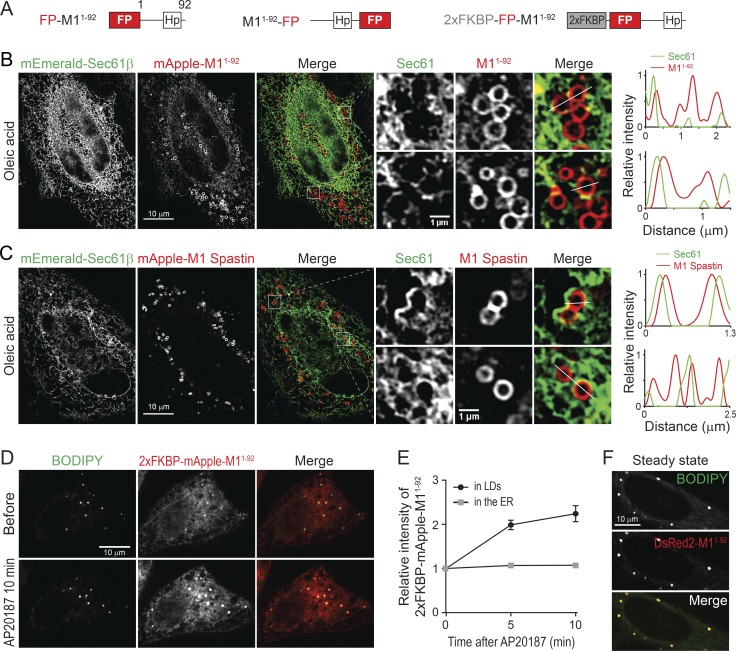
To gain insight into the role of M1 Spastin in LD–peroxisome tethering, we explored how M1 Spastin associates with LDs, focusing on its Hp motif. Two synthetic constructs were generated that contain the amino acids 1–92 of M1 Spastin tagged with mApple, either at its N-terminus (FP-M11–92) or its C-terminus (M11–92-FP; Fig. 3 A). Both mApple-M11–92 and M11–92-mApple labeled the ER and localized to steady-state LDs, oleic acid–induced LDs, and GPAT4152–208-labeled nascent LDs (Fig. S3, A–C; and Video 4). Therefore, similar to M1 Spastin, M11–92 associates with LDs at different stages of LD biogenesis.
Figure 3.
M1 Spastin’s Hp motif directly inserts into the lipid monolayer of LDs. (A) Diagrams of synthetic constructs. Amino acid number and protein domains are indicated as in Fig. 1 A. (B) Localization of mEmerald-Sec61β and mApple-M11–92 monitored by SIM in HeLa cells treated with 300 µM oleic acid for 16 h. Representative images and relative intensity profile are shown. (C) Localization of mEmerald-Sec61β and mApple-M1 Spastin monitored by SIM in HeLa cells treated with oleic acid. Representative images and relative intensity profile are shown. (D) Changes in intensity of 2xFKBP-mApple-M11–92 in BODIPY-493/503–labeled LDs following 1 µM AP20187 treatment in HeLa cells. Representative confocal images are shown. (E) Changes in relative intensity of 2xFKBP-mApple-M11–92 as described in D. Means ± SEM are shown (12 cells from two independent experiments). (F) DsRed2-M11–92 colocalizes with LDs in HeLa cells in steady state. Representative confocal images are shown.
M11–92 could colocalize with LDs either by directly inserting into the lipid monolayer of LDs or by an ER pool of M11–92 wrapping tightly around LDs. To distinguish between these possibilities, we used structured illumination microscopy (SIM), providing a twofold increase in resolution compared with conventional confocal imaging, to visualize ER labeled by mEmerald-Sec61β and mApple-M11–92 in oleic acid–treated cells. SIM imaging coupled with line scan analyses revealed that mApple-M11–92 localizes to LDs rather than to nearby ER tubules (Fig. 3 B), supporting the idea that M11–92 directly inserts into the lipid monolayer of LDs.
Like M11–92, full-length M1 Spastin was also localized on the surface of LDs, showing less colocalization with the ER marker mEmerald-Sec61β (Fig. 3 C). One reason for this difference could relate to full-length M1 Spastin’s ability to oligomerize into hexamers (Pantakani et al., 2008; Roll-Mecak and Vale, 2008), which could cause M1 Spastin to have higher affinity for LDs. To test this possibility, we inserted tandem FKBP (2xFKBP), an AP20187-inducible oligomerization motif (Burnett et al., 2004), into the N-terminus of mApple-M11–92 to generate 2xFKBP-mApple-M11–92 (Fig. 3 A). This protein localized to both LDs and the ER (Fig. 3 D). Shortly following AP20187 treatment, a twofold increase in the relative intensity of 2xFKBP-mApple-M11–92 on LDs occurred (Fig. 3, D and E), suggesting that oligomerization enhances the affinity of M11–92 to LDs. Consistent with this, M11–92 tagged with a dimeric red FP, DsRed2, to enable protein oligomerization also showed primarily LD localization under steady-state conditions (Fig. 3 F). Together, these findings suggest that M1 Spastin directly inserts into the LD lipid monolayer via its Hp motif, with oligomerization enhancing this association.
ATP hydrolysis is required for M1 Spastin to mediate LD–peroxisome contact formation
We systematically surveyed the cytosolic regions of M1 Spastin to determine how it might induce LD–peroxisome contact formation. We found that M1 SpastinΔ1–56 lacking its N-terminal cytosolic region or M1 SpastinHFDD mutant with a defective MIT domain for ESCRT-III binding (Yang et al., 2008) can still localize to LDs and drive LD–peroxisome contact formation (Fig. S3, D–G). Therefore, M1 Spastin’s N-terminal cytosolic region and MIT domain are dispensable for peroxisomal interaction.
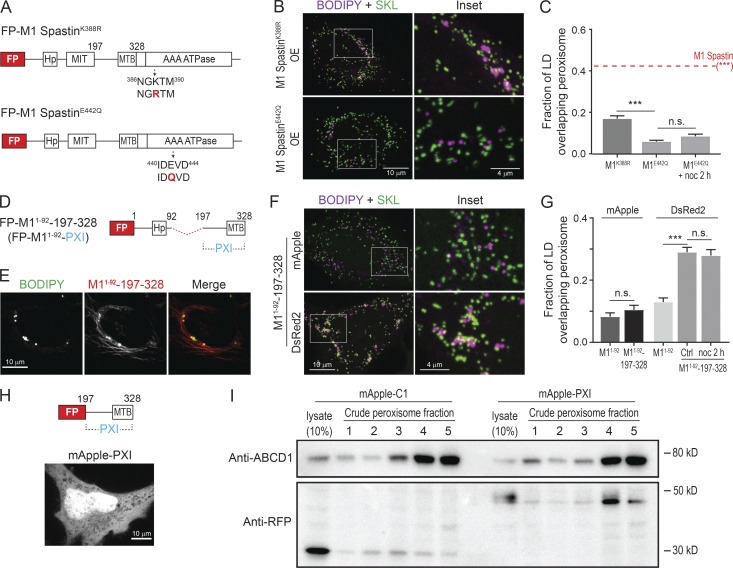
To test whether M1 Spastin’s ATPase domain is important for mediating LD–peroxisome contacts, we generated ATPase-deficient M1 Spastin mutants, M1 SpastinK388R and M1 SpastinE442Q (Fig. 4 A). M1 SpastinK388R and M1 SpastinE442Q both localized to LDs (Fig. S3 H). M1 SpastinE442Q additionally displayed a filamentous pattern. Notably, both mutations significantly abolished the ability of M1 Spastin to promote LD–peroxisome contact formation (Fig. 4, B and C), with greater reduction in the extent of LD–peroxisome contacts in M1 SpastinE442Q–expressing cells than in cells expressing M1 SpastinK388R (Fig. 4 C). As the defect in ATPase activity is more pronounced in M1 SpastinE442Q compared with M1 SpastinK388R (Evans et al., 2005), these results suggested that M1 Spastin’s ATPase activity is important for promoting LD–peroxisome contact formation.
Figure 4.
ATP hydrolysis is required for M1 Spastin to mediate LD–peroxisome contact formation via its PXI region. (A) Diagrams of FP-tagged M1 Spastin mutant constructs. Amino acid number and protein domains are indicated as Fig. 1 A. The mutated residues are labeled in red. (B) Localizations of LDs and peroxisomes in HeLa cells overexpressing mApple-M1 SpastinK388R (top) or mApple-M1 SpastinE422Q (bottom). Representative confocal MIP images are shown. (C) Fraction of LD overlapping peroxisome as described in B and in cells overexpressing mApple-M1 Spastin (red dashed line). mApple-M1 SpastinE422Q-expressing cells were further incubated with nocodazole (noc). Means ± SEM are shown (34–50 cells from at least four independent experiments). ***, P < 0.001; n.s., not significant. (D) Diagrams of a synthetic construct. The red dashed line indicates MIT domain deletion. (E) Colocalization of LDs and mApple-M11–92-197–328 in HeLa cells. Representative confocal images are shown. (F) Association between LDs and peroxisomes in HeLa cells overexpressing mApple- or DsRed2-M11–92-197–328. Representative confocal MIP images are shown. (G) Fraction of LD overlapping peroxisome in HeLa cells overexpressing mApple-M11–92, mApple-M11–92-197–328, DsRed2-M11–92, or DsRed2-M11–92-197–328. DsRed2-M11–92-197–328–expressing cells were further incubated with nocodazole. Means ± SEM are shown (20–37 cells from at least three independent experiments). ***, P < 0.001; n.s., not significant. (H) Diagram of FP-tagged PXI construct (top). mApple-PXI showed cytoplasmic distribution in HeLa cells (bottom). A representative confocal image is shown. (I) Peroxisome purification in HeLa cells transfected with mApple-C1 or mApple-PXI. Crude peroxisome fraction was subjected to density gradient ultracentrifugation to obtain enriched peroxisomes (fractions 4 and 5). Purity of peroxisome and protein levels of overexpressed constructs were assessed by Western blotting using antibodies against ABCD1 and RFP, respectively.
