Abstract
In hydrothermal environments, carbon monoxide (CO) utilisation by thermophilic hydrogenogenic carboxydotrophs may play an important role in microbial ecology by reducing toxic levels of CO and providing H2 for fuelling microbial communities. We evaluated thermophilic hydrogenogenic carboxydotrophs by microbial community analysis. First, we analysed the correlation between carbon monoxide dehydrogenase (CODH)–energy-converting hydrogenase (ECH) gene cluster and taxonomic affiliation by surveying an increasing genomic database. We identified 71 genome-encoded CODH–ECH gene clusters, including 46 whose owners were not reported as hydrogenogenic carboxydotrophs. We identified 13 phylotypes showing > 98.7% identity with these taxa as potential hydrogenogenic carboxydotrophs in hot springs. Of these, Firmicutes phylotypes such as Parageobacillus, Carboxydocella, Caldanaerobacter, and Carboxydothermus were found in different environmental conditions and distinct microbial communities. The relative abundance of the potential thermophilic hydrogenogenic carboxydotrophs was low. Most of them did not show any symbiotic networks with other microbes, implying that their metabolic activities might be low.
Electronic supplementary material
The online version of this article (10.1007/s00203-019-01661-9) contains supplementary material, which is available to authorized users.
Keywords: Microbial community analysis, Next-generation sequencing, Hot spring, Thermophile, Carboxydotroph, Hydrogenogen
Introduction
Hydrothermal systems, where geothermally heated water is expelled through fissures in the Earth’s crust, are located both on land and under the sea. It is now well known that a wide variety of microorganisms, called thermophiles or hyperthermophiles, can prevail and even thrive in such high-temperature environments. The pioneering studies by Brock and his colleagues (Brock 1967; Bott and Brock 1969; Brock and Darland 1970; Brock et al. 1971) at the Yellowstone National Park hot springs established that these organisms grow at near boiling temperatures. Furthermore, a research study led by Pace using molecular phylogenetic techniques demonstrated the high abundance of unidentified thermophilic bacteria and archaea and their remarkable phylogenetic diversity in pink filaments and sediments in the same area (Barns et al. 1994, 1996; Reysenbach et al. 1994; Hugenholtz et al. 1998).
In recent years, microbes that can utilise carbon monoxide (CO) have been found from the hydrothermal area (Sokolova et al. 2009; Techtmann et al. 2009). Although CO is a toxic gas, it can also be a low-potential electron donor and carbon source for many microbes. To date, the list of known thermophilic anaerobic CO-utilizing microorganisms includes acetogenic bacteria (Moorella thermoacetica, for instance), sulfate-reducing bacteria (Desulfotomaculum carboxydivorans), methanogenic archaea (Methanothermobacter thermautotrophicus), and hydrogenogenic bacteria as well as various archaea, such as Carboxydothermus hydrogenoformans, Thermosinus carboxydivorans, and Thermococcus AM4 (Techtmann et al. 2009). Of these, hydrogenogenic bacteria and archaea (collectively designated thermophilic hydrogenogenic carboxydotrophs) are thought to play a key ecological role by virtue of providing a ‘safety valve’ for reducing toxic levels of CO and supplying H2 for fuelling H2-dependent microbial community processes (Techtmann et al. 2009).
In general, the ability of hydrogenogenic carboxydotrophy is linked to the presence of CO dehydrogenase (CODH)–energy-converting hydrogenase (ECH) gene cluster in genomes. This cluster is believed to be horizontally transferred between the representatives of separate taxa (Techtmann et al. 2012). So far, 28 phylogenetically diverse thermophilic anaerobic hydrogenogenic CO-utilizing archaea and bacteria have been reported (Sokolova et al. 2009; Table 2). Most of them (23 species) are members of the phylum Firmicutes.
Table 2.
Prokaryotes possessing CODH–ECH gene clusters
| Organism | Hydrogenogenic carboxydotrophic growth | Isolation source | |
|---|---|---|---|
| Type | References | ||
| Crenarchaeota (thermophilic) | |||
| Thermofilum carboxyditrophus 1505 | Yes (Sokolova et al. 2009) | Water and mud | Sokolova et al. (2009) |
| Euryarchaeota (thermophilic) | |||
| Thermococcus barophilus CH5 | Yes (Kozhevnikova et al. 2016) | Deep-sea hydrothermal fields | Kozhevnikova et al. (2016) |
| Thermococcus barophilus MP | Yes (Kozhevnikova et al. 2016) | Deep-sea hydrothermal fields | Kozhevnikova et al. (2016) |
| Thermococcus guaymasensis DSM 11113 | n.r | Hydrothermal vent sediment | Canganella et al. (1998) |
| Thermococcus onnurineus NA1 | Yes (Bae et al. 2006) | Deep-sea hydrothermal fields | Bae et al. (2006) |
| Thermococcus paralvinellae ES1 | n.r | Active hydrothermal vent chimneys | Hensley et al. (2014) |
| Thermococcus sp. AM4 | Yes (Sokolova et al. 2004b) | Active chimney | Sokolova et al. (2004b) |
| Firmicutes (thermophilic) | |||
| Parageobacillus thermoglucosidasius B4168 | n.r | n.r | n.r |
| Parageobacillus thermoglucosidasius C56-YS93 | n.r | n.r | n.r |
| Parageobacillus thermoglucosidasius DSM 2542a | Yes (Mohr et al. 2018) | n.r | Suzuki et al. (1983) |
| Parageobacillus thermoglucosidasius GT23 | n.r | n.r | n.r |
| Parageobacillus thermoglucosidasius NBRC 107763 | n.r | n.r | n.r |
| Parageobacillus thermoglucosidasius NCIMB 11955 | n.r | n.r | n.r |
| Parageobacillus thermoglucosidasius TG4 | Yes (Inoue et al. 2019b) | Marine sediment | Inoue et al. (2019a) |
| Parageobacillus thermoglucosidasius TM242 | n.r | n.r | n.r |
| Parageobacillus thermoglucosidasius TNO-09.020 | n.r | n.r | n.r |
| Parageobacillus thermoglucosidasius Y4.1MC1 | n.r | n.r | n.r |
| Carboxydocella sp. JDF658 | Yes (Fukuyama et al. 2017) | Open-air stream from a hot spring well | Fukuyama et al. (2017) |
| Carboxydocella sp. ULO1 | Yes (Fukuyama et al. 2017) | Sediment of a maar lake | Fukuyama et al. (2017) |
| Carboxydocella sporoproducens DSM 16521 | Yes (Slepova et al. 2006) | Hot spring | Slepova et al. (2006) |
| Carboxydocella thermautotrophica 019 | Yes (Toshchakov et al. 2018) | Thermal field | Toshchakov et al. (2018) |
| Carboxydocella thermautotrophica 041 | Yes (Sokolova et al. 2002) | Terrestrial hot vent | Sokolova et al. (2002) |
| Desulfosporosinus sp. OL | n.r | n.r | n.r |
| Desulfotomaculum nigrificans CO-1-SRB | Yes (Parshina et al. 2005) | Anaerobic bioreactor sludge | Sokolova et al. (2009) |
| Thermincola ferriacetica Z-0001 | Yes (Zavarzina et al. 2007) | Ferric deposits of a terrestrial hydrothermal spring | Zavarzina et al. (2007) |
| Thermincola potens JR | Yes (Wrighton et al. 2008; Byrne-Bailey et al. 2010) | Thermophilic microbial fuel cell | Wrighton et al. (2008); Byrne-Bailey et al. (2010) |
| Caldanaerobacter subterraneus subsp. pacificus DSM 12653 | Yes (Sokolova et al. 2001; Fardeau et al. 2004) | Oilfields | Fardeau et al. (2004) |
| Caldanaerobacter subterraneus subsp. tengcongensis MB4 | n.r | Oilfields | Fardeau et al. (2004) |
| Caldanaerobacter subterraneus subsp. yonseiensis KB-1 | n.r | Oilfields | Fardeau et al. (2004) |
| Calderihabitans maritimus KKC1 | Yes (Yoneda et al. 2013b) | Submerged marine caldera | Yoneda et al. (2013b) |
| Carboxydothermus hydrogenoformans Z-2901 | Yes (Svetlichny et al. 1991) | Hot swamp | Svetlichny et al. (1991) |
| Carboxydothermus islandicus SET | Yes (Novikov et al. 2011) | Hot spring | Novikov et al. (2011) |
| Moorella glycerini NMP | n.r | Underground gas storage | Slobodkin et al. (1997) |
| Moorella sp. Hama-1 | n.r | Thermophilic anaerobic digestion reactor | Harada et al. (2018) |
| Moorella stamsii DSM 26271 | Yes (Alves et al. 2013) | Anaerobic sludge | Alves et al. (2013) |
| Moorella thermoacetica DSM 21394 | Yes (Jiang et al. 2009) | Anaerobic bioreactors | Jiang et al. (2009) |
| Thermanaeromonas toyohensis ToBE | n.r | Geothermal aquifer in mine | Mori et al. (2002) |
