Abstract.
Acute diarrhea is an important public health issue. Here, we focused on the differences of enteropathogens in acute diarrhea between urban and rural areas in southeast China. Laboratory- and sentinel-based surveillance of acute diarrhea (≥ 3 loose or liquid stools/24 hours) was conducted at 16 hospitals. Fecal specimens were tested for bacterial (Aeromonas sp., Campylobacter sp., diarrheagenic Escherichia coli, Plesiomonas shigelloides, non-typhoidal Salmonella, Shigella sp., Vibrio sp., and Yersinia sp.) and viral (adenovirus, astrovirus, Norovirus, Rotavirus, and Sapovirus) pathogens. Descriptive statistics were used. Between January 1, 2010, and December 31, 2014, 4,548 outpatients with acute diarrhea were enrolled (urban, n = 3,220; rural, n = 1,328). Pathogens were identified in 2,074 (45.6%) patients. Norovirus (25.7%), Vibrio parahaemolyticus (10.2%), enteroaggregative Escherichia coli (EAEC) (8.8%), group A Rotavirus (7.0%), and enterotoxigenic Escherichia coli (ETEC) (5.6%) were the most common pathogens. Enteropathogens were less common in urban than in rural areas (42.0% versus 54.4%, P < 0.001). In urban areas, EAEC and ETEC were more common in high-income than in middle-income regions. Interventions targeting the most common enteropathogens can substantially reduce the burden of acute diarrhea in southeast China.
INTRODUCTION
Diarrhea is a leading cause of morbidity and mortality globally and caused an estimated 1.3 million deaths in 2015.1 China is one of the 15 high-burden countries for diarrhea in the world.2 Understanding the pathogen characteristics of diarrheal diseases is critical to enable the development of more specific disease control strategies.
The differences in pathogen features associated with acute diarrhea among patients of various ages and from different regions have been thoroughly discussed. In addition, previous studies in China have indicated the unique characteristics of some specific enteropathogens associated with acute diarrhea in rural areas.3,4 The enteropathogens of bacterial diarrhea among children was also found to vary in developing and developed regions of China.5,6 However, differences in the features of the enteropathogens of acute diarrhea between urban and rural areas, including both bacteria and viruses, have not been well demonstrated, especially in China.
Our research aimed to reveal the characteristics of enteropathogens associated with acute diarrhea in southeastern China between urban and rural areas. The conclusions drawn in our study provide scientific evidence to support the formulation of appropriate public health policies.
METHODS
Study design and participants.
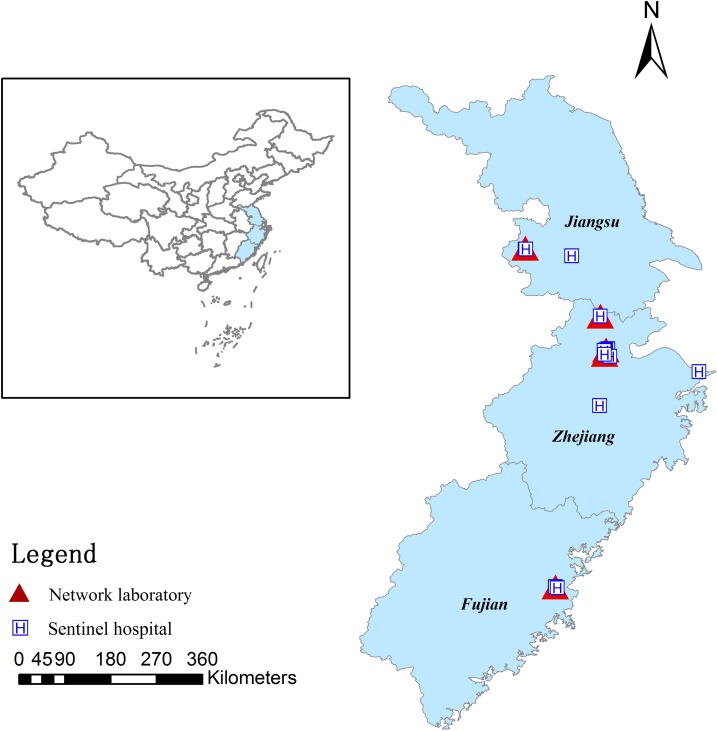
Between January 1, 2010, and December 31, 2014, diarrhea surveillance was conducted at 16 sentinel hospitals in southeast China, covering 25 county-level cities, 59 districts, and 44 counties from the Zhejiang, Jiangsu, and Fujian provinces (Figure 1). The types of hospitals included children’s, general, and urban hospitals, and rural community health service centers.
Figure 1.
The geographic distribution of the five network laboratories and 16 sentinel hospitals. This figure appears in color at www.ajtmh.org.
During each week of the study period, the first 1–5 eligible cases visiting each sentinel hospital were enrolled in our study, with approximately a median of 60 outpatients with acute diarrhea enrolled each year in each hospital; they were primarily enrolled from the enteric, pediatric, infectious disease, emergency, and internal medicine departments. Diarrhea was defined as the passage of three or more loose or liquid stools per day. For breastfed babies (≤ 6 months),7,8 we used the mother’s definition of diarrhea. This study excluded patients with diarrhea lasting more than 14 days,9 patients with comorbid conditions (e.g., hypertension, diabetes mellitus, and cardiovascular disease), patients who had received antibiotics within the preceding 10 days, and patients who had a history of travel (defined as a trip outside of southeast China) in the week preceding the onset of illness.
Information regarding sociodemographic and clinical characteristics was collected using a standardized case reporting form (CRF) during recruitment. Verbal consent was acquired from outpatients or guardians and recorded by the practitioner on the CRF.
Specimen collection and transport.
Fecal specimens were collected and transported to network laboratories for microbiological testing. For each patient, three aliquots of feces were collected under the guidance of a trained nurse at the hospital. For viral testing, 5 g of fresh whole stool was collected in sterilized containers without preservatives and stored at −20°C. For bacterial testing, fresh whole stool was collected using five sterilized cotton swabs and immediately placed in Cary-Blair medium (C-B, Oxoid Ltd., Basingstoke, United Kingdom) at 4°C.
