Abstract
The BBSome, a complex of eight Bardet-Biedl syndrome (BBS) proteins involved in cilia function, has emerged as an important regulator of energy balance, but the underlying cellular and molecular mechanisms are not fully understood. Here, we show that the control of energy homeostasis by the anorexigenic proopiomelanocortin (POMC) neurons and orexigenic agouti-related peptide (AgRP) neurons require intact BBSome. Targeted disruption of the BBSome by Bbs1 gene deletion in POMC or AgRP neurons increases body weight and adiposity. We demonstrate that obesity in mice lacking the Bbs1 gene in POMC neurons is associated with hyperphagia. Mechanistically, we present evidence implicating the BBSome in the trafficking of G protein–coupled neuropeptide Y Y2 receptor (NPY2R) and serotonin 5-hydroxytryptamine (HT)2C receptor (5-HT2CR) to cilia and plasma membrane, respectively. Consistent with this, loss of the BBSome reduced cell surface expression of the 5-HT2CR, interfered with serotonin-evoked increase in intracellular calcium and membrane potential, and blunted the anorectic and weight-reducing responses evoked by the 5-HT2cR agonist, lorcaserin. Finally, we show that disruption of the BBSome causes the 5-HT2CR to be stalled in the late endosome. Our results demonstrate the significance of the hypothalamic BBSome for the control of energy balance through regulation of trafficking of important metabolic receptors.
Introduction
Genetic studies have consistently pointed to the central nervous system as a key player in the regulation of body weight and the development of obesity and its adverse side effects (1,2). This is consistent with the tremendous influence the brain exerts on the processes that control food intake and energy expenditure (3,4). The brain receives afferent signals about the status of body energy reserves, leading to autonomic, humoral, and behavioral adaptations aimed at maintaining body weight and adiposity in a normal range. These neuroendocrine systems are tightly controlled, and their disruption leads to obesity.
The distribution and mechanism of action of the key signals that influence metabolic functions point to a significant role of the hypothalamus in integrating the external and internal signals and in enacting appropriate and consequential metabolic and behavioral responses to maintain energy balance (3,4). Anorexigenic neurons expressing proopiomelanocortin (POMC) and orexigenic neurons expressing agouti-related peptide (AgRP) located in the hypothalamic arcuate nucleus are critically involved in the control of feeding and energy expenditure. These neurons are considered as first-order neurons that mediate the effects of various signals such as leptin, insulin, and serotonin (3,5–7). Despite being the most extensively studied neuronal populations regarding energy balance, our understanding of the molecular processes controlling their activity remains incomplete.
The BBSome is a complex composed of eight Bardet-Biedl syndrome (BBS) proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9, and BBS18) (8,9). The assembly of the BBSome depends on another protein complex that contains three BBS proteins (BBS6, BBS10, and BBS12) (10,11). The discovery that various BBS proteins interact to form complexes that are critical is consistent with the overlapping phenotypes arising from mutations in different BBS genes (11). Indeed, mutation in BBS genes results in human diseases with multiple features, including obesity. In addition, variants of several BBS genes were found to increase susceptibility to obesity in individuals without BBS (12–14). Together, these findings highlight the importance of the BBSome for energy balance and the development of obesity, but the underlying mechanisms are not well understood.
The BBSome is a critical regulator of cilia function by targeting cargos to the ciliary membrane. Primary cilia are important signaling organelles present in virtually every cell type in the body, including neurons (15). A number of G protein–coupled receptors, including somatostatin receptor 3 (SSTR3) (16), neuropeptide Y Y2 receptor (NPY2R) (17), and melanocortin 4 receptor, localize to cilia (18). In addition to its involvement in the trafficking of cargos to cilia, the BBSome has emerged as a key mediator of many cellular processes, including the delivery of receptors to the plasma membrane (19,20).
Here, we reveal an important role of the BBSome in the control of energy balance by POMC and AgRP neurons. We show that selective disruption the BBSome, through Bbs1 gene deletion in POMC or AgRP neurons, led to a significant increase in body weight and adiposity. We also uncover a critical role of the BBSome in the cellular trafficking of serotonin 5-hydroxytryptamine (HT)2C receptor (5-HT2CR), an important receptor for energy homeostasis and the target of the antiobesity drug lorcaserin. Our data demonstrate that the BBSome is required for 5-HT2CR trafficking to the plasma membrane, providing a molecular explanation for the obesity phenotype evoked by disruption of the BBSome.
