Abstract
It is well established that activation of the transcription factor signal transducer and activator of transcription 1 (STAT1) is required for the interferon-γ (IFN-γ)–mediated antiviral response. Here, we found that IFN-γ receptor stimulation also activated Unc-51-like kinase 1 (ULK1), an initiator of Beclin-1-mediated autophagy. Furthermore, the interaction between ULK1 and the mitogen-activated protein kinase kinase kinase MLK3 (mixed lineage kinase 3) was necessary for MLK3 phosphorylation and downstream activation of the kinase ERK5. This autophagy-independent activity of ULK1 promoted the transcription of key antiviral IFN-stimulated genes (ISGs) and was essential for IFN-γ–dependent antiviral effects. These findings define a previously unknown IFN-γ pathway that appears to be a key element of the antiviral response.
Introduction
Interferons (IFNs) are secreted cytokines that act as the first line of defense against viral infections (1). IFN-inducible activation of signal transducer and activator of transcription 1 (STAT1) and STAT2 is important for an antiviral response associated with type I and type III IFNs (2). By contrast, there is little information on the signaling effectors that mediate the antiviral effects of the type II IFN, IFNγ(3). IFNγ initiates its biological activities by binding to its cognate heterodimeric cell surface receptor, composed of two ligand binding IFN-gamma receptor 1 (IFNGR1) subunit chains and two signal-transducing IFNGR2 chains (4). This interaction triggers activation of several downstream signaling events, including the canonical Janus kinase (JAK)-STAT1 pathway, and non-STAT pathways, such as mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), mechanistic target of rapamycin complex 2 (mTORC2), calcium/calmodulin-dependent protein kinase II (CaMKII), and nuclear factor kappa-B (NF-kB) cellular pathways (3,5,6,7,8). Although IFNγ-mediated activation of STAT1 is required for several IFNγ-dependent antiviral responses, some of these activities are STAT1-independent, which implies that there may be activation of alternate and complementary pathways by IFNγ stimulation during a viral infection (3).
The human serine/threonine kinase ULK1 (Unc-51-like kinase 1) is the human homolog of Saccharomyces cerevisiae autophagy-related protein kinase Atg1, which promotes autophagy (9). However, ULK1 activity is required for type I IFN-induced activation of p38 MAPK, and for its antiproliferative effects in normal hematopoiesis and myeloproliferative neoplasms (9). In light of these findings, we questioned whether ULK1 could also be activated downstream of the type II IFNR, IFNGR, and whether ULK1 is required for IFNγ-mediated antiviral activity. We found that IFNγ-dependent engagement of ULK1 promoted activation of downstream IFNγ-dependent signaling events and transcription of key antiviral IFN-stimulated genes (ISGs). We identified that mixed lineage kinase 3 (MLK3) interacted with and was phosphorylated by ULK1 after engagement of the IFNGR. This study defines the molecular mechanism by which ULK1 activates IFNγ-mediated antiviral responses.
Results
IFNγ treatment promotes ULK1-dependent MLK3 activation
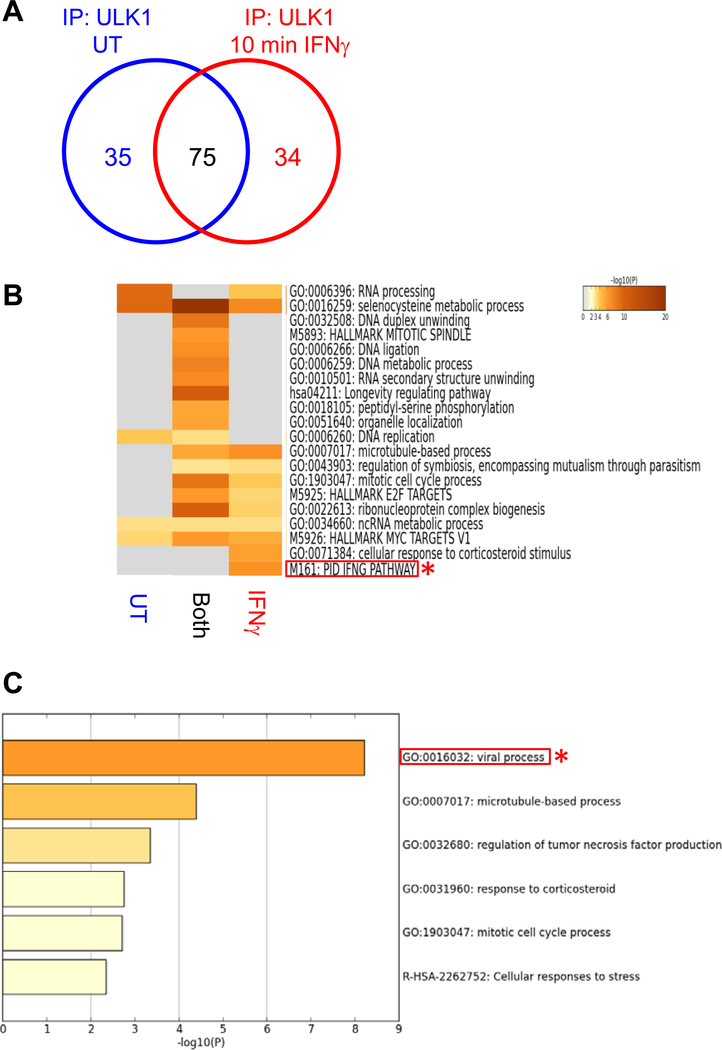
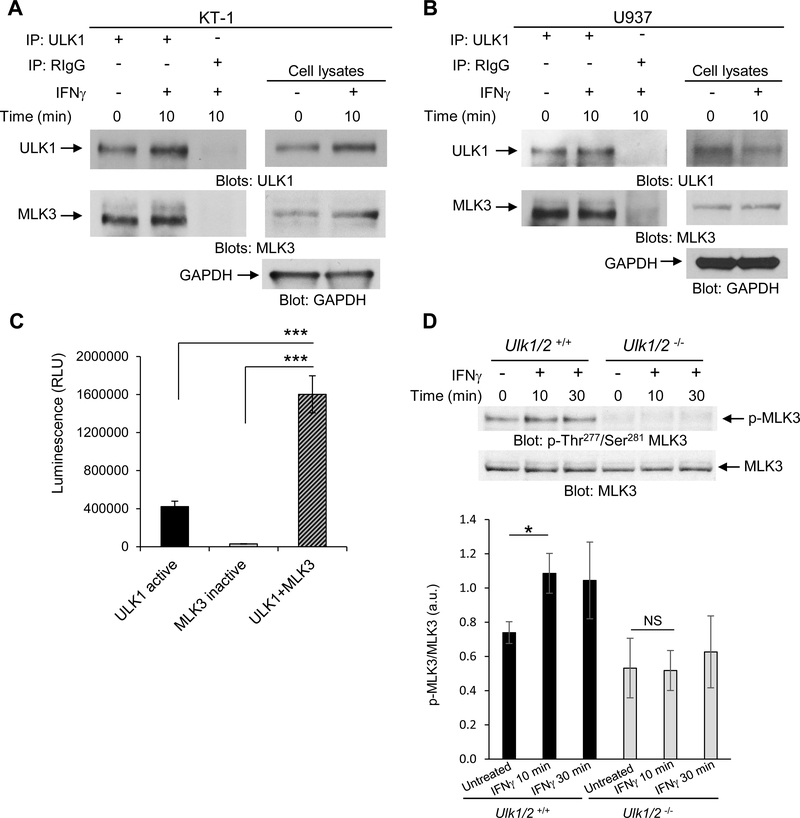
We speculated that IFNγ activation of the IFNGR may engage ULK1, which could in turn interact with other cellular signaling effectors. To identify potential IFNγ-dependent binding partners of ULK1, we performed nano-liquid chromatography (nLC) and tandem mass spectrometry (MS/MS) analysis of endogenous protein-ULK1 complexes immunoprecipitated from untreated and IFNγ-treated KT-1 leukemia cells. We found that ULK1 interacted with 35 proteins under untreated conditions, 75 proteins under both untreated and IFNγ-treated conditions, and 34 unique proteins following IFNγ treatment for 10 minutes (min) (Fig. 1A). Pathway and process enrichment analysis of the putative ULK1 interactors in KT-1 cells identified that 6 known IFNγ signaling pathway-related proteins bound to ULK1 after IFNγ stimulation (Fig. 1B, red asterisk and Table S1). Additionally, a separate pathway and process enrichment analysis of the 34 putative ULK1 interactors after IFNγ treatment revealed enrichment of 20 proteins associated with viral processes (Fig. 1C, red asterisk and Table S2) and indicated that MAP kinase kinase kinase 11 (MAP3K11, also MLK3) was involved in both IFNγ signaling and virus-associated processes (Table S1 and S2). When we probed the binding interaction between ULK1 and MLK3 by co-immunoprecipitation followed by immunoblotting in IFNγ-sensitive KT-1 and U937 cells, we found that ULK1 and MLK3 interacted in both unstimulated and IFNγ-treated cells (Fig. 2, A and B). Furthermore, in vitro kinase reactions between ULK1 and heat-inactivated MLK3 increased the ATP consumption in vitro (Fig. 2C), and loss of Ulk1/2 in mouse embryonic fibroblasts (MEFs) inhibited IFNγ-induced phosphorylation of MLK3 within its activation loop (10) at Thr277 and Ser281 (Fig. 2D). These data suggest that MLK3 is activated downstream of ULK1 during engagement of the IFNGR.
Fig. 1. Putative binding partners of ULK1 after engagement of the IFNGR.
(A to C) Tandem mass spec analysis of protein-ULK1 complexes from untreated (UT) or IFNγ-treated KT-1 cells. Venn diagram (A) indicates the number of proteins that interact with endogenous ULK1 in UT (blue), both (black), and after 10 min’ IFNγ-treatment (red) conditions. Heatmap analysis (B) is of the putative binding partners of ULK1 identified in untreated (UT), IFNγ-treated (IFNγ), and both groups (also Table S1). Ontology analysis (C) is of putative ULK1 interactors identified only after IFNγ stimulation (also Table S2). Note: Red rectangles and asterisks highlight (B) IFNγ signaling pathway and (C) viral process terms. Data are from of one experiment.