The best-characterized function of M1 Spastin’s ATPase domain is microtubule severing (White et al., 2007; Roll-Mecak and Vale, 2008). Thus, we applied nocodazole treatment to disrupt the microtubule network to see if this could rescue the defect in LD–peroxisome contact formation by M1 SpastinE442Q overexpression. Upon nocodazole treatment, no rescue was observed (Fig. 4 C), suggesting that microtubule severing in general is not critical for LD–peroxisome association induced by M1 Spastin. M1 Spastin’s ATPase domain, therefore, must be involved in some other aspect of LD–peroxisome interaction.
Identification of a PXI region on M1 Spastin
To investigate whether the cytosolic region between M1 Spastin’s MIT and ATPase domains plays a role in anchoring LDs to peroxisomes, we engineered a synthetic construct, M11–92-197–328, that contains amino acids 197–328 linked to M11–92 for LD targeting (Fig. 4 D). This synthetic protein successfully localized to LDs (Fig. 4 E). The additional filamentous pattern observed could represent an ER pool of the protein bound to microtubules via its microtubule-binding domain (White et al., 2007).
Overexpression of mApple-M11–92-197–328 did not promote LD–peroxisome contact formation, whereas overexpression of DsRed2-M11–92-197–328, which contains a dimeric red FP, led to significantly increased LD–peroxisome association (Fig. 4, F and G). Consistent with the results with M1 Spastin (Fig. 1 E), nocodazole treatment had no effect on LD–peroxisome contact formation mediated by DsRed2-M11–92-197–328 (Fig. 4 G). We further generated mApple-197–328 without an LD-targeting motif and found that it localized in the cytoplasm (Fig. 4 H). When we purified peroxisomes from HeLa cells overexpressing mApple-C1 or mApple-197–328, we observed that mApple-197–328, but not mApple-C1, copurified with peroxisomes (Fig. 4 I). These results suggested that amino acids 197–328 in M1 Spastin have affinity for peroxisomes, especially when the protein undergoes oligomerization. We have called this region PXI.
ABCD1 forms a tethering complex with M1 Spastin via the PXI region
Some ALD patients with mutations in ABCD1 manifest spasticity symptoms (Maris et al., 1995; Shaw-Smith et al., 2004; Zhan et al., 2013; Koutsis et al., 2015; Balicza et al., 2016), so we tested whether ABCD1 is required for LD–peroxisome contact formation by applying siRNA to knock down ABCD1 (Fig. S2 B). The number of peroxisomes was not affected in siABCD1-transfected cells (Fig. S4 A). Nonetheless, the extent of LD–peroxisome contacts induced by M1 Spastin overexpression was significantly reduced by ABCD1 knockdown (Fig. 5, A and B). Moreover, the enhanced LD–peroxisome contacts following cumyl-OOH or As3+ treatment were decreased in siABCD1-treated cells (Fig. S4 B). This raised the possibility that ABCD1 is required for tethering LDs to peroxisomes.
Figure 5.
ABCD1 forms a tethering complex with M1 Spastin via the PXI region. (A) Localizations of LDs and peroxisomes in mApple-M1 Spastin-overexpressing (OE) HeLa cells cotransfected with siCtrl or siABCD1. Representative confocal MIP images are shown. (B) Fraction of LD overlapping peroxisome as described in A. Means ± SEM are shown (39–44 cells from four independent experiments). ***, P < 0.001. (C) IP of ABCD1 in HeLa cells transfected with mApple-C1 (Ctrl), mApple-M1 Spastin (M1), or mApple-M1 SpastinE442Q (M1EQ). Protein levels of endogenous ABCD1 and overexpressed constructs were assessed by Western blotting using antibodies against ABCD1 and RFP, respectively. (D) Quantification of relative ABCD1 level in the IP as described in C. The value of M1 is set as 1. Means ± SEM from three independent IP experiments are shown. (E) IP of ABCD1 in HeLa cells transfected with mApple-C1 (Control) and mApple-M11–92-PXI. (F) Quantification of relative ABCD1 level in the IP as described in E. The value of mApple-M11–92-PXI is set as 1. Means ± SEM from two independent IP experiments are shown.
To test if ABCD1 forms a complex with M1 Spastin for LD–peroxisome tethering, we performed immunoprecipitation (IP) experiments. Endogenous ABCD1 coimmunoprecipitated with mApple-M1 Spastin (M1) but not with mApple FP (Fig. 5 C, lanes 4 and 5; and Fig. 5 D). M1 Spastin seemed to selectively associate with ABCD1, since we did not detect comparable levels of ACBD5, another peroxisomal membrane protein, in our IP experiments (Fig. S4, C and D). The ability of M1 Spastin to associate with ABCD1 was reduced when an E442Q mutation was introduced into Spastin (M1EQ; Fig. 5 C, lanes 5 and 6; and Fig. 5 D), suggesting that ATPase activity is important for forming the M1 Spastin–ABCD1 complex. We further found that ABCD1 coimmunoprecipitated with overexpressed mApple-M11–92-197–328, namely M11–92-PXI (Fig. 5, E and F). This indicated that the PXI region of M1 Spastin is sufficient to mediate complex formation with ABCD1. Altogether, these data reveal that ABCD1 forms a tethering complex with the PXI region of M1 Spastin.
Monitoring LD-to-peroxisome FA trafficking using 12-(7-nitrobenzofurazan-4-ylamino)dodecanoic acid (NBD-C12)
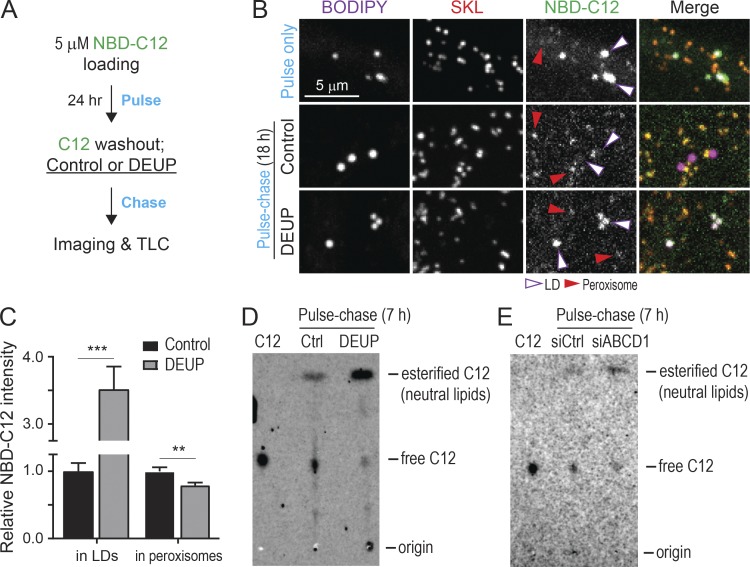
Since ABCD1 is an FA transporter on peroxisomes (Lodhi and Semenkovich, 2014) and appears to form a tethering complex with M1 Spastin at LD–peroxisome contacts, we wondered whether FA trafficking from LDs to peroxisomes could be regulated by M1 Spastin. To monitor LD-to-peroxisome FA trafficking in cells, we used the fluorescence FA analogue NBD-C12 in a pulse-chase assay in which cells were pulse-labeled by incubating with trace amounts of NBD-C12 for 24 h and then chased in label-free medium (Fig. 6 A). Visualization of cells immediately after the pulse labeling revealed that NBD-C12 had incorporated into BODIPY-labeled LDs (Fig. 6 B, top panels). Upon chasing for 18 h, a significant pool of NBD-C12 had now redistributed into peroxisomes (Fig. 6 B, middle panels), suggesting LD-to-peroxisome FA trafficking. Incubation with a pan-lipase inhibitor, diethylumbelliferyl phosphate (DEUP), during the chase abolished the redistribution of NBD-C12 to peroxisomes, coinciding with a substantial accumulation of NBD-C12 in LDs (Fig. 6 B, bottom panels; and Fig. 6 C). Consistent with this, TLC analysis also revealed an accumulation of esterified NBD-C12 (neutral lipids) and a reduction in free NBD-C12 in DEUP-treated cells (Fig. 6 D). Moreover, siABCD1-treated cells displayed decreased levels of free NBD-C12 compared with control siRNA (siCtrl)–treated cells (Fig. 6 E), suggesting that free NBD-C12 is transported to peroxisomes by ABCD1. Altogether, we concluded that release of NBD-C12 from neutral lipids via lipase activities is required for NBD-C12 redistribution from LDs to peroxisomes via ABCD1 transporters.
Figure 6.
Monitoring LD-to-peroxisome FA trafficking using NBD-C12. (A) Schematic diagram representing NBD-C12 pulse-chase assay using with DEUP treatment. (B) Distribution of NBD-C12 in BODIPY-665/676–labeled LDs and mCherry-SKL–labeled peroxisomes after pulse (top) and after pulse-chase in control (middle) or 150 µM DEUP-treated HeLa cells (bottom). Magenta and red arrowheads indicate LDs and peroxisomes, respectively. Representative confocal MIP images are shown. (C) Relative NBC-C12 intensity in LDs or peroxisomes as described in B. Means ± SEM are shown (26–27 cells from three independent experiments). **, P < 0.01; ***, P < 0.001. (D) TLC analysis of NBD-C12 extracts from pulse-chased HeLa cells as described in A. C12, 1 µl of 1 µM NBD-C12 loading for identifying free form of C12 from cell extracts. (E) TLC analysis of NBD-C12 extracts from siCtrl and siABCD1 transfected HeLa cells.
