Abstract
Objective:
To explore the contribution of blood and colon myeloid cells to HIV persistence during antiretroviral therapy (ART).
Design:
Leukapheresis were collected from HIV-infected individuals with undetectable plasma viral load during ART (HIV+ART; n=15) and viremics untreated (HIV+; n=6). Rectal sigmoid biopsies were collected from n=8 HIV+ART.
Methods:
Myeloid cells (total monocytes (Mo), CD16+/CD16- Mo, CD1c+ dendritic cells (DC)) and CD4+ T-cells were isolated by MACS and/or FACS from peripheral blood. Matched myeloid and CCR6+CD4+ T-cells were isolated from blood and rectal biopsies by FACS. Levels of early (RU5 primers), late (Gag primers), and/or integrated HIV-DNA (Alu/HIV primers) were quantified by nested real-time PCR. Replication-competent HIV was amplified by co-culturing cells from HIV+ individuals with CD3/CD28-activated CD4+ T-cells from uninfected donors.
Results:
Early/late but not integrated HIV reverse transcripts were detected in blood myeloid subsets of 4/10 HIV+ART; in contrast, integrated HIV-DNA was exclusively detected in CD4+ T-cells. In rectal biopsies, late HIV reverse transcripts were detected in myeloid cells and CCR6+CD4+ T-cells from 1/8 and 7/8 HIV+ART individuals, respectively. Replication-competent HIV was outgrown from CD4+ T-cells but not from myeloid of untreated/ART-treated HIV+ individuals.
Conclusion:
In contrast to CD4+ T-cells, blood and colon myeloid cells carry detectable HIV only in a small fraction of HIV+ART individuals. This is consistent with the documented resistance of Mo to HIV infection and the rapid turnover of Mo-derived macrophages in the colon. Future assessment of multiple lymphoid and non-lymphoid tissues is required to include/exclude myeloid cells as relevant HIV reservoirs during ART.
Keywords: HIV reservoirs, myeloid cells, CD16+ Mo, CD1c+ DC, antiretroviral therapy
INTRODUCTION
Antiretroviral therapy (ART) controls HIV-1 replication to undetectable plasma levels; however, viral rebound occurs upon ART interruption [1–3]. HIV reservoirs are defined as cells carrying replication-competent integrated HIV-DNA with long-term survival/self-renewal capacity [4]. While CD4+ T-cells fit well into this definition, the contribution of myeloid cells to HIV reservoir persistence during ART remains still a matter of debate [5–7]. This lack of knowledge is explained at least in part by difficulties in accessing myeloid cells residing in vital tissues. This is in contrast to CD4+ T-cells that have the ability to recirculate from the tissues into blood [8, 9].
Monocytes (Mo) are bone marrow-derived myeloid cells that circulate through the blood for a few hours/days [10] before being recruited into tissues, where they differentiate into dendritic cells (DC) or macrophages (MΦ) [11, 12]. Human Mo represent a heterogeneous population that consists of at least three different subsets (classical, intermediate, and non-classical) identified based on their differential expression of CD14 and CD16/FcγRIII and unique functional/transcriptional profiles [13–18]. In healthy individuals, classical CD14+CD16- Mo are predominant (80–85% of blood Mo), whereas intermediate/non-classical CD16+ Mo represent 5–15% of blood Mo. The chemokine receptor CCR2 (CCL2 receptor) is key for Mo to emigrate from the bone marrow into the blood [19, 20]. Classical and intermediate/non-classical Mo use CCR2 and CX3CR1 (CX3CRL1 receptor), respectively, to migrate from blood into various tissues [21–23]. During HIV-1 infection, CD16+ versus CD16- Mo are significantly expanded and produce IL-1 and TNF-α [18, 24–27]. Similar findings were reported for CD16+ Mo in the context of SIV infection [28], with increased CD16+ Mo activation being associated with an increased risk of SIV acquisition in the context of a vaccination trial [29]. Finally, CD16+ and CD16- Mo differentiate into DC and MΦ with unique molecular features that qualify them as key players in HIV pathogenesis [30–32]. While it is well-documented that CD16+ Mo fuel chronic inflammation in HIV/SIV infection, their contribution to HIV reservoir persistence during ART remains unclear.
Infection of myeloid cells may occur at various steps in the differentiation process of Mo into MΦ and/or DC. Early studies documented HIV restriction in blood Mo via post-entry restriction mechanisms [7, 33–35], despite their ability to capture and internalize HIV [22, 30, 31]. It is now established that HIV restriction in Mo, DC and MΦ is mediated by SAMHD1, a restriction factor that limits the efficacy of HIV reverse transcription [36, 37]. Nevertheless, studies by the group of Suzanne Crowe reported that CD16+ versus CD16- Mo are permissive to HIV infection in vitro and harbor Gag HIV-DNA in ART-treated individuals [38, 39]. Mo differentiation into MΦ is associated with increased HIV permissiveness [31, 40], with tissue-resident MΦ being sites of HIV replication in vivo in the absence of ART [6, 7, 41, 42]. Thus, Mo exposed to HIV in the blood or during the process of trans endothelial migration (endothelial cells capture HIV virions [43]) may become sites of productive HIV replication upon differentiation into MΦ and/or DC. Alternatively, myeloid cells may become infected with HIV via the phagocytosis of infected CD4+ T-cells. Indeed, studies demonstrated that MΦ capture and engulf HIV-infected CD4+ T-cells in vitro [44] and SIV-infected T-cells in vivo [45]. Whether MΦ represent HIV/SIV reservoirs during ART remains to be demonstrated [46]. Nevertheless, studies in humanized mouse models point to a direct contribution of myeloid cells to HIV rebound after ART interruption [47, 48]. Finally, although DC are quite resistant to productive HIV infection, they have a unique ability to capture and transmit HIV to CD4+ T-cells [49]. In addition, vaginal epithelial DC were recently identified as an HIV reservoir in ART-treated individuals [50].
In this manuscript, we aimed to shed light into the contribution of myeloid cells versus CD4+ T-cells to HIV reservoir persistence during ART. To reach this objective, we had access to leukapheresis and sigmoid colon biopsies from HIV-infected ART-treated individuals from well-characterized Montreal cohorts. We used polychromatic flow-cytometry cell sorting strategies to insure optimal purity, as well as highly sensitive nested real-time PCR to quantify different forms of cell-associated HIV-DNA and tailored viral outgrowth procedures to detect replication-competent HIV reservoirs. Together, our results support a minor role played by myeloid cells from blood and colon to HIV reservoir persistence during ART relative to CD4+ T-cells, which demonstrated to be predominant viral reservoirs. Our results are consistent with the fact that resistance to HIV infection in Mo, MΦ and DC via post-entry HIV restriction mechanisms is well-documented [7, 33–37], and that myeloid cells from the colon are short-lived as they exhibit high turnover and are mainly replenished by blood Mo [51].
METHODS
Study participants
HIV-infected individuals, untreated (HIV+) and receiving viral-suppressive ART (HIV+ART) (Table 1a–d), and uninfected controls (HIV-) were recruited at the McGill University Health Centre. Leukaphereses were collected, PBMCs isolated by Ficoll and frozen in liquid N2 until use, as we previously described [52, 53]. Plasma viral load was measured using the Amplicor HIV-1 monitor ultrasensitive method (Roche) (detection limit 40 HIV-RNA copies/ml). The date of infection was estimated using clinical and laboratory data. PBMC were isolated and frozen until use, as previously described [54, 55].