| Thermoanaerobacter sp. YS13 | n.r | Geothermal hot spring | Peng et al. (2016) |
| Thermosinus carboxydivorans Nor1 | Yes (Sokolova et al. 2004a) | Hot spring | Sokolova et al. (2004a) |
| Proteobacteria (mesophilic) | |||
| Rhodopseudomonas palustris BisB18 | n.r | River sediment | Oda et al. (2008) |
| Pleomorphomonas carboxyditropha SVCO-16 | n.r | Anaerobic sludge | Esquivel-Elizondo et al. (2018) |
| Pseudovibrio sp. POLY-S9 | n.r | Intertidal marine sponge | Alex and Antunes (2015) |
| Pseudovibrio sp. Tun.PSC04-5.I4 | n.r | n.r | n.r |
| Rhodospirillum rubrum ATCC 11170 | Yes (Kerby et al. 1992) | Fresh water | Munk et al. (2011) |
| Rhodospirillum rubrum F11 | Yes (Singer et al. 2006) | n.r | n.r |
| Desulfovibrio bizertensis DSM 18034 | n.r | Marine sediment | Haouari et al. (2006) |
| Pseudodesulfovibrio piezophilus C1TLV30 | n.r | Wood falls at deep sea | Khelaifia et al. (2011) |
| Geobacter bemidjiensis Bem | n.r | Subsurface sediments | Nevin et al. (2005) |
| Geobacter pickeringii G13 | n.r | Kaolin clays | Shelobolina et al. (2007) |
| Ferrimonas futtsuensis DSM 18154 | n.r | Sediment | Nakagawa et al. (2006) |
| Ferrimonas kyonanensis DSM 18153 | n.r | Alimentary tract of littleneck clams | Nakagawa et al. (2006) |
| Ferrimonas sediminum DSM 23317 | n.r | Coastal sediment | Ji et al. (2013) |
| Shewanella sp. M2 | n.r | n.r | n.r |
| Shewanella sp. R106 | n.r | n.r | n.r |
| Citrobacter amalonaticus Y19 | Yes (Oh et al. 2008) | Anaerobic wastewater sludge digester | Jung et al. (1999) |
| Salmonella enterica subsp. enterica serovar Montevideo 50262 | n.r | n.r | n.r |
| Salmonella enterica subsp. enterica serovar Montevideo 50270 | n.r | n.r | n.r |
| Salmonella enterica subsp. enterica serovar Senftenberg 50263 | n.r | n.r | n.r |
| Salmonella enterica subsp. enterica serovar Senftenberg 50264 | n.r | n.r | n.r |
| Salmonella enterica subsp. enterica serovar Senftenberg 50265 | n.r | n.r | n.r |
| Salmonella enterica subsp. enterica serovar Senftenberg 50271 | n.r | n.r | n.r |
| Salmonella enterica subsp. enterica serovar Senftenberg 50272 | n.r | n.r | n.r |
| Salmonella enterica subsp. enterica serovar Senftenberg SS209 | n.r | n.r | n.r |
| Photobacterium marinum AK15 | n.r | Sediment | Srinivas et al. (2013) |
| Uncultured | n.r | n.r | |
| Candidatus Korarchaeota archaeon MDKW | n.r | Hot springs metagenomes | n.r |
| Clostridiales bacterium DRI-13 | n.r | Subglacial ecosystem | n.r |
| Rhizobiales bacterium AFS016371 | n.r | Soil | n.r |
| Rhizobiales bacterium AFS041951 | n.r | Soil | n.r |
| Rhizobiales bacterium AFS049984 | n.r | Soil | n.r |
| Rhizobiales bacterium AFS089140 | n.r | Soil | n.r |
n.r. not reported
aTwo genomes are available for this strain in the database
In addition to their basic isolation and identification, there are several ecological studies on thermophilic hydrogenogenic carboxydotrophs (Kochetkova et al. 2011; Brady et al. 2015; Yoneda et al. 2015). Notably, a radio isotopic study suggests that the majority of CO is oxidised to CO2 (120 μmol L−1 of sediment day−1) by microbial activities in the hot springs of Uzon Caldera (Kamchatka) (Kochetkova et al. 2011). Thermophilic hydrogenogenic carboxydotrophs of the genera Carboxydocella and Dictyoglomus have also been isolated from the same environment (Kochetkova et al. 2011). A quantitative polymerase chain reaction (qPCR) analysis targeting the CODH gene, which encodes a key enzyme involved in CO oxidation, suggests that the Carboxydothermus species, which is the most studied thermophilic carboxydotrophic species, is widely distributed in a wide range of hydrothermal environments despite its relatively low population size ( ≤ 0.000795% of the total bacterial population) (Yoneda et al. 2015). In addition, using the stable isotope probing (SIP) method by 13CO DNA, Thermincola, Desulfotomaculum, and Carboxydocella species were all detected and enriched at geothermal sites, although they are present at < 1% in the original communities (Brady et al. 2015). While there is evidence for the temporal dominance of the Carboxydothermus species ( ~ 10% of bacterial population) in hydrothermal environments (Yoneda et al. 2013a), thermophilic hydrogenogenic carboxydotrophs are generally considered to occur in low abundance in the environments.
However, these ecological studies on thermophilic hydrogenogenic carboxydotrophs had a few limitations. Because the sequences of CODH genes are highly diverse, it was difficult to design universal primers that could amplify a wide range of CODH genes from different taxa (Yoneda et al. 2013a). SIP is effective for identifying CO-utilizing microbes in the environment (Brady et al. 2015); however, cultivation bias could be observed. In addition, the previous CO-SIP study was limited to a few neutral pH hot springs (Brady et al. 2015). On the other hand, 16S metagenomics is a culture-independent and high-throughput technique, which is applicable for exploring diverse thermophilic hydrogenogenic carboxydotrophs and co-occurring microbes. The number of available microbial genome sequences has vastly increased thanks to recent advances in next-generation sequencing technology, using which CODH genes were detected in some species that had never been reported to show hydrogenogenic carboxydotrophic growth (Mohr et al. 2018; Inoue et al. 2019a). However, the correlation between the presence of CODH–ECH gene cluster and taxonomic affiliation has not been well understood. Here, we performed a comprehensive survey of a current prokaryotic genomic database and revealed the phylogenetic distribution of CODH–ECH gene clusters across prokaryotes. Next, we performed 16S rRNA gene amplicon (V3/V4 region) sequencing analysis on 100 sediment samples from a wide variety of hydrothermal and mesophilic environments in Japan and unveiled the distribution patterns of these “potential hydrogenogenic carboxydotrophs”.
Materials and methods
Sample collection and DNA extraction
We collected a total of 100 sediment samples [17.5 ~ 99.0 °C; pH 2.2 ~ 8.9; oxidation–reduction potential (ORP) − 262 ~ + 449 mV] from terrestrial hydrothermal and mesophilic environments in Japan from May 2014 to March 2017 (Online Resource 1). The sampling sites included 76 on Southern Kyushu Island (Kagoshima prefecture), 14 on Northern Kyushu Island (Oita prefecture), five on the Eastern Izu peninsula (Shizuoka prefecture), and five on the Southern Izu peninsula (Shizuoka prefecture). At the Unagi-onsen hot spring (Southern Kyushu Island), we collected a total of 65 samples in May 2014, May 2015, November 2015, and December 2016 as a previous study suggested that Carboxydothermus species are abundant in this environment (Yoneda et al. 2013a). In addition, we previously isolated the Carboxydocella strains ULO1 and JDF658 at Unagi-ike lake and the Jiunji-onsen hot spring, respectively (Fukuyama et al. 2017). Temperature was measured using a TX10 digital thermometer (Yokogawa, Tokyo, Japan) with a type K temperature probe (Yokogawa, Tokyo, Japan) at each sampling site. The pH and ORP of the sediment pore water were measured using an HM-31P portable pH meter (DKK-TOA, Tokyo, Japan) with pH (GST-2729C; DKK-TOA, Tokyo, Japan) or ORP (PST-2729C; DKK-TOA, Tokyo, Japan) electrodes. Sediment samples were collected using 50 mL plastic tubes filled with pore water, put into plastic bags with AnaeroPouch-Anaero (Mitsubishi Gas Chemical, Tokyo, Japan), and immediately sealed to minimise contact with oxygen. The samples were then packed in a cooler box with ice, transported to the laboratory, and stored at – 80 °C until use. DNA was extracted from 0.5 g of sediment material using an Extrap Soil DNA Kit Plus ver. 2 (Nippon Steel and SUMIKIN Eco-Tech, Tokyo, Japan) following the manufacturer’s instructions. During the homogenising step, we used a bead beater-type homogeniser, Beads Crusher μT-12 (Taitec, Koshigaya, Japan), at a speed of 3200 r min−1 for 60 s. The extracted DNA was stored at – 30 °C until use.
