Abstract
Molecular phylogenetic analysis of 18S rRNA gene sequences of nearly any species of Chytridiomycota has typically challenged traditional classification and triggered taxonomic revision. This has often led to the establishment of new taxa which, normally, appears well supported by zoospore ultrastructure, which provides diagnostic characters. To construct a meaningful and comprehensive classification of Chytridiomycota, the combination of molecular phylogenies and morphological studies of traditionally defined chytrid species is needed. In this work, we have studied morphological and ultrastructural features based on light and transmission electron microscopy as well as molecular phylogenetic analysis of a parasite (strain X-124 CCPP ZIN RAS) morphologically similar to Rhizophydium granulosporum living on the yellowgreen alga Tribonema gayanum. Phylogenetic analysis of the 18S rRNA gene sequence of this strain supports that it represents a new genus and species affiliated to the recently established order Gromochytriales. The ultrastructure of X-124 confirms its phylogenetic position sister to Gromochytrium and serves as the basis for the description of the new genus and species Apiochytrium granulosporum. The 18S rRNA gene of A. granulosporum contains a S943 group I intron that carries a homing endonuclease pseudogene.
Keywords: Chytrids, endonuclease pseudogene, intron, molecular phylogeny, ultrastructure
THE systematics of Chytridiomycota has been under continuous revision after a global molecular phylogenetic tree of fungi based on six gene markers was published some years ago (James et al. 2006a). From that time, the phylogenetic investigation of 18S rRNA gene sequences of nearly any species or strain has triggered the revision of neighboring taxa, often leading to the establishment of new higher rank taxa, for example, family, order and class, and divisions normally supported by zoospore ultrastructure. In the past few years, chytrid taxonomy has experienced considerable changes. For instance, James et al. (2006b) described the new phylum Blastocladiomycota; Letcher et al. (2006) redescribed the Rhizophydium clade (James et al. 2006a) as the order Rhizophydiales; Mozley-Standridge et al. (2009) established the order Cladochytriales for the Cladochytrium clade (James et al. 2006a) and Simmons et al. (2009) redescribed the clade formerly represented in phylogenetic trees by Chytriomyces angularis (James et al. 2006a) as the order Lobulomycetales. New orders have also been proposed: Polychytriales (Longcore and Simmons 2012), Gromochytriales and Mesochytriales (Karpov et al. 2014), and Synchytriales (Longcore et al. 2016). Moreover, some genera with typical chytrid morphology and hence thought to branch early in the fungal tree, such as Olpidium (Sekimoto et al. 2011), Amoeboradix, and Sanchytrium (Karpov et al. 2018), appeared to branch within a clade composed of the Basidiomycota, Ascomycota, and Glomeromycota, out from the Chytridiomycota clade. It is clear that we need molecular data for each traditionally described species of chytrids to construct a meaningful and comprehensive classification of Chytridiomycota (see also Powell 2016).
Transmission electron microscopy (TEM) sometimes reveals peculiarities that can be used as new taxonomic characters or, on the contrary, may show the unimportance of some commonly accepted ultrastructural features. However, zoospore ultrastructure has so far provided useful discriminatory characters for taxonomy in agreement with the molecular phylogeny of fungi. Some chytrids have been found to contain insertions in ribosomal DNA, which may or may not be present in close relatives, but that are rather common in Dikarya and green algae (Karpov et al. 2017).
Here, we present light microscopy observations, ultrastructure, and molecular phylogenetic analysis of strain X- 124 (CCPP ZIN RAS), a parasite of algae isolated from freshwater samples using Tribonema gayanum as a host. Phylogenetic analysis of 18S rRNA gene sequences of this strain supports that it represents a new genus and species affiliated to the recently established order Gromochytriales. The ultrastructure of X-124 confirms its phylogenetic position sister to Gromochytrium and serves as the basis for the description of this new genus and species.
Materials and methods
Collection and isolation
Strain X-124 was isolated by M.A. Mamkaeva in 2012 from the freshwater sample x-22 collected in a small pond in Kuty village, Kingisepp District, Leningrad Province, Russia. A strain was maintained in culture on T. gayanum (strain 20 CALU) as a host as described in Karpov et al. (2016).
Light and transmission electron microscopy Light and DIC microscopy observations of living cultures were carried out on a Zeiss Axioplan microscope (Leica Microsystems, St. Petersburg, Russia) equipped with black and white MRm Axiocam (Leica Microsystems).
Electron microscopy
Infected algal filaments were fixed with 0.3% OsO4 in 0.1 M cacodylate buffer during two minutes, and then, glutaraldehyde was added to 2% final concentration for 30 min in the dark. Then, the fixative was removed, and 2% glutaraldehyde in 0.1 M cacodylate buffer was added for 1 h, rinsed in 0.1 M cacodylate buffer and postfixed in 1% OsO4 in the same buffer for one hour. All fixation stages were carried out on ice. After rinsing in 0.1 M cacodylate buffer for 5 min, the filaments were dehydrated in alcohol series and in propylen oxide and finally embedded in Spurr resin (Sigma Aldrich, St. Louis, MO). Ultrathin sections were prepared using a Leica Ultracut ultratome with a diamond knife. After double staining, the sections were observed using a JEM 1400 microscope (Jeol, Tokyo, Japan) equipped with the digital camera Olympus Veleta (Tokyo, Japan).
