Abstract
Telomere shortening has been associated with multiple age-related diseases such as cardiovascular disease, diabetes, and dementia. However, the biological mechanisms responsible for these associations remain largely unknown. In order to gain insight into the metabolic processes driving the association of leukocyte telomere length (LTL) with age-related diseases, we investigated the association between LTL and serum metabolite levels in 7,853 individuals from seven independent cohorts. LTL was determined by quantitative polymerase chain reaction and the levels of 131 serum metabolites were measured with mass spectrometry in biological samples from the same blood draw. With partial correlation analysis, we identified six metabolites that were significantly associated with LTL after adjustment for multiple testing: lysophosphatidylcholine acyl C17:0 (lysoPC a C17:0, p-value = 7.1 × 10−6), methionine (p-value = 9.2 × 10−5), tyrosine (p-value = 2.1 × 10−4), phosphatidylcholine diacyl C32:1 (PC aa C32:1, p-value = 2.4 × 10−4), hydroxypropionylcarnitine (C3-OH, p-value = 2.6 × 10−4), and phosphatidylcholine acyl-alkyl C38:4 (PC ae C38:4, p-value = 9.0 × 10−4). Pathway analysis showed that the three phosphatidylcholines and methionine are involved in homocysteine metabolism and we found supporting evidence for an association of lipid metabolism with LTL. In conclusion, we found longer LTL associated with higher levels of lysoPC a C17:0 and PC ae C38:4, and with lower levels of methionine, tyrosine, PC aa C32:1, and C3-OH. These metabolites have been implicated in inflammation, oxidative stress, homocysteine metabolism, and in cardiovascular disease and diabetes, two major drivers of morbidity and mortality.
Subject terms: Metabolomics, Telomeres
Introduction
Telomeres are located at the ends of chromosomes and protect against spontaneous DNA damage, thus preserving genomic integrity1,2. The progressive shortening of telomere length with each subsequent cell division underlies the so-called mitotic clock, i.e. the limited replicative capacity of a cell3. Replicative senescence and subsequent cell death occurs when the mean telomere length reaches a critical value and telomere length is therefore seen as a marker for biological age4–6. Short leukocyte telomere length (LTL) has been associated with age5,7–9 and multiple age-related diseases such as cardiovascular disease10–15, diabetes10,16,17 and dementia18,19. Short LTL has also been associated with mortality20–27, although not all studies support this association28–33. However, the biological mechanisms underlying the associations of LTL with age-related diseases and mortality are still largely unknown.
Longevity in humans has previously been associated with various metabolic traits in the elderly, including traits related to blood pressure and lipids, suggesting that changes at the metabolic level are key features in longevity34–36. Metabolic profiles have been associated with age and various age-related diseases, such as type 2 diabetes, atherosclerosis, cancer, and Alzheimer’s Disease37–44. Only a few studies investigated the association of metabolic markers with LTL, all using untargeted metabolomics45–47. One study focused on metabolic markers involved in aging and early development in 6,055 individuals included in the TwinsUK registry45. Although a combined set of 22 metabolites was strongly correlated with age and age-related traits, the individual metabolites were not significantly associated with LTL45. Another study identified 19 metabolites associated with LTL in a small sample of American Indians (n = 423)46. The most recent study was conducted in 3,511 females from the TwinsUK registry, reporting associations of five metabolites with LTL. These include gamma-glutamyltyrosine, gamma-glutamylphenylalanine, 1-stearoylglycerophosphoinositol, 1-palmitoylglycerophosphoinositol, and 4-vinylphenol sulfate47.
In the current study, we used a standardized targeted metabolomics approach to investigate the association between LTL and serum metabolites of key biochemical pathways in the largest sample so far consisting of 7,853 individuals from seven independent population-based cohorts from Europe and Australia. We further aimed to disentangle the metabolic pathways that are represented by the metabolites significantly associated with LTL.
Methods
Cohort descriptions and measurements of LTL and metabolites
The cohorts included in this study are the Cooperative Health Research in the Region of Augsburg (KORA) study, the Netherlands Twin Register (NTR), the Estonian Genome Center University of Tartu (EGCUT) study, the TwinsUK cohort, the Erasmus Rucphen Family (ERF) study, the Leiden Longevity Study (LLS), and the Queensland Institute of Medical Research (QIMR) study, all part of the ENGAGE consortium. Details on the individual cohorts as well as details on the LTL quantitative polymerase chain reaction measurements and the metabolites as measured with the AbsoluteIDQTM p150 kit (BIOCRATES Life Sciences AG, Innsbruck, Austria) are provided in the Supplementary Materials. In summary, both LTL and metabolite concentrations were measured in the same laboratories according to a common protocol, using blood samples taken at the same time point. To ensure good data quality, each metabolite had to meet three criteria for inclusion in the study: 1) coefficient of variation (CV) not exceeding 25%; 2) less than 5% missing values; 3) median of metabolite concentrations above the limit of detection. This quality control was performed per cohort. Supplementary Table 1 describes the reasons for exclusion of metabolites from the analysis for each cohort. The metabolites measured include hexoses (H1), amino acids (AA), acyl-carnitines (AC), sphingomyelins (SMs), diacyl phosphatidylcholines (PC aa), acyl-alkyl-phosphatidylcholines (PC ae) and lysophosphatidylcholines (lysoPC).
Written informed consent was obtained from all study participants. The study protocol was approved by the medical ethics boards of the Helmholtz Zentrum München, VUmc Amsterdam, University of Tartu, St. Thomas’ Hospital London, Erasmus MC Rotterdam, LUMC Leiden, and Queensland Institute of Medical Research and all investigations were carried out in accordance with the Declaration of Helsinki.
Statistical analysis
To standardize LTL measurements across cohorts we Z-transformed the LTL values. Metabolite concentration values were natural log-transformed to attain a better approximation of the normal distribution. We performed partial correlation analysis per cohort, adjusting for age and sex, and if necessary for family relationships (model 1). In the extended model (model 2) we additionally adjusted for body mass index (BMI). We performed a sensitivity analysis to test the robustness of the results and repeated the meta-analysis excluding studies with low sample size or high mean LTL values.
A multiple testing-corrected statistical significance threshold for association of metabolite concentrations with LTL was defined at the meta-analysis level. We accounted for multiple testing by Bonferroni correction based on the effective number of independent variables (VeffLi) in the metabolite concentration data48 (https://neurogenetics.qimrberghofer.edu.au/matSpD/). The VeffLi value was determined using the correlation matrix of the quality controlled and log-transformed metabolomics data in the ERF and NTR cohorts, yielding a VeffLi (representing the number of independent metabolites) of 46 for both cohorts. This resulted in a Bonferroni corrected significance threshold of p-value < 1.1 × 10−3 (=0.05/46).
Pathway analysis of the associated metabolites
For interpretation of the observed associations we followed two bioinformatics approaches in parallel. First, we employed a bioinformatics pipeline based on a workflow management software tool called “Taverna” (http://www.taverna.org.uk)49 to determine if the metabolites associated with LTL share a network space within two reaction steps. We took as input all possible pairs of significantly associated metabolites. In this pipeline, all the reactions within a radius of two steps in the reaction space of the first metabolite were obtained from the KEGG database50. The second metabolite is searched against the substrates and the products of the reactions obtained in the previous step. The presence of the second metabolite in any of the reaction steps is an indication that the two metabolites participate in reactions within two steps of each other. In the final step, the path between the two metabolites is returned to the user. In order to prevent non-specific connections, an intermediate step filters out hub metabolites such as ATP, ADP, and NADP. Next, we used the function “heatmap.2” included in the R package “gplots” (https://CRAN.R-project.org/package=gplots) to prepare a heat map of the correlation of the individual metabolites with LTL in models 1 and 2. For this analysis, default functions for clustering were used (distance measure: euclidean distance). We also derived a correlation matrix for the significantly associated metabolites in ERF and visualized this in a correlogram using the R package “corrplot”51.