16S rRNA gene amplification and sequencing
The V3/V4 region of bacterial and archaeal 16S rRNA genes was amplified with the following prokaryotic universal primer sets (Takahashi et al. 2014): forward (5′-CCTACGGGNBGCASCAG-3′) and reverse (5′-GACTACNVGGGTATCTAATCC-3′) with added overhanging adapter sequences at each 5ʹ-end according to the 16S metagenomic sample preparation guide (https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf). Each sample was amplified with KAPA™ HiFi HotStart ReadyMix (2X) (KAPA Biosystems, South Africa) according to the manufacturer’s instructions. Paired-end (PE, 2 × 300 nucleotides) sequencing was performed with an Illumina MiSeq (MiSeq Reagent kit v2) and followed the manufacturer’s run protocols (Illumina, Inc., San Diego, CA, USA).
16S rRNA gene sequence processing and statistical analyses
Primer-binding regions were removed by trimming 17 and 21 nt sequences from the 5′ ends of the forward and reverse reads without adapter regions, respectively, with VSEARCH ver. 2.6.0 (Rognes et al. 2016). The reads were further processed by trimming low-quality regions from the sequences with Trimmomatic ver. 0.36 (SLIDINGWINDOW: 50:20) (Bolger et al. 2014). Using VSEARCH, the paired-end reads were joined and de-multiplexed, and a further round of quality control was conducted to remove sequences shorter than 200 nt as well as those containing ambiguous bases (N) or bases with a quality score below 20. Chimeric 16S rDNA sequences were detected using the UCHIME algorithm in the USEARCH package implemented within VSEARCH. The SILVA 132 SSU Ref Nr99 (Quast et al. 2013), a comprehensive, quality checked data sets of small subunit rRNA sequences, was used as a reference for chimera detection. Operational taxonomic units (OTUs) were defined as clusters of sequences that were not singletons (unique sequences that are present exactly once in each sample) with 98.7% similarity using VSEARCH. Then, taxonomic classification of individual OTU was performed with the stand-alone SINA ver. 1.2.11 aligner (Pruesse et al. 2012) using the SILVA 132 SSU Ref Nr99 database as a reference. The non-prokaryotic OTUs (i.e., eukaryote and unclassified domain) were then removed. OTU abundance was estimated by adding prokaryotic singleton reads using the global alignment search option of VSEARCH (–usearch_global—id 0.987), to increase sensitivity. Prior to community analysis, samples with less than 10,000 sequences were omitted (leaving 77 samples) in the beta-diversity patterns. The resulting OTU abundance tables were rarefied to an even number of sequences per sample to ensure equal sampling depth (14,146 sequences per sample) using the vegan package (Oksanen et al. 2017) of the R software (R Core Team 2016). Alpha and beta diversity analyses were then performed with the phyloseq (McMurdie and Holmes 2013) and vegan packages of the R software.
Database search for CODH–ECH gene clusters
The amino acid sequences corresponding to CODHs were obtained from the Reference Sequence (RefSeq) Database in National Center for Biotechnology Information (NCBI) (December 2018) through a BLASTp search using C. hydrogenoformans CooSI (ABB14432.1) subunit as a query. Low-scoring and short-length hits (bit score < 200, amino acid length < 550) including HCPs and partial fragments were excluded from the data set. Then, coding sequences (CDS) within 20 CDSs upstream and downstream of the CODH gene locus were annotated by clusters of orthologous groups of proteins (COGs) (Tatusov 2001) through RPS-BLAST search (e value < 10−6) using NCBI Conserved Domain Database (Marchler-Bauer et al. 2002). Of these, we identified CODH genes with ECH small and large subunits (COG3260 nd COG3261, respectively) as CODH–ECH gene clusters.
Phylogenetic analyses
We retrieved the reference 16S rRNA gene sequences that were equal or longer than 1,000 nt and did not include N from the genomes of prokaryotes possessing CODH–ECH gene clusters and those that were classified into the same genera as them via the RefSeq genome database. To obtain a non-redundant data set for phylogenetic analysis, retrieved sequences were trimmed into V3/V4 region identical to the amplicons and clustered with 100% similarity using VSEARCH (the sequences utilised in this analysis are listed in Online Resource 6). The sequences were aligned using MAFFT 7.402 (Katoh and Standley 2013). Maximum-likelihood phylogenetic trees were calculated using FastTree ver. 2.1.9 (Price et al. 2010) with an approximate-maximum-likelihood method using the GTR + GAMMA model. Robustness of the topology of the phylogenetic trees was evaluated by local bootstrap values based on 1000 re-samples. The tree was imported into the iTOL online tool (Letunic and Bork 2016) for visualisation.
Exploring the co-occurrence of thermophilic hydrogenogenic carboxydotrophs and other microbes
Based on the OTU read numbers, a network of phylotype co-occurrence was produced with a minimum Spearman correlation coefficient of 0.8 using R. We retrieved and have presented the smaller networks, including phylotypes, related to the thermophilic hydrogenogenic carboxydotrophs identified in our phylogenetic analysis.
Results and discussion
Sample profiles and overview of 16S rRNA gene amplicon sequencing
We collected 100 sediment samples from geographically distant areas in Japan, including Kyushu Island and the Izu Peninsula (Table 1; additional data are provided in Online Resource 1). Except for a single sample from Unagi-ike lake, which has a moderate environment (17.5 °C; pH 7.37; ORP, + 75 mV), all the samples were collected from geothermally heated hydrothermal environments (33.8–99.0 °C). Although the in situ environmental conditions of the sampling sites were variable, the hot springs on Kyushu Island had an acidic pH [average pH 4.1 ± 1.1 (sd); measurable sites, n = 82], whereas those on the Izu Peninsula were neutral or weakly alkaline (pH 8.3 ± 0.4; n = 10).
Table 1.
Summary of samples
| Sampling area and time point | Sampling date | Numbers of samples | Temperature (°C) | pH | ORP (mV) | Salinity (%) |
|---|---|---|---|---|---|---|
| 1405_Unagi | May 2014 | 13 | 41.4 ~ 99.0 | 2.2 ~ 4.9 | − 218 ~ + 426 | n.m |
| 1505_Unagi | May 2015 | 15 | 33.8 ~ 95.8 | 4.4 ~ 5.8 | − 174 ~ + 277 | n.m |
| 1511_Unagi | November 2015 | 19 | 41.2 ~ 96.2 | 3.3 ~ 5.6 | − 130 ~ + 449 | n.m |
| 1612_Unagi | December 2016 | 18 | 35.5 ~ 96.9 | 2.6 ~ 5.9 | − 262 ~ + 164 | n.m |
| 1612_Kirishima | December 2016 | 10 | 63.4 ~ 88.7 | 2.4 ~ 4.1 | − 179 ~ + 310 | 0 |
| 1703_Komatsu | March 2017 | 14 | 61.1 ~ 80.9 | 2.2 ~ 5.6 | − 179 ~ + 286 | n.m |
| 1501_Eastern_Izu | January 2015 | 5 | 68.2 ~ 80.1 | 8.4 ~ 8.5 | − 22 ~ + 189 | 0.0 ~ 0.2 |
| 1501_Southern_Izu | January 2015 | 5 | 60.1 ~ 78.5 | 7.7 ~ 8.9 | − 30 ~ + 259 | 0.0 ~ 2.4 |
| 1612_Unagi-ike_lake | December 2016 | 1 | 17.5 | 7.37 | 75 | n.m |
n.m. not measured
Our 16S rRNA gene amplicon sequencing analysis generated 8,531,132 bacterial and archaeal quality-controlled sequences from the 100 samples, with a range of 107–398,919 sequences (average, 85,311 sequences) per sample (Online Resource 1). A total of 9,394 prokaryotic OTUs were defined at the 98.7% similarity level, and 23–4,737 OTUs (average, 299 OTUs) were observed in each sample (Online Resource 2). Diversity analysis using rarefied 77 samples with equal or greater than 10,000 sequences revealed that microbial communities in the sampled hot springs showed much lower alpha diversity than those in the moderate environment (Unagi-ike lake; Online Resource 3), indicating that high temperature imposed constraints on community properties as observed in other studies (Sharp et al. 2014).
