Abstract
Four strains of Penicillium and Talaromyces species are described and illustrated in an inventory of fungal species belonging to Eurotiales. The strains, CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31, were isolated from soil and freshwater samples from South Korea. Based on their morphological characteristics and sequence analyses by the combined β-tubulin and calmodulin gene, the CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31 isolates were identified as Penicillium pasqualense, Penicillium sanguifluum, Talaromyces apiculatus, and Talaromyces liani, respectively. The designated strains were found to represent a previously undescribed species of Korean fungal biota. In this study, detailed morphological descriptions and phylogenetic relationships of these species are provided.
Keywords: Eurotiales; Penicillium pasqualense, Penicillium sanguifluum, Talaromyces apiculatus, Talaromyces liani
1. Introduction
Soil and freshwater provide unique environments for the growth of fungi. Fungal communities perform essential functions in the biogeochemical cycles of these ecosystems. In soils, they act as agents governing soil carbon cycling, plant nutrition, and pathology [1]. Freshwater nourishes diverse habitats such as fallen leaves, plant litter, decaying wood, aquatic plants and insects, and soils for fungi. Fungi act in the energy flow, food-web, nutrient transformations, and self-purification in the freshwater ecosystem [2].
The genera Penicillium and Talaromyces (Trichocomaceae, Eurotiales), belonging to the Eurotiomycetes, are of high environmental and biotechnological relevance [3]. These genera are abundant in contaminated foods, fruits, indoor environments, air, soil, and water [4–6]. They also produce a variety of useful secondary metabolites including anticancer and antifungal compounds, extracellular enzymes, and harmful mycotoxins [7–9]. Previously, Talaromyces was thought to be related to teleomorphic Penicillium subgenus Biverticillium and other genera, but recently both have been classed as separate genera containing both sexual and asexual species with the introduction of single name nomenclature [10,11]. The species belonging to both these genera were identified on the recommendations of the polyphasic species concept, using morphological and molecular phylogenetic analyses [11,12].
The genus Penicillium was first described by Link and included asexual fungi bearing Penicillium-like fruiting bodies [13]. New species have since been added to the already 354 accepted Penicillium species [8,14,15]. Members of this genus were divided into 26 sections using the polyphasic approach [16]. To the best of our knowledge, over 100 Penicillium species have been reported from Korea [17,18]. Some of the new species discovered in Korea were isolated from various environmental samples such as P. daejeonium from grape and Schisandra fruit, P. koreense from soil, P. samsonianum from stems and leaves of Viscum album var. coloratum, P. jejuense from marine environments of Jeju Island, P. punicae from pomegranate (Punica granatum) fruit, P. aquaticum and P. acidum from freshwater [16,19–22]. Section Citrina represents one of these sections clade and are abundant worldwide [23]. Among the specified 39 species and 17 new species accepted in the section Citrina, only 11 species have been reported from Korea [24,25].
The genus Talaromyces was introduced by Benjamin for teleomorphic Penicillium species with T. vermiculatus as the type species [26]. These species were characterized taxonomically by their sexual morphology, having cleistothecial or gymnothecial ascomata, unitunicate 8-spored asci, and unicellular ascospores with or without equatorial crests. Currently, the genus accepts 110 species which are divided into seven sections, i.e., Bacillispori, Heici, Islandici, Purpurei, Subinflati, Talaromyces, and Trachyspermi [6]. Recently, several new Talaromyces species were discovered from various environmental habitats [6,15,27,28]. To date, only 21 Talaromyces species have been reported from Korea [17,29–31]. Among the reported species, only six have been well described in Korea, these species are not reported before [30]. Talaromyces apiculatus and Talaromyces liani are classified in section Talaromyces. The diversity of the genus Penicillium and Talaromyces in Korea remains unknown using both molecular and morphological analyses.
During investigation of the fungal species inhabiting soil and freshwater, two Penicillium and two Talaromyces species were identified. The objective of this study was to perform morphological and molecular analyses to characterize the undescribed Penicillium and Talaromyces species in Korea: P. pasqualense, P. sanguifluum, T. apiculatus and T. liani belonging to the Eurotiales.
2. Materials and methods
2.1. Isolation of fungal strains from water and soil samples
Soil samples were collected from Dongdo (eastern islet) of Dokdo Island (37°14′21.3″ N, 131°52′04.4″ E) and under the rhizosphere of a pine tree located at Geumgol Mountain, Jin Island (Jindo). Freshwater samples were collected from Yeosu stream in a small falcon tube. Serial-dilution plating methods were employed using potato dextrose agar (PDA; Becton, Dickinson and Co., Sparks, MD) and malt extract agar (MEA; Becton). Pure isolates were obtained by picking individual colonies of varied morphologies and transferring them to PDA plates and subculturing until pure mycelia were obtained. All pure isolates were maintained in PDA slant tubes and in 20% glycerol at −80 °C at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea.
P. pasqualense, P. sanguifluum, T. apiculatus, and T. liani strains isolated in our study were designated CNUFC-DDS17-1 and CNUFC-DDS17-2, CNUFC-DDS27-1, and CNUFC-DDS27-2, CNUFC-PTM72-1 and CNUFC-PTM72-2, CNUFC-YJW3-31 and CNUFC-YJW3-32, respectively. The isolates, CNUFC-DDS17-1, -DDS17-2, -DDS27-1, and -DDS27-2 were isolated from soil of Dokdo Island; CNUFC-PTM72-1 and -PTM72-1 from soil of Geumgol Mountain; CNUFC-YJW3-31 and -YJW3-32 from freshwater.
2.2. DNA extraction, PCR, and purification
Genomic DNA was extracted using the Solg TM Genomic DNA Prep Kit (Solgent Co. Ltd., Daejeon, Korea). The internal transcribed spacer (ITS)-rDNA region, β-tubulin gene (BenA), and calmodulin (CaM) gene were amplified with the primer pairs ITS1/ITS4 [32], Bt2a/Bt2b [33], and Cmd5/Cmd6 [5], respectively. The PCR amplification reaction mixture was set up according to Nguyen et al. [30]. An Accuprep PCR Purification Kit (Bioneer Corp., Daejeon, Korea) was employed for PCR purification. DNA sequencing was performed using an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA).
