Significance
More than 80% of eukaryotic proteins and 67% of prokaryotic proteins contain multiple domains. Due to the technical difficulties in structural biology, however, 65.3% of solved proteins in the Protein Data Bank contain only single-domain structures. Similarly, most computational approaches are optimized for single-domain structure predictions. We propose a pipeline for assembling domain models into full-length structures with interdomain orientations constrained by an analogous structure alignment search. Large-scale benchmark tests showed an unprecedented ability of DEMO in modeling multidomain structures, with a success rate significantly beyond that of the state-of-the-art approaches built on linker modeling. This development helps bridge the significant gap between the increasingly improved ability of individual domain structure determination and the extremely high demands of community for multidomain structure models.
Keywords: protein structure prediction, domain assembly, multidomain protein, multidomain template recognition
Abstract
Most proteins exist with multiple domains in cells for cooperative functionality. However, structural biology and protein folding methods are often optimized for single-domain structures, resulting in a rapidly growing gap between the improved capability for tertiary structure determination and high demand for multidomain structure models. We have developed a pipeline, termed DEMO, for constructing multidomain protein structures by docking-based domain assembly simulations, with interdomain orientations determined by the distance profiles from analogous templates as detected through domain-level structure alignments. The pipeline was tested on a comprehensive benchmark set of 356 proteins consisting of 2–7 continuous and discontinuous domains, for which DEMO generated models with correct global fold (TM-score > 0.5) for 86% of cases with continuous domains and for 100% of cases with discontinuous domain structures, starting from randomly oriented target-domain structures. DEMO was also applied to reassemble multidomain targets in the CASP12 and CASP13 experiments using domain structures excised from the top server predictions, where the full-length DEMO models showed a significantly improved quality over the original server models. Finally, sparse restraints of mass spectrometry-generated cross-linking data and cryo-EM density maps are incorporated into DEMO, resulting in improvements in the average TM-score by 6.3% and 12.5%, respectively. The results demonstrate an efficient approach to assembling multidomain structures, which can be easily used for automated, genome-scale multidomain protein structure assembly.
Protein in cells frequently consist of multiple domains, each representing a compact and independent folding unit. Although protein domains often perform their functions separately, appropriate interdomain organization is critical to facilitate the implementation of multiple (and often related) functions in a cooperative way. Meanwhile, many biological functions rely on the interaction of different domains; for example, the protease activity of a chymotrypsin is mediated by the active sites buried at the interface between their domains (1). Statistics has shown that at least 80% of eukaryotic proteins and 67% of prokaryotic proteins contain more than 2 domains (2). Determining full-length structure of multidomain proteins is thus a crucial step in elucidating their functions and designing new drugs to regulate these functions.
However, due to the technical difficulties in structural biology, to date most of the multidomain proteins have only single domain structures solved. Currently, only 34.7% of the 608,044 protein chains solved in the Protein Data Bank (PDB) contain multiple domains. Similarly, almost all of the advanced protein structure prediction methods, such as I-TASSER (3–5), QUARK (6), and Rosetta (7, 8), are optimized for modeling single-domain proteins in both force field design and conformational search. These methods result in a significant gap between the (increasingly improved) ability of single-domain structure determination and the high demand of the biomedical community for high-resolution multidomain protein structures.
Despite the importance of multidomain structure modeling, there have been very few methods dedicated to this approach. Among them, AIDA (9) and Rosetta (10) focus mainly on construction of the linker models with domain orientations loosely constrained by some physical terms from generic hydrophobic interactions; this leaves the domain structures largely randomly oriented in the final model. Different from the linker-based methods, in a rigid-body docking approach, which we present here, the domain structures are assembled by sampling the degrees of freedom of interdomain interactions instead of the domain linkers.
Two major issues/questions should be considered in the rigid-body domain assembly approaches. First, can we deduce correct domain orientations from other known multidomain proteins, as template-based tertiary structure modeling approaches typically do? Second, can we assemble multidomain models with improved quality over the models built directly from full-length modeling simulations?
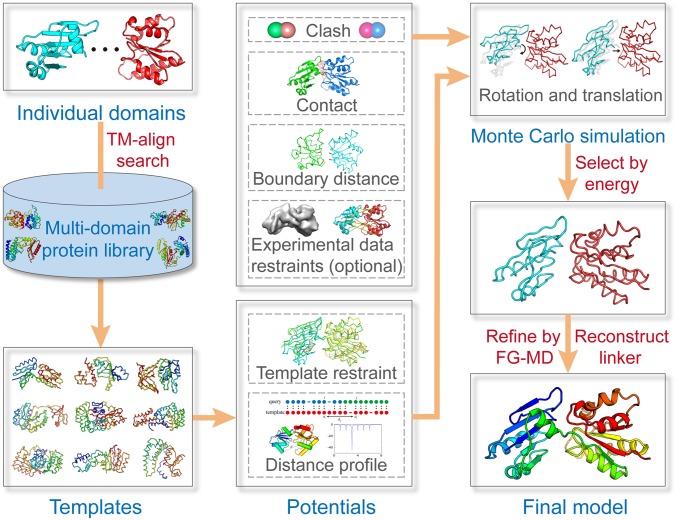
To address these questions, we have developed a domain-enhanced modeling (DEMO) approach to construct multidomain protein models by rigid-body assembling of single-domain models, with the interdomain orientations constrained by the distance profiles from templates detected through local and global structural alignments (Fig. 1). To carefully examine the strengths and weaknesses of this approach, we tested DEMO on 2 large sets of multidomain proteins containing various numbers of continuous and discontinuous domains. The results show that the analogy template-guided domain assembly approach can generate more accurate models for the majority of proteins compared with the state-of-the-art linker-based approaches. It can also create full-length structural models for proteins with partially aligned templates with a quality significantly beyond that of traditional full-length folding simulations. DEMO can be used to assemble both experimentally solved and computationally predicted domain structures. The procedure for DEMO is fully automated, and the online server and stand-alone program, together with all datasets used in the study, are freely available at https://zhanglab.ccmb.med.umich.edu/DEMO/.
Fig. 1.
Flowchart of DEMO for rigid-body protein domain assembly. Starting from individual domain structures, templates are first identified by structurally threading the domains through a nonredundant multidomain structural library using TM-align (11). Replica-exchange Monte Carlo simulations are then used to assemble the domain structures under the guidance of template-based distance profiles (as well as CL and cryo-EM data if available), with models with lowest energy selected for linker reconstruction and side chain refinement. For 3 or more domain assemblies, a global structural refinement simulation is performed following the consecutive pairwise domain assembly.
Results and Discussion
Completeness of Multidomain Protein Structure Library.
To infer initial domain orientation in DEMO, a nonredundant structure library was constructed by collecting all multidomain proteins from the PDB (12) with a 70% pairwise sequence identity cutoff. The domain structures are primarily recognized by DomainParser (13), where multidomain proteins defined in CATH 4.1 (14) and SCOPe 2.06 (15) are also included if they have a sequence identity <70% or a TM-score <0.5 (16) to the DomainParser proteins (details in SI Appendix, Text S1). This resulted in a total of 15,293 template proteins in the library.
To examine the completeness of the library in the domain space, we collected a comprehensive set of 2,269 target proteins from the library at the 30% sequence identity cutoff and structurally matched them through the library using TM-align (11). SI Appendix, Fig. S1 summarizes TM-scores and alignment coverages of the templates identified for all of the target proteins, with close templates with sequence identity >30% to the targets excluded. It is shown that >80% of target proteins have at least 1 template with a TM-score >0.5 and alignment coverage >90%, confirming that most interdomain orientations can be inferred from the template library.