M1 Spastin controls FA trafficking from LDs to peroxisomes
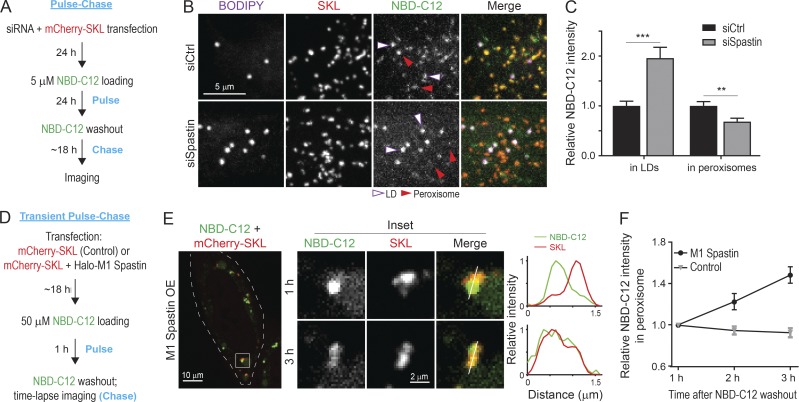
To test if endogenous Spastin is required for transfer of FAs from LDs to peroxisomes, we performed the NBD-C12 pulse-chase assay in Spastin knockdown cells (Fig. 7 A). NBD-C12 accumulated in LDs in siSpastin cells, coinciding with weaker peroxisomal NBD-C12 signal compared with siCtrl cells (Fig. 7, B and C). This suggested that endogenous Spastin is important for FA trafficking from LDs to peroxisomes.
Figure 7.
M1 Spastin controls LD-to-peroxisome FA trafficking. (A) Schematic diagram representing NBD-C12 pulse-chase assay in cells transfected with siRNAs and mCherry-SKL. (B) Distribution of NBD-C12 in BODIPY-665/676–labeled LDs and mCherry-SKL–labeled peroxisomes in HeLa cells transfected with siCtrl or siSpastin. Representative confocal MIP images are shown. (C) Relative NBC-C12 intensity in LDs or peroxisomes as described in B. Means ± SEM are shown (26–30 cells from three independent experiments). **, P < 0.01; ***, P < 0.001. (D) Schematic diagram representing transient NBD-C12 pulse-chase assay in cells transfected with mCherry-SKL alone or mCherry-SKL and Halo-M1 Spastin. (E) Redistribution of NBD-C12 to peroxisomes in Halo-M1 Spastin overexpressing HeLa cells following pulse-chase described in D. Line scans of relative intensity profile are shown (right panels). Representative confocal mages are shown. (F) Relative NBC-C12 intensity in peroxisomes as described in D and E. Means ± SEM are shown (12–13 cells from three independent experiments).
We next examined whether M1 Spastin overexpression could enhance LD-to-peroxisome FA trafficking. We modified the above pulse-chase assay by pulsing cells with NBD-C12 for 1 h before chasing to monitor dynamic FA distribution to peroxisomes (Fig. 7 D). NBD-C12 was successfully incorporated into LDs after pulse labeling, as its signal accumulated in round-shaped structures (Fig. 7 E, left panel). After 1-h chase, most NBD-C12 fluorescence remained in LDs, with minimal levels of NBD-C12 in nearby peroxisomes, as shown by line scan analysis (Fig. 7 E, middle and right panels). After 3-h chase, however, significant levels of NBD-C12 had accumulated in peroxisomes. Quantification showed a steady increase in NBD-C12 intensity in peroxisomes in M1 Spastin–overexpressing cells, while no increase was observed in control cells (Fig. 7 F). These data indicated that M1 Spastin overexpression enhances FA trafficking from LDs to peroxisomes.
M1 Spastin recruits ESCRT-III proteins IST1 and CHMP1B to LDs
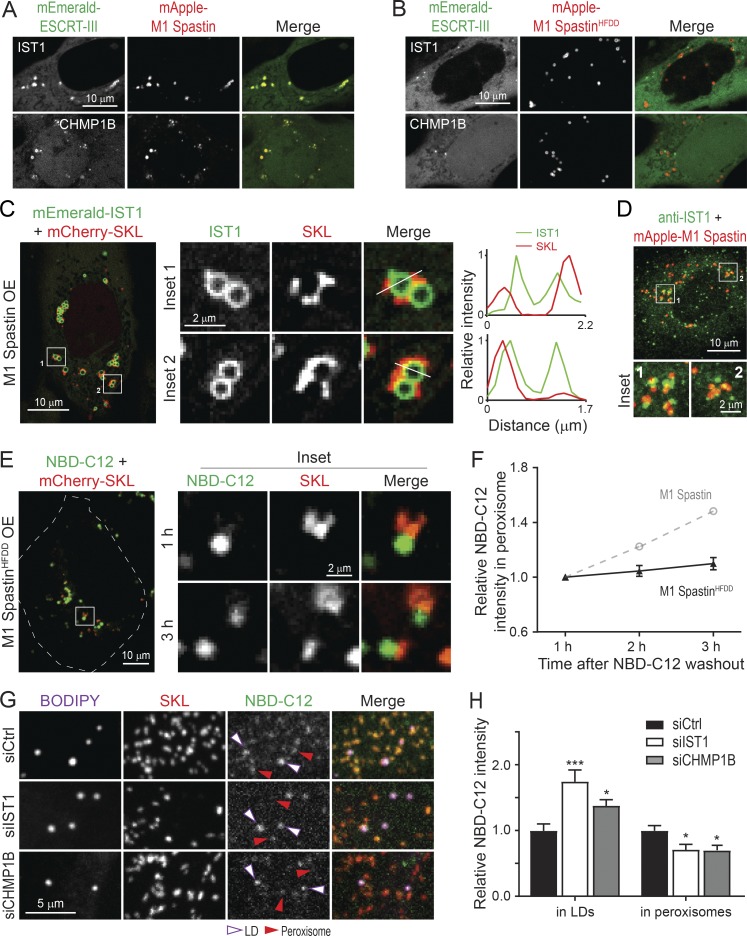
M1 Spastin’s MIT domain interacts with ESCRT-III proteins IST1 and CHMP1B (Reid et al., 2005; Agromayor et al., 2009; Renvoisé et al., 2010). We tested, therefore, whether M1 Spastin could recruit IST1 and CHMP1B onto LDs. We found that FP-tagged IST1 and CHMP1B labeled bright loci that colocalized with mApple-M1 Spastin (Fig. 8 A); these loci disappeared in cells coexpressing the M1 SpastinHFDD mutant unable to bind ESCRT-III (Fig. 8 B). Examining IST1 loci by Airyscan microscopy revealed that IST1 was restricted to the surface of round-shaped LDs and not to nearby peroxisomes (Fig. 8 C). We further observed a partial colocalization of mApple-M1 Spastin and endogenous IST1 by immunostaining (Fig. 8 D). These results indicated that IST1 and CHMP1B are recruited to LDs by M1 Spastin’s MIT domain.
Figure 8.
M1 Spastin recruits IST1 and CHMP1B to LDs to support LD-to-peroxisome FA trafficking. (A and B) Subcellular localization of mEmerald-IST1 (top panels) and mEmerald-CHMP1B (bottom panels) in HeLa cells overexpressing mApple-M1 Spastin (A) or mApple-M1 SpastinHFDD (B). Representative confocal images are shown. (C) mEmerald-IST1 is recruited onto LDs in HeLa cells coexpressing Halo-M1 Spastin, monitored by Airyscan microscopy. Representative images and relative intensity profile are shown. (D) Partial colocalization of immunostained IST1 and mApple-M1 Spastin in HeLa cells. Representative confocal MIP images are shown. (E) Distribution of NBD-C12 in Halo-M1 SpastinHFDD and mCherry-SKL overexpressing HeLa cells following pulse-chase described in Fig. 7 D. Representative confocal images are shown. (F) Relative NBC-C12 intensity in peroxisomes as described in E. Means ± SEM are shown (19 cells from three independent experiments). Gray dashed line is a replica from Fig. 7 F. (G) Distribution of NBD-C12 in BODIPY-665/676–labeled LDs and mCherry-labeled peroxisomes in siCtrl-, siIST1-, or siCHMP1B-treated HeLa cells following NBD-C12 pulse-chase described in Fig. 7 A. Representative confocal MIP images are shown. (H) Relative NBC-C12 intensity in LDs or peroxisomes as described in G. Means ± SEM are shown (25–31 cells from three independent experiments). *, P < 0.05; ***, P < 0.001 compared with siCtrl.