The collected specimens were packed and transported in ice boxes to network laboratories in accordance with the UN3373 transportation requirements within 24 hours for bacterial tests and within 48 hours for viral tests. When the samples arrived, the network laboratories inspected and recorded the quality of the specimens, and unqualified specimens (specimen volume < 5 g or swabs not preserved in Cary-Blair medium) were rejected. In cases of rejected samples, new samples were requested for resubmission.
Microbiological testing.
The network laboratories consisted of the Fujian Provincial Center for Disease Control and Prevention, Huzhou Central Hospital in Zhejiang Province, Jiangsu Provincial Center for Disease Control and Prevention, Zhejiang University, and Zhejiang Provincial Center for Disease Control and Prevention.
A panel of enteropathogens was assayed, including viral (adenovirus, astrovirus, Norovirus, Rotavirus, and Sapovirus) and bacterial (Aeromonas sp., Campylobacter sp., diarrheagenic Escherichia coli (DEC), Plesiomonas shigelloides (P. shigelloides), non-typhoidal Salmonella (NTS), Shigella sp., Vibrio sp., and Yersinia sp.) pathogens. All network laboratories adopted a uniform study protocol, including standardized test methods and operational procedures.10
For Rotavirus testing, an enzyme-linked immunosorbent assay (ProSpecT™ Rotavirus kit, Oxoid Ltd., Basingstoke, United Kingdom) was used to confirm the presence of group A Rotavirus antigens, and reverse transcription–polymerase chain reaction with random primers was used to genotype the ELISA-positive specimens.11 For the other viruses, viral DNA or RNA was extracted from specimens, and the first strand cDNAs were synthesized from the extracted viral RNAs. The multiplex PCR with two sets of specific primers was performed to detect adenovirus, astrovirus, Norovirus (GI and GII), Rotavirus (groups B and C), and Sapovirus.12,13
To isolate Campylobacter sp., the specimens were inoculated on Skirrow selective medium, which added blood and incubated at 42°C in microaerophilic environment for 2–3 days. The suspicious strains were identified following the oxidase, catalase, and hippurate hydrolysis tests.6 To isolate DEC, the specimens were inoculated onto MacConkey (MAC) agar and incubated at 37°C for 16–24 hours. The suspicious colonies were selected to perform biochemical reactions by Kligler iron agar (KIA), motility indole urea (MIU) semisolid medium, and indole/methyl red/Voges-Proskauer/citrate test to identify suspicious presumptive Escherichia coli strains. Multiplex PCR was performed to detect the virulence genes of suspicious presumptive Escherichia coli strains, and the main pathotypes of DEC included enteropathogenic Escherichia coli (EPEC), enteroaggregative Escherichia coli (EAEC), enterotoxigenic Escherichia coli (ETEC), enteroinvasive Escherichia coli (EIEC), and Shiga toxin-producing Escherichia coli (STEC).14 To isolate NTS, the specimens were placed into selenite brilliant green sulfa enrichment broth and incubated at 37°C for 16 hours. Then, the inoculum was placed onto the Salmonella Shigella (SS) agar at 37°C overnight, and the suspicious colonies were selected to conduct ortho-nitrophenyl-beta-D-galactopyranoside test. Finally, the strains were confirmed by Api20E (bioMérieux, France).15 To isolate Shigella sp., specimens were streaked onto the SS agar, MAC agar, or xylose lysine deoxycholate agar, incubated at 37°C for 16–24 hours. The suspicious colonies were chosen to test biochemical reactions by KIA and MIU. The strains were identified and serotyped by the antisera of Shigella. To isolate Vibrio sp., Aeromonas sp., and P. shigelloides, the specimens were cultured by alkaline peptone water at 37°C for 6–8 hours and then inoculated on thiosulfate citrate bile salts sucrose agar, MAC agar, and blood plate. The suspected colonies were tested for oxidase activity, and positive isolates were identified by Api20E/NE. To isolate Yersinia sp., enrichment was performed by using peptone sorbitol bile broth at 4°C for 10–20 days. Then, the strains were inoculated onto Yersinia selective agar (cefsulodin irgasan novobiocin agar) and incubated at 25°C for 24 hours. Suspicious colonies were selected by KIA and MIU, and then identified by Api20E.16
Statistical analysis.
According to the Statistic Provisions for Dividing Urban and Rural Areas from the National Bureau of Statistics and the present addresses of patients, we classified patients residing in cities and towns into urban areas, and those residing in townships and villages into rural areas.17 The income levels of different regions were divided into two categories, high and middle, by adopting the criteria of the World Bank and using the Atlas method.18 Detailed descriptions of the criteria were provided in Supplemental Tables 1 and 2, respectively. The enrolled outpatients were divided into five age groups: < 5, 5–24, 25–44, 45–64, and ≥ 65 years. The onset date of cases was divided into four seasons: spring (March–May), summer (June–August), autumn (September–November), and winter (December–February). As not all specimens underwent a full-range assay of 13 enteropathogens, the prevalence of each pathogen (the proportion of cases that tested positive) was calculated by dividing the number of positive samples by the total number of samples tested for that pathogen. The exact 95% confidence interval (CI) for the prevalence was calculated using a binomial distribution.
The chi-squared test or Fisher’s exact test was used to compare proportions as appropriate. A two-sided P-value < 0.05 was considered statistically significant. All statistical tests were performed using the Statistical Package for Social Science (SPSS, version 13.0, SPSS Inc., Chicago, IL) and Microsoft Excel 2013 (version 15.0, Microsoft Inc., Redmond, WA). A geographic map was processed using ArcGIS (version 9.3, ESRI Inc., Redlands, CA).
Ethics approval and consent to participate.
This study was approved by the Ethics Committee of the Chinese Center for Disease Control and Prevention. Verbal consent was acquired from outpatients or guardians and recorded by the practitioner on the CRF.