Overall Results of DEMO Modeling on Experimental Domains.
To test DEMO, we collected a comprehensive set of benchmark proteins by separately clustering the proteins with different domain types and structures from the template library with a 30% sequence identity cutoff. This benchmark set contained 356 proteins, including 166 2-domain (2dom), 69 3-domain (3dom), 40 ≥ 4-domain (m4dom), and 81 discontinuous-domain (2dis) proteins. Here a discontinuous domain was defined as that containing 2 or more segments from separate regions of the query sequence (SI Appendix, Fig. S10). The maximum number of domains in m4dom was 7. In addition, 425 nonredundant proteins with a sequence identity <30% to the benchmark proteins were selected from the library to train the DEMO potential (Methods); this training set included 197 2dom, 111 3dom, 66 m4dom, and 51 2dis proteins.
In this test, we reassembled the individual domain structures partitioned from the experimental structure according to the domain boundary defined by DomainParser, CATH, or SCOPe. The initial domain structure was randomly rotated and translated before assembly, with templates with a sequence identity >30% to the query excluded. Table 1 summarizes the DEMO modeling results, showing average TM-scores of 0.78 for 2dom, 0.67 for 3dom, and 0.53 for m4dom proteins. These results demonstrate DEMO’s ability to assemble domains with a smaller number of domains. This is understandable, because it is usually harder to find a global template that covers all individual domains when the number of domains increases. In addition, the degrees of freedom increase for proteins of more domains, which increases the search space of domain assembly simulations. Nevertheless, the overall quality of the multidomain models was acceptable, with an average TM-score of 0.62 for proteins with 3 or more domains, 65% of which had a TM-score >0.5.
Table 1.
Summary of domain structure assembly using experimentally solved domains on 356 test proteins
| Domain | Method | TM-score | RMSD, Å | iRMSD, Å | No.of clashes |
| 2dom (N = 166) | DEMO | 0.78 | 7.3 | 5.5 | 0.59 |
| AIDA | 0.68 | 12.3 | 9.2 | 5.09 | |
| Modeller | 0.63 | 13.3 | 11.3 | 5.08 | |
| 3dom (N = 69) | DEMO | 0.67 | 10.7 | 6.2 | 1.60 |
| AIDA | 0.52 | 18.1 | 11.8 | 9.17 | |
| Modeller | 0.49 | 19.9 | 14.6 | 3.70 | |
| m4dom (N = 40) | DEMO | 0.53 | 16.9 | 12.8 | 3.79 |
| AIDA | 0.42 | 23.6 | 16.7 | 14.51 | |
| Modeller | 0.38 | 28.3 | 20.9 | 16.78 | |
| 2dis (N = 81) | DEMO | 0.84 | 5.5 | 3.8 | 1.18 |
| AIDA | 0.71 | 9.7 | 7.4 | 8.11 | |
| Modeller | 0.67 | 11.2 | 8.7 | 5.56 | |
| All (N = 356) | DEMO | 0.74 | 8.6 | 6.1 | 1.28 |
| AIDA | 0.63 | 14.1 | 10.1 | 7.63 | |
| Modeller | 0.58 | 15.8 | 12.4 | 6.24 |
Bold result indicates the best one. N, number of proteins.
Interestingly, the TM-score for discontinuous domains, the structures of which are generally considered difficult to model, had the highest average TM-score (0.84) of all categories. This is probably due to the greater interdomain distance restraints used for guiding the assembly simulations in these cases, as proteins of discontinuous domains contain tighter interdomain coupling from experimental structures compared with proteins of continuous domains. Meanwhile, the connectivity of linker regions between the discontinuous and inserted domains provides additional anchor restraints on the domain orientations (SI Appendix, Fig. S10). Table 1 also lists the RMSD of the full-length model and the RMSD of interface residues (iRMSD), which have a Cα distance between different domains of <10 Å. Similar trends were observed for iRMSD and RMSD; that is, proteins with a smaller number of domains (or with discontinuous domains) have lower iRMSD and RMSD compared with proteins with a larger number of domains (or with continuous domains).
As a control, Table 1 also shows the model results constructed by AIDA (9) and Modeller (17) from the same set of domain structures. Here AIDA is the only program available for multidomain assembly. In Modeller, the full-length models are created using experimental domain structures as the input templates. Generally, DEMO models have a higher TM-score and lower iRMSD/RMSD than the models from the control methods in all categories of domain structures. Overall, the TM-score of the DEMO models (0.74) is 17.5% higher than that produced by AIDA (0.63) and 27.6% higher than that produced by Modeller (0.58). The respective P values on Student’s t test of 4.1E-38 and 4.9E-53 indicate statistically significant differences between the methods. In addition, as an examination of local quality, DEMO models have on average much fewer interdomain steric clashes (1.28) than either AIDA (7.63) or Modeller (6.24).
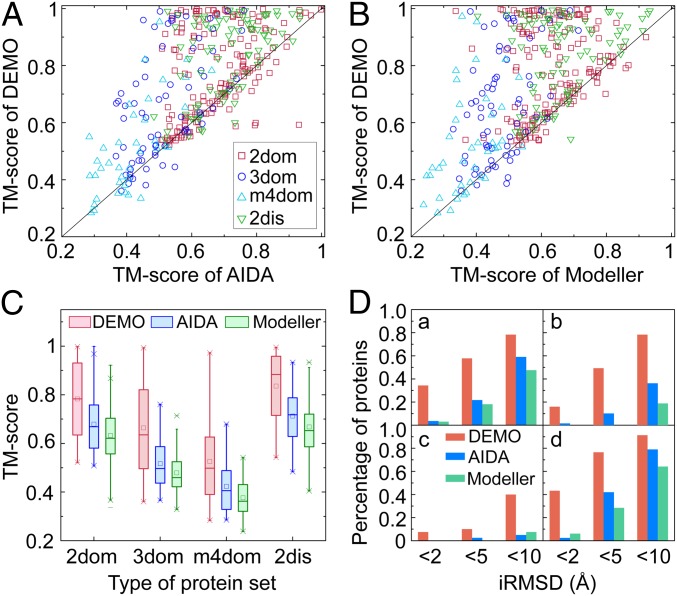
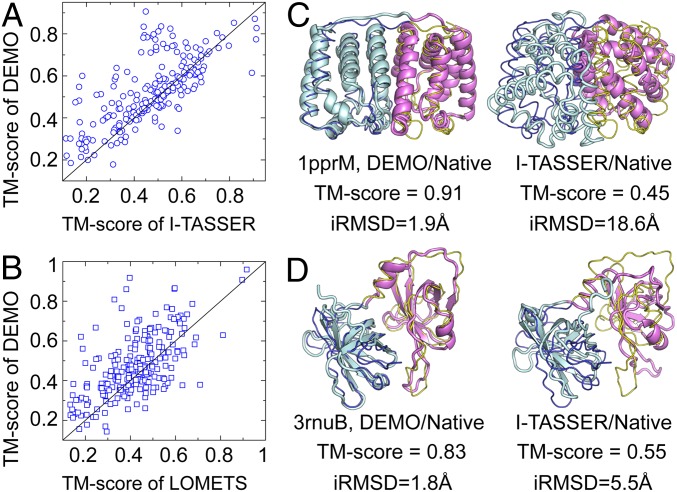
Fig. 2 A and B show the scattering points for the TM-score of DEMO versus that of AIDA and Modeller, respectively. It can be seen that DEMO achieves a higher TM-score than AIDA and Modeller in 279 and 302 cases, respectively, while AIDA and Modeller achieve the highest TM-score in only 77 cases and 54 cases, respectively. Accordingly, in 171 cases, DEMO has a TM-score >0.75, a rate 117% higher than that of AIDA and 317% higher than that of Modeller. Fig. 2 C and D also show the average TM-score and iRMSD histograms in separate categories, which again show that DEMO is able to assemble more accurate full-length models for proteins of different types of domains.
