Abstract
Background
Flavonoid 3′-hydroxlase (F3’H) is an important enzyme in determining the B-ring hydroxylation pattern of flavonoids. In monocots, previous studies indicated the presence of two groups of F3’Hs with different enzyme activities. One F3’H in rice was found to display novel chrysoeriol-specific 5′-hydroxylase activity. However, the evolutionary history of monocot F3’Hs and the molecular basis for the observed catalytic difference remained elusive.
Results
We performed genome-wide survey of 12 common monocot plants, and identified a total of 44 putative F3’H genes. The results showed that F3’H gene family had underwent volatile lineage-specific gene duplication and gene loss events in monocots. The expansion of F3’H gene family was mainly attributed to dispersed gene duplication. Phylogenetic analyses showed that monocot F3’Hs have evolved into two independent lineages (Class I and Class II) after gene duplication in the common ancestor of monocot plants. Evolutionary dynamics analyses had detected positive natural selection in Class II F3’Hs, acting on 7 specific amino acid sites. Protein modelling showed these selected sites were mainly located in the catalytic cavity of F3’H. Sequence alignment revealed that Class I and Class II F3’Hs displayed amino acid substitutions at two critical sites previously found to be responsible for F3’H and flavonoid 3′5’-hydroxylase (F3’5’H) activities. In addition, transcriptional divergence was also observed for Class I and Class II F3’Hs in four monocot species.
Conclusions
We concluded that monocot F3’Hs have evolved into two independent lineages (Mono_F3’H Class I and Class II), after gene duplication during the common ancestor of monocot plants. The functional divergence of monocot F3’H Class II has been affected by positive natural selection, which acted on specific amino acid sites only. Critical amino acid sites have been identified to have high possibility to affect the substrate specificity of Class II F3’Hs. Our study provided an evolutionary and protein structural explanation to the previously observed chrysoeriol-specific 5′-hydroxylation activity for CYP75B4 in rice, which may also be true for other Class II F3’Hs in monocots. Our study presented clear evidence of plant-environmental interaction at the gene evolutionary level, and would guide future functional characterization of F3’Hs in cereal plants.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1947-z) contains supplementary material, which is available to authorized users.
Keywords: Flavonoids, Flavonoid 3′-hydroxylase, Anthocyanin, Cereal crops, Evolutionary dynamics, Gene evolution, Positive natural selection, Plant environmental interactions
Background
Flavonoids including anthocyanins, flavones and flavonols are ubiquitous secondary metabolites present in all organs and tissues of plants [1, 2]. During the past few decades, enormous research attention has been drawn toward their biological functions in monocot cereal crops [1, 3, 4], such as wheat, barley, rice and maize, which are the major sources of human food. From the biological perspective, flavonoid biosynthesis plays important roles in plant’s defence mechanism to various abiotic and biotic stress factors including UV-radiation, heat, heavy metal ions, drought, pathogen and microbial invasion et al. [5–7]. Flavonoid pigments in flower and seed are visible signals to attract insects and animals for pollination and seed dispersal [8, 9]. In addition, flavonoids have been shown to be involved in pollen germination [10, 11], and could also function as developmental regulators in auxin transport and catabolism [12, 13]. Flavonoids such as anthocyanin accumulation in cereal grains has been shown to affect seed dormancy and prevent preharvest sprouting [3, 14], which assists plant’s survival in unfavourable environmental conditions. From the food consumption perspective, flavonoid compounds, due to their antioxidant properties, also have demonstrated great health benefits in the protection of degenerative diseases such as coronary heart disease and cancer [15–17].
The molecular mechanisms of flavonoid biosynthesis has been well established in monocot plants [1]. As the starting point, phenylalanine was transformed via the phenylpropanoid pathway into 4-coumaroyl-CoA, which then enters the flavonoid biosynthesis pathway [18]. Chalcone synthase (CHS) and chalcone isomerase (CHI) are the first two enzymes in the flavonoid pathway, leading to the sequential production of chalcones and naringenin, which act as the precursors for all flavonoid classes [19]. Based on the hydroxylation pattern of the flavonoid B-ring, flavonoid biosynthesis can diverge into three different directions, resulting in the final production of one-hydroxy (pelargonidin-type), two-hydroxy (cyaniding-type) and three-hydroxy (delphinidin-type) anthocyanins [20]. The hydroxylation pattern of the flavonoid B-ring is controlled by two key enzymes flavonoid 3-hydroxylase (F3’H) and flavonoid 3′5’-hydroxylase (F3’5’H). F3’H belongs to the CYP75B subfamily in the cytochrome P45O-dependent monooxygenase superfamily, while F3’5’H belongs to the CYP75A subfamily and represents a lateral functional divergence from F3’H [21]. In the flavonoid pathway, F3’H catalyses the hydroxylation of naringenin and dihydrokampferol at the 3′-position, leading to the final production of cyanidin-based anthocyanins. Instead, F3’5’H is able to hydroxylate the flavonoid B-ring at both 3′ and 5′ position, which is responsible for the delphinidin-based anthocyanin production. The F3’H activity, together with the F3’5’H activity, compete with the central flavonoid pathway without hydroxylation, and have led to the great diversification of the flavonoid biosynthesis pathway [20]. This metabolic diversification has been suggested to play a critical role in plant’s adaption to the diverse environmental conditions during evolution.
Gene duplication is a widespread phenomenon in plant genomes. It generates the raw genetic material for environmental selection to act upon, playing a central role in plant diversification, thus facilitating their environmental adaptation [22, 23]. Following duplication, the gene duplicates could be either lost or retained, depending on whether beneficial function could arise or not at either the protein structural level and/or the gene transcriptional level. The retention of species-specific gene copy number has often been proposed to assist different plants to meet their specific environmental challenges. A recent study reported that 2 copies of F3’Hs (F3’H-1 & F3’H-2) were present in barley genome, which were resulted from a duplication event before the divergence of Triticeae tribe [24]. A tissue-specific expression profile was also observed for F3’H-1 and F3’H-2. In another earlier study, 3 and 2 copies of F3’Hs has been identified in barley and rice, respectively [25]. Interestingly, one of the two F3’Hs in rice (CYP75B4) was proven to have recruited novel 5′-hydroxlase activity on chrysoeriol, which comprised a critical step in tricin biosynthesis [25]. Preliminary phylogeny analysis showed CYP75B4 belonged to an independent phylogenetic group divergent from the normal monocot F3’Hs. CYP75B3 and CYP75B4 in rice were also shown to have different substrate specificity [26]. These observations suggested a potential functional divergence among monocot F3’Hs. However, no systematic and comprehensive evolution analyses have been performed on F3’H gene family in monocot. The protein structural basis underlying the 5′-hydroxylase activity of CYP75B4 remained to be characterised.
In this study, we investigated the conservation of putative F3’H genes in the major cereal plants, for which the genomic data is available in the public databases. The evolutionary history of F3’H genes in monocot plants was characterized by comprehensive phylogenetic and natural selection analyses. We found clear evidence of plant-environmental interaction during the evolution of monocot F3’H gene subfamily. Our study consolidated the flavonoid biosynthesis pathway as a model to investigate plant and environment interaction, and would also serve as a guide for future functional study on the F3’H gene subfamily in monocot plants.
Results
Identification of F3’H in monocot plants
To identify the genuine F3’H genes, a comprehensive Neighbour Joining tree was developed based on the amino acid sequence alignment of the retrieved F3’H homologs. Two distinct branches encompassing the previously characterised F3’Hs and F3’5’Hs, respectively, were identified. Those homologous proteins in the F3’H branch were considered genuine F3’Hs and were selected for further analyses. As summarised in Table 1, a total of 44 putative F3’H genes were identified from 12 monocots. At least two copies of F3’H were present in each species. The highest number of F3’H occurred in Triticum aestivum (9), followed by Triticum dicoccoides (7) in the Triticeae subfamily. All the other Triticeae crops including Hordeum vulgare, Aegilops tauschii and Secale cereale contain 3 copies of F3’H genes, with the exception of Triticum urartu, which has 2 copies instead. Brachypodium distachyon, a close relative to Triticeae, contained 4 copies of F3’Hs. In addition, most plants in the Panicoideae lineage, including Setaria italica and Panicum hallii, retain 2 F3’Hs, whilst Zea mays and Sorhum bicolor have exceptionally 3 and 5 copies, respectively. Notably, the other important crop Oryta sativa also contained 2 F3’Hs.
Table 1.