Results
General characteristics of the study populations are provided in Table 1. The study covers a wide age range, with the mean age of the participants ranging from 18.4 to 62.9 years. Most studies had approximately equal numbers of males and females, except for NTR (33% female) and TwinsUK (only females). Mean LTL ranged from 1.44 (LLS) to 3.58 (TwinsUK). BMI was on average between 25.2 and 27.6 kg/m2, but was unavailable at the time of metabolite and LTL assessment in the QIMR study.
Table 1.
General characteristics of the study populations.
| n | n_BMI | % female | LTL | Age | BMI | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | SD | 95% CI | mean | SD | 95% CI | mean | SD | 95% CI | ||||
| KORA | 3003 | 2988 | 51.8 | 1.85 | 0.33 | 1.84 - 1.86 | 56.08 | 13.25 | 55.61 - 56.55 | 27.61 | 4.80 | 27.44 - 27.78 |
| NTR | 1314 | 1307 | 33.3 | 2.54 | 0.47 | 2.51 - 2.57 | 50.60 | 14.13 | 49.84 - 51.36 | 25.97 | 3.80 | 25.76 - 26.18 |
| EGCUT | 1084 | 1084 | 50.3 | 1.90 | 0.30 | 1.88 - 1.92 | 37.78 | 15.70 | 36.85 - 38.71 | 25.16 | 4.56 | 24.89 - 25.43 |
| TwinsUK | 810 | 810 | 100.0 | 3.58 | 0.64 | 3.54 - 3.62 | 53.72 | 10.76 | 53.10 - 54.34 | 26.44 | 5.35 | 26.13 - 26.75 |
| ERF | 806 | 806 | 53.7 | 1.79 | 0.37 | 1.76 - 1.82 | 47.76 | 13.97 | 46.80 - 48.72 | 27.17 | 4.81 | 26.84 - 27.50 |
| LLS | 643 | 643 | 50.1 | 1.44 | 0.27 | 1.42 - 1.46 | 62.91 | 6.61 | 62.40 - 63.42 | 26.65 | 4.01 | 26.34 - 26.96 |
| QIMR | 193 | 0 | 48.2 | 3.43 | 0.56 | 3.35 - 3.51 | 18.44 | 12.65 | 16.66 - 20.22 | NA | NA | NA |
Abbreviations: n = number of participants with data available on metabolites, telomere length, age, and sex; n_BMI = number of participants with data available on metabolites, telomere length, age, sex, and BMI; LTL = leukocyte telomere length; SD = standard deviation. LTL as a ratio of telomere repeat length to copy number of the single copy gene 36B4; Age in years; BMI in kg/m2.
Out of the 131 metabolites that passed quality control, 27 showed at least nominally significant correlation (p-value < 0.05) with LTL when adjusting for age and sex in model 1 (Table 2). Six metabolites surpassed the multiple-testing corrected significance threshold (p-value < 1.1 × 10−3). Five of these metabolites were consistently associated with LTL in the same direction in at least five out of seven studies: lysophosphatidylcholine acyl C17:0 (lysoPC a C17:0, r = 0.05, p-value = 7.1 × 10−6) and phosphatidylcholine acyl-alkyl C38:4 (PC ae C38:4, r = 0.04, p-value = 9.0 × 10−4) were positively associated with LTL, while methionine (Met, r = −0.04, p-value = 9.2 × 10−5), tyrosine (Tyr, r = −0.04, p-value = 2.1 × 10−4), and phosphatidylcholine diacyl C32:1 (PC aa C32:1, r = −0.04, p-value = 2.4 × 10−4) were negatively associated with LTL. Although hydroxypropionylcarnitine (C3-OH, r = −0.10, p-value = 2.6 × 10−4) was also found negatively associated with LTL, this effect was only based on two out of seven studies. Additionally adjusting for BMI in model 2 had limited effect on the correlation coefficients of the six significant metabolites in model 1 and all metabolites except PC ae C38:4 remained significantly associated with LTL (Table 2). The summary statistics for all metabolite-LTL correlations for both models can be found in Supplementary Table 2. Study-specific results for the age- and sex-adjusted model are provided in Supplementary Table 3.
Table 2.
Partial correlation meta-analysis results of LTL and metabolites (p-value in model 1 < 0.05).
| Metabolite | Model 1: age + sex | Model 2: age + sex + BMI | Metabolite full name | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | direction* | r | p-value | FDR | n | direction* | r | p-value | FDR | ||
| lysoPC a C17:0 | 7853 | ++−++++ | 0.05 | 7.1 × 10 −6 | 9.3 × 10−4 | 7638 | ++−+++ | 0.04 | 4.7 × 10 −4 | 6.9 × 10−3 | Lysophosphatidylcholine acyl C17:0 |
| Met | 7852 | −−−−−−+ | −0.04 | 9.2 × 10 −5 | 6.0 × 10−3 | 7637 | −−−−−− | −0.05 | 7.5 × 10 −5 | 9.3 × 10−4 | Methionine |
| Tyr | 7047 | −−−−?−+ | −0.04 | 2.1 × 10 −4 | 6.9 × 10−3 | 6832 | −−−−?− | −0.04 | 8.9 × 10 −4 | 6.9 × 10−3 | Tyrosine |
| PC aa C32:1 | 7851 | −−−−+−+ | −0.04 | 2.4 × 10 −4 | 6.9 × 10−3 | 7636 | −−−−++ | −0.04 | 3.4 × 10 −4 | 6.9 × 10−3 | Phosphatidylcholine diacyl C32:1 |
| C3-OH | 1449 | ????−−? | −0.10 | 2.6 × 10 −4 | 6.9 × 10−3 | 1449 | ????−− | −0.10 | 2.7 × 10 −4 | 6.0 × 10−3 | Hydroxypropionylcarnitine |
| PC ae C38:4 | 7853 | +−+−+++ | 0.04 | 9.0 × 10 −4 | 2.0 × 10−2 | 7638 | +−+−++ | 0.03 | 4.7 × 10−3 | 3.1 × 10−2 | Phosphatidylcholine acyl-alkyl C38:4 |
| PC ae C40:3 | 7853 | +++++++ | 0.04 | 1.6 × 10−3 | 3.0 × 10−2 | 7638 | ++++++ | 0.03 | 8.5 × 10−3 | 3.8 × 10−2 | Phosphatidylcholine acyl-alkyl C40:3 |
| PC ae C40:5 | 7853 | +−+++++ | 0.04 | 1.9 × 10−3 | 3.1 × 10−2 | 7638 | +−++++ | 0.03 | 1.9 × 10−2 | 1.0 × 10−1 | Phosphatidylcholine acyl-alkyl C40:5 |
