Abstract
The expression systems of the mitochondrial genes are derived from their bacterial ancestors, but have evolved many new features in their eukaryotic hosts. Mitochondrial RNA splicing is a complex process regulated by families of nucleus-encoded RNA-binding proteins, few of which have been characterized in maize (Zea mays L.). Here, we identified the Zea mays small kernel 3 (Zmsmk3) candidate gene, which encodes a mitochondrial transcription termination factor (mTERF) containing two mTERF motifs, which is conserved in monocotyledon; and the target introns were also quite conserved during evolution between monocotyledons and dicotyledons. The mutations of Zmsmk3 led to arrested embryo and endosperm development, resulting in small kernels. A transcriptome of 12 days after pollination endosperm analysis revealed that the starch biosynthetic pathway and the zein gene family were down-regulated in the Zmsmk3 mutant kernels. ZmSMK3 is localized in mitochondria. The reduced expression of ZmSmk3 in the mutant resulted in the splicing deficiency of mitochondrial nad4 intron1 and nad1 intron4, causing a reduction in complex I assembly and activity, impairing mitochondria structure and activating the alternative respiratory pathway. So, the results suggest that ZmSMK3 is required for the splicing of nad4 intron 1 and nad1 intron 4, complex I assembly and kernel development in maize.
Keywords: mitochondrial, embryo, endosperm, intron splicing, Zea mays
The expression systems of mitochondrial genes are derived from their prokaryotic ancestors, but they have evolved many new features in their eukaryotic hosts (Barkan 2011; Hammani and Barkan 2014). These new features are involved in RNA transcription, RNA editing, intron splicing, RNA maturation and RNA degradation, which are far more complex than those of their prokaryotic progenitors and regulated by a plethora of nucleus-encoded proteins. Although these families of nucleus-encoded proteins evolved independently, they seem to share a common structural organization, which is consisted of similar repeated helical motifs and can form the solenoid structures of chromatin (Hammani and Barkan 2014). These families include the pentatricopeptide repeat (PPR) proteins, the mitochondrial transcription termination factors (mTERF), the half-a-tetratricopeptide (HAT) proteins and the octotricopeptide repeat (OPR) proteins.
The mTERF proteins are widely distributed in metazoans and plants, and green alga, but apparently absent in fungi and prokaryotes (Roberti et al. 2009). The mTERF proteins were originally identified and characterized in the metazoans. In metazoans, There are four members of mTERF proteins (mTERF1- mTERF4), which are encoded by nuclear genes and consist of multiple mTERF motifs, each of which contains about 32 amino acids that form three X3LX3 leucine zipper-like elements (Zhao et al. 2014). All of mTERFs are targeted in mitochondria, and they could regulate transcription, translation and DNA replication of mitochondria genes in metazoans.(Chen et al. 2005; Roberti et al. 2009; Cámara et al. 2011). MOC1 (mterf-like gene of Chlamydomonas1) is the first mTERF characterized in a photosynthetic organism, green alga Chlamydomonas reinhardtii (Schönfeld et al. 2004). MOC1 is a mitochondrial protein, which binds specifically to a sequence within the mitochondrial ribosomal RNA (rRNA)-coding module S3 (Wobbe and Nixon 2013). The loss of MOC1 causes perturbed mitochondrial DNA (mtDNA) expression and sensitivity to high light.
In plants, there are more members of mTERFs, however, only a few mTERFs have been well functionally characterized, all of which encode chloroplast or mitochondrial proteins (Quesada 2016). BELAYA SMERT/RUGOSA2 (BSM/RUG2) is a dual-targeted mTERF protein that affects the expression levels in both the mitochondria and the chloroplasts. The bsm mutation causes arrested embryo development and a significant retardation in plant growth and development (Babiychuk et al. 2011). MTERF DEFECTIVE IN Arabidopsis1 (MDA1) functions in chloroplast development and abiotic stress responses; mda1 mutants display pale-green pigmentation and growth retardation, enhanced salt, osmotic stress tolerance, altered sugar responses, and reduced abscisic acid (ABA) sensitivity (Robles et al. 2012a; Robles et al. 2012b). Suppressor of hot1-4 1 (SHOT1) encodes a mitochondrial mTERF protein that suppresses the phenotype of the hot1-4 mutation in Arabidopsis thaliana; the loss of SHOT1 function reduces plant growth and enhances thermos tolerance in the absence of HSP101 (Kim et al. 2012). Singlet oxygen-linked death activator 10 (SOLDAT10) encodes a mitochondrial mTERF protein that suppresses the flu mutation; the soldat10 mutants decrease levels of plastid-specific rRNA and attenuate protein synthesis in plastids (Meskauskiene et al. 2009). TWR-1/MTERF9 is a chloroplast protein, and the loss of the TWR-1/MTERF9 results in defective chloroplast development, reduced mesophyll cell numbers, paleness, and stunted growth (Robles et al. 2015). Arabidopsis mTERF6 is localized in both chloroplasts and mitochondria, which is required for maturation of the chloroplast transfer RNAIle(GAU); the loss of mTERF6 perturbs plastid development and results in seedling lethality (Romani and Manavski 2015). Arabidopsis mTERF15 is a mitochondrial protein required for mitochondrial nad2 intron 3 splicing and complex I activity (Hsu et al. 2014). The null homozygous T-DNA mterf15 mutants display substantial retardation of both vegetative and reproductive development (Hsu et al. 2014). Zm-mTERF4 is the ortholog of the Arabidopsis protein BSM/RUG2. Zm-mTERF4 localizes to the chloroplast stroma and is required for group II intron splicing and the accumulation of plastid ribosomes in maize (Zea mays) chloroplasts (Hammani and Barkan 2014). The Zm-mterf4-1, the null mutant, displays an ivory leaf phenotype, while Zm-mterf4-2 mutant displays a pale yellow-green phenotype. These findings reveal that the plant mTERF genes are required for the regulation of gene expression in chloroplasts and mitochondria.
Here, we identified and characterized a candidate gene ZmSmk3, which is responsible for kernel development in maize. ZmSmk3 encodes an mTERF protein targeted to mitochondria, which contains two mTERF motifs conserved in monocotyledon. The Zmsmk3 mutation arrested the splicing of mitochondrial nad4 intron 1 and nad1 intron 4, reduced the assembly and activity of complex I, impaired mitochondrial structure, and increased the expression levels of the alternative oxidases AOX2 and AOX3, which resulted in small kernels. In summary, we present a new mitochondrial mTERF protein required for mitochondrial intron splicing, complex I assembly and kernel development in maize.
Materials and methods
Plant materials
The Zmsmk3 mutant was isolated from the UniformMu collection stocks, No. UFMu-06341 (McCarty et al. 2005). WT kernels in a segregating ear were used for the evaluation of the kernel phenotypes. For the genotype analysis and the co-segregation analysis, total DNA was extracted from the leaves of each individual using the modified CTAB method.
