Abstract
Allelic differences in A and B mating-type loci are a prerequisite for the progression of mating in the genus Pleurotus eryngii; thus, the crossing is hampered by this biological barrier in inbreeding. Molecular markers linked to mating types of P. eryngii KNR2312 were investigated with randomly amplified polymorphic DNA to enhance crossing efficiency. An A4-linked sequence was identified and used to find the adjacent genomic region with the entire motif of the A locus from a contig sequenced by PacBio. The sequence-characterized amplified region marker 7-2299 distinguished A4 mating-type monokaryons from KNR2312 and other strains. A BLAST search of flanked sequences revealed that the A4 locus had a general feature consisting of the putative HD1 and HD2 genes. Both putative HD transcription factors contain a homeodomain sequence and a nuclear localization sequence; however, valid dimerization motifs were found only in the HD1 protein. The ACAAT motif, which was reported to have relevance to sex determination, was found in the intergenic region. The SCAR marker could be applicable in the classification of mating types in the P. eryngii breeding program, and the A4 locus could be the basis for a multi-allele detection marker.
Keywords: Mating-type gene, A4 locus, Pleurotus eryngii, SCAR marker, homeodomain
1. Introduction
The sexual development of mushrooms starts with the mating step and progresses through haploid and diploid phases. A dikaryotic mycelium results from the fusion of two genetically distant monokaryons, and the two nuclei coexist during vegetative growth and fruiting body formation until the onset of the maturation stage, when these nuclei fuse to form basidia and then produce haploid spores [1,2]. The mating-type loci that control developmental pathways and play a role in maintaining a fertile dikaryon are termed A and B [3]. Similarly, mating compatibility in Pleurotus eryngii is determined by two multiallelic mating-type loci, A and B [4]. The developmental sequence leading to the formation of the dikaryon and further to the binucleate cells that characterize it are present in both sets of mating-type genes [5]. The initiation of A-regulated steps in dikaryon development begins with compatible mating to generate the active transcription factor complex [6], while B genes encode lipopeptide pheromones and pheromone receptors [7]. Previously, Ryu et al. [8] classified mating-type factors for A and B alleles by crossing monokaryons with testers of each allele, thereby a SCAR marker linked to the B3 locus was developed to determine the mating type of P. eryngii.
Since ∼25% of the world’s production of cultivated fungi is the production of edible oyster mushrooms, strain improvement through breeding might ease the production procedure with enhanced knowledge about their mating-type loci [9]. P. eryngii, king oyster mushroom, is among the highly demanded mushrooms in Asia, Europe and the USA, and this species has been grown on a commercial scale in Italy, China, Japan, and Korea [10]. Immunostimulatory, antiproliferative, and antifungal actions against cells make this mushroom medically valuable [11]. P. eryngii species are also famous for their flavor and relatively low-cost cultivation methods. Researchers have focused on obtaining high yielding strains using strain selection and improvements in cultivation technology [12], but limited research has been reported on mating-type specificity [4,8].
The DNA marker approach has benefited modern plant breeding in different ways, such as genotype identification, gene isolation, and the analysis of different agronomic traits. Marker-assisted selection (MAS) has been used in mushroom breeding to distinguish cultivars to protect breeders’ rights [13], and its use in relation to agronomic traits for breeding has made MAS a fast, easy, and inexpensive method [14]. SCAR markers are identified by PCR using specifically designed oligo-nucleotide primers based on sequence data. These markers have been reported as a suitable tool for routine diagnostics [15].
In the present study, we demonstrate the molecular structure of the A4 locus and the development of a SCAR marker specifically linked to this locus.
2. Materials and methods
2.1. Strains and growth conditions
All of the strains used in this study were derived from the previous work by Ryu et al. [8,16] (Supplemental Table S1). Two compatible neohaplonts, P5 and P6, were isolated from dikaryotic P. eryngii KNR2312 by dedikaryotization through homogenization of the protoplast with a lysing enzyme. Mycelial mats were grown in MCM (mushroom complete medium) broth as described by a previous report [8] and were then harvested, freeze-dried, and ground with mortar and pestle for DNA extraction.