Table 1a:
Clinical information on HIV-infected study participants on ART
| ID symbol | Age (years) | Sex | CD4* | CD8* | CD4:CD8 ratio | Plasma viral load# | Time of infection (months) | ART | Time on ART (months) |
|---|---|---|---|---|---|---|---|---|---|
| HIV+ART #1 (●) | 62 | M | 847 | 944 | 0.90 | <40 | 191 | Kivexa Reyataz |
165 |
| HIV+ART #2 (■) | 68 | M | 314 | 793 | 0.40 | <40 | 249 | Darunavir Etravirine Dolutegravir Norvir |
181 |
| HIV+ART #3 (▲) | 23 | M | 277 | 909 | 0.30 | <40 | 11 | Complera | 8 |
| HIV+ART #4 (▼) | 47 | M | 581 | 1,060 | 0.55 | <40 | 96 | Isentress Kivexa | 96 |
| HIV+ART #5 (○) | 21 | M | 796 | 399 | 1.99 | <40 | 7 | Stribild | 4 |
| HIV+ART #6 (□) | 57 | M | 539 | 1,231 | 0.40 | <40 | 252 | Truvada | 84 |
| HIV+ART #7 (◊) | 61 | M | 397 | 238 | 1.67 | <40 | 45 | Stribild | 38 |
| HIV+ART #8 (Δ) | 62 | M | 498 | 531 | 0.94 | <40 | 213 | Raltegravir Intelence Ritonavir |
134 |
| HIV+ART #9 (∇)& | 24 | M | 776 | 478 | 1.62 | <40 | 288 | Complera | 140 |
| HIV+ART #10 (♦) | 31 | M | 824 | 900 | 0.92 | <40 | 58 | Atripla | 47 |
| HIV+ART #11 (×) | 28 | F | 433 | 240 | 1.80 | <40 | 184 | Viracept Truvada |
156 |
| HIV+ART #12 (┼) | 30 | F | 833 | 445 | 1.87 | <40 | 216 | Viracept Truvada |
192 |
ID, participant identity; F, female; M, male;
, cells/µl;
#, HIV-RNA copies/ml; N/A, data not available;
&, Participant ART #9 (∇) was infected at birth.
Table 1d:
Clinical information on HIV-infected ART-treated participants included in the colon biopsy study cohort
| ID symbol | Age (years) | Sex | CD4* | CD8* | CD4:CD8 ratio | Plasma viral load# | Time of infection (months) | ART | Time on ART (months) |
|---|---|---|---|---|---|---|---|---|---|
| HIV+ART #13 | 51 | M | 175 | 1,100 | 0.16 | <40 | 231 | Triumeq | 204 |
| HIV+ART #14 | 50 | M | 453 | 729 | 0.62 | <40 | 233 | Truvada Viramune |
204 |
| HIV+ART #15 | 54 | M | 895 | 1,023 | 0.87 | <40 | 224 | Norvir Reyataz Truvada |
192 |
| HIV+ART #16 | 67 | M | 314 | 793 | 0.40 | <40 | 244 | Darunavir Etravirine Dolutegravir Norvir |
176 |
| HIV+ART #17 | 56 | M | 786 | 1,813 | 0.43 | <40 | 240 | Isentress Truvada |
72 |
| HIV+ART #18 | 58 | M | 718 | 1,079 | 0.67 | <40 | 305 | Tivicay Kivexa |
204 |
| HIV+ART #19 | 55 | M | 513 | 1,095 | 0.47 | <40 | 192 | Kivexa Isentress |
N/A |
| HIV+ART #20 | 56 | M | 288 | 407 | 0.71 | <40 | 216 | Stribild | 216 |
ID, participant identity; M, male;
, cells/µl;
#, HIV-RNA copies/ml; N/A, data not available;
Note: HIV+ART#2 and HIV+ART#16, as well as HIV+ART#6 and HIV+ART#17 represent the same individual, with blood collected at different time points.
Ethics statement
This study using biological samples from HIV-infected and uninfected individuals was conducted in compliance with the principles included in the Declaration of Helsinki and received approval from the Institutional Review Board of the McGill University Health Centre and CHUM-Research Centre. All subjects signed written informed consent for their study participation.
Flow-cytometry analysis
Flow-cytometry analysis was performed using Abs listed in Supplemental Table 1. The viability dye LIVE/DEAD® Fixable Aqua Dead Cell Stain Kit (Invitrogen) was used to exclude dead cells. The BD-LSRII cytometer and FlowJo (©Tree Star, Inc., Ashland, OR) softwares were used for phenotypic analysis. Positivity gates were placed using the Fluorescence minus one (FMO) strategy [56], as we previously described [57].
Magnetic (MACS) and fluorescence activated (FACS) cell sorting
In a first set of experiments (Figure 1), total Mo were purified from peripheral blood by negative selection using magnetic beads (MACS, Pan Monocyte Isolation kit, human, Miltenyi). Highly pure total Mo, CD16- and CD16+ Mo, and matched CD1c+ DC were subsequently sorted by FACS (BD AriaIII) from the MACS-sorted total Mo fraction. Briefly, cells were stained with anti-CD3, CD56, CD19, CD14, CD16, HLA-DR and CD1c Abs (Supplemental Table 1) in a special sorting buffer (PBS 1X with 5% FBS and 25mM Hepes buffer). Mo were identified by their expression of HLA-DR, CD14 and/or CD16 and by the absence of expression of T-cell (CD3, CD8), B cell (CD19), NK cell (CD56) and DC (CD1c) markers. DC were identified as cells with a Lin-HLA-DR+CD1c+ phenotype. Mo/DC purity upon sort was >99%. In parallel, matched total CD4+ T-cells were isolated from PBMC of HIV+ on ART individuals by negative selection using magnetic beads (Miltenyi). In another set of experiments (Figure 2), matched CD16+/CD16- Mo (Lineage-HLA-DR+CD16+ or CD16-), CD1c+ DC (Lineage-HLA-DR+CD1c+) and total CD4+ T-cells (Lineage-CD3+CD4+) were isolated directly from PBMC of HIV+ART individuals by FACS. The cell purity upon sort was >99%.
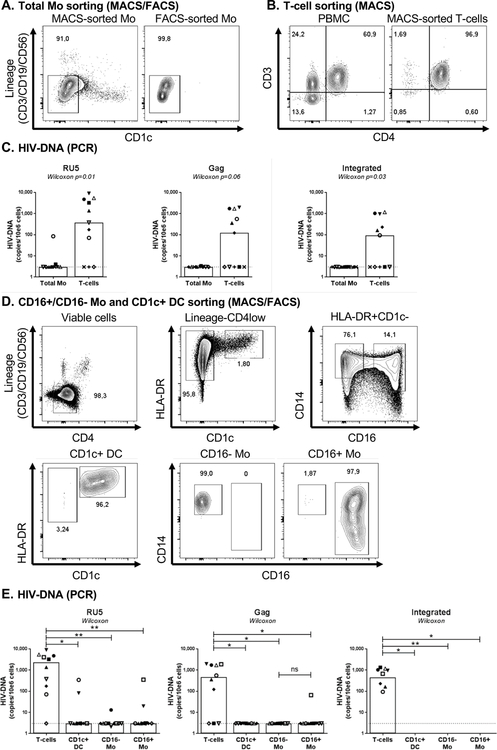
Figure 1. HIV-DNA quantification in blood myeloid cells sorted by MACS and FACS from HIV+ART individuals.
