Abstract
CD4+ regulatory T (Treg) cells have an essential function in maintaining self-tolerance; however, they may also play a detrimental role in antitumor immune responses. The presence of elevated frequencies of Treg cells in tumors correlates with disease progression and poor survival in patients with cancer. The antigen specificity of Treg cells that have expanded in the tumor microenvironment is poorly understood; answering this question may provide important insights for immunotherapeutic approaches. To address this, we used a novel combinatorial approach to characterizing the T cell receptor (TCR) profiles of intratumoral Treg cells from patients with metastatic melanoma, gastrointestinal, and ovarian cancers and elucidated their antigen specificities. The TCR repertoires of tumor-resident Treg cells were diverse yet displayed significant overlap with circulating Treg cells but not with conventional T cells in tumor or blood. TCRs isolated from Treg cells displayed specific reactivity against autologous tumors and mutated neoantigens, suggesting that intratumoral Treg cells act in a tumor antigen–selective manner leading to their activation and clonal expansion in the tumor microenvironment. Tumor antigen–specific Treg-derived TCRs resided in the tumor and in the circulation, suggesting that both Treg cell compartments may serve as a source for tumor-specific TCRs. These findings provide insights into the TCR specificity of tumor-infiltrating human Treg cells that may have potential implications for cancer immunotherapy.
INTRODUCTION
Human CD4+ regulatory T (Treg) cells comprise a small subset of circulating CD4+ T cells with potent suppressive function in vitro and in vivo (1). They play a vital role in regulating immune responses and maintaining self-tolerance; however, they also impede antitumor immunity [reviewed in (2, 3)]. Human Treg cells express high levels of the interleukin-2 receptor α chain (CD25) and the forkhead winged-helix transcription factor (FOXP3), which is pivotal for their development and function [reviewed in (4)]. Elevated frequencies of Treg cells have been reported in many types of tumors, including melanoma (5), breast (6), lung (7), and ovarian carcinoma (8), and their high frequencies correlate with poor prognosis [reviewed in (9)]. In contrast to circulating Treg cells, tumor-resident Treg cells display an activated profile (5–7). Given that T cell receptor (TCR) stimulation is required for the activation and acquisition of suppressive function in Treg cells (10–12), the activated profile of intratumoral Treg cells suggests that antigen stimulation may play an important role in the activation and accumulation of Treg cells in the tumor microenvironment.
The antigen specificity of tumor-infiltrating Treg cells has thus far remained largely unexplored. Lack of an exclusive cell surface marker to unequivocally distinguish activated Treg cells from conventional T (Tconv) cells in tumors forms a major obstacle to isolate viable Treg cells, as staining for intracellular FOXP3 renders the cells nonviable. Using an antigen-specific tetramer against the cancer germline antigen MAGE-A3, Francois et al. (13) isolated and clonally expanded circulating human T cells with phenotypic and functional attributes of Treg cells. Additional studies also identified suppressive CD4 T cells from the peripheral blood (PBL) of patients with cancer with reactivity against nonmutated tumor antigens after stimulation with overlapping peptide libraries (14, 15). Cloning of tumor-infiltrating lymphocytes from melanoma tumors identified CD4+ T cell clones specific for the cancer germline antigen LAGE1 protein (16) that were attributed to be Treg based on their phenotypic and functional characteristics of the clones. All these studies used T cell cloning and expansion techniques that could potentially alter the initial phenotypic and functional status of T cells. Furthermore, the frequency and dominance of these Treg-attributed clones in the tumor and circulation were not reported. Comparison of TCR repertoire of Treg and Tconv cells in humans has been limited to PBL (17, 18), and little is known about the TCR repertoire of intratumoral Treg cells in patients with cancer.
We hypothesized that the elevated frequency of intratumoral Treg cells in human cancers may be due to oligoclonal expansion upon tumor antigen encounter. To explore this hypothesis, we studied the TCR repertoire of tumor-resident Treg cells in human metastatic melanoma, gastrointestinal, and ovarian cancers and elucidated their antigen specificity. We found that the TCR repertoire of intratumoral Treg cells was distinct from Tconv cells in the tumor and PBL of patients; however, it overlapped significantly with circulating Treg cells. Furthermore, the most dominant TCRs derived from intratumoral Treg cells were shown to be tumor reactive and recognized mutated cancer neoantigens. The identified tumor antigen–specific Treg cells were also found in the circulation, suggesting that PBL may be used as an additional source of tumor-specific TCRs. These findings provide insights into the TCR specificity of tumor-infiltrating human Treg cells.
RESULTS
The TCRB repertoires of FOXP3+ Treg cells were distinct from FOXP3− Tconv cells in tumors
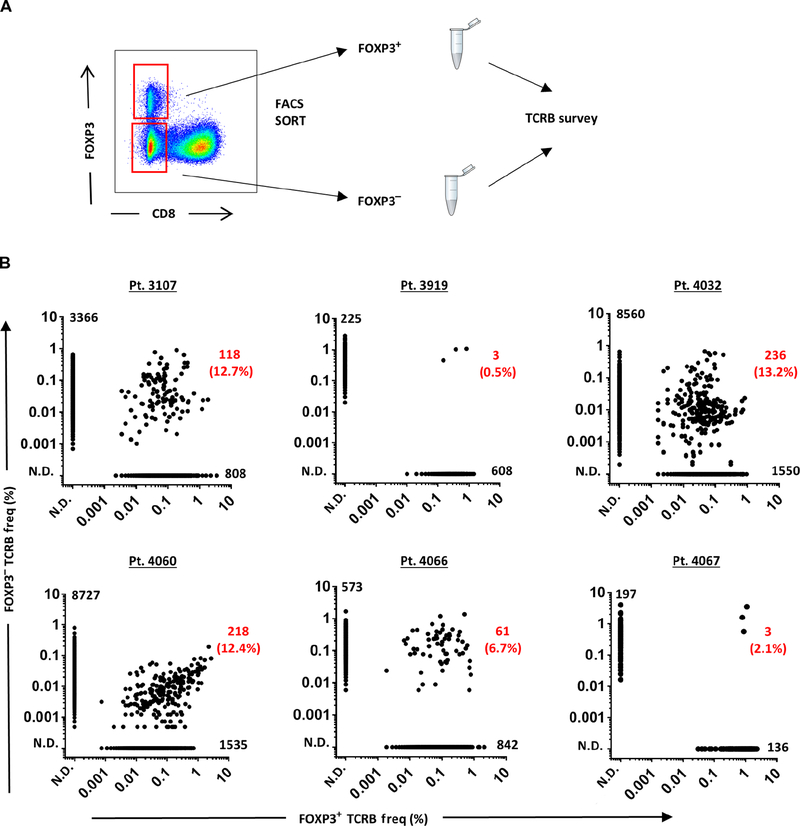
To study the TCR clonotypic repertoire of intratumoral Treg cells of patients with cancer, we performed TCR β (TCRB) chain deep sequencing of Treg cells isolated by flow cytometric sorting based on the expression of FOXP3. Because the enzymatic digestion of tumor samples diminished direct staining for CD4 coreceptor, samples were stained for CD8 and CD3 cell surface markers, followed by intracellular staining for FOXP3 as previously performed (5), and FOXP3+ and FOXP3− CD4 T cell subsets were sorted from CD8−CD3+ T cell populations (Fig. 1A). Previous studies have shown that FOXP3 expression was confined to CD4 Treg cells in vivo, and in vitro activation of Tconv cells can lead to up-regulation of FOXP3 in non-Treg cells in both CD4 and CD8 T cells (19–21); thus, our strategy for the isolation of bona fide intratumoral Treg cells included staining and sorting cells immediately after thawing to not alter the expression of FOXP3. Lack of FOXP3 expression in the intratumoral CD3+CD8+ T cells ex vivo served as a negative control and assurance that the expression of FOXP3 by a subset of intratumoral CD4 T cells was likely confined to CD4 Treg cells as previously reported (5). Functional and epigenetic analyses could not be performed on the sorted intratumoral FOXP3+ cells due to lack of cell viability upon intracellular staining for FOXP3. Moreover, these analyses would be limited on a bulk population and would not be informative for individual cells expressing TCRs of interest.
