Abstract
Limited evidence exists that humans mount a mutation-specific T cell response to epithelial cancers. We used a whole-exomic-sequencing-based approach to demonstrate that tumor-infiltrating lymphocytes (TIL) from a patient with metastatic cholangiocarcinoma contained CD4+ T helper 1 (TH1) cells recognizing a mutation in erbb2 interacting protein (ERBB2IP) expressed by the cancer. After adoptive transfer of TIL containing about 25% mutation-specific polyfunctional TH1 cells, the patient achieved a decrease in target lesions with prolonged stabilization of disease. Upon disease progression, the patient was retreated with a >95% pure population of mutation-reactive TH1 cells and again experienced tumor regression. These results provide evidence that a CD4+ T cell response against a mutated antigen can be harnessed to mediate regression of a metastatic epithelial cancer.
The human immune system has evolved to recognize and eliminate cells expressing foreign, nonself antigens. All malignant tumors harbor nonsynonymous mutations or other genetic alterations (1), some of which may generate neo-”nonself” epitopes that could potentially trigger an antitumor T cell response. Indeed, mutation-reactive T cells can frequently be found infiltrating human melanomas (2) and likely play a critical role in the clinical efficacy of adoptive cell therapy (ACT) and other immunotherapies in melanoma (3–7). However, limited evidence exists demonstrating that the human immune system can mount an endogenous, mutation-specific T cell response against epithelial cancers that comprise over 80% of all human malignancies (8–11), and it is unclear whether this response can be harnessed to develop effective personalized cancer immunotherapies (12). Moreover, epithelial cancers often contain fewer mutations than melanoma (1), which may decrease the probability of eliciting a mutation-specific T cell response. We thus first set out to determine whether tumor-infiltrating lymphocytes (TIL) recognizing patient-specific mutations can be identified in patients with metastatic gastrointestinal (GI) cancers.
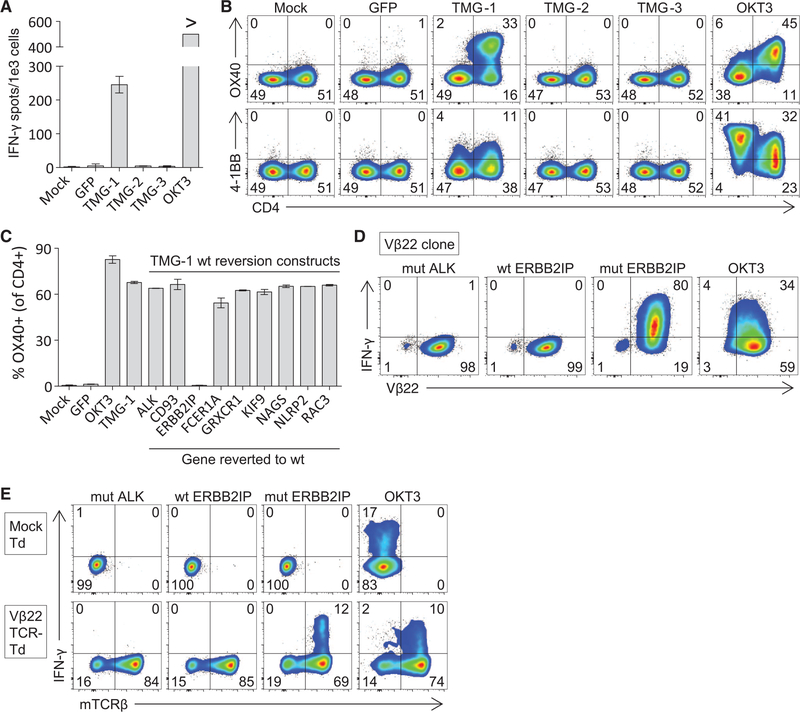
To this end, a 43-year-old woman with widely metastatic cholangiocarcinoma (patient 3737, table S1) who progressed through multiple chemotherapy regimens was enrolled in a TIL-based ACT protocol for patients with GI cancers () (13). Lung metastases were resected and used as a source for whole-exomic sequencing and generation of T cells for treatment. Whole-exomic sequencing revealed 26 nonsynonymous mutations (table S2). To test whether the patient’s TIL recognized any of these mutations, we used a minigene approach. Briefly, for each mutation we designed a minigene construct that encoded for the mutated amino acid flanked on each side by 12 amino acids from the endogenous protein (fig. S1). Multiple minigenes were synthesized in tandem to generate tandem minigene (TMG) constructs (fig. S1 and table S3), which were then used as templates for the generation of in vitro transcribed (IVT) RNA (13). Each of these IVT TMG RNAs was then individually transfected into autologous antigen-presenting cells, followed by a coculture with TIL to determine whether any of the processed and presented mutated antigens were recognized by TIL. We observed reactivity of 3737-TIL to a mutated antigen present in TMG-1 but not TMG-2 or TMG-3 (Fig. 1A). Moreover, the reactivity predominated in the CD4+ T cell population, as demonstrated by up-regulation of the activation markers OX40 and 4–1BB (Fig. 1B). Although some 4–1BB up-regulation was observed in the CD4-negative (CD8+) T cell population, we sorted these cells and found no reactivity against the TMG. To determine which of the nine mutations in TMG-1 was being recognized by 3737-TIL, we synthesized nine additional TMG-1 constructs, each one containing a reversion of one of the mutations back to the wild-type sequence. Reactivity of 3737-TIL to TMG-1 was abrogated only when the erbb2 interacting protein (ERBB2IP) mutation was reverted back to the wild-type sequence, indicating that the TIL specifically recognized the ERBB2IPE805G mutation (Fig. 1C). The response was restricted by HLA-DQB1*0601, and the minimal epitope was located within the 13–amino acid sequence NSKEETGHLENGN (where E is Glu; G, Gly; H, His; K, Lys; L, Leu; N, Asn; S, Ser; and T, Thr) (fig. S2).
Fig. 1. Patient 3737–TIL harbor ERBB2IP mutation–specific CD4+ T cells.
(A to C) Patient 3737–TIL were cocultured with dendritic cells (DCs) transfected with nothing (Mock), green fluorescent protein (GFP) RNA, or the indicated TMG construct encoding the various mutations identified by whole-exomic sequencing.(A) IFN-γ enzyme-linked immunosorbent spot assay at 20 hours. “>“ denotes greater than 500 spots per 1e3 cells. (B and C) Flow-cytometric analysis at 24 hours. Data are gated on live CD3+ cells and, for (C), further gated on CD4+ cells. (D) A CD4+ Vβ22+ clone was co-cultured for 6 hours with autologous B cells pulsed overnight with wild-type (wt) ERBB2IP, mutated (mut) anaplastic lymphoma receptor tyrosine kinase (ALK), or mut ERBB2IP 25-amino acid- long peptides. Flow cytometry was used to detect intracellular IFN-γ production in the CD4+ population. Data are representative of two clones sharing the same TCR-Vβ sequence. (E) Autologous open-repertoire peripheral blood T cells were transduced (Td) with nothing (Mock) or the TCR derived from the Vβ22+ clone and then assessed for reactivity as described in (D). The endogenous Vβ22+ TCR constant regions were swapped with mouse constant regions, allowing for the detection of the introduced TCR using antibodies against the mouse TCRβ constant region (mTCRβ). Plate-bound OKT3 was used as a control in all assays. All data are representative of at least two independent experiments. Error bars are SD.
