Abstract
Algicidal bacteria are important players regulating the dynamic changes of plankton assemblages. Most studies on these bacteria have focused on the effect on single algal species in simple incubation experiments. Considering the complexity of species assemblages in the natural plankton, such incubations represent an oversimplification and do not allow making further reaching conclusions on ecological interactions. Here, we describe a series of co‐incubation experiments with different level of complexity to elucidate the effect of the algicidal bacterium Kordia algicida on mixed cultures of a resistant and a susceptible diatom. The growth of the resistant diatom Chaetoceros didymus is nearly unaffected by K. algicida in monoculture, while cells of the susceptible diatom Skeletonema costatum are lysed within few hours. Growth of C. didymus is inhibited if mixed cultures of the two diatoms are infected with the bacterium. Incubations with filtrates of the infected cultures show that the effects are chemically mediated. In non‐contact co‐culturing we show that low concentrations of the lysed algae support the growth of C. didymus, while higher concentrations trigger population decline. Complex cascading effects of algicidal bacteria have thus to be taken into account if their ecological role is concerned.
Keywords: algicidal bacteria, co‐cultures, community ecology, diatoms, phytoplankton, Plankton interactions
1. INTRODUCTION
Microalgae of the phytoplankton can form spatially and temporally limited blooms within the pelagic environment. Such blooms can be limited by abiotic factors, such as light or nutrient availability. However, a recent survey reflects that environmental factors are insufficient predictors of community structures (Lima‐Mendez et al., 2015). Additional biotic interactions can lead to species dominance and bloom decline. Mechanisms mediating decline of algal populations include grazing by herbivores and viral infection as well as allelopathic interactions in which growth of microalgae is chemically suppressed by competitors (Bratbak, Egge, & Heldal, 1993; Brussaard et al., 1995; Pohnert, 2010). Cell‐lysis caused by algicidal bacteria is another factor that can substantially influence the plankton community (Bidle, 2015; Meyer, Bigalke, Kaulfuss, & Pohnert, 2017). The heterotrophic bacteria can utilize resources excreted by phytoplankton cells or resources released after algal cell death and lysis (Bidle & Falkowski, 2004; Meyer et al., 2017). Due to their universal distribution, algicidal bacteria are of major interest for the understanding of dynamic species successions in the ocean (Teeling et al., 2016; van Tol, Amin, & Armbrust, 2017). However, research on this class of organisms is mainly limited to investigations of bilateral interactions of one bacterial isolate with a specific phytoplankton species (Meyer et al., 2017). In nature, the situation is clearly more complex since multiple players form complex interaction networks that can be disturbed by lytic bacteria. Bacteria might, for example, target one single species within a community by specific lysis or eliminate multiple members of the phytoplankton thereby making room for successive blooms (Meyer et al., 2017). Algae that show resistance against such generalist algicidal bacteria might, therefore, have a substantial competitive advantage that could boost their performance in succession of a lytic event.
To study such potential cascading effects, we set up a tripartite interaction network including a resistant and a susceptible diatom species that were exposed to the algicidal bacterium Kordia algicida. We focused on the two widely distributed diatoms Skeletonema costatum, which is susceptible to bacterial lysis and the resistant Chaetocerous didymus (Paul & Pohnert, 2011). Both diatoms are globally distributed and co‐occur as succeeding bloom forming algae in the environment (Kooistra et al., 2008; Li et al., 2017). It has been recently established in our lab that the algicidal activity of K. algicida is based on the release of proteases that are under the control of quorum sensing mediators (Paul & Pohnert, 2011). The resistance of C. didymus involves the induced release of a presumably counteracting protease (Paul & Pohnert, 2013) confirming that chemical cues are the primary language used by marine organisms (Hay, 2009). To investigate how chemically mediated interactions might be involved in further cascading effects following algal lysis we address here mixed co‐cultures, as well as the activity of filtrates of infected and non‐infected cultures on the respective interaction partners. Additionally, induction of responses that are triggered by diffusible chemical mediators was investigated in non‐contact co‐culturing experiments that allow the free diffusion of chemical signals in between culture compartments containing the respective partners (Paul, Mausz, & Pohnert, 2012). We clearly establish that combinations of only two species in simple laboratory setups do not allow to predict algal performance upon infection in more complex settings. Despite the intrinsic problematic transfer from laboratory to field scenarios our results suggest that in nature lytic bacteria might cause unexpected cascading effects. We also suggest that care has to be taken when planning to use such bacteria in the control of harmful algal blooms since these unpredictable events might lead to even more harmful scenarios.
2. MATERIALS AND METHODS
2.1. Diatom and bacteria culturing
S. costatum (RCC75) was obtained from the Roscoff Culture Collection France and C. didymus (Na20B4) was obtained from Wiebe Kooistra who isolated it in the Gulf of Naples, Italy (LTER sample station Marechiara). Both non‐axenic diatoms were grown in artificial sea water (SW) prepared according to Maier and Calenberg at a pH of 7.8 (Maier & Calenberg, 1994) under a 14/10 hr light/dark cycle with 50–60 µmol m−2 s−1 at 13°C. The initial nutrient concentrations were 620 mM nitrate, 14.5 mM phosphate, and 320 mM silicate. Algal growth was determined either by measuring the relative in vivo chlorophyll a fluorescence on a Mithras LB 940 plate reader (Berthold Technologies, Bad Wildbad, Germany) using 200 µl of each culture in dark 96‐well plates or by cell counting after fixation with Lugol´s iodine solution using Fuchs‐Rosenthal counting chambers under an upright microscope (Leica DM 200, Wetzlar, Germany).
The Gram‐negative marine bacterium K. algicida strain OT‐1 was obtained from the NITE Biological Resource Center (NBRC 100336) and stored at −80°C in 20 vol% glycerol. Growth of K. algicida was maintained at 23°C and controlled by measuring the optical density (OD) at a wavelength of 550 nm (Specord M42 UV‐vis spectrophotometer by Carl Zeiss, Jena Germany). Bacterial inoculation densities were set to a final OD550 of 0.02 for all the experiments. For experiments 1–3, K. algicida was grown on solid full medium plates (3.74% w/v marine broth and 1.5% w/v agar) for three days before harvesting. The inoculation solution for treatments in the experiments was obtained by washing off the bacterial cells from the plates with SW. Inoculation procedure for experimental controls was identical but plates without bacterial cells were used instead.
2.2. Experiment 1
We initially tested how the bacterial lysed S. costatum affects the success of the resistant diatom C. didymus in mixed cultures. Two independent fully replicated sets of experiments were carried out with C. didymus and S. costatum bialgal cultures at an initial cell density of 1×103 and 1×105 cells/ml, respectively, in presence or absence of the algicidal bacterium. Eighty milliliters of algal dilutions were cultivated as described above in cell culture flasks (T‐75, Sarstedt, Nümbrecht, Germany). Monoalgal controls were conducted in the second experiment. K. algicida was cultivated and inoculated as described above. Data points were obtained by cell counts resulting in total replicates of four to seven from both experiments. We used non‐parametric Mann–Whitney Rank Sum tests (U‐test) to obtain significant differences in the contact cultures (Table A1a).
2.3. Experiment 2
We assessed the effects of bacterial lysed S. costatum on the growth of C. didymus by exposing exponential precultured C. didymus to filtrates of S. costatum in declining phase and S. costatum lysed by K. algicida at three successive time points after bacterial inoculation. S. costatum monocultures (100 ml) were inoculated (n = 6) into cell culture flasks (T‐75, Sarstedt). S. costatum growth was assessed as in vivo chlorophyll a fluorescence (RFU) and K. algicida was introduced at the beginning of declining phase of S. costatum as described in “diatoms and bacteria culturing”. Filtrates were obtained directly after K. algicida inoculation (day 0) and after five and 10 days, respectively, by processing the cultures in the following order: gentle centrifugation (3 min.; 570g; Hermle Z400, Wehingen, Germany), filtration through membranes with a pore size of 5 µm (13 mm, Nucleopore Track‐Etch membrane, Whatman, Kent, UK) placed in a syringe filter holder (13 mm, Swinnex®, Merck, Darmstadt, Germany). Subsamples of 5 µm filtrates were further sterile filtered (syringe filter unit Filtropur S 0.2 µm with a polyethersulfone (PES) membrane, Sarstedt).
Two hundred and fifty microliters of either 5 µm filtrates, 0.2 µm filtrates or sea water (control) were immediately added to 250 µl C. didymus cultures (each in independent triplicates) and incubated in 24‐well plates. C. didymus growth was followed by measuring in vivo chlorophyll a fluorescence (RFU). These triplicates were averaged before data processing. The average of sea water control was used as reference value (see below). C. didymus growth rate was calculated from day 0 to day 3: µ = ((ln (n3/n0))/t), where n3 and n0 refers to chlorophyll a fluorescence at day 3 and day 0, respectively. The effects of filtrates obtained from S. costatum cultures on C. didymus were normalized as per cent growth relative to control: (µ[trt]/µ[ctrl]) * 100 where µ[trt] is the growth rate of C. didymus exposed to the 5 µm or 0.2 µm‐filtrates of the competing diatom and µ[ctrl] is the growth rate of C. didymus exposed to the respective sea water control.