Research Design and Methods
Animals
All animal testing was approved by the University of Iowa Animal Care and Use Committee. Mice were housed in groups of three to five per cage and maintained on a 12-h light-dark cycle, with lights on at 6:00 a.m. Room temperature was maintained at 22°C. Food and water were available ad libitum.
To obtain selective deletion of the Bbs1 gene in POMC or AgRP neurons, male transgenic POMCCre or AgRPCre mice (stock #005965 and 012899, respectively; The Jackson Laboratory) were crossed with Bbs1fl/fl female mice (20). POMCCre/Bbs1fl/wt and AgRPCre/Bbs1fl/wt offspring were subsequently crossed with Bbs1fl/wt mice to generate POMCCre/Bbs1fl/fl and AgRPCre/Bbs1fl/fl mice, respectively. The genotype of mice was determined by PCR analysis of tail DNA (20) using the primer sequences provided in Supplementary Table 1. A group of diet-induced obese C57BL/6J mice fed a high-fat diet (D12492; Research Diets) for 12 weeks and controls fed normal chow were obtained from The Jackson Laboratory.
Metabolic Studies
Mice were weighed once weekly, beginning at weaning (4 weeks of age) until 25 weeks of age. Body composition was determined by MRI under anesthesia (91 mg/kg ketamine and 9.1 mg/kg xylazine, i.p.) using a Varian Unity/Inova 4.7 T small-bore MRI system (Varian). To measure food intake, mice were housed individually in regular cages and allowed to acclimate for at least 3 days before measurements of 24-h food intake for 5 days. Resting metabolic rate was measured by push-pull respirometry at thermoneutrality during the daytime, as described previously (20).
Food intake and body weight response to lorcaserin were compared between control animals, POMCCre/Bbs1fl/fl mice, and diet-induced obese mice. Baseline body weight and food intake of individually housed mice were measured daily for 5 consecutive days before beginning treatment with vehicle or lorcaserin (1 μg/μL/g body wt i.p., twice daily at 8:00 a.m. and 5:00 p.m., for 4 days).
Glucose and Insulin Tolerance Tests and Insulin Measurements
Glucose tolerance tests (GTTs) and insulin tolerance tests (ITTs) were performed as described previously. Mice were fasted overnight (GTT) or for 5 h (ITT) in a clean cage. Basal blood glucose measurements were taken before injection of glucose (2 g/kg i.p.) or insulin (1 unit/kg i.p.). Blood glucose measurements were assessed multiple times during the 2-h test. Plasma insulin levels were measured using an ELISA kit (RayBiotech).
Generation of the Mouse IMCD3 Bbs1 Knockout Cell Line
The human codon-optimized Cas9 plasmid pX459 (#48139) was obtained from Addgene. Single guide (sg)RNAs were designed and constructed as described previously. Briefly, target 20 base pair (bp) sequences starting with guanine and preceding the protospacer adjacent motif (5′-NGG-3′) were selected from the mouse Bbs1 exon 4. Potential off-target effects of sgRNA candidates were analyzed using the online tool CRISPR Design (http://crispr.mit.edu/). One sgRNA in exon 4 (Supplementary Table 1) of the mouse Bbs1 gene was selected for transfection to create knockout cell lines. Clonal genomic DNA was extracted and used for PCR and Sanger sequencing with the forward primer and the reverse primer (Supplementary Table 1). This resulted in cell lines with homozygous 1 bp T insertions and homozygous 2 bp deletions (cytosine and thymine) in exon 4.
Mouse kidney epithelial cells (mIMCC-3; #CRL-2123, ATCC) were cultured in DMEM/F12 (Life Technologies) supplemented with 10% FCS. Cells were seeded in a 6-well plate at 1 × 106 cells/well. Transfection was performed with Lipofectamine 2000 using recommended conditions. Briefly, 2 µg plasmid containing an sgRNA targeting Bbs1 exon 4 and 6 µL Lipofectamine 2000 were diluted in 100 µL Opti-MEM, mixed 1:1, and added to cells after 5 min of incubation. After 24 h, the cells were passed at low density (2,000 cells/well) in 10-cm culture dishes and selected with puromycin at 1.5 µg/mL for clonal expansion. The media were changed every 2 days until colonies were harvested using cloning cylinders.