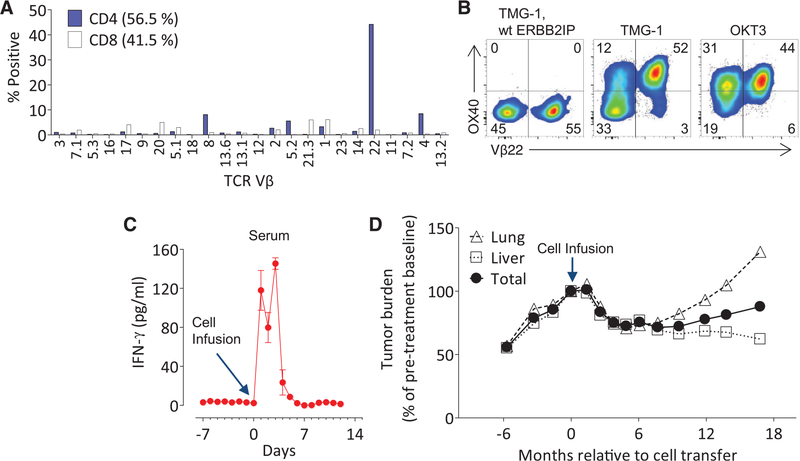
We next characterized the clonality of the mutated ERBB2IP-specific CD4+ T cells by sorting them after antigen-specific activation, using OX40 as a marker of activation (fig. S3A). These cells were then bulk-expanded and cloned by limiting dilution. A flow cytometry–based survey of the T cell receptor–Vβ (TCR-Vβ) repertoire demonstrated that the bulk-expanded population was >95% Vβ22+ and that 10/11 clones assessed were purely Vβ22+ (fig. S3, B and C). TCR sequence analysis revealed the same TCRβ variable, diversity, joining (V-D-J) sequence in 6/6 Vβ22+ clones tested (table S4), suggesting that the majority of the ERBB2IP mutation–reactive T cells was composed of a dominant Vβ22+ T cell clone. T cell clones expressing this Vβ22 TCR specifically produced the cytokine interferon (IFN)–γ upon stimulation with the mutated ERBB2IP peptide (Fig. 1D). Moreover, autologous open repertoire peripheral blood T cells genetically modified with this TCR-Vβ22 chain matched with its alpha chain (table S5) conferred specific reactivity to the mutated ERBB2IP peptide (Fig. 1E), demonstrating that this TCR specifically recognized the ERBB2IPE805G mutation.
The patient underwent adoptive transfer of 42.4 billion TIL containing CD4+ ERBB2IP mutation-reactive T cells followed by four doses of interleukin (IL)–2 to enhance T cell proliferation and function (fig. S4). Flow cytometry analysis demonstrated that about 25% of the entire 3737-TIL product administered was composed of the Vβ22+, mutation-reactive T cells (Fig. 2, A and B), equating to the infusion of over 10 billion ERBB2IP mutation–specific CD4+ T cells. Elevated levels of IFN-γ were detected in the patient’s serum for the first 5 days after cell infusion (Fig. 2C). Although the patient had clear evidence of progressive disease before the cell infusion, tumor regression was observed by the 2-month follow-up, and all target lung and liver lesions continued to regress, reaching a maximum reduction of 30% at 7 months post-treatment (Fig. 2D). The patient experienced disease stabilization for about 13 months after cell infusion, after which disease progression was observed only in the lungs but not liver.
Fig. 2. Adoptive transfer ofTILcontainingERBB2IP mutation–reactive T cells.
(A) Flow-cytometric analysis of the TCR-Vβ repertoire of 3737-TIL, gated on live CD4+ or CD8+ T cells. (B) Patient 3737–TIL were cocultured with DCs transfected with TMG-1 or TMG-1 encoding the wt ERBB2IP reversion, and flow cytometry was used to assess OX40 and Vβ22 expression on CD4+ T cells at 24 hours post-stimulation. Plate-bound OKT3 stimulation was used as a positive control. (A) and (B) are representative of at least two independent experiments. (C) IFN-γ enzyme-linked immunosorbent assay on patient 3737 serum samples pre- and post-adoptive cell transfer of 3737-TIL. Error bars are SEM. (D) Tumor growth curves [Response Evaluation Criteria in Solid Tumors (RECIST), sum of maximum diameters] before and after infusion of 3737-TIL. Data are expressed as a change in percent from pretreatment baseline and stratified on lung, liver, and total tumors.