Bacterial algicidal activity (Paul & Pohnert, 2011) was indirectly assessed by protease activity of the sterile filtrates measured immediately at each medium sampling time point using a commercial Protease Assay Kit (EnzChek™, Invitrogen, Carlsbad CA, USA) which is based on the conversion of a casein dye to a fluorescent product (Jones et al., 1997). The assay was performed according to manufactures instructions and as described previously (Paul & Pohnert, 2011). Briefly, 10 µl of cell‐free filtrates were diluted in 100 µl digestion buffer and 100 µl of the dye at a concentration of 10 µg per ml were added. Samples were incubated in the dark at room temperature for one hour prior to fluorescence read out on a Mithras LB plate reader with an excitation wavelength of 470 ± 5 nm and an emission wavelength of 510 ± 20 nm.
We used unpaired two‐sided t tests to obtain significant differences in chlorophyll a units (RFU) for S. costatum growth comparison and relative growth rate comparisons (%) for C. didymus (Table A2aa,c). Protease activities (RFU) were compared using One way Analysis Of Variance (ANOVA) followed by Holm–Sidak post hoc test when significant differences occurred (Table A2ab).
2.4. Experiment 3
Experiment 3 was conducted with co‐cultivation chambers in a non‐contact situation to further determine whether the ability to alter C. didymus success is chemically mediated or relies on the contact of C. didymus with S. costatum and its associated bacteria. The co‐culture setup consists of two glass vessels separated by a 0.22 µm hydrophilic polyvinylidene fluoride (PVDF) membrane (Durapore, Millipore, Billerica, MA, USA) which allows exchange of small molecules but blocks the passage for all cells (Paul et al., 2012). Modifying the procedure from (Paul et al., 2012) we used a modified co‐cultivation setup with size‐reduced chambers leading to ten times less inoculation volume per vessel. Each vessel can hold 50 ml, has a 43 mm flat edge opening and a 15 mm opening for filling and sampling. Both vessels are fitted together by four screws fixing two fastening rings one at each chamber. Each co‐culture contains both, 50 ml of late exponential or early stationary precultured S. costatum (Sc) at either initially 7 × 104 cells/ml (low‐Sc) or 2 × 105 cells/ml (high‐Sc) concentration in one compartment and 50 ml of exponential precultured C. didymus (Cd) at initial cell densities of 1 × 104 cell/ml in the other. K. algicida (Ka) was introduced as described under “diatoms and bacteria culturing” into each of the diatom containing vessels. All setups were cultivated under conditions described in “diatoms and bacteria culturing”, however with constant slow shaking. Growth was measured as in vivo chlorophyll a fluorescence (RFU) in five (low‐Sc/Cd, high‐Sc/Cd, low‐Sc+Ka/Cd+Ka, high‐Sc+Ka/Cd+Ka) replicates, respectively. We used an unpaired two‐sided t test to test for significant differences in algal growth (RFU) (Table A3).
2.5. Experiment 4
Directly following the last measurement of the co‐culture experiment (day 11), the cultures were tested for growth of K. algicida, the strain used for inoculation at the beginning of the co‐cultivation. Total remaining volumes of each co‐culture (both compartment) were gently vacuum filtered through a 1.2 µm GF/C, 47‐mm filter (Whatman) before sterile filtration (syringe filter unit Filtropur S 0.2, Sarstedt). Two filtrates of “high‐Sc/Cd” replicates were accidentially discarded during processing and not used for K. algicida inoculations. Three (high‐Sc/Cd) and five (low‐Sc+Ka/Cd+Ka, high‐Sc+Ka/Cd+Ka, and low‐Sc/Cd) replicates corresponding to the replicates during co‐cultivation were used for bacterial inoculations. Twelve milliliters of the obtained sterile solutions were transferred to culture flasks and inoculated with a K. algicida culture (final OD550 of 0.02) previously starved for three days in SW after stationary growth in liquid minimal medium, containing 10 amino acids (aspartic acid, alanine, glutamic acid, glycine, isoleucine, leucine, methionine, phenylalanine, proline, and valine in a final concentration of each 0.08% w/v) to reduce carry over from the full medium. Control media for the experiment used were SW and a minimal medium (each n = 1) containing six amino acids (aspartic acid, glutamic acid, glycine, leucine, methionine, and valine in a final concentration of each 0.08% w/v). Growth of K. algicida was maintained at 23°C during constant shaking and was followed over eight days by optical density (550 nm) measurements. We used non‐parametric Mann–Whitney Rank Sum tests (U‐test) to obtain significant differences in treatments from the co‐culture system (Table A4).
2.6. Data analysis
Statistical analysis was conducted using SigmaPlot 13.0 software (Systat Software Inc., London, UK). Levels of significance are given as *p < 0.05, **p < 0.01, and ***p < 0.001. p > 0.05 is considered as not significant.
3. RESULTS
3.1. Effect of K. algicida‐induced lysis of S. costatum on the growth of C. didymus in mixed cultivation (Experiment 1)
Before inoculation with bacteria, the initial cell abundance ratio of S. costatum to that of C. didymus was 100:1 to compensate the higher growth rate and larger cell volumes of C. didymus. At these cell densities, S. costatum growth is lower in co‐cultures with C. didymus compared to the control (Figure 1a). This effect was significant on day 8 (p = 0.038). In the presence of K. algicida, S. costatum is quantitatively lysed on day 1 and the (co‐)cultures do not recover. Both K. algicida treatments have significant negative effects on S. costatum compared to the respective controls from day 1 onwards (Table A1aa). The presence of C. didymus did not affect the overall lysis, indicating that no protective effect in co‐cultures can be observed (p > 0.1 at all days after inoculation except day 4 Table A1aa). Growth responses of C. didymus in mixed cultivation with S. costatum and/or K. algicida are shown in Figure 1b. S. costatum in the co‐culture delayed the growth of C. didymus from the fifth day onwards (p ≤ 0.038, Table A1ab); however, this effect was not significant on the last day of the experiment (p = 0.257). C. didymus is delayed only slightly (significant only on day 4 and 6, Table A1ab) in the presence of K. algicida and reaches comparable cell counts at day 8, indicating resistance of the algae. This was confirmed in an independent experiment where no significant effect of K. algicida on C. didymus was observed (Figure A1). C. diymus growth is fully inhibited when the co‐cultured S. costatum was lysed by the algicidal bacterium (p = 0.001 at day 8 compared to cell abundances in bialgal cultivation; p ≤ 0.042 from day 5 onward compared to C. didymus +K. algicida). Taken together, both diatoms exhibit an inhibitory effect on each other in co‐culture. K. algicida quantitatively lyses S. costatum and the combination of lysed cells and K. algicida arrests growth of C. didymus.
Figure 1.
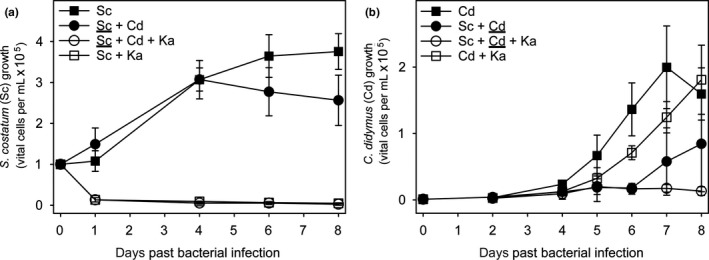
Diatom growth in tripartite mixed cultivation. Cell numbers of (a) S. costatum (Sc) and (b) C. didymus (Cd) with or without additions of K. algicida (Ka) in mono‐ or bialgal contact co‐cultivation. Results are expressed as the mean of four to seven replicates ± SD. Non‐parametric statistical tests were used for comparisons (Data about the statistical evaluation are given in Table A1aa,b). The underlined species in mixed cultivation is the one for which the cell numbers are given
3.2. Effects of filtrates of S. costatum cultures that were lysed with K. algicida on growth of C. didymus (Experiment 2)
K. algicida lyses stationary to declining S. costatum cultures within one day (p = 0.018) that then remain significantly suppressed until day 10 (p < 0.001, Table A2aa) (Figure 2a). Five and 10 days after K. algicida‐induced lysis of S. costatum, protease activity is significantly increased compared to the non‐infected but declining S. costatum cultures (both p < 0.001, Table A2ab) and also compared to K. algicida controls (both p ≤ 0.006) (Figure 2b).
Figure 2.
