Abstract
Fecal pollution of surface waters presents a global human health threat. New molecular indicators of fecal pollution have been developed to address shortcomings of traditional culturable fecal indicators. However, there is still little information on their fate and transport in the environment. The present study uses spatially and temporally extensive data on traditional (culturable enterococci, cENT) and molecular (qPCR-enterococci, qENT and human-associated marker, HF183/BacR287) indicator concentrations in marine water surrounding highly-urbanized San Francisco, California, USA to investigate environmental and anthropogenic processes that impact fecal pollution. We constructed multivariable regression models for fecal indicator bacteria at 14 sampling stations. The human marker was detected more frequently in our study than in many other published studies, with detection frequency at some stations as high as 97%. The odds of cENT, qENT, and HF183/BacR287 exceeding health-relevant thresholds were statistically elevated immediately following discharges of partially treated combined sewage, and cENT levels dissipated after approximately 1 day. However, combined sewer discharges were not important predictors of indicator levels typically measured in weekly monitoring samples. Instead, precipitation and solar insolation were important predictors of cENT in weekly samples, while precipitation and water temperature were important predictors of HF183/BacR287 and qENT. The importance of precipitation highlights the significance of untreated storm water as a source of fecal pollution to the urban ocean, even for a city served by a combined sewage system. Sunlight and water temperature likely control persistence of the indicators via photoinactivation and dark decay processes, respectively.
1. Introduction
Globally, it is estimated that each year exposure to coastal waters polluted with human waste causes more than 120 million gastrointestinal and 50 million severe respiratory illnesses.1 Collectively, illness caused by exposure to recreational waters is referred to as recreational waterborne illness (RWI). In an effort to reduce RWI, the US uses densities of fecal indicator bacteria (FIB) Escherichia coli and enterococci to assess risk. Similar monitoring programs are in place around the globe, guided by recommendations from the World Health Organization.2,3 Although FIB are not usually etiologic agents of RWI, they are used to evaluate coastal water quality, as FIB densities in coastal waters have been linked quantitatively to swimmer illness in epidemiology studies.4 When FIB densities exceed threshold values, beach advisories or closures are issued warning swimmers that exposure could lead to illness.
There are a number of shortcomings associated with using FIB to assess risk.5 In the USA, enterococci alone are recommended by the United States Environmental Protection Agency (USEPA) to assess RWI risk in marine waters owing to their good correlation with adverse health outcomes in epidemiology studies conducted in salt water.6 Enterococci enumeration methods require incubation of bacteria for 24 hours.7–9 This means that beach advisories and closures that warn the public of poor water quality conditions can be issued no sooner than one day after sampling. This is problematic because enterococci concentrations in beach waters are known to vary over time scales shorter than 24 hours, and therefore next-day beach notifications may not accurately reflect the water quality conditions to which bathers are exposed.5
An additional shortcoming concerns the source of FIB. Enterococci occur not only in wastewater and human feces, but also in the feces of other mammals and birds10,11 and naturally in the environment.12 Epidemiology studies that tie enterococci to adverse health outcomes in swimmers, which represent the cornerstone of beach water quality criteria, were conducted at beaches where FIB were believed to come from treated human wastewater. Since exposure to water containing a given concentration of enterococci generally presents a greater threat to human health if the enterococci are derived from human rather than animal sources,13 the risk presented by enterococci of unknown origin is uncertain. Therefore methods that can identify the source of fecal pollution in recreational waters would be expected to improve protection of human health.5
Recent methodological advances have sought to address these shortcomings. To shorten the time required to assess microbiological water quality, rapid molecular assays based on quantitative polymerase chain reaction (qPCR) for enterococci (qENT) have been developed. A drawback of qPCR, however, is that it may perform poorly if sample matrix constituents interfere with the polymerase chain reaction.6 Because of this, it is encouraged to conduct site-specific evaluations of qENT assays over a range of environmental conditions prior to adopting qENT-based recreational water criteria.6 Furthermore, the source concentrations and environmental fate of qENT and culturable enterococci (cENT) may be distinct. For example, mesocosm experiments suggest that the persistence of culturable and molecular fecal indicators may be differentially impacted by sunlight,14,15 and other research suggests that cENT : qENT ratios may vary by source.16,17 To date, there is limited work comparing the spatio-temporal variation of qENT and cENT in the field over large spatial and temporal scales to elucidate factors that differentially affect their concentrations.
At the same time, new microbial source tracking assays have been developed that aim to detect bacteria that come specifically from human feces.13 The performance of human- associated markers has been studied in multi-laboratory trials.18 Studies have examined the occurrence of the most commonly employed marker, which targets the HF183 16S rRNA gene cluster of the Bacteroides genus (HF183/BacR287, henceforth “HF183”), in storm water,19 wastewater,20,21 fresh-water,22,23 and marine water.24–28 However, there is still little observational data available on the occurrence and cooccurrence of HF183 with qENT and cENT in shoreline marine waters over extended temporal and spatial scales. Furthermore, to our knowledge, there are no published studies seeking to understand how multiple environmental factors modulate HF183 fate and transport in surface waters.
To address these knowledge gaps, we measured cENT, qENT, and HF183 (collectively called “indicators”) over two years at 14 stations spanning approximately 40 km of shoreline marine waters around the peninsula city of San Francisco, California, USA (SF). These waters surround several highly-urbanized watersheds serviced by combined wastewater and storm water systems, and are subject to occasional partially treated combined sewer discharges (CSDs). San Francisco has a population over 800 000,29 65% impervious land cover,30 and a Mediterranean climate31 with rain events and CSDs occurring primarily from November to March.
Specifically, this study sought to answer several fundamental and applied research questions. First, we aimed to understand how prevalent these three indicators are and what that suggests about their sources. Second, we aimed to understand how using qENT in lieu of cENT for routine, state-mandated monitoring would affect the number of beach advisories in the region. Third, we aimed to understand how environmental factors and anthropogenic inputs such as CSDs are associated with indicator levels. The results of this study provide valuable information on the source of microbial pollution in this region, and important insights on how various environmental processes affect the fate and transport of traditional (cENT) and more novel indicators (qENT and HF183).
2. Methods
2.1. Sample collection
500 mL of marine water was collected in 2014 and 250 mL in 2016 from 14 routinely monitored ocean and bay stations (“monitoring stations”) surrounding the perimeter of San Francisco, California where recreational water contact may occur (Fig. 1). Samples were collected weekly on the same day of the week (Monday) from January to November in 2014 and from March to November in 2016, yielding a total of 1050 samples (75 weekly samples at each of 14 monitoring stations). Weekly monitoring samples were typically collected in the morning (11 of 1050 were collected after noon) and transported to the laboratory on ice for immediate processing. The time of sample collection was noted for each sample.
Fig. 1.
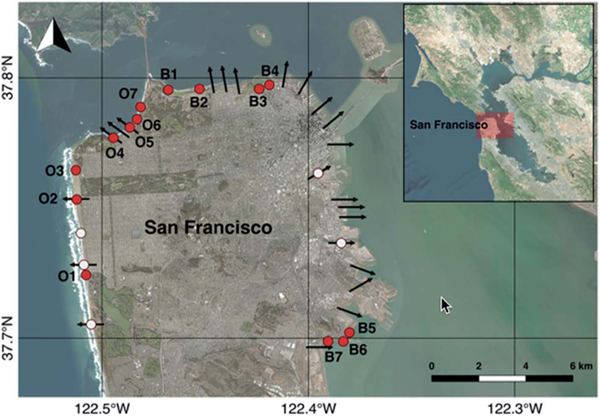
Sampling stations and CSD locations around San Francisco. Red circles represent weekly monitoring stations. White circles represent stations sampled only when a CSD occurs. Arrows represent locations where CSDs may occur. Arrows are symbolic and arrow lengths do not correspond to CSD volumes or distance offshore of discharge. Map made in QGIS open source software (http://qgis.osgeo.org) using Bing satellite imagery.