2.3. Molecular analysis
Fungal sequences were aligned using Clustal_X version 2.1 [34] and edited with Bioedit version 7.2.6.0 [35]. Maximum likelihood (ML) was constructed using MEGA 6 software [36]. Sequences of CNUFC-DDS17-1, CNUFC-DDS17-2, CNUFC-DDS27-1, CNUFC-DDS27-2, CNUFC-PTM72-1, CNUFC-PTM72-2, CNUFC-YJW3-31, and CNUFC-YJW3-32 were deposited in the NCBI database under the accession numbers shown in Table 1.
Table 1.
Taxa, collection numbers, sequences, and GenBank accession numbers used in this study.
| GenBank accession no. |
|||
|---|---|---|---|
| Taxon name | Collection no. (isolate no.) | BenA | CaM |
| Penicllium aurantiacobrunneum | CBS 126228 (T) | JN606702 | JN606522 |
| P. cairnsense | CBS 124325 (T) | JN606693 | JN606512 |
| P. chrzaszii | CBS 217.28 (T) | JN606758 | JN606423 |
| P. citrinum | CBS 139.45 (T) | GU944545 | GU944638 |
| P. cosmopolitanum | CBS 126995 (T) | JN606733 | JN606472 |
| P. decaturense | CBS 117509 (T) | JN606685 | JN606413 |
| P. godlewskii | CBS 215.28 (T) | JN606768 | JN606443 |
| P. manginii | CBS 253.31 (T) | JN606651 | JN606381 |
| P. miczynskii | CBS 220.28 (T) | JN606706 | JN606526 |
| P. neomiczynskii | CBS 126231 (T) | JN606705 | JN606523 |
| P. nothofagi | CBS 130383 (T) | JN606732 | JN606507 |
| P. pancosmium | CBS 276.75 (T) | JN606790 | JN606446 |
| P. pasqualense | CBS 126330 (T) | JN606673 | JN606394 |
| P. pasqualense | CBS 124327 | JN606672 | JN606392 |
| P. pasqualense | CBS 122402 | JN606674 | JN606393 |
| P. pasqualense | CNUFC-DDS17-1 | MK204815 | MK204817 |
| P. pasqualense | CNUFC-DDS17-2 | MK204816 | MK204818 |
| P. quebecense | CBS 101623 (T) | JN606700 | JN606509 |
| P. roseopurpureum | CBS 266.29 (T) | JN606838 | JN606556 |
| P. sanguifluum | CBS 127032 (T) | JN606819 | JN606555 |
| P. sanguifluum | CBS 118024 | JN606833 | JN606537 |
| P. sanguifluum | CBS 643.73 | JN606853 | JN606576 |
| P. sanguifluum | CNUFC-DDS27-1 | MK204819 | MK204821 |
| P. sanguifluum | CNUFC-DDS27-2 | MK204820 | MK204822 |
| P. sizovae | CBS 413.69 (T) | GU944535 | GU944618 |
| P. sucrivorum | CBS 135116 (T) | JX141015 | JX141506 |
| P. sumatrense | CBS 281.36 (T) | JN606639 | JN606368 |
| P. ubiquetum | CBS 126437 (T) | JN606800 | JN606460 |
| P. waksmanii | CBS 230.28 (T) | JN606779 | JN606431 |
| P. westlingii | CBS 231.28 (T) | JN606718 | JN606500 |
| Talaromyces aculeatus | CBS 289.48 (T) | KF741929 | KF741975 |
| T. apiculatus | CBS 101366 | KF741910 | KF741932 |
| T. apiculatus | CBS 312.59 (T) | KF741916 | KF741950 |
| T. apiculatus | CNUFC-PTM72-1 | MK204823 | MK204825 |
| T. apiculatus | CNUFC-PTM72-2 | MK204824 | MK204826 |
| T. aurantiacus | CBS 314.59 (T) | KF741917 | KF741951 |
| T. australis | CBS 137102 (T) | KF741922 | KF741971 |
| T. bacillisporus | CBS 296.48 (T) | AY753368 | KJ885262 |
| T. derxii | CBS 412.89 (T) | JX494305 | KF741959 |
| T. duclauxii | CBS 322.48 (T) | JX091384 | KF741955 |
| T. flavovirens | CBS 102801 (T) | JX091376 | KF741933 |
| T. francoae | CBS 113134 (T) | KX011489 | KX011501 |
| T. funiculosus | CBS 272.86 (T) | JX091383 | KF741945 |
| T. liani | CBS 118434 | KM066139 | KP453744 |
| T. liani | CBS 225.66 (T) | JX091380 | KJ885257 |
| T. liani | CNUFC-YJW3-31 | MK204811 | MK204813 |
| T. liani | CNUFC-YJW3-32 | MK204812 | MK204814 |
| T. kendrickii | CBS 136666 (T) | KF741921 | KF741967 |
| T. marneffei | CBS 388.87 (T) | JX091389 | KF741958 |
| T. purpurogenus | CBS 286.36 (T) | JX315639 | KF741947 |
| T. stellenboschiensis | CBS 135665 (T) | JX091605 | JX140683 |
| T. stollii | CBS 408.93 (T) | JX315633 | JX315646 |
| T. verruculosus | CBS 388.48 (T) | KF741928 | KF741974 |
| T. viridis | CBS 114.72 (T) | JX494310 | KF741935 |
| T. viridulus | CBS 252.87 (T) | JX091385 | KF741943 |
Bold letters indicate isolates and accession numbers described in our study.
CBS: Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands); CNUFC: Chonnam National University Fungal Collection (Gwangju, South Korea); T: ex-type strain.