2.2. Genomic DNA extraction, RAPD analysis, and genome sequencing
Genomic DNA was extracted from the lyophilized mycelia using GenEx plant plus! (GeneAll, Seoul, Korea). PCR was performed with RAPD to screen the mating-type-specific bands according to the method in a previous report [17] with minor modifications. Four gDNAs of monokaryons with the same mating types (A3B3, A3B4, A4B3, and A4B4) derived from P. eryngii KNR2312 were bulked and adjusted to 30 ng/ul for bulked segregant analysis (BSA). Monokaryons in bulked gDNA showing mating-type-specific bands with random primers were subjected to PCR for individual monokaryons with the same primers. PCRs were conducted in a 10 μL mixture containing 30 ng of template genomic DNA, 0.2 mM dNTPs, 0.25 U of e-Taq DNA polymerase (SolGent, Daejeon, Korea), 1× buffer containing 2.5 mM Tris-HCl (pH 8.2) and 1.5 mM MgCl2, and 0.25 pmol of each 10-mer random oligonucleotides including L series (Operon Technologies, Alameda, CA). Amplification conditions were set as follows: initial denaturation for 4 min at 95 °C, followed by 35 cycles of a 1 min denaturation step at 94 °C, 1 min annealing at 37 °C, and 1 min and 30-sec extension at 72 °C. PCR products were run on 3% (w/v) agarose gels in TAE buffer (400 mM Tris, 200 mM sodium acetate, and 20 mM EDTA, pH 8.3), stained Safeview classic (iNtRON Biotechnology, eongnam, Korea) and visualized under ultraviolet light. The 100 bp plus DNA ladder (Bioneer, Daejeon, Korea) was used as a standard size marker.
We prepared gDNA samples from KNR2312P6 (neohaplont) into SMRTbell DNA template libraries of 20-kb average insert size according to the manufacturer’s specification, and Covaris G-tubes were used for fragmentation. SMRT sequencing was carried out on the PacBio RS II platform according to standard protocols, with the XL binding kit used in conjunction with the C4 sequencing kit. All runs were performed with diffusion-based loading and analyzed using standard primary data analysis. Contigs were assembled using CANU v1.7 with long reads.
2.3. SCAR marker development and data analysis
For isolation and identification of DNA in the gel, the mating-type-specific bands were cut out and purified by Gel Elution Kit (Solgent) and directly sequenced on an ABI 377 sequencer (Macrogen, Seoul, Korea) using the Big-Dye cycle sequencing kit (Applied Biosystems, Carlsbad, CA). The A4 mating-type-specific sequence, designated 7-2, was used to find the 7-2 adjacent genomic region and the entire motif of the A4 locus. The SCAR primer was manually designed using the 7-2 sequence and a flanked region (∼1 kb) obtained from a contig of KNR2312P6 sequenced by PacBio. The developed primer sequences were F: 5′-AATCACGGGAAGATCTGGTG-3′ and loci-R: 5′-GTGGTAGGGTTCCCGCCT-3′. PCR conditions for the SCAR marker were optimized as follows: initial denaturation at 98 °C for 30 sec, followed by 35 cycles of a 10 sec denaturation at 98 °C, 15 sec annealing at 70 °C, and 10 sec extension at 72 °C, and a final extension of 10 min at 72 °C. The primer set was applied to determine the mating type of monokaryons from KNR2312 and other strains whose mating types were different (Supplemental Table S1) in addition to the Pleurotus genus.