| SM C20:2 | 7853 | +−+−+++ | 0.03 | 2.4 × 10−3 | 3.5 × 10−2 | 7638 | +−+−++ | 0.03 | 2.7 × 10−3 | 2.0 × 10−2 | Sphingomyeline C20:2 |
| C9 | 5262 | +??−++ ? | 0.04 | 2.9 × 10−3 | 3.8 × 10−2 | 5247 | +??−++ | 0.04 | 1.1 × 10−2 | 7.5 × 10−2 | Nonaylcarnitine |
| PC ae C40:4 | 7853 | +0+++++ | 0.03 | 4.1 × 10−3 | 4.9 × 10−2 | 7638 | +−++++ | 0.03 | 2.8 × 10−2 | 1.1 × 10−1 | Phosphatidylcholine acyl-alkyl C40:4 |
| PC aa C38:3 | 7852 | −−−−+++ | −0.03 | 7.0 × 10−3 | 7.5 × 10−2 | 7637 | −−−−++ | −0.02 | 3.5 × 10−2 | 1.3 × 10−1 | Phosphatidylcholine diacyl C38:3 |
| PC ae C36:1 | 7852 | +−+−+++ | 0.03 | 7.4 × 10−3 | 7.5 × 10−2 | 7637 | +−+−++ | 0.02 | 5.8 × 10−2 | 2.1 × 10−1 | Phosphatidylcholine acyl-alkyl C36:1 |
| PC aa C36:1 | 7850 | −−−−+++ | −0.03 | 9.3 × 10−3 | 8.3 × 10−2 | 7635 | −−−−++ | −0.03 | 4.0 × 10−3 | 3.0 × 10−2 | Phosphatidylcholine diacyl C36:1 |
| PC ae C40:6 | 7853 | +−+−+++ | 0.03 | 9.5 × 10−3 | 8.3 × 10−2 | 7638 | +−+−++ | 0.02 | 8.5 × 10−2 | 2.4 × 10−1 | Phosphatidylcholine acyl-alkyl C40:6 |
| SM (OH) C16:1 | 7047 | +−+−?++ | 0.03 | 1.1 × 10−2 | 9.4 × 10−2 | 6832 | +−+−?+ | 0.02 | 5.3 × 10−2 | 1.6 × 10−1 | Hydroxysphingomyeline C16:1 |
| C2 | 7853 | −−+−−−+ | −0.03 | 1.4 × 10−2 | 1.0 × 10−1 | 7638 | −−+−−− | −0.03 | 1.5 × 10−2 | 8.3 × 10−2 | Acetylcarnitine |
| PC ae C36:2 | 7853 | ++−−+++ | 0.03 | 1.4 × 10−2 | 1.0 × 10−1 | 7638 | +−−−++ | 0.02 | 1.3 × 10−1 | 2.4 × 10−1 | Phosphatidylcholine acyl-alkyl C36:2 |
| PC ae C38:3 | 7852 | +++−+++ | 0.03 | 1.7 × 10−2 | 1.1 × 10−1 | 7637 | +++−++ | 0.02 | 3.6 × 10−2 | 1.3 × 10−1 | Phosphatidylcholine acyl-alkyl C38:3 |
| PC aa C42:0 | 7853 | +++++++ | 0.03 | 2.0 × 10−2 | 1.3 × 10−1 | 7638 | +++−++ | 0.02 | 8.9 × 10−2 | 2.4 × 10−1 | Phosphatidylcholine diacyl C42:0 |
| PC aa C32:0 | 7853 | −−−−+++ | −0.03 | 2.1 × 10−2 | 1.3 × 10−1 | 7638 | −−−−++ | −0.03 | 5.9 × 10−3 | 3.5 × 10−2 | Phosphatidylcholine diacyl C32:0 |
| PC aa C40:5 | 7849 | −−−−+++ | −0.03 | 2.2 × 10−2 | 1.3 × 10−1 | 7634 | −−−−++ | −0.03 | 1.7 × 10−2 | 8.3 × 10−2 | Phosphatidylcholine diacyl C40:5 |
| PC aa C38:1 | 836 | ?????++ | 0.08 | 2.6 × 10−2 | 1.5 × 10−1 | 643 | ?????+ | 0.05 | 1.7 × 10−1 | 2.4 × 10−1 | Phosphatidylcholine diacyl C38:1 |
| PC aa C36:2 | 7853 | −−−−−++ | −0.02 | 3.0 × 10−2 | 1.6 × 10−1 | 7638 | −−−−−+ | −0.03 | 1.0 × 10−2 | 4.9 × 10−2 | Phosphatidylcholine diacyl C36:2 |
| PC aa C34:1 | 7852 | −−−−+++ | −0.02 | 3.1 × 10−2 | 1.6 × 10−1 | 7637 | −−−−++ | −0.03 | 1.3 × 10−2 | 7.5 × 10−2 | Phosphatidylcholine diacyl C34:1 |
| PC ae C42:4 | 7853 | +−+++++ | 0.02 | 3.5 × 10−2 | 1.8 × 10−1 | 7638 | +−++++ | 0.01 | 2.1 × 10−1 | 2.5 × 10−1 | Phosphatidylcholine acyl-alkyl C42:4 |
| SM C26:0 | 5454 | +??−+++ | 0.03 | 4.3 × 10−2 | 2.1 × 10−1 | 5246 | +??−++ | 0.02 | 2.3 × 10−1 | 2.6 × 10−1 | Sphingomyeline C26:0 |
Abbreviations: n = number of participants, r = correlation coefficient, BMI = body mass index, FDR = false discovery rate.
*Order of cohorts in direction column: KORA, NTR, EGCUT, TwinsUK, ERF, LLS, QIMR; Direction of effect represented by − (negative correlation) + (positive correlation) or ? (not included) for each study. Bold p-values: associations surpassing significance threshold (p-value < 1.1 × 10-3).
We next conducted a sensitivity analysis to determine whether the analyses were driven by a single cohort. We removed two cohorts from the analysis: the TwinsUK cohort, which had a high mean LTL () and the QIMR cohort, which had a small sample size (N = 193) and was on average younger than the other cohorts. After excluding the TwinsUK cohort from the meta-analysis, all metabolites remained significantly associated with LTL, except for PC aa C32:1 (p-value = 1.1 × 10−3) (Supplementary Table 4). All metabolites, except for PC ae C38:4 (p-value = 1.8 × 10−3), remained significantly associated with LTL after excluding the QIMR cohort from the meta-analysis (Supplementary Table 5).
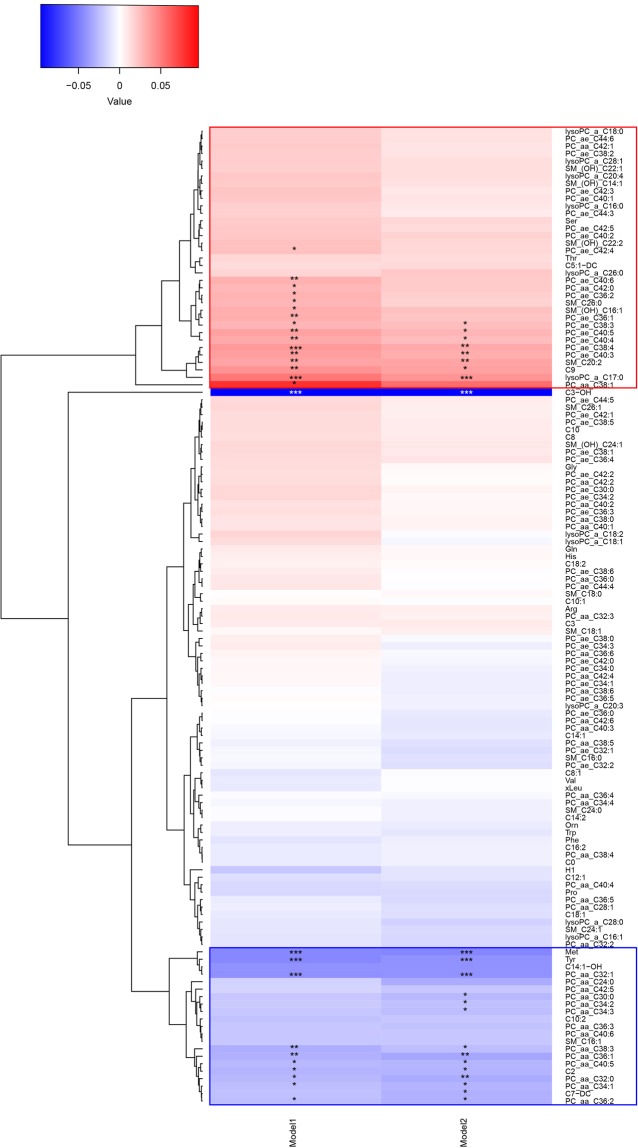
To explore to which extent the various metabolites cluster, we constructed a heat map based on the correlation of each individual metabolite with LTL in both model 1 and model 2 (Fig. 1). The heat map shows two clusters of which one (hereafter referred to as “cluster 1”) includes lysoPC a C17:0, PC ae C38:4, and a series of PC ae metabolites positively associated with LTL, while the second cluster (hereafter referred to as “cluster 2”) includes methionine, tyrosine, PC aa C32:1, and a series of PC aa metabolites negatively associated with LTL. Figure 1 further shows that C3-OH is relatively dissimilar from all other metabolites. A correlogram of the six metabolites associated with LTL after correction for multiple testing is presented in Fig. 2 and shows a positive correlation of methionine with the three PC metabolites. LysoPC a C17:0 and PC ae C38:4 (cluster 1) are most strongly correlated in Fig. 2. Methionine and tyrosine are highly correlated with each other and both amino acids are correlated to PC aa C32.1 (Fig. 2).