IST1 and CHMP1B are required for M1 Spastin–regulated FA trafficking
Membrane curvature contributes to lipid transfer between membranes (Lev, 2012). Since IST1 and CHMP1B are curvature generators (McCullough et al., 2015), we examined if recruitment of IST1 and CHMP1B to LDs by M1 Spastin is important for FA trafficking. To address this question, we used the 1-h FA pulse-chase assay described in Fig. 7 D to assess LD-to-peroxisome FA trafficking in cells expressing the M1 SpastinHFDD mutant unable to bind ESCRT-III. M1 SpastinHFDD overexpression did not affect NBD-C12 incorporation into LDs after 1 h of labeling (Fig. 8 E). However, only a small amount of NBD-C12 appeared in peroxisomes after a 3-h chase in these cells (Fig. 8, E and F), even though M1 SpastinHFDD enhances LD–peroxisome contact formation to the same extent as M1 Spastin (Fig. S3, E and F). As M1 SpastinHFDD lacks a functional MIT domain to recruit ESCRT-III to the surface of LDs, the data support the idea that ESCRT-III is involved in FA trafficking from LDs to peroxisomes. Consistently, knockdown of IST1 or CHMP1B also led to a reduction of peroxisomal NBD-C12 labeling compared with siCtrl cells in pulse-chase experiments described in Fig. 7 A (Fig. 8, G and H; and Fig. S2, C and D). Overall, these results suggested that M1 Spastin controls LD-to-peroxisome FA trafficking by recruiting IST1 and CHMP1B to LDs.
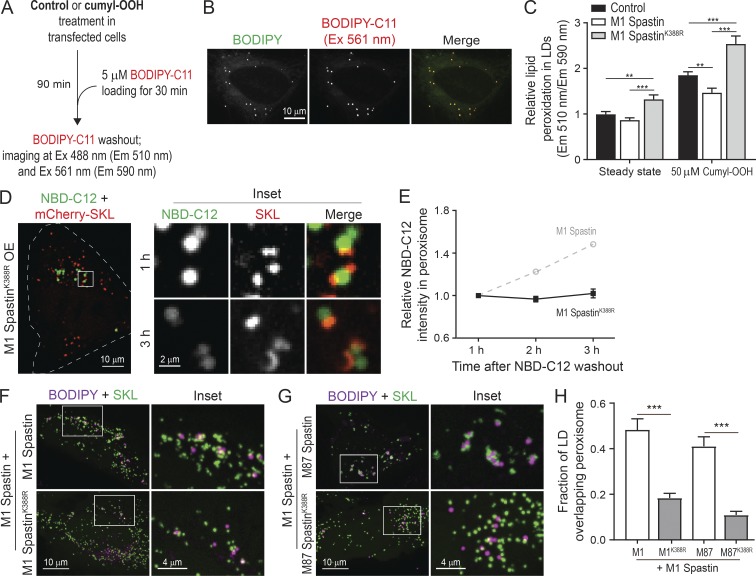
The pathogenic SpastinK388R mutant disrupts LD–peroxisome contact formation, leading to peroxidated lipid accumulation in LDs
As LD–peroxisome contacts are up-regulated following oxidative stress (Fig. 2, A and B), we wondered if FA trafficking at LD–peroxisome contacts may influence lipid peroxidation in LDs. We designed an imaging-based approach using LD-localized BODIPY-C11, a FA analogue that shifts its emission from 590 to 510 nm after cumyl-OOH-induced lipid peroxidation, as a sensor for measuring peroxidated lipids in LDs (Fig. 9, A and B). In control cells, we observed an increase in lipid peroxidation in LDs upon cumyl-OOH treatment (Fig. 9 C). By contrast, overexpression of M1 Spastin significantly reduced the levels of LD peroxidation caused by cumyl-OOH treatment (Fig. 9 C). Given M1 Spastin’s role in mediating LD-to-peroxisome FA trafficking, these findings suggested that the reduced levels of LD peroxidation by M1 Spastin overexpression were due to peroxidated FAs being transferred to peroxisomes.
Figure 9.
The pathogenic SpastinK388R mutant disrupts LD–peroxisome contact formation, leading to peroxidated lipid accumulation in LDs. (A) Schematic diagram representing lipid peroxidation assay using BODIPY-C11 to monitor LD peroxidation. (B) Colocalization of BODIPY-C11– and BODIPY-665/676–labeled LDs in HeLa cells. Representative confocal MIP images are shown. (C) Relative lipid peroxidation in LDs in control and in Halo-M1 Spastin– or Halo-M1 SpastinK388R–overexpressing HeLa cells in the absence or presence of cumyl-OOH. Means ± SEM are shown (32–56 cells from at least three independent experiments). **, P < 0.01; ***, P < 0.001. (D) Distribution of NBD-C12 in Halo-M1 SpastinK388R and mCherry-SKL–overexpressing HeLa cells following pulse-chase described in Fig. 7 D. Representative confocal images are shown. (E) Relative NBC-C12 intensity in peroxisomes as described in D. Means ± SEM are shown (15 cells from three independent experiments). Gray dashed line is a replica from Fig. 7 F. (F and G) Association between LDs and peroxisomes in HeLa cells coexpressing mApple-M1 Spastin and mTagBFP2-M1 Spastin or mTagBFP2-M1 SpastinK388R (F), or coexpressing mApple-M1 Spastin and mTagBFP2-M87 Spastin or mTagBFP2-M87 SpastinK388R (G). Representative confocal MIP images are shown. (H) Fraction of LD overlapping with peroxisome as described in F and G. Means ± SEM are shown (13–22 cells from two independent experiments). ***, P < 0.001.
We next examined how the HSP pathogenic mutant M1 SpastinK388R impacts LD-to-peroxisome FA trafficking and LD peroxidation. M1 SpastinK388R was incapable of promoting LD–peroxisome contact formation (Fig. 4, B and C), and no LD-to-peroxisome FA trafficking was observed in the transient NBD-C12 pulse-chase assay in cells overexpressing M1 SpastinK388R (Fig. 9, D and E). M1-SpastinK388R overexpression led to a significant increase in LD peroxidation at steady state (Fig. 9 C). This suggests that M1 SpastinK388R disrupts the endogenous LD-to-peroxisome FA trafficking pathway. Consistent with this, following cumyl-OOH treatment, M1 SpastinK388R–expressing cells showed a nearly twofold increase in LD peroxidation compared with M1 Spastin–expressing cells (Fig. 9 C).
The K388R mutation in Spastin is an autosomal dominant mutation for HSP (Blackstone, 2018), suggesting that SpastinK388R might exert a dominant-negative effect on M1 Spastin–mediated LD–peroxisome contact formation. Consistent with this possibility, we found a significant reduction in LD–peroxisome contacts in M1 Spastin–expressing cells cotransfected with M1 SpastinK388R or M87 SpastinK388R (Fig. 9, F–H). This dominant negative effect on LD–peroxisome tethering might explain the elevated LD peroxidation observed in M1 SpastinK388R–overexpressing cells.
Discussion
Frequent LD–peroxisome contact sites have been shown in professional FA-handling cells, such as adipocytes and hepatocytes, as well as other cell types including yeast and plant cells (Novikoff and Novikoff, 1973; Novikoff et al., 1973, 1980; Hayashi et al., 2001; Schrader, 2001; Binns et al., 2006; Thazar-Poulot et al., 2015). These studies demonstrated that LD–peroxisome contacts are evolutionarily conserved interorganelle structures and prompted speculation that directed transfer of FAs occurs at these contact sites. It has been a mystery, however, how peroxisomes and LDs coordinate with each other to facilitate this metabolic crosstalk.
We reasoned that some type of protein complex should tether LDs to peroxisomes and modulate their FA trafficking. Seeking such an LD–peroxisome tethering complex, we identified LD-associated M1 Spastin and peroxisomal FA transporter ABCD1 in conjunction with ESCRT-III proteins IST1 and CHMP1B to support FA trafficking between LDs and peroxisomes. Below, we discuss the basis for these findings, their significance for interorganelle FA trafficking, and their relevance to certain types of HSP and other diseases with defective peroxisome functions.
We found that M1 Spastin overexpression dramatically increased the extent of LD–peroxisome contacts within cells, while depletion of Spastin by siRNA significantly reduced LD–peroxisome contacts. Indeed, CLEM and FIB-SEM analyses showed that peroxisomes become tightly arrayed along the surface of the LDs upon M1 Spastin overexpression. By systematically exploring regions of M1 Spastin that promote LD–peroxisome tethering, we identified a PXI region of M1 Spastin for interacting with peroxisomes. Significantly, the PXI region in M1 Spastin forms a complex with ABCD1 in peroxisomes for LD–peroxisome tethering. We further discovered that M1 Spastin’s MIT domain recruits ESCRT-III IST1 and CHMP1B to LDs to facilitate LD-to-peroxisome FA trafficking. Despite these findings, a recent proximity labeling study of LDs in U-2 OS and Huh7 cells did not identify Spastin or ABCD1 (Bersuker et al., 2018). It is possible that endogenous Spastin or ABCD1 was expressed at too-low levels in these cells to be detected by proximity labeling or that other tethering complexes maintained LD–peroxisome contacts in these cells. Related to the latter possibility, other proteomic studies have identified a series of potential tethers between LDs and peroxisomes, including a HSP-related protein called Spartin and ABCD3 (Pu et al., 2011; Young et al., 2018).
