Abstract
Fungi produce an abundance of bioactive secondary metabolites which can be utilized as antibiotics and pharmaceutical drugs. The genes encoding secondary metabolites are contiguously arranged in biosynthetic gene clusters (BGCs), which supports co-regulation of all genes required for any one metabolite. However, an ongoing challenge to harvest this fungal wealth is the finding that many of the BGCs are ‘silent’ in laboratory settings and lie in heterochromatic regions of the genome. Successful approaches allowing access to these regions - in essence converting the heterochromatin covering BGCs to euchromatin - include use of epigenetic stimulants and genetic manipulation of histone modifying proteins. This review provides a comprehensive look at the chromatin remodeling proteins which have been shown to regulate secondary metabolism, the use of chemical inhibitors used to induce BGCs, and provides future perspectives on expansion of epigenetic tools and concepts to mine the fungal metabolome.
Keywords: Chromatin, Natural products, Methylation, Acetylation, Histone, Activation, Repression, Cryptic, Genome mining
1. Introduction
Fungi are a major source of secondary metabolites, also termed natural products, compounds which are an incredibly fruitful source for the development of pharmaceutical drugs (Schueffler and Anke, 2014). The bioactivity of secondary metabolites make them potent drug candidates (Harvey et al., 2015; Newman and Cragg, 2016). Secondary metabolites differ from their primary metabolite counter parts in both the timing of their production, as well as their dispensability for fungal growth under laboratory conditions. In the environment however, these metabolites are important in combating abiotic and biotic stresses (Keller, 2018 (accepted) Fungal Secondary Metabolism: Regulation, function and drug discovery. Nature Reviews Microbiology).
The biosynthetic enzymes that produce secondary metabolites are encoded by genes which are physically clustered in the genome, and are called biosynthetic gene clusters (BGCs). This contiguous arrangement coupled with the conserved sequence of synthases (e.g. polyketide synthase) and synthetases (e.g. non-ribosomal peptide synthetase) encoded in the genome has allowed for the generation of algorithms able to predict the BGC number of any sequenced genome (Khaldi et al., 2010; Medema et al., 2011). For example, analysis of the genomes of 19 Aspergillus species showed a range of 21–66 BGCs in each species (de Vries et al., 2017). In fact there are some species (e.g. Aspergillus westerdijkiae) containing upwards of 80 BGCs (Han et al., 2016). Considering the estimate that there may be between 3 and 5 million species of fungi (Blackwell, 2011; O’Brien et al., 2005), many of which containing equivalent numbers of BGCs as the Aspergilli, the number of uncharacterized BGCs is likely astounding. However, many of these BGCs are silent under laboratory conditions. This impediment has led to the development of various strategies to awaken these BGCs and characterize their cryptic products.
The most straightforward method to activate BGC expression is by culturing a fungus on various growth medias (Frisvad, 2012). This can also include the co-incubation of the fungus of interest with another microbe to induce expression of BGCs (Marmann et al., 2014). These two strategies have been successful in the identification of secondary metabolites, but will not awaken all clusters. At the genetic level, overexpression of transcription factors, both BGC specific or global regulators, has also been fruitful in activating some silent BGCs (Brakhage, 2013; Lim and Keller, 2014). However, overexpression of transcriptional regulators do not always result in BGC expression, which is in part due to epigenetic regulation. Epigenetics is the study of heritable phenotypes, such as production of secondary metabolites, that do not involve changes in the DNA sequence. The “epi” represents features “on top of” or “in addition to” traditional genetic inheritance. The proteins which are involved in the inheritance of gene expression do so, in part, through manipulating chromatin. There has been a movement to induce the expression of these silent BGCs through manipulating chromatin regulators to prevent the formation of silencing chromatin modification over BGCs. This strategy has been very successful in the identification of novel compounds with bioactivities of interest. This review summarizes the findings over the past decade regarding chromatin regulation of secondary metabolism, and poses questions that, once answered, may result in new chromatin regulation strategies to unlock the drug producing potential of fungi.
2. Chromatin structure and regulation
In order to protect, maintain, and organize the millions of nucleotides that make up eukaryotic genomes, DNA is wound around nucleosomes to form chromatin. The nucleosome core molecules are octamers composed of two copies of four histone subunits; H2A, H2B, H3, and H4. Histones are very basic, small proteins (11–15 kDa) which are highly conserved throughout eukaryotic species, each with an N-terminal “histone tail” which extends outwards from the nucleosome. These tails makes up about 25% of the mass of the core histones, and their individual amino acid residues can be highly modified (Cutter and Hayes, 2015). Lastly, association with an additional linker histone, H1, completes the nucleosome organizing approximately 200 base pairs of DNA (Cutter and Hayes, 2015; McGinty and Tan, 2015).
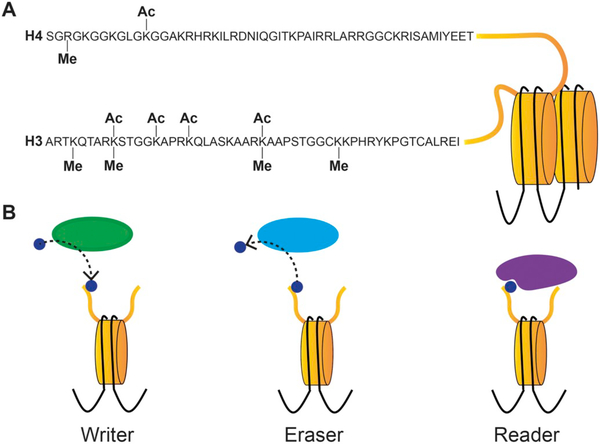
The strength of interactions between the nucleosomes and DNA, and higher order chromatin structures such as heterochromatin or euchromatin, can be controlled through modifications to the chromatin. This can be to the DNA itself, such as the addition of methyl groups to cytosines (Jin et al., 2011), or as post-translational modifications (PTMs) to histone tails. Loosely compact chromatin is more transcriptionally active and is called euchromatin, whereas the tightly compact chromatin is termed heterochromatin. Genes in heterochromatic regions, including BGCs, are typically silent. In particular, PTMs to histone tails are the main target for chromatin regulation of secondary metabolism (Bannister and Kouzarides, 2011; Gacek and Strauss, 2012; Strauss and Reyes-Dominguez, 2011). There are a wide array of PTMs that can be placed on many residues in the histone tail, such as acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, ADP ribosylation, and deimination (Bannister and Kouzarides, 2011). This review focuses on methylation and acetylation of histone tails (Fig. 1), which are the best characterized throughout eukaryotes. There is evidence of some of these other modifications regulating secondary metabolism, such as sumolyation (Szewczyk et al., 2008), however it is not known if it is sumoylation of histone tails or other proteins that leads to the change. The shorthand for writing a histone PTM is by listing the histone, the one letter code for the residue, the number of residues from the end of the histone tail that it is occurring on, followed by the modification. For example, acetylation of the ninth lysine on histone H3 is written as H3K9ac.
Fig. 1.
Histone tail modifications and the enzymes that regulate them. A) Representation of two nucleosomes, and the sequences of the tails of histones H3 and H4 whose post translational modifications (PTM) have been best studied in regards to secondary metabolism. PTM have been placed on residues that have been demonstrated to regulate secondary metabolism. Ac- acetylation. Me-methylation. B) The actions of the three classes of epigenetic machinery. Each protein is acting at a nucleosome, with just two histone tails being shown instead of the eight for simplicity. “Writers” (green) place modifications on histone tails, as represented by the blue dot. “Erasers” (light blue) remove the PTM, illustrated by the blue dot moving in the opposite direction as seen in the “writer”. Lastly, “readers” (blue) recognize the PTM on histone tails, bringing the writers and/or erasers to the correct genomic loci to act at. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The current hypothesis is that these histone PTMs can either act as signals or binding sites for downstream transcriptional processes, or act by influencing the structure of the local chromatin. This hypothesis is called the histone code, suggesting that various combinations of histone modifications will lead to distinct biological outcomes. A criticism to the histone code hypothesis is that it has the potential to be immensely complex, however this complexity has not be seen in vivo (reviewed in Rando, 2012). Most likely there are several patterns which will comprise the histone code, and that not all combinations of modifications occur biologically. Histone PTMs, which make up this histone code, are controlled and interpreted by three types of proteins: proteins which place or “write” modifications on histone tails, proteins which remove or “erase” those modifications, and proteins who interpret or “read” the modifications and mediate the response to that signal (Fig. 1B). Examples of each of these types of proteins and their relationship to secondary metabolism are described in “Writing the Code” (Section 4), “Erasing the Code” (Section 5), and “Reading the Code” (Section 6) below. The strategy for activation of cryptic BGCs has been inhibition, deletion or overexpression of chromatin modifying enzymes, to prevent formation of heterochromatin over BGCs (Fig. 2).
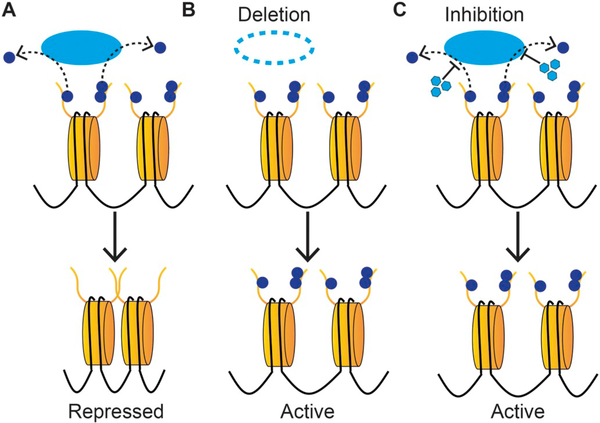
Fig. 2.
Strategies for interfering with chromatin regulation. A) Action of an epigenetic eraser under wild-type conditions. This enzyme removes the activating modifications represented by the blue dots, which leads to more condensed, repressed chromatin where BGC are often found. B) Deletion of the eraser prevents the removal of the activating modifications, and the chromatin remains open and active, allowing for expression of genes which are typically repressed. C) Adding chemical inhibitors (represented by the light blue hexagons) which prevent the eraser from removing the activating PTM. This leads to a similar outcome as deletion of the enzyme, and allows for expression of genes which are typically repressed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Techniques used to study chromatin and secondary metabolism
The majority of work done to study chromatin modifications and their relationship to secondary metabolism have primarily used two major techniques. The first method is assessing the global levels of histone modifications, typically through western blotting. The most successful western blotting protocols will enrich for histones, whether that is through nuclei extraction and purification (Soukup and Keller, 2013), or through acid extraction (Jourquin and Géli, 2017). These extracts are run on a high percentage PAGE gel, and utilize antibodies specific to a particular histone modification. As a loading control, an antibody to the C-terminus (which is unmodified) of H3 or H4 is typically used. Histone extractions can be difficult, and may require optimization for the fungus of interest. With the improvement of mass spectrometry techniques and methods, it is also possible to measure levels of histone PTMs via mass spectrometer (MS) (Krautkramer et al., 2015). This requires a very pure histone preparation, but can yield more information (~62 unique modifications in human cell line) and does not require the purchase of many antibodies. This technique has not been fully utilized in filamentous fungi, so it is hard to say how many unique modifications that can be measured by MS. However, a modification to the protocol demonstrated the MS was able to detect few changes in a few histone PTMs in A. nidulans (Gacek-Matthews et al., 2016, 2015).
While western blotting and MS will inform of the changes in global levels of histone modifications, they do not reveal the histone PTMs at specific loci. Rather, chromatin immunoprecipitation (ChIP) technology is used to identify levels of specific histone modifications at specific loci, as well as determine the binding of proteins of interest directly or indirectly bound to DNA (Orlando, 2000). ChIP typically involves the crosslinking of protein and DNA, shearing of DNA through mechanical means (sonication) or enzymes (micrococcal nuclease), a DNA/protein pull-down using an antibody specific to the protein of interest (e.g. histone PTM such as H3K9me3), and then purification of DNA. DNA can then be quantified via quantitative PCR (qPCR) if only a few genomic loci are of interest, or examined on a genome wide scale through microarray technology (ChIP on chip), or more recently, next generation sequencing technology (ChIP-seq) (Boedi et al., 2012). In combination with RNA-seq, one can study the changes in histone modifications throughout the genome and correlate modifications with transcriptomics.
The advancements in ChIP technology, such as the transition from microarrays to next generation sequencing, has also occurred with the analytical chemistry techniques used. The pioneering paper describing the relationship between histone PTM and secondary metabolism was published in 2007, using thin-layer chromatography (TLC) to measure the differences in production of metabolite (Shwab et al., 2007). High performance liquid chromatography (HPLC) paired with a variety of detectors yields more information about metabolite extracts of various chromatin mutants, and is used now instead of TLC. The use of MS detectors paired with HPLC (Pfannenstiel et al., 2018) or techniques such as 2D NMR spectroscopy (Forseth et al., 2011) give the most thorough analysis of the whole metabolome, and greatly aids the search for novel compounds. This is important to note, as early work may have concluded that secondary metabolism was not impacted by genetic manipulation of chromatin regulators, however the use less sensitive instrumentation could have obscured differences in secondary metabolism.