(A) Monocytes (Mo) were purified from the peripheral blood monucleated cells (PBMC) of HIV-infected on ART individuals (HIV+ART; Table 1a) by negative selection using magnetic associated cell sorting (MACS). To insure high purity of the Mo population with no T cells contamination, total Mo identified as cells with a CD3-CD19-CD56-CD1c- phenotype were subsequently sorted by FACS upon staining with CD3, CD56, CD19, and CD1c Abs. Post-sort purity was >96%. (B) Total CD4+ T-cells were isolated by MACS from matched donors. (C) Levels of early (RU primers; left panel), late (Gag primers, middle panel) HIV reverse transcripts and integrated HIV-DNA (Alu/HIV primers; right panel) were quantified by nested real-time PCR. (D) Highly pure CD14+CD16- and CD14+/lowCD16+ Mo (CD3-CD19-CD56-HLA-DR+CD1c-) and CD1c+ DC (CD3-CD19-CD56- HLA-DR+CD1c+) were sorted by FACS upon staining of the total Mo fraction with CD3, CD4, CD56, CD19, CD14, CD16, HLA-DR and CD1c Abs (upper panels). Post-sort purity of Mo and DC was >96% (lower panels). (E) Levels of early (left panel), late (middle panel) HIV reverse transcripts and integrated HIV-DNA (right panel) were quantified by nested real-time PCR. The lines indicate the limit of detection: 3 HIV-DNA or CD3 copies per PCR replicate. Each PCR reaction was performed in 3–10 replicate wells and median values were calculated. Each symbol represents one different HIV+ART individual (n=10). The limit of detection (3 HIV-DNA copies/test), indicated by the dotted lines, was assigned to any undetectable sample. Wilcoxon paired non-parametric test p-values are indicated on the graphs in panels C and E.
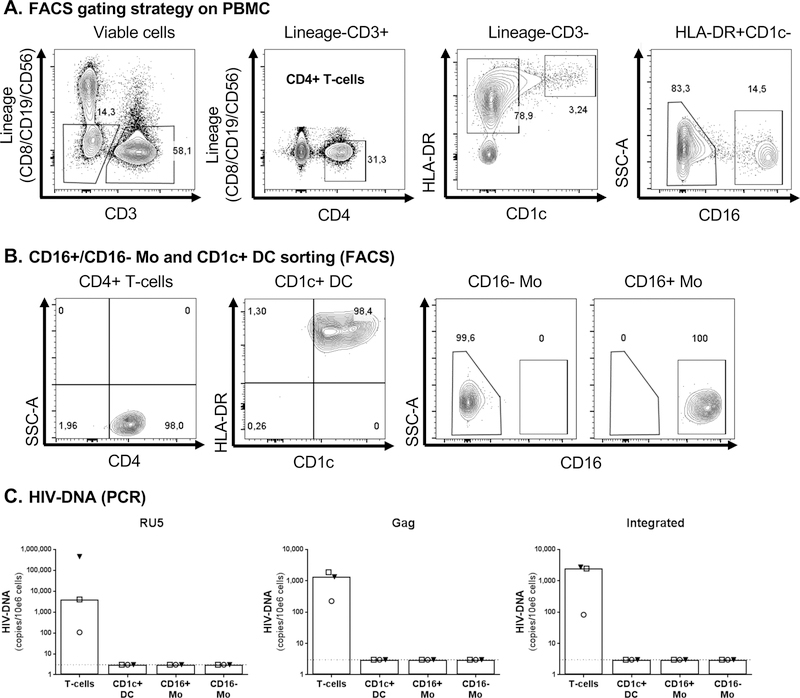
Figure 2. HIV-DNA quantification in myeloid cells sorted by FACS directly from PBMC of HIV+ART individuals.
Matched CD16- Mo, CD16+ Mo, CD1c+ DC, and CD4+ T-cells were sorted directly by FACS from PBMC of HIV+ART individuals (Table 1a) upon staining with CD3, CD4, CD8, CD56, CD19, CD16, HLA-DR and CD1c Abs, using the gating strategy illustrated in panel A. Post-sort cell purity was >98%, as depicted in panel B. Levels of early (left panel), late (middle panel) HIV reverse transcripts and integrated HIV-DNA (right panel) were quantified using nested real-time PCR (C), as detailed in Figure 1 legend. The limit of detection (3 HIV-DNA copies/test), indicated by the dotted lines, was assigned to any undetectable sample. Each PCR reaction was performed in triplicates of 105 cells per test. Each symbol represents one different HIV+ART individual (n=3).
HIV-DNA quantification
The quantification of early (RU5), total (Gag) and integrated HIV-DNA was performed by nested real-time PCR using specific HIV and/or Alu primers, as we previously described[57]. For normalization, CD3 and HIV regions were amplified together during the first amplification and separated for the second amplification. ACH2 cells containing one copy of proviral HIV-DNA was used for standard using 1:10 serial dilutions from 3×105 to 3 cells. The sensitivity of the assay was 3 HIV/CD3 copies/test.
Viral outgrowth assay
Total myeloid cells (Lineage-CD3-HLA-DR+CD33+), mainly including Mo and a small fraction of CD1c+ DC, were sorted by MACS and then by FACS from PBMC of HIV+ individuals before and/or after ART initiation (Table 1b–c). Matched total CD4+ T-cells (Lineage-CD3+CD4+) from HIV+ individuals were isolated by MACS. In parallel, memory CD4+ T-cells of one HIV-uninfected individual were sorted by MACS. T-cells from HIV+/HIV- individuals were activated via the TCR using immobilized CD3 (clone UCHT1, BD) and soluble CD28 Abs (clone CD28.2, BD) (1 µg/ml) for 3 days prior co-culture. Freshly isolated myeloid cells and TCR-activated T-cells (1×106) from HIV+ individuals were cultured with TCR-activated T-cells from the HIV- individual (1×106) at a 1:1 ratio in 48-well plates. Media (RPMI, 10% FBS, 1% penicillin/streptomycin, IL-2) was refreshed and each well was split into two new wells every three days, as previously reported [57, 58]. Levels of HIV-p24 in cell-culture supernatant were quantified by ELISA, as previously reported [57, 58].
Table 1b:
Clinical information on HIV-infected study participants before and after ART
| ID symbol | Age (years) | Sex | CD4* | CD8* | CD4:CD8 ratio | Plasma viral load# | Time of infection (months) | ART | Time on ART (months) |
|---|---|---|---|---|---|---|---|---|---|
| HIV+ #1 | 39 | M | 946 | 1,356 | 0.70 | 8,809 | 9 | - | 0 |
| HIV+ #2 | 24 | M | 316 | 691 | 0.46 | 9,496 | 55 | - | 0 |
| HIV+ #3 | 40 | M | 644 | 931 | 0.69 | 7,556 | 4 | - | 0 |
| HIV+ #1 ART | 42 | M | 795 | 753 | 1.06 | <40 | 46 | Tenofovir Rilpivirine Dolutegravir |
33 |
| HIV+ #2 ART | 27 | M | 437 | 555 | 0.79 | <40 | 86 | Tenofovir Emtricitabine Alafenamide |
12 |
| HIV+ #3 ART | 42 | M | 899 | 497 | 1.81 | <40 | 32 | Abacavir Dolutegravir Iamivudine |
26 |
ID, participant identity; M, male;
, cells/µl;
#, HIV-RNA copies/ml
Table 1c:
Clinical information on HIV-infected untreated study participants
| ID symbol | Age (years) | Sex | CD4* | CD8* | CD4:CD8 ratio | Plasma viral load# | Time of infection (months) | ART | Time on ART (months) |
|---|---|---|---|---|---|---|---|---|---|
| HIV+ #4 | 44 | M | 254 | 812 | 0.31 | 24,729 | 222 | - | 0 |
| HIV+ #5 | 30 | M | 642 | 848 | 0.76 | 6,077 | 36 | - | 0 |
| HIV+ #6 | 41 | M | 795 | 1,062 | 0.75 | 320,573 | 7 | - | 0 |
ID, participant identity; M, male;
, cells/µl;
#, HIV-RNA copies/ml
Cell isolation from blood and colon biopsies
Matched blood (20 ml/donor) and colon biopsies (≈32 biopsies/donor) were collected and processed from a cohort of HIV+ART individuals (Table 1d; n=8), as previously reported [53, 57, 59]. Briefly, biopsies were washed with media (RPMI, 100U/mL penicillin, 100 µg/mL streptomycin, 2 mM L-glutamine), enzymatically digested using Liberase (50 µg/mL), and mechanically disrupted. This procedure was repeated three times. In parallel, PBMC were isolated from fresh blood by Ficoll. Blood/colon cell suspensions were stained with CD3, CD326, CD8, CD19, CD66b, CD64 and HLA-DR Abs (Supplemental Table 1) in FACS sorting buffer (PBS, 5% FBS, 25mM Hepes buffer), as previously described [53, 57]. Matched blood/colon myeloid cells (CD3-CD8-CD19-CD66b-CD326-HLA-DR+) and CD4+ T-cells (CD3+CD8-CD19-CD66b-CD326-) were sorted in parallel by FACS (BDAriaIII, BD).