Figure 1.
Heat map showing the results of the cluster analysis of metabolite correlations with LTL. The two statistical models used in our analysis are shown on the horizontal axis and all individual metabolites are depicted on the vertical axis. In model 1 we investigated the association of the metabolites with LTL adjusting for age and sex and in model 2 we additionally adjusted for BMI. A blue color indicates a negative partial correlation point estimate, while a red color indicates a positive partial correlation point estimate. Cluster 1 is shown by a red rectangle and cluster 2 by a blue rectangle. The stars represent the significance: *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001.
Figure 2.
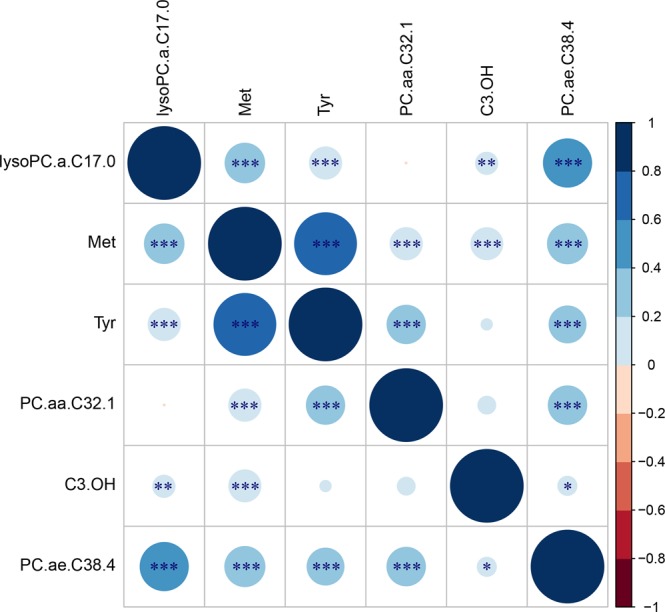
Correlogram of the six metabolites associated with LTL after correction for multiple testing in the first model using ERF data. Positive correlations are displayed in blue and negative correlations in red. Color intensity and the size of the circle are proportional to the correlation coefficients, with larger circles indicating higher correlation point estimates. Abbreviations: lysoPC.a.C17.0 = lysophosphatidylcholine acyl C17:0; Met = Methionine; Tyr = Tyrosine; PC.aa.C32.1 = phosphatidylcholine diacyl C32:1; C3.OH = hydroxypropionylcarnitine; PC.ae.C38.4 = phosphatidylcholine acyl-alkyl C38:4. *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001.
Pathway analysis using the Taverna workflow showed that phosphatidylcholines (lysoPC a C17:0, PC aa C32:1, and PC ae C38:4) and methionine are involved in homocysteine metabolism. Homocysteine is the intermediate product in the conversion of the amino acid methionine to cysteine, a precursor of the antioxidant glutathione (Fig. 3). Briefly, PC is a precursor of choline which is oxidized to betaine. Betaine is used to convert homocysteine to methionine. Methionine is first converted to S-adenosylmethionine followed by demethylation to S-adenosylhomocysteine (SAH). Next, hydrolysis of SAH forms homocysteine, which can either be re-methylated into methionine (transmethylation cycle) or metabolized to cysteine (transsulfuration pathway) as shown in Fig. 3.
Figure 3.
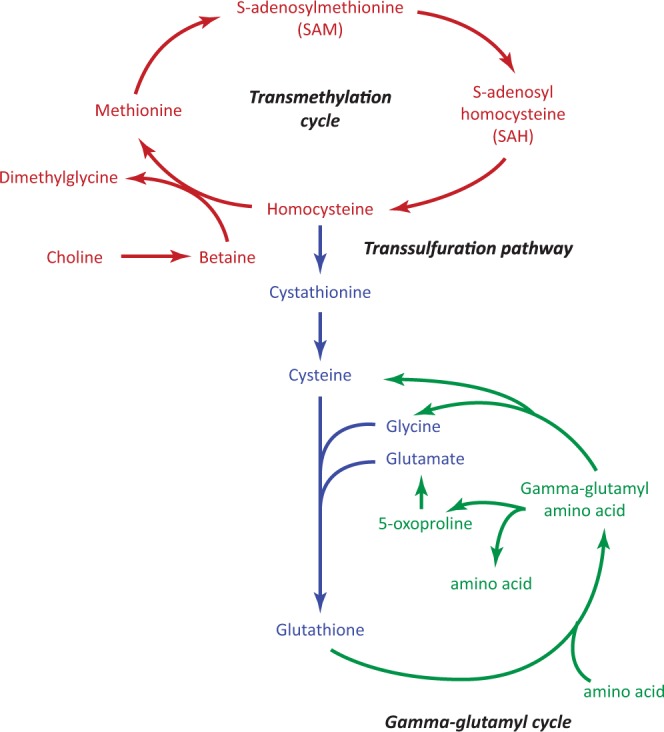
Methionine metabolic pathways. Indicated in red are the metabolites of the transmethylation pathway, in blue those of the transsulfuration pathway, and in green those involved in the gamma-glutamyl cycle. Modified figure from Dash et al.113.
Discussion
When adjusting for false positive findings due to multiple testing, this study identified significant associations between LTL and six metabolites, which form two extended clusters. These metabolites include three phosphatidylcholines (lysoPC a C17:0, PC aa C32:1, PC ae C38:4), two amino acids (methionine, tyrosine), and one acylcarnitine (C3-OH). Longer LTL was associated with higher levels of lysoPC a C17:0 and PC ae C38:4, and with lower levels of methionine, tyrosine, PC aa C32:1, and C3-OH. Pathway analysis highlighted a key role of the homocysteine metabolism.
A problem when comparing our findings to those in earlier studies is that the metabolites are platform-specific and thus we cannot replicate directly previous findings43,44. However, the PCs significantly associated with LTL in our study belong to the same metabolite family of glycerophospholipids52 as two metabolites significantly associated with LTL in the study of Zierer et al.47: 1‐stearoylglycerophosphoinositol and 1‐palmitoylglycero-phosphoinositol. These metabolites are involved in fatty acid metabolism and particularly membrane composition in biological aging. The small study (N = 423) performed in American Indians from the Strong Family Heart Study also found associations of glycerophospholipids (e.g. glycerophosphoethanolamines, glycero-phosphocholine, and glycerophosphoglycerol) with LTL46.
The metabolite most significantly associated with LTL in our study was lysoPC a C17:0. LysoPCs are formed through hydrolysis of PCs by phospholipase A253 and have pro-atherogenic and pro-inflammatory effects through impairment of endothelium-dependent vascular relaxation54, monocyte recruitment and macrophage proliferation55,56, and increased expression of adhesion molecules57. Previously, negative associations of lysoPC a C17:0 levels with high-sensitivity C-reactive protein (hsCRP), interleukin-6, insulin, and myocardial infarction have been found58,59. These results indicate the involvement of inflammation58,59. Inflammation and cardiovascular disease are related to telomere shortening60,61 and our study brings to surface lysoPC a C17:0 as a novel key player.
We further identified a negative association of the two highly correlated amino acids methionine and tyrosine with LTL. Methionine is an essential amino acid and involved in multiple important biological processes necessary for normal growth and development in mammals, including protein synthesis, methylation, the transsulfuration pathway, and homocysteine metabolism62. Previous studies have shown that a methionine-restricted diet increased lifespan in rodents63–67. Tyrosine is a non-essential amino acid and a precursor for several catecholamines, including dopamine, as well as thyroid hormones (T3 and T4)68,69. Low thyroid hormone levels have been associated with increased lifespan in multiple animal models70–72 and in humans73–75. Moreover, a role of tyrosine as developmental regulator and modulator of longevity has been described in Caenorhabditis elegans76. Tyrosine can also form a dipeptide with gamma-glutamate called gamma-glutamyltyrosine (http://www.hmdb.ca/metabolites/HMDB0011741), which was negatively associated with LTL in the TwinsUk cohort47. It is involved in the gamma-glutamyl cycle (as shown in Fig. 3) and indicates involvement of increased oxidative stress47, a factor related to LTL shortening77,78. Elevated blood levels of the amino acid tyrosine are seen in obese individuals79,80, and were found to be a novel risk factor for the development of diabetes38,81. Type 2 diabetes has been associated with shorter LTL10,16,17.
