Abstract
The secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA), formed by gut microbiota from primary bile acids via a multi-step 7α-dehydroxylation reaction, have wide-ranging effects on host metabolism and play an important role in health and disease. A few 7α-dehydroxylating strains have been isolated, where bile acid-inducible (bai) genes were organized in a gene cluster and encoded major enzymes involved. However, only little is known on diversity and abundance of intestinal bacteria catalysing DCA/LCA formation in the human gut in situ. In this study, we took the opportunity to screen metagenome-assembled genomes (MAGs) from sequence data of stool samples provided by two recent studies along with newly available gut-derived isolates for the presence of the bai gene cluster. We revealed in total 765 and 620 MAGs encoding the potential to form DCA/LCA that grouped into 21 and 26 metagenomic species, respectively. The majority of MAGs (92.4 and 90.3%) were associated with a Ruminococcaceae clade that still lacks an isolate, whereas less MAGs belonged to Lachnospiraceae along with eight new isolates (n total = 11) that contained the bai genes. Only a few MAGs were linked to Peptostreptococcaceae. Signatures for horizontal transfer of bai genes were observed. This study gives a comprehensive overview of the diversity of bai-exhibiting bacteria in the human gut highlighting the application of metagenomics to unravel potential functions hidden from current isolates. Eventually, isolates of the identified main MAG clade are required in order to prove their capability of 7α-dehydroxylating primary bile acids.
Keywords: Bile acids, Gut microbiota, Microbiome, Metagenomics, 7α-dehydroxylation, Systems biology
1. Introduction
The primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) are produced from cholesterol in the liver and are subsequently conjugated to taurine or glycine residues. After their excretion into the duodenum they enable absorption of dietary lipids, cholesterol and fat-soluble vitamins that are essential for lipid metabolism and host health [1]. Additionally, bile acids act as signaling molecules regulating their own synthesis, uptake, transportation, and detoxification, and are involved in overall lipid, glucose and energy metabolisms via binding on nuclear and G-protein-coupled bile acid receptors that are expressed throughout the body [2]. The majority of secreted bile (95%) is reabsorbed along the entire gut by active transportation and passive diffusion, subsequently reconjugated in the liver and again secreted into the duodenum, which is referred to as the enterohepatic circulation [1]. Gut microbiota directly act on bile acids substantially modifying the composition of the bile acid pool. As a first step, bacteria initiate their deconjugation via bile salt hydrolases rendering bile acids susceptible to various subsequent bacterial transformations including 7α-dehydroxylation, dehydrogenation, and epimerization that lead to the generation of secondary bile acids [1,3]. Deoxycholic acid (DCA) and lithocholic acid (LCA) comprise the majority of secondary bile acids and are formed from CA and CDCA, respectively, via 7α-dehydroxylation, a multi-step process that primarily occurs in the colon. Upon reabsorption DCA is reconjugated, but not rehydroxylated, which leads to its accumulation in the bile acid pool comprising a substantial part of total bile (around 25%, with large interindividual variations) [4]. In contrast, LCA is reconjugated and additionally sulfonated in the liver promoting its excretion from the body.
Secondary bile acids have wide-ranging effects on host health. On the one hand they promote disease with high levels being cytotoxic and associated with an increased risk of cholesterol gallstone disease and colon cancer [5]. Furthermore, a recent study demonstrated their role in hepatocellular carcinoma via modulating the immune system [6]. On the other hand, numerous studies described their antimicrobial effects highlighting their ability to provide colonization resistance against Clostridioides difficile [c.f. 7].
Enzymes involved in 7α-dehydroxylation are encoded by bile acid inducible (bai) genes that were previously identified in a few strains of Lachnospiraceae and Peptostreptococcaceae [1,8]. However, despite their pivotal role for host physiology, only little is known on diversity of 7α-dehydroxylating bacteria in the human gut in situ. Our recent, extensive survey on publicly available metagenomic/metatranscriptomic datasets suggested that the bai gene cluster is present and expressed in most individuals, yet only in a small fraction (<1%) of total intestinal bacteria [8]. Its main representative was an uncultivated member of the order Clostridiales, namely, Firmicutes bacterium CAG:103 that recruited 63.9% of all bai-associated reads, which displayed high amino acid sequence similarities to the reference. Only a minor portion was linked to Lachnospiraceae (4.7%) and Peptostreptococcaceae (1.9%), which include the species Clostridium scindens and Clostridium hiranonis, respectively.