Statistical analysis
Statistical analysis was performed using the GraphPad Prism 7 software; details are included in Figure legends.
RESULTS
Quantification of early, late and integrated HIV reverse transcripts in myeloid cells from the blood of HIV+ART individuals.
One major limitation for studying the contribution of myeloid cells to HIV reservoir persistence during ART is the purity of sorted samples for optimal exclusion of contaminating CD4+ T-cells. In order to reach maximum purity, total Mo were enriched from PBMC of HIV+ART individuals with undetectable plasma viral load (Table 1a), first by negative selection using magnetic beads (MACS) and then by flow-cytometry (FACS) (Figure 1A). In parallel, highly pure CD4+ T-cells were sorted by MACS negative selection from matched donors (Figure 1B). By using this strategy, sufficient numbers of total Mo (range: 1–9×106 cells from 50–300×106 PBMC) and CD4+ T-cells (range: 7–12×106 cells from 20–100×106 PBMC) were sorted for HIV-DNA PCR quantification. PCR reactions in T-cells and myeloid cells were performed in 3–10 replicates on similar numbers of cells (Suppl. Figure 1A–C). Specific PCR primers were used to quantify early (RU5) and late (Gag), as well as integrated (Alu/LTR) HIV reverse transcripts. RU5 HIV-DNA was detected in total Mo from 2/11 individuals and in CD4+ T-cells of 8/11 individuals (Figure 1C, left panel). Gag HIV-DNA was detected in total Mo from 1/11 individuals and in CD4+ T-cells of 6/11 individuals (Figure 1C, middle panel). Finally, integrated HIV-DNA was detected only in CD4+ T-cells (Figure 1C, right panel). Thus, although early/late reverse transcripts, likely indicative of recent HIV exposition, are detected in blood Mo, these cells do not carry integrated HIV during ART, as it is the case for CD4+ T-cells in the majority of HIV+ART individuals tested.
CD16+ Mo represent a more advanced stage of Mo differentiation [10, 60] prone to contribute to HIV replication/dissemination [30, 31, 38, 39]. Therefore, in a second set of experiments, highly pure CD16+ and CD16- Mo were enriched by FACS from the total Mo fraction enriched by MACS from the blood of n=10 HIV+ART individuals (Table 1a). In parallel, CD1c+ DC (cDC2), another important subset of myeloid cells present in the blood [61], were also sorted by FACS (Figure 1D). By using this strategy, sufficient numbers of CD1c+ DC (range: 0.45–1.5×106), CD16- Mo (range: 8–11×106), and CD16+ Mo (range: 0.8–4.3×106) were sorted from 0.8–3×109 PBMC for HIV-DNA PCR quantification. Each PCR reaction was performed on minimum 104 cells in 3–7 replicates (Supplemental Figure 1D–F). Matched CD4+ T-cells sorted by MACS (Figure 1B) were used as controls. RU5 HIV-DNA was detected in CD1c+ DC of 2/10 individuals (340 and 316 HIV-DNA copies/106 cells), in CD16+ Mo of 2/10 individuals (19 and 349 HIV-DNA copies/106 cells), and in CD16- Mo of 1/10 individuals (11 HIV-DNA copies/106 cells) (Figure 1E, left panel). Gag HIV-DNA was detected in CD16+ Mo of 1/10 individuals (66 HIV-DNA copies/106 cells), while levels were undetectable in CD16- Mo and CD1c+ DCs of n=10 individuals (Figure 1E, middle panel). Integrated HIV-DNA was undetectable in CD16+Mo, CD16- Mo and CD1c+ DC from n=10 individuals (Figure 1E, right panel). Thus, CD4+ T-cells but not CD16+/CD16- Mo nor CD1c+ DC carry integrated HIV-DNA in the blood of HIV+ART individuals.
To further validate the results obtained on myeloid cells sorted by FACS from MACS-enriched Mo/DC fractions, we used a second sorting strategy in which CD1c+ DC, CD16+/CD16- Mo, and CD4+ T-cells were directly sorted by FACS from PBMC (Figure 2A–B). Experiments were performed on n=3 HIV+ART individuals selected based on the presence of integrated HIV-DNA in CD4+ T-cells (Figure 1; Table 1a). The purity of each population sorted was >98% (Figure 2B). Sufficient numbers of CD4+ T-cells (range: 2.8–5.7×106), CD1c+ DC (range: 0.4–1×106), CD16- Mo (range: 2.7–5.9×106) and CD16+ Mo (range: 0.4–2.8×106) were sorted from 120–150×106 PBMC for HIV-DNA PCR amplification; each PCR reaction was performed on minimum 105 cells in 3–6 replicates (Supplemental Figure 2A–C). Relatively high levels of RU5, Gag and integrated HIV-DNA were detected in CD4+ T-cells of 3/3 individuals, while levels were undetectable in CD1c+ DC, CD16- and CD16+ Mo (Figure 2C).
Together, these results demonstrate that integrative HIV infection is undetectable in CD16- and CD16+ Mo subsets, as well as CD1c+ DC isolated from the blood of HIV+ART individuals, while traces of likely abortive HIV exposure, reflected by RU5/Gag HIV-DNA detection, are present in a minor fraction of study participants.
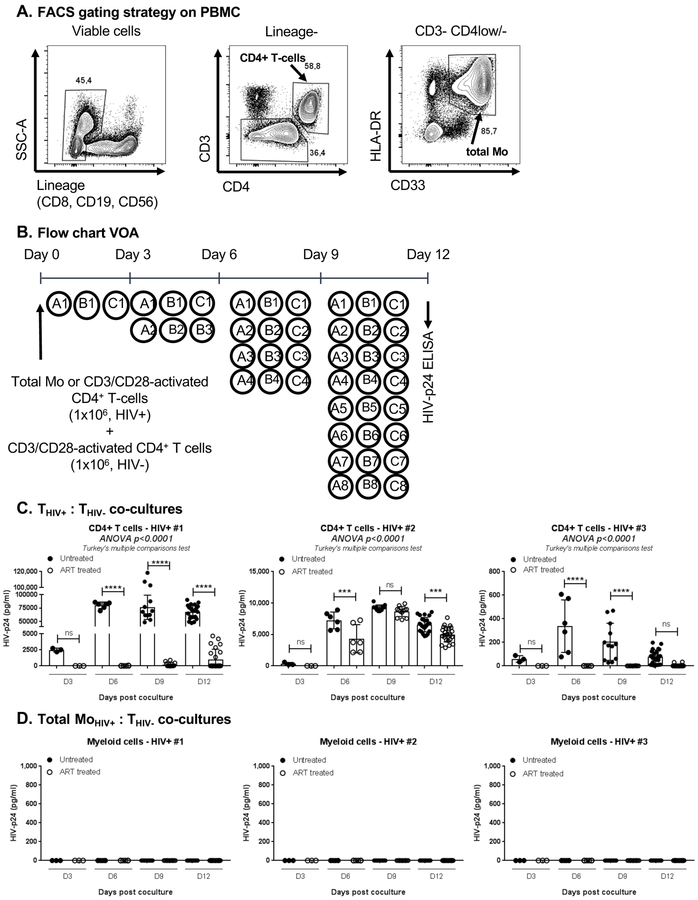
Blood CD4+ T-cells but not Mo carry replication-competent HIV in the absence of ART.