Fig. 1. TCRB repertoire of intratumoral FOXP 3+ Treg cells was primarily distinct from FOXP3− Tconv cells.
(A) Representation of sorting strategy to isolate FOXP3+ Treg cells and FOXP3− Tconv cells from freshly thawed single-cell suspension of a patient’s (3107) tumor digest. The dot plot was gated on CD3+ lymphocytes. The TCRB immunosequencing was performed on each sorted population to determine the rank and frequency of TCRB clonotypes. (B) The frequency of all productive TCRB sequences in FOXP3+ and FOXP3− subsets for each patient (pt.) was plotted along the x axis and y axis, respectively. Each dot represents a unique TCRB clonotype. The number of overlapping clonotypes and the percentage per total FOXP3+ clonotypes are indicated in red. The number of unique TCRB clonotypes for FOXP3+ (x axis) and FOXP3− (y axis) is indicated next to each axis, respectively. N.D., not detected.
A summary of cell numbers, total productive reads, unique TCRB sequences, and TCRB clonality for each sorted FOXP3 subsets from tumor and PBL is listed in table S2. The median values for the number of sorted cells for FOXP3+ TUM, FOXP3− TUM, FOXP3+ PBL, and FOXP3− PBL were 10,500 cells (range, 3000 to 22,000), 30,000 cells (range, 5000 to 50,000), 55,000 cells (range, 10,000 to 100,000), and 500,000 cells (range, 300,000 to 1,500,000), respectively. We collected all the possible events for the FOXP3+ subsets for each sample because this population was limiting both in the tumor and PBL. The TCR repertoire of each sorted population was analyzed using productive reads (in-frame and no stop codons). The total number of productive reads was not statistically different among the sorted populations (Fig. S1), verifying that each population received a comparable sequencing coverage. Although both FOXP3 subsets in the tumor had lower numbers of unique TCRB sequences than their counterparts in PBL, their differences were not statistically significant (Fig. S2). This difference is likely associated with the lower number of cells sorted from the tumor as compared with PBL.
The TCRB repertoire of intratumoral Treg cells (FOXP3+ TUM) appeared diverse and exhibited a distinct and unique TCRB clonotypic repertoire compared with Tconv cells (FOXP3− TUM) for all the six patients studied (Fig. 1B). Only a small fraction of the TCRB clonotypes was shared between the FOXP3+ and FOXP3− subsets, accounting for 0.5 to 13.2% (mean of 7.9%, n = 6 patients) of the FOXP3+ population, consistent with a previous report on the comparison of circulating Treg and Tconv subsets in the PBL of healthy adults (18). These findings reveal that the TCRB repertoires of intratumoral FOXP3+ Treg cells were primarily distinct from intratumoral Tconv cells with low clonal overlaps, consistent with the findings in the PBL (17, 18), suggesting that the accumulation of intratumoral Treg cells might be mediated by antigen-specific clonal expansion in the tumor microenvironment.
The TCRB clonotypes of intratumoral FOXP3+ Treg cells overlapped with circulating FOXP3+ Treg cells
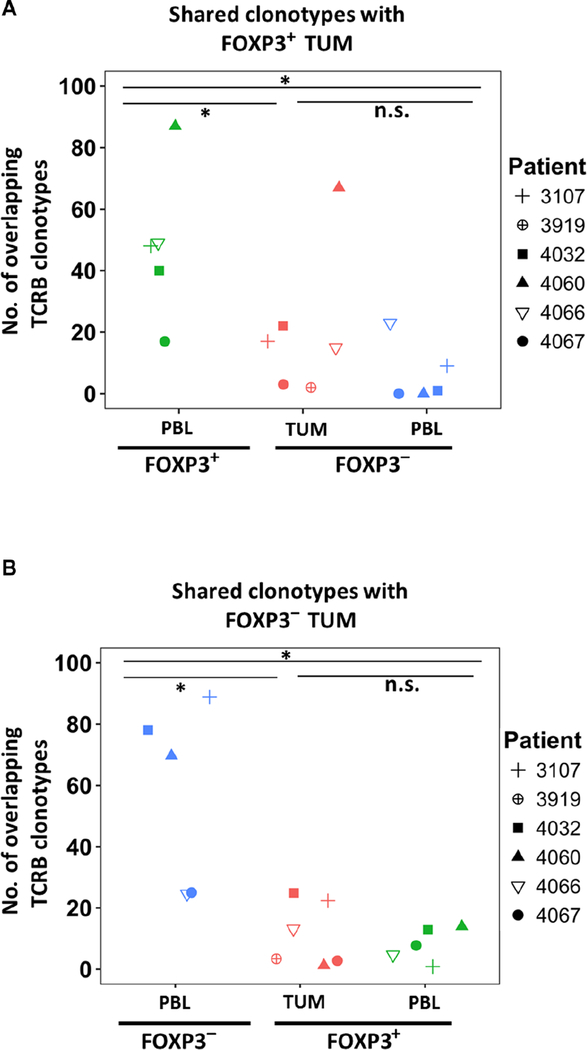
Because the TCRB repertoire analyses of intratumoral FOXP3+ Treg cells revealed that they were principally distinct compared with intratumoral Tconv cells, we subsequently compared the intratumoral FOXP3+ Treg cells repertoire with circulating Treg cells. The most dominant TCRB clonotypes of intratumoral Treg cells (FOXP3+ TUM) overlapped significantly (P < 0.05) with circulating Treg cells (FOXP3+ PBL) but not with FOXP3− Tconv subsets in the tumor (FOXP3− TUM) or in the circulation (FOXP3− PBL; Fig. 2A). There were no overlapping clonotypes detected between FOXP3+ TUM and FOXP3− PBL in two patients (4067 and 4060; Fig. 2A). In contrast, FOXP3− TUM displayed a significant (P < 0.05) overlap with FOXP3− PBL and a minimal overlap with FOXP3+ subsets in the tumor and PBL (Fig 2B). These findings indicate that the most dominant TCRB repertoire of tumor-resident FOXP3+ Treg cells resembles the circulating Treg cells rather than the Tconv cells.
Fig. 2. The dominant TCRB clonotypes in intratumoral FOXP 3+ Treg cells significantly overlapped with circulating FOXP3+ Treg cells.