RESULTS
Characteristics of the study participants.
Between January 1, 2010, and December 31, 2014, 4,548 outpatients with acute diarrhea were enrolled, including 3,220 patients from urban areas and 1,328 patients from rural areas. Although gender, age, and the receipt of oral rehydration before treatment were similar between the patients in urban and rural areas, other characteristics (e.g., income level, season, and the percentages of vomiting, fever, and dehydration) differed (Table 1).
Table 1.
Sociodemographic and clinical characteristics of outpatients with acute diarrhea in southeast China, 2010–2014
| Characteristic | All patients, n = 4,548 | Urban, n = 3,220 | Rural, n = 1,328 | P-value |
|---|---|---|---|---|
| Gender | 0.059 | |||
| Male | 2,439 (53.6) | 1,698 (52.7) | 741 (55.8) | |
| Female | 2,109 (46.4) | 1,522 (47.3) | 587 (44.2) | |
| Age (years) | 0.808 | |||
| < 5 | 1,302 (28.6) | 916 (28.4) | 386 (29.1) | |
| 5–24 | 636 (14.0) | 442 (13.7) | 194 (14.6) | |
| 25–44 | 1,340 (29.5) | 961 (29.8) | 379 (28.5) | |
| 45–64 | 889 (19.5) | 626 (19.4) | 263 (19.8) | |
| ≥ 65 | 381 (8.4) | 275 (8.5) | 106 (8.0) | |
| Income level* | < 0.001 | |||
| High | 3,532 (79.9) | 2,748 (85.3) | 784 (65.2) | |
| Middle | 890 (20.1) | 472 (14.7) | 418 (34.8) | |
| Season of illness onset | < 0.001 | |||
| Spring | 698 (15.3) | 402 (12.5) | 296 (22.3) | |
| Summer | 2,165 (47.6) | 1,628 (50.6) | 537 (40.4) | |
| Autumn | 1,211 (26.6) | 875 (27.2) | 336 (25.3) | |
| Winter | 474 (10.4) | 315 (9.8) | 159 (12.0) | |
| Clinical symptoms/signs | ||||
| Vomiting | 1,054 (23.2) | 671 (20.8) | 383 (28.8) | < 0.001 |
| Fever | 430 (9.5) | 356 (11.1) | 74 (5.6) | < 0.001 |
| Dehydration | 148 (3.3) | 121 (3.8) | 27 (2.0) | 0.003 |
| Oral rehydration before treatment* | 77 (3.7) | 37 (3.1) | 40 (4.6) | 0.076 |
The results in the table are presented as the no (%). Bold characters indicate significant (P < 0.05) values.
* The numbers in the column were not summated for a total because of missing data.
The prevalence of enteropathogens.
Overall, 2,074 (45.6%) outpatients were positive for at least one pathogen, including 1,878 patients with mono-infections and 196 patients with coinfections. Among the 13 identified enteropathogens, Norovirus was the most prevalent (25.7%, 787/3,061), followed by Vibrio parahaemolyticus (V. parahaemolyticus) (10.2%, 328/3,222), EAEC (8.8%, 245/2,788), group A Rotavirus (7.0%, 225/3,194), ETEC (5.6%, 155/2,788), NTS (3.5%, 115/3,266), Aeromonas sp. (3.2%, 102/3,200), EPEC (2.2%, 62/2,788), Sapovirus (1.5%, 47/3,060), P. shigelloides (0.7%, 23/3,199), Vibrio flurialis (0.7%, 15/2,200), Vibrio cholerae (0.6%, 20/3,221), Shigella sp. (0.6%, 19/3,238), group B Rotavirus (0.6%, 18/3,111), adenovirus (0.6%, 17/3,068), astrovirus (0.5%, 16/3,062), group C Rotavirus (0.5%, 14/3,076), STEC (0.3%, 8/2,788), Yersinia sp. (0.2%, 6/3,201), Campylobacter sp. (0.1%, 3/3,177), and EIEC (0.1%, 2/2,788).
In total, 1878 (43.2%) of 4,352 patients were positive for mono-infections, and the major pathogens included Norovirus (23.3%, 690/2,964), V. parahaemolyticus (9.2%, 294/3,188), EAEC (8.0%, 197/2,474), ETEC (5.6%, 134/2,411), and group A Rotavirus (5.2%, 163/3,132) (Table 2).
Table 2.
