Abstract
Mitochondria are essential for the viability of eukaryotic cells, perform crucial functions in bioenergetics, metabolism and signaling, and have been linked to numerous diseases. Recent functional and proteomic studies revealed a remarkable complexity of mitochondrial protein organization. Protein machineries with diverse functions such as protein translocation, respiration, metabolite transport, membrane architecture and quality control interact with each other in dynamic networks. Here we discuss that the mitochondrial protein import machinery forms a housekeeping system that plays a central role in organizing the mitochondrial protein networks. The preprotein translocases not only deliver newly synthesized proteins to their proper intramitochondrial destination, but are also directly involved in establishing dynamic networks. Translocases form building blocks that cooperate with numerous mitochondrial protein complexes. Understanding mitochondrial protein organization requires an integrative view of organelle biogenesis and protein network formation.
Introduction
Mitochondria, the double membrane-bounded powerhouses, are a hallmark of eukaryotic cells. Derived from an α-proteobacteria-related ancestor, mitochondria have retained the high capacity and system of bacteria to synthesize ATP via oxidative phosphorylation, but are also deeply integrated into the metabolism and signaling pathways of their eukaryotic host cells.
Mitochondria consist of two membranes and two aqueous compartments (FIG. 1). The mitochondrial inner membrane possesses a several-fold larger surface than the outer membrane, resulting in an invagination of so-called cristae membranes that harbor the oxidative phosphorylation system, including the respiratory complexes I to IV and the F1FO-ATP synthase for production of ATP. A small set of proteins are encoded by the mitochondrial genome, including 13 proteins in humans and eight proteins in the model organism baker’s yeast. The mitochondrially encoded proteins include highly hydrophobic proteins that form core parts of the oxidative phosphorylation complexes of the mitochondrial inner membrane. Approximately 99% of mitochondrial proteins are encoded by nuclear genes, including originally prokaryotic genes that were transferred to the nucleus as well as genes coding for novel mitochondrial proteins developed by eukaryotic cells. Nuclear encoded mitochondrial proteins are equipped with specific targeting signals that direct the newly synthesized proteins from the cytosol to mitochondrial surface receptors and subsequently into the proper mitochondrial subcompartments1–4.
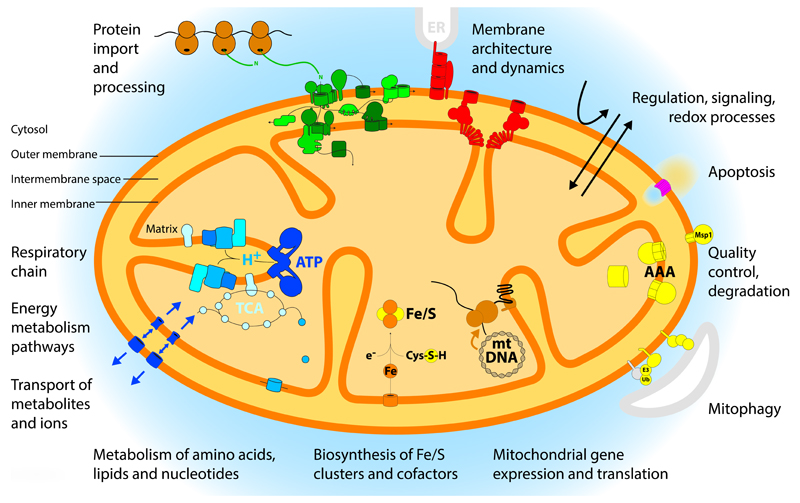
Figure 1. Overview of mitochondria and their functions.
Mitochondria consist of four compartments: outer membrane, intermembrane space, inner membrane and matrix. A large variety of functions have been assigned to mitochondrial proteins and protein complexes and are indicated in the figure: energy metabolism with respiration and synthesis of ATP; metabolism of amino acids, lipids and nucleotides; biosynthesis of iron-sulfur (Fe/S) clusters and cofactors; expression of the mitochondrial genome; quality control and degradation processes including mitophagy and apoptosis; signaling and redox processes; membrane architecture and dynamics; and the import and processing of precursor proteins that are synthesized on cytosolic ribosomes. AAA, ATP-dependent proteases of the inner membrane; E3, ubiquitin ligase; Msp1, mitochondrial sorting of proteins, extracts mistargeted proteins; TCA, tricarboxylic acid cycle; Ub, ubiquitin.
Whereas research in previous decades mainly focused on mitochondrial bioenergetics, studies in the past 10-15 years revealed an unexpected complexity and versatility of mitochondrial activities, integrating mitochondrial energetics with protein biogenesis, metabolic pathways, cellular signaling, stress response and apoptosis. It is becoming more and more evident that mitochondrial protein machineries do not function as independent units, but protein complexes of diverse function are physically and functionally connected. It is currently discussed how these complex networks are formed and maintained and what are the ordering principles behind them.
In this Review, we first discuss recent proteomic studies that showed a large spectrum of mitochondrial proteins and functions. Quantitative proteomics revealed for the first time absolute numbers for the abundance of all important mitochondrial machineries under different growth conditions, unveiling the protein import machinery as a housekeeping system under respiratory and non-respiratory conditions. We then address how preprotein translocases build core parts of dynamic protein networks that link organelle biogenesis, energy metabolism, membrane morphology and dynamics. Based on this integrative view, we discuss the connection of the mitochondrial protein import machinery to mitochondrial stress response, quality control and diseases.
Multifunctional mitochondria
Systematic analyses of the mitochondrial proteome provided a comprehensive overview not only of the mitochondrial protein complement, but also of the huge variety of functions performed by mitochondria, summarized in FIG. 1. We present an overview of the various functions of mitochondria and then discuss how absolute quantification of the mitochondrial proteome considerably shaped our view of mitochondrial activities.
Numerous functions of mitochondria
The typical textbook knowledge about mitochondria comprises the respiratory complexes and the F1FO-ATP synthase in the inner membrane cristae, transporters and channels for metabolites and ions in both mitochondrial membranes, and numerous metabolic pathways mainly localized to the matrix and inner membrane5 (FIG. 1). Major metabolic pathways concern the energy metabolism, such as the tricarboxylic acid cycle (TCA) also known as citric acid cycle or Krebs cycle, and the metabolism of amino acids, lipids and nucleotides. Electrons derived from oxidation of metabolites are fed into the respiratory chain that generates an electrochemical gradient by pumping protons from the matrix to the intermembrane space side. The proton gradient is then used to drive ATP synthesis by the F1FO-ATP synthase, as well as the import of precursor proteins and the transfer of some metabolites across the inner membrane.
Mitochondria perform two tasks that are essential for cell viability under all growth conditions. (i) They contain a complete system for the biosynthesis of iron-sulfur-clusters inside the organelle and provide a not yet identified essential compound for the cytosolic biosynthesis of iron-sulfur-clusters6. (ii) Mitochondrial import and maturation of precursor proteins are essential processes and thus a considerable number of protein import, processing and folding components are essential for cell viability1–4.
The mitochondrial matrix contains a complete genetic system, including the mitochondrial genome, numerous factors for maintaining, regulating and expressing the genome, and the mitochondrial ribosomes that differ in size and composition from cytosolic ribosomes7. Proteins encoded by the mitochondrial genome are typically inserted into the inner membrane in a co-translational mechanism by coupling translating ribosomes to the membrane integrated insertase, termed oxidase assembly (OXA)1,2,4,8.
Several machineries have been identified that control mitochondrial membrane architecture and dynamics. This includes protein factors that mediate fusion or fission of the mitochondrial membranes, in particular dynamin-related GTPases located at the outer and inner membranes, and membrane-shaping components. Mitochondria typically form a dynamic network in most cell types that is continuously remodeled by fusion and fission of the organelles9–12. The mitochondrial contact site and cristae organizing system (MICOS) is critical for maintaining the characteristic shape of inner membrane cristae13–15. MICOS and several further protein complexes are involved in forming contact sites between the mitochondrial outer and inner membranes to promote the transfer of proteins, lipids and metabolites13,14,16,17.
An intensively studied area includes mitochondrial signaling processes, regulation and quality control. Numerous cytosolic signaling cascades target mitochondria under physiological and pathophysiological conditions. The metabolic activity of the organelle serves as a measure for mitochondrial fitness and quality18, and elaborate pathways for mitochondrial stress responses, selective degradation of damaged mitochondria by mitophagy (autophagy)19 and programmed cell death (apoptosis) via mitochondria have been identified20,21. Mitochondria contain a set of internal proteases that are also involved in quality control and turnover of mitochondrial proteins22. In addition, mitochondria are a major site of cellular production of reactive oxygen species and contain numerous redox pathways23.
Quantitative analysis of the mitochondrial proteome
The large number of mitochondrial activities is overwhelming and has led to differential views of mitochondrial organization. Three stages of understanding of mitochondrial functions can be seen. (i) The original research focus on metabolism and ATP production established energetics and metabolism as hallmarks of mitochondria. (ii) The initiation of systematic proteomic studies on mitochondria about 15 years ago led to the identification of numerous new mitochondrial proteins. We currently estimate that mitochondria contain more than 1,000 (yeast) to 1,500 different proteins (humans)24–42. The functional classification of the mitochondrial proteome, based on the number of different proteins, suggested an unexpected perspective30,36. Only up to 15% of the different mitochondrial proteins were directly involved in energy metabolism, including the energy metabolizing pathways and all structural subunits of the oxidative phosphorylation system. However, 20-25% of the mitochondrial proteome were required to maintain, regulate and express the mitochondrial genome that codes for only ~1% of mitochondrial proteins. In addition, numerous components involved in signaling, regulation and membrane dynamics were identified. Thus, regulatory proteins emerged as important factors in mitochondria, suggesting a considerable revision of the textbook view of mitochondria as cellular powerhouses. (iii) The recent systematic quantification of the majority of the mitochondrial proteome yielded the absolute copy numbers of mitochondrial proteins per one cell30,43–47. As detailed in BOX 1, proteins involved in energy metabolism form by far the most abundant protein classes in respiring yeast mitochondria. The ~15% different mitochondrial proteins with a direct role in energy metabolism and respiration mentioned above represent more than half of the mitochondrial protein mass under respiratory conditions, reinforcing the original view of mitochondria as cellular powerhouses. Taking the various metabolic processes, oxidative phosphorylation and metabolite carriers/channels together, ~75% of the protein mass of respiring mitochondria are dedicated to metabolism and bioenergetics30.
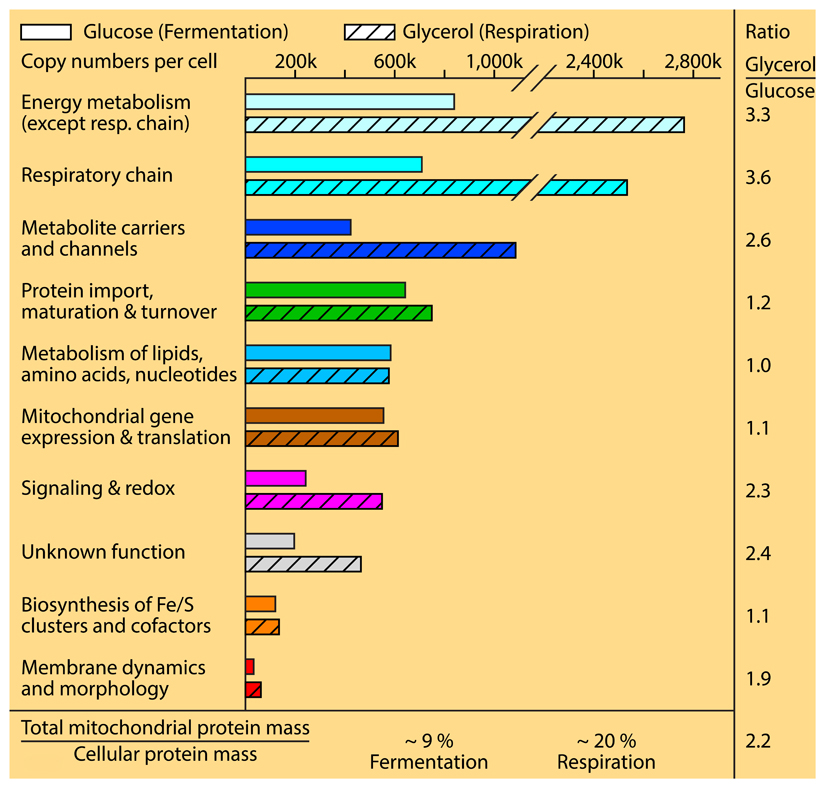
Box 1. The mitochondrial proteome: from fermentation to respiration.
The analysis of the mitochondrial proteome developed from the identification of individual proteins to the systematic determination of most mitochondrial proteins and the quantification of the relative abundance of mitochondrial proteins24–42. Recent studies determined the absolute copy numbers of proteins per cell30,43–47, providing unprecedented insight into the composition and functional organization of mitochondria. Proteins involved in energy metabolism, including enzymes, respiratory chain complexes, F1FO-ATP synthase and metabolite carriers/channels, represent by far the most abundant classes of mitochondrial proteins30. The abundance of these proteins is strongly regulated by the growth conditions. When shifting baker’s yeast from fermentation to respiratory conditions, their protein levels are increased about three-fold or more. The levels of proteins involved in signaling, redox processes, membrane dynamics and morphology are increased about two-fold from fermentation to respiration, comparable to the increase of the total mitochondrial protein mass. However, the two essential processes of mitochondria, protein import & maturation and biosynthesis of iron-sulfur clusters, are only mildly affected by different growth conditions; e.g., the absolute copy numbers per yeast cell of the proteins functioning in protein import, maturation and turnover are only changed from ~640,000 to 750,000 (i.e. a factor of 1.2) when cells are shifted from fermentation to respiration30. These essential machineries are thus identified as housekeeping systems that are present and active under all growth conditions. The protein classes involved in mitochondrial gene expression & translation and in the metabolism of lipids, amino acids and nucleotides are only mildly affected in their overall abundance by different growth conditions, however, differences can be observed for individual groups. For example, proteins involved in lipid metabolism are considerably more expressed under respiratory conditions, whereas proteins involved in amino acid metabolism are more strongly expressed under fermentable conditions30.
