Abstract
Nanoparticle based siRNA formulations often suffer from aggregation and loss of function during storage. We in this study report a frozen targeted RGD-polyethylene glycol (PEG)-ECO/siβ3 nanoparticle formulation with a prolonged shelf life and preserved nanoparticle functionality. The targeted RGD-PEG-ECO/siβ3 nanoparticles are formed by step-wised self-assembly of RGD-PEG-maleimide, ECO, and siRNA. The nanoparticles have a diameter of 224.5 ± 9.41 nm and a zeta potential to 45.96 ± 3.67 mV in water and a size of 234.34 ± 3.01 nm and a near neutral zeta potential in saline solution. The addition of sucrose does not affect their size and zeta potential and substantially preserves the integrity and biological activities of frozen and lyophilized formulations of the targeted nanoparticles. The frozen formulation with as low as 5% sucrose retains nanoparticle integrity (90% siRNA encapsulation), size distribution (polydispersity index [PDI] ≤20%), and functionality (at least 75% silencing efficiency) at −80°C for at least 1 year. The frozen RGD-PEG-ECO/siβ3 nanoparticle formulation exhibits excellent biocompatibility, with no adverse effects on hemocompatibility and minimal immunogenicity. As RNAi holds the promise in treating the previously untreatable diseases, the frozen nanoparticle formulation with the low sucrose concentration has the potential to be a delivery platform for clinical translation of RNAi therapeutics.
Keywords: siRNA nanoparticles, ECO, biocompatibility, long-term stability, RNAi
Introduction
The development of RNAi-based therapies for human disease holds significant promise for the treatment of previously incurable diseases. Currently, there are several RNAi-based therapies undergoing clinical trials to treat cancer, hypercholesterolemia, and viral infections such as Zika virus [1–3]. One such therapy, Patisiran (ONPATTRO™), became the first Food and Drug Administration (FDA)-approved lipid nanoparticle formulation for RNAi-based therapy of hereditary transthyretin amyloidosis [4]. In light of this recent advancement, overcoming delivery barriers is imperative to further facilitate the clinical application of RNAi-based therapies. The systemic administration of RNAi-based therapies is currently limited to targeting diseases associated with the liver, due to its efficient uptake of nanosized therapeutics [5]. The broad application of RNAi-based therapies is still limited by the need of targeted, safe, efficient, and clinically translatable delivery of siRNAs [6,7]. Overcoming these delivery barriers is necessary to facilitate the clinical application of RNAi-based therapies for the treatment of human diseases [6,8].
Various delivery systems, including lipids, polymers, chemically modified siRNAs, and siRNA conjugates, have been developed and tested for RNAi-based therapies [4,9–11]. Cationic lipid siRNA nanoparticles have emerged as an advantageous siRNA delivery system due to their simplicity and ability to mediate safe and efficient siRNA delivery [12]. The clinical application of RNAi therapeutics requires a prolonged shelf-life without compromising therapeutic function. However, currently reported cationic lipid siRNA nanoparticles suffer from various drawbacks, including inconsistent particle formulation, poor stability in aqueous environments, aggregation, and consequent loss in transfection efficiency [13,14]. These drawbacks present significant challenges to satisfy the requirements for the nanoparticle formulations of the regulatory agencies, including consistent and reproducible formulation, as well as acceptable stability and shelf life, before proceeding to clinical studies [15,16]. Therefore, the ability to reproduce cationic lipid siRNA nanoparticles with long-term formulation stability and functionality preservation is essential for the clinical development of RNAi-based therapies.
The aim of this study was to develop a reproducible targeted amino lipid siRNA nanoparticle formulation with long-term stability and good biocompatibility as a delivery platform for the systemic administration of therapeutic siRNAs. We have developed a multifunctional amino lipid based carrier, 1-aminoethylimino[bis(N-oleoylcysteinyl-aminoethyl)propionamide] (ECO), for cytosolic delivery of nucleic acids. ECO forms stable nanoparticles with unmodified siRNAs through self-assembly and mediates efficient gene silencing in tumors through systemic delivery [17,18]. Targeted ECO/siRNA nanoparticles can be readily developed by incorporating a targeting agent through a biocompatible polyethylene glycol (PEG) spacer to the nanoparticles during self-assembly nanoparticle formulation. The targeted RGD-PEG-ECO/siRNA nanoparticles have been tested for successful silencing of multiple oncogenic targets to effectively alleviate primary tumor burden, metastatic potential, and drug resistance of triple negative breast cancer in vivo [19–21]. In this study, we developed a reproducible and stable targeted ECO/siRNA nanoparticle formulation using a siRNA specific to β3 integrin (siβ3) as a model therapeutic siRNA and RGD peptide as a targeting agent for clinical translation. Optimal conditions were established for the formulation of targeted RGD-PEG-ECO/siβ3 nanoparticles by adding sucrose, a common excipient used clinically to aid the stability of lipid nanoparticles [22,23]. The physicochemical properties, including size, zeta potential, siRNA entrapment, stability, and biological properties, including gene silencing efficiency, biocompatibility, and immunogenicity of the formulation, were assessed at different storage conditions up to a year.
Materials and Methods
Cell lines and reagents
BT549 cells were obtained from ATCC (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Sigma). Cells were maintained in a humidified incubator kept at 37°C and 5% CO2. The following siRNAs were ordered from Integrated DNA Technologies (Coralville, IA): human siRNA β3 (sense: 5′-GCUCAUCUGGAAACUCCUCAUCACC-3′; antisense: 5′-GGUGAUGAGGAGUUUCCAGAUGAGCUC-3′); negative control siRNA (sense: 5′-UUAGCGUAGAUGUAAUGUGdTdT-3′; antisense, 5′-CACAUUACAUCUACGCUAA-3′).
Human research statement
Studies were performed according to the amended Declaration of Helsinki. Blood was collected from healthy volunteers under Institutional Review Board approved protocol number OH99-C-N046-C for the National Cancer Institute-Frederick Research Donor Program. All blood donations were voluntary, with signed consent, and donor identities were blinded.
Preparation of ECO/siβ3 nanoparticles
Preparation of the cationic lipid ECO (MW = 1,023) and the targeting ligand RGD-PEG-maleimide (MAL) (PEG, 3.4k; Creative PEGWorks, Durham, NC) were conducted as described previously [18–20]. ECO was dissolved in 100% ethanol to a concentration of 50 mM. siβ3 was dissolved in nuclease-free water to a concentration of 25 μmol/L. Fresh RGD-PEG-MAL was dissolved in nuclease-free water to a concentration of 0.625 mmol/L. To prepare ECO/siβ3 nanoparticles, ECO and siβ3 were mixed at predetermined volumes so that the ratio of protonable amines (N) to phosphates on the siRNA backbone equals 8. In addition, nuclease-free water was added so that the ratio of ethanol:water was fixed at 1:20. To prepare targeted RGD-PEG-ECO/siβ3 nanoparticles, RGD-PEG-MAL was allowed to react with ECO in nuclease-free water for 30 min under gentile agitation. siβ3 in nuclease-free water was subsequently added in a drop-wise manner under gentile vortexing. Particles were subsequently mixed under gentile vortexing for an additional 30 min.
Stability testing
Stability of RGD-PEG-ECO/siβ3 nanoparticles was tested under storage at −80°C. Freshly prepared RGD-PEG-ECO/siβ3 nanoparticles were mixed with sucrose such that excipient concentrations were 5%, 10%, and 20% w/v. Samples were flash-frozen in liquid nitrogen and stored at −80°C for 1 day, 1 week, 3 months, 6 months, and 1 year, respectively. Data represent three independently conducted experiments. Stability of RGD-PEG-ECO/siβ3 nanoparticles was assessed postlyophilization. Freshly prepared RGD-PEG-ECO/siβ3 nanoparticles were mixed with sucrose such that excipient concentrations were 5%, 10%, and 20% w/v and, subsequently, flash-frozen in liquid nitrogen and lyophilized according to standard lyophilization procedure.
