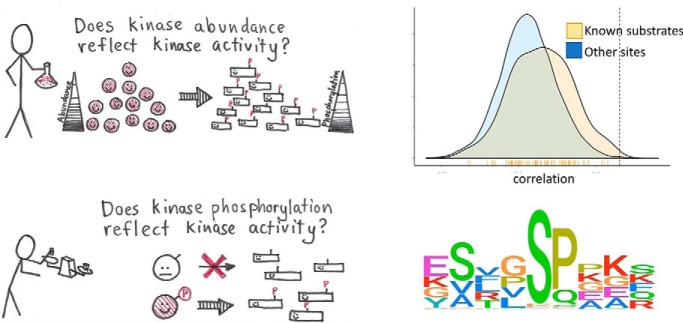
The relationships of kinase levels and activity have been investigated using large, high quality proteomic and phosphoproteomic data sets from tumors. Results show that the protein levels of some kinases correlate with their activity and that activation of kinases is a complex process. This study provides the first analysis of kinase activity in cancer integrating proteomic and phosphoproteomic data.
Keywords: Ovarian cancer, Breast cancer, Cancer Biology, Computational Biology, iTRAQ, Phosphoproteome, Phosphorylation
Graphical Abstract
Highlights
Integration of proteomics and phosphoproteomics data to understand kinase activity.
The abundance of some kinases correlates with activity.
Kinase activity does not necessarily reflect phosphorylation of regulatory sites.
Correlation patterns can be used to extend kinase substrate repertoire.
Abstract
Phosphorylation of proteins is a key way cells regulate function, both at the individual protein level and at the level of signaling pathways. Kinases are responsible for phosphorylation of substrates, generally on serine, threonine, or tyrosine residues. Though particular sequence patterns can be identified that dictate whether a residue will be phosphorylated by a specific kinase, these patterns are not highly predictive of phosphorylation. The availability of large scale proteomic and phosphoproteomic data sets generated using mass-spectrometry-based approaches provides an opportunity to study the important relationship between kinase activity, substrate specificity, and phosphorylation. In this study, we analyze relationships between protein abundance and phosphopeptide abundance across more than 150 tumor samples and show that phosphorylation at specific phosphosites is not well correlated with overall kinase abundance. However, individual kinases show a clear and statistically significant difference in correlation among known phosphosite targets for that kinase and randomly selected phosphosites. We further investigate relationships between phosphorylation of known activating or inhibitory sites on kinases and phosphorylation of their target phosphosites. Combined with motif-based analysis, this approach can predict novel kinase targets and show which subsets of a kinase's target repertoire are specifically active in one condition versus another.
In cellular systems, function is largely carried out by proteins. Regulation of protein function is essential for appropriate cellular function, and dysfunction of regulation can lead to disease states such as cancer (1–3). Though one level of protein regulation is through regulating the amount of protein present to accomplish the function, there are multiple other levels of functional regulation including localization, degradation, and post-translational modification (PTM)1 (4). There are many different forms of PTM utilized by cellular machinery, but phosphorylation is among the most prevalent and best understood (5, 6). Phosphorylation can lead to structural changes affecting activity, changes in affinity for substrates or protein binding, degradation, or changes in localization (7). Phosphorylation is employed in signaling cascades from pathways that link cell-surface receptors to transcription factors in the nucleus and regulate cell differentiation, growth, and migration, among others (8, 9).
Protein kinases modify specific residues on proteins with a phosphate group, which leads to functional changes in the protein in a large number of studied cases (10). Though there are some sequence-based preferences for how kinases select their target proteins for action, sequence alone does not provide enough information to be able to predict what proteins a kinase targets (11). Databases have been compiled of known kinase-target site relationships (12–14), but these are limited in coverage (15).
The growth of mass-spectrometry assisted proteomics recently has allowed rapid determination of phosphorylated residues in thousands of proteins at once (16–18). These data sets have revealed a large number of sites on proteins that can be phosphorylated where there is no functional information about the kinase that is affecting this phosphorylation and/or the functional effect of the phosphorylation. Several recent studies have examined relationships between kinase activity and specific phosphorylation, for example, of phosphorylation of kinase substrates and kinase activity (10, 19, 20), in the context of developing predictive methods for kinase specificity. However, there remains a great need to understand the complex relationships among kinase abundance, phosphorylation of activating sites and the activity of kinases. These relationships are important to the understanding of dysfunctional signaling pathways in cancer and to identify novel therapeutic treatments aimed at kinases and their downstream targets.
The Cancer Genome Atlas (TCGA) recently characterized a large number of ovarian high-grade serous carcinoma (HGSC) tumors (21) and breast cancer tumors (22). Previously we reported the first large-scale proteomic and phosphoproteomic characterizations of subsets of these tumors (23, 24) by the Clinical Proteomic Tumor Analysis Consortium (CPTAC) (25). In our studies, we analyzed 69 HSGC and 83 breast cancer tumors using mass-spectrometry assisted proteomics to acquire quantitative measurements for more than 10,000 proteins and used phosphosite enrichment to identify and quantify the abundance of over 25,000 phosphorylated peptides mapping to phosphosites. Our previous analysis showed that tumors from short and long surviving patients were well-separated by phosphoproteomics when summarized at the pathway level but not as well by the protein or transcript abundance, indicating that phosphorylation levels are an effective measure of pathway activity.
In the current study, we have leveraged the deep proteomic data sets generated by CPTAC to evaluate the relationship between protein abundance and the phosphorylation of cognate phosphosites. We investigate the ability of global proteome-wide correlation analysis of kinase protein expression measurements and phosphopeptide quantifications to pair phosphorylation sites with protein kinases. Integrated analyses of the proteome and phosphoproteome profiles is used to identify potential kinase-target phosphosite interactions in ovarian cancer. Our exploration of the association among protein abundance, phosphorylation and function indicate the complexity of such relationships in cancer.