Both methionine and tyrosine are correlated to PC aa C32:1, which is the fourth metabolite significantly associated with LTL. PCs are the major phospholipids in cell membranes and lipoproteins82,83. They consist of a glycerol backbone with different fatty acid combinations that are linked by ester (a) or ether (e) bonds, resulting in either diacyl (aa) or acyl-alkyl (ae) PCs84. We observed a cluster of metabolites (cluster 2) negatively associated with LTL, including methionine, tyrosine, PC aa C32:1 and multiple other diacyl PCs. Various metabolites of this cluster including PC aa C32:1, PC aa C36:1, PC aa C38:3, and PC aa C40:5, have been associated with increased risk of type 2 diabetes40. The other PC that surpassed the significance threshold in model 1 was PC ae C38:4. However, PC ae C38:4 was nominally significant after including BMI in the model (model 2) and after excluding the younger QIMR study from the meta-analysis. PC ae C38:4 showed a positive association with LTL and clustered with lysoPC a C17:0 and a series of PC ae metabolites (cluster 1) that also show consistent effect across cohorts such as PC ae C40:3, PC ae C40:4, and PC ae C40:5 (FDR < 0.05). In line with this finding, PC ae’s have been shown to have antioxidant properties, protecting lipids from oxidation85,86, and the metabolites in this cluster showed a reduced risk of type 2 diabetes40.
Although also C3-OH was found to be associated with LTL when adjusting for multiple testing, the association with LTL was only observed in ERF and LLS. In the other five studies, this metabolite did not pass the quality control. Therefore, this finding and other findings based on data of two studies only, such as PC aa C38:1, should be interpreted with care and more research, including alternative assessments of these metabolites, is needed. C3-OH is a metabolite of interest for further investigation as it is an acylcarnitine and involved in lipid transport as well as lipid and fatty acid metabolism (http://www.hmdb.ca/metabolites/HMDB0013125). Carnitine is mainly absorbed from the diet but can also be synthesized from the amino acids lysine and methionine87 and is essential for energy metabolism as it transports fatty acids from the cytosol into the mitochondrion for β-oxidation88,89. Figure 2 shows indeed that C3-OH is correlated to methionine, as predicted87. Carnitine insufficiency has been implicated as a common trait of insulin-resistant states, including advanced age, genetic diabetes, and diet-induced obesity90. However, when clustering the correlations of the metabolites to LTL, we find that C3-OH does not cluster with other metabolites (Fig. 1).
Pathway analysis using the Taverna workflow revealed that both methionine and PCs are part of homocysteine metabolism. Our results give novel metabolic insights into the findings of previous studies that describe an increase in plasma homocysteine with age and shortening of LTL with increasing homocysteine levels91,92. Our study suggests that lysoPC a C17:0, PC aa C32:1, PC ae C38:4 as well as methionine and tyrosine are key metabolites in the link between the homocysteine pathway and telomere length. Homocysteine metabolism has been implicated in a wide range of age-related diseases, such as cardiovascular diseases93,94, dementia95,96, Alzheimer’s disease96,97, diabetes and its associated vascular complications98–100, and in mortality101–104. Taking together the findings of our study with that of previous studies, the endothelium may be the tissue of interest. There is substantial evidence that homocysteine and lysoPC are involved in endothelial dysfunction77,78 caused by inflammation and oxidative stress105–109. In cultured endothelial cells, homocysteine was also shown to accelerate telomere shortening and endothelial senescence92,110.
A major strength of this study is that both LTL and metabolites were measured centrally, using a standard protocol and blood samples taken at the same time point. Metabolite levels were quantified using the AbsoluteIDQ p150 kit (Biocrates Life Sciences, Innsbruck, Austria) that detects biologically relevant metabolites from four compound classes: acylcarnitines, amino acids, hexoses, and phosho- and sphingolipids. This method has been proven to be in conformance with FDA Guideline ‘Guidance for Industry—Bioanalytical Method Validation (May 2001)’111, which implies proof of reproducibility within a given error range. At the same time, measuring metabolites with this specific platform may be considered also a limitation of our study as other metabolites might also be related to LTL.
In conclusion, using data from 7,853 individuals from seven independent cohorts, we found longer LTL associated with higher levels of lysoPC a C17:0 and PC ae C38:4, and with lower levels of methionine, tyrosine, PC aa C32:1, and C3-OH. These metabolites form two clusters, one including lysoPC a C17:0, PC ae C38:4, and a series of PC ae metabolites positively associated with LTL, while the second cluster includes methionine, tyrosine, PC aa C32:1, and a series of PC aa metabolites. These metabolites have been implicated in cardiovascular disease and diabetes, two major drivers of morbidity and mortality. The functional role of these metabolites involves inflammation and oxidative stress. Our pathway analysis links the metabolites to homocysteine metabolism, a pathway linked to cardiovascular disease, diabetes and many other age-related diseases.
Supplementary information
Acknowledgements
KORA: KORA was financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany and supported by grants from the German Federal Ministry of Education and Research (BMBF). Part of this work was financed by the German National Genome Research Network (NGFN; NGFNPlus, project number 01GS0834) and supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. Telomere assays were funded by the ENGAGE consortium.
NTR: We thank all participants in the Netherlands Twin Register. Research was funded by the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW grants 904-61-090, 985-10-002,904-61-193,480-04-004, 400-05-717, Addiction-31160008 Middelgroot-911-09-032, Spinozapremie 56-464-14192), Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL, 184.021.007), the VU University’s Institute for Health and Care Research (EMGO+), the European Community’s Seventh Framework Program (FP7/2007-2013), ENGAGE (HEALTH-F4-2007-201413) and the European Science Council (ERC - 230374 and ERC-284167). Harmen H.M. Draisma is funded by the Wellcome Trust (WT205915).
EGCUT: EGCUT was supported by Estonian Research Council [IUT20-60, IUT24-6 and PUT1665 to K.F.]; European Union Horizon 2020 [692145 and European Union through the European Regional Development Fund [2014-2020.4.01.15-0012 GENTRANSMED]. We also thank all participants of the Estonian Biobank cohort. EGCUT studies were financed by University of Tartu (grant “Center of Translational Genomics”), by Estonian Goverment (grant #SF0180142s08, grant #ETF9353) and by European Commission through the European Regional Development Fund in the frame of grant “Centre of Excellence in Genomics” and Estonian Research Infrastructure’s Roadmap and through FP7 grant #313010.
TwinsUK: The study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007–2013), ENGAGE project grant agreement (HEALTH-F4-2007-201413). The study also receives support from the Dept of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London. TDS is an NIHR senior Investigator and is holder of an ERC Advanced Principal Investigator award. Genotyping was performed by The Wellcome Trust Sanger Institute, support of the National Eye Institute via an NIH/CIDR genotyping project.
ERF: The ERF study has received funding from the Centre for Medical Systems Biology (CMSB) and Netherlands Consortium for Systems Biology (NCSB), both within the framework of the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO). ERF study is also a part of EUROSPAN (European Special Populations Research Network) (FP6 STRP grant number 018947 (LSHG-CT-2006-01947)); European Network of Genomic and Genetic Epidemiology (ENGAGE) from the European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F4-2007-201413; “Quality of Life and Management of the Living Resources” of 5th Framework Programme (no. QLG2-CT-2002-01254); FP7 project EUROHEADPAIN (nr 602633), the Internationale Stichting Alzheimer Onderzoek (ISAO); the Hersenstichting Nederland (HSN); and the JNPD under the project PERADES (grant number 733051021, Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease using multiple powerful cohorts, focused Epigenetics and Stem cell metabolomics). A first version of the manuscript has been published in the PhD thesis of Linda Broer “(Genetic) Epidemiology of Aging”112. We are grateful to all general practitioners for their contributions, to P. Veraart for her help in genealogy, J. Vergeer for the supervision of the laboratory work and P. Snijders for his help in data collection.
