Significance
Lactobacillus crispatus primarily inhabits 2 ecological niches: the human vagina and the poultry intestine. The predominance of this commensal species is indicative of a healthy status, and this species has become intriguing as an emerging probiotic. Historically, there has been a paucity of functional studies of L. crispatus due to its recalcitrance to transformation and limited molecular biology tools available. Here, we show how the endogenous type I-E CRISPR-Cas system of L. crispatus can be harnessed for flexible and efficient genetic engineering encompassing insertions, deletions, and single base substitutions. These findings expand the CRISPR toolbox with abundant and diverse type I CRISPR-Cas systems, and illustrate how endogenous systems can be harnessed to develop next-generation probiotics for human and animal health.
Keywords: CRISPR, Cas3, type I-E, genome editing, probiotic
Abstract
CRISPR-Cas systems are now widely used for genome editing and transcriptional regulation in diverse organisms. The compact and portable nature of class 2 single effector nucleases, such as Cas9 or Cas12, has facilitated directed genome modifications in plants, animals, and microbes. However, most CRISPR-Cas systems belong to the more prevalent class 1 category, which hinges on multiprotein effector complexes. In the present study, we detail how the native type I-E CRISPR-Cas system, with a 5′-AAA-3′ protospacer adjacent motif (PAM) and a 61-nucleotide guide CRISPR RNA (crRNA) can be repurposed for efficient chromosomal targeting and genome editing in Lactobacillus crispatus, an important commensal and beneficial microbe in the vaginal and intestinal tracts. Specifically, we generated diverse mutations encompassing a 643-base pair (bp) deletion (100% efficiency), a stop codon insertion (36%), and a single nucleotide substitution (19%) in the exopolysaccharide priming-glycosyl transferase (p-gtf). Additional genetic targets included a 308-bp deletion (20%) in the prophage DNA packaging Nu1 and a 730-bp insertion of the green fluorescent protein gene downstream of enolase (23%). This approach enables flexible alteration of the formerly genetically recalcitrant species L. crispatus, with potential for probiotic enhancement, biotherapeutic engineering, and mucosal vaccine delivery. These results also provide a framework for repurposing endogenous CRISPR-Cas systems for flexible genome targeting and editing, while expanding the toolbox to include one of the most abundant and diverse systems found in nature.
Clustered regularly interspaced short palindromic repeats (CRISPR) and associated proteins (Cas) provide adaptive immunity in prokaryotes against invasive nucleic acids (1). CRISPR-Cas systems are widespread in bacteria (46%) and archaea (90%), although distribution and classification vary greatly within and across phylogenetic clades (2). Currently, 2 major CRISPR-Cas system classes have been described, encompassing 6 types and 34 subtypes (3). Class 1 includes types I, III, and IV, which are defined by the presence of a multiprotein effector complex, such as the CRISPR-associated complex for antiviral defense (Cascade). In contrast, class 2 systems are composed of types II, V, and VI, which rely on single effector nucleases, such as Cas9, Cas12, or Cas13 (3). Despite these distinctions, all types carry out DNA-encoded, RNA-mediated nucleic acid targeting (4, 5), but they vary in their mechanisms of action, molecular targets (DNA or RNA), and specific sequence biases as determined by the protospacer adjacent motif (PAM) (6–8). Exogenous class 2 effector nucleases, such as Cas9 and Cas12, are widely exploited for genome editing in a plethora of eukaryotes (9, 10), hinging on the programmable nature of synthetic guide RNA technology (11–13). Remarkably, few native systems have been harnessed for in situ editing in bacteria, which is somewhat perplexing, given their widespread distribution. More specifically, this applies to type I CRISPR-Cas systems and the signature Cas3 helicase nuclease (14), which constitutes the most abundant and widespread CRISPR-Cas system in bacteria and archaea (2). Presumably, the limited usage of this system is explained by the relative paucity of published studies detailing the activity of CRISPR-Cas systems in their native hosts and lack of fundamental understanding regarding type I CRISPR arrays, accompanying Cas proteins, and corresponding guide CRISPR RNAs (crRNAs) and targeting PAMs necessary for molecular tool development (15). To date, only a handful of type I CRISPR-Cas systems have been characterized, most notably the model type I-E CRISPR-Cas system from Escherichia coli, which was actually the first observed CRISPR locus over 3 decades ago (16) and has been used more recently to demonstrate the dependency of CRISPR immunity on crRNA targeting (17, 18). The Cascade complex, encompassing the crRNA and Cas proteins, constitutes double-stranded DNA recognition machinery that drives the selective nucleotide base-pairing between the crRNA and the cDNA strand (target strand), looping out the nontarget strand generating the “R-loop” structure (19–21). Then, the Cas3 helicase nuclease is recruited by Cascade to unwind and degrade the nontarget strand in a 3′-to-5′ direction (22, 23) via nuclease- and helicase-dependent activities (14, 24). This processive single-stranded DNA degradation, combined with inefficient DNA repair mechanisms, renders self-targeting lethal in bacteria (25) unless a repair template is provided to drive RecA-dependent recombination (26).
The microbiome composition, complexity, and diversity have been the focus of extensive studies over the past decade to understand its impact on health and disease in humans (27, 28) and animals (29, 30). The human vaginal microbiome is dominated by lactobacilli with Lactobacillus crispatus as one of the predominant species (31), which also plays a key role in poultry intestinal health (29) and has been implicated in the maintenance of a healthy status, whereas its absence is correlated with a higher risk of infectious disease (32, 33). Moreover, L. crispatus has become an emerging probiotic for women’s and poultry health due to its ability to fend off invasive pathogenic bacteria through competitive exclusion, production of antimicrobial compounds and exopolysaccharides (34–36), and elicitation of a beneficial host immune response (37). However, the genetic basis of the L. crispatus probiotic features remains unknown due to the lack of molecular tools available for this genetically refractory species.
Here, we characterized a type I-E CRISPR-Cas system in the genetically recalcitrant L. crispatus species in a strain isolated from a healthy human endoscopy. We show how the endogenous Cas machinery can be repurposed in the native host for genome editing by providing engineered CRISPR arrays with self-targeting spacers in combination with various repair templates to generate a variety of genome editing outcomes.
Results
Occurrence and Diversity of CRISPR-Cas Systems in L. crispatus.
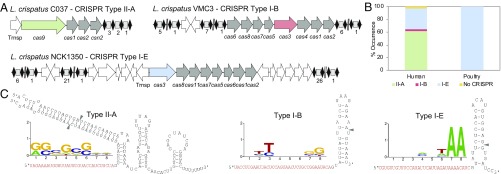
We first investigated the occurrence of CRISPR-Cas systems in 52 available genomes of L. crispatus (SI Appendix, Table S1) and characterized the architecture of the CRISPR loci using in silico analyses. Overall, we identified CRISPR loci in 51 of the 52 genomes (98% occurrence rate) and found types I-B, I-E, and II-A CRISPR-Cas systems (Fig. 1 and SI Appendix, Table S2). This is a rather high level of occurrence and diversity, even for the CRISPR-enriched Lactobacillus genus, in which CRISPR loci occur in ∼63% of genomes (38). The widespread abundance of type I systems, and ∼15% occurrence of type II systems reflect their relative amounts in bacteria (39). A total of 30 CRISPR-Cas systems were identified in the 24 human-associated strains, with 19 type II-A loci, 10 type I-E loci, and a unique type I-B locus (SI Appendix, Table S2). In poultry isolates, all type I-E loci seemed complete, with CRISPR arrays typically accompanied by a canonical set of cas genes (40), whereas only 3 human isolates (DSM20584, NCK1350, and VMC3) displayed a complete system. Interestingly, strains with degenerate type I-E systems did harbor complete type II-A systems (C037, FB049-03, OAB24-B, VMC1, VMC5, and VMC6), except for DISK12 (SI Appendix, Table S2). Noteworthy, all strains with complete type I-E systems carried multiple CRISPR arrays, typically 2 arrays located upstream of the cas locus and a third array located downstream (Fig. 1A). A single type I-B system was also detected in human strain VMC3, which also carried a complete type I-E system. In many incomplete sets, we observed the occurrence of transposases, which have been previously observed in CRISPR loci (41, 42). The coexistence of several CRISPR-Cas systems in the same genome has been previously described in several gut lactobacilli and bifidobacteria (41, 43), as well as in Streptococcus thermophilus starter cultures (44, 45). Overall, we determined widespread occurrence of CRISPR-Cas systems in L. crispatus, notably complete type I-E systems (Fig. 1B).