Measurement of Intracellular Ca2+
IMCD3 and IMCD3-Bbs1−/− cells were transiently transfected with 5-HT2CR tagged with green fluorescent protein (GFP). The cells were loaded with fura-2 48 h after transfection using the acetoxymethyl ester form of the indicator (fura-2/AM) (2 μmol/L for 30 min) to monitor changes in the intracellular Ca2+ concentration ([Ca2+]i) in response to 5 μmol/L serotonin and then to 10 μmol/L ATP. The cells were placed in a chamber for flow-through perfusion, mounted on an inverted IX-71 microscope (Olympus), and perfused with HEPES-buffered Hanks’ balanced salt solution composed of 140 mmol/L NaCl, 5 mmol/L KCl, 1.3 mmol/L CaCl2, 0.4 mmol/L MgSO4, 0.5 mmol/L MgCl2, 0.4 mmol/L KH2PO4, 0.6 mmol/L NaHPO4, 3 mmol/L NaHCO3, 10 mmol/L glucose, and 10 mmol/L HEPES, pH 7.4, with NaOH (310 mOsm/kg with sucrose).
Fluorescence was excited sequentially at 340 nm (12-nm bandpass) and 380 nm (12-nm bandpass) by using a Polychrome V monochromator (TILL Photonics) and focused on the cells via a 20×, 0.75 numerical aperture objective (Olympus). Fluorescence emission was measured at 540 nm (50-nm bandpass) using an IMAGO charge-coupled device camera (640 × 480 pixels; TILL Photonics). Two-by-two binning was used for acquisition (1 pixel, ∼500 nm). A series of 340-nm and 380-nm images was acquired every 2 s. ATP (10 μmol/L) was also used as a positive control. [Ca2+]i data were analyzed using TILLvisION (TILL Photonics) software and presented as F340-to-F380 ratio, where F340 and F380 correspond to the fluorescence intensity at excitation wavelength 340 and 380 nm, respectively. The F340 and F380 values were corrected for background, which was determined for each experiment by measuring fluorescence at 340 and 380 nm in an area free of cells.
Electrophysiological Recording
HEK 293 cells were used to measure serotonin-induced membrane potential. Briefly, the cells were cotransfected with control shRNA or Bbs1-shRNA with GFP-tagged 5-HT2cR. At 72 h after transfection, cells were placed in a perfusion chamber mounted on the stage of an upright Zeiss Axioskop FS2 microscope (Carl Zeiss) equipped with a Hamamatsu C2741-60 charged-coupled device camera (Hamamatsu Photonics). Epifluorescence was briefly used to target fluorescent cells, after which the light source was switched to infrared differential interference contrast imaging to allow visualization while obtaining whole-cell patch clamp recordings. Membrane potentials were recorded in current clamp mode using an Axopatch 700B amplifier (Molecular Devices), low-pass filtered at 1 kHz, digitized at 10 kHz, and analyzed offline on a personal computer with pCLAMP (Molecular Devices) and Origin software (OriginLab). Recording electrodes had resistances of 2.5–5 MΩ when filled with the K-gluconate pipette solution, which contained 120 mmol/L K-gluconate, 10 mmol/L KCl, 10 mmol/L HEPES, 5 mmol/L EGTA, 1 mmol/L CaCl2, 1 mmol/L MgCl2, and 5 mmol/L Mg-ATP adjusted to pH 7.3.
Biochemical Analysis
To detect cell surface receptor expression, cell surface biotinylation assays were performed in HEK 293 cells transiently transfected with the pcDNA3-Flag-5-HT2CR receptor (Flag-5-HT2CR), with Bbs2-shRNA or control shRNA on an LKO1 plasmid with X-tremeGENE 9 transfection reagent (Roche). Silencing efficacy of Bbs2-shRNA was reported previously (20).