In addition, samples were collected from stations on days when CSDs occurred. The CSD stations used in these analyses included 10 of the 14 routinely monitored stations (“monitoring/CSD stations”), as well as 5 stations that were only sampled when CSDs occurred (“CSD-only stations”). Sample times were recorded and occurred as late as 16:00, depending on the time the CSD occurred. Samples collected within 1 km of, and on the same day as, a CSD were denoted “CSD samples” whether they were collected at monitoring/CSD stations or CSD-only stations. Whenever cENT concentrations in CSD samples exceeded the California single sample maximum criterion (104 most probable number (MPN)/100 mL), follow-up samples were collected daily until concentrations dropped below that threshold. Follow-up samples were also generally collected in the afternoon, as their collection depended on lab results from the previous day’s sample.
2.2. CSD occurrence
During our study period, 20 CSDs occurred that were co-located with our sampling stations (considering both monitoring stations and CSD-only stations). CSD flow volumes were estimated by SFPUC staff from infrastructure water level measurements, and are publically available online (https://www.waterboards.ca.gov/ciwqs/publicreports.shtml). Reported discharge volumes ranged from 3.4 × 103 L to 1.6 × 108 L. To assess the impact of these CSDs on fecal indicator values, we mapped samples collected within 1 km of and on the day of a CSD to their respective CSDs. 1 km was selected as the distance threshold because beaches in SF are posted with beach advisories in the event of a CSD within approximately 1 km. Since in some cases multiple stations were located within 1 km of CSD discharge points, this resulted in 34 samples corresponding to these 20 CSDs.
2.3. Culture analysis
Culturable enterococci (cENT) were enumerated using Enter-olert (IDEXX Laboratories, Westbrook, ME) by performing a 1 : 10 dilution using 10 mL of sample diluted into 90 mL of phosphate buffer dilution water per manufacturer’s protocol. Concentrations were determined using an MPN table provided by IDEXX.
2.4. Membrane filtration and DNA extraction
In 2014, 100 mL of water sample was filtered through a 0.4 μm pore size (47 mm in diameter) polycarbonate membrane filter (GE Osmonics, Minnetonka, MA). In 2016, only 50 mL of water sample was filtered to minimize clogging of the filters. One water sample filtration blank (method blank) was processed using 20 mL of sterile phosphate buffered saline before, after, and between every six water samples. All filters were placed into 2 mL screw cap tubes filled with 0.3 g of sterilized glass beads and processed according to USEPA method 1609.1.32 A 5-fold dilution using 20 μL of crude DNA extract was performed for use as template in the qENT assay. The remaining 380 μL of crude DNA extract was purified for HF183 analysis using the DNA-EZ RW02 kit (GeneRite LLC, North Brunswick, NJ) following manufacturer’s instructions.
2.5. qPCR amplification
Real time qPCR assays used in this study are shown in Table 1. The Enterococcus E1a assay, herein referred to as “qENT” for qPCR-Enterococcus, was amplified following the reaction conditions given in USEPA method 1609.1.32 The human-associated Bacteroides assay, HF183/BacR287, was amplified as described in Green et al.20 Briefly, to measure the HF183 target, a total reaction volume of 25 μL, which consisted of 1× TaqMan® Environmental PCR Master Mix (Thermo Fisher Scientific, Grand Island, NY), 0.2 mgmL−1 bovine serum albumin, 1 μM of each primer, 80 nM of each probe, 100 copies of IAC DNA, and 2 μL of DNA sample extract. Reaction conditions for all targets consisted of an initial incubation at 50 °C for 2 min, then 95 °C for 10 min, followed by 40 PCR cycles of 95 °C for 15 s and 60 °C for 1 min. All reactions were carried out in a StepOnePlus™ thermal cycler (Thermo Fisher Scientific, Grand Island, NY).
Table 1.
qPCR assays
2.6. Standards and controls
Calibrator cells of Enterococcus faecalis (ATCC# 29212) were prepared as previously described.34 Approximately 2.66 × 105 E. faecalis cells were present in each calibrator sample. Genomic DNA extracts of E. faecalis were serially diluted to yield estimated target sequences per reaction of 1.0 × 101, 3.5 × 101,1.2 × 102, 3.9 × 102 and 4.3 × 103 copies. A master standard curve for the HF183 assay consisted of diluted plasmid DNA with estimated target sequences of 4.0 × 101, 1.0 × 102, 4.0 × 102, × 103 and 4.0 × 104 copies per reaction. A sample processing control (SPC), consisting of salmon DNA spiked into samples prior to DNA extraction, was used to detect sample matrix interference and assess DNA recovery.33 Extracts showing salmon DNA amplification greater than three cycle quantification units (Cq) above the mean of the corresponding salmon DNA calibrator Cq were reanalyzed after an additional five-fold dilution of the extract. If results still failed the SPC criterion, that sample was recorded as “failed SPC” and was excluded from further analyses. Additionally, plasmid constructs containing target primers (for qENT and HF183, respectively) and universal probes were used as an internal amplification control (IAC) to indicate amplification interference. Samples with IAC Cq exceeding the amplification threshold, which was the mean of the IAC no template control plus three standard deviations, were not used in further analysis. The template sequences for the IACs were constructed as described in USEPA Method 1609.1 (ref. 32) and Green et al.,20 respectively, and then synthesized in vitro and ligated with a pIDTSMART-KAN plasmid vector (Integrated DNA Technologies, Coralville, IA). All qPCR measurements were run in duplicate. Filter blanks and no-template controls were analyzed with each batch of unknown samples.
2.7. Target detection and quantification
Enterococcus faecalis calibrator cell equivalents (CCE)/100 mL of water sample were estimated by the ΔΔCt comparative Ct calibration model.33 The HF183 genetic marker densities were estimated using a master standard curve.35 Theoretical lowest detectable concentrations (LDCs), defined as the sample concentration corresponding to detecting one unit of target (MPN, CCE, or copy), were calculated for each quantification assay by accounting for sample dilution and filtration volume, DNA eluate dilution and volume, and calibrator cell to copy number conversion factors, where applicable. The LDC for cENT was 10 MPN/100 mL. For qENT and HF183, LDCs differed in 2014 and 2016 due to different filtration volumes. LDCs for qENT were 27 and 53 CCE/100 mL in 2014 and 2016, respectively, and 53 and 105 copies/100 mL for HF183 in 2014 and 2016, respectively. A sample was considered positive if at least one analytical replicate exceeded the LDC.
2.8. Hypothesis tests and correlation
All data analysis was performed in R version 3.3.3.36 Three distinct subsets of data were utilized for different analyses. Specifically, to estimate the duration of contamination produced by CSDs, CSD samples and their daily follow-up samples were analyzed. Since daily follow-up samples were collected on a conditional basis - conditional on the knowledge that the previous day’s cENT concentration exceeded 104 MPN/ 100 mL - these specific samples were not considered to be statistically random samples and were thus excluded from all analyses other than duration of contamination estimate. To assess whether CSDs had a statistically significant impact on indicator levels, only samples from monitoring/CSD stations were utilized. For all other analyses, only weekly monitoring samples from all 14 monitoring stations were used.