18S rRNA gene sequencing and molecular phylogeny
Cells from infected cultures were pelleted by centrifugation, and their DNA was purified using the PowerSoil DNA purification kit from MoBio (Carlsbad, CA). 18S rRNA gene fragments were then amplified by PCR using the fungal specific primers Fun_UF1 (50-CGAATCGCATGG CCTTG; Kappe et al. 1996) and Fun_AU4 (50- RTCTCACTAAGCCATTC; Vandenkoornhuyse et al. 2002). PCRs were carried out in a final volume of 25 ll. The reaction mix contained 1 ll of the eluted DNA, 1.5 mM MgCl2, dNTPs (10 nmol each), 20 pmol of each primer, and 0.2 U TaqPlatinum DNA polymerase (Invitrogen, Carlsbad, CA). After a first denaturation step at 94 °C for 2 min, the PCR consisted of 35 cycles of the following iterative cycle: denaturation at 94 °C for 15 s, annealing at 50 °C for 30 s, and polymerization at 72 °C for 2 min. A final elongation at 72 °C for 7 min was carried out. We cloned the resulting 18S rRNA amplicons to identify individual fungal sequences using the Topo TA Cloning System (Invitrogen) following the manufacturer’s instructions. Clone inserts were then amplified by PCR using flanking vector primers, and the inserts of expected sizes were sequenced bidirectionally with vector primers (Beckman Coulter Genomics, Takeley, U.K.). Two closely related chytrid sequences were identified and compared with sequences in the GenBank nr database using BLAST (Altschul et al. 1990). The new sequences and those retrieved by BLAST were aligned using MAFFT with the accurate ‘-linsi’ option (Katoh et al. 2002). The multiple alignment was trimmed with TRIMAL (Capella-Gutiérrez et al. 2009), and a Bayesian phylogenetic tree was reconstructed using MrBayes (Ronquist et al. 2012) applying the GTR + G + I model with four chains and 10,000,000 generations. After checking for convergence, the first 15,000 trees were eliminated as burn-in, and a consensus tree was constructed by sampling every 100 trees. The new Apiochytrium granulosporum strain X-124 18S rRNA sequences have been deposited in GenBank under accession numbers X-124a - MK179157, X-124b - MK179158.
Intron S943 phylogeny
Intron localization was defined by comparing the assembled contig with other sequences from the NCBI nr database using BLAST. Insertion positions were identified using the 16S gene reference of Escherichia coli (Cannone et al. 2002; http://www.rna.icmb.utexas.edu/SAE/2C/rRNA_Introns). Additional SSU sequences containing intron S943 were downloaded from GenBank. Sequences were aligned with MUSCLE (Edgar 2004) and manually adjusted in BioEdit (Hall 1999). Bayesian inference was calculated with MrBayes applying the GTR + G + I model with four chains and 10,000,000 generations. After checking for convergence, the first 50% trees were eliminated as burnin, and a consensus tree was constructed. The resulting Bayesian tree was visualized using the iTOL web tool (Letunic and Bork 2016).