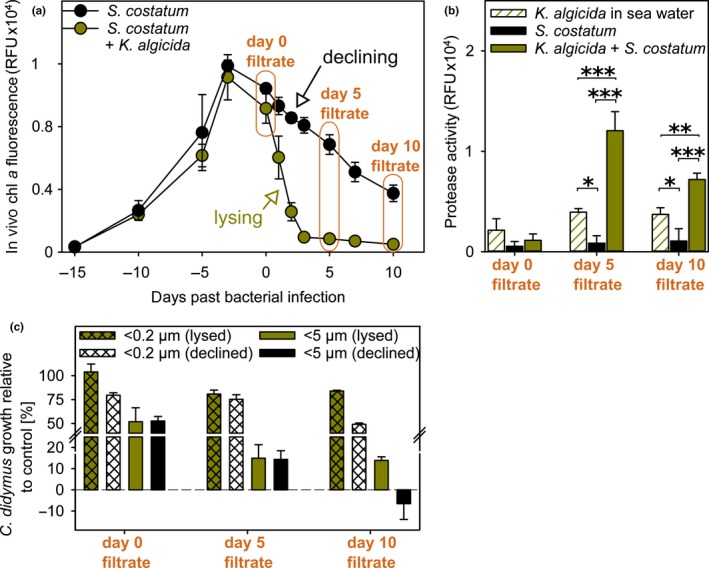
Impact of declining and bacterial‐lysed S. costatum culture filtrate on C. didymus. (a) Growth curve of S. costatum (± K. algicida infection at day 0) (both n = 3 ± SD). Orange sections indicate the time when filtrate was harvested and further processed by filtration (5 µm and 0.2 µm). Unpaired t tests were performed to obtain significant differences in growth (Table A2aa). (b) Protease activity of K. algicida infected S. costatum filtrates (n = 3) compared to filtrates of K. algicida in sea water (technical triplicates, one duplicate) and declining S. costatum cultures (n = 3). Background level of protease activity (sea water control) were substracted before plotting and statistical analysis (One way of variance analysis (ANOVA), Table A2ab). Asterisks show significant differences compared to the controls. Levels of significance are given as *p < 0.05, **p < 0.01, and ***p < 0.001. (c) Inhibitory activity of filtrates from S. costatum and infected S. costatum on C. didymus growth. Relative growth of C. didymus (% RFU of respective control) after three days of incubation (n = 3 ± SD) was tested for significant differences using unpaired t tests (Table A2ac)
Filtrates of K. algicida/S. costatum co‐cultures as well as of pure bacteria and diatom cultures were obtained on the day of inoculation (day 0 filtrate), as well as on day 5 (day 5 filtrate) and day 10 (day 10 filtrate) after inoculation. Two types of filtrates using different pore‐sized filters (5 µm and 0.2 µm, respectively) were generated and administered to C. didymus cultures. The growth of C. didymus was monitored after three days and is given as growth (%) relative to unialgal C. didymus controls in sea water (Figure 2c).
Filtrate (<5 µm) from declining S. costatum cultures inhibits C. didymus growth already at the start of the experiment (Figure 2c, gray). At later days of the experiment with progressing decline of the culture, this effect becomes more pronounced. Filtrate from day 10 of non‐infected S. costatum inhibits C. didymus growth fully. Throughout the experiment, the <5 µm filtrates were more active compared to the <0.2 µm filtrates (p ≤ 0.001 for all incubations, Table A2ac). Filtrates (<5 µm) from S. costatum cultures that were lysed by K. algicida caused also pronounced inhibition of C. didymus growth (Figure 2c, green). Again the <5 µm filtrate was more active compared to the <0.2 µm filtrates (p ≤ 0.006 for all incubations, Table A2ac). Comparison of the impact of S. costatum filtrates in declining phase to filtrates from bacterial‐ lysed cultures shows that the diatom was generally less affected when the bacterium was present, which was significant with both types of day 10 filtrates (significant for both incubations, Table A2ac) and with <0.2 µm day 0 filtrate (p = 0.008). The increased protease activity in K. algicida lysed cultures (Figure 2b) has thus no negative effect on C. didymus. In summary, filtrate from declining S. costatum cultures, whether bacterial‐lysed or not, exhibits a strong negative allelopathic effect on C. didymus. K. algicida alleviates this effect partially.
3.3. Effect of K. algicida‐induced lysis of S. costatum on the growth of C. didymus in non‐contact co‐cultivation (Experiment 3)
Experiment 2 demonstrated that the interaction of C. didymus with S. costatum is at least partly chemically mediated. Testing filtrates, as in Experiment 2 does not allow to conclude about additional dynamic mechanisms influencing the interaction. These could include the induction of chemical responses by signaling molecules or by modulated resource activity. To learn more about these aspects, we conducted co‐cultivation experiments where the diatoms are physically separated by a membrane that allows the diffusion of chemical signals (Figure 3). We included treatments with comparable S. costatum cell counts to those in Experiment 1 and 2 and lower concentrated S. costatum inoculations. The initial cell abundance ratio of S. costatum to that of C. didymus was therefore adjusted to 20:1 (high‐Sc) and 7:1 (low‐Sc). The low‐Sc treatment led to similar initial biomass for both diatoms, when considering different sizes of the cells (Harrison, Conway, Holmes, & Davis, 1977; Menden‐Deuer, Lessard, & Satterberg, 2001) whereas S. costatum is dominant in the high‐Sc co‐culturing setups. Lysis of S. costatum by K. algicida occurred with similar kinetics compared to the above experiments and growth responses were significantly reduced from day 1 after inoculation with the bacteria (p ≤ 0.003 for all data points from day 1 onwards at days with equal variance, Table A3). Growth of C. didymus did not differ in the high‐ and low‐Sc treatments in the absence of K. algicida (at all data points P values exceeded 0.219, Table A3). K. algicida strongly modulated the outcome of the co‐culturing. Dependent on the initial S. costatum concentration, C. didymus growth was either promoted (low‐Sc+Ka) or inhibited (high‐Sc+Ka) in the presence of K. algicida. These effects manifested from day 7 onward (day 7: p = 0.045, day 10: p = 0.029 for growth support) and p = 0.017 for growth inhibition at day 11. pH was monitored throughout the co‐culturing and no changes were observed (data not shown). The results support a complex interaction pattern once the three partners can chemically interact.
Figure 3.
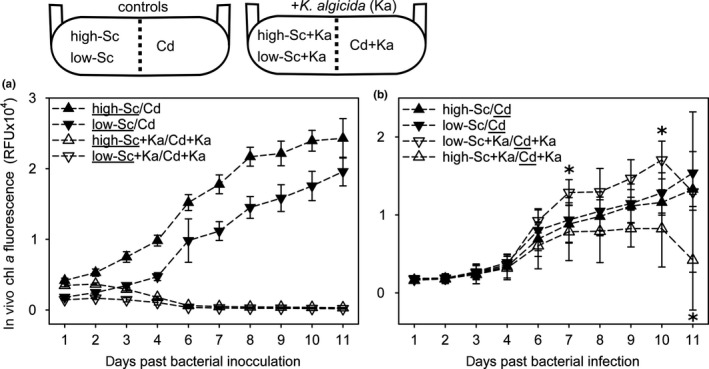
Diatom growth in tripartite non‐contact co‐cultivation. Growth development of (a) S. costatum (Sc) and (b) C. didymus (Cd) with or without K. algicida (Ka) infection in non‐contact co‐cultivations. The underlined species in mixed cultivation is the one for which the cell numbers are given. Results expressed as mean of five replicates ± SD with asterisk indicating significant differences (p < 0.05, unpaired t test, Table A3)
3.4. Effect of co‐culture exudates on the growth of K. algicida (Experiment 4)
K. algicida grew in <0.2 µm filtrates from both treatments of the co‐culture experiment in which bacterial lysis was induced (low‐Sc+Ka/Cd+Ka and high‐Sc+Ka/Cd+Ka) as well as in the minimal medium serving as positive control (Figure 4). However, bacterial cultures after eight days of incubation with the co‐culture filtrates appear whitish whereas K. algicida fully gained its typical yellowish phenotype in the minimal medium. Sea water and both <0.2 µm filtrates of co‐culturing experiments that were not infected with K. algicida did not support the growth of K. algicida.
Figure 4.
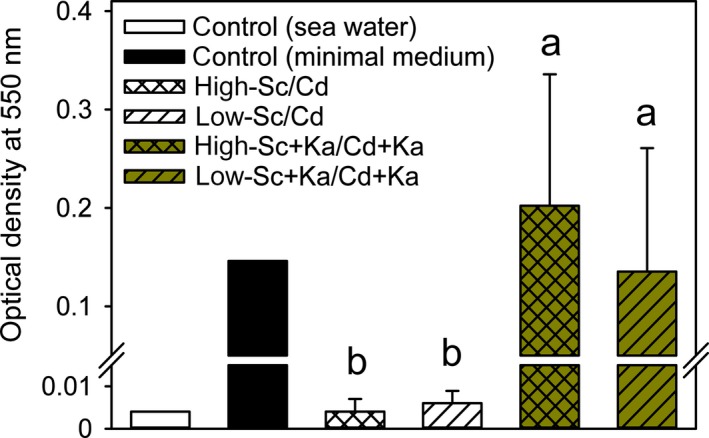
Growth of K. algicida in filtered spent medium obtained from the co‐culture systems. Optical density (OD) of K. algicida was recorded after eight days in filtrates of the co‐cultures from Figure 3 at day 11. Standard deviation (±SD) represents the mean of five replicates except for high‐Sc/Cd (n = 3). Means with letters were compared (non‐parametric U‐test) and different letters are significant different (Table A4)
4. DISCUSSION
Only a few studies on algicidal bacteria have focused on their effects on phytoplankton communities (Jung, Kim, Katano, Kong, & Han, 2008; Pokrzywinski, Place, Warner, & Coyne, 2012) and to the best of our knowledge, cascading effects in defined assemblages that give mechanistic insights have not been addressed. Results from this study clearly document that the effect of algicidal bacteria in common bilateral incubation experiments represent an oversimplification compared to the situation in consortia. A cascading effect of algicidal bacteria in mixed algal assemblages can lead to fundamentally different outcomes after bacterial infection compared to mono cultures. In this and previous studies, the pure culture of C. didymus was entirely resistant against K. algicida (Paul & Pohnert, 2011). In mixed cultures, however, the bacterial lysis of the competitor resulted in concentration dependent effect on the performance of the resistant alga. At high concentration of the lysed competitor growth arrest was observed (Figure 1) while low concentrations of lysed cells supported its growths (Figure 3b). We demonstrate that chemical factors mediate these concentration–dependent interactions in a dynamic manner (Figures 2 and 3). Furthermore we illustrate that the effect of algicidal bacteria and that of allelopathic interactions between two competing algae cannot be fully untangled. Despite the fact that cell densities in our co‐culturing setups exceed those in the natural environment we conclude that overlaying multi–process interaction can also be expected to occur in natural plankton where mixed phyto‐ and bacterioplankton communities prevail. In these natural scenarios close contact interactions with locally enhanced metabolites within the diffusion limited zone (phycosphere) around the producing organism will prevail (Seymour, Amin, Raina, & Stocker, 2017). These steep concentration gradients are not covered in the study here but were averaged over the culture, thus only indirect conclusions about processes in natural waters can be given. Since both diatom genera under investigation frequently dominate phytoplankton blooms and co‐exist and phycospheres will encounter in the oceans, our observations have consequences for natural populations (Kooistra et al., 2008; Li et al., 2017).