Each symbol represents a patient’s total number of overlapping TCRB clonotypes among the top 100 TCRB sequences from intratumoral FOXP3+ (A) or FOXP3− (B) subset compared with the other T cell compartments. The total samples were n = 6 and n = 5 for tumor and for PBL, respectively. The dominant TCRB clonotypes of FOXP3+ TUM did not share any clonotypes with FOXP3− PBL for patients 4060 and 4067 (A). *P < 0.05 using Wilcoxon signed-rank test; n.s., nonsignificant values.
Clonal expansion of tumor-infiltrating FOXP3+ Treg cells
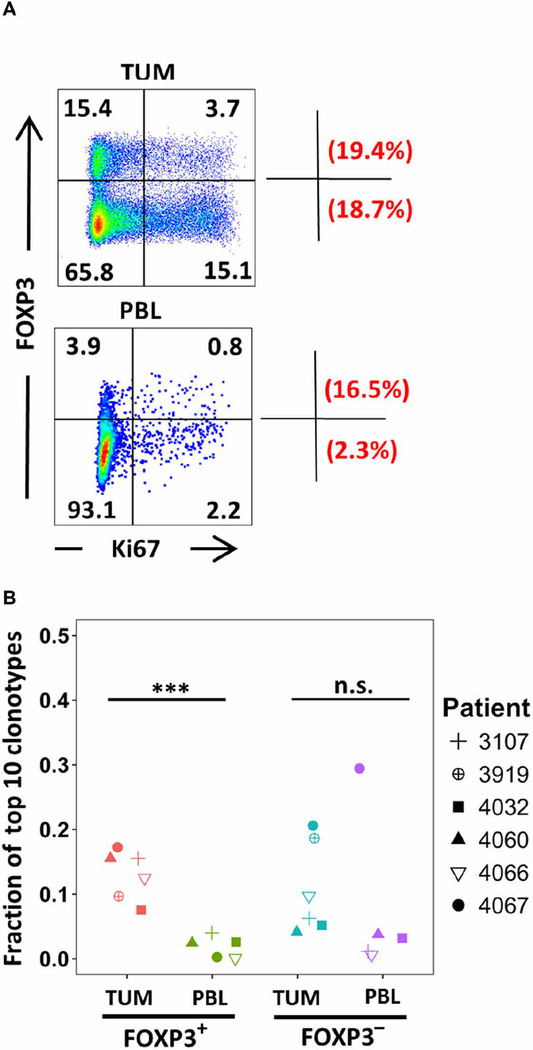
In contrast to circulating Treg cells, intratumoral Treg cells were previously shown to exhibit phenotypic and functional characteristics of activated Treg cells particularly with higher expression level of CTLA-4, OX40, TIGIT, 4–1BB, and CD45RO (5–7, 22). Consistent with these previous studies, the frequency of FOXP3+ Treg was higher in tumors (TUM) than in circulation (PBL) by several folds (Fig. 3A). Directly ex vivo, a fraction (19.4%) of intratumoral FOXP3+ Treg cells expressed Ki67, a marker for recently dividing cells, indicating that intratumoral Treg such as Tconv (FOXP3−, lower quadrants) cells were actively dividing within the tumor microenvironment (Fig. 3A, top panel). Intratumoral Treg cells were reported to be more proliferative than Tconv cells in breast tumors (6). We also compared the clonality for each sorted population in the tumor and blood (Fig. S3 and table S2) as previously reported (23). The median values of clonality for FOXP3+ TUM, FOXP3− TUM, FOXP3+ PBL, and FOXP3− PBL were 0.088 (range, 0.049 to 0.161), 0.080 (range, 0.063 to 0.104), 0.059 (range, 0.054 to 0.092), and 0.068 (range, 0.035 to 0.184), respectively. No significant difference was found in the clonality of the entire sorted population among FOXP3 subsets isolated from tumor or PBL. In contrast, the top 10 TCRB clonotypes in the intratumoral Treg (FOXP3+ TUM) subset constituted a significantly (P < 0.005) higher fraction than the circulating T (FOXP3+reg PBL) subset in all studied patients, regardless of their tumor histology (Fig. 3B). In contrast to FOXP3+ TUM, no significant difference was detected in the frequency of the top 10 TCRB clonotypes between FOXP3− subsets in the tumor and PBL (Fig. 3B). Overall, these findings suggest that the oligoclonality observed within the intratumoral FOXP3+ population may be the result of clonal expansion in response to tumor antigen stimulation.
Fig. 3. Clonal expansion of tumor-infiltrating FOXP 3+ Treg cells.
(A) A single-cell suspension of tumor digest and PBL from the same patient (3107) was stained for CD3, CD8, FOXP3, and Ki67; the dot plots were gated on CD8−CD3+ T cells. The values within each quadrant represent the percentage of cells in that quadrant. The fraction of dividing cells within the Treg (Ki67+FOXP3+/total FOXP3+) and Tconv (Ki67+FOXP3−/total FOXP3−) is depicted as percentage values in the upper corner and lower corner outside the dot plots, respectively. The quadrants were set on the basis of negative control. (B) The fraction of the top 10 TCRB clonotypes was calculated by taking the sum of their TCRB frequencies divided by the total TCRB frequencies per each FOXP3+ or FOXP3− subset in tumor or PBL for each patient. Each symbol represents one patient. The total samples were n = 6 and n = 5 for tumor and for PBL, respectively. ***P < 0.0005 using Wilcoxon signed-rank test.
Tumor reactivity of the most dominant FOXP3+ Treg-derived TCRs in the tumor
To determine the antigen specificity of the most frequent FOXP3+ Treg cells in tumors of patients, we identified the paired TCRB and TCR α (TCRA) chain sequences from the top-ranking TCRB clonotypes in tumors using pairSEQ, a statistical model for pairing TCRA and TCRB sequences (24). These paired TCRA and TCRB sequences were used to reconstruct TCRs, which were subsequently cloned into retroviral vectors (23, 25). Next, retroviral supernatants generated from Treg-derived TCRs were used to transduce autologous PBL (patients 3107 and 4066) or human leukocyte antigen (HLA) class II–matched donor PBL (patient 3919) when PBLs were not available. Subsequently, Treg-derived TCR-transduced T cells were examined for T cell reactivity against autologous and allogeneic tumor cell (TC) lines using interferon-γ (IFN-γ) production assays and up-regulation of the T cell activation marker CD137 (4–1BB) by flow cytometry.