Nanoparticle characterization
Through dynamic light scattering using a Litesizer 500 from Anton Paar (Austria), the nanoparticle hydrodynamic diameter was determined, along with the polydispersity index and the intensity weight distribution. Measurements were performed with a 40 mW laser diode light (λ = 658 nm) at 22 ± 0.1°C, with scattered light collected at 90°. Nanoparticles were diluted 1:20 in nuclease-free water, 10 mM sodium chloride saline solution (pH = 7.2), and Dulbecco's phosphate-buffered saline (DPBS). Data represent three independently conducted experiments.
The zeta potential of nanoparticles was determined in nuclease-free water, 10 mM sodium chloride saline solution (pH = 7.2), and DPBS through electrophoretic light scattering by the Continuously-Monitored Phase-Analysis Light Scattering (cmPALS, patent EP2735870) technology from Austria. Measurements were taken using a 40 mW laser diode light (λ = 658 nm) at 15° detection angle. Data represent three independently conducted experiments.
Transmission electron microscopy
Nanoparticle diameter was confirmed using transmission electron microscopy (TEM) as described previously [24]. Briefly, nanoparticles were loaded onto a 300-mesh copper grid containing a carbon film (20 nm). Samples were stained with 3 μL of 2% uranyl acetate solution, dried, and imaged using TEM.
Gel electrophoresis for siRNA loading
Agarose gel electrophoresis was performed as described previously [18]. Briefly, 20 μL of nanoparticles were mixed with 4 μL of loading dye purchased from Roche (Basel, Switzerland) and loaded onto a 1% agarose gel containing ethidium bromide. The gel was subsequently submerged in 0.5× Tris/Borate/ethylenediaminetetraacetic acid containing ethidium bromide and ran at 100 V for 30 min. Free siRNA was used as a control. Gels were visualized using a ChemiDoc XRS system from BioRad (Hercules, CA).
Entrapment efficiency
siRNA entrapment efficiency was quantified using a Ribogreen assay from Thermo Fisher (Waltham, MA) [25]. Free siRNA following nanoparticle formation was quantified in reference to a linear standard, using a SpectraMax microplate reader from Molecular Devices (San Jose, CA) with an excitation of 500 nm and emission of 525 nm. siRNA entrapment efficiency was calculated by the following equation:
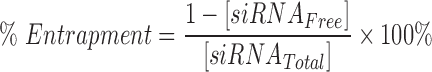 |
Immunoblotting analyses
BT549 cells (400,000 cells/well) were seeded onto a six well plate and allowed to adhere overnight. Cells were incubated with RGD-PEG-ECO/siNS (negative control) and RGD-PEG-ECO/siβ3 nanoparticles for 48 h [N/P = 8, (siRNA) = 100 nM] in complete growth medium. Cell pellets were harvested, washed in DPBS, and boiled in 1× Laemmli sample buffer with 2× protease inhibitor solution from Roche (Basel, Switzerland) for 10 min, vortexing every 5 min. After cooling for 2 min, samples were centrifuged at 15,000 g for 15 min. Protein extracts were subsequently quantified using Bradford/Lowry assay with bovine serum albumin used as a standard. Thirty micrograms of proteins were loaded onto a 4%–20% gradient gel and separated using sodium dodecyl sulfate/polyacrylamide gel electrophoresis. Separated proteins were transferred onto a nitrocellulose membrane using 1× Tris/Glycine buffer containing 5% methanol. Nitrocellulose membrane was incubated overnight with anti-β3 antibody from Cell Signaling (Danvers, MA; 1:1,000) and loading control anti-β-actin (Cell Signaling; 1:1,000). Gels were visualized using a ChemiDoc XRS system from BioRad.
Hemolysis
The analysis of the hemolytic properties of RGD-PEG-ECO/siβ3 nanoparticles was done according to the NCL protocol ITA-1, Analysis of Hemolytic Properties of Nanoparticles (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_ITA-1.pdf). Briefly, freshly drawn human blood was anticoagulated with lithium heparin and diluted in phosphate-buffered saline(PBS) to a final concentration of 10 mg/mL total blood hemoglobin. Diluted whole blood was subsequently incubated with RGD-PEG-ECO/siβ3 nanoparticles at concentrations of 0.01, 0.06, 0.3, and 3.3 μg/mL siβ3 for 3 h at 37°C. Afterward, cell-free supernatants were prepared using centrifugation and analyzed for the presence of plasma-free hemoglobin and its metabolites through the detection of cyanmethemoglobin by measuring the absorbance at 540 nm, against hemoglobin standards.
Platelet aggregation
The effect of long-term, −80°C stored RGD-PEG-ECO/siβ3 nanoparticles on human platelets was evaluated using NCL protocol ITA-2, Analysis of Platelet Aggregation (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_ITA-2.2.pdf). Platelet aggregation was monitored through changes in sample turbidity, as well as ATP release using ChronoLum reagent. In short, platelet rich plasma (PRP) and plasma poor plasma (PPP) were prepared from freshly drawn whole blood and pooled from three donors. PRP was incubated with RGD-PEG-ECO/siβ3 nanoparticles at concentrations of 0.01, 0.06, 0.3, and 3.3 μg/mL siβ3, as well as ChronoLum reagent. Sample turbidity and ATP release were analyzed using a ChronoLog aggregometer. Collagen and PPP were used as the positive and background controls, respectively.
Plasma coagulation times
Herein, the NCL protocol ITA-12, Coagulation Assay (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_ITA-12.pdf) was used to analyze the impact of long-term −80°C stored RGD-PEG-ECO/siβ3 nanoparticles on plasma coagulation time in vitro. Specifically, plasma coagulation was evaluated by examining prothrombin time (PT), activated partial thromboplastin time (APTT), and thrombin time (TT). Plasma from three donors was pooled from freshly drawn human blood and incubated with RGD-PEG-ECO/siβ3 nanoparticles at concentrations of 0.01, 0.06, 0.3, and 3.3 μg/mL siβ3 for 30 min at 37°C. Plasma coagulation initiation reagents (neoplastin, CaCl2, or thrombin) were subsequently added; coagulation times were measured using a STArt4 coagulometer (Diagnostica Stago). Normal and abnormal plasma are used as negative and positive controls, respectively.
Complement activation
Quantitative determination of complement activation was analyzed using an enzyme immunoassay outlined in NCL protocol ITA-5.2 (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_ITA-5.2.pdf). Briefly, plasma was pooled from three donors using freshly drawn human blood. Plasma was incubated with long-term −80°C stored RGD-PEG-ECO/siβ3 nanoparticles at concentrations of 0.01, 0.06, 0.3, and 3.3 μg/mL siβ3 and positive control cobra venom factor (CVF; Quidel Corp., A006) for 30 min at 37°C. Afterward, the samples were analyzed for the complement component iC3b using a MicroVue iC3b EIA Kit (Quidel Corp., A006) according to manufacturer's instructions.
Leukocyte proliferation assay
To assess the effects of long-term, −80°C stored RGD-PEG-ECO/siβ3 nanoparticles on the immunologic function of human leukocytes, the NCL protocol ITA-6 was used (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_ITA-6.pdf). In this study, human leukocytes were isolated from human blood and anti-coagulated with Li-heparin. Isolated leukocytes were incubated with or without positive control phytohemaglutinin-M (PHA-M), in the presence or absence of RGD-PEG-ECO/siβ3 nanoparticles at concentrations of 0.01, 0.06, 0.3, and 3.3 μg/mL siβ3 for 72 h at 37°C. Afterward, cells were incubated with MTT agent (5 mg/mL) for 4 h and analyzed spectrophotometrically using a plate reader at 570 nm.
Leukocyte procoagulant activity
The ability of long-term, −80°C stored RGD-PEG-ECO/siβ3 nanoparticles to induce leukocyte procoagulant activity in vitro was evaluated using NCL protocol ITA-17 (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_ITA-17.pdf). In this study, normal leukocytes represented by peripheral blood mononuclear cells (PBMCs) were isolated from human whole blood from three donors and anti-coagulated with Li-heparin. Cells were incubated with RGD-PEG-ECO/siβ3 nanoparticles at concentrations of 0.01, 0.06, 0.3, and 3.3 μg/mL siβ3, as well as positive control lipopolysaccharide (LPS) and negative control PBS for 24 h at 37°C. Cells were washed and incubated with autologous plasma to initiate coagulation. Coagulation was measured using the STart4 coagulometer (Diagnostica Stago).
