SUMMARY
Type 1 CD8α+ conventional dendritic cells (cDC1s) are required for CD8+ T cell priming but, paradoxically, promote splenic Listeria monocytogenes infection. Using mice with impaired cDC2 function, we ruled out a role for cDC2s in this process and instead discovered an interleukin-10 (IL-10)-depen-dent cellular crosstalk in the marginal zone (MZ) that promoted bacterial infection. Mice lacking the guanine nucleotide exchange factor DOCK8 or CD19 lost IL-10-producing MZ B cells and were resistant to Listeria. IL-10 increased intracellular Listeria in cDC1s indirectly by reducing inducible nitric oxide synthase expression early after infection and increasing intracellular Listeria in MZ metallo-philic macrophages (MMMs). These MMMs trans-in-fected cDC1 s, which, in turn, transported Listeria into the white pulp to prime CD8+ T cells. However, this also facilitated bacterial expansion. Therefore, IL-10-mediated crosstalk between B cells, macrophages, and cDC1 s in the MZ promotes both Listeria infection and CD8+ T cell activation.
Graphical Abstract
In Brief
Splenic dendritic cells (DCs), IL-10, and marginal zone (MZ) B cells each promote Listeria monocytogenes infection. Liu et al. show that these paradoxical responses are linked. IL-10 production from MZ B cells enhances the intracellular Listeria burden in DCs by inhibiting bacterial killing in MZ metal lo philic macrophages. This crosstalk in the MZ promotes CD8+ T cell activation but also Listeria infection.
INTRODUCTION
Listeria monocytogenes (Lm) is a Gram-positive facultative intracellular bacterium and the causative agent of listeriosis, which has a mortality rate of 20%–3Q% (de Noordhout et al., 2014), Upon infection, bacteria quickly disseminate to the spleen and liver (Cossart, 2011). The spleen plays a protective role in host resistance to many infections. Therefore, splenectomy substantially impairs the host defense against bacterial invasion, particularly encapsulated organisms (Cheslyn-Curtis et al., 1988; Cull-ingford et al., 1991). Contrary to this, splenectomized hosts are resistant to Listeria infection (Skamene and Chayas ir isobhon, 1977) because Listeria manipulates the microenvironment of the spleen to favor its survival; however, the cellular mechanisms of this are only partially understood.
The spleen is a peripheral lymphoid organ and the largest filter of blood. Functionally and morphologically, the spleen can be divided into two distinct compartments: the red and white pulp. The white pulp (WP) is demarcated in mice by the marginal zone (MZ), which contains a number of immune cell types, including MZ B cells, granulocytes, macrophages, and XCR1+ CD8α+ type 1 conventional dendritic cells (cDC1 s) (Eisenbarth, 2019). CD8α+ cDC1 s transport Listeria to the WP to initiate antigen presentation to CD8+ T cells, which are crucial for clearance of the pathogen (Qiu et al., 2018); however, the migration of CD8α+ cDC1 s also promotes Listeria proliferation and pathology (Aoshi et al., 2008). Elimination of cDC1s in Baff3-deficient mice results in resistance to Listeria infection but also impaired CD8+ T cell priming (Edelson et al., 2011; Neuenhahn et al., 2006; Theisen and Murphy, 2017), whereas increased numbers of CD8α+ cDC1s cause more severe disease (Alaniz et al., 2004). In this way, Listeria uses CD8α+ cDC1s as a “Trojan horse” to migrate to the sanctuary of the splenic WP to escape killing in the MZ by phagocytes. The second conventional DC subset in the spleen, CD4+33D1+CD11 b+ type 2 conventional dendritic cells (cDC2s) (Guilliams et al., 2014), are also found at the border of the MZ but reside in a distinct region called the bridging channel. The role of cDC2s in regulating infection or T cell responses to Listeria has not been tested.
DOCK8 (dedicator of cytokinesis 8) is an atypical guanine nucleotide exchange factor that plays an important role in controlling many cellular functions, including migration of immune cells (Calabro et al., 2016b; Harada et al., 2012; Zhang et al., 2014). Patients with mutations in DOCK8 are immuno-deficient and susceptible to bacterial infections (Zhang et al., 2009). Mice lacking DOCK8 have impaired interstitial DC migration (Harada et al., 2012; Krishnaswamy et al., 2015), but only in the cDC2 subset, leaving splenic cDC1 migration intact (Calabro et al., 2016b; Krishnaswamy et al., 2017). This results in loss of CD4+ but not CD8+ T cell activation to protein antigens in the circulation (Calabro et al., 2016a; Calabro et al., 2016b).
Consistent with our previous studies, cDC2 but not cDC1 migration was impaired in the spleen of Dock8−/− mice after Listeria infection. We found that, although CD8+ T cell responses to heat-killed (HK) Listeria were normal, Dock8−/− mice demonstrated impaired CD8+ T cell responses to live Listeria infection. Although cDC1 s migrated normally to the WP in Dock8−/− mice during Listeria infection, the load of cDC1s with intracellular Listeria was significantly reduced, and, therefore, the mice were resistant to Listeria infection. Using different cell-specific DOCK8-deficient mice, we confirmed that bacterial resistance was not due to a DC-intrinsic effect but, rather, a B cell-intrinsic defect, resulting in loss of MZ B cells. These innate-like B cells co-localized with cDC1s in the MZ during the early stages of infection and could potentially modify the function of cDC1s. Using two models of MZ B cell loss, Dock8−/− and Cd19−/− mice, along with cell-specific deletion of interleukin-10 (IL-10) or the IL-10 receptor (IL-1 OR), we showed that IL-10 production by MZ B cells promoted the cDC1 intracellular Listeria burden by regulating the adjacent MZ metallophilic macrophages (MMMs); ultimately, this propagates infection in the WP after cDC1 migration. Our work uncovers a crosstalk between MZ B cells, MMMs, and cDC1s within the MZ that promotes DC bacterial load and thereby enhances Listeria infection but also promotes CD8+ T cell activation.
RESULTS
Dock8−/− Mice Have Intact CD8+ T Cell Responses to Heat-Killed, but Not Live, Listeria
CD8α+ cDC1 s are the primary cross-presenting conventional DC subset and are essential for CD8+ T cell activation to multiple systemic antigens (Theisen and Murphy, 2017), However, cDC1s play a detrimental role during Listeria infection by harboring and transporting Listeria into the WP. How cDC2s affect Listeria infection or the CD8+ T cell response to these intracellular bacteria has not been studied. To investigate the role of cDC2s in Listeria infection, we took advantage of Dock8−/− mice, which have a selective migration defect in cDC2s and, therefore, impaired CD4+T cell responses to systemic antigen in the spleen (Calabro et al., 2016a, 2016b; Krishnaswamy et al., 2015). We first confirmed that Dock8-deficient mice have a cDC2, but not cDC1, intra-splenic migration defect during live Listeria infection using in vivo labeling (Figure S1A). This finding is similar to our prior results using lipopolysaccharide and allogeneic red blood cells (Calabro et al., 2016a) and consistent with intact CD8+ T cell activation to intravenous (i.v.) ovalbumin (OVA) immunization (Calabro et al., 2016b).
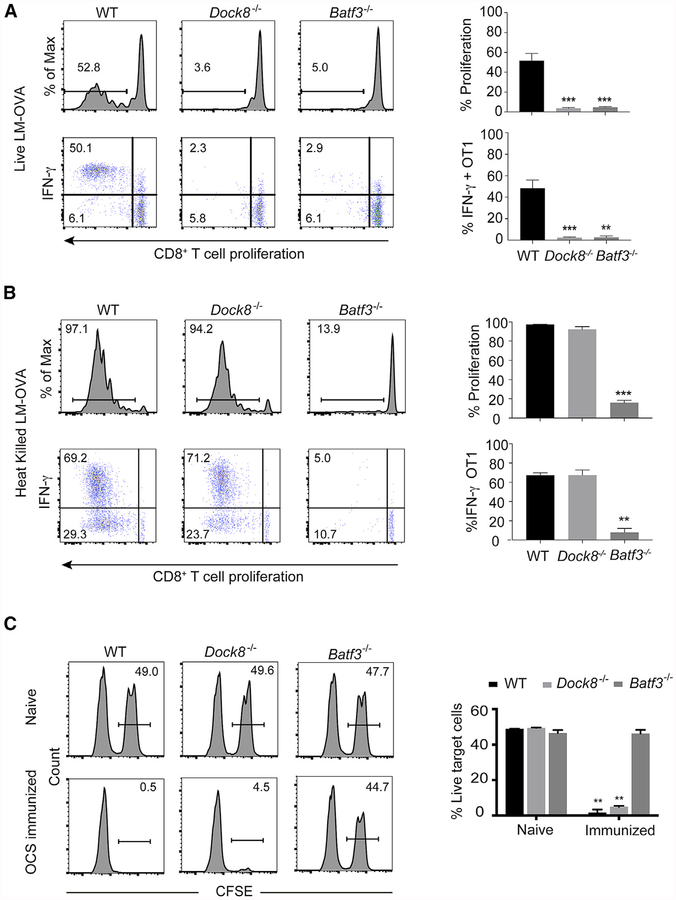
To test the T cell response to Listeria, we adoptively transferred OVA-specific carboxyfluorescein succinimidyl ester (CFSE)-labeled OT-1 CD8+ T cells into wild-type (WT), Dock8−/−, and Batf3−/− mice and infected recipient mice with 103 live recombinant Listeria expressing OVA (rLm-OVA). On day 3 after infection, WT, but not cDC1-deficient, Batf3−/− mice demonstrated intact CD8+ T cell priming (Figure 1A), consistent with a previous study (Edelson et al., 2011). OT-1 T cell proliferation and cytokine production were also impaired in Dock8−/− mice (Figure 1A). Using a second mouse strain with selective loss of cDC2 development, CD11ccre-Irf4fl/fl (DC-Irf4−/−) mice (Bajaña et al., 2012), we did not observe a defect in OT-1 proliferation (Figure S1B), indicating that the loss of cDC2 migration in Dock8−/− mice was not likely to account for the CD8+T cell activation defect. Therefore, we tested whether Listeria viability determined the outcome of the T cell response to OVA in Dock8−/− mice. We transferred CFSE-labeled OT-1 and OT-2 T cells into WT, Dock8−/−, and Batf3−/− mice, followed by i.v. immunization with heat-killed rLm-OVA (HKLm-OVA), Again, OT-1 T cell proliferation and cytokine production were normal in WT but not Batf3−/− mice (Figure 1 B). In contrast, OT-2 activation to HKLm-OVA was normal in both Batf3−/− and WT mice but defective in Dock8−/− mice (Figure S1C). This is consistent with our previous findings using OVA immunization and supports the division-of-labor paradigm between cDC1 and cDC2 priming of CD8+versus CD4+T cells, respectively. CD8+ T cell proliferation and interferon y (IFNγ) production to HKLm-OVA were also intact in Dock8−/− mice (Figure 1B), suggesting that CD8α+ cDC1s are indeed functional in the absence of DOCK8 but that the immune response to a live infection is altered.
Figure 1. Dock8−/− Mice Have an Impaired CD8+ T Cell Response to Live Listeria Infection but Intact CTL Effector Responses to Inert Antigens.
(A and B) 1 × 106 CFSE-labeled OT-1 T cells were adoptively transferred into WT, Dock8−/−, and Batf3−/− mice. One day later, the recipient mice were i.v. infected with 103 live rLm-OVA (A) or with 108 HKLm-OVA (B). After 3 days, the mice were sacrificed, and the spleens were collected for OT-1 cell proliferation and IFNγ production analysis by flow cytometry. Results are representative of five independent experiments with n = 3 or 4/group.
(C) Wild-type (WT), Dock8−/−, and Batf3−/− mice were i.v. injected with 107 irradiated OVA-coated MHC class l-deficient splenocytes. 5 days later, these mice were i.v. injected with 107 OVA peptide pulsed target cells (CFSEhi) and 107 non-pulsed cells (CFSEI0). 16 h after target cell injection, the percentage of CFSEhl cells in the spleen was evaluated by flow cytometry. Results are representative of three independent experiments with n = 3 or 4/group.
Numbers indicate the percentage of proliferating cells or cytokine producers in the indicated gates. Data are means ± SD. **p < 0.01, ***p < 0.001 (compared with the WT in A and B and Batf3−/− in C). See also Figure S1.
To test cytotoxic CD8+ T cell function directly, we used an in vivo killing assay (Prajeeth et al., 2010). In this assay, WT, Dock8−/−, and Batf3−/− mice were i.v. injected i.v. with pre-irra-diated ovalbumin-coated splenocytes (OCSs) from major histocompatibility complex (MHC) class l-deficient mice. 5 days later, these mice were i.v. injected with unmodified or OVA peptide-pulsed target cells. Cytotoxic T lymphocytes (CTLs) generated in Dock8−/− mice were as efficient as in WT mice at killing target cells, whereas Batf3−/− mice failed to generate effective CTLs (Figure 1C). Thus, Dock8-deficient mice have a selective defect in CD8+ T cell priming to live but not inert antigens.
Dock8−/−Mice Are Resistant to Live Listeria Infection
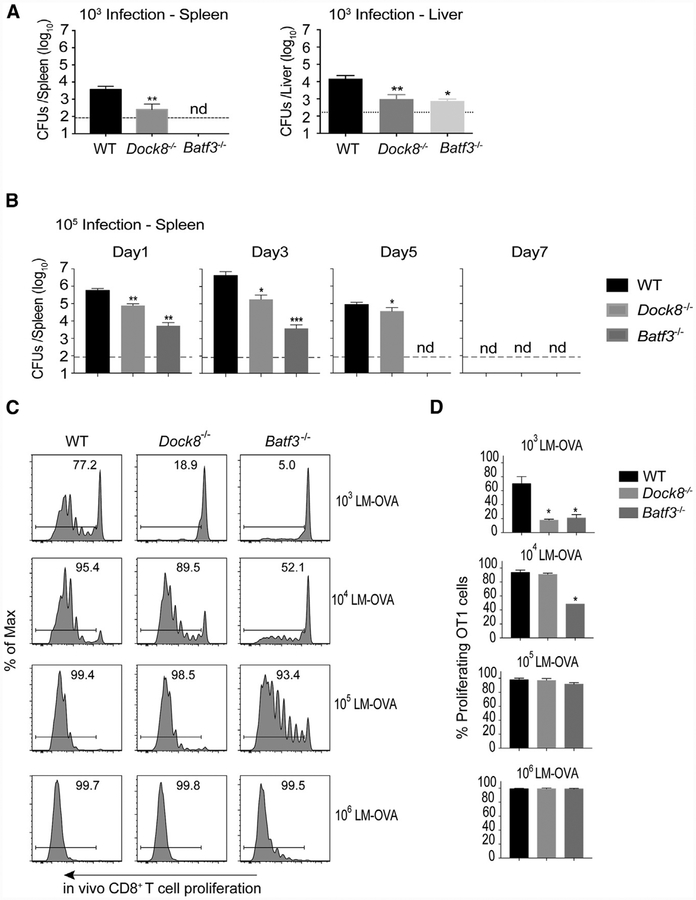
CD8+ T cell activation to Listeria is proportional to the bacterial burden (Edelson et al., 2011). Therefore, loss of cDC1s in Saff3-deficient mice impairs CD8+ T cell activation not only because they are the primary antigen-presenting cells (APCs) for CTLs but also because they promote the bacterial burden. We therefore asked whether loss of CD8+ T cell activation in Dock8−/− mice with live but not HK Listeria was due to a reduced Listeria burden. We analyzed the bacterial burden in both the spleen and liver of WT, Dock8−/−, and Batf3−/− mice 3 days after Listeria infection at the dose used to analyze T cell activation and at a higher dose that facilitates Listeria colony-forming unit (CFU) quantitation. Similar to Batf3−/− mice, Dock8−/− mice were highly resistant to Listeria infection in both the spleen and liver throughout the course of infection (Figures 2A and 2B; Figure S2). Therefore, Batf3−/− mice had a defect in CD8+ T cell activation during both live and dead Listeria infection because of the combined defect in cDC1 antigen presentation and a reduced bacterial load. In contrast, Dock8−/− mice had a defect in bacterial burden but not antigen presentation to CD8+ T cells and, therefore, only demonstrated defects during a live but not dead Listeria challenge (Figures 1 and 2).
Figure 2. Dock8−/− Mice Are Resistant to Live Listeria Infection.