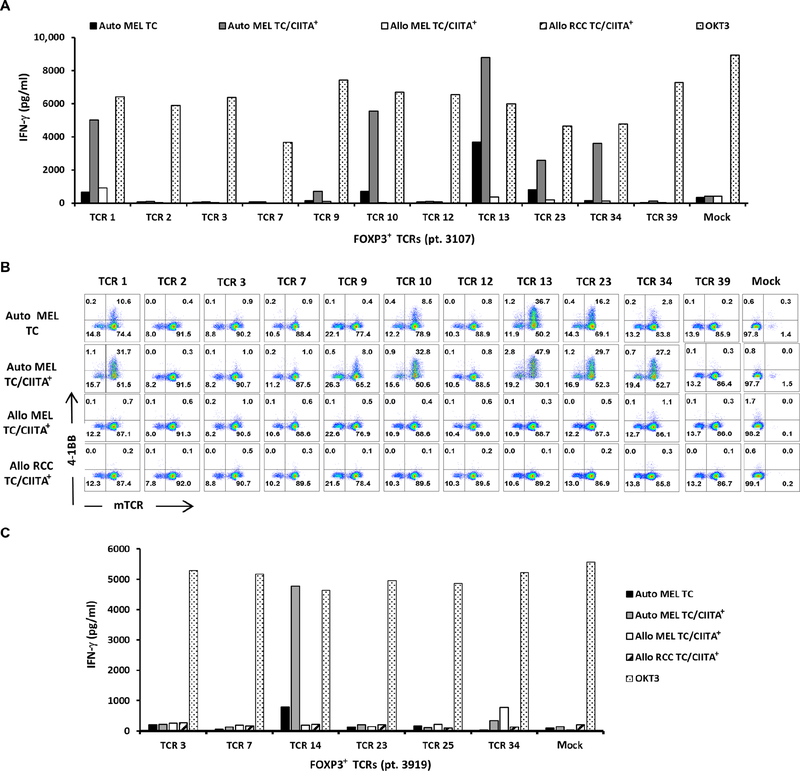
Eleven TCRs were constructed from the most dominant intratumoral Treg cells from a metastatic melanoma tumor (patient 3107), ranging from ranks 1 to 39. Six of these TCRs (TCRs 1, 9, 10, 13, 23, and 34) exhibited specific tumor recognition of the autologous TC as measured by IFN-γ production (Fig. 4A) and up-regulation of 4–1BB on TCR-transduced cells (Fig. 4B). The production of IFN-γ was more profound against TC transduced with class II major histocompatibility complex transactivator (CIITA), presumably due to the higher expression levels of HLA class II molecules on TC (Fig. S4). No or minimal recognition was detected of CIITA-transduced allogeneic melanoma (MEL) and renal cell carcinoma (RCC) TC (Fig. 4, A and B). Overall, 6 of 11 intratumoral Treg-derived TCRs from patient 3107 exhibited specific tumor recognition, and 3 of these TCRs (TCRs 1, 9, and 10) ranked among the top 10 clonotypes in the intratumoral Treg cell population.
Fig. 4. FOXP 3+ Treg-derived TCRs exhibited tumor reactivity.
(A) The most dominant TCR clonotypes derived from the intratumoral FOXP3+ Treg cells (patient 3107) were transduced into autologous PBL and subsequently cocultured overnight with autologous (Auto MEL) or allogenic (Allo MEL or Allo RCC) TC lines, and the IFN-γ in the supernatant was quantified by ELISA. (B) The cocultured cells were also stained with anti-CD3, anti-CD4, anti-mTCRB, and anti−4–1BB antibodies to quantify the percentage of 4–1BB up-regulation on mTCRB+ T cells by FACS. The dot plots were gated on CD4+CD3+ propidium iodine (PI)− T cells. (C) The most dominant TCR clonotypes derived from the intratumoral FOXP3+ Treg cells (patient 3919) were transduced into HLA class II–matched donor PBL and cocultured overnight with autologous or allogenic TC, and the IFN-γ in the supernatant was quantified by ELISA. Data are representative of at least two independent experiments.
We also constructed nine TCRs from FOXP3− Tconv cells isolated from the same metastatic melanoma tumor (patient 3107), ranging from ranks 1 to 20. Seven of these TCRs exhibited specific tumor recognition against the autologous TC, as demonstrated by the production of IFN-γ (Fig. S5A) and up-regulation of 4–1BB (Fig. S5B). The recognition of autologous TC was enhanced by CIITA transduction. In addition to the reactivity against autologous MEL-3107, FOXP3− TCR 20 recognized allogeneic RCC-1764 and not MEL-2630. Because the HLA class II expression is partially matched among these TCs (table S3), it is not clear whether this recognition pattern by TCR 20 reflects either on the reactivity against a shared antigen expressed by RCC-1764 and MEL-3107 and not MEL-2630 or on the lack of expression of the HLA restriction element of TCR 20 by MEL-2630. Overall, seven of nine intratumoral Tconv-derived TCRs exhibited tumor reactivity, and three of seven ranked among the top 10 clonotypes in the intratumoral Tconv cell population.
TCRs from intratumoral FOXP3+ Treg cells from two additional patients with metastatic melanoma (patients 4066 and 3919) were also evaluated using a similar approach. For patient 4066, all the top 10 FOXP3+ TCRs were constructed and screened; however, TCR-transduced T cells showed no discernible specific reactivity against the autologous TC or the mutated neoantigens tested. For patient 3919, six FOXP3+ TCRs were constructed, ranging from ranks 3 to 34 (TCRs 3, 7, 14, 23, 25, and 34), and screened against autologous TC. One of these six FOXP3+ TCRs (TCR 14) exhibited tumor reactivity specific to autologous TC (Fig. 4C). Three FOXP3− TCRs (TCRs 1, 17, and 25) were also reconstructed, cloned, and screened. None of these three TCRs recognized autologous TC (Fig. S5C). In summary, TCRs from intratumoral Treg cells in the two evaluable patients displayed specific tumor reactivity.
Neoantigen reactivity of the most dominant FOXP3+ TCRs in the tumor
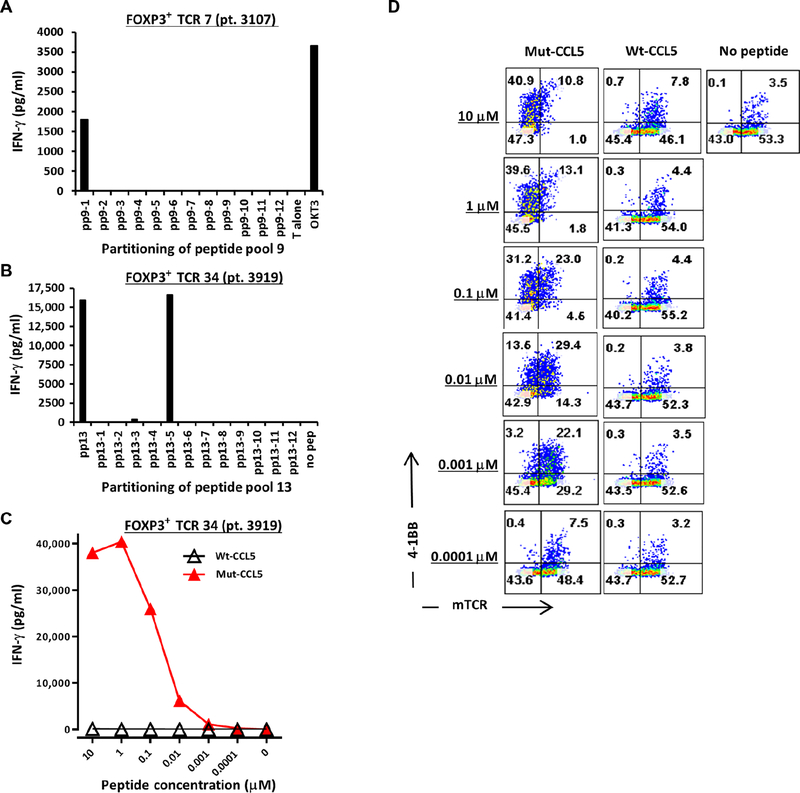
We also asked whether FOXP3+ Treg TCRs were specific for patient-specific mutated neoantigens. For patient 3107, tumor-reactive TCRs were screened for reactivity against 163 identified somatic mutations identified from the patient’s tumor using whole-exome and RNA sequencing (RNA-seq). Autologous dendritic cells (DCs) were pulsed with pools of mutant peptides, with each peptide representing one mutated neoantigen. One of 11 FOXP3+ TCRs screened exhibited reactivity to peptide pool (pp) 9 (pp9) and not to any of the other 13 peptide pools, as assessed by the up-regulation of OX40 and 4–1BB (Fig. S6). Subsequent screening of individual mutant peptides within pp9 revealed reactivity to mutated annexin A1 (ANXA1; Fig. 5A). For patient 3919, FOXP3+ TCR 34 exhibited recognition of DC pulsed with pp13 (Fig. 5B). The sequences of mutant and wild-type peptides are shown in table S4. Subsequent screening of individual mutant peptides within pp13 identified reactivity to mutated CCL-5 (CC chemokine ligand 5, also known as RANTES; Fig. 5B). The other remaining five FOXP3+ TCRs and three FOXP3− TCRs did not display reactivity against any of the screened peptide pools. The reactivity of FOXP3+ TCR 34 was specific to mutated CCL5, as assessed by IFN-γ production (Fig. 5C) and the up-regulation of 4–1BB (Fig. 5D). Recognition of as low as 1 nM of mutated CCL5 peptide was observed, indicating high functional avidity (Fig. 5C). No or minimal