Cytokines
NCL protocol ITA-10 (https://ncl.cancer.gov/sites/default/files/protocols/NCL_Method_ITA-10.pdf) was used to evaluate the ability of long-term, −80°C stored RGD-PEG-ECO/siβ3 nanoparticles to induce pro-inflammatory cytokines. Whole blood from healthy donor volunteers was anticoagulated using Li-heparin, diluted in RPMI-1640 medium (Invitrogen). From there, diluted whole blood was incubated with RGD-PEG-ECO/siβ3 nanoparticles at concentrations of 0.01, 0.06, 0.3, and 3.3 μg/mL siβ3, as well as positive control LPS and PHA-M for 24 h at 37°C. Following incubation, samples were centrifuged and supernatants were analyzed for the presence of inflammatory cytokines and interferons using multiplex ELISA Kits (Quansys, Logan, UT).
Statistical analysis
All experiments were independently replicated at least thrice unless otherwise stated. Statistical significance was calculated using GraphPad software. Statistical significance between two groups was calculated using an unpaired t-test. Data between three groups were compared using one-way analysis of variance with P < 0.05 being statistically significant.
Results
Formulation and stability of RGD-PEG-ECO/siβ3 nanoparticle in aqueous solutions
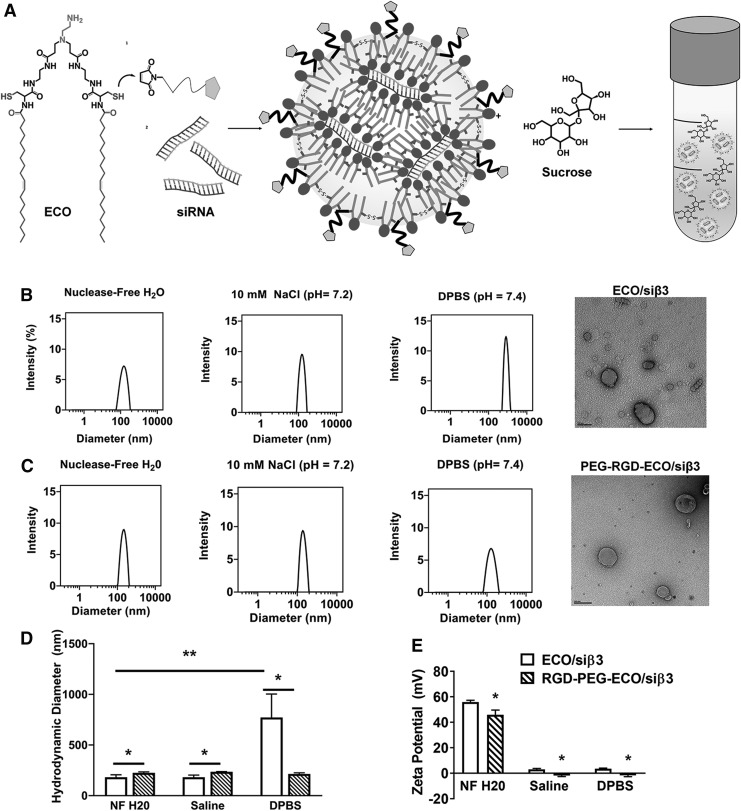
The targeted RGD-PEG-ECO/siβ3 nanoparticles were formulated by first mixing ECO with RGD-PEG-MAL (2.5 mol%), followed by self-assembly with unmodified siRNA in nuclease-free water (Fig. 1A). The targeting agent with the PEG spacer (MW = 3,400 Da) was designed to achieve cell-specific binding, improve stability in aqueous media, and minimize nonspecific tissue uptake of the nanoparticles. The formation of ECO/siβ3 and RGD-PEG-ECO/siβ3 nanoparticles was demonstrated using dynamic light scattering and TEM (Fig. 1B, C). Unmodified ECO/siβ3 nanoparticles had a uniform distribution with an average particle diameter of 181.46 ± 24 nm and zeta potential of 55.97 ± 1.36 mV in nuclease-free water and maintained an average size of 180.77 ± 21.73 nm in saline solution (10 mM, pH 7.2) with a decrease in zeta potential to 2.93 ± 0.74 mV (Fig. 1B, D, and E). Dilution of ECO/siβ3 nanoparticles in DPBS resulted in a significant increase in particle diameter to 772.25 ± 231.2 nm and a decrease in zeta potential to 3.56 ± 0.4 mV. This size increase is likely due to particle aggregation in buffered solution, which could affect intracellular siRNA delivery. In contrast, RGD-PEG-ECO/siβ3 nanoparticles maintained uniform size distributions in all tested media (Fig. 1C, D). Modification with RGD-PEG-MAL resulted in a slight increase of the nanoparticle diameter to 224.5 ± 9.41 nm and decreased zeta potential to 45.96 ± 3.67 mV compared with unmodified ECO/siβ3 nanoparticles. RGD-PEG-ECO/siβ3 nanoparticles had a near neutral zeta potential and a size of 234.34 ± 3.01 nm and 213 ± 11.42 nm when diluted in saline solution and DPBS, respectively. RGD-PEG-ECO/siβ3 nanoparticles maintained a similar nanoparticle size with no aggregation in buffered solution.
FIG. 1.
Characterization of freshly prepared ECO/siβ3 nanoparticles with and without MAL-PEG-RGD in aqueous solutions and in the presence or absence of sucrose. (A) Schematic of experimental design. Representative intensity peaks of ECO/siβ3 nanoparticles (B) and RGD-PEG-ECO/siβ3 nanoparticles (C) in nuclease-free water, 10 mM NaCl (pH = 7.2), and DPBS (CaCl2: 0.9 mM, MgCl2: 0.5 mM, KCl: 2.67 mM, KH2PO4: 1.47 mM, NaCl: 137.9 mM, Na2HPO4: 8 mM). Transmission electron microscopy images of ECO/siβ3 nanoparticles (B) and RGD-PEG-ECO/siβ3 nanoparticles (C). Comparison of the hydrodynamic diameters (D) and zeta potential (E) of ECO/siβ3 nanoparticles and RGD-PEG-ECO/siβ3 nanoparticles in aqueous solutions. (F) Representative intensity peaks of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose in nuclease-free water. Hydrodynamic diameter, polydispersity index (G), and zeta potential (H) of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose in nuclease-free water. (I) Agarose gel retardation of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose compared to free siβ3. (J) RiboGreen assay quantifying siRNA entrapment of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose. (K) Western blot analysis of β3 integrin expression (indicated by the arrowhead) in BT549 cells 48 h after RGD-PEG-ECO/siβ3 nanoparticle treatment (error bars denote SEM, *P < 0.05, **P < 0.01). DPBS, Dulbecco's phosphate-buffered saline; MAL, maleimide; PEG, polyethylene glycol; SEM, standard error of the mean.
We further investigated RGD-PEG-ECO/siβ3 nanoparticle stability, siRNA entrapment, and silencing efficiency in the presence of an excipient sucrose. The addition of various sucrose concentrations to freshly made RGD-PEG-ECO/siβ3 nanoparticles had no significant effect in nanoparticle diameter, size distribution, and zeta potential (Fig. 1F–H). Agarose gel electrophoresis revealed robust entrapment of siRNA in RGD-PEG-ECO/siβ3 nanoparticles (Fig. 1I). The addition of sucrose to RGD-PEG-ECO/siβ3 nanoparticles slightly improved siRNA entrapment compared with the no sucrose control, as confirmed by the quantitative Ribogreen assay, where 70%–90% siRNA entrapment was observed with increasing sucrose concentrations (Fig. 1I, J).