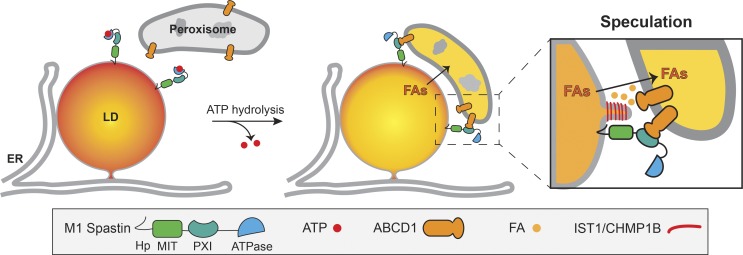
We propose the following model to conceptualize the functions elicited by M1 Spastin at LD–peroxisome contact sites (Fig. 10). Tethering of LDs to peroxisomes would be initiated when M1 Spastin’s PXI domain is made available to form a tethering complex with ABCD1 on peroxisome membranes. We speculate that ATP hydrolysis by M1 Spastin’s ATPase domain may trigger a conformational change that favors PXI region and ABCD1 complex formation. At LD–peroxisome contacts, FAs would be transferred from LDs to peroxisomes with the assistance of IST1 and CHMP1B. One crucial step for FA trafficking is FA release from LDs, which requires lipases to gain access to the lipids within LDs and is likely facilitated by increasing LD membrane curvature (Leff and Granneman, 2010). Given that IST1 and CHMP1B are capable of modifying lipid bilayers (McCullough et al., 2015), we speculate that IST1 and CHMP1B recruitment onto LDs by M1 Spastin could tubulate the lipid monolayer to facilitate FA trafficking (Fig. 10, inset). In yeast, high-curvature peroxisomal protrusions enriched in FA β-oxidation enzymes were found to extend into the lipid core of LDs at LD–peroxisome contacts (Binns et al., 2006). Therefore, altering membrane morphology at LD–peroxisome contacts seems to be a common mechanism to facilitate FA trafficking. Because ESCRT-III proteins require VPS4 ATPase for assembly/disassembly cycles, one can ask whether VPS4 or possibly the ATPase activity from M1 Spastin is important for the remodeling of IST1 and CHMP1B at LD–peroxisome contacts to regulate FA trafficking.
Figure 10.
Model of M1 Spastin in conjunction with ABCD1, IST1, and CHMP1B regulating LD–peroxisome contact formation and LD-to-peroxisome FA trafficking.
This model may be instructive for understanding the pathogenesis of HSP diseases. Many HSP-associated mutations affect proteins that impact FA allocation in LDs. For instance, HSP-associated proteins involved in LD biogenesis such as Atlastin-1, REEP1, and Seipin are responsible for FA incorporation in LDs (Klemm et al., 2013; Falk et al., 2014; Renvoisé et al., 2016). Furthermore, HSP-associated lipase DDHD2 and its related proteins, DDHD1 and PNPLA6, may contribute to the release of FAs from LDs (Rainier et al., 2008; Tesson et al., 2012; Inloes et al., 2014). As demonstrated in this work, M1 Spastin and ABCD1 appear to have an important role in FA trafficking from LDs into peroxisomes. Together, these data suggest that rather than impacting disparate pathways associated with HSP, the mutations giving rise to HSP are all affecting different steps of a single pathway involving FA incorporation into LDs, FA release from LDs, and delivery to peroxisomes. When this pathway is dysfunctional, as occurs in certain types of HSP as well as diseases associated with peroxisomal defects, LDs are unable to deliver FAs into peroxisomes, causing major downstream consequences for lipid detoxification, FA/lipid homeostasis, and energy production.
Materials and methods
Reagents
BODIPY 493/503, BODIPY 665/676, BODIPY 581/591-C11, cumyl-OOH, anti-CHMP1B antibody (PA5-44773), and Dulbecco’s PBS (DPBS) were purchased from Thermo Fisher Scientific. NBD-C12, anti-ABCD1 antibody (ab197013), anti-PMP70 antibody (ab109448), anti-catalase antibody (ab110292), and anti-RFP antibody (ab124754) were obtained from Abcam. Oleic acid-BSA complex, nocodazole, anti-Spastin antibody (ABN368), anti-ACBD5 antibody (HPA012145), and DEUP were purchased from Sigma-Aldrich. Anti-GAPDH antibody (2118) was purchased from Cell Signaling. Anti-IST1 antibody (GTX101972) was obtained from GeneTex. PFA and glutaraldehyde (GA) were obtained from Electron Microscopy Science. AP20187 (B/B homodimerizer) was purchased from Takara. The recombinant anti-M1 Spastin rabbit monoclonal antibody, clone RM346 (31-1232-00: RevMAb Biosciences USA), was generated using RevMAb Biosciences’ proprietary B cell cloning technology against M1-specific sequences.
Cell culture and transfection
HeLa cells (CCL-2) and U-2 OS (HTB-96) cells were purchased from ATCC. HeLa and U-2 OS cells were maintained in Eagle’s minimal essential medium (ATCC) and McCoy’s 5A medium (ATCC), respectively, with 10% FBS (Corning) and 1× penicillin/streptomycin solution (Corning). MRC-5 fibroblasts were kindly provided by Dr. Evan Reid and cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and 1× penicillin/streptomycin solution. For imaging experiments, DNA plasmids (15–50 ng) and siRNAs (∼20 nM) were transfected into HeLa cells with TransIT-LT1 reagent for 16–20 h and TransIT-TKO reagent for 64–68 h, respectively (Muris). For mEmerald-SKL or mCherry-SKL and siRNA cotransfection, HeLa cells were transfected simultaneously with 10 ng plasmid and 20 nM siRNA using both TransIT-LT1 and TransIT-TKO following the manufacturer’s instructions. For plasmids and siRNA transfection in Fig. 5, HeLa cells were first transfected with siRNA using TransIT-TKO for 48 h and subsequently transfected with 15 ng mEmerald-SKL and 50 ng mApple-M1 Spastin using TransIT-X2 for ∼16 h. U-2 OS and MRC-5 cells were transfected with 15–50 ng DNA plasmids using TransIT-X2.
Constructs and siRNAs
mEmerald-Sec61β, mEmerald-SKL, and mCherry-SKL were provided by the Molecular Biology Core at Janelia Research Campus. PA-GFP-SKL was generated by replacing the mEmerald portion of mEmerald-SKL with PA-GFP using AgeI and BsrGI restriction sites. FP-M11–92 plasmids were generated by inserting a PCR fragment containing amino acids 1–92 of human M1 Spastin (GenBank accession number NM_014946) into FP-C1 vectors using EcoRI and BamHI restriction sites. M11–92-mApple plasmid was generated by inserting M11–92 PCR fragment into mApple-N1 vector using BglII and HindIII restriction sites. 2xFKBP-mApple-M11–92 was generated by inserting 2xFKBP PCR fragment into mApple-M11–92 plasmid using NheI and AgeI sites. FP-M1 SpastinWT and FP-M87 Spastin plasmids were constructed the same way as FP-M11–92 with PCR fragments containing amino acids 1–616 and 87–616 of M1 Spastin, respectively. We then introduced an M-to-A mutation at the 87th amino acid in FP-M1 SpastinWT plasmids via site-direct mutagenesis to disrupt the second start codon, and we used these constructs (FP-M1 Spastin) for subsequent cloning and mutagenesis. FP-M1 SpastinΔ1–56 was generated the same way as FP-M11–92 with PCR fragments containing amino acids 57–616 of M1 Spastin. FP-M1 SpastinHFDD, FP-M1 SpastinK388R, and FP-M1 SpastinE442Q were generated using site-directed mutagenesis. For generating mApple-M11–92-197–328, PCR fragments containing M1 Spastin amino acids 197–328 was inserted into the C-terminus of FP-M11–92 linearized by BamHI enzyme using In-Fusion cloning kit (Takara). DsRed-M11–92-197–328 was constructed by replacing mApple in mApple-M11–92-197–328 with DsRed2 using NheI and EcoRI restriction sites. mApple-197–328 was generated by inserting PCR fragments containing amino acids 197–328 of M1 Spastin into mApple-C1 vector using EcoRI and BamHI restriction sites. mEmerald-GPAT4152–208 was constructed by inserting a PCR fragment containing amino acids 152–208 of human GPAT4 (NM_178819) into mEmerald-C1 vector using EcoRI and BamHI restriction sites. mEmerald-IST1 and mEmerald-CHMP1B were constructed by inserting PCR fragments of full-length IST1 (NM_001270975) and CHMP1B (NM_020412.4), respectively, into mEmerad-C1 vector using SalI and KpnI restriction sites. All oligonucleotides used in plasmid generation are listed in Table S1. Homemade siRNAs used in this study were generated by giardia dicer as previously described (Liou et al., 2005; Guiley et al., 2012). All oligonucleotides used in siRNA generation are listed in Table S1.
Fluorescence microscopy imaging
All cells were grown and transfected on Lab-Tek II chambered #1.5 coverglasses (Thermo Fisher Scientific) or MatTek dishes with #1.5 coverslip (MatTek Corporation). To label LDs, BODIPY 493/503 or BODIPY 665/676 was added to cells at 500 ng/ml 5 min or 16 h before imaging, respectively, and was present during imaging. To disrupt microtubule networks, cells were placed on ice for 15 min and subsequently incubated with 10 µM nocodazole at 37°C for 2–3 h. To induce lipid peroxidation, cells were treated with 50 µM cumyl-OOH or 500 µM As3+ for indicated periods. Treated cells were imaged in media without cumyl-OOH or As3+. Confocal microscopy was performed on a custom-built Nikon Eclipse Ti microscope with a Yokogawa CSU-X1 Spinning Disk Confocal unit using SR Apochromat total internal reflection fluorescence 100×/1.49 or Plan Apochromat λ 60×/1.40 objectives. Airyscan and SIM imaging were performed using Plan Apochromat 63×/1.4 objective on Zeiss LSM 880 with Airyscan and Zeiss ELYRA Superresolution Microscopy, respectively. Live-cell confocal and Airyscan experiments were conducted with cells incubated in phenol red-free medium at 37°C with 5% CO2 and humidified air. For SIM experiments, transfected cells were washed with DPBS once and fixed with 4% PFA and 0.1% GA for 20 min at room temperature. Fixed cells were then quenched with 100 mM glycine in DPBS followed by DPBS wash twice and subjected to SIM imaging at room temperature. Photoactivation was performed on the custom-built Nikon microscope equipped with Bruker photoactivation module. HeLa cells expressing PA-GFP-SKL or coexpressing PA-GFP-SKL and mApple-M1 Spastin were subjected to 405-nm laser pulse at maximal intensity in 3.14-µm2 circular areas containing LDs labeled with BODIPY-665/676 and imaged every 20 s for 10–20 min.