Furthermore, our beta diversity analysis revealed apparent differences between the acidic hot springs on Kyushu Island and the neutral or weak alkaline environments on Izu Peninsula and Unagi-ike lake (Fig. 1). At the domain level, microbial communities in the acidic hot springs were dominated by archaea, whereas those in the neutral or weak alkaline environments were dominated by bacteria (Online Resource 4). The phylotypes that shared 100% identity with Vulcanisaeta souniana (phylum Crenarchaeota; OTU_1) and Thermus thermophilus (phylum Deinococcus-Thermus; OTU_20) were notably prominent in the acidic hot springs and neutral or weak alkaline environments, respectively. V. souniana is a heterotrophic anaerobic hyperthermophilic crenarchaeote found in hot springs that grows optimally at 85–90 °C and pH 4.0–4.5 (Itoh et al. 2002). In contrast, T.thermophilus is an extremely thermophilic bacterium also found in hot springs, but its optimal growth occurs at 65–72 °C and pH 7.5 (Oshima and Imahori 1974). Although the major phylotypes were the same in each acidic and neutral or weak alkaline environments, our non-metric multidimensional scaling analysis using rarefied 77 samples with greater equal than 10,000 sequences shows that microbial community compositions vary across each sampling sites (Fig. 1).
Fig. 1.
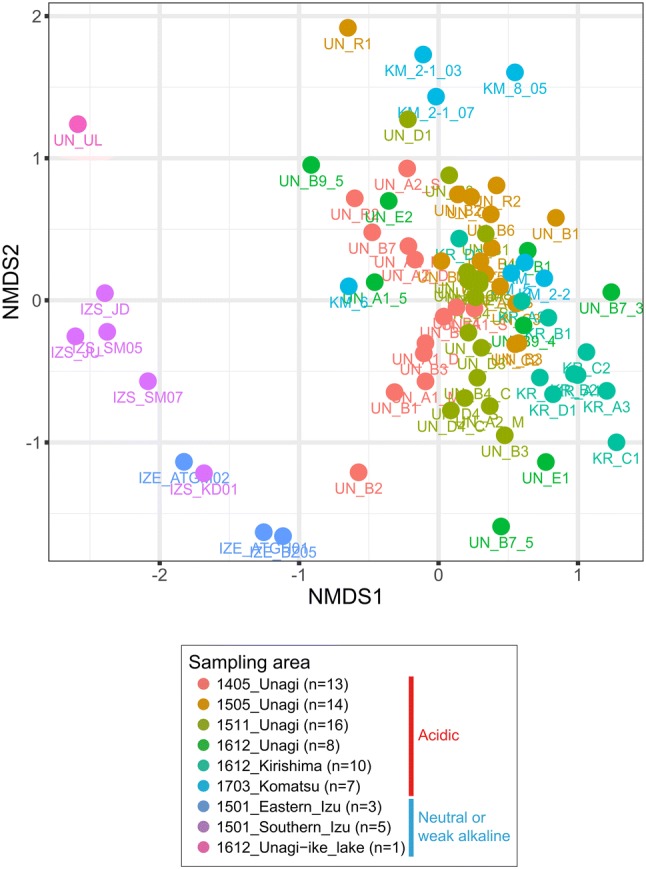
Non-metric multidimensional scaling analysis based on Bray–Curtis dissimilarity. Plot colours illustrate sampling area and period
CODH–ECH gene clusters found in prokaryotic genomes
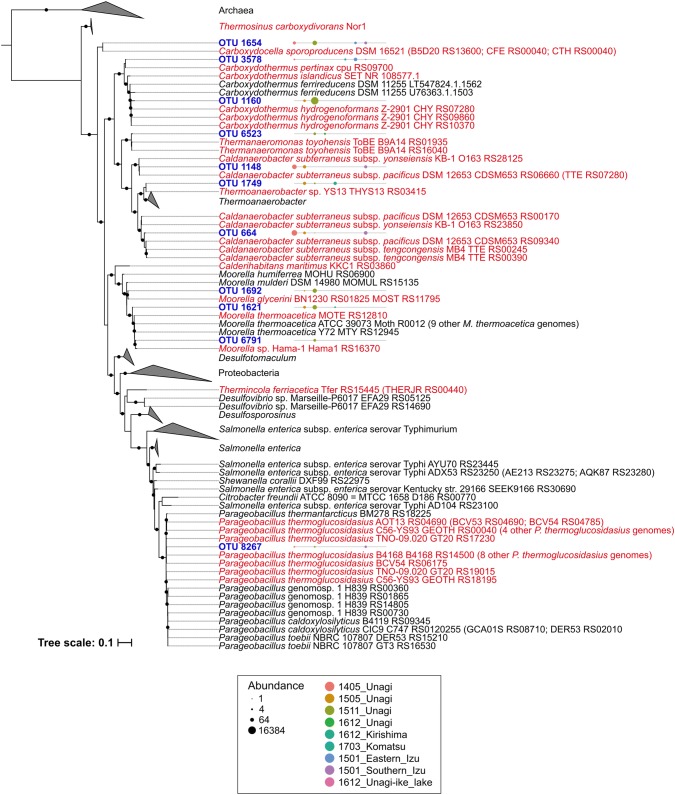
A previous study examined CODHs and their genomic context in 2887 microbial genomes and revealed 185 genomes that encoded at least one CODH gene (Techtmann et al. 2012). Of these, 12 genomes possessed CODH–ECH gene clusters. However, by December 2018, the number of sequenced microbial genome entries in the RefSeq genome database had reached 142,909, and novel thermophilic hydrogenogenic carboxydotrophs had been reported. Therefore, we searched CODH–ECH gene clusters in the current RefSeq database and examined their taxonomic information. We identified 71 genomes encoding CODH–ECH gene clusters, which include 40 thermophile genomes (14 genera), 25 mesophile genomes (12 genera), and six unclassified microbial genomes (Table 2; additional data are listed in Online Resource 5). All mesophilic members were classified into the phylum Proteobacteria, which included phototrophic bacteria or sulfate-reducing bacteria, whereas thermophilic members were phylogenetically diverse and classified into the phylum Crenarchaeota, Euryarchaeota, and Firmicutes. Of these 71 genomes, 46 have never been reported as hydrogenogenic carboxydotrophs (Table 2), and the presence of CODH–ECH gene clusters in 22 genomes was reported for the first time in this study (Online Resource 5).