(A) WT, Dock8−/−, and Batf3−/− mice were i.v. infected with 103 live rLm-OVA. The burdens of rLm-OVA in the spleen and liver were determined by colony-forming units (CFUs) on day 3 after infection. Dashed lines indicate detection limit. Results are representative of two independent experiments with n = 4 or 5/group.
(B) WT, Dock8−/−, and Batf3−/− mice were i.v. infected with 10s live rLm-OVA. The burden of rLm-OVA in the spleen was determined by CFUs at the indicated time points after infection. Results are representative of two independent experiments with n = 4 or 5/group.
(C and D) 1 × 106CFSE-Iabeled OT-1 T cells were adoptively transferred into WT, Dock8−/−, and Batf3−/− mice. 1 day later, recipient mice were i.v. infected with the indicated dose of live rLm-OVA. 3 days later, T cell proliferation was analyzed by flow cytometry. Representative flow cytometry plots (C) and summary bar graphs (D) are shown. Results are representative of three independent experiments with n = 2 or 3/group.
Data are means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 (compared with the WT); nd, not detectable. See also Figure S2.
To quantitatively examine the contribution of Listeria burden to CD8+ T cell priming, we adoptively transferred OT-1 T cells and infected the recipient mice with different doses of live rLm-OVA. Increasing the dose of Listeria infection restored CD8+ T cell priming in Dock8−/− mice (Figures 2C and 2D). Impaired CD8+ T cell priming in Batf3−/− mice could also be restored with increasing Listeria doses, indicating that BATF3-independent APCs (Seillet et al., 2013) could compensate for CD8+T cell activation at higher antigen doses (Figures 2C and 2D), consistent with previous findings (Edelson et al., 2011).
Reduced Intracellular Listeria in Dock8-Deficient CD8α+ cDC1s Is Due to a DC-Extrinsic Effect
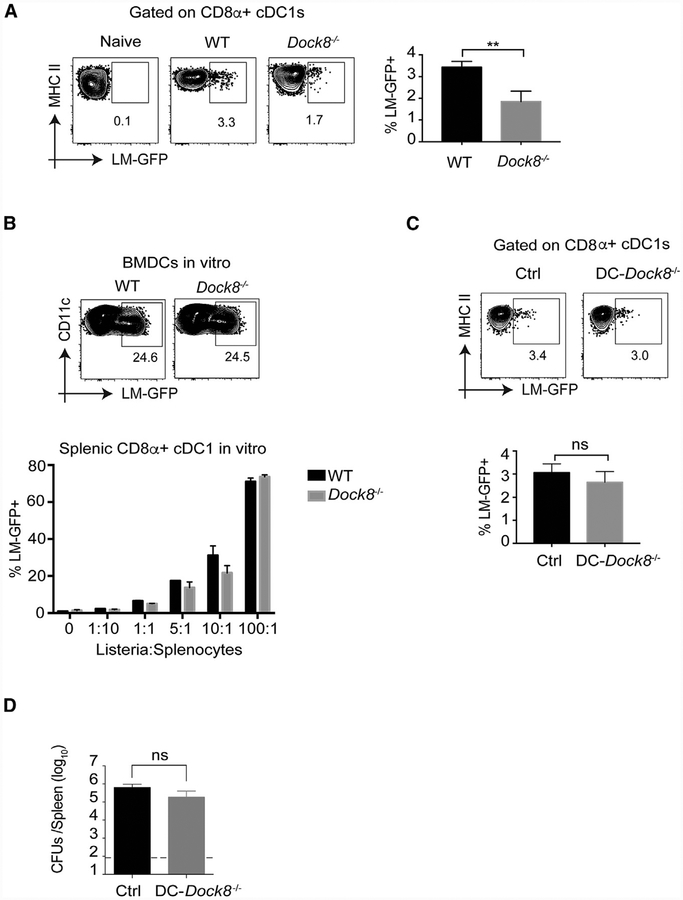
CD8α+ splenic cDC1 s are crucial for transporting Listeria from the red pulp (RP) to the WP, which promotes Listeria proliferation and the bacterial burden (Aoshi et al., 2008; Neuenhahn et al., 2006). Because cDC1s in Dock8−/− mice migrated normally into the WP (Figure S1A), we asked whether they transported an equivalent amount of bacteria to the WP. Using GFP-expressing Listeria (Lm-GFP), we quantitated intracellularListeria in DCs (Figures S3A and S3B). First, we wanted to know whether splenic cDC1s and cDC2s take up equivalent amounts of Listeria. To test this, we in vitro infected WT splenocytes with live immunized Lm-GFP at an MOI. of 5 for 2.5 h. Flow cytometric analysis of cDC1s and cDC2s demonstrated that a small but equivalent fraction of both subsets contained intracellular Listeria (Figure S3B), suggesting that cDC1s and cDC2s did not have intrinsic differences that alter Listeria uptake. However, this in vitro infection model lacks the splenic architecture and potential cell-cell interactions present in vivo. To test the DC Listeria load in vivo, we i.v. infected both WT and Dock8−/− mice with Lm-GFP and analyzed intracellular Lm-GFP in CD8α+ cDC1s at 4 h post-infection. Although the total burden of bacteria in the spleen was only minimally different between WT and Dock8−/− mice at this early time point (Figure S3C), the amount of Listeria in CD8α+ cDC1s in Dock8−/− mice was significantly less than in cDC1s from WT mice (Figure 3A). This was not due to impaired phagocytic ability of Dock8−/− cDC1s because Dock8−/− bone-marrow-derived DCs (BMDCs) and primary splenic cDC1s contained an equivalent number of intracellular Listeria upon in vitro infection (Figure 3B).
Figure 3. CD8α+ cDC1s Contain Less Listeria in Dock8−/−Mice but Not Because of a DC-Intrinsic Defect.
(A) WT and Dock8−/− mice were i.v. infected with 5–10 × 108 Lm-GFP. 4 h later, the mice were sacrificed to measure intracellular Listeria in CD8α+ cDC1s. **p < 0.01. Results are representative of four independent experiments with n = 3 or 5/group.
(B) Bone-marrow-derived dendritic cells (BMDCs) generated from WT or Dock8−/− mice were in vitro infected with Lm-GFP at MOI 5, and the BMDC Listeria load was analyzed based on GFP expression by flow cytometry (top dot plots). Freshly prepared splenocytes from naive WT or Dock8−/− mice were in vitro infected with Lm-GFP at different MOIs, and the CD8α+ cDC1 bacterial load was gauged by GFP expression (bottom graph). Results are representative of three independent experiments.
(C) DC-Dock8−/− (CD11ccre-Dock8fl/fl) and (Dock8fl/fl) Cre control mice were i.v. infected with 5–10 × 108 Lm-GFP. 4 h later, the mice were sacrificed to measure intracellular Listeria in CD8α+ cDC1s. Results are representative of two independent experiments with n = 3 or 5/group.
(D) DC-Dock8−/− (CD11ccre-Dock8fl/fl) and Cre− control mice were i.v. infected with 10s live rLm-OVA. The burdens of rLm-OVA in the spleen were determined by CFUs 3 days after infection, ns, not significant. Results are representative of three independent experiments with n = 4 or 5/group. Data are means ± SD. See also Figure S3.
To test whether DC-intrinsic loss of DOCK8 affected resistance to Listeria infection, we generated mice with isolated Dock8 deficiency in DCs by crossing CD11ccre mice with Dock8m1 mice (Krishnaswamy et al., 2017). CD8α+ cDC1s contained equivalent intracellular Listeria in CD11ccre-Dock8fl/fl (IDC-Dock8−/−) mice 4 h after infection (Figure 3C), and the splenic bacterial burden was normal in CD11ccre-Dock8fl/fl mice 3 days after infection (Figure 3D). Using a second model of cDC2 loss, CD11ccre-Irf4fl/fl mice, we further confirmed that cDC2s play no role in bacterial burden (Figure S3D). These data indicate that the reduced Listeria burden in cDC1s of Dock8-deficient mice is due to a DC-extrinsic mechanism.
Marginal Zone B Cells Quantitatively Promote Listeria Infection and Are Lost in Dock8-Deficient Mice
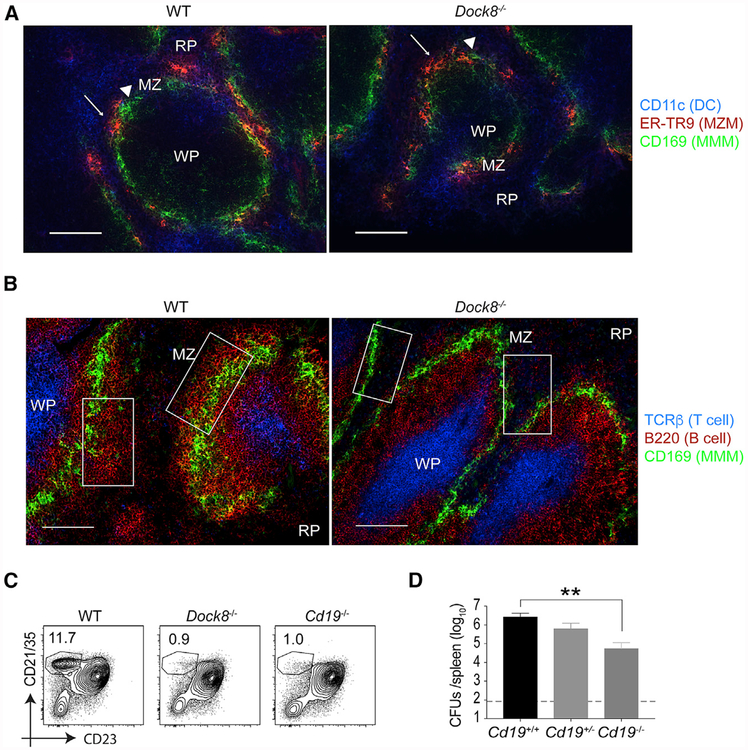
A recent study suggests that CD169+ MMMs trans-infect CD8α+ cDC1 s (Perez et al., 2017). MMMs and SIGN-R1+ marginal zone macrophages (MZMs) are the primary cellular site for Listeria entry into the spleen (Qiu et al., 2018). We therefore examined the MZ for these cellular immune constituents by immunofluores cence staining. Both MMMs and MZMs were present and situated normally in the MZ of Dock8-deficient mice, and the splenic architecture was grossly intact (Figure 4A; Figures S4A and S4B). However, we did observe a slight reduction in CD169 staining on macrophages by flow cytometry (data not shown); the reason for this is not clear but is reminiscent of the reported loss of SIGN-R1 expression on MZMs in mice lacking MZ B cells (You et al., 2011).
Figure 4. Loss of MZ B Cells Enhances Resistance to Listeria Infection.
(A and B) Fluorescence images of a spleen from a naive WT mouse (left) and a Dock8−/− mouse (right).
(A) Dendritic cells (DCs; CD11c in blue), marginal zone macrophages (MZMs; ER-TR9 in red, white arrow), and metallophilic macrophages (MMMs; CD169 [MOMA-1] in green, white triangles). Scale bars, 100 μm.
(B) T cell zone (T cell receptor β [TCRβ] in blue), B cells (B220 in red), and MM Ms (CD169 in green). Scale bars, 100 urn. White squares in the panels indicate MZ B cell areas. A representative spleen from 4 different mice/group is shown.
(C) Naive WT, Dock8−/− and Cd19−/− mice were analyzed for the percentages of the MZ B cell population among total B cells in the spleens by flow cytometry.
(D) WT (Cd19+/+), Cd19+/−, and Cdt9−/− mice were i.v. infected with 105 live rLm-OVA. The burdens of rLm-OVA in the spleen were determined by CFUs on day 3 after infection. **p < 0.01. Results are representative of two or three independent experiments with n = 4 or 5/group. RP, red pulp; WP, white pulp; MZ, marginal zone. Data are means ± SD. See also Figure S4.
MZ B cells (Lopes-Carvalho and Kearney, 2004), another major constituent of the splenic MZ, have also been reported to regulate Listeria burden through an IL-10-dependent mechanism (Lee and Kung, 2012). Dock8 deficiency causes loss of MZ B cell development (Figures 4B and 4C; Krishnaswamy et al., 2015; Randall et al., 2009) for unclear reasons. Cd19-deficient mice also fail to develop MZ B ceils (Lopes-Carvalho and Kearney, 2004), presumably because of loss of phosphatidylinositol 3-kinase (PI3K) signaling pathways downstream of CD19 co-ligation (Anzelon et al., 2003). DOCK8 has been reported to regulate Cd19 transcription (Sun et al., 2018). Cd19−/− mice are resistant to Listeria infection through loss of IL-10 production (Horikawa et al., 2013), although the mechanism by which IL-10 promotes infection has not been established. We confirmed that the Cd19−/− mice phenocopied Dock8−/− mice and further showed a quantitative correlation between Listeria burden and the MZ B cell frequency (Figures 4D, S4C, and S4D). Consistent with previous studies, these data suggest that MZ B cells are detrimental to the host during Listeria infection and that loss of this B cell population in both Dock8−/− and Cd19−/− mice enhances bacterial resistance.
WT Marginal Zone B Cells Restore Listeria Susceptibility in Dock8−/− Mice
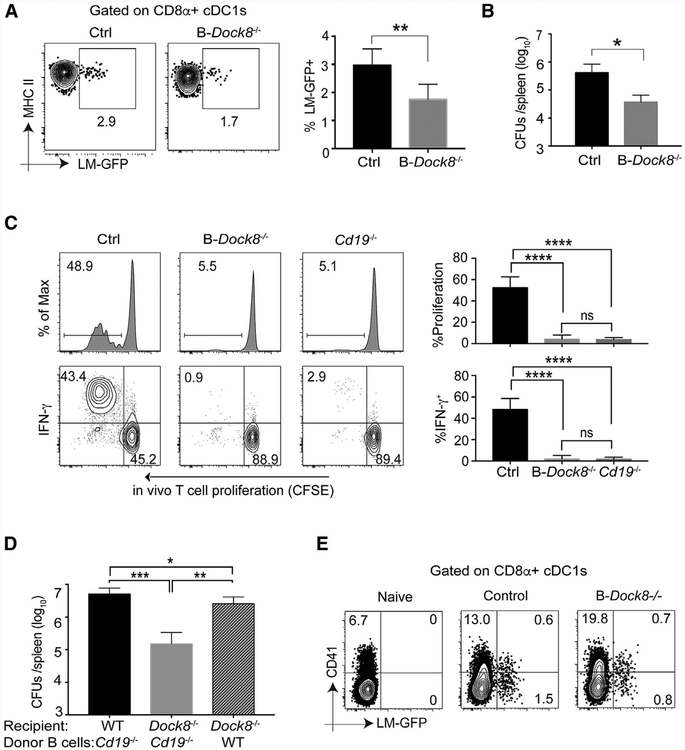
To confirm the B cell-intrinsic role of DOCK8 in the MZ B-dependent Listeria burden, we generated mice with conditional deletion of Dock8 in B cells by crossing Mb1cre mice with Dock8fl/fl mice. These Mb1cre-Dock8fl/fl (B-Dock8−/− ) mice had normal T cell and DC subset frequencies (Figure S5A) but almost complete loss of MZ B cells (Figure S5B), demonstrating that DOCK8 regulates MZ B cell development in a cell-intrinsic manner. In vivo Lm-GFP tracking experiments demonstrated that Mb1cre-Dock8fl/fl mice phenocopied the reduced intracellular Listeria in cDC1 s found in global Dock8 knockout mice (Figure 5A). We i.v. infected Mb1cre-Dock8fl/fl and control mice with 1Q5 live rLm-OVA. 3 days later, the Listeria burden in the spleen was evaluated. Mb1cre-Dock8fl/fl mice had significantly lower burdens than control mice (Figure 5B). Accordingly, adoptively transferred OT-1 cells failed to proliferate in Mb1cre-Dock8fl/fl mice (Figure 5C). These data indicate that B cell-intrinsic loss of DOCK8 regulates the intracellular bacterial burden in CD8α+ cDC1s, likely through loss of MZ B cells. Using a second model of MZ B cell deficiency, Cd19−/− mice, we again observed impaired CD8+ T cell activation to Listeria (Figure 5C). Therefore, impaired CD8+ T cell activation to Listeria is likely due to bacterial resistance afforded by MZ B cell elimination.
Figure 5. Conditional Deletion of Dock8 in B Cells Phenocopies Dock8−/− Mice, and WT B Cell Reconstitution Restores the Listeria Susceptibility of Dock8−/− Mice.