The gene silencing efficiency of RGD-PEG-ECO/siβ3 nanoparticles in the presence of sucrose was determined in BT549 breast cancer cells using western blotting. As previously reported, RGD-PEG-ECO/siβ3 nanoparticles reduced β3 integrin expression by ∼75% compared with cells treated with RGD-PEG-ECO/siNS nanoparticles containing a negative control siRNA [19]. Cells treated with RGD-PEG-ECO/siβ3 nanoparticles containing 5% and 10% sucrose exhibited a similar silencing efficiency of β3 integrin compared to the no sucrose control (Fig. 1K). Interestingly, formulations containing 20% sucrose were not able to reduce β3 integrin expression as effectively as the no sucrose control, indicating that high concentrations of sucrose may hinder silencing efficiency and therefore treatment efficacy. Together, the results indicate that surface modification with a targeting agent and a PEG spacer prevents aggregation and improves nanoparticle stability. In addition, the presence of sucrose did not affect the formation of RGD-PEG-ECO/siβ3 nanoparticles, but rather improved siRNA entrapment in the nanoparticles. Sucrose had no effect on nanoparticle gene silencing efficiency until its concentration reached 20% on the final nanoparticle formulation.
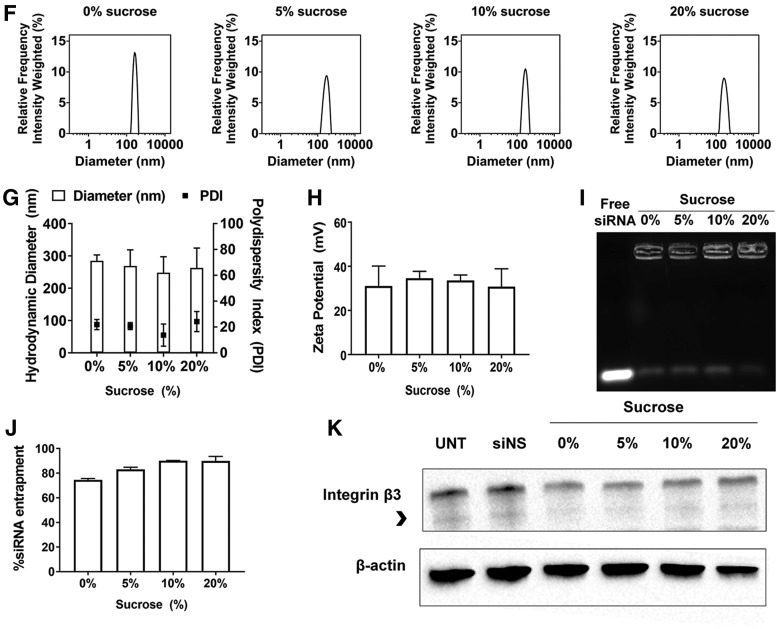
Sucrose stabilizes RGD-PEG-ECO/siβ3 nanoparticle formulation in −80°C
We sought to develop a stable frozen RGD-PEG-ECO/siβ3 formulation with sucrose as a stabilizing excipient for storage at −80°C. RGD-PEG-ECO/siβ3 nanoparticles aggregated significantly when stored in −80°C, as exhibited by the significant increase in nanoparticle diameter (1170.44 ± 590.88 nm) and polydispersity (27.97 ± 3.76%) (Fig. 2A, B). The addition of different concentrations of sucrose prevented RGD-PEG-ECO/siβ3 nanoparticle aggregation, as exhibited by the uniform size distribution and polydispersity with no significant changes in zeta potential (Fig. 2A–C). Agarose gel electrophoresis showed that sucrose had little effect on siRNA entrapment in RGD-PEG-ECO/siβ3 nanoparticles, while quantitative Ribogreen assay revealed slightly improved siRNA entrapment in frozen nanoparticle formulations in the presence of sucrose (Fig. 2D, E).
FIG. 2.
Sucrose improves −80°C storage of RGD-PEG-ECO/siβ3 nanoparticles. (A) Representative intensity peaks of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose poststorage in −80°C for 1 week. Comparison of the hydrodynamic diameters, polydispersity index, (B) and zeta potential (C) of RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose in nuclease-free water. (D) Agarose gel retardation of −80°C stored RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose compared with free siβ3. (E) RiboGreen assay quantifying siRNA entrapment of −80°C stored RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose. (F) Western blot analysis of β3 integrin (indicated by the arrowhead) expression in BT549 cells 48 h after treatment with −80°C stored RGD-PEG-ECO/siβ3 nanoparticles (error bars denote SEM, *P < 0.05 compared to formulations containing 0% sucrose).
Next, we investigated the silencing efficiency of the frozen RGD-PEG-ECO/siβ3 formulations. RGD-PEG-ECO/siβ3 formulations containing 5% and 10% sucrose exhibited an over 75% silencing efficiency of β3 integrin, denoted by the lower band as indicated, in BT549 cells compared to 60% with the formulation without sucrose (Fig. 2F). The preservation of silencing efficiency of the formulation with 5 of 10% sucrose is likely due to their stable small nanoparticle diameter, which is essential for efficient cellular uptake. Again, the formulation containing 20% sucrose showed lower β3 integrin silencing than those containing 5% and 10% sucrose. The results suggest that the addition of low amounts of sucrose (up to 10%) improved the stability of frozen RGD-PEG-ECO/siβ3 formulations at −80°C and maintained their high silencing efficiency.
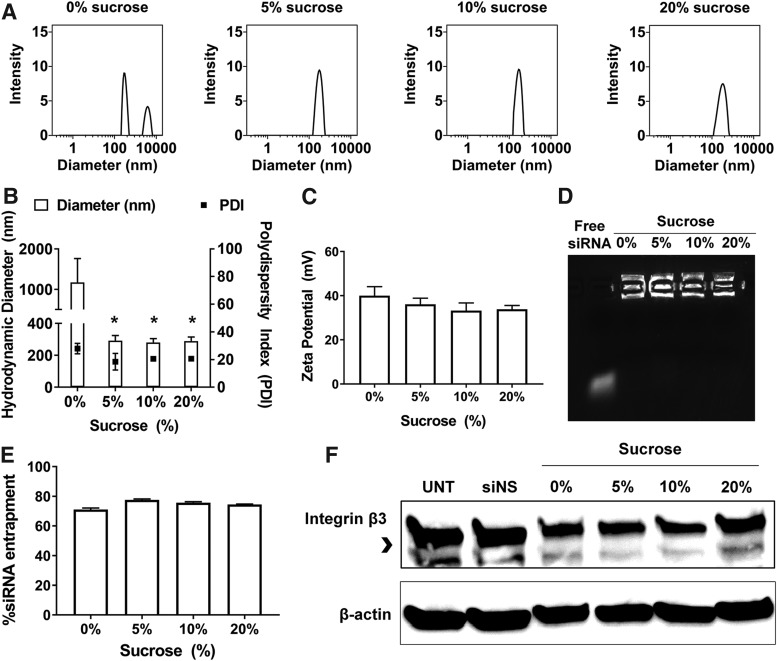
Lyophilized formations of RGD-PEG-ECO/siβ3 nanoparticles
We next sought to develop a lyophilized formulation of RGD-PEG-ECO/siβ3 nanoparticles. Solid formulations of RGD-PEG-ECO/siβ3 nanoparticles with and without sucrose were obtained by lyophilization. The formulations were reconstituted into a predetermined volume with nuclease-free water to assess their stability. After lyophilization, significant aggregation of RGD-PEG-ECO/siβ3 nanoparticles was observed, as exhibited by an increase in nanoparticle diameter and polydispersity (Fig. 3A, B). The addition of sucrose showed a concentration dependent reduction of particle aggregation upon lyophilization. Higher sucrose concentrations (up to 20%) were better in preventing nanoparticle aggregation (Fig. 3A, B). Lyophilization did not cause significant changes in zeta potential with or without sucrose (Fig. 3C). Gel electrophoresis showed the lack of siRNA bands with the lyophilized nanoparticles without sucrose, indicating nanoparticle destabilization (Fig. 3D). In contrast, the presence of sucrose maintained robust siRNA entrapment in the nanoparticles, as exhibited by gel electrophoresis and quantitative Ribogreen assay (Fig. 3D, E). Significant gene silencing (>70% silencing efficiency) was observed for the lyophilized RGD-PEG-ECO/siβ3 formulations containing 5% and 10% sucrose in BT549 cells, while the formulations of without sucrose and with 20% sucrose exhibited low silencing efficiency. Together, the addition of sucrose significantly improved the stability of lyophilized formulations of RGD-PEG-ECO/siβ3 and maintained a good gene silencing efficiency, especially with 10% sucrose.