Conservation patterns of CODH–ECH gene clusters were different in each genus (Fig. 2). We classified these genera into three groups: (1) the CODH–ECH gene clusters and the hydrogenogenic carboxydotrophy ability were well conserved; (2) a portion of members conserved the CODH–ECH gene clusters; and (3) genera that we could not classify into (1) nor (2) because of inadequate availability of genomic information. Thermincola, Carboxydocella, Carboxydothermus, and Caldanaerobacter were classified into the group (1). In most cases, the phylogeny of CODH genes was corresponding to their taxonomic phylogeny in this group (Adam et al. 2018; Fukuyama et al. 2018; Toshchakov et al. 2018), suggesting that the CODH–ECH gene clusters descended from the common ancestors of each genus. The genus Carboxydothermus has been one of the most studied models of thermophilic carboxydotrophy, and the members of this genus possess four or five CODH genes (Fukuyama et al. 2018). A comparative genomic analysis in Carboxydothermus revealed that the CODH–ECH gene clusters were conserved in the members except for C. pertinax, which lacked only the CODH (CODH-I) unit of CODH–ECH gene cluster and Carboxydothermus ferrireducens, which lacked the whole CODH–ECH gene cluster (Fukuyama et al. 2018). C. ferrireducens can grow carboxydotrophically, but is not hydrogenogenic (Slobodkin et al. 2006). On the other hand, C.pertinax can grow by hydrogenogenic carboxydotrophy (Yoneda et al. 2012), and it is suggested that C. pertinax could couple alternative CODH (CODH-II) to the distal ECH (Fukuyama et al. 2018). C. pertinax was the only isolate that could grow by hydrogenogenic carboxydotrophy without the CODH–ECH gene cluster. Caldanaerobacter subterraneus subspecies can oxidise CO and possess CODH–ECH gene clusters, whose structures are very similar (Sant’Anna et al. 2015). However, phylogenetic reconstruction of CODH genes revealed that CODH genes from C. subterraneus have distinct evolutionary histories. It is suggested that replacement of CODH gene occurred by a horizontal gene transfer event in C. subterraneus subsp. tengcongensis and C. subterraneus subsp. yonseiensis (Sant’Anna et al. 2015). Thermococcus, Thermofilum, Thermoanaerobacter, Moorella, Desulfotomaculum, Desulfosporosinus, Parageobacillus, and members of the phylum Proteobacteria were classified into group (2). Because most species of Thermococcus, Thermofilum, Thermoanaerobacter, Desulfotomaculum, and Desulfosporosinus did not possess the CODH–ECH gene clusters, it was suggested that CODH–ECH gene clusters might have been obtained by a portion of the members in a horizontal gene transfer event. In fact, this cluster is believed to be horizontally transferred between the representatives of separate taxa (Techtmann et al. 2012). In the genus Moorella, Moorella stamsii and Moorella glycerini possessed identical CODHs that were flanked by ECH gene clusters. Moorella sp. Hama-1 and Moorella thermoacetica DSM 21394, which formed a different subclade from M. stamsii and M. glycerini, also possessed a similar CODH–ECH gene cluster. However, it was revealed that the other 11 M. thermoacetica strains did not possess the CODH–ECH gene cluster (Online Resource 5). M. thermoacetica might be an acetogenic carboxydotroph rather than being hydrogenogenic, as reported previously (Pierce et al. 2008; Schuchmann and Müller 2014), and only strain DSM 21394 might be hydrogenogenic. Parageobacillus thermoglucosidasius is the only facultative anaerobic bacillus among the thermophilic hydrogenogenic carboxydotrophic species (Mohr et al. 2018). Although other Parageobacillus species did not possess the CODH–ECH gene cluster, P. thermoglucosidasius possesses a CODH–ECH gene cluster that is phylogenetically related to those of Moorella and Caldanaerobacter (Mohr et al. 2018). Unlike M. thermoacetica, all 10 genomes of P. thermoglucosidasius have conserved the CODH–ECH gene clusters (Online Resource 5), and hydrogenogenic carboxydotrophy might be an important trait for this species. The other species, Thermanaeromonas toyohensis, Thermosinus carboxydivorans, Calderihabitans maritimus, and uncultured archaea and bacteria (Candidatus Korarchaeota archaeon MDKW, Clostridium bacterium DRI-13, and Rhizobiales bacterium) were classified into the group (3).
Fig. 2.
Phylogenetic reconstruction of potential thermophilic hydrogenogenic carboxydotrophic phylotypes of the Phylum Firmicutes. The 16S rRNA sequences used in this analysis are listed in Online Resource 6. Other but identical sequences to their leaves are shown in parenthesis (only one sequence per genome are shown). The phylotype sequences obtained in this study are expressed by ‘operational taxonomic unit (OTU)’ prefix. Microbes possessing CODH − ECH gene clusters and Carboxydothermus pertinax (cpu_RS09700) are shown in red font. Nodes supported by a bootstrap value greater than 80% are indicated by black circles. The bubble plots which are shown at the right of OTUs display the distribution pattern of each phylotype. Abundance is indicated by the number of amplicon reads in each sample
Diversity and distribution of thermophilic hydrogenogenic carboxydotrophs
In the 16S amplicon sequencing analysis, we revealed that the representative sequences of 13 phylotypes showed > 98.7% identity with known thermophilic hydrogenogenic carboxydotrophs or microbes possessing CODH–ECH gene clusters, and 10 phylotypes were members of the phylum Firmicutes (Fig. 2, Online Resource 7). Of these, the representative sequences of OTU_1654 and OTU_3578 were identical to Carboxydocella species and C. pertinax, respectively, and OTU_664 and OTU_1148 showed 98.8% and 99.5% identities with C. subterraneus subspecies, respectively. They were members of group (1). It should be noted that the abundant phylotype OTU_1160 showed 97.7% identity with Carboxydothermus species, all of which possess multi CODH gene clusters. The phylotypes that were close to Thermofilum carboxyditrophus 1505 (OTU_1051, identity = 99%), M. thermoacetica DSM 21394 (OTU_1621, identity = 98.8%; OTU_6791, identity = 99.1%), M. glycerini DSM 26271 or M. stamsii NMP (OTU_1692, identity = 99.3%), Thermoanaerobacter sp. YS13 (OTU_1749, identity = 100%), Thermococcus barophilus (OTU_1816, identity = 99%), T. toyohensis ToBE (OTU_6523, identity = 99.3%), and P. thermoglucosidasius (OTU_8267, identity = 100%), were members of group (2) hydrogenogenic carboxydotrophs, suggesting that these phylotypes are also potential thermophilic hydrogenogenic carboxydotrophs. We also found that OTU_1000 showed 99% identity with Candidatus Korarchaeota archaeon MDKW, whose genome was assembled from Washburn Hot Spring metagenome.
The 13 phylotypes of potential thermophilic hydrogenogenic carboxydotrophs were detected in 45 samples (Fig. 2, Online Resource 7). Of these, OTU_1654 (Carboxydocella), OTU_664 (C. subterraneus), OTU_1148 (C. subterraneus), OTU_3578 (C. pertinax), and OTU_8267 (P. thermoglucosidasius) were detected in 7 to 21 samples and widely distributed in geographically distinct areas (both Kyushu Island and the Izu Peninsula) that showed different environmental conditions and microbial community structures (Fig. 2, Online Resource 7). OTU_1000, uncultured archaeon phylotype, was also detected widely from 11 samples. The distribution of OTU_1051 (T. carboxyditrophus), OTU_1692 (M. glycerini or M. stamsii), OTU_1749 (Thermoanaerobacter sp. YS13), OTU_6523 (T. toyohensis), OTU_1621 (M. thermoacetica DSM 21394), and OTU_6791 (M. thermoacetica DSM 21394) was limited to hot springs in Kyushu Island (mainly in Unagi-onsen in May 2015, November 2015, and December 2016). OTU_1816, the phylotype of T. barophilus that was isolated from a deep-sea hydrothermal vent (Marteinsson et al. 1999), was uniquely detected in the saline hot springs in the Izu Peninsula (Online Resource 7).
In most cases, the phylotypes of potential thermophilic hydrogenogenic carboxydotrophs showed a relative abundance of < 0.1%. Previous studies also suggested that Firmicutes carboxydotroph abundance in hydrothermal environments is usually low (Brady et al. 2015; Yoneda et al. 2015). However, the phylotypes of C. subterraneus (OTU_664), Carboxydocella (OTU_1654), C. pertinax (OTU_3578), and Carboxydothermus phylotype (OTU_1160) exhibited a relative abundance of > 0.1% in nine samples (Online Resource 7). In particular, we found that the relative abundance of OTU_1654 reached 8.47% per sample at the 1511_UN_A2_D site (70.9 °C, pH 4.68). OTU_1160 was abundant in Unagi-onsen in November 2015, and its relative abundance reached 7.75% and 11% at the 1511_UN_A2_D and 1511_UN_B4_C (94.9 °C, pH 3.65) sites, respectively. However, we could not identify whether the phylotypes, whose relative abundance exceeded 0.1% were growing in these environments, because six of the nine sites showed higher temperature or lower pH than the growth conditions for the isolates of C. subterraneus subspecies (50–80 °C, pH 4.5–9.0) (Fardeau et al. 2004), Carboxydocella species (40–70 °C, pH 6.2–8.0) (Sokolova et al. 2002; Slepova et al. 2006; Toshchakov et al. 2018), and Carboxydothermus species (40–78 °C, pH 4.6 − 8.6) (Svetlichny et al. 1991; Novikov et al. 2011; Yoneda et al. 2012) (Online Resource 8). The other three sites including 1511_UN_A2_D showed moderate environmental conditions, where the growth could occur (Online Resource 8), but the DNA yields from these sites were low ( < 15 ng/g sediment). Firmicutes members of Carboxydothermus, Carboxydocella, and Caldanaerobacter are reported to be able to form endospore (Kim et al. 2001; Wu et al. 2005; Slepova et al. 2006). Notably, these groups possessed the genes for endospore formation. It was speculated that DNAs of these phylotypes might persist in such environments longer than those of non-spore-forming prokaryotes.