2.4. Morphological studies
The isolated strains CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31 were cultured on Czapek yeast autolysate agar (CYA), Blakeslee’s MEA, and yeast extract sucrose agar (YES) [5]. The plates were incubated at 25 °C in the dark for one week. To observe fine fungal structures, the isolates were fixed in 2.5% paraformaldehyde-glutaraldehyde in 0.05 M phosphate buffer (pH 7.2) for 2 h, and then washed with cacodylate buffer (Junsei Chemical Co. Ltd., Kyoto, Japan). Cellular membranes were preserved by fixing the samples in 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA) diluted in cacodylate buffer for 1 h. Samples were then washed again in cacodylate buffer, dehydrated in graded ethanol (Emsure, Darmstadt, Germany) and isoamyl acetate (Junsei Chemical Co. Ltd.), and dried in a fume hood. Finally, samples were sputter-coated with gold and observed under a Hitachi S4700 field emission scanning electron microscope at the Korea Basic Science Institute, Gwangju, Korea. Samples were also observed under an Olympus BX51 microscope with DIC optics (Olympus, Tokyo, Japan) by mounting in a lactophenol solution (Junsei Chemical Co. Ltd.).
3. Results
3.1. Molecular phylogenetic analysis
The results constructed by ML analyses based on combined BenA and CaM sequences analysis of the isolates respectively belonging to Penicillium and Talaromyces are shown in Figures 1 and 2. A BLASTn search was conducted using the ITS, BenA, and CaM sequences of CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31 isolates. The ITS region of isolates CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31 show 99.3% (543/547 bp), 98.9% (520/526 bp), 98.6% (348/353 bp), and 100% (537/537 bp) sequence similarities with P. pasqualense CBS 126330 (JN617676), P. sanguifluum CV1856 (JX140865), T. apiculatus CBS 101366 (KF741977), and T. liani CBS 225.66 (MH858781). BLASTn analysis of BenA of CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31 showed similarities of 97.8% (402/411 bp), 100% (413/413 bp), 98.9% (373/377 bp), and 98.4% (358/364 bp), respectively, with P. pasqualense CBS122402 (JN606674), P. sanguifluum CBS 643.73 (JN606853), T. apiculatus CBS 312.59 (JX091378), and T. liani CBS 225.66 (JX091380). Similarly, BLASTn analysis of CaM of CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31 showed similarities of 98.1% (418/426 bp), 99.8% (504/505 bp), 98.7% (441/447 bp), and 99.3% (446/449 bp) with P. pasqualense CBS 122402 (JN606393), P. sanguifluum CBS 118020 (JN606536), T. apiculatus CBS 101366 (KF741932), and T. liani CBS 225.56 (KJ885257), respectively.
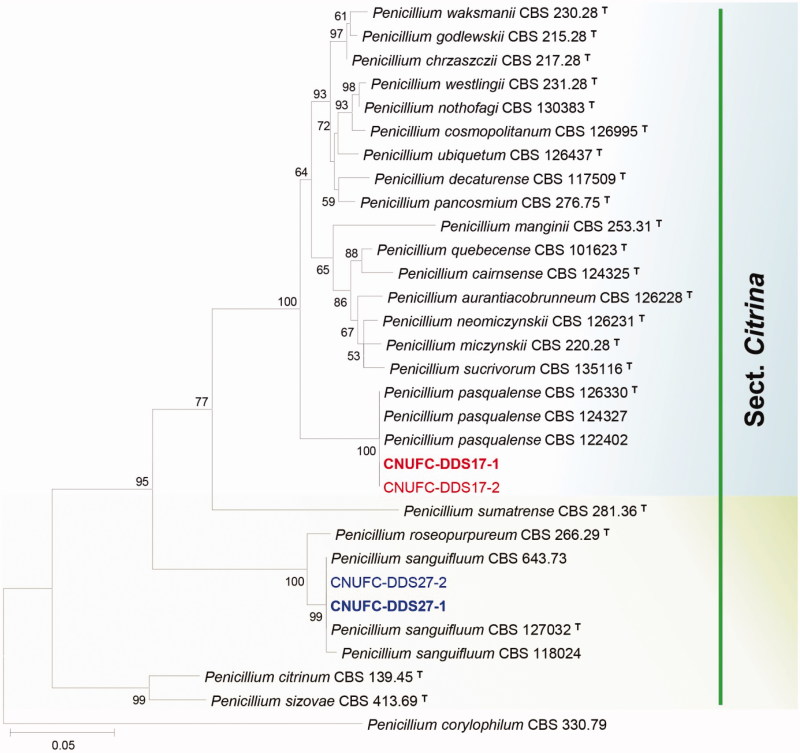
Figure 1.
Phylogenetic tree of Penicillium pasqualense CNUFC-DDS17-1 and CNUFC-DDS17-2, Penicillium sanguifluum CNUFC-DDS27-1 and CNUFC-DDS27-2, and related species based on maximum likelihood analysis of the combined datasets for BenA and CaM. The sequence of Penicillium corylophilum was used as an out group. Numbers at the nodes indicate the bootstrap values (>50%) from 1000 replicates. The bar indicates the number of substitutions per nucleotide. The study isolates are shown in bold red and blue.
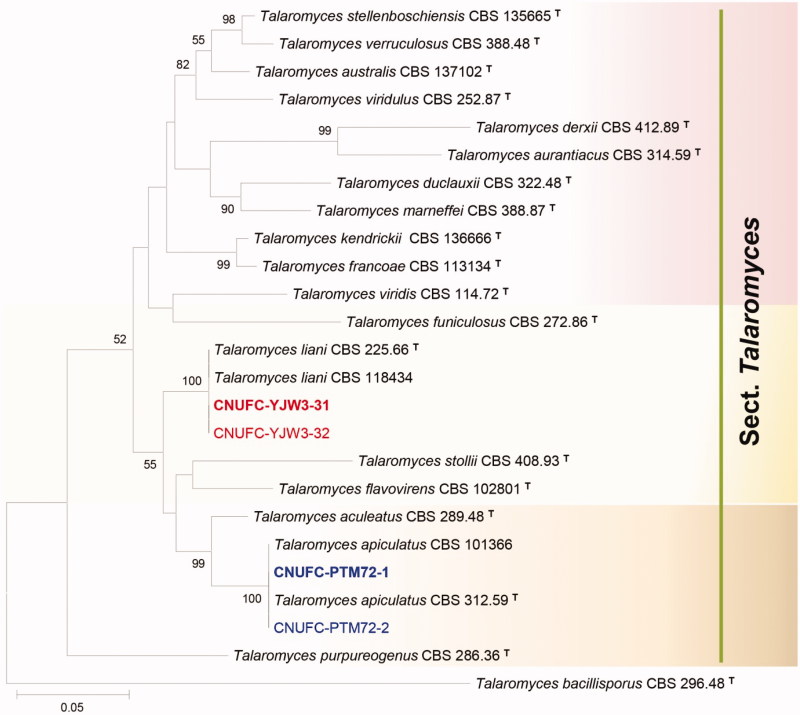
Figure 2.