(A) B-Dock8−/− (Mb1cre-Dock8fl/fl) and Cre− control mice were infected with 5–10 × 108 Lm-GFP. 4 h later, the Listeria load in CD8α+ cDC1s was measured by flow cytometry. Results are representative of three independent experiments with n = 3/group.
(B) B-Dock8−/− (Mb1cre-Dock8fl/fl) and control mice were i.v. infected with 105 live rLm-OVA. The burdens of rLm-OVA in the spleen were determined by CFUs on day 3 after infection. Results are representative of three independent experiments with n = 4 or 5/group.
(C) 1 × 106 CFSE+ OT-1 T cells were adoptively transferred into WT, B-Dock8−/−, and Cd19−/− mice, and 1 day later, the recipients were i.v. infected with 103 live rLm-OVA. 3 days later, the mice were sacrificed, and the spleens were collected for analyses of T cell proliferation and IFNγ production by flow cytometry. Results are representative of three independent experiments with n = 3/group.
(D) Sub-lethally irradiated WT and Dock8−/− mice were reconstituted with 2 × 107 enriched B cells from Cd19−/− or WT mice. 8–10 weeks after transfer, the recipient mice were i.v. infected with 105 live rLm-OVA. The burdens of rLm-OVA in the spleen were determined by CFUs on day 3 after infection. Results are representative of two independent experiments with n = 4 or 5/group.
(E) B-Dock8−/− (Mb1cre-Dock8fl/fl) and control mice were i.v. infected with 109 live CFSE-labeled Lm-GFP. 4 h later, the mice were sacrificed, and splenocytes were surface stained, fixed, and permeabilized for intracellular platelet marker CD41 staining in CD8α+ cDC1s. Results are representative of two independent experiments with n = 4 or 5/group.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are means ± SD. See also Figure S5.
We used adoptive transfer experiments to test whether the Listeria-resistant phenotype of Dock8−/− mice can be reversed by MZ B cell reconstitution. WT and Dock8−/− mice were sub-lethally irradiated, and enriched B cells from Cd19−/− or WT mice were adoptively transferred into recipient mice (Figure S5C). 8–10 weeks later, recipient mice were i.v. infected with 105 live rLm-OVA. Dock8−/− mice that received WT B cells had significantly higher burdens than Dock8−/− mice reconstituted with MZ B cell-deficient B cells from Cd19−/− donors (Figure 5D), indicating that WT MZ B cells could restore the Listeria susceptibility of Dock8−/− mice. These results demonstrate that Dock8 deficiency in B cells resulted in loss of MZ B cells, which is responsible for the enhanced resistance and impaired T cell priming in Dock8−/− mice. Previous work identified a platelet-based “delivery mechanism” of Listeria to CD8α+ cDC1s (Verschoor et al., 2011). However, we did not see a difference in platelet-associated GFP+ Listeria in cDC1s between control and Mb1cre-Dock8fl/fl mice (Figure 5E), suggesting that MZ B cells regulate cDC1 infectivity through another pathway.
Marginal Zone B Cell-Derived IL-10 Enhances the Intracellular Listeria Burden in CD8α+ cDCts
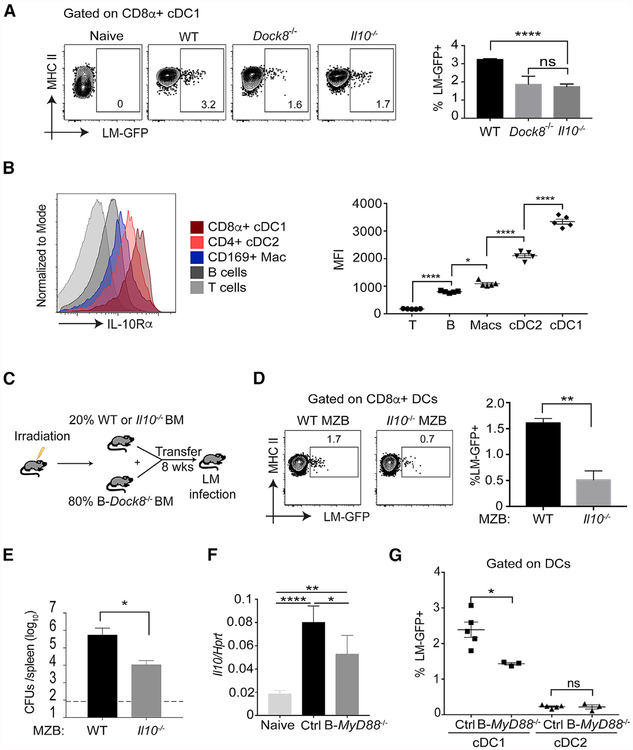
II-10−/− mice are highly resistant to Listeria infection (Dai et al., 1997). It has been demonstrated that MZ B cells are the major producers of IL-10 upon Listeria infection (Lee and Kung, 2012). It has been presumed that B cell-derived IL-10 impairs macrophage clearance of Listeria in the RP and thereby promotes the splenic bacterial load (Horikawa et al., 2013). We hypothesized instead that an IL-10 response by MZ B cells to Listeria infection acts on neighboring cells in the MZ, enhancing the intracellular bacterial burden in cDC1s, ultimately promoting infection through the Trojan horse effect. To test this, we infected WT, Dock8−/−, and II10−/− mice with Lm-GFP (Figure 6A). The amount of Listeria in CD8α+ cDC1s in both II10−/− and Dock8−/− mice was reduced significantly (Figure 6A).
Figure 6. Loss of Marginal Zone B Cell-Derived IL-10 Impairs Acquisition of Listeria by CD8α+ cDC1s.
(A) Naive, WT, Dock8−/−, and II10−/− mice were infected with 5–10 × 108 Lm-GFP. 4 h later, the mice were sacrificed to measure the Listeria burden in CD8α+ cDC1 s. Results are representative of two independent experiments with n = 4 – 5/group.
(B) Surface expression of IL-10Ra on different cell subsets in the spleen of naive WT mice were analyzed by flow cytometry and expressed as mean fluorescence intensity (MFI). Results are representative of two independent experiments with n = 2–5/group.
(C) Cartoon illustration of mixed BM chimeric mice. 2 × 105 BM cells from either WT or II10−/− mice and 8 × 105 BM cells from B-Dock8−/− (Mb1cre-Dockfl/fl) mice were adoptively transferred into lethally irradiated CD45.1 B6 recipient mice. 8 weeks later, the intracellular cDC1 Listeria and splenic bacterial burdens were analyzed.
(D) CD8α+ cDC1 Listeria load of mixed BM chimeric mice with either WT or II10−/− MZ B cells 4 h after Lm-GFP infection. Results are representative of two independent experiments with n = 3–6/group.
(E) Mixed BM chimeric mice were i.v. infected with 105 live rLm-OVA. The burden of rLm-OVA in the spleen was determined by CFUs on day 3 after infection.
(F) B-Myd88−/− (Mb1cre-Myd88fl/fl) and (Myd88fl/fl) Cre− control mice were infected with 5–10 × 108 Lm-GFP. 4 h later, B cells were enriched and used for II-10 mRNA analysis byqPCR.
(G) B-MyD88−/− and Cre− control mice were infected with 5–10 × 108 Lm-GFP. 4 h later, the Listeria load in CD8cz+ cDC1s and CD11b+ cDC2s was measured by flow cytometry. Results are representative of two independent experiments with n = 3–5/group.
*p < 0.05, **p < 0.01, ****p < 0.0001. Data are means ± SD. See also Figure S6.
To find out whether IL-10 can act directly on DCs, we measured the surface expression of IL-10R« on different cell subsets in the spleen of naive WT mice. CD8α+ cDC1s expressed the highest amounts of IL-10R«, including other DC and macrophage populations (Figure 6B). This expression pattern is maintained even after Listeria infection (Figure S6A). To confirm that MZ B cells are the relevant source of IL-10 that affects cDC1s, we generated mixed bone marrow (BM) chimeric mice to isolate IL-10 deficiency to MZ B cells (Figures 6C and S6B). Transfer of 20% BM cells from either WT or II-10−/− mice with 80% BM cells from Mb1cre-Dock8fl/fl mice into lethally irradiated CD45.1 B6 recipient mice generated “WT MZ B” (IL-10-sufficient MZ B) or “II-10−/− MZ B.” Eight weeks later, recipient mice were infected with Lm-GFP. CD8α+ cDC1s from II-10−/− MZ B mice had significantly lower Listeria burdens than those from WT MZ B mice (Figure 6D). When mixed BM chimeric mice were i.v. infected with 105 live rLm-OVA, II-10−/− MZ B mice were also resistant to infection (Figure 6E) and had impaired CD8+ T cell activation (Figure S6C). These data suggest that MZ B cell-derived IL-10 acts, either directly or indirectly, on CD8α+cDC1s to promote intracellular Listeria and that disruption of this axis results in bacterial resistance and impaired CD8+ T cell activation.
MyD88 Signaling in Marginal Zone B Cells Induces IL-10 and Promotes Intracellular Listeria Burden in CD8α+ cDCfs
We next asked which signal induces IL-10 production from MZ B ceils. Listeria is a Gram-positive, flagellated rod with multiple virulence factors that activate numerous pattern recognition receptors (Zenewicz and Shen, 2007). In addition, Listeria coating by complement can facilitate its uptake by innate immune cells but also facilitates spleen colonization (Verschoor et al., 2011). Because MZ B cells express high levels of complement receptors (Cerutti et al., 2013), it is possible that complement-coated Listeria interacts with MZ B cells and that this triggers early IL-10 production. Listeria infection induces II-10 mRNA in B cells within 4 h of infection (Figure 6F). This is significantly impaired when B cells lack the Toll-like receptor signaling adaptor MyD88 (Figure 6F) but not in mice lacking the central complement component C3 (Figure S6D). Indeed, in vitro activation of B cells by the Toll-like receptor 2 (TLR2) ligand Pam3CSK4 significantly induced II-10, whereas one of the other primary Toll-like receptor (TLR) ligands expressed by Listeria, flagellin, failed to do so (data not shown), possibly because of the low expression of mRNA for Tir5 in MZ B cells (Heng et al., 2008). Mice with B cell-specific Myd88 deletion also demonstrated a significant reduction in intracellular Listeria in cDC1s (Figure 6G) but not within cDC2s (Figure 6G) or MZ B cells (Figure S6E). These data suggest that MZ B cells recognize Listeria via a TLR, most likely TLR2, and that this induces IL-10, which selectively promotes the cDC1 bacterial load.
IL-10 Acts on trans-Infecting Macrophages in the Marginal Zone to Promote the cDC1 Listeria Load
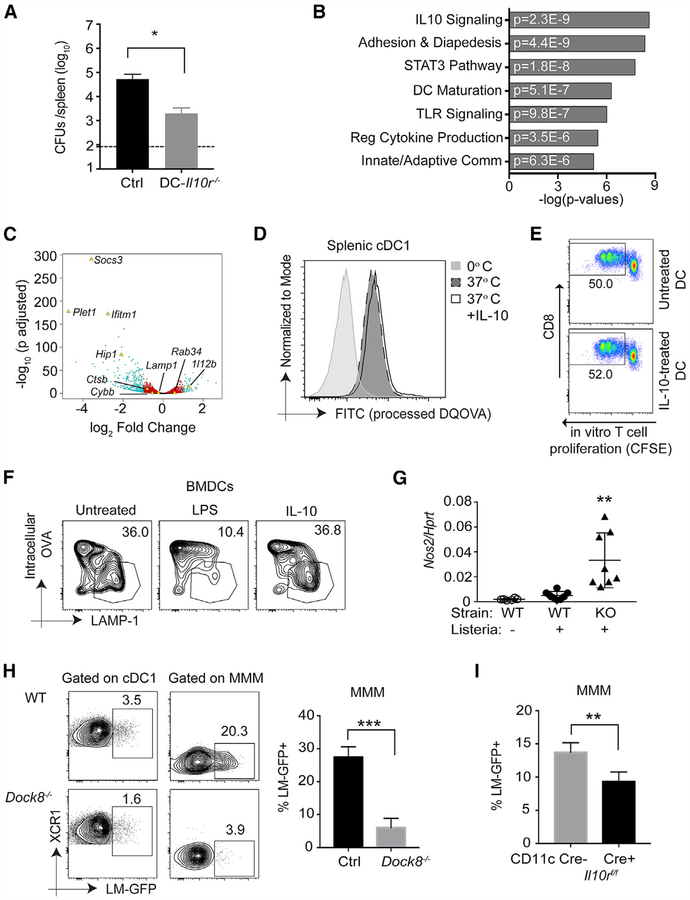
To directly test whether IL-10 alters the handling of intracellular Listeria by DCs, we generated mice in which the IL-10 receptor was selectively deficient on cells expressing CD11c, which includes all conventional DCs but also some macrophage populations (Probst et al., 2005) and some activated B cells. These Cd11ccre-II10rfl/fl (DC-II10rfl/fl) mice indeed had a lower bacterial burden in the spleen, mirroring mice in which IL-10 or MZ B cells are lost (Figure 7A). This difference was most pronounced in female mice, consistent with the enhanced susceptibility of female mice to Listeria because of elevated IL-10 expression (Pasche et al., 2005). If IL-10 was directly acting on DCs to promote intracellular Listeria, then we hypothesized that it could act in three ways: enhance the cellular machinery for phagocytosis, promote Listeria escape from the phagosome into the cytosol, or reduce killing of phagocytosed bacteria by blocking acidification and lysosomal fusion of phagosomes. We found no difference in uptake of labeled inert antigens such as beads in IL-10-treated DCs nor enhanced escape of Listeria into the cytosol using RFP reporter bacteria (data not shown). It is known that CD8α+ cDC1s are specialized at cross-presentation and have developed specific strategies in their endocytic pathway to promote preservation of phagocytosed antigen and transport to the cytosol (Joffre et al., 2012; Schuette and Burgdorf, 2014). Therefore, we focused on the last mechanism of antigen processing and degradation.
Figure 7. IL-10 Enhances the Intracellular Bacterial Load Directly in Marginal Zone Metallophilic Macrophages but Not cDC1s.
(A) DC-II10−/− (Cd11ccre-II10rfl/fl) and (II10rfl/fl) Cre− control female mice were i.v. infected with 105 live rLm-OVA. The burdens of rLm-OVA in the spleen were determined by CFUs on day 3 after infection. Results are representative of two independent experiments with n = 4 or 5/group.
(B and C) RNA-seq analyses of live CD11c+ MHChlCD11b−CD8+ splenic cDC1s stimulated with medium or 200 ng/mL IL-10 for 4 h.
(B) Ingenuity Pathway Analysis (IPA) showing selected significant canonical pathways.
(C) Volcano plot showing statistical significance against fold change between control-and IL-10-treated samples. Teal dots, adjusted p value (p adj.) ≤ 0.05 and fold change of ≥ 2; red dots, p adj. < 0.05 and fold change of < 2; black dots, p adj. > 0.05 and fold change of < 2. Genes of interest (some significant and some not) are indicated with labels and yellow triangles.
(D) Primary splenic DCs were incubated with 200 ng/mL IL-10 for 16 h and then incubated with 100 μg/mL DQ OVA for 20 min at 0°C or 37°C. After thorough washing, DC populations and processed DQ OVA (fluorescein isothiocyanate [FITC]) were identified and quantified by flow cytometry. Data shown are representative of two independent experiments.
(E) Enriched splenic WT DCs were incubated in the presence or absence of 200 ng/mL recombinant murine IL-10 (rmlL-10) for 4 h at 37°C. The pretreated DCs were further pulsed with 100 jxg/mL OVA for 1 h at 37°C, and free OVA was removed by washing. Pulsed DCs were then co-cultured with CFSE-labeled purified OT-1 T cells for 72 h. OT-1 proliferation was assessed by measuring CFSE dilution by flow cytometry. Numbers indicate the percentage of proliferating cells in the indicated gates. Shown is one representative of two independent experiments.
(F) WT BMDCs were incubated in the presence of 100 ng/mL lipopolysaccharide (LPS) (16 h) or 200–500 ng/mL IL-10 (4–16 h) before bead-bound OVA was phagocytosed. Phagosome maturation was analyzed by flow organellocytometry to assess phagosomal OVA degradation in addition to phagosomal acquisition of LAMP-1 after a 120-min chase period. The data displayed here are representative of three independent experiments.