Microbiological findings of outpatients with acute diarrhea
| Enteropathogens | No. positive/no. tested (%) | P-value | ||
|---|---|---|---|---|
| All patients, n = 4,548 | Urban, n = 3,220 | Rural, n = 1,328 | ||
| Mono-infection | 1,878/4,352 (43.2) | 1,245/3,114 (40.0) | 633/1,238 (51.1) | < 0.001 |
| Bacterial | 954/3,428 (27.8) | 600/2,469 (24.3) | 354/959 (36.9) | < 0.001 |
| Diarrheagenic Escherichia coli | 415/2,692 (15.4) | 259/1,764 (14.7) | 156/928 (16.8) | 0.146 |
| EAEC | 197/2,474 (8.0) | 116/1,621 (7.2) | 81/853 (9.5) | 0.041 |
| EHEC | 5/2,282 (0.2) | 4/1,509 (0.3) | 1/773 (0.1) | 0.855 |
| EIEC | 0/2,277 (0.0) | 0/1,505 (0.0) | 0/772 (0.0) | / |
| EPEC | 47/2,324 (2.0) | 25/1,530 (1.6) | 22/794 (2.8) | 0.065 |
| ETEC | 134/2,411 (5.6) | 96/1,601 (6.0) | 38/810 (4.7) | 0.187 |
| Untyped | 32/2,309 (1.4) | 18/1,523 (1.2) | 14/786 (1.8) | 0.243 |
| Non-typhoidal Salmonella | 107/3,258 (3.3) | 60/2,266 (2.6) | 47/992 (4.7) | 0.002 |
| Shigella sp. | 13/3,232 (0.4) | 11/2,254 (0.5) | 2/978 (0.2) | 0.386 |
| Aeromonas sp. | 71/3,169 (2.2) | 47/2,192 (2.1) | 24/977 (2.5) | 0.583 |
| Plesiomonas shigelloides | 14/3,190 (0.4) | 9/2,203 (0.4) | 5/987 (0.5) | 0.922 |
| Campylobacter sp. | 1/3,175 (0.0) | 0/2,192 (0.0) | 1/983 (0.1) | 0.310 |
| Vibrio sp. | 322/2,239 (14.4) | 206/1,713 (12.0) | 116/526 (22.1) | < 0.001 |
| V. cholerae (serogroup O1 and O139) | 1/3,202 (0.0) | 1/2,211 (0.0) | 0/991 (0.0) | 1.000 |
| V. cholerae (serogroup non-o1/o139) | 14/3,215 (0.4) | 11/2,221 (0.5) | 3/994 (0.3) | 0.631 |
| V. parahaemolyticus | 294/3,188 (9.2) | 184/2,211 (8.3) | 110/977 (11.3) | 0.008 |
| V. flurialis | 13/2,198 (0.6) | 10/1,692 (0.6) | 3/506 (0.6) | 1.000 |
| Yersinia sp. | 4/3,199 (0.1) | 2/2,205 (0.1) | 2/994 (0.2) | 0.781 |
| Other bacteria* | 7/1,532 (0.5) | 6/1,028 (0.6) | 1/504 (0.2) | 0.517 |
| Viral | 924/3,398 (27.2) | 645/2,514 (25.7) | 279/884 (31.6) | 0.001 |
| Rotavirus (groups A, B, and C) | 167/3,013 (5.5) | 98/2,268 (4.3) | 69/745 (9.3) | < 0.001 |
| Group A rotavirus | 163/3,132 (5.2) | 95/2,369 (4.0) | 68/763 (8.9) | < 0.001 |
| Group B rotavirus | 3/3,096 (0.1) | 3/2,305 (0.1) | 0/791 (0.0) | 0.575 |
| Group C rotavirus | 1/3,063 (0.0) | 0/2,283 (0.0) | 1/780 (0.1) | 0.255 |
| Norovirus | 690/2,964 (23.3) | 500/2,223 (22.5) | 190/741 (25.6) | 0.079 |
| G I | 48/2,322 (2.1) | 34/1,757 (1.9) | 14/565 (2.5) | 0.430 |
| G II | 642/2,916 (22.0) | 466/2,189 (21.3) | 176/727 (24.2) | 0.100 |
| Sapovirus | 34/3,047 (1.1) | 20/2,261 (0.9) | 14/786 (1.8) | 0.039 |
| Astrovirus | 8/3,054 (0.3) | 3/2,270 (0.1) | 5/784 (0.6) | 0.047 |
| Adenovirus | 11/3,062 (0.4) | 10/2,273 (0.4) | 1/789 (0.1) | 0.357 |
| Other viruses† | 14/2,993 (0.5) | 14/2,226 (0.6) | 0/767 (0.0) | 0.058 |
| Coinfection | 196/2,670 (7.3) | 106/1,975 (5.4) | 90/695 (12.9) | < 0.001 |
| Viral–viral | 54/2,528 (2.1) | 32/1,901 (1.7) | 22/627 (3.5) | 0.006 |
| Bacterial–bacterial | 63/2,537 (2.5) | 36/1,905 (1.9) | 27/632 (4.3) | 0.001 |
| Viral–bacterial | 79/2,553 (3.1) | 38/1,907 (2.0) | 41/646 (6.3) | < 0.001 |
| Total | 2,074/4,548 (45.6) | 1,351/3,220 (42.0) | 723/1,328 (54.4) | < 0.001 |
EAEC = enteroaggregative Escherichia coli; EHEC = enterohaemorrhagic Escherichia coli; EIEC = enteroinvasive Escherichia coli; EPEC = enteropathogenic Escherichia coli; ETEC = enterotoxigenic Escherichia coli. Bold characters indicate significant (P < 0.05) values.
* The full range of eight bacteria tested (n = 1,532) was used to calculate the prevalence of other bacteria (e.g., Vibrio vulnificus; Sphingomonas paucimobilis).
† The full range of five viruses tested (n = 2,993) was used to calculate the prevalence of other viruses (e.g., Enterovirus 71, poliovirus).
In total, 196 (7.3%) of 2,670 patients were positive for coinfection, and the main combinations of pathogens involved were Norovirus and DEC coinfection (1.4%, 35/2,553), Norovirus and group A Rotavirus coinfection (1.1%, 27/2,528), DEC and V. parahaemolyticus coinfection (0.5%, 12/2,537), and DEC and Aeromonas sp. coinfection (0.4%, 11/2,537).
The detection rate of at least one pathogen-positive specimen was lower in urban areas than in rural areas (42.0% versus 54.4%, P < 0.001), including mono-infections (40.0% versus 51.1%, P < 0.001) and coinfections (5.4% versus 12.9%, P < 0.001). More specifically, in instances of mono-infection, some enteropathogens differed between urban and rural areas, including EAEC (7.2% versus 9.5%, P = 0.041), NTS (2.6% versus 4.7%, P = 0.002), V. parahaemolyticus (8.3% versus 11.3%, P = 0.008), group A Rotavirus (4.0% versus 8.9%, P < 0.001), Sapovirus (0.9% versus 1.8%, P = 0.039), and astrovirus (0.1% versus 0.6%, P = 0.047) (Table 2).