Considering the detection of early/late HIV reverse transcripts in myeloid cells from a fraction of HIV+ART individuals (Figure 1E), and the capacity of myeloid cells to capture and transmit HIV to CD4+ T-cells in vitro [31, 49], we further investigated whether blood myeloid cells carry replication-competent HIV in vivo. Total HLA-DR+CD33+ myeloid cells (including mainly Mo and a small fraction of CD1c+ DC) and CD4+ T-cells were sorted by FACS (Figure 3A). A viral outgrowth assay (VOA) was developed, in which matched freshly-isolated Mo and TCR-activated CD4+ T-cells isolated from n=3 HIV+ individuals before/after ART initiation (Table 1b) were co-cultured with TCR-activated CD4+ T-cells isolated from an HIV-uninfected donor (Figure 3B). Replication-competent HIV was robustly detected in CD4+ T-cells before treatment, with ART initiation leading to a significant reduction in viral outgrowth from T-cells ex vivo (Figure 3C). In contrast, the viral outgrowth was undetectable when co-cultures were performed with Mo from the same HIV+ individuals, before/after ART initiation (Figure 3D). One particularity of these three HIV+ individuals is their relatively low plasma viral load before ART initiation (<10,000 HIV-RNA copies/ml; Table 1b). To expand these investigations, further VOA were performed with matched T-cells and Mo isolated from three other untreated HIV+ individuals with high, intermediate, and low plasma viral loads (Table 1c). HIV-DNA was detected in T-cells from 2/3 HIV+ individuals, but not in Mo ex vivo (Supplemental Figure 3B). Similar to results in Figure 3, HIV outgrowth was detected when co-cultures were performed with T-cells but not Mo from HIV+ individuals (Supplemental Figure 3C–D). These results demonstrate that CD4+ T-cells but not Mo carry replication-competent HIV in the blood of viremic untreated and aviremic ART-treated HIV+ individuals.
Figure 3. Blood CD4+ T-cells but not myeloid cells from untreated and ART-treated HIV-infected individuals carry replication-competent HIV.
(A) Highly pure matched total myeloid (Lin-CD3-HLA-DR+CD33+) and total CD4+ T-cells were isolated by FACS from n=3 untreated HIV-infected individuals with detectable plasma viral load in the absence of ART (HIV+) (Table 1b). (B) A viral outgrowth assay (VOA) was performed by co-culturing myeloid cells (MoHIV+) and CD3/CD28-activated CD4+ T-cells (THIV+) from HIV+ individuals with CD3/CD28-activated CD4+ T-cells from one same uninfected individual (THIV-). Co-cultures were performed in 48-well plates at a MoHIV+:THIV- or THIV+:THIV- ratios of 1:1 and a final cell density of 2×106 cells/well, in the presence of IL-2 for 12 days. Cells were split every 3 days in 2 new wells for optimal density. The presence of replication-competent HIV was measured by ELISA at day 3, 6, 9 and 12 post co-culture (detection limit of 5 pg/ml for a dilution of cell-culture supernatant of 1/10–1/500 for T-cells and ½ for Mo). (C-D) Shown are results on HIV reactivation in CD4+ T-cells (C) and myeloid cells (D) from n=3 HIV+ individuals before and after ART initiation (Table 1b). Bar graphs in C-D indicate mean±SD values or replicate wells, with each symbol representing one replicate value per time point. One-way Anova and Turkey’s multiple comparison test p-values are indicated on the graphs in panel C.
HIV-DNA is rarely detected in blood and colon myeloid cells during ART.
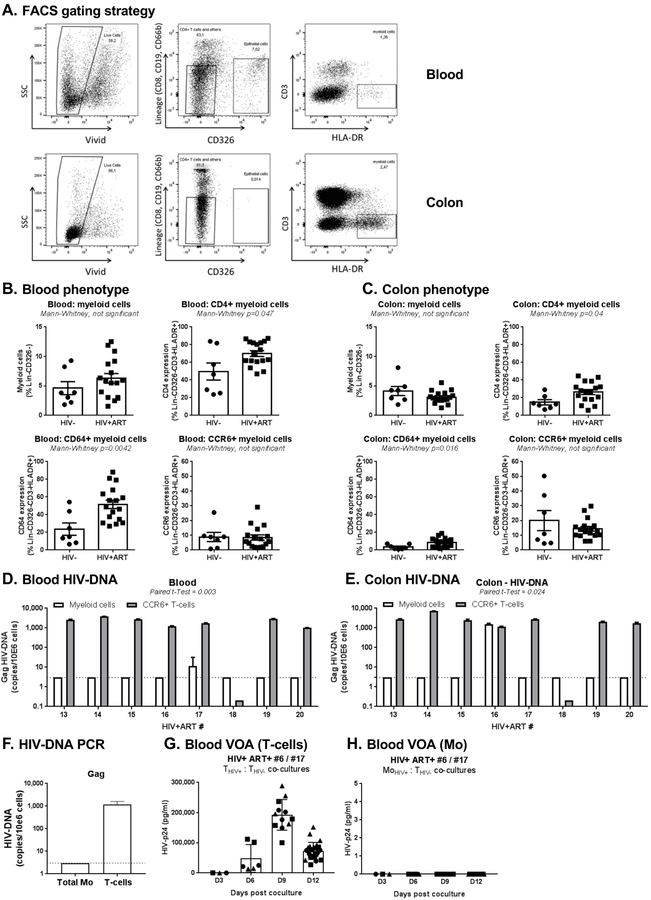
The gut-associated lymphoid tissues (GALT) represent major sites of viral replication early after HIV-infection [62, 63] and during ART-treated chronic HIV infection [53, 64, 65]. Because HIV permissiveness increases with Mo differentiation into MΦ [31, 34], we hypothesized that myeloid cells from the colon, a site rich in CD4+ T-cells carrying HIV reservoirs [53], are more likely to carry HIV-DNA of HIV+ART individuals. To test this hypothesis, we had access to matched blood and sigmoid colon biopsies from HIV+ART (Table 1d) and HIV-, from a previously described cohort [53]. Myeloid cells from blood and colon with a CD8-CD19-CD66b-CD326-CD3-HLA-DR+ phenotype were identified as illustrated in Figure 4A. The frequency of myeloid cells, as well as their expression of the HIV receptor CD4 [3], the MΦ marker CD64/FcγRI [66], and the DC marker CCR6 [67], were compared between HIV+ART (n=17) and uninfected (n=7) individuals in blood and colon. In blood and colon, the frequency of myeloid cells expressing CD4 and CD64 was significantly increased in HIV+ART versus HIV- individuals, with no significant differences observed in the frequency of myeloid cells and CCR6 expression (Figure 4B–C).The FACS sorting strategy illustrated in Figure 4A allowed the simultaneous sorting of myeloid cells from blood (median±SD: 113,140 ± 216,930; range: 0.1 – 0.3×106) and colon (median±SD: 43,608 ± 20,734; range: 0.3–0.8×105) from 8–20×106 total cells extracted from 30 biopsies per donor (n=8 HIV+ART individuals). The presence of Gag HIV-DNA was measured by nested real-time PCR. Each PCR reaction was performed on 0.04–0.3×105 colon and 0.06–0.45×105 blood myeloid cells (Supplementary Fig 4A–B). In these n=8 HIV+ART individuals, Gag HIV-DNA was detected in blood myeloid cells from 1/8 individuals (11.58 HIV-DNA copies/106 cells) and colon myeloid cells of another 1/8 individuals (1,562 HIV-DNA copies/106 cells) (Figure 4D–E). As previously reported [53], blood and colon memory CCR6+CD4+ T-cells isolated from 7/8 donors carried relatively high levels of Gag HIV-DNA (range: 303–7,124 HIV-DNA copies/106 cells) (Figure 4D–E).