MATERIALS AND METHODS
Data Description
Proteomic and phosphoproteomic profiles for high grade serous ovarian tumors were obtained from the Clinical Proteomic Tumor Analysis Consortium (CPTAC). Briefly, 69 HGSC samples previously characterized by the TCGA (21) were characterized using isobaric tags for relative and absolute quantitation (iTRAQ) (26). A portion of the sample was characterized with extensive high-pH reversed phase liquid chromatography (RPLC) prefractionation (27) and high-resolution tandem mass spectrometry (MS). The remainder of the isobarically labeled samples was subjected to immobilized metal affinity chromatography (iMAC) enrichment for phosphopeptide analysis. A “universal reference” representing a pool of all tumor samples in each iTRAQ experiment was used to provide relative peptide quantitation. A total of 9923 proteins and 20,732 unique phosphorylation sites contained in 4100 proteins were identified in the previous analysis. 16,788 phosphospetides mapping to 2096 proteins found to be affected by warm ischemia were removed from further analysis in the previous study (23, 28). In the current analysis, we consider all phosphopeptides because the effects of varying ischemic time are assumed to be random across tumors.
Additionally, we utilized our recently published data set that used very similar methods to measure protein and phosphopeptide abundance in 83 breast cancer tumors (24). This analysis identified 10,599 proteins and 31,017 unique phosphosites localized to 5,898 proteins.
Analysis of Protein Phosphosite and Intraphosphosite Correlation
We first sought to examine the association between protein abundance and phosphorylation. Spearman correlation was calculated in each cancer type between the abundance of each protein in the proteomic data set with each of its corresponding cognate phosphosites (i.e. phosphosites on the same protein) in the phosphoproteomic data set. To obtain reliable correlations between protein and cognate phosphosites, only those protein-phosphosite pairs having both protein and phosphosite abundances observed in at least 20% of samples (7067 and 25,396 protein-phosphosite pairs in ovarian and breast cancer respectively) were included in the analysis. We then analyzed the degree of coordination of phosphorylation of phosphosites on the same protein. For proteins with more than one phosphosite, Spearman correlation was determined between the phosphorylation levels of the phosphosites on the same protein for which the phosphorylation level was known in at least 20% of samples (16,158 and 176,426 pairs of phosphosites in ovarian and breast cancer respectively).
Analysis of Kinase Substrate Relationships
A compilation of known kinase-substrate interactions was obtained from the PhosphoSitePlus database (12). This reference database was used to quantify the extent to which substrate phosphorylation levels are determined by the abundance of their respective kinases. For each given kinase-substrate pair in the database for which both kinase abundance values in the proteomic data set and phosphorylation levels of the known target substrate phosphorylation sites were available in at least 20% of samples, we calculated the Spearman correlation for these known kinase-substrate relationships. In our ovarian and breast cancer data, we found 849 and 1718 known kinase-substrate relationships respectively (corresponding to 123 and 175 unique kinases) where we could accurately calculate correlation (less than 80% missing data in either component). To determine kinase-substrate interactions, the correlation was then calculated for each protein kinase in the database represented in the proteomic data set with every phosphosite in the phosphoproteomic data set, not just known target substrates keeping the same 20% threshold of kinase-phosphosite pairs with sufficient non-missing data for each component. The dimensions of the kinase-phosphosite matrices computed from the ovarian and breast data sets were 7645 × 243 and 30,519 × 281 respectively with entry i,j being the computed correlation between phosphosite i and kinase j.
Kinase Activity Network Construction
Kinase and phosphopeptide correlations from proteomic measurements of protein abundance and phosphorylation were used to infer kinase substrate regulatory relationships and construct a kinase-target interaction network in ovarian cancer. The kinase substrate network is a bipartite graph with directed edges from protein kinases to their putative target substrate phosphosites. This activity network was constructed by filtering the set of correlations of all kinase-phosphosite pairs (as represented by a kinase-phosphosite correlation matrix computed above) formed by a kinase in the proteomic data set with a phosphosite observed in the phosphoproteomic data set to keep pairs above a correlation cutoff of 0.65. The target set of the kinase was defined to be the set of phosphosites that paired with the kinase in the filtered set. For those phosphosites that had a correlation above the cutoff with more than one kinase, they were mapped to the kinase with which the correlation was highest so that each phosphosite was assigned to a unique kinase. The constructed kinase substrate network was visualized using Cytoscape (29).
Kinase Phosphorylation Motif Analysis
We used the Gibbs Motif Sampler (30) available as a web server at http://ccmbweb.ccv.brown.edu/gibbs/gibbs.html for motif discovery from the predicted target substrate phosphosites for the kinases. To search for a motif in the substrate set of a kinase, the peptide sequences comprising residues in the vicinity of each phosphorylation site were input to the program. Default parameters were used for motif searching from the peptide sequences centered at each phosphorylation site in the target set of the kinase and flanked by the amino acids immediately upstream and downstream of the phosphosite. Motifs with a positive maximum a posteriori probability value were used to identify consensus sequences significantly different from a random background (31). The cognate position weight matrix of a motif obtained from the Gibb's Motif Sampler was used to create a sequence logo representation of the consensus sequence using ggseqlogo (32). Functional enrichment analysis for the predicted substrates in the kinase activity network was carried out using clusterProfiler (33) with an FDR of 0.05.
Analysis of Relationship Between Kinase Phosphorylation and Substrate Phosphorylation
We examined the relationship between known kinase activating (and inhibitory) sites and kinase activity as defined by the levels of known substrates. Spearman correlation was calculated for activating/inhibitory phosphosites of kinases for which such functional annotation was available in PhosphositePlus with known substrate levels. For both cancer data sets, analysis was limited to those kinase functional sites with non-missing values in at least one fifth of samples in addition to at least ten observed substrates.
Computing
All analyses were carried out using custom scripts in the R programming language and environment for statistical computing (34) (version 3.4.1) along with the packages Biostrings (35) (version 2.46.0), tidyverse (36) (version 1.2.1) and qdapTools (37) (version 1.3.2). Figures were created with the help of the packages ggplot2 (38) (version 3.0.0), ggrepel (39) (version 0.8.0), cowplot (40) (version 0.9.3), ggpubr (41) (version 0.1.7) and ggridges (42) (version 0.5.0).