Fig. 2.
Summary of full-length models assembled from experimental domain structures by different methods. (A) TM-scores of models by DEMO and by AIDA. (B) TM-scores of models by DEMO and by Modeller. (C) Boxplot for TM-score of models by DEMO, AIDA, and Modeller. The square and solid vertical lines represent the mean and median TM-scores, and crosses on the top and bottom are the maximum and minimum TM-scores, respectively. (D) Percentage of proteins at different iRMSD thresholds. Red, blue, and green columns represent the results of DEMO, AIDA, and Modeller, respectively. (a)–(d) 2dom, 3dom, m4dom, and 2dis proteins, respectively.
DEMO Improves Domain Orientations of Initial Templates from Structural Alignments.
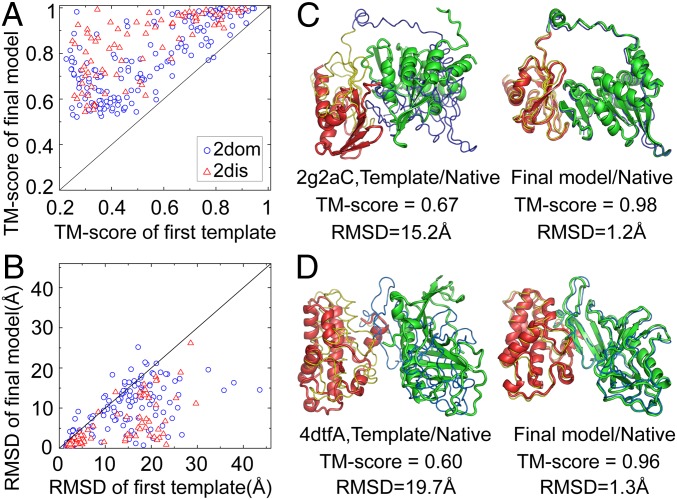
Since DEMO assembles multidomain structures under guidance of the templates identified by TM-align, it is of interest to examine whether DEMO has the ability to draw the initial templates closer to the native structure. Fig. 3 A and B presents TM-scores and RMSDs of the first DEMO models and the top-ranked template, respectively. Because assemblies of proteins with 3 or more domains are treated as consecutive 2-domain protein assemblies in DEMO, the figure shows results only for 2dom and 2dis proteins.
Fig. 3.
Comparison of final DEMO models and initial templates identified by TM-align. (A) TM-scores of the first DEMO model vs. the top-rank template. (B) RMSDs of the first DEMO model vs. the top-rank template in the same aligned regions. (C and D) Representative examples showing improvement of the DEMO models over the templates. The thin lines are experimental structures, and cartoons represent initial TM-align templates or DEMO models, with different domains signified by different colors.
Fig. 3A shows that 99% of proteins were improved by DEMO for both 2dom and 2dis proteins in terms of TM-score. When considering the RMSD in the same aligned regions, DEMO improved the initial models in 92% of cases. This improvement had an RMSD >1 Å in 74% of the cases and of 5 Å in 30% (Fig. 3B).
This significant improvement in the templates may be attributed in part to the fact that input domains were from an experimental solution, while domains in the templates were from nonnative proteins. To eliminate this effect, we constructed a hybrid model set by superimposing the native domain structures onto the initial templates. The results show that the average TM-score of the DEMO models is still significantly higher than that of the hybrid models (0.80 vs. 0.75; P = 3.2E-14). Meanwhile, the DEMO models have a lower number of clashes (0.78) and iRMSD (5.0 Å) compared with the hybrid models (4.1 and 11.8 Å, respectively) (SI Appendix, Fig. S2B).
The second and likely more dominant reason for the improvement is the use of consensus distance profiles collected from multiple templates, which often have greater accuracy compared with the individual templates (18). Fig. 3 C and D presents illustrative examples from the VirB11 ATPase (PDB ID code 2gzaC), a 2dom protein, and VgrG Vibrio cholerae toxin (PDB ID code 4dtfA), a 2dis protein, respectively. The average error of the interdomain distances extracted from multiple templates is 7.2 Å for 2gzaC and 8.0 Å for 4dtfA, significantly lower values than extracted from the top-rank templates (11.6 Å and 14.6 Å, respectively). Accordingly, the TM-score of the templates was increased by 46.3% and 60.0%, respectively, and RMSD was reduced by >13 Å in the 2 examples.
Assembly of Predicted Domain Models from I-TASSER.
Because most proteins do not have experimentally solved domains, it is important to examine the ability of DEMO to assemble domains from low-resolution protein structure prediction. Table 2 summarizes the full-length model results using domain structures predicted by I-TASSER (5). Here, when generating the I-TASSER domain models, all homologous templates with a sequence identity >30% to the query have been excluded. For discontinuous domains that contain multiple segments (e.g., Domain-I in SI Appendix, Fig. S10), an artificial continuous sequence is formed by sequentially connecting the sequences of all segments and then inputting them to I-TASSER for modeling, where residues are reindexed to match the original atomic order after modeling is completed. The original DEMO test dataset involves a total of 81 discontinuous domains, in which the average TM-score of the I-TASSER models is 0.57, generally comparable to that of the continuous domains (0.61), showing the feasibility of I-TASSER for modeling the discontinuous domain structures. To clearly check the effect of domain assembly and rule out the negative impact from incorrect domain models, the data focus only on cases with all domain folds correctly predicted by I-TASSER with a TM-score >0.5; these include 116 2dom proteins, 47 3dom proteins, 24 m4dom proteins, and 41 2dis proteins. The average TM-score for these I-TASSER domain models is 0.77.
Table 2.
Summary of domain structure assembly using domain models by I-TASSER prediction on 228 proteins
| Domain | Method | TM-score | RMSD, Å | iRMSD, Å | No. of clashes |
| 2dom (N = 116) | DEMO | 0.67 | 9.8 | 8.0 | 0.56 |
| AIDA | 0.55 | 13.4 | 10.8 | 3.69 | |
| Modeller | 0.51 | 19.0 | 11.7 | 66.30 | |
| 3dom (N = 47) | DEMO | 0.55 | 12.8 | 9.7 | 1.70 |
| AIDA | 0.43 | 18.9 | 13.0 | 16.80 | |
| Modeller | 0.38 | 24.3 | 14.9 | 159.57 | |
| m4dom (N = 24) | DEMO | 0.47 | 19.0 | 14.2 | 4.47 |
| AIDA | 0.38 | 24.8 | 17.4 | 13.39 | |
| Modeller | 0.32 | 31.8 | 22.5 | 382.25 | |
| 2dis (N = 41) | DEMO | 0.58 | 12.4 | 8.9 | 2.36 |
| AIDA | 0.52 | 14.8 | 10.7 | 12.66 | |
| Modeller | 0.49 | 19.1 | 11.7 | 55.61 | |
| All (N = 228) | DEMO | 0.61 | 11.9 | 9.1 | 1.53 |
| AIDA | 0.50 | 16.0 | 11.9 | 9.03 | |
| Modeller | 0.46 | 21.5 | 13.5 | 116.86 |
Bold result indicates the best one. N, number of proteins.
