Significance
Mitochondrial DNA (mtDNA) mutations have been associated with common metabolic and degenerative disease phenotypes, implying a bioenergetic etiology for these diseases. For example, the mtDNA tRNALeu(UUR) m.3243A > G mutation manifests as diabetes, neurodegenerative disease, or lethal pediatric disease, depending on the percentage of mutant mtDNAs within the cell (heteroplasmy). Cultured cybrid cell lines harboring 3243G heteroplasmy levels corresponding to the different clinical phenotypes have distinct transcriptional profiles. Exhaustive metabolomic and histone posttranscriptional modification analysis of these 3243G cybrids revealed that changes in mtDNA heteroplasmy cause changes in mitochondrial intermediates and redox state, which result in distinctive histone modification changes. Thus, changes in the mitochondrial genotype change mitochondrial metabolism, which change the epigenome and transcriptome, which induce distinct clinical phenotypes.
Keywords: mitochondria, epigenetics, metabolism, transcription, common diseases
Abstract
Diseases associated with mitochondrial DNA (mtDNA) mutations are highly variable in phenotype, in large part because of differences in the percentage of normal and mutant mtDNAs (heteroplasmy) present within the cell. For example, increasing heteroplasmy levels of the mtDNA tRNALeu(UUR) nucleotide (nt) 3243A > G mutation result successively in diabetes, neuromuscular degenerative disease, and perinatal lethality. These phenotypes are associated with differences in mitochondrial function and nuclear DNA (nDNA) gene expression, which are recapitulated in cybrid cell lines with different percentages of m.3243G mutant mtDNAs. Using metabolic tracing, histone mass spectrometry, and NADH fluorescence lifetime imaging microscopy in these cells, we now show that increasing levels of this single mtDNA mutation cause profound changes in the nuclear epigenome. At high heteroplasmy, mitochondrially derived acetyl-CoA levels decrease causing decreased histone H4 acetylation, with glutamine-derived acetyl-CoA compensating when glucose-derived acetyl-CoA is limiting. In contrast, α-ketoglutarate levels increase at midlevel heteroplasmy and are inversely correlated with histone H3 methylation. Inhibition of mitochondrial protein synthesis induces acetylation and methylation changes, and restoration of mitochondrial function reverses these effects. mtDNA heteroplasmy also affects mitochondrial NAD+/NADH ratio, which correlates with nuclear histone acetylation, whereas nuclear NAD+/NADH ratio correlates with changes in nDNA and mtDNA transcription. Thus, mutations in the mtDNA cause distinct metabolic and epigenomic changes at different heteroplasmy levels, potentially explaining transcriptional and phenotypic variability of mitochondrial disease.
The mtDNA is maternally inherited and codes for 13 essential subunits of the mitochondrial energy generating system, oxidative phosphorylation (OXPHOS), plus the 2 rRNAs for the mitochondrial, chloramphenicol (CP)-sensitive, ribosome and the 22 tRNAs required for mitochondrial mRNA translation. Mutations in the mtDNA tRNAs impair the biogenesis of all mtDNA-encoded OXPHOS enzyme complexes, but particularly compromise complex I (NADH:CoQ oxidoreductase), which incorporates 7 of the 13 mtDNA encoded polypeptides (1).
The overall prevalence of mtDNA mutations is about 1 in 5,000 (2, 3). This high prevalence of mtDNA diseases results from the high mtDNA mutation rate, which is reflected in 1 in 200 cord bloods harboring 1 of the 10 most common pathogenic mtDNA mutations (4, 5) and 1 in 8 adults harboring a disease-associated mutation at ≥1% heteroplasmy (6).
One of the most common pathogenic mtDNA mutations is the tRNALeu(UUR) m.3243 A > G mutation, which causes starkly different clinical phenotypes depending on the heteroplasmy levels (1). At 20% to 30% m.3243G mutant, patients commonly present with type 1 or type 2 diabetes or manifest autism. At 50% to 80% mutant, patients present with myopathy, cardiomyopathy, lactic acidosis, and stroke-like episodes, presenting separately or when in aggregate referred to as the MELAS syndrome. At 90% to 100% mutant, individuals manifest perinatal lethal diseases such as Leigh syndrome.
To investigate the molecular basis of the relationship between the mtDNA heteroplasmy levels and shifts in patient phenotype, we prepared transmitochondrial cybrids with the same nucleus, but with different heteroplasmic percentages of the tRNALeu(UUR) m.3243G mutation. Cell biological and biochemical characterization of these cybrid cell lines revealed that the physiological parameters of the cybrids mirrored those previously observed in the patients with the same ranges of m.3243G heteroplasmy. Transcriptional analysis by RNA sequencing revealed that cybrids with similar heteroplasmy levels corresponding to a set of clinical phenotypes (e.g., 50% to 80% m.3243G associated with neurodegenerative disease) had a similar transcriptional profile, which was strikingly different from transcriptional profiles of cybrids with different heteroplasmy (SI Appendix, Fig. S1). Therefore, the marked shifts in the patient phenotypes associated with the different levels of m.3243G heteroplasmy correlate with transcriptional phase shifts in the nuclear gene expression (7).