Carboxydotrophs have been suggested to be functionally important, because they mediate a ‘currency exchange’ between CO and hydrogen in hydrothermal environments (Techtmann et al. 2009). For example, symbiotic interactions have been observed between C. hydrogenoformans and thermophilic sulfate reducers in culture, wherein the carboxydotroph provides protection from CO toxicity, whereas H2 is removed by sulfate reduction, thus reducing end-product inhibition (Parshina et al. 2005). We investigated the co-occurrence of the potential thermophilic hydrogenogenic carboxydotrophs and other microbes using non-parametric Spearman correlations of phylotype presence/absence across all sampling sites. Among the phylotypes present in at least seven sites, networks between OTU_664 and four uncultured microbes, and between OTU_1000 and two uncultured bacteria were identified with a Spearman correlation coefficient > 0.8 (Online Resource 9). There seem to be no specific symbiotic interactions between most of the potentially hydrogenogenic carboxydotrophic phylotypes and other microbes at these sampling sites.
A microbial population whose relative abundance is < 0.1% is called ‘rare biosphere’ and contributes to a persistent microbial seed bank, which is a collection of dormant microorganisms that can respond to favourable environmental conditions (Lynch and Neufeld 2015). Endospore formation has an important role for dormancy as well as microbial dispersal (Hubert et al. 2009; Müller et al. 2013; Zeigler 2014; Lynch and Neufeld 2015). It was considered that Firmicutes members of the potential thermophilic hydrogenogenic carboxydotrophs found in a variety of hot springs (in most case, as rare biosphere) might form endospores in extreme environmental conditions and have a strategy of microbial seed bank dynamics. The result that most of the potential hydrogenogenic carboxydotrophs did not show any symbiotic networks with other microbes also might support the speculation that metabolic activities of these members are low in extreme environments.
Conclusion
This study explored the distribution, diversity, and ecology of thermophilic carboxydotrophs across various hydrothermal environments using microbial community analysis. First, we searched CODH–ECH gene clusters in the current microbial genomic database and revealed 71 genomes encoding CODH–ECH gene clusters. Of these, 46 were genomes whose carriers have never been reported as hydrogenogenic carboxydotrophs. In a microbial community analysis, we identified 13 phylotypes that showed > 98.7% identity with thermophilic members of these taxa. Of these, 10 phylotypes were members of the phylum Firmicutes, and Parageobacillus, Carboxydocella, Caldanaerobacter, and Carboxydothermus phylotypes were found across geographically distant hot springs with different environmental conditions, wherein distinct microbial community structures were formed. Although the relative abundance of the Carboxydothermus and Carboxydocella phylotypes was greater than 1% at some sites, most of the potentially thermophilic hydrogenogenic carboxydotrophs were usually rare biospheres, whose relative abundances were < 0.1%. They might be in dormant states in extreme environmental conditions. Although symbiotic interactions between hydrogenotrophic microbes and hydrogenogenic carboxydotrophs have been suggested (Parshina et al. 2005), no symbiotic interaction was identified between most of these phylotypes and other microbes in our study, leading to the speculation that thermophilic hydrogenogenic carboxydotrophic species might not be active in these environments. However, the previous sediment incubation and cultivation studies have shown that Carboxydothermus and Carboxydocella species respond to the presence of CO and actively grow (Kochetkova et al. 2011; Yoneda et al. 2012 , 2015; Brady et al. 2015). There is also evidence that an unusually high-density population (equivalent to 9.45 × 105 cells g sediment−1) of Carboxydothermus is present in Unagi-onsen hot springs (Yoneda et al. 2013a), suggesting that they are viable in the environment. While further studies such as transcription analysis are needed to better understand the ecological function of thermophilic hydrogenogenic carboxydotrophs, the present study provides essential information concerning their distribution and diversity in a variety of volcanic environments.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Eitaro Ikeda, Tatsuki Oguro, and Shin Fujiwara for their assistance during field sampling. We thank Masao Inoue and Issei Nakamoto for helping with computational analysis. Part of the computational analysis was completed at the Supercomputer System at the Institute for Chemical Research, Kyoto University.
Funding
This study was supported by Grants-in-Aid for Scientific Research (A) 25252038, and (S) 16H06381 from The Ministry of Education, Culture, Sports, Science and Technology (MEXT) and by a Grant-in-Aid from the Japan Society for the Promotion of Science (JSPS) Fellows (16J11269).
Compliance with ethical standards
Conflict of interest
None declared.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adam PS, Borrel G, Gribaldo S. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc Natl Acad Sci. 2018;115:E1166–E1173. doi: 10.1073/pnas.1716667115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex A, Antunes A. Whole genome sequencing of the symbiont Pseudovibrio sp. from the intertidal marine sponge Polymastia penicillus revealed a gene repertoire for host-switching permissive lifestyle. Genome Biol Evol. 2015;7:3022–3032. doi: 10.1093/gbe/evv199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves JI, van Gelder AH, Alves MM, et al. Moorella stamsii sp. nov., a new anaerobic thermophilic hydrogenogenic carboxydotroph isolated from digester sludge. Int J Syst Evol Microbiol. 2013;63:4072–4076. doi: 10.1099/ijs.0.050369-0. [DOI] [PubMed] [Google Scholar]
- Bae SS, Kim YJ, Yang SH, et al. Thermococcus onnurineus sp. nov., a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent area at the PACMANUS field. J Microbiol Biotechnol. 2006;16:1826–1831. [Google Scholar]
- Barns SM, Fundyga RE, Jeffries MW, Pace NR. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns SM, Delwiche CF, Palmer JD, Pace NR. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott TL, Brock TD. Bacterial growth rates above 90 °C in Yellowstone hot springs. Science. 1969;164:1411–1412. doi: 10.1126/science.164.3886.1411. [DOI] [PubMed] [Google Scholar]
- Brady A, Sharp C, Grasby S, Dunfield P. Anaerobic carboxydotrophic bacteria in geothermal springs identified using stable isotope probing. Front Microbiol. 2015;6:897. doi: 10.3389/fmicb.2015.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TD. Life at high temperatures. Science. 1967;158:1012–1018. doi: 10.1126/science.158.3804.1012. [DOI] [PubMed] [Google Scholar]
- Brock TD, Darland GK. Limits of microbial existence: temperature and pH. Science. 1970;169:1316–1318. doi: 10.1126/science.169.3952.1316. [DOI] [PubMed] [Google Scholar]
- Brock TD, Brock ML, Bott TL, Edwards MR. Microbial life at 90 °C: the sulfur bacteria of Boulder Spring. J Bacteriol. 1971;107:303–314. doi: 10.1128/jb.107.1.303-314.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne-Bailey KG, Wrighton KC, Melnyk R, et al. Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J Bacteriol. 2010;192:4078–4079. doi: 10.1128/JB.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canganella F, Jones WJ, Gambacorta A, Antranikian G. Thermococcus guaymasensis sp. nov. and Thermococcus aggregans sp. nov., two novel thermophilic archaea isolated from the Guaymas Basin hydrothermal vent site. Int J Syst Evol Microbiol. 1998;48:1181–1185. doi: 10.1099/00207713-48-4-1181. [DOI] [PubMed] [Google Scholar]