It is shown again that the average TM-score of the DEMO models decreases with an increasing number of domains, due to the increased degrees of freedom and searching space in domain orientations. However, different from that on experimental domains, the model quality for 2dom targets is better than that of 2dis proteins when I-TASSER models are used. This is probably because the I-TASSER models can have various errors in the domain structures (including the tail and linker regions). Therefore, including additional restraints from the noisy interdomain linkers and anchor points does not improve the models for proteins of discontinuous domains.
Nevertheless, the DEMO models compare favorably with the models by the control methods in all categories of proteins, with the average TM-score of 0.61 being 22.0% higher than that of AIDA (0.50) and 32.6% higher than that of Modeller (0.46); the P value on Student’s test is 2.6E-36 and 3.2E-49, respectively, relative to AIDA and Modeller, showing statistically significant differences. Overall, there are 154 cases in which DEMO models have a TM-score >0.5, compared with 110 such cases for AIDA and 76 for Modeller (SI Appendix, Fig. S3). Meanwhile, average iRMSD (or RMSD) and the number of steric clashes of the DEMO models are also lower than those of the AIDA and Modeller models, showing better local qualities achieved by DEMO compared with the control methods.
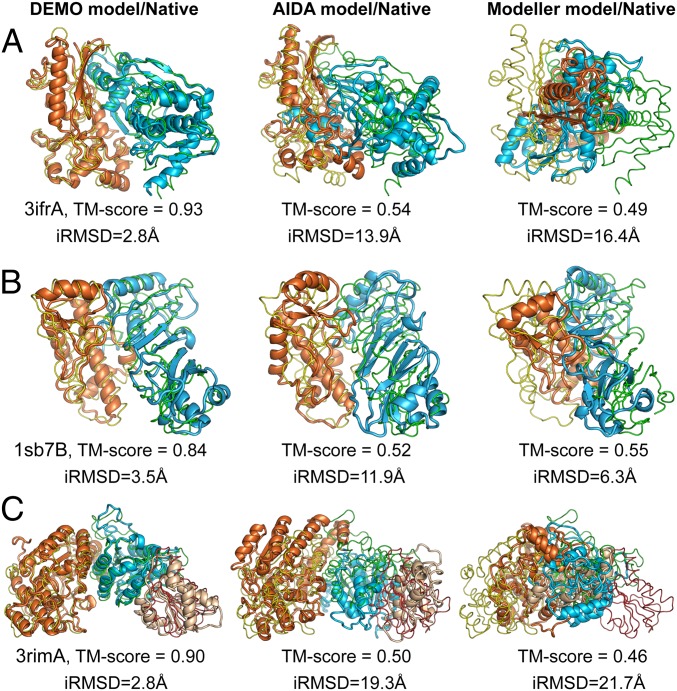
Fig. 4 presents 3 representative examples from the xylulose kinase (PDB ID code 3ifrA), a 2dom protein; pseudouridine synthase TruD (PDB ID code 1sb7B), a 2dis protein; and Mycobacterium tuberculosis Transketolase (PDB ID: 3rimA), a 3dom protein. Although the domain models from I-TASSER have correct fold (TM-score >0.5), there are various local errors along the models, with an average RMSD of 6.8 Å, 10.9 Å, and 16.6 Å, respectively. However, the TM-align search based on individual domain structure alignments identifies correct multidomain templates with a reasonable TM-score—0.91, 0.75, and 0.81, respectively, for the 3 targets respectively. This is probably due to the inherent correlation between the structural similarity of local domains and that of global structures. Under the restraints of these templates, DEMO constructed final full-length models of TM-scores 0.93, 0.84 and 0.90, which are significantly higher than those of AIDA (0.54, 0.52, and 0.50) and Modeller (0.49, 0.55, and 0.46). The success shown in these examples was due mainly to the identification of correct global templates, the quality of which was further improved through local domain structure packing and refinement.
Fig. 4.
Illustrative examples of domain assembly results with domain structure predicted by I-TASSER. The thin lines represent the experimental structures, and cartoons are the final models by DEMO (Left), AIDA (Center), and Modeller (Right), with different colors indicating different domains. (A) 3ifrA. (B) 1sb7B. (C) 3rimA.
Domain Assembly Outperforms Whole-Chain–Based Modeling.
A classical problem in template-based multidomain protein structure prediction is that threading alignment often cannot identify homologous templates that cover all domain regions, although it may recognize single-domain templates well (19). Thus, structural models need to be created for individual domains and then assembled into full-length models. To examine DEMO’s ability to address problems in such cases, we collected a separate set of 228 2dom proteins that have at least 1 domain with alignment coverage <30% in the whole-chain–based threading alignment by HHsearch (20). Proteins in this dataset are nonredundant, with a sequence identity <30% to one another, and are also nonredundant with the 425 training proteins used to train DEMO.
We first use I-TASSER to generate 3D structure models for each of the individual models, excluding all close templates with a sequence identity >30% to the target. This results in an average TM-score of 0.64 for all 456 (228 2) domains, 350 of which have a TM-score >0.5. Table 3 summarizes the results for the full-length models assembled by DEMO from the I-TASSER–predicted single-domain models. The DEMO models have an average TM-score of 0.53 and the global fold is correct, with 136 cases with a TM-score >0.5. This compares favorably with the full-length models built directly by I-TASSER simulation, which have an average TM-score of 0.47 but with only 104 cases with a TM-score >0.5 (Fig. 5A).
Table 3.
Modeling on 228 2dom proteins with at least 1 domain missed in whole-chain threading alignment
| Criterion | Domain-based assembly | Whole-chain folding | ||
| DEMO | AIDA | Modeller | I-TASSER | |
| TM-score | 0.53 | 0.48 | 0.46 | 0.47 |
| RMSD, Å | 14.1 | 16.3 | 20.1 | 17.8 |
| iRMSD, Å | 9.8 | 11.8 | 11.2 | 12.1 |
| No. of clashes | 1.73 | 8.89 | 72.59 | 1.31 |
Bold indicates the best result in each category.
Fig. 5.
Comparison of individual domain assembly and whole-chain structure prediction. (A) TM-scores of models assembled by DEMO vs. models assembled by whole-chain I-TASSER prediction. (B) TM-scores of templates identified by domain-based structural alignment using TM-align vs. those identified by full-chain threading using LOMETS. (C and D) Illustrative examples from 1pprM and 3rnuB, where thin lines are native structures and cartoons are DEMO and I-TASSER models. with different colors representing different domains.
There are 2 main reasons for the superior performance of the DEMO-based pipeline. First, the domain split allows I-TASSER to detect domain-specific templates and thus construct better models for individual domains, because many proteins in the PDB were solved as individual domains. When looking at the first templates of the 456 domains, the average TM-score identified by LOMETS (21) from single-domain sequences is 0.56, which is 115% higher than that identified by LOMETS from full-length sequences. Furthermore, the I-TASSER force field is optimized for folding single-domain proteins, as many energy terms (e.g., solvation and radius-gyration constraints) are designed for single globular domains. Thus, the individual domain models built by domain-based I-TASSER have a higher average TM-score (0.64) compared with models built by whole-chain–based I-TASSER modeling (0.50). In fact, starting from the individual domain models, a simple domain assembly approach from AIDA and Modeller can generate full-length structures with average TM-scores (0.46 and 0.48, respectively) comparable to that achieved by whole-chain I-TASSER modeling (0.47), although the former has a higher number of steric clashes (Table 3).