Identification of putative F3’H genes in cereal plants. The F3’H class was classified based on the phylogeny analyses. The gene duplication pattern was determined using the MCScanX tool. NA stands for “not applicable”
| Species | Chr | F3’H gene ID | F3’H class | Duplication pattern | Start position | End position | |
|---|---|---|---|---|---|---|---|
| Triticeae | H. vulgare | 1H | HORVU1Hr1G094880 | Class I | Dispersed duplication | 556,691,949 | 556,693,904 |
| 6H | HORVU6Hr1G002400 | Class II | Dispersed duplication | 6,328,532 | 6,330,799 | ||
| 7H | HORVU7Hr1G095900.47 | Class II | Dispersed duplication | 585,061,615 | 585,071,468 | ||
| A. tauschiii | Aet1 | AET1Gv21041800 | Class I | Dispersed duplication | 499,122,364 | 499,126,756 | |
| Aet6 | AET6Gv20027200 | Class II | Dispersed duplication | 5,657,246 | 5,659,238 | ||
| Aet7 | AET7Gv20999800 | Class II | Dispersed duplication | 523,485,446 | 523,487,617 | ||
| T. turgidum | Tt1A | TRIDC1AG064860 | Class I | NA | 590,437,663 | 590,440,408 | |
| Tt1B | TRIDC1BG074020 | Class I | 685,939,174 | 685,944,027 | |||
| Tt2B | TRIDC2BG088640 | Class I | 792,785,227 | 792,787,184 | |||
| Tt6A | TRIDC6AG001340 | Class II | 4,624,395 | 4,644,086 | |||
| Tt6B | TRIDC6BG002010 | Class II | 11,399,578 | 11,402,254 | |||
| Tt7A | TRIDC7AG057400 | Class II | 599,327,799 | 599,328,465 | |||
| Tt7B | TRIDC7BG049820 | Class II | 562,791,603 | 562,792,604 | |||
| T. aestivum | Ta1A | TraesCS1A01G442300.1 | Class I | NA | 590,995,642 | 590,997,413 | |
| Ta1B | TraesCS1B01G476400.1 | Class I | 685,231,562 | 685,233,491 | |||
| Ta1D | TraesCS1D01G450100.1 | Class I | 492,534,241 | 492,538,011 | |||
| Ta2B | TraesCS2B01G613200.1 | Class I | 792,677,476 | 792,679,409 | |||
| Ta6A | TraesCS6A01G012600.1 | Class II | 5,861,572 | 5,863,417 | |||
| Ta6B | TraesCS6B01G018800.1 | Class I | 11,574,703 | 11,578,097 | |||
| Ta6D | TraesCS6D01G015200.1 | Class II | 6,319,419 | 6,321,226 | |||
| Ta7A | TraesCS7A01G411700.1 | Class II | 602,804,667 | 602,806,415 | |||
| Ta7D | TraesCS7D01G404900.1 | Class II | 522,502,518 | 522,504,208 | |||
| S. cereale | Sc1 | Sc1Loc01465431 | Class I | NA | Lo7_v2_contig_2871825 | ||
| Sc2 | Sc2Loc01684522 | Class I | Lo7_v2_contig_326626 | ||||
| Sc7 | Sc7Loc01952123 | Class II | Lo7_v2_contig_61986 | ||||
| T. urartu | Tu1 | TuG1812G0100004862 | Class I | Dispersed duplication | 581,402,997 | 581,404,997 | |
| Tu7 | TuG1812G0700004460 | Class II | Dispersed duplication | 590,301,744 | 590,303,739 | ||
| B. distachyon | Bd1 | Bradi1g17180 | Class I | Dispersed duplication | 13,787,434 | 13,789,806 | |
| Bd1 | Bradi1g24840 | Class II | Dispersed duplication | 20,108,563 | 20,112,348 | ||
| Bd3 | Bradi3g04750 | Class I | Dispersed duplication | 3,260,666 | 3,262,706 | ||
| Bd4 | Bradi4g16560 | Class II | Dispersed duplication | 17,368,956 | 17,372,786 | ||
| O. sativa indica | Os10 | LOC_Os10g17260 | Class I | Proximal duplication | 8,679,309 | 8,681,284 | |
| Os10 | LOC_Os10g16974 | Class II | Proximal duplication | 8,494,247 | 8,504,329 | ||
| Panicoideae | Z. mays | Zm4 | Zm00008a016611 | Class I | WGD/Segmental | 131,908,959 | 131,910,431 |
| Zm5 | Zm00008a022212 | Class I | WGD/Segmental | 177,289,973 | 177,292,210 | ||
| Zm8 | Zm00008a031477 | Class II | Dispersed duplication | 116,235,840 | 116,239,418 | ||
| S. bicolor | Sb4 | Sobic.004G200800 | Class I | Tandem duplication | 55,221,098 | 55,224,686 | |
| Sb4 | Sobic.004G200833 | Class I | Tandem duplication | 55,225,513 | 55,227,179 | ||
| Sb4 | Sobic.004G200900 | Class I | Proximal duplication | 55,233,582 | 55,236,702 | ||
| Sb4 | Sobic.004G201100 | Class I | Proximal duplication | 55,261,682 | 55,264,545 | ||
| Sb9 | Sobic.009G162500 | Class II | Dispersed duplication | 51,943,204 | 51,948,939 | ||
| S. italica | Si9 | Seita.9G244600 | Class II | Dispersed duplication | 19,091,837 | 19,094,929 | |
| Si9 | Seita.9G242900 | Class I | Dispersed duplication | 18,990,913 | 18,992,801 | ||
| P. hallii | Ph8 | Pahal.H01052 | Class II | Dispersed duplication | 31,682,928 | 31,687,178 | |
| Ph9 | Pahal.I03232 | Class I | Dispersed duplication | 16,748,267 | 16,750,212 | ||
Phylogeny inference
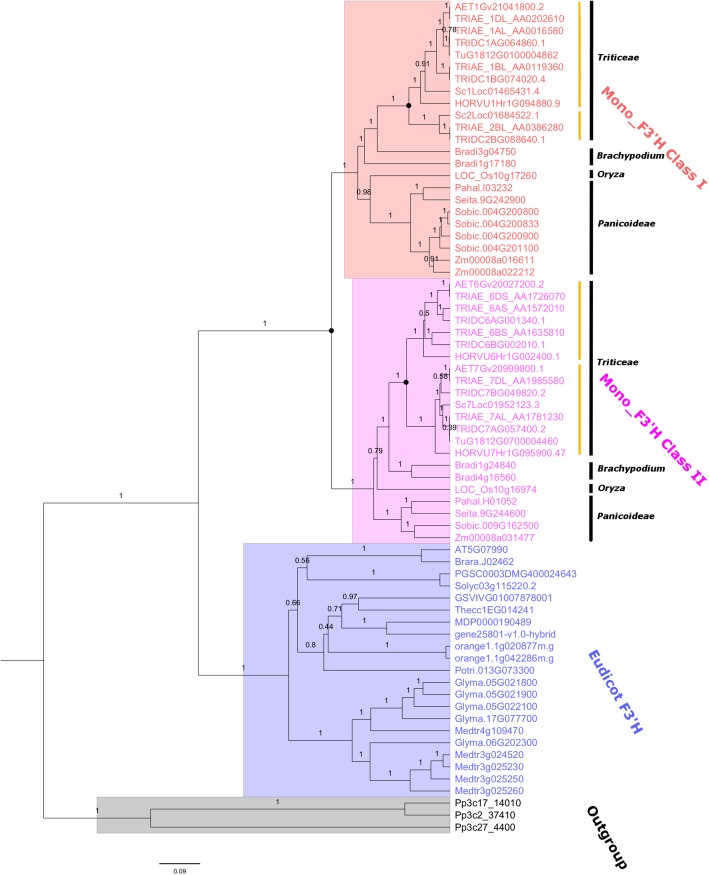
To investigate the evolutionary history of the F3’H gene family in monocot plants, a Bayesian phylogeny was developed based on the coding domain sequence (CDS) alignment of the identified F3’Hs. Eudicot F3’Hs and the remote F3’H homologs from the lower plant Physcomitrella patens were included as the out-group. Overall, the phylogenetic tree demonstrated a strong topology support, indicating the resolved phylogeny was highly reliable. As shown in Fig. 1, the target F3’Hs were grouped into two major clusters, corresponding to eudicot F3’H and monocot F3’H, respectively. The monocot F3’Hs further separated into two distinct lineages, which were classified here as Mono_F3’H Class I and Class II, respectively. Noteworthy, each of Mono_F3’H Class I and Class II covered all the monocot species included in the present study, suggesting the divergence occurred before the species diversification. A closer inspection on the phylogeny showed that these two lineages shared the same evolutionary pattern that resembles the species phylogeny of monocot plants. Specifically, within both Mono_F3’H Class I and Class II, Panicoideae plants including Z. mays, S. bicolor and S. italica diverged firstly, followed by O. sativa, which represents an evolutionary intermediate between Panicoideae and Triticeae. For the other plants, B. distachyon diverged before Triticeae. F3’Hs retrieved from Triticeae plants were clustered together with strong support. These results indicated that the Class I and Class II F3’Hs have evolved vertically within monocot plants, providing further support that the divergence between Class I and Class II F3’Hs have occurred during the common ancestor of monocot crops. The universal conservation of Class I and Class II F3’Hs among monocots indicated that both F3’H classes are essential for the normal growth of these plants.
Fig. 1.
Phylogenetic tree displaying the evolutionary history of plant F3’Hs. The phylogeny was developed using Bayesian method. F3’H homologs from P. patens was included as outgroup. Posterior support was displayed above each branch. The deduced duplication event was indicated by solid black dot symbol. Yellow vertical lines indicated the division of sub-branches of F3’Hs in Triticeae
The evolution of F3’Hs in monocots displayed a clear Class- and species-specific profile. Whilst only a single copy of both Class I and Class II F3’Hs were conserved within Panicoideae plants S. italic and P. hallii, S. bicolor and Z. mays contained 4 and 2 copies of Class I F3’Hs, respectively. In addition, lineage-specific expansion of F3’H were also observed in B. distachyon, which has 2 copies of both Class I and Class II F3’Hs. O. sativa resembled S. italic and P. hallii with one Class I and one Class II F3’H. For Triticeae, the evolution of F3’H seems to be independent from the above plants. Within the Mono_F3’H Class II lineage, Triticeae F3’Hs could be further divided into two sub-branches (yellow line; Fig. 1), which covered all the Triticeae plants included in this study. This indicated that Class II F3’Hs have underwent an extra round of duplication during the common ancestor of Triticeae, but after the divergence of B. distachyon. Noteworthy, a similar expansion pattern could be observed for Triticieae Class I F3’Hs, which had also evolved into two distinct sub-branches (yellow line; Fig. 1), indicating a duplication event predating the Triticeae diversification as well. One of the two sub-branches of Class I F3’H (the upper yellow line) covered all Triticeae plants, whilst the other is preserved only in T. turgidum, T. aestivum and S. cereale, but absent in H. vulgare, A. tauschii. The absence of the secondary sub-branch might be due to gene loss after duplication. Noteworthy, the identified duplication events for Class I and Class II F3’Hs in Triticeae may point to a shared genome-wide duplication event in the common ancestor of Triticeae. Further investigation is needed to verify this hypothesis. In addition to Triticeae, Class I and Class II F3’Hs in B. distachyon tended to have evolved independently as separate lineages.