- Esquivel-Elizondo S, Krajmalnik-Brown R, Maldonado J. Anaerobic carbon monoxide metabolism by Pleomorphomonas carboxyditropha sp. nov., a new mesophilic hydrogenogenic carboxydotroph. FEMS Microbiol Ecol. 2018;94:fiy056. doi: 10.1093/femsec/fiy056. [DOI] [PubMed] [Google Scholar]
- Fardeau M-L, Salinas MB, L’Haridon S, et al. Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: reassignment of T. subterraneus, Thermoanaerobacter yonseiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum to Caldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies. Int J Syst Evol Microbiol. 2004;54:467–474. doi: 10.1099/ijs.0.02711-0. [DOI] [PubMed] [Google Scholar]
- Fukuyama Y, Oguro T, Omae K, et al. Draft genome sequences of two hydrogenogenic carboxydotrophic bacteria, Carboxydocella sp. strains JDF658 and ULO1, isolated from two distinct volcanic fronts in Japan. Genome Announc. 2017;5:e00242–17. doi: 10.1128/genomeA.00242-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama Y, Omae K, Yoneda Y, et al. Insight into energy conservation via alternative carbon monoxide metabolism in Carboxydothermus pertinax revealed by comparative genome analysis. Appl Environ Microbiol. 2018;84:e00458–18. doi: 10.1128/AEM.00458-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouari O, Fardeau M-L, Casalot L, et al. Isolation of sulfate-reducing bacteria from Tunisian marine sediments and description of Desulfovibrio bizertensis sp. nov. Int J Syst Evol Microbiol. 2006;56:2909–2913. doi: 10.1099/ijs.0.64530-0. [DOI] [PubMed] [Google Scholar]
- Harada J, Yamada T, Giri S, et al. Draft genome sequence of Moorella sp. Strain Hama-1, a novel acetogenic bacterium isolated from a thermophilic digestion reactor. Microbiol Resour Announc. 2018;6:e00517. doi: 10.1128/genomeA.00517-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley SA, Jung J-H, Park C-S, Holden JF. Thermococcus paralvinellae sp. nov. and Thermococcus cleftensis sp. nov. of hyperthermophilic heterotrophs from deep-sea hydrothermal vents. Int J Syst Evol Microbiol. 2014;64:3655–3659. doi: 10.1099/ijs.0.066100-0. [DOI] [PubMed] [Google Scholar]
- Hubert C, Loy A, Nickel M, et al. A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science. 2009;325:1541–1544. doi: 10.1126/science.1174012. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Nakamoto I, Omae K, et al. Structural and phylogenetic diversity of anaerobic carbon-monoxide dehydrogenases. Front Microbiol. 2019;9:3353. doi: 10.3389/fmicb.2018.03353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Tanimura A, Ogami Y, et al. Draft genome sequence of strain TG4, a hydrogenogenic carboxydotrophic bacterium isolated from a marine sediment. Microbiol Resour Announc. 2019;8:e01666–18. doi: 10.1128/MRA.01666-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Suzuki K, Nakase T. Vulcanisaeta distributa gen. nov., sp. nov., and Vulcanisaeta souniana sp. nov., novel hyperthermophilic, rod-shaped crenarchaeotes isolated from hot springs in Japan. Int J Syst Evol Microbiol. 2002;52:1097–1104. doi: 10.1099/00207713-52-4-1097. [DOI] [PubMed] [Google Scholar]
- Ji S, Zhao R, Li Z, et al. Ferrimonas sediminum sp. nov., isolated from coastal sediment of an amphioxus breeding zone. Int J Syst Evol Microbiol. 2013;63:977–981. doi: 10.1099/ijs.0.042408-0. [DOI] [PubMed] [Google Scholar]
- Jiang B, Henstra A-M, Paulo PL, et al. Atypical one-carbon metabolism of an acetogenic and hydrogenogenic Moorella thermoacetica strain. Arch Microbiol. 2009;191:123–131. doi: 10.1007/s00203-008-0435-x. [DOI] [PubMed] [Google Scholar]
- Jung GY, Kim JR, Jung HO, et al. A new chemoheterotrophic bacterium catalyzing water–gas shift reaction. Biotechnol Lett. 1999;21:869–873. doi: 10.1023/A:1005599600510. [DOI] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerby RL, Hong SS, Ensign S, et al. Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J Bacteriol. 1992;174:5284–5294. doi: 10.1128/jb.174.16.5284-5294.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelaifia S, Fardeau M-L, Pradel N, et al. Desulfovibrio piezophilus sp. nov., a piezophilic, sulfate-reducing bacterium isolated from wood falls in the Mediterranean Sea. Int J Syst Evol Microbiol. 2011;61:2706–2711. doi: 10.1099/ijs.0.028670-0. [DOI] [PubMed] [Google Scholar]
- Kim BC, Grote R, Lee DW, et al. Thermoanaerobacter yonseiensis sp. nov., a novel extremely thermophilic, xylose-utilizing bacterium that grows at up to 85 °C. Int J Syst Evol Microbiol. 2001;51:1539–1548. doi: 10.1099/00207713-51-4-1539. [DOI] [PubMed] [Google Scholar]
- Kochetkova TV, Rusanov II, Pimenov NV, et al. Anaerobic transformation of carbon monoxide by microbial communities of Kamchatka hot springs. Extremophiles. 2011;15:319–325. doi: 10.1007/s00792-011-0362-7. [DOI] [PubMed] [Google Scholar]
- Kozhevnikova DA, Taranov EA, Lebedinsky AV, et al. Hydrogenogenic and sulfidogenic growth of Thermococcus archaea on carbon monoxide and formate. Microbiology. 2016;85:400–410. doi: 10.1134/S0026261716040135. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MDJ, Neufeld JD. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015;13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker B, et al. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteinsson VT, Birrien JL, Reysenbach AL, et al. Thermococcus barophilus sp. nov., a new barophilic and hyperthermophilic archaeon isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1999;49:351–359. doi: 10.1099/00207713-49-2-351. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr T, Aliyu H, Küchlin R, et al. CO-dependent hydrogen production by the facultative anaerobe Parageobacillus thermoglucosidasius. Microb Cell Fact. 2018;17:108. doi: 10.1186/s12934-018-0954-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Hanada S, Maruyama A, Marumo K. Thermanaeromonas toyohensis gen. nov., sp. nov., a novel thermophilic anaerobe isolated from a subterranean vein in the Toyoha Mines. Int J Syst Evol Microbiol. 2002;52:1675–1680. doi: 10.1099/00207713-52-5-1675. [DOI] [PubMed] [Google Scholar]
- Müller AL, de Rezende JR, Hubert CRJ, et al. Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J. 2013;8:1153–1165. doi: 10.1038/ismej.2013.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk AC, Copeland A, Lucas S, et al. Complete genome sequence of Rhodospirillum rubrum type strain (S1T) Stand Genomic Sci. 2011;4:293–302. doi: 10.4056/sigs.1804360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Iino T, Suzuki K, Harayama S. Ferrimonas futtsuensis sp. nov. and Ferrimonas kyonanensis sp. nov., selenate-reducing bacteria belonging to the Gammaproteobacteria isolated from Tokyo Bay. Int J Syst Evol Microbiol. 2006;56:2639–2645. doi: 10.1099/ijs.0.64399-0. [DOI] [PubMed] [Google Scholar]
- Nevin KP, Holmes DE, Woodard TL, et al. Geobacter bemidjiensis sp. nov. and Geobacter psychrophilus sp. nov., two novel Fe(III)-reducing subsurface isolates. Int J Syst Evol Microbiol. 2005;55:1667–1674. doi: 10.1099/ijs.0.63417-0. [DOI] [PubMed] [Google Scholar]
- Novikov AA, Sokolova TG, Lebedinsky AV, et al. Carboxydothermus islandicus sp. nov., a thermophilic, hydrogenogenic, carboxydotrophic bacterium isolated from a hot spring. Int J Syst Evol Microbiol. 2011;61:2532–2537. doi: 10.1099/ijs.0.030288-0. [DOI] [PubMed] [Google Scholar]
- Oda Y, Larimer FW, Chain PSG, et al. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc Natl Acad Sci. 2008;105:18543–18548. doi: 10.1073/pnas.0809160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y-K, Kim H-J, Park S, et al. Metabolic-flux analysis of hydrogen production pathway in Citrobacter amalonaticus Y19. Int J Hydrogen Energy. 2008;33:1471–1482. doi: 10.1016/j.ijhydene.2007.09.032. [DOI] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M et al (2017) Vegan: community ecology package. R package version 2.4–3. https://CRAN.R-project.org/package=vegan
- Oshima T, Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol. 1974;24:102–112. doi: 10.1099/00207713-24-1-102. [DOI] [Google Scholar]