Second, DEMO exploits a procedure to identify global templates through domain-based structural alignments, which can help detect reasonable whole-chain templates even when homologous whole-chain templates do not exist in the PDB. Fig. 5B shows a head-to-head comparison of the whole-chain templates identified by TM-align vs. those identified by whole-chain threading using LOMETS, where TM-align detects templates with a higher TM-score in 151 of the 228 cases, while LOMETS does so in only 77 cases. On average, the TM-score by TM-align is 12.9% higher than that by LOMETS (0.473 vs. 0.419; P = 1.7E-07, Student’s t test). This advantage of whole-chain template identification is the major reason why DEMO can generate better domain orientations, with an average TM-score at least 10% higher than those generated by all other domain-assembly or whole-chain–based model prediction protocols (Table 3).
Fig. 5 C and D shows 2 illustrative case examples from peridinin-chlorophyll protein (PDB ID code 1pprM) and IFI16 receptor (PDB ID code 3rnuB). For 1pprM (Fig. 5C), the I-TASSER–based full-length model has only 1 domain modeled correctly due to the lack of full-length structure templates (with average TM-scores of 0.78 and 0.29 for the N- and C-terminal domains, respectively). After the split of the domains, I-TASSER correctly predicted individual domain structures, with an average TM-score of 0.86 for both the N- and C-domains, where the reassembly by DEMO resulted in a full-length model with an average TM-score of 0.91 and iRMSD of 1.9 Å.
For 3rnuB, a full-length model computed directly by I-TASSER has both domains correctly folded (with TM-scores of 0.84 and 0.79 for the N- and C-terminal domains, respectively), probably due to I-TASSER’s ability to combine multiple templates that can have the different templates aligned on different domains. However, the orientation of the domains was not correctly predicted, resulting in a marginal TM-score of 0.55 with an iRMSD of 5.5 Å. Under the guidance of the TM-align templates, DEMO created a first-rank model of much better overall quality (average TM-score of 0.83 and iRMSD of 1.8 Å). These examples highlight both aspects of advantages of individual domain folding and the analogous template-guided domain assembly.
Multidomain Structure Assembly on CASP Targets.
In this test, we collect all of the multidomain protein targets from the most recent CASP12 and CASP13 experiments. As listed in SI Appendix, Table S1, these include 41 targets, with 20 2dom proteins, 4 3dom proteins, 4 m4dom proteins, 9 2dis proteins, 2 3dom proteins with 1 discontinuous domain, 1 3dom protein with 2 discontinuous domains, and 1 5dom protein with 1 discontinuous domain. As a control, we select models from 3 top servers: Zhang-Server, QUARK (22), and Baker-Rosetta (23). While all 3 servers modeled individual domains separately, Zhang-Server and QUARK assembled full-length models by docking domain structures with the full-length I-TASSER models (22), and Baker-Rosetta used linker-based domain docking guided by contact-map prediction (10).
Table 4 compares the original models by the top server with models reassembled by DEMO based on the domain structures excised from the original full-length models. Here all domain structures are randomly rotated and translated before reassembly, with PDB templates released after May 1, 2016, for CASP12 and after May 1, 2018, for CASP13 excluded from the TM-align search. It is observed that DEMO reassembly results in an obvious improvement in the domain orientations, with the average TM-score increased by 3%–4% and iRMSD (or RMSD) decreased by 1–2 Å compared with the original models, although with a slight increase in the number of clashes (all <2).
Table 4.
Comparison of domain reassembly results and the original models from 3 top servers on 41 multidomain targets in CASP12 and CASP13 experiments
| Server | Criterion | Original | DEMO | AIDA | Modeller |
| Zhang-Server | TM-score | 0.454 | 0.469 | 0.429 | 0.415 |
| RMSD, Å | 20.4 | 18.6 | 22.7 | 23.9 | |
| iRMSD, Å | 15.1 | 13.5 | 16.2 | 18.9 | |
| No. of clashes | 1.27 | 1.90 | 9.24 | 3.43 | |
| QUARK | TM-score | 0.453 | 0.467 | 0.417 | 0.405 |
| RMSD, Å | 19.4 | 17.7 | 23.2 | 24.1 | |
| iRMSD, Å | 14.3 | 12.9 | 16.1 | 19.2 | |
| No. of clashes | 0.71 | 1.23 | 11.22 | 1.84 | |
| Baker-Rosetta | TM-score | 0.412 | 0.428 | 0.395 | 0.389 |
| RMSD, Å | 21.6 | 19.6 | 23.1 | 25.3 | |
| iRMSD, Å | 17.6 | 15.8 | 17.2 | 18.9 | |
| No. of clashes | 0.07 | 1.68 | 7.51 | 8.10 |
Bold indicates the best result in each category.
SI Appendix, Fig. S4 shows an example from T0920, for which individual domain structures were correctly predicted by all 3 servers, with average TM-scores of 0.90/0.86, 0.88/0.86, and 0.90/0.83 for the N-/C- terminal domains by Zhang-Server, QUARK, and Baker-Rosetta, respectively, but the orientation of the domains was incorrect, resulting in full-length models with low TM-scores (0.61, 0.61, and 0.59, respectively). On the other hand, starting from the server domain models, TM-align identifies a correct interdomain template structure with a TM-score of 0.95, whereas the full-length models reassembled by DEMO have a TM-score >0.85 and an iRMSD <3 Å in all cases (i.e., TM-score/iRMSD of 0.93/2.8 Å, 0.92/2.7 Å, and 0.87/2.9 Å starting from Zhang-Server, QUARK, and Rosetta domain models, respectively).
As a control, Table 4 also presents the model results by AIDA and Modeller from the same domain structures, showing significantly poorer performance compared with both DEMO and the original server models. These data demonstrate again the important impact of modeling method on the quality of multiple domain structure assembly.
Cross-Linking and Cryo-EM Data-Assisted Domain Assembly.
In addition to template-based restraints, DEMO also allows for integration of data from mass spectrometry generated cross-linking (CL) and cryogenic electron microscopy (cryo-EM) experiments, which are calculated as contact and density-map correlation restraints, respectively (Fig. 1 and SI Appendix, Eqs. S9 and S10). Table 5 summarizes the modeling results by the data-assisted DEMO pipelines, DEMO-CL and DEMO-EM, compared with the default DEMO without using experimental data. For brevity, we focus only on the 2dom and 2dis proteins; the assembly of proteins with 3 or more domains is a straightforward extension of the 2-domain assembly process.
Table 5.
Comparison of DEMO domain assembly models with or without experimental data assistance
| Domain | Method | TM-score | RMSD, Å | iRMSD, Å | No. of clashes |
| Starting with experimentally solved domain structures | |||||
| 2dom (N = 166) | DEMO | 0.78 | 7.3 | 5.5 | 0.59 |
| DEMO-CL | 0.85 | 3.6 | 2.0 | 0.45 | |
| DEMO-EM | 0.89 | 3.9 | 2.7 | 0.23 | |
| 2dis (N = 81) | DEMO | 0.84 | 5.5 | 3.8 | 1.18 |
| DEMO-CL | 0.86 | 3.7 | 2.4 | 1.02 | |
| DEMO-EM | 0.92 | 3.3 | 2.9 | 0.62 | |
| All (N = 247) | DEMO | 0.80 | 6.7 | 5.0 | 0.78 |
| DEMO-CL | 0.85 | 3.7 | 2.1 | 0.64 | |
| DEMO-EM | 0.90 | 3.7 | 2.8 | 0.36 | |
| Starting with I-TASSER predicted domain models | |||||
| 2dom (N = 116) | DEMO | 0.67 | 9.8 | 8.0 | 0.56 |
| DEMO-CL | 0.70 | 8.0 | 5.3 | 1.49 | |
| DEMO-EM | 0.72 | 7.2 | 6.7 | 0.60 | |
| 2dis (N = 41) | DEMO | 0.58 | 12.4 | 8.9 | 2.36 |
| DEMO-CL | 0.63 | 10.8 | 8.1 | 2.31 | |
| DEMO-EM | 0.66 | 10.2 | 8.3 | 2.45 | |
| All (N = 157) | DEMO | 0.65 | 10.5 | 8.2 | 1.03 |
| DEMO-CL | 0.68 | 8.7 | 6.0 | 1.70 | |
| DEMO-EM | 0.71 | 8.0 | 7.1 | 1.08 | |
Bold indicates the best result in each category. N, number of proteins.