A complex interaction network between the three investigated species was observed for mixed cultures in Experiment 1. If both diatoms were inoculated within the same culture vessel the growth C. didymus was delayed and S. costatum entered the declining phase earlier (Figure 1). Thus, both algae in direct interaction negatively affect the growth of the partner. Allelochemical interactions would be a possible explanation for this phenomenon but microalgae in direct contact might influence the respective growth using alternative mechanisms. These include inhibition by shading effects that reduce light availability to the competitors, concurrent consumption of nutrients, or direct interactions mediated by cell‐cell contact (Dunker, Althammer, Pohnert, & Wilhelm, 2017). If K. algicida is added as third interaction partner, fast lysis of S. costatum is observed, which is in full agreement with experiments using isolated cultures (Paul & Pohnert, 2011). The lysis indicates that quorum sensing up‐regulation of algicidal activity as observed in Paul and Pohnert (2011) is active in our experimental setups. Interestingly, growth of C. didymus that is fully resistant against bacterial lysis in bilateral interactions was entirely suppressed in the tripartite co‐culturing. A possible explanation for this observed inhibition is that exudates, released from declining or perished cells of S. costatum are inhibiting the growth of the competitor (Imada, Kobayashi, Tahara, & Oshima, 1991). This is also supported by the observation that the growth arrest by lysed cells is concentration dependent (see below). Alternatively, the bacteria might up‐regulate their algicidal activity in the presence of both partners, thereby resulting in an increased infectivity even against the otherwise resistant algae.
To evaluate if chemical signals meditate this interaction, we conducted Experiment 2 with filtrates of incubations where either the algae or both, algae and bacteria were removed using different pore sized filters (Figure 2). These experiments confirmed that in fact the “smell of death” of the declining or lysed S. costatum population is responsible for the observed effect on C. didymus (Figure 2c). To exclude potentially overlaying effects of nutrient starvation we exposed freshly transferred C. didymus to the filtrates thereby guaranteeing sufficient access to nutrients during three days of the experiment. The inhibitory effect on C. didymus was observed in both, filtrates from declining and bacterially lysed S. costatum and is thus most likely caused by chemical mediators from this diatom. This main or exclusive contribution of S. costatum derived mediators is also supported by the notion that algicidal proteases that are up‐regulated in S. costatum infections (Figure 2b) are not affecting C. didymus (Figure 2c) (Paul & Pohnert, 2013). Also the role of proteases released during death of diatoms in response to abiotic stress as observed by Berges and Falkowski (1998) can be excluded since no such increase was observed in the declining S. costatum culture (Figure 2b). Qualitatively, the Skeletonema‐effect is in agreement with previous studies demonstrating growth–inhibiting effects of spent medium of diatoms from the end of their growth phase (Imada et al., 1991; Vidoudez & Pohnert, 2008). Negative influence on co‐occurring phytoplankton has been repeatedly reported for Skeletonema spp. (Wang et al., 2017; Yamasaki et al., 2011) while only few reports on positive allelopathy of this alga are known (Paul, Barofsky, Vidoudez, & Pohnert, 2009). The metabolite class of polyunsaturated aldehydes (PUAs) are often brought forward as examples for negative allelopathic exudates of Skeletonema sp. (Fontana, d'Ippolito, Cutignano, Miralto et al., 2007; Fontana, d'Ippolito, Cutignano, Romano et al., 2007; Ianora, Bentley et al., 2011; Ianora, Romano et al., 2011; Sieg, Poulson‐Ellestad, & Kubanek, 2011; Wichard et al., 2005). These metabolites are released during the late exponential phase of growth (Vidoudez & Pohnert, 2008) but cell lysis triggers an even more pronounced production (Ribalet et al., 2014). Other compound classes, such as sterol sulfates from Skeletonema marinoi can mediate cell death (Gallo, d'Ippolito, Nuzzo, Sardo, & Fontana, 2017). While these sterols have been made responsible for an auto induced cell lysis it can be envisaged that that they might also alter the physiology of other diatoms, particularly when suddenly released after lysis (Xu, Tang, Qin, Duan, & Gobler, 2015). The fact that sterile filtered culture supernatants had consistently lower effects on C. didymus compared to 5 µm filtrates that still contained bacteria and other particulate organic matter suggests that in addition to diffusible chemicals additional factors are involved in the interaction (Figure 2c). We did not undertake further elucidation of the involved chemical triggers but rather focused on functional aspects of the interaction.
In direct contact co‐cultures the growth of C. didymus was inhibited in a more pronounced way in the presence of S. costatum and K. algicida compared to the presence of S. costatum alone (Figure 1b). In contrast, filtrates from the co‐culture at day 10 suppressed growth less compared to those from declining cultures (Figure 2c). Here, the bacterium apparently reduces the harmful effect, which could be due to recycling of diatom‐derived organic matter during the prolonged time of the experiment or due to nutrient exchange that is often the basis for positive algal/bacterial interactions (Amin, Parker, & Armbrust, 2012; Orellana, Pang, Durand, Whitehead, & Baliga, 2013). It is also possible that K. algicida metabolizes toxins released by lysed S. costatum within the 10 days required to manifest the effect. Such detoxification by bacteria has already been documented in cyanobacteria (Yamada, Murakami, Kawamura, & Sakakibara, 1994).
To obtain a more refined picture about the mechanism of interaction we carried out a non‐contact co‐cultivation Experiment 3 (Figure 3). In the experimental setup we ensure that chemical mediators can reach all interaction partners within the system, while the two competing algal species remain spatially separated (Paul et al., 2009). At comparable cell concentrations to the experiments described above, exudates from the lysed S. costatum resulted in significantly inhibited growth of C. didymus. This effect manifested most pronouncedly toward the end of the experiment (Figure 3b). Mediators released from the association of K. algicida with the dead dense S. costatum culture are thus freely diffusible and have the capacity to reduce C. didymus that is otherwise resistant to the lytic bacterium. The pH remained constant throughout the experiments thereby excluding this potential cause for reduced performance of C. didymus.
The situation is entirely reversed if S. costatum cell counts in the co‐culturing chamber are reduced. In this case, C. didymus performance is increased after day 7 higher abundance of this alga is observed compared to the control (Figure 3b). It has been demonstrated for the diatom Thalassiosira weissflogii that it performs better in co‐culture with Skeletonema sp. and such a support is obviously also occurring in the mixed culture under study in this investigation (Paul et al., 2009). Since C. didymus is performing better if S. costatum is lysed by the bacterium it might be envisaged that it benefits metabolites or nutrients released from disrupted cells. Use of organic substrates would require the capability of mixotrophic growth that has been documented in diatoms previously (Shishlyannikov, Klimenkov, Bedoshvili, Mikhailov, & Gorshkov, 2014; Villanova et al., 2017). Nutrient release can also be envisaged since diatoms are known to store internally nutrients that might be released during bacterial lysis (De La Rocha, Terbruggen, Volker, & Hohn, 2010). Alternatively, metabolites such as PUA, released during diatom wounding might be priming defence capabilities in the co‐cultured algae, a phenomenon that has been described for PUA treated monocultures of the diatom Phaeodactylum tricornutum (Vardi et al., 2006).
Looking at the third interaction partner we could show K. algicida benefits from the lysis of the diatoms. By exposing it to sterile filtrates of the co‐cultivations we observed that it grew effectively on exudates from setups with lysed S. costatum cells compared to those that contained only the exudates of the co‐cultures without bacterial lysis (Figure 4). From macroscopic investigations of cell pellets of grown bacterial cultures we recognized that cultures after growth on algal‐derived lysis products appear whitish compared to the yellowish controls in artificial bacterial media. Obviously, the algal exudates remaining in the co‐culture are not sufficient for the full development of the phenotype but support efficient growth in the range of that observed in minimum medium. The observed increased bacterial growth on lysis products fully agrees with field data that heterotrophic bacteria follow the bloom of phytoplankton (Teeling et al., 2012).
5. CONCLUSION
Despite the already complex outcome of the experiments, the situation in the plankton are undoubtedly more complex, since additional associated microorganism might modulate the chemical mediators by metabolic transformations (Margulis, 1990). However, our study illustrates that the chemical interaction within even a simple model community makes the outcome hard to predict. This calls for caution if algicidal bacteria are released into the environment to control for example harmful algal blooms.
CONFLICT OF INTERESTS
The authors declare no conflict of interests.