Phylogenetic tree of Talaromyces apiculatus CNUFC-PTM72-1 and CNUFC-PTM72-2, Talaromyces liani CNUFC-YJW3-31 and CNUFC-YJW3-32, and related species based on maximum likelihood analysis of the combined datasets for BenA and CaM. Sequence of Talaromyces bacillisporus was used as an out group. Numbers at the nodes indicate the bootstrap values (>50%) from 1000 replicates. The bar indicates the number of substitutions per nucleotide. The study isolates are shown in bold red and blue.
In the combined BenA and CaM sequence tree, the isolates CNUFC-DDS17-1, CNUFC-DDS27-1, CNUFC-PTM72-1, and CNUFC-YJW3-31 were identical to P. pasqualense, P. sanguifluum, T. apiculatus, and T. liani, respectively (Figures 1 and 2).
3.2. Taxonomy
3.2.1. Taxonomy of CNUFC-DDS17-1
Penicillium pasqualense Houbraken, Frisvad & Samson, Studies in Mycology 70: 108 (2011) (Table 2, Figure 3).
Table 2.
Morphological characteristics of CNUFC-DDS17-1 compared with the reference strain of Penicillium pasqualense.
| Character | CNUFC-DDS17-1 | P. pasqualensea |
|---|---|---|
| Conidiophores | Biverticillate, triverticillate conidiophores, measured 100–300 μm long, 4.5–5 μm wide | Predominantly symmetrically biverticillate, 200–400 μm, smooth, 2.5–3.0 μm wide |
| Metuale | Few metuale, 10–18.9 × 2.5–3.2 μm | Metulae in a divergent vertical terminal, 11–17 × 2.5–3.5 μm, branches longer up to 25 μm |
| Phialides | Ampulliform, 5.9−10.2 × 2.5−3.5 μm | Ampulliform, 7.5–10 × 2.5–3.5 μm |
| Conidia | Globose to subglobose, 2.3–3.1 µm in diameter | Globose to subglobose, spinose, 2.5–3.5 µm in diameter |
| Sclerotia | Absent | Orange or brown |
From the description by Houbraken et al. [24].
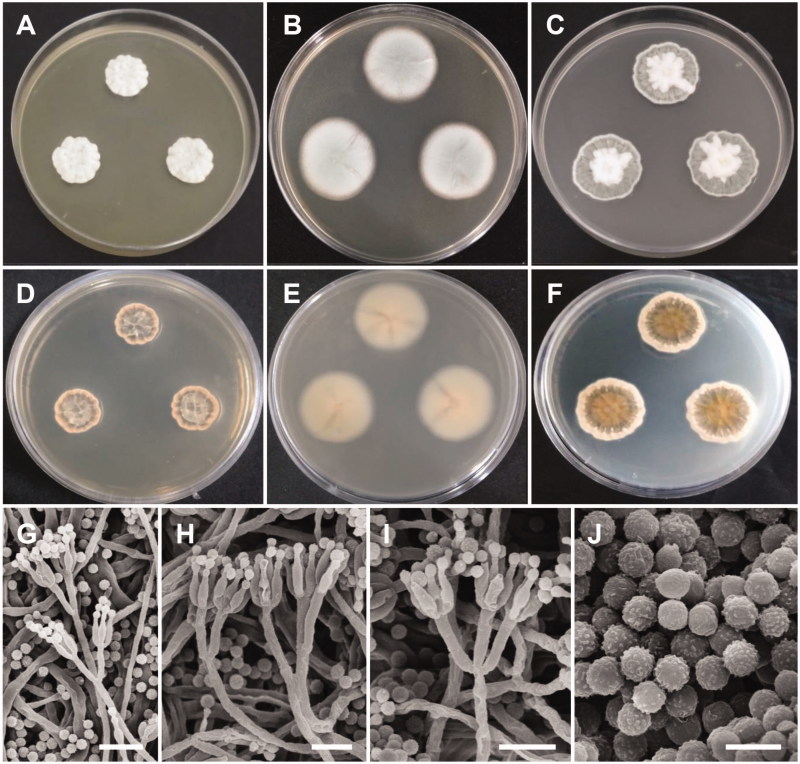
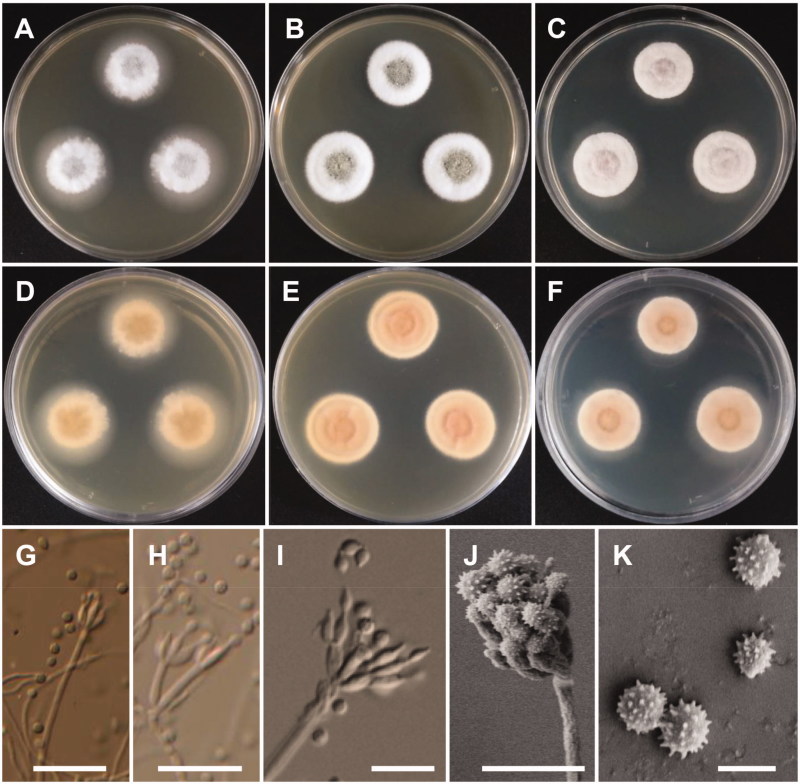
Figure 3.