2.4. Gene prediction and sequence alignment
The sequence including the A4 allele was obtained from a contig of KNR2312P6 using a sequence search with DNAMAN (Lynnon Biosoft, Quebec, Canada) with an A4-specific genomic sequence. A4 sequence has been deposited in the National Center for Biotechnology Information (NCBI) database and assigned the Accession Number MK522809. Sequences were analyzed using FGENESH (http://www.softberry.com/) and NCBI Conserved Domain Search [18] to predict the genes present and their functions. COILS (window width 14 and MTIDK matrix) [19] and NLStradamus (prediction cutoff, 0.5) [20] were adapted for coiled-coil domains (putative dimerization motif) and nuclear localization signals, respectively. Genomic data of A mating-type genes for P. eryngii KNR2312P5, P. eryngii ATCC90797, P. tuoliensis CCMSSC00489 and CCMSSC00485, and P. ostreatus PC15 were obtained from an edible mushroom DB (http://112.220.192.2/per/)[21], NCBI’s GenBank and Joint Genomic Institute (https://genome.jgi.doe.gov/portal/). Alignment analysis was performed with DNAMAN (Lynnon Biosoft) with default parameters to determine the nucleotide frequency, including the transcription initiation site.
3. Results
3.1. A4 mating-type-linked SCAR marker
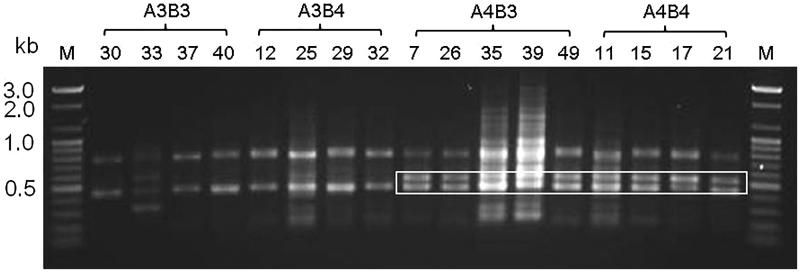
Among 200 primers, several primers were segregated with mating types (data not shown), including the OPL-07 primer specifying A4Bx by an ∼500 bp PCR product (designated 7-2) (Figure 1). All RAPD markers showing mating-type specificity were converted into SCAR markers based on their sequences. Most SCAR markers showed low or no mating-type specificity (data not shown). The marker from 7-2 was unstable and weak; thus, a more accurate marker was designed based on the flanked sequences of 7-2 obtained from a contig that was one of the 498 contigs (∼50 Mb) of KNR2312P6 sequenced by PacBio. Cosegregation of the specific 299 bp bands with the A4Bx locus was observed in KNR2312-derived monokaryons with the SCAR marker (designated 7-2299). A critical annealing temperature of 70 °C was determined to best identify the mating type. Thus, to observe the strong linkage of 7-2299 with the A4Bx locus, 17 monokaryons from KNR2312 were tested. Eight monokaryotic isolates showed a specific band for mating-type A4B3, A4B4, and P6 (A4B3), while no amplification was detected for A3B3, A3B4, and P5 (A3B4) (Figure 2). All 98 monokaryons from KNR2312 [8] showed consistent results of the 299 bp genotype coinciding with the A4Bx phenotype at all times (data not shown).
Figure 1.
RAPD patterns for 16 monokaryons of Pleurotus eryngii KNR2312 with OPL-07 (10-mer). Box shows the A4 specific band. M: 100 bp ladder; 30, 33, 37, 40: A3B3; 12, 25, 29, 32: A3B4; 7, 26, 35, 39, 49: A4B3; 11, 15, 17, 21: A4B4.
Figure 2.