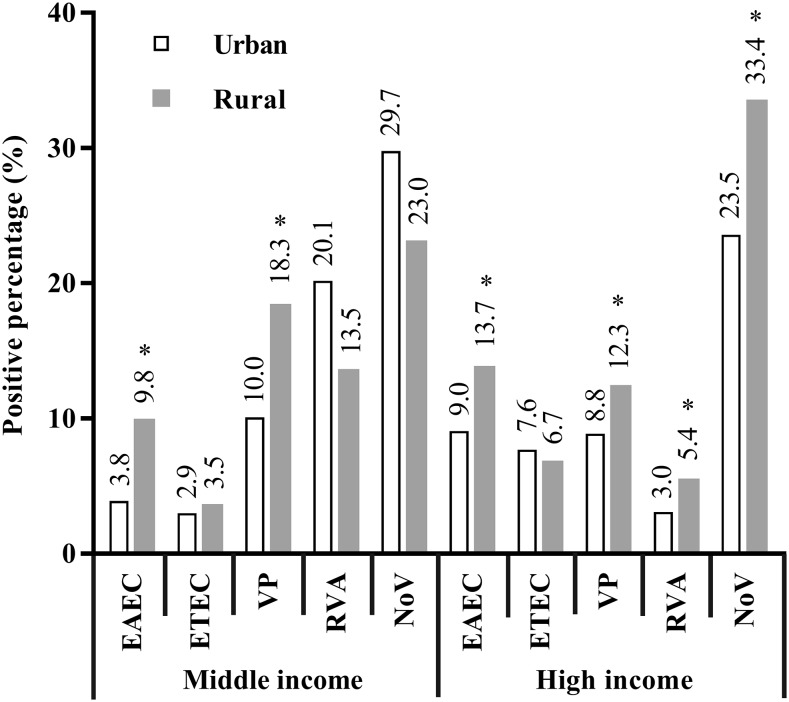
In regions with middle-income level, the percentage of EAEC-infected patients was lower in urban areas than in rural areas, and the same pattern was observed for V. parahaemolyticus–infected patients. In regions with high-income level, the percentages of EAEC, V. parahaemolyticus, group A Rotavirus, and Norovirus were lower in urban areas than in rural areas (Figure 2). Moreover, the percentage of EAEC-positive patients was lower in regions with middle-income level than in regions with high-income level (urban: 3.8% versus 9.0%, P = 0.012), and the same pattern was observed for ETEC (urban: 2.9% versus 7.6%, P = 0.014) and Norovirus (rural: 23.0% versus 33.4%, P = 0.008).
Figure 2.
Positive percentage (%) of the main enteropathogens between middle- and high-income levels. *P < 0.05. EAEC = enteroaggregative Escherichia coli; ETEC = enterotoxigenic Escherichia coli; NoV = Norovirus; RVA = group A Rotavirus; VP = Vibrio parahaemolyticus.
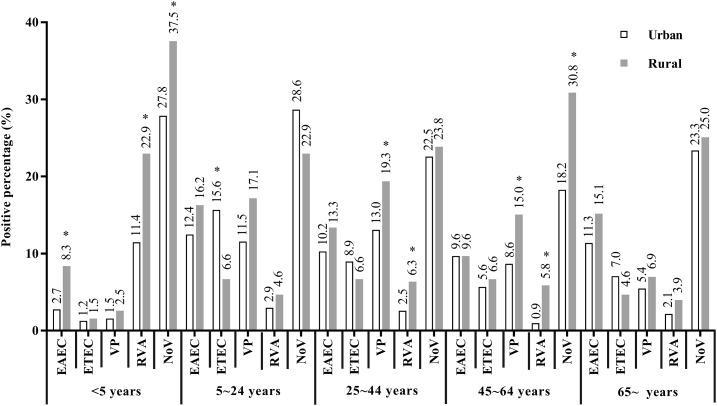
In both urban and rural areas, Norovirus was the leading pathogen among all age groups. In patients aged < 5 years, the percentage of EAEC-positive cases was lower in urban areas than in rural areas (2.7% versus 8.3%, P < 0.001), and the same pattern was observed for group A Rotavirus (11.4% versus 22.9%, P < 0.001) and Norovirus (27.8% versus 37.5%, P = 0.002). In patients aged 25–44 years, the percentage of V. parahaemolyticus infections was lower in urban areas than in rural areas (13.0% versus 19.3%, P = 0.009), and the same pattern was observed for group A Rotavirus (2.5% versus 6.3%, P = 0.011). In patients aged 45–64 years, the percentage of V. parahaemolyticus infections was lower in urban areas than in rural areas (8.6% versus 15.0%, P = 0.010), and the same pattern was observed for group A Rotavirus (0.9% versus 5.8%, P = 0.002) and Norovirus (18.2% versus 30.8%, P = 0.003). However, in patients aged 5–24 years, the percentage of ETEC-positive cases was higher in urban areas than in rural areas (15.6% versus 6.6%, P = 0.015) (Figure 3).
Figure 3.
Positive percentage (%) of main enteropathogens among different age groups. *P < 0.05. EAEC = enteroaggregative Escherichia coli; ETEC = enterotoxigenic E. coli; NoV = Norovirus; RVA = group A Rotavirus; VP = Vibrio parahaemolyticus.
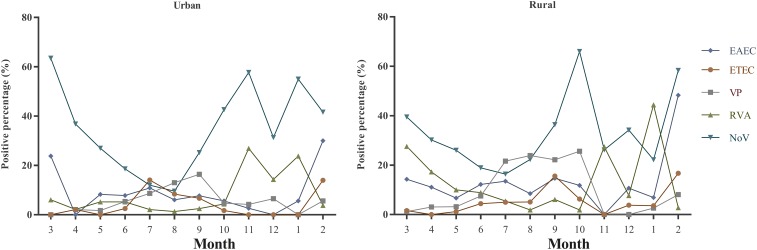
In urban areas, Norovirus was the most prevalent pathogen during all seasons, except for July (ETEC) and August (V. parahaemolyticus). In rural areas, aside from July (V. parahaemolyticus), August (V. parahaemolyticus), November (group A Rotavirus), and January (group A Rotavirus), the most prevalent pathogen was Norovirus (Figure 4). The percentage of EAEC-positive patients in April was lower in urban areas than in rural areas (0.0% versus 11.1%, P = 0.049), the same pattern was observed for ETEC (September: 6.7% versus 15.6%, P = 0.016), V. parahaemolyticus (July: 8.7% versus 21.6%, P < 0.001; August: 13.0% versus 23.9%, P < 0.001; October: 4.7% versus 25.6%, P < 0.001), group A Rotavirus (January: 23.8% versus 44.4%, P = 0.043; March: 6.1% versus 27.6%, P < 0.001; April: 2.2% versus 17.3%, P = 0.004), and Norovirus (August: 9.6% versus 22.3%, P < 0.001; October: 42.6% versus 66.0%, P = 0.003). However, the opposite pattern was observed in ETEC (July: 14.1% versus 5.0%, P = 0.007) and Norovirus (January: 55.0% versus 22.2%, P = 0.002; March: 63.5% versus 39.5%, P = 0.004; November: 57.7% versus 26.2%, P < 0.001) (Figure 4).