Thus, both seemingly controversial views, mitochondria as powerhouses vs. mitochondria as organelles with a huge number of different functions, have to be combined to understand the cellular significance of mitochondria. Based on absolute protein mass, metabolism and bioenergetics indeed represent the quantitatively major tasks of mitochondria and the term powerhouses is justified. However, many additional functions are of central importance for mitochondrial fitness, cellular growth and development. A striking example is the system for biosynthesis of iron-sulfur clusters that comprises less than 1% of the protein mass of respiring mitochondria, but is essential for the viability of eukaryotic cells6. Similarly, the machineries for mitochondrial membrane morphology and dynamics represent less than 1% of the mitochondrial protein mass30, although they play crucial roles in mitochondrial architecture, fusion and fission and thus are critical for maintaining the mitochondrial network in cells and for remodeling mitochondria under different growth conditions9–12. Mitochondria can thus be seen as super powerhouses that in addition to their predominant metabolic and energetic functions, are deeply integrated into cellular signaling, biosynthesis and dynamics by performing a multitude of functions. We discuss below that these functions are not independent tasks, but the various machineries and proteins are physically connected in large, dynamic networks.
Plasticity of the mitochondrial proteome
The mitochondrial content of a cell can vary considerably under different growth conditions and between different organisms and tissues25,28,30–32,48. The systematic analysis of yeast mitochondria revealed that the total mitochondrial protein mass represented ~9% of the cellular protein mass under fermentable conditions, where less activity of mitochondria is required. Under respiratory growth conditions, the mitochondrial protein mass is more than doubled to ~20% of the cellular protein mass30. As outlined in BOX 1, the changes from fermentation to respiration are quite different for mitochondrial proteins belonging to different functional classes. The strongest increase toward respiration is observed for proteins directly involved in energy metabolism and respiration with a more than three-fold increase of the absolute protein copy numbers of these proteins per cell. Proteins functioning in signaling, redox processes and membrane dynamics are increased about two-fold, like the overall increase of mitochondrial mass30.
Remarkably, the protein classes responsible for the two mitochondrial processes, which are essential for cell viability in all cell types and growth conditions, protein biogenesis & folding and biosynthesis of iron-sulfur clusters, are only moderately altered in their overall copy numbers upon shift of yeast cells from fermentation to respiration30. Thus, these systems are very well equipped already under non-respiratory conditions and are only slightly increased in their abundance during respiration. It seems that these systems are so central to mitochondrial and cellular behavior that their protein complement is largely present under all conditions, although their activity has to be strongly increased under respiratory conditions. The protein import machinery has to import more than double the amount of proteins during respiration and thus it is evident that the machinery is working at considerably lower yield under fermentation. We conclude that the protein import machinery and the system for biosynthesis of iron-sulfur clusters are central housekeeping systems of yeast mitochondria and are not or only moderately regulated by their copy numbers. Indeed, studies on the phosphorylation of the main protein entry gate, the translocase of the outer membrane (TOM) complex, revealed that at least four different cytosolic signaling systems regulate the TOM activity by phosphorylation, leading to a sophisticated pattern of stimulatory and inhibitory effects depending on the kinase and TOM subunits involved49–52. The advantage of a stable presence of housekeeping systems and their regulation by reversible modification such as phosphorylation is the rapid response to changing conditions. When an increased capacity of the protein import machinery would require an increased gene expression, translation and import, the system would be rather slow in adapting to different requirements. Similarly, inhibitory effects are faster when directly executed by protein modification than by the slower copy number reduction. For example, upon fermentable growth, where less translocation of metabolites into and out of mitochondria is needed, the activity of the TOM receptor of 70 kDa (Tom70), which is critical for targeting of metabolite carriers, is inhibited by phosphorylation, leading to an immediate decrease of carrier import into mitochondria52. Therefore, the presence of essential housekeeping systems with largely stable protein copy numbers and the regulation of these systems by post-translational modification will provide a high flexibility of mitochondrial biogenesis and plasticity.
Protein assembly and functional networks
The analysis of protein import from the cytosol into mitochondria was originally based on the assumption that one central pathway is responsible for translocating the 1,000-1,500 different proteins to their mitochondrial destination, however, the characterization of precursor proteins carrying different targeting signals revealed a higher complexity. We first present that mitochondria use at least five major protein import pathways, each one directed by a different type of targeting signal. Then we discuss the next level of complexity as preprotein translocases do not operate as isolated units, but are connected to numerous mitochondrial protein complexes that belong to different, at first glance unrelated functional categories.
Five major import pathways of precursor proteins into mitochondria
The presequence pathway is the best known mitochondrial protein import pathway responsible for the transport of ~60% of all mitochondrial proteins53. The precursor proteins carry amino-terminal targeting signals, termed presequences, which form positively charged amphipathic α-helices. These presequences are typically recognized by the TOM receptors of ~20 and ~22 kDa (Tom20 and Tom22)54,55 on the mitochondrial surface and direct translocation through the main protein translocation channel of the outer membrane of ~40 kDa (Tom40)2,4,56,57 (FIG. 2). Upon translocation across the outer membrane, the preproteins are engaged by the presequence translocase of the inner membrane (TIM23 complex) that directs their transfer across the inner membrane58–61. The membrane potential (Δψ) across the inner membrane (negative on the inside) activates the Tim23 channel and drives the positively charged presequences toward the matrix62–65. The presequence translocase-associated motor (PAM) contains the mitochondrial heat shock protein 70 (mtHsp70) as central ATP-driven chaperone66. Together with five co-chaperones, PAM promotes translocation of the entire polypeptide chain into the matrix67–72. The presequences are removed by the mitochondrial processing peptidase (MPP)2,4,73. Additional processing enzymes in the matrix are involved in quality control functions as mitochondrial proteins can be degraded according to the N-end rule pathway, depending on the presence of stabilizing or destabilizing amino acid residues at the amino-termini of proteins53,74. The intermediate cleaving peptidase of 55 kDa (Icp55) and the octapeptidyl peptidase (Oct1) remove destabilizing amino acid residues, which can be located at the amino-termini of imported proteins after cleavage by MPP, and thus generate stable amino-termini that are less prone to degradation by matrix proteases53,75,76. With the help of mtHsp70 and further chaperones such as Hsp60-Hsp1077,78, the proteins are then folded to their active form.
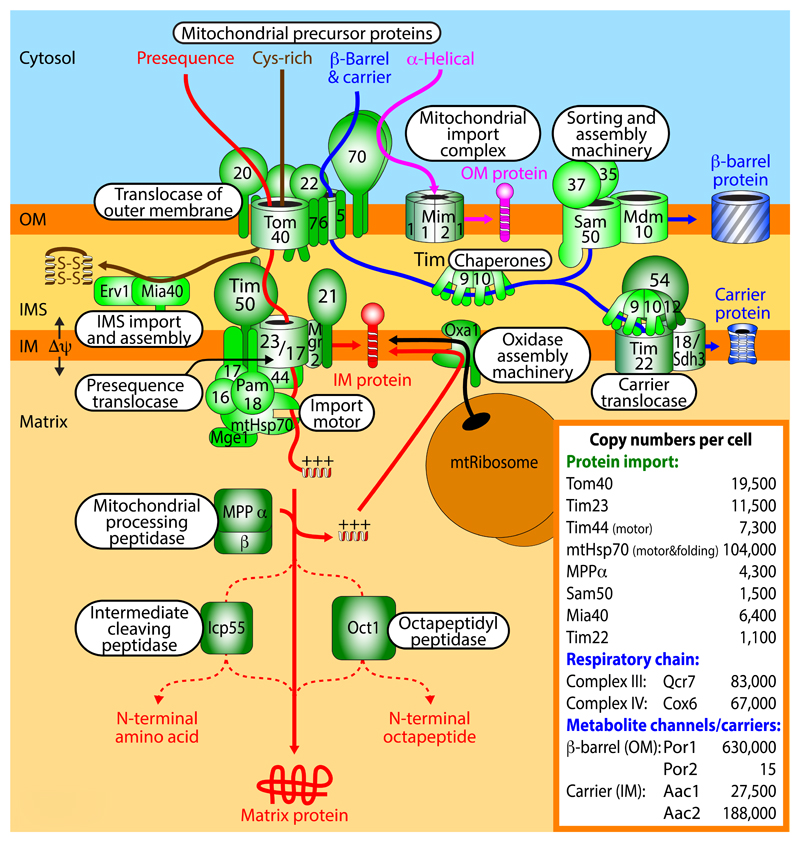
Figure 2. Protein import pathways into mitochondria.
Five major pathways of mitochondrial protein import have been identified. The protein import machineries have been well conserved from fungi (shown in this figure) to mammals (shown in BOX 4). First, the presequence pathway transports presequence-carrying cleavable preproteins through the translocase of the outer membrane (TOM) and the presequence translocase of the inner membrane (TIM23) with the presequence translocase-associated motor (PAM). The membrane potential Δψ across the inner membrane (IM) activates the TIM23 channel and drives translocation of the positively charged presequences into the matrix. The presequences are removed by the mitochondrial processing peptidase (MPP) and additional proteolytic processing can occur by the intermediate cleaving peptidase (Icp55) or the octapeptidyl peptidase (Oct1). Cleavable IM proteins are either laterally released from the TIM23 complex or are transported via the matrix and inserted into the IM by the oxidase assembly (Oxa1) insertase. IM proteins synthesized on mitochondrial ribosomes are also inserted by Oxa1. Second, cysteine-rich proteins destined for the intermembrane space (IMS) are imported through the TOM complex and are recognized by the mitochondrial IMS import and assembly protein (Mia40) that functions as oxidoreductase to insert disulfide bonds into the imported proteins. The sulfhydryl oxidase Erv1 forms a disulfide relay with Mia40, transferring disulfides from Erv1 to Mia40 to imported proteins. Third, the precursors of non-cleavable IM proteins such as the carrier proteins are imported by the TOM complex, followed by transfer to the small TIM chaperones in the IMS and insertion into the IM by the TIM22 carrier translocase. Fourth, the precursors of outer membrane (OM) β-barrel proteins use the TOM complex and small TIM chaperones and are inserted into the OM by the sorting and assembly machinery (SAM). Fifth, many OM proteins with α-helical transmembrane segments are inserted into the membrane by the mitochondrial import (MIM) complex. α-helical OM proteins typically do not use the Tom40 channel, but Tom70 can be involved in their recognition. Inset, assessment of absolute copy numbers of mitochondrial proteins in a respiring yeast cell30. The porin isoform Por1 of the OM is one of the most abundant mitochondrial proteins, whereas the isoform Por2 is one of the least abundant proteins.
Presequence-carrying precursors that become integrated into the inner membrane follow two distinct routes (FIG. 2). A number of presequence-carrying preproteins possess a hydrophobic sorting signal behind the matrix targeting signal. This sorting signal arrests translocation in the TIM23 complex and the lateral gatekeeper Mgr2 permits a release of the protein into the inner membrane (stop transfer pathway)79. Other inner membrane proteins are first transported into the matrix and are translocated into the inner membrane by the OXA insertase that has been conserved from bacteria to mitochondria and is also used by mitochondrially synthesized proteins (conservative sorting)8,80–83.
Most other protein import pathways also use the TOM channel for preprotein translocation across the outer membrane84,85 (FIG. 2), although the dependence on the three TOM receptors Tom20, Tom22 and Tom70 and the mode of delivery from the cytosol to TOM can differ86–88. The carrier pathway is dedicated to the import of hydrophobic multi-spanning inner membrane proteins, which do not possess amino-terminal presequences, but distinct types of internal targeting signals that are not fully defined, yet include hydrophobic elements. Cytosolic chaperones of the Hsp90 and Hsp70 classes deliver the hydrophobic precursor proteins to the receptor Tom7089. The precursors are released from the chaperones, translocated through the Tom40 channel in a loop formation90,91 and engaged by small TIM chaperones of the intermembrane space to prevent aggregation of the hydrophobic precursors92–94 (FIG. 2). The small TIM chaperones guide the carrier precursors to the carrier translocase of the inner membrane (TIM22 complex) that operates in a Δψ-dependent manner to drive membrane insertion of the multi-spanning proteins95–99.
Many proteins of the mitochondrial intermembrane space contain characteristic cysteine motifs that become oxidized to form stabilizing disulfide bonds in the mature proteins. The mitochondrial intermembrane space import and assembly (MIA) system consists of two main components: the oxidoreductase with disulfide isomerase activity and intermembrane space receptor of ~40 kDa, Mia40100,101, and the sulfhydryl oxidase essential for respiration and viability, Erv1102 (FIG. 2). Upon translocation of the precursors through the Tom40 channel, Mia40 recognizes the precursors that contain a mitochondrial intermembrane space sorting signal, typically consisting of a hydrophobic element flanked by a cysteine residue103–105. Mia40 forms a transient disulfide bond with the precursor protein and then transfers disulfide bonds to the imported protein, leading to an oxidation and stabilization of the protein by intramolecular disulfide bonds106,107. Upon each transfer of a disulfide bond to an imported protein, cysteines of Mia40 become reduced and are re-oxidized by Erv1. In this disulfide relay, disulfide bonds are thus transferred from Erv1 to Mia40 to imported intermembrane space proteins.