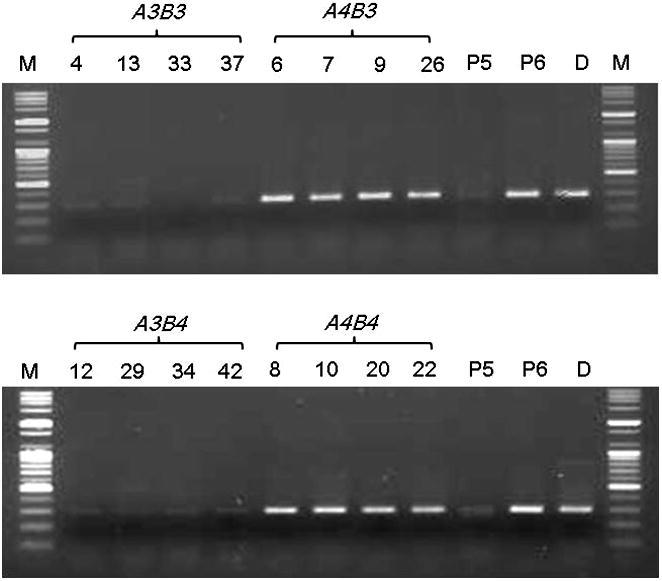
Amplification patterns with 7-2299 (A4 specific marker) in different sets of monokaryons derived from Pleurotus eryngii KNR2312. M: 100 bp ladder; Upper: KNR2312(3)-4, 13, 33, 37: A3B3; 6, 7, 8, 26: A3B4. Lower: 12, 29, 34, 42: A4B3; 8, 10, 20, 22: A4B4; P5: A3B4; P6: A4B3; D: dikaryon (P5 × P6).
3.2. Application of the SCAR marker (7-2299) to other strains of P. eryngii and other species
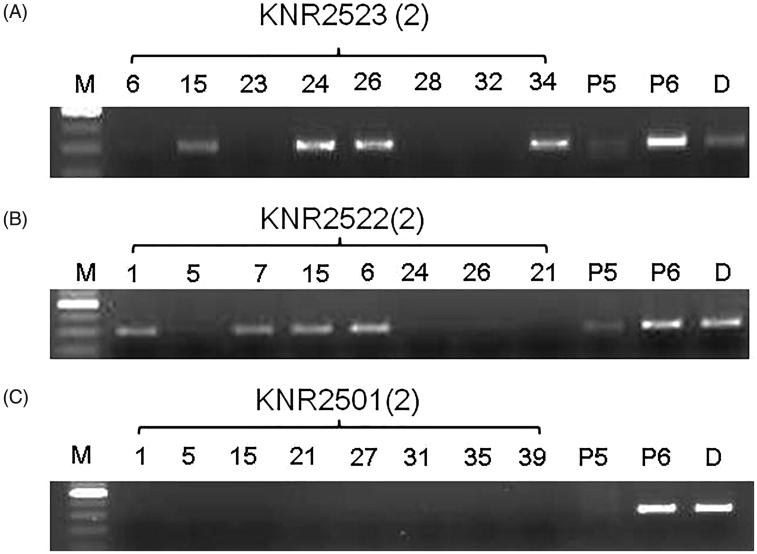
Different monokaryons derived from KNR2501, KNR2522, and KNR2523 were used to evaluate the suitability of the 7-2299 marker. Because the A4 allele was shared in KNR2523 and KNR2522 but not in KNR2501 [8], 8 monokaryons, two from each mating type, were selected for the new marker. The 7-2299 marker amplified unique bands from KNR2523(2)-15, 34 (A4B3) and KNR2523(2)-24, 26 (A4B12) but not from the remaining strains (KNR2523(2)-6 and -28 for A12B3 and KNR2523(2)-23 and -32 for A12B12). Similarly, a unique band of 299 bp was observed from KNR2522(2)-1, 6 (A4B12) and KNR2522(2)-7, 15 (4B3). No band was shown in the strains carrying the A12B12 and A12B3 mating types in addition to the KNR2501(2)-derived monokaryons with A3B5, A5B5, A3B4, and A5B4 (Figure 3). A 299 bp band was amplified from all monokaryons with A4Bx and P6 (A4B3), which indicated its suitability for the detection of the A4Bx locus in other strains of P. eryngii. Some other mushroom fungi of the same genus, such as P. florida, P. sajor-caju, P. salmoneostramineus, P. cornucopiae, and P. ostreatus, were also used to examine the presence of the A4Bx locus with the 7-2299 marker, but none of them were positive for the A4Bx locus (data not shown).