Fig. 2. ULK1 interacts with and phosphorylates MLK3 during engagement of the IFNGR.
(A and B) Co-immunoprecipitation analysis of ULK1 interaction with MLK3 in KT-1 (A) or U937 (B) cells left untreated, or treated with IFNγ for 10 min, as indicated. Blots are representative of 3 independent experiments. (C) ADP-Glo kinase assay analysis of ADP concentration produced by in vitro kinase reaction of recombinant human ULK1 and heat-inactivated MLK3 alone, or in combination. Data are means ± SEM of 3 independent experiments performed in triplicates. (D) Western blot analysis of pMLK3 in lysates from Ulk1/2+/+ and Ulk1/2−/− MEFs treated with IFNγ for 10 or 30 min, as indicated. Blots (top) are representative of 5 independent experiments. Quantified data (bottom) are means ± SEM pooled from all experiments. *P<0.05, **P<0.01, ***P< 0.001, and NS; not significant by one-way ANOVA followed by Tukey’s multiple comparisons test (C) or two-way ANOVA using Bonferroni correction (D).
IFNγ-dependent signaling events require ULK1/2 activity
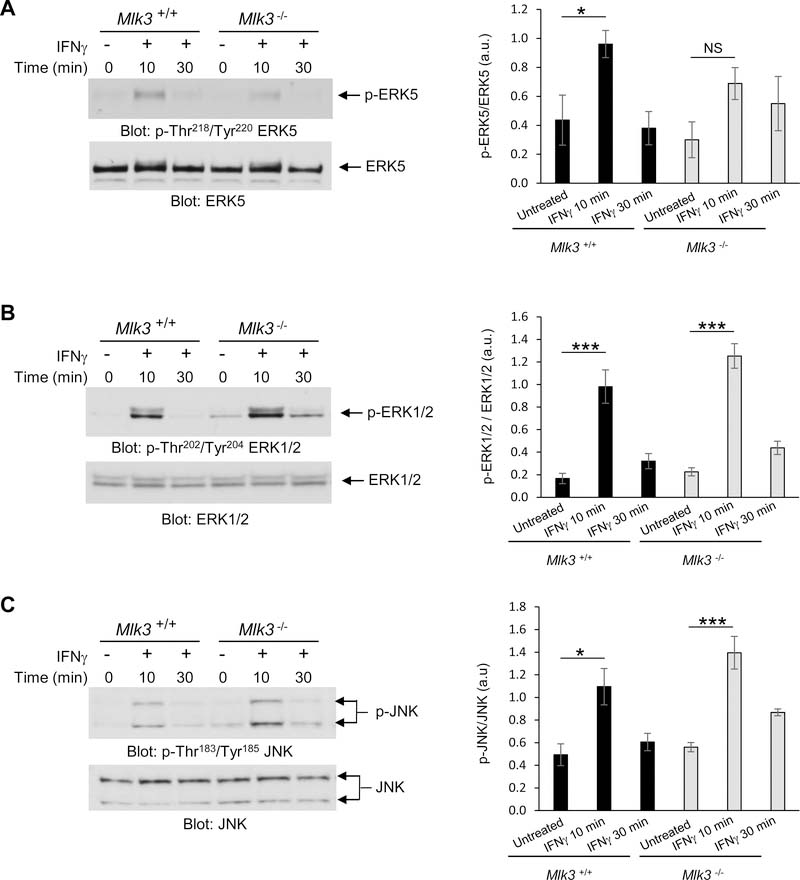
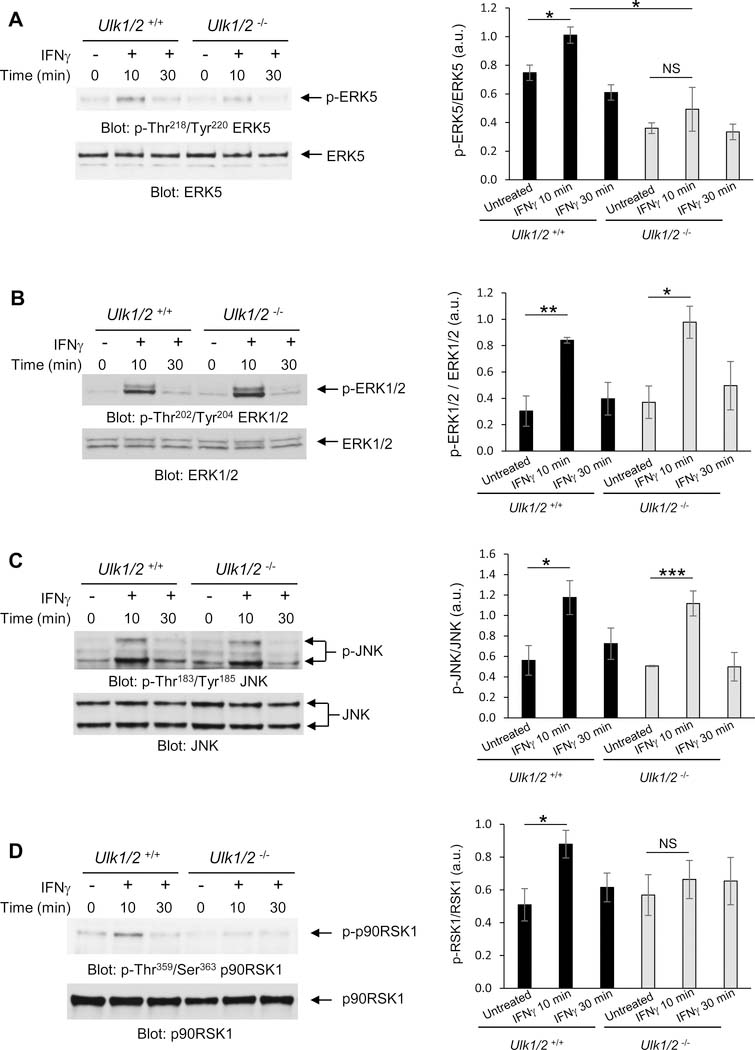
MLK3, a member of the MAP3K family, is a Ser/Thr protein kinase essential for activation of several MAPK signaling cascades in response to different stimuli (11,12,13). Specifically, in response to IFNγ MLK3 is required for the activation of the transcription factor CCAAT/enhancer-binding protein-β (C/EBP-β) (14). However, it is not known whether MLK3 promotes activation of MAP kinases after engagement of the IFNGR. Thus, we evaluated the importance of MLK3 for IFNγ-induced phosphorylation of the MAP kinases ERK5, ERK1/2 and JNK, using Mlk3+/+ and Mlk3−/− mouse embryonic fibroblasts (MEFs) (12). IFNγ-mediated activation of ERK5 by phosphorylation of the Thr-Glu-Tyr motif within the activation loop of its kinase domain (15) (Thr218/Tyr220) was defective in Mlk3−/− MEFs when compared to Mlk3+/+ MEFs (Fig. 3A). In contrast, IFNγ-induced phosphorylation of ERK1/2 and JNK were unaffected in Mlk3−/− cells (Fig. 3, B and C). When we examined whether ULK1 is required for IFNγ-dependent phosphorylation of ERK5 kinase in Ulk1/2+/+ and Ulk1/2−/− MEFs (16), we found that ERK5 phosphorylation in MEFs is defective in the absence of Ulk1/2 (Fig. 4A). However, there was some increased baseline phosphorylation of ERK5 in Ulk1/2+/+ MEFs compared to Ulk1/2−/− MEFs (Fig. 4A). Furthermore, IFNγ stimulation resulted in phosphorylation of both ERK1/2 and JNK kinases, as well as STAT1, independently of Ulk1/2 presence (Fig. 4, B and C and fig. S1). Together, our results suggest that ULK1/2 activity is required for IFNγ-mediated phosphorylation of MLK3, which in turn is required for IFNγ-induced activation of ERK5.
Fig. 3. IFNγ stimulates MLK3-mediated activation of ERK5, but not ERK1/2 and JNK.
(A to C) Western blot analysis of pERK5 (A), pERK1/2 (B), and pJNK (C) in lysates from Mlk3+/+ and Mlk3−/− MEFs treated with IFNγ for 10 or 30 min, as indicated. Blots (left) are representative of 3 (C) or 4 (A,B) independent experiments. Quantified data (right) are means ± SEM pooled from all experiments. *P<0.05, ***P< 0.001, and NS; not significant by two-way ANOVA using Bonferroni’s correction.
Fig. 4. ULK1/2 promotes IFNγ-induced phosphorylation of ERK5 and p90RSK1.
(A to D) Western blot analysis of pERK5 (A), pERK1/2 (B), pJNK (C), and p-p90RSK1 (D) in lysates from Ulk1/2+/+ and Ulk1/2−/− MEFs treated with IFNγ for 10 or 30 min, as indicated. Blots (left) are representative of 3 independent experiments. Quantified data (right) are means ± SEM pooled from all experiments. *P<0.05, **P< 0.01, ***P< 0.001, and NS; not significant by two-way ANOVA using Bonferroni’s correction.
Once activated, ERK5 plays a critical role as a master regulator of transcription by promoting the activation of many transcription factors and protein kinases, including p90 ribosomal S6 kinase 1 (p90RSK1) (16,17). The p90RSK1 Ser/Thr kinase modulates several biological processes by increasing both transcription and mRNA translation of specific genes (18,19,20). IFNγ-induced phosphorylation of p90RSK1 was defective in Ulk1/2−/− MEFs compared to Ulk1/2+/+ MEFs (Fig. 4D). Together these results suggest that ULK1/2 may play a role in the regulation of IFNγ-induced activation of ERK5 and p90RSK1.