4. Writing the code
In the analogy of histone modifying enzymes, those which are responsible for placing the highly dynamic, reversible modifications on chromatin are called “writers”. Writers can be placed into several groups (e.g. methyltransferases, acetyltransferases), depending upon the type of modifications they are responsible for placing on histone tails, and further divided based upon their protein domains which perform these actions. When considering filamentous fungi and the regulation of secondary metabolism, there are two main enzymes which are “writing” on chromatin, those which write with methyl groups (histone methyltransferases) and those write with acetyl groups (histone acetyltransferases) (Table 1).
Table 1:
Epigenetic writers, erasers, and readers which have been genetically shown to regulate secondary metabolism.
| Class | Action | Complex/Protein | Modification | Organism |
|---|---|---|---|---|
| Writer | Methyltransferase | Bre2/Ash2/CclA (COMPASS) | H3K4me2/3 | A. nidulans |
| A. oryzae | ||||
| F. fujikuroi | ||||
| F. graminearum | ||||
| P. fici | ||||
| Set1 (COMPASS) | H3K4me1/2/3 | F. graminearum | ||
| F. vertilicillioides | ||||
| M. orzyae | ||||
| Clr4/Dim-5 | H3K9me3 | A. nidulans | ||
| E. festucae | ||||
| F. vertilicillioides | ||||
| Kmt6 (PRC2) | H3K27me3 | E. festucae | ||
| F. graminearum | ||||
| F. fujikuroi | ||||
| Set2 | H3K36me3 | F. fujikuroi | ||
| F. vertilicillioides | ||||
| Ash1 | H3K36me3 | F. fujikuroi | ||
| Dot1 | H3K79me3 | A. flavus | ||
| Hmt1 | H4R3me2 | A. flavus | ||
| F. graminearum | ||||
| Acetyltransferase | Gcn5 (SAGA/Ada) | H3K9K14K18K27ac | A. nidulans | |
| F. fujikuroi | ||||
| F. graminearum | ||||
| T. atroviride | ||||
| U. maydis | ||||
| Esa1 | H4K12ac | A. nidulans | ||
| Sas3 | H3ac | F. graminearum | ||
| M. robertsii | ||||
| P. microspora | ||||
| Hat1 | H3/H4ac | P. microspora | ||
| Eraser | Demethylase | Rph1 | H3K36me3 | A. nidulans |
| F. fujikuroi | ||||
| Jhd2 | H3K4me3 | A. flavus | ||
| A. nidulans | ||||
| Deacetylase | Rpd3 | H3/H4ac | A. nidulans | |
| Hos2 | H3/H4ac | A. nidulans | ||
| F. fujikuroi | ||||
| Hda1 | H3/H4ac | A. nidulans | ||
| C. arbuscular | ||||
| F. fujikuroi | ||||
| F. asiaticum | ||||
| M. orzyae | ||||
| P. chrysogenum | ||||
| P. fici | ||||
| P. microspora | ||||
| Hos3 | H3/H4ac | F. fujikuroi | ||
| Hst1 | H4K16ac | A. nidulans | ||
| Hst4 | H3K9K18K56ac | A. nidulans | ||
| A. orzyae | ||||
| Reader | Ssp1 (COMPASS) | H3K4me3 | A. orzyae | |
| HP1 | Heterochromatin | A. nidulans | ||
| F. graminearum | ||||
| Snt2 | H3K4me3 H3ac | A. nidulans | ||
| A. flavus |
4.1. Methyltransferases
Histone methyltransferases add a level of complexity to the histone code, as the presence or absence of methylation does not always lead to the same output. Unlike histone acetylation which is generally associated with transcriptional activation (Eberharter and Becker, 2002), the effect of methylation depends on which particular residue in a histone tail is targeted. In addition, there are three methylation levels of lysines (mono-, di-, and tri-), which can each have their own biological significance. Methylation of H3K4, K36, or K79 is associated with active chromatin, whereas methylation of H3K9, K27, or H4K20 is typically associated with repressed chromatin (Martin and Zhang, 2005). Arginine residues are targets of methylation as well, and can lead to activation or repression (Blanc and Richard, 2017). Several complexes of methyltransferases have been characterized as secondary metabolite regulators, and are listed separately below and in Table 1.
4.1.1. COMPASS
The Complex of proteins associated with Set1 (COMPASS) is a highly conserved heteromeric complex from yeast to humans. Set1 (YHR119W) is a methyltransferase responsible for the methylation of the fourth lysine of Histone 3 (H3K4) (Krogan et al., 2002). H3K4me3 is typically associated with transcriptional activation, and is found in areas of active transcription (Sims and Reinberg, 2006). COMPASS is also associated with transcriptional silencing near telomeres (Schneider et al., 2005). Considering that many BGCs are located in sub-telomeric regions and are silenced, one way to induce the expression of these was by manipulating the COMPASS complex in A. nidulans (Bok et al., 2009). Bre2/ASH2 homolog cclA (AN9399), which is required for H3K4 methyltransferase activity, was deleted and subsequent metabolomics identified the production of emodin, monodictyphenone, and its derivatives which were at the time not known to be produced by A. nidulans. Deletion of suspect backbone genes identified the cluster responsible for these metabolites, and led to the identification of F9775A/ B polyketides (Bok et al., 2009). Chromatin precipitation of two genes within the cluster and one gene that flanked the BGC revealed a decrease in H3K4me2/3 in all three genes, and an increase in H3K9me2/3 in the two genes within the cluster but not the flanking gene in the ΔcclA strain indicating the involvement of the COMPASS complex at this genomic locus. Similarly, in A. fumigatus, deletion of cclA results in the global loss of H3K4me2/3, and regulation of several secondary metabolites as seen by thin layer chromatography (Palmer et al., 2013).
In Pestalotiopsis fici, PfcclA (PFICI_05127) was deleted and assessed for novel metabolite production (Wu et al., 2016). This led to the characterization of several compounds such as pestaloficiols and ficipyrone. It was shown that media composition (rice or PDA) was also a factor in the regulation of compounds, indicating that BGC activation and nutrient availability can be interdependent and should both be considered when mining for novel natural products.
In A. oryzae, a transcription factor and chromatin remodeling deletion library was screened that identified two gene deletions within the COMPASS complex, cclA (AO090124000076) and an additional gene which will be discussed in “Reading the Code” (Section 6) of this review, sspA (AO090003001570). Deletion of either of these two genes eliminates H3K4 methyltransferase activity, and resulted in loss of H3K4me3, and an increase in various astellolide compounds (Shinohara et al., 2016).
The cclA homolog CCL1 was deleted in the plant pathogens Fusarium fujikuroi (FFUJ_04433) and F. graminearum (FGSG_09564), and was demonstrated to be responsible for H3K4me3 in these species as well (Liu et al., 2015; Studt et al., 2017). Despite H3K4me3 being an activating modification, there were observed changes in both the loss and increase in various secondary metabolites. Interestingly, metabolites which are known virulence factors (e.g. gibberellic acid and deoxynivalenol also known as vomitoxin) were found to be decreased when grown on laboratory media, and yet production was unchanged during infection, indicating the fact that environmental factors can compensate for losses in chromatin machinery (Studt et al., 2017). An important observation from this study was apparent disconnect between CCL1 disruption, H3K4me3 levels and transcription of the BGCs of interest, strongly suggesting the misregulation of secondary metabolism may be indirect, and through some additional cis–/trans- acting factors (Studt et al., 2017).
Set1, for which the complex is named, has additionally been studied for its role in secondary metabolism. Unlike deletion of Bre2/Ash2/CclA/Ccl1 which leads to a decrease in H3K4me3, deletion of Set1 leads to a decrease in mono-, di-, and trimethylated H3K4 (Liu et al., 2015). This was demonstrated in F. graminearum (FGSG_07445), which resulted in the loss of the mycotoxin deoxynivalenol. Interactions between the COMPASS complex members identified from S. cerevisiae was confirmed to be similar in F. graminearum as well. ChIP-qPCR demonstrated that there were significant decreases in H3K4me, H3K4me2, and H3K4me3 levels at various deoxynivalenol biosynthetic genes, partially explaining the decrease in transcription of these genes (Liu et al., 2015). Similarly, in F. verticillioides FvSet1 (FVEG_07811) is required for fumonisin B1 production and transcription, although changes in histones modifications were not assessed (Gu et al., 2017b). Although their production was not directly assessed, Set1 (MGG_15053) in Magnaporthe orzyae regulates BCGs transcriptionally (Pham et al., 2015).
4.1.2. Clr4
The highly conserved methyltransferase known as Clr4 in Shizzosaccharomyces pombe, DIM-5 in Neurospora crassa, and Su(var)3–9 in Drosphila, is responsible for the trimethylation of H3K9. H3K9me3 is a repressive mark, and is recognized by heterochromatin protein 1 (HP1) which is a key component of heterochromatin, leading to heterochromatin formation (Allshire and Madhani, 2017). Following naming conventions in A. nidulans, the homolog named clrD (AN1170), has been shown to regulate secondary metabolism. ClrD is required for H3K9me3 in A. nidulans and the loss of this protein leads to an increase in expression of several BGCs, including the sterigmatocystin BGC (Reyes-Dominguez et al., 2010). The loss of H3K9me3 also led to a loss of HepA (HP1 homolog, AN1905) occupancy at the sterigmatocystin BGC, illustrating a requirement for HepA in proper heterochromatin formation. Using ChIP-qPCR, it was shown that HepA and H3K9me3 levels typically decrease through primary growth phase and are replaced by H3K9K14ac, which leads to activation of the gene cluster and production of sterigmatocystin, and demonstrated how facultative heterochromatin is involved in the regulation of secondary metabolism.
In the plant endophyte Epichloë festucae, H3K9me3 was also shown to repress several alkaloid BGCs in axenic culture, and these levels were reduced in plant infected tissue (Chujo and Scott, 2014). Similar to A. nidulans, the clrD homolog (EfM3.062280) is responsible for H3K9me3. The loss of H3K9me3 only partially derepressed the alkaloids produced by E. festucae, with the kmt6 homolog (discussed in Section 4.1.3) also playing a role in the heterochromatin formation and repression of the alkaloid BGCs. Similar to the previous two examples, in F. vertilicillioides the Clr4/Dim-5/Su(var)3–9 homolog Fvdim5 (FVEG_08911), is responsible for the majority of H3K9me3. Fvdim5 represses fumonisin B1 production, and expression of melanin biosynthetic enzymes (Gu et al., 2017a). Taken together, removal of H3K9me3 through deletion of the Clr4/Dim-5/Su(var)3–9 homologs in fungi can remediate some of the repression from heterochromatin formation, but it may take knocking out several heterochromatin histone modifications in order to awaken silent BGCs in some fungi.
4.1.3. Polycomb repressive complex 2
Lysine(k) methyltransferase 6 (Kmt6) is part of the polycomb group (PcG) called the polycomb repressive complex 2 (PRC2) first characterized in Drosophila (Jürgens, 1985). Kmt6 has been shown in two Fusarium species to be responsible for the presence of the repressive mark H3K27me3. H3K27me3 is heavily enriched at telomeric/subtelomeric regions where many predicted BGCs are located, and thus possibly explaining why BGC are typically silent in these regions. Kmt6 is an essential protein in F. fujikorui (FFUJ_00719) but not in F. graminearum (FGSG_15795). The knock down of KMT6 led to the induction of the recently characterized beauvericin BGC in F. fujikuroi (Niehaus et al., 2016). The repressive H3K27me modification at the beauvericin cluster was replaced with the activating H3K27ac, and in fact H3K27ac modification was increased globally. Interestingly, H3K27me3 forms several foci that are spread throughout the nucleus, however this distribution is disrupted in the knock down of KMT6, demonstrating that histone modifications may be playing several biological functions (Niehaus et al., 2016).
Characterization of Kmt6 has particularly insightful in presenting differences in epigenetic regulation in various fungal species. For example, in Neurospora crassa 5–8% of the genome is covered in H3K27me3 whereas about a third of the Fusarium genomes are covered in this mark. Loss of H3K27me3 led to expression of 14% of the silent genes in a wild-type strain of Fusarium, while 20% of the genome remained silent indicating that loss of this PTM is not enough to activate the entire genome and that there are additional regulatory elements that keep these gene silent. The multiple layers of repressive elements has been demonstrated in E. festucae, where H3K27me3 was present in higher levels at gene promoters whose expression was repressed in axenic culture (Chujo and Scott, 2014). Deletion of the kmt6 homolog ezhB (EfM3.069800) which is responsible for the H3K27me3 in the cell, relieved some of the repression of the examined BGC. However, it was only in a double deletion of ezhB and the H3K9me3 methyltransferase clrD, that these BGC were fully derepressed (Chujo and Scott, 2014).