Domain assembly with CL data.
The CL data are simulated from the target structures by randomly picking up restraints from all interdomain residue pairs that have a Cα distance <10 Å and separation |i − j| > 5, where , or , or . Here it is noted that can differ depending on the shape and size of interdomain interfaces. In fact, 7% (or 34%) of the test proteins have (or ) due to the small interfaces. To add noise, false restraints with Cα >10 Å are selected at random and included in the CL pool. Thus, up to 20 CL restraints are collected with at least 5% of false-positive pairs included. Table 5 presents the results of DEMO-CL starting with the experimentally solved domain structure, where the CL data increase the TM-score of DEMO from 0.78 to 0.85 for 2dom proteins and from 0.84 to 0.86 for 2dis proteins; this corresponds to P values of 2.7E-12 and 9.3E-09, respectively, on Student’s t test, showing that the improvement is statistically significant. SI Appendix, Fig. S5A presents a head-to-head TM-score comparison of DEMO and DEMO-CL, showing significantly more cases with a higher TM-score by DEMO-CL than that by DEMO. Accordingly, the average RMSD and iRMSD of DEMO-CL is reduced by 83.3% and 134.0%, respectively, compared with the values of DEMO, and with fewer clashes (Table 5).
Table 5 also presents the modeling results using I-TASSER–predicted domain models. A similar positive effect of CL restraints was observed in which DEMO-CL achieved an average TM-score 4.6% higher than that of DEMO, corresponding to P = 1.2E-06 on Student’s t test. There were 87% of cases with a TM-score >0.5, 10.2% more compared with those of DEMO (SI Appendix, Fig. S5B). The average RMSD and iRMSD were 20.5% and 36.4% lower, respectively, in DEMO-CL compared with DEMO. However, the DEMO-CL models contained slightly more clashes (1.70 vs. 1.03), probably due to the conflict of I-TASSER model errors with the CL data.
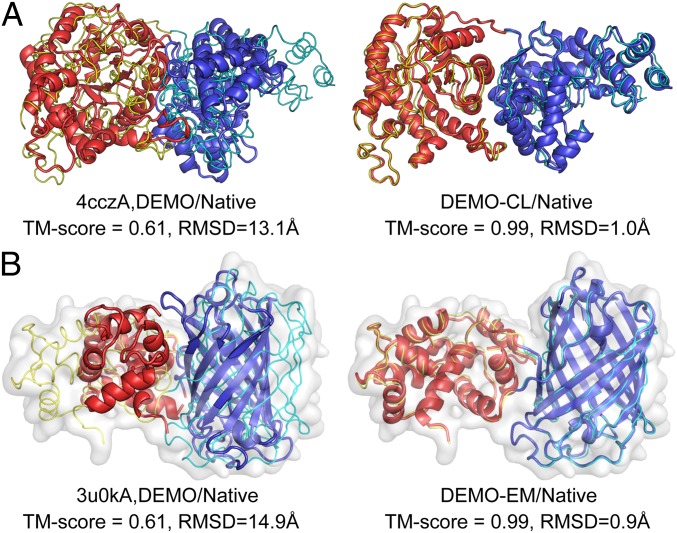
Fig. 6A shows an illustrative example starting with the target domains from Chain-A of the methionine synthase (PDB ID code 4cczA), a 2dom protein, in which DEMO generates a medium-quality model with TM-score of 0.61 and RMSD of 13.1 Å. The inclusion of CL restraints (17 positive and 1 negative) resulted in a significantly improved model with an average TM-score of 0.99 and RMSD of 1.0 Å. SI Appendix, Fig. S6A shows another example from Cys-Gly metallodipeptidase (PDB ID code 4g1pA), a 2dis protein, which starts with the I-TASSER–predicted domains (with TM-scores of 0.86 and 0.84 for the N- and C-terminal domains, respectively). The DEMO model was also drastically improved by the CL data, with increases in TM-score from 0.74 to 0.89 and in RMSD from 6.2 Å to 3.8 Å. In both examples, the fraction of satisfied CL pairs is low in the original DEMO models (5.9% for 4cczA and 20% for 4g1pA) and significantly increased in the DEMO-CL models (94.1% and 80%, respectively). In fact, the CL satisfaction rate was improved in nearly all the test cases, with the average rate increased from 0.53/0.32 (by DEMO) to 0.84/0.65 (DEMO-CL) using the experimental/I-TASSER domain structures. These data again demonstrate the impact of the cross-linking data in domain assembly experiments.
Fig. 6.
Illustrative examples of domain assembly by DEMO starting from the target domain structures when assisted by experimental restraints. The thin lines are experimental structures, and cartoons are full-length models assembled by the DEMO programs, with different colors representing different domains. (A) 4cczA assisted with CL data. (B) 3u0kA assisted with cryo-EM data.
Domain assembly with cryo-EM density maps.
To examine the effect of cryo-EM data on DEMO, we simulate the density maps from the experimental structures using the IMP program (24) with resolution of 10 Å and voxel size of 2 Å. Table 5 summarizes the models generated by integrating the cryo-EM data with DEMO, showing that a 12.5% higher TM-score by DEMO-EM than by DEMO (0.90 vs. 0.80) starting from the target domain structures, which corresponds to P = 3.2E-21 on Student’s t test. When starting from the I-TASSER–predicted domain structures, the percentage of TM-score increase by DEMO-EM is slightly reduced (by 9.2%; 0.71 vs. 0.65) but still statistically significant, with P = 2.4E-09 on Student’s t test. As shown in SI Appendix, Fig. S5 C and D, there are a significantly greater number of targets with a higher TM-score by DEMO-EM than by DEMO: 75% of cases when starting from the target domain and 69% of cases when using the I-TASSER prediction domain structures.
Fig. 6B and SI Appendix, Fig. S6B show 2 examples from calcium indicator protein (PDB ID code 3u0kA), a 2dis protein, starting from experimental domains, and poliovirus 3CD protein (PDB ID code 2ijd1), a 2dom protein, starting from the I-TASSER–predicted domain model. In both cases, the original DEMO models do not fit well with the cryo-EM density map, where DEMO-EM with density map correlation restraints significantly improved the model and density map matches, resulting in a much improved global model with increases in average TM-score from 0.61 to 0.99 and from 0.67 to 0.89 and decreases in RMSD from 14.9 to 0.9 Å and from 13.2 to 3.7 Å in the 2 examples. These data clearly demonstrate the impact of the cryo-EM data on the domain assembly process in DEMO.