Fig. 1.
CRISPR-Cas systems in L. crispatus. (A) Architecture of the CRISPR loci II-A, I-B, and I-E detected in L. crispatus strains, with the signature cas genes in green (Cas9, type II-A), red (Cas3, type I-B), and blue (Cas3, type I-E) and cas genes in gray. Repeats are represented as black diamonds and spacers as gray squares, with the number of total spacers in each CRISPR array indicated below. Trnsp, transposase. (B) Occurrence and diversity of CRISPR-Cas systems in L. crispatus strains from human (gut and urogenital tract) and poultry (gut) isolates. (C) PAM prediction and representation using the frequency plot of WebLogo for each CRISPR subtype. The crRNA/tracrRNA-predicted interaction in the type II-A system with the RNase III-predicted processing sites indicated with gray arrowheads (Left) and the crRNA-predicted structure for type I-B (Center) and type I-E (Right) with the putative Cas6 processing site indicated with a gray arrowhead are shown.
PAM and Guide RNA Characterization.
Once we determined the occurrence and diversity of CRISPR-Cas systems in L. crispatus and selected type I-E as the most widespread and promising candidate, we next determined the sequences that guide Cas nucleases, namely, the PAM and the crRNAs. By nature, CRISPR spacers represent a vaccination record of immunization events over time. Therefore, we first analyzed CRISPR spacer sequences to elucidate the flanking protospacer sequences in their matching targets so as to predict the PAM, which is essential for target DNA recognition and binding (6, 8). In silico analysis of the CRISPR spacers revealed sequence homology to plasmids, phages, and bacterial chromosomes (SI Appendix, Tables S3–S5), allowing us to identify 5′-AA-3′ as a conserved PAM upstream of the protospacer for the type I-E L. crispatus CRISPR-Cas3 (Fig. 1C).
Using NUPACK to depict the predicted guides (46, 47), we determined the consensus repeat sequence for each CRISPR subtype and predicted the crRNA sequence and structure for type I and crRNA/trans-activating crRNA (tracrRNA) for type II, using previously established molecular rules about guide RNA composition and complementarity (48) (Fig. 1C). Variations in repeat sequences did not alter the predicted crRNA structures, since polymorphisms occurred in predicted bulges (SI Appendix, Table S2).
The Native Type I-E System Is Active in L. crispatus NCK1350.
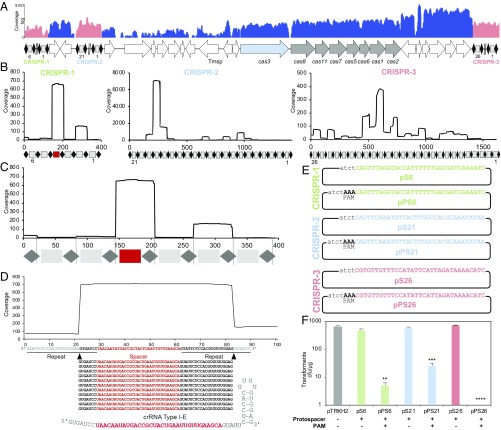
Once we established the widespread occurrence of complete type I-E CRISPR-Cas systems in L. crispatus and predicted the necessary guide RNA and targeting PAM, we selected a human endoscopy isolate, NCK1350, to validate our predictions and test the functionality of the endogenous system. RNA-sequencing (RNA-seq) data revealed constitutive expression of the cas genes encompassing a monocistronic transcript for cas3 and polycistronic expression for cascade (Fig. 2A), while the small RNA (smRNA)-seq analyses probed the transcription profiles of all 3 associated CRISPR arrays (Fig. 2B), enabling the determination of mature crRNA composition (Fig. 2 C and D). The mature crRNA structure is unique, with a 5′ handle consisting of 7 nucleotides (Fig. 2D), which differs from the canonical crRNA processing by Cas6 generating a 5′ handle of 8 nucleotides (49, 50).
Fig. 2.
CRISPR locus expression and functionality. (A) RNA-seq coverage displaying the transcriptional profile of the CRISPR locus type I-E in L. crispatus NCK1350, with mRNA in blue and smRNA for the 3 CRISPR arrays in red. Trnsp, transposase. The smRNA-seq expression profiles of the CRISPR arrays display the coverage for each spacer in each array (B), and a detailed representation of CRISPR-1 displays the coverage for each spacer repeat (C). (D) smRNA-seq displayed the crRNA maturation with the generation of the 5′ handle consisting of 7 nucleotides (5′GUGAUCC-tag). The crRNA boundaries with the terminal hairpin at the 3′ end were manually depicted. (E) Protospacer corresponding to the most recently acquired spacer of each CRISPR array was cloned into the shuttle vector pTRKH2, with and without the PAM 5′-AAA-3′, for plasmid interference assays. The lowercase sequence displays the plasmid sequence upstream of the protospacer. (F) Interference assays with a reduction of between 2 and 3 log units compared with the vector pTRKH2 or the non–PAM-containing plasmids. Bar graphs represent the mean of 3 independent biological replicates, and the error bars represent the SD. **P < 0.01, ***P < 0.001, ****P < 0.001 after Welch’s t test to compare each sample with the non–PAM-containing control.
Next, we used a plasmid interference assay to test the ability of the native system to prevent uptake of a plasmid carrying a sequence complementary to a native CRISPR spacer, flanked by the predicted PAM. Analysis of the NCK1350 spacer matches revealed 5′-AAA-3′ (an extension of the aforementioned 5′-AA-3′ PAM) as the likely PAM (SI Appendix, Table S5). We tested all 3 endogenous CRISPR loci, using a protospacer corresponding to the most recently acquired spacer within each CRISPR array (5′ end of the array, closest to the leader sequence), by cloning the corresponding protospacer into the shuttle vector pTRKH2 with or without a flanking predicted PAM (Fig. 2E and SI Appendix, Table S6). Results showed that all 3 CRISPR loci can drive interference against plasmids that carry a target protospacer flanked by the predicted PAM. Specifically, the transformation efficiency was reduced by 10-fold, 100-fold, and over 1,000-fold for loci 2, 1, and 3, respectively (Fig. 2F), reflecting high activity and specificity of this type I-E system. Overall, these results validated the predicted PAM 5′-AAA-3′, determined the guide RNA sequences, and confirmed activity of the native system in standard laboratory conditions.
Repurposing the Endogenous Type I-E CRISPR-Cas3 System for Genome Editing.
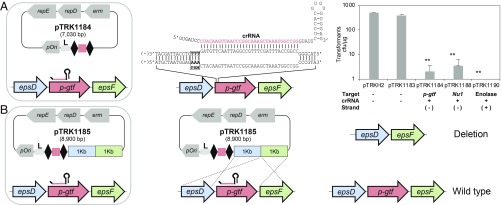
Once the functionality of the endogenous type I-E CRISPR-Cas was demonstrated in L. crispatus NCK1350, we next repurposed this endogenous system for genome editing by codelivering a self-targeting CRISPR array with editing templates. We first surveyed the L. crispatus NCK1350 genome (∼2.0 Mbp) for potential PAM sequences and found 56,591 instances of the 5′-AAA-3′ motif and 181,672 occurrences of 5′-AA-3′ on the coding strand as well as 55,061 instances of 5′-AAA-3′ and 182,194 occurrences of 5′-AA-3′ on the noncoding strand. This high frequency of PAM sequences within the NCK1350 genome suggests that the endogenous type I-E can be used to target and potentially alter every single gene in the genome, with a canonical PAM occurring, on average, every 35 nucleotides, virtually enabling widespread genome editing across this chromosome.