Figure 3.
SCAR marker amplification patterns for the A4 locus in diverse phenotypes. Amplifications were performed with SCAR (7-2299) primers using monokaryons derived from KNR2523, KNR2522, and KNR2501. Two monokaryons were selected from each mating type. (A) monokaryons from KNR2523(2) (6, 28: A12B3; 15, 34: A4B3; 23, 32: A12B12; and 24, 26: A4B12), P5: A3B4; P6: A4B3; D: dikaryon (P5 × P6); (B) monokaryons from KNR2522(2) (1, 6: A4B12; 5, 21: A12B12; 24, 26: A12B3; and 7, 15: A4B3); (C) monokaryons from KNR2501(2) (15, 35: A3B4; 1, 31: A3B5; 21, 39: A5B4; and 5, 27: A5B5).
3.3. Molecular structure of the A4 locus
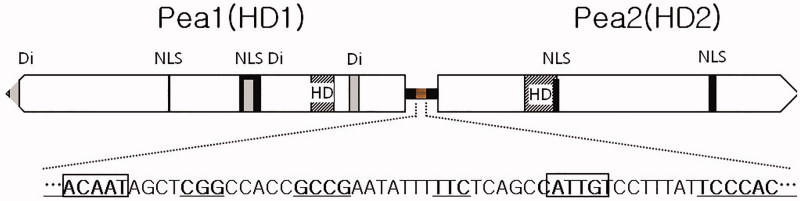
The BLASTp search of the flanked sequence of the A4-specific marker identified two homeodomains (HD1, designated Pea1, and HD2, designated Pea2), located 200 kb apart from the marker. The two genes were located side-by-side, and the direction of transcription was outward from an intergenic region for both genes, similar to other basidiomycetes [22,23]. The two HD genes were located across 4.6 kb, HD1 was assumed to be 2197 bp gDNA and 694 a.a. with two introns, while HD2 was likely 2062 bp gDNA and 627 a.a with 3 introns (Supplemental Figure S1). The two HD genes were considered a general feature of HD in Pleurotus [23,24]; however, P. eryngii ATCC90797 and P. ostreatus PC15 contained three HD genes (Supplemental Figure S2). Pea1 (HD1) showed high similarity, ranging from 51.9–70.4% with those of other Pleurotus, while Pea2 (HD2) had high similarity with HD2 of Pleurotus, ranging from 69.0–49.6% (Supplemental Table S2). The intergenic region between Pea1 (HD1) and Pea2 (HD2) was 163 bp long, which was similar to those of other Pleurotus species ranging from 166 to 178 bp (Supplemental Figure S3). Alignment of the intergenic regions among Pleurotus showed several conserved sequences, including two ACAAT motifs (one was reversed) (Figure 4), GCCG and TCCCAC. Pea1 contained two NLS and three dimerization motifs (coiled-coil domain). Both genes had homeodomains located at the 129–168th a.a. (Pea1, 39 a.a. in length) and 150–206th a.a. (Pea2, 56 a.a. in length). Some NLS domains overlapped with coiled-coil motifs and homeodomains (Figure 4). Interestingly, coiled coils were not detected at high levels in Pea2 (three with 0.021 as the highest value were detected). Multiple alignment with HD mating-type proteins from the genus Pleurotus showed two sectors: one had highly diverse sequences, and the other had highly similar sequences. Diverged sequences were terminated at the beginning of the homeodomain (Supplemental Figure S2).
Figure 4.