RESULTS
Data Sources
The Cancer Genome Atlas (TCGA) is a large-scale effort for the genomic characterization of multiple tumor types across large patient cohorts. A companion consortium, the Clinical Proteomics Tumor Analysis Consortium (CPTAC), has profiled a subset of tumors using global proteomics and phosphoproteomics. As part of CPTAC, we have recently conducted the first extensive analysis of TCGA high grade serous ovarian cancer samples using iTRAQ (isobaric tag for relative and absolute quantitation) (26) labeling in conjunction with extensive fractionation to provide comprehensive measurements on both the proteome and phosphoproteome (23). In addition, CPTAC investigators have also measured protein and phosphopeptide abundance in 83 breast cancer tumors (24). The analyses in this manuscript are conducted on these large-scale ovarian and breast cancer mass spectrometry proteomic and phosphoproteomic data sets.
Relationship Between Protein and Phosphorylation Abundance
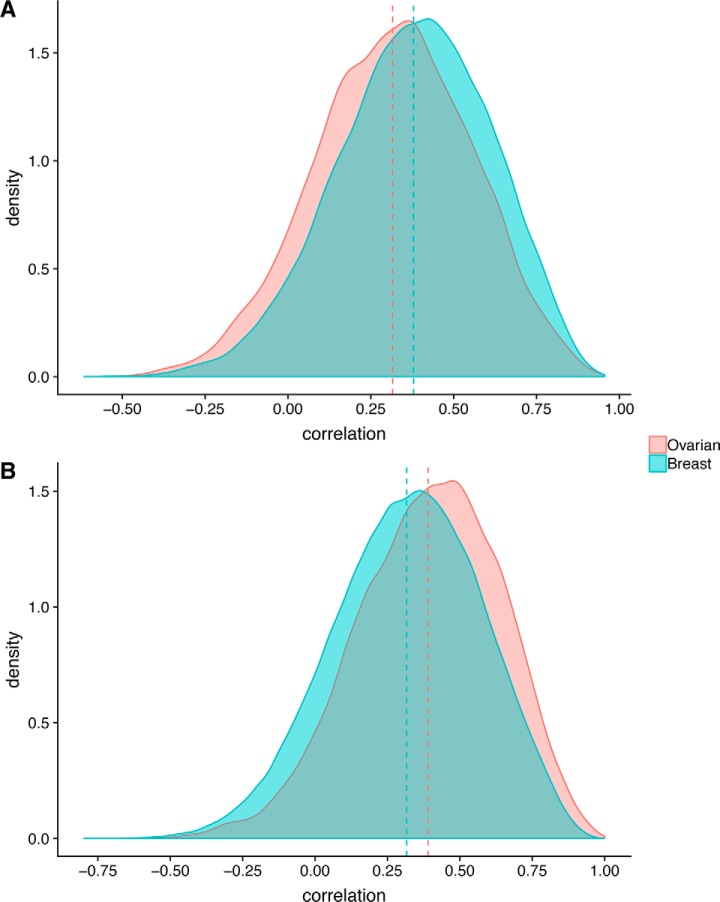
We previously reported a low concordance of mRNA levels to protein levels across ovarian tumors (23), similar to that observed in colorectal (43) and breast cancers (24). This observation highlights the levels of post-transcriptional regulation occurring that impact protein levels and presumably, function. It is clear that similar mechanisms exist in regulation of PTMs for proteins, but the extent and nature of this regulation are unclear. We looked at whether all proteins are phosphorylated to the same extent, regardless of changes in protein abundance. To do so, we calculated correlation between each protein and phosphopeptides that mapped to the same protein (cognate phosphopeptides) (Fig. 1A). We found that the correlation between protein abundance and phosphopeptide abundance was positive but modest (mean correlation 0.32 and 0.38 in ovarian and breast tumors, respectively). This suggests, unsurprisingly, that protein abundance does not dictate relative levels of phosphorylation overall. However, shown in Fig. 1A are a broad range of correlations with individual phosphosites.
Fig. 1.
Protein phosphosite and intraphosphosite correlation. Distribution of correlations between (A) protein and cognate phosphosite abundance and (B) phosphorylation of phosphosites (co-phosphorylation) on the same protein, for ovarian (red) and breast (blue) tumors. Dashed vertical lines indicate the means of the distributions.
Relationship Among Phosphorylation Levels of Protein Phosphosites
Although it is well understood that phosphorylation at different sites on the same protein can lead to dramatically different functional outcomes (44, 45), it is unclear as to the extent to which the phosphorylation of different sites might be coordinated on a global level. We explored whether phosphorylation at different sites on the same protein would be at similar levels leading to high correlation among sites. To do so, we calculated the correlation of phosphopeptide abundance between different sites on the same protein for those proteins with two or more observed phosphosites across all samples (Fig. 1B). We found that the mean intra-protein phosphosite correlation (i.e. correlation of phosphosites on the same protein) was 0.39 in the ovarian and 0.32 in the breast cancer data set. These qualitatively low levels of correlation indicate that individual phosphosites on the same protein are poorly coordinated in their phosphorylation, however they are still significantly (p < 0.001 Wilcoxon test) more correlated than phosphosites belonging to different proteins (mean inter-protein phosphosite correlation of phosphosites on distinct proteins is 0.10).
We speculated that the proteins with phosphosites most correlated with their abundance would also have the most correlated phosphosites but found that this was not the case and the trend was opposite (supplemental Fig. S1). This is an interesting finding and if the lack of correlation of phosphosites with cognate protein abundance is an indication of a higher level of regulation at the kinase level (less of the variation in the phosphosites can be explained by protein abundance) this may indicate that regulation of phosphosites occurs in a coordinated fashion for proteins in general.