(G) WT and Dock8−/− female mice were or were not infected with 5–10 × 108 Lm-GFP. 4 h later, Nox2 mRNA analysis was performed by qPCR on total splenocytes. Results are pooled from two independent experiments. **p < 0.01 compared with the WT with or without Listeria infection.
(H) WT and Dock8−/− female mice were i.v. infected with 5–10 × 108 Lm-GFP. 4 h later, the mice were sacrificed to measure intracellular Listeria in XCR1+ cDC1 s and CD169+ MMMs. ***p < 0.001. Results are representative of two independent experiments with n = 3–5/group.
(I) WT mice receiving BM from DC-II10r−/− (Cd11ccre-H10rfl/fl) or Cre− control female mice were i.v. infected with 5–10 × 108 Lm-GFP. 4 h later, intracellular Listeria in CD169+ MMMs was measured by flow cytometry. **p < 0.01. Results are representative of two independent experiments with n = 4/group. Data are means ± SD. See also Figure S7.
To understand how IL-10 might be promoting the bacterial load in DCs by altering antigen handling, we performed RNA sequencing (RNA-seq) analysis on primary splenic cDC1s (Mach et al,. 2000) treated or not treated with IL-10 for 4 h (a time frame analogous to when differences are observed in vivo). As expected, IL-10 signaling was the pathway most affected in treated DCs (Figure 7B). Consistent with previous studies, IL-10 rapidly inhibited particular pro-inflammatory anti-gen-presenting DC pathways, including II-12 and Cd80 induction (De Smedt et al., 1997; Teitz-Tennenbaum et al., 2018), and induced the inhibitory molecule Socs3 (Figures 7B and 7C; Mittal and Roche, 2015). In macrophages, IL-10 has been shown to suppress expression of cathepsins, hydrolytic enzymes, and membrane proteins that facilitate phagolysosomal fusion (Hop et al., 2018), but we found no difference in vesicular trafficking molecules such as Lamp1 or Lamp2, hydrolytic enzymes such as cathepsins and N-acetyl-β-D-hexosaminidases (HEXs), or other molecules relevant for phagosomal maturation, such as Cybb, Fscn1, or Tfeb (Figure 7C and data not shown), nor did we find evidence of changes in antigen presentation genes such Cd74, B2m, and Ciita or individual MHC genes. Therefore, we did not observe a significant effect of IL-10 on the RNA expression of machinery that handles antigens in DCs. Because these processes could be regulated post-transcriptionally, we tested IL-10-treated DCs for antigen presentation alterations relevant to intracellular bacterial survival. TLR signaling in DCs restrains phago-lysosome fusion to promote cross-presentation (Alloatti et al., 2015); we tested whether IL-10 similarly inhibited DC phago-lysosome fusion and thereby allowed Listeria-enhanced survival in cDC1s, Using multiple assays to interrogate antigen processing, phago-lysosomal fusion, and antigen crosspresentation (Hoffmann et al., 2016; Li et al., 2001), we did not observe an effect of IL-10 on cDC1s (Figures 7D–7F), consistent with the RNA-seq data.
Because of CD11c expression by some macrophages (Probst et at., 2005), we next asked whether the cDC1 phenotype we observed was indirectly mediated through an IL-10 effect on macrophages. IL-10 is known to affect handling of phagocytosed pathogens in both human and mouse macrophages across a wide spectrum of tissues, resulting in enhanced survival of intracellular pathogens, including Listeria (Fleming et al., 1999: Hop et al., 2018; O’Leary et al., 2011); this effect is in part mediated by delaying phagosome maturation, inhibiting ¡NOS production, and blocking IFNγ-induced macrophage activation. Indeed, in vitro, IL-10 enhanced intracellular Listeria in macrophages but not in DCs (Figures S7A and S7B). Four hours after infection, before the influx and differentiation of TNF and iNOS-producing DCs (TIP-DCs) (Kang et al., 2008), we observed induction of Nox2 (iNOS) in the spleen of infected mice; this was significantly elevated in the absence of MZ B cells (Dock8-deficient mice [Figure 7G] and Cd19-deficient mice [data not shown]) or when IL-1 OR was lost on CD11c-expressing myeloid cells (Figures S7E).
The metallophilic macrophage population in the MZ has the ability to promote cross-presentation by splenic cDC1s by transferring antigen and, during Listeria infection, “trans-infects” cDC1s with live bacteria (Backer et al., 2010; Perez et al., 2017). In multiple mouse lines lacking MZ B cells, we found a significant reduction in intracellular Listeria in MMMs (Figures 7H and S7C). Consistent with expression of CD11 c by these macrophages, CD11c-Cre mice crossed to IL-10R floxed mice also demonstrated a reduction in MMM IL-10 receptor staining (Figure S7D) and, accordingly, a reduction in intracellular Listeria (Figure 7I). Therefore, our data support a model in which MZ B cell-derived IL-10 acts on neighboring MMMs but not cDC1s to enhance intracellular bacterium survival, likely via inhibition of phagosomal maturation and nitrous oxide (NO) production in macrophages. We propose, based on work from others, that IL-10-exposed MMMs then deliver a greater load of bacteria to more cDC1s, which ultimately migrate into the WP and promote infection but also CTL priming. This intricate cellular relay in the MZ might augment the induction of a robust cDC1-mediated CTL response but is vulnerable to manipulation by pathogens, as in the case of Listeria.
DISCUSSION
Listeria is a Gram-positive bacterium that is commonly associated with gastrointestinal infections through the consumption of contaminated food (Qiu et al., 2018). Upon infection of gastrointestinal epithelial cells, Listeria can break the intestinal barrier and disseminate to the spleen and liver. In the spleen, Listeria activates both innate and adaptive immunity (Zenewicz and Shen, 2007), resulting in potent antigen-specific CD8+ and CD4+ T cell responses (Pamer, 2004; Qiu et al., 2018). Given these immunostimulatory properties, Listeria has been developed as a vaccine vector in pre-clinical studies (Gunn et al., 2001; Paterson and Maciag, 2005; Shahabi et al., 2008; Singh et al., 2005). Recently, genetically attenuated Listeria vectors have been administered i.v. to boost anti-cancer vaccines in clinical trials (Qiu et al., 2018). Understanding the complex innate cellular immune response to Listeria in the spleen, which ultimately regulates the adaptive immune response, will be important to further these efforts and uncover fundamental rules governing antigen presentation to CD8+ T cells. Although a tremendous amount of knowledge has been gained in the mouse system regarding the orchestration of the innate and adaptive immune response to systemic Listeria, how this translates to the human splenic immune system remains unknown. This is largely due to the dearth of knowledge about the structure-function of the border between the RP and WP in the human spleen (Lewis et al., 2019). Some of the cell types identified in the mouse MZ have been observed in a distinct region of the human spleen called the perifollicular zone, and the nature of B cells in the MZ of mouse and human are quite distinct (Cerutti et al., 2013; Lewis et al., 2019). Until these discrepancies are understood, it is difficult to predict how similar or different the innate immune response to Listeria would be between mouse and human.
DC migration is crucial for immune surveillance and induction of protective immune responses (Eisenbarth, 2019). This is true both in the periphery from a tissue to a draining lymph node and within the spleen (Calabro et al., 2016b). Accordingly, redistribution of Listeria from the MZ to the WP is important for the initiation of protective CD8+T cell responses but, paradoxically, also facilitates escape of Listeria from sterilizing phagocytes in the MZ. CD8α+ cDC1 s harbor and transport Listeria from the MZ and RPtothe WP and thereby aid establishment of a productive infection (Edelson et al., 2011; Neuenhahn et al., 2006). At the same time, cDC1s are also the primary APCs for the induction of protective primary and memory CD8+ T cell responses during Listeria infection (Alexandre et al., 2016). We showed that cDC2s also contained intracellular Listeria early during infection and migrated into the WP. However, loss of cDC2 development or migration within the spleen did not affect the course of Listeria infection or CD8+ T cell responses. It is possible that cDC2s have enhanced destruction of ingested Listeria and therefore fail to propagate the bacteria in the WP. This is supported by data demonstrating less viable Listeria recovered from cDC2s than from cDC1s (Neuenhahn et al., 2006). cDC2s are also less adept at cross-presentation and, therefore, potentially cannot prime CD8+ T cells unless the antigen dose is high (Edelson et al., 2011). Although CD8+ T cell priming is largely intact in the absence of functional cDC2s, CD4+ T cell activation is impaired. Generation of optimal CD4+ T cell responses promotes the development of CD8+ memory T cells (Shedlock and Shen, 2003; Sun and Bevan, 2003); therefore, it is possible that loss of functional cDC2s does, in fact, impair CD8+ T cell-mediated protection, but only during re-infection.
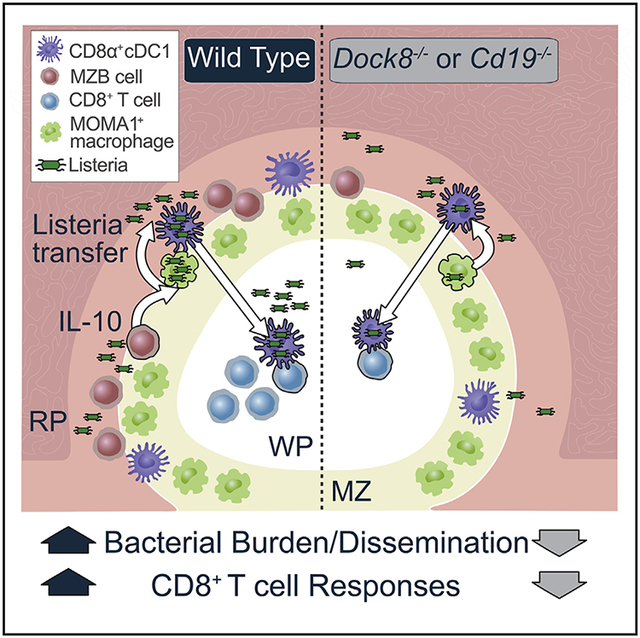
We used a model of impaired cDC2 migration to test the role of cDC2s during Listeria infection: Dock8 deficiency. Although we ruled out a role of cDC2 function in the early stages of the CTL response to Listeria, we discovered an unexpected DC-macro-phage-B cell (cDC1-MMM-MZ B) cell interaction in the MZ of the spleen that ultimately promotes the adaptive immune response to Listeria as well as the propagation of Listeria infection. We found that Dock8-deficient mice failed to activate CD8+ T cells to live but not dead Listeria because of bacterial resistance. Using mice with cell-specific deletion of Dock8, we demonstrated that B cell-specific loss of DOCK8 resulted in loss of IL-10-producing MZ B cells and enhanced resistance to Listeria. MZ B cells likely recognize Listeria via TLR2, and this induces II-10; specific loss of MZ B cell IL-10 production impairs the Listeria load in cDC1s, the bacterial burden in the spleen, and CD8+ T cell responses.
This crosstalk between CD8α+ cDC1 s and MZ B cells provides a model to explain previous findings that the absence of MZ B cells or IL-10 results in enhanced resistance to Listeria (Horikawa et al., 2013; Lee and Kung, 2012). We wanted to understand how IL-10 regulates bacteria in cDC1s. IL-10 is classically thought of as an immunosuppressive cytokine, including for DCs; however, it can also promote particular immune responses, such as memory CD8+T cell differentiation (Foulds et al., 2006; Laidlaw et al., 2015). Although there is ample evidence that IL-10 inhibits phagosome maturation in macrophages, there is little direct evidence that the same is true for bona fide DCs. I n fact, most in vitro work demonstrating IL-10-induced inhibition of human or mouse DC activation has either been done with BMDCs, which contain significant macrophage populations (Helft et al., 2015), or monocytes (D’Amico et al., 2000; Matsumura et al., 2013; Westcott et al., 2010). In support of a model in which I L-10 has selective effects on macrophages but not DCs, we compared known lysosomal degradative molecules that are suppressed in macrophages 4 h after IL-10 treatment with the mRNA profile of IL-10-treated cDC1s after 4 h and found little correlation. We showed here that, early after IL-10 stimulation of cDC1s, II-12 is indeed suppressed, consistent with previous DC work; however, we found no evidence to support a role of IL-10 in directly modulating DC antigen handling or cross-presentation, nor did we observe a decrease in Ccr7 or Fascini expression in IL-10-treated DCs (D’Amico et al., 2000; Yamakita et al., 2011), consistent with intact WP homing (Calabro et al., 2016b) observed in Dock8−/− mice after Listeria infection. Our data show instead that IL-10 acts primarily on MMMs to augment the intracellular bacterial burden. These macrophages pass off their internalized bacteria to cDC1 s, which can, after migration to the WP, present this antigen to T cells but also allow Listeria to escape early innate immune control in the MZ. In light of this model, evaluation of whether IL-10-induced alterations in CD8+ T cell memory could, in part, work via effects on antigen delivery to cDC1 s warrants further investigation.
Unlike DCs, macrophages do not immediately digest and present phagocytosed antigen; instead, a signal from other leukocytes through IFNγ stimulates phago-lysosomal fusion and surface MHC antigen presentation. IL-10 actually blocks this process, and the two cytokines act antagonistically (Fleming et al., 1999; Mittal and Roche, 2015). Phagosome maturation and iNOS induction, which are crucial steps in intracellular pathogen destruction, also degrade antigen, making it less available for presentation. We propose that this IL-10-lFNγ cytokine switch might act as a molecular timer in macrophages for retaining phagocytosed antigens and promoting sampling of ingested contents by endosomal pattern recognition receptors (Wu et al., 2015). For a few hours after phagocytosis, IL-10 pauses macrophage phagosomal degradation. Then, either through downregulation of IL-10 production, departure of MZ B cells into the follicles, or refractory sensing of IL-10 by macrophages, the endocytosed material is ultimately degraded, likely initiated by unimpeded IFNγ signaling.
Possibly in concert, this also provides intact antigen to cDC1 s from MMMs via pathways that remain unknown (Backer et al., 2010; Perez et al., 2017). Multiple mechanisms exist by which live Listeria could be transferred from macrophages to DCs. Cytosolic cargo, including live intracellular pathogens such as Listeria, can be transferred between viable or damaged macrophages via trogocytosis or efferocytosis, respectively (Czucz-man et al., 2014; Steele et al., 2016). Whether these or other cellular processes are employed by cDC1 s to sample macrophage cargo and, in the process, acquire Listeria remains to be determined. Unfortunately, this IL-10-IFNγ cytokine timer presents a window of opportunity for intracellular pathogens to escape killing in macrophages and, in the case of Listeria, find safe passage into the WP harbored in cDC1 s. Altogether, these data define a previously unknown three-innate-cell relay in the MZ between MZ B cells, MMMs, and cDC1s that might have evolved to facilitate cross-presentation but ultimately promotes Listeria infection.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stephanie C. Eisenbarth (stephanie.eisenbarth@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains
All mouse strains used here are on a C57BL/6 background. C57BL/6 and congenie C57BL/6-Ly5.1 [B6.SJL-PtprcaPepcb/BoyCrI] WT mice were purchased from Charles River Laboratories (Wilmington, MA). CD11ccre (Itgax-Cre) [B6.Cg-Tg(ltgax-Cre)1–1 Reiz/J], Mb1cre [B6.C(Cg)-Cd79atm1(Cre)Reth/EhobJ], CD19−/− (CD19cre/cre), Batf3−/− [B6.129S(C)-Batf3tm1Kmm/J], MHC Class I−/− [B6.129P2-B2mtm1Unc/J], Irf4fl [B6.129S1-Irf4tm1Rdf/J], Myd88fl/fl [B6.129P2(SJL)-Myd88tm1 Defr/J], IL-10Rαfl/fl [B6(SJL)-II10-ratm1.1 Tlg/J], C3−/− [B6.129S4-C3tm1Crr/J], II10−/− [B6.129P2-II10tm1Cgn/J], OT-1 [C57BlV6-Tg(TcraTcrb)110OMjb/J] and OT-2 [B6.Cg-Tg(TcraTcrb)425Cbn/J] mice were purchased from Jackson Laboratories (Bar Harbor, ME). OT-1 and OT-2 mice were crossed onto the CD45.1 mice. Dock8−/− and Dock8fl/fl mice were generated as described previously (Krishnaswamy et al., 2015; Williams et al., 2016). Conditional Dock8fl/fl mice were crossed with CD11ccre or Mb1cre mice, to generate CD11ccre-Dock8lx/lx mice or Mb1cre-Dock8lx/lx mice, respectively. Myd88fl/fl mice were crossed with Mb1cre mice to generate Mb1cre-/Wyd88, lx/lx mice. Conditional IL-10Rαfl/fl mice were crossed with CD11ccre mice to generate Mb1cre-IL-10Rαfl/fl mice. We used age-and sex-matched (male and female) mice that were between 6 and 16 weeks of age in all experiments. All protocols used in this study were approved by the Institutional Animal Care and Use Committee at the Yale University School of Medicine.