The cells were incubated 48 h after transfection with cleavable EZ-Link Sulfo-NHS-SS-Biotin (Thermo Scientific) for 30 min on ice, and biotinylated proteins were recovered by using streptavidin-agarose beads (GE Healthcare). Beads were resuspended in Laemmli buffer, and bound proteins were separated by SDS-PAGE and detected by immunoblotting using mouse M2 anti-Flag (F1804; Sigma-Aldrich) to detect 5-HT2CR and mouse anti-transferrin receptor (Invitrogen) monoclonal antibody.
To detect interaction between BBS1 or other BBSome proteins and CD63, HEK 293 cells were transiently transfected with hemagglutinin (HA)-tagged BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, or BBS9 plasmids with control shRNA or Bbs2-shRNA. The cells were lysed 48 h after transfection with lysate buffer (50 mmol/L HEPES, pH 7.5, 150 mmol/L NaCl, 1 mmol/L MgCl2, 1 mmol/L CaCl2, 10 mmol/L NaF, 5 mmol/L EDTA, 1% Triton, 2 mmol/L sodium orthovanadate, and Roche cocktail protease inhibitor tablet). Cellular protein samples (800 μg) were subjected to immunoprecipitation assay with 5 μg anti-HA antibody (11815016001; Roche Applied Science). Immunocomplex was separated on a 9% SDS-PAGE gel, transferred to polyvinylidene fluoride membrane, and probed with anti-CD63 antibody (1:1,000) (EXOAB-CD63A-1; SBI System Biosciences), followed by a secondary anti-rabbit horse radish peroxidase goat antibody (1:10,000) (7074; Cell Signaling) or a goat anti-mouse horseradish peroxidase (1:10,000) (7076; Cell Signaling). Protein expression was visualized with the ECL detection kit (GE Healthcare) and exposed to film.
Gene Expression Analysis
Mice were sacrificed by CO2 asphyxiation, and the brain was quickly isolated and put on ice. The mediobasal hypothalamus was carefully dissected and total RNA isolated using the RNeasy Plus Mini Kit (Qiagen). Total RNA (2 μg in final volume of 40 μL) was used to synthesize first-strand cDNAs with the Super-Script preamplification system. Then, 10 μL cDNA and 0.4 mmol/L primers were added in a final volume of 25 μL PCR mixture (iQ SYBR Green supermix, Bio-Rad) and amplified in a iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad). The PCR conditions for all genes were as follows: denaturation for 5 min at 95°C, 40 cycles for 30 s at 95°C, and 30 s at 60°C. S18 rRNA expression was used as internal control to normalize mRNA expression of these genes. Primer sequences are provided in Supplementary Table 1.
RNAscope
To detect Bbs1 and POMC mRNA, the brain was freshly harvested and put into optimum cutting temperature solution. The brain was sectioned with a cryostat, and the brain sections were hybridized with specific probes purchased from Advanced Cell Diagnostics (491121 for Bbs1 and 314081-C2 for POMC). Processed brain sections were mounted using VectaShield mounting medium with DAPI. Images were visualized using confocal microscopy (LSM710; Zeiss) and analyzed using ImageJ software.
Immunofluorescence
Mice anesthetized with ketamine and xylazine were perfused with 50 mL PBS (5 mL/min), followed by 4% paraformaldehyde/HistoChoice Tissue Fixative (Amresco) in 50 mL PBS (2.5 mL/min) using a Harvard PHD 22/2000 Syringe Pump. The entire brain was excised and incubated in the same fixative overnight at 4°C. Fixed brains were washed three times with PBS and incubated in 30% sucrose/PBS overnight with one change of solution after 4–6 h of initial incubation. Brains were vibratome sectioned with 30-μm thickness. Immunochemistry was performed on brain sections using antibodies against POMC (1:250) (H-029-30; Phoenix Pharmaceuticals), phosphorylated Stat3 (1:1,000) (#9131S; Cell Signaling), 5-HT2CR (1:250) (RA24505; Neuromics), NPY2R (1:250) (14112; Neuromics), or adenylyl cyclase type III (1:250 dilution) (sc-32113 [N-14]; Santa Cruz Biotechnology), as described previously (20,21). For phosphorylated Stat3 staining, mice were fasted overnight and treated with leptin (2 mg/kg, i.p.) 1 h before sacrifice. The anti–5-HT2CR antibody was validated using cells expressing a GFP-tagged 5-HT2CR and brains of 5-HT2CR−/− mice (provided by Dr. J. Elmquist, University of Texas Southwestern Medical Center). Processed brain sections were mounted using VectaShield mounting medium with DAPI. Images were visualized using confocal microscopy (LSM710; Zeiss) and analyzed using ImageJ software.