Relationship Between Kinase Abundance and the Phosphorylation Levels of Known Substrates
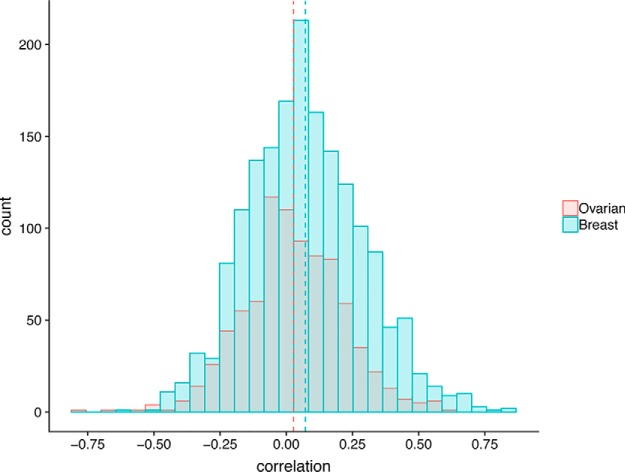
Signaling through cellular pathways via phosphorylation is important to cellular function, including aberrant cancer-associated functions. In heterogeneous samples such as human tumors, we hypothesized that the abundance of a kinase would reflect its activity and should be roughly correlated with phosphorylation levels of known target phosphosites. We used the PhosphoSitePlus database (12) that contains known kinase-substrate relationships to calculate the correlation of kinase protein abundance with known substrate phosphorylation levels. Our results showed that, overall, kinase abundance and target phosphosite abundance are uncorrelated (mean r = 0.03 and 0.07 for ovarian and breast data sets respectively; Fig. 2), though in each case this slight positive correlation is significantly higher than the background correlation.
Fig. 2.
Kinase known substrate correlation. Frequency distribution of correlations of kinases with their known substrates in ovarian (red) and breast (blue) cancer. Dashed lines indicate means.
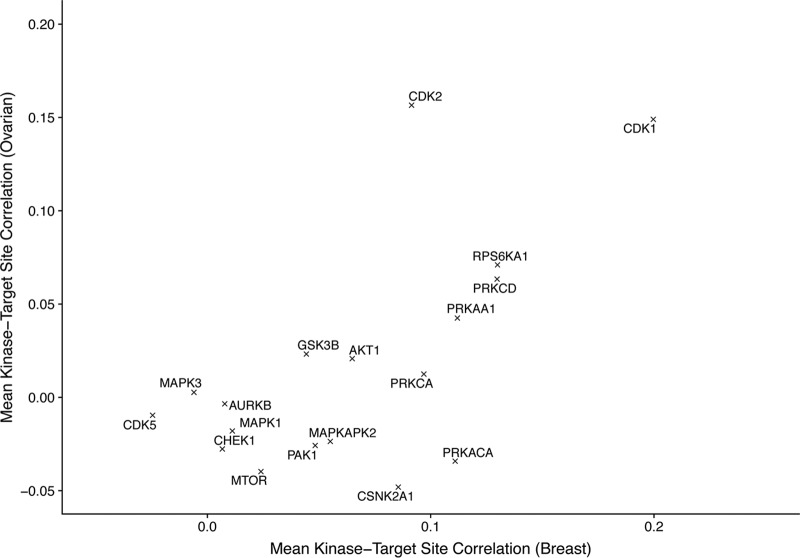
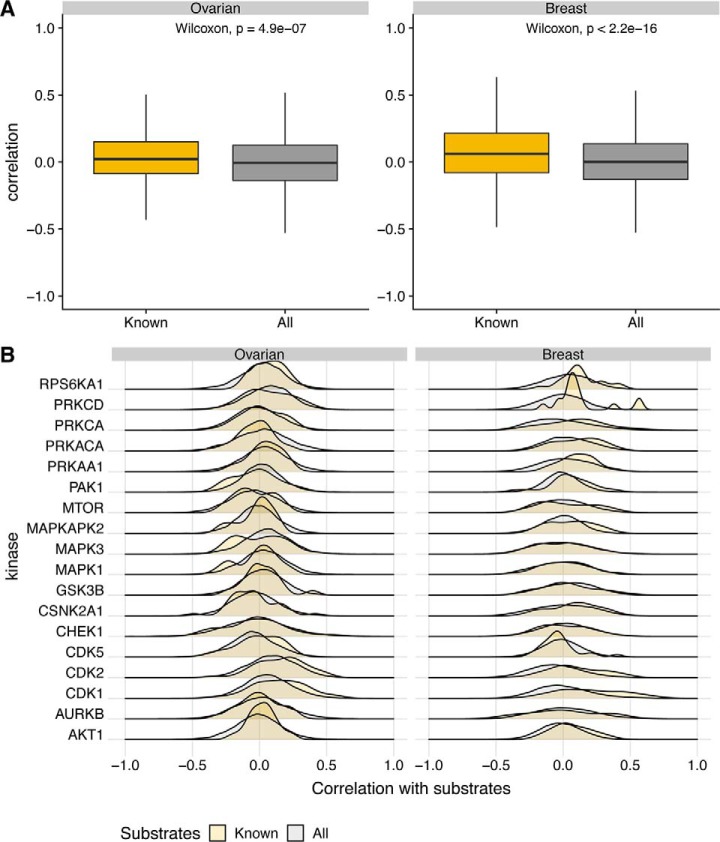
However, analysis of individual kinases revealed that some kinases were correlated with their known substrate phosphosites (e.g. CDK1, CDK2, PRKCD, RPS6KA1) whereas many were not (Fig. 3). Comparing results from ovarian and breast cancer data sets revealed a strong correspondence between the correlation in these two data sets (with a correlation of 0.63 across the mean kinase-substrate correlations in the two tumor types). These results suggest that for some kinases, high correlation between kinase abundance and phosphosite abundance is a reasonable predictor that there may be a real interaction, though this is difficult to assess completely because of the sparsity of known kinase-substrate relationships and the likely presence of many true relationships that are not yet known.
Fig. 3.
Kinase mean substrate correlation. Mean correlation of kinase abundance with known substrate abundance for ovarian and breast tumors.