Morphology of Penicillium pasqualense. (A,D) Colonies on yeast extract sucrose agar (YES); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on Czapek yeast autolysate agar (CYA); (A–C: obverse view, D–F: reverse view); (G–I) Conidiophores; (J) Conidia (Scale bars: G = 15 μm, H–I = 10 μm, J = 5 μm).
Description: Colonies on YES were dark beige with white mycelium, no soluble pigment, weak to moderate sporulation, and reached 20–26 mm in diameter after 7 d at 25 °C. Colonies on MEA were pale green, with good sporulation, and velvety to floccose texture. Colonies grew slow, reaching 25–35 mm in diameter after 7 d at 25 °C. Colonies were dark brown, with no soluble pigment, entire margins, and floccose colony texture at the center on CYA, reaching 25–30 mm in diameter. There were no formations of asci or ascospores. The conidiophores were predominantly symmetrically biverticillate, triverticillate (few) with additional branches and divergent terminal verticals of metulae, measured 10–18.9 × 2.5–3.2 μm. Phialides were ampulliform and measured 5.9–10.2 × 2.5–3.5 μm in diameter. Conidia were dark green or dark-blue green in color, globose to subglobose, and spinose and measured 2.3–3.1 μm in diameter.
3.2.2. Taxonomy of CNUFC-DDS27-1
Penicillium sanguifluum (Sopp) Biourge, La Cellule 33: 105 (1923) (Table 3, Figure 4).
Table 3.
Morphological characteristics of CNUFC-DDS27-1 compared with the reference strain of Penicillium sanguifluum.
| Character | CNUFC-DDS27-1 | P. sanguifluuma |
|---|---|---|
| Conidiophores | Monoverticillate, 17–50 μm | Monoverticillate, 15–50 µm |
| Metuale | Not observed | Branched-like metulae scarcely formed, strongly vesiculate (10–) 13–18 × 2.0–3.0 μm at base |
| Phialides | Ampulliform, 5.5–8.3 × 2.0–3.0 μm | Ampulliform, terminally, and subterminally formed, 6.5–8 × 2–3 μm |
| Conidia | Globose to subglobose, smooth to finely roughened, 2.0–2.5 µm in diameter | Globose to subglobose, smooth to finely roughened, 2.0–2.5 μm in diameter |
| Sclerotia | Absent | Absent |
From the description by Houbraken et al. [24].
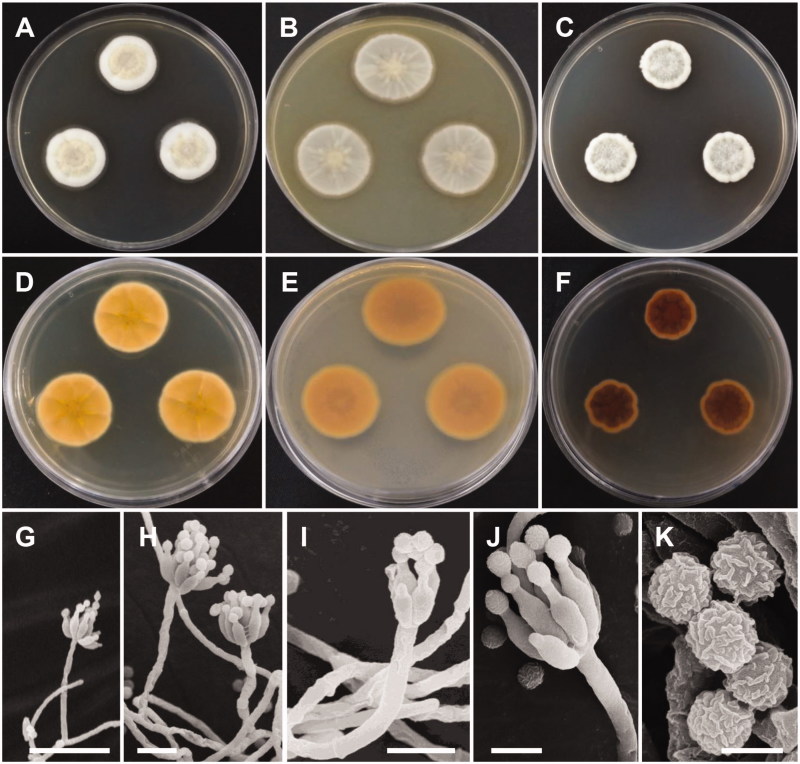
Figure 4.
Morphology of Penicillium sanguifluum. (A,D) Colonies on yeast extract sucrose agar (YES); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on Czapek yeast autolysate agar (CYA); (A–C: obverse view, D–F: reverse view); (G–J) Conidiophores; (K) Conidia (Scale bars: G = 20 μm, H–I = 10 μm, J = 5 μm, K = 2 μm).
≡Citromyces sanguifluus Sopp, Skrifter udgivne af Videnskabs-Selskabet i Christiania. Mathematisk-Naturvidenskabelig Klasse 11: 115 (1912).
Description: Colonies on YES were pale yellow with white mycelium, sparse sporulation, reaching 25–35 mm in diameter after 7 d at 25 °C. On MEA, colonies were muted greenish-blue at the center to white mycelium, colony texture floccose, sporulation sparse, and reaching 25–30 mm in diameter after 7 d at 25 °C. Colonies on CYA grew slow, reaching 25–30 mm in diameter at 25 °C, were pale beige to grey green at the center, had absent to moderate sporulation, and an entire or irregular margin. Sclerotia absent. Conidiophores were monoverticillate with short stipes and measured to be 17–50 μm. Phialides were ampulliform and measured 5.5–8.3 × 2.0–3.0 μm. Conidia were grey-green in color, globose to subglobose and measured 2.0–2.5 μm in diameter.