Cytological observation
Kernels at 6, 12, and 21 DAP were harvested from the selfed +/Zmsmk3 heterozygotes and fixed overnight in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA), dehydrated in an ethanol gradient series (30%, 50%, 70%, 85%, 95%, and 100% ethanol), and embedded in Paraplast Plus (Sigma-Aldrich). The samples were sectioned into 8–12 µm slices using a Leica RM2265 microtome (Leica Microsystems, Wetzlar, Germany) and stained with 0.5% toluidine blue O. Images were captured using a Leica MZFLIII microscope (Leica Microsystems).
For the TEM analysis, the 10-DAP endosperms of the WT and Zmsmk3 kernels were fixed, washed, dehydrated, embedded, and cut into ultrathin sections, as previously described (Ren et al. 2017). Ultra-thin sections were obtained using a Leica EM UC7 ultra microtome (Leica Microsystems). The sections were stained with uranyl acetate and subsequently with lead citrate, then imaged using a Tecnai G2 20 TWIN transmission electron microscope. These procedures were performed by Pei Zhang (Core Facility and Technical Support, Wuhan Institute of Virology, China).
RNA extraction and gene expression analysis
The developing tissues, including the root, stem, leaf, tassel, ear, silk, ovary, endosperm, and embryo, were collected to analyze the expression pattern of ZmSmk3. The total RNA of these tissues was extracted using the Ambion Pure Link Plant RNA Reagent (Thermo Fisher Scientific, Waltham, MA, USA), then reverse transcribed using M-MLV reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. RT-qPCR was performed for the expression pattern analysis of ZmSmk3 using the primer pairs ZmSmk3-qPCR-F and ZmSmk3-qPCR-R (primer sequences listed in Supplemental Table 2). The RT-qPCR was performed using the SYBR Select Master kit (Thermo Fisher Scientific), following the manufacturer’s instructions, with three biological replicates. The maize actin gene (ZmActin; GRMZM2G126010) was used as the internal control, and the relative expression levels were calculated using the comparative Ct method.
For the transcriptional analysis of the mitochondrial and AOX genes, the total RNAs were extracted from 12-DAP kernels of the WT and the Zmsmk3 mutant after the pericarp was removed. The RNAs were treated with RNase-free DNase I, then their abundances were normalized to both the total RNA level and the abundance of the ZmActin transcripts.
The primers used for the amplification of the mitochondrial genes, RT-qPCRs, and RT-PCRs in Figure 6 and Figure S4 were previously published by Xiu et al. (2016).
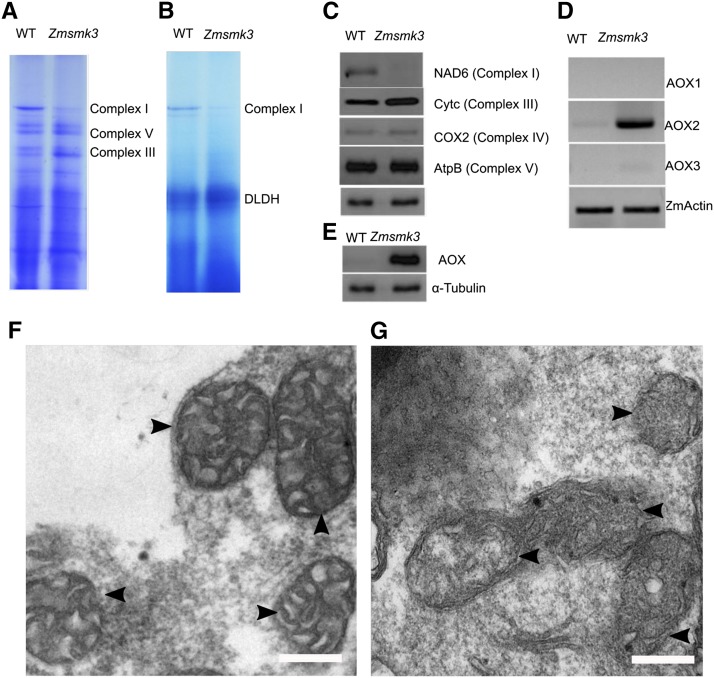
Figure 6.
The deficiency of complex I assembly and disordered mitochondrial structure in Zmsmk3. (A) Comparison of the accumulation of complex I in the wild type (WT) and Zmsmk3 mutant. Mitochondrial complexes of 12-DAP maize kernels without pericarps were separated using 3–12.5% blue native polyacrylamide gel electrophoresis (BN-PAGE). Complexes I, III, and IV are indicated. (B) Comparison of the NADH dehydrogenase activity of complex I in the WT and Zmsmk3. Darker staining indicates higher levels of activity. Dihydrolipoamide dehydrogenase (DLDH) was used as a loading control. (C) Comparison of the abundance of respiratory enzymes in the WT and Zmsmk3. Crude mitochondrial extracts from 12-DAP kernels with the pericarp removed were used for a western blot analysis using antibodies against Nad6 (subunit of complex I), Cytc (subunit of complex III), COX2 (subunit of complex IV), AtpB (subunit of complex V) and α-tubulin as a sample loading control. (D) RT-PCR analysis of AOX1, AOX2, and AOX3 expression in the WT and Zmsmk3 mutant. Total RNA was extracted from 12-DAP kernels with the pericarp removed. The expression level was normalized against ZmActin. (E) Western blot analysis using antibodies against AOX (alternative oxidase) and α-tubulin as a sample loading control. (F-G) Morphological analysis of mitochondria in wild-type (F) and Zmsmk3 (G) endosperms at 10-DAP. Mitochondria are indicated by the arrows. Scale bar = 0.5 µm.
RNA in situ hybridization
WT kernels at 12 DAP were used for the mRNA in situ hybridization assay. The kernels were fixed in 4% paraformaldehyde, following the procedures for cytological observation outlined above. The probes used to detect the ZmSmk3 transcripts corresponded to the full length of ZmSmk3 cDNA, and were constructed using the ZmSmk3-CDS-F/R primers. The T7 and Sp6 polymerases were used to produce sense and antisense RNA probes for in vitro transcription, which were then labeled with digoxigenin-UTP (Roche, Basel, Switzerland). Nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche) were used to detect the digoxigenin signal, following the manufacturer’s instructions. A Nikon Eclipse 80i differential interference contrast microscope (Nikon Instruments, Tokyo, Japan) was used to the capture images, following the methods described by Ren et al. (2017).
Subcellular localization of ZmSMK3
The full-length coding sequence of ZmSmk3 without the termination codon was amplified using the ZmSmk3-Kpn1-F and ZmSmk3-Xba1-R primers, which are listed in Supplemental Table 2. The PCR product was purified, verified by sequencing, then inserted into the pM999-GFP vector to generate a fusion construct. The construct was introduced into maize protoplasts derived from seedling leaves using a polyethylene glycol (PEG)/calcium-mediated transformation (Yoo et al. 2007). MitoTracker Red (Thermo Fisher Scientific) was used as the mitochondrion marker.