The correlation among m.3243G mutant load, transcriptional profile, and clinical phenotype suggests that nDNA transcription responds to signals generated from the mitochondrion. One likely source of such signals are the high-energy mitochondrial intermediates that are the substrates for the posttranslational histone-modifying enzymes. These include ATP for phosphorylation, NAD+ for sirtuin-mediated deacetylation, folates for S-adenosylmethionine (SAM) synthesis for methylation, and tricarboxylic acid (TCA) cycle intermediates including acetyl-CoA for histone acetylation and α-ketoglutarate (αKG) for histone demethylation (8–10).
To determine whether mtDNA variation affects the nuclear epigenome, we correlated mtDNA tRNALeu(UUR) m.3243 A > G cybrids’ metabolic and redox status with changes in the complete array of nuclear histone modifications. We found multiple associations between metabolites and histone modifications as the level of m.3243 A > G heteroplasmy increased, of which some were observed only at a particular heteroplasmy level. Thus, mtDNA genotype regulates the epigenome through mitochondrial metabolism.
Results
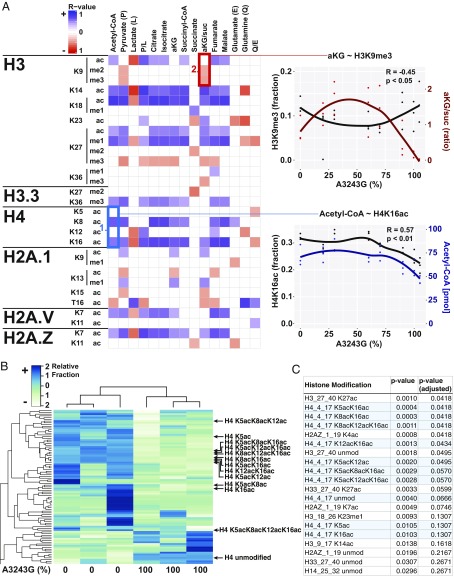
To determine the mitochondrial signals that may regulate the epigenome, we quantified TCA cycle intermediates across our 7 mtDNA tRNALeu(UUR) nt 3243 A > G patient-derived cybrid lines with heteroplasmy levels of ∼0%, 20%, 30%, 60%, 70%, 90%, and 100% m.3243G mutant, but with the same nuclear DNA. We then correlated the cybrid metabolite levels with all nuclear histone modifications assessed by liquid chromatography mass spectrometry (SI Appendix, Fig. S2). The result was an extensive matrix of statistically significant correlations between mitochondrially generated metabolites and histone modifications across the heteroplasmy spectrum (Fig. 1A), confirming broad spectrum control of nuclear histone modifications by mitochondrial metabolism.
Fig. 1.
MtDNA heteroplasmy modulates metabolites and histone modifications. (A) Seven mtDNA m.3243A > G heteroplasmic cell lines plus the ρ0 parent cells were cultured in parallel for metabolite (j = 15) and histone modification (i = 107) quantification by liquid chromatography tandem mass spectrometry. Spearman correlation coefficient was calculated for each histone modification (unmod = unmodified peptide) against each metabolite (blue = positive, red = negative, and blank = no correlation), with the color intensity corresponding to correlation strength (significance = P < 0.05). (Example #1, blue box) Positive association between acetyl-CoA levels and histone H4 acetylation of lysines 8 (K8) and 16 (K16) across heteroplasmy levels. (Example #2, red box) Negative association between αKG/succinate ratio and histone H3 lysine 9 di- (me2) and trimethylation (me3) across heteroplasmy levels. Each point represents a measurement and lines represent local regression of the mean (R-software loess), n = 3. (B) Heat map of histone acetylation modifications of 0% and 100% m.3243G lines (n = 3), showing a dramatic effect of mtDNA genotype on the histone epigenome. (C) List of top 20 histone modifications ranked by P value, showing the strong effect on mtDNA genotype on H4K5, K8, K12, and K16 acetylation (n = 3). Although it can be argued that modification level changes at a particular site are not independent of either other changes at that site or overall changes in the associated peptide, we also include P values with an arbitrary correction factor, allowing a false discovery rate of 1% (adjusted P value).
To test whether the differential abundance of the mitochondrial metabolites causes the changes in the histone modifications, we focused on 2 relationships: a positive association between acetyl-CoA and histone H4 lysines 8 (H4K8) and 16 (H4K16) acetylation (Fig. 1A, blue square #1), and a negative association between the αKG/succinate ratio and histone H3 lysine 9 (H3K9) methylation (Fig. 1A, red square #2). The first association allowed direct examination of acetyl-CoA effect on histone acetylation, as histone H4 tail residues (K5/K8/K12/K16) cannot be methylated. The second association permitted analyzing the effect of αKG level on methylation, as αKG/succinate ratio correlated with H3K9 di- and trimethylation (H3K9me2/3), but not H3K9 acetylation.