Substitution of values below LDCs was avoided for hypothesis tests and correlation.37 Maximum likelihood estimation assuming common statistical distributions (e.g., lognormal, negative binomial, gamma) yielded poor fits to indicator concentration data. Thus, non-parametric statistics capable of handling censored data (since many values were below LDCs) from the ‘NADA’38 and ‘interval’39 packages in R were used. Standard errors of medians were computed with 1000 bootstrap samples. To compare 3 or more groups of concentration data, the Generalized Wilcoxon (GW) test for interval-censored data was used. To test rank-order correlation between indicators, intervals were treated as left-censored, and Kendall’s tau- a correlation coefficient for left-censored data (hereafter, KTC), which is expected to be about 0.15 units smaller than Spearman’s coefficient for the same degree of correlation,40 was used. The Wilcoxon Signed-Rank (WSR) test was used to assess differences in mean ranks between paired groups of correlation coefficients, and Fisher scoring was used to test differences between pairs of correlation coefficients. The Wilcoxon Rank Sum (WRS) was used to assess differences between independent groups of non-censored data. Hypothesis tests of independence of categorical variables were performed using Chi-square (χ2) tests or, for tests stratified by station, Cochran-Mantel-Haenszel (CMH) tests. McNemar’s test was used to test difference of proportion between paired categorical data. Exact tests were used when contingency tables contained expected values less than 5. To test for the degree of association between indicator levels and CSD occurrence while controlling for the effect of precipitation, indicator concentrations were dichotomized at health-relevant thresholds (see Results) and regressed against CSD occurrence and 72 hour cumulative precipitation using binary logistic generalized linear mixed effects models (GLMM). GLMMs were performed using the R package ‘lme4’41 and accounted for differences between sampling stations by treating station as a random effect. Further details on these statistical analyses are provided in ESI.†
2.9. Multivariate regression models
Multivariate binary logistic regression models incorporated environmental variables expected to be important in controlling indicator concentrations. Each monitoring station was modeled separately, with indicator concentrations dichotomized at the median value at each station (Table 2). Models were fit to data from weekly monitoring samples of cENT, qENT, and HF183, respectively, from all 14 monitoring stations, yielding a total of 42 models. LASSO regularization was used for variable selection and coefficient estimation,42 implemented in the R package ‘glmnet’.43
Table 2.
Indicator summary statisticsa
| cENT | qENT | HF183 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N |
Median (SE) |
N |
Median (SE) |
N |
Median (SE) |
||||
| Station | [−] | [CFU/100 mL] | [−] | [CCE/100 mL] | [−] | [Copies/100 mL] | |||
| O1 | 75 | <10 | (NA) | 75 | <53 | (NA) | 75 | <105 | (NA) |
| O2 | 75 | <10 | (NA) | 74 | <53 | (NA) | 75 | 217 | (40.9) |
| O3 | 75 | <10 | (NA) | 75 | <53 | (NA) | 75 | 204 | (43.1) |
| O4 | 75 | <10 | (NA) | 74 | <53 | (NA) | 75 | 230 | (44.9) |
| O5 | 75 | <10 | (NA) | 75 | <53 | (NA) | 75 | 284 | (78.7) |
| O6 | 75 | 20 | (5.5) | 75 | 109 | (21.7) | 75 | 224 | (64.3) |
| O7 | 75 | <10 | (NA) | 74 | <53 | (NA) | 75 | 362 | (90.6) |
| B1 | 75 | <10 | (NA) | 75 | 78 | (15.4) | 75 | 635 | (89.5) |
| B2 | 75 | 10 | (4.0) | 75 | 122 | (27.7) | 75 | 689 | (101.9) |
| B3 | 75 | <10 | (NA) | 75 | 190 | (31.6) | 75 | 1530 | (303.6) |
| B4 | 75 | <10 | (NA) | 75 | 150 | (18.5) | 75 | 1197 | (234.8) |
| B5 | 75 | 10 | (2.0) | 62 | 232 | (38.2) | 75 | 170 | (86.6) |
| B6 | 75 | 41 | (7.6) | 67 | 357 | (86.3) | 75 | <105 | (NA) |
| B7 | 75 | 31 | (8.4) | 65 | 457 | (84.4) | 75 | <105 | (NA) |
| Global | 1050 | <10 | (NA) | 1016 | 80 | (3.7) | 1050 | 293 | (24.3) |
Interval censored values were assigned the squared mean root of the Interval censored values were assigned the squared mean root of the interval bounds. See Methods for details. SEs are based on 1000 bootstrap samples. Lower N for qENT is due to failing sample processing control procedures.
Predictor variables considered in the models were CSD occurrence (CSD occurred on day of sample within 1 km of station), precipitation (cumulative precipitation over the prior 72 hours), time since solar noon (equal to time of sampling minus time of solar noon, in hours), tide level (at time of sampling), tidal gradient (at time of sampling), daily tidal range, significant wave height (at time of sampling), average daily solar insolation, average daily water temperature, and average daily wind speed (Table 3). These variables were chosen because previous modeling work has identified these variables as potentially important predictors of indicator concentrations.44–46 All daily variables pertain to the day on which the sample was taken (these values were highly correlated with day-before sample values, data not shown). Further regression modeling details are provided in ESI.†
Table 3.
Description of predictor variables used in regression models
| Predictor variable | Description | Type | Original unitsa |
Source |
|---|---|---|---|---|
| CSD occurrence | Combined sewer discharge occurred (=1) on day of sampling within 1 km of station, or did not (=0) | Binary | – | SFPUC |
| Precipitation, 72 h | Cumulative precipitation over 72 hours prior to sampling; square-root transformed | Continuous | cm | NOAA47 |
| Solar insolation, daily | Mean solar insolation on day of sampling | Continuous | kW h m −2 | NASA48 |
| Tidal gradient | Time rate of change of tide level at time of sampling (6 minute resolution) | Continuous | m h −1 | Derived |
| Tidal range, daily | Maximum tide level minus minimum tide level that occurred on day of sampling | Continuous | m | Derived |
| Tide level | Tidal level at time of sampling (6 minute resolution) | Continuous | m | NOAA49 |
| Time since solar noon | Time recorded by sampler minus time of solar noon for that day | Continuous | h | NOAA50 |
| Water temperature, daily | Mean water temperature on day of sampling | Continuous | °C | UC Davis Bodega Marine Laboratory51 |
| Wave height | Signi cant wave height: mean of the highest one-third of all wave heights during the 20 minute measurement window closest to the time of sampling | Continuous | m | NOAA52 |
| Wind speed, daily | Mean wind speed on day of sampling | Continuous | m s −1 | NASA48 |
All continuous variables were z-standardized for coefficient estimation.
3. Results
3.1. Performance of microbiological assays
Of 1352 total samples collected, 55 samples failed the SPC criterion. Five-fold dilution did not cause any samples that initially failed the SPC criterion to pass the SPC criterion. Nearly all SPC-failed samples were taken from sites B5, B6, and B7 (Table 2). These sites are typically shallow and muddy, and water samples taken from them are frequently more turbid than those from the other sites (data not shown). None of the samples failed the IAC criterion. The amplification efficiencies of the qENT and HF183 assays were 98% and 97%, respectively. Method blanks and no template controls showed no signs of contamination, as Cq values were undetermined for these reactions.
3.2. Indicator detection frequency
Across all 14 monitoring stations, the mean cENT detection frequency was 42% (global n = 1050; range among stations = 17%, 79%) (Table 4). Mean qENT detection frequency was 75% (n = 1016; 49%, 99%), and mean HF183 detection frequency was 77% (n = 1050; 41%, 97%). qENT was detected more frequently than cENT at all stations, while HF183 was detected less frequently than cENT at 2 stations. In 5% (53/1016) of samples, cENT was detected and qENT was not. Detection frequency of each indicator varied by station (χ2 test for each indicator, p < 0.05). Aggregating data across stations, cENT and qENT were detected more frequently on the bay than the ocean side (χ2 test, p < 0.05) of the peninsula. HF183 detection frequency did not vary between stations located on the bay versus ocean side (χ2 test, p > 0.05). The null hypothesis that cENT detection frequency was independent of qENT detection frequency (stratified by station) was rejected (CMH exact test, p < 0.05). The null hypothesis that cENT detection frequency was independent of HF183 detection frequency (stratified by station) was not rejected (p > 0.05). The null hypothesis that qENT detection frequency was independent of HF183 detection frequency (stratified by station) was rejected (p < 0.05). These three CMH tests were Bonferroni corrected for three comparisons.
Table 4.