ULK1/2 activity is essential for transcription of key IFNγ-inducible antiviral genes
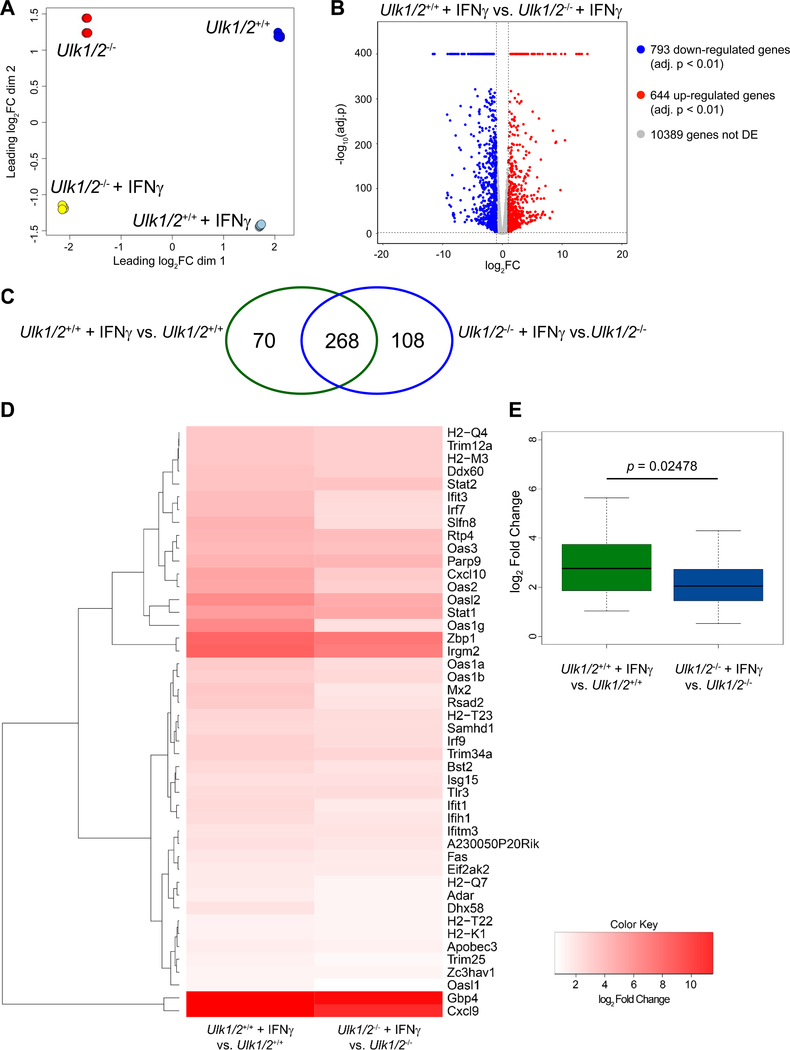
To determine whether ULK1/2 activity was required for IFNγ-dependent gene transcription, we performed high-throughput single-end RNA-sequencing of total RNA isolated from untreated and IFNγ-treated Ulk1/2+/+ and Ulk1/2−/− MEFs. When we used Multidimensional scaling (MDS) on the expression of the top 500 genes in all samples to determine how distinct and reproducible the samples were, we found that all four biological replicates in each group clustered together (fig. 5A). We identified those genes that were differentially expressed after IFNγ treatment using edgeR (21). This analysis revealed that the expression of 793 genes was decreased and that of 644 genes was increased in IFNγ-treated Ulk1/2+/+ MEFs compared to IFNγ-treated Ulk1/2−/− MEFs (Fig. 5B). Moreover, IFNγ treatment altered the expression of 338 genes in Ulk1/2+/+ MEFs (Fig. 5C, green ellipse, and fig. S2A), and 376 genes in Ulk1/2−/− MEFs (Fig. 5C, blue ellipse, and fig. S2B). From these, 268 common genes were differentially expressed in both groups after IFN-γ treatment (Fig. 5C, fig. S3, and Table S3). We also identified 70 IFN-γ-stimulated genes that were expressed only in Ulk1/2+/+ MEFs (Fig. 5C, fig. S4, and Table S4), and 108 IFNγ-stimulated genes that were expressed only in Ulk1/2−/− cells (Fig. 5C, fig. S5, and Table S5). Gene ontology analyses were performed to classify the differentially expressed genes detected following IFNγ treatment in both genotypic groups, those only in Ulk1/2+/+ cells, and those only in Ulk1/2−/− cells among ontology clusters (fig. S6A–C and Table S6–S8). The 268 commonly differentially expressed genes are mainly associated with processes involved in the defense response to virus infection, namely the innate immune response and antigen processing and presentation (fig. S6A and Table S6). The 70 genes differentially expressed only in Ulk1/2+/+ MEFs are predominantly associated with processes involved in immune cell differentiation, regulation of the JAK-STAT cascade, and regulation of inflammatory responses (fig. S6B and Table S7). By contrast, the 108 genes differentially expressed only in Ulk1/2−/− MEFs were predominantly associated with regulation of leukocyte proliferation and migration and positive regulation of the MAPK cascade (fig. S6C and Table S8). To closely evaluate whether the absence of Ulk1/2 affects IFNγ-mediated expression of antiviral genes, we selected the IFNγ-inducible genes depicted in our gene ontology analysis (Table S6–S8) associated with antiviral processes (total 66 genes) (22,23). From these genes, we found that expression of 46 genes was increased by IFNγ more in Ulk1/2+/+ MEFs than Ulk1/2−/− MEFs (Fig. 5D) and overall this difference was statistically significant (Fig. 5E).
Fig. 5. Ulk1/2 augments IFNγ-mediated transcription of ISGs.
(A to E) RNASeq analysis of transcript expression in Ulk1/2+/+ and Ulk1/2−/− MEFs untreated or treated with mouse IFNγ for 6 hours, as indicated. Multidimensional (MDS) plot of all groups (A) and Volcano plot of differentially expressed genes after IFNγ treatment in Ulk1/2+/+ versus Ulk1/2−/− MEFs (B). Venn diagram (C) indicates the gene expression overlap existing between differentially expressed genes after IFNγ treatment in Ulk1/2+/+ (green ellipse) and Ulk1/2−/− (blue ellipse) MEFs. Heatmap analysis (D) of antiviral genes induced in Ulk1/2+/+ MEFs compared to Ulk1/2−/− MEFs by IFNγ treatment. Box plot (E) of log2FC distribution of IFNγ-inducible antiviral genes in Ulk1/2+/+ and Ulk1/2−/− MEFs. Data are from 4 biological replicates per group (see also fig. S2–S6, and Table S3–S8). Statistical analyses were performed using Wilcoxon unpaired rank sum test.
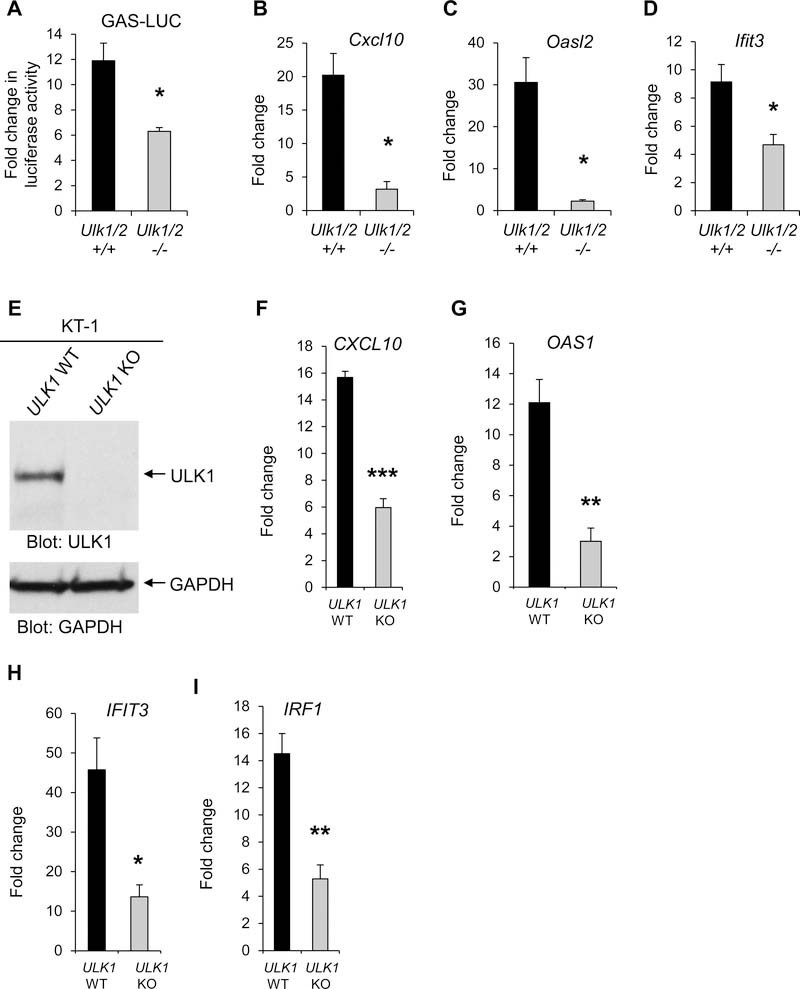
To evaluate whether ULK1/2 activity was required for optimal IFNγ-dependent transcriptional activation of ISGs, we generated Ulk1/2+/+ and Ulk1/2−/− MEFs stably expressing an IFNγ activation site (GAS) element-luciferase reporter gene (GAS-LUC) and performed luciferase reporter assays. IFNγ-induced transcriptional activation of GAS elements was significantly reduced in Ulk1/2−/− cells compared to Ulk1/2+/+ cells (Fig. 6A). Additionally, when we examined the specific role of ULK1/2 on the expression of select IFNγ-inducible antiviral genes Cxcl10 (24,25), Oasl2 and Ifit3 (26,27) by qRT-PCR, we found that expression of these genes after IFNγ treatment was significantly decreased in the absence of Ulk1/2 (Fig. 6, B to D). Although the activation of ULK1 promotes induction of autophagy (28), treatment of Ulk1/2+/+ MEFs with the autophagy inhibitor, chloroquine, did not significantly affect IFNγ-mediated mRNA expression of Cxcl10, Oasl2 and Ifit3 (fig. S7, A to C). Moreover, siRNA-mediated knockdown of VPS34 (PIK3C3) and Beclin-1 (BECN1), which are necessary for ULK1-mediated autophagy (29), did not reduce IFNγ-induced expression of antiviral ISGs in KT-1 cells (fig. S7, D to I). These results suggest that the key role of ULK1/2 related to IFNγ-mediated transcription of antiviral genes is independent of the activation of autophagy.