Listeria
rLM-OVA (Listeria monocytogenes strain 10403s expressing OVA) was originally from Hao Shen (Foulds et al., 2002) and kindly provided by Lauren A. Zenewicz (The University of Oklahoma Health Sciences Center), LM-GFP (Listeria monocytogenes strain 10403S expressing GFP) Listeria was a gift from Herve Agaisse (Chong et al., 2009) (University of Virginia School of Medicine). Pre-titered and aliquoted Listeria stocks were stored in PBS at −80°C and diluted with sterile PBS for i.v. infection.
METHOD DETAILS
In vivo and in vitro Listeria Infection
To determine the organ Listeria burden, spleens and livers from infected mice were homogenized separately in PBS then 1:1 mixed with 0.1% Triton X-10Q (Sigma). Serial dilutions of homogenate were plated on brain heart infusion (BHI) agar plates, and bacterial CFUs were assessed after 24–48 hr growth at 37°C, with detection limits of 83 CFUs for spleen and 167 CFUs for liver. HKLM-OVA were prepared by suspending at 1Q10 CFU/mL live rLM-OVA in PBS and incubated in a 80°C water bath for 1 h. The heat-killed rLM-OVA stocks were aliquoted and stored at −80°C. The absence of live bacteria was confirmed by lack of overnight growth on BHI agar plates at 37°C.
For analyzing in vivo DC or macrophage infectivity, WT, Dock8−/−, II10−/− and CD19−/− mice were infected with 5–10×108 GFP expressing Listeria (LM-GFP) by i.v. injection. 4 h later the mice were sacrificed and the spleens were minced using razor blades in ice cold PBS in the presence of 5 μg/ml of Gentamycin, before passing through a 70 μm cell strainer to achieve a single cell suspension. After RBC lysis and washing with cold PBS, cells were stained with fluorescence conjugated antibodies for flow cytometric analysis. The evaluation of infectivity of Listeria was based on the GFP expression on gated CD8α+ cDC1s, For quantification LM-GFP in CD169+ MMM, mechanically disrupted spleens were additionally digested in RPMI containing fetal bovine serum (2%; Sigma), collagenase IV (0.5mg/mL; Sigma), DNase I (100units/mL; Sigma) and 5 ng/ml of Gentamycin for 30 minutes at 37°C. Lin-(B220, TCRβ, Ly6G) CD11b+CD169+ cells were identified as CD169+MMM.
T cell proliferation and cytokine assay
OVA-specific CD4+ or CD8+ T cells were prepared from spleen and lymph nodes of OT-2 or OT-1 TCR transgenic mice by negative selection using the EasySep™ CD4+ or CD8+ T Cell Isolation Kit (StemCell Technologies, Inc.) according to the manufacturer’s instructions. For T cell proliferation assays, OT-2 CD4+ or OT-1 CD8+ T cells were labeled with 2 μM CFSE before transfer. 106 cells purified OT-2 CD4+ or OT-1 CD8+ T cells were transferred into mice by retro orbital (r.o.) injection. 24 h later, mice were i.v. infected with indicated doses of live rLM-OVA or HKLM-OVA. Spleens were harvested 3 days post infection, and single-cell suspensions were prepared and restimulated with 1 ¡ig/ml of OVA 257–264 peptides (SIINFEKL) for 4 h at 37°C in the presence of 10 ng/ml Brefeldin A. The cells were surface stained and fixed, and then permeabilized for intracellular cytokine staining. Stained cells were analyzed on a MACSQuant flow cytometer (Miltenyi Biotec).
Marginal zone B cell reconstitution
Enriched B cells from WT or CD19−/− mice were adoptively transferred into sub-lethally irradiated WT or Dock8−/− mice. 8 weeks later, some mice were sacrificed to check the reconstitution of MZ B cells before Listeria infection.
Mixed bone marrow chimera generation
WT CD45.1 mice were irradiated with 2 doses of 650 rad 3 h apart. 2 h after the second irradiation, 2×105 bone marrow cells from WT or II10−/− mice mixed with 8×105 bone marrow cells from Mb1cre-Dock8fl/fl mice were adoptively transferred by i.v. injection into irradiated naive wild-type recipient mice. All experiments with bone marrow chimeric mice were performed 8–12 weeks after bone marrow transplant.
Immunofluorescence Analysis
Splenic tissue from WT and Dock8−/− mice were fixed in 4% paraformaldehyde overnight at 4° C. After washing in PBS, tissues were dehydrated through sequential exposure to solutions of 10%, 20% and 30% sucrose, mounted in acryomold with O.C.T. Compound (Tissue-Tek, Sakura), and stored at −80°C prior to sectioning (7 μm) and staining. The following antibodies were used for staining different cell subsets: CD11c (N418), CD169 (MOMA), TCRβ (H57–597), F4/80 (BM8, Invitrogen), Fibroblast Marker (ER-TR7, Santa Cruz Biotechnology), SIGN-R1 (ER-TR9), B220 (RA3–6B2). The images were acquired immediately after stain with the Nikon eclipse Ti microscope using 20x objectives.
Intravascuiar/marginal zone antibody labeling
Mice were i.v. infected with 108 rLM-OVA, 6 h later, 1.5 μg of Anti-CD11 c-APC were injected intravenously. 3 m later, the mice were sacrificed and spleens were collected in cold PBS. To obtain optimal DC staining for injected antibodies, single splenocyte suspension was prepared by mechanically dissociating spleens.
In vivo CTL killing assay
Single cell suspension of spleen cells obtained from MHC class I-deficient (b2M−/−) mice was Xray-irradiated (1500 Rads) and splenocytes were re-suspended and incubated in complete RPM11640 medium (GIBCO) containing 10 mg/m L of OVA (Sigma-Aid rich) for 10 m at 37°C. After washing twice with cold PBS, these OCS were suspended in cold PBS at 108/ml (containing 5 μg/ml CpG ODN1668) and retro-orbital (r.o.) injected into mice as a source of cellular antigen (2 × 107 cells/mouse) (Li et al., 2001). After 5 days of OCS priming, splenocytes from WT naive B6 mice were equally split into two parts. One part was pulsed with 1 ng/mL of OVA257–264 peptides (SIINFEKL) for 1 h at 37°C and then labeled with a higher concentration (2 μM) of CFSE (CFSEhi). The other part was labeled with a lower concentration (0.2 μM) of CFSE (CFSEIo). Equal amounts of cells from each part were mixed and a total of 2 × 107 cells were adoptively transferred by i.v. injection into OCS primed mice. 16 h later, mice were sacrificed and splenocytes suspensions were analyzed by flow cytometry and each population was distinguished by CFSE intensity. Percent OVA-specific CTL killing was determined by loss of the peptide-pulsed CFSEhl population.
BMDC culture and in vitro Listeria infection
Murine bone marrow cells were isolated from WT and Dock8−/− mice and cultured on non-tissue culture treated 10 cm dishes at a concentration of 2×105 cells/mL in 10 mL of RPMI1640 (GIBCO) supplemented with 1% fetal bovine serum (GE Healthcare Life Sciences), 1% Penicillin Streptomycin (GIBCO), 1% L-Glutamine (GIBCO), 1% HEPES (GIBCO), 1% Sodium Pyruvate (GIBCO), 50 μM β-Mercaptoethanol (Sigma) in presence of 20 μg/mL of recombinant murine GM-CSF(PeproTech). 10 mL of the differentiation medium was added on day 3 and half of the media was replaced on day 6. On day 7, loosely adherent WT and DOCK8-deficient BMDCs were harvested from dishes and plated in triplicates in non-tissue culture treated 24 well plates at a concentration of 5×105 cells/mi. 2.5×106 LM-GFP was added to each well. In some experiments, splenocytes from WT or DOCK8-deficient mice were plated in nontissue culture treated 24 well plates at a concentration of 5–10×106 cells/ml and cocultured with LM-GFP at different M.O.I.s. Plates were centrifuged at 600 g for 10 m at room temperature to synchronize the infection of cells and then were incubated at 37°C for 25 m. After incubation, gentamicin (Sigma) is added to reach final concentration of 5 ng/mL. This concentration kills any remaining extracellular Listeria but does not impact growth of intracellular bacteria. Plates were kept at 37°C for an additional 2 h and infection was determined by the percentages of GFP+ BMDCs or splenocytes.
Analyze platelet binding to Listeria
For analysis of in vivo platelet binding to Listeria (Verschoor et al., 2011), LM-GFP were incubated for 30 m at 37°C in PBS containing 5 n-M CSFE. After washing twice with cold PBS, 109 LM-GFP i.v. injected into recipient mice and followed the procedure mentioned above. Surface stained splenocytes were fixed with BDfix/perm solution for 30 m, then permeabilized and stained intracellularly with anti-CD41. The evaluation of platelet binding to Listeria was based on the GFP and CD41 double positive population within gated CD8α+ cDC1s.
Flow cytometry Analysis and Antibodies
Single cell suspensions of spleen were acquired either with LSR II (BD), or MACSQuant (Miltenyi Biotec) flow cytometers and analyzed using F low Jo software (Tree Star). The following antibodies were used for staining different cell subsets (from BioLegend unless specified): TCRβ (H57–597), B220 (RA3–6B2), MHC II (M5/114.15.2), CD11c (N418), 33D1 (33D1), XCR1 (ZET), Va2 (B20.1), CD4 (GK1.5), CD8 (53–6.7), CD19 (6D5), CD23 (B3B4), CD21/35 (7E9), F4/80 (CI:A3–1), CD11 b (M1/70), CD41 (MWReg30, BD Bioscience), IFN-γ (XMG1.2), IL-10Rα (1B1.3a), CD169(SER-4, eBioscience), Ly6G(1A8).
Flow Organellocytometry
Flow organellocytometry was carried out as in (Hoffmann et al., 2016) with the following modifications. In brief, cells were treated with 200 ng/mL IL-10 (BioLegend) for 4–16 h or 100 ng/mL LPS (invivogen) for 16 h prior to the procedure. Following 30 m incubation at 16°C of bead-bound OVA (Polysciences and Sigma) with BMDCs (day 9) or splenic DCs at a particle-to-cell ratio of 10:1 and cell density of 20 × 106 cells/mL, cells were repeatedly washed with cold PBS to remove floating particles. Phagosomal antigen degradation was then allowed to occur for different time points at 37°C, or halted immediately by placing the samples on ice. Samples were incubated with 200 ng/mL IL-10 or PBS during pulse and chase periods. Non-specific binding sites were blocked by incubation with CD16/32 antibody (BD Biosciences) and external beads were labeled to be excluded from analysis (rabbit anti-chicken egg albumin; Sigma and AF 647 donkey anti-rabbit IgG; BioLegend clone Poly4064). Samples were subsequently resuspended in homogenization buffer and mechanically disrupted with a 22 G needle fitted to a 3 mL syringe (BD Biosciences) to release cytosol and cell organelles. Centrifugation was used to separate the post-nuclear supernatant for labeling of phagosomal OVA (rabbit anti-chicken egg albumin, Sigma and PE donkey anti-rabbit IgG; BioLegend clone Poly4064) and LAMP-1 (Biotin anti-mouse CD107a; BioLegend, clone 1D4B and BV 421 Strepavidin; BD Biosciences) on ice. Samples were measured by flow cytometry using on the MACSQuant (Miltenyi Biotec) flow cytometer and analyzed by FlowJo software (Tree Star).
In vitro antigen processing analysis
BMDCs (day 7) or splenic DCs were treated with 200 ng/mL IL-10 (BioLegend) or PBS for 16 h before being incubated with 100 |ig/mL DO Ovalbumin (ex. 505 nm, em. 515 nm; Molecular Probes) and 200 ng/mL IL-10 (BioLegend) or PBS for 20 m at 37°C or 0°C. Samples were then extensively washed with cold PBS + 2% FBS, stained in PBS + 2% FBS + 1 mM EDTA for 20 m on ice, and DC populations and DO OVA were identified and quantified by flow cytometry using the MACSQuant (Miltenyi Biotech) flow cytometer and analyzed by FlowJo software (Tree Star, Inc). The DQ-OVA was quantified using the FITC channel. The following antibodies were used for staining DC subsets (from BioLegend unless specified): rat anti-mouse CD16/32 (BD Biosciences), LIVE/DEAD Fixable Aqua (Invitrogen), Pacific Blue anti-mouse I-A/I-E (M5/114.15.2), PerCP/Cy5.5 anti-mouse/human CD11 b (M1/70), PE/Cy7 anti-mouse TCRβ (H57–597), PE/Cy7 anti-mouse/human CD45R/B220 (RA3–6B2), APC anti-mouse/rat XCR1 (ZET), APC/Fire 750 anti-mouse CD11 c (N418).
RT-PCR and primers
RNA from cells was isolated using TRIzol (Life Technologies) and chloroform (CHCI3). Cel Is were suspended in TRIzol and centrifuged at 375 g for 15 s before incubation at room temperature for 5 m. 200 μL chloroform was added per 1 mL TRIzol, followed by vortexing for 5 m and another room temperature incubation for 2 m. The samples were then centrifuged for 15 m at 12,000 g at 4°C. The aqueous phase was transferred to a fresh tube and an equal volume of 2-propanol was added. Samples were vortexed for 5 s, followed by a 10 m incubation at room temperature and then centrifugation at 12,000 g for 10 m at 4°C. Supernatant was decanted and samples were washed with 500 μL 75% ethanol. Samples were then air-dried for 5 m, resuspended in RNase-free water, and incubated at 57°Cfor 10 m. cDNAwas prepared using Oligo(dT) (Sigma), dNTP Mix (Lamda Biotech), and SMART MMLV Reverse Transcriptase Kit (Clontech Laboratories), in accordance with the manufacturer’s protocol. Real-time PCR was performed using KAPA SYBR Fast Master Mix (Kapa Biosystems) and Low ROX (Kapa Biosystems) and run on the QuantStudio3 (Applied Biosystems). cDNA expression was analyzed by the △Ct (change in cycle threshold) method normalized to values of Hprt obtained in parallel reactions during each cycle. The following primers were used: Hprt: forward 5′-CTGGTGAAAAGGACCTCTCG & reverse 5′-TGAAG TACTCATTATAGTCAAGGGCA; 1110: forward 5′- G GTTG CC AAG CCTT AT CG G A & reverse 5′- ACCT G CT CCACT G CCTT G CT; Tnf: forward 5′- TCCCAGGTTCTCTTCAAGGGA, reverse 5′-GGTGAGGAGCACGTAGTCGG; Nox2: forward 5′-TTCACCCAGTTGTG CATCGACCTA & reverse 5′- TCCATGGTCACCTCCAACACAAGA
Primary splenic DC isolation
B16 Flt3L mouse melanoma cells were cultured in DMEM (GIBCO) supplemented with 10% fetal bovine serum (GE Healthcare Life Sciences), 1 % Sodium Pyruvate (GIBCO), 1 % HEPES (GIBCO), 1 % L-Glutamine (GIBCO), 1 % Penicillin Streptomycin (GIBCO), and 50 μM β-mercaptoethanol (Sigma). At 80% confluency, cells were washed with PBS and 5 × 106 cells were resuspended in 200 μL PBS. Following anesthetization with 30% isoflurane, 8–12 week C57BLÍ6 mice were subcutaneously injected with 5 × 106 murine Flt3L-secreting B16 melanoma cells on the dorsal side in order to expand CD11c+ DCs in the spleen. Tumors were left to grow for 14 days or until the tumor reached ~1cm3 in size, as in (Mach et al., 2000). Spleens were isolated from individual mice and digested in RPMI containing fetal bovine serum (2%; Sigma), collagenase IV (0.5 mg/mL; Sigma), collagenase D (0.5 mg/mL; Roche), 1 mM EDTA and DNase I (100 units/mL; Sigma) for 45 mat 37° C in aC02 incubator. After digestion, EDTA(10 mM final concentration) was added for another 5 m to disrupt DC:T ceil conjugates. Undigested material was removed by filtering through a 70 μm cell strainer. DCs were enriched by negative selection using the EasySep Mouse Pan-DC Enrichment Kit (StemCell Technologies, Inc.) according to the manufacturer’s instructions.