Interestingly, the magnitude of TM-score improvement by the cryo-EM data is greater than that by the CL data in both tests (i.e., 12.5% vs. 6.3% when starting from the target domain and 9.2% vs. 4.6% when starting from the I-TASSER domain structures). This is probably because the cryo-EM density map provides 3D structural shape matches, which are more informative (or provide more stringent constraints on the domain orientations) than the contact-like CL data that are essentially 1D binary constraints. Nevertheless, the effect of the CL data also depends on the data quality. If we increase the number of CL restraints up to L/5 (L = length of the target) with shorter distance (<8 Å), the magnitude of TM-score increase can reach a similar level as that obtained using cryo-EM restraints in this experiment. If we simply implement the CL and cryo-EM restraints simultaneously, the model quality can be further improved (e.g., with an increase in average TM-score to 0.96 starting from the target domains).
Concluding Remarks
Modeling of multidomain protein structures is an important problem that is largely ignored by mainstream computational biology compared with the extensive effort and rapid progress made in single-domain tertiary structure folding. This is partly due to the difficulty in modeling interdomain orientations, as multidomain proteins have a much higher degree of freedom in domain orientation, and the stability of multidomain structures often involves interactions with other protein cofactors. Meanwhile, the complex interplay of various levels of continuous and discontinuous domain assembly and linker refinement makes the method development difficult to automate.
In this work, we tried to meet the aforementioned challenges and developed a pipeline, termed DEMO, to automatically assemble full-length protein structures with both continuous and discontinuous domain architectures. Considering that purely physics-based approaches have difficulty modeling domain–domain interactions, which usually have small interfaces and often involve interactions with other protein cofactors, the method starts with recognition of analogous whole-chain templates by structurally aligning the component domains with known proteins from a nonredundant multidomain structure library. Interdomain distance profiles are then extracted from the templates, which are combined with the physics-based steric potential and used to guide the Monte Carlo domain assembly simulations.
DEMO was tested in a comprehensive set of 356 proteins containing various levels of continuous and discontinuous domain structures. Starting with domain models predicted by I-TASSER (5), which have an average TM-score of 0.77, the structure-alignment–based search could detect correct whole-chain templates with a TM-score >0.5 in 113 of the 228 cases. After DEMO docking reassembly, this number was increased to 154, with the average TM-score increased from 0.52 to 0.61. Due to the increased degrees of freedom and searching space in domain assembly, the model quality by DEMO generally decreases with the number of domains involved. Meanwhile, proteins with discontinuous domain structures are more difficult to model than those with continuous domain structures; however, this is not the case when using experimental domain structures, because restraints from interdomain linkers provide additional information to facilitate the domain assembly simulations. Meanwhile, the overall TM-score and iRMSD values were improved in all domain types when using experimentally solved high-resolution domain structures, demonstrating the impact of domain quality on the full-length structure models.
Overall, DEMO demonstrated significant advantages over the state-of-the-art linker- or homology-based domain assembly methods in terms of both global domain orientation modeling and reduction of interdomain steric clashes. The superior performance of DEMO stems mainly from the fact that the structure-based global template identification can provide a promising initial orientation of domains due to the inherent correlation of local domain and global structures in natural proteins. Second, the Monte Carlo simulations, as guided by the composite energy function combining consensus template-based restraints and physical terms, help refine the domain docking structures and identify models with domain orientations closer to the native than the individual templates. Here we note that we also tried to combine TM-align with the homology-based threading alignments for whole-chain template identification, but found that the addition from threading is modest (SI Appendix, Text S7). This result probably suggests that the structure-based domain matches can effectively cover both analogous and homologous interdomain orientation spaces, and thus we stick to the DEMO pipeline, with only structural alignment for briefness and efficiency of method implementation.
As an application, DEMO was used to assemble predicted domains for the hard proteins that have at least 1 domain missed in the threading alignments. Out of the 228 2dom proteins, DEMO correctly predicted the full-length structure with a TM-score >0.5 in 136 cases, which is 31% higher than that of the models built directly with full-length structure modeling simulations. The improvement is mainly due to the fact that domain-based threading can identify specific tertiary templates and thus result in better domain models. Second, the structure-based alignments can help detect more reasonable global templates to guide the domain assembly simulations; this is particularly true for cases in which homologous templates do not exist but analogous templates can be detected by domain-level structure alignments.
As a further illustration, DEMO was applied to reassemble the multidomain models generated by the top servers in the most recent CASP experiments. Although these servers have built the domain models individually, the DEMO domain reassembly achieved considerable improvements in full-length model quality, with the average TM-score increased by 3% and iRMSD decreased by >1 Å, which demonstrates the importance of domain-level structure assembly.
Finally, we tested the ability of DEMO to integrating data from CL and cryo-EM experiments. It was shown that the full-length models assembled by DEMO when starting from experimentally solved domain structures, are significantly improved by the CL and cryo-EM restraints, with average TM-score increases from 0.80 to 0.85 and 0.90, respectively. When starting with the low-resolution I-TASSER–predicted domain models, the full-length models with corrected orientation (TM-score > 0.5) are increased by 10.2% and 12.1% with CL and cryo-EM restraints, respectively. These results show that the DEMO domain assembly process can benefit significantly from the sparse experimental data.
Despite the successes documented herein, DEMO could be further improved in several aspects. Currently, domain structures are kept rigid during the DEMO simulations, which cannot appropriately account for binding-induced structural changes. In addition, the predicted domains often have low resolution, and thus introducing backbone flexibility to the domain assembly simulation provides the potential for local domain structure refinement. Second, coevolution and deep-learning–based contact predictions have recently demonstrated advantages in protein 3D structure prediction (25). Similarly, sequence-based interdomain contact and distance predictions can be introduced into DEMO to help refine domain orientations. Work along these lines is in progress. With continuous development, we expect that DEMO will become an efficient tool for solving a significant problem that despite its extreme importance has not received sufficient attention by the field of computational structural biology.
Methods
DEMO involves 4 steps: global template identification, initial template superposition, Monte Carlo domain assembly simulation, and atomic-level linker reconstruction and refinement. A flowchart is displayed in Fig. 1.
Analogous Structure Template Identification.
Whole-chain templates are identified from the multidomain structure library by a 2-step procedure based on TM-align (11) (SI Appendix, Fig. S7). In the first step, individual domains of the query are structurally aligned with a template, regardless of the overlap between the alignments of different domains. The average TM-score of all domains is defined as the local score (L-score) for the template, where the top 500 templates with highest L-scores are selected. In the second step, domains are aligned on each of the 500 selected templates twice, once from the N to C terminal and once from the C to N terminal, with no overlap allowed in the alignments of different domains (SI Appendix, Fig. S7D). The average TM-score of all domains is defined as the global score (G-score), and alignments with the highest G-scores are selected for the next steps of initial model construction and interdomain distance profile deduction.
Initial Model Construction.
The initial conformation in DEMO is constructed from the top templates ranked by G-score. Since the domain alignments are performed separately, the aligned regions of 2 domains may be far away from each other. When this happens, we use a sliding-window based procedure to recreate domain alignments so that neighboring domains have the initial structure constructed from the neighboring regions of the template (SI Appendix, Fig. S8). In this procedure, the N domain is superposed with every position along the template, where at each position, the C domain is allowed to superpose in the remaining regions of the template at a maximum of 10 residues away from the N domain. The alignment with the highest average TM-score is finally selected to construct the initial full-length model for the query sequence.
Replica-Exchange Monte Carlo Simulation for Domain Assembly.
Starting from the initial structure, replica-exchange Monte Carlo simulations (26) are performed for domain assembly (SI Appendix, Section S3). The movements contain rigid-body rotation and translation of the smaller domain, where the larger domain is fixed during the simulation. The force field of DEMO contains 5 energy items: (i) Cα clashes between domains, (ii) a generic interdomain contact potential, (iii) interdomain distance restraints derived from templates (SI Appendix, Fig. S9), (iv) domain boundary connectivity (SI Appendix, Fig. S10), and (v) local domain distance restraint to the initial domain-template superposition. Details of these energy items are given in SI Appendix, Section S2.