Fig. 6. ULK1/2 is required for IFNγ-dependent transcriptional induction of antiviral ISGs.
(A) Luciferase reporter assay analysis of GAS promoter activity in Ulk1/2+/+ and Ulk1/2−/− cells untreated or treated with IFNγ for 6 hours. Data are means ± SEM from 4 independent experiments. (B to D) qRT-PCR analysis of Cxcl10 (B), Oasl2 (C), and Ifit3 (D) mRNA expression in Ulk1/2+/+ and Ulk1/2−/−MEFs untreated or treated with IFNγ for 6 hours. Data are means ± SEM from 3 (B) or 4 (C and D) independent experiments. (E) Western blot analysis of ULK1 in lysate from KT-1 ULK1 KO cells generated by CRISPR/Cas9 genome editing. Blots are representative of 3 independent experiments. (F to I) qRT-PCR analysis of CXCL10 (F), OAS1 (G), IFIT3 (H), and IRF1 (I) mRNA expression in KT-1 ULK1 WT and KT-1 ULK1 KO cells were incubated in the presence or absence of IFNγ for 6 hours. Data are means ± SEM from 3 (F) or 4 (G to I) independent experiments. *P< 0.05, **P< 0.01, ***P< 0.001 by Unpaired t test (two-tailed) with Welch’s correction.
To determine whether ULK1 activity was also required for IFNγ-induced transcription of select antiviral genes in human cells, we generated KT-1 ULK1 knockout (KO) cells using CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 genomic editing. Plasmids encoding Cas9 and single-guide RNA (sgRNA) targeting the ULK1 gene (fig. S8) were transduced into KT-1 cells, and stably-transduced cells were selected with blasticidin and puromycin antibiotics. The complete absence of ULK1 protein was confirmed using western blot analysis and by comparing ULK1 abundance between the KT-1 parental cell line (ULK1 wild-type [WT]) and KT-1 ULK1 KO cells (Fig. 6E). We verified that IFNγ-induced gene expression of the antiviral effectors CXCL10, OAS1, IFIT3, and IRF1 was significantly reduced in ULK1 KO cells compared to ULK1 WT cells by qRT-PCR analysis (Fig. 6, F to I). Together, our data suggest that ULK1 promotes antiviral-ISG transcription.
ULK1/2 kinase activity is required for IFNγ-induced antiviral responses
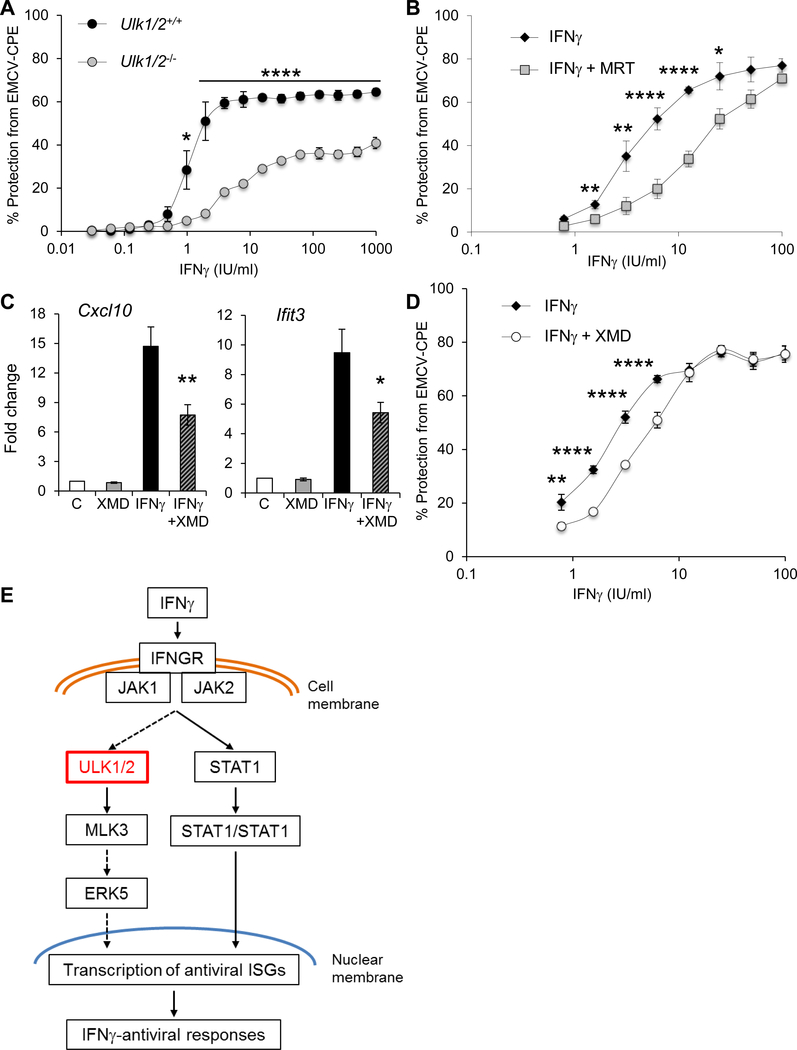
Because ULK1/2 expression was required for the optimal transcription of select IFNγ-induced antiviral genes, we tested whether ULK1 was required for IFNγ-stimulated antiviral responses. For this, we compared the protective effects of mouse IFNγ on protection from encephalomyocarditis virus (EMCV)-induced cell death in Ulk1/2+/+ and Ulk1/2−/− MEFs. We found that cells that lacked Ulk1/2 were less responsive to IFNγ treatment than WT cells (Fig. 7A). Moreover, human fibrosarcoma 2fTGH cells were less sensitive to the antiviral effects of human IFNγ treatment against EMCV infection when treated with the ULK1 kinase inhibitor MRT68921 (30) (Fig. 7B). Similarly, selective inhibition of ERK5 activity with XMD8–92 (31) resulted in a reduction of IFNγ-induced expression of the antiviral genes Cxcl10 and Ifit3 in Ulk1/2+/+ MEFs (Fig. 7C) and of IFNγ-induced antiviral effects in human 2fTGH cells (Fig. 7D). Together, these data suggest that engagement of the ULK1/2-ERK5 pathway augments IFNγ-dependent antiviral responses (Fig. 7E).
Fig. 7. Requirement of ULK1/2 for the generation of IFNγ-dependent antiviral effects.
(A) Crystal violet viability analysis of EMCV-induced cytopathic effects in Ulk1/2+/+ and Ulk1/2−/− MEFs pretreated with IFNγ at the indicated doses for 16 hours and subsequently challenged with encephalomyocarditis virus (EMCV) for 24 hours. Data are means ± SEM of quadruplicate assays from 3 independent experiments. (B) Crystal violet viability analysis of EMCV-induced cytopathic effects in human fibrosarcoma 2fTGH cells pretreated for 2 hours with the ULK1 kinase inhibitor MRT68921 (MRT) as indicated, then exposed to IFNγ for 16 hours before challenge with EMCV for 24 hours. Data are means ± SEM of quadruplicate assays from 3 independent experiments. (C) qRT-PCR analysis of Cxcl10 (left) and Ifit3 (right) mRNA expression in Ulk1/2+/+ MEFs were treated with DMSO (vehicle-control, C), XMD8–92 (XMD), and/or IFNγ, as indicated. Data are means ± SEM from 4 independent experiments. (D) Crystal violet viability analysis of EMCV-induced cytopathic effects in human fibrosarcoma 2fTGH cells pretreated for 2 hours with the ERK5 inhibitor XMD8–92 (XMD) as indicated, then exposed to IFNγ for 16 hours before challenge with EMCV for 24 hours. Data are means ± SEM of quadruplicate assays from 3 independent experiments. (E) Schematic illustration of the potential role of ULK1/2 in IFNγ signaling. *P<0.05, **P< 0.01, ****P< 0.0001 by one-way ANOVA analysis followed by Tukey’s multiple comparisons test (C) or two-way ANOVA with Post-hoc t-tests (A, B and D).
Discussion
IFNγ is a critical immunomodulator, regulating the innate and adaptive immune responses to pathogen infection (32). Indeed, both IFNγ KO and IFNGR KO mice are more susceptible to bacterial and viral infections than their WT counterparts (33,34,35,36). Moreover, mice injected with a monoclonal antibody against IFNγ succumb to Toxoplasma gondii infection (37) and Listeria monocytogenes at higher rates (38). Distinct from the ability of IFNγ to influence the differentiation and activation of specific immune cell subsets required for pathogen clearance, IFNγ promotes apoptosis of pathogen-infected cells and induces the expression of type I IFNs and of several antiviral and antimicrobial genes (39,40). However, a comprehensive understanding of the signaling pathways activated downstream of the IFNGR and how these impact IFNγ-driven responses remains unclear. Herein we provide evidence that IFNγ-induced engagement of ULK1 was required to inhibit virus infection. We found that lack of Ulk1/2 genes or pharmacological inhibition of the ULK1/2 kinase activity abrogated IFNγ-induced antiviral activity. This correlated with defective IFNγ-mediated transcription of antiviral genes in the absence of ULK1/2 activity, although we cannot exclude that additional pathways may participate in the induction of antiviral responses. Our work suggests that IFNγ-signaling involves the interaction of ULK1 with MLK3, which was required for IFNγ-dependent phosphorylation/activation of MLK3. These data suggest that ULK1 likely acts upstream, either as a kinase or a scaffold protein, of IFNγ-activated MAPK signaling cascades. In human fibroblasts, MLK3 is required for both cytokine- and mitogen-induced activation of several MAPK signaling cascades (ERK1/2, JNK, and p38 MAPK) (41). Moreover, tumor necrosis factor (TNF)-induced stimulation of JNK, but not p38 MAPK or ERK1/2, is defective in Mlk3−/− MEFs when compared to WT cells (12). Similarly, MLK3 is required for IFNγ-driven gene expression, independently of ERK1/2 activation (15). Here we showed in MEFs that both Ulk1/2 and Mlk3 were required for IFNγ-mediated phosphorylation of ERK5, but not JNK and ERK1/2. These data suggest that ULK1 and MLK3 may act upstream of MEKK2, MEKK3 or MEK5 kinases, which are known to activate ERK5 in IFNγ-stimulated cells (42). Our results support a model in which IFNγ-dependent engagement of ULK1 leads to phosphorylation of MLK3 and ERK5, which promotes transcription of antiviral ISGs and consequent IFNγ-induced antiviral responses that may complement the antiviral effects induced by the canonical JAK1/JAK2-STAT signaling.