Strikingly, these 2 associations show different behaviors across the m.3243G mutant heteroplasmy spectrum. Acetyl-CoA levels and H4K16 acetylation were stable from 0% to 70% mutant mtDNAs, but then declined together at 90% to 100% mutant (R = 0.57; Fig. 1 A, Bottom Right). This decline was confirmed for all H4 residues in independent experiments (Fig. 1 B and C and SI Appendix, Fig. S3 A–C). In contrast, the αKG/succinate ratio was low at 0% mutant, increased in 30% to 70% mutant, and declined again in 90% to 100% mutant cells. The H3K9 di- and trimethylation showed the inverse pattern, being high at 0% mutant, low at 50% to 70% mutant, and high at 90% to 100% mutant (R = −0.45; Fig. 1 A, Top Right). αKG alone showed a similar pattern (SI Appendix, Fig. S3D). The inverse αKG-histone methylation association is expected, as Jumonji C (JmjC) histone demethylases known to be active on H3K9 require αKG to demethylate histones and are inhibited by succinate (11).
Because acetyl-CoA was significantly decreased in 100% mutant cells (Fig. 1A #1 and SI Appendix, Fig. S4 C, Right), but can be generated from multiple sources (12), we sought to determine the contribution of mitochondrial metabolism of glucose in the generation of acetyl-CoA and acetylation of histone H4. We incubated 0% and 100% mutant cells with uniformly 13C -labeled glucose and traced the label through pyruvate, mitochondrial generation and exportation to the cytosol of citrate, conversion of cytosolic citrate into acetyl-CoA and oxaloacetate by ATP-citrate lyase (13), and use of the mitochondrially derived acetyl-CoA for histone acetylation (Fig. 2A). We found that about 70% of acetyl-CoA was derived from 13C-labeled glucose, and that this was reduced to 45% in the 100% mutant cell line (Fig. 2B), as in 100% mutant cells, much of the pyruvate was metabolized to lactate, thus limiting acetyl-CoA production (SI Appendix, Fig. S4 A–C).
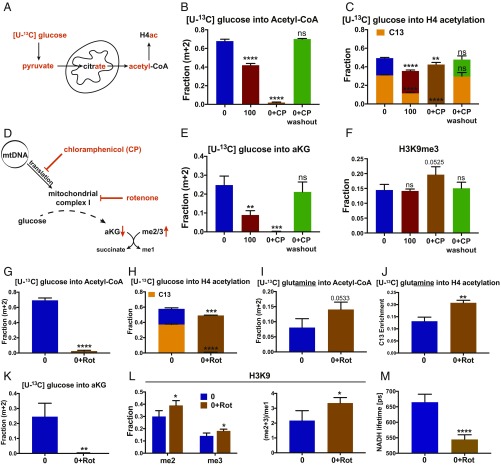
Fig. 2.
Mitochondria as key supplier of epigenetic metabolites. (A) Mitochondrial metabolism of 13C glucose to produce nuclear/cytosolic acetyl-CoA. (B) Conversion of 13C-glucose into acetyl-CoA in 0% (0) and 100% (100) m.3243G cells and in 0% cells treated with CP (0+CP) without or with CP washout. (C) Steady-state (total) histone 4 acetylation (bar height) and 13C-glucose H4 acetylation (orange shading) in 0% and 100% heteroplasmy cells and 0% cells with CP. (D) Effect of CP and rotenone (Rot) on complex I, the TCA cycle, αKG levels, and histone JmjC demethylation. (E) Conversion of 13C-glucose to αKG, using the same labeling protocols as B). (F) Steady-state (total) histone 3 lysine 9 trimethylation, using the protocols of B). (G) 13C-glucose incorporation into acetyl-CoA in 0% mutant cells, without and with rotenone (Rot) inhibition. (H) Steady-state (total) (bar height) and 13C-glucose derived (orange bar) H4 acetylation without and with Rot inhibition. (I) 13C-glutamine incorporation into acetyl-CoA without and with Rot inhibition. (J) 13C-glutamine incorporation into H4 acetylation without and with Rot inhibition. (K) Conversion of 13C-glucose into αKG without and with Rot inhibition. (L) H3K9 di- and trimethylation (Left) and poly/monomethylation ratio (Right) of 0% cells without and with Rot inhibition. (M) Mitochondrial NAD+/NADH ratio determined by NADH lifetime in 0% cells without and with Rot inhibition; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant; n = 3.
To confirm that decreased incorporation of glucose-derived 13C into acetyl-CoA was a result of the mitochondrial protein synthesis defect, we treated the 0% m.3243G cybrid cells for 7 d with CP to completely block mitochondrial protein synthesis of the 0% mutant cells. CP reduced the transfer of glucose-derived 13C into acetyl-CoA to ∼2%. Therefore, mitochondria are the major source of acetyl-CoA, and impaired mitochondrial protein synthesis limits mitochondrial acetyl-CoA production. This was confirmed by wash out of CP, resulting in the complete restoration of the incorporation of glucose-derived 13C into acetyl-CoA (Fig. 2B).
In parallel with the reduced mitochondrial incorporation of glucose-derived 13C into acetyl-CoA, we assessed the effects of 100% mutant and CP on the acetylation of histone H4 (Fig. 2C). In the bicolored bar graphs, the total bar height reflects total histone acetylation levels. The different colored subbars represent the time-limited incorporation of the 13C from the labeled precursor. Both the total steady state histone H4 and 13C-glucose-derived H4 acetylation were decreased from 50% and 35% in 0% mutant cells to 37% and 15% in 100% mutant cells, respectively (Fig. 2C). When 0% mutant cells were treated with CP, steady state H4 acetylation was decreased to 40%, but the 13C-glucose-derived acetylation was eliminated. Following washout of CP, the total steady state H4 acetylation returned to 50% and the glucose-derived 13C H4 acetylation returned to 35% (Fig. 2C). Thus, impaired mitochondrial protein synthesis caused by either 100% 3243G mutant mtDNAs or CP inhibition resulted in the depletion of glucose-derived acetyl-CoA and in the reduction of histone H4 acetylation. Mitochondrially derived acetyl-CoA is thus the primary substrate for histone acetylation.