Results
Morphology
Zoospores
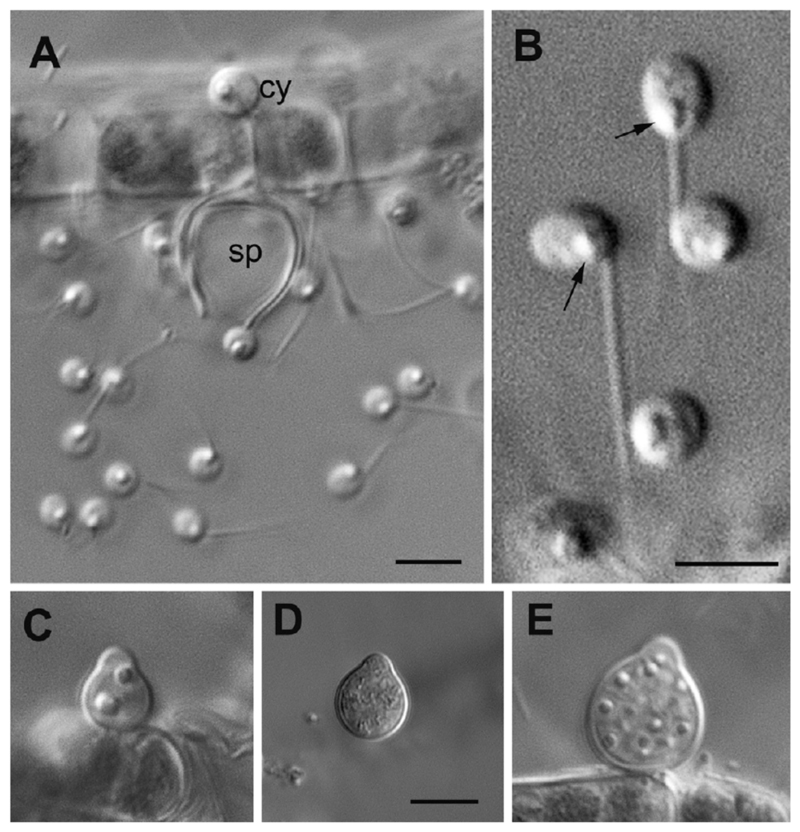
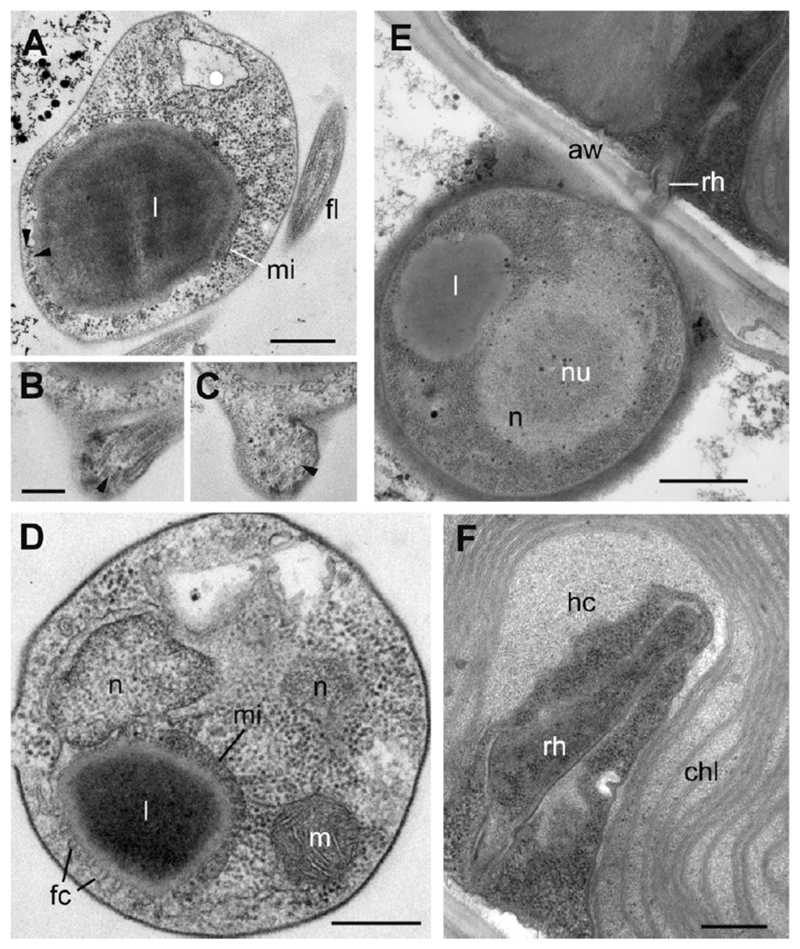
This fungus produces slightly elongated or spherical zoospores 2.3–2.7 lm in diameter with a posterior single lipid globule and a comparatively short flagellum about 8– 9 lm in length having a short acroneme of approximately 1 lm (Fig. 1A, B). In released zoospores, the single prominent lipid globule locates at a posterior position, near the point of flagellum emergence (Fig. 2A–D). The lipid globule is associated with a flat microbody and covered with fenestrated cisternae facing the plasma membrane (Fig. 2D). The elongated nucleus is slightly curved and oriented in posterior–anterior direction. Its posterior end is located near the microbody-lipid complex (MLC; Fig. 2D), and the lateral cavity of the nucleus is filled with small vesicles and rare cisternae of the Golgi apparatus (not shown). Mitochondrial profiles with flat cristae are scattered in the cytoplasm, and the ribosomes do not aggregate (Fig. 2A, D). Mature intrasporangial zoospores are predominantly of amoeboid appearance and in general exhibit the same location of MLC, nuclear cavity, Golgi apparatus, dispersed ribosomes, and mitochondria (Fig. 4B–D). A nonfunctional kinetosome (centriole) is located at sharp angle (ca. 60°) with the flagellar kinetosome and connected to it with a broad fibrillar bridge (Fig. 4C, D). The flagellar transition zone contains a spiral fiber or cylinder (Fig. 2B, C). A short posterior root of two microtubules passes, between kinetosome and centriole, to the plasma membrane (Fig. 4D). Traces of one more root (probably anterior) of 2–3 microtubules can be found between the plasma membrane and the fenestrated cisternae (Fig. 2A), but its origin could not be fully tracked.
Figure 1.
DIC images of living zoospores (A, B) and sporangia (C–E) of Apiochytrium granulosporum sp. nov. (X-124 CCPP ZIN RAS). A—released zoospores, cyst (cy), and empty sporangium (sp), B—zoospores at high magnification. Arrows on B show posterior lipid globule. C, D—young sporangia, E—premature sporangium. Scale bars: A—5 µm, B—3 µm, C–E—5 µm.
Figure 2.
Transmission electron micrographs of Apiochytrium granulosporum sp. nov. (X-124 CCPP ZIN RAS). (A–D) sections of free-swimming zoospores. Arrowheads on A show microtubules of the anterior root. (B, C) consecutive sections of flagellar transition zone with spiral filament (arrowheads), (E) encysted zoospore penetrating the algal cell wall (aw) with a rhizoid (rh), (F) rhizoid (rh) inside the alga. aw = algal cell wall; chl = chloroplast; fc = fenestrated cisterna; fl = flagellum; hc = host cytoplasm; l = lipid globule; m = mitochondrion; mi = microbody; n = nucleus. Scale bars: A—500 nm, B, C—200 nm, D—300 nm, E—800 nm, F—400 nm.