Autophagy is a degradative process that aids the host immune response against viral infections, including induction of type I IFN production and activation of IFNγ-induced JAK-STAT signaling (43,44). Assembly of the nondegradative Atg5-Atg12/Atg16L1 autophagy protein complex is also required for IFNγ-dependent antiviral activity (45). In contrast, ULK1-dependent autophagy activates a different pathway of phagophore nucleation that requires phosphorylation of Beclin-1 and activation of vacuolar protein sorting 34 (VPS34) (46). Using pharmacologic and genetic inhibition we found that autophagy is not required for the effects of ULK1 on IFNγ-dependent gene expression. Thus, multicellular organisms may have developed ways to use autophagy-related proteins for additional immune functions. In support of this, here we report a nondegradative role of ULK1 that is required for transcription of IFNγ inducible antiviral genes.
The precise mechanisms by which ULK1 or MLK3 may stimulate ERK5 phosphorylation remain undetermined. MEK5 is the direct upstream activator of ERK5 (47), and MEKK2/3 (48,49,50), Cot/TPL-2 (51) and MLTK (52) are MAP3Ks that activate ERK5 in response to multiple stimuli. Additionally, another MAP3K, transforming growth factor β-activated kinase 1 (TAK1), interacts with MEKK3 and promotes its autophosphorylation in TFNα-signaling (53). Similarly, ULK1 and/or MLK3 could act as scaffold proteins, or MAP3Ks of ERK5 upstream kinases after IFNγ treatment. Although p38 MAPK activity is not required for IFNγ-induced transcription of ISGs in MEFs (54), it is involved in the generation of IFNγ-inducible biological effects in different human cell types (55,56,57,58). Given that ULK1 promotes p38 MAPK activity downstream of the type I IFNR (9), in future studies, it would be of interest to evaluate whether ULK1-MLK3 interactions are required for IFNγ-dependent activation of p38 MAPK in human cells. Viewed altogether our studies identify an IFN-γ−stimulated signaling cascade that is critical for expression of IFNγ-inducible ISGs and antiviral responses. These findings may suggest new points of therapeutic intervention for the treatment of several diseases where IFNγ has proven clinical activity (59,60,61) or how to enhance immune checkpoint blockade where IFNγ responses are involved (62,63).
Materials and Methods
Cell culture
KT-1 (64) and U937 (CRL-1593.2; ATCC) cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. 2fTGH cells (65) were kindly provided by Dr. George R. Stark (Cleveland Clinic) and were cultured in DMEM high glucose medium supplemented with 10% FBS and antibiotics. The immortalized Ulk1/2+/+ and Ulk1/2−/− MEFs (17) were kindly provided by Dr. Craig B. Thompson (Memorial Sloan-Kettering Cancer Center) and were cultured in DMEM supplemented with 10% FBS and antibiotics. Primary Mlk3+/+ and Mlk3−/− MEFs were described previously (12) and were cultured in DMEM high glucose medium supplemented with 10% bovine growth serum, 1x L-glutamine, 1x penicillin/streptomycin solution and 8 μl of 2-mercaptoethanol per liter of medium. All experiments were performed using primary MEFs between passages 2 and 4. The other cell lines were frozen at low passage in liquid nitrogen and were kept in culture for no longer than 8 passages. All cells were cultured at 37°C and 5% CO2. All cells were tested for mycoplasma contamination using MycoAlert PLUS mycoplasma detection kit following the manufacturer’s instructions (Lonza).
Reagents
IFNγ recombinant human (# PHC4033) and mouse (# PMC4033) proteins were from Gibco (Life Technologies). MRT68921 dual autophagy kinase ULK1/2 inhibitor was purchased from Selleckchem (# S7949). Chloroquine (CQ) was purchased from Sigma-Aldrich. XMD8–92 was purchased from Tocris (# 4132). The Edit-R human lentiviral ULK1 sgRNAs (# VSGH10142–246477203) and Edit-R Lentiviral hEF1α-Blast-Cas9 nuclease plasmid DNA (# CAS10138) were purchased from GE Healthcare Dharmacon. ON-TARGETplus Non-targeting Control Pool siRNA (# D-001810–10-05), ON-TARGETplus PIK3C3 siRNA (SMARTpool: # L-005250–00-0005) and ON-TARGETplus BECN1 siRNA (SMARTpool: # L-010552–00-0005) were purchased from Dharmacon. Antibodies against ULK1 (D8H5) (#8054), MLK3 (#2817), phospho-ERK5 (Thr218/Tyr220) (#3371), ERK5 (D3I5V) (#12950), phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (D13.14.4E) (#4370), p44/42 MAPK (ERK1/2) (#9102), phospho-SAPK/JNK (Thr183/Tyr185) (81E11) (#4668), SAPK/JNK (#9252), phospho-STAT1 (Ser727) (D3B7) (#8826), phospho-STAT1 (Tyr701) (D4A7) (#7649), phospho-p90RSK (Thr359/Ser363) antibody (#9344), and RSK1 (D6D5) (#8408) were purchased from Cell Signaling. Anti-MLK3 (phospho Thr277 + Ser281) antibody (#ab191530) was purchased from abcam and STAT1 p84/p91 antibody (E-23) (#sc-346) was from Santa Cruz Biotechnology. Antibody against GAPDH (6C5) (#MAB374) was purchased from EMD Millipore.
Mass spectroscopy analyses
To identify proteins that interact with endogenous ULK1, KT-1 cells were left untreated or were treated with 5×103 IU/ml of human IFNγ for 10 min and then lysed in NP-40 buffer (20mM HEPES pH7.4, 180mM KCl, 0.2mM EGTA, 0.1% NP-40) supplemented with protease and phosphatase inhibitors. Three mg of protein (total cell lysates) from untreated and IFNγ-treated samples were used for immunoprecipitation (IP) of endogenous protein-ULK1 complexes using ULK1 (D8H5) rabbit monoclonal antibody-conjugated to magnetic beads (Custom order, Cell signaling). As control, the same procedure was followed for IFNγ-treated lysates, but using Rabbit (DA1E) monoclonal antibody IgG XP Isotype control-conjugated to magnetic beads (#8726, Cell Signaling) instead of the ULK1 antibody. After incubating the samples overnight with rotation at 4°C, the beads were washed two times with NP-40 buffer and one time with washing buffer (20mM HEPES pH7.4, 180mM KCl, 0.2mM EGTA). Protein-ULK1 complexes were eluted from the beads by incubation with lane marker reducing sample buffer (Pierce) at 95°C for 10 min and proteomic analyses were performed in the Northwestern Proteomics Core Facility (Northwestern University, Chicago). IP-eluted proteins were initially separated using SDS-PAGE and cut into ten equivalent height bands prior to standard in-gel digestion (66). Resulting peptides were extracted from the gel pieces and desalted using solid phase extraction on a Pierce C18 Spin column, prior to elution in 40 μL of 80% acetonitrile in 0.2% formic acid. After lyophilization, peptides were reconstituted with 0.1% formic acid in water and injected onto a trap column (150 μm ID × 3 cm) coupled with a nanobore analytical column (75 μm ID × 15 cm, both ReproSil C18aq, 3 μm). Samples were separated using a linear gradient of solvent A (95% water, 5% acetonitrile, 0.1% formic acid) and solvent B (5% water, 95% acetonitrile, 0.1% formic acid) over 60 min. nLC-MS/MS data were obtained on a Velos Orbitrap (Thermo) mass spectrometer. Data were searched using Mascot (Matrix Science) 2.5 against the human SwissProt database and results were reported at 1% FDR in Scaffold 4.5 (Proteome Software). Proteins identified by nLC-MS/MS analysis in the control group (Rabbit IgG) were excluded from our data analysis.
Protein function enrichment analysis
Protein lists identified in untreated and/or IFNγ-treated groups were converted to gene lists that were submitted to the Metascape database (67,68), a gene annotation and analysis resource (http://metascape.org/), for pathway and process enrichment analysis. For each given gene list, Metascape carries pathway and process enrichment analysis using the following ontology sources: GO Biological Processes, KEGG Pathway and Reactome Gene Sets. All genes in the genome were used as the enrichment background. Terms with p-value < 0.01, minimum count 3, and enrichment factor > 1.5 are collected and grouped into clusters based on their membership similarities. More specifically, p-values are calculated based on accumulative hypergeometric distribution, q-values are calculated using the Banjamini-Hochberg procedure to account for multiple testing. Kappa scores were used as the similarity metric when performing hierarchical clustering on the enriched terms and then sub-trees with similarity > 0.3 are considered a cluster. The most statistically significant term within a cluster is chosen as the one representing the cluster.