Correlation Between Kinase Abundance and Phosphorylation Levels Can Identify Novel Kinase Target Substrates
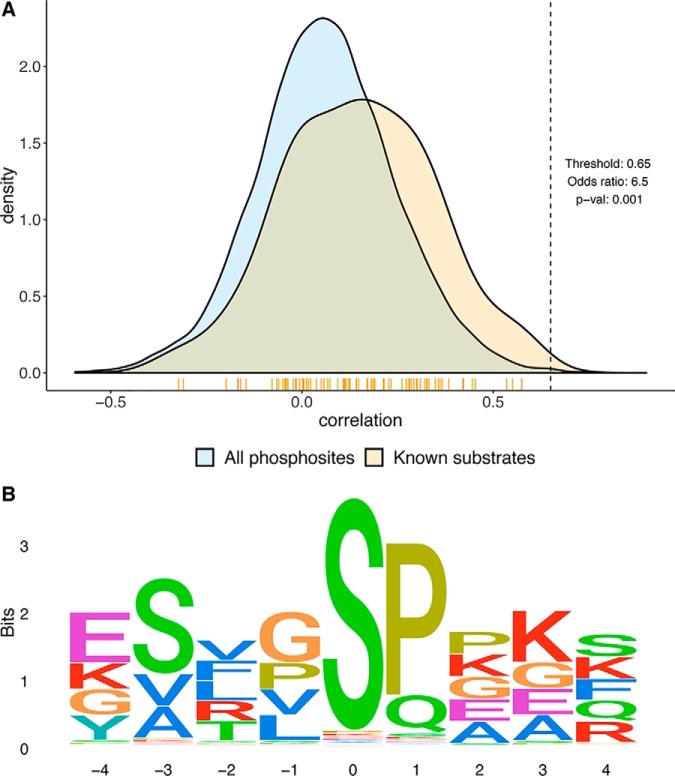
To extend this analysis to expand the potential repertoire of individual kinases, we determined the correlation between kinases and all phosphosites observed in the data, not just the known phosphosites. Fig. 4 shows the distribution of the correlation of kinases with known substrates against the complete set of phosphorylation sites profiled. For certain kinases, the distribution of the correlation of the kinase with known substrates was like that with all substrates whereas for others the two distributions were distinct. Because there were significant differences between known substrates and other phosphosites for some proteins we used a simple threshold to predict novel phosphosite targets for kinases. The threshold used was 0.65, which provided good p values and odds ratios (p value below 0.05 and odds ratio above 4) when tested against correlation with known phosphosites. This threshold should yield several high-confidence predictions for novel phosphosites for these kinases. The predicted kinase-substrate network for ovarian cancer is shown in supplemental Fig. S2.
Fig. 4.
Kinase phosphosite correlation - known versus all. Comparison of correlations of kinase abundance with the phosphorylation of known substrates against the complete set of phosphorylation sites profiled. A, Boxplots comparing the correlations of the entire kinase set with their known substrates against the background of correlations of the kinases with all of the phosphosites in the data. B, Distributions of correlations for individual kinases with known substrate target sites against all phosphosites in the data.
We searched for kinase protein phosphorylation motifs surrounding the predicted phosphosites for individual kinases using the Gibbs Motif Sampler (30), a method for discovering motifs in DNA or protein sequences. As an indication of the potential utility of this approach, the phosphorylation motif extracted from the predicted substrates for the kinase CDK1 (a kinase responsible for driving the eukaryotic cell cycle) using this methodology is shown in Fig. 5. The consensus sequence indicates a conservation of proline and lysine residues at positions 1 and 3 respectively which agrees with the global analysis of CDK1 phosphorylation sites in (46). Further, functional enrichment analysis for the predicted substrates indicated an enrichment for multiple processes including mitotic centrosome separation and the regulation of mitotic cell cycle (supplemental Fig. S3).
Fig. 5.
Prediction of CDK1 target phosphosites. An example of kinase-specific target substrate prediction for the kinase CDK1 by correlation analysis of kinase protein abundance with phosphorylation levels of phosphosites. The top panel shows the distributions of correlations of the kinase CDK1 with known kinase substrate phosphosites against all phosphosites in the data. The threshold is used for new phosphosite prediction from the data. Phosphosites above the threshold were used to predict kinase targets. The vertical yellow bars below the density plot mark where the known substrate phosphosites line up. The bottom panel is a sequence logo representation of the identified phosphorylation motif from the predicted target substrate phosphosites of the kinase CDK1. Position zero indicates the phosphorylation site.
As means of validation, we repeated the motif-based analysis using the same procedure independently on the breast cancer data sets (supplemental Fig. S4). The phosphorylation motif identified is very similar to that in ovarian cancer indicating that in some cases this approach could be used to identify putative substrates.