Indicator detection frequency by station. Cells are color- coded to represent relative detection frequency for each indicator. Means were calculated by collapsing across the appropriate category (ocean/bay or all stations)
| Detection frequency |
||||
|---|---|---|---|---|
| Region | Station | cENT | qENT | HF183 |
| Ocean | O1 | 25% | 49% | 53% |
| O2 | 25% | 58% | 73% | |
| O3 | 24% | 52% | 80% | |
| O4 | 17% | 57% | 88% | |
| O5 | 24% | 59% | 81% | |
| O6 | 65% | 81% | 80% | |
| O7 | 35% | 64% | 84% | |
| Mean | 31% | 60% | 77% | |
| Bay | B1 | 33% | 77% | 91% |
| B2 | 51% | 88% | 89% | |
| B3 | 49% | 91% | 97% | |
| B4 | 28% | 93% | 97% | |
| B5 | 61% | 89% | 67% | |
| B6 | 79% | 99% | 51% | |
| B7 | 73% | 95% | 41% | |
| Mean | 54% | 90% | 76% | |
|
Grand Mean |
42% | 75% | 77% | |
3.3. Indicator concentration trends
Median concentrations (units of MPN, CCE, and copies/100 mL, respectively) across all monitoring samples were <10 (standard error (se) = not applicable (NA) because median not precisely known; n = 1050), 80 (3.7; 1016), and 293 (24.3; 1050) for cENT, qENT, and HF183, respectively (Table 2). Concentrations of all three indicators varied across the 14 stations (GW test,p < 0.05), with median concentrations ranging from <10 (se = NA) to 41 (7.6) for cENT, <53 (NA) to 457 (84.4) for qENT, and <105 (NA) to 1530 (303.6) for HF183. Indicator concentrations aggregated across stations varied by month (Fig. 2), with median concentrations of all three indicators lowest in dry summer months (cENT <10 in April-September and November, qENT = <53 in June and August, HF183 = <104 in August) and highest in wet winter months (cENT = 20 (14.8), qENT = 219 (110.1), HF183 = 1746 (355.7), all in January). Indicator correlations between stations showed no clear trend as a function of distance between stations.
Fig. 2.
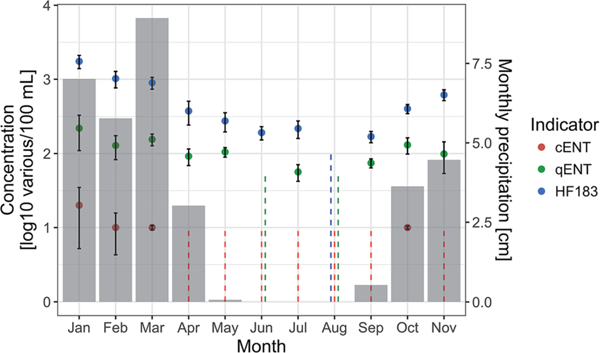
Median indicator values by month, aggregated over years 2014 and 2016. Grey bars represent mean monthly precipitation over years 2014 and 2016. Error bars represent bootstrapped standard errors, based on 1000 bootstrap samples. Dashed lines represent the interval containing the median for the months in which medians were below the highest LDC for each indicator, i.e., <10 MPN/100 mL, <53 CCE/ 100 mL, and <105 copies/100 mL for cENT, qENT, and HF183, respectively. Note that due to this log 10 representation, the dashed line intervals only extend to 0 log 10 (equal to 1 in original concentration units), while the true intervals extend to include 0 in original concentration units.
3.4. Correlation between indicators
cENT and qENT were significantly positively correlated (KTC, p < 0.05) at 9 of 14 monitoring stations: O3, O4, O6, B1, B2, B3, B5, B6, and B7 (Fig. 3). Median and maximum correlation coefficients between cENT and qENT across all stations were 0.17 and 0.46, respectively. cENT and HF183 were significantly positively correlated at 3 of 14 stations (stations O2, O3, and O4). The other eleven stations showed no significant correlation between cENT and HF183. Median correlation between cENT and HF183 was 0.05 and maximum was 0.14. qENT and HF183 were significantly positively correlated (KTC, p < 0.05) at 2 of 14 stations: B3 and B7. Median correlation between qENT and HF183 across all stations was 0.12, and maximum was 0.20.
Fig. 3.
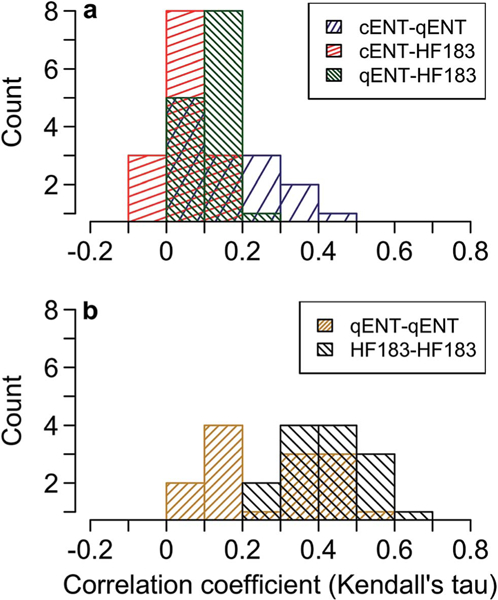
Histograms of correlations between indicators at each station (14 stations with n = 75 at each station, except for stations at which qENT failed SPC measures, see Table 2). (a) Depicts correlations between different indicators, and (b) depicts correlations between analytical duplicates of each molecular assay: the qENT assay (”qENT-qENT”) and the HF183 assay (“HF183-HF183”), respectively. Correlation coefficient is Kendall’s tau-a for left-censored data.
To compare the variability of the qENT and HF183 molecular assays, correlations between analytical duplicates of qENT (“qENT-qENT”) and between analytical duplicates of HF183 (“HF183-HF183”) were also computed (Fig. 3). Analytical duplicates of qENT were significantly positively correlated at 11 of 14 monitoring stations. Median and maximum qENT-qENT correlations across all stations were 0.28 and 0.53, respectively. Analytical duplicate HF183 measurements were significantly positively correlated with each other at all monitoring stations, with median and maximum correlations of 0.41 and 0.66, respectively. Duplicate HF183 measurements were more highly correlated with each other than duplicate qENT measurements were with each other (p < 0.05).
Correlations between indicators were higher in CSD samples, with KTC = 0.72 (n = 28), 0.42 (n = 34), and 0.52 (n = 28) for cENT-qENT, cENT-HF183, and qENT-HF183, respectively, than in all monitoring samples taken together (p < 0.05). To test whether the increased correlation following CSDs is merely a product of indicator levels being elevated, these correlation coefficients were compared to correlations between indicators in all other samples containing high cENT levels ($104 MPN/100 mL). Correlation coefficients between indicators in these “high-cENT” samples were 0.29 (n = 58), −0.05 (n = 68), and 0.13 (n = 58), for cENT-qENT, cENT-HF183, and qENT-HF183, respectively. Correlations between cENT-qENT and cENT-HF183 were statistically greater (p < 0.05) in CSD samples than in all high-cENT samples, and qENT-HF183 showed a trend toward correlations being greater in CSD samples than high-cENT samples (p = 0.07).
3.5. qENT versus cENT for beach posting decisions
We compared all available paired cENT and qENT measurements from monitoring stations (n = 1016) to the cENT California single sample criterion (104 MPN/100 mL) and the qENT statistical threshold value (STV) set forth in the USEPA 2012 Recreational Water Quality Criteria corresponding to a risk threshold of 32 illnesses per 1000 primary contact recreators (1280 CCE/100 mL) (Fig. 4). These criteria are used to post beaches as unfit for swimming. Overall, cENT and qENT measurements led to the same beach posting decision (post or no-post) in 95% of samples. Global comparisons yielded fewer excursions for qENT (32 excursions, 3% of all samples) than for cENT (59 excursions, 6% of all samples), with both indicators simultaneously exceeding their respective STVs in 21 samples (2% of all samples). The null hypothesis that cENT and qENT were equally likely to exceed their respective standards was rejected (McNemar’s test, p < 0.05).
Fig 4.