Genomic structure analyses
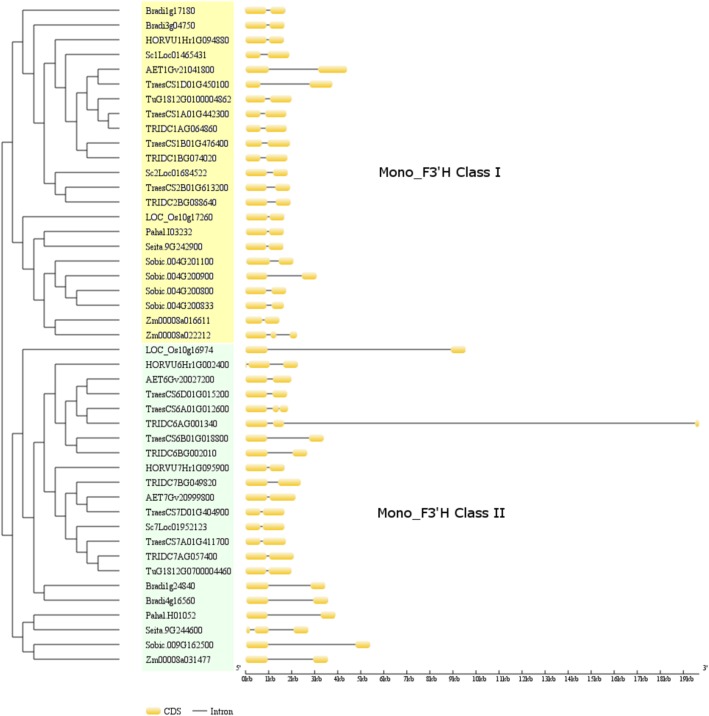
To characterise the gene structural profiles of F3’H and their potential relation with the evolution history of monocot F3’H gene subfamily, the gene structures of monocot F3’Hs were analysed based on the developed phylogeny. As shown in Fig. 2, majority (41/45) of monocot F3’Hs contained two exons, regardless of the phylogeny groups. The other four putative F3’Hs, corresponding to Zm00008a022212 in Class I and TraesCS6A01G012600, TRIDC6AG001340, Seita.9G244600 in Class II, retained three exons. The intron length tends to be conserved for most F3’Hs from different phylogeny branches, with the exception of non-Triticeae Class II F3’Hs, which displayed a clear and universal increase in intron length. In addition to the non-Triticeae Class II F3’Hs, several other F3’Hs from both Class I and Class II also showed an increase in intron length, which corresponded to AET1Gv21041800, TraesCS1D01G450100, Sobic.004G200900 (Class I) and TraesCS6B01G018800, TRIDC6BG002010 (Class II). Of these five genes, AET1Gv21041800/TraesCS1D01G450100 and TRIDC6BG002010/TraesCS6B01G018800 are close homolog pairs and may have reflected the origin of T. aestivum D and B subgenomes from A. tauschii and T. turgidum, respectively. It should be noted that the gene structure data presented here is based on the gene annotation in the plant genome databases. Laboratory gene cloning and sequencing in respective species are needed to further validate these results. We also refrained to compare the putative promoter regions including the 5’UTR and 3’UTR of F3’Hs due to the lack of experimental information.
Fig. 2.
Genomic structures of monocot F3’Hs. The F3’H genes were clustered based on the developed phylogeny in the present study. Two features: CDS and Intron were displayed for each F3’H based data extracted from public database
Duplication pattern and synteny analyses
Gene duplicates arising from different mechanisms can be divided into four categories: Whole genome duplication (WGD)/segmental duplication, tandem duplication, proximal duplication and dispersed duplication. To further investigate the evolutionary history of F3’H family, gene duplication pattern were determined for F3’Hs in 9 monocot species (Table 1). T. aestivum, T. turgidum and S. cereale were excluded for this analysis due to either their multi-ploidity or the lack of fine genome annotation information. As shown in Table 1, all of the F3’Hs in Triticeae plants were identified as dispersed duplicates. The same observation was made with the F3’Hs in B. distacyon (4 copies), all of which had arisen from dispersed gene duplication. F3’Hs in rice (2 copies), located close to each other on chromosome 10, were found to be proximal duplicates. Interestingly, 2 F3’Hs (Zm00008a016611, Zm00008a022212) from Z. mays were found to have originated from whole-genome or segmental duplication, reflecting a different evolution origin for F3’Hs in this species. The other F3’H (Zm00008a031477) in Z. mays was identified as a dispersed duplicate as well. In addition, 4 F3’Hs on chromosome 4 in S. bicolor were identified as tandem duplication or proximal duplication, which was quite unusual compared to other species. The other F3’H (Sobic.009G162500) in S. bicolor was found as a dispersed duplicate. Unlike Z. mays and S. bicolor, all of the F3’Hs from S. italic and P. hallii in the Panicoideae tribe were found to have resulted from dispersed duplication.
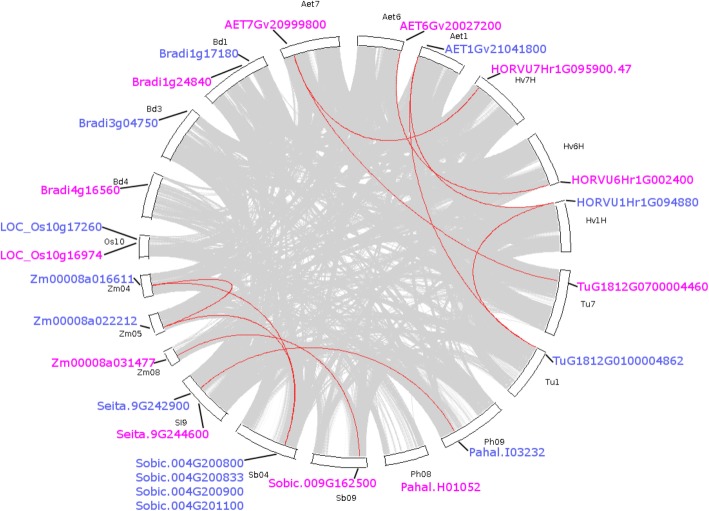
To investigate the syntenic conservation of F3’Hs across monocot plants, collinear F3’H gene pairs were identified. As shown in Fig. 3, a total of 12 collinear F3’H pairs have been identified for F3’Hs in 9 monocot species. These gene pairs could be divided into two clusters: Triticeae-specific and Panicoideae-specific. For Triticeae, F3’Hs located on chromosome Hv1H, Hv6H and Hv7H in H. vulgare were found to be collinear with F3’Hs on chromosome Aet1, Aet6 and Aet7 in A. tauschii, respectively. Noteworthy, TuG1812G0100004862 on chromosome Tu1 in T. urartu was located in a collinear region with HORVU1Hr1G094880 and AET1Gv21041800, TuG1812G0100004862 on chromosome Tu7 was collinear with AET7Gv20999800, which also formed collinear pair with HORVU7Hr1G095900.47. Interestingly, all these gene pairs occurred within the same class of F3’Hs. No inter-class F3’H pair had been observed in Triticeae. Taken together, the identified F3’H collinear pairs reflected the close relationship among Triticeae species and also demonstrated the strict conservation of F3’Hs in these plants. Noteworthy, no F3’H has been found on chromosome Tu6 in T. urartu, which may have been lost in this species.
Fig. 3.
Displays the collinear F3’H gene pairs across monocot plants. The circle plot was created by MCScanX tool. Identified colinear genes were linked by red curved lines. Hv, Bd, Aet, Tu, Os, Zm, SI, Sb and Ph represent H. vulgare, B. distachyon, A. tauschii, T. urartu, O. sativa, Z. mays, S. italica, S. bicolor and P. hallii, respectively. Only the chromosomes containing F3’H genes were included for this analysis. Class I and Class II F3’Hs were labelled in blue and pink colours, respectively
The other identified collinear F3’H pairs were found in Panicoideae only (Fig. 3). No collinearity could be identified for the F3’Hs in O. sativa and B. distacyon, which may reflect a divergent genomic location for the F3’Hs in these two species. For Panicoideae including Z. mays, S. italic, S. bicolor and P. hallii, collinearity was mainly found for Class II F3’Hs, with one intro-species pair (Zm00008a016611-Zm00008a022212) identified in Z. mays. In addition, only one collinear pair (Zm00008a031477-Sobic.009G162500) was found for Class I F3’Hs in Panicoideae. No collinearity could be observed for the Class I F3’Hs in S. italica (Seita.9G244600) and P. hallii (Pahal.H01052). Again, no inter-class collinearity could be identified between Class I and Class II F3’Hs in Panicoideae.