The mitochondrial outer membrane contains different classes of membrane proteins: single-spanning and multi-spanning proteins with α-helical transmembrane segments; and β-barrel proteins. The precursors of β-barrel proteins are initially translocated by the TOM complex to the intermembrane space and interact with small TIM chaperones like the carrier precursors108 (FIG. 2). Insertion of β-barrel precursors into the outer membrane is mediated by the sorting and assembly machinery (SAM)109–111 in a step-wise process that involves translocation into the SAM channel and lateral release into the lipid phase of the membrane112. The carboxy-terminal β-strand of the precursor functions as β-signal that directs insertion via SAM113. The β-signal and the central channel-forming protein Sam50 have been conserved from bacteria to humans114. α-Helical outer membrane proteins follow distinct import routes that in most cases do not involve the Tom40 channel. The sorting signal is typically contained within the α-helical transmembrane segment(s) and flanking positively charged amino acid residues. Single-spanning proteins with amino-terminal membrane anchor (signal-anchored proteins) as well as multi-spanning outer membrane proteins can use the mitochondrial import (MIM) channel for membrane insertion, assisted by the receptor Tom70 at least in the case of multi-spanning proteins115–120. In case of single-spanning proteins with carboxy-terminal membrane anchor (tail-anchored proteins) and some multi-spanning proteins, the lipid composition of the membrane seems to be important, yet the exact molecular mechanism is unknown121–124. The views reach from a protein-independent insertion directly into the phospholipid membrane to MIM complex-assisted insertion or to the possible involvement of an as yet unknown proteinaceous insertase of the outer membrane.
Abundance and versatility of import machineries
The inset of FIG. 2 shows the absolute copy numbers of characteristic translocase components in a respiring yeast cell, and for comparison also the abundance of respiratory complexes, metabolite channels and carriers of the mitochondrial membranes30. The TOM complex is the most abundant translocase consistent with its role in feeding precursors into at least four distinct down-stream translocase systems. The abundance of the TIM23 presequence translocase fits to the major role of the presequence import pathway. Interestingly, the central motor component mtHsp70 is about ten times more abundant than Tim23 and other motor subunits such as Tim44. mtHsp70 plays a dual role: a fraction of mtHsp70 molecules act in the TIM23-associated motor PAM to drive preprotein import, whereas the major fraction of mtHsp70 is dedicated to protein folding in the mitochondrial matrix. Sam50 and Tim22 are present in quite low amounts, reflecting their more specialized role in protein sorting. A comparison with the abundance of their major substrates, however, underscores the importance and activity of the translocases. The outer membrane β-barrel metabolite channel porin (isoform Por1) is one of the most abundant mitochondrial proteins and also the inner membrane metabolite carriers are of high abundance.
The TOM complexes can form different types of dynamic supercomplexes: a TOM-SAM supercomplex for efficient transfer of β-barrel precursors and a two membrane-spanning TOM-TIM23-preprotein supercomplex125–130. TOM also interacts with the small TIM chaperones, and in mammals TIM29 of the TIM22 complex was found to associate with TOM85,131,132. The differential abundance of the translocases (FIG. 2, inset) indicates that indeed sufficient TOM complexes are available to form the different supercomplexes. It is currently discussed whether separate pools of TOM complexes exist for different import pathways or whether the TOM complexes are freely interchangeable in one large dynamic pool. The translocation of several intermembrane space precursor proteins across the outer membrane depends on Tom40 but does not require the TOM receptor domains133,134 and competition experiments suggest that intermembrane space precursors and presequence-carrying precursors do not use the same TOM complexes133, supporting the view of distinct forms of the main protein entry gate. We speculate that in addition to the full-size TOM complex, which contains Tom40 channels, all three receptors and three small Tom proteins85, simplified forms of TOM complexes may exist. These simplified TOM complexes may just contain the Tom40 channel and possibly some of the small Tom subunits and may be dedicated to the import of e.g. intermembrane space precursors, which are recognized by the receptor Mia40 and do not depend on classical TOM receptors. The differential phosphorylation of TOM complexes by cytosolic signaling cascades also contributes to a heterogeneity of TOM complexes and thus favors the engagement in distinct import routes in a signal-controlled manner49–52.
In metazoans, the presequence translocase exists in two forms with a differential tissue distribution, containing either the stably expressed housekeeping subunit TIM17B (skeletal muscle) or the stress-regulated subunit TIM17A (brain)135. Under stress conditions, the levels of TIM17A are decreased by two means, decreased synthesis and increased degradation by the ATP-dependent AAA protease of the inner membrane that is exposed to the intermembrane space (i-AAA protease) (FIG. 3), promoting the induction of a mitochondrial unfolded protein response136. Thus, whereas the overall abundance of the mitochondrial protein import machinery of rapidly growing uni-cellular organisms like yeast is quite stable under different metabolic conditions, the abundance of tissue-specific isoforms can be regulated in metazoans, suggesting an additional regulatory level in multi-cellular organisms135–137.
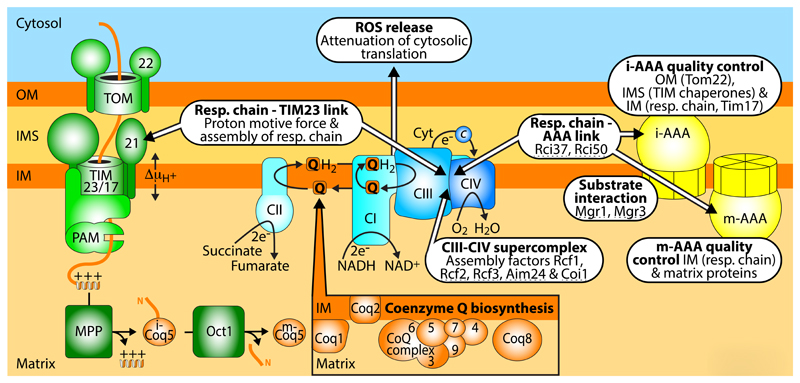
Figure 3. Interaction network of respiratory complexes, biogenesis and quality control machineries.
Supercomplexes of the mitochondrial respiratory chain are integrated into functional networks with the presequence translocase TIM23 (see BOX 2) and the AAA proteases of the inner membrane (IM). The ATP-dependent AAA proteases not only degrade several IM proteins, but also selected proteins of the matrix, intermembrane space (IMS) and outer membrane (OM), functioning as a quality control system of mitochondria. Several respiratory chain-AAA linker proteins, AAA-substrate adapter proteins, and assembly factors for respiratory supercomplexes were identified in fungi (indicated by dashed lines). The coenzyme Q (CoQ, Q) biosynthetic complex on the matrix side of the IM provides CoQ for the respiratory chain and further enzymes. The precursors of the CoQ complex are imported by the TOM and TIM machineries. Proteolytic processing in the matrix can involve two steps like for the precursor of Coq5. The mitochondrial respiratory chain is a main source for the generation of ROS that can exert harmful effects but also function in signaling. Targeting of the cytosolic translation machinery by ROS leads to a decreased protein synthesis, providing a link between the status of the respiratory chain and protein biogenesis.
Elements of different import pathways can be combined for the creation of new import pathways. For example, the single-spanning outer membrane protein Om45 contains a large domain in the intermembrane space. The precursor is first transported by the presequence pathway through the TOM complex and interacts with the TIM23 complex, followed by escape into the intermembrane space and insertion into the outer membrane by a reverse action of the MIM complex138,139. Further examples of the versatility of the mitochondrial import machinery are cleavable carboxy-terminal targeting signals that are imported via the presequence pathway and are removed by matrix or intermembrane space peptidases, followed by differential sorting to intramitochondrial destinations140–142. Thus, the diversity and versatility of mitochondrial protein import pathways go substantially beyond the known five major pathways and we expect that the systematic analysis of the large number of substrates will reveal the existence of further import routes and possibly also of further translocases.
Respiratory chain interactions link bioenergetics, biogenesis and quality control
The respiratory chain complexes of the mitochondrial inner membrane form the center of a large network that connects bioenergetics to biogenesis, regulation and turnover processes (FIG. 3). The respiratory complexes I (NADH:ubiquinone oxidoreductase), III (cytochrome c reductase) and IV (cytochrome c oxidase) assemble into large supercomplexes. The I-III-IV supercomplexes are also termed respirasomes143–147. The existence of respiratory supercomplexes is now generally accepted, however, different views exist about the functions of these large assemblies144,146. The supercomplexes may influence the assembly and stability of respiratory complexes, regulate the activity of the complexes and/or reduce the formation of reactive oxygen species (ROS). Various factors involved in the formation of respiratory supercomplexes have been reported (FIG. 3), yet it is currently discussed if they mainly function in the assembly of individual respiratory complexes or in the formation of supercomplexes144,146,148–151.
Preprotein translocases are not only in crosstalk with each other, but form physical contacts with other mitochondrial machineries, including machineries involved in mitochondrial energy metabolism. The TIM23 presequence translocase forms a hub in the sorting of preproteins at the inner membrane and cooperates with a remarkable number of different protein complexes. In addition to the complexes directly involved in preprotein import (TOM and PAM), the TIM23 complex forms supercomplexes with the respiratory complexes III and IV as well as with the ADP/ATP carrier (FIG. 3; BOX 2). These interactions of the TIM23 complex support protein import under energy-limiting conditions152–154; and in human mitochondria, TIM23-respiratory chain interactions have been shown to promote assembly of respiratory complexes155–157 (BOX 2). The respiratory complexes as well as the ADP/ATP carrier are several-fold more abundant than the TIM23 complex30 (FIG. 2, inset) and thus only a fraction of them are engaged in the interaction with TIM23. Respiratory complexes are preferentially located in cristae membranes, yet a smaller fraction is found in the inner boundary membrane, which is adjacent to the outer membrane158, and can thus interact with the TIM23 complexes. The interaction is likely not permanent, but TIM23 and respiratory complexes form dynamic supercomplexes152,153.
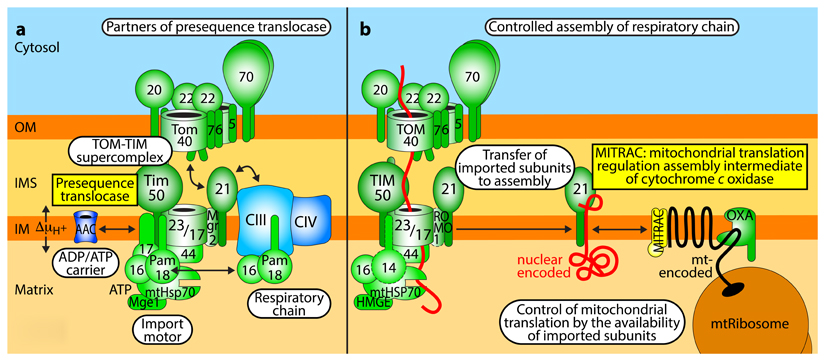
Box 2. Mitochondrial presequence translocase and respiratory chain assembly and function.
a. Interaction network of presequence translocase
The TIM23 complex of the inner membrane (IM) is a central junction in the presequence import pathway and interacts with several partner complexes in a dynamic manner: with the TOM complex during preprotein transfer from the outer membrane (OM) to the IM, forming a TOM-TIM-preprotein supercomplex; with the ATP-driven presequence translocase-associated motor (PAM); with the respiratory chain complexes III and IV that generate an electrochemical proton gradient (ΔμH+) driving preprotein insertion152,153; and with the ADP/ATP carrier that also supports preprotein translocation154,229. The translocase subunit Tim21 functions as a dynamic coupling factor that interacts with TOM and the respiratory supercomplex III-IV in an alternating manner. In fully active, respiring yeast mitochondria, the activity of the presequence translocase drives preprotein import efficiently. However, when the respiratory activity is decreased, coupling of the translocase to machineries involved in bioenergetics is beneficial to maintain the energy-dependent action of the translocase152–154. Preprotein translocases in the immediate vicinity of proton pumping respiratory complexes likely experience an increased proton motive force (localized proton gradients) and thus under energylimiting conditions preprotein insertion into the IM is still possible152,153, ensuring that the biogenesis of respiratory complexes is ongoing e.g. under limited food supply.
b. Coupling of presequence translocase to respiratory chain assembly
The characterization of respiratory chain biogenesis in human mitochondria revealed a further level of cooperation between the protein import machinery and the respiratory chain, mediated by the mitochondrial translation regulation assembly intermediate of cytochrome c oxidase (MITRAC)155–157. MITRAC comprises several assembly intermediate complexes of the respiratory chain and plays a dual role. (i) It links the presequence translocase to respiratory chain assembly intermediates via the TIM21-mediated transfer of imported proteins from TIM23 to MITRAC156. (ii) MITRAC cooperates with the machineries for mitochondrial protein synthesis and insertion by adapting the efficiency of mitochondrial translation to the import of nuclear encoded partner proteins (translational plasticity). MITRAC assembly factors bind to partially synthesized/membrane inserted proteins such as the highly hydrophobic COX1 protein and delay their synthesis till the appropriate partner proteins have been imported, thus promoting a proper balance of nuclear and mitochondrially encoded subunits157.
OXA, insertion machinery (oxidase assembly). The role of human TIM14/PAM18 and ROMO1/MGR2 in the TIM23 complex has not been defined so far (indicated by dashed borders).
A further link between the mitochondrial respiratory chain and protein biogenesis was found by analyzing the effects of mitochondrially generated ROS. The mitochondria respiratory chain is a major source for ROS. Stress conditions and dysfunction of the respiratory chain can lead to increased ROS production with oxidative damage of proteins, DNA and membranes. ROS can also perform various signaling functions and thus mitochondrially produced ROS can signal the functional state of mitochondria23,146,159. ROS was found to target redox-sensitive cysteine residues (redox switches) of the cytosolic translation apparatus, including the ribosome and translation factors159. When mitochondrial dysfunction leads to increased ROS production, the efficiency of translation is decreased in a reversible manner (FIG. 3). The respiratory chain thus participates in controlling cytosolic protein synthesis to decrease the protein load under mitochondrial stress conditions159.