FIG. 3.
Sucrose improves RGD-PEG-ECO/siβ3 nanoparticle stability postlyophilization. RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose were flash-frozen and lyophilized overnight. Formulations were reconstituted in their original volume of nuclease-free water. (A) Representative intensity peaks of reconstituted RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose. Comparison of the hydrodynamic diameters, polydispersity index, (B) and zeta potential (C) of reconstituted RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose in nuclease-free water. (D) Agarose gel retardation of reconstituted RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose compared to free siβ3. (E) RiboGreen assay quantifying siRNA entrapment of reconstituted RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% sucrose. (F) Western blot analysis of β3 integrin (indicated by the arrowhead) expression in BT549 cells 48 h after treatment with reconstituted RGD-PEG-ECO/siβ3 nanoparticles (error bars denote SEM, *P < 0.05 compared to formulations containing 0% sucrose).
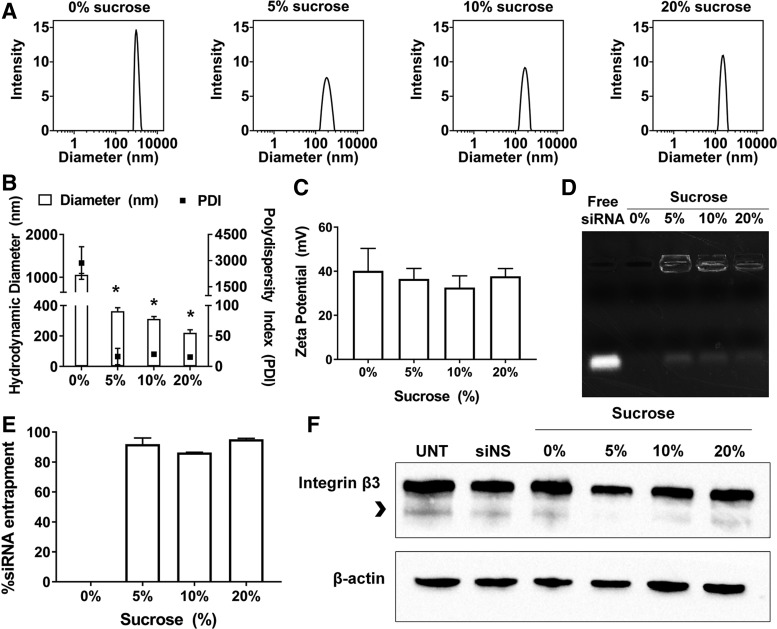
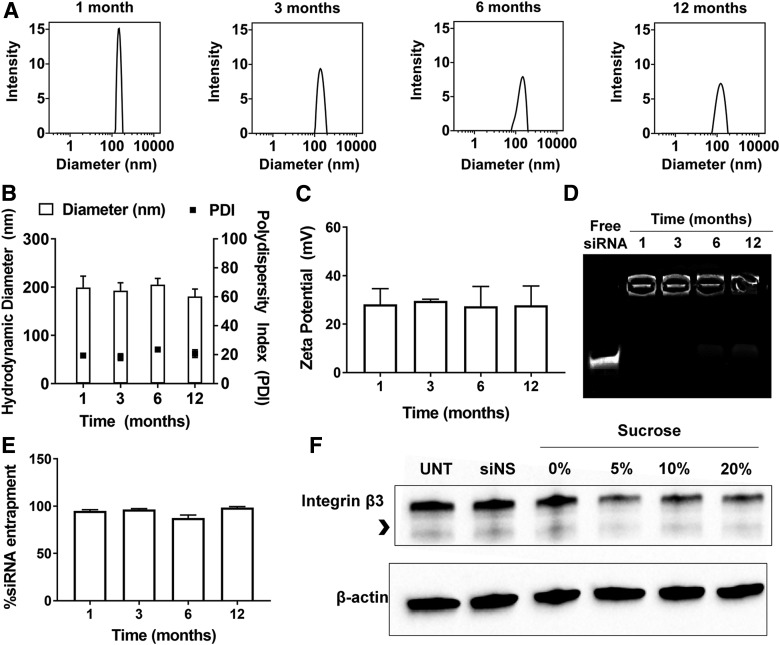
Long-term stability of frozen RGD-PEG-ECO/siβ3 nanoparticle formulations
Maintaining a long-term stability for the siRNA nanoparticle formulations is essential for their clinical translation and application. It appears that the frozen formulation of RGD-PEG-ECO/siβ3 with 5% sucrose is a promising formulation with the best stability and gene silencing efficiency. We next determined the stability of the frozen RGD-PEG-ECO/siβ3 formulation containing 5% sucrose in −80°C for up to 1 year. The frozen RGD-PEG-ECO/siβ3 formulation exhibited no significant change in particle diameter, polydispersity, and zeta potential at −80°C for up to 12 months, indicating good stability of the formulation during long-term storage (Fig. 4A–D). No significant change in siRNA entrapment was observed after 1 year of storage, with siRNA entrapment ranging from 85% to 90% compared to naked siRNA controls (Fig. 4D, E). The long-term stability of the formulations containing 10% and 20% sucrose was also investigated for up to 12 months; no significant change of the parameters was observed for the formulations during the storage (Supplementary Fig. S1).
FIG. 4.
Sucrose improves long-term storage of RGD-PEG-ECO/siβ3 nanoparticles. RGD-PEG-ECO/siβ3 nanoparticles containing 5% sucrose were flash-frozen and stored in −80°C for 1, 3, 6, and 12 months. (A) Representative intensity peaks of long-term stored RGD-PEG-ECO/siβ3 nanoparticles containing 5% sucrose in nuclease-free water. Comparison of the hydrodynamic diameters, polydispersity indexes (B), and zeta potential (C) of RGD-PEG-ECO/siβ3 nanoparticles stored for 1, 3, 6, and 12 months in nuclease-free water. (D) Agarose gel retardation of RGD-PEG-ECO/siβ3 nanoparticles stored for 12 months compared to free siβ3. (E) RiboGreen assay quantifying siRNA entrapment of RGD-PEG-ECO/siβ3 nanoparticles stored for 1, 3, 6, and 12 months. (F) Western blot analysis of β3 integrin expression (indicated by the arrowhead) in BT549 cells 48 h after treatment with RGD-PEG-ECO/siβ3 nanoparticles containing 0%, 5%, 10%, and 20% stored for 12 months (error bars denote SEM).
The frozen formulation with 5% and 10% sucrose maintained gene silencing efficiency after storage at −80°C for 12 months, with ∼75% silencing efficiency (Fig. 4F). Interestingly, the formulation containing 20% sucrose exhibited 65% silencing efficiency, higher than the freshly prepared formulations. The formulation without sucrose maintained only 37% silencing compared to BT549 cells treated with RGD-PEG-ECO/siNS nanoparticles, a substantial decrease from 75% silencing efficiency of freshly made particles. These results show together that the frozen RGD-PEG-ECO/siβ3 nanoparticle formulation with sucrose possesses long-term stability with consistent nanoparticle diameter, polydispersity, and preserved biological functions.