AUTHORS CONTRIBUTION
GP and AB conceived the study, AB performed the experiments, AB and GP wrote, edited, and approved the manuscript.
ETHICS STATEMENT
Not required.
ACKNOWLEDGEMENTS
This work was funded by the German Research Foundation within the CRC ChemBioSys, CRC1127.
APPENDIX 1.
Table A1a.
Cell counts of S. costatum in co‐cultures and mono‐cultures ±bacterial infection were compared using U‐tests
| Comparisons | Size | Median (cells/mL) | 25% percentile | 75% percentile | T | p (exact) |
|---|---|---|---|---|---|---|
| Day1: Sc+Cd versus Sc+Cd+Ka | 4;4 | 150,768;10,837 | 110,128;10,519 | 186,628;18,567 | 26.00 | 0.029* |
| Day4: Sc+Cd versus Sc+Cd+Ka | 6;7 | 305,044;4,144 | 277,153;3,825 | 339,469;7,969 | 63.00 | 0.001** |
| Day6: Sc+Cd versus Sc+Cd+Ka | 4;4 | 276,356;3,825 | 224,878;2,948 | 330,703;9,244 | 26 | 0.029* |
| Day8: Sc+Cd versus Sc+Cd+Ka | 6;7 | 257,231;2,231 | 195,234;638 | 319,866;4,781 | 63 | 0.001** |
| Day1: Sc+Ka versus Sc+Cd+Ka | 4;4 | 10,838;10,838 | 10,519; 10,519 | 18,567;18,567 | 18 | 1 |
| Day4: Sc+Ka versus Sc+Cd+Ka | 4;7 | 9,244;4,144 | 8,766;3,825 | 9,961;7,969 | 36 | 0.024* |
| Day6: Sc+Ka versus Sc+Cd+Ka | 4;4 | 5,897;3,825 | 5,100; 2,948 | 8,367; 9,244 | 22 | 0.343 |
| Day8: Sc+Ka versus Sc+Cd+Ka | 4;7 | 4,941;2,231 | 3,586;638 | 5,817;4,781 | 33 | 0.109 |
| Day1: Sc versus Sc+Ka | 4;4 | 112,040;10,838 | 82,397;10,519 | 129,492; 18,567 | 26 | 0.029* |
| Day4: Sc versus Sc+Ka | 4;4 | 325,125;9,244 | 258,506;8,766 | 336,759;9,961 | 26 | 0.029* |
| Day6: Sc versus Sc+Ka | 4;4 | 356,044;5,897 | 320,344;5,100 | 417,084;8,367 | 26 | 0.029* |
| Day8: Sc versus Sc+Ka | 4;4 | 391,744;4,941 | 329,428;3,586 | 405,769;5,817 | 26 | 0.029* |
| Day1: Sc versus Sc+Cd | 4;4 | 112,040; 150,768 | 82,397; 110,128 | 129,492; 186,628 | 12 | 0.114 |
| Day4: Sc versus Sc+Cd | 4;6 | 325,125; 305,043 | 258,506; 277,153 | 336,759; 339,469 | 23 | 0.914 |
| Day6: Sc versus Sc+Cd | 4;4 | 356,043; 276,356 | 320,344; 224,878 | 417,084; 330,703 | 23 | 0.200 |
| Day8: Sc versus Sc+Cd | 4;6 | 391,743; 257,231 | 329,428; 195,234 | 405,769; 319,866 | 32 | 0.038* |
Level of significances:
p < 0.05
p < 0.01
p < 0.0001.
Table A1b.
Cell counts of C. didymus in co‐cultures and mono‐cultures ±bacterial infection were compared using U‐tests
| Comparisons | Size | Median (cells/ml) | 25% percentile | 75% percentile | T | p (exact) |
|---|---|---|---|---|---|---|
| Day2: Sc+Cd versus Sc+Cd+Ka | 6;6 | 3,028; 2071 | 1913;0 | 7,438;5,844 | 52 | 0.181 |
| Day4: Sc+Cd versus Sc+Cd+Ka | 6;7 | 6,295; 6,693 | 5,220;2,550 | 23,375;16,044 | 45.5 | 0.628 |
| Day5: Sc+Cd versus Sc+Cd+Ka | 6;7 | 5,657; 17,956 | 4,821;13,706 | 44,758;19,656 | 34 | 0.295 |
| Day6: Sc+Cd versus Sc+Cd+Ka | 4;4 | 18,009; 14,343 | 15,380;10,678 | 20,639;25,181 | 21 | 0.486 |
| Day7: Sc+Cd versus Sc+Cd+Ka | 6;7 | 34,584; 17,850 | 18,966;13,069 | 121,603;20,400 | 55 | 0.073 |
| Day8: Sc+Cd versus Sc+Cd+Ka | 6;7 | 47,653; 14,343 | 29,803;6,163 | 176,083;19,763 | 63 | 0.001** |
| Day2: Cd+Ka versus Sc+Cd+Ka | 4;7 | 3,188;2072 | 1952;0 | 3,586;5,844 | 28 | 0.527 |
| Day4: Cd+Ka versus Sc+Cd+Ka | 4;7 | 12,511;6,694 | 9,363;2,550 | 16,137;16,044 | 31 | 0.230 |
| Day5: Cd+Ka versus Sc+Cd+Ka | 4;7 | 27,253;17,956 | 20,798;13,706 | 49,486;19,656 | 35 | 0.042* |
| Day6: Cd+Ka versus Sc+Cd+Ka | 4;4 | 66,141;14,344 | 65,105;10,678 | 81,759;25,181 | 26 | 0.029* |
| Day7: Cd+Ka versus Sc+Cd+Ka | 4;7 | 131,963;17,850 | 99,530;13,069 | 141,684;20,400 | 38 | 0.006** |
| Day8: Cd+Ka versus Sc+Cd+Ka | 4;7 | 188,700;14,344 | 128,217;6,163 | 225,755;19,763 | 38 | 0.006** |
| Day2: Cd versus Cd+Ka | 4;4 | 3,585;3,188 | 2072;1952 | 4,263;3,586 | 20.5 | 0.486 |
| Day4: Cd versus Cd+Ka | 4;4 | 23,348;12,511 | 18,248;9,363 | 29,285;16,137 | 26 | 0.029* |
| Day5: Cd versus Cd+Ka | 4;4 | 73,471;27,253 | 34,664;20,798 | 92,198;49,486 | 24 | 0.114 |
| Day6: Cd versus Cd+Ka | 4;4 | 132,600;66,141 | 102,080;65,105 | 174,117;81,759 | 26 | 0.029* |
| Day7: Cd versus Cd+Ka | 4;4 | 192,365;131,963 | 143,677;99,530 | 262,809;141,684 | 24 | 0.114 |
| Day8: Cd versus Cd+Ka | 4;4 | 148,856;188,700 | 130,130;128,217 | 199,378;225,755 | 16 | 0.686 |
| Day2: Cd versus Sc+Cd | 4;6 | 3,585;3,028 | 2072;3,028 | 4,263;7,438 | 22 | 1 |
| Day4: Cd versus Sc+Cd | 4;6 | 23,348;6,295 | 18,248;5,220 | 29,285;23,375 | 29 | 0.171 |
| Day5: Cd versus Sc+Cd | 4;6 | 73,471;5,657 | 34,664;4,821 | 92,198;44,758 | 32 | 0.038* |
| Day6: Cd versus Sc+Cd | 4;4 | 132,600;18,009 | 102,080;15,380 | 174,117;20,639 | 26 | 0.029* |
| Day7: Cd versus Sc+Cd | 4;6 | 192,365;34,584 | 143,677;18,966 | 262,809;121,603 | 34 | 0.010* |
| Day8: Cd versus Sc+Cd | 4;6 | 148,856;47,653 | 130,130;29,803 | 199,378;176,083 | 28 | 0.257 |
Level of significances:
p < 0.05,
p < 0.01,
p < 0.0001.
Table A2a.
Comparison of S. costatum with and without K. algicida, the infection happened at day 0
| Comparisons (S. costatum vs. S. costatum +K. algicida) | Size | Mean (RFU) | Std Dev | t | df | p |
|---|---|---|---|---|---|---|
| Day −15 | 3;3 | 325;348 | 18.4;19.9 | −1.443 | 4 | 0.223 |
| Day −10 | 3;3 | 2,656;2,392 | 631.4;130.8 | 0.708 | 4 | 0.518 |
| Day −5 | 3;3 | 7,624;6,156 | 2,421.4;718.0 | 1.006 | 4 | 0.371 |
| Day −3 | 3;3 | 11,876;11,148 | 369.5;1,441.5 | 0.846 | 4 | 0.445 |
| Day 0 | 3;3 | 10,436;9,155 | 251.4;935.7 | 2.290 | 4 | 0.084 |
| Day 1 | 3;3 | 9,320;6,035 | 550.3;1,367 | 3.861 | 4 | 0.018* |
| Day 2 | 3;3 | 8,556;2,573 | 318.0;575.8 | 15.754 | 4 | <0.001*** |
| Day 3 | 3;3 | 8,087;949 | 496.8;72.6 | 24.622 | 4 | <0.001*** |
| Day 5 | 3;3 | 6,858;852 | 627.2;59.7 | 16.511 | 4 | <0.001*** |
| Day 7 | 3;3 | 5,107;703 | 616.3;72.1 | 12.293 | 4 | <0.001*** |
| Day 10 | 3;3 | 3,747;499 | 526.4;77.1 | 10.572 | 4 | <0.001*** |
Values are based on in vivo chlorophyll a fluorescence (RFU). T test performed with two‐tailed p‐values.