DC RNA-Seq analysis
For RNA-seq, the enriched DCs were stained with DAPI, CD11c, CD11 b, CD8α and MHC class II monoclonal antibodies for 30 m on ice, washed and sorted on BD Aria cell sorter. Purified CD8α+ DCs from each mouse were split in two equal parts and were in vitro cultured at 2 million cells/mL with or without recombinant murine IL-10 (200 ng/mL) for 4 h. 2×106 cells were resuspeneded in 100 uL of RLT (QIAGEN) and snap frozen. These cells were thawed and volume was added to 350 uL of RLT lysis buffer (QIAGEN) with 1 % β-mercaptoethanol (β-ME; Sigma). The lysates were homogenized by vortexing followed by incubating at room temperature for 5 min to ensure complete lysis of the cells. RNA was purified using RNeasy mini kit (QIAGEN) as per manufacturer’s instructions. For each RNA sample, on column DNA digestion was performed. Purified total RNA was eluted in nuclease-free water. RNA-Seq libraries were prepared with KAPA Stranded mRNA-Seq kit (Roche) according to manufacturer’s instructions. First, polyA RNA was isolated from 300 ng total RNA using oligo-dT magnetic beads. Purified RNA was then fragmented at 85°Cfor 6 m, targeting fragments ranging 250–300 bp. Fragmented RNA was reverse transcribed with an incubation of 25°C for 10 m, 42°C for 15 m and an inactivation step at 70°C for 15 m. This was followed by second strand synthesis at 16°C, 60 m. Double stranded cDNA fragments were purified using Ampure XP beads (Beckman). The ds cDNA were then A-tailed, and ligated with illumina unique UDI adapters. Adaptor-ligated DNA was purified using Ampure XP beads. This was followed by 10 cycles of PCR amplification. The final library was cleaned up using AMpure XP beads. Sequencing was performed on Illumina HiSeq4000 platform generating paired end reads of 76 bp. FastQ files were checked with FastQC (version 0.11.5), before tim-ming with Trim Galore (version 0.4.0). Fragments were quasi-mapped to the mouse transcriptome mm10 using salmon (Patroetal., 2017) (version 0.7.2) atthe gene level and differential expression analysis between IL-10-treated and control cells was performed using DESeq2 package (Love et al., 2014) in R (version 3.3.1). Differentially expressed genes with adjusted p values ≤ 0.001 and foldchange ≥ 2 were used for the pathway analysis with I PA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using Prism 7 (GraphPad Software). Data were analyzed with the unpaired t test using Welch’s correction or one-way ANOVA with Tukey correction. Data are presented as mean ± SD. *p < 0.05; **p < 0.01; ****p < 0.0001; ns, not significant.
DATA AND CODE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is Gene Expression Omnibus (GEO): GSE124771. All software used in the analysis is listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| APC/Cyanine7 anti-mouse CD11c Antibody (N418) | BioLegend | Cat# 117324, RRID:AB_830649 |
| PE anti-mouse CD169 (Siglec-1) Antibody (SER-4) | eBioscience | Cat# 12-5755-82, RRID:AB_2572625 |
| Alexa Fluor® 647 anti-mouse/rat XCR1 Antibody (ZET) | BioLegend | Cat# 148214, RRID:AB_2564369 |
| PE/Cy7 anti-mouse/human CD45R/B220 Antibody (RA3–6B2) | BioLegend | Cat# 103222, RRID:AB_313005 |
| PE/Cy7 anti-mouse Ly-6G Antibody (1A8) | BioLegend | Cat# 127618, RRID:AB_1877261 |
| PE/Cy7 anti-mouse TCR β chain Antibody (H57–597) | BioLegend | Cat# 109222, RRID:AB_893625 |
| PE/Cy7 anti-mouse CD49b (pan-NK cells) Antibody (DX5) | BioLegend | Cat# 108922, RRID:AB_2561460 |
| PE anti-mouse/human CD11b Antibody (M1/70) | BioLegend | Cat# 101208, RRID:AB_312791 |
| APC anti-mouse CD8a Antibody (53–6.7) | BioLegend | Cat# 100712, RRID:AB_312751 |
| Biotin anti-mouse CD210 (IL-10 R) antibody (1B1.3a) | BioLegend | Cat# 112704, RRID:AB_313517 |
| Anti-chicken egg albumin (OVA antibody) | Sigma | Cat# C6534, RRID:AB_258953 |
| Purified rat anti-mouse CD16/CD32 (mouse Fc block) (2.4G2) | BD Biosciences | Cat# 553142, RRID:AB_394657 |
| Anti-mouse CD107a (LAMP-1) antibody (1D4B) | BioLegend | Cat# 121603, RRID:AB_572002 |
| PE Donkey anti-rabbit IgG (minimal x-reactivity) Antibody (Poly4064) | BioLegend | Cat# 406421, RRID:AB_2563484 |
| Alexa Fluor® 647 Donkey anti-rabbit IgG (minimal x-reactivity) Antibody(Poly4064) | BioLegend | Cat# 406414, RRID:AB_2563202 |
| Alexa Fluor® 647 anti-mouse TCR β chain Antibody (H57–597) | BioLegend | Cat# 109218, RRID:AB_493346 |
| Alexa Fluor® 488 anti-mouse TCR β chain Antibody (H57–597) | BioLegend | Cat# 109215, RRID:AB_493344 |
| APC anti-mouse CD11c Antibody (N418) | BioLegend | Cat# 117310, RRID:AB_313779 |
| FITC anti-mouse CD169 (Siglec-1) Antibody (3D6.112) | BioLegend | Cat# 142406, RRID:AB_2563107 |
| eFluor 570 F4/80 Monoclonal Antibody (BM8) | Invitrogen | Cat# 41-4801-80, RRID:AB_2573610 |
| Fibroblast Marker Antibody (ER-TR7) Alexa Fluor® 647 | Santa Cruz Biotechnology | Cat# sc-73355 AF647 |
| Alexa Fluor® 647 anti-mouse/human CD45R/B220 Antibody | BioLegend | Cat# 103229, RRID:AB_492875 |
| Brilliant Violet 510™ anti-mouse I-A/I-E Antibody | BioLegend | Cat# 107636, RRID:AB_2734168 |
| Biotin anti-mouse DC Marker (33D1) Antibody | BioLegend | Cat# 124904, RRID:AB_1186159 |
| Pacific Blue™ anti-mouse TCR Vα2 Antibody (B20.1) | BioLegend | Cat# 127816, RRID:AB_10613647 |
| PE anti-mouse CD4 Antibody (GK1.5) | BioLegend | Cat# 100408, RRID:AB_312693 |
| APC/Cyanine7 anti-mouse CD19 Antibody (6D5) | BioLegend | Cat# 115530, RRID:AB_830707 |
| PE anti-mouse CD23 Antibody (B3B4) | BioLegend | Cat# 101608, RRID:AB_312833 |
| APC anti-mouse CD21/CD35 (CR2/CR1) Antibody (7E9) | BioLegend | Cat# 123412 |
| Pacific Blue™ anti-mouse F4/80 Antibody (CI:A3–1) | BioLegend | Cat# 123124, RRID:AB_893475 |
| PerCP/Cy5.5 anti-mouse CD45.1 Antibody | BioLegend | Cat# 110728, RRID:AB_893346 |
| Brilliant Violet 510™ anti-mouse CD45.2 Antibody | BioLegend | Cat# 109838, RRID:AB_2650900 |
| PE Rat Anti-Mouse CD41 (MWReg30) | BD Bioscience | Cat# 558040, RRID:AB_397004 |
| APC anti-mouse IFN-γ Antibody (XMG1.2) | BioLegend | Cat# 505810, RRID:AB_315404 |
| Bacterial and Virus Strains | ||
| Listeria monocytogenes strain 10403s expressing OVA | Dr. Lauren A. Zenewicz | The University of Oklahoma Health Sciences Center |
| Listeria monocytogenes strain 10403S expressing GFP | Dr. Herve Agaisse | University of Virginia School of Medicine |
| Listeria monocytogenes strain 10403S expressing RFP | Dr. Eric G. Pamer | Sloan Kettering Institute |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Albumin from chicken egg white (OVA) | Sigma | Cat# A5503 |
| OVA257–264 peptides (SIINFEKL) | InvivoGen | Cat# vac-sin |
| CpG ODN1668) | InvivoGen | Cat# tlrl-1668–1 |
| Recombinant mouse IL-10 (carrier-free) | BioLegend | Cat# 575802 |
| BV421 streptavidin | BD BioSciences | Cat# 563259 |
| PE Streptavidin | BioLegend | Cat# 405204 |
| RPMI1640 | Corning | Cat# 10040CV |
| 0.5M EDTA, pH 8.0 | Invitrogen | Cat# 15-575-020 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 |
| Penicillin-Streptomycin | Gibco | Cat# 15140122 |
| Deoxyribonuclease I | Sigma-Aldrich | Cat# D5025 |
| Collagenase type IV | Sigma-Aldrich | Cat# C5138 |
| Fetal Bovine Serum | ATLANTA biologicals | Cat# S11150 |
| Sodium Pyruvate (100 mM) | Gibco | Cat# 11360070 |
| RBC Lysis Buffer (10X) | BioLegend | Cat# 420301 |
| HEPES, 1M buffer solution | AmericanBIO | Cat# AB06021–00100 |
| L-Glutamine (200 mM) | Gibco | Cat# 25030081 |
| GolgiStop™ Protein Transport Inhibitor (containing Monensin) | BD Bioscience | Cat# 554724 |
| Fixation/Permeabilization Solution Kit | BD Bioscience | Cat# 54714 |
| CellTrace™ CFSE Cell Proliferation Kit | Invitrogen | Cat# C34554 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Invitrogen | Cat# L34957 |
| TRITON X-100 | AmericanBIO | Cat# AB02025 |
| BBL™ Brain Heart Infusion | BD Bioscience | Cat# 211059 |
| Bacto™ Dehydrated Agar | BD Bioscience | Cat# 214010 |
| Amine-modified polystyrene microspheres, 3 μm diamer | Polysciences | Cat# 17145–5 |
| CO2-independent medium | Gibco | Cat# 18045–088 |
| Iscove’s modified Dulbecco’s medium (IMDM) | Gibco | Cat# 12440053 |
| Bovine serum albumin (BSA) | AmericanBio | Cat# AB01088 |
| Glycine | Sigma | Cat# G7126 |
| Imidazole | Sigma | Cat# I-0250 |
| Sucrose | AmericanBio | Cat# AB01900 |
| Phenylmethanesulfonyl fluoride (PMSF) | Sigma | Cat# P7626 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11836170001 |
| Dithiothreitol (DTT) | Sigma | Cat# 646563 |
| Trypan blue solution, 0.4% | Gibco | Cat# 15250061 |
| Recombinant Murine GM-CSF | PEPROTECH | Cat# 315–03 |
| Lipopolysaccharides from Escherichia coli O111:B4 | Sigma | Cat# L3012 |
| DQ Ovalbumin | Molecular Probes | Cat# D-12053 |
| Glutaraldehyde, EM grade, 50% | Polysciences | Cat# 18428 |
| TRIzol™ Reagent | Invitrogen | Cat# 15596018 |
| SMART® MMLV Reverse Transcriptase | Clontech | Cat# 639524 |
| Chloroform | JT Baker | Cat# 9180–01 |
| 2-Propanol (Isopropyl Alcohol) | JT Baker | Cat# 9084–01 |
| dNTPs | Lamda BIOTECH | Cat# D107 |
| KAPA SYBR® FAST qPCR Kits | Kapa Biosystems | Cat# KK4605 |
| Gentamycin | Sigma | Cat# G1264 |
| Critical Commercial Assays | ||
| EasySep™ Mouse CD4+ T Cell Isolation Kit | StemCell Technologies | Cat# 19852 |
| EasySep™ Mouse CD8+ T Cell Isolation Kit | StemCell Technologies | Cat # 19853 |
| EasySep™ Mouse Pan-DC Enrichment Kit | StemCell Technologies | Cat # 19763 |
| EasySep™ Mouse Pan-B Cell Isolation Kit | StemCell Technologies | Cat # 19844 |
| Deposited Data | ||
| Splenic cDC1s RNA-Seq | This paper | GEO: GSE124771 |
| Experimental Models: Cell Lines | ||
| B16 Flt3L mouse melanoma cells | Dr. Richard A. Flavell | RRID:CVCL_IJ12 |
| L929 cells | ATCC | Cat# CCL-1, RRID:CVCL_0462 |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT: C57BL/6NCrl | Charles River Laboratories | Cat# CRL:27, RRID:IMSR_CRL:27 |
| Mouse:WT:C57BL/6-Ly5.1[B6.SJL-PtprcaPepcb/BoyCrl] | Charles River Laboratories | Cat# CRL:494, RRID:IMSR_CRL:494) |
| Mouse: CD11ccre (Itgax-Cre) [B6.Cg-Tg(Itgax-Cre)1–1Reiz/J] | The Jackson Laboratory | Cat# JAX:008068, RRID:IMSR_JAX:008068 |
| Mouse: Mb1cre [B6.C(Cg)-Cd79atm1(Cre)Reth/EhobJ] | The Jackson Laboratory | Cat# JAX:020505, RRID:IMSR_JAX:020505 |
| Mouse: CD19−/-(CD19cre/cre) [B6.129P2(C)-Cd19tm1(cre)Cgn/J] | The Jackson Laboratory | Cat# JAX:006785, RRID:IMSR_JAX:006785 |
| Mouse: Batf3−/- [B6.129S(C)-Batf3tm1Kmm/J] | The Jackson Laboratory | Cat# JAX:013755, RRID:IMSR_JAX:013755 |
| Mouse: MHC Class I−/- [B6.129P2-B2mtm1Unc/J] | The Jackson Laboratory | Cat# JAX:002087, RRID:IMSR_JAX:002087 |
| Mouse: Irf4fl [B6.129S1-Irf4tm1Rdf/J] | The Jackson Laboratory | Cat# JAX:009380, RRID:IMSR_JAX:009380 |
| Mouse: Myd88fl [B6.129P2(SJL)-Myd88tm1Defr/J] | The Jackson Laboratory | Cat# JAX:008888, RRID:IMSR_JAX:008888 |
| Mouse: IL-10Rαfl [B6(SJL)-Il10ratm1.1Tlg/J] | The Jackson Laboratory | Cat# JAX:028146, RRID:IMSR_JAX:028146 |
| Mouse: C3−/- [B6.129S4-C3tm1Crr/J] | The Jackson Laboratory | Cat# JAX:029661, RRID:IMSR_JAX:029661 |
| Mouse: Il10−/- [B6.129P2-Il10tm1Cgn/J] | The Jackson Laboratory | Cat# JAX:002251, RRID:IMSR_JAX:002251 |
| Mouse: OT-1 [C57BL/6-Tg(TcraTcrb)1100Mjb/J] | The Jackson Laboratory | Cat# JAX:003831, RRID:IMSR_JAX:003831 |
| Mouse: OT-2 [B6.Cg-Tg(TcraTcrb)425Cbn/J] |
The Jackson Laboratory | Cat# JAX:004194, RRID:IMSR_JAX:004194 |
| Mouse: Dock8−/- | Krishnaswamy et al., 2015 | N/A |
| Mouse: Dock8fl/fl | Krishnaswamy et al., 2017 | N/A |
| Oligonucleotides | ||
| hypoxanthine guanine phosphoribosyl transferase (Hprt) forward 5’-CTGGTGAAAAGGACCTCTCG |
Sigma | N/A |
| Hprt reverse 5’-TGAAGTACTCATTATAGTCAAGGGCA | Sigma | N/A |
| Interleukin 10 (IL-10) forward 5’-GGTTGCCAAGCCTTATCGGA | Sigma | N/A |
| IL-10 reverse 5’- ACCTGCTCCACTGCCTTGCT | Sigma | N/A |
| NADPH oxidase (Nox2) forward 5’-TTCACCCAGTTGTGCATCGACCTA | Sigma | N/A |
| Nox2 reverse 5’- TCCATGGTCACCTCCAACACAAGA | Sigma | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| GraphPad Prism 7 | Graphpad Inc | RRID: SCR_002798 |
| Flowjo v10 | Treestar Inc | RRID: SCR_008520 |
| Adobe Photoshop | https://www.adobe.com/products/photoshop.html | RRID:SCR_014199 |
| Adobe Illustrator | https://www.adobe.com/products/illustrator.html | RRID:SCR_010279) |
| IPA | QIAGEN Bioinformatics | RRID:SCR_008653 |
| Trim Galore version 0.4.0 | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | RRID:SCR_011847 |
| FastQC version 0.11.5 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 |
| Salmon version 0.7.2 | https://combine-lab.github.io/salmon/ | RRID: SCR_017036 |
| R version 3.3.1 libraries “tximport”, “DESeq2”, “pheatmap”, “RColorBrewer”, “ggplot2” | https://www.r-project.org/ | RRID:SCR_001905 |
| Other | ||
Supplementary Material
Highlights.