4.1.4. Set2
Set2 (YJL168C) is the only H3K36 methyltransferase identified in S. cerevisiae, and is required for the mono-, di-, and trimethylation of H3K36 (Strahl et al., 2002). H3K36 methylation is often associated with transcriptional activation. FvSET2 (FVEG_06937) has been deleted in F. verticillioides, and was shown to be required for H3K36me3, similar to what has been seen in N. crassa and S. cerevisiae (Adhvaryu et al., 2005; Strahl et al., 2002). Set2 was similarly required for normal levels of the mycotoxin fumonisin B1, demonstrating the requirement of H3K36me3 for production of at least one secondary metabolite, however the study did not determine if this modification is present at the BGC itself (Gu et al., 2017c). Set2 (FFUJ_08690) is also required for distributing the majority of H3K36me3 modifications in F. fujikuroi, and plays a role in regulating various secondary metabolites such as fusarins, fusarubins, and gibberellic acids (Janevska et al., 2018). Set1 H3K36me3 was found mostly within euchromatic regions in F. fujiuroi, and overlaps with the pattern of H3K4me2/3.
4.1.5. Ash1
Ash1 (FFUJ_05655) is a H3K36 methyltransferase, responsible for a minority of H3K36me3 in F. fujikouroi. The other H3K36 methyltransferase in F. fujikouroi is Set2 (FFUJ_08690) (Section 4.1.4), and in a double deletion there is a complete loss of H3K36me3. These two methyltransferases appear to target different chromosomal regions, and Ash1 mediated H3K36me was typically found in facultative heterochromatin located in subtelomeric regions (Janevska et al., 2018). These regions overlap with H3K27me3, a silencing mark placed by Kmt6 activity (Section 4.1.3). This suggests a very different role for K36 methylation for filamentous fungi than what is seen in other species including yeasts and higher eukaryotes (Janevska et al., 2018). Deletion of Ash1 led to either an increase or decrease of several metabolites such as gibberellic acids, bikaverin, fusarins, fusarubins, and fusaric acid which are primarily located in facultative heterochromatin. This work strongly suggests that the location and the chromatin context can strongly influence the function of a modification.
4.1.6. DOT1
Dot1 is a H3K79 methyltransferase in yeast, and has not been studied in filamentous fungi heavily. A dot1 homolog (AFLA_093140) in A. flavus has been deleted, and shown to transcriptionally regulate the aflatoxin BGC (Liang et al., 2017). However, it remains unclear if this homolog does in fact regulate H3K79 methylation, or how prevalent this modification is in filamentous fungi.
4.1.7. Hmt1
There are only two examples of protein arginine methyltransferases (PRMT) studied in the context of secondary metabolism, both are homologs of the S. cerevisiae gene Hmt1 (YBR034C). Methylation of arginine can be activating or repressing, depending on whether the methylation is symmetrical (one methyl group on each of the terminal nitrogens) or asymmetrical (two methyl groups to one terminal nitrogen) (Di Lorenzo and Bedford, 2011). The Hmt1 homolog arginine methyltransferase A (RmtA, AN10526) has been shown to have H4R3 specificity in A. nidulans (Trojer et al., 2004). There was no general growth phenotype observed in the deletion mutant, and secondary metabolite production was not tested, however the deletion of this homolog, amt1 (FGSG_01134) in F. graminearum led to a decrease in radial growth and production of the mycotoxin deoxynivalenol (Bauer et al., 2010; Wang et al., 2012). Similar to F. graminearum, the Hmt1 homolog in A. flavus RmtA (AFLA_127370) positively regulates the production of the mycotoxin aflatoxin (Satterlee et al., 2016). Although not demonstrated to contain the arginine methylation ability in either F. graminearum or A. flavus, the high homology to A. nidulans suggests that impact is due to the loss of H4R3 methyltransferase activity. However, there is no evidence whether this modification present at the deoxynivalenol or aflatoxin BGCs, so it is not known if the PTM presents a direct or indirect relationship.
4.2. Acetyltransferases
Histone acetylation has been proposed and subsequently found to be associated with transcriptional activation since the 1960s (Allfrey et al., 1964; Pogo et al., 1966). Histone acetyltransferases (HAT) will transfer the acetyl group from acetyl-CoA to lysines in the histone tails, and can be very promiscuous in their specificity, and not just targeting a single lysine (Struhl, 1998). The general hypothesis is that the acetylation of a lysine will negate the positive charge of that amino acid, decreasing the interaction between the histone and DNA. The weaker the interaction between these two bodies, the more open the DNA is, allowing for higher rates of transcription (Berger, 2007; Workman and Kingston, 1998). Outside of genetic manipulation of HATs, acetylation has been shown to be an important natural process of progressive gene cluster activation in A. flavus (Roze et al., 2007). In addition, mutation of histone residues which can no longer be acetylated showed a decrease in secondary metabolite production (Nützmann et al., 2013). Although only one complex is mentioned below, HATs are often part of larger complexes and require these binding partners to carry out their respective functions in the cell (Lee and Workman, 2007). The HATs which have been shown to regulate secondary metabolism are described below.
4.2.1. SAGA/Ada
The Spt-Ada-Gcn5 acetyltransferase (SAGA) complex is a highly conserved global chromatin regulating complex, impacting 13% of the genome in S. cerevisiae (Lee et al., 2000), and ~10.9% in A. nidulans (Cánovas et al., 2014). This complex in S. cerevisiae is responsible for the acetylation of histones, as well as the deubiquitination of the H2B tail (Georgakopoulos et al., 2013; Shukla et al., 2006). The best studied SAGA complex in regards to secondary metabolism is in A. nidulans, where it was found to mediate the fungal secondary metabolite response to co-culture with Streptomyces rapamycinicus (Fischer et al., 2018; Nützmann et al., 2011). The complex in A. nidulans lacks the deubiquitination components of the yeast SAGA/Ada complex, and suggests that this type of chromatin remodeling is mediated from another complex (Georgakopoulos et al., 2013). GcnE, a histone acetyltransferase and member of the SAGA complex, has also been shown to interact with the light regulated protein, LreA, mediating histone acetylation in response to light (Hedtke et al., 2015). The role of GcnE has been heavily studied in A. nidulans, and is responsible for acetylation of H3K9 and H3K14 residues at many genomic loci such as at BGCs (e.g. orsellinic acid BGC), nitrogen utilization genes, and developmental regulators (Bok et al., 2013; Cánovas et al., 2014; Fischer et al., 2018; Nützmann et al., 2011; Reyes-Dominguez et al., 2008). The SAGA/Ada complex is the bacterial target in an additional co-culture interaction between the bacterium Pseudomonas piscium, found in the wheat head microbiome, and the wheat pathogen F. graminearum (Chen et al., 2018). In this interaction, P. piscium produces a metabolite which inhibits FgGcn5 (FGRRES_00280), which impacts histone acetylation of many residues including H3K14, H3K18, H3K27, and H2BK11 acetylation. This and one other study reported that FgGcn5 is required for deoxynivalenol production (Chen et al., 2018; Kong et al., 2018). Together, these papers provide a mechanism of how co-culture can control production of secondary metabolites through chromatin regulation, and how this is an integral aspect of microbial communication.
The SAGA complex has also been identified in the rice pathogen F. fujikuroi, whose protein composition matched that of A. nidulans (Rösler et al., 2016). Deletion of Ffgcn5 (FFUJ_00382) and two other components of the SAGA complex, Ffada2 (FFUJ_08807) and Ffada3 (FFUJ_00496), all resulted in alterations in secondary metabolism production including gibberellic acids, fusarubin, fusaric acid, and bikaverin. Unlike what is seen in A. nidulans, there is no global change in acetylation levels of H3K14, however there are global loses of acetylation levels at H3K4, H3K9, H3K18, and H3K27 when the HAT gcnE is deleted in F. fujikuroi (Rösler et al., 2016). The Gcn5 (TRIATDRAFT_47901) homolog in the mycoparasite Trichoderma atroviride similarly regulates secondary metabolism genes, however the metabolites themselves were not quantified (Gómez-Rodríguez et al., 2018). The changes in expression patterns did not alter T. atroviride parasitism of the phytopathogenic fungus Rhizoctonia solani. In A. flavus, AflgcnE is required for aflatoxin production (Lan et al., 2016). Finally, an examination of the gcn5 deletion mutant in Ustilago maydis provides one of the few studies to show an impact of chromatin modifiers on secondary metabolism in the Basidiomycete taxon of fungi (Martínez-Soto et al., 2015).
4.2.2. Esa1
The only S. cerevisiae Esa1 (YOR244W) homolog that has been studied in relation to secondary metabolism in filamentous fungi is EsaA (AN10956) a MYST (named for its founding members; MOZ, Ybf2/Sas3, Sas2, and Tip60) type acetyltransferase. Esa1 is a member of the NuA4 complex, and acetylates many lysine residues of H4, including K5, K8, and K12, as well as H2BK5 (Smith et al., 1998; Suka et al., 2001). EsaA was identified from a suppressor screen of the deletion of the global regulator of secondary metabolism laeA in A. nidulans (Soukup et al., 2012). In a ΔlaeA strain, overexpression of esaA was able to restore production of sterigmatocystin. The overexpression of EsaA also led to the increase in production of several other secondary metabolites (penicillin, xanthones, and emericellin). Although there are several lysines targeted by EsaA homologs, in A. nidulans its upregulation of secondary metabolite BGCs seems to come from its acetylation of H4K12 as assessed by ChIP-qPCR.
4.2.3. Sas3
The second MYST type acetyltransferases to be studied for their control of secondary metabolism are the something about silencing (Sas3) yeast homologs. Sas3 (YBL052C) is part of the NuA3 histone acetyltransferase complex, and has been shown to regulate secondary metabolism in two species. In Pestalotiopsis microspora the Sas3 homolog mst2 (KX268363) is a positive regulator of pestalotiollide B and the melanin associated polyketide synthase (Zhang et al., 2018). Conversely, deletion of the homolog in Metarhizium robertsii (HAT1, MAA_02282) was used to discover several novel natural products which were highly produced in the deletion mutant, including meromusides A-I, and meromutides A and B (Fan et al., 2017). There is a 50% decrease in histone acetylation of H3 when HAT1 is deleted, indicating it is responsible for a significant amount of the H3 acetylation in M. robertsii. Finally, FgSas3 (FGRRES_08481) positively transcriptionally regulates the production of deoxynivalenol in F. graminearum (Kong et al., 2018).
4.2.4. Hat1
There are two major classes of histone acetyltransferases, type A and B (Ruiz-García et al., 1998), categorized on the basis of where they are located and what they acetylate. Type A acetylates chromatin associated histones in the nucleus while Type B is cytoplasmic and acetylates free histones. Of these type B histone acetyltransferases, there is one in particular which has been shown to regulate secondary metabolism, Hat1 (ANS59908) (Zhang et al., 2016). Hat1 was deleted in P. microspora, and was shown to be required for production of several secondary metabolites including pestalotiollide B. The exact mechanism of how Hat1 functions has not been defined, so whether this is a direct or indirect relationship of Hat1 to secondary metabolite BGCs is yet to be determined.
5. Erasing the code
The antagonists to the epigenetic writing machinery are the epigenetic erasers. These enzymes catalyze the removal of the histone PTMs that are written on histone tails. It is this balance between ‘erasers’ and ‘writers’ that makes chromatin regulation a dynamic process. There are two groups of eraser enzymes which have been studied in relation to secondary metabolism, those that remove the acetyl-PTM from histone tails (histone deacetylases, HDAC) and those which remove the methyl groups (demethylases) (Table 1).
5.1. Histone demethylases
As discussed in Section 4.1, histone tail lysines are the primary residues modified by histone methyltransferase. The reversible methylation mark can be removed by lysine demethylases, of which there are two reported classes; those which are amino oxidase homologs of KDM1 (KIAA0601), and those which contain the Jumonji C (JmjC) domain (D’Oto et al., 2016). However, only the class which contains a JmjC domain which can remove all three methylation states of a lysine (Klose et al., 2006; Tsukada et al., 2006), and are discussed below.
5.1.1. Rph1
The yeast homolog of Rph1, lysine(k) demethylase A (KdmA, AN1060), has been shown to have K36 demethylase activity, and is a transcriptional regulator of primary and secondary metabolism genes (Gacek-Matthews et al., 2015). The deletion of kdmA in A. nidulans did not lead to an increase in global H3K36me3/2 levels but did impact methylation at specific loci. For example, the local levels of H3K36me3 increased at the penicillin and sterigmatocystin BGCs in the deletion mutant. Expression of several other BGCs were altered (both increased and decreased) in the A. nidulans kdmA deletion mutant, however whether this is a direct or indirect relationship is to be determined. The homolog of KdmA in F. fujikuroi (KDM4, FFUJ_01769), similarly did not lead to any noticeable changes in global H3K36me3 levels (Janevska et al., 2018). In addition, there were no differences in the four secondary metabolites measured, providing additional evidence that chromatin regulation varies between species of fungi.