Immunostaining
All procedures were performed at room temperature unless otherwise indicated, and all washing steps were done with DPBS for 5 min. HeLa cells transfected with mApple-M1 Spastin were rinsed with DPBS and fixed with 4% PFA and 0.1% GA for 20 min. Fixed cells were quenched with 100 mM glycine in DPBS, washed twice, and incubated with 0.3% Triton X-100 in DPBS for 20 min for permeabilization. Permeabilized cells were then blocked with 5% normal donkey serum (Jackson ImmunoResearch Laboratories) in DPBS for 1 h followed by incubation with anti-catalase antibody (1:200 dilution), anti-PMP70 antibody (1:200 dilution), or anti-IST1 antibody (1:1,000 dilution) in DPBS with 1% BSA at 4°C overnight. After three washes, the cells were incubated with fluorescent secondary antibody (1:2,000 dilution) for 1 h. The stained samples were washed three times and imaged with confocal microscopy at room temperature.
Image analysis
All image analyses were performed using ImageJ (National Institutes of Health) unless otherwise indicated. All intensity analyses were subjected to background subtraction. To obtain relative intensity profiles, the intensity values from different conditions were normalized to that at the first time point or in control groups. Colocalization of LDs and peroxisomes was quantified by measuring the Mander’s overlap coefficient of confocal maximum-intensity projection (MIP) images using the JACoP plugin. For analysis of relative duration of LD contacting peroxisome, LDs making contact with peroxisomes at least once during 10-min imaging were included. The frames in which LD contacted peroxisomes were then divided by total imaging frames (31 frames in 10 min) to obtain the relative duration. For intensity analysis of 2xFKBP-mApple-M11–92, regions of interest (ROIs) in LDs and the ER were generated using oval selection and polygon selection, respectively. Mean gray values of 2xFKBP-mApple-M11–92 intensity in LDs were obtained and then averaged. For NBD-C12 intensity analyses in pulse-chase experiments, ROIs of LDs and peroxisomes were generated using their respective confocal MIP images. These ROIs were then applied to NBD-C12 confocal MIP images to obtain the mean gray values of NBD-C12 in LDs and peroxisomes. Similar analyses were applied to obtain NBD-C12 intensity in peroxisomes in transient pulse-chase assay with control cells or with M1 SpastinK388R–overexpressing cells. Due to the close association of LDs and peroxisomes in M1 Spastin or M1 SpastinHFDD–overexpressing cells, ROIs generated from peroxisome mask showed a substantial overlap with LDs. Thus, we manually selected peroxisome ROIs without overlapping LDs to analyze NBD-C12 intensity in transient pulse-chase assay with cells overexpressing M1 Spastin or M1 SpastinHFDD. For analyses of lipid peroxidation in LDs, ROIs of LDs were generated using oval selection. Mean gray values of ROIs from both green (emission 510 nm) and red (emission 590 nm) channels were extracted. Relative lipid peroxidation was derived from the ratio of emission 510 to 590 nm.
CLEM
HeLa cells grown on 25-mm photoetched coverslips (Electron Microscopy Sciences) were transfected with 250 ng mApple-M1 Spastin and 100 ng mEmerald-SKL plasmids for ∼16 h in the presence of 15 µM oleic acid. Cells were then washed with DPBS and fixed with 4% PFA, 0.1% GA, and 2 mM CaCl2 in 0.08 M sodium cacodylate buffer. Fixative was present during light microscopy image acquisition. M1 Spastin and peroxisome fluorescence images of cells of interest and their corresponding etched number on the coverslips were acquired by Zeiss LSM 880 with Airyscan. Subsequently, the fixative for light microscopy was removed, and fresh 2% GA and 2 mM CaCl2 in 0.08 M sodium cacodylate buffer, pH 7.2, was added. Cells were kept in fixative at 4°C for 16 h, postfixed in 2% osmium tetroxide/1.25% potassium ferrocyanide in 0.1 M cacodylate buffer for 30 min followed by 2% osmium in the same buffer for another 30 min, and processed for epon embedding. Cells imaged by Airyscan were localized on the grid (imprinted in the epon block). Ultrathin sections (60 nm) from the imaged cells were cut and stained with uranyl acetate/lead citrate and imaged in a Tecnai 12 electron microscope (FEI) operating at 80 kV equipped with an Ultrascan 4000 digital camera (Gatan).
FIB-SEM
Sample preparation
HeLa cells grown on sapphire coverslips (3-mm diameter, 0.05-mm thickness; Nanjing Co-Energy Optical Crystal Co.) were transfected with mApple-M1 Spastin plasmid for ∼16 h in the presence of 15 µM oleic acid. Wide-field fluorescence and differential interference contrast images were collected before high-pressure freezing for subsequent identification of cells of interest. The sapphire coverslips were clamped between two aluminum planchettes (Technotrade International) using 25% dextran as a filler. High-pressure freezing was performed in a Wohlwend Compact 01 high-pressure freezer (Wohlwend), followed by freeze-substitution (FS). Coverslips were released from the planchettes and transferred to cryotubes containing FS medium (2% OsO4, 40 mM imidazole, 40 mM 1,2,4-triazole, 0.1% uranyl acetate, and 4% water in acetone) under liquid N2. FS was performed using an automated FS machine (AFS2, Leica Microsystems) with FS schedule adapted from Buser and Walther (2008) as follows: −140°C to −90°C for 2 h, −90°C for 24 h, −90°C to 0°C for 12 h, 0 to 22°C for 1 h, and 22°C for 90 min. Resin embedding was performed immediately after FS. Samples were removed from the FS machine, washed three times in anhydrous acetone for a total of 10 min, and embedded in Eponate 12 as follows: acetone/Eponate 12 (2:1) for 1 h, acetone/Eponate 12 (1:1) for 1 h, acetone/Eponate 12 (1:2) for 1 h, Eponate 12 for 2 h, Eponate 12 2 h to overnight, and Eponate 12 for 2 h. Coverslips with cells side up were then placed in the slots of a flat embedding silicone mold filled with Eponate 12 and incubated at 60°C for 2 d to allow Eponate 12 polymerization. The epoxy was removed using a razor blade to expose the sapphire coverslip surface not containing cells. The coverslip was separated by sequential dipping into liquid N2 and hot water. The exposed surface was immediately reembedded in Durcupan, which helps reduce the streaks during FIB-SEM imaging (Xu et al., 2017). X-rays of the entire reembedded block were taken using XRadia 510 Versa micro X-ray system (Zeiss). X-ray images were then overlaid with fluorescence and differential interference contrast images to identify cells of interest. The sample block was then remounted using Durcupan on a copper stud (Xu et al., 2017) and trimmed using an ultramicrotome (EM UC7; Leica Microsystems). Finally, the sample was sputter coated with 10 nm of Au and 100 nm of carbon in a sputter-coating system (PECS 682; Gatan).
FIB-SEM imaging
FIB-SEM imaging was performed using a customized Zeiss Merlin crossbeam system previously described (Xu et al., 2017). The Zeiss Capella FIB column was repositioned at 90° to the SEM column. The block face was imaged by a 2-nA electron beam with 1.2-keV landing energy at 500 kHz. The x-y pixel resolution was set at 8 nm. A subsequently applied focused Ga+ beam of 15 nA at 30 keV strafed across the top surface and ablated away 4 nm of the surface. The newly exposed surface was then imaged again. The ablation–imaging cycle continued about once every 30 s for 1 wk. The sequence of acquired images formed a raw imaged volume, followed by postprocessing of image registration and alignment using a Scale Invariant Feature Transform–based algorithm. The aligned stack was binned by a factor of 2 along z to form a final isotropic volume of 100 × 7 × 80 µm3 with 8 × 8 × 8-nm3 voxels, which can be viewed in arbitrary orientations. Organelle segmentation and 3D rendering were manually annotated using Amira software (Thermo Fisher Scientific).
IP and Western blotting
HeLa cells were cultured on 6-cm dishes and transfected with 1 µg of plasmid DNA for ∼18 h in the presence of 15 µM oleic acid. Cells were then washed with DPBS before lysis with IP lysis buffer (Thermo Fisher Scientific) supplemented with 1× Halt protease inhibitor cocktail (Thermo Fisher Scientific) on ice for 30 min. The lysates were subjected to centrifugation at 18,000 g for 15 min at 4°C, and the clear lysates (supernatants) were collected. The clear lysates were mixed with RFP-nAb agarose resins (Allele Biotechnology) and incubated with tumbling at 4°C for 4 h. The immunoprecipitated proteins were eluted with NuPAGE LDS sample buffer (Thermo Fisher Scientific) after washing the RFP-nAb agarose resins twice with IP lysis buffer and twice with 10 mM Tris buffer containing 500 mM NaCl. To determine endogenous protein expression levels following siRNA treatment, cells grown in 12-well plates were transfected with siRNAs (∼20 nM) for ∼70 h, and the cell lysates were collected as described above. The eluted proteins of IP experiments and lysates for siRNA experiments were analyzed by Western blotting. HRP-conjugated secondary antibodies were used to visualize protein bands, and chemiluminescence was detected by ChemiDoc Imaging System (Bio-Rad). Quantification of IP was done by densitometry analysis using ImageJ.