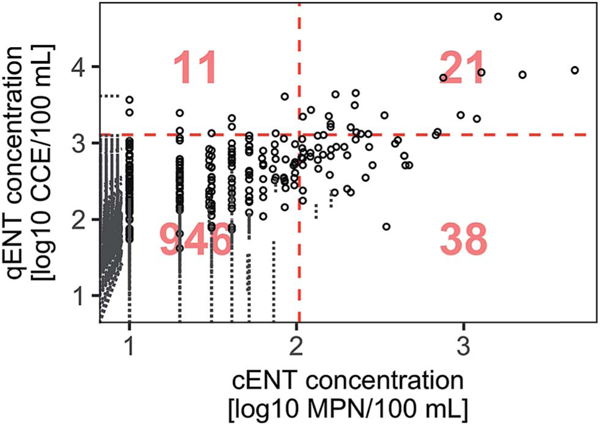
All paired cENT and qENT measurements (n = 1016). Red dashed lines represent thresholds for posting beaches based on a single sample measured at weekly intervals. The vertical red dashed line is the California cENT single sample maximum (104 MPN/100 mL). The horizontal dashed line is the USEPA qENT statistical threshold value (1280 CCE/100 mL).6 Numbers inside the plot represent the number of points in each quadrant. Gray dotted lines correspond to censored values (at least one replicate below LDC), and represent the interval spanning the possible values. Diagonal dotted lines appear for samples where both cENT and qENT were censored. Note that due to this log10 representation, the dashed line intervals only extend to the edge of the plot area, while the true intervals extend to include 0 in original concentration units.
3.6. Associations between CSDs and indicator levels
During our study period, 20 distinct CSD events occurred that were co-located with our sampling stations, resulting in 34 CSD samples (see Methods). Reported discharge volumes ranged from 3.4 × 103 L to 1.6 × 108 L. Of 34 CSD samples, 14 samples had cENT levels below 104 MPN/100 mL. These 14 samples corresponded to CSD events with statistically smaller flow volumes (WRS, p < 0.05), with median volume = 57 × 103 L, than the other 18 CSDs, with median flow volume = 54 × 106 L. In the event of a CSD, water samples were collected daily until cENT levels dropped below 104 MPN/100 mL. On average, it took 1.1 days (95% CI: 0.5, 1.6) from the date of the CSD for cENT concentrations to drop below 104 MPN/100 mL, although in one case a CSD was followed by 8 consecutive exceedance days of cENT over 104 MPN/100 mL. As described in Methods, mixed effects models were used to test the degree of association between indicator levels and CSD occurrence using samples from the ten monitoring stations where at least one CSD occurred during the study period. GLMMs controlled for the effect of precipitation, accounted for differences between sampling stations, and excluded daily follow-up samples. On average, CSD occurrence was associated with a 20-fold (GLMM, 95% CI: 6, 66) increase in the odds of measuring high cENT (cENT exceeding the CA single sample maximum criterion 104 MPN/100 mL) in a sample, a 10-fold (95% CI: 3, 39) increase in the odds of measuring high qENT (qENT exceeding the USEPA STV of 1280 CCE/100 mL), and a 3-fold (95% CI: 1, 11) increase in the odds of measuring high HF183 (HF183 exceeding the risk-based benchmark estimated in a previous QMRA study of 4200 copies/100 mL (ref. 53)).
3.7. Multivariate regression models
Regression results from all 42 models (one model for each of the three indicators at fourteen stations) are summarized in Table 5. Predictor variables are ranked by the total number of models in which they were selected by the LASSO method, i.e., explained a statistically important amount of variance in the odds of the outcome variable being high as opposed to low (above or below median).
Table 5.
Summary of 42 regression models. The number in each cell represents the number of times a predictor variable was selected by the LASSO method for inclusion in a model out of a total of 14 models (one for each station). Selected variables can be thought of as explaining a statistically important amount of variance in the outcome variable, and were selected via 10-fold cross validation. All models are binary logistic models of indicators dichotomized at the median value for each station. All quantitative predictor variables were standardized for coefficient estimation. Cells are colored to reflect the relative selection frequency of each predictor, with a maximum possible selection frequency of 14. Note that wave height was only included for ocean (not bay) stations
| Outcome variable |
||||
|---|---|---|---|---|
| Predictor | cENT | qENT | HF183 | Sum |
| Precipitation, 72 h | 2 | 3 | 9 | 14 |
| Water temperature, mean daily | 0 | 3 | 5 | 8 |
| Solar insolation, mean daily | 3 | 2 | 1 | 6 |
| Tidal range, daily | 0 | 0 | 2 | 2 |
| Time since solar noon | 0 | 1 | 1 | 2 |
| Significant wave height | 0 | 1 | 0 | 1 |
| Tidal gradient | 0 | 1 | 0 | 1 |
| Tide level | 0 | 0 | 1 | 1 |
| CSD occurrence | 0 | 0 | 0 | 0 |
| Wind speed, mean daily | 0 | 0 | 0 | 0 |
Mean daily solar insolation and 72 h precipitation were the only two predictors selected in cENT models. When selected, insolation was always negatively related to cENT and precipitation was always positively related. At ten of fourteen stations, the null model was selected for cENT (i.e., including any predictors did not improve model performance per the cross-validated LASSO method).
Mean daily solar insolation (coefficient always negative) and 72 h precipitation (coefficient always positive) were relatively important predictors in qENT models, as was mean daily water temperature (coefficient always negative). Time since solar noon (positive coefficient), significant wave height (positive coefficient), and tidal gradient (negative coefficient) were each chosen in one model, and the null model was the best performing at eight of fourteen stations.
For HF183, 72 h precipitation (coefficient always positive) was selected at nine of fourteen stations and water temperature (coefficient always negative) was selected at five stations. Tidal range was selected at two stations (coefficient always positive), and solar insolation (negative coefficient), time since solar noon (positive coefficient), and tide level (positive coefficient) were each selected once. The null model was the best performing at three of fourteen stations for HF183.
Notably, CSD occurrence and mean daily wind speed were the only two predictors not selected in any model for any indicator. To confirm the null effect of CSDs identified in these models, all models were run again twice: once without the precipitation variable to check whether co-linearity with the CSD variable was obscuring a CSD effect, and once with CSD volume instead of CSD occurrence. Neither scenario changed the number of models (zero) in which CSD was selected as an important predictor. Correlations between the precipitation variable and the water temperature and solar insolation variables (Pearson’s correlation = −0.36 and −0.27, respectively), were within the range of correlations between predictors employed in previous models.44 A summary of alternative models is given in Table S1,† and coefficient estimates of the final model with precipitation and CSD occurrence are given in Table S2.†
Predictors were not chosen consistently across indicators at individual stations. For example, precipitation was not selected for all three indicators at any one station. Precipitation was selected for cENT at stations O3 and B1, for qENT at stations O4, B4, and B5, and for HF183 at stations O2, O4, O5, O6, O7, B1, B3, B4, and B6. No clear geographic patterns of predictors emerged. See Table S2† for regression results broken out by station.