Mitochondrial complex activity assay and western blotting assay
Crude and intact mitochondria were isolated from maize embryos and endosperms at 15-DAP, as described by Ren et al. (2017). The mitochondrial suspension was used for a BN-PAGE using a Native PAGE sample prep kit (Thermo Fisher Scientific). The in-gel complex I activity assay was performed as described previously (Meyer et al. 2009). Gel strips were loaded with extracts from 150 µg maize mitochondria.
Crude mitochondrial extracts from 15-DAP kernels with the pericarp removed were used for the western blot analyses using antibodies (from Agrisera, Vännäs, Sweden) against Nad6 (subunit of complex I), Cytc (subunit of complex III), COX2 (subunit of complex IV), AtpB (subunit of complex V), AOX (alternative oxidase), and α-tubulin as a loading control.
RNA-Seq analysis
Total RNA was extracted from three biological samples of 15-DAP WT and Zmsmk3 seeds with the pericarp removed. Library construction was performed according to the standard instructions provided by Illumina (San Diego, CA, USA). FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic (Bolger et al. 2014) were used to obtain clean reads. The RNA-Seq data were analyzed using TopHat and Cufflinks (Ghosh and Chan 2016) and normalized as fragments per kilobase of transcript per million mapped reads (FPKM). A hypergeometric distribution was used for the statistical analysis.
Data availability
There are two files (one excel file and one word file) in the Supplemental Material, File S1 and File S2. File S1 contains two supplementary tables. Table S1 includes Gene ontology classifications of DEGs with functional annotation. Table S2 shows Primers used in this study. File S2 contains four supplementary figures. Figure S1 showed aborted development in Zmsmk3 mutants. Figure S2 showed identification of ZmSmk3 by TAIL-PCR. Figure S3 showed phylogenetic analysis of ZmSMK3. Figure S4 showed RT-PCR analysis of mitochondrial tRNA and rRNA encoded genes transcriptional levels in the wild type and the Zmsmk3 mutant. Supplemental material available at FigShare: https://doi.org/10.6084/m9.figshare.8217473.
Results
Embryo and endosperm development are arrested in Zmsmk3
The Zmsmk3 is a small kernel (smk) mutant identified from the Uniform Mu population of W22 plants mutated with Mutator transposons, stock No. UFMu-06341 (McCarty et al. 2005). The self-pollinated ZmSmk3 heterozygotes segregated into normal and small kernels at a ratio of approximately 3:1 (1651:526, χ2-test, P > 0.05; Figure 1A), indicating that Zmsmk3 is a monogenic recessive nuclear mutant allele. Compared with the kernels of wild type (WT), the tops of the mutant kernels were shrunken and the starchy endosperm did not fill the pericarp (Figure 1B and 1C). The 100-kernel weight of the mutant was about 1/4 of that of the WT (Figure 1D). During sectioning of the mature kernels, some of the mutant embryos only had a root apical meristems (RAM), (Figure S1A and S1B), some embryos were even necrotic (Figure S1C), and only part of embryos were well-structured with smaller size. (Figure 1C). The germination rate and a seedling survival rate of Zmsmk3 were sharply decreased. About 75% of Zmsmk3 could germinate, however, some of which developed into abnormal seedlings (Figure S1E), resulting in only 30% well-structured seedlings (Figure S1D), which showed much slower growth than the WT (Figure S1F). These results indicated that the mutation of ZmSmk3 affected both kernel and plant development.
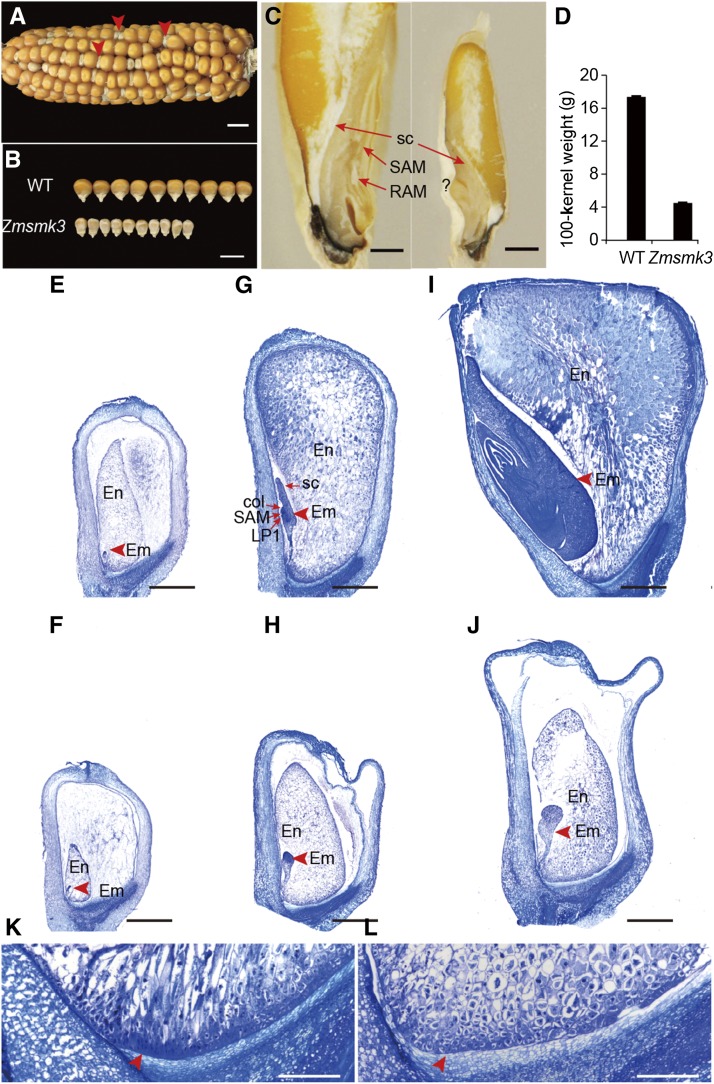
Figure 1.
The Zmsmk3 mutants are arrested in embryogenesis and endosperm development. (A) A mature segregating ear of maize. Arrows indicate the Zmsmk3 mutant kernels. (B) A size comparison between the wild-type (WT) and Zmsmk3 mutant kernels. (C) Dissection of mature WT (left) and Zmsmk3 (right) kernels. RAM, root apical meristem; SAM, shoot apical meristem; sc, scutellum, (D) Comparison of WT and Zmsmk3 kernel weights. Sections of developing kernels at 6 days after pollination (DAP; E-F), 12 DAP (G-H), and 21 DAP (I-J), and the basal endosperm transfer layer (BETL) at 21 DAP (K-L). (E, G, I, K) Wild-type (WT) kernels, (F, H, J, L) Zmsmk3 kernels. col, coleoptile; em, embryo; en, endosperm; LP1, first leaf primordium; SAM, shoot apical meristem; sc, scutellum. Arrows in K and L indicate the BETL. Values are the means with SE; n = 3 individuals. Scale bar = 1 cm in (A, B), 1 mm in (C, E, F, G, H, I, J) and 0.2 mm in (K–L).