Co-immunoprecipitation of protein-ULK1 complexes
To confirm the physical interaction between ULK1 and MLK3 identified by nLC-MS/MS analysis, KT-1 cells were left untreated or were treated with human IFNγ (5×103 IU/ml) for 10 min and U937 cells were cultured overnight in serum-free RPMI 1640 medium and then were left untreated or were treated with human IFNγ (5×103 IU/ml) for 10 min. After treatment, cell pellets were lysed in NP-40 buffer (20mM HEPES pH7.4, 180mM KCl, 0.2mM EGTA, 10% glycerol, 0.1% NP-40) supplemented with protease and phosphatase inhibitors. For immunoprecipitation of endogenous protein-ULK1 complexes, 200 μg of protein (total cell lysates) from each sample were incubated overnight at 4°C with rotation with ULK1 (D8H5) rabbit monoclonal antibody (1:100) (#8054, Cell signaling), followed by incubation for 1 hour at 4°C with rotation with protein G Sepharose 4 Fast Flow beads (GE Healthcare Life Sciences). As control, the same procedure was followed using Rabbit (DA1E) monoclonal antibody IgG XP Isotype control (#3900, Cell Signaling) instead of ULK1 antibody. After immunoprecipitation, the beads were washed three times with NP-40 buffer without glycerol. Protein-ULK1 complexes were eluted from the beads by incubation with lane marker reducing sample buffer (Pierce) at 95°C for 10 min. Eluates were resolved by SDS-PAGE and processed for immunoblotting analyses.
In vitro kinase assay
Recombinant human MLK3 active protein (#M19–11G, SignalChem) was heat inactivated at 65°C for 20 min in a sonicating bath. To confirm heat inactivation of MLK3 recombinant protein, kinase reactions between MLK3 heat inactive protein (1 μg) and MBP substrate (5 μg) (#M42–51N, SignalChem) were performed. To determine whether ULK1 can directly phosphorylate MLK3, kinase reactions between recombinant human ULK1 active protein (100 ng) (#U01–11G, SignalChem) and MLK3 heat inactive protein (1.66 μg) were performed. As controls, the same kinase reactions were carried out, but using each recombinant protein alone. For each kinase reaction we used DTT (#D86–09B, SignalChem), ATP (#V915A, Promega), and 5x kinase buffer (#K03–09, SignalChem) following the manufacturer’s instructions. Kinase mixtures were incubated for 40 min shaking at 300 rpm and 30°C. The ADP formed from the kinase reactions was measured using the ADP-Glo Kinase Assay Kit (#V6930, Promega) following the manufacturer’s instructions.
MEFs treatment and lysis for immunoblotting analyses
Prior to IFNγ treatment, MEFs were either cultured in 10% FBS-containing medium (for detection of phosphorylation of ERK5, STAT1 and ERK1/2) or starved overnight (for detection of phosphorylation of JNK, MLK3, and p90RSK1). MEFs were then treated with 5×103 IU/ml of mouse IFNγ for 10 or 30 min, as indicated. After treatment, cell pellets were lysed with lysis buffer (50 mM HEPES pH 7.3, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA pH 8.0, 100 μM sodium fluoride, 10 μM sodium pyrophosphate, 0.5% Triton X-100, and 10% glycerol) supplemented with protease and phosphatase inhibitors.
Immunoblotting analyses
Equal amounts of total cell lysates were resolved by SDS-PAGE and transferred to a Immobilon-P PVDF membrane (Millipore) using the Trans-Blot Turbo transfer system (Bio-Rad). For immunoblotting analyses, the membranes were probed with primary antibodies, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies, and antibody binding was detected by enhanced chemiluminescence using Amersham ECL prime western blotting detection reagent (GE Healthcare Life Sciences). Bands corresponding to protein of interest were scanned and quantified by densitometry using ImageJ software.
Library Construction and RNA-Sequencing (RNA-Seq)
Ulk1/2+/+ and Ulk1/2−/− MEFs were plated as four biological replicates and were either left untreated or were treated for 6 hours with 2.5×103 IU/ml of mouse IFNγ. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN), following the manufacturer’s instructions. Library construction and stranded mRNA sequencing were conducted at the NUSeq Core Facility of Northwestern University. Briefly, RNA quality and quantity were first determined with the Agilent Bioanalyzer 2100 and Qubit fluorometer, and all samples presented a RNA integrity number of 10. Sequencing libraries were prepared from 1μg of high-quality RNA samples using Illumina TruSeq Stranded mRNA Library Preparation Kit (Illumina) as per the manufacturer’s instructions. This procedure includes mRNA purification and fragmentation, cDNA synthesis, 3’ end adenylation, Illumina adapter ligation, library PCR amplification and validation. An Illumina NextSeq 500 Sequencer was used to sequence the libraries with the production of single-end, 75 bp reads.
RNA-Seq reads mapping and transcript abundance estimation
Raw reads in the fastq format were controlled for quality using Trimmomatic (69). Trimmed reads were then aligned to the mouse genome (UCSC mm9) using TopHat v2.1.0 (70). Only uniquely mapped reads with a maximum of two mismatches over the whole length of the gene were considered for ensuing analyses. Gene annotations came from Ensembl release 75. Exonic reads were assigned to specific genes from Ensembl release 75 using the htseq-count script from HTSeq-0.6.1 (71).
Differential analysis of gene regulation at transcript resolution
Gene counts computed by HTSeq were used as input for edgeR (version 3.18.1) (72), which was used to calculate the differential expression and analyze the significances of observed changes between two groups using the standard edgeR workflow. Genes with Benjamini-Hochburg adjusted p-values less than 0.01 were considered to be differentially expressed unless otherwise specified.
Gene clustering
RNA-Seq results are shown as a heatmap and include transcripts whose expression were the most significantly altered by IFNγ treatment in Ulk1/2+/+ and Ulk1/2−/− MEFs. Colored rectangles represent normalized total reads per million (RPM) mRNA abundance of the transcript. Hierarchical clustering was performed with R version 3.4.0 software using the heatmap.2 function and distance correlation. The intensity of the color is proportional to the RPM values from each RNA-Seq measurement, as indicated on the bar next to the heatmap image.
Gene Set Enrichment analyses
The gene list of the differentially induced genes by IFNγ found only in Ulk1/2+/+ MEFs was submitted to the Metascape database (68,69) (http://metascape.org). Metascape carries pathway and process enrichment analysis using the following ontology sources: GO Biological Processes, KEGG Pathway and Reactome Gene Sets. Gene ontology functional analysis was conducted using default parameters.
Generation of Ulk1/2+/+ and Ulk1/2−/− cell lines stably expressing GAS luciferase reporter gene elements
pGF1-GAS-LUC, which expresses a puromycin selection cassette and firefly luciferase reporter under the control of a minimal CMV promoter followed by four tandem consensus GAS elements (5′-AGTTTTCATATTACTCTAAATC-3′) was purchased from System Biosciences (SBI). Reporter vector carrying viral particles was produced by co-transfection of the lentiviral plasmid and the packaging vectors into Lenti-X 293T cell line (Clontech). Ulk1/2+/+ and Ulk1/2−/− MEFs were infected with virus-containing fresh supernatant using Transdux reagent (SBI). Stably-transduced cells were selected using 2 μg/ml of puromycin (Gibco, Life Technologies).
Luciferase assays
Ulk1/2+/+ and Ulk1/2−/− MEFs stably expressing pGF1-GAS-LUC were plated in a 96-well plate (three or five replicates of 5000 cells per well) and 24 hours later were maintained in serum-free RPMI 1640 medium overnight. Serum-starved cells were left untreated (control) or were treated with 2.5×103 IU/ml of mouse IFNγ for 6 hours and then lysed using 1x Reporter Lysis Buffer (Promega). Luciferase assay substrate buffer (Luciferase Assay System # E4030, Promega) was used per the manufacturer’s instructions and luciferase activities were measured using a Cytation 3 cell imaging multi-mode microplate reader (BioTek).
Quantitative RT-PCR (qRT-PCR) analyses
To determine the effect of ULK1/2 on IFNγ-induced expression of antiviral ISGs, Ulk1/2+/+ and Ulk1/2−/− MEFs or KT-1 WT and KT-1 ULK1 KO cells were left untreated or were treated for 6 hours with 2.5×103 IU/ml of mouse or human IFNγ, respectively. To determine whether ERK5 is required for IFNγ-induced expression of antiviral ISGs, Ulk1/2+/+ MEFs were pre-incubated for 1 hour with 0.5μM of the ERK5 inhibitor XMD8–92 followed by 6 hours of co-treatment with 2.5×103 IU/ml of mouse IFNγ. Cells were treated with vehicle-control (DMSO) or each compound alone (controls). To define whether inhibition of autophagy would affect antiviral ISG expression, KT-1 cells were transfected with control, PIK3C3 (VPS34) or BECN1 (Beclin-1) siRNAs using Amaxa Cell Line Nucleofector Kit V (Lonza) following the manufacturer’s instructions and, 18 hours later, cells were either left untreated or were treated with 2.5×103 IU/ml of human IFNγ for 6 hours. Additionally, Ulk1/2+/+ MEFs were pre-incubated for 1 hour with 6μM of chloroquine (CQ) followed by six hours of co-treatment with 2.5×103 IU/ml of mouse IFNγ. As controls, cells were left untreated or were treated with each compound alone. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) following the manufacturer’s instructions. 2μg of total cellular mRNA was reverse-transcribed into cDNA using the Omniscript RT kit (QIAGEN) and oligo(dT)12–18 primers (Life Technologies). Quantitative RT-PCR was carried out using an ABI7500 sequence detection system (Applied Biosystems) or a Bio-Rad CFX96 Real Time System (Bio-Rad) using commercially-available FAM-labeled primer/probe sets (Thermo Fisher) to determine mouse Cxcl10 (Mm00445235_m1), Ifit3 (Mm01704846_s1), Oasl2 (Mm00496187_m1), and human CXCL10 (Hs01124252_g1), OAS1 (Hs00973635_m1), IFIT3 (Hs01922752_s1), IRF1 (Hs00971965_m1), PIK3C3 (Hs00176908_m1), and BECN1 (Hs01007018_m1) mRNA expression. Mouse Gapdh (Mm99999915_g1) and human GAPDH (Hs02758991_g1) were used for normalization. The mRNA amplification was calculated as previously (9), and the data were plotted as the increase of fold change as compared with control samples.