Natural selection test
To investigate the evolutionary dynamics in monocot F3’H gene family, natural selection tests were performed on the developed F3’H phylogeny. The ratio (ω) of non-synonymous and synonymous substitution is an important parameter to assess the selection pressure on evolving genes, whereby ω < 1, ω = 1 and ω > 1 indicate purifying selection, neutral evolution and positive selection, respectively. Branch and amino acid site specific ω values were calculated for Monocot F3’H Class I and Class II under different hypotheses. Eudicot F3’H was used as the reference. For the branch specific models, three hypotheses (Table 2) were tested. Likelihood-Ratio Tests (LRTs) showed that the two-ratio models ω[eudi] = ω[mono1] ≠ ω[mono2] and ω[eudi] = ω[mono2] ≠ ω[mono1], which specified divergent ω values for Monocot F3’H Class I and Class II, respectively, were both significantly better (df = 1, p < 0.0001; df = 1, p = 0.0444) than the one ratio model ω[eudi] = ω[mono1] = ω[mono2]. In addition, the three ratio model ω[eudi] ≠ ω[mono1] ≠ ω[mono2], specifying different ω values for all the three branches, fit the dataset significantly (df = 1, p < 0.0001) better than ω[eudi] = ω[mono2] ≠ ω[mono1], but not better (df = 1, p = 1.0) than ω[eudi] = ω[mono1] ≠ ω[mono2]. These calculations indicated that ω[mono2] was significantly different from ω[eudi] and ω[mono1], whilst ω[eudi] and ω[mono2] were not significantly different from one another. This suggested that Monocot Class II F3’Hs had underwent significantly different selection pressure compared to Monocot Class II F3’Hs and Eudicot F3’Hs. Under the best-fitting model ω[eudi] = ω[mono1] ≠ ω[mono2], ω[mono2] was calculated as 0.82808, while ω[eudi] and ω[mono1] equalled 0.11857. These calculations indicated that Monocot Class I F3’Hs and Eudicot F3’Hs were under strong purifying selection, whilst Monocot Class II F3’Hs were relatively more divergent.
Table 2.
Natural selection tests on plant F3’Hs. “np” stands for the number of parameters. ln(Likelihood) refers the log value of the likelihood
| Model | np | ln(Likelihood) | Estimates of parametersa (ω = dN: dS; P – percentage of site) |
Positively selected sitesb |
|---|---|---|---|---|
| One-ratio | ||||
| ω[eudi] = ω[mono1] = ω[mono2] | 1 | −29,872.87 | ω[eudi] = ω[mono1] = ω[mono2] = 0.12094 | Not Allowed (NA) |
| Branch-specific models | ||||
| ω[eudi] = ω[mono1] ≠ ω[mono2] | 2 | −29,862.42 |
ω[eudi] = ω[mono1] = 0.11857, ω[mono2] = 0.82808 |
NA |
| ω[eudi] = ω[mono2] ≠ ω[mono1] | 2 | −29,870.85 |
ω[eudi] = ω[mono2] = 0.11990, ω[mono1] = 0.43835 |
NA |
| ω[eudi] ≠ ω[mono1] ≠ ω[mono2] | 3 | − 29,862.42 | ω[eudi] = 0.11853, ω[mono2] = 0.78501, ω[mono1] = 0.12479 | NA |
| Site-specific models | ||||
| Neutral M1 (2 site classes) | 2 | −29,413.91 |
P0 = 0.87805 (P1 = 1-P0 = 0.12195); ω0 = 0.09334 (ω1 = 1- ω0 = 1.0) |
NA |
| Selection M1 (3 site classes | 3 | −29,413.91 |
P0 = 0.87805, P1 = 0.01421, P2 = 1-P0-P1 = 0.10774; ω0 = 0.09334 (ω1 = 1.0), ω2 = 1.0 |
NA |
| Branch-site models | ||||
| Model A Null (Class I F3’H) | 3 | −29,408.41 |
P0 = 0, P1 = 0.0, P2 + P3 = 1; ω0 = 0.09152, ω1 = 1.0, ω2 = 1.0 |
|
| Model A (Class I F3’H) | 4 | −29,408.41 |
P0 = 0.00005, P1 = 0.00001, P2 + P3 = 0.99994; ω0 = 0.09152, ω1 = 1.0, ω2 = 1.0 |
None |
| Model A Null (Class II F3’H) | 3 | −29,394.83 |
P0 = 0.51238, P1 = 0.07209, P2 + P3 = 0.41553; ω0 = 0.09046, ω1 = 1.0, ω2 = 1.0 |
NA |
| Model A (Class II F3’H) | 4 | −29,392.04 |
P0 = 0.68799, P1 = 0.09608, P2 + P3 = 0.21593; ω0 = 0.09035, ω1 = 1.0, ω2 = 8.19737 |
108R,222A, 265 V, 274 T, 355Q, 447S, 449 L (p < 0.05) |
aIn the site-specific model M1, two site classes were specified: highly conserved sites (ω0) and neutral sites (ω1 = 1). For the site-specific model M2, there were three site classes: highly conserved sites (ω0), neutral sites (ω1 = 1) and positively selected sites (ω2). In Model A, four site classes were specified. The first two classes had ω ratios of ω0 and ω1 respectively, corresponding to highly conserved sites and neutral sites across all lineages. In the other two site classes, the background lineages had ω0 or ω1 while the foreground lineages had ω2. bPositively selected amino acids at P-value ≤0.05 are numbered according to HORVU6Hr1G002400.1, excluding the first 34 amino acids predicted as membrane targeting signal
To further characterise the evolutionary dynamics of monocot F3’Hs, the site-specific model, which allows the ω value to vary along different amino acid sites, were applied to the same dataset. Results (Table 2) showed that Selection M1 was not better (df = 1, p = 1.0) than the neutral M1. No amino acid site could be identified to be under positive selection in the selection model. To test whether ω may vary at specific amino acid sites in specific branches, branch-site models were also tested (Table 2). When Monocot Class II F3’H was set as the foreground branch, the selection Model A revealed that 7 amino acid sites (108R, 222A, 265 V, 274 T, 355Q, 447S, 449 L) in the Monocot Class II F3’H branch were under positive selection (ω 2 = 8.19737; p < 0.05). LRTs showed that Model A was significantly better than its null hypothesis Model A Null, which specified ω 2 = 1.0. Comparison of Model A with Neutral M1 (df = 2, p < 0.0001) also supported that these amino acid sites were under positive selection. In contrast, when Monocot Class I F3’H was set as the foreground branch, no amino acid site could be identified to be under positive selection at the significant level (Table 1). In this case, the selection Model A did not fit the dataset better (df = 1, p = 1.0) than its null hypothesis Model A Null. Taken together, natural selection assessments indicated that Monocot Class II F3’Hs were under significantly positive selection, which had been detected only in Monocot Class II F3’Hs, affecting specific amino acid sites in this branch.
Sequence alignment and protein modelling analyses
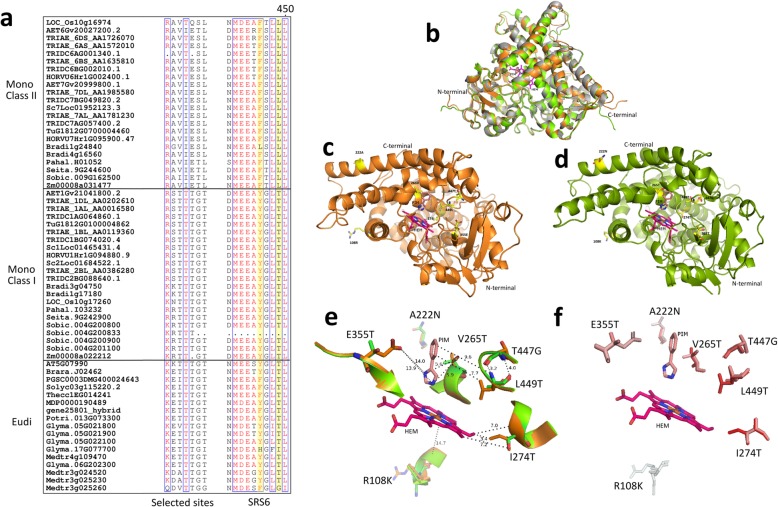
The amino acid substitutions at the positively selected sites between Class I and Class II F3’Hs were analysed by sequence alignment. As shown in Fig. 4a, monocot Class I and Class II F3’Hs displayed clear amino acid substitutions at 6 of the 7 selected sites. For all of these sites, Class I F3’Hs resembled eudicot F3’Hs, which differed from Class II F3’Hs, suggesting a closer relationship between Class I F3’Hs and eudicot F3’Hs. The enzyme activity of F3’Hs could be affected by critical amino acid sites. Previous studies have identified 6 substrate recognition sites (SRS1-SSR6) for Cytochrome P450s proteins. Sequence alignment showed that 4 of the 7 selected amino acid sites were located in SRS4 (265 V, 274 T) and SRS6 (447S, 449 L), suggesting direct potential to affect the enzyme activity of Class II F3’Hs. Moreover, two amino acid sites (yellow highlight in Fig. 4a) in SRS6 have been shown to be critical to determine the activity of F3’H and F3’5’H. As shown in Fig. 4a, most Class II F3’Hs contained 446F-449L, whilst Class I F3’Hs displayed 446Y-449 T. Interestingly, the T446F substitution has previously been shown to be able to enable 5′-hydroxylation activity in some eudicot F3’Hs [27]. The conservation of Thr at position 449 was also considered critical for the 3′-hydroxylation activity in F3’Hs [27]. It should also be noted that the amino acids at position 447 and 449 were identified to be affected by positive natural selection in Class II F3’Hs. Taken together, these observations indicated a strong potential of functional divergence for Class II F3’Hs. In addition, SRS6 was found to be missing in Sobic.004G200833 from S. bicolor, indicating that this Class I F3’H may not be functional.
Fig. 4.