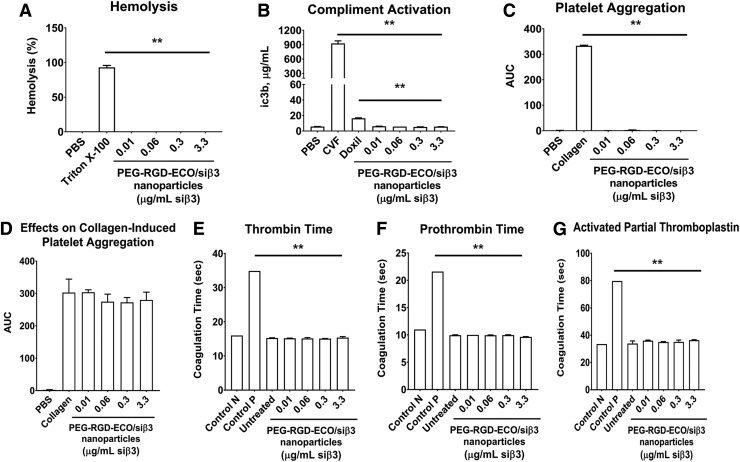
Hemocompatibility of frozen RGD-PEG-ECO/siβ3 formulation
The hemocompatibility of the frozen RGD-PEG-ECO/siβ3 formulation with 5% sucrose was assessed in vitro with standard procedures. Incubation of the formulation with erythrocytes did not induce hemolysis at all tested concentrations (Fig. 5A). Since systemic administration of nanoparticles may activate the complement system, we analyzed complement activation by measuring reactive thioester iC3b in human plasma exposed to RGD-PEG-ECO/siβ3 nanoparticles. Compared to positive control CVF, human plasma incubated with RGD-PEG-ECO/siβ3 nanoparticles showed no activation of the human complement system in vitro at all tested concentrations, as shown by the low expression iC3b similar to negative control PBS (Fig. 5B). Interestingly, RGD-PEG-ECO/siβ3 nanoparticles showed significantly lower expression of iC3b than Doxil, an FDA approved nanoparticle formulation.
FIG. 5.
(A) Hemolytic activity of frozen RGD-PEG-ECO/siβ3 nanoparticles compared to negative control PBS and positive control Triton X-100. (B) Complement activation of frozen RGD-PEG-ECO/siβ3 nanoparticles compared to positive control cobra venom factor, negative control PBS, and liposomal formulation Doxil. The effect of frozen RGD-PEG-ECO/siβ3 nanoparticles on platelet aggregation (C) and collagen-induced platelet aggregation (D) was evaluated compared to positive control collagen and negative control PBS. Human plasma coagulation was evaluated comparing thrombin time (E), prothrombin time (F), and activated partial thromboplastin (G) in response to treatment with varying concentrations of frozen RGD-PEG-ECO/siβ3 nanoparticles. Controls used were normal (control N) and abnormal (control P) plasma standards, as well as untreated plasma (error bars denote SEM, **P < 0.01). PBS, phosphate-buffered saline.
We next investigated the effects of RGD-PEG-ECO/siβ3 nanoparticles on platelet aggregation and anticoagulation in vitro. At all tested concentrations of RGD-PEG-ECO/siβ3, no significant platelet aggregation was observed upon incubation with PRP compared to negative control, while significant aggregation was observed for positive control, collagen (Fig. 5C). Incubation of PRP with collagen and RGD-PEG-ECO/siβ3 nanoparticles showed no significant change in platelet aggregation induced by collagen (Fig. 5D). The effect of RGD-PEG-ECO/siβ3 nanoparticles on coagulation time was also evaluated using three specialized assays: TTT (final common pathway), PT (extrinsic pathway), and APTT assay (intrinsic pathway). At all tested concentrations, incubation of RGD-PEG-ECO/siβ3 nanoparticles with human plasma did not significantly affect coagulation time compared to normal (control N) plasma standards in all three tests (Fig. 5E–G). Together, these results indicate good hemocompatibility of the frozen RGD-PEG-ECO/siβ3 nanoparticle formulation.
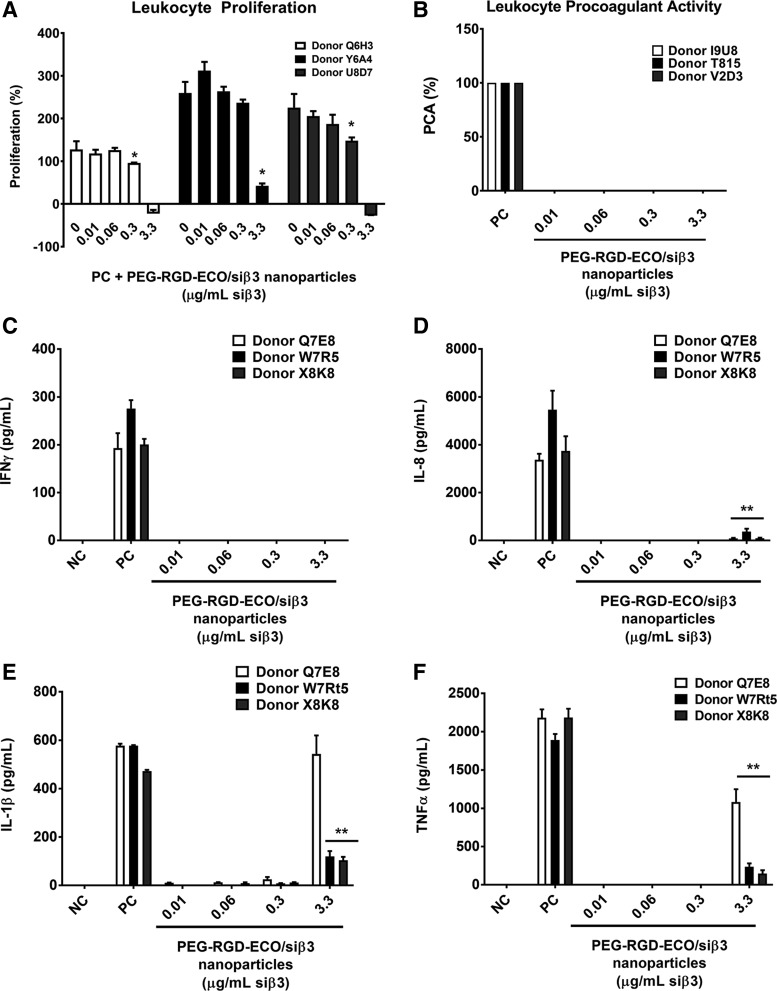
Immunogenicity of the frozen RGD-PEG-ECO/siβ3 formulation
The immunogenicity of the frozen RGD-PEG-ECO/siβ3 formulation with 5% sucrose was evaluated based on leukocyte activation, procoagulant activity, and cytokine secretion. To assess the effects of RGD-PEG-ECO/siβ3 nanoparticles on leukocyte proliferation, the nanoparticles were incubated for 72 h with human leukocytes in the presence of PHA-M, a lectin mucoprotein used for the stimulation of significant leukocyte proliferation. RGD-PEG-ECO/siβ3 nanoparticles inhibited PHA-M induced leukocyte proliferation at the siβ3 concentration of 0.3 μg/mL in two donors and 3.3 μg/mL in three donors. No inhibition was observed at lower siβ3 concentrations (Fig. 6A). To assess leukocyte procoagulant activity, RGD-PEG-ECO/siβ3 nanoparticles were incubated with normal leukocytes (PBMCs) from healthy donor volunteers. RGD-PEG-ECO/siβ3 nanoparticles did not induce leukocyte procoagulant activity in vitro at all tested concentrations (Fig. 6B).
FIG. 6.
(A) The effect of frozen RGD-PEG-ECO/siβ3 nanoparticles on PHA-M proliferation using three different donors at various concentrations compared to positive control PHA-M. (B) Leukocyte procoagulant activity of RGD-PEG-ECO/siβ3 nanoparticles compared to positive control Escherichia coli K12 lipopolysaccharide treated peripheral blood mononuclear cells. The effect of RGD-PEG-ECO/siβ3 nanoparticle treatment on pro-inflammatory cytokines IFNγ (C), IL-8 (D), IL-1β (E), and TNFα (F) was evaluated in whole blood cultures of healthy donor volunteers (n = 3) (error bars denote SEM, *P < 0.05, **P < 0.01). PHA-M, phytohemaglutinin-M; IL, interleukin; IFN, interferon; TNFα, tumor necrosis factor alpha.