Coenzyme Q (CoQ), also termed ubiquinone, is a central molecule for the function of the respiratory chain160,161. CoQ is a redox-active lipid that mediates electron transfer from respiratory complexes I and II to complex III (FIG. 3) and functions as cofactor of many further enzymes. Several enzymes of CoQ biosynthesis were only identified recently, using systematic mass spectrometry profiling at the proteomic, lipidomic and metabolomic level30,75,160,161. The CoQ biosynthetic complex located at the matrix side of the mitochondrial inner membrane contains numerous enzymes involved in CoQ biosynthesis as well as lipids and cofactors. All protein subunits of this dynamic CoQ biosynthetic complex are encoded by nuclear genes and have to be imported by the TOM and TIM machineries160,161. The precursor of the methyltransferase Coq5 is processed twice. The first processing by MPP generates an instable intermediate and thus a second processing by the octapeptidyl peptidase is required to generate the stable mature Coq5 enzyme (FIG. 3). Disturbance of the processing by Oct1 leads to CoQ deficiency and respiratory defects75. Import and specific processing of the Coq precursor proteins thus functionally link the mitochondrial protein import machinery and the CoQ biosynthetic complex.
The mitochondrial inner membrane carries two large ATP-dependent protease complexes, the i-AAA protease and the matrix-exposed m-AAA protease22,162–164. These proteases are main elements of a quality control system for protein turnover and processing in mitochondria. AAA proteases not only cleave/degrade inner membrane proteins such as some subunits of respiratory complexes and preprotein translocases, but are also involved in the quality control and turnover of selected matrix, intermembrane space and outer membrane proteins (FIG. 3). In fungi, the adapter proteins Mgr1 and Mgr3 associate with the i-AAA protease and promote substrate recognition by the protease162,164. A recent proteomic study of yeast mitochondria identified the respiratory chain interacting proteins Rci37 and Rci50 and demonstrated that they also interacted with the AAA proteases, Rci37 with the m-AAA protease and Rci50 with the i-AAA protease30, revealing specific connections between respiratory complexes III and IV and the inner membrane quality control system.
The functional network of the mitochondrial respiratory chain thus includes respiratory supercomplexes and machineries for protein biogenesis, cofactor biosynthesis, regulation and quality control.
Mitochondrial membrane architecture and protein interactions
Contact sites between the mitochondrial outer and inner membranes and between the outer membrane and the endoplasmic reticulum (ER) are crucial elements of a large network that functions in protein and lipid biogenesis, membrane architecture and dynamics, metabolite/ion transport, mitochondrial dynamics and inheritance (FIG. 4). Preprotein translocases form central building blocks of this ER-mitochondria organizing network (ERMIONE)165. The TOM and SAM complexes of the outer membrane interact with the large MICOS morphology complex of the inner membrane13–15,17,166–169. MICOS is enriched at crista junctions170,171, the tubular entry gates into the cristae lumen. The largest MICOS subunit, Mic60, plays an important role in the formation of outer-inner membranes contact sites. In addition, Mic60 transiently interacts with the receptor and oxidoreductase Mia40 of the intermembrane space assembly machinery13. MICOS thus helps to position the down-stream machineries MIA and SAM close to the main protein import channel TOM and promotes the efficient import of cysteine-rich precursors into the intermembrane space and of β-barrel precursors into the outer membrane13,166.
Figure 4. Mitochondrial organizing network.
The mitochondrial contact site and cristae organizing system (MICOS) of the inner membrane (IM) and the protein translocases TOM and SAM of the outer membrane (OM) form the core of a large ER-mitochondria organizing network (ERMIONE) that includes multiple dynamic interactions: to the ER-mitochondria encounter structure (ERMES); to further ER-mitochondria contact sites that involve the receptor Tom70 and IP3 receptors or the lipid transfer protein Lam6/Ltc1, as well as to vacuole-mitochondria contact sites (including Tom40 and the bridging protein Vps39/Vam6); to the kinase PINK1 and the metabolite channel VDAC (porin); to the mitochondrial intermembrane space (IMS) protein import and assembly system (Mia40); to respiratory chain complexes, the F1FO-ATP synthase, and the fusion protein OPA1 of the IM; and to mtDNA nucleoids (with the mtDNA packaging factor, termed mitochondrial transcription factor A, TFAM)256 of the matrix. Most components shown have been functionally conserved from yeast to humans; proteins that have been characterized in fungi only are indicated by a dashed border, whereas proteins that have been characterized in metazoans so far are bordered in red. In sum, ERMIONE forms a membrane-spanning system for the coordination of protein and lipid biogenesis, energetics, inheritance and quality control of mitochondria.
The MICOS-SAM-TOM core of ERMIONE undergoes multiple interactions with further mitochondrial machineries. FIG. 4 gives an overview of this huge network. Most interactions of ERMIONE partners have been observed by biochemical means via direct physical associations or by genetic means, i.e. synthetic growth defects of double mutants. Analyses in vivo and in organello provided evidence for functional crosstalks. The molecular mechanisms governing this network and the regulation of mitochondrial functions by interaction of diverse machineries are currently the subject of intensive research. (i) In the inner membrane, Mic10, a core component of MICOS172,173, and its partner protein Mic27 are in dynamic contact with the dimeric F1FO-ATP synthase that shapes cristae rims, leading to a crosstalk between the two major membrane-shaping machineries of the inner membrane, MICOS and F1FO-ATP synthase147,174,175. Assembly of the Mic10-containing subcomplex of MICOS is linked to respiratory complexes and the mitochondria-specific dimeric phospholipid cardiolipin176. In addition, MICOS is connected to the machineries for mitochondrial fusion, including the inner membrane fusion protein optic atrophy 1 (OPA1, termed mitochondrial genome maintenance 1 [Mgm1] in yeast)147,177,178. (ii) By mutant studies, MICOS has been functionally linked to nucleoid aggregation and inheritance of mtDNA179,180. The underlying molecular mechanisms will require further analysis. (iii) At the outer membrane, the SAM complex not only directly interacts with a fraction of TOM complexes in TOM-SAM supercomplexes, but is also in exchange with the ER-mitochondria encounter structure (ERMES) that links the mitochondrial outer membrane to the ER (FIG. 4). The outer membrane β-barrel protein mitochondrial distribution and morphology 10 (Mdm10) is a subunit of both SAM, where it functions in TOM biogenesis, and ERMES, where it contributes to lipid transfer and maintenance of mitochondrial morphology181–184. The shuttling of Mdm10 between SAM and ERMES is regulated by Tom7. The small protein Tom7 has a dual localization. It is mainly located in the TOM complex, but additional Tom7 molecules act as regulatory factors that promote Mdm10 transfer to ERMES185–188. Non-assembled Tom7 retards TOM assembly by shifting Mdm10 from the SAM-form to the ERMES-form, representing a direct regulatory mechanism when an excess of non-assembled TOM subunits accumulate in mitochondria. (iv) The major outer membrane metabolite channel porin, also termed voltage-dependent anion channel (VDAC), interacts with MICOS as well as the TOM complex14,189, linking metabolite transport to ERMIONE. (v) Tom70 performs receptor functions in the TOM complex, but is also found in pools outside of the TOM complex and plays crucial roles in forming ER-mitochondria contact sites (FIG. 4). Tom70 and its isoform Tom71 interact with the lipid transfer protein anchored at membrane contact sites termed Lam6/Ltc1, which contains lipid binding sites and regulates contact sites between mitochondria, ER and further organelles190,191. Mammalian TOM70 also interacts with inositol trisphosphate (IP3) receptors of the ER to promote Ca++ transfer from the ER to mitochondria192. The TOM complex with is central component Tom40 participates in the formation of vacuole-mitochondria contact sites, termed vacuole and mitochondria patch (vCLAMP), involving the vacuolar GTPase Ypt7 and the bridging protein Vps39/Vam6190,193. (vi) Moreover, Tom70/Tom71 have been linked to mitochondrial quality control and degradation systems. Vesicles can be released from the mitochondrial outer membrane that may direct selected cargo to degradation194–196 and Tom70/71 were found to be required for the formation of at least some mitochondria-derived vesicles197. The outer membrane ATPase Msp1 promotes the extraction of mistargeted proteins198,199. Upon accumulation of non-imported precursor proteins, Msp1 is recruited to Tom70 via the peripheral membrane protein Cis1, leading to removal of non-imported proteins and their degradation by the proteasome200. (vii) Finally, as discussed in the following chapter, TOM and MICOS are involved in the accumulation of the PTEN-induced putative kinase 1 (PINK1) at the outer membrane, under conditions of mitochondrial dysfunction that lead to removal of damaged mitochondria by mitophagy19,201–203.
The mitochondrial membranes thus contain at least two large protein networks, both containing TOM complexes: the TOM-TIM23-respiratory chain-AAA network coupling protein import to bioenergetics and quality control, and the MICOS-SAM-TOM-ER network (ERMIONE) that links protein biogenesis to membrane contact sites and membrane morphology. Whereas MICOS is enriched at crista junctions, TOM-TIM23-preprotein supercomplexes are preferentially found in a distance of ~30-60 nm away from crista junctions127. At present, it is open if these two large networks function independently of each other or if the exchange of TOM complexes and further components between the networks may provide a dynamic coordination between protein biogenesis, energetics, membrane morphology and quality control. Though substantial future work will be required for understanding ERMIONE on a molecular and functional level, the identification of these networks clearly demonstrates that mitochondrial machineries do not function as stand-alone units, but are intimately linked to each other.
Protein import and pathophysiology
The efficiency of protein import into mitochondria is a sensitive indicator of the energetic state and the fitness of mitochondria. Various disorders of mitochondrial respiration and metabolism lead to a reduction of the inner membrane potential204,205. Since the membrane potential is crucial for protein translocation into and across the inner membrane, the import of preproteins is diminished1–4. Defects of protein homeostasis in the mitochondrial matrix by the accumulation of misfolded proteins also lead to a reduced protein import, likely by disturbing the mtHsp70 import motor206. An impaired activity of the mitochondrial protein import machinery under stress conditions or in mitochondrial diseases is a direct indicator of impaired mitochondrial functions and can induce several responses from rescuing stress responses to removal of damaged mitochondria by mitophagy (BOX 3).
Box 3. Mitochondrial protein import and processing in cellular quality control.
Mitochondrial protein import machinery in stress response and quality control
Mitochondrial protein import machinery in stress response and quality control Mitochondrial unfolded protein response (UPRmt): The activating transcription factor associated with stress ATFS-1/ATF5 contains mitochondrial and nuclear localization signals. The factor is imported into healthy mitochondria and degraded. When mitochondrial import is impaired, the transcription factor accumulates in the cytosol, is translocated into the nucleus and induces expression of chaperones, proteases and further factors to promote recovery of impaired mitochondria207,208.
Unfolded protein response activated by mistargeted mitochondrial proteins (UPRam), also termed mitochondrial precursor over-accumulation stress (mPOS): Upon disturbance of mitochondrial protein import, precursor proteins accumulating in the cytosol trigger a stress response that reduces the efficiency of cytosolic protein synthesis and increases the activity of the proteasome, thus reducing the accumulation of mistargeted proteins in the cytosol209,210.
PINK1/parkin: The mitochondrial kinase PINK1 has been identified in familial cases of Parkinson’s disease. In healthy mitochondria, PINK1 is imported by the presequence pathway and processed by MPP and the presenilin-associated rhomboid-like protease PARL, followed by release into the cytosol and degradation by the proteasome. When protein import or processing by the presequence pathway are disturbed, unprocessed PINK1 accumulates at the TOM complex221–223 (FIG. 4), where it phosphorylates ubiquitin and the E3 ubiquitin ligase parkin, triggering the removal of damaged mitochondria by mitophagy.
Mitochondria as guardian in cytosol (MAGIC): Some aggregation-prone or misfolded cytosolic proteins may be imported into mitochondria and degraded215, suggesting a role of mitochondria in cytosolic proteostasis.
Mitochondrial preprotein processing, turnover and membrane dynamics
After processing by MPP, the matrix peptidases Icp55 and octapeptidyl peptidase can remove destabilizing amino acid residues from the amino-termini of imported yeast mitochondrial proteins (FIG. 2), generating proteins with stabilizing amino-termini that are less susceptible to degradation by matrix proteases53,75,76.
The inner membrane fusion protein OPA1 is present in long and short isoforms. Upon import, MPP performs the first cleavage to generate the long isoform. A second processing by inner membrane-bound proteases such as AAA proteases (FIG. 3) and OMA1 in mammals generates short isoforms216–218. The efficiency of the second processing is influenced by stress and energetic conditions (ATP, membrane potential), leading to different ratios between long and short isoforms that modulate mitochondrial fusion and fragmentation.
A mild disturbance of mitochondrial protein import can trigger the activation of the mitochondrial unfolded protein response (UPRmt) by impairing mitochondrial import of the transcription factor ATFS-1/ATF5, leading to its transport into the nucleus207,208, where it induces a mitochondrial stress response to rescue partially damaged mitochondria (BOX 3). The stress-regulated decrease of the levels of TIM17A also leads to decreased mitochondrial protein import and promotes the induction of an UPRmt136. In addition, accumulation of mitochondrial precursor proteins in the cytosol leads to an attenuation of cytosolic protein synthesis and activation of the proteasome to clear the mistargeted proteins from the cytosol209–212. This process is termed unfolded protein response activated by mistargeted mitochondrial proteins (UPRam) or mitochondrial precursor over-accumulation stress (mPOS).