LLS: We thank all participants of the Leiden Longevity Study. The research leading to these results has received funding from the European Union’s Seventh Framework Programme (FP7/2007–2011) under grant agreement n° 259679. This study was supported by a grant from the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology, Biobanking and Biomolecular Resources Research Infrastructure, (BBMRI-NL, 184.021.007) and the Netherlands Consortium for Healthy Ageing (grant 050-060-810), all in the framework of the Netherlands Genomics Initiative, Netherlands Organization for Scientific Research (NWO), and by Unilever Colworth. J. Deelen was financially supported by the Alexander von Humboldt Foundation.
QIMR: We thank Marlene Grace and Ann Eldridge for twin recruitment and data collection, Lisa Bardsley for preparation of DNA samples, David Smyth for IT/database support, and the twins and their families for their participation. Data collection was supported by grants to NGM and MJW from the Australian Research Council and Australian National Health and Medical Research Council (NHMRC). Telomere length assessment was co-funded by the European Community’s Seventh Framework Programme (FP7/2007–2013), ENGAGE project, grant agreement HEALTH-F4-2007-201413 and NHMRC-European Union Collaborative Research Grant 496739. GWM and DRN were supported by the NHMRC Fellowship (619667) and ARC Future Fellowship (FT0991022) schemes, respectively.
Author Contributions
L.B., H.H.M.D., R.P., D.I.B., C.M.D. and C.G. designed the study. A.S., L.B., H.H.M.D., R.P., E.A., M.B., M.M., M.R., D.R.N., H.K.D., V.C., J.D. and A.A.M.V. performed the analyses. L.B., M.B., M.M., V.C., N.A., E.J.C.G., A.D., I.Y., K.F., T.H., A.K.H., A.I., S.E.M., G.W.M., S.P.M., K.S., H.E.D.S., D.H., R.W.S., J.B.W., G.W. and M.J.W. were involved in data collection and/or provided data. N.G.M., N.J.S., A.M., P.E.S., T.D.S., D.I.B., C.M.D. and C.G. were involved in supervision of individual cohorts. A.S., L.B., H.H.M.D., R.P., D.I.B., C.M.D. and C.G. interpreted the results and A.S., L.B., C.M.D. and C.G. wrote the manuscript. All authors critically reviewed and approved the manuscript.
Data Availability
All results generated during this study are included in this published article and its Supplementary Materials. The datasets analysed for each individual cohort can be requested by contacting the responsible Principal Investigator. Because of restrictions based on privacy regulations and informed consent of the participants, data cannot be made freely available in a public repository. For the Rotterdam Study data, requests should be directed towards the management team of the Rotterdam Study (secretariat.epi@erasmusmc.nl), which has a protocol for approving data requests.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ashley van der Spek, Linda Broer, Harmen H. M. Draisma and René Pool contributed equally.
Dorret I. Boomsma, Cornelia M. van Duijn and Christian Gieger jointly supervised this work.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47282-6.
References
- 1.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 3.Vaziri H, et al. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc. Natl. Acad. Sci. USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdallah P, et al. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/S0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 6.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 7.Lindsey J, McGill NI, Lindsey LA, Green DK, Cooke HJ. In vivo loss of telomeric repeats with age in humans. Mutat. Res. 1991;256:45–48. doi: 10.1016/0921-8734(91)90032-7. [DOI] [PubMed] [Google Scholar]
- 8.Slagboom PE, Droog S, Boomsma DI. Genetic determination of telomere size in humans: a twin study of three age groups. Am. J. Hum. Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- 9.Broer L, et al. Meta-analysis of telomere length in 19,713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet. 2013;21:1163–1168. doi: 10.1038/ejhg.2012.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzpatrick AL, et al. Leukocyte telomere length and cardiovascular disease in the cardiovascular health study. Am. J. Epidemiol. 2007;165:14–21. doi: 10.1093/aje/kwj346. [DOI] [PubMed] [Google Scholar]
- 11.Maubaret CG, et al. Telomeres are shorter in myocardial infarction patients compared to healthy subjects: correlation with environmental risk factors. J. Mol. Med. (Berl.) 2010;88:785–794. doi: 10.1007/s00109-010-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehkopf DH, et al. Leukocyte Telomere Length in Relation to 17 Biomarkers of Cardiovascular Disease Risk: A Cross-Sectional Study of US Adults. PLoS Med. 2016;13:e1002188. doi: 10.1371/journal.pmed.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brouilette S, Singh RK, Thompson JR, Goodall AH, Samani NJ. White cell telomere length and risk of premature myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 2003;23:842–846. doi: 10.1161/01.ATV.0000067426.96344.32. [DOI] [PubMed] [Google Scholar]
- 14.Brouilette SW, et al. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet. 2007;369:107–114. doi: 10.1016/S0140-6736(07)60071-3. [DOI] [PubMed] [Google Scholar]
- 15.Haycock PC, et al. Leucocyte telomere length and risk of cardiovascular disease: systematic review and meta-analysis. BMJ. 2014;349:g4227. doi: 10.1136/bmj.g4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeanclos E, et al. Shortened telomere length in white blood cells of patients with IDDM. Diabetes. 1998;47:482–486. doi: 10.2337/diabetes.47.3.482. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Miao K, Wang H, Ding H, Wang DW. Association between telomere length and type 2 diabetes mellitus: a meta-analysis. PLoS One. 2013;8:e79993. doi: 10.1371/journal.pone.0079993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honig LS, Kang MS, Schupf N, Lee JH, Mayeux R. Association of shorter leukocyte telomere repeat length with dementia and mortality. Arch. Neurol. 2012;69:1332–1339. doi: 10.1001/archneurol.2012.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Ruiz C, et al. Telomere length predicts poststroke mortality, dementia, and cognitive decline. Ann. Neurol. 2006;60:174–180. doi: 10.1002/ana.20869. [DOI] [PubMed] [Google Scholar]
- 20.Fitzpatrick AL, et al. Leukocyte telomere length and mortality in the Cardiovascular Health Study. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:421–429. doi: 10.1093/gerona/glq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marioni Riccardo E, Harris Sarah E, Shah Sonia, McRae Allan F, von Zglinicki Thomas, Martin-Ruiz Carmen, Wray Naomi R, Visscher Peter M, Deary Ian J. The epigenetic clock and telomere length are independently associated with chronological age and mortality. International Journal of Epidemiology. 2016;45(2):424–432. doi: 10.1093/ije/dyw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakaysa SL, et al. Telomere length predicts survival independent of genetic influences. Aging Cell. 2007;6:769–774. doi: 10.1111/j.1474-9726.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- 23.Cawthon RM, Smith KR, O’Brien E, Sivatchenko A, Kerber RA. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet. 2003;361:393–395. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- 24.Ehrlenbach S, et al. Influences on the reduction of relative telomere length over 10 years in the population-based Bruneck Study: introduction of a well-controlled high-throughput assay. Int. J. Epidemiol. 2009;38:1725–1734. doi: 10.1093/ije/dyp273. [DOI] [PubMed] [Google Scholar]
- 25.Kimura M, et al. Telomere length and mortality: a study of leukocytes in elderly Danish twins. Am. J. Epidemiol. 2008;167:799–806. doi: 10.1093/aje/kwm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deelen J, et al. Leukocyte telomere length associates with prospective mortality independent of immune-related parameters and known genetic markers. Int. J. Epidemiol. 2014;43:878–886. doi: 10.1093/ije/dyt267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rode L, Nordestgaard BG, Bojesen SE. Peripheral blood leukocyte telomere length and mortality among 64,637 individuals from the general population. J. Natl. Cancer Inst. 2015;107:djv074. doi: 10.1093/jnci/djv074. [DOI] [PubMed] [Google Scholar]