To assess the impact of the Zmsmk3 mutation on the development of the embryo and endosperm, we examined different stages after pollination of the mutant and WT kernels in the same segregating ears using paraffin sectioning and microscopy. At six days after pollination (DAP), the size of Zmsmk3 kernels were similar to those of the WT; however, the embryos and endosperms were smaller in Zmsmk3 kernels (Figure 1E, F). At 12 DAP, the WT embryos had progressed to the late embryogenesis stage and had differentiated the first leaf primordium, coleoptile and SAM, and the starchy endosperm had filled the pericarp (Figure 1G). However, in the 12-DAP Zmsmk3 kernels, the embryos remained at the transition stage and there was an obvious gap between the endosperm and the pericarp, which persisted in the later stages (Figure 1H). At 21 DAP, the WT kernels had developed a mature embryo with a scutellum, four leaf primordia, and a starch-filled endosperm (Figure 1I), while the development of the Zmsmk3 kernels remained arrested (Figure 1J). We further compared the differentiation of the basal endosperm transfer layer (BETL) in the WT and Zmsmk3 endosperm at 21 DAP. In the WT, there were cell wall ingrowths in the BETL cell layers, which could increase the surface area of the BETL plasma membrane (Figure 1K). In Zmsmk3, the BETL cell structure was diminished, which might impede the nutrient transport from the maternal placenta to the developing endosperm (Figure 1L). These results indicated that both embryogenesis and endosperm development were severely arrested in Zmsmk3 mutant.
The deposition of storage substances during seed filling is defective in Zmsmk3
We analyzed the starch deposition in the endosperm of the WT and Zmsmk3 kernels. At 12 DAP, there were many starch granules in the WT endosperm (Figure 2A) but fewer in Zmsmk3 mutants (Figure 2B). At 25 DAP, the starch granules in the outer region of the WT endosperm were tightly embedded in a proteinaceous matrix, and endosperm cell structure had already disappeared (Figure 2C); however, in the mutant, the protein matrix was diminished and the endosperm cell structure was still retained (Figure 2D).
Figure 2.
Storage substances is reduced in the Zmsmk3 kernels. Starch accumulation in (A, B) 12 DAP kernels, (C, D) 25-DAP kernels, (A, C) wild-type kernels, and (B, D) Zmsmk3 kernels. Scale bar = 50 µm. (E) GO analysis of differentially expressed genes (DEGs) between the wild-type and Zmsmk3 kernels.
To better understand the impacts of Zmsmk3, we removed the pericarps from WT and Zmsmk3 kernels harvested from the same segregating ear at 12-DAP and performed a transcriptome analysis using RNA-seq. Among the 44,466 genes expressed in both the WT and Zmsmk3 kernels, we identified 5,113 significantly differentially expressed genes (DEGs) between the two genotypes using a threshold fold change > 2.0 and P < 0.05. Of these, 3,521 genes were upregulated and 1,592 were downregulated in Zmsmk3. The gene ontology (GO) classifications revealed that the DEGs were largely associated with storage filling and energy metabolism (Figure 2E). DEGs related to starch biosynthesis (GO: 00019252, P = 3.47E-07), nutrient reservoir activity (GO: 0045735, P = 2.05E-05), and amyloplast function (GO: 0009501, P = 8.80E-05) were extensively downregulated (Supplemental Table 1). The DEGs related to fructose 6-phosphate metabolism (GO: 0006002, P = 6.73E-06) and carbohydrate phosphorylation (GO: 0046835, P = 9.77E-05) were generally upregulated. These results indicated that the pathways of storage product deposition and energy harvesting were greatly affected in Zmsmk3.
Identification of the candidate gene for Zmsmk3 by TAIL-PCR
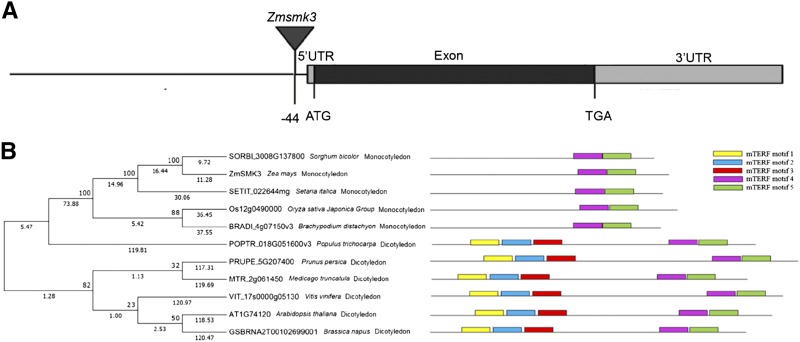
To identify the causal gene of the Zmsmk3 mutant, Genomic DNA extracted from leaves of Zmsmk3/+ and +/+ individuals, whose genotype had been ascertained by selfing, was used for modified thermal asymmetric interlaced PCR (TAIL-PCR) (Settles et al. 2004; Zhang et al. 2013). For this procedure, three arbitrary primers combined with specific Mu-flanking primers were used to amplify larger flanking sequences from Mu element insertions, and the specific DNA binds (Figure S2A-C) for Zmsmk3 were identified and sequenced. The results showed that the Mu element was inserted into the promoter of gene GRMZM2G177019, 44 bp upstream of the predicted translation start site (ATG) in Zmsmk3 mutants. Co-segregation analysis was performed on a total of 500 F2 individuals from the selfing heterozygotes, and the results showed that the Mu element insertion was tightly linked to Zmsmk3 without any recombination (Figure S2D). So GRMZM2G177019 is the key candidate gene for Zmsmk3. GRMZM2G177019 (ZmSmk3) is an intron-free gene encoding an mTERF protein (Figure 3A). Phylogenetic analysis was conducted with 143 homologous proteins of ZmSMK3 from six species (Oryza sativa, Zea mays, Arabidopsis, Sorghum bicolor, Setaria italic, and Brachypodium distachyon), and the results showed that ZmSMK3 was highly homologous to mTERF15 (AT1G74120) (Figure S3), which is required for the splicing of nad2 intron 3 in Arabidopsis thaliana (Hsu et al. 2014). However, when the species for phylogenetic analysis was enlarged to eleven (Figure 3B), including both monocotyledons and dicotyledons, two branches were found in the phylogenetic tree for monocotyledons and dicotyledons respectively. Interestingly, by aligning the homologous proteins of the eleven species, we found that there were five mTERF motifs (http://smart.embl-heidelberg.de/) in the dicotyledonous species; but only two mTERF motifs are included in the monocotyledonous species (Figure 3B).
Figure 3.