Correlation Between Kinase Activation and Substrate Phosphorylation Levels
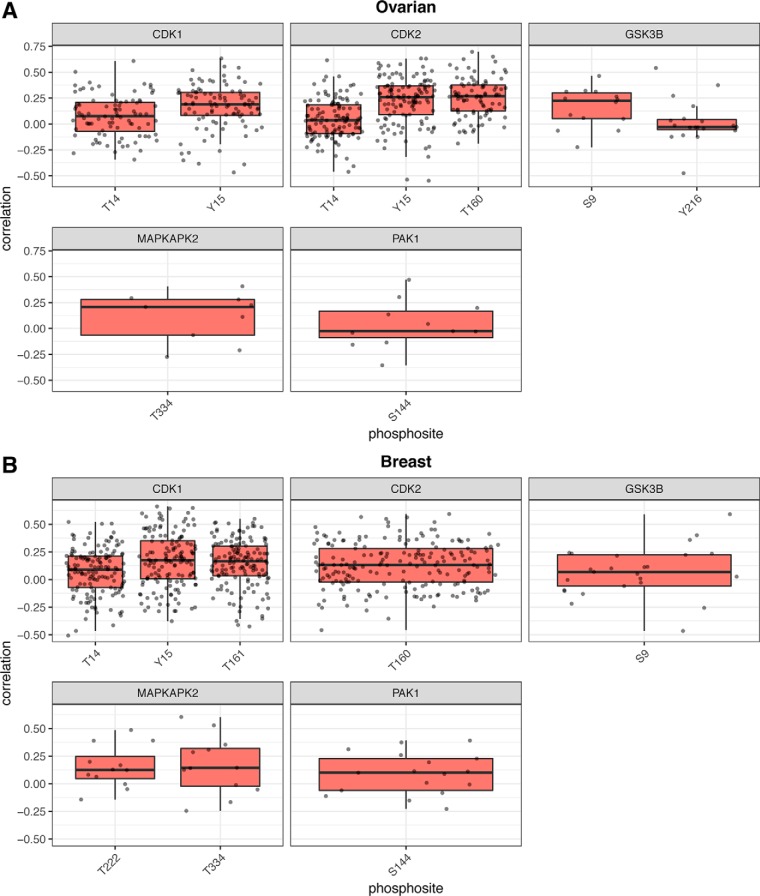
Thus far we have concentrated on relationships between kinase protein abundance and kinase activity, as assessed by phosphorylation of target phosphosites. For a subset of kinases, phosphorylation sites on those kinases are understood to play an activating or inhibitory role for kinase activity. Previously, it was reported that phosphorylation of autophosphorylation activating sites on kinases correlated with the phosphorylation of the kinase's known substrates (10), but this analysis was limited by the overall quality of the data set and did not assess more general activating and inhibitory sites on kinases. Therefore, we assessed the extent to which phosphorylation of known activating or inhibitory sites on the kinase correlate with kinase activity. From information in the PhosphositePlus database, we observed phosphorylation consistently on six activation sites and five inhibitory sites from five kinases (supplemental Table S1). Examining correlation between these sites and the known substrates for each of the kinases, we found variable results (Fig. 6; supplemental Table S1). Chiefly, there did not seem to be a clear relationship between the activation state of a kinase, as indicated by phosphorylation at the activating or inactivating site, and the activity of that kinase, as indicated by phosphorylation of known substrates. The inhibitory sites on CDK1 and CDK2 appear to be as positively correlated with activity as the activating site. In the case of GSK3B, phosphorylation at the inhibitory site is positively correlated with activity whereas the activating site is not. This was a surprising finding, so to determine if this was because of the untargeted nature of the mass-spectrometry measurement, we assessed this relationship using data from reverse-phase protein arrays (RPPA). We calculated the correlation between the inhibitory phosphosite (GSK3B-S9) and known substrates EIF4EBP1-T37 and ESR1-S118 in 418 tumors from the TCGA, which include all the ovarian tumors in the current study. We found that the correlations were 0.513 and 0.205 respectively, the average of which is 0.36, agreeing very well with our mass-spectrometry-based assessment.
Fig. 6.
Kinase functional phosphosite known substrate correlation. Boxplots of distribution of correlations of kinase functional (activating and inhibitory) phosphosites with known substrates in the (A) ovarian and (B) breast cancer data sets.
DISCUSSION
Though kinase phosphorylation and signaling are crucial to the understanding of many biological processes, and mass-spectrometry techniques have advanced rapidly allowing the measurement of the abundance of tens of thousands of phosphosites from one sample, understanding of the basic relationships between kinase activity and phosphorylation remain unclear. We have analyzed deep proteomic and phosphoproteomic data from tumor samples for ovarian and breast tumors (23, 24). In the current study, we show that phosphorylation levels are largely unrelated to the protein abundance of the cognate protein or the phosphorylation of other sites on the same protein, neither of which are surprising observations. Somewhat surprisingly we found that abundance of the kinase is largely uncorrelated with its activity, as assessed by phosphorylation of known substrates. However, we found that using a stringent threshold for this relationship was a reasonable approach for the identification of novel substrates for some kinases.
Finally, we showed that phosphorylation of kinases on their activating or inhibiting sites did not seem to correlate well with their activity. In the case of GSK3B, the reported inhibitory site was positively correlated with activity, directly opposite of the expected relationship. This raises several possibilities. One possibility is that the original information about the site is incorrect. However, a number of publications have reported previously that this site is inhibitory under several conditions (47, 48). Another likely possibility is that the action of the kinase is highly context dependent, with different sets of substrates being targeted under different conditions. It's possible that the ovarian and breast cancer environments, overall, represent a set of conditions under which GSK3B acts differently. A third possibility is that the measurement of phosphorylation on specific sites by either mass spectrometry or RPPA is a population-based measurement, such that the effect of specific phosphorylation events is obscured by population differences.
Previously, some studies have used phosphorylation levels that have been normalized to protein abundance (23) whereas others have used unnormalized phosphoproteomic data (24, 49), and often phosphoproteomic analyses are conducted without gathering corresponding global protein abundance data (50–52). There are differences of opinion about whether to normalize phosphopeptide abundance to the cognate protein abundance and each approach comes with advantages and caveats that must be considered in interpretation. Leaving the data unnormalized means that increases in phosphopeptide abundance (and thus measured phosphorylation of the associated sites) may also reflect changes in protein abundance. However, normalization may obscure information about kinase activity that is inherent in protein abundance.
Previous studies have analyzed relationships within phosphoproteomic data sets to look at kinase activity. Ochoa et al., compiled a large set of phosphoproteomic data from different studies and used this to examine the relationship between cell treatment and kinase activation patterns by assessing overall phosphorylation of known kinase substrates (10). Additionally, they reported that the phosphorylation of one known activating site on AURKA was well-correlated with AURKA activity. However, our findings show that not all known activating or inhibitory sites on kinases behave in such a straightforward manner, with many sites seeming to display behavior indicative of more complicated regulatory processes. Similarly, Petsalaki et al., showed that known substrates of kinases were significantly enriched in groups of correlated phosphosites, showing that this approach could be used to identify candidate kinase-substrate relationships (53). A study by Ayati et al., uses, in part, the same ovarian data set generated by our group to build a predictive method for identifying novel kinase substrates (19). In this study, the authors show that phosphorylation sites known to be targets of a kinase are significantly more correlated with each other than are all phosphosites in the data set. This “co-phosphorylation” is significant, but the effect, like our results, is very small in terms of correlation. This result fits well with our results showing a modest, but significant, correlation between kinase abundance and substrate phosphorylation (see Fig. 4), given that it is likely that multiple phosphosites correlated with the same kinase level would also be correlated with each other.