- 28.Njajou OT, et al. Association between telomere length, specific causes of death, and years of healthy life in health, aging, and body composition, a population-based cohort study. J. Gerontol. A Biol. Sci. Med. Sci. 2009;64:860–864. doi: 10.1093/gerona/glp061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bischoff C, et al. No association between telomere length and survival among the elderly and oldest old. Epidemiology. 2006;17:190–194. doi: 10.1097/01.ede.0000199436.55248.10. [DOI] [PubMed] [Google Scholar]
- 30.Harris SE, et al. The association between telomere length, physical health, cognitive ageing, and mortality in non-demented older people. Neurosci. Lett. 2006;406:260–264. doi: 10.1016/j.neulet.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Ruiz CM, Gussekloo J, van Heemst D, von Zglinicki T, Westendorp RG. Telomere length in white blood cells is not associated with morbidity or mortality in the oldest old: a population-based study. Aging Cell. 2005;4:287–290. doi: 10.1111/j.1474-9726.2005.00171.x. [DOI] [PubMed] [Google Scholar]
- 32.Strandberg TE, et al. Association of telomere length in older men with mortality and midlife body mass index and smoking. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:815–820. doi: 10.1093/gerona/glr064. [DOI] [PubMed] [Google Scholar]
- 33.Houben JM, Giltay EJ, Rius-Ottenheim N, Hageman GJ, Kromhout D. Telomere length and mortality in elderly men: the Zutphen Elderly Study. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:38–44. doi: 10.1093/gerona/glq164. [DOI] [PubMed] [Google Scholar]
- 34.Cheng S, et al. Distinct metabolomic signatures are associated with longevity in humans. Nat Commun. 2015;6:6791. doi: 10.1038/ncomms7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montoliu I, et al. Serum profiling of healthy aging identifies phospho- and sphingolipid species as markers of human longevity. Aging (Albany NY) 2014;6:9–25. doi: 10.18632/aging.100630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez-Covarrubias V, et al. Lipidomics of familial longevity. Aging Cell. 2013;12:426–434. doi: 10.1111/acel.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang-Sattler R, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, et al. Metabolomics based markers predict type 2 diabetes in a 14-year follow-up study. Metabolomics. 2017;13:104. doi: 10.1007/s11306-017-1239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suhre K, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Floegel A, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto T, Kobayashi T, Kamata K. Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr. Med. Chem. 2007;14:3209–3220. doi: 10.2174/092986707782793899. [DOI] [PubMed] [Google Scholar]
- 42.Cao M, Zhao L, Chen H, Xue W, Lin D. NMR-based metabolomic analysis of human bladder cancer. Anal. Sci. 2012;28:451–456. doi: 10.2116/analsci.28.451. [DOI] [PubMed] [Google Scholar]
- 43.Oresic M, et al. Metabolome in progression to Alzheimer’s disease. Transl Psychiatry. 2011;1:e57. doi: 10.1038/tp.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Z, et al. Human serum metabolic profiles are age dependent. Aging Cell. 2012;11:960–967. doi: 10.1111/j.1474-9726.2012.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menni C, et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013;42:1111–1119. doi: 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J, et al. Metabolic profiles of biological aging in American Indians: the Strong Heart Family Study. Aging (Albany NY) 2014;6:176–186. doi: 10.18632/aging.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zierer J, et al. Metabolomics profiling reveals novel markers for leukocyte telomere length. Aging (Albany NY) 2016;8:77–94. doi: 10.18632/aging.100874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb.) 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 49.Hull D, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei, T. & Simko, V. R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available from, https://github.com/taiyun/corrplot (2017).
- 52.Blanco, A. & Blanco, G. Chapter 5: Lipids. In Medical biochemistry 99–119 (Academic Press, 2017).
- 53.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, et al. Oxidative modification of low density lipoprotein in normal and hyperlipidemic patients: effect of lysophosphatidylcholine composition on vascular relaxation. J. Lipid Res. 1997;38:546–553. [PubMed] [Google Scholar]
- 55.Weber C, Erl W, Weber PC. Enhancement of monocyte adhesion to endothelial cells by oxidatively modified low-density lipoprotein is mediated by activation of CD11b. Biochem. Biophys. Res. Commun. 1995;206:621–628. doi: 10.1006/bbrc.1995.1088. [DOI] [PubMed] [Google Scholar]
- 56.Olofsson KE, Andersson L, Nilsson J, Bjorkbacka H. Nanomolar concentrations of lysophosphatidylcholine recruit monocytes and induce pro-inflammatory cytokine production in macrophages. Biochem. Biophys. Res. Commun. 2008;370:348–352. doi: 10.1016/j.bbrc.2008.03.087. [DOI] [PubMed] [Google Scholar]
- 57.Kume N, Cybulsky MI, Gimbrone MA., Jr. Lysophosphatidylcholine, a component of atherogenic lipoproteins, induces mononuclear leukocyte adhesion molecules in cultured human and rabbit arterial endothelial cells. J. Clin. Invest. 1992;90:1138–1144. doi: 10.1172/JCI115932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paapstel K, et al. Inverse relations of serum phosphatidylcholines and lysophosphatidylcholines with vascular damage and heart rate in patients with atherosclerosis. Nutr. Metab. Cardiovasc. Dis. 2018;28:44–52. doi: 10.1016/j.numecd.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 59.Ward-Caviness CK, et al. Improvement of myocardial infarction risk prediction via inflammation-associated metabolite biomarkers. Heart. 2017;103:1278–1285. doi: 10.1136/heartjnl-2016-310789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Donovan A, et al. Cumulative inflammatory load is associated with short leukocyte telomere length in the Health, Aging and Body Composition Study. PLoS One. 2011;6:e19687. doi: 10.1371/journal.pone.0019687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bekaert S, et al. Telomere length and cardiovascular risk factors in a middle-aged population free of overt cardiovascular disease. Aging Cell. 2007;6:639–647. doi: 10.1111/j.1474-9726.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 62.Finkelstein JD. Methionine metabolism in mammals. J. Nutr. Biochem. 1990;1:228–237. doi: 10.1016/0955-2863(90)90070-2. [DOI] [PubMed] [Google Scholar]
- 63.Lee BC, Kaya A, Gladyshev VN. Methionine restriction and life-span control. Ann. N. Y. Acad. Sci. 2016;1363:116–124. doi: 10.1111/nyas.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun L, Sadighi Akha AA, Miller RA, Harper JM. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J. Gerontol. A Biol. Sci. Med. Sci. 2009;64:711–722. doi: 10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J. Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 66.Zimmerman JA, Malloy V, Krajcik R, Orentreich N. Nutritional control of aging. Exp. Gerontol. 2003;38:47–52. doi: 10.1016/S0531-5565(02)00149-3. [DOI] [PubMed] [Google Scholar]
- 67.Miller RA, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.During MJ, Acworth IN, Wurtman RJ. Effects of systemic L-tyrosine on dopamine release from rat corpus striatum and nucleus accumbens. Brain Res. 1988;452:378–380. doi: 10.1016/0006-8993(88)90043-1. [DOI] [PubMed] [Google Scholar]
- 69.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- 71.Ooka H, Fujita S, Yoshimoto E. Pituitary-thyroid activity and longevity in neonatally thyroxine-treated rats. Mech. Ageing Dev. 1983;22:113–120. doi: 10.1016/0047-6374(83)90104-5. [DOI] [PubMed] [Google Scholar]
- 72.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 73.Atzmon G, Barzilai N, Hollowell JG, Surks MI, Gabriely I. Extreme longevity is associated with increased serum thyrotropin. J. Clin. Endocrinol. Metab. 2009;94:1251–1254. doi: 10.1210/jc.2008-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rozing MP, et al. Familial longevity is associated with decreased thyroid function. J. Clin. Endocrinol. Metab. 2010;95:4979–4984. doi: 10.1210/jc.2010-0875. [DOI] [PubMed] [Google Scholar]