Figure 4.
Seasonal patterns of the main enteropathogens. EAEC = enteroaggregative Escherichia coli; ETEC = enterotoxigenic Escherichia coli; NoV = Norovirus; RVA = group A Rotavirus; VP = Vibrio parahaemolyticus. This figure appears in color at www.ajtmh.org.
Clinical symptoms of major enteropathogens.
Norovirus-infected outpatients were younger than noninfected outpatients, but their clinical symptoms were similar, except for higher percentages of patients with vomiting and dehydration among Norovirus-infected outpatients. EAEC-infected outpatients were older than noninfected outpatients, and the clinical characteristics of the outpatients were similar, aside from a lower percentage of fever among EAEC-infected outpatients. The percentage of outpatients with vomiting was higher among V. parahaemolyticus–infected outpatients than among noninfected outpatients. Group A Rotavirus-infected outpatients were significantly younger than noninfected outpatients, and the clinical signs differed between the two groups, including a higher percentage of vomiting, more episodes of vomiting in 24 hours, a higher percentage of fever, and a higher percentage of dehydration among group A Rotavirus-infected outpatients than the corresponding parameters among noninfected outpatients (Table 3).
Table 3.
Ages and clinical characteristics of outpatients infected with major enteropathogens
| Characteristics | EAEC | ETEC | Vibrio parahaemolyticus | Group A rotavirus | Norovirus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative, n = 1,475 | Positive, n = 197 | P-value | Negative, n = 1,475 | Positive, n = 134 | P-value | Negative, n = 1,853 | Positive, n = 294 | P-value | Negative, n = 1,721 | Positive, n = 163 | P-value | Negative, n = 1,622 | Positive, n = 690 | P-value | |
| Age, years, median (IQR) | 30.0 (4.0–51.0) | 33.0 (21.5–55.5) | 0.009 | 30.0 (4.0–51.0) | 33.0 (24.0–51.0) | 0.020 | 32.0 (19.0–52.0) | 32.5 (25.0–49.0) | 0.100 | 26.0 (1.0–48.0) | 1.0 (0.0–17.0) | < 0.001 | 27.0 (1.0–49.0) | 24.0 (1.0–42.0) | < 0.001 |
| Duration of diarrhea, days, median (IQR) | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) | 0.014 | 1.0 (1.0–2.0) | 1.0 (1.0–1.0) | < 0.001 | 1.0 (1.0–2.0) | 1.0 (0.0–1.0) | < 0.001 | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) | 0.961 | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) | < 0.001 |
| No. of stools in 24 hours, median (IQR) | 5.0 (4.0–6.0) | 5.0 (4.0–6.0) | 0.643 | 5.0 (4.0–6.0) | 5.0 (3.0–6.0) | 0.164 | 5.0 (4.0–6.0) | 5.0 (4.0–6.0) | 0.005 | 5.0 (3.0–6.0) | 5.0 (4.0–7.0) | 0.003 | 5.0 (3.0–6.0) | 5.0 (3.0–6.0) | 0.094 |
| Vomiting present, % | 19.8 | 16.2 | 0.236 | 19.8 | 17.2 | 0.462 | 17.7 | 33.3 | < 0.001 | 18.2 | 44.2 | < 0.001 | 17.1 | 34.1 | < 0.001 |
| Duration of vomiting, days, median (IQR) | 1.0 (0.0–1.0) | 1.0 (0.0–1.0) | 0.298 | 1.0 (0.0–1.0) | 1.0 (1.0–1.0) | 0.545 | 1.0 (0.0–1.0) | 1.0 (0.0–1.0) | 0.009 | 1.0 (0.0–1.0) | 1.0 (0.5–2.0) | 0.191 | 1.0 (0.0–1.0) | 1.0 (0.0–1.0) | 0.005 |
| No. of vomiting episodes in 24 hours, median (IQR) | 1.0 (1.0–3.0) | 1.0 (1.0–2.75) | 0.676 | 1.0 (1.0–3.0) | 1.0 (1.0–3.0) | 0.859 | 1.0 (1.0–2.0) | 2.0 (1.0–3.0) | < 0.001 | 1.0 (1.0–3.0) | 3.0 (1.0–4.0) | < 0.001 | 1.0 (1.0–2.0) | 2.0 (1.0–3.0) | < 0.001 |
| Fever present, % | 6.1 | 1.5 | 0.008 | 6.1 | 7.5 | 0.532 | 6.8 | 7.8 | 0.521 | 11.7 | 19.0 | 0.006 | 10.2 | 10.9 | 0.615 |
| Temperature of fever, °C, median (IQR) | 38.3 (38.0–38.8) | 38.5 (38.0–/) | 0.992 | 38.3 (38.0–38.8) | 38.0 (38.0–38.525) | 0.425 | 38.3 (38.0–38.7) | 38.1 (37.7–38.5) | 0.152 | 38.4 (38.0–39.0) | 38.5 (38.0–39.0) | 0.614 | 38.4 (38.0–38.8) | 38.0 (38.0–38.9) | 0.275 |
| Dehydration present, % | 3.5 | 3.0 | 0.765 | 3.5 | 3.0 | 0.968 | 3.0 | 3.4 | 0.687 | 1.2 | 4.3 | 0.004 | 1.0 | 4.2 | < 0.001 |
EAEC = enteroaggregative Escherichia coli; ETEC = enterotoxigenic Escherichia coli. IQR = interquartile range. Bold characters indicate significant (P < 0.05) values.