2. Results & Discussion
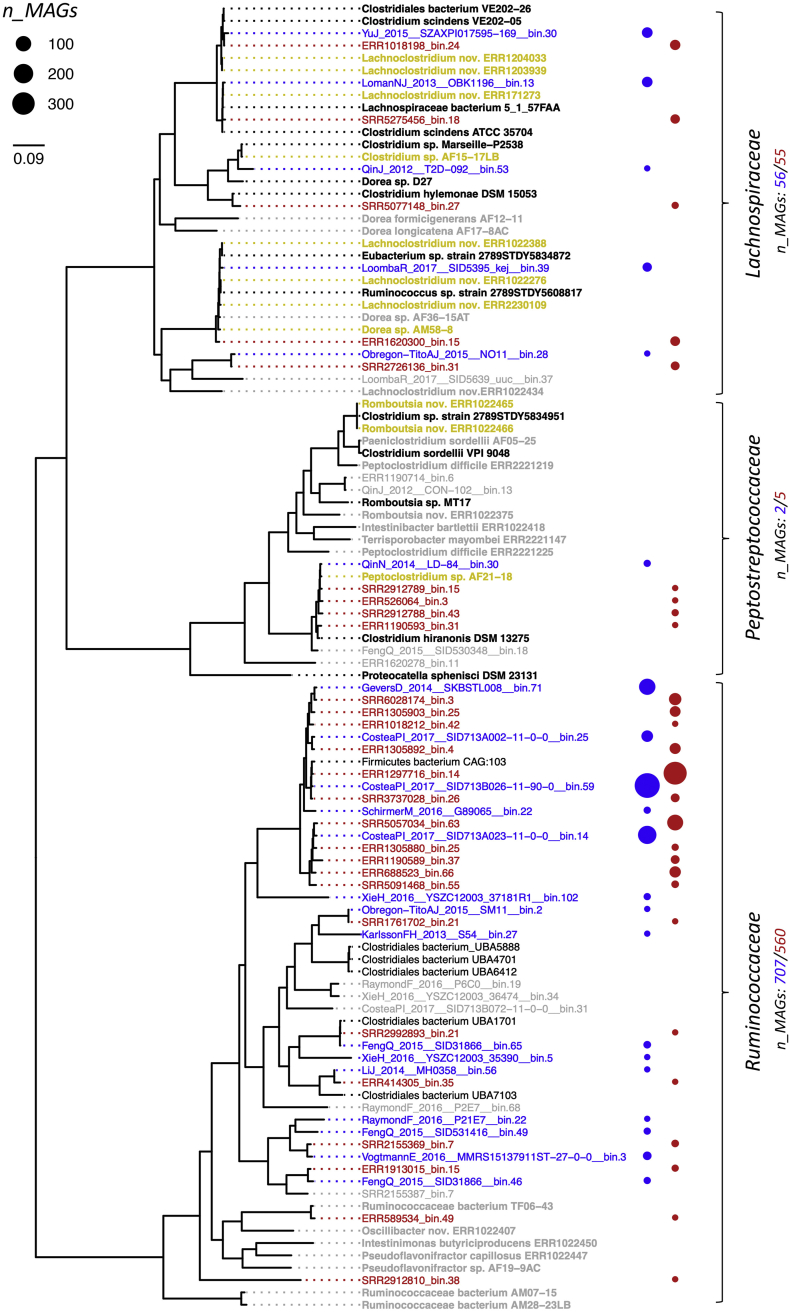
Recently, vast amounts of metagenome-assembled genomes (MAGs) of gut bacteria became available from two signature studies, namely, Pasolli et al. [9] and Almeida et al. [10], which are referred to as study A and B, respectively, in the following text. Furthermore, efforts from Forster et al. [11] and Zou et al. [12] substantially increased cultured representatives of intestinal bacteria. We took the opportunity to screen for bai-containing genomes in those references in order to get a comprehensive overview of bacteria exhibiting this crucial function in the gut environment and to expand current reference sequences. Eleven new isolates containing the gene cluster were identified (Fig. 1). For metagenomic-based data, both studies yielded very similar results with 765 and 620 bai-exhibiting MAGs obtained from study A and B, respectively, which represented 0.51% (A) and 0.73% (B) of all stool-associated MAGs in these studies (excluding infants, which were devoid of bai-containing MAGs), and clustered into 21 (A) and 26 (B) species-level genome bins (SGBs) (Fig. 1). For detailed distribution of bai-containing bacteria in human gut microbiota, i.e., their total abundance and abundances of individual clades, we want to refer the reader to our previous report [8]. Most bai-exhibiting MAGs were associated with Ruminococcaceae – 92.4 and 90.3% of all bai-containing MAGs were associated with this family in study A and B, respectively. The vast majority of these MAGs, 97.5% (A) and 98.2% (B), were closely related to the previously identified metagenome-derived Firmicutes bacterium CAG:103, with the bulk harboring all bai genes (baiA-I). Despite recent isolation efforts, this clade still lacks a cultured representative; most closely related gut isolates were the Ruminococcaceae genera Oscillibacter, Intestinimonas, and Pseudoflavonifractor. BLASTing of selected housekeeping genes from main SGBs against NCBI's non-redundant protein database revealed the same genera as mentioned above as their closest relatives. It should be mentioned that although SGBs, which surround Firmicutes bacterium CAG:103 in the phylogenetic tree shown in Fig. 1, formed a functionally coherent group, we detected several medium quality SGBs, i.e., genome bins with completeness <90% and/or contamination >5%, that were devoid of the bai gene cluster and interleaved with bai-containing SGBs (data not shown). Furthermore, major SGBs contained many non bai-exhibiting MAGs, where, for instance, only 66.2 and 66.1% of total MAGs of the SGBs CosteaPI_2017__SID713B026-11-90-0__bin.59 and CosteaPI_2017__SID713A023-11-0-0__bin.14, respectively, harbored the target genes (no data is available for study B). It is, thus, questionable whether this clade is truly functionally consistent and care should be taken based on analyses using SGBs as