Identification of the candidate gene for Zmsmk3 . (A) Gene structure of the candidate gene for Zmsmk3. The location of the insertion in the candidate gene Zmsmk3 is indicated by the triangle. (B) Phylogenetic analysis and mTERF motifs alignment of candidate gene ZmSMK3 with its orthologs. (Left) Phylogenetic analysis of the candidate gene ZmSMK3. ZmSMK3 and identified homologous proteins in Sorghum bicolor, Setaria italica, Oryza sativa Japonica Group, Brachypodium distachyon, Populus trichocarpa, Prunus persica, Medicago truncatula, Vitis vinifera, Arabidopsis and Brassica napus were aligned by ClustalW. The phylogenetic tree was constructed using MEGA7 by Neighbor-joining method. The numbers at the nodes represent the percentage of 1000 bootstraps. The numbers at the lines represent the branch lengths. (Right) The alignment of the ZmSMK3 protein with its orthologs. mTERF motifs are indicated with different colors.
ZmSmk3 is constitutively expressed and ZmSmk3 protein is targeted to mitochondria
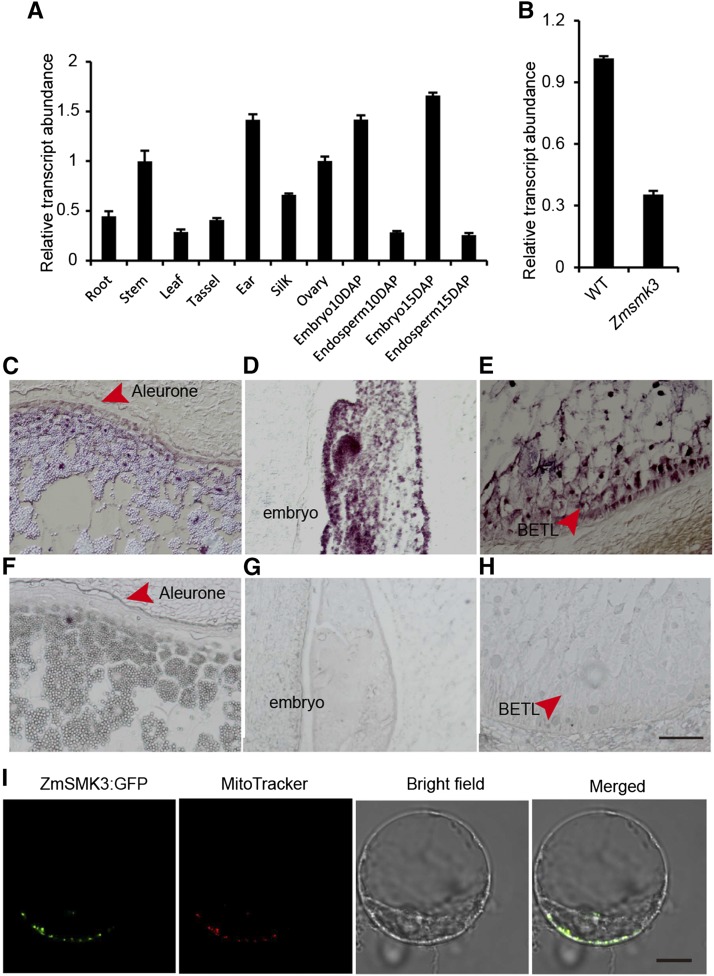
Quantitative real-time PCR (RT-qPCR) was performed to identify the expression pattern of ZmSmk3. As a result, ZmSmk3 was ubiquitously expressed in all tested tissues, with higher expression levels in the stem, ear, ovary, and embryo and lower levels in the root, leaf, tassel, silk, and endosperm (Figure 4A). We next genotyped developing seeds with the pericarp removed. Homozygous Zmsmk3 seeds at 12-DAP with the pericarp removed were used to analyze expression of ZmSmk3 and mitochondrial genes. Transcript levels were reduced in the Zmsmk3 homozygous mutants (Figure 4B). To further characterize ZmSmk3 spatial expression of the developing kernels, an mRNA in situ hybridization was performed in the kernels at 12 DAP. The hybridization signal was detected in the aleurone layer (Figure 4C), the BETL of the endosperm (Figure 4D), and the SAM of the embryo (Figure 4E), with particularly high expression levels in the BETL and SAM. Hybridization to the sense RNA probe did not detect any significant signals (Figure 4F-H). These results suggested that ZmSmk3 was ubiquitously expressed in the developing kernels, but may play an important role in the BETL and SAM of the developing kernels.
Figure 4.
The expression of ZmSmk3 (A) Expression profiles of ZmSmk3 in various tissues. (B) Comparison of ZmSmk3 expression in the wild-type (WT) and Zmsmk3 mutant kernels. RNA was extracted from kernels at 12 days after pollination (DAP) following the removal of the pericarp. For each RNA sample in (A) and (B), three technical replicates were performed. Values are means with SE; n = 3 individuals. (C–H) mRNA in situ hybridization analysis of ZmSmk3 in 12-DAP WT kernels using an antisense probe (C–E) and a sense probe (F–H). (I) Subcellular localization of ZmSMK3. The ZmSMK3:GFP fusion protein was transiently expressed in maize leaf protoplasts. Fluorescent signals from ZmSMK3:GFP are displayed in green and mitochondria stained with MitoTracker are red. Scale bar = 100 µm in (C–H) and 10 µm in (I).
To determine the localization of ZmSMK3, the full-length coding region of the gene was fused to GFP and transiently expressed in maize protoplasts. The GFP signals were detected in small dots that were identified as mitochondria by observing the red fluorescence of the MitoTracker Red dye (Figure 4I). This result indicated that ZmSMK3 was targeted to the mitochondria.
Zmsmk3 exhibits deficiency in the splicing of mitochondrial nad4 intron 1 and nad1 intron 4
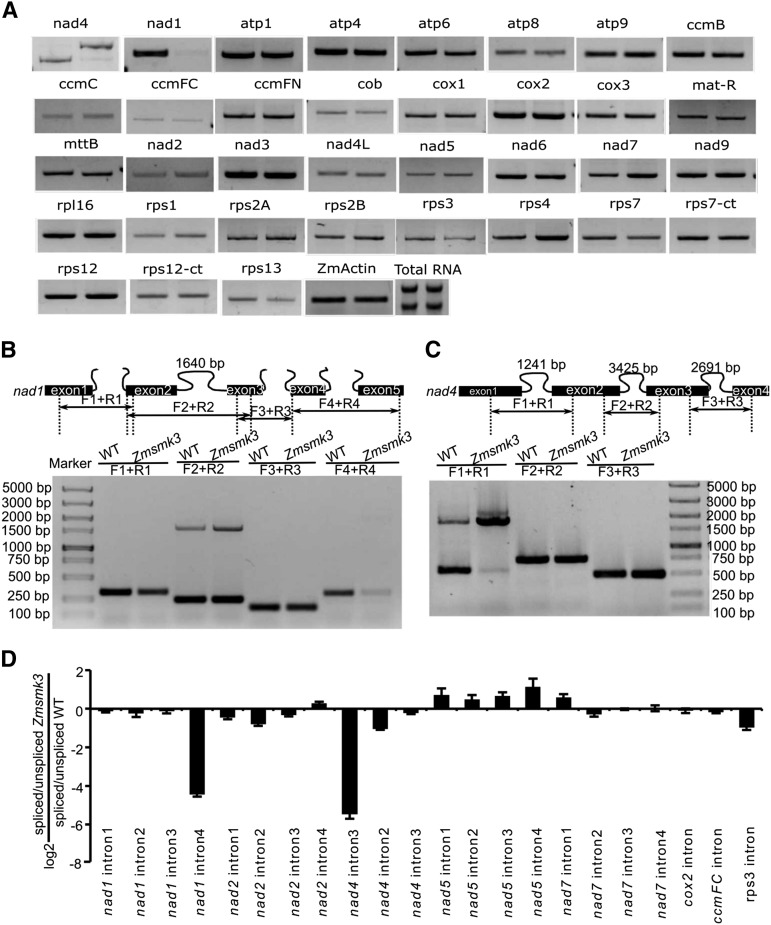
Because ZmSMK3 appeared to target to mitochondria, we investigated whether the Zmsmk3 mutation had any effect on gene expression in this organelle. We analyzed the transcript levels of the maize NB mitochondrial genes in wild-type and Zmsmk3 mutants by RT-PCR. Total RNAs were extracted from 12 DAP kernels of wild-type and Zmsmk3 from the same segregating ear with the pericarp removed. Gene-specific primers were used as Xiu described (2016) to amplify transcripts of mitochondrial genes from the cDNA template. The results showed that the expression level of most mitochondrial genes was not significantly different between wild-type and the Zmsmk3 mutant and some tRNAs were a little higher in Zmsmk3, while the precursor transcript levels of nad4 were high, and the abundance of its mature transcript was dramatically reduced, and the mature nad1 transcripts were also dramatically reduced (Figure 5A and Supplementary Fig. S4).