Impairment of mitochondrial protein synthesis resulting from 100% m.3243G mutant mtDNAs or CP inhibition should impair complex I, and thus the mitochondrial oxidation of NADH. The resulting decrease in NAD+ should stall the TCA cycle, reducing mitochondrial production of αKG. Because αKG is the required substrate for the JmjC histone demethylases (14), histone demethylation should be impaired by a protein synthesis defect resulting in increased histone methylation (Fig. 2D). Both 100% mutant and CP-treated 0% mutant cells showed decreased incorporation of glucose-derived 13C into αKG, which was restored to normal in CP-treated 0% mutant cells after CP washout (Fig. 2E). Moreover, although 100% mutant cells did not show an increase in H3K9 trimethylation, likely as a result of decreased SAM production (SI Appendix, Fig. S4D), the 0% mutant cells treated with CP showed significantly increased levels of H3K9 trimethylation, which returned to normal on CP washout (Fig. 2F). Thus, inhibition of mitochondrial protein synthesis results in reduced αKG and leads to increased histone methylation, presumably as a result of limiting JmjC-mediated histone demethylation.
To further evaluate the role of NADH oxidation and TCA cycle-mediated acetyl-CoA production, we determined the effect of rotenone, a potent complex I inhibitor, on 13C-glucose-derivation of acetyl-CoA. Rotenone completely eliminated the incorporation of glucose-derived 13C into acetyl-CoA (Fig. 2G) and resulted in a significant decrease in the steady state acetylation of H4 histone residues, as well as complete elimination of incorporation of glucose-derived 13C into H4 acetylation (Fig. 2H and SI Appendix, Fig. S5 A–F).
Because acetyl-CoA is essential, depletion of acetyl-CoA by rotenone inhibition of complex I must be compensated. This could occur by the conversion of glutamine to glutamate and then αKG, with αKG being converted to acetyl-CoA by either entering the TCA cycle to generate citrate or by reductive carboxylation (15). When labeling cells with glutamine-derived 13C, 0% mutant cells produced about 8% of the acetyl-CoA from glutamine. However, after rotenone inhibition this rose to about 14% (Fig. 2I). In parallel, glutamine-derived 13C incorporated into H4 acetylation increasing from about 14% in 0% mutant cells to about 21% after rotenone inhibition (Fig. 2J and SI Appendix, Fig. S5 G and H). Thus, glutamine can make a small contribution to acetyl-CoA for histone acetylation.
Rotenone not only depleted glucose-derived 13C incorporation into αKG (Fig. 2K) but also resulted in elevated levels of di- and trimethylation and in the di- and trimethylation to monomethylation ratio of H3K9 (Fig. 2L). Thus, mitochondrially generated αKG must play a role in the activity of the JmjC histone demethylases.
NAD+/NADH ratio is crucial in regulating TCA metabolism, and was reduced in 100% mutant cells (SI Appendix, Fig. S4 C, Bottom Left), but it is also known to vary between cellular compartments. To assess subcellular levels of NAD+/NADH ratios, we employed fluorescence lifetime imaging microscopy (FLIM) to quantify NADH lifetimes in the mitochondria and the nucleus of the cybrid cell lines. By FLIM, decreased NADH lifetime correlates with increased free NADH and decreased NAD+/NADH ratio (16).
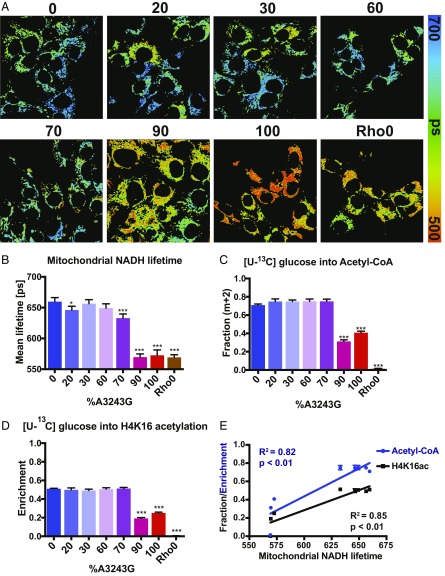
In the mitochondria, the NAD+/NADH ratio was relatively stable from 0% to 60% heteroplasmy. It began to decline at 70% mutant and then dropped precipitously at 90% and 100% mutant (Fig. 3 A and B). Similarly, glucose-derived acetyl-CoA was sustained in 0% to 70% mutant cells, but then dropped precipitously at 90% to 100% m.3243G mutant heteroplasmy (Fig. 3C). In parallel, glucose-derived H4 acetylation was sustained up to 70% mutant, followed by a drop in the 90% to 100% mutant cells (Fig. 3 D and E).
Fig. 3.