Given the highly heterogeneous nature of these samples, tumors representing different genetic backgrounds, environmental histories, and subtypes of ovarian and breast cancer, it is somewhat surprising that we uncovered any relationships at all. Many previous studies of such relationships have been focused on more highly controlled systems with homogenous genetic and environmental backgrounds and rigorously controlled experimental conditions. We recognize that a limitation of our findings is the heterogeneous nature of our data but emphasize that our findings represent a lower bound based on utilization of biologically relevant samples. Our findings are based on sampling the diverse cells in a tumor and will mask the dynamic nature of phosphorylation and signaling. However, our previous results have indicated that the state of phosphorylation in this snapshot of the distribution of dynamic states in tumors is more closely related to phenotype (overall survival) than the proteome, transcriptome, or genetic composition (23).
Data Availability
All raw primary MS data for the tumor samples analyzed in this study is publicly available from the CPTAC Data Coordinating Center (https://cptac-data-portal.georgetown.edu).
Supplementary Material
Acknowledgments
The proteomics work described herein was performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) national scientific user facility located at the Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL01830.
Footnotes
* This work was supported by the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium under grants U01CA214116 and U24CA210955.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- PTM
- post-translational modification
- CPTAC
- Clinical Proteomic Tumor Analysis Consortium
- HGSC
- high grade serous carcinoma
- iMAC
- immobilized metal ion affinity chromatography
- iTRAQ
- isobaric tag for relative and absolute quantitation
- RPLC
- reverse phase liquid chromatography
- TCGA
- The Cancer Genome Atlas.
REFERENCES
- 1. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell. 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 2. Giancotti F. G. (2014) Deregulation of cell signaling in cancer. FEBS Lett. 588, 2558–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gonzalez M. W., and Kann M. G. (2012) Chapter 4: Protein interactions and disease. PLoS Comput. Biol. 8, e1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vogel C., and Marcotte E. M. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Gen. 13, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma K., D'Souza R. C., Tyanova S., Schaab C., Wisniewski J. R., Cox J., and Mann M. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Reports 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 6. Hunter T. (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80, 225–236 [DOI] [PubMed] [Google Scholar]
- 7. Pawson T. (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 [DOI] [PubMed] [Google Scholar]
- 8. Lemmon M. A., and Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beltrao P., Albanese V., Kenner L. R., Swaney D. L., Burlingame A., Villen J., Lim W. A., Fraser J. S., Frydman J., and Krogan N. J. (2012) Systematic functional prioritization of protein posttranslational modifications. Cell 150, 413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ochoa D., Jonikas M., Lawrence R. T., El Debs B., Selkrig J., Typas A., Villen J., Santos S. D., and Beltrao P. (2016) An atlas of human kinase regulation. Mol. Syst. Biol. 12, 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Douglass J., Gunaratne R., Bradford D., Saeed F., Hoffert J. D., Steinbach P. J., Knepper M. A., and Pisitkun T. (2012) Identifying protein kinase target preferences using mass spectrometry. Am. J. Physiol. Cell Physiol. 303, C715–C727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2014) PhosphoSitePlus: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu J., Rho H. S., Newman R. H., Zhang J., Zhu H., and Qian J. (2014) PhosphoNetworks: a database for human phosphorylation networks. Bioinformatics 30, 141–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gnad F., Gunawardena J., and Mann M. (2011) PHOSIDA : the posttranslational modification database. Nucleic Acids Res. 39, D253–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wirbel J., Cutillas P., and Saez-Rodriguez J. (2018) Phosphoproteomics-based profiling of kinase activities in cancer cells. Methods Mol. Biol. 1711, 103–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doll S., and Burlingame A. L. (2015) Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem. Biol. 10, 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olsen J. V., and Mann M. (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell Proteomics 12, 3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Humphrey S. J., Azimifar S. B., and Mann M. (2015) High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 [DOI] [PubMed] [Google Scholar]
- 19. Ayati M, Wiredja D., Schlatzer D., Maxwell S., Li M., Koyuturk M., and Chance M. R. (2019) CoPhosK: A method for comprehensive kinase substrate annotation using co-phosphorylation analysis. PLoS Comput. Biol. 15, e1006678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Domanova W., Krycer J., Chaudhuri R., Yang P., Vafaee F., Fazakerley D., Humphrey S., James D, and Kuncic Z. (2016) Unraveling kinase activation dynamics using kinase-substrate relationships from temporal large-scale phosphoproteomics studies. PLoS ONE 11, e0157763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cancer Genome Atlas Research Network. (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas Research Network. (2012) Comprehensive molecular portraits of human breast tumours, Nature 490, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang H., Liu T., Zhang Z., Payne S. H., Zhang B., McDermott J. E., Zhou J. Y., Petyuk V. A., Chen L., Ray D., Sun S., Yang F., Chen L., Wang J., Shah P., Cha S. W., Aiyetan P., Woo S., Tian Y., Gritsenko M. A., Clauss T. R., Choi C., Monroe M. E., Thomas S., Nie S., Wu C., Moore R. J., Yu K. H., Tabb D. L., Fenyo D., Bafna V., Wang Y., Rodriguez H., Boja E. S., Hiltke T., Rivers R. C., Sokoll L., Zhu H., Shih I. M., Cope L., Pandey A., Zhang B., Snyder M. P., Levine D. A., Smith R. D., Chan D. W., and Rodland K. D. (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., Gatza M. L., Wilkerson M., Perou C. M., Yellapantula V., Huang K. L., Lin C., McLellan M. D., Yan P., Davies S. R., Townsend R. R., Skates S. J., Wang J., Zhang B., Kinsinger C. R., Mesri M., Rodriguez H., Ding L., Paulovich A. G., Fenyo D., Ellis M. J., and Carr S. A. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ellis M. J., Gillette M., Carr S. A., Paulovich A. G., Smith R. D., Rodland K. K., Townsend R. R., Kinsinger C., Mesri M., Rodriguez H., and Liebler D. C. (2013) Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 3, 1108–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., and Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Yang F., Gritsenko M. A., Wang Y., Clauss T., Liu T., Shen Y., Monroe M. E., Lopez-Ferrer D., Reno T., Moore R. J., Klemke R. L., Camp D. G. 2nd, and Smith R. D. (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 11, 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mertins P., Yang F., Liu T., Mani D. R., Petyuk V. A., Gillette M. A., Clauser K. R., Qiao J. W., Gritsenko M. A., Moore R. J., Levine D. A., Townsend R., Erdmann-Gilmore P., Snider J. E., Davies S. R., Ruggles K. V., Fenyo D., Kitchens R. T., Li S., Olvera N., Dao F., Rodriguez H., Chan D. W., Liebler D., White F., Rodland K. D., Mills G. B., Smith R. D., Paulovich A. G., Ellis M., and Carr S. A. (2014) Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell Proteomics 13, 1690–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thompson W., Rouchka E. C., and Lawrence C. E. (2003) Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res. 31, 3580–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson W., McCue L. A., and Lawrence C. E. (2005) Using the Gibbs motif sampler to find conserved domains in DNA and protein sequences. Current Protocols in Bioinformatics Chapter 2, Unit 2.8 [DOI] [PubMed] [Google Scholar]
- 32. Wagih O. (2017) ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647 [DOI] [PubMed] [Google Scholar]
- 33. Yu G., Wang L. G., Han Y., and He Q. Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. R Core Team. (2014) R: A Language and Environment for Statistical Computing. [Google Scholar]
- 35. Pages H., Aboyoun P., Gentleman R., and DebRoy S. (2017) Biostrings: Efficient manipulation of biological strings. R package version 2.46.0 ed [Google Scholar]
- 36. Wickham H. (2017) tidyverse: easily install and load the 'Tidyverse'. R package version 1.2.1 ed [Google Scholar]
- 37. Rinker T. (2015) qdapTools: tools to accompany the qdap package. R package version 1.3.2 ed [Google Scholar]
- 38. Wickham H. (2016) ggplot2: elegant graphics for data analysis. Springer-Verlag; New York [Google Scholar]
- 39. Slowikowski K. (2018) ggrepel: Automatically Position Non-Overlapping Text Labels with 'ggplot2'. R package version 0.8.0 ed [Google Scholar]
- 40. Wilke C. O. (2018) cowplot: streamlined plot theme and plot annotations for 'ggplot2'. R package version 0.9.3 ed [Google Scholar]
- 41. Kassambara A. (2018) ggpubr: 'ggplot2' based publication ready plots. R package version 0.1.7 ed [Google Scholar]
- 42. Wilke C. O. (2018) ggridges: ridgeline plots in 'ggplot2'. R package version 0.5.0 ed [Google Scholar]
- 43. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J., and Liebler D. C. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nishi H., Shaytan A., and Panchenko A. R. (2014) Physicochemical mechanisms of protein regulation by phosphorylation. Frontiers Gen. 5, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nishi H., Demir E., and Panchenko A. R. (2015) Crosstalk between signaling pathways provided by single and multiple protein phosphorylation sites. J. Mol. Biol. 427, 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P., and Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ko H-W., Lee H-H., Huo L., Xia W., Yang C-C., Hsu J. L., Li L-Y., Lai C-C., Chan L-C., Cheng C-C., Labaff A. M., Liao H-W., Lim S-O., Li C-W., Wei Y., Nie L., Yamaguchi H., and Hung M-C. (2016) GSK3β inactivation promotes the oncogenic functions of EZH2 and enhances methylation of H3K27 in human breast cancers. Oncotarget. 7, 57131–57144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xing H. Y., Cai Y. Q., Wang X. F., Wang L. L., Li P., Wang G. Y., and Chen J. H. (2015) The cytoprotective effect of hyperoside against oxidative stress is mediated by the Nrf2-ARE signaling pathway through GSK-3beta inactivation. PLoS ONE 10, e0145183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vasaikar S., Huang C., Wang X., Petyuk V. A., Savage S. R., Wen B., Dou Y., Zhang Y., Shi Z., Arshad O. A., Gritsenko M. A., Zimmerman L. J., McDermott J. E., Clauss T. R., Moore R. J., Zhao R., Monroe M. E., Wang Y. T., Chambers M. C., Slebos R. J. C., Lau K. S., Mo Q., Ding L., Ellis M., Thiagarajan M., Kinsinger C. R., Rodriguez H., Smith R. D., Rodland K. D., Liebler D. C., Liu T., and Zhang B. (2019) Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell 177, 1035–1049.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hosseini M. M., Kurtz S. E., Abdelhamed S., Mahmood S., Davare M. A., Kaempf A., Elferich J., McDermott J. E., Liu T., Payne S. H., Shinde U., Rodland K. D., Mori M., Druker B. J., Singer J. W., and Agarwal A. (2018) Inhibition of interleukin-1 receptor-associated kinase-1 is a therapeutic strategy for acute myeloid leukemia subtypes. Leukemia. 32, 2374–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilkes E. H., Terfve C., Gribben J. G., Saez-Rodriguez J., and Cutillas P. R. (2015) Empirical inference of circuitry and plasticity in a kinase signaling network. Proc. Natl. Acad. Sci. U.S.A. 112, 7719–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beekhof R., van Alphen C., Henneman A. A., Knol J. C., Pham T. V., Rolfs F., Labots M., Henneberry E., Le Large T. Y., de Haas R. R., Piersma S. R., Vurchio V., Bertotti A., Trusolino L., Verheul H. M., and Jimenez C. R. (2019) INKA, an integrative data analysis pipeline for phosphoproteomic inference of active kinases. Mol. Syst. Biol. 15, e8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petsalaki E., Helbig A. O., Gopal A., Pasculescu A., Roth F. P., and Pawson T. (2015) SELPHI: correlation-based identification of kinase-associated networks from global phospho-proteomics data sets. Nucleic Acids Res. 43, W276–W282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw primary MS data for the tumor samples analyzed in this study is publicly available from the CPTAC Data Coordinating Center (https://cptac-data-portal.georgetown.edu).