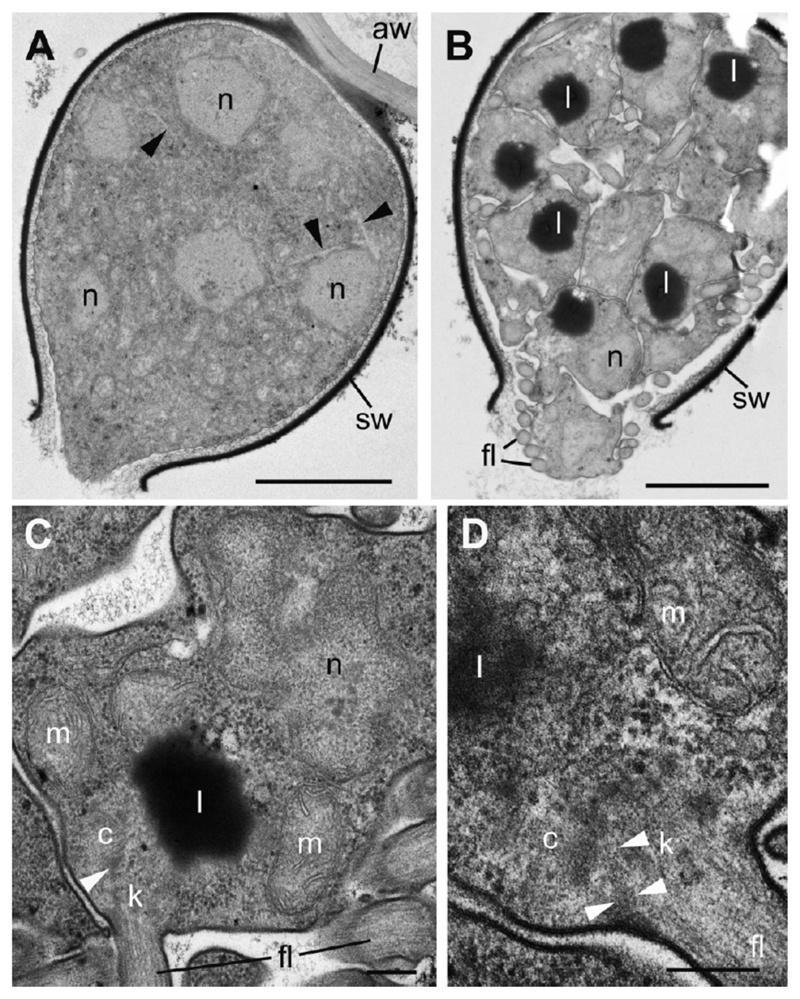
Figure 4.
Transmission electron micrographs of Apiochytrium granulosporum sp. nov. (X-124 CCPP ZIN RAS). (A) immature sporangium with open papilla, several nuclei, and without lipid globule. Arrowheads show developing cell borders, (B) sporangium with mature and releasing zoospores, (C) intrasporangial zoospore. Arrowhead shows a fibrillar bridge between centriole (c) and kinetosome (k). (D) longitudinal section of centriole adjacent to kinetosome which is obliquely sectioned here. Arrowheads show a posterior root of two microtubules. aw = algal cell wall; c = centriole; fl = flagellum; k = kinetosome; l = lipid globule; m = mitochondrion; n = nucleus. Scale bars: A—2.5 µm, B—2 µm, C—200 nm, D—200 nm.
Cyst
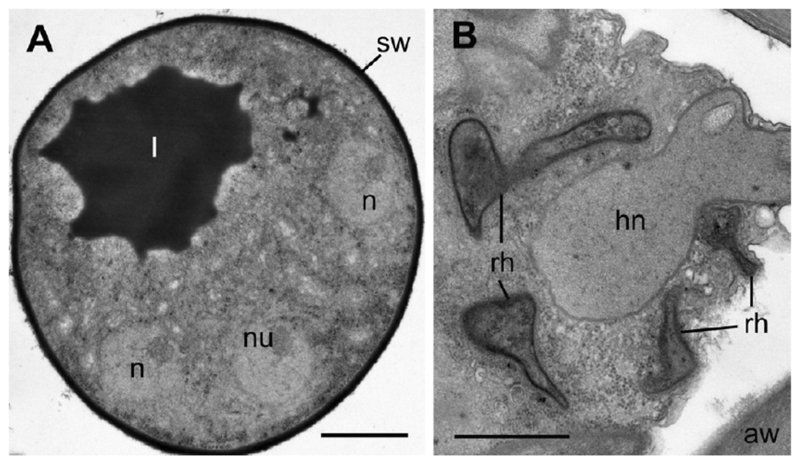
Zoospores encyst on the surface of the host alga and produce a rhizoid penetrating the host cell wall (Fig. 1A, 2E, F). In addition to the chitin wall, the cyst is covered with a rather thick layer of electron dense amorphous material (Fig. 2E). Each cyst contains a nucleus with a large central nucleolus and a lipid globule that can be visible in the thin sections (Fig. 2E). The rhizoids were not visible on the living material, but the ultrastructural observation revealed a very thin rhizoidal stem penetrating the algal cell wall and its thickened part just under the wall of the host (Fig. 2E, F). Such a narrow hole (<200 nm as measured in serial sections) in the algal cell wall for thin rhizoids has been also found in the mature sporangium (not shown). The rhizoid distal part branches inside the cell, sometimes enveloping the host nucleus, which looks deformed (Fig. 3B).
Figure 3.
Transmission electron micrographs of Apiochytrium granulosporum sp. nov. (X-124 CCPP ZIN RAS). (A) young sporangium with several nuclei and a big lipid globule, (B) host nucleus (hn) surrounded by rhizoids (rh). aw = algal cell wall; hn = host nucleus; l = lipid globule; n = nucleus; nu = nucleolus; rh = rhizoids; sw = sporangial wall. Scale bars: A—2 µm, B—1 µm.