Molecular structure of the A4 locus in Pleurotus eryngii KNR2312P6. The A4 locus spans ∼4.6 kb in length and consists of two homeodomain transcription genes with a 164 bp intergenic region. Pea1 and Pea2 consist of 2197 bp and 2062 bp gDNA and 694 aa and 627 aa lengths, respectively. Conserved sequences in the intergenic region are ACAAT, GCCG, and TCCCAC. The gray block in the intergenic region indicates the relative position of the conserved sequence area shown below. Bold indicates conserved sequences revealed by alignment analysis with that of another genus Pleurotus (see M & M). Overlapped conserved domains are adjusted block height for identification of locations. HD: homeodomain; Di: dimerization domain (coiled coil); NLS: nuclear localization sequence.
4. Discussion
4.1. Molecular marker linked to the A4 locus
The development of new desired strains and the methods to distinguish them are some of the main issues in mushroom breeding. Previously, different techniques were employed to verify strains such as small subunit ribosomal DNA (SSU rDNA) [25], multiple nuclear genes [26], or PCR-based DNA fingerprinting methodologies including RAPD, AFLP, and SCAR [27]. These marker techniques save time by identifying the mating type more reliably than traditional methods such as crossing. SCAR markers are comparatively stable and overcome the limitations of RAPD markers in the form of reproducibility. SCARs have been used for identification purposes in Lentinula edodes [14] and Laccaria bicolor [28]. In the present study, polymorphisms were frequently detected in the RAPD products of the A4Bx-specific compatible monokaryons (A4B3 and A4B4) (Figure 1). As SCAR markers can identify single bands instead of complex patterns, they are more accurate than RAPDs. Therefore, a SCAR marker (7-2299) was developed to achieve well-verified and consistent results (Figures 2 and 3). Although the 7-2299 marker is located 200 kb away from the A4 locus, it cosegregated with the A4 locus trait among all monokaryons from KNR2312 and a wide range of other strains. This result suggested that the inter-region between the 7-2299 and A4 loci exerts strong selection pressure. The genomic region around the A locus in the genus Pleurotus was reported to be conserved during long evolutions [24]. Thus, due to the high specificity, consistency, and wide range of applicability of the PCR-based method, the 7-2299 marker might be an efficient tool for distinguishing A4Bx-locus strains in breeding programs for P. eryngii. The newly developed method might be preferred over the traditional method for the identification of mating-type, based on the shorter duration, i.e., 1–3 days instead of 15 days. Rapid colony PCR for mycelium is well established for massive selections [29]; therefore, breeders could save time and materials by mating only compatible strains with SCAR markers in the case of inbreeding. Moreover, P. eryngii KNR2312 has been used as a parent for many cultivars [30–32] due to a narrow genetic pool; thus, this marker will be more effective for breeding using KNR2312-derived cultivars.
4.2. Molecular structure of the A4 locus
The homeodomain proteins HD1 and HD2 play a role in the regulation of sexual development in several fungi [33]. The A4 locus showed a general feature of the A mating locus of the genus Pleurotus reported [23,24]. The two HD genes were located side-by-side and separated by a 163 bp intergenic region. Exceptions were the three HDs in P. eryngii ATCC90797 and P. ostreatus PC15, suggesting that these strains are likely closer to a prototype. However, whether all HDs are linked to the A mating type is unknown. Alignment of the intergenic region revealed conserved sequences. ACAAT motifs were reported to be enhancer elements for eukaryotic transcription factors [34]. Interestingly, some of them were reported to be involved in sex determination in animals, suggesting that the motif has been conserved across kingdoms for sex or sex-related functions. The function of the remaining GCCG and TCCCAC motifs has not been reported. ACAAT motifs were also found in L. edodes, whereas GCGGAGT, found in L. edodes, was not detected, indicating that cognate DNA subsites of transcription factors were likely genus specific. The conserved domain in the HD proteins was homeodomain, coiled-coil, and NLS. A mating-type protein could interact with a dimerized form and in the nucleus. Similarly, an interaction between HD1 and HD2 homeodomains has been reported for sexual development regulation in basidiomycetes [2,35]. Interestingly, Pea2 contained only three coiled-coil domains with a value less than 0.021 for coiled-coil possibility. In this case, an HD protein with a low value was found in the P. tuoliensis CC00489 HD2 protein. Whether these low values are sufficient for dimerization is unknown; however, the dimerization motif in HD2 is reported to be weak due to the existence of additional forces, such as protein-protein interactions [2].