The ability of RGD-PEG-ECO/siβ3 nanoparticles to induce elevated cytokine levels in human whole blood was investigated using enzyme linked immunosorbent assay. Interferon (IFN)-γ was not induced at all tested concentrations of RGD-PEG-ECO/siβ3 nanoparticles (Fig. 6C). No induction of pyrogenic markers interleukin (IL)-8, IL-1β, and tumor necrosis factor alpha (TNFα) was observed until the tested siβ3 concentration reached 3.3 μg/mL in all three human donors (Fig. 6D–F). However, RGD-PEG-ECO/siβ3 nanoparticles at high siβ3 concentration (3.3 μg/mL siβ3) resulted in an increase of TNFα and IL-8 induction, but significantly lower compared to positive control LPS + PHA-M contained in whole blood from all tested donors. RGD-PEG-ECO/siβ3 nanoparticles at 3.3 μg/mL siβ3 resulted in high induction of IL-1β expression in one donor, but significantly lower induction in two other donors compared to positive control. Taken together, these results indicate low immunogenicity of the frozen RGD-PEG-ECO/siβ3 nanoparticle formulation.
Discussion
The broad clinical development of RNAi therapies is hindered by the lack of biocompatible and stable formulations that preserve the integrity and functionality of siRNA nanoparticles in the long term [26]. In this work, we have shown that the targeted ECO/siRNA nanoparticles are able to form stable formulations with preserved gene silencing efficiency when stored for at least 1 year. Compared to other lipid/siRNA nanoparticles, the ECO/siRNA nanoparticles possess several unique features, including formation of stable nanoparticles with nucleic acids without helper lipids and pH-sensitive amphiphilic endosomal escape and reductive cytosolic siRNA release (PERC). Since the lipid carrier ECO has two thiol groups, it forms stable nanoparticles with siRNA at a low N/P ratio by forming reversible disulfide bonds, without the presence of a helper lipid, for example, cholesterol that is commonly used in other lipid siRNA nanoparticles. The surface of ECO/siβ3 nanoparticles can readily be modified with a targeting agent through a PEG spacer for targeted siRNA delivery and for further stabilization of the nanoparticle formulation. Moreover, ECO was previously shown to protect the siRNAs from serum degradation, addressing a main hurdle in siRNA delivery [18]. Other methods can be used to evaluate siRNA stability, such as Förster resonance energy transfer, where fluorescence dequenching is observed upon nucleic acid degradation [27–29]. Additional research is ongoing for evaluating the intracellular uptake of RGD-PEG-ECO/siRNA nanoparticles. Enhanced stability in saline and buffer solutions was achieved with the targeted RGD-PEG-ECO/siβ3 nanoparticles. Besides the targeting agent tested in this study, other targeting agents can also be readily incorporated into the nanoparticles using the same chemistry to target different molecular targets on cell surface.
The stability of frozen and lyophilized solid formulations of RGD-PEG-ECO/siβ3 nanoparticles was further improved by adding a cryoprotectant sucrose. It has been shown that the freezing process could compromise particle stability [30]. Lyophilization could also cause irreversible nanoparticle fusion and particle aggregation [14,30,31]. Various excipients, including sucrose, have been used to reserve nanoparticle integrity, for example, with a concentration as high as 30% for sucrose [14,32]. In this study, we have shown that frozen formulations maintained more consistent physicochemical properties than the lyophilized formulations, especially at low sucrose content. The addition of as little as 5% sucrose was sufficient to prevent particle aggregation in the frozen formulations and alleviate aggregation in the lyophilized formulations. A higher concentration of sucrose (10%–20%) was needed for the lyophilized formulations, likely due to the additional protection needed for water removal upon lyophilization [15]. However, the RGD-PEG-ECO/siβ3 formulation containing 20% sucrose showed reduced silencing compared to the RGD-PEG-ECO/siβ3 formulations with 5% and 10% sucrose. This could be attributed to inhibition of clathrin-dependant endocytosis by the increased sucrose concentration [33]. In addition, increased sucrose concentration has been shown to enhance transforming growth factor-β signaling, which could also attribute to the elevated expression of β3 integrin [34]. Therefore, the frozen formulation with 5% sucrose could be a suitable formulation of the targeted siRNA nanoparticles for further clinical development.
The frozen RGD-PEG-ECO/siβ3 formulation with 5% sucrose exhibited good blood compatibility, which is essential for its further clinical translation. Because RNAi therapies are administered intravenously, they should avoid negative side effects such as hemolysis, compliment activation, cytokine secretion, and leukocyte procoagulation [35]. It has been reported that some siRNA nanoparticles formulated with cationic polymers and liposomes exhibited cytotoxicity, concentration dependent disseminated intravascular coagulation, and induced platelet activation [36–39]. The RGD-PEG-ECO/siβ3 formulation showed no significant effect at all tested concentrations on hemolysis, compliment activation, and platelet and leukocyte coagulation. Specifically for RGD-PEG-ECO/siβ3 nanoparticles, β3 integrin is a platelet receptor that plays an essential role in hemostasis and thrombosis [40,41]. It binds a number of ligands, including fibrinogen, fibrin, von Willebrand factor, and fibronectin, to mediate platelet aggregation [40]. The incubation of platelets with RGD-PEG-ECO/siβ3 nanoparticles did not induce any negative effects on platelet aggregation.
The frozen RGD-PEG-ECO/siβ3 formulation exhibited low immunogenicity. Substantial effect on leukocyte proliferation was observed until reaching the highest testing siβ3 concentration at 3.3 μg/mL. The inhibition of leukocyte proliferation at this high concentration could be attributed to silencing β3 integrin, as β3 integrin is essential for T cell proliferation [42]. The RGD-PEG-ECO/siβ3 formulation did not induce IFN-γ secretion at all concentrations. Induced secretion of TNFα, IL-8, and IL-1β was only observed at the highest siβ3 concentration (3.3 μg/mL). The potential immunogenicity and induced cytokine secretion for in vivo application could be avoided by controlling overall blood concentration during intravenous infusion.
In summary, we have developed a frozen formulation of RGD-PEG-ECO/siβ3 nanoparticles with prolonged stability and preserved biological function for clinical development of RNAi therapeutics. The formulation maintains excellent nanoparticle integrity and effective biological function after storage for at least 1 year. The RGD-PEG-ECO/siβ3 formulation also exhibits excellent hemocompatibility and low immunogenicity. Further studies are needed for comprehensive evaluation of the safety, pharmacokinetics, and pharmacodynamics in animal models before clinical development of the formulation. Since RNAi based therapy has the potential to treat a plethora of diseases, the nanoparticle formulation has the potential to serve as a delivery platform for various RNAi therapeutics for broader clinical applications.
Supplementary Material
Acknowledgments
The work was supported, in part, by the National Cancer Institute, the National Institutes of Health, under award number R01CA194518. The formulation described herein was characterized by the Nanotechnology Characterization Laboratory as part of its free Assay Cascade characterization service for cancer nanomedicines, https://ncl.cancer.gov/working-ncl/ncl-assay-cascade-application-process. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Z.-R.L. is a M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering.
Author Disclosure Statement
Z.-R.L. is a cofounder of Cleveland Theranostics, LLC, a startup company focused on the development of multifunctional pH-sensitive amino lipids for gene therapy.
Z.-R.L. and N.R.A. have a patent interest related to this work. All other authors declare no conflicts of interest.