Figure 4. Myeloid cells from blood and colon rarely harbor HIV-DNA in HIV+ART individuals.
Myeloid cells were identified in matched blood and colon biopsies of HIV+ART (n=16) and HIV- (n=7) individuals (cohorts previously described in [53]), using the gating illustrated in panel A. Briefly, live/dead dye (Vivid; Invitrogen) was used to exclude dead cells from analysis. Staining with CD8, CD19, CD66b, and CD326 Abs allowed the exclusion of CD8+ T-cells, B cells, granulocytes and epithelial cells, respectively. Myeloid cells were identified as Lin-CD326-CD3-HLA-DR+. (B-C) Shown is the frequency of total myeloid cells (upper left panel), as well as the expression of CD4 (upper right panel), CD64 (lower left panel) and CCR6 (lower right panel) on myeloid cells from blood (B) vs colon (C) of HIV- vs. HIV+ART individuals. (D-E) Shown are levels of late HIV reverse transcripts (Gag primers) quantified using nested real-time PCR in myeloid cells from blood (D) and colon (E) of n=8 HIV+ART individuals. For comparison, included are levels of Gag HIV-DNA detected by real-time nested PCR in matched CCR6+CD4+ T-cells sorted by FACS, as previously published [53]. (F-H) Frozen PBMC from the HIV+ART#17 participant (Table 1d), with leukapheresis collected at a previous time point (identified as HIV+ART#6 in Table 1a), were used to sort matched CD4+ T-cells and Mo by MACS and FACS, respectively. Shown are Gag HIV-DNA levels in T-cells and Mo ex vivo (F), as well as levels of replication-competent HIV detected in VOA performed with T-cells (G) and Mo (H) from this participant. Bar graphs in G-H indicate mean±SD values or replicate wells, with each symbol representing one replicate value per time point.
To validate the presence of HIV-DNA in blood Mo of HIV+ART#17 (Table 1d), we repeated the PCR and performed a VOA using total Mo and CD4+ T-cells isolated from a leaukapheresis donated by the same donor (HIV+ART#6; Table 1a). HIV-DNA was detected in T-cells but no longer in Mo (Figure 4F), likely due to the stochastic distribution of HIV reservoirs and/or the distinct composition of Mo isolated from fresh blood versus leukapheresis. Similarly, the VOA revealed the presence of replication-competent HIV in CD4+ T-cells but not in Mo (Figure 4G–H). Thus, in contrast to CD4+ T-cells, blood and colon myeloid cells represent a minor HIV reservoir during ART-treated and untreated infection, regardless of the levels of plasma viral load in the absence of ART.
DISCUSSION
In this study, we demonstrate that infection of myeloid cells from blood and colon of ART-treated HIV-infected individuals is a rare event. This contrasts with the detection of integrated HIV-DNA and/or replication-competent HIV reservoirs in CD4+ T-cells from these two anatomic compartments in the majority of donors tested.
The fact that early/late reverse transcripts were rarely detected in blood CD16+/CD16- Mo and CD1c+ DC of HIV+ individuals with undetectable plasma viral load during ART, but never integrated HIV-DNA, is consistent with the documented resistance to HIV infection of myeloid cells [7, 33–35], as well as the relatively short Mo life-span [10]. There was no correlation between the presence of early/late HIV reverse transcripts and clinical parameters such as age, CD4:CD8 ratio, time since infection, time of ART initiation, or time on ART. Noteworthy, 3–10 PCR replicates were performed for each sample. The fact that few replicates only tested positive is indicative that the infection of myeloid cells is a rare event, compared to CD4+ T-cells. Our results contrast with previous studies reporting that CD16+ Mo carry Gag HIV-DNA in ART-treated individuals [38, 39]. Such discrepancies may be explained by i) the inclusion of HIV+ART individuals with detectable viral load (50–45,400 copies/mL) and ii) potential CD4+ T-cell contaminations of Mo fractions upon magnetic bead isolation. Indeed, one major concern relative to HIV-DNA quantification in myeloid cells is the purity of the samples. In our study, we used a combination of MACS and FACS or only FACS sorting to ensure high purity of samples (>98%).
Despite their well-documented resistance to HIV infection [7, 33–37], Mo exhibit the capacity to capture/transmit intact HIV virions in vivo [30, 31]. To detect replication-competent HIV potentially captured by blood Mo in vivo, we used a myeloid-tailored VOA, where freshly isolated CD33+HLA-DR+ myeloid cells isolated from leukapheresis of HIV+ individuals collected before/after ART initiation were co-cultured with TCR-activated CD4+ T-cells from an HIV-uninfected individual. Matched TCR-activated T-cells were used as positive controls. Replication-competent HIV was recovered from CD4+ T-cells but not HLA-DR+CD33+ myeloid cells of n=3 HIV+ individuals with relatively low plasma viral load in the absence of treatment (<10,000 HIV-RNA copies/ml). After the initiation of viral-suppressive ART, the detection of replication-competent HIV was significantly reduced but still detectable in CD4+ T-cells and remained undetectable in the myeloid fraction. Similar results were obtained with matched myeloid and T-cells isolated from three additional untreated HIV+ individuals with low, intermediate and high plasma viral load. Thus, in contrast to CD4+ T-cells, replication-competent HIV was not detectable in myeloid cells of ART-treated neither viremics untreated HIV+ individuals.
The gut-associated lymphoid tissues (GALT) are enriched in CD4+ T-cells and represent major sites of viral replication early on after infection [62, 63, 68]. MΦ are abundant in the intestine [69]; they are mainly replenished by blood Mo [51] and accumulate in the gut mucosa of ART naive individuals [70]. Although, intestinal MΦ appear to be resistant to HIV infection in vitro [71], the presence of HIV reservoirs in this myeloid cells (as a result of direct infection or phagocytosis of infected T-cells) remains unexplored in the colon. Here, we demonstrate that detection of HIV-DNA in myeloid cells from the colon is a rare event occurring in 1/8 HIV+ART individuals, in contrast to the frequent infection of CD4+ T-cells, and this for similar numbers of cells sampled by PCR. These results raise new questions on the reason why myeloid cells are not detected as being infected in an environment rich in HIV reservoirs [53], likely prone to residual viral transcription [72]. The existence of a deep state of HIV latency in colon-infiltrating T-cells [73], may explain the lack of infection in short-lived myeloid cells. However, we cannot exclude limits in our ability to isolate sufficient HIV-infected myeloid cells from the colon due to their high activation/adherence, which may cause their loss during the flow-cytometry isolation procedure. Such potential limitations emphasize the need for future implementation of in situ visualization approaches, such as HIV DNA/RNAscope [72, 74].