Mitochondrial NADH lifetime regulates glucose incorporation into acetyl-CoA and histone 4 lysine 16 acetylation. (A) Pseudocolored images mitochondrial NADH lifetime analysis using FLIM across cybrid heteroplasmy levels. (B) Quantification of mitochondrial NADH lifetimes (NAD+/NADH ratios) across cybrid heteroplasmy levels. (C) Conversion of 13C-glucose into acetyl-CoA across cybrid heteroplasmy levels. (D) 13C-glucose incorporation into H4K6 acetylation across cybrid heteroplasmy levels. (E) Relationship between NADH lifetime (NAD+/NADH) and 13C-glucose conversion to acetyl-CoA and H4K16 acetylation, showing strong correlation across cybrid heteroplasmy levels (Spearman), n = 3.
When impairment of mitochondrial protein synthesis reaches the level caused by 100% 3243G mutant, respiration is impaired (SI Appendix, Fig. S6B), the mitochondria can no longer oxidize NADH back to NAD+, the TCA cycle is inhibited, mitochondrial production of glucose-derived acetyl-CoA is reduced, and histone acetylation is limited. This limitation is independent of glucose concentration in the media (SI Appendix, Fig. S7).
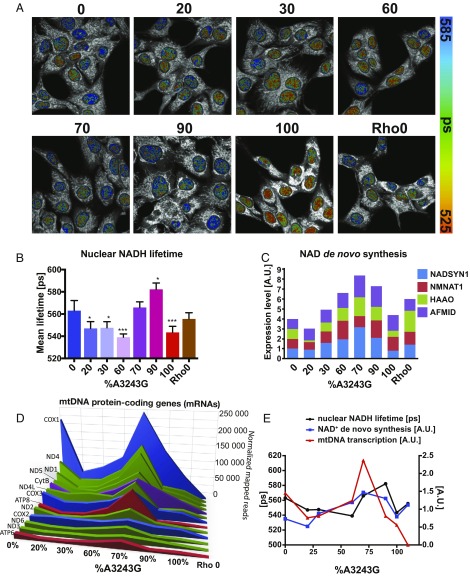
In the nucleus, the NAD+/NADH ratio declined sharply from that of the 0% mutant cells, reaching a minimum in 20% to 60% mutant cells (Fig. 4 A and B). The nuclear NAD+/NADH ratio then rose abruptly between 60% and 70% mutant cells to levels well in excess of those of the 0% mutant cells. Finally, the NAD+/NADH ratio declined again at 100% mutant (Fig. 4B). The sudden rise in NAD+/NADH ratio in the 60% to 70% mutant cells correlated with the induction of genes for de novo NAD+ synthesis, starting at 30% mutant, peaking at 70% mutant, and then declining to approximately the 0% level at 100% mutant (Fig. 4C). The unusual behavior of the nuclear NAD+/NADH ratio exactly corresponds to the striking changes that occurred in the levels of the mtDNA OXPHOS gene mRNAs (Fig. 4 D and E), suggesting that the nuclear NAD+ levels may regulate the activity of the mitochondrial biogenesis genes. Nuclear NAD+ levels also correlated with the phase changes in nDNA gene transcription seen between 20% to 30% versus 60% to 90% mutant cells (SI Appendix, Fig. S1). The induction of NAD+ synthesis may be a final attempt to reestablish a functional NAD+/NADH redox balance and stimulate mitochondrial OXPHOS.
Fig. 4.
Nuclear NADH lifetime correlates with mtDNA transcription and NAD+ biosynthesis gene induction. (A) Pseudocolored images mitochondrial NADH lifetime analysis using FLIM across cybrid heteroplasmy levels. (B) Quantification of nuclear NADH lifetimes (NAD+/NADH ratio) across cybrid heteroplasmy levels. (C) mRNA levels of key NAD+ de novo synthesis enzymes across cybrid heteroplasmy levels (AFMID, Arylformamidase; NADSYN1, NAD Synthetase 1 [rate-limiting]; HAAO, 3-hydroxyanthranilate 3,4-dioxygenase NMNAT1, Nicotinamide Nucleotide Adenylyltransferase 1). (D) mtDNA mRNA levels across cybrid heteroplasmy levels. Reproduced from ref. 7. (E) Parallel fluctuations of Nuclear NADH lifetime (black, left axis), normalized NADH synthesis mRNA levels (blue), and normalized mtDNA mRNA levels (red) across cybrid heteroplasmy levels; n = 3.