Level of significances:
p < 0.05,
p < 0.01,
p < 0.0001.
Table A2b.
Comparison of protease activity in relative fluorescence units
| ANOVA | Post hoc test | |||||
|---|---|---|---|---|---|---|
| Size | Mean | Std Dev | t | p | ||
| Filtrates of day 0 (df = 7; F = 2.967; p = 0.141) | ‐ | |||||
| S. costatum | 3 | 539 | 483.9 | |||
| S. costatum +K. algicida | 3 | 1,134 | 640.4 | |||
| K. algicida in sea water | 2 | 2,142 | 1,145.5 | |||
| Filtrates of day 5 (df = 8; F = 70.613; p < 0.001) | Holm‐Sidak (all pairwise) α 0.05 | |||||
| S. costatum | 3 | 851 | 738.5 | S. costatum versus S. costatum +K. algicida | 11.5 | <0.001*** |
| S. costatum +K. algicida | 3 | 12,057 | 1897.6 | S. costatum +K. algicida versus K. algicida in sea water | 8.334 | <0.001*** |
| K. algicida in sea water | 3 | 3,939 | 352.5 | S. costatum versus K. algicida in sea water | 3.170 | 0.019* |
| Filtrates of day 10 (df = 8; F = 36.111; p < 0.001) | Holm‐Sidak (all pairwise) α 0.05 | |||||
| S. costatum | 3 | 1,064 | 1,226.6 | S. costatum versus S. costatum +K. algicida | 8.473 | <0.001*** |
| S. costatum +K. algicida | 3 | 7,188 | 636.6 | S. costatum +K. algicida versus K. algicida in sea water | 4.810 | 0.006** |
| K. algicida in sea water | 3 | 3,712 | 664.4 | S. costatum versus K. algicida in sea water | 3.663 | 0.011* |
Averaged blank values for protease activity of sea water were subtracted before one way analysis of variance (ANOVA) was carried out. Background subtraction resulted in one negative value (S. costatum with day 5 filtrates) which was set to zero.
Level of significances:
p < 0.05,
p < 0.01,
p < 0.0001.
Table A2c.
Comparison of C. didymus growth response with filtrates of declining S. costatum and bacterial‐lysed S. costatum obtained at three time points based on in vivo chlorophyll a fluorescence values (RFU)
| Comparisons (S. costatum vs. S. costatum+K.algicida) | Size | Mean (RFU) | Std Dev | t | df | p |
|---|---|---|---|---|---|---|
| Day0 filtrate: 0.2 µm (lysed) versus 5 µm (lysed) | 3;3 | 104;52 | 8.2;14.6 | 5.366 | 4 | 0.006** |
| Day5 filtrate: 0.2 µm (lysed) versus 5 µm (lysed) | 3;3 | 81;15 | 4.1;6.4 | 15.008 | 4 | <0.001*** |
| Day10 filtrate: 0.2 µm (lysed) versus 5 µm (lysed) | 3;3 | 84;14 | 0.9;1.7 | 62.645 | 4 | <0.001*** |
| Day0 filtrate: 0.2 µm (declined) versus 5 µm (declined) | 3;3 | 80;53 | 2.6;4.8 | 8.610 | 4 | 0.001** |
| Day5 filtrate: 0.2 µm (declined) versus 5 µm (declined) | 3;3 | 75;14 | 4.8;4.2 | 16.646 | 4 | <0.001*** |
| Day10 filtrate: 0.2 µm (declined) versus 5 µm (declined) | 3;3 | 49;−7 | 1.3;7.4 | 12.821 | 4 | <0.001*** |
| Day0 filtrate: 0.2 µm (declined) versus 0.2 µm (lysed) | 3;3 | 80;104 | 2.6;8.2 | −4.874 | 4 | 0.008** |
| Day5 filtrate: 0.2 µm (declined) versus 0.2 µm (lysed) | 3;3 | 75;81 | 4.8;4.1 | −1.497 | 4 | 0.209 |
| Day10 filtrate: 0.2 µm (declined) versus 0.2 µm (lysed) | 3;3 | 49;84 | 1.3;0.9 | −38.224 | 4 | <0.001*** |
| Day0 filtrate: 5 µm (declined) versus 5 µm (lysed) | 3;3 | 53;52 | 4.8;14.6 | 0.0844 | 4 | 0.937 |
| Day5 filtrate: 5 µm (declined) versus 5 µm (lysed) | 3;3 | 14;15 | 4.2;6.4 | −0.141 | 4 | 0.895 |
| Day10 filtrate: 5 µm (declined) versus 5 µm (lysed) | 3;3 | −7;14 | 7.4;1.7 | −4.649 | 4 | 0.0097** |
T test performed with two‐tailed p‐values.
Level of significances:
p < 0.05,
p < 0.01,
p < 0.0001.
Table A3.
Comparison of diatom growth response in non‐contact co‐cultures (± bacterial infections) were compared using t tests. Significances were obtained as p‐values from two‐tailed tests
| Size | Mean (RFU) | Std Dev | T | df | p–value | |
|---|---|---|---|---|---|---|
| S. costatum (low‐Sc/Cd); S. costatum (low‐Sc+Ka/Cd+Ka) | ||||||
| Day 1 | 5;5 | 1,803; 1,457 | 96; 96 | 5.683 | 8 | <0.001*** |
| Day 2 | 5;5 | 2,480; 1,664 | 323; 179 | 4.935 | 8 | 0.001** |
| Day 3 | 5;5 | 3,467; 1,441 | 308; 297 | 10.580 | 8 | <0.001*** |
| Day 4# | 5;5 | 4,712;1,060 | 560.5;354.1 | 12.318 | 8 | <0.001*** |
| S. costatum (high‐Sc/Cd); S. costatum (high‐Sc+Ka/Cd+Ka) | ||||||
| Day 1 | 5;5 | 4,180; 3,486 | 320; 191 | 4.169 | 8 | 0.003** |
| Day 2 | 5;5 | 5,291; 3,669 | 511; 182 | 6.687 | 8 | <0.001*** |
| Day 3# | 5;5 | 7,499; 2,947 | 748; 224 | 13.032 | 8 | <0.001*** |
| C. didymus (low‐Sc/Cd); C. didymus (high‐Sc/Cd) | ||||||
| Day 1 | 5;5 | 1,847; 1,628 | 321; 228 | 1.243 | 8 | 0.249 |
| Day 2 | 5;5 | 1,878; 1,824 | 558; 504 | 0.159 | 8 | 0.878 |
| Day 3 | 5;5 | 2,700; 2,438 | 889; 1,271 | 0.378 | 8 | 0.715 |
| Day 4 | 5;5 | 3,887; 3,388 | 1,090; 1,485 | 0.606 | 8 | 0.561 |
| Day 6 | 5;5 | 8,024; 6,896 | 2,614; 2,225 | 0.735 | 8 | 0.483 |
| Day 7 | 5;5 | 9,352; 8,827 | 2,867; 2,442 | 0.312 | 8 | 0.763 |
| Day 8 | 5;5 | 10,486; 9,806 | 2,456; 2,480 | 0.436 | 8 | 0.675 |
| Day 9 | 5;5 | 11,459; 11,141 | 2,506; 2,111 | 0.217 | 8 | 0.834 |
| Day 10 | 5;5 | 12,820; 11,604 | 2,585; 1625 | 0.890 | 8 | 0.399 |
| Day 11 | 5;5 | 15,381; 13,285 | 2,738; 2,200 | 1.334 | 8 | 0.219 |
| C. didymus (low‐Sc/Cd); C. didymus (low‐Sc+Ka/Cd+Ka) | ||||||
| Day 1 | 5;5 | 1,847;1,752 | 321;213 | 0.552 | 8 | 0.596 |
| Day 2 | 5;5 | 1,878;1,875 | 558;578 | 0.007 | 8 | 0.995 |
| Day 3 | 5;5 | 2,700;2,586 | 889.1;904.9 | 0.201 | 8 | 0.846 |
| Day 4 | 5;5 | 3,887;3,608 | 1,090;596 | 0.503 | 8 | 0.628 |
| Day 6 | 5;5 | 8,024;9,249 | 2,614;1549 | −0.902 | 8 | 0.394 |
| Day 7 | 5;5 | 9,352;12,870 | 2,867;1657 | −2.376 | 8 | 0.045* |
| Day 8 | 5;5 | 10,486;12,957 | 2,456.4;3,002.9 | −1.424 | 8 | 0.192 |
| Day 9 | 5;5 | 11,459; 14,676 | 2,506; 2,405 | −2.071 | 8 | 0.072 |
| Day 10 | 5;5 | 12,820;17,044 | 2,584.7;2,425.7 | −2.665 | 8 | 0.029* |
| Day 11 | 5;5 | 15,381; 12,939 | 2,738;10,284 | 0.513 | 8 | 0.622 |
| C. didymus (high‐Sc/Cd); C. didymus (high‐Sc+Ka/Cd+Ka) | ||||||
| Day 1 | 5;5 | 1,628;1,619 | 228.2;166.5 | 0.0735 | 8 | 0.943 |
| Day 2 | 5;5 | 1,824;1,800 | 504.4;603.5 | 0.0685 | 8 | 0.947 |
| Day 3 | 5;5 | 2,438;2,371 | 1,270.8;1,204.2 | 0.0086 | 8 | 0.934 |
| Day 4 | 5;5 | 3,388;3,179 | 1,485.4;1,498.0 | 0.222 | 8 | 0.830 |
| Day 6 | 5;5 | 6,895.9;6,038 | 2,224.8;2,951.1 | 0.519 | 8 | 0.618 |
| Day 7 | 5;5 | 8,827;7,845 | 2,442.2;3,710.1 | 0.494 | 8 | 0.634 |
| Day 8 | 5;5 | 9,806;7,908.8 | 2,479.6;4,031.0 | 0.897 | 8 | 0.396 |
| Day 9 | 5;5 | 11,141;8,250 | 2,111.3;2,364.8 | 2.040 | 8 | 0.076 |
| Day 10 | 5;5 | 11,604;8,243 | 1624.5;4,930.7 | 1.448 | 8 | 0.186 |
| Day 11 | 5;5 | 13,285;4,205 | 2,199.9;6,405.3 | 2.998 | 8 | 0.017* |
Level of significances:
No homogeneity of variance was observed after these days.
p < 0.05,
p < 0.01,
p < 0.0001.