Figure 5.
The expression of nad1 and nad4 is dramatically decreased due to the Splicing deficiency of mitochondrial nad4 intron 1 and nad1 intron 4. (A) RT-PCR analysis of 35 mitochondria-encoded transcripts in WT (left) and Zmsmk3 (right) seeds. (B) Schematic structure of the maize mitochondrial nad1 (up) and the splicing efficiency of the four introns by RT-PCR (down). (C) Schematic structure of the maize mitochondrial nad4 (up) and the splicing efficiency of the three introns by RT-PCR (down). (D) Quantitative RT-PCR analysis of 22 intron-containing mitochondrial transcripts. The RNA was isolated from the same ear segregating for WT and Zmsmk3 seeds at 12 DAP. ZmActin (GRMZM2G126010) was served as internal control.
Primers spanning the exons adjacent to the introns of the mature nad1 and nad4 transcripts were further used to examine the intron-splicing efficiency in the Zmsmk3 and its sibling WT kernels. The results revealed that the splicing efficiency of nad1 intron 4 and nad4 intron 1 was decreased in the Zmsmk3 mutant (Figure 5B-C). Furthermore, we examined the ratio of spliced to un-spliced transcripts for 22 of mitochondrial intron in Zmsmk3 and WT using quantitative RT‐PCR (qRT‐PCR). Of the 22 introns examined, splicing efficiency of the nad1 intron 4 and nad4 intron 1 was dramatically decreased in Zmsmk3 compared with that in WT (Figure 5D), but splicing efficiencies of the other introns were not significantly affected in Zmsmk3 (Figure 5D). These results demonstrate that ZmSmk3 is required for the splicing of nad1 intron 4 and nad4 intron 1.
Zmsmk3 exhibits deficiency of mitochondrial complex I assembly and impaired mitochondrial function
To further investigate the assembly of respiratory complexes, mitochondrial proteins isolated from 12 DAP Zmsmk3 and WT endosperm were analyzed by blue native polyacrylamide gel electrophoresis (BN-PAGE). The results showed that Zmsmk3 mutants had a reduced accumulation of complex I (Figure 6A) and a significant reduction in complex I activity (Figure 6B), while the complex III was slightly increased, which might be as a complementary of the deficiency of complex I. Furthermore, we determined the steady‐state level of some subunits by western blotting. Surprisingly, a great difference between WT and Zmsmk3 was revealed using an antibody against Nad6, a subunit of complex I. The accumulation of complexes IV and V subunits (Cox2 and AtpB) were insignificantly different between Zmsmk3 and WT, whereas the accumulation of complexes III subunits (Cytc) was a little higher in Zmsmk3 mitochondria than that in WT (Figure 6C). So the results showed that deficiency of splicing of nad1 and nad4 resulted in disordered assembly and activity of complex I.
We further investigated the morphology of the mitochondria in the endosperm at 10 DAP using transmission electron microscopy. It was discovered that the WT mitochondria showed normally folded cristae structures with distinct inner spaces (Figure 6F), whereas the cristae structure of Zmsmk3 mitochondria was irregular (Figure 6G). Another alternative oxidation pathway was also analyzed by examining the expression of the three alternative oxidase (AOX) genes, AOX1, AOX2 and AOX3. The expression of AOX2 was much higher in the Zmsmk3 mutants than that in the WT, and the expression levels of AOX1 and AOX3 were slightly increased (Figure 6D). Specific antibodies against the alternative oxidation proteins were used for western blotting. As a result, there was a dramatic increase in the levels of AOX proteins in Zmsmk3 mutant (Figure 6E). These results indicated that the deficiency of ETC could cause a dramatic increase of the expression levels of AOX genes.
Discussion
ZmSmk3 is required for kernel development and seedling growth in maize
Here, we identified a candidate gene for Zmsmk3 in maize, which encodes an mTERF protein required for the development of maize kernels and seedling growth. The kernels are composed of endosperm, embryo and pericarp. Endosperm is the main storage organ for nutrients, so the arrested endosperm development in Zmsmk3 resulted in small kernels with reduced weights (Figure 1 and Figure S1). The differentiated endosperm is divided into four major cell types, namely the cells of the embryo surrounding region, the BETL, the aleurone layer, and the starchy endosperm (Olsen 2001). BETL is crucial to transport nutrients from maternal placenta to the endosperm in the development of kernels, and the absence of a properly formed BETL is correlated with reduced rates of grain filling and increased levels of seed abortion (Cai et al. 2017; Ren et al. 2017). Here, ZmSmk3 was found to be highly expressed in the BETL of the endosperm (Figure 4e), suggesting that ZmSmk3 is important for BETL development. Consistent with this finding, BETL differentiation was diminished in the Zmsmk3 mutants (Figure 1), resulting in arrested nutrients enrichment in the Zmsmk3 endosperm (Figure 2). This discovery was further supported by the down regulated expression of starch biosynthesis genes and reduced starch granules in the 12DAP endosperm of Zmsmk3 mutants (Figure 2, Supplemental Table 1). The development of embryo proceeds in three stages in maize, namely the transition, coleoptilar, and late embryogenesis stages (Olsen 2001). In this study, we found that ZmSmk3 was highly expressed in the SAM of embryos (Figure 4). The loss of ZmSmk3 function caused the early halting of embryo development at the transition stage, resulting in an inflated transition stage embryo (Figure 1). However, part of the embryos of Zmsmk3 mutant could germinate and grow into intact seedlings (Figure 1 and Figure S1) with lower germination rates and seedling survival rates, and the survived seedlings grew much slowly (Figure S1). These findings suggest that ZmSmk3 is not only crucial for kernel development but also for the development of the seedlings. However, to finally validate the function of ZmSmk3, complementation and allelism tests would be adopted in the future research.