5.1.2. Jhd2
The second lysine demethylase, KdmB (AN8211) to be studied in A. nidulans was also shown to be a regulator of secondary metabolism and is the homolog of the JmjC domain-containing histone demethylase (JHD2) in S. cerevisiae. KdmB is a specific H3K4me3 demethylase, which is an activating mark. Transcriptomics and ChIPseq identified regions which are regulated directly and indirectly via KdmB (Gacek-Matthews et al., 2016). BGCs and cell structure and function genes were among the most regulated genes in the deletion mutant. Deletion of kdmB led to the loss of sterigmatocystin, emericellamides, and orsellinic acid, and an increase in emodin production. Interestingly even in BGCs, such as the sterigmatocystin BGC, when activated, the H3K4me3 mark is largely absent, indicating that it is either present at very low levels or the BGC activation was indirect. The homolog (Rum1, AFLA_006240) in A. flavus has also been shown to regulate aflatoxin production transcriptionally (Hu et al., 2018).
5.2. Histone deacetylases
Histone deacetylases (HDACs), were the first histone modifying enzymes shown to link secondary metabolism with chromatin remodeling in fungi (Shwab et al., 2007). HDACs are responsible for the removal of acetyl groups, and can be placed into four classes. There are three “classical” HDAC classes; Class I (Rpd3-like), Class II (HdaI-like), and Class IV (Seto and Yoshida, 2014). Class IV enzymes have only been identified in plants and mammals and are not present in fungi (Graessle et al., 2001). These classical HDACs are similar in their requirement for zinc and remove the acetyl group with a molecule of H2O (Grunstein, 1997; Kuo and Allis, 1998), whereas the HDAC Class III (sirtuins) are NAD+ dependent HDACs (Marks and Xu, 2009).
Histone acetylation is typically associated with transcriptional activation. The original hypothesis from the 2007 study was that removal or inhibition of enzymes which erase this particular mark would result in increased gene activation and concomitant increases in secondary metabolite production (Fig. 2). Shwab et al. looked at three particular HDACs, each one belonging to a different class of HDACs, and tested for any regulation of terraquinone, sterigmatocystin, and penicillin in A. nidulans, and found that HdaA was a negative regulator of the two subtelomeric BGCs of sterigmatocystin and penicillin (Shwab et al., 2007). In addition, treatment with the small molecule HDAC inhibitor tricostatin A (TSA) influenced the production of secondary metabolism in both Alternaria alternata and Penicillium expansum (Shwab et al., 2007). Since this study, research on HDACs control over secondary metabolism has exploded. Below we summarize the work performed in each HDAC class, using the nomenclature originally described in S. cerevisiae.
5.2.1. Class I
There are two reported Class I HDACs in S. cerevisiae, Rpd3 (reduced potassium dependency) and Hos2 (HAD one similarity) (Robyr et al., 2002; Rundlett et al., 1996). Rpd3 is often used as the architype of the Class I HDAC. Although not as heavily studied as the Class II HDACs for induction and study of secondary metabolism, Class I HDACs can be utilized to discover novel secondary metabolites. Of note, Rpd3 homologs have been found to be essential in most filamentous fungi in contrast to the dispensable nature of this protein in S. cerevisiae.
5.2.1.1. Rpd3. One of the major members of the Class I HDACs are Rpd3-like enzymes.
Rpd3 (YNL330C) was originally named in S. cerevisiae and is conserved from yeast to humans (Taunton et al., 1996; Vidal et al., 1990). The Rpd3 homolog in A. nidulans (rpdA, AN4493), and in F. fujikuroi (Ffhda3, FFUJ_00456), are both essential genes (Studt et al., 2013; Tribus et al., 2010). The metabolome of an A. nidulans rpdA knockdown strain was analyzed alongside a wild-type strain, and compared to a strain grown with the HDAC chemical inhibitor suberoylanilide hydoxamic acid (SAHA). Interestingly the profile of both the chemical inhibitor as well as the rpdA knockdown showed great similarities in the array of secondary metabolites up and down regulated (Albright et al., 2015). In a follow up study, this knockdown strain was also used to identify novel aspercryptins in A. nidulans (Henke et al., 2016).
5.2.1.2. Hos2.
The S. cerevisiae Hos2 (YGL194C) homolog in F. fujikuroi, Ffhda2 (FFUJ_01551), was shown to be a regulator of secondary metabolism. Ffhda2 was responsible 25% of total HDAC activity in this fungus and shown to be a positive regulator of the secondary metabolites bikaverin, gibberellic acids, fusaric acid, and fusarins (Studt et al., 2013). The A. nidulans homolog (HosA, AN3806) is both a positive and negative transcriptional regulator; repressing the orsellinic acid BGC and activating the penicillin BGC. ChIP-qPCR of several of these upregulated clusters showed they had an enrichment of H3K9ac and H4K16ac in their promoters, demonstrating the role of HosA as a chromatin remodeling agent (Pidroni et al., 2018).
5.2.2. Class II
A prominent member of the Class II HDACs is HDA1 (histone deacetylase 1), one of the best studied histone modifying enzymes in regards to secondary metabolism. The second member from S. cerevisiae in Class II is Hos3, which is not found outside of fungi.
5.2.2.1. Hda1.
The HDA1 ortholog HdaA was shown to be responsible for the majority of HDAC activity in A. nidulans, and appears to be active as a monomer (Tribus et al., 2005). As detailed above, deletion of HdaA (AN8042) in A. nidulans led to increased expression of the subtelomeric BGCs producing penicillin and sterigmatocystin (Shwab et al., 2007). HdaA (Afu5g01980) was also found to regulate some secondary metabolites in A. fumigatus including gliotoxin, as well as other unknown metabolites (Lee et al., 2009). In this species, HdaA positively and negatively regulated several BGCs containing NRPSs. There was no measurable increase in global histone H3 acetylation when hdaA was deleted in A. fumigatus as may have been expected considering that HdaA is responsible for the majority of HDAC activity in A. nidulans (Lee et al., 2009; Tribus et al., 2005).
The HdaA homolog in F. fujikuroi, Ffhda1 (FFUJ_09787), was similarly found to be responsible for the majority of HDAC activity (> 60%). Of the several secondary metabolites measured, Ffhda1 acts as both a positive (bikaverin, fusarubins, gibberellic acids, and fusaric acid) and negative (bikaverin during highly inducing conditions) regulator. Genome wide analysis of H3K9ac was assessed by ChIP-seq, and generally the trend of loss of acetylation coincided with loss of metabolite, and vice versa (Studt et al., 2013). The F. fujikuroi metabolite, beauvericin, observed in the Ffhda1 deletion strain was later assigned to an uncharacterized BGC (Niehaus et al., 2016). This gene cluster was shown to be repressed by the H3K27 methyltransferase Ffkmt6 (Section 4.1.3), and it was shown that the activating H3K274ac modification is enriched both genome wide (western blotting), and at the beauvericin cluster (ChIP-qPCR) (Niehaus et al., 2016).
Several other studies have utilized HdaA homologs to modulate the production of secondary metabolites. P. fici has been examined for metabolite changes in response to PfhdaA (PFICI_08988) deletion (Wu et al., 2016). In the absence of PfhdaA, the metabolites ficiolides and asperpentyn were upregulated, indicating either direct or indirect regulation of secondary metabolism from PfhdaA. Similarly, in Pestalotiopsis microspora the HdaA homolog named hid1 (GenBank: AHE63221. 1), is a negative regulator of pestalotiollide B, and increased the production of this metabolite by two fold even in media conditions that had been optimized for production (Niu et al., 2015). Deletion of the HdaA homolog in Magnaporthe orzyae led to increase in melanin biosynthetic gene expression, and its deletion led to an increase in trichothecenes in Fusarium asiaticum (Maeda et al., 2017). In the endophyte Calcarisporium arbuscular, hdaA deletion drastically modified the secondary metabolite profile, resulting in activation of over 75% of the biosynthetic enzymatic backbone encoding genes, and discovery of several metabolites including arbumycin, arbumelin, arubuscullic acid A and B (Mao et al., 2015). Additional metabolites isolated from the deletion mutant demonstrated inhibition of expression of matrix metalloproteinases, in human breast cancer cells (Bai et al., 2018). Interestingly, previous attempts to induce expression of secondary metabolites from this fungus using epigenetic inducing chemicals or different media were not successful (Mao et al., 2015). Taken together, these studies on C. arbuscular demonstrate the need to examine more than one method to induce metabolite production. The Hda1 homolog in Penicillium chrysogenum, hdaA (EN45_076690), regulated the chryogine and sorbicillinoid BGC, and led to the production of an unknown metabolite (Guzman-Chavez et al., 2018).
5.2.2.2. Hos3.
The third set of fungal HDACs are homologs of Hos3 (YPL116W) originally identified in S. cerevisiae (Rundlett et al., 1996). This subset of Class II HDAC enzymes are unique to fungi, and are resistant to the HDAC inhibitor TSA (Carmen et al., 1999; Trojer et al., 2003). In F. fujikuroi the Hos3 homolog Ffhda4 (FFUJ_08772) was not responsible for a significant amount of the HDAC activity in the fungus, and no changes in secondary metabolite production were observed when the gene was deleted although the mutant did show aberrant development which is often tied in with secondary metabolism (Studt et al., 2013). The A. nidulans Hos3 homolog hosB (AN7019) does show intrinsic HDAC activity, however has not been studied in the context of secondary metabolism (Trojer et al., 2003).
5.2.3. Class III: sirtuins
The last class of HDACs are called the sirtuins which are known as Class III HDACs. Sirtuins erase acetylation, on both histones and other proteins, in a NAD+ dependent manner (Bheda et al., 2016). Sirtuins have been shown to demonstrate different sub cellular localizations, and not all sirtuins have been able to demonstrate deacetylase activity, suggesting sirtuins have several functions (Du et al., 2011; Michishita et al., 2005). Sitruins can be placed into four classes (I-IV). All classes are not present in all organisms, such as prokaryotes which do not contain classes II and IV (Frye, 2000). The A. nidulans class I sirtuin HstA sirtuin (AN11067) was the first sirtuin to be examined for an impact on secondary metabolism where it had no impact on the three metabolites measured in that study (Shwab et al., 2007). However, with the increased panel of known A. nidulans secondary metabolites and advances in mass spectrometry technologies, there may in fact be changes in some metabolites that were unable to be detected previously.
Another A. nidulans sirtuin, SirA (AN10449) the homolog of the class I sirtuin Hst1 in S. cerevisiae, was shown to deacetylate H4K16 in the promoter regions of the penicillin and sterigmatocystin BGCs (Shimizu et al., 2012). A transcriptomic study of the sirA deletion mutant showed both up and down regulated secondary metabolite BGCs, and measured increases in austinol and sterigmatocystin production (Itoh et al., 2017b). Another class I sirtuin SirE (AN1226) has also been studied in A. nidulans, and was found to regulate both primary and secondary metabolism (Itoh et al., 2017a). Deletion of sirE led to an increase in H3K9ac, H3K18ac, as well as H3K56ac during various growth conditions (Itoh et al., 2017a).
Sirtuins have also been studied in A. oryzae, where it was found that the A. nidulans sirE homolog hstD (AO090038000370) regulates development and secondary metabolism, specifically the production of penicillin and kojic acid (Kawauchi et al., 2013). An epistatic relationship was shown between hstD and the global regulator of secondary metabolism laeA, where laeA phenotype was dominant to that of hstD, whether hstD was overexpressed or deleted.
6. Reading the code
The final group of proteins to be discussed is the epigenetic “reading” machinery. Readers can interpret the code, and are required for the writing and erasing enzymes to find the correct genomic loci to modify (Yun et al., 2011). Readers contain protein domains which recognize specific histone modifications and some of these domains can be contained within a writing or erasing protein itself (Gacek-Matthews et al., 2016). There are several domains that can recognize various histone modifications, and each has a particular preference for recognizing a histone modification (e.g. Bromo domain and acetylation, Tudor domain and methylation). One can find many proteins containing reading domains in the genomes of fungi (Table 2). Despite this high number (~83 proteins containing reading domains Table 2) there are only three examples of reading proteins having been studied in the context of secondary metabolism. Thus, reading proteins represent the area with the greatest space for exploration.