Discussion
Our work suggests that mtDNA heteroplasmy can result in discrete cellular transcriptional profiles and clinical phenotypes by simultaneously modulating multiple metabolites key to epigenetic signaling (SI Appendix, Fig. S8). In contrast to wild-type 0% mutant cells (SI Appendix, Fig. S8, State A), in the 20% to 30% tRNALeu(UUR) m.3243 A > G mutant cells, a decline in the nuclear NAD+/NADH ratio is associated with a decline in mtDNA transcripts and a switch from the 0% mutant transcriptional profile to that of the 20% to 30% m.3243 A > G mutant cells (SI Appendix, Figs. S1 and S8, State B). This correlates with the chronic mitochondrial energy deficiency observed in type 2 diabetes (17, 18) and autism (19). In the 60% to 70% m.3243 A > G mutant cells, the rise in nuclear NAD+ levels, corresponding to the induction of NAD+ synthesis, is paralleled by an increase in αKG levels and a decline in histone methylation. This correlates with the hyperexpression of the mtDNA genes and the transition to the next discrete cellular transcriptional profile (SI Appendix, Fig. S1). This transcription profile corresponds to neuromuscular degenerative disease manifestations and the KEGG-identified transcriptional profiles of Alzheimer, Parkinson, and Huntington diseases (Figure 5D in ref. 7 and SI Appendix, Fig. S8, State C). Finally, in the 90% to 100% mtDNA tRNALeu(UUR) m.3243 A > G mutant cells, the drop in acetyl-CoA levels in association with the decline in histone acetylation is associated with a drop in mtDNA transcripts and the final transition in transcriptional profile (SI Appendix, Figs. S1 and S8, State D), this being associated with perinatal lethal disease (7). Thus, increasing severity of the m.3243G mtDNA genetic defect directly affects mitochondrial metabolites and subcellular redox levels. Our data suggest that these changes cause discrete changes in the epigenomic state that correlate with altered gene transcription and associated clinical manifestations.
Methods
The mtDNA tRNALeu(UUR) nt 3243A > G Mutation Cybrids.
Cell lines harboring different percentages of the mtDNA tRNALeu(UUR) nt 3243A > G mutation were prepared by fusion of cytoplasts from a heteroplasmic mtDNA tRNALeu(UUR) nt 3243A > G donor (20) to human 143B(TK−) ρ0 cells. Transmitochondrial cybrid cell lines were maintained in DMEM (4.5 g/L [25 mM] glucose + 110 mg/L sodium pyruvate [Gibco #10569]) supplemented with 50 μg/mL uridine, nonessential amino acids (1:100, Gibco #11140), and 10% FBS, grown at 37 °C in 5% CO2 (21).
Cybrid mtDNA tRNALeu(UUR) nt 3243A > G heteroplasmy levels were quantified by PCR amplification of a 264-nt fragment surrounding the 3243 site and digestion with HaeIII. The cybrid heteroplasmy levels were remarkably stable (SI Appendix, Fig. S9).
Cells for analysis of metabolites and histones were grown to ∼80% confluent in DMEM supplemented with 10% FBS, 1 mM glucose, 2 mM glutamine, and 50 ng/L uridine (experimental media). For isotopic experiments, 13C-uniformly labeled glucose or glutamine and dialyzed serum were employed.
Mitochondrial protein synthesis was inhibited with 100 μg/mL CP for 7 d (22), with inhibition reversed by growth without CP for 3 d. Complex I was inhibited with 1 μM rotenone for 4 h.
Glucose and lactate consumption were analyzed by incubating cells for 4, 8, or 24 h, the medium collected, the glucose and lactate measurement using YSI 2950 Bioanalyzer, and the levels corrected for cell proliferation and media evaporation. Oxygen consumption of digitonin permeabilized cells was analyzed using the Oroboros instrument with 106 cells/mL in 2.2 mL of Miro5 buffer (0.5 mM EGTA, 3 mM MgCl2 0.6 H2O, 60 mM K‐lactobionate, 20 mM Taurine, 10 mM KH2PO4, 20 mM Hepes, 110 mM Sucrose, 1 g/L fatty acid free BSA at pH 7.0 with KOH). Substrates and complex inhibitors were added as in SI Appendix, Fig. S6.
Acetyl-CoA/Free CoA, SAM, and Organic Acid Measurement.
Cells were cultured for 12 h in DMEM with 4.5 g/L glucose, 10% FBS, 50 μg/mL uridine, 1 mM sodium pyruvate, 1× NEAA and 2 mM glutamine; harvested; and frozen for 10 s in liquid nitrogen and stored in −80 °C. The concentration of amino acids was determined by Agilent 1260 HPLC system, using precolumn derivatization with o-phthalaldehyde (23). Short-chain Acyl-CoAs and SAM levels were determined by Agilent LC/MS 6410 Triple Quad system using 872-365 multiple reaction monitoring (MRM) for Benzoyl-CoA (Internal standard), 868-361 MRM for Succinyl-CoA, 854-347 MRM for Malonyl-CoA, 838-331 MRM for Butyryl-CoA, 824-317 MRM for Propionyl-CoA, 810-303 MRM for Acetyl-CoA, 768-261 for free CoA, and 399-250 MRM for SAM. Organic acids levels were determined by the isotope-dilution approach and GC-MS system (24).
Acyl-CoA and Polar Metabolite Quantification and Isotopologue Analysis.