Sporangium
Monocentric epibiotic sessile slightly broaden pear-shaped sporangia up to 10–12 lm in height with a broad apical papilla for zoospore releasing locate on the Tribonema filament (Fig. 1C–E). The immature sporangia have characteristic granulated contents (Fig. 1E) with several nuclei and one big lipid globule (Fig. 3A). At a later stage of sporangium maturation (Fig. 4A), this globule disappears, likely as a consequence of its use as an energy source for zoospore formation. Interestingly, in spite of the fact that the plasmodium division just has started and the flagella are not yet formed, the papilla is already open (Fig. 4A). In the mature sporangium, each newly formed zoospore contains a single big globule immediately prior to zoospore release. Thus, each intrasporangial zoospore already contains a posterior lipid globule near the flagellar base (Fig. 4B–D).
18S rRNA phylogeny
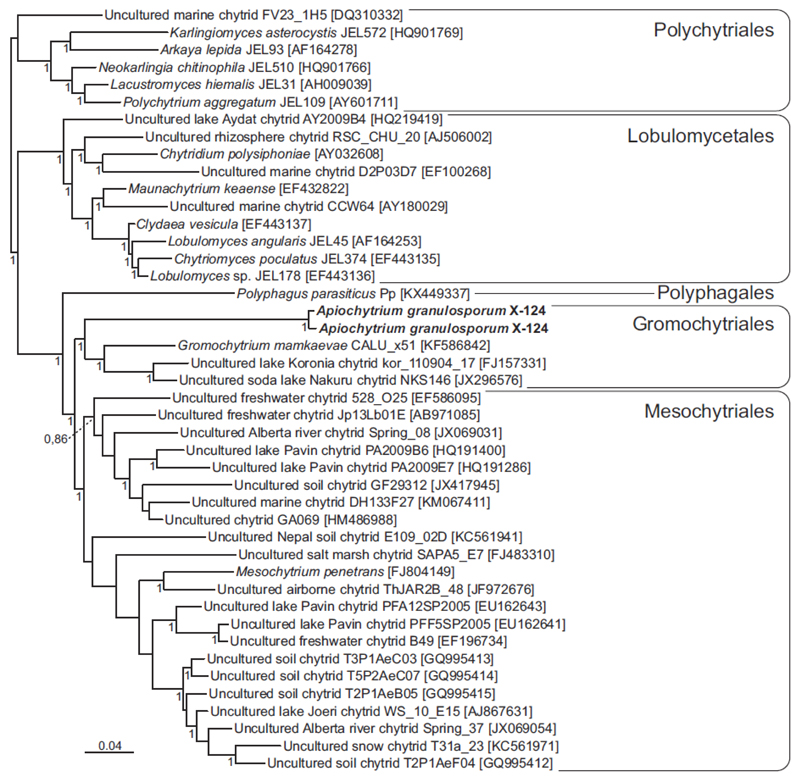
The two 18S rRNA gene sequences of the strain X-124 were very similar (99.4% identical) and formed a clade sister to the cluster containing Gromochytrium mamkaevae and some environmental sequences with high support (Fig. 5). We ignore if the two X-124 sequences correspond to two closely related chytrid strains present in our culture or to two slightly divergent 18S rRNA genes present in a single species. According to our tree, this strain represents a new deep-branching lineage within the order Gromochytriales, clearly distinct from Gromochytrium. In addition, the strain X-124 sequences contains a large (1,476 bp) insertion absent in the other Gromochytriales sequences. The presence of this large insertion may have impacted the whole 18S rRNA gene structure of X-124, which may explain the relatively long branch of this strain compared with the other Gromochytriales. Taking into account its phylogenetic distance, we consider that strain X-124 represents a new genus and species within the order Gromochytriales. Our tree also supports strongly the sister relationship between the Gromochytriales and the Mesochytriales, and the position of the recently described Polyphagales as sister of the Gromochytriales + Mesochytriales cluster (Fig. 5), in agreement with previous analyses (Karpov et al. 2016).
Figure 5.
18S rRNA gene Bayesian phylogenetic tree based on 1,456 conserved sites. The tree is rooted using sequences of Lobulomycetales and Polychytriales. Numbers at branches are Bayesian posterior probabilities (only values higher than 0.7 are shown).
Portrayal of S943 intron of the strain X-124
The 18S rRNA gene of the strain X-124 is noticeably longer than usual because it contains a group I intron (1,476 bp) with an insertion position at S943 according to the E. coli reference (Cannone et al. 2002). The intron carries a homing endonuclease gene that locates on the antisense strand of P8 region and coding His-Cys box protein 244 amino acid residues long. Homing endonuclease genes are rare but widely distributed among S943 introns (Haugen et al. 2004). The predicted homing endonuclease protein of the strain X-124 does not have at its C-terminus the 2nd zinc-binding motif and dimerization tail which are common for His–Cys box family proteins (Chevalier and Stoddard 2001). However, zinc-binding conservative motif can be detected if multiple frame shifts are introduced into the nucleotide sequence. Previously, pseudogenes/ remnants of homing endonuclease genes were found in fungal S943 introns (Haugen et al. 2004). Homing endonuclease genes in 18S rDNAs have been found so far in two chytridiomycete species: Gonapodya sp. JEL612 (Dee et al. 2015; the same intron insertion position S943) and Phlyctochytrium planicorne (James et al. 2006b; another intron insertion position S508). According to tblastn search against nr/nt database of NCBI, the endonuclease from Gonapodya sp. is the most similar to the endonuclease from the strain X-124. This indicates that the chyrid X-124 and monoblepharid Gonapodia sp. JEL612 obtained their homing endonuclease genes either “vertically” from an ancient common ancestor or horizontally from an unknown source.