recognition of wild-type CCL5 peptide was detected (Fig. 5, C and D). FOXP3+ TCR 34 did not display recognition of the autologous TC (Fig. 4C). At the genomic level, mutated CCL5 was detected both in the tumor and in the TC (patient 3919) based on whole-exome sequencing, and TC exhibited loss of heterozygosity at this site. However, RNA-seq revealed that TC lacked expression of CCL5 (table S5), providing a plausible explanation for the lack of TC recognition by FOXP3+ TCR 34 despite its specific reactivity against the mutated CCL5 peptide. Similarly, FOXP3+ TCR 7 recognized mutated ANXA1 peptide pulsed on DC but not the autologous TC. The low expression of mutated ANXA1 by TC (table S5) may explain its lack of recognition by this TCR. Overall, two intratumoral Treg-derived TCRs isolated from two patients displayed reactivity against mutated cancer neoantigens.
Fig. 5. FOXP 3+ Treg-derived TCRs exhibited specific reactivity against mutant neo-antigens.
(A) FOXP3+ TCR 7 (patient 3107) was cocultured overnight with autologous DC pulsed with individual long mutated peptides from pp9, and IFN-γ production was quantified by ELISA. (B) HLA class II–matched donor PBL was transduced with FOXP3+ TCR 34 (patient 3919) and subsequently cocultured overnight with the donor-derived DCs that were pulsed with pp13 or its individual peptides, and IFN-γ production was quantified by ELISA. (C) FOXP3+ TCR 34 was co-cultured with titrated amount of high-performance liquid chromatography (HPLC) purified mutated (mut-) or wild-type (wt-) CCL5 (initially identified as pp13–5), and IFN-γ in the supernatant was quantified by ELISA. (D) The cocultured cells from (B) were stained similarly as in Fig. 4B to assess the up-regulation of 4–1BB on FOXP3+ TCR 34–transduced T cells (mTCRB+ cells) after an overnight coculture with DC pulsed with mut- or wt-CCL5 peptides (HPLC). The dot plots were gated on CD4+CD3+ PI T cells. All data are representative of at least two independent experiments.
Tumor-reactive FOXP3+ TCRs were also found in the circulating Treg population
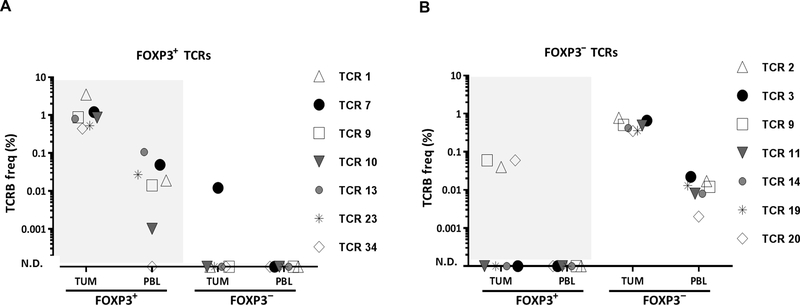
To investigate whether tumor-reactive Treg-derived TCRs identified in patients’ tumors can also be detected in the circulation, we identified TCRB clonotypes for the circulating FOXP3+ and FOXP3− T cell subsets using TCRB deep sequencing from PBL sample collected before the tumor resection. Six of seven tumor-reactive FOXP3+ TCRs (patient 3107), including mutated neoantigen-reactive TCR 7, were also detected in the circulating FOXP3+ Treg population (FOXP3+ PBL) and not in the FOXP3− Tconv cells (FOXP3− PBL) (Fig. 6A). Although TCR 7 was not detected in FOXP3− PBL, it was detected at a very low frequency (0.01%) in FOXP3− TUM. However, this frequency was 100-fold lower than that in FOXP3+ TUM (1.2%) and 5-fold lower than that in FOXP3+ PBL (0.05%). Given that the total productive TCRB reads were 10-fold higher for FOXP3− PBL than that for FOXP3− TUM (table S2) and yet we could not detect this TCR in FOXP3− PBL, its detection at such a low frequency in FOXP3− TUM might be the result of a potential cross-contamination during sorting.
Fig. 6. Tumor-reactive FOXP 3+ TCRs were also found in the circulating Treg population.
(A) The frequency of six tumor-reactive and one neoantigen-reactive FOXP3+ TCRs (patient 3107) was assessed in each FOXP3 subset in the tumor and PBL using TCRB immunosequencing survey. (B) Similarly, the frequency of seven tumor-reactive FOXP3− TCRs (patient 3107) was assessed in each FOXP3 compartment. Each symbol represents a single reactive TCR. The total TCRB clonotypes for FOXP3+ TUM, FOXP3+ PBL, FOXP3− TUM, and for FOXP3− PBL were 926, 3007, 3484, and 50,586 sequences, respectively.
Similar to TCRs isolated from FOXP3+ TUM that were confined to FOXP3+ subsets in tumors and blood, all seven tumor-reactive TCRs isolated from FOXP3− TUM were confined to the circulating Tconv cells (FOXP3− PBL) and not to Treg cells (FOXP3+ PBL) (Fig. 6B). Although four of seven tumor-reactive FOXP3− TCRs were not detected in the FOXP3+ TUM, three of them were found in this subset albeit at low frequency (<0.1%). PBL sample was unavailable to track Treg-derived TCRs in patient 3919. Overall, the finding that intratumoral Treg-derived TCRs with tumor/neoantigen reactivity can be found among the circulating Treg cells suggests that the PBL Treg cells may also be used as a source to identify tumor-reactive and mutated neoantigen-reactive TCRs.
DISCUSSION
Accumulation of FOXP3+ CD4+ Treg cells in human tumors and their expression of markers associated with activation and proliferation (5–7) are potentially driven by antigens presented in the tumor microenvironment (9). In this study, we found that the TCR repertoire of intratumoral Treg cells was distinct from that of intratumoral Tconv cells. Furthermore, the most dominant TCRs derived from intratumoral Treg cells could be tumor-reactive and recognize mutated tumor neoantigens, suggesting that tumor antigens may drive the clonal expansion of intratumoral Treg cells. Last, the TCR repertoire of the dominant intratumoral Treg cells displayed similarity with circulating Treg cells. Collectively, these results reveal tumor- and neoantigen-specific reactivity of Treg-derived TCRs that may provide an opportunity for generating tumor-specific TCR-modified T cells for adoptive T cell therapies for patients with cancer.