A plasmid-based tool was developed to reprogram the endogenous type I-E machinery based on the expression of an artificial and programmable CRISPR array carrying a self-targeting CRISPR spacer. For this purpose, a double-stranded gene block containing a promoter, 2 CRISPR repeats, and a rho-independent terminator was cloned into BglII-SalI–digested pTRKH2 to generate a flexible plasmid, pTRK1183, in which self-targeting spacers can readily be cloned (Table 1 and SI Appendix, Fig. S1). For the promoter, the native leader of the CRISPR-3 array (AT content of ∼70%) was chosen to drive the expression of the artificial CRISPR array. Conveniently, we designed pTRK1183 with 2 BsaI sites between the 2 CRISPR repeats, allowing flexible and easy insertion of spacers (33 base pairs [bp]) as programmable self-targeting guides, using annealing oligonucleotides with overhang ends compatible with the BsaI-digested plasmid (SI Appendix, Fig. S1). Thus, the artificial guide expressed from the plasmid will mimic the native type I-E crRNA from NCK1350. We used this tool to clone various self-targeting spacers close to the target gene start codon (SI Appendix, Fig. S2), redirecting the endogenous Cascade-Cas3 machinery against select chromosomal locations. For this purpose, we engineered the plasmids pTRK1184, pTRK1188, and pTRK1190 targeting, respectively, the nonessential exopolysaccharide priming-glycosyltransferase (p-gtf), the prophage DNA packaging Nu1, and the essential and highly transcribed enolase (Table 1). In all instances, self-targeting was lethal, with constructs killing over 99% of the cells across the 3 target sites (Fig. 3A).
Table 1.
Strains and plasmids used in this study
| Description | |
| Strains | |
| L. crispatus NCK1350 | L. crispatus isolated from a human endoscopy with CRISPR-Cas systems subtype I-E |
| NCK2635 | L. crispatus NCK1350 mutant with the deletion (643 bp) of the exopolysaccharide p-gtf gene |
| NCK2656 | L. crispatus NCK1350 mutant with 3 stop codons inserted (p-gtf15_16::taatagtga) in the p-gtf gene and the protospacer sequence deleted |
| NCK2659 | L. crispatus NCK1350 mutant with a single base substitution altering the PAM sequence (14A > G) (K5R) in the p-gtf gene |
| NCK2662 | L. crispatus NCK1350 mutant with the prophage DNA packaging Nu1 deleted (308 bp) |
| NCK2665 | L. crispatus NCK1350 mutant with the GFP inserted in the chromosome downstream of the enolase |
| Plasmids | |
| pTRKH2 | High-copy gram-positive shuttle vector; Ermr |
| pS6 | Spacer 6 from CRISPR-1 cloned into pTRKH2 |
| pPS6 | PAM + spacer 6 from CRISPR-1 cloned into pTRKH2 |
| pS21 | Spacer 18 from CRISPR-2 cloned into pTRKH2 |
| pPS21 | PAM + spacer 18 from CRISPR-2 cloned into pTRKH2 |
| pS26 | Spacer 26 from CRISPR-3 cloned into pTRKH2 |
| pPS26 | PAM + spacer 26 from CRISPR-3 cloned into pTRKH2 |
| pTRK1183 | Plasmid-based technology with an artificial crRNA (leader + 2 repeats + rho-terminator) cloned into pTRKH2 |
| pTRK1184 | Targeting plasmid on the exopolysaccharide p-gtf gene obtained after cloning with annealing oligonucleotides a 33-nt spacer into pTRK1183 |
| pTRK1185 | Editing plasmid containing the repair template (RTKO) to generate a knockout of the p-gtf gene, cloned into pTRK1184 |
| pTRKH2-RT | Control plasmid containing the repair template (RTKO) used to generate a knockout of the p-gtf gene, cloned into pTRKH2 |
| pTRK1186 | Editing plasmid containing the repair template (RTSTOP) to generate the insertion of 3 stop codons in the p-gtf gene, cloned into pTRK1184 |
| pTRK1187 | Editing plasmid containing the repair template (RTSNP) to perform single nucleotide substitution altering the PAM sequence in the p-gtf gene, cloned into pTRK1184 |
| pTRK1188 | Targeting plasmid on the prophage DNA packaging Nu1 gene obtained after cloning with annealing oligonucleotides a 33-nt spacer into pTRK1183 |
| pTRK1189 | Editing plasmid containing the repair template (RTKO) to generate a knockout of the Nu1 gene, cloned into pTRK1188 |
| pTRK1190 | Targeting plasmid on the enolase gene obtained after cloning with annealing oligonucleotides a 33-nt spacer into pTRK1183 |
| pTRK1191 | Editing plasmid containing the repair template (RTGFP) to generate the chromosomal insertion of the GFP gene, cloned into pTRK1188 |
Ermr, erythromycin resistance; nt, nucleotide.
Fig. 3.
Repurposing the endogenous type I-E CRISPR-Cas system. (A) Artificial crRNA is expressed with a plasmid-based system (SI Appendix, Fig. S1 and Table 1) to repurpose the endogenous type I-E against the desired chromosomal target (Center), causing cell death (Right). (Right) Base pair of the crRNA with the protospacer target located on the negative (−) or positive (+) strand is indicated The bar graphs represent the mean of 3 independent biological replicates, and the error bars represent the SD. **P < 0.01 after Welch’s t test to compare each sample with the control pTRK1183. cfu, colony-forming unit. (B) Cloning a 2-kb homologous repair template in the targeting plasmid (SI Appendix, Figs. S1 and S2) allowed generation of a markerless technology to perform genome editing in L. crispatus NCK1350 with different applications.
To trigger genome editing, we codelivered a repair template cloned into the self-targeting plasmid containing the CRISPR array to enable the host to overcome Cas3-based targeting and damage. First, we used the p-gtf target to generate a knockout, since the mutants will conveniently display a visibly distinct phenotype due to the altered exopolysaccharide content (51–53), which can also lead to altered probiotic features, such as adherence, stress resistance, and modulation of the host immune system (54–57). We designed the repair template to encompass sequences 1 kilobase (kb) upstream and 1 kb downstream of the target protospacer, and cloned into SalI-PvuI–digested pTRK1184 to generate pTRK1185 (SI Appendix, Fig. S2A). All tested transformants generated a smaller PCR product, revealing the 643-bp expected deletion in the NCK2635 mutant (100% efficiency), confirmed by sequencing (Fig. 4A). Similarly, a control plasmid was generated containing the same repair template (RT) but lacking the targeting guide (pTRKH2-RT). Indeed, when this plasmid was transformed into L. crispatus NCK1350, hundreds of transformants were obtained (Fig. 4D) and none of the PCR-screened colonies presented the deletion, indicating low-efficiency recombination without CRISPR selective pressure. This result suggests the deletion mutant NCK2635 was the consequence of Cascade-Cas3 targeting, followed by homology-directed repair based on the repair template provided on the plasmid, rather than naturally occurring homologous recombination (HR). Also, these results confirmed the lethality of Cas3-based DNA damage when a self-targeting array is delivered to repurpose the endogenous system and trigger lethal cleavage without a repair template.
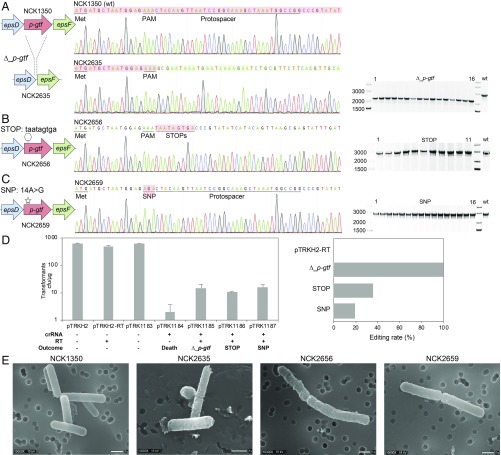
Fig. 4.