Loss of marginal zone (MZ) B cells reduces intracellular Listeria in CD8α+ cDCIs
Listeria stimulates MZ B cells to produce IL-10 via a MyD88-dependent pathway
IL-10 does not directly alter cDC1 handling of bacteria or antigen presentation
IL-10 inhibits iNOS and increases splenic macrophage intracellular bacterial burden
ACKNOWLEDGMENTS
We would like to thank M. Firla for technical assistance, M. Wimsatt for the illustration, L.A. Zenewicz and H. Agaisse for providing strains of Lm, E. Hoffman for technical guidance regarding phagosome analysis, T. Zhang for help with imaging, S. Calabro and J. Krishnaswamy for helpful discussions, and E.G. Pamer for RFP-Lm and critical review of the manuscript. This work was supported by a CIHR postdoctoral fellowship (to D.L.), P01 HL132819 (to S.C.E.), R01 Al 108829 (to S.C.E.), and R21 AI135221 and R21 AI133440 (to A.W.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/l0.1016/j.immuni.2019.05.011.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alaniz RC, Sandali S, Thomas EX., and Wilson CB (2004). Increased dendritic cell numbers impair protective immunity to intracellular bacteria despite augmenting antigen-specific CD8+ T lymphocyte responses. J. Immunol 172, 3725–3735. [DOI] [PubMed] [Google Scholar]
- Alexandre YO, Ghilas S, Sanchez C, Le Bon A, Crozat K, and Dalod M (2016). XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J. Exp. Med 273, 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloatti A, Kotsias F, Pauwels AM, Carpier JM, Jouve M,Timmerman E, Pace L, Vargas P, Maurin M,Gehrmann U, et al. (2015).Toll-like Receptor4 Engagement on Dendritic Cells Restrains Phago-Lysosome Fusion and Promotes Cross-Presentati on of Antigens. Immunity 43,1087–1100. [DOI] [PubMed] [Google Scholar]
- Anzeion AN, Wu H, and Rickert RC (2003). Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat. Immunol 4, 287–294. [DOI] [PubMed] [Google Scholar]
- Aoshi T, Zinseimeyer BH, Konjufca V, Lynch JN, Zhang X, Koide Y, and Miller MJ (2008). Bacterial entry to the splenic white pulp initiates antigen presentation to CD8+ T cells. Immunity 29, 476–486. [DOI] [PubMed] [Google Scholar]
- Backer R, Schwandt T, Greuter M, Oosting M, Jüngerkes F, Tüting T, Boon L, O’Toole T, Kraal G, Ummer A, and den Haan JM (2010). Effective collaboration between marginal metallophilic macrophages and CD8+ dendritic cells in the generation of cytotoxic T cells. Proc. Natl. Acad. Sci. USA 107, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaña S, Roach K, Turner S, Paul J, and Kovats S (2012). IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol 189, 3368–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabro S, Gallman A, Gowthaman U, Liu D, Chen P, Liu J, Krishnaswamy JK, Nascimento MS, Xu L, Patel SR, et al. (2016a). Bridging channel dendritic cells induce immunity to transfused red blood cells. J. Exp. Med 213, 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabro S, Liu D, Gallman A, Nascimento MS, Yu Z, Zhang TT, Chen P, Zhang B, Xu L, Gowthaman U, et al. (2016b). Differential Intrasplenic Migration of Dendritic Cell Subsets Tailors Adaptive immunity. Ceil Rep 16, 2472–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A, Cols M, and Puga I (2013). Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol 13,118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheslyn-Curtis S, Aldridge MC, Biglin JE, Dye J, Chadwick SJ, and Dudley HA (1988). Effect of splenectomy on gram-negative bacterial clearance in the presence and absence of sepsis. Br. J. Surg 75, 177–180. [DOI] [PubMed] [Google Scholar]
- Chong R, Swiss R, Briones G, Stone KL, Gulcicek EE, and Agaisse H (2009). Regulatory mimicry in Listeria monocytogenes actin-based motility. Cell Host Microbe 6, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 108, 19484–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullingford GL, Watkins DN, Watts AD, and Mallon DF (1991). Severe late postsplenectomy infection. Br. J. Surg 78, 716–721. [DOI] [PubMed] [Google Scholar]
- Czuczman MA, Fattouh R, van Rijn JM, Canadien V, Osborne S, Muise AM, Kuchroo VK, Higgins DE, and Brumell JH (2014). Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature 509,230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico G, Frascaroli G, Bianchi G, Transidico P, Doni A, Vecchi A, Sozzani S, Allavena P, and Mantovani A (2000). Uncoupling of inflammatory chemokine receptors by IL-10: generation of functional decoys. Nat. Immunol 1, 387–391. [DOI] [PubMed] [Google Scholar]
- Dai WJ, Köhler G, and Brombacher F (1997). Both innate and acquired immunity to Listeria monocytogenes infection are increased in IL-10-deficient mice. J. Immunol 158, 2259–2267. [PubMed] [Google Scholar]
- de Noordhout CM, Devleesschauwer B, Angulo FJ, Verbeke G, Haagsma J, Kirk M, Havelaar A, and Speybroeck N (2014). The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect. Dis 14, 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smedt T, Van Mechelen M, De Becker G, Urbain J, Leo O, and Moser M (1997). Effect of interleukin-10 on dendritic cell maturation and function. Eur. J. Immunol 27, 1229–1235. [DOI] [PubMed] [Google Scholar]
- Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, Kc W, Belizaire R, Aoshi T, Schreiber RD, Miller MJ, et al. (2011). CD8α(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity 35, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC (2019). Dendritic cell subsets inT cell programming: location dictates function. Nat. Rev. Immunol 19, 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming SD, Leenen PJ, Freed JH, and Campbell PA (1999). Surface interleukin-10 inhibits listericidal activity by primary macrophages. J. Leukoc. Biol 66, 961–967. [DOI] [PubMed] [Google Scholar]
- Foulds KE, Rotte MJ, and Seder RA (2006). IL-10 is required for optimal CD8 T cell memory following Listeria monocytogenes infection. J. Immunol 177,2565–2574. [DOI] [PubMed] [Google Scholar]
- Foulds KE,Zenewicz LA, Shediock DJ, Jiang J,Troy AE, and Shen H (2002). Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J. Immunol 168, 1528–1532. [DOI] [PubMed] [Google Scholar]
- Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E,Tussiwand R, and Yona S (2014). Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol 14, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn GR, Zubair A, Peters C, Pan ZK,Wu TC, and Paterson Y (2001). Two Listeria monocytogenes vaccine vectors that express different molecular forms of human papilloma virus-16 (HPV-16) E7 induce qualitatively different T cell immunity that correlates with their ability to induce regression of established tumors immortalized by HPV-16. J. Immunol 167, 6471–6479. [DOI] [PubMed] [Google Scholar]
- Harada Y, Tanaka Y, Terasawa M, Pieczyk M, Habiro K, Katakai T, Hanawa-Suetsugu K, Kukimoto-Niino M, Nishizaki T, Shirouzu M, et al. (2012). DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses. Blood 119, 4451–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft J, Böttcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, Goubau D, and Reis e Sousa C (2015). GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCil(+) Macrophages and Dendritic Cells. Immunity 42, 1197–1211. [DOI] [PubMed] [Google Scholar]
- Heng TS, and Painter MW; Immunological Genome Project Consortium (2008). The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Hoffmann E, Pauweis AM, Alloatti A, Kotsias F, and Amigorena S (2016). Analysis of Phagosomal Antigen Degradation by Flow Organellocytometry. Bio. Protoc 6, e2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hop HT, Reyes AWB, Huy TXN, Arayan LT, Min W, Lee HJ, Rhee MH, Chang HH, and Kim S (2018). Interleukin 10 suppresses lysosome-mediated killing of Brucella abortus in cultured macrophages. J. Biol. Chem 293, 3134–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa M, Weimer ET, DiUllo DJ, Venturi GM, Spolski R, Leonard W.ü., Heise MT, and Tedder TF (2013). Regulatory B cell (B10 Cell) expansion during Listeria infection governs innate and cellular immune responses in mice. J. immunol 190, 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre OP, Segura E, Savina A, and Amigorena S (2012). Cross-presen-tation by dendritic cells. Nat. Rev. Immunol 12, 557–569. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Liang HE, Reizis B, and Locksley RM (2008). Regulation of hierarchical clustering and activation of innate immune cells by dendritic cells. Immunity 29, 819–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy JK, Gowthaman U, Zhang B, Mattsson J, Szeponik L, Liu D, Wu R, White T, Calabro S, Xu L, et al. (2017). Migratory CD11b+ conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci. Immunol 2, eaam9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy JK, Singh A, Gowthaman U, Wu R, Gorrepati P, Sales Nascimento M, Gallman A, Liu D, Rhebergen AM, Calabro S, et al. (2015). Coincidental loss of DOCK8 function in NLRP10-deficient and C3H/HeJ mice results in defective dendritic cell migration. Proc. Natl. Acad. Sci. USA 112, 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Cui W, Amezquita RA, Gray SM, Guan T, Lu Y, Kobayashi Y, Flavell RA, Kleinstein SH, Craft J, and Kaech SM (2015). Production of IL-10 by CD4(+) regulatory T cells during the resolution of infection promotes the maturation of memory CD8(+) T cells. Nat. Immunol 16, 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, and Kung JT (2012). Marginal zone B cell is a major source of 11–10 in Listeria monocytogenes susceptibility. J. immunol 189, 3319–3327. [DOI] [PubMed] [Google Scholar]
- Lewis SM, Williams A, and Eisenbarth SC (2019). Structure and function of the immune system in the spleen. Sci. Immunol 4, eaau6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Davey GM, Sutherland RM, Kurts C, Lew AM, Hirst C, Carbone FR, and Heath WR (2001). Cell-associated ovalbumin is cross-presented much more efficiently than soluble ovalbumin in vivo. J. Immunol 166, 6099–6103. [DOI] [PubMed] [Google Scholar]
- Lopes-Carvalho T, and Kearney JF (2004). Development and selection of marginal zone B cells. Immunol. Rev 197, 192–205. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N, GiHessen S, Wilson SB, Sheehan C, Mihm M, and Dranoff G (2000). Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage coiony-stimulating factor or Flt3-ligand. Cancer Res 60, 3239–3246. [PubMed] [Google Scholar]
- Matsumura F, Yamakita Y, Starovoytov V, and Yamashiro S (2013). Fascin confers resistance to Listeria infection in dendritic cells. J. Immunol 191, 6156–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal SK, and Roche PA (2015). Suppression of antigen presentation by IL-10. Curr. Opin. Immunol 34, 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenhahn M, Kerksiek KM, Nauerth M, Suhre MH, Schiemann M, Gebhardt FE, Stemberger C, Panthel K, Schröder S, Chakraborty T, et al. (2006). CD8alpha+ dendritic cells are required for efficient entry of Listeria monocytogenes into the spleen. Immunity 25, 619–630. [DOI] [PubMed] [Google Scholar]
- O’Leary S, O’Sullivan MP, and Keane J (2011). IL-10 blocks phagosome maturation in mycobacterium tuberculosis-infected human macrophages. Am. J. Respir. Cell Mol. Biol 45, 172–180. [DOI] [PubMed] [Google Scholar]
- Pamer EG (2004). Immune responses to Listeria monocytogenes. Nat. Rev. Immunol 4, 812–823. [DOI] [PubMed] [Google Scholar]
- Pasche B, Kalaydjiev S, Franz TJ, Kremmer E, Gailus-Durner V, Fuchs H, Hrabé de Angelis M, Lengeiing A, and Busch DH (2005). Sex-depen-dent susceptibility to Listeria monocytogenes infection is mediated by differential interleukin-10 production. Infect. Immun 73, 5952–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson Y, and Maciag PC (2005). Listeria-based vaccines for cancer treatment. Curr. Opin. Mol. Ther 7, 454–460. [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez OA, Yeung ST, Vera-Licona P, Romagnoli PA, Samji T, Ural BB, Maher L, Tanaka M, and Khanna KM (2017). CD169+macrophages orchestrate innate immune responses by regulating bacterial localization in the spleen. Sci. Immunol 2, eaah5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajeeth CK, Jirmo AC, Krishnaswamy JK, Ebensen T, Guzman CA, Weiss S, Constabel H, Schmidt RE, and Behrens GM (2010). The synthetic TLR2 agonist BPPcysMPEG leads to efficient cross-priming against co-administered and linked antigens. Eur. J. Immunol 40, 1272–1283. [DOI] [PubMed] [Google Scholar]
- Probst HC, Tschannen K, Odermatt B, Schwendener R, Zinkernagel RM, and Van Den Broek M (2005). Histological analysis of CD11c-DTR/GFP mice after in vivo depletion of dendritic cells. Clin. Exp. Immunol 141, 398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Khairallah C, and Sheridan BS (2018). Listeria Monocytogenes: A Model Pathogen Continues to Refine Our Knowledge of the CD8 T Cell Response. Pathogens 7, E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall KL, Lambe T, Johnson AL, Treanor B, Kucharska E, Domaschenz H, Whittle B, Tze LE, Enders A, Crockford TL, et al. (2009). Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat. Immunol 10,1283–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette V, and Burgdorf S (2014). The ins-and-outs of endosomal antigens for cross-presentation. Curr. Opin. Immunol 26, 63–68. [DOI] [PubMed] [Google Scholar]
- Seillet C, Jackson JT, Markey KA, Brady HJ, Hill GR, Macdonald KP, Nutt SL, and Beiz GT (2013). CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood 121, 1574–1583. [DOI] [PubMed] [Google Scholar]
- Shahabi V, Reyes-Reyes M, Wailecha A, Rivera S, Paterson Y, and Maciag P (2008). Development of a Listeria monocytogenes based vaccine against prostate cancer. Cancer Immunol. Immunother 57, 1301–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock DJ, and Shen H (2003). Requirement for CD4T cell help in generating functional CD8 T cell memory. Science 300, 337–339. [DOI] [PubMed] [Google Scholar]