Peroxisome purification
Peroxisomes were purified using peroxisome isolation kit (Sigma-Aldrich) following the manufacturer’s protocol. In brief, HeLa cells in 15-cm dishes with ∼70% confluence were transfected with mApple-C1 (5 µg/dish; four dishes total) or mApple-PXI (5 µg/dish; four dishes total) for ∼18 h. Transfected cells were homogenized in peroxisome extraction buffer and subsequently subjected to differential centrifugation to obtain the crude peroxisome fraction. To enrich peroxisomes, the crude peroxisome fraction was further subjected to density gradient ultracentrifugation. Top layers, which mainly consist of the ER, lysosomes, and mitochondria, were carefully removed after ultracentrifugation. The rest of the sample was equally divided into fractions 1–5 (top to bottom). Samples were then boiled in NuPAGE LDS sample buffer for 10 min. Equal volumes of fractions 1–5 and 10% volume of lysate (supernatant after 1,000 g) were analyzed by Western blotting. The purities of peroxisome and the overexpressed protein in different fractions were assessed by ABCD1 antibody and RFP antibody, respectively.
TLC
NBD-C12 pulse-chased HeLa cells were harvested by trypsinization. Lipids were extracted by vortexing cell pellets in chloroform for 1 min (1.5 × 106 cells per 100 µl chloroform). The samples were then centrifuged at 18,000 g for 10 min at 4°C, and the lipids in the lower (organic) phase were obtained. 25 µl of the extracted lipids and 1 µl of 1 µM NBD-C12 solution were spotted onto precoated TLC aluminum plates (Silica gel 60 F254; Millipore) and separated in a solvent system of hexanes/ethyl acetate (1:2; Rambold et al., 2015). NBD-C12 fluorescence on TLC plates were visualized by ChemiDoc Imaging System.
Statistical analyses
Data were statistically analyzed by two-tailed t test or one-way ANOVA with Tukey’s multiple comparisons using Prism (GraphPad Software). Data distribution was assumed to be normal but was not formally tested. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Online supplemental material
Fig. S1 shows that M1 Spastin, but not M87 Spastin, overexpression enhances LD–peroxisome contact formation in various cell lines. Fig. S2 shows reduced protein expression levels following siRNA treatment. Fig. S3 shows the localization of synthetic proteins and M1 Spastin mutants in LD and/or peroxisomes. Fig. S4 shows the effect of ABCD1 knockdown in LD–peroxisome contacts and minimal interaction between ACBD5 and M1 Spastin. Table S1 shows oligonucleotides used in this study. Video 1 shows the FIB-SIM slices and organelle segmentation containing LD–peroxisome contacts. Videos 2 and 3 are time-lapse videos demonstrating dynamic association between LDs and peroxisomes. Video 4 is a time-lapse video showing colocalization of GPAT4158–208 and M11–92.
Supplementary Material
Acknowledgments
We thank Dr. Luke Lavis at the Janelia Research Campus for providing JF Halo ligands; Dr. Claire Deo and Dr. Ya-Cheng Liao at Janelia Research Campus for technical assistance; Dr. Evan Reid at Cambridge Institute for Medical Research, Cambridge, UK, for kindly providing MRC-5 cells; Rick Webb at University of Queensland, Queensland, Australia, for advice on FS; and Victoria Custard for administrative assistance.
This work was supported by Howard Hughes Medical Institute, Janelia Research Campus, and the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: C-L. Chang, C. Blackstone, and J. Lippincott-Schwartz conceived and designed the study. C-L. Chang, M.S. Ioannou, H.A. Pasolli, C.S. Xu, D.R. Peale, G. Shtengel, and M. Freeman performed the experiments. C-L. Chang, A.V. Weigel, and M.S. Ioannou analyzed the data. A.V. Weigel analyzed and segmented the FIB-SEM images. H.F. Hess supervised the FIB-SEM experiment. C-L. Chang and J. Lippincott-Schwartz wrote the manuscript with the input from all coauthors. J. Lippincott-Schwartz supervised the project.
References
- Agromayor M., Carlton J.G., Phelan J.P., Matthews D.R., Carlin L.M., Ameer-Beg S., Bowers K., and Martin-Serrano J.. 2009. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell. 20:1374–1387. 10.1091/mbc.e08-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes M., Gressens P., Baumgart E., Carmeliet P., Casteels M., Fransen M., Evrard P., Fahimi D., Declercq P.E., Collen D., et al. . 1997. A mouse model for Zellweger syndrome. Nat. Genet. 17:49–57. 10.1038/ng0997-49 [DOI] [PubMed] [Google Scholar]
- Balicza P., Grosz Z., Gonzalez M.A., Bencsik R., Pentelenyi K., Gal A., Varga E., Klivenyi P., Koller J., Züchner S., and Molnar J.M.. 2016. Genetic background of the hereditary spastic paraplegia phenotypes in Hungary - An analysis of 58 probands. J. Neurol. Sci. 364:116–121. 10.1016/j.jns.2016.03.018 [DOI] [PubMed] [Google Scholar]
- Bersuker K., Peterson C.W.H., To M., Sahl S.J., Savikhin V., Grossman E.A., Nomura D.K., and Olzmann J.A.. 2018. A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev. Cell. 44:97–112.e117. 10.1016/j.devcel.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D., Januszewski T., Chen Y., Hill J., Markin V.S., Zhao Y., Gilpin C., Chapman K.D., Anderson R.G., and Goodman J.M.. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173:719–731. 10.1083/jcb.200511125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C. 2018. Converging cellular themes for the hereditary spastic paraplegias. Curr. Opin. Neurobiol. 51:139–146. 10.1016/j.conb.2018.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett S.H., Kershen E.J., Zhang J., Zeng L., Straley S.C., Kaplan A.M., and Cohen D.A.. 2004. Conditional macrophage ablation in transgenic mice expressing a Fas-based suicide gene. J. Leukoc. Biol. 75:612–623. 10.1189/jlb.0903442 [DOI] [PubMed] [Google Scholar]
- Buser C., and Walther P.. 2008. Freeze-substitution: the addition of water to polar solvents enhances the retention of structure and acts at temperatures around -60 degrees C. J. Microsc. 230:268–277. 10.1111/j.1365-2818.2008.01984.x [DOI] [PubMed] [Google Scholar]
- Claudiani P., Riano E., Errico A., Andolfi G., and Rugarli E.I.. 2005. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp. Cell Res. 309:358–369. 10.1016/j.yexcr.2005.06.009 [DOI] [PubMed] [Google Scholar]
- Connell J.W., Lindon C., Luzio J.P., and Reid E.. 2009. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 10:42–56. 10.1111/j.1600-0854.2008.00847.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen M., Kemp S., de Visser M., van Geel B.M., Wanders R.J., Aubourg P., and Poll-The B.T.. 2012. X-linked adrenoleukodystrophy (X-ALD): clinical presentation and guidelines for diagnosis, follow-up and management. Orphanet J. Rare Dis. 7:51 10.1186/1750-1172-7-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K.J., Gomes E.R., Reisenweber S.M., Gundersen G.G., and Lauring B.P.. 2005. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 168:599–606. 10.1083/jcb.200409058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J., Rohde M., Bekhite M.M., Neugebauer S., Hemmerich P., Kiehntopf M., Deufel T., Hübner C.A., and Beetz C.. 2014. Functional mutation analysis provides evidence for a role of REEP1 in lipid droplet biology. Hum. Mutat. 35:497–504. 10.1002/humu.22521 [DOI] [PubMed] [Google Scholar]
- Finn P.F., and Dice J.F.. 2006. Proteolytic and lipolytic responses to starvation. Nutrition. 22:830–844. 10.1016/j.nut.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Guiley K.Z., Pratt A.J., and MacRae I.J.. 2012. Single-pot enzymatic synthesis of Dicer-substrate siRNAs. Nucleic Acids Res. 40:e40 10.1093/nar/gkr1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi H.F., and Goodman J.M.. 2015. The life cycle of lipid droplets. Curr. Opin. Cell Biol. 33:119–124. 10.1016/j.ceb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M., Hayashi H., Hara-Nishimura I., and Nishimura M.. 2001. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma. 218:83–94. 10.1007/BF01288364 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Reese M.L., and Goodman J.M.. 2018. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 37:e98947 10.15252/embj.201898947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inloes J.M., Hsu K.L., Dix M.M., Viader A., Masuda K., Takei T., Wood M.R., and Cravatt B.F.. 2014. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl. Acad. Sci. USA. 111:14924–14929. 10.1073/pnas.1413706111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islinger M., Voelkl A., Fahimi H.D., and Schrader M.. 2018. The peroxisome: an update on mysteries 2.0. Histochem. Cell Biol. 150:443–471. 10.1007/s00418-018-1722-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm R.W., Norton J.P., Cole R.A., Li C.S., Park S.H., Crane M.M., Li L., Jin D., Boye-Doe A., Liu T.Y., et al. . 2013. A conserved role for atlastin GTPases in regulating lipid droplet size. Cell Reports. 3:1465–1475. 10.1016/j.celrep.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N., Farese R.V. Jr., and Walther T.C.. 2016. Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol. 26:535–546. 10.1016/j.tcb.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsis G., Lynch D.S., Tucci A., Houlden H., Karadima G., and Panas M.. 2015. A novel ABCD1 mutation detected by next generation sequencing in presumed hereditary spastic paraplegia: A 30-year diagnostic delay caused by misleading biochemical findings. J. Neurol. Sci. 355:199–201. 10.1016/j.jns.2015.05.031 [DOI] [PubMed] [Google Scholar]