S943 introns are widely distributed in different fungi, amoebae, algae, cercozoans, and ciliates. The majority of these taxa include nonintronized and intronized species at the different phylogeny levels. However, S943 intron distribution is mosaic because only 1–5% species in each mentioned group carry this intron. We constructed the Bayesian phylogenetic tree of nucleotide sequences of S943 introns using 279 fungal and nonfungal species. S943 introns of filamentous fungi form a large monophyletic group sister to the strain X-124 and an uncultured chytrid Jp13Ch04E (Fig. S1). Introns of ascomycetes form a large cluster with a few exceptions including one basidiomycete and one zygomycete species. Introns of basidiomycetes form three paraphyletic groups and one group inside the ascomycete cluster. Introns from zoosporic fungi (five monoblepharidomycetes, two uncultured chytrid clones, and the aphelid Amoeboaphelidium occidentale) and nonfungal taxa form a set of poorly resolved lines outside the filamentous fungi.
Discussion
Phylogeny and ultrastructure
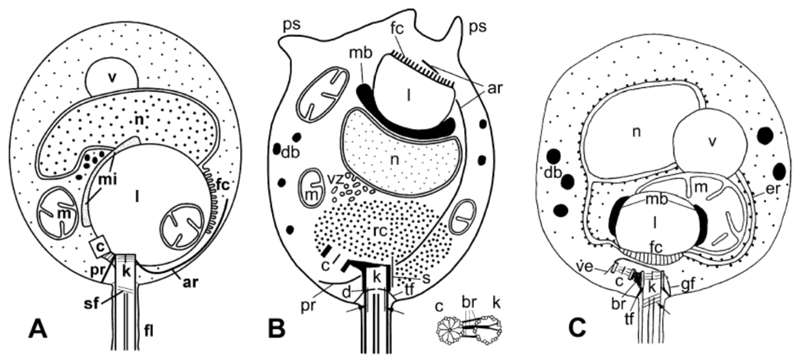
The 18S rRNA gene sequences of the X-124 strain form a monophyletic cluster with some environmental sequences (Fig. 5) that affiliates to the recently described order Gromochytriales (Karpov et al. 2014). This order includes one cultured chytrid, G. mamkaevae, having zoospores with posterior ribosome aggregates unbounded by endoplasmic reticulum and one MLC with anterior location with a rather thick microbody (Fig. 6). By contrast, X-124 shows dispersed ribosomes and a posterior MLC with a thin microbody. Both species exhibit a single lipid globule with fenestrated cisternae facing toward the exterior, posterior, and probably anterior microtubular roots, a cylinder or spiral filament in the flagellar transition zone and several dispersed mitochondria. Gromochytrium mamkaevae has a centriole at an angle of approx. 30° with respect to the kinetosome vs. 60° in X-124 (Fig. 6). We did not observe a centriole in released zoospores; nonetheless, we can propose that the angle of centriole location in the X-124 intrasporangial zoospore may probably be the same after complete maturation in the released zoospore (Fig. 6). Although a centriole-kinetosome bridge was certainly present, our material did not allow studying it in detail. Some zoospore characters of X-124, that is, dispersed ribosomes and posterior MLC, are similar to those of Mesochytrium penetrans (Karpov et al. 2010), which belongs to the closely related order Mesochytriales (Fig. 6). However, other zoospore characters of M. penetrans essentially differ from those of X-124: the ER bounding MLC associated with a single big mitochondrion, fenestrated cisterna facing the flagellar base, and the presence of a vacuole and several fibrils associated with the centriole and kinetosome (Karpov et al. 2010). The flagellar apparatus of the M. penetrans zoospore contains a spiral fiber in the transition zone, but does not have any microtubular roots (Karpov et al. 2010).
Figure 6.
General scheme of zoospore structure in Apiochytrium granulosporum (A), Gromochytrium mamkaevae (B) (Gromochytriales), and Mesochytrium penetrans (C) (Mesochytriales). ar = anterior microtubular root; br = bridge connecting centriole to kinetosome; c = centriole; d = diaphragm; db (vz) = dense bodies (located in the posterior nuclear cavity in A. granulosporum); er = endoplasmic reticulum; fc = fenestrated cisterna; fl = flagellum; k = kinetosome; l = lipid globule; m = mitochondrion; mb (mi) = microbody; n = nucleus; nu = nucleolus; pr = posterior microtubular root; ps = pseudopodia; rc = ribosomal core; sf = spiral filament in flagellar transition zone; tf = transition fibers; ve, s, gf-kinetid fibrillar derivates probably absent in A. granulosporum. B—after: Karpov et al. (2014); C—after: Karpov et al. (2010).
In summary, the zoospore morphology and, in particular, the flagellar apparatus of X-124 are more similar to those of G. mamkaevae than to those of M. penetrans, confirming the affiliation to the Gromochytriales. The partial zoospore similarity of X-124 to M. penetrans is in agreement with a somewhat intermediate phylogenetic position of X- 124 between Gromochytrium and Mesochytrium. In fact, the X-124 18S rRNA sequences branch deep in the Gromochytriales (Fig. 5), which might explain that X-124 has retained some features of the last common ancestor of Gromochytriales and Mesochytriales, sharing characters with both orders.
From a taxonomy perspective, our data provide additional material for revision of the key diagnostic characters defining the order Gromochytriales. However, at this stage of investigation, such a revising is premature. More molecular and ultrastructural data from other representatives of the order will be needed to attempt that task.