3.2.3. Taxonomy of CNUFC-PTM72-1
Talaromyces apiculatus R.A. Samson, N. Yilmaz & J.C. Frisvad, Studies in Mycology 70: 174 (2011) (Table 4, Figure 5).
Table 4.
Morphological characteristics of CNUFC-PTM72-1 compared with reference strain of Talaromyces apiculatus.
| Character | CNUFC-PTM72-1 | Talaromyces apiculatusa |
|---|---|---|
| Conidiophores | Biverticillate, 61–141 × 2.3–3.5 μm | Biverticillate (100–)150–350 × 2.5–3 μm |
| Phialides | Flask-shaped phialides, three to five per metulae, 7.5–10 × 2.3–3.6 μm | Flask-shaped phialides, three to five per metulae, 9–11 × 2.5–3.5 μm |
| Conidia | Rough-walled to echinulate, globose, 3.3–4.6 × 3.3–4.3 μm. | Rough-walled to echinulate, globose, 3–4(–5.5)×3–4(–5) μm |
| Ascomata | Not observed | Not observed |
From the description by Yilmaz et al. [11].
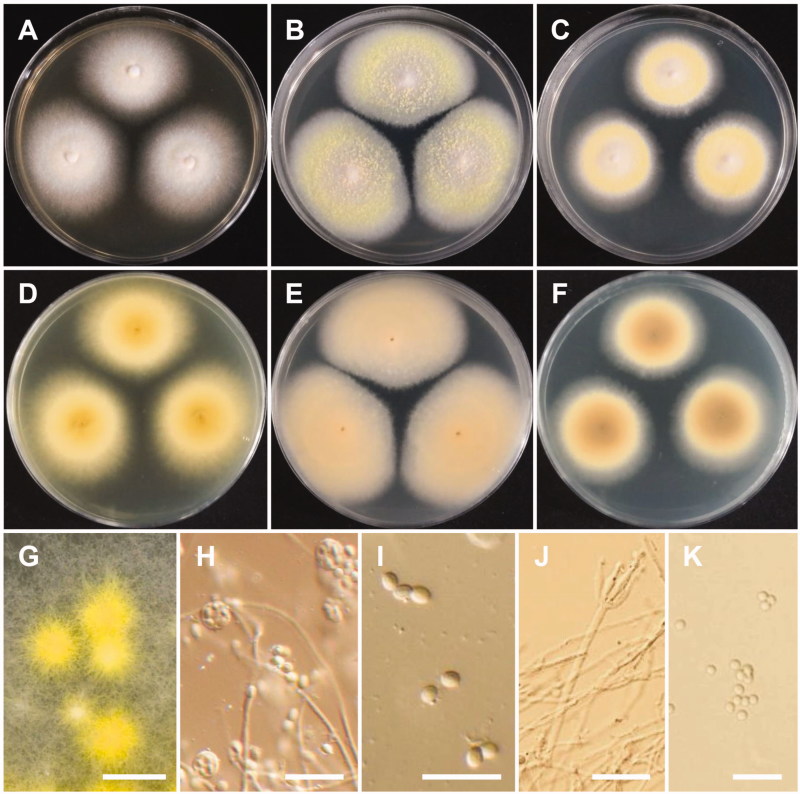
Figure 5.
Morphology of Talaromyces apiculatus. (A,D) Colonies on yeast extract sucrose agar (YES); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on Czapek yeast autolysate agar (CYA); (A–C: obverse view, D–F: reverse view); (G–J) Conidiophores; (K) Conidia (Scale bars: G–J = 20 μm, K = 5 μm).
≡Penicillium aculeatum var. apiculatum Abe, S., J. Gen. Appl. Microbiol., Tokyo 2: 124 (1956).
Description: Colonies on CYA were moderately deep, had radial, low margins, white mycelia, floccose texture, absent to sparse sporulation, no soluble pigment, and reached 22–23 mm in diameter after 7 d at 25 °C. On MEA, colonies were moderately deep, slightly raised at the center, had white mycelia, floccose texture, no soluble pigment, their reverse was brownish orange, and they reached 24–25 mm in diameter. On YES, colonies were moderately deep, low margins, white mycelia, no sporulation, no soluble pigment, and reaching 30–32 mm in diameter. Conidiophores were monoverticillate and measured to be 61–141 × 2.3–3.5 μm. Phialides were flask-shaped and measured 7.5–10 × 2.3–3.6 μm. Conidia were globose with rough echinulate walls and measured 3.3–4.6 × 3.3–4.3 μm in diameter.
3.2.4. Taxonomy of CNUFC-YJW3-31
Talaromyces liani (Kamyschko) N. Yilmaz, J.C. Frisvad & R.A. Samson, Studies in Mycology 78: 266 (2014) (Table 5, Figure 6).
Table 5.
Morphological characteristics of CNUFC-YJW3-31 compared with reference strain of Talaromyces liani.
| Character | CNUFC-YJW3-31 | Talaromyces liania |
|---|---|---|
| Conidiophores | Monoverticillate and biverticillate | Monoverticillate and biverticillate |
| Phialides | Acerose, three to six per metula, 10–17.5 × 2–3.5 μm | Acerose, three to six per metulae, 9–20 × 2–3.5 μm |
| Conidia | Globose to ellipsoidal, 2.5–4.0×2–3.5 μm. | Ellipsoidal, 2.5–4(–4.5)×2–3.5 μm. |
| Ascomata | Yellow, globose to subglobose, 110–429.5 × 109–428.5 μm | Yellow to orange red, globose to subglobose, 150–550 × 150–545 μm. |
| Asci | Globose to subglobose or slightly ellipsoidal, 9.5–11.5 × 8.0–10.0 μm | 9–13 × 7.5–11 μm |
| Ascospores | Ellipsoidal, spiny, 3.5 –5.5 × 3.0–4.5 μm | Ellipsoidal, spiny, 4–6 × 2.5–4 μm |
From the description by Yilmaz et al. [11].
Figure 6.
Morphology of Talaromyces liani. (A,D) Colonies on yeast extract sucrose agar (YES); (B,E) Colonies on Blakeslee’s malt extract agar (MEA); (C,F) Colonies on Czapek yeast autolysate agar (CYA); (A–C: obverse view, D–F: reverse view); (G) Ascomata on MEA; (H) Asci and ascopsores; (I) Ascopsores; (J) Conidiophore; (K) Conidia (Scale bars: G = 100 μm, H–K = 20 μm).