representatives for bai-containing bacteria, since this might lead to overestimations in their abundance. Only 55 (A) and 56 (B) MAGs were associated with Lachnospiraceae, whereas the majority of new isolates belonged to this family. MAGs from the Lachnospiraceae formed two main clades intermitted by non bai-exhibiting Dorea species. A tiny fraction of MAGs was associated with Peptostreptococcaceae. Phylogenetic analysis of bai genes showed signatures of horizontal gene transfer, where Lachnospiraceae-associated bai genes related to those identified in Dorea sp. AM58-8 grouped with sequences of the main Ruminococcaceae clade that contained the majority of MAGs (Fig. S1). Bai genes of the Peptostreptococcaceae related to those of C. hiranonis clustered in-between genes of the Lachnospiraceae species C. scindens and C. hylemonae, respectively. Genes of members associated with C. sordelli formed an outgroup. While baiA-I catalyze the oxidative arm of 7α-dehydroxylation, enzymes of the reductive arm are largely unknown [1]. Recently, a flavoprotein (baiN) isolated from C. scindens was suggested to play a role in the reductive reactions [13]. Screening for baiN in the present study did yield hits in most genomes, however, amino acid identities were often low, even for C. hiranonis, a verified 7α-dehydroxylating bacterium (Table S1). Furthermore, also non baiA-I-containing taxa exhibited similar genes, which was already shown in the original publication [13]. We could, hence, not convincingly point baiN homologous gene sequences outside of the main Lachnospiraceae clade that includes C. scindens and C. hylemonae and more detailed investigations, including biochemical testing, needs to be performed in order to unravel enzymes encoding the reductive arm in baiA-I-exhibiting bacteria revealed in this study.
Fig. 1.
Approximately-Maximum-Likelihood tree of bai-gene-containing reference genomes based on 92 housekeeping genes. In blue, metagenome-derived species-level genome bins (SGBs) of study A are depicted, whereas SGBs from study B are shown in red. Abundances, i.e., number of bai-gene-containing metagenome-assembled genomes (MAGs) associated with individual SGBs, are shown on the right. Names of isolates are displayed in bold with those highlighted in gold derived from recent isolation efforts. Names shown in grey represent gut-derived isolates and SGBs not exhibiting the bai gene cluster (only high quality SGBs were considered). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In conclusion, this study gives an extensive overview of the diversity of bai-exhibiting bacteria in the human gut and highlights the application of metagenomics to unravel potential functions hidden from current isolates. Obtained reference sequences will assist guided isolation of target bacteria enabling in vitro experiments to validate 7α-dehydroxylation of primary bile acids.