DISCUSSION
The results of this diarrhea surveillance study on the epidemiology of enteropathogens in southeast China showed differences in characteristics of enteropathogens between urban and rural areas, and indicated a higher positive percentage of enteropathogens in rural areas than in urban areas. Norovirus, V. parahaemolyticus, EAEC, group A Rotavirus, and ETEC were the most common enteropathogens in this study.
Overall, 45.6% of the enrolled patients were positive for at least one pathogen, which is similar to the results of a diarrhea surveillance study in Shanghai, China.19 In contrast to most studies that focused on patients aged < 5 years, this surveillance study enrolled patients in all age groups. Thus, Norovirus was the predominant pathogen in this study rather than rotavirus, as identified in other studies12,20–23; however, group A Rotavirus was still a leading pathogen among patients aged < 5 years in this study. Norovirus plays an important role in sporadic diarrheal cases across all age groups.24–27 Moreover, previous studies have reported that the most prevalent enteropathogen in adults was Norovirus.6,22,28 DEC is a main cause of bacterial diarrhea in developing countries,29–31 and in this study, EAEC was predominant among the pathotypes of DEC, which is in line with the results of previous studies.32,33 In total, 7.3% of patients with acute diarrhea were positive for more than one pathogen; this detection rate is much lower than those reported previously.34
In patients infected with Norovirus, V. parahaemolyticus, and group A Rotavirus, the percentage of patients with vomiting was higher than the corresponding percentage of noninfected patients, suggesting that if vomiting were included in the “case definition,” the positive percentages of specific pathogens may improve. Some studies have incorporated this idea in the surveillance of Norovirus and Rotavirus.35
Unlike other major pathogens, in urban areas, EAEC and ETEC exhibited a higher positive percentage in regions with high-income level than in those with middle-income level. The specific reason for this observation requires further investigation, especially in the context of rapid urbanization.
LIMITATIONS OF THIS STUDY
However, this study had several limitations. First, this study lacked a case–control design. Hospital-based surveillance was not representative of the overall population. Because of the limited capacity of laboratories, most specimens were not assayed for the full range of enteropathogens. For example, unlike the previously reported positive percentage (7.1%) in Shanghai, China,36 Campylobacter sp. in this study may have been underestimated. Therefore, some stool specimens should be tested again with improved technology or appropriate selective enrichment. Alternatively, the negative results of some stool specimens may have been due to the absence of the enteropathogens in our surveillance scheme. Some “new” agents of diarrhea have been described, including bacterial (Klebsiella oxytoca,37 enterotoxigenic Bacteroides fragilis,38 and Laribacter hongkongensis39) and viral (Parechovirus40 and bocavirus41) pathogens. Moreover, the information regarding the subtypes of enteropathogens was not complete. Finally, long-term trends were not observed in this study as the period of 5 years was too short. Further continuous surveillance will be able to clarify such trends.
CONCLUSION
In conclusion, precise interventions targeting the five most common pathogens (Norovirus, V. parahaemolyticus, EAEC, group A Rotavirus, and ETEC) can substantially reduce the burden of acute diarrhea in southeast China. The differences between urban and rural areas should be emphasized in future surveillance and intervention efforts. Moreover, further studies are needed to explore the risk factors for enteropathogen infection and acute diarrhea.
Supplementary Material
Acknowledgments
We would like to thank the patients, nurses, and clinicians who participated in this study and the laboratory and administration staff at all sites for their assistance.
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1.GBD Diarrhoeal Diseases Collaborators , 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the global burden of disease study 2015. Lancet Infect Dis 17: 909–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O’Brien KL, Campbell H, Black RE, 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381: 1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiao N, et al. 2017. Variation analysis of Norovirus among children with diarrhea in rural Hebei Province, North of China. Infect Genet Evol 53: 199–205. [DOI] [PubMed] [Google Scholar]
- 4.Wang XY, et al. 2005. Incidence of diarrhea caused by Rotavirus infections in rural Zhengding, China: prospective, population-based surveillance. J Infect Dis 192 (Suppl 1): S100–S105. [DOI] [PubMed] [Google Scholar]
- 5.Qin S, Duan R, Jing HQ, Wang X, 2018. Etiology of bacterial diarrhea in large cities, mid-sized/small cities and rural areas of China. Zhonghua Liu Xing Bing Xue Za Zhi 39: 651–655. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Wang J, Sun H, Xia S, Duan R, Liang J, Xiao Y, Qiu H, Shan G, Jing H, 2015. Etiology of childhood infectious diarrhea in a developed region of China: compared to childhood diarrhea in a developing region and adult diarrhea in a developed region. PLoS One 10: e0142136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Section on Breastfeeding , 2012. Breastfeeding and the use of human milk. Pediatrics 129: e827–e841. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization , 2018. Infant and Young Child Feeding. Geneva, Switzerland: WHO. [Google Scholar]
- 9.Shane AL, et al. 2017. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 65: 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Management Office of National Science and Technology Major Project of China , 2011. Diarrheal Syndrome Surveillance Protocol (2012 Version). Beijing, China: Chinese Center for Disease Control and Prevention. [Google Scholar]
- 11.Yan H, Nguyen TA, Phan TG, Okitsu S, Li Y, Ushijima H, 2004. Development of RT-multiplex PCR assay for detection of adenovirus and group A and C rotaviruses in diarrheal fecal specimens from children in China. Kansenshogaku Zasshi 78: 699–709. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, et al. 2015. Etiological epidemiology of viral diarrhea on the basis of sentinel surveillance in children younger than 5 years in Gansu, northwest China, 2009–2013. J Med Virol 87: 2048–2053. [DOI] [PubMed] [Google Scholar]