- 75.Rozing MP, et al. Low serum free triiodothyronine levels mark familial longevity: the Leiden Longevity Study. J. Gerontol. A Biol. Sci. Med. Sci. 2010;65:365–368. doi: 10.1093/gerona/glp200. [DOI] [PubMed] [Google Scholar]
- 76.Ferguson AA, et al. TATN-1 mutations reveal a novel role for tyrosine as a metabolic signal that influences developmental decisions and longevity in Caenorhabditis elegans. PLoS Genet. 2013;9:e1004020. doi: 10.1371/journal.pgen.1004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.von Zglinicki T, Saretzki G, Docke W, Lotze C. Mild hyperoxia shortens telomeres and inhibits proliferation of fibroblasts: a model for senescence? Exp. Cell Res. 1995;220:186–193. doi: 10.1006/excr.1995.1305. [DOI] [PubMed] [Google Scholar]
- 78.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem. Sci. 2002;27:339–344. doi: 10.1016/S0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 79.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moore SC, et al. Human metabolic correlates of body mass index. Metabolomics. 2014;10:259–269. doi: 10.1007/s11306-013-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skipski VP, et al. Lipid composition of human serum lipoproteins. Biochem. J. 1967;104:340–352. doi: 10.1042/bj1040340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cole LK, Vance JE, Vance DE. Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim. Biophys. Acta. 2012;1821:754–761. doi: 10.1016/j.bbalip.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 84.Magnusson CD, Haraldsson GG. Ether lipids. Chem. Phys. Lipids. 2011;164:315–340. doi: 10.1016/j.chemphyslip.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 85.Engelmann, B. Plasmalogens: targets for oxidants and major lipophilic antioxidants. Biochem. Soc. Trans. 32, 147–150, doi:10.1042/ (2004). [DOI] [PubMed]
- 86.Zoeller RA, et al. Plasmalogens as endogenous antioxidants: somatic cell mutants reveal the importance of the vinyl ether. Biochem. J. 1999;338(Pt 3):769–776. doi: 10.1042/bj3380769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vaz FM, Wanders RJ. Carnitine biosynthesis in mammals. Biochem. J. 2002;361:417–429. doi: 10.1042/bj3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fritz IB, Yue KT. Long-Chain Carnitine Acyltransferase and the Role of Acylcarnitine Derivatives in the Catalytic Increase of Fatty Acid Oxidation Induced by Carnitine. J. Lipid Res. 1963;4:279–288. [PubMed] [Google Scholar]
- 89.McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 90.Noland RC, et al. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J. Biol. Chem. 2009;284:22840–22852. doi: 10.1074/jbc.M109.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richards JB, et al. Homocysteine levels and leukocyte telomere length. Atherosclerosis. 2008;200:271–277. doi: 10.1016/j.atherosclerosis.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 92.Xu D, Neville R, Finkel T. Homocysteine accelerates endothelial cell senescence. FEBS Lett. 2000;470:20–24. doi: 10.1016/S0014-5793(00)01278-3. [DOI] [PubMed] [Google Scholar]
- 93.Homocysteine Studies C. Homocysteine and risk of ischemic heart disease and stroke: a meta-analysis. JAMA. 2002;288:2015–2022. doi: 10.1001/jama.288.16.2015. [DOI] [PubMed] [Google Scholar]
- 94.Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. BMJ. 2002;325:1202. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith AD, et al. Homocysteine-lowering by B vitamins slows the rate of accelerated brain atrophy in mild cognitive impairment: a randomized controlled trial. PLoS One. 2010;5:e12244. doi: 10.1371/journal.pone.0012244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith AD, et al. Homocysteine and Dementia: An International Consensus Statement. J. Alzheimers Dis. 2018;62:561–570. doi: 10.3233/JAD-171042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morris MS. Homocysteine and Alzheimer’s disease. Lancet Neurol. 2003;2:425–428. doi: 10.1016/S1474-4422(03)00438-1. [DOI] [PubMed] [Google Scholar]
- 98.Smulders YM, et al. Fasting and post-methionine homocysteine levels in NIDDM. Determinants and correlations with retinopathy, albuminuria, and cardiovascular disease. Diabetes Care. 1999;22:125–132. doi: 10.2337/diacare.22.1.125. [DOI] [PubMed] [Google Scholar]
- 99.Buysschaert M, Dramais AS, Wallemacq PE, Hermans MP. Hyperhomocysteinemia in type 2 diabetes: relationship to macroangiopathy, nephropathy, and insulin resistance. Diabetes Care. 2000;23:1816–1822. doi: 10.2337/diacare.23.12.1816. [DOI] [PubMed] [Google Scholar]
- 100.Joshi MB, et al. Elevated homocysteine levels in type 2 diabetes induce constitutive neutrophil extracellular traps. Sci. Rep. 2016;6:36362. doi: 10.1038/srep36362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bostom AG, et al. Nonfasting plasma total homocysteine levels and stroke incidence in elderly persons: the Framingham Study. Ann. Intern. Med. 1999;131:352–355. doi: 10.7326/0003-4819-131-5-199909070-00006. [DOI] [PubMed] [Google Scholar]
- 102.Dangour AD, et al. Plasma homocysteine, but not folate or vitamin B-12, predicts mortality in older people in the United Kingdom. J. Nutr. 2008;138:1121–1128. doi: 10.1093/jn/138.6.1121. [DOI] [PubMed] [Google Scholar]
- 103.Nygard O, et al. Plasma homocysteine levels and mortality in patients with coronary artery disease. N. Engl. J. Med. 1997;337:230–236. doi: 10.1056/NEJM199707243370403. [DOI] [PubMed] [Google Scholar]
- 104.Vollset SE, et al. Plasma total homocysteine and cardiovascular and noncardiovascular mortality: the Hordaland Homocysteine Study. Am. J. Clin. Nutr. 2001;74:130–136. doi: 10.1093/ajcn/74.1.130. [DOI] [PubMed] [Google Scholar]
- 105.Li JJ, et al. Homocysteine Triggers Inflammatory Responses in Macrophages through Inhibiting CSE-H2S Signaling via DNA Hypermethylation of CSE Promoter. Int. J. Mol. Sci. 2015;16:12560–12577. doi: 10.3390/ijms160612560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Au-Yeung KK, et al. Hyperhomocysteinemia activates nuclear factor-kappaB in endothelial cells via oxidative stress. Circ. Res. 2004;94:28–36. doi: 10.1161/01.RES.0000108264.67601.2C. [DOI] [PubMed] [Google Scholar]
- 107.Topal G, et al. Homocysteine induces oxidative stress by uncoupling of NO synthase activity through reduction of tetrahydrobiopterin. Free Radic. Biol. Med. 2004;36:1532–1541. doi: 10.1016/j.freeradbiomed.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 108.Hoffman M. Hypothesis: hyperhomocysteinemia is an indicator of oxidant stress. Med. Hypotheses. 2011;77:1088–1093. doi: 10.1016/j.mehy.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 109.Pushpakumar S, Kundu S, Sen U. Endothelial dysfunction: the link between homocysteine and hydrogen sulfide. Curr. Med. Chem. 2014;21:3662–3672. doi: 10.2174/0929867321666140706142335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kurz DJ, et al. Chronic oxidative stress compromises telomere integrity and accelerates the onset of senescence in human endothelial cells. J. Cell Sci. 2004;117:2417–2426. doi: 10.1242/jcs.01097. [DOI] [PubMed] [Google Scholar]
- 111.US Department of Health and Human Services, F. A. D. A. Center for Drug Evaluation and Research (CDER) Center for Veterinary Medicine (CVM) Guidance for Industry Bioanalytical Method Validation. (2001).
- 112.Broer, L. (Genetic) Epidemiology of Aging. Erasmus University Rotterdam. Retrieved from, http://hdl.handle.net/1765/50038 (2013).
- 113.Dash PK, et al. Traumatic Brain Injury Alters Methionine Metabolism: Implications for. Pathophysiology. Front. Syst. Neurosci. 2016;10:36. doi: 10.3389/fnsys.2016.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All results generated during this study are included in this published article and its Supplementary Materials. The datasets analysed for each individual cohort can be requested by contacting the responsible Principal Investigator. Because of restrictions based on privacy regulations and informed consent of the participants, data cannot be made freely available in a public repository. For the Rotterdam Study data, requests should be directed towards the management team of the Rotterdam Study (secretariat.epi@erasmusmc.nl), which has a protocol for approving data requests.