The intercompartmental comparison of TCR repertoire of FOXP3 subsets in tumors and PBL in the same patients allowed for a comprehensive analysis that revealed a substantial polyclonal diversity between intratumoral Treg and Tconv cells yet a significant intercompartmental overlap between FOXP3+ Treg population in the tumor and in the circulation. Collectively, these results suggest that accumulation of Treg cells in tumors is likely driven by selective migration of circulating Treg cells into tumors and their subsequent proliferation at that site. Although the differentiation of Tconv into Treg cells at extrathymic sites has been well documented in murine models [reviewed in (26, 27)], the low overlap of TCR repertoire between Treg and Tconv cells isolated from either tumors [the current study and (6)] or PBL (17, 18) provides little support for this conversion in humans. Our findings further suggest that, unlike some murine models that Tconv-derived Treg may also contribute to intratumoral Treg pool [(28); reviewed in (27, 29)], it is not a prominent mechanism for the accumulation of Treg cells in human tumors, although it cannot be exclusively ruled out. The main mechanism for the accrual of intratumoral Treg cells in humans may remain elusive; however, the tumor-specific reactivity of intratumoral Treg-derived TCRs revealed in this study suggests that their TCRs may be exploited for potential antitumor therapeutic approaches.
Using the current strategy, it was not feasible to isolate viable intratumoral Treg cells for functional suppression assays due to intracellular staining for FOXP3. In general, combination of CD25+CD127− phenotype, also shared with recently activated intratumoral effector T cells that lack FOXP3 expression in vivo (5), is used to isolate viable Treg cells from PBL. However, this approach results in the enrichment of a polyclonal population of Treg cells, and identification of tumor-reactive Treg cells in this bulk population might be challenging due to lack of appropriate biological readout for antigen-reactive Treg cells. Although current technical limitations would not permit us to evaluate the functional and epigenetic properties of individual FOXP3+ T cells whose TCRs were functionally characterized in this study, the TCRB repertoire analyses of the bulk FOXP3+ CD4 T cells in tumor and PBL indicate that they were distinct and nonoverlapping with FOXP3− Tconv cells, suggesting that there was a minimal conversation of Tconv cells into FOXP3+ T cells, consistent with a recent study by Rudensky and colleagues (6) in breast tumors. Moreover, the TCRB repertoire comparison among the FOXP3 subsets further allowed us to indirectly assess the integrity of FOXP3+ TCRs that we functionally characterized. Six of seven tumor- and neoantigen-reactive FOXP3+ TCRs in the tumor were found in FOXP3+ PBL and not in the FOXP3− subsets whether in the tumor or PBL, suggesting that they were likely derived from bona fide Treg cells.
In conclusion, we report that human intratumoral Treg cells have a distinct TCR repertoire with minimal overlap with circulating and tumor-resident Tconv cells and are tumor and neoantigen reactive. Accumulation of Treg cells in the tumors can be exploited for the identification and isolation of novel and potent tumor- and neoantigen-reactive TCRs for immunotherapy of patients with cancer. In the current study, TCRs were solely selected on the basis of their frequency in the tumor. Future studies may select TCRs based on the coexpression of FOXP3 and activation markers to further enrich for a subset of activated intratumoral Treg cells. Furthermore, this combinatorial approach has the potential to be exploited for antigen screening of TCRs isolated from other regulatory and nonregulatory T cells with obscure functional readouts. By cloning a TCR into autologous PBL, the intrinsic functional property of original cells can be bypassed to evaluate TCR’s antigen specificity using known effector parameters.
MATERIALS AND METHODS
Patients, PBMCs, and tumor samples
Tumor specimens and PBL samples were collected from patients with metastatic melanoma (n = 3), gastrointestinal (n = 2), and ovarian (n = 1) tumors. Table S1 summarizes the clinical characteristics of patients including their metastatic sites and their prior treatments that might have included two or more of the following treatments: surgery, chemotherapy, immunotherapy, or none of the above. The peripheral blood mononuclear cell (PBMC) samples were prepared over Ficoll-Hypaque (LSM, MP Biomedicals Inc.) gradient and were cryopreserved until analyzed. Tumor specimens were processed by sterile mechanical dissection, followed by enzymatic digestion as previously described (30). The tumor single-cell suspensions were cryopreserved until further analyzed. Patients were not undergoing any therapy at the time when samples were collected. All protocols were approved by the Institutional Review Board of the National Cancer Institute, and informed consents were obtained from the patients.
Flow cytometry, antibodies, and reagents
The following monoclonal antibodies specific for human antigens were used: allophycocyanin (APC)–H7–conjugated anti-CD3 (SK7), fluorescein isothiocyanate (FITC)–conjugated anti-CD8 (SK1), phycoerythrin (PE)–conjugated anti-CD25 (2A3), and APC-conjugated anti-FOXP3 (PCH101) for isolation and sorting of FOXP3 subsets and FITC-conjugated anti-OX40 (ACT35), APC-conjugated anti- 4-1BB (4B4–1), PE-Cy7–conjugated anti-murine TCRβ (H57–597), and PE-conjugated anti-CD4 (SK3). Fluorescence-activated cell sorting (FACS) buffer consisted of phosphate-buffered saline (PBS) supplemented with 3% fetal bovine serum (FBS) with or without 2 mM EDTA (for staining T cells after an overnight coculture). T cells were sorted by flow cytometry using FACSAria (BD Biosciences) and analyzed using FlowJo software v10.
Isolation of FOXP3+ CD4 subsets
Single-cell suspensions from tumor digests or PBMCs were initially stained with anti-CD3 and anti-CD8 for 30 min at 4°C, washed, and subsequently fixed with a 1:20 diluted Fixation/Permeabilization buffer (eBioscience) for 45 min, followed by one wash with FACS buffer and one wash with Permeabilization buffer (eBioscience) according to the manufacturer’s instructions. The Fixation/Permeabilization buffer was titrated to 1:20 rather than 1:4, the dilution recommended by the manufacturer, to minimize DNA damage by paraformaldehyde without hampering FOXP3 staining. The sorted cells were collected into PBS constituted with 5% FBS and 2.5% Hepes, pelleted by centrifuging at 4000 revolutions per minute (rpm) for 20 to 30 min, quick freeze on liquid nitrogen for 1 to 2 min, and stored at −80°C before sending to Adaptive Biotechnologies (Seattle, WA) for TCRB immunosequencing survey.
TCRB immunosequencing survey and matching TCRA-TCRB pairs
TCRB sequencing survey (ImmunoSEQ) was performed by Adaptive Biotechnologies on genomic DNA isolated from sorted FOXP3+ and FOXP3− CD4 T cells from PBL and single-cell suspension of tumor digests. Only productive TCRB rearrangements were used in the calculations of TCRB clonotype frequencies. The number of total productive TCRB reads per sample varied according to the number of cells that was sorted. Unfractionated (10 × 105 cells) and/or an enriched CD4 T cell fraction (0.3 to 1 × 105 cells) from the single-cell tumor digest samples were pelleted in a table top centrifuge at 6000 rpm for 20 to 30 min, resuspended in 200 μl of RNAlater (Invitrogen), snap frozen, and sent to Adaptive Biotechnologies to identify the matching TCRA-TCRB chains by pairSEQ technology, as previously described (24).