Recent studies demonstrated the persistence of HIV reservoirs in MΦ of a fraction (33%) of humanized myeloid-only mice (MoM) during ART and their capacity to sustain HIV replication in absence of T cells [47, 48]. The identity of myeloid cells that contribute to HIV persistence in this MoM model of ART-treated HIV infection remains to be defined. This finding points to the necessity to extend HIV reservoir studies on myeloid cells to other human tissues of ART-treated individuals, such as liver, lungs and brain. Noteworthy, the new understanding of the ontology of tissue-resident myeloid cells [51] suggest potential differences between embryonic/fetal and Mo-derived myeloid cells in their capacity to act as HIV reservoirs during ART.
In conclusion, our results support a minor role played by myeloid cells from blood and colon to HIV reservoir persistence during ART, in contrast to CD4+ T-cells that proved to be major HIV reservoirs in these two compartments. Nevertheless, considering the major paradigm shift that occurred in the last years relative to the ontogeny of myeloid cells [51], access to long-lived self-renewing tissue-resident MΦ from multiple lymphoid/non-lymphoid tissues will be critical in future studies to rationally include/exclude myeloid cells as relevant cellular HIV reservoirs [5–7, 75].
Supplementary Material
Acknowledgements
The authors acknowledge the contribution of Dr. Dominique Gauchat and Mr. Philippe St-Onge (Flow-Cytometry Core Facility, CHUM-Research Centre, Montreal, QC, Canada) for expert technical support with FACS analysis and sorting, Mr. Mario Legault for help with ethical approvals and informed consents, Mrs. Josée Girouard, Mrs. Angie Massicotte, and Mrs. Maria Fraraccio (McGill University Health Centre-Glen site, Montreal, QC, Canada) for critical contribution to blood collection from HIV-infected study participants. Finally, the authors acknowledge all study participants for their precious gift of leukapheresis essential for this study.
Funding
This study was supported by grants from the Canadian Institutes of Health Research (CIHR) (HOP-120239 and MOP-114957) to PA, grants from the CIHR Canadian HIV Trials Network (CTN #247), the Fonds de Recherche du Qubec-Sant (FRQ-S)/AIDS and Infectious Diseases Network, Qubec, Canada, to JPR, and by The Canadian HIV Cure Enterprise (CanCURE) Team Grant from the CIHR in partnership with CANFAR and IAS to PA, EAC and JPR (HIG-133050). JPR holds a Louis Lowenstein Chair in Hematology and Oncology, McGill University. The funding institutions played no role in the design, collection, analysis, and interpretation of data.
Footnotes
Competing interests
The authors declare no financial and non-financial competing interests.
REFERENCES
- 1.Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, et al. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med 2016; 22(8):839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goulder PJ, Lewin SR, Leitman EM. Paediatric HIV infection: the potential for cure. Nat Rev Immunol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barre-Sinoussi F, Ross AL, Delfraissy JF. Past, present and future: 30 years of HIV research. Nat Rev Microbiol 2013; 11(12):877–883. [DOI] [PubMed] [Google Scholar]
- 4.Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012; 37(3):377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton KL, Garcia JV, Clements JE, Walker BD. HIV Infection of Macrophages: Implications for Pathogenesis and Cure. Pathogens & immunity 2017; 2(2):179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson M Role of myeloid cells in HIV-1-host interplay. Journal of neurovirology 2015; 21(3):242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sattentau QJ, Stevenson M. Macrophages and HIV-1: An Unhealthy Constellation. Cell Host Microbe 2016; 19(3):304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Andrian UH, Mackay CR. T-cell function and migration. Two sides of the same coin. N Engl J Med 2000; 343(14):1020–1034. [DOI] [PubMed] [Google Scholar]
- 9.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, et al. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 2012; 490(7419):283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 2017; 214(7):1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science 2010; 327(5966):656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randolph GJ, Beaulieu S, Lebecque S, Steinman RM, Muller WA. Differentiation of monocytes into dendritic cells in a model of transendothelial trafficking. Science 1998; 282(5388):480–483. [DOI] [PubMed] [Google Scholar]
- 13.Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood 2010; 116(16):e74–80. [DOI] [PubMed] [Google Scholar]
- 14.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005; 5(12):953–964. [DOI] [PubMed] [Google Scholar]
- 15.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol 2009; 27:669–692. [DOI] [PubMed] [Google Scholar]
- 16.Ancuta P A slan-based nomenclature for monocytes? Blood 2015; 126(24):2536–2538. [DOI] [PubMed] [Google Scholar]
- 17.Ziegler-Heitbrock L Blood Monocytes and Their Subsets: Established Features and Open Questions. Front Immunol 2015; 6:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wacleche VS, Tremblay CL, Routy JP, Ancuta P. The Biology of Monocytes and Dendritic Cells: Contribution to HIV Pathogenesis. Viruses 2018; 10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol 2008; 26:421–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol 2006; 7(3):311–317. Epub 2006 Feb 2005. [DOI] [PubMed] [Google Scholar]
- 21.Weber C, Belge KU, von Hundelshausen P, Draude G, Steppich B, Mack M, et al. Differential chemokine receptor expression and function in human monocyte subpopulations. J Leukoc Biol 2000; 67(5):699–704. [DOI] [PubMed] [Google Scholar]
- 22.Ancuta P, Rao R, Moses A, Mehle A, Shaw SK, Luscinskas FW, et al. Fractalkine preferentially mediates arrest and migration of CD16+ monocytes. J Exp Med 2003; 197(12):1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 2003; 19(1):71–82. [DOI] [PubMed] [Google Scholar]
- 24.Thieblemont N, Weiss L, Sadeghi HM, Estcourt C, Haeffner-Cavaillon N. CD14lowCD16high: a cytokine-producing monocyte subset which expands during human immunodeficiency virus infection. Eur J Immunol 1995; 25(12):3418–3424. [DOI] [PubMed] [Google Scholar]
- 25.Ancuta P, Kamat A, Kunstman KJ, Kim EY, Autissier P, Wurcel A, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS ONE 2008; 3(6):e2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ansari AW, Meyer-Olson D, Schmidt RE. Selective expansion of pro-inflammatory chemokine CCL2-loaded CD14+CD16+ monocytes subset in HIV-infected therapy naive individuals. Journal of clinical immunology 2013; 33(1):302–306. [DOI] [PubMed] [Google Scholar]
- 27.Dutertre CA, Amraoui S, DeRosa A, Jourdain JP, Vimeux L, Goguet M, et al. Pivotal role of M-DC8(+) monocytes from viremic HIV-infected patients in TNFalpha overproduction in response to microbial products. Blood 2012; 120(11):2259–2268. [DOI] [PubMed] [Google Scholar]
- 28.Moniuszko M, Liyanage NP, Doster MN, Parks RW, Grubczak K, Lipinska D, et al. Glucocorticoid treatment at moderate doses of SIVmac251-infected rhesus macaques decreases the frequency of circulating CD14+CD16++ monocytes but does not alter the tissue virus reservoir. AIDS Res Hum Retroviruses 2015; 31(1):115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaccari M, Fourati S, Gordon SN, Brown DR, Bissa M, Schifanella L, et al. HIV vaccine candidate activation of hypoxia and the inflammasome in CD14(+) monocytes is associated with a decreased risk of SIVmac251 acquisition. Nat Med 2018; 24(6):847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ancuta P, Autissier P, Wurcel A, Zaman T, Stone D, Gabuzda D. CD16+ Monocyte-Derived Macrophages Activate Resting T Cells for HIV Infection by Producing CCR3 and CCR4 Ligands. J Immunol 2006; 176(10):5760–5771. [DOI] [PubMed] [Google Scholar]