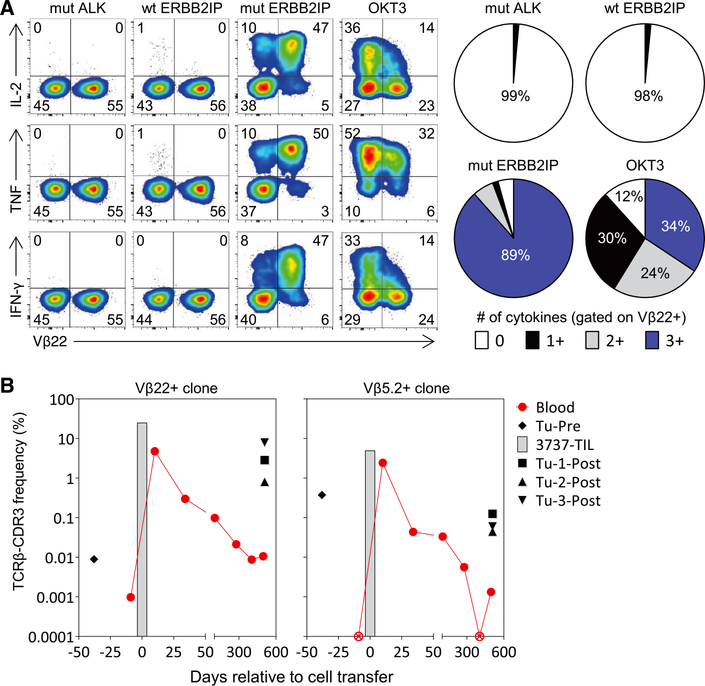
To determine whether there was evidence that the CD4+ ERBB2IP mutation–reactive T cells played a role in the disease stabilization, we evaluated the in vitro phenotype and function and in vivo persistence of these cells. A number of pre-clinical studies (14–20) and one case report in melanoma (21) have demonstrated that T helper 1 (TH1) cells, through a variety of mechanisms including the secretion of the pleiotropic cytokines IFN-γ and tumor necrosis factor (TNF) (22, 23), can mediate tumor regression. Moreover, CD4+ T cells that can simultaneously produce multiple effector cytokines (polyfunctional T cells) have been correlated with effective antitumor T cell responses (19, 24). We found that the Vβ22+ ERBB2IP mutation–reactive CD4+ T cells were polyfunctional TH1 cells, because stimulation with the mutated ERBB2IP peptide induced the robust coexpression of IFN-γ, TNF, and IL-2 (Fig. 3A) but little to no IL-4 or IL-17. Further phenotypic characterization revealed that these cells were predominantly effector memory CD4+ T cells with cytolytic potential (fig. S5, A and B).
Fig. 3. Functional phenotype and persistence of ERBB2IP mutation–specific CD4+ T cells.
(A) Patient 3737–TIL were cocultured for 6 hours with autologous B cells pulsed overnight with wt ERBB2IP, mut ALK, or mut ERBB2IP 25–amino acid–long peptides. Flow cytometry was used to assess expression of Vβ22 and to detect intracellular production of IL-2, TNF, and IFN-γ in the CD4+ population. Pie charts display the percentage of Vβ22+ cells that expressed the indicated number of cytokines. Data are representative of at least three independent experiments. (B) TCR-Vβ deep sequencing of 3737-TIL and blood and tumors of patient 3737 at various times pre- and post-adoptive cell transfer with 3737-TIL. Data show the frequency of the two ERBB2IP mutation–specific TCRβ-CDR3 clonotypes. ⊗, not detected.
There also appeared to be a minor population of polyfunctional Vβ22-negative, ERBB2IP mutation–reactive CD4+ Tcells present in 3737-TIL (Figs. 2B and 3A). Sorting of the Vβ22-negative cells followed by activation of these cells revealed that one or more additional clonotypes reactive to this epitope were present in 3737-TIL (fig. S6, A and B), the most dominant clonotype of which was Vb5.2 (fig. S6, C and D). The majority of the Vβ5.2+ cells produced multiple cytokines in an antigen-specific manner (fig. S6E and table S6). There also appeared to be a minor population of Vβ5.2-negative (and Vβ22-negative) CD4+ T cells that recognized mutated ERBB2IP (fig. S6E). Thus, the TIL used to treat patient 3737 contained at least three different polyfunctional CD4+ Tcell clones that recognized the same mutation in ERBB2IP, suggesting that this mutation was highly immunogenic.