Upon severe damage of mitochondrial protein import, the kinase PINK1 is not imported, processed and degraded, but associates with the TOM complex as full-length protein, initiating a cascade that leads to removal of damaged mitochondria by mitophagy19,201–203 (BOX 3). Since mutations of PINK1 have been linked to Parkinson’s disease, it is discussed that an insufficient mitophagy may be one of the factors in the development of the disease201. Recently, PINK1 and MIC60 of the MICOS complex were found to interact transiently, suggesting a crosstalk between PINK1 accumulation and inner membrane cristae remodeling213,214 (FIG. 4).
It has been suggested that mitochondria may function as guardian of cytosolic proteins (MAGIC) by importing and degrading misfolded cytosolic proteins215. This process will require further studies to define its relevance for cellular protein homeostasis (proteostasis) and its relation to stress responses initiated by a decreased mitochondrial protein import efficiency such as UPRmt, UPRam and PINK1/parkin.
Proteolytic processing of precursor proteins also plays a role in mitochondrial quality control (BOX 3). One mechanism is the removal of destabilizing amino-terminal amino acid residues by the processing enzymes Icp55 or octapeptidyl peptidase to stabilize imported proteins against proteolytic degradation53,75,76 (FIG. 2). Another mechanism concerns the differential processing of imported proteins, yielding two or more isoforms with distinct amino-termini, exemplified with the inner membrane fusion protein OPA1. OPA1 is first processed by MPP, yielding a long isoform. Further processing by inner membrane-proteases generates short isoforms, depending on stress conditions and the energetic state of the inner membrane. The balance between long and short isoforms that is important for membrane fusion and fission is thus modulated by stress and mitochondrial activity216–218.
An impaired processing of preproteins has been linked to mitochondrial dysfunctions in Alzheimer’s disease219. The matrix peptidasome degrades presequences and other peptides such as Alzheimer linked amyloid β-peptides. Upon accumulation of amyloid β-peptides in mitochondria, the degradation of presequences is slowed down competitively, leading to an inhibition of processing peptidases. As a consequence, proteins imported into mitochondria are retained in precursor or intermediate forms that cannot fold properly and are prone to rapid degradation. The accumulation of amyloid β-peptides thus causes numerous changes in mitochondrial protein composition, providing possible explanations for a wide variety of mitochondrial alterations observed in Alzheimer’s disease.
Studies in recent years provided increasing evidence for the involvement of mitochondrial protein import and processing in the pathogenesis of human diseases. At present, different views exist if mitochondrial dysfunctions are directly or indirectly involved in the development of major neurodegenerative diseases such as Parkinson’s disease and Alzheimer’s disease220. In BOX 4, we provide an overview of more rare diseases and disorders that have been linked to specific components of the mitochondrial machineries for protein import and maturation, suggesting an involvement in disease pathogenesis. The diseases mostly affect the nervous system and other tissues with a high energy demand such as heart, muscles and kidney. On a mechanistic level, defects in preprotein targeting, presequence pathway, processing and folding, MIA pathway and carrier pathway have been observed (BOX 4).
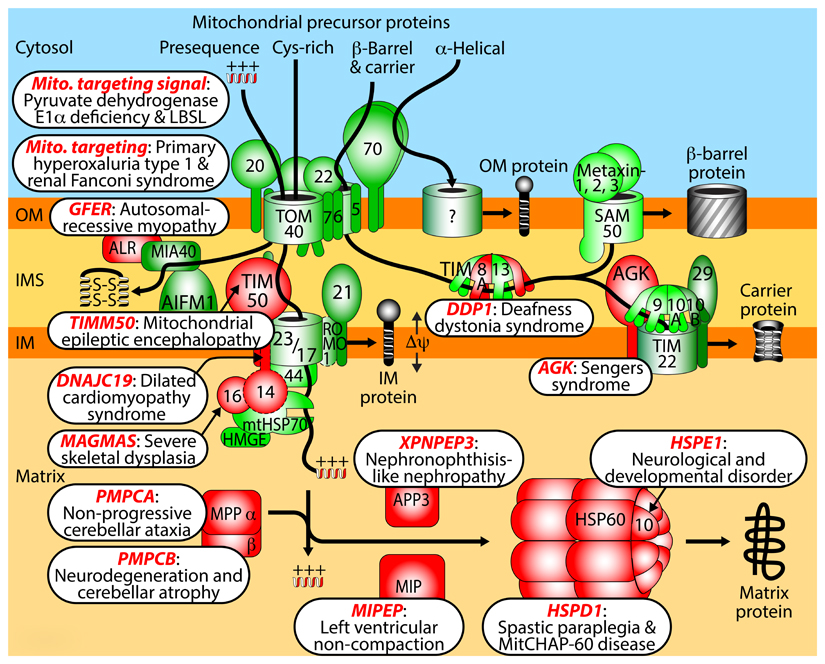
Box 4. Disorders and diseases linked to distinct steps of human mitochondrial protein import and maturation.
Mitochondrial targeting signal: mutations of targeting signals can impair import of individual proteins, causing pyruvate dehydrogenase E1α deficiency230 or mitochondrial aspartyl-tRNA synthetase import defect linked to leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation (LBSL)231.
Mitochondrial targeting: L-alanine:glyoxylate aminotransferase resides in peroxisomes in humans, however, mutations can generate a mitochondrial targeting signal, leading to mistargeting to mitochondria and primary hyperoxaluria type 1232,233. Similarly, in a form of renal Fanconi syndrome, a mutation generates a mitochondrial targeting signal in a peroxisomal protein involved in fatty acid oxidation, causing its mistargeting to mitochondria and disturbance of mitochondrial energy production in the proximal tubule234.
Growth factor, augmenter of liver regeneration ERV1 homolog (GFER/ALR): a mutation in the gene for the disulfide relay component ERV1/GFER/ALR causes impairment of FAD cofactor binding resulting in myopathy with cataract and combined respiratory chain deficiency235,236.
TIMM50: mutations of the presequence translocase receptor lead to epileptic encephalopathy, 3-methylglutaconic aciduria and variable complex V deficiency237.
DnaJ domain containing protein DNAJC19: mutations in the gene for the human PAM18/TIM14 homolog cause dilated cardiomyopathy with ataxia, anemia and testicular dysgenesis238,239. Since DNAJC19 is mainly associated with prohibitin complexes affecting cardiolipin metabolism, cardiolipin alteration is likely involved in disease pathogenesis240. DNAJC15/MCJ (methylation-controlled J-protein), a further human PAM18/TIM14 homolog241, has been linked to tumorigenesis. DNAJC19 and DNAJC15 have been connected to distinct presequence translocase forms135, yet their exact relevance for protein import needs further analysis (indicated by a dashed border).
Mitochondria-associated granulocyte macrophage colony stimulating factor-signaling molecule (MAGMAS): a mutation in the gene for human PAM16/TIM16 is linked to a severe spondylodysplastic dysplasia242.
Mitochondrial processing peptidase (MPP): mutations in the genes for MPP subunits α (PMPCA) or β (PMPCB) cause defects in preprotein processing, linked to cerebellar ataxia243 or early childhood neurodegeneration with cerebellar atrophy244.
Mitochondrial intermediate peptidase (MIPEP): mutations of octapeptidyl peptidase cause left ventricular non-compaction cardiomyopathy with hypotonia and developmental delay245.
X-prolyl aminopeptidase 3 (XPNPEP3): mutations of the human intermediate cleaving peptidase are linked to nephronophthisis-like cystic kidney disease246,247.
Heat shock protein family D member 1 (HSPD1): mutations of HSP60 lead to neurodegenerative disorders, spastic paraplegia and mitochondrial chaperonin-60 disease248,249.
Heat shock protein family E member 1 (HSPE1): a mutation of the co-chaperonin HSP10 is associated with infantile spasms and developmental delay250.
Deafness/dystonia protein 1 (DDP1): mutations of subunit TIMM8a of small TIM chaperones cause deafness dystonia syndrome, also termed Mohr-Tranebjaerg syndrome251,252.
Acyl glycerol kinase (AGK): mutations in the AGK gene lead to cataracts, cardiomyopathy and skeletal myopathy (Sengers syndrome)253. AGK plays a dual role as lipid kinase and as subunit of the human TIM22 carrier translocase, linking lipid metabolism and protein import to Sengers syndrome254,255.
The elaborate networks between preprotein translocases and other mitochondrial machineries have been mostly studied under physiological conditions. We expect that these networks will play an important role in understanding the mechanistic basis of mitochondrial stress responses and pathogenesis of diseases, exemplified with the role of TOM and MICOS in the accumulation of PINK1 at the outer membrane and the subsequent removal of damaged mitochondria by mitophagy213,221–223.
Conclusions and perspectives
Here, we have discussed that mitochondrial preprotein translocases, respiratory complexes, metabolite transporters, proteases, morphology complexes and membrane contact sites do not function as independent machineries, but are physically and functionally connected in large dynamic networks. The protein translocases represent an essential housekeeping system of mitochondria. The translocases are not only responsible for importing ~1,000-1,500 different proteins, but also form stable building blocks of the mitochondrial protein networks.
The rapid progress in identifying connections between machineries of different functions15,30,147,160,161,224,225 indicates that we have not reached a saturation in the analysis of mitochondrial protein networks. In addition to the experimentally established connections described in this Review, interesting further network candidates include: scaffold protein complexes that locally organize the protein-lipid composition of the inner membrane, such as the prohibitin ring complexes and stomatin-like protein 2 that associates with protease complexes and regulates the processing of PINK1 and OPA115,226; lipid biosynthesis and remodeling enzymes; and cytosolic machineries that are involved in transferring preproteins, lipids or metabolites to mitochondria. Whereas several contact sites between mitochondria and other cell organelles have been identified recently, we have only a limited understanding of the interplay between cytosolic proteins/protein complexes and the mitochondrial outer membrane. This includes the potential involvement of specialized pools of cytosolic ribosomes in protein delivery to mitochondria227, the role of cytosolic chaperones, co-chaperones and potential targeting factors in cytosol-mitochondria crosstalks, and the emerging evidence that numerous mitochondrial proteins possess a dual function and localization30.
Important questions concern the dynamics, regulation and turnover of the protein networks. It is likely that partner complexes in networks are turned over in different rates. Examples are the stress-regulated degradation of the TIM17A isoform of metazoan presequence translocases136 and the selective degradation of the outer membrane proteins Tom22 and porin-associated Om45 by the i-AAA protease164 (FIG. 3), whereas the other subunits of the complexes are turned over by different proteolytic machineries. The differential control of the networks by mitochondrial proteolytic systems, the cytosolic ubiquitin-proteasome system and mitophagy, as well as the role of lipids in establishing and maintaining the networks will become central topics of research.
The large number of distinct functions observed in mitochondrial protein networks may give the initial impression that collaborations of protein machineries have developed in a random manner. The mechanistic studies performed so far, however, indicate that the interactions are highly specialized and specifically regulated, such as between presequence translocase and respiratory supercomplexes, and between MICOS, TOM, SAM and ER-mitochondria contact sites. To date, the studies have been mainly performed in yeast and partially in human mitochondria that both belong to the same supergroup of eukaryotes, opisthokonts, which include fungal and metazoan kingdoms. Since the characterization of the mitochondrial protein import machinery in different supergroups yielded remarkable insight into core machineries and the high variability of transport complexes1,228, a systematic analysis of mitochondrial protein networks in the five eukaryotic supergroups will represent a rich source for defining core principles and variable parts of mitochondrial organization.
Glossary terms.
Oxidative phosphorylation
Oxidation of metabolites liberates energy that is used to synthesize ATP, in mitochondria performed by the respiratory chain that generates a proton gradient across the inner membrane to drive the F1FO-ATP synthase.
Insertase
Membrane-bound machinery that facilitates the insertion of precursor proteins into the lipid phase of a membrane, such as the oxidase assembly (OXA) insertase of the mitochondrial inner membrane.
MICOS
The mitochondrial contact site and cristae organizing system is a large protein complex of the inner membrane with a dual role, formation of contact sites to the outer membrane and maintenance of the cristae architecture of the inner membrane.
TOM complex
The translocase of the outer membrane forms the major mitochondrial entry site for precursor proteins synthesized in the cytosol.
SAM complex
The sorting and assembly machinery inserts β-barrel proteins into the mitochondrial outer membrane and is also called topogenesis of outer membrane β-barrel proteins (TOB).
Heat shock proteins 70
Large family of ATP-dependent molecular chaperones of ~70 kDa that bind loosely folded proteins and prevent their misfolding or aggregation. The major mitochondrial heat shock protein 70 (mtHsp70) has a dual role in driving ATP-dependent protein import into the matrix and assisting in folding of imported proteins.
Respirasomes
Large supercomplexes in the mitochondrial inner membrane consisting of the complexes I, III and IV of the respiratory chain.
ERMES
The endoplasmic reticulum-mitochondria encounter structure is a multi-subunit protein complex that connects endoplasmic reticulum and the mitochondrial outer membrane. ERMES is likely involved in lipid transfer between the organelles and is required for maintaining the morphology of mitochondria.