IMCD3 and IMCD3-Bbs1−/− cells were transfected with GFP-tagged 5-HT2CR, NPY2R, or SSTR3, and cilia were stained with acetylated α-tubulin (sc-23950; Santa Cruz Biotechnology). To detect 5-HT2CR or BBS1 subcellular localization in IMCD3-Bbs1−/− cells, the cells were transfected with GFP-tagged 5-HT2CR or cotransfected with red fluorescent protein (RFP)-Bbs1 with GFP-Rab5, GFP-Rab7, and GFP-Golgi, and then stained with early endosome marker EEA1 (3288; Cell Signaling), Lysosomal marker Lamp1 (9091; Cell Signaling), endosome marker Cav1 (3267; Cell Signaling), Golgi marker RCAS1 (12290; Cell Signaling), or late endosome marker CD63 (EXOAB-CD63A-1; SBI System Biosciences). HEK 293 cells were transiently cotransfected with GFP-tagged 5-HT2CR with control shRNA or Bbs2-shRNA, and the receptor was visualized after 48 h of transfection.
Data Analysis
The data are expressed as means ± SEM and were analyzed using the Student t test or one- or two-way ANOVA. When ANOVA reached significance, a post hoc comparison was made using the Fisher test. A P < 0.05 value was considered statistically significant. Univariate regression modeling with body weight considered a covariate was used to assess resting metabolic rate data using SPSS software.
Results
Selective Deletion of the Bbs1 Gene in POMC Neurons Causes Obesity
In silico reanalysis of available single-cell RNA sequencing data sets from dissociated mouse hypothalamic cells (22) revealed that BBS genes are expressed in both POMC and AgRP neurons (Supplementary Fig. 1A and B). We have confirmed Bbs1 gene expression in POMC neurons using RNAscope (Supplementary Fig. 1C). Strikingly, fasting was found to cause contrasting changes in the expression of several BBS genes in POMC versus AgRP neurons (Supplementary Fig. 1A and B). To determine the relevance of the BBSome in these neuronal populations for energy homeostasis, we generated mice bearing selective deletion of the Bbs1 gene in POMC neurons (POMCCre/Bbs1fl/fl mice) by breeding the conditional Bbs1fl/fl mice with POMCCre mice. Further crossing with a reporter mouse model expressing fluorescent td-Tomato (ROSA), in a Cre-dependent manner, confirmed the Cre-mediated recombination in POMC neurons (Supplementary Fig. 2A and B). It should be noted that absence of the BBS1 protein disrupts BBSome formation and function (20). Furthermore and consistent with previous reports implicating the BBSome in leptin sensitivity and leptin receptor (LepRb) trafficking (20,23), we found that leptin-induced Stat3 activation was blunted in td-Tomato+ cells of the arcuate nucleus of POMCcre/Bbs1fl/fl mice (Supplementary Fig. 2C).
Body weight was comparable between young (4–7 weeks old) POMCCre/Bbs1fl/fl mice and littermate controls (Fig. 1A and B). However, the body weight of both males and females begin to diverge at ∼8 weeks of age, with POMCCre/Bbs1fl/fl mice gaining more weight than the controls. The divergence in body weight continued over time, with POMCCre/Bbs1fl/fl mice weighing 10–17 g more than the control mice at 25 weeks of age (Fig. 1A and B). The increase in body weight in POMCCre/Bbs1fl/fl mice was due to an increase in adiposity, as indicated by the increased fat mass (Fig. 1C and D and Supplementary Fig. 2D and E). Liver mass was elevated in POMCCre/Bbs1fl/fl mice relative to controls (females: 1.74 ± 0.15 vs. 1.12 ± 0.09 g, P < 0.05; males: 2.94 ± 0.3 vs. 1.71 ± 0.08 g, P < 0.05). However, there was no difference in lean mass between POMCCre/Bbs1fl/fl mice and controls (Fig. 1E).