ZmSmk3 affects complex I assembly by modulating the splicing of mitochondrial nad4 intron1 and nad1 intron4 in maize
NAD1 and NAD4 are the core components of complex I, which is located on the mitochondrial membrane (Klodmann et al. 2010; Meyer et al. 2011). There are three trans-introns and 0ne cis-intron in nad1 and three cis-introns in nad4, and the proper splicing of them is crucial for their functions. Some PPR proteins have been reported to function as splicing factors of nad1 and nad4. In Zea mays, the defective kernel 2 (dek2) mutation reduced the splicing efficiency of mitochondrial nad1 intron 1 and severely impeded complex I assembly (Qi et al. 2017). Dek35 and Emp11 encode PPR proteins that affected the cis-splicing of mitochondrial nad4 intron 1 and nad1 introns, respectively, which both influenced seed development in maize (Chen et al. 2017; Ren et al. 2017). In Zmsmk3 mutants, the nad1 intron4 and nad4 intron1 could not be spliced properly, resulting in the decrease of mature transcripts of nad1 and nad4 (Figure 5A-C), while other mitochondrial introns in Zmsmk3 were correctly spliced. So the results suggested that a single Zmsmk3 mutation specifically affected the splicing of nad1 intron4 and nad4 intron1. RNA metabolism in plant mitochondria is a complex process that combines bacteria-like traits with novel features that evolved in the host cell, and is consequently regulated by families of nucleus-encoded factors (Hammani and Giege 2014). Various factors are required for the splicing and processing of mitochondrial introns in plants, including the PPRs (Barkan and Small 2014), mTERFs (Hammani and Barkan 2014), the regulators of chromosome condensation (Kühn et al. 2011), plant organellar RNA recognition proteins (Kroeger et al. 2009; Francs-Small et al. 2012), and chloroplast RNA splicing and ribosome maturation proteins (Zmudjak et al. 2013). To interpret the non-specificity of ZmSmk3 to nad4 intron1 and nad1 intron 4, we assume that ZmSmk3 may play a role in the recognition of precursor nad4 and nad1 mRNA and in the maintenance of the nad4 and nad1 conformation for intron splicing by cooperating with other factors. Consistent with previously reported phenotypes of the complex I-deficient mutants nuclear maturase 1 (nmat1) (Keren et al. 2012), nmat2 (Keren et al. 2009), and nmat4 (Cohen et al. 2014), in Zmsmk3, the defect mature transcripts of nad1 and nad4 affected the assembly and activity of complex I (Figure 6A-C), limiting ATP production. The loss of complex I function results in a defective ETC, which affects the respiratory metabolism and the inner mitochondrial structure (Qi et al. 2017). When the ETC is defective, alternative respiratory enzymes, the AOXs, are activated (Sabar et al. 2000). Plants with partial mitochondrial dysfunction can increase AOX expression to increase their tolerance of energy stress (Vanlerberghe et al. 2016). Similarly, our results found that the mitochondrial complex I was reduced in the mutant, and the expression of AOXs was dramatically increased in the Zmsmk3 mutant (Figure 6D-E). Though it is known that ZmSMK3 functions in the splicing of nad1 and nad4, its remains to be seen whether this factor have additional roles in organellar RNA metabolism.
ZmSMK3 has the structural and functional divergence with its ortholog mTEFRF15 in Arabidopsis
According to the results, ZmSmk3 is involved in the splicing of nad1 intron4 and nad4 intron1. However, in Arabidopsis, mTERF15, the ortholog of ZmSMK3, is required for mitochondrial nad2 intron 3 splicing and complex I activity (Hsu et al. 2014). The structure comparison of the two proteins showed that there are 2 mTERF motifs in maize ZmSMK3, while there are 5 mTERF motifs in Arabidopsis mTERF15 (Figure 3B). More interestingly, the alignment of ZmSMK3 orthologs among 11 species showed that the homologous of ZmSMK3 in the monocotyledons was distinctly different from the homologous in dicotyledons in the number of mTERF motifs, which is exactly identical with the comparison between ZmSMK3 and mTERF15 (Figure 3B). However, the target introns in Zea mays, including nad1 intron4, nad2 intron3 and nad4 intron1, showed considerable similarity to Arabidopsis (data not shown), suggesting that the introns were quite conserved during evolution between the two species. According to the results, the ZmSMK3 and mTERF15 could bind to different introns. Therefore, on one hand, the function of the mTERF proteins is conservative as both ZmSMK3 and AtmTERF15 were participate in intron splicing, on the other hand, the variation of their structure resulted in divergence in target intron splicing.
Acknowledgments
We are grateful to Pei Zhang from The Core Facility and Technical Support, Wuhan Institute of Virology, for her help with producing TEM micrographs. This work was supported by the National Natural Science Foundation of China (91735306) and the National Key Research and Development Program of China (2016YFD0100303), received by FQ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.6084/m9.figshare.8217473.