Many A alleles were found in P. eryngii [8], and thus, the development of more accurate molecular markers is required. The molecular structure of the A4 locus will provide a basis for the development of vast and diverse A-allele detection markers. The newly developed method might be preferred over the traditional method for the identification of mating type due to its shorter duration, i.e., 1–3 days instead of 15 days. Rapid colony PCR for mycelium was well established for massive selections [29]; therefore, breeders could save time and materials by mating only compatible strains with SCAR markers in the case of inbreeding. Moreover, P. eryngii KNR2312 has been used as a parent for many cultivars [30–32] due to a narrow genetic pool; thus, this marker will be more effective for breeding using KNR2312-derived cultivars.
Supplementary Material
Funding Statement
This study was supported by a grant from the Golden seed project [IPET No. 213007-05-3-SBI30], MIFAFF.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kües U. From two to many: multiple mating types in Basidiomycetes. Fungal Biol Rev. 2015;29:126–166. [Google Scholar]
- 3.Raudaskoski M, Kothe E. Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell. 2010;9:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim K-H, Kang YM, Im CH, et al. Identification and functional analysis of pheromone and receptor genes in the B3 mating locus of Pleurotus eryngii. PLoS One. 2014;9:e104693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banham AH, Asante-Owusu RN, Gottgens B, et al. An N-terminal dimerization domain permits homeodomain proteins to choose compatible partners and initiate sexual development in the mushroom Coprinus cinereus. Plant Cell. 1995;7:773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Shea SF, Chaure PT, Halsall JR, et al. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics. 1998;148:1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryu J-S, Kim MK, Ro H-S, et al. Identification of mating type loci and development of SCAR marker genetically linked to the B3 locus in Pleurotus eryngii. J Microbiol Biotechnol. 2012;22:1177–1184. [DOI] [PubMed] [Google Scholar]
- 9.Kothe E. Mating-type genes for basidiomycete strain improvement in mushroom farming. Appl Microbiol Biotechnol. 2001;56:602–612. [DOI] [PubMed] [Google Scholar]
- 10.Estrada AR, Royse D. Yield, size and bacterial blotch resistance of Pleurotus eryngii grown on cottonseed hulls/oak sawdust supplemented with manganese, copper and whole ground soybean. Bioresour Technol. 2007;98:1898–1906. [DOI] [PubMed] [Google Scholar]
- 11.Ngai PH, Ng T. A hemolysin from the mushroom Pleurotus eryngii. Appl Microbiol Biotechnol. 2006;72:1185–1191. [DOI] [PubMed] [Google Scholar]
- 12.Zervakis G, Venturella G. Mushroom breeding and cultivation enhances ex situ conservation of Mediterranean Pleurotus taxa In: Engels JMM, Ramanantha Rao V, Brown AHD, Jackson MT, editors. Managing plant genetic diversity. New York, USA: CABI Publishing; 2002. p. 351–358. [Google Scholar]
- 13.Zhang R, Hu D, Zhang J, et al. Development and characterization of simple sequence repeat (SSR) markers for the mushroom Flammulina velutipes. J Biosci Bioeng. 2010;110:273–275. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka A, Miyazaki K, Murakami H, et al. Sequence characterized amplified region markers tightly linked to the mating factors of Lentinula edodes. Genome. 2004;47:156–162. [DOI] [PubMed] [Google Scholar]
- 15.Scovel G, Ben-Meir H, Ovadis M, et al. RAPD and RFLP markers tightly linked to the locus controlling carnation (Dianthus caryophyllus) flower type. Theor Appl Genet. 1998;96:117–122. [Google Scholar]
- 16.Ryu J, Kim K, Im C, et al., editors Complete genome sequence of Pleurotus eryngii KNR2312 using the next generation sequencing (NGS). 80th Meeting of the Mycological Society of America. The Mycological Society of America, New Haven, CT, USA; 2012.