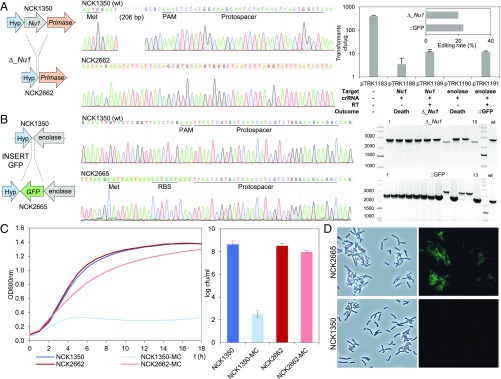
Diversity of genome editing outcomes achieved by repurposing the endogenous type I-E system in L. crispatus NCK1350. Different editing strategies can be achieved based on the repair template cloned in the targeting plasmid (SI Appendix, Figs. S1 and S2A). (A) Deletion of 643 bp in the exopolysaccharide p-gtf gene, with the chromatogram showing the sequence of the NCK1350 wild-type strain (wt) and the deletion mutant NCK2635. (B) Insertion of stop codons while deleting the protospacer region in the p-gtf gene to generate the mutant NCK2656 (eps15_16::taatagtga). (C) Single base editing performed as single base substitution to alter the PAM sequence (14A > G), creating a missense mutation (K5R) in the derivative mutant NCK2659. (D) Transformation efficiencies and editing rates (%). cfu, colony-forming unit. (E) Scanning electron microscopy of the wild-type strain L. crispatus NCK1350 and the derivative mutants NCK2635, NCK2656, and NCK2659 harboring a deletion, interruption, or single base substitution in the exopolysaccharide p-gtf gene, respectively. (Magnification: 10,000× to 13,000×.) (Scale bar: 1 μm.) Images courtesy of North Carolina State University/Claudio Hidalgo-Cantabrana and Valerie Lapham.
We then used a similar strategy to generate other genome editing outcomes to illustrate the versatility of the technology. We used the same targeting plasmid (pTRK1184), in which we cloned different repair templates to perform various editing outcomes within the p-gtf gene (SI Appendix, Fig. S2A). We introduced a stop codon in the p-gtf gene while simultaneously deleting the protospacer region (pTRK1186 in Table 1 and SI Appendix, Fig. S2A). When the plasmid was transformed into L. crispatus NCK1350, 11 transformants were obtained and PCR screening confirmed the insertion of the stop codon at the desired location with 36% efficiency (4 of 11 colonies), generating NCK2656 (Fig. 4B). The other survivors appeared to carry defective plasmids in which the targeting spacer had been excised, presumably by HR between the CRISPR repeats. Next, we carried out a single base substitution (14A > G) yielding a missense mutation (K5R) in the p-gtf target (pTRK1187 in Table 1 and SI Appendix, Fig. S2A). In this case, 16 transformants were obtained and the PCR screening confirmed the genesis of the desired single base substitution in NCK2659 (Fig. 4C) with an efficiency of 19% (3 of 16 colonies). The exopolysaccharide (EPS)-derivative mutants NCK2635 and NCK2656 displayed a rough phenotype due to the p-gtf deletion or interruption, visually distinguishable from the smooth phenotype of the wild-type strain NCK1350, when using scanning electron microscopy (Fig. 4E). The EPS-derivative mutant NCK2659 displayed an intermediate surface phenotype between the parental strain NCK1350 and the deletion mutant NCK2635 (Fig. 4E) as the amino acid change K5R did show features of both the smooth and rough morphologies of L. crispatus. These results showed that this approach can be used to generate deletions, insert stop codons, or precisely mutate a single base efficiently in the p-gtf gene.
Next, to illustrate the versatility of this approach, we targeted another chromosomal location, and deleted the prophage DNA packaging Nu1, to provide a proof of concept for prophage curing. Using the aforementioned vector, we designed a repair template completely ablating the Nu1, cloned it into SalI-PvuI–digested pTRK1188 (pTRK1189 in Table 1 and SI Appendix, Fig. S2B), and generated a 308-nucleotide deletion mutant, NCK2662, with 20% efficiency (2 of 10 colonies) (Fig. 5A). Finally, we targeted a third chromosomal locus to generate a knock-in. We strategically selected the downstream region of the enolase gene as a stable and highly expressed locus, which we previously used for antigen expression in Lactobacillus acidophilus (58) using a upp plasmid-based cloning system (59, 60). We designed a repair template containing the green fluorescent protein (GFP) gene, flanked by 2-kb homologous arms, cloned into SalI-PvuI–digested pTRK1190 to generate pTRK1191 (Table 1 and SI Appendix, Fig. S2C). In this case, PCR screening of the transformants revealed the intended GFP integration (730 bp) with 23% efficiency (3 of 13 colonies) (Fig. 5B). Prophage curing, leading to the enhancement of strain genetic stability, was demonstrated under the selective induction of mitomycin C (0.75 μg/mL), with the deletion mutant NCK2662 being able to grow, whereas cell lysis occurred in the wild-type strain NCK1350 due to prophage excision from the chromosome (Fig. 5C). The fluorescence signal of the chromosomal inserted GFP was detected in the derivative mutant NCK2665 using fluorescence microscopy, enabling monitoring of probiotic strains in future characterization through in vitro and in vivo analyses (Fig. 5D). Overall, these results show that various loci can be targeted by the endogenous type I-E machinery to generate deletions and insertions flexibly and efficiently.
Fig. 5.
Diversity of genome editing loci achieved by repurposing the endogenous type I-E system in L. crispatus NCK1350. (A, Left) Deletion of the prophage DNA packaging Nu1 gene (308 bp), with the chromatogram showing the sequence of the NCK1350 wild-type strain (wt) and the derivative mutant NCK2662. Note that the repair template was designed 206 bp upstream from the PAM to delete the complete gene (SI Appendix, Fig. S2). (A, Right) Transformation efficiencies and the editing rate (%) (Top) and the corresponding gels (Bottom) are shown. (B) Chromosomal insertion of the GFP (730 bp) downstream of the enolase gene, with the chromatogram showing the sequence of the wild-type strain and the derived mutant NCK2665. (C) Growth curve (OD600nm) of NCK1350 and derivative mutant NCK2662 in the presence of mitomycin C (MC) for prophage induction. cfu, colony-forming unit. (D) Fluorescence microscopy of NCK1350 and derivative mutant NCK2665 expressing the GFP inserted in the chromosome, using a white filter (Left) and fluorescein isothiocyanate filter (Right). (Magnification: 40×.)
Discussion
The advent of CRISPR-based technologies has revolutionized genome editing and enabled the alteration of virtually any sequence in any organism of interest. Much of this success is due to the portability, ease of delivery, and accessibility of materials and protocols for genome editing and transcriptional control (61). However, the current toolbox is limited to only a few Cas9, Cas12, and Cas13 effector proteins, predominantly optimized for use in eukaryotes. With thousands of native CRISPR-Cas systems widely occurring in bacteria and archaea, we have the opportunity to repurpose endogenous systems in their native host for genome editing, provided we can characterize their guide RNAs and targeting PAM sequences (15). Harnessing the endogenous machinery enables efficient genome editing simply by delivering a CRISPR array, together with desired repair templates. The development of such a potent tool has the potential to facilitate the engineering of many valuable bacteria that play critical roles in human health (62, 63) and important biological functions in the various habitats and niches they inhabit. Also, this opens new avenues for the functional enhancement of bacterial communities and rational design of beneficial microbes and probiotics to promote host health.