4. Discussion
4.1. Molecular indicators were more frequently detected than the culturable indicator
Overall, qENT was detected in 75% and HF183 in 77% of samples, whereas cENT was detected in 42%. It was expected that qENT would be detected more frequently than cENT because culturable enterococci represent a subset of total Enterococcus 23S rRNA gene content in a sample. However, in a small subset of samples, cENT was detected and qENT was not. This disparity may be attributable to the fact that the LDCs for qENT (27 and 53 CCE/100 mL in 2014 and 2016, respectively) were higher than the LDC for cENT (10 MPN/100 mL), although units differ. It may also be due to variability in 23S rRNA gene copy number across Enterococcus spp.,54 less than 100% efficient DNA extraction,19 or analytical uncertainty in qENT measurements.55 In contrast, as shown in this study, HF183 detection frequency can be substantially greater than or less than that of cENT depending on the station, which may be due to variation in sources of fecal pollution5 or differential persistence of cENT and HF183 in seawater.15
4.2. HF183 was frequently detected
Overall, HF183 was detected in 77% percent of samples. This suggests that humans are a source of fecal pollution along the San Francisco shoreline. Based on these data, it may make sense to conduct additional investigation into the areas contributing to stations B3 and B4, where HF183 was detected in 97% of monitoring samples and median HF183 concentrations (1530 and 1197 copies/100 mL, respectively) were twice as high as the next highest station. HF183 was detected more frequently in our marine shoreline samples than in a study of several urban-impacted recreational beaches in Florida and South Carolina (detection frequency 0% to 20%);46 a study of two non-point source impacted beaches in Southern California (27%);25 a regional study of freshwater coastal drainages in Southern California, in which 21% and 52% of samples were positive for HF183 in dry weather and wet weather, respectively;24 and a study of urban storm water outfalls in Milwaukee (57%).19 These differences in detection frequency may be due to differences in watershed land use (e.g., San Francisco’s watersheds are highly urbanized), rainfall (San Francisco receives more rainfall than Southern California), solar insolation (generally lower in San Francisco than in sites examined in these other studies), and water matrices (seawater versus freshwater or storm water). Differences in DNA extraction and amplification efficiency, as well as detection assays (McQuaig et al., 2012 used a form of endpoint PCR to detect HF183) and definitions of positive detection, may also contribute. The HF183 marker is highly specific to human feces as determined in a large California-based method comparison study.18 However, in some locales, the HF marker has been detected in other mammalian feces albeit at low densities.56
4.3. HF183 detection patterns differed from those of cENT
In fact, global HF183 detection varied independently of cENT detection. While enterococci detection was more frequent at bay stations than ocean stations, HF183 detection was not. Furthermore, the two stations (B6 and B7) with the lowest HF183 detection frequency had the highest enterococci (both cENT and qENT) detection frequency, which may suggest frequent fecal inputs from non-human sources or environmental enterococci. One possible source is gulls, which are observed regularly flocking to waters adjacent to these two stations (data not shown). While relative human fecal contribution may differ by station, using these data to make fecal source attribution is complicated by the fact that cENT and HF183 decay at different rates in marine water15 and that fecal source type - contaminated sediment, raw feces, sewage, or septage - impacts cENT and HF183 decay rates.16,17
4.4. All three indicators showed seasonal variation
When fecal indicator concentrations were averaged across stations for each sample date, a distinct seasonal pattern emerged for cENT, qENT, and HF183, with higher concentrations in winter and lower in summer months. A large body of evidence shows that winter conditions found in San Francisco, including colder water temperatures57 and reduced solar radiation,15,16,58 increase persistence of cENT in marine water. Studies that simultaneously examined the effect of sunlight on qENT and HF183 decay in marine water found a less pronounced effect on qENT than on cENT, and have provided contrasting evidence on the effect of sunlight on HF183.14–16 However, given that cENT, qENT, and HF183 in San Francisco shoreline waters share at least some common sources, such as CSDs59,60 and urban runoff,46,61,62 which themselves show strong seasonality in San Francisco, it is not surprising that all three indicators show seasonality.
4.5. The current CA standard for cENT yielded nearly twice as many exceedances as the qENT standard given in USEPA’s 2012 recreational water quality criteria
This indicates that over our two-year study period, cENT was a more conservative fecal indicator than qENT. Without accompanying site-specific pathogen data or epidemiological data, however, we cannot determine which indicator more accurately reflects risk to human bathers in these waters. Over-all, because most samples contained enterococci well below the cENT and qENT thresholds, these indicators produced the same beach posting decision in the great majority of samples.
4.6. CSDs were associated with significantly increased odds of cENT, qENT, and HF183 exceeding health-relevant thresholds’ controlling for precipitation
This result was obtained using data from the ten monitoring stations where at least one CSD occurred during the study period. It includes samples taken immediately after CSDs occurred (CSD samples), even if those samples were not part of the weekly monitoring routine, and it excludes all daily follow-up samples, as justified in the Methods. CSD flows in San Francisco comprise a mixture of urban runoff and sewage, which receives treatment equivalent to wet-weather primary treatment, and thus are expected to contain high levels of cENT, qENT, and HF183.19,59,61,63 Interestingly, however, in 14 out of 34 CSD samples, cENT concentrations were below 104 MPN/100 mL. All 14 of these samples were taken within 6 hours of the termination of the corresponding CSDs, which ranged in volume from 38 × 103 to 47 × 106 L. This result is not necessarily surprising given our understanding of the complexity of nearshore currents and the patchiness of nearshore contamination.64
At first, this result may seem to contradict the results from the multiple regression analysis that showed CSD was not an important predictor of indicator levels measured during routine, weekly sampling. A likely explanation is that CSDs did not frequently coincide with weekly monitoring samples. Only 8 of 1050 weekly monitoring samples used to construct the regression models were CSD samples.
4.7. Precipitation, solar insolation, and water temperature were the most important predictors in 42 models of weekly monitoring samples
For cENT, precipitation and solar insolation were relatively important predictors. In statistical models presented by two other studies, precipitation is consistently found to be an important predictor of cENT levels at marine beaches, while solar insolation is sometimes significant though less consistently so.46,65 Precipitation and water temperature were the most important predictors of the two molecular indicators, especially HF183. The importance of precipitation for all three indicators highlights the significance of untreated storm water, as a source of fecal pollution to SF shoreline water, even though most of the city is serviced by a combined sewer system. Further study is warranted to understand the mechanisms that mediate the effects of runoff on indicator levels in this system. Likely, sunlight and water temperature control the persistence of indicators once introduced to shoreline waters, acting via photoinactivation and dark decay processes, respectively.
Interestingly, environmental predictors contained more information about HF183 levels than cENT or qENT levels, as the null model was selected for HF183 at only 3 of 14 stations, compared to 10 of 14 for cENT and 8 of 14 for qENT. Tidal variables showed a positive relationship with HF183 at two stations, suggesting that washing of sand66 and other shoreline surfaces or hydraulic pumping of submarine ground water67 may be sources of human fecal pollution at these stations. Given the importance of precipitation and water temperature for HF183, these two measurements in particular could prove useful for predicting HF183 levels in SF shoreline waters. Overall, our results represent the first statistical models connecting field measurements of HF183 in marine waters to these environmental factors.
The three dominant predictors - precipitation, solar insolation, and water temperature - show the same relationship directionality (positive or negative) across all indicators and stations, suggesting that these drivers act similarly in all SF shoreline waters, and that the effects are unlikely to be a result of Type I error. However, given that specific predictors were not consistently selected at individual stations (e.g., precipitation was not selected at the same stations for cENT, qENT, and HF183), it appears that individual models may be required not only to model each station, but also to model each indicator.
4.8. Analytical duplicates of HF183 were more strongly correlated than were analytical duplicates of qENT
These results suggest that duplicate qENT correlations were somewhat low in comparison to duplicate HF183 correlations. This may stem from the fact that the qENT protocol32 employed crude extracts of environmental DNA, while HF183 extracts were purified. Different calibration models for the qENT and HF183 protocols may also contribute to this difference in analytical replicate variability.
5. Conclusions
HF183 was detected more frequently in San Francisco shoreline waters than in many other marine water studies, even though cENT exceeded the CA single sample maximum criterion in only 6% of samples.
Controlling for precipitation, CSDs were associated with elevated odds of cENT, qENT, and HF183 exceeding health-relevant thresholds, even though CSDs were not important predictors of indicator levels typically measured throughout the year.
Regression results suggest that storm water flows are likely an important source of all three indicators to shoreline waters, especially HF183.
Statistical models also suggest that elevated water temperature may be an important driver of decay of molecular indicators, and that solar radiation may be more important for cENT decay.
Overall, cENT was a more conservative indicator of fecal pollution than was qENT, as cENT resulted in 6% and qENT resulted in 3% of samples exceeding recreational water quality criteria.
Supplementary Material
Environmental significance.
This study yields insights into the environmental processes and anthropogenic inputs that impact fecal pollution levels in the urban ocean. We identify factors that differentially impact traditional fecal indicators (culturable enterococci, cENT) and new molecular indicators (qPCR-enterococci, qENT, and a human- specific marker, HF183/BacR287). This paper describes quantitative relationships between these three indicators, the understanding of which is crucial for strategically monitoring recreational waters and identifying areas of highest concern to human health using HF183.