Table 2:
Proteins with epigenetic reading domain in Aspergillus and Fusarium spp.
| PHD | BAH | Bromo | YEATS | Tudor | Chromo | PWWP | 14-3-3 | BRCT | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| A. nidulans | 28 | 3 | 7 | 2 | 6 | 25 | 3 | 3 | 14 | 91 |
| A. flavus | 27 | 3 | 6 | 2 | 5 | 8 | 2 | 3 | 16 | 72 |
| A. orzyae | 27 | 2 | 6 | 2 | 6 | 9 | 3 | 3 | 13 | 71 |
| F. graminearum | 30 | 5 | 9 | 2 | 6 | 15 | 3 | 3 | 17 | 90 |
| F. fujikuroi | 28 | 4 | 9 | 2 | 6 | 17 | 3 | 3 | 17 | 89 |
| F. verticillioides | 29 | 5 | 10 | 2 | 5 | 14 | 3 | 3 | 18 | 89 |
PHD-Plant Homeodomain; BAH- Bromo Adjacent Homology; Bromo-bromodomain; YEATS-Yaf9, ENL, AF9, Taf14, Sas5; Tudor- tudor domain; Chromo- chromodomain; PWWP- “Pro-Trp-Trp-Pro” motif domain; BRCT-BRCA1 C terminus domain.
6.1. Spp1
Deletion of SppA (AO090003001570), homolog of the S. cerevisiae Spp1 (YHR184W), was shown to repress astellolide production in A. oryzae (Shinohara et al., 2016). SppA contains a single plant homeodomain (PHD, referred to as a PHD finger) which is an example of an epigenetic reading domain that typically recognizes lysine methylation (Musselman and Kutateladze, 2009; Sanchez and Zhou, 2011). This reader is a member of the COMPASS complex (4.1.1). The loss of sppA led to an equal reduction in H3K4me3 as the deletion of the COMPASS methyltransferase cclA, however the levels of H3K4me/me2 were consistent as is seen in the cclA deletion, and is unlike what is seen in a set1 deletion (Liu et al., 2015; Shinohara et al., 2016). The Spp1 homolog in F. graminearum however is dispensable for H3K4me3 (Liu et al., 2015). This underlines the importance of a reader; where manipulation can lead to a similar output as deletion of a writer or eraser.
6.2. HP1
Heterochromatin protein 1 (HP1) is an epigenetic reader that has a major role in heterochromatin formation. HP1 and its homologs contain two conserved protein domains, an N-terminal chromo-domain and a C-terminal chromo-shadow domain. The N-terminal chromo-domain is required for recognition of the repressive H3K9me modification, which is required for function (Bannister et al., 2001; Platero et al., 1995). This protein is conserved in many species, however is absent in S. cerevisiae thus most of the fungal work on this particular protein has been in S. pombe and N. crassa (Freitag et al., 2004; Wang et al., 2000).
The HP1 homolog has been studied in A. nidulans (HepA, AN1905) and F. graminearum (HEP1, FG08763) (Reyes-Dominguez et al., 2012, 2010). HEP1 positively regulates the deoxynivalenol BGC and negatively regulates aurofusarin in F. graminearum. The loss of Hep1 led to a decrease in H3K9me at both clusters, despite one being activated and the other repressed, (Reyes-Dominguez et al., 2012). In A. nidulans, ChIP with HepA showed it covered the secondary metabolite BGC of sterigmatocystin, and was removed in a laeA dependent manner (Reyes-Dominguez et al., 2010). The deletion of hepA led to an increase in transcript of several BGCs including sterigmatocystin, penicillin, and terriquinone BGCs. This study was the first demonstration of facultative heterochromatin controlling expression of BGCs.
6.3. Snt2
Aspergillus nidulans SntB (AN9517), named for homology to S. cerevisiae Snt2, was recognized as a regulator of secondary metabolism from a forward genetic screen in A. nidulans (Pfannenstiel et al., 2017). SntB contains five conserved domains that recognize specific histone tail modifications: the SANT domain, three PHD fingers, and one bromoadjacent homology (BAH) domain. The BAH domain contained two mutations which led to amino acid substitutions in the mutant that arose from the screen, indicating that this BAH domain is crucial for function. In A. nidulans, it was shown that deletion of this reading protein led to misregulation of secondary metabolism, with increases (F-9775A/B) and decreases (sterigmatocystin, alternariol, microperfuranone, and cichorine) in various metabolites. This phenotype was conserved in A. flavus (AFLA_029990), where deletion led to the loss of many secondary metabolites (aflavarin, aflatoxin, asparasoneA, aflatrem, and kojic acid) but an increase in the production of the cryptic metabolites ditryptophenline and leporine B (Pfannenstiel et al., 2018). Studies in fission and budding yeasts show that the yeast Snt2 complex contains various readers and writers that differ with Snt2 complexed with RpdA (S. cerevisiae) or KdmB (S. pombe) (Baker et al., 2013; Roguev et al., 2004). Assessment of the global histone modifications in the A. flavus sntB deletion showed an increase in both histone H3 acetylation as well as H3K4me3, suggesting that Aspergillus SntB may form a hybrid complex with these two epigenetic erasers (Pfannenstiel et al., 2018).
7. Chemical inhibition of epigenetic machinery
Although molecular manipulation of epigenetic machinery has greatly added to the quest for increased secondary metabolite synthesis in fungi, this tactic is not possible for all fungi as many remain recalcitrant to genetic transformation. However, it is also possible to alter chromatin modifications utilizing small molecule inhibitors of chromatin regulators (Cole, 2008). The first example that chemical small molecule inhibitors could be used to awaken silent BGCs was the treatment of A. alternata and P. expansum with the HDAC inhibitor and antifungal antibiotic trichostatin A (Shwab et al., 2007). Since this study, the use of chemical treatment to induce expression of silent BGC has grown.
One of the two primary groups of inhibitors utilized inhibit DNA methyltransferases (DNMTs). The DNMT inhibitor 5-azacytidine is the most commonly used chemical which inhibits DNA methylation when incorporated into DNA (Christman, 2002). 5-azacytidine has been used to successfully identify novel metabolites in Alternaria sp. (Sun et al., 2012), A. niger (Fisch et al., 2009), Beauveria bassiana (Yakasai et al., 2011), Cladosporium resinae (Khan et al., 2017), Cladosporium cladosporioides (Williams et al., 2008), Cordyceps indigotica (Asai et al., 2012d), Diatrype sp. (Williams et al., 2008), Muscodor yucatanensis (Qadri et al., 2017), Penicillium citreonigrum (Wang et al., 2010), and Pestalotiopsis crassiuscula (Yang et al., 2014). A second DNMT inhibitor, RG-108, which blocks the active site of DNMTs (Savickiene et al., 2012), was used with success to induce secondary metabolism in Isaria tenuipes (Asai et al., 2012a).
The second class of inhibitors used targets HDACs. Five inhibitors that have met with success in increasing secondary metabolite production in fungi target the Class I and Class II HDACs: octanoylhydroxamic acid (OHA), trichostatin A, sodium valproate (VPA), SAHA (vorinostat), and suberoyl bis-hydroxamic acid (SBHA) (Marks and Xu, 2009). Trichostatin A was the first inhibitor used to show HDAC inhibitors can change secondary metabolite profiles (Shwab et al., 2007). SAHA has been used by several groups to identify novel metabolites in A. nidulans (Albright et al., 2015), A. niger (Henrikson et al., 2009), Chalara sp. (Adpressa et al., 2017), C. resinae (Khan et al., 2017), Daldinia sp. (Du et al., 2014), Drechslera sp. (Siless et al., 2018), Microascus sp. (Vervoort et al., 2011), and M. yucatanensis (Qadri et al., 2017). OHA is an analog of SAHA which has induced production in Drechslera sp. (Siless et al., 2018). SBHA has also been widely used in B. bassiana (Yakasai et al., 2011), Cordyceps annullata (Asai et al., 2012b), C. cladosporoides (Williams et al., 2008), C. indigotica (Asai et al., 2012e), I. tenuipes (Asai et al., 2012a), and Torrubiella luterorostrata (Asai et al., 2011). Lastly, VPA has been used successfully in Drechslera sp. (Siless et al., 2018).
Class III HDAC inhibitors can also be used to alter the secondary metabolite profile of strains. The most common Class III inhibitor that has been used is nicotinamide, which binds to a conserved pocket of the HDAC that is responsible for the NAD+ binding and catalysis (Avalos et al., 2005). Sodium butyrate, sirtinol, and splitomycin have also been used, but not nearly as widely as nicotinamide. Nicotinamide has been successful in inducing metabolite production in Chaetomium canroideum (Asai et al., 2016), Chaetomium mollipilium (Asai et al., 2012c), Graphiopsis chlorocephala (Asai et al., 2013), Monascus ruber (Hu et al., 2017), and Penicillium brevicompactum (El-Hawary et al., 2018). A secondary metabolite produced by Didymobotryum rigidum, 5-methylmellein, was identified as an inhibitor of SirA from A. nidulans, and treatment of this inhibitor led to increased metabolite profiles in A. nidulans (Shigemoto et al., 2018).
8. Future prospects
In the approximate 10 years since the first publication of epigenetic regulation of secondary metabolism in fungi (Shwab et al., 2007), research has exploded in the applications of this concept to mining fungal genomes (Fig. 2). This has expanded to additional fungi, types of histone modifying enzymes, and inhibitors. Despite the growth, there are still several areas which need additional attention as the field continues to study the regulation of BGCs through epigenetic proteins (Fig. 3).
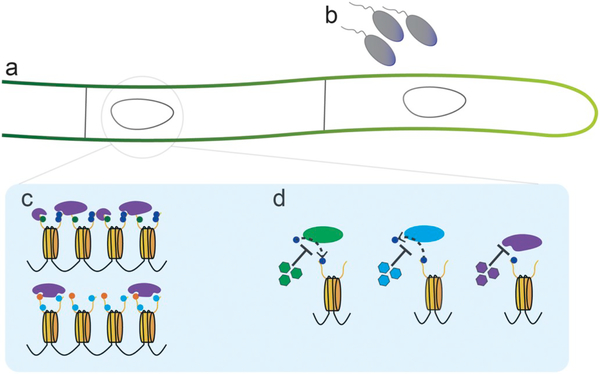
Fig. 3.
Areas of future research. Depiction of a fungal hyphae, which contains two nuclei represented by the ovals. Each letter represents an area that needs further research relating to chromatin regulation of secondary metabolism. “a” is labeling the fungus, because despite the extensive research that has occurred within Aspergillus and Fusarium, very few taxa of fungi have been studied for how their BGC are regulated by chromatin. “b” marks the bacterial interaction which has been demonstrated to regulate secondary metabolite production. “c” a zoomed in look in the nucleus, shows chromatin with various purple reading proteins. This represents an untapped resource of proteins which may be controlling secondary metabolism through recognition of PTM, and recruitment of writing or erasing enzymes. “d” represents the use of chemical inhibitors to prevent the actions of the histone modifying enzymes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The first area that needs to be expanded is the taxa of fungi to be studied (Fig. 3a). As seen in Table 1, the vast majority of research has focused on the Ascomycete genera Aspergillus and Fusarium. The fungi that have been studied outside of these two genera have also been almost exclusively Ascomycetes, whether through genetic manipulation or treatment with chemical inhibitors. Members of the Basidiomycetes and Mucorales, many rich in various secondary metabolites, have largely been ignored in epigenetic mining. Although these fungi are less accessible to genetic manipulations, treatment with chemical inhibitors might stimulate production of cryptic metabolites.
A second area of great interest is characterizing the mechanism of microbial interactions, and how this controls secondary metabolism (Fig. 3b). Currently there have been two phenomenon in a microbial interaction that has controlled secondary metabolism, interestingly both through the SAGA/Ada complex (Chen et al., 2018; Nützmann et al., 2011). Additional interactions should be characterized to see if this is a common theme, or if regulation of secondary metabolism occurs through different epigenetic complexes. For instance, several of the Fusarium metabolites regulated by the histone deacetylase HdaA (bikaverin, gibberellic acid and beauvericin) are induced by confrontations with the bacterium Ralstonia solanacearum (Spraker et al., 2018) possibly an indication of bacterial regulation of these BGCs through affecting HdaA expression.
The types of histone modifying proteins studied has been limited, and should be expanded in order to understand chromatin regulation of BGC (Fig. 3c). Erasers and writers that place and remove phosphorylation, ubiquitylation, sumoylation, ADP ribosylation, and deamination of histone tails have not been assessed. Also there is little known about how epigenetic readers regulate secondary metabolism. Possibly this group of enzymes which may inform us the most as to how secondary metabolite gene clusters are regulated. It is this group of proteins which will tell us how the writers and erasers arrive to the locations that they need to act, as well as what modifications are there based off of the recognition domains within the reading proteins themselves. On several occasions it has been seen that heterochromatic regions flank several well characterized BGCs (Palmer and Keller, 2010). The question of how the BGC chromatin is regulated, may have to do with specific histone modifications that cover the BGC. Along the same vein, the types of chemical inhibitors used in genome mining for secondary metabolites has also been extremely limited, and in order to establish more efficient and complete genome mining efforts, additional chemical inhibitors should be used. On the forefront of epigenetic therapies for cancer and other genetic diseases are inhibitors of epigenetic reader domains (Greschik et al., 2017; Zaware, 2017). These types of inhibitors are untested when it comes to fungi, and represent a novel manner in chromatin perturbation to induce expression of silent BGC (Fig. 3d).