- 17.Michelmore RW, Paran I, Kesseli R. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88:9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. [DOI] [PubMed] [Google Scholar]
- 20.Ba ANN, Pogoutse A, Provart N, et al. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im CH, Park Y-H, Hammel KE, et al. Construction of a genetic linkage map and analysis of quantitative trait loci associated with the agronomically important traits of Pleurotus eryngii. Fungal Genet Biol. 2016;92:50–64. [DOI] [PubMed] [Google Scholar]
- 22.Au CH, Wong MC, Bao D, et al. The genetic structure of the A mating-type locus of Lentinula edodes. Gene. 2014;535:184–190. [DOI] [PubMed] [Google Scholar]
- 23.Zhao M, Zhang J, Chen Q, et al. The famous cultivated mushroom Bailinggu is a separate species of the Pleurotus eryngii species complex. Sci Rep. 2016;6:33066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James TY, Liou S-R, Vilgalys R. The genetic structure and diversity of the A and B mating-type genes from the tropical oyster mushroom, Pleurotus djamor. Fungal Genet Biol. 2004;41:813–825. [DOI] [PubMed] [Google Scholar]
- 25.Berbee ML, Taylor JW. Detecting morphological convergence in true fungi, using 18S rRNA gene sequence data. Biosystems. 1992;28:117–125. [DOI] [PubMed] [Google Scholar]
- 26.Hansen K, LoBuglio KF, Pfister DH. Evolutionary relationships of the cup-fungus genus Peziza and Pezizaceae inferred from multiple nuclear genes: RPB2, beta-tubulin, and LSU rDNA. Mol Phylogenet Evol. 2005;36:1–23. [DOI] [PubMed] [Google Scholar]
- 27.Lopandic K, Molnár O, Prillinger H. Application of ITS sequence analysis, RAPD and AFLP fingerprinting in characterising the yeast genus Fellomyces. Microbiol Res. 2005;160:13–26. [DOI] [PubMed] [Google Scholar]
- 28.Weber J, Díez J, Selosse M-A, et al. SCAR markers to detect mycorrhizas of an American Laccaria bicolor strain inoculated in European Douglas-fir plantations. Mycorrhiza. 2002;12:19–27. [DOI] [PubMed] [Google Scholar]
- 29.Izumitsu K, Hatoh K, Sumita T, et al. Rapid and simple preparation of mushroom DNA directly from colonies and fruiting bodies for PCR. Mycoscience. 2012;53:396–401. [Google Scholar]
- 30.Im CH, Kim M-K, Kim K-H, et al. Breeding of king oyster mushroom, Pleurotus eryngii with a high yield and earliness of harvest trait and its sensory test. Kor J Mycol. 2013;41:91–96. [Google Scholar]
- 31.Shin P, Park Y, Yoo Y, et al. Characteristics and breeding of a new cultivar Pleurotus eryngii, Song-A. J Mushroom Sci Prod. 2011;9:59–62. [Google Scholar]
- 32.Lee Y, Kim Y, Seuk S, et al. Mband characterization of a cultivar “DanBi 5Ho” with a long shelf life. J Mushrooms. 2016;14:64–69. [Google Scholar]
- 33.Burglin T. A comprehensive classification of homeobox genes In: Duboule D, editor. Guidebook to the homeobox genes. Oxford: Oxford University Press; 1994. p. 25–27. [Google Scholar]
- 34.Narayana N, Weiss MA. Crystallographic analysis of a sex-specific enhancer element: sequence-dependent DNA structure, hydration, and dynamics. J Mol Biol. 2009;385:469–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kües U, Göttgens B, Stratmann R, et al. A chimeric homeodomain protein causes self‐compatibility and constitutive sexual development in the mushroom Coprinus cinereus. EMBO J. 1994;13:4054–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.