Communicating editor: M. Hufford
Literature Cited
- Babiychuk E., Vandepoele K., Wissing J., Garcia-Diaz M., De Rycke R. et al. , 2011. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. USA 108: 6674–6679. 10.1073/pnas.1103442108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., 2011. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532. 10.1104/pp.110.171231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., and Small I., 2014. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442. 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Li S., Sun F., Sun Q., Zhao H. et al. , 2017. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J. 91: 132–144. 10.1111/tpj.13551 [DOI] [PubMed] [Google Scholar]
- Cámara Y., Asin-Cayuela J., Park C. B., Metodiev M. D., Shi Y. et al. , 2011. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 13: 527–539. 10.1016/j.cmet.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Chen X., Feng F., Qi W., Xu L., Yao D. et al. , 2017. Dek35 Encodes a PPR Protein that Affects cis-Splicing of Mitochondrial nad4 Intron 1 and Seed Development in Maize. Mol. Plant 10: 427–441. 10.1016/j.molp.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhou G., Yu M., He Y., Tang W. et al. , 2005. Cloning and functional analysis of human mTERFL encoding a novel mitochondrial transcription termination factor-like protein. Biochem. Biophys. Res. Commun. 337: 1112–1118. 10.1016/j.bbrc.2005.09.164 [DOI] [PubMed] [Google Scholar]
- Cohen S., Zmudjak M., Colas des Francs-Small C., Malik S., Shaya F. et al. , 2014. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78: 253–268. 10.1111/tpj.12466 [DOI] [PubMed] [Google Scholar]
- Francs-Small C. C., Kroeger T., Zmudjak M., Ostersetzer-Biran O., Rahimi N. et al. , 2012. A PORR domain protein required for rpl2 and ccmF(C) intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 69: 996–1005. 10.1111/j.1365-313X.2011.04849.x [DOI] [PubMed] [Google Scholar]
- Ghosh S., and Chan C. K., 2016. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 1374: 339–361. 10.1007/978-1-4939-3167-5_18 [DOI] [PubMed] [Google Scholar]
- Hammani K., and Barkan A., 2014. An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res. 42: 5033–5042. 10.1093/nar/gku112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., and Giege P., 2014. RNA metabolism in plant mitochondria. Trends Plant Sci. 19: 380–389. 10.1016/j.tplants.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Hsu Y. W., Wang H. J., Hsieh M. H., Hsieh H. L., and Jauh G. Y., 2014. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS One 9: e112360 10.1371/journal.pone.0112360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I., Bezawork-Geleta A., Kolton M., Maayan I., Belausov E. et al. , 2009. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15: 2299–2311. 10.1261/rna.1776409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I., Tal L., des Francs-Small C. C., Araujo W. L., Shevtsov S. et al. , 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 71: 413–426. [DOI] [PubMed] [Google Scholar]
- Kim M., Lee U., Small I., des Francs-Small C. C., and Vierling E., 2012. Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell 24: 3349–3365. 10.1105/tpc.112.101006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J., Sunderhaus S., Nimtz M., Jansch L., and Braun H. P., 2010. Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810. 10.1105/tpc.109.073726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger T. S., Watkins K. P., Friso G., van Wijk K. J., and Barkan A., 2009. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 106: 4537–4542. 10.1073/pnas.0812503106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K., Carrie C., Giraud E., Wang Y., Meyer E. H. et al. , 2011. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J. 67: 1067–1080. 10.1111/j.1365-313X.2011.04658.x [DOI] [PubMed] [Google Scholar]
- McCarty D. R., Settles A. M., Suzuki M., Tan B. C., Latshaw S. et al. , 2005. Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. 10.1111/j.1365-313X.2005.02509.x [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Wursch M., Laloi C., Vidi P. A., Coll N. S. et al. , 2009. A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses (1)O(2)-induced cell death. Plant J. 60: 399–410. 10.1111/j.1365-313X.2009.03965.x [DOI] [PubMed] [Google Scholar]
- Meyer E. H., Solheim C., Tanz S. K., Bonnard G., and Millar A. H., 2011. Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 286: 26081–26092. 10.1074/jbc.M110.209601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E. H., Tomaz T., Carroll A. J., Estavillo G., Delannoy E. et al. , 2009. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 151: 603–619. 10.1104/pp.109.141770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O. A., 2001. ENDOSPERM DEVELOPMENT: Cellularization and Cell Fate Specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 233–267. 10.1146/annurev.arplant.52.1.233 [DOI] [PubMed] [Google Scholar]
- Qi W., Yang Y., Feng X., Zhang M., and Song R., 2017. Mitochondrial Function and Maize Kernel Development Requires Dek2, a Pentatricopeptide Repeat Protein Involved in nad1 mRNA Splicing. Genetics 205: 239–249. 10.1534/genetics.116.196105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., 2016. The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol. Plant. 157: 389–399. 10.1111/ppl.12416 [DOI] [PubMed] [Google Scholar]
- Ren X., Pan Z., Zhao H., Zhao J., Cai M. et al. , 2017. EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J. Exp. Bot. 68: 4571–4581. 10.1093/jxb/erx212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti M., Polosa P. L., Bruni F., Manzari C., Deceglie S. et al. , 2009. The MTERF family proteins: mitochondrial transcription regulators and beyond. Biochim. Biophys. Acta 1787: 303–311. 10.1016/j.bbabio.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Robles P., Micol J. L., and Quesada V., 2012a Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS One 7: e42924 10.1371/journal.pone.0042924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles P., Micol J. L., and Quesada V., 2012b Unveiling plant mTERF functions. Mol. Plant 5: 294–296. 10.1093/mp/sss016 [DOI] [PubMed] [Google Scholar]
- Robles P., Micol J. L., and Quesada V., 2015. Mutations in the plant-conserved MTERF9 alter chloroplast gene expression, development and tolerance to abiotic stress in Arabidopsis thaliana. Physiol. Plant. 154: 297–313. 10.1111/ppl.12307 [DOI] [PubMed] [Google Scholar]
- Romani I., and Manavski N., 2015. A Member of the Arabidopsis Mitochondrial Transcription Termination Factor Family Is Required for Maturation of Chloroplast Transfer RNAIle(GAU). 169: 627–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M., De Paepe R., and de Kouchkovsky Y., 2000. Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 124: 1239–1250. 10.1104/pp.124.3.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfeld C., Wobbe L., Borgstädt R., Kienast A., Nixon P. J. et al. , 2004. The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J. Biol. Chem. 279: 50366–50374. 10.1074/jbc.M408477200 [DOI] [PubMed] [Google Scholar]
- Settles A. M., Latshaw S., and McCarty D. R., 2004. Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res. 32: e54 10.1093/nar/gnh052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G. C., Martyn G. D., and Dahal K., 2016. Alternative oxidase: a respiratory electron transport chain pathway essential for maintaining photosynthetic performance during drought stress. Physiol. Plant. 157: 322–337. 10.1111/ppl.12451 [DOI] [PubMed] [Google Scholar]
- Wobbe L., and Nixon P. J., 2013. The mTERF protein MOC1 terminates mitochondrial DNA transcription in the unicellular green alga Chlamydomonas reinhardtii. Nucleic Acids Res. 41: 6553–6567. 10.1093/nar/gkt313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu Z., Sun F., Shen Y., Zhang X., Jiang R. et al. , 2016. EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J 85: 507–519. 10.1111/tpj.13122 [DOI] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., and Sheen J., 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zhang Y. F., Hou M. M., and Tan B. C., 2013. The requirement of WHIRLY1 for embryogenesis is dependent on genetic background in maize. PLoS One 8: e67369 10.1371/journal.pone.0067369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cai M., Zhang X., Li Y., Zhang J. et al. , 2014. Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS One 9: e94126 10.1371/journal.pone.0094126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmudjak M., Colas des Francs-Small C., Keren I., Shaya F., Belausov E. et al. , 2013. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytol. 199: 379–394. 10.1111/nph.12282 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are two files (one excel file and one word file) in the Supplemental Material, File S1 and File S2. File S1 contains two supplementary tables. Table S1 includes Gene ontology classifications of DEGs with functional annotation. Table S2 shows Primers used in this study. File S2 contains four supplementary figures. Figure S1 showed aborted development in Zmsmk3 mutants. Figure S2 showed identification of ZmSmk3 by TAIL-PCR. Figure S3 showed phylogenetic analysis of ZmSMK3. Figure S4 showed RT-PCR analysis of mitochondrial tRNA and rRNA encoded genes transcriptional levels in the wild type and the Zmsmk3 mutant. Supplemental material available at FigShare: https://doi.org/10.6084/m9.figshare.8217473.