- 13.Zheng S, et al. 2016. Enteropathogens in children less than 5 years of age with acute diarrhea: a 5-year surveillance study in the Southeast Coast of China. BMC Infect Dis 16: 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller D, Greune L, Heusipp G, Karch H, Fruth A, Tschape H, Schmidt MA, 2007. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol 73: 3380–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner F, Villar R, Angulo F, Tauxe R, Swaminathan B, 2000. Salmonella nomenclature. J Clin Microbiol 38: 2465–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Cui Z, Jin D, Tang L, Xia S, Wang H, Xiao Y, Qiu H, Hao Q, Kan B, 2009. Distribution of pathogenic Yersinia enterocolitica in China. Eur J Clin Microbiol Infect Dis 28: 1237–1244. [DOI] [PubMed] [Google Scholar]
- 17.National Bureau of Statistics of China , 2008. Statistic Provisions for Dividing Urban and Rural Areas. Beijing, China: National Bureau of Statistics of China. [Google Scholar]
- 18.The World Bank , 2018. World Bank Country and Lending Groups. Washington, DC: The World Bank. [Google Scholar]
- 19.Li J, Pan H, Xiao WJ, Gong XH, Zhuang Y, Kuang XZ, Wu HY, Yuan ZA, 2017. Epidemiological and etiological surveillance study of infectious diarrhea in Shanghai in 2013–2015. Zhonghua Yu Fang Yi Xue Za Zhi 51: 1113–1117. [DOI] [PubMed] [Google Scholar]
- 20.Chen C-J, Wu F-T, Huang Y-C, Chang W-C, Wu H-S, Wu C-Y, Lin J-S, Huang F-C, Hsiung CA, 2015. Clinical and epidemiologic features of severe viral gastroenteritis in children: a 3-year surveillance, multicentered study in Taiwan with partial Rotavirus immunization. Medicine 94: e1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Operario DJ, et al. 2017. Etiology of severe acute watery diarrhea in children in the global Rotavirus surveillance network using quantitative polymerase chain reaction. J Infect Dis 216: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Podkolzin AT, et al. 2009. Hospital-based surveillance of Rotavirus and other viral agents of diarrhea in children and adults in Russia, 2005–2007. J Infect Dis 200 (Suppl 1): S228–S233. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, et al. 2015. Rotavirus-specific and overall diarrhea mortality in Chinese children younger than 5 years: 2003 to 2012. Pediatr Infect Dis J 34: e233–e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA, 2014. Global prevalence of Norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14: 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glass RI, Parashar UD, Estes MK, 2009. Norovirus gastroenteritis. New Engl J Med 361: 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien SJ, Donaldson AL, Iturriza-Gomara M, Tam CC, 2016. Age-specific incidence rates for Norovirus in the community and presenting to primary healthcare facilities in the United Kingdom. J Infect Dis 213 (Suppl 1): S15–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel MM, Hall AJ, Vinje J, Parashar UD, 2009. Noroviruses: a comprehensive review. J Clin Virol 44: 1–8. [DOI] [PubMed] [Google Scholar]
- 28.Zhang ZK, Lai SJ, Yu JX, Geng QB, Yang WQ, Chen Y, Wu JG, Jing HQ, Yang WZ, Li ZJ, 2017. Etiology of acute diarrhea in the elderly in China: a six-year observational study. Plos One 12: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canizalez-Roman A, Flores-Villaseñor HM, Gonzalez-Nuñez E, Velazquez-Roman J, Vidal JE, Muro-Amador S, Alapizco-Castro G, Díaz-Quiñonez JA, León-Sicairos N, 2016. Surveillance of diarrheagenic Escherichia coli strains isolated from diarrhea cases from children, adults and elderly at Northwest of Mexico. Front Microbiol 7: 1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, et al. 2018. Bacterial pathogen spectrum of acute diarrheal outpatients in an urbanized rural district in Southwest China. Int J Infect Dis 70: 59–64. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhu X, Hou H, Lu Y, Yu J, Mao L, Mao L, Sun Z, 2018. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: a hospital based study. BMC Infect Dis 18: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, et al. 2014. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in Southeastern China. Clin Microbiol Infect 20: 52–58. [DOI] [PubMed] [Google Scholar]
- 33.Nataro JP, Mai V, Johnson J, Blackwelder WC, Heimer R, Tirrell S, Edberg SC, Braden CR, Morris JG, Hirshon JM, 2006. Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin Infect Dis 43: 402–407. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S-X, et al. 2016. Impact of co-infections with enteric pathogens on children suffering from acute diarrhea in southwest China. Infect Dis Poverty 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satter SM, Gastanaduy PA, Islam K, Rahman M, Rahman M, Luby SP, Heffelfinger JD, Parashar UD, Gurley ES, 2017. Hospital-based surveillance for Rotavirus gastroenteritis among young children in Bangladesh: defining the potential impact of a Rotavirus vaccine program. Pediatr Infect Dis J 36: 168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, et al. 2014. Nontyphoidal Salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis 11: 200–206. [DOI] [PubMed] [Google Scholar]
- 37.Gorkiewicz G, 2009. Nosocomial and antibiotic-associated diarrhoea caused by organisms other than Clostridium difficile. Int J Antimicrob Agents 33: S37–S41. [DOI] [PubMed] [Google Scholar]
- 38.Sears CL, 2009. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev 22: 349–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo PC, Lau SK, Teng JL, Que T-L, Yung RW, Luk W-K, Lai RW, Hui W-T, Wong SS, Yau H-H, 2004. Association of Laribacter hongkongensis in community-acquired gastroenteritis with travel and eating fish: a multicentre case-control study. Lancet 363: 1941–1947. [DOI] [PubMed] [Google Scholar]
- 40.Guo Y, Duan Z, Qian Y, 2013. Changes in human parechovirus profiles in hospitalized children with acute gastroenteritis after a three-year interval in Lanzhou, China. PLoS One 8: e68321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lasure N, Gopalkrishna V, 2017. Molecular epidemiology and clinical severity of human bocavirus (HBoV) 1–4 in children with acute gastroenteritis from Pune, Western India. J Med Virol 89: 17–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.