Table A4.
Differences in growth of K. algicida in filtered spent medium obtained from co‐cultures evaluated by U‐tests (Mann–Whitney Rank Sum Test)
| Comparisons | Size | Median (cells/ml) | 25% percentile | 75% percentile | T | p (exact) |
|---|---|---|---|---|---|---|
| high‐Sc+Ka/Cd+Ka versus low‐Sc+Ka/Cd+Ka | 5;5 | 0.220;0.145 | 0.090;0.011 | 0.305;0.255 | 33 | 0.310 |
| high‐Sc+Ka/Cd+Ka versus high‐Sc/Cd | 5;3 | 0.220;0.004 | 0.090;0.001 | 0.305;0.007 | 6 | 0.036* |
| high‐Sc+Ka/Cd+Ka versus low‐Sc/Cd | 5;5 | 0.220;0.005 | 0.090;0.004 | 0.305;0.008 | 40 | 0.008** |
| low‐Sc+Ka/Cd+Ka versus high‐Sc/Cd | 5;3 | 0.145;0.004 | 0.011;0.001 | 0.255;0.007 | 6 | 0.036* |
| low‐Sc+Ka/Cd+Ka versus low‐Sc/Cd | 5;5 | 0.145;0.005 | 0.011;0.004 | 0.255;0.008 | 38.5 | 0.016* |
| high‐Sc/Cd versus low‐Sc/Cd | 3;5 | 0.004;0.005 | 0.001;0.004 | 0.007;0.008 | 11 | 0.571 |
Level of significances:
p < 0.05,
p < 0.01,
p < 0.0001.
Figure A1.
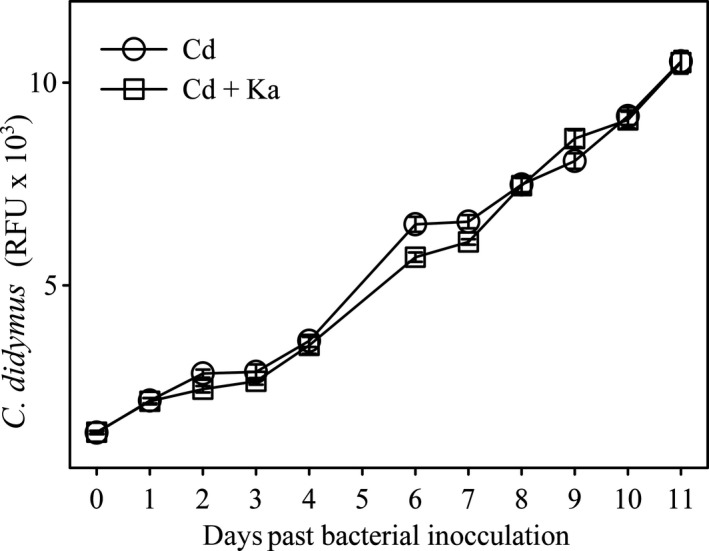
Growth of C. didymus (Cd) with and without added K. algicida (Ka). Results are expressed as the mean of five replicates ± SD. Conditions were otherwise as in Figure 1 of the main manuscript
Bigalke A, Pohnert G. Algicidal bacteria trigger contrasting responses in model diatom communities of different composition. MicrobiologyOpen. 2019;8:e818 10.1002/mbo3.818
DATA ACCESSIBILITY
Underlying data for this publication will be made available upon reasonable request.
REFERENCES
- Amin, S. A. , Parker, M. S. , & Armbrust, E. V. (2012). Interactions between diatoms and bacteria. Microbiology and Molecular Biology Reviews, 76, 667–684. 10.1128/MMBR.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges, J. A. , & Falkowski, P. G. (1998). Physiological stress and cell death in marine phytoplankton: Induction of proteases in response to nitrogen or light limitation. Limnology and Oceanography, 43, 129–135. 10.4319/lo.1998.43.1.0129 [DOI] [Google Scholar]
- Bidle, K. D. (2015). The molecular ecophysiology of programmed cell death in marine phytoplankton. Annual Review of Marine Science, 7, 341–375. 10.1146/annurev-marine-010213-135014 [DOI] [PubMed] [Google Scholar]
- Bidle, K. D. , & Falkowski, P. G. (2004). Cell death in planktonic, photosynthetic microorganisms. Nature Reviews Microbiology, 2, 643–655. 10.1038/nrmicro956 [DOI] [PubMed] [Google Scholar]
- Bratbak, G. , Egge, J. K. , & Heldal, M. (1993). Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Marine Ecology Progress Series, 93, 39–48. 10.3354/meps093039 [DOI] [Google Scholar]
- Brussaard, C. P. D. , Riegman, R. , Noordeloos, A. A. M. , Cadée, G. C. , Witte, H. , Kop, A. J. , … Bak, R. P. M. (1995). Effects of grazing, sedimentation and phytoplankton cell lysis on the structure of a coastal pelagic food web. Marine Ecology Progress Series, 123, 259–271. 10.3354/meps123259 [DOI] [Google Scholar]
- De La Rocha, C. L. , Terbruggen, A. , Volker, C. , & Hohn, S. (2010). Response to and recovery from nitrogen and silicon starvation in Thalassiosira weissflogii: Growth rates, nutrient uptake and C, Si and N content per cell. Marine Ecology Progress Series, 412, 57–68. 10.3354/meps08701 [DOI] [Google Scholar]
- Dunker, S. , Althammer, J. , Pohnert, G. , & Wilhelm, C. (2017). A fateful meeting of two phytoplankton species: Chemical vs. cell‐cell‐interactions in co‐cultures of the green algae Oocystis marsonii and the cyanobacterium Microcystis aeruginosa . Microbial Ecology, 74, 22–32. 10.1007/s00248-016-0927-1 [DOI] [PubMed] [Google Scholar]
- Fontana, A. , d'Ippolito, G. , Cutignano, A. , Miralto, A. , Ianora, A. , Romano, G. , & Cimino, G. (2007). Chemistry of oxylipin pathways in marine diatoms. Pure and Applied Chemistry, 79, 481–490. [Google Scholar]
- Fontana, A. , d'Ippolito, G. , Cutignano, A. , Romano, G. , Lamari, N. , Massa Gallucci, A. , … Ianora, A.. (2007). LOX‐induced lipid peroxidation mechanism responsible for the detrimental effect of marine diatoms on zooplankton grazers. ChemBioChem, 8, 1810–1818. [DOI] [PubMed] [Google Scholar]
- Gallo, C. , d'Ippolito, G. , Nuzzo, G. , Sardo, A. , & Fontana, A. (2017). Autoinhibitory sterol sulfates mediate programmed cell death in a bloom‐forming marine diatom. Nature Communications, 8, 1292 10.1038/s41467-017-01300-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P. J. , Conway, H. L. , Holmes, R. W. , & Davis, C. O. (1977). Marine diatoms grown in chemostats under silicate or ammonium limitation. III. Cellular chemical composition and morphology of Chaetoceros debilis, Skeletonema costatum, and Thalassiosira gravida . Marine Biology, 43, 19–31. 10.1007/BF00392568 [DOI] [Google Scholar]
- Hay, M. E. (2009). Marine chemical ecology: Chemical signals and cues structure marine populations, communities, and ecosystems. Annual Review of Marine Science, 1, 193–212. 10.1146/annurev.marine.010908.163708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianora, A. , Romano, G. , Carotenuto, Y. , Esposito, F. , Roncalli, V. , Buttino, I. , & Miralto, A. (2011). Impact of the diatom oxylipin 15S‐HEPE on the reproductive success of the copepod Temora stylifera . Hydrobiologia, 666, 265–275. [Google Scholar]
- Ianora, A. , Bentley, M. G. , Caldwell, G. S. , Casotti, R. , Cembella, A. D. , Engstrom‐Ost, J. , … Paldavičienë, A. (2011). The relevance of marine chemical ecology to plankton and ecosystem function: An emerging field. Marine Drugs, 9, 1625–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada, N. , Kobayashi, K. , Tahara, K. , & Oshima, Y. (1991). Production of an autoinhibitor by Skeletonema costatum and its effect on the growth of other phytoplankton. Nippon Suisan Gakkaishi, 57, 2285–2290. [Google Scholar]
- Jones, L. J. , Upson, R. H. , Haugland, R. P. , Panchuk‐Voloshina, N. , Zhou, M. , & Haugland, R. P. (1997). Quenched BODIPY dye‐labeled casein substrates for the assay of protease activity by direct fluorescence measurement. Analytical Biochemistry, 251, 144–152. 10.1006/abio.1997.2259 [DOI] [PubMed] [Google Scholar]
- Jung, S. W. , Kim, B. H. , Katano, T. , Kong, D. S. , & Han, M. S. (2008). Pseudomonas fluorescens HYK0210‐SK09 offers species‐specific biological control of winter algal blooms caused by freshwater diatom Stephanodiscus hantzschii . Journal of Applied Microbiology, 105, 186–195. [DOI] [PubMed] [Google Scholar]
- Kooistra, W. H. , Sarno, D. , Balzano, S. , Gu, H. , Andersen, R. A. , & Zingone, A. (2008). Global diversity and biogeography of Skeletonema species (Bacillariophyta). Protist, 159, 177–193. 10.1016/j.protis.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Boonprakob, A. , Gaonkar, C. C. , Kooistra, W. H. C. F. , Lange, C. B. , Hernández‐Becerril, D. , … Lundholm, N. (2017). Diversity in the globally distributed diatom genus Chaetoceros (Bacillariophyceae): three new species from warm‐temperate waters. PLoS ONE, 12, e0168887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima‐Mendez, G. , Faust, K. , Henry, N. , Decelle, J. , Colin, S. , Carcillo, F. , …… Raes, J. (2015). Ocean plankton. Determinants of community structure in the global plankton interactome. Science, 348, 1262073 10.1126/science.1262073 [DOI] [PubMed] [Google Scholar]