Supplementary Material
References
- 1. Kulkarni JA, Cullis PR. and van der Meel R. (2018). Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther 28:146–157 [DOI] [PubMed] [Google Scholar]
- 2. Gaudet D, Drouin-Chartier JP. and Couture P. (2017). Lipid metabolism and emerging targets for lipid-lowering therapy. Can J Cardiol 33:872–882 [DOI] [PubMed] [Google Scholar]
- 3. Barrett ADT. (2018). Current status of Zika vaccine development: Zika vaccines advance into clinical evaluation. NPJ Vaccines 3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adams D, Gonzalez-Duarte A, O'Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, et al. (2018). Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med 379:11–21 [DOI] [PubMed] [Google Scholar]
- 5. Lorenzer C, Dirin M, Winkler AM, Baumann V. and Winkler J. (2015). Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics. J Control Release 203:1–15 [DOI] [PubMed] [Google Scholar]
- 6. Tatiparti K, Sau S, Kashaw SK. and Iyer AK. (2017). siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials (Basel) 7:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gujrati M, Vaidya A. and Lu ZR. (2016). Multifunctional pH-sensitive amino lipids for siRNA delivery. Bioconjug Chem 27:19–35 [DOI] [PubMed] [Google Scholar]
- 8. Chakraborty C, Sharma AR, Sharma G, Doss CGP. and Lee SS. (2017). Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids 8:132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garba AO. and Mousa SA. (2010). Bevasiranib for the treatment of wet, age-related macular degeneration. Ophthalmol Eye Dis 2:75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lusthaus JA. and Goldberg I. (2016). Investigational and experimental drugs for intraocular pressure reduction in ocular hypertension and glaucoma. Expert Opin Investig Drugs 25:1201–1208 [DOI] [PubMed] [Google Scholar]
- 11. Wittrup A. and Lieberman J. (2015). Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet 16:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leung RK. and Whittaker PA. (2005). RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol Ther 107:222–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R. and Wagner E. (1998). The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther 5:1425–1433 [DOI] [PubMed] [Google Scholar]
- 14. Ball RL, Bajaj P. and Whitehead KA. (2017). Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int J Nanomed 12:305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdelwahed W, Degobert G, Stainmesse S. and Fessi H. (2006). Freeze-drying of nanoparticles: formulation, process and storage considerations. Adv Drug Deliv Rev 58:1688–1713 [DOI] [PubMed] [Google Scholar]
- 16. Andersen MO, Howard KA, Paludan SR, Besenbacher F. and Kjems J. (2008). Delivery of siRNA from lyophilized polymeric surfaces. Biomaterials 29:506–512 [DOI] [PubMed] [Google Scholar]
- 17. Malamas AS, Gujrati M, Kummitha CM, Xu R. and Lu ZR. (2013). Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J Control Release 171:296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gujrati M, Malamas A, Shin T, Jin E, Sun Y. and Lu ZR. (2014). Multifunctional cationic lipid-based nanoparticles facilitate endosomal escape and reduction-triggered cytosolic siRNA release. Mol Pharm 11:2734–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parvani JG, Gujrati MD, Mack MA, Schiemann WP. and Lu ZR. (2015). Silencing beta3 integrin by targeted ECO/siRNA nanoparticles inhibits EMT and metastasis of triple-negative breast cancer. Cancer Res 75:2316–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gujrati M, Vaidya AM, Mack M, Snyder D, Malamas A. and Lu ZR. (2016). Targeted dual pH-sensitive lipid ECO/sirna self-assembly nanoparticles facilitate in vivo cytosolic sieIF4E delivery and overcome paclitaxel resistance in breast cancer therapy. Adv Healthc Mater 5:2882–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X, Jiang H, Sun D, Scheidt J, Qian V, et al. (2019). Systemic delivery of tumor-targeting siRNA nanoparticles against an oncogenic lncRNA facilitates effective triple-negative breast cancer therapy. Bioconjug Chem 30:907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, Janke O, Endruschat J, Durieux B, et al. (2008). Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res 68:9788–9798 [DOI] [PubMed] [Google Scholar]
- 23. Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O, Lange C, Giese K, Kaufmann J, Khan M. and Drevs J. (2014). First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol 32:4141–4148 [DOI] [PubMed] [Google Scholar]
- 24. Sun D, Sahu B, Gao S, Schur RM, Vaidya AM, Maeda A, Palczewski K. and Lu ZR. (2017). Targeted multifunctional lipid ECO plasmid DNA nanoparticles as efficient non-viral gene therapy for Leber's congenital amaurosis. Mol Ther Nucleic Acids 7:42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones LJ, Yue ST, Cheung CY. and Singer VL. (1998). RNA quantitation by fluorescence-based solution assay: RiboGreen reagent characterization. Anal Biochem 265:368–374 [DOI] [PubMed] [Google Scholar]
- 26. Suzuki Y, Hyodo K, Tanaka Y. and Ishihara H. (2015). siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo. J Control Release 220:44–50 [DOI] [PubMed] [Google Scholar]
- 27. Gupta K, Afonin KA, Viard M, Herrero V, Kasprzak W, Kagiampakis I, Kim T, Koyfman AY, Puri A, et al. (2015). Bolaamphiphiles as carriers for siRNA delivery: from chemical syntheses to practical applications. J Control Release 213:142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gupta K, Mattingly SJ, Knipp RJ, Afonin KA, Viard M, Bergman JT, Stepler M, Nantz MH, Puri A. and Shapiro BA. (2015). Oxime ether lipids containing hydroxylated head groups are more superior siRNA delivery agents than their nonhydroxylated counterparts. Nanomedicine (Lond) 10:2805–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim T, Afonin KA, Viard M, Koyfman AY, Sparks S, Heldman E, Grinberg S, Linder C, Blumenthal RP. and Shapiro BA. (2013). In silico, in vitro, and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Mol Ther Nucleic Acids 2:e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kasper JC, Troiber C, Kuchler S, Wagner E. and Friess W. (2013). Formulation development of lyophilized, long-term stable siRNA/oligoaminoamide polyplexes. Eur J Pharm Biopharm 85:294–305 [DOI] [PubMed] [Google Scholar]
- 31. Kundu AK, Chandra PK, Hazari S, Ledet G, Pramar YV, Dash S. and Mandal TK. (2012). Stability of lyophilized siRNA nanosome formulations. Int J Pharm 423:525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chacon M, Molpeceres J, Berges L, Guzman M. and Aberturas MR. (1999). Stability and freeze-drying of cyclosporine loaded poly(d,l lactide-glycolide) carriers. Eur J Pharm Sci 8:99–107 [DOI] [PubMed] [Google Scholar]
- 33. Hansen SH, Sandvig K. and van Deurs B. (1993). Clathrin and HA2 adaptors: effects of potassium depletion, hypertonic medium, and cytosol acidification. J Cell Biol 121:61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen CL, Hou WH, Liu IH, Hsiao G, Huang SS. and Huang JS. (2009). Inhibitors of clathrin-dependent endocytosis enhance TGFbeta signaling and responses. J Cell Sci 122:1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dobrovolskaia MA. and McNeil SE. (2013). Understanding the correlation between in vitro and in vivo immunotoxicity tests for nanomedicines. J Control Release 172:456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dobrovolskaia MA, Patri AK, Simak J, Hall JB, Semberova J, De Paoli Lacerda SH. and McNeil SE. (2012). Nanoparticle size and surface charge determine effects of PAMAM dendrimers on human platelets in vitro. Mol Pharm 9:382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ilinskaya AN. and Dobrovolskaia MA. (2013). Nanoparticles and the blood coagulation system. Part II: safety concerns. Nanomedicine (Lond) 8:969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dobrovolskaia MA, Patri AK, Potter TM, Rodriguez JC, Hall JB. and McNeil SE. (2012). Dendrimer-induced leukocyte procoagulant activity depends on particle size and surface charge. Nanomedicine (Lond) 7:245–256 [DOI] [PubMed] [Google Scholar]
- 39. Juliano RL, Hsu MJ, Peterson D, Regen SL. and Singh A. (1983). Interactions of conventional or photopolymerized liposomes with platelets in vitro. Exp Cell Res 146:422–427 [DOI] [PubMed] [Google Scholar]
- 40. Durrant TN, van den Bosch MT. and Hers I. (2017). Integrin alphaIIbbeta3 outside-in signaling. Blood 130:1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nieswandt B, Varga-Szabo D. and Elvers M. (2009). Integrins in platelet activation. J Thromb Haemost 7 (Suppl. 1):206–209 [DOI] [PubMed] [Google Scholar]
- 42. Neto EH, Coelho AL, Sampaio AL, Henriques M, Marcinkiewicz C, De Freitas MS. and Barja-Fidalgo C. (2007). Activation of human T lymphocytes via integrin signaling induced by RGD-disintegrins. Biochim Biophys Acta 1773:176–184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.