Acknowledgements
This work was supported by the European Research Council (ERC) Consolidator Grant No. 648235, the Excellence Initiative of the German federal and state governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School), the Deutsche Forschungsgemeinschaft (PF 202/8-1 and 202/9-1; WA 1598/5-1), and the Sonderforschungsbereiche 746 and 1140.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 2.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 3.Kutik S, Stroud DA, Wiedemann N, Pfanner N. Evolution of mitochondrial protein biogenesis. Biochim Biophys Acta. 2009;1790:409–415. doi: 10.1016/j.bbagen.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Wiedemann N, Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 5.van der Bliek AM, Sedensky MM, Morgan PG. Cell Biology of the Mitochondrion. Genetics. 2017;207:843–871. doi: 10.1534/genetics.117.300262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lill R. Function and biogenesis of iron–sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 7.Ott M, Amunts A, Brown A. Organization and Regulation of Mitochondrial Protein Synthesis. Annu Rev Biochem. 2016;85:77–101. doi: 10.1146/annurev-biochem-060815-014334. [DOI] [PubMed] [Google Scholar]
- 8.Hell K, Neupert W, Stuart RA. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraus F, Ryan MT. The constriction and scission machineries involved in mitochondrial fission. J Cell Sci. 2017;130:2953–2960. doi: 10.1242/jcs.199562. [DOI] [PubMed] [Google Scholar]
- 10.Wai T, Langer T. Mitochondrial Dynamics andMetabolic Regulation. Trends Endocrinol Metab. 2016;27:105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Labbé K, Murley A, Nunnari J. Determinants and Functions of Mitochondrial Behavior. Annu Rev Cell Dev Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 12.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 13.von der Malsburg K, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Harner M, et al. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoppins S, et al. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [References 13, 14 and 15 report the identification of the mitochondrial contact site and cristae organizing system (MICOS), a multi-subunit complex that links outer and inner membranes and is crucial for the maintenance of crista junctions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aaltonen MJ, et al. MICOS and phospholipid transfer by Ups2–Mdm35 organize membrane lipid synthesis in mitochondria. J Cell Biol. 2016;213:525–534. doi: 10.1083/jcb.201602007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ott C, et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat Rev Mol Cell Biol. 2018;19:109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 19.Pickles S, Vigié P, Youle RJ. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinou J-C, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tait SWG, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 22.Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sena LA, Chandel NS. Physiological Roles of Mitochondrial Reactive Oxygen Species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Gaucher SP, et al. Expanded Coverage of the Human Heart Mitochondrial Proteome Using Multidimensional Liquid Chromatography Coupled with Tandem Mass Spectrometry. J Proteome Res. 2004;3:495–505. doi: 10.1021/pr034102a. [DOI] [PubMed] [Google Scholar]
- 27.Hung V, et al. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefort N, et al. Proteome profile of functional mitochondria from human skeletal muscle using one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Journal of Proteomics. 2009;72:1046–1060. doi: 10.1016/j.jprot.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald T, et al. Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol Cell Proteomics. 2006;5:2392–2411. doi: 10.1074/mcp.T500036-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Morgenstern M, et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017;19:2836–2852. doi: 10.1016/j.celrep.2017.06.014. [Systematic quantitative analysis of the proteome of yeast mitochondria, revealing the absolute copy numbers of most mitochondrial protein machineries under fermentable and respiratory growth conditions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohlmeier S, Kastaniotis AJ, Hiltunen JK, Bergmann U. The yeast mitochondrial proteome, a study of fermentative and respiratory growth. J Biol Chem. 2004;279:3956–3979. doi: 10.1074/jbc.M310160200. [DOI] [PubMed] [Google Scholar]
- 32.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prokisch H, et al. Integrative analysis of the mitochondrial proteome in yeast. Plos Biol. 2004;2:e160. doi: 10.1371/journal.pbio.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 35.Rhee H-W, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sickmann A, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith AC, Robinson AJ. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016;44:D1258–D1261. doi: 10.1093/nar/gkv1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor SW, et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 39.Vögtle FN, et al. Landscape of submitochondrial protein distribution. Nature Communications. 2017;8:290. doi: 10.1038/s41467-017-00359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vögtle FN, et al. Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics. 2012;11:1840–1852. doi: 10.1074/mcp.M112.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahedi RP, et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, et al. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 2008;8:1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Godoy LMF, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 44.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 45.Beck M, et al. The quantitative proteome of a human cell line. Molecular Systems Biology. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 47.Wiśniewski JR, Hein MY, Cox J, Mann M. A ‘proteomic ruler’ for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteomics. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulo JA, et al. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. Journal of Proteomics. 2016;148:85–93. doi: 10.1016/j.jprot.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harbauer AB, et al. Mitochondria. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science. 2014;346:1109–1113. doi: 10.1126/science.1261253. [DOI] [PubMed] [Google Scholar]
- 50.Gerbeth C, et al. Glucose-induced regulation of protein import receptor Tom22 by cytosolic and mitochondria-bound kinases. Cell Metabolism. 2013;18:578–587. doi: 10.1016/j.cmet.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Rao S, et al. Biogenesis of the preprotein translocase of the outer mitochondrial membrane: protein kinase A phosphorylates the precursor of Tom40 and impairs its import. Mol Biol Cell. 2012;23:1618–1627. doi: 10.1091/mbc.E11-11-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt O, et al. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [This study reports that biogenesis and activity of the TOM complex are regulated by cytosolic kinases, revealing the main protein import site of mitochondria as a major target for cytosolic signaling pathways.] [DOI] [PubMed] [Google Scholar]
- 53.Vögtle FN, et al. Global Analysis of the Mitochondrial N-Proteome Identifies a Processing Peptidase Critical for Protein Stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 54.Abe Y, et al. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 55.van Wilpe S, et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- 56.Kuszak AJ, et al. Evidence of Distinct Channel Conformations and Substrate Binding Affinities for the Mitochondrial Outer Membrane Protein Translocase Pore Tom40. J Biol Chem. 2015;290:26204–26217. doi: 10.1074/jbc.M115.642173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melin J, et al. Presequence recognition by the Tom40 channel contributes to precursor translocation into the mitochondrial matrix. Mol Cell Biol. 2014;34:3473–3485. doi: 10.1128/MCB.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohret TA, Jensen RE, Kinnally KW. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J Cell Biol. 1997;137:377–386. doi: 10.1083/jcb.137.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bauer MF, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- 60.Dekker PJ, et al. Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett. 1993;330:66–70. doi: 10.1016/0014-5793(93)80921-g. [DOI] [PubMed] [Google Scholar]
- 61.Demishtein-Zohary K, Marom M, Neupert W, Mokranjac D, Azem A. GxxxG motifs hold the TIM23 complex together. FEBS J. 2015;282:2178–2186. doi: 10.1111/febs.13266. [DOI] [PubMed] [Google Scholar]
- 62.Truscott KN, et al. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat Struct Biol. 2001;8:1074–1082. doi: 10.1038/nsb726. [DOI] [PubMed] [Google Scholar]
- 63.Malhotra K, et al. Cardiolipin mediates membrane and channel interactions of the mitochondrial TIM23 protein import complex receptor Tim50. Sci Adv. 2017;3:e1700532. doi: 10.1126/sciadv.1700532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denkert N, et al. Cation selectivity of the presequence translocase channel Tim23 is crucial for efficient protein import. eLife. 2017;6:e28324. doi: 10.7554/eLife.28324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ramesh A, et al. A disulfide bond in the TIM23 complex is crucial for voltage gating and mitochondrial protein import. J Cell Biol. 2016;214:417–431. doi: 10.1083/jcb.201602074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang PJ, et al. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 67.Demishtein-Zohary K, et al. Role of Tim17 in coupling the import motor to the translocation channel of the mitochondrial presequence translocase. eLife. 2017;6:e22696. doi: 10.7554/eLife.22696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ting S-Y, Yan NL, Schilke BA, Craig EA. Dual interaction of scaffold protein Tim44 of mitochondrial import motor with channel-forming translocase subunit Tim23. eLife. 2017;6:e23609. doi: 10.7554/eLife.23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banerjee R, Gladkova C, Mapa K, Witte G, Mokranjac D. Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein. eLife. 2015;4:e11897. doi: 10.7554/eLife.11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sikor M, Mapa K, von Voithenberg LV, Mokranjac D, Lamb DC. Real-time observation of the conformational dynamics of mitochondrial Hsp70 by spFRET. EMBO J. 2013;32:1639–1649. doi: 10.1038/emboj.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schendzielorz AB, et al. Two distinct membrane potential-dependent steps drive mitochondrial matrix protein translocation. J Cell Biol. 2017;216:83–92. doi: 10.1083/jcb.201607066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schulz C, Rehling P. Remodelling of the active presequence translocase drives motor-dependent mitochondrial protein translocation. Nature Communications. 2014;5:4349. doi: 10.1038/ncomms5349. [DOI] [PubMed] [Google Scholar]
- 73.Fukasawa Y, et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veling MT, et al. Multi-omic Mitoprotease Profiling Defines a Role for Oct1p in Coenzyme Q Production. Mol Cell. 2017;68:970–977. doi: 10.1016/j.molcel.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vögtle FN, et al. Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell. 2011;22:2135–2143. doi: 10.1091/mbc.E11-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng MY, et al. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989;337:620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- 78.Ostermann J, Horwich AL, Neupert W, Hartl FU. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989;341:125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 79.Ieva R, et al. Mgr2 functions as lateral gatekeeper for preprotein sorting in the mitochondrial inner membrane. Mol Cell. 2014;56:641–652. doi: 10.1016/j.molcel.2014.10.010. [Identification of a lateral gatekeeper protein at the TIM23 complex to control the proper release of preproteins with hydrophobic sorting signal into the inner membrane.] [DOI] [PubMed] [Google Scholar]
- 80.Stiller SB, et al. Mitochondrial OXA Translocase Plays a Major Role in Biogenesis of Inner-Membrane Proteins. Cell Metabolism. 2016;23:901–908. doi: 10.1016/j.cmet.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park K, Botelho SC, Hong J, Osterberg M, Kim H. Dissecting Stop Transfer versus Conservative Sorting Pathways for Mitochondrial Inner Membrane Proteins in Vivo. J Biol Chem. 2013;288:1521–1532. doi: 10.1074/jbc.M112.409748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bohnert M, et al. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr Biol. 2010;20:1227–1232. doi: 10.1016/j.cub.2010.05.058. [DOI] [PubMed] [Google Scholar]
- 83.Herrmann JM, Neupert W, Stuart RA. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bausewein T, et al. Cryo-EM Structure of the TOM Core Complex from Neurospora crassa. Cell. 2017;170:693–700. doi: 10.1016/j.cell.2017.07.012. [Cryoelectron microscopy structure of the TOM core complex of the mitochondrial outer membrane. Two translocation pores formed by Tom40 β-barrels are connected by two copies of the central receptor Tom22.] [DOI] [PubMed] [Google Scholar]
- 85.Shiota T, et al. Molecular architecture of the active mitochondrial protein gate. Science. 2015;349:1544–1548. doi: 10.1126/science.aac6428. [Mapping of the TOM complex architecture by site-specific crosslinking in the native membrane, revealing different translocation pathways for hydrophilic and hydrophobic precursor proteins through the Tom40 channel.] [DOI] [PubMed] [Google Scholar]
- 86.Backes S, et al. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J Cell Biol. 2018;217:1369–1382. doi: 10.1083/jcb.201708044. [This study reports a role of Tom70 in the import of presequence-containing preproteins that contain additional internal targeting signals. Whereas Tom20/22 bind the amino-terminal presequences, Tom70 binds the internal signals to prevent aggregation of the preproteins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Melin J, et al. A presequence-binding groove in Tom70 supports import of Mdl1 into mitochondria. Biochim Biophys Acta. 2015;1853:1850–1859. doi: 10.1016/j.bbamcr.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 88.Yamamoto H, et al. Roles of Tom70 in Import of Presequence-containing Mitochondrial Proteins. J Biol Chem. 2009;284:31635–31646. doi: 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 90.Wiedemann N, Pfanner N, Ryan MT. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Curran SP, Leuenberger D, Schmidt E, Koehler CM. The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J Cell Biol. 2002;158:1017–1027. doi: 10.1083/jcb.200205124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vial S, et al. Assembly of Tim9 and Tim10 into a functional chaperone. J Biol Chem. 2002;277:36100–36108. doi: 10.1074/jbc.M202310200. [DOI] [PubMed] [Google Scholar]
- 93.Koehler CM, et al. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 1998;17:6477–6486. doi: 10.1093/emboj/17.22.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sirrenberg C, et al. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- 95.Okamoto H, Miyagawa A, Shiota T, Tamura Y, Endo T. Intramolecular disulfide bond of Tim22 protein maintains integrity of the TIM22 complex in the mitochondrial inner membrane. J Biol Chem. 2014;289:4827–4838. doi: 10.1074/jbc.M113.543264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wrobel L, Trojanowska A, Sztolsztener ME, Chacinska A. Mitochondrial protein import: Mia40 facilitates Tim22 translocation into the inner membrane of mitochondria. Mol Biol Cell. 2013;24:543–554. doi: 10.1091/mbc.E12-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rehling P, et al. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 2003;299:1747–1751. doi: 10.1126/science.1080945. [DOI] [PubMed] [Google Scholar]