Sequence alignment and protein modelling analyses. a Amino acid sequence alignment of the selected sites and SRS6 in plant F3’Hs. b The overall superimposition of the LOC_Os10g16974 (orange) and LOC_Os10g17260 (green) models with CYP76AH1 (grey; PDB: 5YM3); c The spatial location of the positively selected sites in LOC_Os10g16974; d The spatial location of the positively selected sites in LOC_Os10g17260; e The potential interaction of the positively selected sites with the superimposed substrates; f The hydrophobicity changes between LOC_Os10g16974 and LOC_Os10g17260 at the positively selected amino acid residues. Red and white colours represent the most hydrophobic and the most hydrophilic, respectively. HEM stands for heme. PIN refers enzyme inhibitor in PDB 5YM3
To further investigate the potential effects of natural selection on the enzyme function of Class II F3’Hs, 3D structural models of the Class II (LOC_Os10g16974) and Class I (LOC_Os10g17260) F3’Hs in rice were developed by homology modelling. A recently determined CYP76AH1 crystal structure (PDB: 5YM3) was identified as a close homolog (~ 35% amino acid identity) to F3’H and was used as the template. The rice homologs were chosen due to the strict conservation of a single copy F3’H for each class in this species. As shown in Fig. 4b, the majority of the full length rice F3’Hs could be reliably modelled, with the exception of the short fragments at the N and C terminals. The un-modelled N terminal peptides were predicted as hydrophobic membrane binding domain and do not have direct effect on the enzyme function. The spatial location of the 7 amino acid sites were analysed, as displayed in Fig. 4c & d. Five out of the 7 amino acid sites were found to be located in the catalytic region (Fig. 4e), forming part of the catalytic cavity. The other two amino acid sites belonged to the N terminal region and were located on the exterior surface. Specifically, the side-chains of the 5 amino acid sites in the catalytic region were positioned toward the bound substrates heme and PIM with close distance (3.2 Å ~ 14.0 Å) and may have direct effect on the cavity volume and substrate binding. Structural superimposition revealed amino acid substitutions between LOC_Os10g16974 (Class I) and LOC_Os10g17260 (Class II) F3’Hs at all of the 7 amino acid sites. Noteworthy, four amino acid sites in the catalytic region of LOC_Os10g17260 were found to have the Threonine residue (neutral side chain), which were replaced with Valine (hydrophobic), Isoleucine (hydrophobic), Glutamic acid (charged side chain) and Leucine (hydrophobic). The hydrophobicity profiles of the selected sites were displayed in Fig. 4f. The amino acid substitutions V265 T, I274T, T447G and L449 T not only caused clear hydrophobicity changes between LOC_Os10g16974 and LOC_Os10g17260, but also affect the size of the catalytic cavity due to differences in the side chains. These observations strongly indicated a potential functional divergence after the split of the two classes of F3’Hs in monocot plants, which has been driven by positive natural selection.
In addition, based on the CYP76AH1 structure, the catalytic sites of monocot F3’Hs were identified by selecting the amino acid residues within a close distance (< 5 Å) to the bound heme molecule and enzyme inhibitor PIM. As a result, a total of 39 amino acid sites were identified (Additional file 1). Sequence alignment showed that, the majority of these putative catalytic sites were strictly conserved among all plant F3’Hs. However, an extra amino acid substitution between Class I and Class II F3’Hs was identified at position 84, which was proximal to the enzyme substrate.
Transcriptional analyses
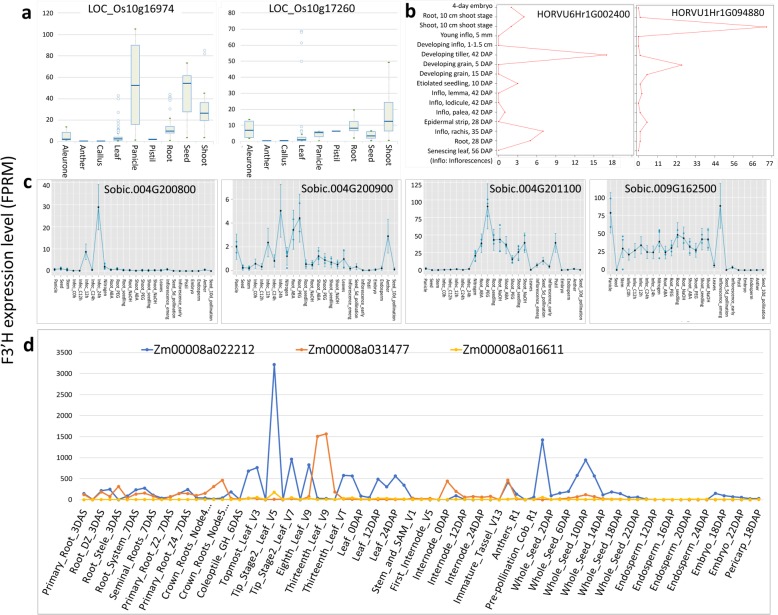
To explore the potential transcriptional divergence between Class I and Class II F3’Hs, the transcriptional data for monocot F3’Hs were searched in public databases. In rice (Fig. 5a), the Class I F3’H LOC_Os10g16974 was highly expressed in panicle, seed and shoot, relatively lower in root, and was barely expressed in other tissues such as anther, callus, leaf and pistil. Compared to LOC_Os10g16974, the Class II F3’H LOC_Os10g17260 generally displayed much lower expression in all tissues, except the pistil tissue, in which LOC_Os10g17260 had relatively higher transcription. The highest expression of LOC_Os10g17260 was found in shoot and root. It should be noted that, while the median expression level of LOC_Os10g17260 was very low in leaf, its expression could reach exceptionally high in some conditions. In barley, a clear transcriptional divergence for Class I and Class II F3’Hs was also observed (Fig. 5b). The Class I F3’H HORVU6Hr1G002400 was found to be mainly expressed in the developing tillers. Slight expression of HORVU6Hr1G002400 was also observed in inflorescences rachis and seedling root. In contrast, the Class II F3’H HORVU1Hr1G094880 was predominantly expressed in seedling shoots, followed by the developing grain at the early stage. Little expression of HORVU1Hr1G094880 was found in other tissues. An interesting observation with F3’Hs in sorghum (Fig. 5c) is that, the sorghum Class I F3’H Sobic.009G162500 was barely transcribed in all the reproductive tissues including anther, seeds, endosperm, embryo, pistil and the early inflorescence, with the exception of the emerging inflorescence. Instead, abundant expression of Sobic.009G162500 was found in the vegetative tissues panicle, stem, root and shoot. In contrast, Class II F3’Hs Sobic.004G201100 displayed moderate expression in pistil and young seeds. Significant transcription of Sobic.004G201100 was also found in vegetative tissues root and shoot. The other two Class II F3’Hs Sobic.004G200800 and Sobic.004G200900, which are tandem duplicates, had very low or barely no expression in all tissues studied. Noteworthy, although Sobic.004G200800 was barely expressed in sorghum, a dramatic increase of its expression was observed in leaves upon pathogen infection. In maize, the Class I and Class II F3’Hs also displayed a clear transcriptional divergence (Fig. 5d). The highest expression of Zm00008a031477 (Class I F3’H) was found in leaves, followed by moderate expression in root, internode and meiotic tassel. In contrast, Zm00008a022212 (Class II F3’H) displayed more widespread expression abundant in leave tips, silks and whole seeds at 10 days after pollination (DAP), and moderate in pooled leaf, topmost leaf and mature leave. Interestingly, the other Class II F3’H Zm00008a016611 was barely expressed in all tissues studied.
Fig. 5.
Transcriptional profiles of monocot F3’Hs. a LOC_Os10g16974 (Class II) and LOC_Os10g17260 (Class I) in rice; b HORVU6Hr1G002400 (Class II) and HORVU1Hr1G094880 (Class I) in barley; c Sobic.004G200800, Sobic.004G200833, Sobic.004G200900, Sobic.004G201100 (Class I) and Sobic.009G162500 (Class II) in Sorghum; d Zm00008a016611, Zm00008a022212 (Class I) and Zm00008a031477 (Class II) in maize
Discussions
The evolution of complex metabolic pathways such as flavonoid biosynthesis has been indicated to play a critical role in plant evolution, helping plants adapt to various biotic and abiotic stressors [28, 29]. The flavonoid biosynthetic pathway has been extensively studied in di-cotyledon plants, whilst only moderate attention has been paid toward monocot cereal crops [1]. This observation is staggering, considering the fact that cereal plants such as wheat, rice, barley and maize comprise the most economically important food and feed sources for human and animals. Due to the ubiquitous presence of flavonoids in plant tissues and flavonoids being potent antioxidant, a close relationship between flavonoid biosynthesis and environmental adaptation has also been established in cereal plants [3]. Previous studies on the adaptive role of flavonoids in cereals have mainly been reported in barley [14, 30–35], wheat [36–40] and rice [41–43], maize [44–47]. These studies generally can be divided into two categories: the measurement of flavonoid content changes and the transcriptional responses of the flavonoid biosynthetic gene under biotic or abiotic stress conditions. Given the constant selection pressure confronting cereal plants during evolution, the evidence of environmental adaptation at the gene molecular level should theoretically also be prevalent in the flavonoid biosynthetic pathway. In the present study, we aim to explore the potential interaction between cereal plants and the environment from the gene evolutionary dynamics perspective. We focused on the F3’H gene subfamily, which encoded a critical enzyme that controls the hydroxylation patterns of the flavonoid B-ring.