Recent studies have established L. crispatus as a key commensal species for women’s health and poultry intestinal health (29, 31–33), although it is unclear what the genetic basis of those probiotic features is. Furthermore, research in this species has been limited by the paucity of molecular tools available for functional studies, as well as limited transformation efficiencies in this genetically recalcitrant species (64, 65). Indeed, the lack of molecular tools for L. crispatus represents a bottleneck for a more comprehensive understanding of its physiology and further enhancement of its probiotic features through genome editing. The methods we used to edit various chromosomal loci in L. crispatus NCK1350 using the native CRISPR-Cas3 system illustrate how endogenous CRISPR-Cas systems can be easily repurposed for precise genome editing encompassing insertions, deletions, and single base alterations. Similar approaches have been used previously for transcriptional control in the model bacterium E. coli (66, 67) and in archaea (68), for genome editing in archaea (69, 70), and also for genome engineering of a bacteriophage (71) and Clostridium (72, 73). Here, we used the endogenous type I-E CRISPR-Cas system for efficient and precise genome editing in lactobacilli. The only unique tool available previously was based on the heterologous expression of Streptococcus pyogenes Cas9 in Lactobacillus reuteri, Lactobacillus casei, and Lactobacillus plantarum (74–76). While Cas3-based exonucleolytic activity can be toxic to bacterial cells (25, 77), the widespread HR machinery mediated by RecBCD resects DNA ends. Subsequently, RecA is recruited to drive recombination (26, 78) or RecA is recruited via the RecF pathway with RecFOR at the initial steps (79) to assist with DNA repair and genesis of the desired genome editing outcomes encoded on the repair template. In this study, we show that providing an adequately designed repair template (2-kb size) in the targeting plasmid constitutes an efficient means to carry out various editing outcomes, even in a recalcitrant species such as L. crispatus. The flexible genetic manipulation of the commensal L. crispatus uncovers tremendous potential to develop next-generation probiotics for women’s health and poultry health, including, but not limited to, enhancing the probiotic features or the development of vaccines against infectious diseases and sexually transmitted diseases. These findings also open new avenues for engineering other Lactobacillus species by repurposing their endogenous active CRISPR-Cas systems (80, 81) to enhance bacterial applications, microbiome targeting, and modulation in humans and animals. Indeed, this technology relies on the use of a single plasmid conveniently designed for easy cloning, thus enabling potent CRISPR targeting and programmable genome editing without the necessity of a large heterologous Cas nuclease, which usually requires complex plasmid engineering leading to stability artifacts.
Overall, this study provides a framework to characterize endogenous CRISPR-Cas systems, based on in silico examination, transcriptomic analyses, and plasmid interference assays. We have demonstrated how endogenous type I CRISPR-Cas systems can be repurposed for efficient genome editing of bacteria in situ, opening new avenues for next-generation engineering of industrial workhorses, commensal microbes, and beneficial probiotic bacteria for the development of engineered biotherapeutics. Additionally, this research may serve as a framework for future studies that aim at developing next-generation CRISPR-Cas tools.
Methods
CRISPR-Cas System Detection and Characterization in Silico.
The 52 L. crispatus genomes (SI Appendix, Table S1) available in the National Center for Biotechnology Information (NCBI) GenBank in December 2017 were mined to determine the occurrence and diversity of CRISPR-Cas systems in this species. The in silico analyses were performed using Cas proteins (Cas1, Cas3, and Cas9), previously identified in other lactobacilli species (38), as queries, using BLAST (82) to retrieve the Cas proteins among L. crispatus strains. Then, the putative CRISPR array(s) of each genome were identified using the CRISPR Recognition Tool (83) implemented in Geneious 10.0.6 software (84). Thereafter, the CRISPR-Cas systems of each strain were manually curated and annotated. The CRISPR subtypes were designated based on the occurrence of signature Cas proteins (Cas9-typeII and Cas3-typeI) and associated proteins as previously reported (39).
Spacer Analyses, PAM Prediction, and Guide RNA Identification.
CRISPR spacers represent an iterative vaccination record for bacteria. Bioinformatic analyses were performed to predict the PAM sequence based on spacer–protospacer match, using the spacers of each L. crispatus strain and the web server CRISPRTarget (85). The WebLogo server was used to represent the PAM sequence based on a frequency chart where the height of each nucleotide represents the conservation of that nucleotide at each position (86).
In type I systems, the crRNA represents the guide RNA that interacts with the Cascade complex to define the complementary sequence. The crRNA encompasses the repeat-spacer pair, so a repeat-spacer nucleotide sequence was used to predict the structure of the crRNA of type I-B and type I-E using the NUPACK web server (87), and then manually depicted. In type II systems, the tracrRNA has a complementary region to the CRISPR repeat sequence of the crRNA, allowing creation of the duplex crRNA/tracrRNA. Therefore, the repeat sequence of type II-A was used to identify the tracrRNA in the CRISPR locus as previously described (15), and the interaction between crRNA and tracrRNA was then predicted using the NUPACK web server and depicted manually.
Bacterial Strains and Growth Conditions.
L. crispatus NCK1350 and derivative strains used in this study (Table 1) were propagated in De Man, Rogosa, and Sharpe (MRS; Difco) broth or on MRS agar (1.5% wt/vol) plates at 37 °C under anaerobic conditions. E. coli DH10B and MC1061 were used as cloning hosts. E. coli strains were grown in brain heart infusion (BHI; Difco) broth at 37 °C with aeration (250 rpm) or on BHI agar plates at 37 °C aerobically. Transformants were selected in the presence of erythromycin (Erm) at 150 μg⋅mL−1 for E. coli or 2.5 μg⋅mL−1 for L. crispatus.
Genome Sequencing and Assembly.
Total DNA of L. crispatus NCK1350 was isolated using an UltraClean Microbial DNA Isolation Kit (MOBIO), and whole-genome sequencing was performed using a MiSeq System (Illumina) at the Roy J. Carver Biotechnology Centre of the University of Illinois (Urbana–Champaign, IL) following the supplier’s protocol. Libraries were prepared with the Hyper Library construction kit from Kapa Biosystems. The libraries were pooled as instructed, quantitated by qPCR, and sequenced on 1 lane per pool for 301 cycles from each end of the fragments on a MiSeq flowcell using a MiSeq 600-cycle sequencing kit (version 3). Fastq files of the pair-end reads were generated and demultiplexed with bcl2fastq Conversion Software (v2.17.1.14, Illumina). The adaptors were trimmed from the sequencing reads, and sequences were quality-retained. The fastq files of the pair-end reads were used as input for the genome assembly through the PATRIC web server (https://www.patricbrc.org) and also for the protein-encoding open reading frame prediction and annotation. Then, the genome annotations were manually curated in Geneious 11.0.5.
RNA Extraction and RNA Sequencing Analysis.
Total RNA of L. crispatus NCK1350 was isolated from a 10-mL MRS culture, with 2 independent biological replicates grown under anaerobic conditions to an optical density at 600 nm (OD600nm) ∼ 0.6. Cells were harvested by centrifugation (3,200 × g, 10 min, 4 °C), and the cell pellets were flash-frozen and stored at −80 °C until RNA extraction was performed. Total RNA was isolated using a Zymo Direct-Zol RNA Miniprep Kit (Zymo Research), following the protocol previously described (88). The messenger RNA (mRNA) and smRNA library preparation and sequencing were performed at the Roy J. Carver Biotechnology Centre, and data analysis was performed as previously described (88). Finally, the RNA-seq reads were mapped onto the L. crispatus NCK1350 genome using Geneious 11.0.5 software (84) with default settings, and the expression level for each coding sequence was calculated based on the normalized transcripts per million (89).
DNA Manipulations.
Chromosomal DNA from L. crispatus was isolated using the UltraClean Microbial DNA Isolation Kit, and plasmid DNA from E. coli was obtained using a QIAprep Spin Miniprep Kit (Qiagen) following the manufacturer’s instructions. PCR primers, double-stranded synthetic DNA for plasmid interference assays, and single-stranded DNA for annealing oligonucleotides were synthesized by Integrated DNA Technologies (IDT; Morrisville, NC). Synthetic DNA for the target-specific crRNA was synthesized by Genewiz (Suzhou, China). PCR amplicons for colony screening were generated using standard PCR protocols and Taq blue DNA polymerase (Denville Scientific). Q5 Hot Start High-Fidelity Polymerase (New England Biolabs [NEB]) was used to PCR-amplify DNA for cloning purpose. PCR products were analyzed on 0.8 to 1.5% agarose gels. DNA sequencing was performed by Genewiz to confirm sequence content. Restriction digestions were performed with 1 μg of plasmid DNA in a final volume of 50 μL, at 37 °C for 1 h, using high-fidelity restriction enzymes (NEB). Purification of digested products for ligation was performed using a Monarch PCR & DNA Cleanup Kit (NEB) or Monarch DNA Gel Extraction Kit (NEB). Ligation reactions were performed at a 3:1 insert/vector ratio using 50 ng of vector in a final volume of 10 μL, using Instant Sticky-end Ligase Master Mix (NEB) based on the manufacturer’s instructions.