The in vivo persistence of adoptively transferred T cells in the blood at 1 month post–cell transfer is correlated with improved outcomes in melanoma patients treated with ACT (25). We thus evaluated the in vivo persistence of the ERBB2IP mutation–reactive CD4+ T cell clones in patient 3737. TCR-Vβ deep sequencing revealed that these clonotypes were rare or not detectable in the peripheral blood before ACT (Fig. 3B). Ten days after ACT, both clones were present at greater than 2% of the total T cells in the peripheral blood, but declined to less than 0.3% by day 34 post–cell infusion (Fig. 3B). Three lung metastases, which were resected nearly a year and a half after ACT, were infiltrated by the ERBB2IP mutation–reactive T cells (Fig. 3B), suggesting that these cells contributed to the cancer regression and disease stabilization. The Vβ22+ ERBB2IP mutation–reactive clone was the most frequent clone detected in tumor nodule–3 (Tu-3-Post) and represented nearly 8% of total T cells in the tumor (Fig. 3B), whereas this clone was the second and twelfth most frequent in tumor nodules−1 and −2, respectively. The Vβ5.2+ ERBB2IP mutation- reactive clone was also enriched compared with its frequency in blood in all three tumor nodules (Fig. 3B). Thus, patient 3737 experienced tumor regression with stabilization of disease for more than 1 year after receiving over 10 billion ERBB2IP mutation–specific polyfunctional TH1 cells that infiltrated and persisted in the metastatic lesions.
We observed relatively high levels of ERBB2IP expression in both the original and the recurrent lung lesions as determined by quantitative reverse transcription polymerase chain reaction (fig. S7A), and Sanger sequencing validated the presence of the ERBB2IP mutation in all tumor lesions (fig. S7B). The T cell infiltrate and major histocompatibility complex expression of the tumors in situ are summarized in tables S7 and S8, respectively. The presence of T cells reactive to the ERBB2IP mutation in progressing tumors that expressed this mutation suggests the presence of immunosuppressive influences at the tumor site that may need to be overcome to increase the anti-cancer effects of the transferred cells.
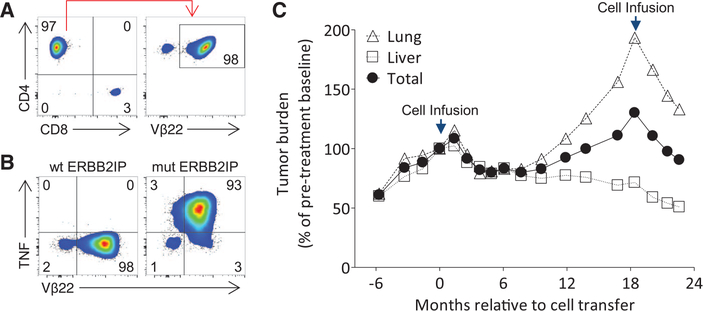
To specifically evaluate the contribution of mutation-reactive TH1 cells to the antitumor response in vivo, we generated and adoptively transferred a TIL product that was comprised of >95% of the Vβ22+ ERBB2IP mutation–reactive TH1 cells (Fig. 4, A and B) (13). Again, the patient experienced a decrease in target lesions, but, unlike the first treatment, tumor regression was observed even at the first-month follow-up and continues as of the last follow-up at 6 months (Fig. 4C).
Fig. 4. Evidence of tumor regression after treatment with a highly pure population of Vβ22+ ERBB2IP mutation–reactive CD4+ T cells.