TCR reconstruction, cloning of TCRs into a retroviral vector, retrovirus production, and retroviral transduction of T cells
Reconstruction of full-length TCRs was performed as previously described (23). For generation of TCRs, full-length TCRA V-J regions were fused to the mouse TCRA constant chain and the TCRB V-D-J regions to the mouse TCRB constant chain (31). The murine constant region was modified to allow preferential pairing of the TCR chains of interest and to enhance its surface expression and functionality (32, 33). Furthermore, the TCRA and TCRB chains were separated by a RAKR-SGSG P2A linker to ensure a comparable expression efficiency of the two chains (34). Last, the full-length TCRA and TCRB chains, separated by the above described linker, were synthesized and cloned in the TCRB-TCRA orientation into the pMSGV1 retroviral vector (GenScript).
Autologous PBMCs (or HLA class II–matched PBL for patient 3919) were transduced with a retroviral vector encoding the TCR, as previously described (23, 25). Retroviral supernatant was used to transduce autologous pretreatment PBMCs that were stimulated with soluble anti-CD3 (50 ng/ml; OKT3, Miltenyi Biotec) and rhIL2 (300 IU/ml; Chiron) for 2 days before retroviral transduction. Transduced T cells were used at 10 to 15 days after transduction or cryopreserved until used in coculture assays. Transduction efficiency was determined by flow cytometric analysis using the anti-murine TCRB (clone H57–597) antibody.
Target cell preparation
Melanoma TC lines (TC lines 3107, 3919, and 4066) were established from tumor fragments or from mechanically or enzymatically separated TCs and cultured in RPMI 1640 plus 10% FBS (Sigma-Aldrich) supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C in 5% CO2. TCs were also retrovirally transduced with CIITA plasmid (carrying puromycin-resistant gene) using vesicular stomatitis virus envelope glycoprotein (VSVG) and 293GP packaging cell line. Stably transduced TCs were selected after puromycin (5 μg/ml) selection for 24 to 48 hours. Autologous (patients 3107 and 4066) and HLA class II–matched (patient 3919) DCs were prepared as previously described (35, 36). Nonsynonymous mutations identified by whole-exome sequencing and RNA-seq were each synthesized as long peptides (25 mers) by GenScript. Antigen-presenting cells were pulsed with long peptides overnight at concentrations of 10 μg/ml or lower (as indicated) and washed once before the overnight coculture with TCR-transduced T cells.
Target cell recognition and functional assay
Briefly, TCR-transduced or untransduced (mock) T cells (50,000 cells per well) were cocultured with TC (50,000 cells per well) or peptide-pulsed DC (50,000 to 75,000 cells per well) in 96-well round-bottom plates overnight. The supernatant was collected the next day to quantify the amount of IFN-γ by enzyme-linked immunosorbent assay (ELISA). Cocultured cells were stained with anti-CD3, anti-CD4, anti-CD137, and anti-murine TCRB antibodies after the overnight coculture (16 to 20 hours) and acquired by BD FACSCanto II (BD Biosciences). The coexpression of murine TCRB constant chain (identified as mTCR) and CD137 was used to assess the frequency of TCR-transduced antigen-reactive T cells (to be considered reactive, the CD137 up-regulation had to be greater than 1%, three times the background). FACS results were analyzed using FlowJo software.
Whole-exome sequencing and RNA-seq of tumors
Whole-exome sequencing was performed by Personal Genome Diagnostics (Baltimore, MD) or at Surgery Branch facility, as previously described (37, 38). Sequencing was done on a fresh tumor fragment embedded in optimum cutting temperature (O.C.T.) compound and/or TC lines and matched normal PBL sample. The data were aligned to genome build hg 18. An mRNA sequencing library was prepared from fresh tumors using an Illumina TruSeq RNA library prep kit, as previously described (39).
Statistical analysis
Differences in the frequencies of TCRB clonotypes from different CD4 T cell subsets were analyzed using Wilcoxon signed-rank test. Significance values are indicated as *P < 0.05 and ***P < 0.001.
Supplementary Material
Fig. S1. Number of total productive TCRB reads for FOXP3+ and FOXP3− subsets sorted from tumor or PBL.
Table S3. Summary of HLA typing of TC lines.
Table S4. The amino acid sequences of CCL5 and ANXA1 mutated and wild-type peptides.
Table S5. Genomic analysis of CCL5 and ANAX1 neoantigens.
Table S6. Raw data from figures.
Fig. S2. Number of total unique TCRB reads for FOXP3+ and FOXP3− subsets sorted from tumor or PBL.
Fig. S3. Clonality of FOXP3+ and FOXP3− subsets sorted from tumor or PBL.
Fig. S4. High level expression of HLA class II alleles on TC lines after transduction by CIITA.
Fig. S5. FOXP3− Tconv-derived TCRs exhibited tumor reactivity.
Fig. S6. FOXP3+ TCR 7 exhibited specific reactivity to pp9.
Table S1. Clinical characteristics of patients.
Table S2. Summary of sorted FOXP3 populations in tumor and PBL.
Acknowledgments:
We thank A. Mixon and S. Farid for flow cytometry support, T. Prickett and J. Gartner for sequencing analysis, Y. Li and S. Ray for technical support, M. Parkhurst for peptide synthesis, and K. Hanada and Y. C. Lu for providing reagents.
Funding: This research was supported by the Intramural Research Program of NIH.
Footnotes
SUPPLEMENTARY MATERIALS
immunology.sciencemag.org/cgi/content/full/4/31/eaao4310/DC1
Competing interests: Authors declare that they have no competing financial interests.
Data and materials availability: All the FASTQ files related to this study are available through the National Center for Biotechnology Information Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) under BioProject Accession PRJNA498668.