Lineage-specific evolution is an important mechanism for gene evolution via duplication. It is a common observation during plant evolution and diversification. Facilitated by the latest whole genome sequencing data of barley, rye, wheat and other wheat relatives, this study presented a comprehensive genome-wide survey and a systematic phylogeny analysis on F3’H genes in 12 monocot species. We found that the distribution of putative F3’Hs was highly unbalanced among monocots. For Triticeae, it ranges from 2 copies in T. urartu to 9 copies in T. aestivum. A high volatility of F3’H number was also observed for non-Triticeae, with S. bicolor and B. distachyon retaining 5 and 4 copies, respectively. In this study, we identified 3 F3’Hs in barley genome, in contrast to the study by Vikhorev [24], which reported only 2 copies of F3’Hs. In comparison, we identified an extra F3’H (HORVU7Hr1G095900.47) on chromosome 7H of barley, which was collinear with AET7Gv20999800 and TuG1812G0700004460, located on chromosomes Aet7 and Tu7 of A. tauschii and T. urartu, respectively. Our results are consistent with the study by Lam [25], who also identified 3 F3’Hs in barley. The expansion of F3’Hs in monocots was found to be mainly attributed to dispersed gene duplication. This finding is consistent with the observations made with another three gene families (MYB, MYC and F3’5’H) from the anthocyanin biosynthetic pathway in monocot plants [48]. Noteworthy, we found that tandem duplication and proximal duplication have contributed to the expansion of F3’Hs specifically in S. bicolor and O. sativa. In addition, one WGD/Segmental duplication was observed for Zm00008a016611 and Zm00008a022212 in Z. mays. These observations may have reflected the species-specific evolutionary history of F3’Hs in these three plants. Notably, despite 5 F3’Hs were present in S. bicolor genome, one F3’H Sobic.004G200833 may not be functional due to the loss of the critical substrate recognition sites SRS6.
Phylogeny analyses in the present study revealed that monocot F3’Hs have evolved into two independent lineages: Mono_F3’H Class I and Mono_F3’H Class II, with the previously characterised CYP75B4 in rice classified as a Class II F3’H. This finding is consistent with the report by Lam [25], which found that CYP75B4 belonged to separate phylogeny clade, and had obtained novel 5′-hydroxylase activity on chrysoeriol. Here, we showed that the divergence between Class I and Class II F3’Hs has originated from a duplication event predicating the species diversification of monocots. In addition, an additional duplication event could also be proposed for both Class I and Class II F3’Hs during the common ancestor of Triticeae, leading to the formation of 2 sub-branches in each F3’H class in Triticeae. The identified duplication events for Class I and Class II F3’Hs in Triticeae may point to a shared genome-wide duplication event in the common ancestor of Triticeae. Interestingly, the conservations of Class I/Class II F3’Hs and the different sub-branches in Triticeae both displayed unequal distribution among different species. Whilst the 2 sub-branches of Class II F3’Hs were both preserved in all Triticeae species included in the present study, the secondary sub-branch of Class I was found to be absent in H. vulgare and A. tauschii. This gene absence might be due to gene loss after duplication, which is a common observation in plant genomes [22, 23]. Taken together, these findings showed that F3’Hs in monocot have underwent volatile lineage-specific gene duplication and gene loss events.
As the focus of the present study, evolutionary dynamic analyses showed that monocot Class II F3’Hs had been clearly affected by positive natural selection. The detection of positive selection in Class II F3’H within this study presented direct evidence that the evolution of flavonoid biosynthetic pathway has been affected by environmental selection. F3’H and F3’5’H are close homologs in the cytochrome P450 superfamily, responsible for the production of red and blue anthocyanins, respectively. A similar observation has been made on monocot F3’5’H gene subfamily [48], in which positive selection has been shown to drive the emergence of a separate F3’5’H lineage responsible for the accumulation of blue anthocyanins in Triticeae grains. The selection on monocot F3’5’H was suggested to have resulted from plants’ adaptation to strong light or heat stresses. The detection of positive selection in both F3’H and F3’5’H subfamilies lend support to our earlier hypothesis that the evidence of plant-environmental interaction at the gene evolutionary level should be prevalent in the flavonoid biosynthetic pathway. Unlike monocot F3’5’H, which displayed selection for increased protein thermostability, the divergence between Class I and Class II F3’Hs seems to be more related with enzyme function at the protein structural level. This is supported by the results from the sequence alignment and protein modelling analyses in the present study. The detection of positive selection in both F3’H and F3’5’H families is corroborated by a recent report which showed that light environment may induce differences in photoprotective phenolic compounds during long-term photoacclimation [49].
F3’H is among the poorly understood flavonoid biosynthetic genes in monocot plants. Characterization of F3’H has only been reported for maize Pr1 [50, 51], controlling the red aleurone colour; for rice CYP75B3 and CYP75B4 [25, 26], underlying the 3′-hydroxylated flavonoids and tricin formation; and for sorghum [52–54], involved in 3-deoxyanthocyanidins biosynthesis. Interestingly, CYP75B3 and CYP75B4 in rice, classified as Class I and Class II, respectively, have been shown to have divergent enzyme activity [25, 26]. In particular, the Class II F3’H CYP75B4 was shown to display novel chrysoeriol-specific 5′-hydroxylase activity and played an indispensable role in tricin biosynthesis [25]. The recruitment of 5′-hydroxylase activity for CYP75B4 was suggested to have contributed to the prevalence of tricin-derived metabolites in grasses and monocots, which have important function in plant-defence mechanisms. These observations suggested that CYP75B4 may obtained a novel biological role due to protein functional divergence. Indeed, Class II F3’Hs in monocots including CYP75B4 were found to be affected by positive natural selection, acting 7 specific amino acid sites. Protein modelling and amino acid property analyses showed that majority of these selected sites were located at the catalytic cavity of F3’H, and may have a direct effect on substrate specificity. These findings may provide an evolutionary and protein structural explanation to the observed enzyme activity differences between CYP75B3 and CYP75B4 [25, 26]. Notably, the molecular basis underlying the functional difference between F3’H and F3’5’H has been well-characterised [27]. Amino acid substitutions at two critical sites in SRS6 were shown to control the 3′- and 5′- hydroxylase activities. Interestingly, sequence alignment in our study showed that Class I and Class II F3’Hs displayed distinct amino acid substitutions at these two sites. Intriguingly, Class II F3’Hs contained a Phe at position 446, which was commonly observed for F3’5’Hs. In contrast, Class I F3’Hs retained Tyr at this position, consistent with previously characterised F3’Hs. These results are consistent with the observed 5′-hydroxylase activity for CYP75B4, a Class II F3’H. Instead, no 5′-hydroxylase activity has been observed for CYP75B3 (Class I F3’H) [25]. Our analyses provided a protein structural explanation for the 5′-hydroxylase activity in CYP75B4. The results presented here resembled a similar observation made in Asteraceae, in which some CYP75B proteins were identified to be clustered together with F3’Hs but displayed F3’5’H activities [55]. Our study indicated that the whole monocot Class II F3’Hs may have obtained the novel chrysoeriol-specific 5′-hydroxylase function.
In addition to the divergence at the protein structural level, functional divergence after gene duplication could also occur at the transcriptional level. In fact, gene expression analyses have been widely used to study many flavonoid biosynthetic genes in regard to their responses to various biotic and abiotic stressors. Among these studies, three differentially expressed F3’Hs (Class II) in sorghum leave under cutting stress [53, 54], two tissue-specific F3’Hs (Class I and Class II) in barley [24] and two F3’Hs (Class I and Class II) in rice [25] have been reported. Interestingly, in all cases, Class I and Class II displayed divergent expression profiles. This observation is consistent with the results from our transcriptional analyses, which also covered the three F3’H paralogs from maize. By combining these data together, although a common expression pattern for Class I and Class II F3’Hs across these monocot species can not be drawn at the moment, a transcriptional divergence for the two F3’H lineages may be proposed. The observation of gene functional divergence at both protein structural and gene transcriptional level have been reported in many plant gene families [56–58], and consolidated the theory of gene evolution via duplication [22, 23]. In this study, the expression of different F3’H paralogs may have evolved a species-specific profile, meeting the particular environmental challenges faced by different plants. For example, we found one of the Class I F3’Hs identified from sorghum displayed a clear transcriptional response to pathogen infection but was not expressed at all under normal growing condition. This result is corroborated by previous studies on F3’Hs in sorghum [52–54]. In addition to monocot plants, transcriptional divergence has also been reported for paralogous F3’Hs and F3’5’Hs in several eudicot plants, such as F3’Hs in tea tree leave [59], which further confirmed that functional divergence at the gene transcriptional level is a common observation during gene evolution via duplication. An evolutionary explanation for this observation would be that, the development of the complex expression profile for flavonoid biosynthetic genes resulted from the environmental selection pressure acting on different plants, and also have in turn improved plants’ survivability in nature.
Conclusions
Based on the results from the genome-wide survey, phylogeny, evolutionary dynamics and protein structural modelling analyses, we found that monocot F3’Hs had underwent volatile lineage-specific gene duplication and gene loss events in monocot plants. We concluded that monocot F3’Hs have evolved into two independent lineages (Mono_F3’H Class I and Class II), after gene duplication during the common ancestor of monocot plants. The functional divergence of monocot F3’H Class II has been affected by positive natural selection, acting on several specific amino acid sites. The amino acid substitutions at these selected sites and other sites in SRS6 displayed high potential to affect the substrate binding of F3’Hs, and may have contributed to the recruitment of chrysoeriol-specific 5′-hydroxylation activity in F3’H Class I, as evidenced by CYP75B4 in rice. In addition, transcriptional divergence between F3’H Class I and Class II have also been observed. Taken together, our study revealed clear evidence of plant-environmental interaction for the flavonoid biosynthetic pathway at the gene evolutionary level.