CRISPR/Cas9 approach to generate ULK1 knockout KT-1 cells
KT-1 cells were transduced with Edit-R Lentiviral hEF1α-Blast-Cas9 Nuclease Plasmid DNA and Edit-R Human Lentiviral ULK1 sgRNA (#CAS10138 and #VSGH10142–246477203, GE Healthcare Dharmacon). Transduction was carried out by spinoculation (750g) for 90 min at RT in the presence of Transdux reagent (SBI) followed by 48 hours of incubation. Stably-transduced cells were selected using blasticidin (5 μg/ml) and puromycin (2.5 μg/ml) antibiotics. One week later, antibiotic-resistant cells were separated from dead cells using a BD FACSAria 5-Laser Cell Sorter (BD Biosciences) and performed at the Northwestern University Flow Cytometry Core Facility. Lack of ULK1 protein in KT-1 ULK1 KO cells was confirmed using western blot analysis.
Antiviral assays
Ulk1/2+/+ and Ulk1/2−/− MEFs were seeded in quadruplicate in 96-well plates overnight and then treated with the indicated doses of mouse IFNγ for 16 hours. Cells were subsequently challenged with EMCV and EMCV-induced cytopathic effects (CPEs) were determined 24 hours later. Briefly, medium was aspirated and cells were fixed in 95% ethanol for 30 minutes, then stained with 0.1% crystal violet in 2% ethanol for a further 30 minutes. Cells were then destained in 0.5M NaCl in 50% ethanol for 1 hour, then absorbance readings at 570nm were taken to determine the extent of uninfected, viable cells. For those assays where the ULK1 pharmacological inhibitor MRT68921 or the ERK5 inhibitor XMD8–92 were included, human fibrosarcoma 2fTGH cells were seeded in quadruplicate in 96-well plates and then treated with either human IFNγ for 16 hours or pretreated for 2 hours with MRT68921 (1 μM) or with XMD8–92 (5 μM), then the medium removed and the cells treated with MRT68921 or XMD8–92 and human IFNγ, as indicated, for a further 16 hours. Cells were subsequently challenged with EMCV and EMCV-induced CPEs were determined 24 hours later, as described above.
Statistical Analyses
PRISM v6.0 software (GraphPad Software) and SAS 9.4. software (SAS Institute Inc.) were used for statistical analyses. Unpaired two-tailed t test with Welch’s correction was used for comparison of means between two groups, one-way analysis of variance (ANOVA) was used to compare means among more than two independent groups followed by Tukey’s multiple comparisons test, and two-way ANOVA with Bonferroni-corrected post-hoc t-tests were used to compare the mean differences among groups classified by two factors. Differences were considered statistically significant when p values were less than 0.05.
Supplementary Material
Table S1. Gene ontology analysis of putative ULK1 interactors in untreated versus IFNγ-treated KT-1 cells identified by mass spectrometry analysis.
Table S2. Gene ontology analysis of putative ULK1 interactors identified after IFNγ stimulation by mass spectrometry analysis.
Table S3. Genes differentially expressed in both Ulk1/2+/+ and Ulk1/2−/− MEFs after IFNγ treatment.
Table S4. Genes differentially expressed only in Ulk1/2+/+ MEFs after IFNγ treatment.
Table S5. Genes differentially expressed only in Ulk1/2−/− MEFs after IFNγ treatment.
Table S6. Gene ontology analysis of differentially expressed genes identified in both Ulk1/2+/+ and Ulk1/2−/− MEFs after IFNγ treatment.
Table S7. Gene ontology analysis of differentially expressed genes identified only in Ulk1/2+/+ MEFs after IFNγ treatment.
Table S8. Gene ontology analysis of differentially expressed genes identified only in Ulk1/2−/− MEFs after IFNγ treatment.
Fig. S1. ULK1/2 activity is not required for IFNγ-induced phosphorylation of STAT1.
Fig. S2. Effects of targeted disruption of Ulk1/2 gene expression on IFNγ-dependent gene transcription.
Fig. S3. Genes differentially expressed in both Ulk1/2+/+ and Ulk1/2−/− MEFs after IFNγ treatment..
Fig. S4. Genes differentially expressed only in Ulk1/2+/+ MEFs after IFNγ treatment.
Fig. S5. Genes differentially expressed only in Ulk1/2−/− MEFs after IFNγ treatment.
Fig. S6. Gene ontology analysis of IFNγ-dependent differentially expressed genes.
Fig. S7. Inhibition of autophagy does not affect transcription of antiviral ISGs.
Fig. S8. Genomic location for the ULK1 single-guide RNA in the human ULK1 open reading frame.
Acknowledgments
We thank Dr. Craig B. Thompson (Memorial Sloan-Kettering Cancer Center) for the Ulk1/2+/+ and Ulk1/2−/− MEFs. We thank Dr. Ali Shilatifard (Northwestern University, Feinberg School of Medicine) for helping with the analysis of the RNA-Seq data.
Funding: This work was supported in part by NIH grants CA161196, CA77816, and CA155566 and by grant I01CX000916 from the Department of Veterans Affairs. A.D.A. was supported in part by NIH/NCI grant T32 CA070085, and D.S. was supported in part by NIH/NCI grant T32 CA080621. ENF is a Tier 1 Canada Research Chair in Women’s Health and Immunobiology. Proteomic services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. Flow Cytometry Cell Sorting was performed on a BD FACSAria SORP system, purchased through the support of NIH 1S10OD011996-01 and this work was supported by the Northwestern University – Flow Cytometry Core Facility supported by Cancer Center Support Grant (NCI CA060553). Sequencing of RNA libraries were performed by the NUSeq Core of Northwestern University.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw RNA-Seq data have been uploaded to the NIH GEO web portal and the accession number is GSE118766. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009018. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Lin FC, Young HA, Interferons: Success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 25, 369–376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann HH, Schneider WM, Rice CM, Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 36, 124–138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gough DJ, Levy DE, Johnstone RW, Clarke CJ, IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 19, 383–394 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Fish EN, Platanias LC, Interferon receptor signaling in malignancy: a network of cellular pathways defining biological outcomes. Mol Cancer Res. 12, 1691–1703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platanias LC, Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat Rev Immunol. 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Stark GR, How cells respond to interferons revisited: from early history to current complexity. Cytokine Growth Factor Rev. 18, 419–423 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroczynska B, Rafidi RL, Majchrzak-Kita B, Kosciuczuk EM, Blyth GT, Jemielity J, Warminska Z, Saleiro D, Mehrotra S, Arslan AD, Fish EN, Platanias LC, Interferon γ (IFNγ) Signaling via Mechanistic Target of Rapamycin Complex 2 (mTORC2) and Regulatory Effects in the Generation of Type II Interferon Biological Responses. J Biol Chem. 291, 2389–2396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroczynska B, Blyth GT, Rafidi RL, Majchrzak-Kita B, Xu L, Saleiro D, Kosciuczuk EM, Jemielity J, Su B, Altman JK, Eklund EA, Fish EN, Platanias LC, Central Regulatory Role for SIN1 in Interferon γ (IFNγ) Signaling and Generation of Biological Responses. J Biol Chem. 292 4743–4752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleiro D, Kosciuczuk EM, Platanias LC, Beyond autophagy: New roles for ULK1 in immune signaling and interferon responses. Cytokine Growth Factor Rev. 29, 17–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fey D, Croucher DR, Kolch W, Kholodenko BN, Crosstalk and signaling switches in mitogen-activated protein kinase cascades. Front Physiol. 3, 355 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brancho D, Ventura JJ, Jaeschke A, Doran B, Flavell RA, Davis RJ, Role of MLK3 in the regulation of mitogen-activated protein kinase signaling cascades. Mol. Cell Biol 25, 3670–3681 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arthur JS, Ley SC, Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 13, 679–692 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Keshet Y, Seger R, The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 661, 3–38 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Roy SK, Shuman JD, Platanias LC, Shapiro PS, Reddy SP, Johnson PF, Kalvakolanu DV, A role for mixed lineage kinases in regulating transcription factor CCAAT/enhancer-binding protein-{beta}-dependent gene expression in response to interferon-{gamma}. J. Biol. Chem 280, 24462–24471 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Nithianandarajah-Jones GN, Wilm B, Goldring CE, Müller J, Cross MJ, ERK5: structure, regulation and function. Cell Signal. 24, 2187–2196 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB, Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl Acad. Sci. USA 108, 11121–11126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranganathan A, Pearson GW, Chrestensen CA, Sturgill TW, Cobb MH, The MAP kinase ERK5 binds to and phosphorylates p90 RSK. Arch Biochem Biophys. 449, 8–16 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Anjum R, Blenis J, The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 9, 747–758 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Kroczynska B, Kaur S, Katsoulidis E, Majchrzak-Kita B, Sassano A, Kozma SC, Fish EN, Platanias LC, Interferon-dependent engagement of eukaryotic initiation factor 4B via S6 kinase (S6K)- and ribosomal protein S6K-mediated signals. Mol Cell Biol. 29, 2865–2875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroczynska B, Joshi S, Eklund EA, Verma A, Kotenko SV, Fish EN, Platanias LC, Regulatory effects of ribosomal S6 kinase 1 (RSK1) in IFNλ signaling. J Biol Chem. 286, 1147–1156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neill LA, Bowie AG, Sensing and signaling in antiviral innate immunity. Curr Biol. 20, 328–333 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Randall RE, Goodbourn S, Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen Virol. 89, 1–47 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Christensen JE, de Lemos C, Moos T, Christensen JP, Thomsen AR, CXCL10 is the key ligand for CXCR3 on CD8+ effector T cells involved in immune surveillance of the lymphocytic choriomeningitis virus-infected central nervous system. J Immunol. 176, 4235–4243 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Wuest TR, Carr DJ, Dysregulation of CXCR3 signaling due to CXCL10 deficiency impairs the antiviral response to herpes simplex virus 1 infection. J Immunol. 