Mosaic rDNA introns distribution
Group I introns localized in the same insertion sites usually group together at the phylogenetic tree of introns. S943 introns are one of the most common rDNA introns (Karpov et al. 2017) so they can be used as a model of the group I intron evolution. S943 intron of the strain X-124 raises two previously observed issues (Haugen et al. 2004). The first one is an evolutionary trend of homing endonuclease gene transformation into pseudogene and its subsequent loss. Multiple frameshifts in the homing endonuclease coding sequence from the strain X-124 intron correspond to this trend. The second corresponds to the controversial predominance of vertical inheritance of group I introns. The filamentous fungi form a monophyletic clade at the broad taxon sampling S943 introns tree (Fig. S1). Likewise, myxomycetes, green algae, and ciliates form monophyletic or nearly monophyletic (a couple of long branches of amoebae located among green algae) groups (Fig. S1). In other words, all the variety of S943 introns arose most likely from one source in each listed group (filamentous fungi, myxomycetes, green algae, ciliates), and then, they were vertically inherited and diverged along with species divergence in the evolution. However, a mosaic distribution of introns within each group remains a difficulty of such interpretation. Indeed, it seems unlikely that common ancestors of different fungi taxon ranks remained the S943 intron in their 18S rRNA genes throughout their evolutionary radiation while only 1–5% of recent species share this trait. In this case, there should be a recent massive intron loss event happened after taxon radiation. Another hypothesis solving the mosaic intron distribution issue is horizontal transfer. But this hypothesis explains the observed grouping of closely related filamentous fungi only if the horizontal transfer was limited to closely related species. According to the accepted phylogeny of the fungal species, we expected that S943 introns of the chytridiomycetes would form a sister clade to the filamentous fungi. However, the evolutionary path of S943 intron heredity in zoosporic fungi is still under question because of poor resolution of this part of the tree and lack of representative data. Similar ambiguity appears if we interpret the distribution of homing endonuclease genes, rather than the nucleotide sequences of S943 introns. Only two distantly related species of zoosporic fungi have a homing endonuclease gene in their S943 introns, and they are the most similar to each other among all the available homing endonuclease sequences in the NCBI database. If this similarity is due to vertical inheritance from a common ancestor, then the question arises why the closest relatives of each of these species did not inherit homing endonuclease gene as well. If the similarity is due to horizontal transfer from a common source, then the question is what is the source and why we still have no sequence data about it. Despite the insufficient amount of zoosporic fungi data, we can consider that the model of group I intron heredity in 18S rRNA gene is more complicated than it seems and might involve both vertical or horizontal transfer.
Taxonomy
According to light microscopic observations, strain X-124 is similar to Rhizophydium granulosporum Scherff., a parasite of Tribonema with a broadly pyriform sporangium of close dimensions (7–14 lm), delicate rhizoids arising from a short main axis and ovoid zoospores of nearly the same dimensions (3 lm long and 2 lm diameter) with a large colorless globule (Letcher and Powell 2012; Sparrow 1960). The globule in Rh. granulosporum zoospore is also posterior as in X- 124, but the flagellum of Rh. granulosporum seems to be longer than in X-124, although the flagellar length was not shown in the diagnosis of Rh. granulosporum. The resting spore was not found in X-124.
Thus, zoospores of X-124 strain do not differ essentially from those of Rh. granulosporum. In this case based on the shape of sporangium, LM characters of zoospores and similar dimensions we can use Rh. granulosporum Scherff. as a basyonym for description of new genus and species which belongs to the order Gromochytriales on the base of molecular phylogeny and zoospore ultrastructure.
Apiochytrium Karpov et Moreira gen. nov
MycoBank MB 827963
Monocentric, epibiotic, endogenously developing chytrid on an algal substrate. Zoospores contain posterior MLC with single lipid globule and flat microbody near the flagellar base, dispersed ribosomes, and several scattered mitochondria. Kinetid structure as for the order.
Etymology: Apio (Greek)—pear, referring to the pyriform sporangium.
Type species: A. granulosporum Karpov et Moreira sp. nov.
Apiochytrium granulosporum Karpov et Moreira sp. nov
MycoBank MB 827964
Monocentric epibiotic sessile pyriform sporangia up to 12 lm in height and 9 lm broad with 1 broad apical papilla for zoospore releasing. Branching rhizoid is invisible on the living material. Elongated to spherical zoospores 2.3–2.7 lm in diameter or up to 3 lm in length with posterior single lipid globule and flagellum 8–9 lm in length. A nonfunctional kinetosome (centriole) is located at sharp angle to the flagellar kinetosome. Parasite of yellow-green alga T. gayanum.
Etymology: after the name Rh. granulosporum—most similar to the strain X-124.
GenBank Accession Numbers X-124a - MK179157, X-124b - MK179158. Type: Fig. 1. Karpov et al. this publication. RUSSIA, Leningrad Province, Kingisepp District, small pond in Kuty village (59°48062″N, 28°94066″E). Sample collected by Maria Mamkaeva in April 2012. Ex type culture deposited in Culture Collection of Parasitic Protists of Zoological Institute of Russian Academy of Science (CCPP ZIN RAS; Malysheva et al. 2016) under No: X-124.