Methods
Sequence retrieval and genuine F3’H homolog identification
Due to the close homology between F3’5’H and F3’H, genuine F3’H homologs were identified by a method as described in a previous study [48]. Twelve monocot crops and twelve eudicot species were included. The amino acid sequence of the previously characterised CYP75B3 homolog in barley was used as queries for BLASTP (E-value threshold: 1e-30) against public databases of monocot plant genomes. Remote F3’H homologs were also retrieved from lower plant moss (P. pattens). The overall process is as following: all homologs identified above were included for the development of a preliminary Neighbour Joining tree using MEGA7.0 software [60]. Sequence alignment was performed using Muscle [61]. The substitution model used is P-distance. 1000 times bootstrapping iteration was performed for tree assessment. The distinct phylogeny branch containing the previously reported F3’Hs (CYP75B3 & CYP75B4) in rice was selected as the genuine F3’Hs and was used for further analyses.
Phylogeny reconstruction
The CDS sequences of the above identified F3’Hs and the remote F3’H homologs in patten were used for phylogeny reconstruction. Codon-based sequence alignment of the CDS sequences was performed using Muscle with 8 iterations [61]. The resulted sequence alignment was checked manually to remove significant alignment gaps and 5′ signal peptides. The phylogeny was searched by Bayesian simulations implemented in BEAST2 [62] under strict molecular clock assumption. The unlinked substitution model Yule + G (5 categories) was used. A single Markov Chain - Monte Carlo Chain was run for 1000,000 generations. Trees were sampled every 100 generations with 1, 000 pre burn-in until convergence. The final phylogenetic tree was inferred by TreeAnnotator with the first 1000 trees discarded. All phylogenetic trees in the present study were annotated using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Gene structural analyses
The CDS and genomic sequences for F3’Hs were downloaded from the corresponding genome database for the target monocot species. The gene structural diagram was constructed using the GSDS v2.0 online tool (http://gsds.cbi.pku.edu.cn/). The phylogenetic subtree for monocot F3’Hs developed in the present study was also used as an input to cluster the gene structures based on phylogeny relationship.
Gene duplication pattern and colinearity analyses
The MCScanX package [63] was used to characterise the gene duplication pattern. The original genomic data was downloaded from public database and was further processed to generate the input files for MCScanX. Intra- and inter-species genome comparisons were performed using the standalone NCBI-BLAST-2.2.29 tool with an E-value threshold of 1e-05. Intra-genome all-vs-all BLAST was performed for gene duplication pattern identification. For colinear gene pair identification, genome dataset from different species were combined for all-vs-all BLAST.
Sequence alignment and protein modelling
Amino acid sequence alignment and annotation were carried out using ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/index.php) Homologous structure template was identified by BLASTp against the PDB database using the amino acid sequences of rice F3’Hs as queries. The protein structure of CYP76AH1 (PDB: 5YM3) with the highest amino acid identity was used for the model development. The protein models of F3’Hs were created by homology modelling using the Modeller server. 5 structural models were generated for each protein. The best model was selected based on the lowest Discrete Optimized Protein Energy (DOPE) values and GA 341 score of 1, which indicate reliability of these models. The final model was validated by Ramachandran plot analysis using PROCHECK (http://www.ebi.ac.uk/thornton-srv/software/PROCHECK). Molecular visualizations were performed using PyMOL (Version 1.3r1. Schrodinger, LLC).
Transcriptional data mining
The transcriptional data for the target F3’H genes was extracted from individual databases: for rice (http://expression.ic4r.org/index), barley (https://apex.ipk-gatersleben.de/apex/f?p=284:10), maize (https://www.maizegdb.org/) and sorghum (http://sorghum.riken.jp/morokoshi/Home.html).
Additional file
Amino acid sequence alignment of plant F3’Hs. (PDF 50 kb)
Acknowledgements
We acknowledge the plant research community for making the genomic data and transcriptional data available to the public.
Abbreviations
- CDS
Coding domain sequence
- CHI
Chalcone isomerase
- CHS
Chalcone synthase
- DAP
Days after pollination
- DOPE
Discrete Optimized Protein Energy
- F3’H
Flavonoid 3′-hydroxylase
- F3′5’H
Flavonoid 3’5’-hydroxylase
- LRTs
Likelihood Ratio Tests
- SRS
Substrate recognition sites
- WGD
Whole genome duplication
Authors’ contributions
CL & YX supervised the project and provided valuable comments on manuscript development. YJ conceived the study and developed the manuscript. YJ & BL retrieved the gene sequences. BL & YJ are responsible for the gene structural analysis. YJ performed phylogeny, evolutionary dynamic and protein modelling analyses. YJ, YZ & XZ performed gene duplication, synteny and gene expression data mining analyses. All authors read and approved the final manuscript.
Funding
This project is supported by the Grains Research & Development Corporation (GRDC) project “Improved adaptation of barley to acid soils” (Project ID: UMU00046).
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Figshare repository at https://figshare.com/s/a811638eb6fcd3496d2f.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yong Jia and Bo Li contributed equally to this work.
Contributor Information
Yong Jia, Email: y.jia@murdoch.edu.au.
Bo Li, Email: 294828485@qq.com.
Yujuan Zhang, Email: Yujuan.Zhang@murdoch.edu.au.
Xiaoqi Zhang, Email: Xiao-Qi.Zhang@murdoch.edu.au.
Yanhao Xu, Email: xyh09@yangtzeu.edu.cn.
Chengdao Li, Email: c.li@murdoch.edu.au.
References
- 1.Tohge T, de Souza LP, Fernie AR. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J Exp Bot. 2017;68(15):4013–4028. doi: 10.1093/jxb/erx177. [DOI] [PubMed] [Google Scholar]
- 2.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126(2):485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khlestkina EK. The adaptive role of flavonoids: emphasis on cereals. Cereal Res Commun. 2013;41(2):185–198. [Google Scholar]
- 4.Dykes L, Rooney LW. Phenolic compounds in cereal grains and their health benefits. Cereal Food World. 2007;52(3):105–111. [Google Scholar]
- 5.Agati G, Brunetti C, Di Ferdinando M, Ferrini F, Pollastri S, Tattini M. Functional roles of flavonoids in photoprotection: new evidence, lessons from the past. Plant Physiol Bioch. 2013;72(11):35–45. doi: 10.1016/j.plaphy.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Guo J, Han W, Wang MH. Ultraviolet and environmental stresses involved in the induction and regulation of anthocyanin biosynthesis: a review. Afr J Biotechnol. 2008;7(25):4966–4972. [Google Scholar]
- 7.Gould KS. Nature's Swiss army knife: the diverse protective roles of anthocyanins in leaves. J Biomed Biotechnol. 2004;2004(5):314–320. doi: 10.1155/S1110724304406147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradshaw HD, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426(6963):176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- 9.Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends Plant Sci. 1998;3(6):212–217. [Google Scholar]
- 10.Mo YY, Nagel C, Taylor LP. Biochemical complementation of Chalcone synthase mutants defines a role for Flavonols in functional pollen. P Natl Acad Sci USA. 1992;89(15):7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor LP, Hepler PK. Pollen germination and tube growth. Annu Rev Plant Phys. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- 12.Friml J, Jones AR. Endoplasmic reticulum: the rising compartment in auxin biology. Plant Physiol. 2010;154(2):458–462. doi: 10.1104/pp.110.161380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BSJ, Muday GK. Auxin and ethylene induce Flavonol accumulation through distinct transcriptional networks. Plant Physiol. 2011;156(1):144–164. doi: 10.1104/pp.111.172502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himi E, Yamashita Y, Haruyama N, Yanagisawa T, Maekawa M, Taketa S. Ant28 gene for proanthocyanidin synthesis encoding the R2R3 MYB domain protein (Hvmyb10) highly affects grain dormancy in barley. Euphytica. 2012;188(1):141–151. [Google Scholar]
- 15.Hyun JW, Chung HS. Cyanidin and malvidin from Oryza sativa cv. Heugjinjubyeo mediate cytotoxicity against human monocytic leukemia cells by arrest of G(2)/M phase and induction of apoptosis. J Agr Food Chem. 2004;52(8):2213–2217. doi: 10.1021/jf030370h. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C, Giusti MM, Malik M, Moyer MP, Magnuson BA. Effects of commercial anthocyanin-rich extracts on colonic cancer and nontumorigenic colonic cell growth. J Agr Food Chem. 2004;52(20):6122–6128. doi: 10.1021/jf049517a. [DOI] [PubMed] [Google Scholar]
- 17.Tsuda T, Horio F, Osawa T. Cyanidin 3-O-beta-D-glucoside suppresses nitric oxide production during a zymosan treatment in rats. J Nutr Sci Vitaminol. 2002;48(4):305–310. doi: 10.3177/jnsv.48.305. [DOI] [PubMed] [Google Scholar]
- 18.Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3(1):2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 19.Ferreyra MLF, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Yoshikazu, Brugliera Filippa. Flower colour and cytochromes P450. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368(1612):20120432. doi: 10.1098/rstb.2012.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bak S, Beisson F, Bisshop G, Hamberger B, Hofer R, Paquette SM, Reichhart DW. Cytochromes P450. Arabidopsis Book. 2011;9:e0144. doi: 10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang JZ. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18(6):292–298. [Google Scholar]