After 16 h growth in normal media, cells were incubated in experimental media containing labeled metabolites for 4 h. For acyl-CoA quantification, the attached cells were fixed by the addition of 1 mL ice-cold 10% (wt/vol) trichloroacetic acid, harvested by scrapping, disrupted by pulse-sonication (sonic dismembrator, Fisher Scientific), and the protein sedimented at 17,000 rcf for 10 min at 4 °C. Supernatants were purified using Oasis HLB 1-mL (30 mg) solid-phase extraction columns (Waters), conditioned with methanol and water. The supernatants were applied to the columns, the columns washed with 1 mL H2O, the analytes eluted with methanol containing 25 mM ammonium acetate, dried with N2 gas, and resuspended in 50 µl of 5% 5-sulfosalicylic acid. For quantification, 0.1 mL of internal standard was added containing [13C315N1]-labeled acyl-CoAs generated in pan6-deficient yeast culture (25). All other polar analytes were quantified after cell extraction with 80:20 methanol:water at −80 °C, sonication, protein removal, drying with N2 gas, and resuspension in 5-sulfosalicylic acid. Samples were analyzed using an Ultimate 3000 Quaternary UHPLC coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) operating in the positive ion mode (26, 27). For 13C tracing experiments, isotopic enrichment was calculated using the FluxFix online calculator (28).
Histone Extraction and Western Blot Analysis.
For histone extraction, cells were switched to experimental media, washed with ice-cold PBS, and 0.5 mL of nuclei isolation buffer (NIB-250) added and incubated for 5 min. NIB-250 is 15 mM Tris⋅HCl at pH 7.5, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, and 250 mM sucrose, to which was added 1 mM DTT, 10 mM sodium butyrate, 1× Protein Inhibitor Mixture (Sigma), and 0.1% Nonidet P-40 (Sigma). Cells were harvested by scrapping, pelleted at 600 rcf for 5 min at 4 °C, washed twice with NIB-250 without Nonidet P-40, and pelleted. Histones were extracted in 0.8 mL 0.4 N H2SO4 for 2 h at 4 °C, the insoluble fraction pelleted at 11,000 rcf for 10 min in 4 °C, and the soluble fraction containing histones precipitated with 0.2 mL 100% TCA at 4 °C overnight. The film-like pellet of histone was sedimented at 11,000 rcf for 10 min at 4 °C, washed with ice-cold 1 mL acetone with 0.1% HCl plus 2 washes in ice-cold pure acetone, air-dried, and stored at −80 °C for Western blotting or mass spectrometry analysis.
For Western blotting, 0.5 to 1 μg of histone extract was electrophoresed on a gradient 4% to 12% Bis-Tris gel at 120 V for 60 min, and the proteins transferred onto a 0.2 cellulose membrane in methanol buffer overnight. Membranes were blocked in 5% dry milk, incubated in primary antibody (H4K5ac, Millipore 07-327; H4, ab10158; H3, ab1791) for 2 h, washed 3 times incubated with secondary horseradish-peroxidase conjugated antibody for 1 h, and peroxidase stained (Biorad Gel-Doc XR).
Histone Modification Quantification and Isotopologue Analysis.
Histones were extracted from cells incubated in experimental media containing labeled metabolites for 4 h. For mass spectrometer analysis, histones were derivatized using propionic anhydride, digested using trypsin, and the new N-termini were also derivatized. The resulting samples were stage-tipped and subjected to mass spectrometer analysis, using 0.1% formic acid in water and 0.1% formic acid in acetonitrile as buffers, with separation carried out on a C18 column (29). Data acquisition used high-resolution DIA and the data processed using EpiProfile software (30).
NADH FLIM.
FLIM microscopy of NADH (NADH FLIM) (16) was performed on cells grown in glass-bottom dishes (Greiner, 627870) with the medium changed to Tyrodes buffer (pH 7.4) and the cells incubated for 4 h at 37 °C in room air. The cultures were then placed in the microscope chamber 15 min before image acquisition, with FLIM imaged used a Zeiss LSM 710 laser scanning microscope equipped with a femtosecond pulsed 2-photon laser (80 MHz, Coherent). NADH was excited at 730 nm with a laser power of <5 mW and fluorescence monitored using time-correlated single photon counting (HPM-100-40, Becker and Hickl) through a 680-nm short-pass and 460/50 bandpass filter. Data were recorded using SPCM 9.77 and subsequently analyzed using SPCImage 6.2 assuming a biexponential decay with fixed τ1 = 400 ps and τ2 = 2,500 ps for free and protein-bound NADH. Proper fitting of the lifetime curve was evaluated using χ2, and the mean lifetime (τmean) was calculated.
Total Cellular NAD+/NADH Ratio Measurement.
NAD+/NADH levels were analyzed for cells grown for 12 h in DMEM with 4.5 g/L glucose, 10% FBS, 50 μg/mL uridine, 1 mM sodium pyruvate, 1× NEAA and 2 mM glutamine, then collected and frozen for 10 s in liquid nitrogen and stored in −80 °C. Frozen pellets were resuspended in Tris buffer (pH = 8) and half extracted with perchloric acid and half with alkaline buffer. After filtration, samples were loaded onto a HPLC system, using an YMC-Pack ODS-A column (5 μm, 4.6 × 250 mm) preceded by a guard column. NAD+ and NADH peak areas were integrated by the Shimadzu Lab Solution software with standard curves normalized to protein (31).
RNA-Sequencing Data Analysis.
RNA-sequence data were used to determine expression levels of the NAD+ biosynthetic genes (7), reported as reads per kilobase per million reads normalized to 0% m.3243G.
Data Analysis and Statistical Methods.
R software (3.4.2) was used for metabolite-histone correlations; Spearman correlation for correlating dependent variables across heteroplasmy spectrum; and unpaired, 2-tailed, t test (Prism software) for individual comparisons. For hierarchical clustering, P values were adjusted using the FDR method with false-positive discovery rate of 1%.