(A) Flow-cytometric analysis of the TIL product used for retreatment. Data are gated on live CD3+ T cells (left) and further gated on CD4+ cells (right).(B) Retreatment TIL were cocultured for 6 hours with autologous B cells pulsed overnight with wt or mut ERBB2IP 25–amino acid–long peptides. Flow cytometry was used to detect intracellular TNF production in the CD4+ population. (A) and (B) are representative of at least two independent experiments. (C) Tumor growth curves (sum of maximum diameters) of patient 3737 before and after first and second adoptive cell transfers. The first infusion product consisted of 42.4 billion T cells containing about 25% (10 billion) Vβ22+ ERBB2IP mutation–reactive TH1 cells. The second infusion product consisted of 126 billion T cells containing over 95% (120 billion) Vβ22+ ERBB2IP mutation–reactive TH1 cells. Data are expressed as a change from baseline and stratified on lung, liver, and total tumors. Some target lesions selected in (C) differ from Fig. 2D because only lesions that were present throughout both treatments were selected for measurement.
Mapping of the mutational landscape of human cancers is occurring at a rapid pace, but strategies to exploit this information for clinical benefit remain to be fully realized. Here, we used a whole-exome sequencing approach to demonstrate that the human immune system can mount an endogenous TH1 response against a mutation expressed by an epithelial cancer, and we provide evidence that this response can be harnessed for therapeutic benefit. Given that a major hurdle for the success of immunotherapies for GI and other cancers is the apparent low frequency of tumor-reactive T cells (26), the strategies reported here could be used to generate a T cell product that is highly enriched in mutation-reactive T cells for use in ACT. The ability to immunologically target unique mutations in cancers can potentially extend highly personalized immunotherapies to patients with epithelial cancers, which account for about 90% of cancer deaths in the United States.
Supplementary Material
Acknowledgments:
We thank the Milstein Family Foundation for their generous support; C.-C. R. Lee, Y. F. Li, A. Mixon,S. Farid, and J. Gartner for helpful technical support and reagents; and the Surgery Branch clinical team for outstanding patient care. The data presented in this manuscript are tabulated in the main paper and the supplementary materials. The raw whole-exome sequence data are available on the Sequence Read Archive database: Bioproject PRJNA243084. This research was supported by the Intramural Research Program of the NIH and NCI.
Footnotes
Supplementary Materials
References and Notes
- 1.Vogelstein B et al. , Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Bruggen P, Stroobant V, Vigneron N,Van den Eynde B, “Tumor antigens resulting from mutations,” Cancer Immunity (2013), http://cancerimmunity.org/peptide/mutations/. [PMC free article] [PubMed] [Google Scholar]
- 3.Robbins PF et al. , Nat. Med 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Rooij N et al. , J. Clin. Oncol 31, e439–e442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu YC et al. , J. Immunol 190, 6034–6042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbière V et al. , Cancer Res. 71, 1253–1262 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Huang J et al. , J. Immunol 172, 6057–6064 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echchakir H et al. , Cancer Res. 61, 4078–4083 (2001). [PubMed] [Google Scholar]
- 9.Mandruzzato S, Brasseur F, Andry G, Boon T,van der Bruggen P, J. Exp. Med 186, 785–793 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogan KT et al. , Cancer Res. 58, 5144–5150 (1998). [PubMed] [Google Scholar]
- 11.Karanikas V et al. , Cancer Res. 61, 3718–3724 (2001). [PubMed] [Google Scholar]
- 12.Wedén S et al. , Int. J. Cancer 128, 1120–1128 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Materials and methods are available as supplementary materials on Science Online.
- 14.Mumberg D et al. , Proc. Natl. Acad. Sci. U.S.A. 96, 8633–8638 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corthay A et al. , Immunity 22, 371–383 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Quezada SA et al. , J. Exp. Med 207, 637–650 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Y et al. , J. Exp. Med 207, 651–667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg PD, Kern DE, Cheever MA, J. Exp. Med 161, 1122–1134 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding ZC et al. , Blood 120, 2229–2239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shafer-Weaver KA et al. , Cancer Res. 69, 6256–6264 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Hunder NN et al. , N. Engl. J. Med 358, 2698–2703 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braumüller H et al. , Nature 494, 361–365 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Qin Z, Blankenstein T, Immunity 12, 677–686 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Yuan J et al. , Proc. Natl. Acad. Sci. U.S.A. 105, 20410–20415 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg SA et al. , Clin. Cancer Res. 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turcotte S et al. , J. Immunol 191, 2217–2225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.