Single-stranded DNA oligonucleotides were resuspended in IDT Duplex Buffer to a final concentration of 100 μM. Then, equal amounts (2 μg) of each strand (A + B) were mixed, and the final volume was adjusted to 50 μL with IDT Duplex Buffer. Both strands were annealed at 95 °C for 2 min, followed by incubation at 25 °C for 45 min. All annealed oligonucleotides were stored at −20 °C.
Construction of Interference Plasmids.
The pTRKH2 plasmid (90), a replicating shuttle vector for E. coli and Lactobacillus, was used for all plasmid constructions. The interference plasmids were constructed by ligation of the synthetic double-stranded DNA protospacers, with or without the PAM, into BglII-SalI–digested pTRKH2 (SI Appendix, Table S6). The constructs were transformed into rubidium chloride-treated competent E. coli DH10B cells using heat shock at 42 °C for 1 min, followed by another 2-min incubation on ice. Cells were recovered in 900 μL of SOC medium (NEB) at 37 °C aerobically for 3 h and then plated on BHI with 150 μg⋅mL−1 Erm. The resulting interference plasmids were PCR-screened in E. coli transformants with M13 primers (SI Appendix, Table S6) for the presence of the insert and sequenced to confirm sequence content.
Construction of the CRISPR-Based Editing Vector pTRK1183 to Repurpose the Endogenous Type I-E System in L. crispatus NCK1350.
The plasmid-based technology pTRK1183 was constructed by ligation of the synthetic double-stranded gene block that represents the artificial crRNA of NCK1350 into BglII-SalI–digested pTRKH2 (Table 1). The artificial crRNA contains a promoter that is the native leader of the CRISPR-3 array of L. crispatus NCK1350, together with 2 repeats and a rho-independent terminator (BBa_B1006, registry of standard biological parts) (SI Appendix, Fig. S1). The ligation was transformed in rubidium chloride competent E. coli DH10B cells as described above. The resulting pTRK1183 plasmid was isolated from E. coli transformants, checked by PCR with M13 primers (SI Appendix, Table S6) for the presence of the insert, and sequenced to confirm sequence composition.
Two BsaI sites are located between the 2 direct repeats of the artificial crRNA in pTRK1183 to allow the insertion of spacers (targets) using annealing oligonucleotides. The pTRK1183 plasmid was isolated from E. coli, digested with BsaI, and ligated with the annealing oligonucleotides carrying overhang ends. The constructs were transformed in rubidium chloride competent E. coli DH10B cells as described above. The resulting plasmid is a pTRK1183 derivative containing a spacer to target the exopolysaccharide gene p-gtf (Enzyme Commission [EC] 2.7.8.6) generating the plasmid pTRK1184, a spacer to target the prophage DNA packaging gene Nu1 generating pTRK1188, or a spacer to target the enolase gene (EC 4.2.1.11) generating the plasmid pTRK1190 (Table 1). The resulting plasmids were isolated from E. coli transformants, checked by PCR with M13 primers (SI Appendix, Table S6) for the presence of the insert, and sequenced to confirm sequence content.
pTRK1183 and derived targeting plasmids (pTRK1184, pTRK1188, and pTRK1190) present a SalI-PvuI restriction site ideal to clone a designed repair template to perform genome editing repurposing the endogenous type I-E system in L. crispatus NCK1350. For this purpose, a double-stranded DNA synthetic gene block containing 2-kb homologous arms to the p-gtf gene (SI Appendix, Fig. S2A) was PCR-amplified with primers p-gtf_RTKO_SalI_F and p-gtf_RTKO_PvuI_R (SI Appendix, Table S6) and cloned into SalI-PvuI–digested pTRK1184 generating the plasmid pTRK1185 (Table 1) that contains both the crRNA guide to target the gene and the repair template to perform a deletion of 643 bp. The same repair template was cloned into SalI-PvuI–digested pTRKH2 generating the plasmid pTRKH2-RT (Table 1), containing the repair template but not the targeting CRISPR array, that serves as a control plasmid for spontaneous HR.
Similarly, a different gene block (2 kb) designed to introduce 3 stop codons in the p-gtf gene (SI Appendix, Fig. S2A) was amplified by PCR using the primers p-gtf_RTSTOP_PvuI_R and p-gtf_RTSTOP_PvuI_R and cloned into SalI-PvuI–digested pTRK1184 generating the plasmid pTRK1186.
Another repair template (2 kb) was designed to introduce a single base substitution in the p-gtf gene to alter the PAM. In this case, the primers p-gtf_RTSNP_Up_SalI_F and p-gtf_RTSNP_Up_R were used to perform a chromosomal amplification of the upstream homologous arm introducing the mutation in the repair template, and the primers p-gtf_RTSNP_Dw_SOE-PCR_F and p-gtf_RTSNP_Dw_PvuI_R amplified the downstream region. Then, both repair templates were overlapped using splicing by overlapping extension (SOE)-PCR with the primers p-gtf_RTSNP_Up_SalI_F and gtf_RTSNP_Dw_PvuI_R to generate the final 2-kb repair template that was cloned into SalI-PvuI–digested pTRK1184 generating plasmid pTRK1187.
To delete the prophage DNA packaging gene Nu1, a double-stranded DNA synthetic gene block containing 2-kb homologous arms (SI Appendix, Fig. S2B) was amplified using the primers Nu1_RTKO_SalI_F and Nu1_RTKO_SalI_R and cloned into SalI-PvuI–digested pTRK1188 generating plasmid pTRK1189.
To perform the chromosomal insertion of the GFP at the 3′ end of the enolase gene, a repair template containing 730 bp corresponding to the GFP and 2-kb homologous arms to the enolase gene region was designed (SI Appendix, Fig. S2C). For this purpose, the enolase downstream region was amplified using the primers enolase_RTGFP_Dw_SalI_F and enolase_RTGFP_Dw_R, the upstream region was amplified using the primers enolase_RTGFP_Up_F and enolase_RTGFP_Up_PvuI_R, and the gene block containing the GFP was amplified using the primers RTGFP_GFP_SOE-PCR_F and RTGFP_GFP_SOE-PCR_R. Then, the 3 PCR fragments were overlapped using SOE-PCR with the primers enolase_RTGFP_Dw_SalI_F and enolase_RTGFP_Up_PvuI_R, and the resulting amplicon (2.73 kb) was cloned into SalI-PvuI–digested pTRK1190 generating plasmid pTRK1191.
The final plasmid constructs were PCR-screened using the general M13_F and lacZ_Rev primers or M13_F and 253_R primers (SI Appendix, Table S6) to check plasmid content.
Transformation of L. crispatus NCK1350.
The transformation of L. crispatus NCK1350 was optimized based on a slight modification of a previously described transformation protocol for lactobacilli (60). Stationary cells grown anaerobically were inoculated (1% vol/vol) into MRS broth previously reduced to anaerobic conditions and grown until OD600nm ∼ 0.3 was achieved. At this point, penicillin G was added to a final concentration of 10 μg⋅mL−1 and cells were incubated for another hour. Then, cells were harvested by centrifugation (3,200 × g, 10 min, 4 °C) and washed 3 times with electroporation buffer containing 1 M sucrose and 3.5 mM MgCl2. Finally, cells were resuspended in 1 mL of electroporation buffer and aliquoted in 200 μL for direct use. For each transformation, 2 μg of plasmid was combined with 200 μL of cells, and 2-mm cuvettes were used for electrotransformation under 2.5-kV, 25-μF, and 400-Ω conditions. Cells were recovered in 1 mL of MRS broth previously reduced to anaerobic conditions and incubated at 37 °C in anaerobic conditions for 18 h. Transformants were selected on MRS plates with 2.5 μg⋅mL−1 Erm for 48 to 72 h.