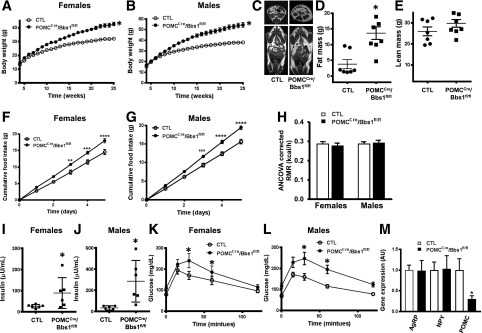
Figure 1.
Mice lacking the Bbs1 gene in POMC neurons develop obesity. Weekly body weights of female (A) and male (B) mice (n = 12–16/group: 12 males and 15 females for control [CTL] mice, 16 males and 16 females for POMCcre/Bbs1fl/fl mice). C–E: Fat mass and lean mass, measured by MRI (n = 7/group: three males and four females in each group). F and G: Cumulative daily food intake (n = 6–13/group: 6 males and 12 females for control and 6 males and 13 females for POMCcre/Bbs1fl/fl mice). H: Resting metabolic rate (RMR) (n = 4–6/group: six males and four females in each group), analyzed by univariate regression modeling (model, P < 0.001; body mass, P < 0.001; genotype, P = 0.912; sex, P = 0.501; interaction P = 0.453; body mass evaluated at 34.263 g; R2 = 0.772). I and J: Plasma insulin levels. K and L: GTT (n = 10–12/group: 5 males and 5 females for controls and 6 males and 6 females for POMCcre/Bbs1fl/fl mice). M: Hypothalamic mRNA levels of metabolic neuropeptides (n = 6/group: three males and three females in each group). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 vs. control group.
Consistent with the obesity phenotype, POMCCre/Bbs1fl/fl mice exhibited elevated food intake (Fig. 1F and G). In contrast, resting metabolic rate, corrected for body weight by univariate regression modeling (model P < 0.001, body weight P < 0.001), was comparable between POMCCre/Bbs1fl/fl mice and controls (Fig. 1H) (genotype P = 0.912, sex P = 0.501, interaction P = 0.453). These findings indicate that obesity in mice lacking the Bbs1 gene in POMC neurons is driven by hyperphagia.
Circulating insulin levels were elevated in POMCCre/Bbs1fl/fl mice (Fig. 1I and J). A significant increase in fasting blood glucose was also noted in POMCCre/Bbs1fl/fl mice (114.7 ± 7.7 mg/dL) compared with controls (77.1 ± 6.8 mg/dL, P < 0.05). Moreover, POMCCre/Bbs1fl/fl mice displayed glucose intolerance (Fig. 1K and L) and tended to be insulin resistant (Supplementary Fig. 2F and G).
Interestingly, disruption of the BBSome in POMC neurons led to a significant decrease in the expression of the hypothalamic anorexigenic Pomc gene without altering the expression of orexigenic Agrp and Npy genes (Fig. 1M). Of note, there was no difference in the expression pattern of these genes between male and female POMCCre/Bbs1fl/fl mice. These findings highlight the relevance of the BBSome in POMC neurons for the mechanisms that underlie metabolic regulation.
Mice Lacking the Bbs1 Gene in AgRP Neurons Are Obese
We previously reported that global BBS mice and mice lacking the Bbs1 gene in the nervous system or LepRb-expressing cells have normal expression of hypothalamic Agrp and Npy genes. This led us to hypothesize that loss of the BBSome in AgRP neurons does not contribute to the obesity phenotype observed in these mice. To test this directly, we generated mice lacking the Bbs1 gene selectively in AgRP neurons by breeding the Bbs1fl/fl mice with AgRPCre mice. As above, we used the td-Tomato (ROSA) reporter mouse model to confirm that AgRPCre drives Cre-recombinase, indicated by td-Tomato protein expression (Supplementary Fig. 3A).