- 31.Ancuta P, Kunstman KJ, Autissier P, Zaman T, Stone D, Wolinsky SM, et al. CD16+ monocytes exposed to HIV promote highly efficient viral replication upon differentiation into macrophages and interaction with T cells. Virology 2006; 344(2):267–276. . [DOI] [PubMed] [Google Scholar]
- 32.Wacleche SW, Cattin A, Fonseca do Rosario N, Goulet JP, Gauchat D, Cleret-Buhot A, et al. CD16+ monocytes give rise to CD103+RALDH2+TCF4+ dendritic cells with unique transcriptional and immunological features. Blood Advances 2018; In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonza S, Maerz A, Deacon N, Meanger J, Mills J, Crowe S. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J Virol 1996; 70(6):3863–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triques K, Stevenson M. Characterization of restrictions to human immunodeficiency virus type 1 infection of monocytes. J Virol 2004; 78(10):5523–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaushik R, Zhu X, Stranska R, Wu Y, Stevenson M. A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe 2009; 6(1):68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011; 474(7353):654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 2012; 13(3):223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, et al. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol 2007; 178(10):6581–6589. [DOI] [PubMed] [Google Scholar]
- 39.Jaworowski A, Kamwendo DD, Ellery P, Sonza S, Mwapasa V, Tadesse E, et al. CD16+ monocyte subset preferentially harbors HIV-1 and is expanded in pregnant malawian women with plasmodium falciparum malaria and HIV-1 infection. J Infect Dis 2007; 196(1):38–42. [DOI] [PubMed] [Google Scholar]
- 40.Rich EA, Chen IS, Zack JA, Leonard ML, O’Brien WA. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1). J Clin Invest 1992; 89(1):176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science 1997; 276(5320):1857–1861. [DOI] [PubMed] [Google Scholar]
- 42.Fischer-Smith T, Croul S, Sverstiuk AE, Capini C, L’Heureux D, Regulier EG, et al. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. Journal of neurovirology 2001; 7(6):528–541. [DOI] [PubMed] [Google Scholar]
- 43.Bobardt MD, Saphire AC, Hung HC, Yu X, Van der Schueren B, Zhang Z, et al. Syndecan captures, protects, and transmits HIV to T lymphocytes. Immunity 2003; 18(1):27–39. [DOI] [PubMed] [Google Scholar]
- 44.Baxter AE, Russell RA, Duncan CJ, Moore MD, Willberg CB, Pablos JL, et al. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe 2014; 16(6):711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calantone N, Wu F, Klase Z, Deleage C, Perkins M, Matsuda K, et al. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity 2014; 41(3):493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiNapoli SR, Hirsch VM, Brenchley JM. Macrophages in Progressive Human Immunodeficiency Virus/Simian Immunodeficiency Virus Infections. J Virol 2016; 90(17):7596–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, et al. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 2017; 23(5):638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, et al. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed Z, Kawamura T, Shimada S, Piguet V. The role of human dendritic cells in HIV-1 infection. The Journal of investigative dermatology 2015; 135(5):1225–1233. [DOI] [PubMed] [Google Scholar]
- 50.Pena-Cruz V, Agosto LM, Akiyama H, Olson A, Moreau Y, Larrieux JR, et al. HIV-1 replicates and persists in vaginal epithelial dendritic cells. J Clin Invest 2018; 128(8):3439–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 2014; 14(6):392–404. [DOI] [PubMed] [Google Scholar]
- 52.Boulassel MR, Spurll G, Rouleau D, Tremblay C, Edwardes M, Sekaly RP, et al. Changes in immunological and virological parameters in HIV-1 infected subjects following leukapheresis. J Clin Apher 2003; 18(2):55–60. [DOI] [PubMed] [Google Scholar]
- 53.Gosselin A, Wiche Salinas TR, Planas D, Wacleche VS, Zhang Y, Fromentin R, et al. HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS 2017; 31(1):35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, et al. Peripheral blood CCR4+ CCR6+ and CXCR3+ CCR6+ CD4+ T cells are highly permissive to HIV-1 infection. J Immunol 2010; 184(3):1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, et al. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J Immunol 2011; 186(8):4618–4630. [DOI] [PubMed] [Google Scholar]
- 56.Roederer M Compensation in flow cytometry. Current protocols in cytometry / editorial board, Robinson J Paul, managing editor [et al. ] 2002; Chapter 1:Unit 1 14. [DOI] [PubMed] [Google Scholar]
- 57.Planas D, Zhang Y, Monteiro P, Goulet JP, Gosselin A, Grandvaux N, et al. HIV-1 selectively targets gut-homing CCR6+CD4+ T cells via mTOR-dependent mechanisms. JCI Insight 2017; 2(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wacleche VS, Goulet JP, Gosselin A, Monteiro P, Soudeyns H, Fromentin R, et al. New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology 2016; 13(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shacklett BL, Critchfield JW, Lemongello D. Isolating mucosal lymphocytes from biopsy tissue for cellular immunology assays. Methods in molecular biology 2009; 485:347–356. [DOI] [PubMed] [Google Scholar]
- 60.Ancuta P, Liu KY, Misra V, Wacleche VS, Gosselin A, Zhou X, et al. Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16- monocyte subsets. BMC genomics 2009; 10(1):403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017; 356(6335). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med 2004; 200(6):761–770. Epub 2004 Sep 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 2004; 200(6):749–759. Epub 2004 Sep 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macal M, Sankaran S, Chun TW, Reay E, Flamm J, Prindiville TJ, et al. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol 2008; 1(6):475–488. [DOI] [PubMed] [Google Scholar]
- 65.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol 2016; 14(1):55–60. [DOI] [PubMed] [Google Scholar]
- 66.Ancuta P, Weiss L, Haeffner-Cavaillon N. CD14+CD16++ cells derived in vitro from peripheral blood monocytes exhibit phenotypic and functional dendritic cell-like characteristics. Eur J Immunol 2000; 30(7):1872–1883. [DOI] [PubMed] [Google Scholar]
- 67.Dieu MC, Vanbervliet B, Vicari A, Bridon JM, Oldham E, Ait-Yahia S, et al. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J Exp Med 1998; 188(2):373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veazey RS, Lackner AA. Getting to the guts of HIV pathogenesis. J Exp Med 2004; 200(6):697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hume DA. The mononuclear phagocyte system. Curr Opin Immunol 2006; 18(1):49–53. Epub 2005 Dec 2009. [DOI] [PubMed] [Google Scholar]
- 70.Allers K, Fehr M, Conrad K, Epple HJ, Schurmann D, Geelhaar-Karsch A, et al. Macrophages accumulate in the gut mucosa of untreated HIV-infected patients. J Infect Dis 2014; 209(5):739–748. [DOI] [PubMed] [Google Scholar]
- 71.Shen R, Richter HE, Clements RH, Novak L, Huff K, Bimczok D, et al. Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. J Virol 2009; 83(7):3258–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 2017; 23(11):1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Telwatte S, Lee S, Somsouk M, Hatano H, Baker C, Kaiser P, et al. Gut and blood differ in constitutive blocks to HIV transcription, suggesting tissue-specific differences in the mechanisms that govern HIV latency. PLoS Pathog 2018; 14(11):e1007357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moysi E, Estes JD, Petrovas C. Novel Imaging Methods for Analysis of Tissue Resident Cells in HIV/SIV. Curr HIV/AIDS Rep 2016; 13(1):38–43. [DOI] [PubMed] [Google Scholar]
- 75.Rodrigues V, Ruffin N, San-Roman M, Benaroch P. Myeloid Cell Interaction with HIV: A Complex Relationship. Front Immunol 2017; 8:1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.