The transformants obtained were PCR-screened and sequenced to confirm the presence of desired mutations. For the exopolysaccharide gene p-gtf, the primers KO_p-gtf_F and KO_p-gtf_R were used for the chromosomal PCR amplification (2.8 kb in wild type and 2.2 kb in deletion mutant), and the primers p-gtf_F and p-gtf_R were used to sequence the p-gtf region for the 3 different editing outcomes performed in this target. For the prophage DNA packaging Nu1 gene, the primers KO_Nu1_F and KO_Nu1_R were used for the chromosomal PCR amplification (2.8 kb in wild type and 2.5 kb in deletion mutant), and the primers Nu1_F and Nu1_R were used for sequencing. To check the insertion of the GFP in the enolase region, the primers GFP_Insertion_F and GFP_Insertion_R were used for PCR amplification (2.4 kb in wild type and 3.1 kb in insertion mutant) of the chromosomal location, and the primers GFP_F and GFP_R were used to check the sequence.
Scanning Electron Microscopy.
The L. crispatus NCK1350 and derived exopolysaccharide mutants (NCK2635, NCK2656, and NCK2659) were grown for 16 h as described above. Bacterial cells from 10 mL of culture were harvested by centrifugation (10 min, 2,500 rpm), resuspended in 10 mL of 3% glutaraldehyde in 0.1 M Na cacodylate buffer (pH 5.5), and stored at 4 °C until processed. Bacterial suspensions were filtered using a 0.4-μm pore polycarbonate Nucleopore filter. Filters containing bacteria were washed 3 times with 30-min changes of 0.1 M Na cacodylate buffer (pH 5.5), dehydrated with a graded series of ethanol to 100% ethanol, and then critical point-dried (Tousimis Samdri-795; Tousimis Research Corp.) in liquid CO2. Dried filters were mounted on stubs with double-stick tape and silver paint and sputter-coated (Hummer 6.2 sputtering system; Anatech USA) with 50 Å of Au/Pd. Samples were held in a vacuum desiccator until viewed using a JEOL JSM-5900LV scanning electron microscope. Images were acquired at a resolution of 1,280 × 960 pixels. Sample preparation and scanning electron microscopy were performed at the College of Agriculture and Life Sciences Center for Electron Microscopy at North Carolina State University.
Prophage Induction.
L. crispatus NCK1350 and the NCK2662 mutant lacking the prophage DNA packaging Nu1 (Table 1) were grown for 16 h as described above. Then, 10 mL of fresh broth was inoculated (1%), and mitomycin C (Sigma) was added (0.75 μg/mL) when the cultures reached OD600nm = 0.2 to 0.3. Bacterial growth was monitored (OD600nm) over 18 h, and cell counts where performed on regular media at the final time point. Three independent biological replicates were performed, with 2 technical replicates in each experiment.
Fluorescence Microscopy.
The L. crispatus NCK1350 and NCK2665 derivative mutants expressing the chromosomal inserted GFP were grown for 16 h as described above. Then, bacterial cells were washed, placed on a microscope slide, and covered with a coverslip (Fisher Scientific). The preparations were observed with a Nikon Eclipse E600 microscope using 40× magnification. The fluorescein isothiocyanate filter (excitation = 480, emission = 585) was used for visualization of the GFP signal.
Statistical Analyses.
In all figures, the bar graphs represent the mean of 3 independent biological replicates and the error bars represent the SD. Data distribution was analyzed with Welch’s t test, used to compare 2 unpaired groups (sample vs. control) under the hypothesis that the 2 groups contain equal means. Comparisons with a P value <0.05 were considered statistically significant. The statistical analyses were performed in R studio, v1.1.463.
Accession Numbers.
The chromosomal sequence and the RNA-seq data of L. crispatus NCK1350 reported in this paper have been deposited in the NCBI genome database and Short Read Archive database, respectively, under the BioProject ID PRJNA521996 (91). The whole-genome sequence has been deposited under the accession number SGWL00000000. The mRNA sequences have been deposited under the accession numbers SRR8568636 to SRR8568637, and the smRNA sequences have been deposited under the accession numbers SRR8568722 to SRR8568723.
Supplementary Material
Acknowledgments
We thank Valerie Lapham of the College of Agriculture and Life Sciences Center for Electron Microscopy (North Carolina State University) for her electron microscopy assistance and technical expertise, as well as Courtney Klotz for editorial support. We acknowledge funding by North Carolina State University and the North Carolina Agricultural Foundation.
Footnotes
Conflict of interest statement: R.B. and C. H.-C. are inventors on several patents related to CRISPR-Cas systems and their uses. R.B. is a shareholder of Caribou Biosciences, Intellia Therapeutics, Locus Biosciences, and Inari Ag, and a cofounder of Intellia Therapeutics and Locus Biosciences. C.H.-C. is an advisor and cofounder of Microviable Therapeutics.
Data deposition: The chromosomal sequence and RNA-sequencing data of Lactobacillus crispatus NCK1350 have been deposited in the National Center for Biotechnology database under BioProject ID PRJNA521996. The whole-genome sequence has been deposited under accession number SGWL00000000. The messenger RNA sequences have been deposited under accession numbers SRR8568636–SRR8568637, and the small RNA sequences have been deposited under accession numbers SRR8568722–SRR8568723.
See Profile on page 15754.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905421116/-/DCSupplemental.