≡Penicillium liani Kamyschko, Not. Syst. Crypt. Inst. Bot. Acad. Sci. USSR 86: 115 (1962).
Description: Colonies were light yellow, with sparse sporulation, no soluble pigment, and their reverse was pastel yellow on CYA after 7 d at 25 °C. On MEA, colonies formed abundant yellow ascomata, had sparse sporulation, no soluble pigments, and their reverse was greyish yellow. Colonies were deep yellow on their reverse, had poor sporulation, white mycelium, and no soluble pigments on YES after 7 d at 25 °C. Conidiophores were monoverticillate and biverticillate, 2.0–4.5 μm wide. Phialides were acerose, three to six per metula, and measured 10–17.5 × 2–3.5 μm. Conidia were globose to ellipsoidal and measured 2.5–4.0 × 2–3.5 μm. Ascomata occurred within 7 d on MEA at 25 °C, were yellow in color and from globose to subglobose shape, and measured 110–429.5 × 109–428.5 μm. Asci were globose to subglobose or slightly ellipsoidal and measured 9.5–11.5 × 8.0–10.0 μm. Ascospores were ellipsoidal, spiny, and measured 3.5 –5.5 × 3.0–4.5 μm.
4. Discussion
The use of molecular study based on DNA sequences has dramatically increased the identification of fungal species [37]. DNA sequences, especially those of ITS-5.8S rDNA, have become important features for the rapid identification of fungi [38]. Although ITS sequences are well-accepted barcodes and place Penicillium species into their respective sections, more than 50% of the species could be erroneously linked as many species share the same ITS sequences. This led to the use of secondary markers, such as BenA and CaM, for the correct identification of Penicillium species in section Citrina. These markers act as a hub covering larger databases for the diverse genus Penicillium. Similarly, phylogenetic analyses of BenA genes are imperative for identification of Talaromyces species [11]. Here, we discuss the phylogeny and morphological characteristics of Penicillium and Talaromyces species and compare them to the most closely related species.
There were differences observed in the colony diameter for P. pasqualense and P. sanguifluum in comparison to previous descriptions. Colony diameter on MEA and YES media was shown to be different from the previously described P. pasqualense species (YES: 25–35 mm; MEA: 25–30) and (YES: 40–45 mm; MEA: 30–40) [8,24]. However, cultures on the other media were similar to the described species. In comparison to P. sanguifluum described by Visagie et al. [8], colony growth was shown to be restricted on CYA to 15–26 (vs. 7–16) mm at 25 °C. Also, no sclerotia production was observed for P. pasqualense [8,24]. These differences could be attributed to different soil conditions, seasons of sample collection, or different climate conditions. Another reason could be that the present isolated strain is a sub-cultured strain, because most species produce sclerotia in their first isolated strains [24].
Species of section Citrina have a cosmopolitan distribution and are commonly isolated from soil, but are also isolated from foods, leaf litter, and indoor air. Previously P. pasqualense and P. sanguifluum were reported to be isolated from various habitats; soils, indoor air environments, ants, and chestnuts [8,24]. Members belonging to Penicillium section Citrina are valuable owing to their ability to produce a variety of metabolites including citrinins and citreoviridins [23]. Many specific extrolites were extracted and identified from P. pasqualense and P. sanguifluum such as pyrenocines, indol alkaloids, PAS, bisanthrons, roseopurpurin, β -hydroxycurvularin, dehydrocurvularin, curvularin, FOSI, FYKS, SNIT, TIDL, and VERN [24]. The members of section Citrina can grow at temperatures ranging from 23 to 26 °C [24]. In this study, the two isolates had slow growth at lower temperatures (5–10 °C) and optimum growth at 25–30 °C, whereas growth was restricted at higher temperatures 35–37 °C for P. pasqualense and P. sanguifluum.
In comparison to T. apiculatus [11], the present isolate CNUFC-PTM72-1 shows similar morphological characteristics with small length conidiophores. The morphological characteristics of the T. liani isolate studied were generally similar to those previously described by Yilmaz et al. [11]. However, our observations showed that the size of asci and ascomata of isolate CNUFC-YJW3-31 were smaller than previously reported. Sexual reproduction structures were only observed on MEA. There were previously only reports of T. apiculatus and T. liani from soil samples [11,39]. This is the first isolation of T. liani from freshwater. T. apiculatus produces various bioactive extrolites such as macrocyclic polylactones (apiculides) NG-011, NG-012 (potentiators of nerve growth factors); BK223-A (=NG-012), BK-223B, BK-223C, 15G256B, and 15G256a-2 (act as antifungal) and bioxanthracene B, whereas no available information on the metabolites produced by T. liani yet [11].
Four species were identified, namely P. pasqualense, P. sanguifluum, T. apiculatus, and T. liani, successfully combining molecular and morphological analyses in this study. The main focus was to describe the diversity of Penicillium and Talaromyces species from soil and freshwater in Korea. The soil ecosystem has a tremendous impact on environmental sustainability in forestry, agriculture, and horticulture. Therefore, it is necessary to identify fungal species from soil and to characterize their role in the environment. Thus, our future studies would include studying the species ecological roles, such as the production of extracellular enzyme activity, antibacterial and antifungal activities, as well as the production of secondary metabolites.