- 23.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183(3):557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 24.Vikhorev AV, Strygina KV, Khlestkina EK. Duplicated flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes in barley genome. Peerj. 2019;7:e6266. doi: 10.7717/peerj.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam PY, Liu HJ, Lo C. Completion of Tricin biosynthesis pathway in Rice: cytochrome P450 75B4 is a unique Chrysoeriol 5 '-hydroxylase. Plant Physiol. 2015;168(4):1527–1536. doi: 10.1104/pp.15.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park Sangkyu, Choi Min, Lee Jong, Kim Jae, Ha Sun-Hwa, Lim Sun-Hyung. Molecular and Biochemical Analysis of Two Rice Flavonoid 3’-Hydroxylase to Evaluate Their Roles in Flavonoid Biosynthesis in Rice Grain. International Journal of Molecular Sciences. 2016;17(9):1549. doi: 10.3390/ijms17091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seitz C, Ameres S, Forkmann G. Identification of the molecular basis for the functional difference between flavonoid 3 '-hydroxylase and flavonoid 3 ',5 '-hydroxylase. FEBS Lett. 2007;581(18):3429–3434. doi: 10.1016/j.febslet.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 28.Weng JK. The evolutionary paths towards complexity: a metabolic perspective. New Phytol. 2014;201(4):1141–1149. doi: 10.1111/nph.12416. [DOI] [PubMed] [Google Scholar]
- 29.Mouradov A, Spangenberg G. Flavonoids: a metabolic network mediating plants adaptation to their real estate. Front Plant Sci. 2014;5:620. doi: 10.3389/fpls.2014.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skadhauge B, Thomsen KK, von Wettstein D. The role of the barley testa layer and its flavonoid content in resistance to Fusarium infections. Hereditas. 1997;126(2):147–160. [Google Scholar]
- 31.Christensen AB, Gregersen PL, Olsen CE, Collinge DB. A flavonoid 7-O-methyltransferase is expressed in barley leaves in response to pathogen attack. Plant Mol Biol. 1998;36(2):219–227. doi: 10.1023/a:1005985609313. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz-Hoerner R, Weissenbock G. Contribution of phenolic compounds to the UV-B screening capacity of developing barley primary leaves in relation to DNA damage and repair under elevated UV-B levels. Phytochemistry. 2003;64(1):243–255. doi: 10.1016/s0031-9422(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 33.Lee SH, Lee KW, Kim KY, Choi GJ, Yoon SH, Ji HC, Seo S, Lim YC, Ahsan N. Identification of salt-stress induced differentially expressed genes in barley leaves using the annealing-control-primer-based GeneFishing technique. Afr J Biotechnol. 2009;8(7):1326–1331. [Google Scholar]
- 34.Lachman J, Dudjak J, Miholova D, Kolihova D, Pivec V. Effect of cadmium on flavonoid content in young barley (Hordeum sativum L.) plants. Plant Soil Environ. 2005;51(11):513–516. [Google Scholar]
- 35.Tattini M, Galardi C, Pinelli P, Massai R, Remorini D, Agati G. Differential accumulation of flavonoids and hydroxycinnamates in leaves of Ligustrum vulgare under excess light and drought stress. New Phytol. 2004;163(3):547–561. doi: 10.1111/j.1469-8137.2004.01126.x. [DOI] [PubMed] [Google Scholar]
- 36.Giovanini MP, Puthoff DP, Nemacheck JA, Mittapalli O, Saltzmann KD, Ohm HW, Shukle RH, Williams CE. Gene-for-gene defense of wheat against the hessian fly lacks a classical oxidative burst. Mol Plant Microbe In. 2006;19(9):1023–1033. doi: 10.1094/MPMI-19-1023. [DOI] [PubMed] [Google Scholar]
- 37.Li XL, Lv X, Wang XH, Wang LH, Zhang MS, Ren MJ. Effects of abiotic stress on anthocyanin accumulation and grain weight in purple wheat. Crop Pasture Sci. 2018;69(12):1208–1214. [Google Scholar]
- 38.Himi E, Mares DJ, Yanagisawa A, Noda K. Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. J Exp Bot. 2002;53(374):1569–1574. doi: 10.1093/jxb/erf005. [DOI] [PubMed] [Google Scholar]
- 39.Ma DY, Sun DX, Wang CY, Li YG, Guo TC. Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol Bioch. 2014;80:60–66. doi: 10.1016/j.plaphy.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 40.Gondor OK, Janda T, Soos V, Pal M, Majlath I, Adak MK, Balazs E, Szalai G. Salicylic acid induction of flavonoid biosynthesis pathways in wheat varies by treatment. Front Plant Sci. 2016;7:1447. doi: 10.3389/fpls.2016.01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hao ZN, Wang LP, He YP, Liang JG, Tao RX. Expression of defense genes and activities of antioxidant enzymes in rice resistance to rice stripe virus and small brown planthopper. Plant Physiol Bioch. 2011;49(7):744–751. doi: 10.1016/j.plaphy.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Ithal N, Reddy AR. Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 2004;166(6):1505–1513. [Google Scholar]
- 43.Chutipaijit S, Cha-Um S, Sompornpailin K. Differential accumulations of proline and flavonoids in Indica Rice varieties against salinity. Pakistan J Bot. 2009;41(5):2497–2506. [Google Scholar]
- 44.Casati P, Walbot V. Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol. 2003;132(4):1739–1754. doi: 10.1104/pp.103.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christie PJ, Alfenito MR, Walbot V. Impact of low-temperature stress on general Phenylpropanoid and anthocyanin pathways - enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta. 1994;194(4):541–549. [Google Scholar]
- 46.Marrs KA, Walbot V. Expression and RNA splicing of the maize glutathione S-transferase Bronze2 gene is regulated by cadmium and other stresses. Plant Physiol. 1997;113(1):93–102. doi: 10.1104/pp.113.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolra R, Barcelo J, Poschenrieder C. Constitutive and aluminium-induced patterns of phenolic compounds in two maize varieties differing in aluminium tolerance. J Inorg Biochem. 2009;103(11):1486–1490. doi: 10.1016/j.jinorgbio.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 48.Jia Y, Selva C, Zhang Y, Li B, Lee MA, Broughton S, Xiaoqi Z, Sharon W, Penghao W, Cong T et al: Uncovering the evolutionary origin of blue anthocyanins in cereal grains. Under review, to be published. 2019.
- 49.Oksana S, Marek Z, Susanne N, Peyman MT, Marian B. Precultivation of young seedlings under different color shades modifies the accumulation of phenolic compounds in Cichorium leaves in later growth phases. Environ Exp Bot. 2019;165:30–38. [Google Scholar]
- 50.Larson R, Bussard JB, Coe EH. Gene-Dependent Flavonoid 3′-Hydroxylation in Maize. Biochem Genet. 1986;24(7–8):615–624. doi: 10.1007/BF00504338. [DOI] [PubMed] [Google Scholar]
- 51.Sharma M, Chai CL, Morohashi K, Grotewold E, Snook ME, Chopra S. Expression of flavonoid 3 '-hydroxylase is controlled by P1, the regulator of 3-deoxyflavonoid biosynthesis in maize. BMC Plant Biol. 2012;12:196. doi: 10.1186/1471-2229-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boddu J, Svabek C, Sekhon R, Gevens A, Nicholson RL, Jones AD, Pedersen JF, Gustine DL, Chopra S. Expression of a putative flavonoid 3 '-hydroxylase in sorghum mesocotyls synthesizing 3-deoxyanthocyanidin phytoalexins. Physiol Mol Plant P. 2004;65(2):101–113. [Google Scholar]
- 53.Shih CH, Chu IK, Yip WK, Lo C. Differential expression of two flavonoid 3 '-hydroxylase cDNAs involved in biosynthesis of anthocyanin pigments and 3-deoxyanthocyanidin phytoalexins in sorghum. Plant Cell Physiol. 2006;47(10):1412–1419. doi: 10.1093/pcp/pcl003. [DOI] [PubMed] [Google Scholar]
- 54.Mizuno H, Yazawa T, Kasuga S, Sawada Y, Kanamori H, Ogo Y, Hirai MY, Matsumoto T, Kawahigashi H. Expression of flavone synthase II and flavonoid 3 '-hydroxylase is associated with color variation in Tan-colored injured leaves of Sorghum. Front Plant Sci. 2016;7:1718. doi: 10.3389/fpls.2016.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G. Cloning, functional identification and sequence analysis of flavonoid 3 '-hydroxylase and flavonoid 3 ',5 '-hydroxylase cDNAs reveals independent evolution of flavonoid 3 ',5 '-hydroxylase in the Asteraceae family. Plant Mol Biol. 2006;61(3):365–381. doi: 10.1007/s11103-006-0012-0. [DOI] [PubMed] [Google Scholar]
- 56.Jia Y, Wong DCJ, Sweetman C, Bruning JB, Ford CM. New insights into the evolutionary history of plant sorbitol dehydrogenase. BMC Plant Biol. 2015;15:101. doi: 10.1186/s12870-015-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehti-Shiu MD, Uygun S, Moghe GD, Panchy N, Fang L, Hufnagel DE, Jasicki HL, Feig M, Shiu SH. Molecular evidence for functional divergence and decay of a transcription factor derived from whole-genome duplication in Arabidopsis thaliana. Plant Physiol. 2015;168(4):1717–1734. doi: 10.1104/pp.15.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panchy N, Lehti-Shiu M, Shiu SH. Evolution of gene duplication in plants. Plant Physiol. 2016;171(4):2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei K, Wang LY, Zhang CC, Wu LY, Li HL, Zhang F, Cheng H. Transcriptome analysis reveals key flavonoid 3 '-hydroxylase and flavonoid 3 ',5 '-hydroxylase genes in affecting the ratio of Dihydroxylated to Trihydroxylated Catechins in Camellia sinensis. PLoS One. 2015;10(9):e0137925. doi: 10.1371/journal.pone.0137925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang YP, Tang HB, DeBarry JD, Tan X, Li JP, Wang XY, Lee TH, Jin HZ, Marler B, Guo H, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment of plant F3’Hs. (PDF 50 kb)
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Figshare repository at https://figshare.com/s/a811638eb6fcd3496d2f.