This review has shown that there is no one perfect way to manipulate chromatin regulators to induce expression of all silent BGC. It will be a combination of multiple strategies through molecular means of those fungi which are genetically amenable, as well as the use of multiple chemical inhibitors of these proteins which will increase the efficacy of genome mining efforts. Ideally the knowledge gleaned from this research will increase the push for utilizing fungal secondary metabolites as new pharmaceutical drugs and antibiotics.
Acknowledgements
The authors would like to thank Dr. Claudio Greco for assistance in proofreading. BTP was supported by the Predoctoral Training Program in Genetics, funded by the National Institutes of Health (5T32 GM007133-40). This work was also funded in part by NIHR01GM112739-01 to NPK.
References
- Adhvaryu KK, Morris SA, Strahl BD, Selker EU, 2005. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell 4, 1455–1464. 10.1128/EC.4.8.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adpressa DA, Stalheim KJ, Proteau PJ, Loesgen S, 2017. Unexpected biotransformation of the HDAC inhibitor vorinostat yields aniline-containing fungal metabolites. ACS Chem. Biol 12, 1842–1847. 10.1021/acschembio.7b00268. [DOI] [PubMed] [Google Scholar]
- Albright JC, Henke MT, Soukup AA, McClure RA, Thomson RJ, Keller NP, Kelleher NL, 2015. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol 10, 1534–1541. 10.1021/acschembio.5b00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE, 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A 51, 786–794. 10.1073/PNAS.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Madhani HD, 2017. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol 19, 229–244. 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Yamamoto T, Oshima Y, 2011. Histone deacetylase inhibitor induced the production of three novel prenylated tryptophan analogs in the entomopathogenic fungus Torrubiella luteorostrata. Tetrahedron Lett. 52, 7042–7045. 10.1016/J.TETLET.2011.10.020. [DOI] [Google Scholar]
- Asai T, Chung Y-M, Sakurai H, Ozeki T, Chang F-R, Yamashita K, Oshima Y, 2012a. Tenuipyrone, a novel skeletal polyketide from the entomopathogenic fungus, Isaria tenuipes, cultivated in the presence of epigenetic modifiers. Org. Lett 14, 513–515. 10.1021/ol203097b. [DOI] [PubMed] [Google Scholar]
- Asai T, Luo D, Obara Y, Taniguchi T, Monde K, Yamashita K, Oshima Y, 2012b. Dihydrobenzofurans as cannabinoid receptor ligands from Cordyceps annullata, an entomopathogenic fungus cultivated in the presence of an HDAC inhibitor. Tetrahedron Lett. 53, 2239–2243. 10.1016/J.TETLET.2012.02.088. [DOI] [Google Scholar]
- Asai T, Morita S, Shirata N, Taniguchi T, Monde K, Sakurai H, Ozeki T, Oshima Y, 2012c. Structural diversity of new C 13 -polyketides produced by Chaetomium mollipilium cultivated in the presence of a NAD + −dependent histone deacetylase inhibitor. Org. Lett 14, 5456–5459. 10.1021/ol302539s. [DOI] [PubMed] [Google Scholar]
- Asai T, Yamamoto T, Chung Y-M, Chang F-R, Wu Y-C, Yamashita K, Oshima Y, 2012d. Aromatic polyketide glycosides from an entomopathogenic fungus Cordyceps indigotica. Tetrahedron Lett. 53, 277–280. 10.1016/J.TETLET.2011.10.013. [DOI] [Google Scholar]
- Asai T, Yamamoto T, Oshima Y, 2012e. Aromatic polyketide production in Cordyceps indigotica, an entomopathogenic fungus, induced by exposure to a histone deacetylase inhibitor. Org. Lett 14, 2006–2009. 10.1021/ol3005062. [DOI] [PubMed] [Google Scholar]
- Asai T, Otsuki S, Sakurai H, Yamashita K, Ozeki T, Oshima Y, 2013. Benzophenones from an endophytic fungus, Graphiopsis chlorocephala, from Paeonia lactiflora cultivated in the presence of an NAD + −dependent HDAC inhibitor. Org. Lett 15, 2058–2061. 10.1021/ol400781b. [DOI] [PubMed] [Google Scholar]
- Asai T, Morita S, Taniguchi T, Monde K, Oshima Y, 2016. Epigenetic stimulation of polyketide production in Chaetomium cancroideum by an NAD + −dependent HDAC inhibitor. Org. Biomol. Chem 14, 646–651. 10.1039/C5OB01595B. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C, 2005. Mechanism of sirtuin inhibition by Nicotinamide: altering the NAD+ cosubstrate specificity of a Sir2 Enzyme. Mol. Cell 17, 855–868. 10.1016/J.MOLCEL.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Bai J, Mu R, Dou M, Yan D, Liu B, Wei Q, Wan J, Tang Y, Hu Y, 2018. Epigenetic modification in histone deacetylase deletion strain of Calcarisporium arbuscula leads to diverse diterpenoids. Acta Pharm. Sin. B 8, 687–697. 10.1016/J.APSB.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LA, Ueberheide BM, Dewell S, Chait BT, Zheng D, Allis CD, 2013. The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxidemediated oxidative stress. Mol. Cell. Biol 33, 3735–3748. 10.1128/MCB.00025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T, 2011. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T, 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bauer I, Graessle S, Loidl P, Hohenstein K, Brosch G, 2010. Novel insights into the functional role of three protein arginine methyltransferases in Aspergillus nidulans. Fungal Genet. Biol 47, 551–561. 10.1016/J.FGB.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Berger SL, 2007. The complex language of chromatin regulation during transcription. Nature 447, 407–412. 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bheda P, Jing H, Wolberger C, Lin H, 2016. The substrate specificity of sirtuins. Annu. Rev. Biochem 85, 405–429. 10.1146/annurev-biochem-060815-014537. [DOI] [PubMed] [Google Scholar]
- Blackwell M, 2011. The Fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot 98, 426–438. 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Blanc RS, Richard S, 2017. Arginine methylation: the coming of age. Mol. Cell 65, 8–24. 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Boedi S, Reyes-Dominguez Y, Strauss J, 2012. Chromatin Immunoprecipitation Analysis in Filamentous Fungi, in: Fungal Secondary Metabolism. Humana Press, Totowa, NJ, pp. 221–236. 10.1007/978-1-62703-122-6_16. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang Y-M, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP, 2009. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol 5, 462–464. 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Soukup AA, Chadwick E, Chiang Y-M, Wang CCC, Keller NP, 2013. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol. Microbiol 89, 963–974. 10.1111/mmi.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA, 2013. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol 11, 21–32. 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Cánovas D, Marcos AT, Gacek A, Ramos MS, Gutiérrez G, Reyes-Domínguez Y, Strauss J, 2014. The histone acetyltransferase GcnE (GCN5) plays a central role in the regulation of Aspergillus asexual development. Genetics 197, 1175–1189. 10.1534/GENETICS.114.165688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen AA, Griffin PR, Calaycay JR, Rundlett SE, Suka Y, Grunstein M, 1999. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. U. S. A 96, 12356–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang J, Yang N, Wen Z, Sun X, Chai Y, Ma Z, 2018. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat. Commun 9, 3429 10.1038/s41467-018-05683-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman JK, 2002. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495. 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Chujo T, Scott B, 2014. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte-plant symbiosis. Mol. Microbiol 92, 413–434. 10.1111/mmi.12567. [DOI] [PubMed] [Google Scholar]
- Cole PA, 2008. Chemical probes for histone-modifying enzymes. Nat. Chem. Biol 4, 590–597. 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AR, Hayes JJ, 2015. A brief review of nucleosome structure. FEBS Lett. 589, 2914–2922. 10.1016/j.febslet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo A, Bedford MT, 2011. Histone arginine methylation. FEBS Lett. 585, 2024–2031. 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Oto A, Tian Q-W, Davidoff AM, Yang J, 2016. Histone demethylases and their roles in cancer epigenetics. J. Med. Oncol. Ther 1, 34–40. [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H, 2011. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809. 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, King JB, Cichewicz RH, 2014. Chlorinated polyketide obtained from a Daldinia sp. treated with the epigenetic modifier suberoylanilide hydroxamic acid. J. Nat. Prod 77, 2454–2458. 10.1021/np500522z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB, 2002. Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep. 3, 224–229. 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hawary S, Sayed A, Mohammed R, Hassan H, Zaki M, Rateb M, Mohammed T, Amin E, Abdelmohsen U, 2018. Epigenetic modifiers induce bioactive phenolic metabolites in the marine-derived fungus Penicillium brevicompactum. Mar. Drugs 16, 253 10.3390/md16080253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan A, Mi W, Liu Z, Zeng G, Zhang P, Hu Y, Fang W, Yin W-B, 2017. Deletion of a histone acetyltransferase leads to the pleiotropic activation of natural products in Metarhizium robertsii. Org. Lett 19, 1686–1689. 10.1021/acs.orglett.7b00476. [DOI] [PubMed] [Google Scholar]
- Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Wägele H, Cichewicz RH, 2009. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J. Ind. Microbiol. Biotechnol 36, 1199–1213. 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- Fischer J, Müller SY, Netzker T, Jäger N, Gacek-Matthews A, Scherlach K, Stroe MC, García-Altares M, Pezzini F, Schoeler H, Reichelt M, Gershenzon J, Krespach MK, Shelest E, Schroeckh V, Valiante V, Heinzel T, Hertweck C, Strauss J, Brakhage AA, 2018. Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites. Elife. 7 10.7554/eLife.40969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forseth RR, Fox EM, Chung D, Howlett BJ, Keller NP, Schroeder FC, 2011. Identification of cryptic products of the gliotoxin gene cluster using NMR-based comparative metabolomics and a model for gliotoxin biosynthesis. J. Am. Chem. Soc 133, 9678–9681. 10.1021/ja2029987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU, 2004. HP1 is essential for DNA methylation in Neurospora. Mol. Cell 13, 427–434. 10.1016/S1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, 2012. Media and Growth Conditions for Induction of Secondary Metabolite Production, in: Fungal Secondary Metabolism. Humana Press, Totowa, NJ, pp. 47–58. 10.1007/978-1-62703-122-6_3. [DOI] [PubMed] [Google Scholar]
- Frye RA, 2000. Phylogenetic classification of Prokaryotic and Eukaryotic Sir2-like Proteins. Biochem. Biophys. Res. Commun 273, 793–798. 10.1006/BBRC.2000.3000. [DOI] [PubMed] [Google Scholar]
- Gacek A, Strauss J, 2012. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol 95, 1389–1404. 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacek-Matthews A, Noble LM, Gruber C, Berger H, Sulyok M, Marcos AT, Strauss J, Andrianopoulos A, 2015. KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans. Mol. Microbiol 96, 839–860. 10.1111/mmi.12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacek-Matthews A, Berger H, Sasaki T, Wittstein K, Gruber C, Lewis ZA, Strauss J, 2016. KdmB, a Jumonji histone H3 demethylase, regulates genome-wide H3K4 trimethylation and is required for normal induction of secondary metabolism in Aspergillus nidulans. PLoS Genet. 12, e1006222 10.1371/journal.pgen.1006222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos P, Lockington RA, Kelly JM, 2013. The Spt-Ada-Gcn5 Acetyltransferase (SAGA) complex in Aspergillus nidulans. PLoS ONE 8, e65221 10.1371/journal.pone.0065221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Rodríguez EY, Uresti-Rivera EE, Patrón-Soberano OA, Islas-Osuna MA, Flores-Martínez A, Riego-Ruiz L, Rosales-Saavedra MT, Casas-Flores S, 2018. Histone acetyltransferase TGF-1 regulates Trichoderma atroviride secondary metabolism and mycoparasitism. PLoS ONE 13, e0193872 10.1371/journal.pone.0193872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessle S, Loidl P, Brosch G, 2001. Histone acetylation: plants and fungi as model systems for the investigation of histone deacetylases. Cell. Mol. Life Sci 58, 704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greschik H, Schüle R, Günther T, 2017. Selective targeting of epigenetic reader domains. Expert Opin. Drug Discov 12, 449–463. 10.1080/17460441.2017.1303474. [DOI] [PubMed] [Google Scholar]
- Grunstein M, 1997. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352. 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Gu Q, Ji T, Sun X, Huang H, Zhang H, Lu X, Wu L, Huo R, Wu H, Gao X, 2017a. Histone H3 lysine 9 methyltransferase FvDim5 regulates fungal development, pathogenicity and osmotic stress responses in Fusarium verticillioides. FEMS Microbiol. Lett 364 10.1093/femsle/fnx184. [DOI] [PubMed] [Google Scholar]