- 98.Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol. 1997;139:1663–1675. doi: 10.1083/jcb.139.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- 100.Koch JR, Schmid FX. Mia40 Combines Thiol Oxidase and Disulfide Isomerase Activity to Efficiently Catalyze Oxidative Folding in Mitochondria. J Mol Biol. 2014;426:4087–4098. doi: 10.1016/j.jmb.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 101.Chacinska A, et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mesecke N, et al. A Disulfide Relay System in the Intermembrane Space of Mitochondria that Mediates Protein Import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 103.Milenkovic D, et al. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell. 2009;20:2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sideris DP, et al. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J Cell Biol. 2009;187:1007–1022. doi: 10.1083/jcb.200905134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peleh V, Cordat E, Herrmann JM. Mia40 is a trans-site receptor that drives protein import into the mitochondrial intermembrane space by hydrophobic substrate binding. eLife. 2016;5:e16177. doi: 10.7554/eLife.16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neal SE, et al. Mia40 Protein Serves as an Electron Sink in the Mia40-Erv1 Import Pathway. J Biol Chem. 2015;290:20804–20814. doi: 10.1074/jbc.M115.669440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kojer K, Peleh V, Calabrese G, Herrmann JM, Riemer J. Kinetic control by limiting glutaredoxin amounts enables thiol oxidation in the reducing mitochondrial intermembrane space. Mol Biol Cell. 2015;26:195–204. doi: 10.1091/mbc.E14-10-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jores T, et al. Characterization of the targeting signal in mitochondrial β-barrel proteins. Nature Communications. 2016;7 doi: 10.1038/ncomms12036. 12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klein A, et al. Characterization of the insertase for β-barrel proteins of the outer mitochondrial membrane. J Cell Biol. 2012;199:599–611. doi: 10.1083/jcb.201207161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wiedemann N, et al. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 111.Paschen SA, et al. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 112.Höhr AIC, et al. Membrane protein insertion through a mitochondrial β-barrel gate. Science. 2018;359 doi: 10.1126/science.aah6834. eaah6834. [Mapping of the membrane insertion pathway of β-barrel precursors through the SAM channel, signal-induced opening of the lateral gate of Sam50, and release of the folded β-barrel protein into the outer membrane.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kutik S, et al. Dissecting Membrane Insertion of Mitochondrial β-Barrel Proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 114.Ulrich T, Rapaport D. Biogenesis of beta-barrel proteins in evolutionary context. Int J Med Microbiol. 2015;305:259–264. doi: 10.1016/j.ijmm.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 115.Krüger V, et al. Identification of new channels by systematic analysis of the mitochondrial outer membrane. J Cell Biol. 2017;216:3485–3495. doi: 10.1083/jcb.201706043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dimmer KS, et al. A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J Cell Sci. 2012;125:3464–3473. doi: 10.1242/jcs.103804. [DOI] [PubMed] [Google Scholar]
- 117.Papic D, Krumpe K, Dukanovic J, Dimmer KS, Rapaport D. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J Cell Biol. 2011;194:397–405. doi: 10.1083/jcb.201102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hulett JM, et al. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol. 2008;376:694–704. doi: 10.1016/j.jmb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 119.Popov-Čeleketić J, Waizenegger T, Rapaport D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol. 2008;376:671–680. doi: 10.1016/j.jmb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 120.Becker T, et al. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J Cell Biol. 2011;194:387–395. doi: 10.1083/jcb.201102044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dukanovic J, Rapaport D. Multiple pathways in the integration of proteins into the mitochondrial outer membrane. Biochim Biophys Acta. 2011;1808:971–980. doi: 10.1016/j.bbamem.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 122.Keskin A, Akdoğan E, Dunn CD. Evidence for Amino Acid Snorkeling from a High-Resolution, In Vivo Analysis of Fis1 Tail-Anchor Insertion at the Mitochondrial Outer Membrane. Genetics. 2017;205:691–705. doi: 10.1534/genetics.116.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vögtle FN, et al. The fusogenic lipid phosphatidic acid promotes the biogenesis of mitochondrial outer membrane protein Ugo1. J Cell Biol. 2015;210:951–960. doi: 10.1083/jcb.201506085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sauerwald J, et al. Genome-Wide Screens in Saccharomyces cerevisiae Highlight a Role for Cardiolipin in Biogenesis of Mitochondrial Outer Membrane Multispan Proteins. Mol Cell Biol. 2015;35:3200–3211. doi: 10.1128/MCB.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Albrecht R, et al. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 2006;7:1233–1238. doi: 10.1038/sj.embor.7400828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chacinska A, et al. Distinct Forms of Mitochondrial TOM-TIM Supercomplexes Define Signal-Dependent States of Preprotein Sorting. Mol Cell Biol. 2010;30:307–318. doi: 10.1128/MCB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gold VAM, et al. Visualizing active membrane protein complexes by electron cryotomography. Nature Communications. 2014;5:4129. doi: 10.1038/ncomms5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qiu J, et al. Coupling of Mitochondrial Import and Export Translocases by Receptor-Mediated Supercomplex Formation. Cell. 2013;154:596–608. doi: 10.1016/j.cell.2013.06.033. [Identification of a TOM-SAM supercomplex that facilitates the efficient transfer of β-barrel precursors from the TOM import channel to the SAM membrane insertion sites of the mitochondrial outer membrane.] [DOI] [PubMed] [Google Scholar]
- 129.Waegemann K, Popov-Celeketic D, Neupert W, Azem A, Mokranjać D. Cooperation of TOM and TIM23 complexes during translocation of proteins into mitochondria. J Mol Biol. 2015;427:1075–1084. doi: 10.1016/j.jmb.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 130.Wenz L-S, et al. Sam37 is crucial for formation of the mitochondrial TOM-SAM supercomplex, thereby promoting β-barrel biogenesis. J Cell Biol. 2015;210:1047–1054. doi: 10.1083/jcb.201504119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Callegari S, et al. TIM29 is a subunit of the human carrier translocase required for protein transport. FEBS Lett. 2016;590:4147–4158. doi: 10.1002/1873-3468.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kang Y, et al. Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. eLife. 2016;5:e17463. doi: 10.7554/eLife.17463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gornicka A, et al. A discrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol Biol Cell. 2014;25:3999–4009. doi: 10.1091/mbc.E14-06-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wiedemann N, et al. Biogenesis of yeast mitochondrial cytochrome c: a unique relationship to the TOM machinery. J Mol Biol. 2003;327:465–474. doi: 10.1016/s0022-2836(03)00118-9. [DOI] [PubMed] [Google Scholar]
- 135.Sinha D, Srivastava S, Krishna L, D'Silva P. Unraveling the intricate organization of mammalian mitochondrial presequence translocases: existence of multiple translocases for maintenance of mitochondrial function. Mol Cell Biol. 2014;34:1757–1775. doi: 10.1128/MCB.01527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rainbolt TK, Atanassova N, Genereux JC, Wiseman RL. Stress-Regulated Translational Attenuation Adapts Mitochondrial Protein Import through Tim17A Degradation. Cell Metabolism. 2013;18:908–919. doi: 10.1016/j.cmet.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Opalińska M, Parys K, Murcha MW, Janska H. The plant i-AAA protease controls the turnover of an essential mitochondrial protein import component. J Cell Sci. 2018;131 doi: 10.1242/jcs.200733. jcs200733. [DOI] [PubMed] [Google Scholar]
- 138.Wenz L-S, et al. The presequence pathway is involved in protein sorting to the mitochondrial outer membrane. EMBO Rep. 2014;15:678–685. doi: 10.1002/embr.201338144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Song J, Tamura Y, Yoshihisa T, Endo T. A novel import route for an N-anchor mitochondrial outer membrane protein aided by the TIM23 complex. EMBO Rep. 2014;15:670–677. doi: 10.1002/embr.201338142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee CM, Sedman J, Neupert W, Stuart RA. The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J Biol Chem. 1999;274:20937–20942. doi: 10.1074/jbc.274.30.20937. [DOI] [PubMed] [Google Scholar]
- 141.Ieva R, et al. Mitochondrial inner membrane protease promotes assembly of presequence translocase by removing a carboxy-terminal targeting sequence. Nature Communications. 2013;4:2853. doi: 10.1038/ncomms3853. [DOI] [PubMed] [Google Scholar]
- 142.Sinzel M, et al. Mcp3 is a novel mitochondrial outer membrane protein that follows a unique IMP-dependent biogenesis pathway. EMBO Rep. 2016;17:965–981. doi: 10.15252/embr.201541273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Acin-Perez R, Fernández-Silva P, Peleato ML, Pérez-Martos A, Enríquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 144.Melber A, Winge DR. Inner Secrets of the Respirasome. Cell. 2016;167:1450–1452. doi: 10.1016/j.cell.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of Mammalian Respiratory Supercomplex I1III2IV1. Cell. 2016;167:1598–1609. doi: 10.1016/j.cell.2016.11.012. [Cryo-electron microscopy structure of the 1.7 MDa respiratory supercomplex at near-atomic resolution, revealing the arrangement of complexes I, III and IV, and the position of cofactors and phospholipids.] [DOI] [PubMed] [Google Scholar]
- 146.Milenkovic D, Blaza JN, Larsson N-G, Hirst J. The Enigma of the Respiratory Chain Supercomplex. Cell Metabolism. 2017;25:765–776. doi: 10.1016/j.cmet.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 147.Schweppe DK, et al. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci U S A. 2017;114:1732–1737. doi: 10.1073/pnas.1617220114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen Y-C, et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metabolism. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vukotic M, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metabolism. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 151.Singhal RK, et al. Coi1 is a novel assembly factor of the yeast complex III-complex IV supercomplex. Mol Biol Cell. 2017;28:2609–2622. doi: 10.1091/mbc.E17-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van der Laan M, et al. A Role for Tim21 in Membrane-Potential-Dependent Preprotein Sorting in Mitochondria. Curr Biol. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 153.Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol. 2007;179:1115–1122. doi: 10.1083/jcb.200709087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mehnert CS, et al. The mitochondrial ADP/ATP carrier associates with the inner membrane presequence translocase in a stoichiometric manner. J Biol Chem. 2014;289:27352–27362. doi: 10.1074/jbc.M114.556498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dennerlein S, et al. MITRAC7 Acts as a COX1-Specific Chaperone and Reveals a Checkpoint during Cytochrome c Oxidase Assembly. Cell Rep. 2015;12:1644–1655. doi: 10.1016/j.celrep.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 156.Mick DU, et al. MITRAC Links Mitochondrial Protein Translocation to Respiratory-Chain Assembly and Translational Regulation. Cell. 2012;151:1528–1541. doi: 10.1016/j.cell.2012.11.053. [DOI] [PubMed] [Google Scholar]
- 157.Richter-Dennerlein R, et al. Mitochondrial Protein Synthesis Adapts to Influx of Nuclear-Encoded Protein. Cell. 2016;167:471–483. doi: 10.1016/j.cell.2016.09.003. [Identification of a mechanism how MITRAC assembly factors adjust the efficiency of mitochondrial synthesis of membrane-integrated respiratory chain subunits to the import of nuclear encoded partner proteins, termed mitochondrial translational plasticity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stoldt S, et al. Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat Cell Biol. 2018;20:528–534. doi: 10.1038/s41556-018-0090-7. [DOI] [PubMed] [Google Scholar]
- 159.Topf U, et al. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nature Communications. 2018;9:324. doi: 10.1038/s41467-017-02694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Floyd BJ, et al. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stefely JA, et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat Biotechnol. 2016;34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gerdes F, Tatsuta T, Langer T. Mitochondrial AAA proteases--towards a molecular understanding of membrane-bound proteolytic machines. Biochim Biophys Acta. 2012;1823:49–55. doi: 10.1016/j.bbamcr.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 163.Puchades C, et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science. 2017;358 doi: 10.1126/science.aao0464. eaao0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wu X, Li L, Jiang H. Mitochondrial inner-membrane protease Yme1 degrades outer-membrane proteins Tom22 and Om45. J Cell Biol. 2018;217:139–149. doi: 10.1083/jcb.201702125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van der Laan M, Bohnert M, Wiedemann N, Pfanner N. Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol. 2012;22:185–192. doi: 10.1016/j.tcb.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 166.Bohnert M, et al. Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol Biol Cell. 2012;23:3948–3956. doi: 10.1091/mbc.E12-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zerbes RM, et al. Role of MINOS in mitochondrial membrane architecture: cristae morphology and outer membrane interactions differentially depend on mitofilin domains. J Mol Biol. 2012;422:183–191. doi: 10.1016/j.jmb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 168.Körner C, et al. The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria. Mol Biol Cell. 2012;23:2143–2155. doi: 10.1091/mbc.E11-10-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein Mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- 170.Rabl R, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jans DC, et al. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc Natl Acad Sci U S A. 2013;110:8936–8941. doi: 10.1073/pnas.1301820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Barbot M, et al. Mic10 Oligomerizes to Bend Mitochondrial Inner Membranes at Cristae Junctions. Cell Metabolism. 2015;21:756–763. doi: 10.1016/j.cmet.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 173.Bohnert M, et al. Central Role of Mic10 in the Mitochondrial Contact Site and Cristae Organizing System. Cell Metabolism. 2015;21:747–755. doi: 10.1016/j.cmet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 174.Rampelt H, et al. Mic10, a Core Subunit of the Mitochondrial Contact Site and Cristae Organizing System, Interacts with the Dimeric F1Fo-ATP Synthase. J Mol Biol. 2017;429:1162–1170. doi: 10.1016/j.jmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 175.Eydt K, Davies KM, Behrendt C, Wittig I, Reichert AS. Cristae architecture is determined by an interplay of the MICOS complex and the F1FO ATP synthase via Mic27 and Mic10. Microb Cell. 2017;4:259–272. doi: 10.15698/mic2017.08.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Friedman JR, Mourier A, Yamada J, McCaffery JM, Nunnari J. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 2015;4:e07739. doi: 10.7554/eLife.07739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Barrera M, Koob S, Dikov D, Vogel F, Reichert AS. OPA1 functionally interacts with MIC60 but is dispensable for crista junction formation. FEBS Lett. 2016;590:3309–3322. doi: 10.1002/1873-3468.12384. [DOI] [PubMed] [Google Scholar]