Supplementary Material
Acknowledgements
The isolation, cultivation, morphological study of strain X- 124, and the manuscript writing have been supported by RSF grant no. 16-14-10302, and the molecular analysis by ERC Advanced Grant No. 322669 “ProtistWorld.” We also thank for support the RFBR grants No. 15-29-02734 and 18-04-01210; the scholarship program “Jean d’Alembert” of Paris-Saclay University; the program of the RAS Presidium “Evolution of the organic world. The role and significance of planetary processes”; the Research Resource Center for Molecular and Cell Technologies (RRC MCT) at St. Petersburg State University (SPbSU) for access to the EM facilities; ZIN RAS program AAAA-A17-117030310322-3. The investigated strain has been cultivated at the Center for Culturing Collection of Microorganisms of Research park of St. Petersburg State University.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, M€uller KM, Pande N, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/ intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee J, Mollicone MRN, Longcore JE, Roberson RW, Berbee M. Cytology and molecular phylogenetics of Monoblepharidomycetes provide evidence for multiple independent origins of the hyphal habit in the Fungi. Mycologia. 2015;107:710–728. doi: 10.3852/14-275. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 1999;41:95–98. [Google Scholar]
- Haugen P, Reeb V, Lutzoni F, Bhattacharya D. The evolution of homing endonuclease genes and group I introns in nuclear rDNA. Mol Biol Evol. 2004;21:129–140. doi: 10.1093/molbev/msh005. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsh HT, et al. Reconstructing the early evolution of the fungi using a six-gene phylogeny. Nature. 2006a;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, Griffith GW, Vilgalys R. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota) Mycologia. 2006b;98:860–871. doi: 10.3852/mycologia.98.6.860. [DOI] [PubMed] [Google Scholar]
- Kappe R, Fauser C, Okeke CN, Maiwald M. Universal fungus-specific primer systems and group-specific hybridization oligonucleotides for 18S rDNA. Mycoses. 1996;39:25–30. doi: 10.1111/j.1439-0507.1996.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Karpov SA, Kobzeva AA, Mamkaeva MA, Mamkaeva KA, Mikhailov KV, Mirzaeva GS, Aeoshin VV. Gromochytrium mamkaevae gen. et sp. nov. and two new orders: Gromochytriales and Mesochytriales (Chytridiomycetes) Persoonia. 2014;32:115–126. doi: 10.3767/003158514X680234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov SA, Letcher PM, Mamkaeva MA, Mamkaeva KA. Phylogenetic position of the genus Mesochytrium (Chytridiomycota) based on zoospore ultrastructure and 18S and 28S rRNA gene sequences. Nowa Hedwigia. 2010;90:81–94. [Google Scholar]
- Karpov SA, Lopez-Garcia P, Mamkaeva MA, Tcvetkova VS, Vishnyakov AE, Klimov VI, Moreira D. The chytrid- like parasites of algae Amoeboradix gromovi gen. et sp. nov. and Sanchytrium tribonematis belong to a new fungal lineage. Protist. 2018;169:122–140. doi: 10.1016/j.protis.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov SA, Lopez-Garcia P, Mamkaeva MA, Vishnyakov A, Moreira D. Chytridiomycete Polyphagus parasiticus: molecular phylogeny supports the erection of a new chytridiomycete order. Mikol Fitopatol. 2016;50:362–366. [Google Scholar]
- Karpov SA, Mamanazarova KS, Popova OV, Aleoshin VV, James TY, Mamkaeva MA, Tcvetkova VS, Vishnyakov AE, Longcore JE. Monoblepharidomycetes diversity includes new parasitic and saprotrophic species with highly intronized rDNA. Fungal Biol. 2017;21:729–741. doi: 10.1016/j.funbio.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher PM, Powell MJ. A taxonomic summary and revision of Rhizophydium (Rhizophydiales, Chytridiomycota) Alabama University; Tuscaloosa, AL: 2012. Printing, No. 1. Imprint. [Google Scholar]
- Letcher PM, Powell MJ, Churchill PF, Chambers JG. Ultrastructural and molecular phylogenetic delineation of a new order, the Rhizophydiales. Mycol Res. 2006;110:898–915. doi: 10.1016/j.mycres.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longcore JE, Simmons DR. The Polychytriales ord. nov. contains chitinophilic members of the rhizophlyctoid alliance. Mycologia. 2012;104:276–294. doi: 10.3852/11-193. [DOI] [PubMed] [Google Scholar]
- Longcore JE, Simmons DR, Letcher PM. Synchytrium microbalum sp. nov. is a saprobic species in a lineage of parasites. Fungal Biol. 2016;120:1156–1164. doi: 10.1016/j.funbio.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Malysheva MN, Mamkaeva MA, Kostygov AY, Frolov AO, Karpov SA. Culture collection of parasitic protists at the Zoological Institute RAS (CCPP ZIN RAS) Protistology. 2016;10:26–42. [Google Scholar]
- Mozley-Standridge SE, Letcher PM, Longcore JE, Porter D, Simmons DR. Cladochytriales – a new order in Chytridiomycota. Mycol Res. 2009;113:498–507. doi: 10.1016/j.mycres.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Powell MJ. Chytridiomycota. In: Archibald JM, Simpson AGB, Slamovits C, editors. Handbook of the Protists. Springer International Publishing; Cham, Switzerland: 2016. pp. 1–36. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Heohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto S, Rochon A, Lon JE, Dee JM, Berbee ML. A multigene phylogeny of Olpidium and its implications for early fungal evolution. BMC Evol Biol. 2011;11:331. doi: 10.1186/1471-2148-11-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DR, James TY, Meyer AF, Longcore JE. Lobulomycetales, a new order in the Chytridiomycota. Mycol Res. 2009;113:450–460. doi: 10.1016/j.mycres.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Sparrow FK. Aquatic phycomycetes. The University of Michigan Press; Ann Arbor, MI: 1960. [Google Scholar]
- Vandenkoornhuyse P, Husband R, Daniell TJ, Watson IJ, Duck JM, Fitter AH, Young JP. Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol Ecol. 2002;11:1555–1564. doi: 10.1046/j.1365-294x.2002.01538.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.