- Gu Q, Tahir HAS, Zhang H, Huang H, Ji T, Sun X, Wu L, Wu H, Gao X, 2017b. Involvement of FvSet1 in fumonisin B1 biosynthesis, vegetative growth, fungal virulence, and environmental stress responses in Fusarium verticillioides. Toxins 9 10.3390/toxins9020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Wang Z, Sun X, Ji T, Huang H, Yang Y, Zhang H, Tahir HAS, Wu L, Wu H, Gao X, 2017c. FvSet2 regulates fungal growth, pathogenicity, and secondary metabolism in Fusarium verticillioides. Fungal Genet. Biol 107, 24–30. 10.1016/J.FGB.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Guzman-Chavez F, Salo O, Samol M, Ries M, Kuipers J, Bovenberg RAL, Vreeken RJ, Driessen AJM, 2018. Deregulation of secondary metabolism in a histone deacetylase mutant of Penicillium chrysogenum. Microbiology, e00598 10.1002/mbo3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chakrabortti A, Zhu J, Liang Z-X, Li J, 2016. Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae. BMC Genomics 17, 633 10.1186/s12864-016-2974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey AL, Edrada-Ebel R, Quinn RJ, 2015. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov 14, 111–129. 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- Hedtke M, Rauscher S, Röhrig J, Rodríguez-Romero J, Yu Z, Fischer R, 2015. Light-dependent gene activation in Aspergillus nidulans is strictly dependent on phytochrome and involves the interplay of phytochrome and white collar-regulated histone H3 acetylation. Mol. Microbiol 97, 733–745. 10.1111/mmi.13062. [DOI] [PubMed] [Google Scholar]
- Henke MT, Soukup AA, Goering AW, McClure RA, Thomson RJ, Keller NP, Kelleher NL, 2016. New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans. ACS Chem. Biol 11, 2117–2123. 10.1021/acschembio.6b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH, 2009. A chemical epigenetics approach for engineering the in situbiosynthesis of a cryptic natural product from Aspergillus niger. Org. Biomol. Chem 7, 435–438. 10.1039/B819208A. [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhou Y, Mao Z, Li H, Chen F, Shao Y, 2017. NAD+−dependent HDAC inhibitor stimulates Monascus pigment production but inhibit citrinin. AMB Express 7, 166 10.1186/s13568-017-0467-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yang G, Zhang D, Liu Y, Li Y, Lin G, Guo Z, Wang S, Zhuang Z, 2018. The PHD transcription factor Rum1 regulates morphogenesis and aflatoxin biosynthesis in Aspergillus flavus. Toxins 10 10.3390/toxins10070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh E, Odakura R, Oinuma K-I, Shimizu M, Masuo S, Takaya N, 2017a. Sirtuin E is a fungal global transcriptional regulator that determines the transition from the primary growth to the stationary phase. J. Biol. Chem 292, 11043–11054. 10.1074/jbc.M116.753772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh E, Shigemoto R, Oinuma K-I, Shimizu M, Masuo S, Takaya N, 2017b. Sirtuin a regulates secondary metabolite production by Aspergillus nidulans. J. Gen. Appl. Microbiol 63, 228–235. 10.2323/jgam.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Janevska S, Baumann L, Sieber CMK, Münsterkötter M, Ulrich J, Kämper J, Güldener U, Tudzynski B, 2018. Elucidation of the two H3K36me3 histone methyltransferases Set2 and Ash1 in Fusarium fujikuroi unravels their different chromosomal targets and a major impact of Ash1 on genome stability. Genetics 208, 153–171. 10.1534/genetics.117.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD, 2011. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2, 607–617. 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourquin F, Géli V, 2017. Histone purification from Saccharomyces cerevisiae, in. Methods Mol. Biol 69–73. 10.1007/978-1-4939-6630-1_5. (Clifton, N.J.). [DOI] [PubMed] [Google Scholar]
- Jürgens G, 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316, 153–155. 10.1038/316153a0. [DOI] [Google Scholar]
- Kawauchi M, Nishiura M, Iwashita K, 2013. Fungus-specific sirtuin HstD coordinates secondary metabolism and development through control of LaeA. Eukaryot. Cell 12, 1087–1096. 10.1128/EC.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, 2018. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND, 2010. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol 47, 736–741. 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Bacha N, Ahmad B, Bakht J, Lutfullah G, Ali J, 2017. Synthesis of secondary metabolites by Cladosporium resinae (NRL-6437) under different growth media and chemical inducers and their pharmaceutical activity. Pak. J. Pharm. Sci 30, 1617–1624. [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y, 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet 7, 715–727. 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Kong X, van Diepeningen AD, van der Lee TAJ, Waalwijk C, Xu J, Xu J, Zhang H, Chen W, Feng J, 2018. The Fusarium graminearum histone acetyltransferases are important for morphogenesis, DON biosynthesis, and pathogenicity. Front. Microbiol 9, 654 10.3389/fmicb.2018.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkramer KA, Reiter L, Denu JM, Dowell JA, 2015. Quantification of SAHA-dependent changes in histone modifications using data-independent acquisition mass spectrometry. J. Proteome Res 14, 3252–3262. 10.1021/acs.jproteome.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A, 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem 277, 10753–10755. 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- Kuo M-H, Allis CD, 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays 20, 615–626. . [DOI] [PubMed] [Google Scholar]
- Lan H, Sun R, Fan K, Yang K, Zhang F, Nie XY, Wang X, Zhuang Z, Wang S, 2016. The Aspergillus flavus histone acetyltransferase AflGcnE regulates morphogenesis, aflatoxin biosynthesis, and pathogenicity. Front. Microbiol 7, 1324 10.3389/fmicb.2016.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL, 2007. Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol 8, 284–295. 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FCP, Shen W-C, Hannett N, Jennings EG, Winston F, Green MR, Young RA, 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405, 701–704. 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Lee I, Oh J-H, Shwab EK, Dagenais TRTT, Andes D, Keller NP, 2009. HdaA, a class 2 histone deacetylase of Aspergillus fumigatus affects germination and secondary metabolite production. Fungal Genet. Biol 46, 782–790. 10.1016/j.fgb.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Liu Y, Yang K, Lin G, Xu Z, Lan H, Wang X, Wang S, Liang L, Liu Y, Yang K, Lin G, Xu Z, Lan H, Wang X, Wang S, 2017. The putative histone methyltransferase DOT1 regulates aflatoxin and pathogenicity attributes in Aspergillus flavus. Toxins 9, 232 10.3390/toxins9070232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim FY, Keller NP, 2014. Spatial and temporal control of fungal natural product synthesis. Nat. Prod. Rep 31, 1277–1286. 10.1039/c4np00083h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu N, Yin Y, Chen Y, Jiang J, Ma Z, 2015. Histone H3K4 methylation regulates hyphal growth, secondary metabolism and multiple stress responses in Fusarium graminearum. Environ. Microbiol 17, 4615–4630. 10.1111/1462-2920.12993. [DOI] [PubMed] [Google Scholar]
- Maeda K, Izawa M, Nakajima Y, Jin Q, Hirose T, Nakamura T, Koshino H, Kanamaru K, Ohsato S, Kamakura T, Kobayashi T, Yoshida M, Kimura M, 2017. Increased metabolite production by deletion of an HDA1-type histone deacetylase in the phytopathogenic fungi, Magnaporthe oryzae (Pyricularia oryzae) and Fusarium asiaticum. Lett. Appl. Microbiol 65, 446–452. 10.1111/lam.12797. [DOI] [PubMed] [Google Scholar]
- Mao X-M, Xu W, Li D, Yin W-B, Chooi Y-H, Li Y-Q, Tang Y, Hu Y, 2015. Epigenetic genome mining of an endophytic fungus leads to the pleiotropic biosynthesis of natural products. Angew. Chem. Int. Ed 54, 7592–7596. 10.1002/anie.201502452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Xu W-S, 2009. Histone deacetylase inhibitors: potential in cancer therapy. J. Cell. Biochem 107, 600–608. 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmann A, Aly A, Lin W, Wang B, Proksch P, Marmann A, Aly AH, Lin W, Wang B, Proksch P, 2014. Co-cultivation—a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 12, 1043–1065. 10.3390/md12021043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y, 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol 6, 838–849. 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Martínez-Soto D, González-Prieto JM, Ruiz-Herrera J, 2015. Transcriptomic analysis of the GCN5 gene reveals mechanisms of the epigenetic regulation of virulence and morphogenesis in Ustilago maydis. FEMS Yeast Res. 15, fov055 10.1093/femsyr/fov055. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Tan S, 2015. Nucleosome structure and function. Chem. Rev 115, 2255–2273. 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R, 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I, 2005. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635. 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Kutateladze TG, 2009. PHD fingers: epigenetic effectors and potential drug targets. Mol. Interv 9, 314–323. 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM, 2016. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod 79, 629–661. 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Niehaus E-M, Studt L, von Bargen KW, Kummer W, Humpf H-U, Reuter G, Tudzynski B, 2016. Sound of silence: the beauvericin cluster in Fusarium fujikuroi is controlled by cluster-specific and global regulators mediated by H3K27 modification. Environ. Microbiol 18, 4282–4302. 10.1111/1462-2920.13576. [DOI] [PubMed] [Google Scholar]
- Niu X, Hao X, Hong Z, Chen L, Yu X, Zhu X, 2015. A putative histone deacetylase modulates the biosynthesis of pestalotiollide B and conidiation in Pestalotiopsis microspora. J. Microbiol. Biotechnol 25, 579–588. [DOI] [PubMed] [Google Scholar]
- Nützmann H-W, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schümann J, Hertweck C, Strauss J, Brakhage AA, 2011. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Adamediated histone acetylation. Proc. Natl. Acad. Sci. U. S. A 108, 14282–14287. 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H-WW, Fischer J, Scherlach K, Hertweck C, Brakhagea AA, Brakhage AA, 2013. Distinct amino acids of Histone H3 control secondary metabolism in Aspergillus nidulans. Appl. Environ. Microbiol 79, 6102–6109. 10.1128/AEM.01578-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien HE, Parrent JL, Jackson JA, Moncalvo J-M, Vilgalys R, 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol 71, 5544–5550. 10.1128/AEM.71.9.5544-5550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci 25, 99–104. [DOI] [PubMed] [Google Scholar]
- Palmer JM, Keller NP, 2010. Secondary metabolism in fungi: does chromosomal location matter? Curr. Opin. Microbiol 13, 431–436. 10.1016/j.mib.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JM, Bok JW, Lee S, Dagenais TRT, Andes DR, Kontoyiannis DP, Keller NP, 2013. Loss of CclA, required for histone 3 lysine 4 methylation, decreases growth but increases secondary metabolite production in Aspergillus fumigatus. Peer J. 1, e4 10.7717/peerj.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannenstiel BT, Zhao X, Wortman J, Wiemann P, Throckmorton K, Spraker JE, Soukup AA, Luo X, Lindner DL, Lim FY, Knox BP, Haas B, Fischer GJ, Choera T, Butchko RAE, Bok J-W, Affeldt KJ, Keller NP, Palmer JM, 2017. Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. MBio 8 10.1128/mBio.01246-17. (e01246–17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannenstiel BT, Greco C, Sukowaty AT, Keller NP, 2018. The epigenetic reader SntB regulates secondary metabolism, development and global histone modifications in Aspergillus flavus. Fungal Genet. Biol 120, 9–18. 10.1016/J.FGB.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham KTM, Inoue Y, Van Vu B, Nguyen HH, Nakayashiki T, Ikeda K, Nakayashiki H, 2015. MoSET1 (histone H3K4 methyltransferase in Magnaporthe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet. 11, e1005385 10.1371/journal.pgen.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidroni A, Faber B, Brosch G, Bauer I, Graessle S, 2018. A Class 1 histone deacetylase as major regulator of secondary metabolite production in Aspergillus nidulans. Front. Microbiol 9, 2212 10.3389/fmicb.2018.02212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero JS, Hartnett T, Eissenberg JC, 1995. Functional analysis of the chromo domain of HP1. EMBO J. 14, 3977–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogo BG, Allfrey VG, Mirsky AE, 1966. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl. Acad. Sci. U. S. A 55, 805–812. 10.1073/PNAS.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadri M, Nalli Y, Jain SK, Chaubey A, Ali A, Strobel GA, Vishwakarma RA, Riyaz-Ul-Hassan S, 2017. An insight into the secondary metabolism of Muscodor yucatanensis: small-molecule epigenetic modifiers induce expression of secondary metabolism-related genes and production of new metabolites in the endophyte. Microb. Ecol 73, 954–965. 10.1007/s00248-016-0901-y. [DOI] [PubMed] [Google Scholar]