181, 7985–7993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider WM, Chevillotte MD, Rice CM, Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoggins JW, Rice CM, Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1 519–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Kundu M, Viollet B, Guan KL, AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL, ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 15 741–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petherick KJ, Conway OJ, Mpamhanga C, Osborne SA, Kamal A, Saxty B, Ganley IG, Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem. 290, 11376–11383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Q, Deng X, Lu B, Cameron M, Fearns C, Patricelli MP, Yates JR 3rd, Gray NS, Lee JD, Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell. 18, 258–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroder K, Hertzog PJ, Ravasi T, Hume DA, Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 75 163–189 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M, Immune response in mice that lack the interferon-gamma receptor. Science. 259, 742–1745 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Kamijo R, Le J, Shapiro D, Havell EA, Huang S, Aguet M, Bosland M, Vilcek J, Mice that lack the interferon-gamma receptor have profoundly altered responses to infection with Bacillus Calmette-Guérin and subsequent challenge with lipopolysaccharide. J Exp Med. 178, 1435–1440 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Broek MF, Müller U, Huang S, Zinkernagel RM, Aguet M, Immune defence in mice lacking type I and/or type II interferon receptors. Immunol Rev. 148:5–18 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Pearl JE, Saunders B, Ehlers S, Orme IM, Cooper AM, Inflammation and lymphocyte activation during mycobacterial infection in the interferon-gamma-deficient mouse. Cell Immunol. 211, 43–50 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Suzuki Y, Orellana MA, Schreiber RD, Remington JS, Interferon-gamma: the major mediator of resistance against Toxoplasma gondii. Science. 240, 516–518 (1988). [DOI] [PubMed] [Google Scholar]
- 38.Buchmeier NA, Schreiber RD, Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc Natl Acad Sci USA. 82, 7404–7408 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D, Gene modulation and immunoregulatory roles of interferon gamma. Cytokine. 50, 1–14 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Diamond MS, Farzan M, The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 13, 46–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chadee DN, Kyriakis JM, MLK3 is required for mitogen activation of B-Raf, ERK and cell proliferation. Nat Cell Biol. 6, 770–776 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Drew BA, Burow ME, Beckman BS, MEK5/ERK5 pathway: the first fifteen years. Biochim Biophys Acta. 1825, 37–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi Y, Bowman JW, Jung JU, Autophagy during viral infection - a double-edged sword. Nat Rev Microbiol. [Epub ahead of print] (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang YP, Tsai CC, Huang WC, Wang CY, Chen CL, Lin YS, Kai JI, Hsieh CY, Cheng YL, Choi PC, Chen SH, Chang SP, Liu HS, Lin CF, Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation. J Biol Chem. 285, 28715–28722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, Green KY, Lopez-Otin C, Xavier RJ, Thackray LB, Virgin HW, Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 11, 397–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dikic I, Elazar Z, Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 19, 349–364 (2018) [DOI] [PubMed] [Google Scholar]
- 47.Zhou G, Bao ZQ, Dixon JE, Components of a new human protein kinase signal transduction pathway. J Biol Chem. 270, 12665–12669 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Chao TH, Hayashi M, Tapping RI, Kato Y, Lee JD, MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J Biol Chem. 274, 36035–36038 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Chayama K, Papst PJ, Garrington TP, Pratt JC, Ishizuka T, Webb S, Ganiatsas S, Zon LI, Sun W, Johnson GL, Gelfand EW, Role of MEKK2-MEK5 in the regulation of TNF-alpha gene expression and MEKK2-MKK7 in the activation of c-Jun N-terminal kinase in mast cells. Proc Natl Acad Sci USA. 98, 4599–4604 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun W, Kesavan K, Schaefer BC, Garrington TP, Ware M, Johnson NL, Gelfand EW, Johnson GL, MEKK2 associates with the adapter protein Lad/RIBP and regulates the MEK5-BMK1/ERK5 pathway. J Biol Chem. 276, 5093–5100 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Chiariello M, Marinissen MJ, Gutkind JS, Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol Cell Biol. 20, 1747–1758 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gotoh I, Adachi M, Nishida E, Identification and characterization of a novel MAP kinase kinase kinase, MLTK. J Biol Chem. 276, 4276–4286 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Blonska M, Shambharkar PB, Kobayashi M, Zhang D, Sakurai H, Su B, Lin X, TAK1 is recruited to the tumor necrosis factor-alpha (TNF-alpha) receptor 1 complex in a receptor-interacting protein (RIP)-dependent manner and cooperates with MEKK3 leading to NF-kappaB activation. J Biol Chem. 280, 43056–43063 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Sassano A, Majchrzak B, Deb DK, Levy DE, Gaestel M, Nebreda AR, Fish EN, Platanias LC, Role of p38alpha Map kinase in Type I interferon signaling. J Biol Chem. 279, 970–979 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Verma A, Deb DK, Sassano A, Kambhampati S, Wickrema A, Uddin S, Mohindru M, Van Besien K, Platanias LC, Cutting edge: activation of the p38 mitogen-activated protein kinase signaling pathway mediates cytokine-induced hemopoietic suppression in aplastic anemia. J Immunol. 168, 5984–5988 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Valledor AF, Sánchez-Tilló E, Arpa L, Park JM, Caelles C, Lloberas J, Celada A, Selective roles of MAPKs during the macrophage response to IFN-gamma. J Immunol. 180, 4523–4529 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Hardy PO, Diallo TO, Matte C, Descoteaux A, Roles of phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase in the regulation of protein kinase C-alpha activation in interferon-gamma-stimulated macrophages. Immunology. 128, 652–660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuzawa T, Kim BH, Shenoy AR, Kamitani S, Miyake M, Macmicking JD, IFN-γ elicits macrophage autophagy via the p38 MAPK signaling pathway. J Immunol. 189, 813–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marciano BE, Wesley R, De Carlo ES, Anderson VL, Barnhart LA, Darnell D, Malech HL, Gallin JI, Holland SM, Long-term interferon-gamma therapy for patients with chronic granulomatous disease. Clin Infect Dis. 39, 692–699 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Jarvis JN, Meintjes G, Rebe K, Williams GN, Bicanic T, Williams A, Schutz C, Bekker LG, Wood R, Harrison TS, Adjunctive interferon-γ immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS. 26, 1105–1113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu YJ, Cai WM, Li Q, Liu Y, Shen H, Mertens PR, Dooley S, Weng HL Long-term antifibrotic action of interferon-γ treatment in patients with chronic hepatitis B virus infection. Hepatobiliary Pancreat Dis Int. 10, 151–157 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, Cucolo L, Lee DS, Pauken KE, Huang AC, Gangadhar TC, Amaravadi RK, Schuchter LM, Feldman MD, Ishwaran H, Vonderheide RH, Maity A, Wherry EJ, Minn AJ, Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. 167, 1540–1554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Topalian SL, Drake CG, Pardoll DM, Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanagisawa K, Yamauchi H, Kaneko M, Kohno H, Hasegawa H, Fujita S, Suppression of cell proliferation and the expression of a bcr-abl fusion gene and apoptotic cell death in a new human chronic myelogenous leukemia cell line, KT-1, by interferon-alpha. Blood. 91, 641–648 (1998). [PubMed] [Google Scholar]
- 65.McKendry R, Pellegrini S, Kerr IM, Stark GR, Constitutive production of alpha and beta interferons in mutant human cell lines. J Virol. 68, 4057–4062 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M, In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Soonthornvacharin S, Rodriguez-Frandsen A, Zhou Y, Galvez F, Huffmaster NJ, Tripathi S, Balasubramaniam VR, Inoue A, de Castro E, Moulton H, Stein DA, Sánchez-Aparicio MT, De Jesus PD, Nguyen Q, König R, Krogan NJ, García-Sastre A, Yoh SM, Chanda SK, Systems-based analysis of RIG-I-dependent signalling identifies KHSRP as an inhibitor of RIG-I receptor activation. Nat Microbiol. 2, 17022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, Yángüez E, Andenmatten D, Pache L, Manicassamy B, Albrecht RA, Gonzalez MG, Nguyen Q, Brass A, Elledge S, White M, Shapira S, Hacohen N, Karlas A, Meyer TF, Shales M, Gatorano A, Johnson JR, Jang G, Johnson T, Verschueren E, Sanders D, Krogan N, Shaw M, König R, Stertz S, García-Sastre A, Chanda SK, Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 18, 723–735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolger AM, Lohse M, Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14,R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anders S, Pyl PT, Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarthy JD, Chen Y, Smyth GK, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Gene ontology analysis of putative ULK1 interactors in untreated versus IFNγ-treated KT-1 cells identified by mass spectrometry analysis.
Table S2. Gene ontology analysis of putative ULK1 interactors identified after IFNγ stimulation by mass spectrometry analysis.
Table S3. Genes differentially expressed in both Ulk1/2+/+ and Ulk1/2−/− MEFs after IFNγ treatment.
Table S4. Genes differentially expressed only in Ulk1/2+/+ MEFs after IFNγ treatment.
Table S5. Genes differentially expressed only in Ulk1/2−/− MEFs after IFNγ treatment.
Table S6. Gene ontology analysis of differentially expressed genes identified in both Ulk1/2+/+ and Ulk1/2−/− MEFs after IFNγ treatment.
Table S7. Gene ontology analysis of differentially expressed genes identified only in Ulk1/2+/+ MEFs after IFNγ treatment.
Table S8. Gene ontology analysis of differentially expressed genes identified only in Ulk1/2−/− MEFs after IFNγ treatment.
Fig. S1. ULK1/2 activity is not required for IFNγ-induced phosphorylation of STAT1.
Fig. S2. Effects of targeted disruption of Ulk1/2 gene expression on IFNγ-dependent gene transcription.
Fig. S3. Genes differentially expressed in both Ulk1/2+/+ and Ulk1/2−/− MEFs after IFNγ treatment..
Fig. S4. Genes differentially expressed only in Ulk1/2+/+ MEFs after IFNγ treatment.
Fig. S5. Genes differentially expressed only in Ulk1/2−/− MEFs after IFNγ treatment.
Fig. S6. Gene ontology analysis of IFNγ-dependent differentially expressed genes.
Fig. S7. Inhibition of autophagy does not affect transcription of antiviral ISGs.
Fig. S8. Genomic location for the ULK1 single-guide RNA in the human ULK1 open reading frame.