- Parshina SN, Kijlstra S, Henstra AM, et al. Carbon monoxide conversion by thermophilic sulfate-reducing bacteria in pure culture and in co-culture with Carboxydothermus hydrogenoformans. Appl Microbiol Biotechnol. 2005;68:390–396. doi: 10.1007/s00253-004-1878-x. [DOI] [PubMed] [Google Scholar]
- Peng T, Pan S, Christopher LP, et al. Growth and metabolic profiling of the novel thermophilic bacterium Thermoanaerobacter sp. strain YS13. Can J Microbiol. 2016;62:762–771. doi: 10.1139/cjm-2016-0040. [DOI] [PubMed] [Google Scholar]
- Pierce E, Xie G, Barabote RD, et al. The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum) Environ Microbiol. 2008;10:2550–2573. doi: 10.1111/j.1462-2920.2008.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2016. [Google Scholar]
- Reysenbach A, Wickham GS, Pace NR. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna F, Lebedinsky A, Sokolova T, et al. Analysis of three genomes within the thermophilic bacterial species Caldanaerobacter subterraneus with a focus on carbon monoxide dehydrogenase evolution and hydrolase diversity. BMC Genom. 2015;16:757. doi: 10.1186/s12864-015-1955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchmann K, Müller V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol. 2014;12:809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- Sharp CE, Brady AL, Sharp GH, et al. Humboldt’s spa: microbial diversity is controlled by temperature in geothermal environments. ISME J. 2014;8:1166–1174. doi: 10.1038/ismej.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelobolina ES, Nevin KP, Blakeney-Hayward JD, et al. Geobacter pickeringii sp. nov., Geobacter argillaceus sp. nov. and Pelosinus fermentans gen. nov., sp. nov., isolated from subsurface kaolin lenses. Int J Syst Evol Microbiol. 2007;57:126–135. doi: 10.1099/ijs.0.64221-0. [DOI] [PubMed] [Google Scholar]
- Singer SW, Hirst MB, Ludden PW. CO-dependent H2 evolution by Rhodospirillum rubrum: Role of CODH:CooF complex. Biochim Biophys Acta Bioenerg. 2006;1757:1582–1591. doi: 10.1016/j.bbabio.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Slepova TV, Sokolova TG, Lysenko AM, et al. Carboxydocella sporoproducens sp. nov., a novel anaerobic CO-utilizing/H2-producing thermophilic bacterium from a Kamchatka hot spring. Int J Syst Evol Microbiol. 2006;56:797–800. doi: 10.1099/ijs.0.63961-0. [DOI] [PubMed] [Google Scholar]
- Slobodkin A, Reysenbach AL, Mayer F, Wiegel J. Isolation and characterization of the homoacetogenic thermophilic bacterium Moorella glycerini sp. nov. Int J Syst Bacteriol. 1997;47:969–974. doi: 10.1099/00207713-47-4-969. [DOI] [PubMed] [Google Scholar]
- Slobodkin AI, Sokolova TG, Lysenko AM, Wiegel J. Reclassification of Thermoterrabacterium ferrireducens as Carboxydothermus ferrireducens comb. nov., and emended description of the genus Carboxydothermus. Int J Syst Evol Microbiol. 2006;56:2349–2351. doi: 10.1099/ijs.0.64503-0. [DOI] [PubMed] [Google Scholar]
- Sokolova TG, González JM, Kostrikina NA, et al. Carboxydobrachium pacificum gen. nov., sp. nov., a new anaerobic, thermophilic, CO-utilizing marine bacterium from Okinawa Trough. Int J Syst Evol Microbiol. 2001;51:141–149. doi: 10.1099/00207713-51-1-141. [DOI] [PubMed] [Google Scholar]
- Sokolova TG, Kostrikina NA, Chernyh NA, et al. Carboxydocella thermautotrophica gen. nov., sp. nov., a novel anaerobic, CO-utilizing thermophile from a Kamchatkan hot spring. Int J Syst Evol Microbiol. 2002;52:1961–1967. doi: 10.1099/00207713-52-6-1961. [DOI] [PubMed] [Google Scholar]
- Sokolova TG, González JM, Kostrikina NA, et al. Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park. Int J Syst Evol Microbiol. 2004;54:2353–2359. doi: 10.1099/ijs.0.63186-0. [DOI] [PubMed] [Google Scholar]
- Sokolova TG, Jeanthon C, Kostrikina NA, et al. The first evidence of anaerobic CO oxidation coupled with H2 production by a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Extremophiles. 2004;8:317–323. doi: 10.1007/s00792-004-0389-0. [DOI] [PubMed] [Google Scholar]
- Sokolova TG, Henstra A-M, Sipma J, et al. Diversity and ecophysiological features of thermophilic carboxydotrophic anaerobes. FEMS Microbiol Ecol. 2009;68:131–141. doi: 10.1111/j.1574-6941.2009.00663.x. [DOI] [PubMed] [Google Scholar]
- Srinivas TNR, Bhaskar YV, Bhumika V, Kumar PA. Photobacterium marinum sp. nov., a marine bacterium isolated from a sediment sample from Palk Bay India. Syst Appl Microbiol. 2013;36:160–165. doi: 10.1016/j.syapm.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kishigami T, Inoue K, et al. Bacillus thermoglucosidasius sp. nov., a new species of obligately Thermophilic Bacilli. Syst Appl Microbiol. 1983;4:487–495. doi: 10.1016/S0723-2020(83)80006-X. [DOI] [PubMed] [Google Scholar]
- Svetlichny VA, Sokolova TG, Gerhardt M, et al. Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir Island. Syst Appl Microbiol. 1991;14:254–260. doi: 10.1016/S0723-2020(11)80377-2. [DOI] [Google Scholar]
- Takahashi S, Tomita J, Nishioka K, et al. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS One. 2014;9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techtmann SM, Colman AS, Robb FT. “That which does not kill us only makes us stronger”: the role of carbon monoxide in thermophilic microbial consortia. Environ Microbiol. 2009;11:1027–1037. doi: 10.1111/j.1462-2920.2009.01865.x. [DOI] [PubMed] [Google Scholar]
- Techtmann SM, Lebedinsky AV, Colman AS, et al. Evidence for horizontal gene transfer of anaerobic carbon monoxide dehydrogenases. Front Microbiol. 2012;3:1–16. doi: 10.3389/fmicb.2012.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov SV, Lebedinsky AV, Sokolova TG, et al. Genomic insights into energy metabolism of Carboxydocella thermautotrophica coupling hydrogenogenic CO oxidation with the reduction of Fe(III) minerals. Front Microbiol. 2018;9:1759. doi: 10.3389/fmicb.2018.01759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Agbo P, Warnecke F, et al. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008;2:1146–1156. doi: 10.1038/ismej.2008.48. [DOI] [PubMed] [Google Scholar]
- Wu M, Ren Q, Durkin S, et al. Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 2005;1:e65. doi: 10.1371/journal.pgen.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y, Yoshida T, Kawaichi S, et al. Carboxydothermus pertinax sp. nov., a thermophilic, hydrogenogenic, Fe(III)-reducing, sulfur-reducing carboxydotrophic bacterium from an acidic hot spring. Int J Syst Evol Microbiol. 2012;62:1692–1697. doi: 10.1099/ijs.0.031583-0. [DOI] [PubMed] [Google Scholar]
- Yoneda Y, Yoshida T, Daifuku T, et al. Quantitative detection of carboxydotrophic bacteria Carboxydothermus in a hot aquatic environment. Fundam Appl Limnol. 2013;182:161–170. doi: 10.1127/1863-9135/2013/0374. [DOI] [Google Scholar]
- Yoneda Y, Yoshida T, Yasuda H, et al. A thermophilic, hydrogenogenic and carboxydotrophic bacterium, Calderihabitans maritimus gen. nov., sp. nov., from a marine sediment core of an undersea caldera. Int J Syst Evol Microbiol. 2013;63:3602–3608. doi: 10.1099/ijs.0.050468-0. [DOI] [PubMed] [Google Scholar]
- Yoneda Y, Kano SI, Yoshida T, et al. Detection of anaerobic carbon monoxide-oxidizing thermophiles in hydrothermal environments. FEMS Microbiol Ecol. 2015;91:fiv093. doi: 10.1093/femsec/fiv093. [DOI] [PubMed] [Google Scholar]
- Zavarzina DG, Sokolova TG, Tourova TP, et al. Thermincola ferriacetica sp. nov., a new anaerobic, thermophilic, facultatively chemolithoautotrophic bacterium capable of dissimilatory Fe(III) reduction. Extremophiles. 2007;11:1–7. doi: 10.1007/s00792-006-0004-7. [DOI] [PubMed] [Google Scholar]
- Zeigler DR. The Geobacillus paradox: why is a thermophilic bacterial genus so prevalent on a mesophilic planet? Microbiology. 2014;160:1–11. doi: 10.1099/mic.0.071696-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.