The weighting parameters of different energy items are optimized by maximizing the energy–RMSD correlations of decoy structure (27), based on a separate set of 425 training proteins that are nonredundant with the test proteins reported in this study. For a target, 5 simulation runs are performed, each run with 30 replicas and each replica with 10,000 movement iterations. The decoy conformation with the lowest energy is selected for the next step of linker-based refinements.
Experimental Data Integration.
When CL and cryo-EM data are available, DEMO provides options for integrating such data as additional restraints to guide the domain assembly simulations. For cross-linking data, the restraints are implemented as a contact potential (SI Appendix, Eq. S9), with other parts of the DEMO pipeline remaining unchanged. For cryo-EM data, DEMO first fits the initial structure generated from the template into the density map by performing a quick Metropolis Monte Carlo simulation with 500 steps. This fitting simulation is guided only by the density-map correlation energy as defined by SI Appendix, Eq. S10, with movements including rigid-body rotation and translation of the full-length model after matching the centers of the structure and the global density map. Following the fitting, the normal replica-exchange Monte Carlo simulations are performed to optimize the orientation of domains with the combined cryo-EM restraints and inherent DEMO energy force field.
Domain Connectivity and Linker Refinement.
Linkers between 2 domains may be disconnected after the domain assembly simulations. To connect the domain structures, the residues from the domains are gradually released until the Cα distance between the 2 unclosed residues is <3.5(l + 1) Å, where l is the number of released residues. We note that the number of released residues is usually low for reasonable domain assembly conformations, in which only 4.7 residues on average are required to release in our benchmark tests.
Following the linker residue release, an initial Cα linker model is constructed through self-avoid random walks from the anchor of the N-terminal domain to that of the C-terminal domain, where at each walk step, a new Cα atom must satisfy the conditions that the distance between the unclosed termini is <3.5(l + 1) Å and no clash exists between the new atom and other atoms. Next, Metropolis Monte Carlo simulations are performed to refine the linker models, with movements consisting of rotations of a randomly selected segments around the axis connecting the 2 ending atoms of the segment. Following each movement, the backbone atoms (N, C, and O) and side chain centers are added to the Cα traces according to the average coordinates derived from the statistics of the PDB structures at a local Cartesian system. The energy of each decoy is evaluated by a potential containing 4 items (SI Appendix, Text S5): (i) statistical torsion-angle potential from Ramachandran plots (6, 28), (ii) Cα clashes between the linker and domain structures, (iii) statistical N-Cα-C bond angle potential, and (iv) orientation-dependent side chain contact potential (SI Appendix, Fig. S11) (27). A total of 30,000 movements are performed for each linker, with the linker model with the lowest energy function selected. Finally, the side chain conformations are added and refined, together with the full-chain model, by the FG-MD program (29).
Assembly Refinement Simulations for 3 or More Domains.
The DEMO simulation was designed for assembling 2 neighboring domains. For proteins of 3 or more domains, full-length model is constructed by iterative implementations of the 2-domain assembly simulations, where in each iteration the structure assembled from the last step is treated as a rigid-body virtual N domain.
After all iterations are completed, a domain-level global refinement is performed through a short Metropolis Monte Carlo simulation, guided by the energy of the sum of all pairwise domain interactions in previous iterations (SI Appendix, Text S6). A total of 10,000 movements are performed, consisting of small rotation and translation of individual domains, where the conformation with the lowest total energy is selected. This simulation is designed to fine-tune the arrangement of all domains to avoid possible local structural traps from the consecutive domain assembly iterations.
Supplementary Material
Acknowledgments
We thank Biao Zhang and Xi Zhang for helpful discussions on cryo-EM data processing. This work was supported in part by the National Natural Science Foundation of China (61773346, to G.Z.), the National Institute of General Medical Sciences (GM083107 and GM116960, to Y.Z.), the National Institute of Allergy and Infectious Diseases (AI134678, to Y.Z.), and the National Science Foundation (NBI1564756, to Y.Z.). This work used the Extreme Science and Engineering Discovery Environment, which is supported by the National Science Foundation (ACI1548562).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905068116/-/DCSupplemental.
References
- 1.Blevins R. A., Tulinsky A., Comparison of the independent solvent structures of dimeric alpha-chymotrypsin with themselves and with gamma-chymotrypsin. J. Biol. Chem. 260, 8865–8872 (1985). [PubMed] [Google Scholar]
- 2.Chothia C., Gough J., Vogel C., Teichmann S. A., Evolution of the protein repertoire. Science 300, 1701–1703 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J., et al. , The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu D., Zhang Y., Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R., Baker D., Macromolecular modeling with rosetta. Annu. Rev. Biochem. 77, 363–382 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Bradley P., Misura K. M., Baker D., Toward high-resolution de novo structure prediction for small proteins. Science 309, 1868–1871 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Xu D., Jaroszewski L., Li Z., Godzik A., AIDA: Ab initio domain assembly for automated multi-domain protein structure prediction and domain-domain interaction prediction. Bioinformatics 31, 2098–2105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wollacott A. M., Zanghellini A., Murphy P., Baker D., Prediction of structures of multidomain proteins from structures of the individual domains. Protein Sci. 16, 165–175 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Skolnick J., TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westbrook J., Feng Z., Chen L., Yang H., Berman H. M., The Protein Data Bank and structural genomics. Nucleic Acids Res. 31, 489–491 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Xu D., Gabow H. N., Protein domain decomposition using a graph-theoretic approach. Bioinformatics 16, 1091–1104 (2000). Correction in: Bioinformatics17, 290 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Lam S. D., et al. , Gene3D: Expanding the utility of domain assignments. Nucleic Acids Res. 44, D404–D409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandonia J.-M., Fox N. K., Brenner S. E., SCOPe: Manual curation and artifact removal in the structural classification of proteins—Extended database. J. Mol. Biol. 429, 348–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Skolnick J., Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 1–32 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 18, 342–348 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kryshtafovych A., et al. , Evaluation of the template-based modeling in CASP12. Proteins 86 (suppl. 1), 321–334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Söding J., Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Wu S., Zhang Y., LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C., Mortuza S. M., He B., Wang Y., Zhang Y., Template-based and free modeling of I-TASSER and QUARK pipelines using predicted contact maps in CASP12. Proteins 86 (suppl. 1), 136–151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovchinnikov S., Park H., Kim D. E., DiMaio F., Baker D., Protein structure prediction using Rosetta in CASP12. Proteins 86 (suppl. 1), 113–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russel D., et al. , Putting the pieces together: Integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abriata L. A., Dal Peraro M., Assessment of FM and FM/TBM modeling in CASP13 (2018). http://predictioncenter.org/casp13/doc/presentations/Assessment_FM_Abriata_DalPeraro.pdf. Accessed 17 July 2019.
- 26.Swendsen R. H., Wang J.-S., Replica Monte Carlo simulation of spin glasses. Phys. Rev. Lett. 57, 2607–2609 (1986). [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Kolinski A., Skolnick J., TOUCHSTONE II: A new approach to ab initio protein structure prediction. Biophys. J. 85, 1145–1164 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran G. T., Sasisekharan V., Conformation of polypeptides and proteins. Adv. Protein Chem. 23, 283–438 (1968). [DOI] [PubMed] [Google Scholar]
- 29.Zhang J., Liang Y., Zhang Y., Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.