- 178.Glytsou C, et al. Optic Atrophy 1 Is Epistatic to the Core MICOS Component MIC60 in Mitochondrial Cristae Shape Control. Cell Rep. 2016;17:3024–3034. doi: 10.1016/j.celrep.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Itoh K, Tamura Y, Iijima M, Sesaki H. Effects of Fcj1-Mos1 and mitochondrial division on aggregation of mitochondrial DNA nucleoids and organelle morphology. Mol Biol Cell. 2013;24:1842–1851. doi: 10.1091/mbc.E13-03-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li H, et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 2016;23:380–392. doi: 10.1038/cdd.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [Identification of the endoplasmic reticulum-mitochondria encounter structure. ERMES-mediated contact sites between ER and mitochondria are involved in lipid transfer and maintenance of mitochondrial morphology.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Meisinger C, et al. The Mitochondrial Morphology Protein Mdm10 Functions in Assembly of the Preprotein Translocase of the Outer Membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 183.Yamano K, Tanaka-Yamano S, Endo T. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 2010;11:187–193. doi: 10.1038/embor.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Flinner N, et al. Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim Biophys Acta. 2013;1833:3314–3325. doi: 10.1016/j.bbamcr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 185.Ellenrieder L, et al. Separating mitochondrial protein assembly and endoplasmic reticulum tethering by selective coupling of Mdm10. Nature Communications. 2016;7 doi: 10.1038/ncomms13021. 13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Meisinger C, et al. Mitochondrial protein sorting: differentiation of beta-barrel assembly by Tom7-mediated segregation of Mdm10. J Biol Chem. 2006;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- 187.Stroud DA, et al. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011;413:743–750. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 188.Yamano K, Tanaka-Yamano S, Endo T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem. 2010;285:41222–41231. doi: 10.1074/jbc.M110.163238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Müller CS, et al. Cryo-slicing Blue Native-Mass Spectrometry (csBN-MS), a Novel Technology for High Resolution Complexome Profiling. Mol Cell Proteomics. 2016;15:669–681. doi: 10.1074/mcp.M115.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Elbaz-Alon Y, et al. Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep. 2015;12:7–14. doi: 10.1016/j.celrep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Murley A, et al. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J Cell Biol. 2015;209:539–548. doi: 10.1083/jcb.201502033. [References 190 and 191 report that the lipid transfer protein Lam6/Ltc1 is located at and regulates contact sites between ER, mitochondria and further organelles. Lam6/Ltc1 interacts with the receptor Tom70 in ER-mitochondria contact sites..] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Filadi R, et al. TOM70 Sustains Cell Bioenergetics by Promoting IP3R3-Mediated ER to Mitochondria Ca2+ Transfer. Curr Biol. 2018;28:369–382. doi: 10.1016/j.cub.2017.12.047. [The receptor TOM70 of the mitochondrial outer membrane interacts with inositol trisphosphate receptors of the endoplasmic reticulum, supporting the formation of contact sites for Ca2+ transfer to mitochondria.] [DOI] [PubMed] [Google Scholar]
- 193.González Montoro A, et al. Vps39 Interacts with Tom40 to Establish One of Two Functionally Distinct Vacuole-Mitochondria Contact Sites. Dev Cell. 2018;45:621–636. doi: 10.1016/j.devcel.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 194.McLelland G-L, Lee SA, McBride HM, Fon EA. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J Cell Biol. 2016;214:275–291. doi: 10.1083/jcb.201603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, McBride HM. Reconstitution of Mitochondria Derived Vesicle Formation Demonstrates Selective Enrichment of Oxidized Cargo. PLoS ONE. 2012;7:e52830. doi: 10.1371/journal.pone.0052830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Soubannier V, et al. A Vesicular Transport Pathway Shuttles Cargo from Mitochondria to Lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [References 195 and 196 report that stress conditions can induce the formation of mitochondria-derived vesicles that transport selected cargo such as oxidized proteins to lysosomes, as part of a mitochondrial quality control system.] [DOI] [PubMed] [Google Scholar]
- 197.Hughes AL, Hughes CE, Henderson KA, Yazvenko N, Gottschling DE. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. eLife. 2016;5:e13943. doi: 10.7554/eLife.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen Y-C, et al. Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 2014;33:1548–1564. doi: 10.15252/embj.201487943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Okreglak V, Walter P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc Natl Acad Sci U S A. 2014;111:8019–8024. doi: 10.1073/pnas.1405755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Weidberg H, Amon A. MitoCPR—A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360 doi: 10.1126/science.aan4146. eaan4146. [References 198, 199 and 200 report that the AAA-type ATPase Msp1 extracts mistargeted or non-imported proteins from the mitochondrial outer membrane for degradation by the proteasome in the cytosol.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sekine S, Youle RJ. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 2018;16:2. doi: 10.1186/s12915-017-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Müller M, Lu K, Reichert AS. Mitophagy and mitochondrial dynamics in Saccharomyces cerevisiae. Biochim Biophys Acta. 2015;1853:2766–2774. doi: 10.1016/j.bbamcr.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 203.Okamoto K. Quality control: Organellophagy: Eliminating cellular building blocks via selective autophagy. J Cell Biol. 2014;205:435–445. doi: 10.1083/jcb.201402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nunnari J, Suomalainen A. Mitochondria: In Sickness and in Health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Frazier AE, Thorburn DR, Compton AG. Mitochondrial energy generation disorders: genes, mechanisms and clues to pathology. J Biol Chem. 2017 doi: 10.1074/jbc.R117.809194. jbc.R117.809194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Melber A, Haynes CM. UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial Import Efficiency of ATFS-1 Regulates Mitochondrial UPR Activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [Impaired mitochondrial protein import activates an unfolded protein response. The transcription factor ATFS-1 is normally imported into mitochondria and degraded. Disturbance of mitochondrial import leads to cytosolic accumulation of ATFS-1 and translocation into the nucleus, where it induces expression of chaperones and further rescue factors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fiorese CJ, et al. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr Biol. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wrobel L, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- 210.Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [References 209 and 210 identify a stress response that is induced by accumulation of mitochondrial precursor proteins in the cytosol. The response leads to a decrease of cytosolic protein synthesis and an increased proteasomal activity to reduce the accumulation of toxic mistargeted proteins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bragoszewski P, et al. Retro-translocation of mitochondrial intermembrane space proteins. Proc Natl Acad Sci U.S.A. 2015;112:7713–7718. doi: 10.1073/pnas.1504615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bragoszewski P, Gornicka A, Sztolsztener ME, Chacinska A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol Cell Biol. 2013;33:2136–2148. doi: 10.1128/MCB.01579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Akabane S, et al. PKA Regulates PINK1 Stability and Parkin Recruitment to Damaged Mitochondria through Phosphorylation of MIC60. Mol Cell. 2016;62:371–384. doi: 10.1016/j.molcel.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 214.Tsai P-I, et al. PINK1 Phosphorylates MIC60/Mitofilin to Control Structural Plasticity of Mitochondrial Crista Junctions. Mol Cell. 2018;69:744–756. doi: 10.1016/j.molcel.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 215.Ruan L, et al. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 2017;543:443–446. doi: 10.1038/nature21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Anand R, et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ehses S, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mossmann D, et al. Amyloid-β peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metabolism. 2014;20:662–669. doi: 10.1016/j.cmet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 220.Schapira AH. Mitochondrial diseases. Lancet. 2012;379:1825–1834. doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- 221.Okatsu K, Kimura M, Oka T, Tanaka K, Matsuda N. Unconventional PINK1 localization to the outer membrane of depolarized mitochondria drives Parkin recruitment. J Cell Sci. 2015;128:964–978. doi: 10.1242/jcs.161000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bertolin G, et al. The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance. Autophagy. 2013;9:1801–1817. doi: 10.4161/auto.25884. [DOI] [PubMed] [Google Scholar]
- 223.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 Binding to the TOM Complex and Alternate Intracellular Membranes in Recruitment and Activation of the E3 Ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [This study shows that upon dissipation of the mitochondrial membrane potential, the kinase PINK1 accumulates at the mitochondrial outer membrane in a complex with the translocase of the outer membrane (TOM).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Liu F, Rijkers DTS, Post H, Heck AJR. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods. 2015;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 225.Huttlin EL, et al. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545:505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wai T, et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 2016;17:1844–1856. doi: 10.15252/embr.201642698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Segev N, Gerst JE. Specialized ribosomes and specific ribosomal protein paralogs control translation of mitochondrial proteins. J Cell Biol. 2018;217:117–126. doi: 10.1083/jcb.201706059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Harsman A, Schneider A. Mitochondrial protein import in trypanosomes: Expect the unexpected. Traffic. 2017;18:96–109. doi: 10.1111/tra.12463. [DOI] [PubMed] [Google Scholar]
- 229.Dienhart MK, Stuart RA. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol Biol Cell. 2008;19:3934–3943. doi: 10.1091/mbc.E08-04-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Takakubo F, et al. An amino acid substitution in the pyruvate dehydrogenase E1 alpha gene, affecting mitochondrial import of the precursor protein. Am J Hum Genet. 1995;57:772–780. [PMC free article] [PubMed] [Google Scholar]
- 231.Messmer M, et al. A human pathology-related mutation prevents import of an aminoacyl-tRNA synthetase into mitochondria. Biochem J. 2011;433:441–446. doi: 10.1042/BJ20101902. [DOI] [PubMed] [Google Scholar]
- 232.Purdue PE, Allsop J, Isaya G, Rosenberg LE, Danpure CJ. Mistargeting of peroxisomal L-alanine:glyoxylate aminotransferase to mitochondria in primary hyperoxaluria patients depends upon activation of a cryptic mitochondrial targeting sequence by a point mutation. Proc Natl Acad Sci U S A. 1991;88:10900–10904. doi: 10.1073/pnas.88.23.10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Danpure CJ, Cooper PJ, Wise PJ, Jennings PR. An enzyme trafficking defect in two patients with primary hyperoxaluria type 1: peroxisomal alanine/glyoxylate aminotransferase rerouted to mitochondria. J Cell Biol. 1989;108:1345–1352. doi: 10.1083/jcb.108.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Klootwijk ED, et al. Mistargeting of Peroxisomal EHHADH and Inherited Renal Fanconi's Syndrome. N Engl J Med. 2014;370:129–138. doi: 10.1056/NEJMoa1307581. [DOI] [PubMed] [Google Scholar]
- 235.Di Fonzo A, et al. The mitochondrial disulfide relay system protein GFER is mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am J Hum Gen. 2009;84:594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ceh-Pavia E, Ang SK, Spiller MP, Lu H. The disease-associated mutation of the mitochondrial thiol oxidase Erv1 impairs cofactor binding during its catalytic reaction. Biochem J. 2014;464:449–459. doi: 10.1042/BJ20140679. [DOI] [PubMed] [Google Scholar]
- 237.Shahrour MA, et al. Mitochondrial epileptic encephalopathy, 3-methylglutaconic aciduria and variable complex V deficiency associated with TIMM50 mutations. Clinical Genetics. 2017;91:690–696. doi: 10.1111/cge.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ojala T, et al. New mutation of mitochondrial DNAJC19 causing dilated and noncompaction cardiomyopathy, anemia, ataxia, and male genital anomalies. Pediatr Res. 2012;72:432–437. doi: 10.1038/pr.2012.92. [DOI] [PubMed] [Google Scholar]
- 239.Davey KM, et al. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Gen. 2006;43:385–393. doi: 10.1136/jmg.2005.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Richter-Dennerlein R, et al. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metabolism. 2014;20:158–171. doi: 10.1016/j.cmet.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 241.Schusdziarra C, Blamowska M, Azem A, Hell K. Methylation-controlled J-protein MCJ acts in the import of proteins into human mitochondria. Hum Mol Genet. 2013;22:1348–1357. doi: 10.1093/hmg/dds541. [DOI] [PubMed] [Google Scholar]
- 242.Mehawej C, et al. The impairment of MAGMAS function in human is responsible for a severe skeletal dysplasia. PLoS Genet. 2014;10:e1004311. doi: 10.1371/journal.pgen.1004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Jobling RK, et al. PMPCA mutations cause abnormal mitochondrial protein processing in patients with non-progressive cerebellar ataxia. Brain. 2015;138:1505–1517. doi: 10.1093/brain/awv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Vögtle FN, et al. Mutations in PMPCB Encoding the Catalytic Subunit of the Mitochondrial Presequence Protease Cause Neurodegeneration in Early Childhood. Am J Hum Gen. 2018;102:557–573. doi: 10.1016/j.ajhg.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Eldomery MK, et al. MIPEP recessive variants cause a syndrome of left ventricular non-compaction, hypotonia, and infantile death. Genome Med. 2016;8:106. doi: 10.1186/s13073-016-0360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Otto EA, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Gen. 2011;48:105–116. doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.O’Toole JF, et al. Individuals with mutations in XPNPEP3, which encodes a mitochondrial protein, develop a nephronophthisis-like nephropathy. J Clin Invest. 2010;120:791–802. doi: 10.1172/JCI40076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Magen D, et al. Mitochondrial Hsp60 Chaperonopathy Causes an Autosomal-Recessive Neurodegenerative Disorder Linked to Brain Hypomyelination and Leukodystrophy. Am J Hum Gen. 2008;83:30–42. doi: 10.1016/j.ajhg.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hansen JJ, et al. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70:1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bie AS, et al. Effects of a Mutation in the HSPE1 Gene Encoding the Mitochondrial Co-chaperonin HSP10 and Its Potential Association with a Neurological and Developmental Disorder. Front Mol Biosci. 2016;3:e874. doi: 10.3389/fmolb.2016.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Koehler CM, et al. Human deafness dystonia syndrome is a mitochondrial disease. Proc Natl Acad Sci U S A. 1999;96:2141–2146. doi: 10.1073/pnas.96.5.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Roesch K, Curran SP, Tranebjaerg L, Koehler CM. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum Mol Genet. 2002;11:477–486. doi: 10.1093/hmg/11.5.477. [DOI] [PubMed] [Google Scholar]
- 253.Mayr JA, et al. Lack of the mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am J Hum Gen. 2012;90:314–320. doi: 10.1016/j.ajhg.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kang Y, et al. Sengers Syndrome-Associated Mitochondrial Acylglycerol Kinase Is a Subunit of the Human TIM22 Protein Import Complex. Mol Cell. 2017;67:457–470. doi: 10.1016/j.molcel.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 255.Vukotic M, et al. Acylglycerol Kinase Mutated in Sengers Syndrome Is a Subunit of the TIM22 Protein Translocase in Mitochondria. Mol Cell. 2017;67:471–483. doi: 10.1016/j.molcel.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 256.Kukat C, et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A. 2015;112:11288–11293. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]