- Rando OJ, 2012. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr. Opin. Genet. Dev 22, 148–155. 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Narendja F, Berger H, Gallmetzer A, Fernandez-Martin R, Garcia I, Scazzocchio C, Strauss J, 2008. Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryot. Cell 7, 656–663. 10.1128/EC.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J, 2010. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol 76, 1376–1386. 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Boedi S, Sulyok M, Wiesenberger G, Stoppacher N, Krska R, Strauss J, 2012. Heterochromatin influences the secondary metabolite profile in the plant pathogen Fusarium graminearum. Fungal Genet. Biol 49, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M, 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109, 437–446. 10.1016/S0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Roguev A, Shevchenko A, Schaft D, Thomas H, Stewart AF, Shevchenko A, 2004. A comparative analysis of an orthologous proteomic environment in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol. Cell. Proteomics 3, 125–132. 10.1074/mcp.M300081-MCP200. [DOI] [PubMed] [Google Scholar]
- Rösler SM, Kramer K, Finkemeier I, Humpf H-U, Tudzynski B, 2016. The SAGA complex in the rice pathogen F usarium fujikuroi : structure and functional characterization. Mol. Microbiol 102, 951–974. 10.1111/mmi.13528. [DOI] [PubMed] [Google Scholar]
- Roze LV, Arthur AE, Hong S-Y, Chanda A, Linz JE, 2007. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol 66, 713–726. 10.1111/j.1365-2958.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- Ruiz-García AB, Sendra R, Galiana M, Pamblanco M, Pérez-Ortín JE, Tordera V, 1998. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem 273, 12599–12605. 10.1074/JBC.273.20.12599. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M, 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. U. S. A 93, 14503–14508. 10.1073/PNAS.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou M-M, 2011. The PHD finger: a versatile epigenome reader. Trends Biochem. Sci 36, 364–372. 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee T, Cary JW, Calvo AM, 2016. RmtA, a putative arginine methyltransferase, regulates secondary metabolism and development in Aspergillus flavus. PLoS ONE 11, e0155575 10.1371/journal.pone.0155575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savickiene J, Treigyte G, Jazdauskaite A, Borutinskaite V, Navakauskiene R, 2012. DNA methyltransferase inhibitor RG108 and histone deacetylase inhibitors cooperate to enhance NB4 cell differentiation and E-cadherin re-expression by chromatin remodelling. Cell Biol. Int 36, 1067–1078. 10.1042/CBI20110649. [DOI] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee J-S, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A, 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 19, 849–856. 10.1016/J.MOLCEL.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Schueffler A, Anke T, 2014. Fungal natural products in research and development. Nat. Prod. Rep 31, 1425–1448. 10.1039/C4NP00060A. [DOI] [PubMed] [Google Scholar]
- Seto E, Yoshida M, 2014. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol 6, a018713 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto R, Matsumoto T, Masuo S, Takaya N, 2018. 5-Methylmellein is a novel inhibitor of fungal sirtuin and modulates fungal secondary metabolite production. J. Gen. Appl. Microbiol 2018 (01), 001 10.2323/jgam.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Masuo S, Fujita T, Doi Y, Kamimura Y, Takaya N, 2012. Hydrolase controls cellular NAD, sirtuin, and secondary metabolites. Mol. Cell. Biol 32, 3743–3755. 10.1128/MCB.00032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y, Kawatani M, Futamura Y, Osada H, Koyama Y, 2016. An overproduction of astellolides induced by genetic disruption of chromatin-remodeling factors in Aspergillus oryzae. J. Antibiot. (Tokyo) 69, 4–8. 10.1038/ja.2015.73. [DOI] [PubMed] [Google Scholar]
- Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR, 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol 26, 3339–3352. 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP, 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6, 1656–1664. 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siless GE, Gallardo GL, Rodriguez MA, Rincón YA, Godeas AM, Cabrera GM, 2018. Metabolites from the dark septate endophyte Drechslera sp. evaluation by LC/MS and principal component analysis of culture extracts with histone deacetylase inhibitors. Chem. Biodivers 15, e1800133 10.1002/cbdv.201800133. [DOI] [PubMed] [Google Scholar]
- Sims RJ, Reinberg D, 2006. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 20, 2779–2786. 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD, 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. U. S. A 95, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup A, Keller NP, 2013. Western Analysis of Histone modifications (Aspergillus nidulans). Bio-protocol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup AA, Chiang Y-M, Bok JW, Reyes-Dominguez Y, Oakley BR, Wang CCC, Strauss J, Keller NP, 2012. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol 86, 314–330. 10.1111/j.1365-2958.2012.08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spraker JE, Wiemann P, Baccile JA, Venkatesh N, Schumacher J, Schroeder FC, Sanchez LM, Keller NP, 2018. Conserved responses in a war of small molecules between a plant-pathogenic bacterium and fungi. MBio 9 10.1128/mBio.00820-18. (e00820–18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun Z-W, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD, 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol 22, 1298–1306. 10.1128/MCB.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Reyes-Dominguez Y, 2011. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol 48, 62–69. 10.1016/J.FGB.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Studt L, Schmidt FJ, Jahn L, Sieber CMK, Connolly LR, Niehaus E-M, Freitag M, Humpf H-U, Tudzynski B, 2013. Two histone deacetylases, FfHda1 and FfHda2, are important for Fusarium fujikuroi secondary metabolism and virulence. Appl. Environ. Microbiol 79, 7719–7734. 10.1128/AEM.01557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt L, Janevska S, Arndt B, Boedi S, Sulyok M, Humpf H-U, Tudzynski B, Strauss J, 2017. Lack of the COMPASS component Ccl1 reduces H3K4 trimethylation levels and affects transcription of secondary metabolite genes in two plant–-pathogenic Fusarium species. Front. Microbiol 07, 2144 10.3389/fmicb.2016.02144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M, 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8, 473–479. [DOI] [PubMed] [Google Scholar]
- Sun J, Awakawa T, Noguchi H, Abe I, 2012. Induced production of mycotoxins in an endophytic fungus from the medicinal plant Datura stramonium. Bioorg. Med. Chem. Lett 22, 6397–6400. 10.1016/J.BMCL.2012.08.063. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Chiang Y-M, Oakley CE, Davidson AD, Wang CCC, Oakley BR, 2008. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl. Environ. Microbiol 74, 7607–7612. 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL, 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411. 10.1126/SCIENCE.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tribus M, Galehr J, Trojer P, Brosch G, Loidl P, Marx F, Haas H, Graessle S, 2005. HdaA, a major class 2 histone deacetylase of Aspergillus nidulans, affects growth under conditions of oxidative stress. Eukaryot. Cell 4, 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribus M, Bauer I, Galehr J, Rieser G, Trojer P, Brosch G, Loidl P, Haas H, Graessle S, 2010. A novel motif in fungal class 1 histone deacetylases is essential for growth and development of Aspergillus. Mol. Biol. Cell 21, 345–353. 10.1091/mbc.e09-08-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Brandtner EM, Brosch G, Loidl P, Galehr J, Linzmaier R, Haas H, Mair K, Tribus M, Graessle S, 2003. Histone deacetylases in fungi: novel members, new facts. Nucleic Acids Res. 31, 3971–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Dangl M, Bauer I, Graessle S, Loidl P, Brosch G, 2004. Histone methyltransferases in Aspergillus nidulans: evidence for a novel enzyme with a unique substrate specificity. Biochemistry 43, 10834–10843. 10.1021/BI049626I. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y, 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Vervoort HC, Drašković M, Crews P, 2011. Histone deacetylase inhibitors as a tool to up-regulate new fungal biosynthetic products: isolation of EGM-556, a cyclodepsipeptide, from Microascus sp. Org. Lett 13, 410–413. 10.1021/ol1027199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Buckley AM, Hilger F, Gaber RF, 1990. Direct selection for mutants with increased K+ transport in < i > Saccharomyces cerevisiae. Genetics 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, Anderluh G, Asadollahi M, Askin M, Barry K, Battaglia E, Bayram Ö, Benocci T, Braus-Stromeyer SA, Caldana C, Cánovas D, Cerqueira GC, Chen F, Chen W, Choi C, Clum A, Dos Santos RAC, de Damásio ARL, Diallinas G, Emri T, Fekete E, Flipphi M, Freyberg S, Gallo A, Gournas C, Habgood R, Hainaut M, Harispe ML, Henrissat B, Hildén KS, Hope R, Hossain A, Karabika E, Karaffa L, Karányi Z, Kraševec N, Kuo A, Kusch H, LaButti K, Lagendijk EL, Lapidus A, Levasseur A, Lindquist E, Lipzen A, Logrieco AF, MacCabe A, Mäkelä MR, Malavazi I, Melin P, Meyer V, Mielnichuk N, Miskei M, Molnár ÁP, Mulé G, Ngan CY, Orejas M, Orosz E, Ouedraogo JP, Overkamp KM, Park H-S, Perrone G, Piumi F, Punt PJ, Ram AFJ, Ramón A, Rauscher S, Record E, Riaño-Pachón DM, Robert V, Röhrig J, Ruller R, Salamov A, Salih NS, Samson RA, Sándor E, Sanguinetti M, Schütze T, Sepčić K, Shelest E, Sherlock G, Sophianopoulou V, Squina FM, Sun H, Susca A, Todd RB, Tsang A, Unkles SE, van de Wiele N, van Rossen-Uffink D, de Oliveira JVC, Vesth TC, Visser J, Yu J-H, Zhou M, Andersen MR, Archer DB, Baker SE, Benoit I, Brakhage AA, Braus GH, Fischer R, Frisvad JC, Goldman GH, Houbraken J, Oakley B, Pócsi I, Scazzocchio C, Seiboth B, VanKuyk PA, Wortman J, Dyer PS, Grigoriev IV, 2017. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 18, 28 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ma A, Chow CM, Horsley D, Brown NR, Cowell IG, Singh PB, 2000. Conservation of heterochromatin protein 1 function. Mol. Cell. Biol 20, 6970–6983. 10.1128/MCB.20.18.6970-6983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sena Filho JGJG, Hoover AR, King JB, Ellis TK, Powell DR, Cichewicz RH, 2010. Chemical epigenetics alters the secondary metabolite composition of guttate excreted by an Atlantic-forest-soil-derived Penicillium citregonigrum. J. Nat. Prod 73, 942–948. 10.1021/np100142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wang C, Hou R, Zhou X, Li G, Zhang S, Xu J-R, 2012. The AMT1 arginine methyltransferase gene is important for plant infection and normal hyphal growth in Fusarium graminearum. PLoS ONE 7, e38324 10.1371/journal.pone.0038324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH, 2008. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem 6, 1895 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE, 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem 67, 545–579. 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Wu G, Zhou H, Zhang P, Wang X, Li W, Zhang W, Liu X, Liu H-W, Keller NP, An Z, Yin W-B, 2016. Polyketide production of pestaloficiols and macrodiolide ficiolides revealed by manipulations of epigenetic regulators in an endophytic fungus. Org. Lett 18, 1832–1835. 10.1021/acs.orglett.6b00562. [DOI] [PubMed] [Google Scholar]
- Yakasai AA, Davison J, Wasil Z, Halo LM, Butts CP, Lazarus CM, Bailey AM, Simpson TJ, Cox RJ, 2011. Nongenetic reprogramming of a fungal highly reducing polyketide synthase. J. Am. Chem. Soc 133, 10990–10998. 10.1021/ja204200x. [DOI] [PubMed] [Google Scholar]
- Yang X-L, Huang L, Ruan X-L, 2014. Epigenetic modifiers alter the secondary metabolite composition of a plant endophytic fungus Pestalotiopsis crassiuscula obtained from the leaves of Fragaria chiloensis. J. Asian Nat. Prod. Res 16, 412–417. 10.1080/10286020.2014.881356. [DOI] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, Li B, 2011. Readers of histone modifications. Cell Res. 21, 564–578. 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaware N, 2017. Chemical modulators for epigenome reader domains as emerging epigenetic therapies for cancer and inflammation. Curr. Opin. Chem. Biol 39, 116–125. 10.1016/J.CBPA.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Chen L, Yu X, Liu H, Akhberdi O, Pan J, Zhu X, 2016. A B-type histone acetyltransferase Hat1 regulates secondary metabolism, conidiation, and cell wall integrity in the taxol-producing fungus Pestalotiopsis microspora. J. Basic Microbiol 56, 1380–1391. 10.1002/jobm.201600131. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Akhberdi O, Wei D, Chen L, Liu H, Wang D, Hao X, Zhu X, 2018. A MYST histone acetyltransferase modulates conidia development and secondary metabolism in Pestalotiopsis microspora, a taxol producer. Sci. Rep 8, 8199 10.1038/s41598-018-25983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]