Funding Statement
This work was in part supported by the Graduate Program for the Undiscovered Taxa of Korea, and in part by the Project on Survey and Discovery of Indigenous Fungal Species of Korea funded by NIBR and Project on Discovery of Fungi from Freshwater and Collection of Fungarium funded by NNIBR of the Ministry of Environment (MOE), and in part carried out with the support of the Cooperative Research Program for Agriculture Science and Technology Development (PJ013744), Rural Development Administration, Republic of Korea. This work was in part supported by the BK21 plus program through the National Research Foundation (NRF) funded by the Ministry of Education of Korea. This work was also supported by grants from National Institute of Biological Resources.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Warcup JH. Soil-steaming: a selective method for the isolation of ascomycetes from soil. Trans Br Mycol Soc. 1951;34:515–518. [Google Scholar]
- 2.Gerbersdorf SU, Hollert H, Brinkmann M, et al. . Anthropogenic pollutants affect ecosystem services of freshwater sediments: the need for a “triad plus x” approach. J Soils Sediments. 2011;11:1099–1114. [Google Scholar]
- 3.Taniwaki MH, Pitt JI, Iamanaka BT, et al. . Penicillium excelsum sp. nov from the Brazil nut tree ecosystem in the Amazon Basin. PLOS ONE. 2015;10:e0143189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frisvad JC, Samson RA. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol. 2004;49:1–174. [Google Scholar]
- 5.Samson RA, Houbraken J, Thrane U, et al. Food and Indoor Fungi. CBS laboratory manual series 2. Utrecht: CBS KNAW Fungal Biodiversity Centre. 2010. p. 390. [Google Scholar]
- 6.Guevara-Suarez M, Sutton DA, Gené J, et al. . Four new species of Talaromyces from clinical sources. Mycoses. 2017;60:651–662. [DOI] [PubMed] [Google Scholar]
- 7.Frisvad JC, Filtenborg O, Samson RA, et al. . Chemotaxonomy of the genus Talaromyces. Antonie Van Leeuwenhoek. 1990;57:179–189. [DOI] [PubMed] [Google Scholar]
- 8.Visagie CM, Seifert KA, Houbraken J, et al. . Diversity of Penicillium section Citrina within the fynbos biome of South Africa, including a new species from a Protea repens infructescence. Mycologia. 2014;106:537–552. [DOI] [PubMed] [Google Scholar]
- 9.Schafhauser T, Kirchner N, Kulik A, et al. . The cyclochlorotine mycotoxin is produced by the nonribosomal peptide synthetase CctN in Talaromyces islandicus (‘Penicillium islandicum’). Environ Microbiol. 2016;18:3728–3741. [DOI] [PubMed] [Google Scholar]
- 10.Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011;70:1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yilmaz N, Visagie CM, Houbraken J, et al. . Polyphasic taxonomy of the genus Talaromyces. Stud Mycol. 2014;78:175–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houbraken J, de Vries RP, Samson RA. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol. 2014;86:199–249. [DOI] [PubMed] [Google Scholar]
- 13.Link HF. Observationes in ordines plantarum naturales. Dissertatio 1ma. Mag Ges Naturf Freunde Berlin. 1809;3:3–42. [Google Scholar]
- 14.Wang XC, Chen K, Zeng ZQ, et al. . Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci Rep. 2017;7:8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbosa RN, Bezerra JDP, Souza-Motta CM, et al. . New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek. 2018;111:1883–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houbraken J, Wang L, Lee HB, et al. . New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia. 2016;36:299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HJ, Kim JS, Cheon KH, et al. . Species list of Aspergillus, Penicillium and Talaromyces in Korea, Based on ‘one fungus one name’ system. Kor J Mycol. 2016;44:2017–2219. [Google Scholar]
- 18.Park MS, Lee S, Lim YW. A new record of four Penicillium species isolated from Agarum clathratum in Korea. J Microbiol. 2017;55:237–246. [DOI] [PubMed] [Google Scholar]
- 19.Sang H, An TJ, Kim CS, et al. . Penicillium daejeonium sp. nov., a new species isolated from a grape and schisandra fruit in Korea. J Microbiol. 2013;51:536–539. [DOI] [PubMed] [Google Scholar]
- 20.You YH, Cho HS, Song J, et al. . Penicillium koreense sp. nov., isolated from various soils in Korea. J Microbiol Biotechnol. 2014;24:1606–1608. [DOI] [PubMed] [Google Scholar]
- 21.Tibpromma S, Hyde KD, Jeewon R, et al. . Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017;83:1–602. [Google Scholar]
- 22.Wanasinghe DN, Phukhamsakda C, Hye KD, et al. . Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89:1–236. [Google Scholar]
- 23.Houbraken J, Frisvad JC, Samson RA, et al. . Taxonomy of Penicillium citrinum and related species. Fungal Divers. 2010;44:117–133. [Google Scholar]
- 24.Houbraken J, Frisvad JC, Samson RA. Taxonomy of Penicillium section Citrina. Stud Mycol. 2011;70:53–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park MS, Eom JE, Fong JJ, et al. . New record and enzyme activity of four species in Penicillium section Citrina from marine environments in Korea. J Microbiol. 2015;53:219–225. [DOI] [PubMed] [Google Scholar]
- 26.Benjamin CR. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47:669–687. [Google Scholar]
- 27.Yilmaz N, López-Quintero CA, Vasco-Palacios AM, et al. . Four novel Talaromyces species isolated from leaf litter from Colombian Amazon rain forests. Mycol Progress. 2016;15:1041–1056. [Google Scholar]
- 28.Jiang XZ, Yu ZD, Ruan YM, et al. . Three new species of Talaromyces sect. Talaromyces discovered from soil in China. Sci Rep. 2018;8:4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seok H, Ko JH, Shin I, et al. . Disseminated Talaromyces marneffei and Mycobacterium intracellulare coninfection in an HIV-infected patient. Int J Infect Dis. 2015;38:86–88. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen TTT, Paul NC, Lee HB. Characterization of Paecilomyces variotii and Talaromyces amestolkiae in Korea based on the morphological characteristics and multigene phylogenetic analyses. Mycobiology. 2016;44:248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryu MR, Yoo IY, Song DJ, et al. . Penicillium species other than Talaromyces marneffei producing red pigment from clinical specimens: isolation of Talaromyces albobiverticillius. Lab Med Online. 2017;7:211–214. [Google Scholar]
- 32.White TJ, Bruns T, Lee S, et al. . Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. Cambridge (MA): Academic Press; 1990. p. 315–322. [Google Scholar]
- 33.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Gibson TJ, Plewniak F, et al. . The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai YC, Cui BK, Si J, et al. . Dynamics of the worldwide number of fungi with emphasis on fungal diversity in China. Mycol Prog. 2015;14:62. [Google Scholar]
- 38.Schoch CL, Seifert KA, Huhndorf S, et al. . Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samson RA, Yilmaz N, Houbraken J, et al. . Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70:159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]