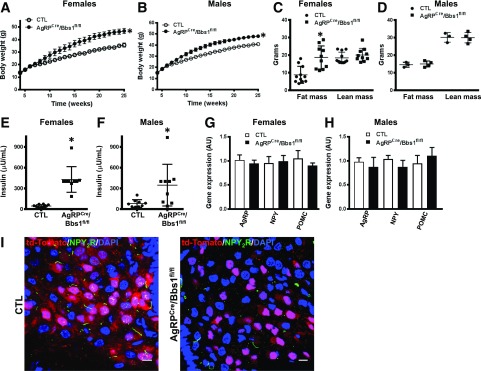
Surprisingly and in contrast to our expectation, mice lacking expression of the Bbs1 gene in AgRP neurons (AgRPCre/Bbs1fl/fl) developed obesity, as indicated by the significantly increased body weight, starting at ∼6 weeks of age (Fig. 2A and B). Notably, this increase in body weight was more pronounced in the females relative to the males, suggesting that loss of the BBSome in AgRP neurons may contribute to the sex difference in the development of obesity in BBS mice (24,25). Fat mass was increased in female but not male AgRPcre/Bbs1fl/fl mice (Fig. 2C and D and Supplementary Fig. 3B and C) without alteration in lean mass (Fig. 2C and D). Consistent with body weight, the increase in liver mass was more noticeable in female than in male AgRPcre/Bbs1fl/fl mice (females: 2.74 ± 0.46 vs. 1.58 ± 0.12 g, P < 0.05; males: 3.41 ± 0.58 vs. 2.45 ± 0.22 g, P = 0.06). Of note, plasma insulin levels were elevated in female and male AgRPcre/Bbs1fl/fl mice relative to control littermates (Fig. 3E and F).
Figure 2.
Bbs1 gene deletion in AgRP neurons causes obesity. Weekly body weights of female (A) and male (B) mice (n = 10–23/group: 19 males and 23 females for controls [CTL] and 15 males and 10 females for AgRPcre/Bbs1fl/fl mice). C and D: Fat and lean masses. E and F: Plasma insulin levels. G and H: Hypothalamic mRNA levels of metabolic neuropeptides (n = 6/group). I: Selective loss of ciliary localization of NPY2R in td-Tomato+ neurons of the hypothalamic arcuate nucleus of AgRPcre/Bbs1fl/fl mice. *P < 0.05 vs. control group. Scale bars, 10 µm.
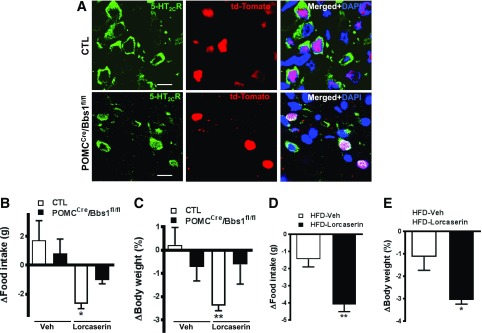
Figure 3.
The BBSome mediates surface expression of the 5-HT2CR in POMC neurons. A: Reduced membrane localization of the 5-HT2CR in td-Tomato+ cells of POMCCre/Bbs1fl/fl mice. B and C: Blunted lorcaserin-induced decrease in food intake and body weight in POMCCre/Bbs1fl/fl mice. CTL, control; Veh, vehicle. Lorcaserin decreases food intake (D) and body weight (E) in high-fat diet (HFD)–induced obese mice. *P < 0.05 and **P < 0.01 vs. control group. Scale bars, 10 µm.
There was no significant change in the expression of hypothalamic anorexigenic and orexigenic genes in AgRPcre/Bbs1fl/fl mice compared with control littermates, although expression of AgRP tended to be lower in AgRPcre/Bbs1fl/fl mice (Fig. 2G). These findings indicate that different molecular mechanisms may underlie the obesity phenotype of AgRPcre/Bbs1fl/fl mice versus POMCcre/Bbs1fl/fl mice.
Consistent with the previous report (17), localization of NPY2R in primary cilia of td-Tomato+ cells was substantially reduced in AgRPcre/Bbs1fl/fl mice (Fig. 2H). Of note, there was no difference in Npy2r gene expression between AgRPcre/Bbs1fl/fl mice (1.4 ± 0.4 arbitrary units [AU]) and controls (1.0 ± 0.3 AU). These findings confirm the importance of the BBSome for the trafficking of key metabolic receptors in hypothalamic neurons.