Supplementary Material
Acknowledgments
We thank Yuanchao Zhang for help in algorithm development for data analysis; Kierstin Keller for experimental suggestions; and Y. Daikhin, O. Horyn, and Ilana Nissim for performing the analysis of amino and organic acids, acyl-CoAs, and SAM levels in the Metabolomics Core Facility at the Children’s Hospital of Philadelphia. This work was supported by Howard Hughes Medical Institute Research Fellowship and Children’s Hospital of Philadelphia Foerderer Award awarded to P.K.K., Perelman School of Medicine’s Institute for Translational Medicine and Advanced Therapeutics Human Maturational Biology Grant (P.K.K., D.C.W., and B.A.G.), as well as National Institutes of Health grants NS021328, MH108592, and OD010944 and US Department of Defense grant W81XWH-16-1-0401 (to D.C.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Histone Post-Translational Modification Data: The current Chorus mass spectrometry database is labeled project 1569. It is publicly shared at the URL https://chorusproject.org/pages/dashboard.html#/projects/public/1569/experiments.
See Commentary on page 15763.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906896116/-/DCSupplemental.
References
- 1.Wallace D. C., Mitochondrial genetic medicine. Nat. Genet. 50, 1642–1649 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gorman G. S., et al. , Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol. 77, 753–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lightowlers R. N., Taylor R. W., Turnbull D. M., Mutations causing mitochondrial disease: What is new and what challenges remain? Science 349, 1494–1499 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Elliott H. R., Samuels D. C., Eden J. A., Relton C. L., Chinnery P. F., Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet. 83, 254–260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnery P. F., Elliott H. R., Hudson G., Samuels D. C., Relton C. L., Epigenetics, epidemiology and mitochondrial DNA diseases. Int. J. Epidemiol. 41, 177–187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebolledo-Jaramillo B., et al. , Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 111, 15474–15479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picard M., et al. , Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl. Acad. Sci. U.S.A. 111, E4033–E4042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace D. C., Fan W., Energetics, epigenetics, mitochondrial genetics. Mitochondrion 10, 12–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace D. C., Fan W., Procaccio V., Mitochondrial energetics and therapeutics. Annu. Rev. Pathol. 5, 297–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verdin E., NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Carey B. W., Finley L. W., Cross J. R., Allis C. D., Thompson C. B., Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S., et al. , ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep. 17, 1037–1052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellen K. E., et al. , ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukada Y., et al. , Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Wallace D. C., Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer P. M., et al. , Mitochondrial matrix pH as a decisive factor in neurometabolic imaging. Neurophotonics 4, 045004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen K. F., Dufour S., Befroy D., Garcia R., Shulman G. I., Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 350, 664–671 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen K. F., et al. , Mitochondrial dysfunction in the elderly: Possible role in insulin resistance. Science 300, 1140–1142 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalkia D., et al. , Association between mitochondrial DNA haplogroup variation and autism spectrum disorders. JAMA Psychiatry 74, 1161–1168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heddi A., Stepien G., Benke P. J., Wallace D. C., Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J. Biol. Chem. 274, 22968–22976 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Trounce I. A., Kim Y. L., Jun A. S., Wallace D. C., Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 264, 484–509 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Wallace D. C., Bunn C. L., Eisenstadt J. M., Cytoplasmic transfer of chloramphenicol resistance in human tissue culture cells. J. Cell Biol. 67, 174–188 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissim I., et al. , The molecular and metabolic influence of long term agmatine consumption. J. Biol. Chem. 289, 9710–9729 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg J. M., Venkatachalam M. A., Roeser N. F., Nissim I., Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc. Natl. Acad. Sci. U.S.A. 97, 2826–2831 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder N. W., et al. , Production of stable isotope-labeled acyl-coenzyme A thioesters by yeast stable isotope labeling by essential nutrients in cell culture. Anal. Biochem. 474, 59–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frey A. J., et al. , LC-quadrupole/Orbitrap high-resolution mass spectrometry enables stable isotope-resolved simultaneous quantification and 13C-isotopic labeling of acyl-coenzyme A thioesters. Anal. Bioanal. Chem. 408, 3651–3658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo L., et al. , Diisopropylethylamine/hexafluoroisopropanol-mediated ion-pairing ultra-high-performance liquid chromatography/mass spectrometry for phosphate and carboxylate metabolite analysis: Utility for studying cellular metabolism. Rapid Commun. Mass Spectrom. 30, 1835–1845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trefely S., Ashwell P., Snyder N. W., FluxFix: Automatic isotopologue normalization for metabolic tracer analysis. BMC Bioinf. 17, 485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karch K. R., Sidoli S., Garcia B. A., Identification and quantification of histone PTMs using high-resolution mass spectrometry. Methods Enzymol. 574, 3–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Z. F., et al. , EpiProfile 2.0: A computational platform for processing epi-proteomics mass spectrometry data. J. Proteome Res. 17, 2533–2541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormack S., et al. , Pharmacologic targeting of sirtuin and PPAR signaling improves longevity and mitochondrial physiology in respiratory chain complex I mutant Caenorhabditis elegans. Mitochondrion 22, 45–59 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.