References
- 1.Barrangou R., et al. , CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Crawley A. B., Henriksen J. R., Barrangou R., CRISPRdisco: An automated pipeline for the discovery and analysis of CRISPR-cas systems. CRISPR J 1, 171–181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarova K. S., Wolf Y. I., Koonin E. V., Classification and nomenclature of CRISPR-Cas systems: Where from here? CRISPR J 1, 325–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasiunas G., Sinkunas T., Siksnys V., Molecular mechanisms of CRISPR-mediated microbial immunity. Cell. Mol. Life Sci. 71, 449–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abudayyeh O. O., et al. , C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mojica F. J., Díez-Villaseñor C., García-Martínez J., Almendros C., Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Marraffini L. A., Sontheimer E. J., Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deveau H., et al. , Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrangou R., Doudna J. A., Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Zetsche B., et al. , Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M., et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P., et al. , RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinkunas T., et al. , Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidalgo-Cantabrana C., Goh Y. J., Barrangou R., Characterization and repurposing of type I and type II CRISPR-Cas systems in bacteria. J. Mol. Biol. 431, 21–33 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A., Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brouns S. J., et al. , Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marraffini L. A., Sontheimer E. J., CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11, 181–190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jore M. M., et al. , Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18, 529–536 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Xiao Y., et al. . Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas System. Cell 170, 48–60.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinkunas T., et al. , In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 32, 385–394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeff L., Brouns S. J. J., Joo C., Repetitive DNA reeling by the Cascade-Cas3 complex in nucleotide unwinding steps. Mol. Cell. 70, 385–394.e3. [DOI] [PubMed] [Google Scholar]
- 23.Mulepati S., Bailey S., In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 288, 22184–22192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huo Y., et al. , Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol. 21, 771–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomaa A. A., et al. , Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio 5, e00928-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selle K., Barrangou R., Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 23, 225–232 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Human Microbiome Project Consortium , Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Integrative HMP (iHMP) Research Network Consortium , The integrative human microbiome project: Dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 16, 276–289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei S., Morrison M., Yu Z., Bacterial census of poultry intestinal microbiome. Poult. Sci. 92, 671–683 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Dec M., Nowaczek A., Stępień-Pyśniak D., Wawrzykowski J., Urban-Chmiel R., Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 18, 168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravel J., et al. , Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 1), 4680–4687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M. B., et al. , Diverse vaginal microbiomes in reproductive-age women with vulvovaginal candidiasis. PLoS One 8, e79812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arokiyaraj S., Seo S. S., Kwon M., Lee J. K., Kim M. K., Association of cervical microbial community with persistence, clearance and negativity of human papillomavirus in Korean women: A longitudinal study. Sci. Rep. 8, 15479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnarumma G., et al. , Lactobacillus crispatus L1: High cell density cultivation and exopolysaccharide structure characterization to highlight potentially beneficial effects against vaginal pathogens. BMC Microbiol. 14, 137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nardini P., et al. , Lactobacillus crispatus inhibits the infectivity of Chlamydia trachomatis elementary bodies, in vitro study. Sci. Rep. 6, 29024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parolin C., et al. , Lactobacillus crispatus BC5 interferes with Chlamydia trachomatis infectivity through integrin modulation in cervical cells. Front. Microbiol. 9, 2630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizzo A., et al. , Lactobacillus crispatus mediates anti-inflammatory cytokine interleukin-10 induction in response to Chlamydia trachomatis infection in vitro. Int. J. Med. Microbiol. 305, 815–827 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Sun Z., et al. , Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 6, 8322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koonin E. V., Makarova K. S., Zhang F., Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarova K. S., et al. , An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hidalgo-Cantabrana C., Crawley A. B., Sanchez B., Barrangou R., Characterization and exploitation of CRISPR loci in Bifidobacterium longum. Front. Microbiol. 8, 1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horvath P., et al. , Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131, 62–70 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Briner A. E., et al. , Occurrence and diversity of CRISPR-cas systems in the genus Bifidobacterium. PLoS One 10, e0133661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath P., et al. , Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190, 1401–1412 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magadán A. H., Dupuis M. E., Villion M., Moineau S., Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One 7, e40913 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chylinski K., Le Rhun A., Charpentier E., The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 10, 726–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briner A. E., Henriksen E. D., Barrangou R., Prediction and validation of native and engineered Cas9 guide sequences. Cold Spring Harb. Protoc. 2016, pdb.prot086785. [DOI] [PubMed] [Google Scholar]
- 48.Briner A. E., Barrangou R., Guide RNAs: A glimpse at the sequences that drive CRISPR-cas systems. Cold Spring Harb. Protoc. 2016, pdb.top090902 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Li H., Structural principles of CRISPR RNA processing. Structure 23, 13–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang R., Preamplume G., Terns M. P., Terns R. M., Li H., Interaction of the Cas6 riboendonuclease with CRISPR RNAs: Recognition and cleavage. Structure 19, 257–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lebeer S., et al. , Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 75, 3554–3563 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fanning S., et al. , Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. U.S.A. 109, 2108–2113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Kranenburg R., Vos H. R., van Swam I. I., Kleerebezem M., de Vos W. M., Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: Complementation, expression, and diversity. J. Bacteriol. 181, 6347–6353 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hidalgo-Cantabrana C., et al. , Immune modulation capability of exopolysaccharides synthesised by lactic acid bacteria and bifidobacteria. Probiotics Antimicrob. Proteins 4, 227–237 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Castro-Bravo N., Wells J. M., Margolles A., Ruas-Madiedo P., Interactions of surface exopolysaccharides from Bifidobacterium and Lactobacillus within the intestinal environment. Front. Microbiol. 9, 2426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hidalgo-Cantabrana C., et al. , A single mutation in the gene responsible for the mucoid phenotype of Bifidobacterium animalis subsp. lactis confers surface and functional characteristics. Appl. Environ. Microbiol. 81, 7960–7968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawaharada Y., et al. , Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312 (2015). [DOI] [PubMed] [Google Scholar]
- 58.O’Flaherty S., Klaenhammer T. R., Multivalent chromosomal expression of the Clostridium botulinum serotype A neurotoxin heavy-chain antigen and the Bacillus anthracis protective antigen in Lactobacillus acidophilus. Appl. Environ. Microbiol. 82, 6091–6101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Douglas G. L., Klaenhammer T. R., Directed chromosomal integration and expression of the reporter gene gusA3 in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 77, 7365–7371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goh Y. J., et al. , Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 75, 3093–3105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LaManna C. M., Barrangou R., Enabling the rise of a CRISPR World. CRISPR J 1, 205–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goh Y. J., Barrangou R., Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli. Curr. Opin. Biotechnol. 56, 163–171 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Klotz C., Barrangou R., Engineering components of the Lactobacillus S-layer for biotherapeutic applications. Front. Microbiol. 9, 2264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beasley S. S., Takala T. M., Reunanen J., Apajalahti J., Saris P. E., Characterization and electrotransformation of Lactobacillus crispatus isolated from chicken crop and intestine. Poult. Sci. 83, 45–48 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Heravi R. M., Nasiraii L. R., Sankian M., Kermanshahi H., Varasteh A. R., Optimization and comparison of two electrotansformation methods for lactobacilli. Biotechnology (Faisalabad) 11, 50–54 (2012). [Google Scholar]
- 66.Chang Y., Su T., Qi Q., Liang Q., Easy regulation of metabolic flux in Escherichia coli using an endogenous type I-E CRISPR-Cas system. Microb. Cell Fact. 15, 195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo M. L., Mullis A. S., Leenay R. T., Beisel C. L., Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 43, 674–681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stachler A. E., Marchfelder A., Gene repression in haloarchaea using the CRISPR (clustered regularly interspaced short palindromic repeats)-cas I-B system. J. Biol. Chem. 291, 15226–15242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y., et al. , Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 44, e34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng F., et al. , Harnessing the native type I-B CRISPR-Cas for genome editing in a polyploid archaeon. J. Genet. Genomics 44, 541–548 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Kiro R., Shitrit D., Qimron U., Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 11, 42–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pyne M. E., Bruder M. R., Moo-Young M., Chung D. A., Chou C. P., Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 6, 25666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J., Zong W., Hong W., Zhang Z. T., Wang Y., Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab. Eng. 47, 49–59 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Leenay R. T., et al. , Genome editing with CRISPR-cas9 in Lactobacillus plantarum revealed that editing outcomes can vary across strains and between methods. Biotechnol. J. 14, e1700583 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Oh J. H., van Pijkeren J. P., CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 42, e131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song X., Huang H., Xiong Z., Ai L., Yang S., CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei. Appl. Environ. Microbiol. 83, e01259-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vercoe R. B., et al. , Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9, e1003454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wigley D. B., Bacterial DNA repair: Recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11, 9–13 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Morimatsu K., Kowalczykowski S. C., RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol. Cell 11, 1337–1347 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Crawley A. B., Henriksen E. D., Stout E., Brandt K., Barrangou R., Characterizing the activity of abundant, diverse and active CRISPR-Cas systems in lactobacilli. Sci. Rep. 8, 11544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanozky-Dawes R., Selle K., O’Flaherty S., Klaenhammer T., Barrangou R., Occurrence and activity of a type II CRISPR-Cas system in Lactobacillus gasseri. Microbiology 161, 1752–1761 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bland C., et al. , CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8, 209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kearse M., et al. , Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Biswas A., Gagnon J. N., Brouns S. J., Fineran P. C., Brown C. M., CRISPRTarget: Bioinformatic prediction and analysis of crRNA targets. RNA Biol. 10, 817–827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zadeh J. N., et al. , NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Theilmann M. C., et al. , Lactobacillus acidophilus metabolizes dietary plant glucosides and externalizes their bioactive phytochemicals. MBio 8, e01421-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wagner G. P., Kin K., Lynch V. J., Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012). [DOI] [PubMed] [Google Scholar]
- 90.O’Sullivan D. J., Klaenhammer T. R., High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137, 227–231 (1993). [DOI] [PubMed] [Google Scholar]
- 91.Hidalgo-Cantabrana C., et al. , Lactobacillus crispatus NCK1350. BioProject. Available at https://www.ncbi.nlm.nih.gov/bioproject/521996. Deposited 12 February 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.