Acknowledgements
The authors would like to acknowledge C. Chrisman at SFDPH for sample collection assistance, personnel of the SFPUC Natural Resources and Water Quality Divisions, especially L. Delpuerto, for their assistance in sample collection and bacterial analyses, and E. Natesan at SFPUC for valuable comments on the manuscript. W. C. J. was supported by a U.S. National Science Foundation Graduate Research Fellowship.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7em00594f
Conflicts of interest
The authors have no conflicts of interest in conducting or reporting this research. The findings and conclusions in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency or the San Francisco Public Utilities Commission. Mention of trade names, products or services does not constitute endorsement or recommendation for use by the US Government, the U.S. EPA, or SFPUC.
References
- 1.Shuval H, Estimating the global burden of thalassogenic diseases: human infectious diseases caused by wastewater pollution of the marine environment, J. Water Health, 2003, 1(2), 53–64. [PubMed] [Google Scholar]
- 2.Bartram J and Rees G, Monitoring bathing waters: A practical guide to the design and implementation of assessments and monitoring programme, London: E & FN Spon, 2000. [Google Scholar]
- 3.World Health Organization, Guidelines for safe recreational water environments. Volume 1: Coastal and fresh waters October 17 2017, http://www.who.int/water_sanitation_health/publications/srwe1/en/. [PubMed]
- 4.Boehm AB and Soller J, Recreational water risk: Pathogens and fecal indicators, in Environmental Toxicology: Selected Entries from the Encyclopedia of Sustainability Science and Technology, 2013, pp. 441–59. [Google Scholar]
- 5.Boehm AB, Ashbolt NJ, Colford JM, Dunbar LE, Fleming LE, Gold MA, et al. A sea change ahead for recreational water quality criteria, J. Water Health, 2009, 7(1), 9–20. [DOI] [PubMed] [Google Scholar]
- 6.USEPA, Recreational water quality criteria, Office of Water, United States Environmental Protection Agency, 2012. [Google Scholar]
- 7.Boehm AB, Grant SB, Kim JH, Mowbray SL, McGee CD, Clark CD, et al. Decadal and shorter period variability of surf zone water quality at Huntington Beach, California, Environ. Sci. Technol, 2002, 36(18), 3885–3892. [DOI] [PubMed] [Google Scholar]
- 8.Whitman RL, Nevers MB, Korinek GC and Byappanahalli MN, Solar and temporal effects on Escherichia coli concentration at a Lake Michigan swimming beach, Appl. Environ. Microbiol, 2004, 70(7), 4276–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, Grant SB, McGee CD, Sanders BF and Largier JL, Locating sources of surf zone pollution: A mass budget analysis of fecal indicator bacteria at Huntington Beach, California, Environ. Sci. Technol, 2004, 38(9), 2626–2636. [DOI] [PubMed] [Google Scholar]
- 10.Harwood VJ, Whitlock J and Withington V, Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: Use in predicting the source of fecal contamination in subtropical waters, Appl. Environ. Microbiol, 2000, 66(9), 3698–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Layton BA, Walters SP and Boehm AB, Distribution and diversity of the enterococcal surface protein (esp) gene in animal hosts and the Pacific coast environment, J. Appl. Microbiol, 2009, 106(5), 1521–1531. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson DM, Griffith JF, McGee CD, Weisberg SB and Hagedorn C, Comparison of enterococcus species diversity in marine water and wastewater using Enterolert and EPA Method 1600, J. Environ. Public Health, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE and Ashbolt NJ, Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination, Water Res, 2010, 44(16), 4674–4691. [DOI] [PubMed] [Google Scholar]
- 14.Bae S and Wuertz S, Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide, Water Res, 2009, 43(19), 4850–4859. [DOI] [PubMed] [Google Scholar]
- 15.Walters SP, Yamahara KM and Boehm AB, Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: Implications for their use in assessing risk in recreational waters, Water Res, 2009, 43(19), 4929–4939. [DOI] [PubMed] [Google Scholar]
- 16.Wanjugi P, Sivaganesan M, Korajkic A, Kelty CA, McMinn B, Ulrich R, et al. Differential decomposition of bacterial and viral fecal indicators in common human pollution types, Water Res, 2016, 105, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson KL, Whitlock JE and Harwood VJ, Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments, Appl. Environ. Microbiol, 2005, 71(6), 3041–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehm AB, Van De Werffiorst LC, Griffith JF, Holden PA, Jay JA, Shanks OC, et al. Performance of forty-one microbial source tracking methods: A twenty-seven lab evaluation study, Water Res, 2013, 47(18), 6812–6828. [DOI] [PubMed] [Google Scholar]
- 19.Sauer EP, VandeWalle JL, Bootsma MJ and McLellan SL, Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment, Water Res, 2011, 45(14), 4081–4091. [DOI] [PubMed] [Google Scholar]
- 20.Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, et al. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples, Appl. Environ. Microbiol, 2014, 80(10), 3086–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed W, Sidhu JPS, Smith K, Beale DJ, Gyawali P and Toze S, Distributions of fecal markers in wastewater from different climatic zones for human fecal pollution tracking in Australian surface waters, Appl. Environ. Microbiol, 2015, 82(4), 1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staley C, Reckhow KH, Lukasik J and Harwood VJ, Assessment of sources of human pathogens and fecal contamination in a Florida freshwater lake, Water Res, 2012, 46(17), 5799–5812. [DOI] [PubMed] [Google Scholar]
- 23.Staley ZR, Vogel L, Robinson C and Edge TA, Differential occurrence of Escherichia coli and human Bacteroidales at two Great Lakes beaches, J. Great Lakes Res, 2015, 41(2), 530–535. [Google Scholar]
- 24.Cao Y, Raith MR, Smith PD, Griffith JF, Weisberg SB, Schriewer A, et al. Regional assessment of human fecal contamination in southern California coastal drainages, Int. J. Environ. Res. Public Health, 2017, 14(8), 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McQuaig S, Griffith J and Harwood VJ, Association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution, Appl. Environ. Microbiol, 2012, 78(18), 6423–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Symonds EM, Young S, Verbyla ME, McQuaig-Ulrich SM, Ross E, Jimenez JA, et al. Microbial source tracking in shellfish harvesting waters in the Gulf of Nicoya, Costa Rica, Water Res, 2017, 111, 177–184. [DOI] [PubMed] [Google Scholar]
- 27.Wade C, Otero E, Poon-Kwong B, Rozier R and Bachoon D, Detection of human-derived fecal contamination in Puerto Rico using carbamazepine, HF183 Bacteroides, and fecal indicator bacteria, Mar. Pollut. Bull, 2015, 101(2), 872–877. [DOI] [PubMed] [Google Scholar]
- 28.Santoro AE and Boehm AB, Frequent occurrence of the human-specific Bacteroides fecal marker at an open coast marine beach: relationship to waves, tides and traditional indicators, Environ. Microbiol, 2007, 9(8), 2038–2049. [DOI] [PubMed] [Google Scholar]
- 29.Anonymous. The largest US cities: Cities ranked 1 to 100 [Internet]. Available from: http://www.citymayors.com/gratis/uscities_100.html.
- 30.US Geological Survey. Impervious surface - conterminous United States 100 meter resolution Albers projection. The National Map. 2013. Available from: http://nationalmap.gov/small_scale/atlasftp.html.