- Singh R, Dominiecki ME, Jaffee EM, and Paterson Y (2005). Fusion to Listerioiysin O and delivery by Listeria monocytogenes enhances the immuno-genicity of HER-2/neu and reveals subdominant epitopes in the FVB/N mouse. J. Immunol 175, 3663–3673. [DOI] [PubMed] [Google Scholar]
- Skamene E, and Chayasirisobhon W (1977). Enhanced resistance to Listeria monocytogenes in splenectomized mice. Immunology 33, 851–858. [PMC free article] [PubMed] [Google Scholar]
- Steele S, Radlinski L, Taft-Benz S, Brunton J, and Kawula TH (2016). Trogocytosis-associated cell to cell spread of intracellular bacterial pathogens. eLife 5, e10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, and Bevan MJ (2003). Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300, 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang J, Qin T, Zhang Y, Huang L, Niu L, Bai X, Jing Y, Xuan X, Miller H, et al. (2018). Dock8 regulates BCR signaling and activation of memory B cells via WASP and CD19. Blood Adv 2, 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitz-Tennenbaum S, Vigliarti SP, Roussey JA, Levitz SM, Olszewski MA, and Osterholzer JJ (2018). Autocrine IL-10 Signaling Promotes Dendritic Cell Type-2 Activation and Persistence of Murine Cryptococcal Lung Infection. J. Immunol 201, 2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen D, and Murphy K (2017). The role of cDC1s in vivo: CD8 T cell priming through cross-presentation. F1000Res 6, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschoor A, Neuenhahn M, Navarini AA, Graef P, Plaumann A, Seidlmeier A, Nieswandt B, Massberg S, Zinkernagel RM, Hengartner H, and Busch DH (2011). A platelet-mediated system for shuttling blood-borne bacteria to CD8α+ dendritic cells depends on glycoprotein GPIb and complement C3. Nat. Immunol 12, 1194–1201. [DOI] [PubMed] [Google Scholar]
- Westcott MM, Henry CJ, Amis JE, and Hiltbold EM (2010). Dendritic cells inhibit the progression of Listeria monocytogenes intracellular infection by retaining bacteria in major histocompatibility complex class ll-rich phagosomes and by limiting cytosolic growth. Infect. Immun 78,2956–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Henao-Mejia J, and Flaveil RA (2016). Editing the Mouse Genome Using the CRISPR-Cas9 System. Cold Spring Harb. Protoc Published online February 1, 2016. 10.1101/pdb.top087536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Gowda NM, and Gowda DC (2015). Phagosomal Acidification Prevents Macrophage Inflammatory Cytokine Production to Malaria, and Dendritic Cells Are the Major Source at the Early Stages of Infection: IMPLICATION FOR MALARIA PROTECTIVE IMMUNITY DEVELOPMENT. J. Biol. Chem 290, 23135–23147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakita Y, Matsumura F, Lipscomb MW, Chou PC, Werlen G, Burkhardt JK, and Yamashiro S (2011). Fascini promotes cell migration of mature dendritic cells. J. Immunol 186, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Myers RC, Freeberg L, Foote J, Kearney JF, Justement LB, and Carter RH (2011). Marginal zone B cells regulate antigen capture by marginal zone macrophages. J. Immunol 186, 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenewicz LA, and Shen H (2007). Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect 9, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Davis JC, Lamborn IT, Freeman AF, Jing H, Favreau AJ, Matthews HF, Davis J, Turner ML, Uzel G, et al. (2009). Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med 361, 2046–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Dove CG, Hör JL, Murdock HM, Strauss-Albee DM, Garcia JA., Mandl JN, Grodick RA, Jing H, Chandler-Brown DB, et al. (2014). DOCK8 regulates lymphocyte shape integrity for skin antiviral immunity. J. Exp. Med 211, 2549–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is Gene Expression Omnibus (GEO): GSE124771. All software used in the analysis is listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| APC/Cyanine7 anti-mouse CD11c Antibody (N418) | BioLegend | Cat# 117324, RRID:AB_830649 |
| PE anti-mouse CD169 (Siglec-1) Antibody (SER-4) | eBioscience | Cat# 12-5755-82, RRID:AB_2572625 |
| Alexa Fluor® 647 anti-mouse/rat XCR1 Antibody (ZET) | BioLegend | Cat# 148214, RRID:AB_2564369 |
| PE/Cy7 anti-mouse/human CD45R/B220 Antibody (RA3–6B2) | BioLegend | Cat# 103222, RRID:AB_313005 |
| PE/Cy7 anti-mouse Ly-6G Antibody (1A8) | BioLegend | Cat# 127618, RRID:AB_1877261 |
| PE/Cy7 anti-mouse TCR β chain Antibody (H57–597) | BioLegend | Cat# 109222, RRID:AB_893625 |
| PE/Cy7 anti-mouse CD49b (pan-NK cells) Antibody (DX5) | BioLegend | Cat# 108922, RRID:AB_2561460 |
| PE anti-mouse/human CD11b Antibody (M1/70) | BioLegend | Cat# 101208, RRID:AB_312791 |
| APC anti-mouse CD8a Antibody (53–6.7) | BioLegend | Cat# 100712, RRID:AB_312751 |
| Biotin anti-mouse CD210 (IL-10 R) antibody (1B1.3a) | BioLegend | Cat# 112704, RRID:AB_313517 |
| Anti-chicken egg albumin (OVA antibody) | Sigma | Cat# C6534, RRID:AB_258953 |
| Purified rat anti-mouse CD16/CD32 (mouse Fc block) (2.4G2) | BD Biosciences | Cat# 553142, RRID:AB_394657 |
| Anti-mouse CD107a (LAMP-1) antibody (1D4B) | BioLegend | Cat# 121603, RRID:AB_572002 |
| PE Donkey anti-rabbit IgG (minimal x-reactivity) Antibody (Poly4064) | BioLegend | Cat# 406421, RRID:AB_2563484 |
| Alexa Fluor® 647 Donkey anti-rabbit IgG (minimal x-reactivity) Antibody(Poly4064) | BioLegend | Cat# 406414, RRID:AB_2563202 |
| Alexa Fluor® 647 anti-mouse TCR β chain Antibody (H57–597) | BioLegend | Cat# 109218, RRID:AB_493346 |
| Alexa Fluor® 488 anti-mouse TCR β chain Antibody (H57–597) | BioLegend | Cat# 109215, RRID:AB_493344 |
| APC anti-mouse CD11c Antibody (N418) | BioLegend | Cat# 117310, RRID:AB_313779 |
| FITC anti-mouse CD169 (Siglec-1) Antibody (3D6.112) | BioLegend | Cat# 142406, RRID:AB_2563107 |
| eFluor 570 F4/80 Monoclonal Antibody (BM8) | Invitrogen | Cat# 41-4801-80, RRID:AB_2573610 |
| Fibroblast Marker Antibody (ER-TR7) Alexa Fluor® 647 | Santa Cruz Biotechnology | Cat# sc-73355 AF647 |
| Alexa Fluor® 647 anti-mouse/human CD45R/B220 Antibody | BioLegend | Cat# 103229, RRID:AB_492875 |
| Brilliant Violet 510™ anti-mouse I-A/I-E Antibody | BioLegend | Cat# 107636, RRID:AB_2734168 |
| Biotin anti-mouse DC Marker (33D1) Antibody | BioLegend | Cat# 124904, RRID:AB_1186159 |
| Pacific Blue™ anti-mouse TCR Vα2 Antibody (B20.1) | BioLegend | Cat# 127816, RRID:AB_10613647 |
| PE anti-mouse CD4 Antibody (GK1.5) | BioLegend | Cat# 100408, RRID:AB_312693 |
| APC/Cyanine7 anti-mouse CD19 Antibody (6D5) | BioLegend | Cat# 115530, RRID:AB_830707 |
| PE anti-mouse CD23 Antibody (B3B4) | BioLegend | Cat# 101608, RRID:AB_312833 |
| APC anti-mouse CD21/CD35 (CR2/CR1) Antibody (7E9) | BioLegend | Cat# 123412 |
| Pacific Blue™ anti-mouse F4/80 Antibody (CI:A3–1) | BioLegend | Cat# 123124, RRID:AB_893475 |
| PerCP/Cy5.5 anti-mouse CD45.1 Antibody | BioLegend | Cat# 110728, RRID:AB_893346 |
| Brilliant Violet 510™ anti-mouse CD45.2 Antibody | BioLegend | Cat# 109838, RRID:AB_2650900 |
| PE Rat Anti-Mouse CD41 (MWReg30) | BD Bioscience | Cat# 558040, RRID:AB_397004 |
| APC anti-mouse IFN-γ Antibody (XMG1.2) | BioLegend | Cat# 505810, RRID:AB_315404 |
| Bacterial and Virus Strains | ||
| Listeria monocytogenes strain 10403s expressing OVA | Dr. Lauren A. Zenewicz | The University of Oklahoma Health Sciences Center |
| Listeria monocytogenes strain 10403S expressing GFP | Dr. Herve Agaisse | University of Virginia School of Medicine |
| Listeria monocytogenes strain 10403S expressing RFP | Dr. Eric G. Pamer | Sloan Kettering Institute |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Albumin from chicken egg white (OVA) | Sigma | Cat# A5503 |
| OVA257–264 peptides (SIINFEKL) | InvivoGen | Cat# vac-sin |
| CpG ODN1668) | InvivoGen | Cat# tlrl-1668–1 |
| Recombinant mouse IL-10 (carrier-free) | BioLegend | Cat# 575802 |
| BV421 streptavidin | BD BioSciences | Cat# 563259 |
| PE Streptavidin | BioLegend | Cat# 405204 |
| RPMI1640 | Corning | Cat# 10040CV |
| 0.5M EDTA, pH 8.0 | Invitrogen | Cat# 15-575-020 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 |
| Penicillin-Streptomycin | Gibco | Cat# 15140122 |
| Deoxyribonuclease I | Sigma-Aldrich | Cat# D5025 |
| Collagenase type IV | Sigma-Aldrich | Cat# C5138 |
| Fetal Bovine Serum | ATLANTA biologicals | Cat# S11150 |
| Sodium Pyruvate (100 mM) | Gibco | Cat# 11360070 |
| RBC Lysis Buffer (10X) | BioLegend | Cat# 420301 |
| HEPES, 1M buffer solution | AmericanBIO | Cat# AB06021–00100 |
| L-Glutamine (200 mM) | Gibco | Cat# 25030081 |
| GolgiStop™ Protein Transport Inhibitor (containing Monensin) | BD Bioscience | Cat# 554724 |
| Fixation/Permeabilization Solution Kit | BD Bioscience | Cat# 54714 |
| CellTrace™ CFSE Cell Proliferation Kit | Invitrogen | Cat# C34554 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Invitrogen | Cat# L34957 |
| TRITON X-100 | AmericanBIO | Cat# AB02025 |
| BBL™ Brain Heart Infusion | BD Bioscience | Cat# 211059 |
| Bacto™ Dehydrated Agar | BD Bioscience | Cat# 214010 |
| Amine-modified polystyrene microspheres, 3 μm diamer | Polysciences | Cat# 17145–5 |
| CO2-independent medium | Gibco | Cat# 18045–088 |
| Iscove’s modified Dulbecco’s medium (IMDM) | Gibco | Cat# 12440053 |
| Bovine serum albumin (BSA) | AmericanBio | Cat# AB01088 |
| Glycine | Sigma | Cat# G7126 |
| Imidazole | Sigma | Cat# I-0250 |
| Sucrose | AmericanBio | Cat# AB01900 |
| Phenylmethanesulfonyl fluoride (PMSF) | Sigma | Cat# P7626 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat# 11836170001 |
| Dithiothreitol (DTT) | Sigma | Cat# 646563 |
| Trypan blue solution, 0.4% | Gibco | Cat# 15250061 |
| Recombinant Murine GM-CSF | PEPROTECH | Cat# 315–03 |
| Lipopolysaccharides from Escherichia coli O111:B4 | Sigma | Cat# L3012 |
| DQ Ovalbumin | Molecular Probes | Cat# D-12053 |
| Glutaraldehyde, EM grade, 50% | Polysciences | Cat# 18428 |
| TRIzol™ Reagent | Invitrogen | Cat# 15596018 |
| SMART® MMLV Reverse Transcriptase | Clontech | Cat# 639524 |
| Chloroform | JT Baker | Cat# 9180–01 |
| 2-Propanol (Isopropyl Alcohol) | JT Baker | Cat# 9084–01 |
| dNTPs | Lamda BIOTECH | Cat# D107 |
| KAPA SYBR® FAST qPCR Kits | Kapa Biosystems | Cat# KK4605 |
| Gentamycin | Sigma | Cat# G1264 |
| Critical Commercial Assays | ||
| EasySep™ Mouse CD4+ T Cell Isolation Kit | StemCell Technologies | Cat# 19852 |
| EasySep™ Mouse CD8+ T Cell Isolation Kit | StemCell Technologies | Cat # 19853 |
| EasySep™ Mouse Pan-DC Enrichment Kit | StemCell Technologies | Cat # 19763 |
| EasySep™ Mouse Pan-B Cell Isolation Kit | StemCell Technologies | Cat # 19844 |
| Deposited Data | ||
| Splenic cDC1s RNA-Seq | This paper | GEO: GSE124771 |
| Experimental Models: Cell Lines | ||
| B16 Flt3L mouse melanoma cells | Dr. Richard A. Flavell | RRID:CVCL_IJ12 |
| L929 cells | ATCC | Cat# CCL-1, RRID:CVCL_0462 |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT: C57BL/6NCrl | Charles River Laboratories | Cat# CRL:27, RRID:IMSR_CRL:27 |
| Mouse:WT:C57BL/6-Ly5.1[B6.SJL-PtprcaPepcb/BoyCrl] | Charles River Laboratories | Cat# CRL:494, RRID:IMSR_CRL:494) |
| Mouse: CD11ccre (Itgax-Cre) [B6.Cg-Tg(Itgax-Cre)1–1Reiz/J] | The Jackson Laboratory | Cat# JAX:008068, RRID:IMSR_JAX:008068 |
| Mouse: Mb1cre [B6.C(Cg)-Cd79atm1(Cre)Reth/EhobJ] | The Jackson Laboratory | Cat# JAX:020505, RRID:IMSR_JAX:020505 |
| Mouse: CD19−/-(CD19cre/cre) [B6.129P2(C)-Cd19tm1(cre)Cgn/J] | The Jackson Laboratory | Cat# JAX:006785, RRID:IMSR_JAX:006785 |
| Mouse: Batf3−/- [B6.129S(C)-Batf3tm1Kmm/J] | The Jackson Laboratory | Cat# JAX:013755, RRID:IMSR_JAX:013755 |
| Mouse: MHC Class I−/- [B6.129P2-B2mtm1Unc/J] | The Jackson Laboratory | Cat# JAX:002087, RRID:IMSR_JAX:002087 |
| Mouse: Irf4fl [B6.129S1-Irf4tm1Rdf/J] | The Jackson Laboratory | Cat# JAX:009380, RRID:IMSR_JAX:009380 |
| Mouse: Myd88fl [B6.129P2(SJL)-Myd88tm1Defr/J] | The Jackson Laboratory | Cat# JAX:008888, RRID:IMSR_JAX:008888 |
| Mouse: IL-10Rαfl [B6(SJL)-Il10ratm1.1Tlg/J] | The Jackson Laboratory | Cat# JAX:028146, RRID:IMSR_JAX:028146 |
| Mouse: C3−/- [B6.129S4-C3tm1Crr/J] | The Jackson Laboratory | Cat# JAX:029661, RRID:IMSR_JAX:029661 |
| Mouse: Il10−/- [B6.129P2-Il10tm1Cgn/J] | The Jackson Laboratory | Cat# JAX:002251, RRID:IMSR_JAX:002251 |
| Mouse: OT-1 [C57BL/6-Tg(TcraTcrb)1100Mjb/J] | The Jackson Laboratory | Cat# JAX:003831, RRID:IMSR_JAX:003831 |
| Mouse: OT-2 [B6.Cg-Tg(TcraTcrb)425Cbn/J] |
The Jackson Laboratory | Cat# JAX:004194, RRID:IMSR_JAX:004194 |
| Mouse: Dock8−/- | Krishnaswamy et al., 2015 | N/A |
| Mouse: Dock8fl/fl | Krishnaswamy et al., 2017 | N/A |
| Oligonucleotides | ||
| hypoxanthine guanine phosphoribosyl transferase (Hprt) forward 5’-CTGGTGAAAAGGACCTCTCG |
Sigma | N/A |
| Hprt reverse 5’-TGAAGTACTCATTATAGTCAAGGGCA | Sigma | N/A |
| Interleukin 10 (IL-10) forward 5’-GGTTGCCAAGCCTTATCGGA | Sigma | N/A |
| IL-10 reverse 5’- ACCTGCTCCACTGCCTTGCT | Sigma | N/A |
| NADPH oxidase (Nox2) forward 5’-TTCACCCAGTTGTGCATCGACCTA | Sigma | N/A |
| Nox2 reverse 5’- TCCATGGTCACCTCCAACACAAGA | Sigma | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| GraphPad Prism 7 | Graphpad Inc | RRID: SCR_002798 |
| Flowjo v10 | Treestar Inc | RRID: SCR_008520 |
| Adobe Photoshop | https://www.adobe.com/products/photoshop.html | RRID:SCR_014199 |
| Adobe Illustrator | https://www.adobe.com/products/illustrator.html | RRID:SCR_010279) |
| IPA | QIAGEN Bioinformatics | RRID:SCR_008653 |
| Trim Galore version 0.4.0 | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | RRID:SCR_011847 |
| FastQC version 0.11.5 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 |
| Salmon version 0.7.2 | https://combine-lab.github.io/salmon/ | RRID: SCR_017036 |
| R version 3.3.1 libraries “tximport”, “DESeq2”, “pheatmap”, “RColorBrewer”, “ggplot2” | https://www.r-project.org/ | RRID:SCR_001905 |
| Other | ||