3. Materials and Methods
Metagenome-assembled genomes (MAGs) from study A [9] and B [10] as well as genomes of isolates from Zou et al. [12] were downloaded, subjected to prokka (v. 1.13.3, default mode) [14], and screened for individual bai genes (baiA-I) using Hidden Markov Chain Models (HMM) as described previously [8]. Protein sequence score cut-offs were set at 50% of the lowest protein reference from our previous database [8] and all genomes exhibiting ≥4 genes in synteny (defined as being separated by ≤10 genes based on locus tag) were selected as candidates. Manual inspections were performed for all genes based on phylogenetic trees. Study A provided association of each MAG with its representative species-level genome bin (SGB) representing all MAGs spanning a 95% genetic similarity. For study B, genetic distances of all bai-exhibiting MAGs were calculated using Mash (v. v. 2.1.1, option "-s 1e4" for sketching) [15] that were subsequently clustered into SGBs (≥ 95% genetic similarity) using hierarchical clustering in R (v. 3.5.2, function stats::hclust followed by function stats::cutree). Raw data of new isolates from Forster et al. (2019) [11] were downloaded, quality filtered using fastp (v. 0.20.0, options “-5 20 -3 20 -l 70”) [15], and assembled via SPAdes on paired-end read mode (v. 3.13.0, option “--careful”) [16]. Contigs were then subjected to prokka before screening with HMMs. All reference gene sequences are available at https://www.pathofunctions.com. The tree shown in Fig. 1 was constructed from 92 housekeeping genes using UBCG (v. 3.0, default mode) [17], whereas concatenated sequences of baiCD,E,H, that were found in most genomes, were used to construct the tree in Fig. S1 applying FastTree (v. 2.1.10, default mode) [18]. Both trees were visualized with ggtree (v. 1.14.6) [19]. Screening for baiN was performed by BLASTing reference sequences (EDS08212.1, ZP_03776912.1) against all genomes shown in Fig. 1 using DIAMOND (v. 0.9.24) [20]; only top hits were recorded.
The following are the supplementary data related to this article.
Approximately-Maximum-Likelihood tree of references based on the genes baiCD,E,H. In blue, metagenome-derived species-level genome bins (SGBs) of study A are depicted, whereas SGBs from study B are shown in red. Organization of individual bai genes is given on the right. Names of isolates are displayed in bold with those highlighted in gold derived from recent isolation efforts.
Search for baiN homologs in genomes. Taxa are arranged according to the tree shown in Fig. 1, where those highlighted in green exhibit enzymes involved in the oxidative arm of 7α-dehydroxylation.
Declaration of Competing Interest
There is no conflict of interest.
Acknowledgements
This work was funded by RESIST (Excellence Cluster), 1.7.2019-30.6.2022, and by the Helmholtz Association's Initiatives on Personalized Medicine (iMed) and Aging and Metabolic Programming (AMPro).
References
- 1.Ridlon J.M., Harris S.C., Bhowmik S., Kang D.J., Hylemon P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li T., Chiang J.Y.L. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foley M.H., O'Flaherty S., Barrangou R., Theriot C.M. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridlon J.M., Kang D.-J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Wells J.E., Hylemon P.B. Identification and characterization of a bile acid 7a-dehydroxylation operon in clostridium sp. strain TO-931, a highly active 7a-dehydroxylating strain isolated from human feces. Appl Environ Microbiol. 2000;66:1107–1113. doi: 10.1128/aem.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma C., Han M., Heinrich B., Fu Q., Zhang Q., Sandhu M. Gut microbiome – mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360:1–9. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buffie C.G., Bucci V., Stein R.R., McKenney P.T., Ling L., Gobourne A. Precision microbiome reconstitution restores bile acid mediated resistance to clostridium difficile. Nature. 2014;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rath S., Rud T., Karch A., Pieper D.H., Vital M. Pathogenic functions of host microbiota. Microbiome. 2018;6(174):1–13. doi: 10.1186/s40168-018-0542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A. A new genomic blueprint of the human gut microbiota. Nature. 2019:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forster S.C., Kumar N., Anonye B.O., Almeida A., Viciani E., Stares M.D. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou Y., Xue W., Luo G., Deng Z., Qin P., Guo R. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37:179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris S.C., Devendran S., Alves J.M.P., Mythen S.M., Hylemon P.B., Ridlon J.M. Identification of a gene encoding a flavoprotein involved in bile acid metabolism by the human gut bacterium Clostridium scindens ATCC 35704. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:276–283. doi: 10.1016/j.bbalip.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. [Google Scholar]
- 15.Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., Koren S. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:1–14. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Na S.I., Kim Y.O., Yoon S.H., Ha S., Baek I., Chun J. UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol. 2018;56:281–285. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 18.Price M.N., Dehal P.S., Arkin A.P. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.Y. Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 20.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Approximately-Maximum-Likelihood tree of references based on the genes baiCD,E,H. In blue, metagenome-derived species-level genome bins (SGBs) of study A are depicted, whereas SGBs from study B are shown in red. Organization of individual bai genes is given on the right. Names of isolates are displayed in bold with those highlighted in gold derived from recent isolation efforts.
Search for baiN homologs in genomes. Taxa are arranged according to the tree shown in Fig. 1, where those highlighted in green exhibit enzymes involved in the oxidative arm of 7α-dehydroxylation.