- Leff T., and Granneman J.G.. 2010. Molecular Mechanisms of Adipocyte Lipolysis. In Adipose Tissue in Health and Disease. Wiley-VCH Verlag, Weinheim, Germany. 83–96. [Google Scholar]
- Lev S. 2012. Nonvesicular lipid transfer from the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4:a013300 10.1101/cshperspect.a013300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E. Jr., and Meyer T.. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15:1235–1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi I.J., and Semenkovich C.F.. 2014. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19:380–392. 10.1016/j.cmet.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris T., Androulidakis E.J., Tzagournissakis M., Papavassiliou S., Moser H., and Plaitakis A.. 1995. X-linked adrenoleukodystrophy presenting as neurologically pure familial spastic paraparesis. Neurology. 45:1101–1104. 10.1212/WNL.45.6.1101 [DOI] [PubMed] [Google Scholar]
- McCullough J., Clippinger A.K., Talledge N., Skowyra M.L., Saunders M.G., Naismith T.V., Colf L.A., Afonine P., Arthur C., Sundquist W.I., et al. . 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 350:1548–1551. 10.1126/science.aad8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.B., Louie S.M., Daniele J.R., Tran Q., Dillin A., Zoncu R., Nomura D.K., and Olzmann J.A.. 2017. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell. 42:9–21. e5. 10.1016/j.devcel.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff A.B., and Novikoff P.M.. 1973. Microperoxisomes. J. Histochem. Cytochem. 21:963–966. 10.1177/21.11.963 [DOI] [PubMed] [Google Scholar]
- Novikoff A.B., Novikoff P.M., Rosen O.M., and Rubin C.S.. 1980. Organelle relationships in cultured 3T3-L1 preadipocytes. J. Cell Biol. 87:180–196. 10.1083/jcb.87.1.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff P.M., Novikoff A.B., Quintana N., and Davis C.. 1973. Studies on microperoxisomes. 3. Observations on human and rat hepatocytes. J. Histochem. Cytochem. 21:540–558. 10.1177/21.6.540 [DOI] [PubMed] [Google Scholar]
- Pantakani D.V., Swapna L.S., Srinivasan N., and Mannan A.U.. 2008. Spastin oligomerizes into a hexamer and the mutant spastin (E442Q) redistribute the wild-type spastin into filamentous microtubule. J. Neurochem. 106:613–624. 10.1111/j.1471-4159.2008.05414.x [DOI] [PubMed] [Google Scholar]
- Papadopoulos C., Orso G., Mancuso G., Herholz M., Gumeni S., Tadepalle N., Jüngst C., Tzschichholz A., Schauss A., Höning S., et al. . 2015. Spastin binds to lipid droplets and affects lipid metabolism. PLoS Genet. 11:e1005149 10.1371/journal.pgen.1005149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Zhu P.P., Parker R.L., and Blackstone C.. 2010. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 120:1097–1110. 10.1172/JCI40979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Antonenkov V.D., Glumoff T., and Hiltunen J.K.. 2006. Peroxisomal beta-oxidation--a metabolic pathway with multiple functions. Biochim. Biophys. Acta. 1763:1413–1426. 10.1016/j.bbamcr.2006.08.034 [DOI] [PubMed] [Google Scholar]
- Pol A., Gross S.P., and Parton R.G.. 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204:635–646. 10.1083/jcb.201311051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Ha C.W., Zhang S., Jung J.P., Huh W.K., and Liu P.. 2011. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2:487–496. 10.1007/s13238-011-1061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier S., Bui M., Mark E., Thomas D., Tokarz D., Ming L., Delaney C., Richardson R.J., Albers J.W., Matsunami N., et al. . 2008. Neuropathy target esterase gene mutations cause motor neuron disease. Am. J. Hum. Genet. 82:780–785. 10.1016/j.ajhg.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold A.S., Cohen S., and Lippincott-Schwartz J.. 2015. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell. 32:678–692. 10.1016/j.devcel.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos O., Carrizales L., Yáñez L., Mejía J., Batres L., Ortíz D., and Díaz-Barriga F.. 1995. Arsenic increased lipid peroxidation in rat tissues by a mechanism independent of glutathione levels. Environ. Health Perspect. 103:85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E., Connell J., Edwards T.L., Duley S., Brown S.E., and Sanderson C.M.. 2005. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 14:19–38. 10.1093/hmg/ddi003 [DOI] [PubMed] [Google Scholar]
- Renvoisé B., Parker R.L., Yang D., Bakowska J.C., Hurley J.H., and Blackstone C.. 2010. SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol. Biol. Cell. 21:3293–3303. 10.1091/mbc.e09-10-0879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvoisé B., Malone B., Falgairolle M., Munasinghe J., Stadler J., Sibilla C., Park S.H., and Blackstone C.. 2016. Reep1 null mice reveal a converging role for hereditary spastic paraplegia proteins in lipid droplet regulation. Hum. Mol. Genet. 25:5111–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., and Vale R.D.. 2008. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 451:363–367. 10.1038/nature06482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg H.H., Richardson E.P., Johnson P.C., Cohen R.B., Powers J.M., and Raine C.S.. 1972. Schilder’s disease. Sex-linked recessive transmission with specific adrenal changes. Arch. Neurol. 27:458–460. 10.1001/archneur.1972.00490170090014 [DOI] [PubMed] [Google Scholar]
- Schrader M. 2001. Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J. Histochem. Cytochem. 49:1421–1429. 10.1177/002215540104901110 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., and Bohnert M.. 2017. A different kind of love - lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862:1188–1196. 10.1016/j.bbalip.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C.J., Lewis S.J., and Reid E.. 2004. X-linked adrenoleukodystrophy presenting as autosomal dominant pure hereditary spastic paraparesis. J. Neurol. Neurosurg. Psychiatry. 75:686–688. 10.1136/jnnp.2003.022970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson C., Nawara M., Salih M.A., Rossignol R., Zaki M.S., Al Balwi M., Schule R., Mignot C., Obre E., Bouhouche A., et al. . 2012. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am. J. Hum. Genet. 91:1051–1064. 10.1016/j.ajhg.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazar-Poulot N., Miquel M., Fobis-Loisy I., and Gaude T.. 2015. Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. USA. 112:4158–4163. 10.1073/pnas.1403322112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger R.H., Clark G.O., Scherer P.E., and Orci L.. 2010. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim. Biophys. Acta. 1801:209–214. 10.1016/j.bbalip.2009.10.006 [DOI] [PubMed] [Google Scholar]
- van der Kraaij A.M., de Jonge H.R., Esterbauer H., de Vente J., Steinbusch H.W., and Koster J.F.. 1990. Cumene hydroperoxide, an agent inducing lipid peroxidation, and 4-hydroxy-2,3-nonenal, a peroxidation product, cause coronary vasodilatation in perfused rat hearts by a cyclic nucleotide independent mechanism. Cardiovasc. Res. 24:144–150. 10.1093/cvr/24.2.144 [DOI] [PubMed] [Google Scholar]
- Wali G., Sutharsan R., Fan Y., Stewart R., Tello Velasquez J., Sue C.M., Crane D.I., and Mackay-Sim A.. 2016. Mechanism of impaired microtubule-dependent peroxisome trafficking and oxidative stress in SPAST-mutated cells from patients with Hereditary Spastic Paraplegia. Sci. Rep. 6:27004 10.1038/srep27004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Chung J., and Farese R.V. Jr. 2017. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R.J. 2013. Peroxisomes in human health and disease: metabolic pathways, metabolite transport, interplay with other organelles and signal transduction. Subcell. Biochem. 69:23–44. 10.1007/978-94-007-6889-5_2 [DOI] [PubMed] [Google Scholar]
- Welte M.A. 2015. Expanding roles for lipid droplets. Curr. Biol. 25:R470–R481. 10.1016/j.cub.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.R., Evans K.J., Lary J., Cole J.L., and Lauring B.. 2007. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol. 176:995–1005. 10.1083/jcb.200610072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. . 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.H., Gatta A.T., and Levine T.P.. 2019. Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 20:85–101. 10.1038/s41580-018-0071-5 [DOI] [PubMed] [Google Scholar]
- Xu C.S., Hayworth K.J., Lu Z., Grob P., Hassan A.M., García-Cerdán J.G., Niyogi K.K., Nogales E., Weinberg R.J., and Hess H.F.. 2017. Enhanced FIB-SEM systems for large-volume 3D imaging. eLife. 6:e25916 10.7554/eLife.25916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C., and Hurley J.H.. 2008. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15:1278–1286. 10.1038/nsmb.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P.A., Senkal C.E., Suchanek A.L., Grevengoed T.J., Lin D.D., Zhao L., Crunk A.E., Klett E.L., Füllekrug J., Obeid L.M., and Coleman R.A.. 2018. Long-chain acyl-CoA synthetase 1 interacts with key proteins that activate and direct fatty acids into niche hepatic pathways. J. Biol. Chem. 293:16724–16740. 10.1074/jbc.RA118.004049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Z.X., Liao X.X., Du J., Luo Y.Y., Hu Z.T., Wang J.L., Yan X.X., Zhang J.G., Dai M.Z., Zhang P., et al. . 2013. Exome sequencing released a case of X-linked adrenoleukodystrophy mimicking recessive hereditary spastic paraplegia. Eur. J. Med. Genet. 56:375–378. 10.1016/j.ejmg.2013.04.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.