- Maier, I. , & Calenberg, M. (1994). Effect of extracellular Ca2+ and Ca2+‐antagonists on the movement and chemoorientation of male gametes of Ectocarpus siliculosus (Phaeophyceae). Botanica Acta, 107, 451–460. [Google Scholar]
- Margulis, L. (1990). Words as battle cries: Symbiogenesis and the new field of endocytobiology. BioScience, 40, 673–677. 10.2307/1311435 [DOI] [PubMed] [Google Scholar]
- Menden‐Deuer, S. , Lessard, E. J. , & Satterberg, J. (2001). Effect of preservation on dinoflagellate and diatom cell volume and consequences for carbon biomass predictions. Marine Ecology Progress Series, 222, 41–50. 10.3354/meps222041 [DOI] [Google Scholar]
- Meyer, N. , Bigalke, A. , Kaulfuss, A. , & Pohnert, G. (2017). Strategies and ecological roles of algicidal bacteria. FEMS Microbiology Reviews, 41, 880–899. 10.1093/femsre/fux029 [DOI] [PubMed] [Google Scholar]
- Orellana, M. V. , Pang, W. L. , Durand, P. M. , Whitehead, K. , & Baliga, N. S. (2013). A role for programmed cell death in the microbial loop. PLoS ONE, 8, e62595 10.1371/journal.pone.0062595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, C. , Barofsky, A. , Vidoudez, C. , & Pohnert, G. (2009). Diatom exudates influence metabolism and cell growth of co‐cultured diatom species. Marine Ecology Progress Series, 389, 61–70. 10.3354/meps08162 [DOI] [Google Scholar]
- Paul, C. , Mausz, M. A. , & Pohnert, G. (2012). A co‐culturing/metabolomics approach to investigate chemically mediated interactions of planktonic organisms reveals influence of bacteria on diatom metabolism. Metabolomics, 9, 349–359. 10.1007/s11306-012-0453-1 [DOI] [Google Scholar]
- Paul, C. , & Pohnert, G. (2011). Interactions of the algicidal bacterium Kordia algicida with diatoms: Regulated protease excretion for specific algal lysis. PLoS ONE, 6, e21032 10.1371/journal.pone.0021032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, C. , & Pohnert, G. (2013). Induction of protease release of the resistant diatom Chaetoceros didymus in response to lytic enzymes from an algicidal bacterium. PLoS ONE, 8, e57577 10.1371/journal.pone.0057577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohnert, G. (2010). Chemical noise in the silent ocean. Journal of Plankton Research, 32, 141–144. 10.1093/plankt/fbp118 [DOI] [Google Scholar]
- Pokrzywinski, K. L. , Place, A. R. , Warner, M. E. , & Coyne, K. J. (2012). Investigation of the algicidal exudate produced by Shewanella sp. IRI‐160 and its effect on dinoflagellates. Harmful Algae, 19, 23–29. 10.1016/j.hal.2012.05.002 [DOI] [Google Scholar]
- Ribalet, F. , Bastianini, M. , Vidoudez, C. , Acri, F. , Berges, J. , Ianora, A. , … Casotti, R. (2014). Phytoplankton cell lysis associated with polyunsaturated aldehyde release in the Northern Adriatic Sea. PLoS ONE, 9, e85947 10.1371/journal.pone.0085947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour, J. R. , Amin, S. A. , Raina, J. B. , & Stocker, R. (2017). Zooming in on the phycosphere: The ecological interface for phytoplankton‐bacteria relationships. Nature Microbiology, 30, 17065. [DOI] [PubMed] [Google Scholar]
- Shishlyannikov, S. M. , Klimenkov, I. V. , Bedoshvili, Y. D. , Mikhailov, I. S. , & Gorshkov, A. G. (2014). Effect of mixotrophic growth on the ultrastructure and fatty acid composition of the diatom Synedra acus from Lake Baikal. Journal of Biological Research‐Thessaloniki, 21, 8 10.1186/2241-5793-21-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieg, R. D. , Poulson‐Ellestad, K. L. , & Kubanek, J. (2011). Chemical ecology of the marine plankton. Natural Products Reports, 28, 388–399. 10.1039/C0NP00051E [DOI] [PubMed] [Google Scholar]
- Teeling, H. , Fuchs, B. M. , Becher, D. , Klockow, C. , Gardebrecht, A. , Bennke, C. M. , … Amann, R. (2012). Substrate‐controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science, 336, 608–611. 10.1126/science.1218344 [DOI] [PubMed] [Google Scholar]
- Teeling, H. , Fuchs, B. M. , Bennke, C. M. , Kruger, K. , Chafee, M. , Kappelmann, L. , … Amann, R. I. (2016). Recurring Patterns in Bacterioplankton Dynamics during Coastal Spring Algae Blooms. Elife, 5, e11888 10.7554/eLife.11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tol, H. M. , Amin, S. A. , & Armbrust, E. V. (2017). Ubiquitous marine bacterium inhibits diatom cell division. ISME Journal, 11, 31–42. 10.1038/ismej.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi, A. , Formiggini, F. , Casotti, R. , De Martino, A. , Ribalet, F. , Miralto, A. , & Bowler, C. (2006). A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biology, 4, e60 10.1371/journal.pbio.0040060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidoudez, C. , & Pohnert, G. (2008). Growth phase‐specific release of polyunsaturated aldehydes by the diatom Skeletonema marinoi . Journal of Plankton Research, 30, 1305–1313. 10.1093/plankt/fbn085 [DOI] [Google Scholar]
- Villanova, V. , Fortunato, A. E. , Singh, D. , Dal Bo, D. , Conte, M. , Obata, T. , … … J. (2017). Investigating mixotrophic metabolism in the model diatom Phaeodactylum tricornutum . Philosophical Transactions of the Royal Society B: Biological Sciences, 372, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Wang, J. T. , Xue, Q. N. , Sha, X. Y. , Tan, L. J. , & Guo, X. (2017). Allelopathic interactions between Skeletonema costatum and Alexandrium minutum . Chemical Ecology, 33, 485–498. [Google Scholar]
- Wichard, T. , Poulet, S. A. , Halsband‐Lenk, C. , Albaina, A. , Harris, R. , Liu, D. , & Pohnert, G. (2005). Survey of the chemical defence potential of diatoms: Screening of fifty one species for α, β, γ, δ‐ unsaturated aldehydes. Journal of Chemical Ecology, 31, 949–958. [DOI] [PubMed] [Google Scholar]
- Xu, N. , Tang, Y. Z. , Qin, J. L. , Duan, S. S. , & Gobler, C. J. (2015). Ability of the marine diatoms Pseudo‐nitzschia multiseries and P. pungens to inhibit the growth of co‐occurring phytoplankton via allelopathy. Aquatic Microbial Ecology, 74, 29–41. 10.3354/ame01724 [DOI] [Google Scholar]
- Yamada, N. , Murakami, N. , Kawamura, N. , & Sakakibara, J. (1994). Mechanism of an early lysis by fatty acids from axenic Phormidium tenue (musty odor‐producing cyanobacterium) and its growth prolongation by bacteria. Biological &/and Pharmaceutical Bulletin, 17, 1277–1281. 10.1248/bpb.17.1277 [DOI] [PubMed] [Google Scholar]
- Yamasaki, Y. , Ohmichi, Y. , Shikata, T. , Hirose, M. , Shimasaki, Y. , Oshima, Y. , & Honjo, T. (2011). Species‐specific allelopathic effects of the diatom Skeletonema costatum . Thalassas, 27, 21–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Underlying data for this publication will be made available upon reasonable request.