REFERENCES AND NOTES
- 1.Baecher-Allan C, Viglietta V, Hafler DA, Human CD4+CD25+ regulatory T cells. Semin. Immunol 16, 89–98 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Yamaguchi T, Nomura T, Ono M, Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Josefowicz SZ, Lu L-F, Rudensky AY, Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudensky AY, Regulatory T cells and Foxp3. Immunol. Rev. 241, 260–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmadzadeh M, Felipe-Silva A, Heemskerk B, Powell DJ Jr., Wunderlich JR, Merino MJ, Rosenberg SA, FOXP3 expression accurately defines the population of intratumoral regulatory T cells that selectively accumulate in metastatic melanoma lesions. Blood 112, 4953–4960 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EV, Chudakov DM, Rudensky AY, Regulatory T cells exhibit distinct features in human breast cancer. Immunity 45, 1122–1134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Simone M, Arrigoni A, Rossetti G, Gruarin P, Ranzani V, Politano C, Bonnal RJP, Provasi E, Sarnicola ML, Panzeri I, Moro M, Crosti M, Mazzara S, Vaira V, Bosari S, Palleschi A, Santambrogio L, Bovo G, Zucchini N, Totis M, Gianotti L, Cesana G, Perego RA, Maroni N, Ceretti AP, Opocher E, De Francesco R, Geginat J, Stunnenberg HG, Abrignani S, Pagani M, Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity 45, 1135–1147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, Zhu Y, Wei S, Kryczek I, Daniel B, Gordon A, Myers L, Lackner A, Disis ML, Knutson KL, Chen L, Zou W, Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa H, Sakaguchi S, Regulatory T cells in tumor immunity. Int. J. Cancer 127, 759–767 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Levine AG, Arvey A, Jin W, Rudensky AY, Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vahl JC, Drees C, Heger K, Heink S, Fischer JC, Nedjic J, Ohkura N, Morikawa H, Poeck H, Schallenberg S, Rieß D, Hein MY, Buch T, Polic B, Schönle A, Zeiser R, Schmitt-Gräff A, Kretschmer K, Klein L, Korn T, Sakaguchi S, Schmidt-Supprian M, Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity 41, 722–736 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Li MO, Rudensky AY, T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 16, 220–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francois V, Ottaviani S, Renkvist N, Stockis J, Schuler G, Thielemans K, Colau D, Marchand M, Boon T, Lucas S, van der Bruggen P, The CD4+ T-cell response of melanoma patients to a MAGE-A3 peptide vaccine involves potential regulatory T cells. Cancer Res. 69, 4335–4345 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Bonertz A, Weitz J, Pietsch D-H, Rahbari NN, Schlude C, Ge Y, Juenger S, Vlodavsky I, Khazaie K, Jaeger D, Reissfelder C, Antolovic D, Aigner M, Koch M, Beckhove P, Antigen-specific Tregs control T cell responses against a limited repertoire of tumor antigens in patients with colorectal carcinoma. J. Clin. Invest. 119, 3311–3321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vence L, Palucka AK, Fay JW, Ito T, Liu Y-J, Banchereau J, Ueno H, Circulating tumor antigen-specific regulatory T cells in patients with metastatic melanoma. Proc. Natl. Acad. Sci. U.S.A. 104, 20884–20889 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang HY, Lee DA, Peng G, Guo Z, Li Y, Kiniwa Y, Shevach EM, Wang RF, Tumor-specific human CD4+ regulatory T cells and their ligands: Implications for immunotherapy. Immunity 20, 107–118 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Lei H, Kuchenbecker L, Streitz M, Sawitzki B, Vogt K, Landwehr-Kenzel S, Millward J, Juelke K, Babel N, Neumann A, Reinke P, Volk HD, Human CD45RA− FoxP3hi memory-type regulatory T cells show distinct TCR repertoires with conventional T cells and play an important role in controlling early immune activation. Am. J. Transplant. 15, 2625–2635 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Golding A, Darko S, Wylie WH, Douek DC, Shevach EM, Deep sequencing of the TCR-β repertoire of human forkhead box protein 3 (FoxP3)+and FoxP3-T cells suggests that they are completely distinct and non-overlapping. Clin. Exp. Immunol. 188, 12–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmadzadeh M, Antony PA, Rosenberg SA, IL-2 and IL-15 each mediate de novo induction of FOXP3 expression in human tumor antigen-specific CD8 T cells. J. Immunother. 30, 294–302 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavin MA, Torgerson TR, Houston E, DeRoos P, Ho WY, Stray-Pedersen A, Ocheltree EL, Greenberg PD, Ochs HD, Rudensky AY, Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc. Natl. Acad. Sci. U.S.A. 103, 6659–6664 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran DQ, Ramsey H, Shevach EM, Induction of FOXP3 expression in naive human CD4+FOXP3− T cells by T-cell receptor stimulation is transforming growth factor-β–dependent but does not confer a regulatory phenotype. Blood 110, 2983–2990 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, Nagase H, Nishimura J, Yamamoto H, Takiguchi S, Tanoue T, Suda W, Morita H, Hattori M, Honda K, Mori M, Doki Y, Sakaguchi S, Two FOXP3+CD4+ T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 22, 679–684 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Pasetto A, Gros A, Robbins PF, Deniger DC, Prickett TD, Matus-Nicodemos R, Douek DC, Howie B, Robins H, Parkhurst MR, Gartner J, Trebska-McGowan K, Crystal JS, Rosenberg SA, Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol. Res. 4, 734–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie B, Sherwood AM, Berkebile AD, Berka J, Emerson RO, Williamson DW, Kirsch I, Vignali M, Rieder MJ, Carlson CS, Robins HS, High-throughput pairing of T cell receptor α and β sequences. Sci. Transl. Med. 7, 301ra131 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Stevanovic S, Pasetto A, Helman SR, Gartner JJ, Prickett TD, Howie B, Robins HS, Robbins PF, Klebanoff CA, Rosenberg SA, Hinrichs CS, Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 356, 200–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shevach EM, Thornton AM, tTregs, pTregs, and iTregs: Similarities and differences. Immunol. Rev. 259, 88–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilate AM, Lafaille JJ, Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu. Rev. Immunol. 30, 733–758 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Zhou G, Levitsky HI, Natural regulatory T cells and de novo-induced regulatory T cells contribute independently to tumor-specific tolerance. J. Immunol. 178, 2155–2162 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Savage PA, Leventhal DS, Malchow S, Shaping the repertoire of tumor-infiltrating effector and regulatory T cells. Immunol. Rev. 259, 245–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gros A, Turcotte S, Wunderlich JR, Ahmadzadeh M, Dudley ME, Rosenberg SA, Myeloid cells obtained from the blood but not from the tumor can suppress T-cell proliferation in patients with melanoma. Clin. Cancer Res. 18, 5212–5223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA, Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 66, 8878–8886 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen CJ, Li YF, El-Gamil M, Robbins PF, Rosenberg SA, Morgan RA, Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 67, 3898–3903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haga-Friedman A, Horovitz-Fried M, Cohen CJ, Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J. Immunol. 188, 5538–5546 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DAA, Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–594 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, Rosenberg SA, Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y-C, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y, Rosenberg SA, Robbins PF, Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin. Cancer Res. 20, 3401–3410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran E, Ahmadzadeh M, Lu Y-C, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, Wunderlich JR, Somerville RP, Rosenberg SA, Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones S, Wang T-L, Shih I-M, Mao T-L, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr., Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N, Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran E, Robbins PF, Lu Y-C, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, Kriley IR, Rosenberg SA, T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Number of total productive TCRB reads for FOXP3+ and FOXP3− subsets sorted from tumor or PBL.
Table S3. Summary of HLA typing of TC lines.
Table S4. The amino acid sequences of CCL5 and ANXA1 mutated and wild-type peptides.
Table S5. Genomic analysis of CCL5 and ANAX1 neoantigens.
Table S6. Raw data from figures.
Fig. S2. Number of total unique TCRB reads for FOXP3+ and FOXP3− subsets sorted from tumor or PBL.
Fig. S3. Clonality of FOXP3+ and FOXP3− subsets sorted from tumor or PBL.
Fig. S4. High level expression of HLA class II alleles on TC lines after transduction by CIITA.
Fig. S5. FOXP3− Tconv-derived TCRs exhibited tumor reactivity.
Fig. S6. FOXP3+ TCR 7 exhibited specific reactivity to pp9.
Table S1. Clinical characteristics of patients.
Table S2. Summary of sorted FOXP3 populations in tumor and PBL.