- 31.Largier JL, Hollibaugh JT and Smith SV, Seasonally hypersaline estuaries in Mediterranean-climate regions, Estuarine, Coastal Shelf Sci, 1997, 45(6), 789–797. [Google Scholar]
- 32.USEPA, Method 1609.1: Enterococci in water by TaqMan quantitative polymerase chain reaction (qPCR) with internal amplification control (IAC) assay, U.S. Environmental Protection Agency, 2015. [Google Scholar]
- 33.Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L and Shanks OC, Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR, Syst. Appl. Microbiol, 2010, 33(6), 348–357. [DOI] [PubMed] [Google Scholar]
- 34.Chern EC, Brenner KP, Wymer L and Haugland RA, Comparison of fecal indicator bacteria densities in marine recreational waters by qPCR, Water Qual Expo Health, 2009, 1(3–4), 203–214. [Google Scholar]
- 35.Sivaganesan M, Haugland RA, Chern EC and Shanks OC, Improved strategies and optimization of calibration models for real-time PCR absolute quantification, Water Res, 2010, 44(16), 4726–4735. [DOI] [PubMed] [Google Scholar]
- 36.R Core Team, R: A language and environment for statistical computing, Vienna, Austria: R Foundation for Statistical Computing, 2017, https://www.R-project.org/. [Google Scholar]
- 37.Helsel DR, Statistics for censored environmental data using Minitab and R. 2nd edn, Wiley, NJ, USA, 2012. [Google Scholar]
- 38.Lee L. NADA: Nondetects and data analysis for environmental data, 2017, https://CRAN.R-project.org/package=NADA.
- 39.Fay MP and Shaw PA, Exact and asymptotic weighted logrank tests for interval censored data: The interval R package, J. Stat. Softw, 2010, 36(2), 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helsel DR and Hirsch RM, Statistical methods in water resources, in US Geological Survey, Techniques of Water-Resources Investigations Book 4, Chapter A3, U.S. Geological Survey, 2002. [Google Scholar]
- 41.Bates D, Maechler M, Bolker B and Walker S, Fitting Linear Mixed-Effects Models Using lme4, J. Stat. Softw., 2015, 67(1), 1–48. [Google Scholar]
- 42.Tibshirani R, Regression shrinkage and selection via the LASSO, J. Roy. Stat. Soc. B Stat. Meth, 1994, 58, 267–288. [Google Scholar]
- 43.Friedman JH, Hastie T and Tibshirani R, Regularization paths for generalized linear models via coordinate descent, J. Stat. Softw, 2010, 33(1), 1–22. [PMC free article] [PubMed] [Google Scholar]
- 44.Thoe W, Gold M, Griesbach A, Grimmer M, Taggart ML and Boehm AB, Predicting water quality at Santa Monica Beach: Evaluation of five different models for public notification of unsafe swimming conditions, Water Res, 2014, 67(Supplement C), 105–117. [DOI] [PubMed] [Google Scholar]
- 45.Nevers MB and Whitman RL, Nowcast modeling of Escherichia coli concentrations at multiple urban beaches of southern Lake Michigan, Water Res, 2005, 39(20), 5250–5260. [DOI] [PubMed] [Google Scholar]
- 46.Molina M, Hunter S, Cyterski M, Peed LA, Kelty CA, Sivaganesan M, et al. Factors affecting the presence of human-associated and fecal indicator real-time quantitative PCR genetic markers in urban-impacted recreational beaches, Water Res, 2014, 64, 196–208. [DOI] [PubMed] [Google Scholar]
- 47.NOAA, Climate data online. National Centers for Environmental Information, https://www.ncdc.noaa.gov/cdo-web/datasets/LCD/stations/WBAN:23234/detail.
- 48.NASA. Prediction of worldwide energy resource, https://power.larc.nasa.gov/cgi-bin/cgiwrap/solar/timeseries.cgi.
- 49.NOAA. Tides and currents, https://Tidesandcurrents.noaa.gov/waterlevels.html?id=9414290&units=metric&bdate=20140101&edate=20140131&timezone=LST/LDT&datum=MLLW&interval=6&action=data.
- 50.NOAA. NOAA solar calculator, Earth System Research Laboratory, https://www.esrl.noaa.gov/gmd/grad/solcalc/. [Google Scholar]
- 51.UC Davis Bodega Marina Laboratory. Sf Bay Fort Point water time series, http://bmlsc.ucdavis.edu:8080/erddap/tabledap/FPT_WTS.html.
- 52.NOAA. Station 46026. National Data Buoy Center, http://www.ndbc.noaa.gov/station_history.php?station=46026. [Google Scholar]
- 53.Boehm AB and Soller JA, Shanks OC. Human-Associated Fecal Quantitative Polymerase Chain Reaction Measurements and Simulated Risk of Gastrointestinal Illness in Recreational Waters Contaminated with Raw Sewage, Environ. Sci. Technol. Lett, 2015, 2(10), 270–275. [Google Scholar]
- 54.Wang D, Yamahara KM, Cao Y and Boehm AB, Absolute quantification of Enterococcal 23S rRNA gene using digital PCR, Environ. Sci. Technol. , 2016, 50(7), 3399–3408. [DOI] [PubMed] [Google Scholar]
- 55.Whitman RL, Ge Z, Nevers MB, Boehm AB, Chern EC, Haugland RA, et al. Relationship and variation of qPCR and culturable Enterococci estimates in ambient surface waters are predictable, Environ. Sci. Technol, 2010, 44(13), 5049–5054. [DOI] [PubMed] [Google Scholar]
- 56.Harwood VJ, Staley C, Badgley BD, Borges K and Korajkic A, Microbial source tracking markers for detection of fecal contamination in environmental waters: Relationships between pathogens and human health outcomes, FEMS Microbiol. Rev, 2014, 38(1), 1–40. [DOI] [PubMed] [Google Scholar]
- 57.Noble R. t., Lee I. m. and Schiff K. c., Inactivation of indicator micro-organisms from various sources of faecal contamination in seawater and freshwater, J. Appl. Microbiol, 2004, 96(3), 464–472. [DOI] [PubMed] [Google Scholar]
- 58.Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR and Harwood VJ, Enterococci in the environment, Microbiol. Mol. Biol. Rev, 2012, 76(4), 685–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boehm AB and Sassoubre LM, Enterococci as indicators of environmental fecal contamination, in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, ed. Gilmore MS, Clewell DB, Ike Y and Shankar N, Boston, Massachusetts Eye and Ear Infirmary, 2014, http://www.ncbi.nlm.nih.gov/books/NBK190421/. [PubMed] [Google Scholar]
- 60.Mayer RE, Bofill-Mas S, Egle L, Reischer GH, Schade M, Fernandez-Cassi X, et al. Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution, Water Res, 2016, 90(Supplement C), 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant SB, Sanders BF, Boehm AB, Redman JA, Kim JH, Mrse RD, et al. Generation of enterococci bacteria in a coastal saltwater marsh and its impact on surf zone water quality, Environ. Sci. Technol, 2001, 35(12), 2407–2416. [DOI] [PubMed] [Google Scholar]
- 62.Whitman RL and Nevers MB, Summer E. coli patterns and responses along 23 Chicago beaches, Environ. Sci. Technol, 2008, 42(24), 9217–9224. [DOI] [PubMed] [Google Scholar]
- 63.Chern EC, Brenner K, Wymer L and Haugland RA, Influence of wastewater disinfection on densities of culturable fecal indicator bacteria and genetic markers, J. Water Health, 2014, 12(3), 410–417. [DOI] [PubMed] [Google Scholar]
- 64.Boehm AB, Enterococci concentrations in diverse coastal environments exhibit extreme variability, Environ. Sci. Technol, 2007, 41(24), 8227–8232. [DOI] [PubMed] [Google Scholar]
- 65.Thoe W, Gold M, Griesbach A, Grimmer M, Taggart ML and Boehm AB, Sunny with a Chance of Gastroenteritis: Predicting Swimmer Risk at California Beaches, Environ. Sci. Technol, 2015, 49(1), 423–431. [DOI] [PubMed] [Google Scholar]
- 66.Yamahara KM, Layton BA, Santoro AE and Boehm AB, Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters, Environ. Sci. Technol, 2007, 41(13), 4515–4521. [DOI] [PubMed] [Google Scholar]
- 67.Boehm AB, Shellenbarger GG and Paytan A, Groundwater discharge: Potential association with fecal indicator bacteria in the surf zone, Environ. Sci. Technol, 2004, 38(13), 3558–3566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