Plasma Membrane Localization of 5-HT2CR in POMC Neurons Required the BBSome
Next, we asked whether the BBSome is involved in the trafficking of other receptors that are important for energy homeostasis, such as the serotonin 5-HT2CR, which is present in POMC neurons (7,26). Using an antibody that recognizes specifically the 5-HT2CR, as confirmed by Western blot and immunohistochemistry in cells and arcuate nucleus of 5-HT2CR−/− null mice (Supplementary Fig. 4A–C), we found that in the arcuate nucleus of control mice, 5-HT2CR is predominantly present in the plasma membrane of td-Tomato+ cells as well as in some td-Tomato− cells (Fig. 3A). Notably, in POMCcre/Bbs1fl/fl mice, the localization of 5-HT2CR is altered selectively in td-Tomato+ cells, appearing stalled in the cytoplasm (Fig. 3A). Of note, 5-HT2CR does not localize in cilia of td-Tomato+ or td-Tomato− cells of the hypothalamic arcuate nucleus of control and POMCcre/Bbs1fl/fl mice. In contrast, the NPY2R localizes predominantly in neuronal cilia, and POMCcre/Bbs1fl/fl mice displayed reduced ciliary localization of this receptor in td-Tomato+ cells of the arcuate nucleus (Supplementary Fig. 5A and B). Hypothalamic mRNA levels of 5-HT2CR and NPY2R were not altered in POMCcre/Bbs1fl/fl mice (0.9 ± 0.1 and 1.2 ± 0.3 AU, respectively) relative to littermate controls (1.0 ± 0.1 AU for both). AgRPcre/Bbs1fl/fl mice also exhibit normal hypothalamic mRNA levels of 5-HT2CR (0.9 ± 0.1 AU) compared to controls (1.0 ± 0.2 AU). Interestingly, deletion of the Bbs1 gene does not appear to affect cilia formation, as indicated by the relatively normal cilia, stained with adenylyl cyclase type III, in the arcuate nucleus td-Tomato+ cells of POMCcre/Bbs1fl/fl mice (Supplementary Fig. 5B).
Based on the evidence above, we predicted that the metabolic effects evoked by activation of 5-HT2CR would be blunted in POMCcre/Bbs1fl/fl mice. Thus, we assessed the anorectic and weight-reducing responses induced by the 5-HT2CR agonist, lorcaserin. In control mice, lorcaserin decreases food intake and body weight (Fig. 3B and C). However, the responses evoked by lorcaserin are significantly blunted in POMCcre/Bbs1fl/fl mice. The reduced responses to lorcaserin in POMCcre/Bbs1fl/fl mice does not appear due to obesity but rather caused by loss of the BBSome. This is supported by the decrease in food intake and body weight in diet-induced obese mice treated with lorcaserin (Fig. 3D and E). Taken together, these data demonstrate the importance of the BBSome in POMC neurons in underlying serotonin-mediated control of energy homeostasis.
BBSome Deficiency Hinders Serotonin-Evoked Cellular Responses
To facilitate the analysis of the trafficking of the 5-HT2CR in vitro, we generated a GFP-tagged receptor (5-HT2CR–GFP). First, we used confocal microscopy to investigate the membrane localization of the 5-HT2CR in the IMCD3 cell line. Although 5-HT2CR–GFP was present in the plasma membrane, we were unable to detect 5-HT2CR–GFP in cilia (labeled with acetylated α-tubulin) (Fig. 4A), indicating that the 5-HT2CR is not targeted to the cilium. In contrast, we found that NPY2R and SSTR3 localize almost exclusively to cilia in IMCD3 cells (Supplementary Fig. 6A and B).
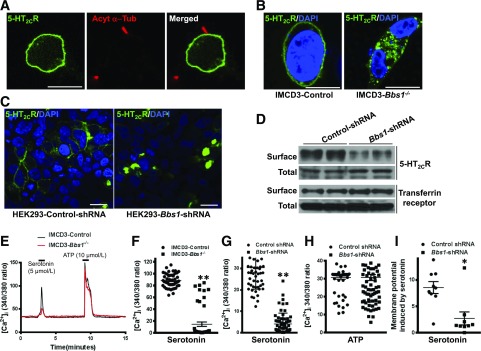
Figure 4.
The BBSome is required for the cellular responses evoked by the 5-HT2CR. A: Lack of localization of 5-HT2CR in cilia (labeled with acetylated α-tubulin [Acyt α-Tub]) in IMCD3 cells. B: Reduced membrane localization of 5-HT2CR in IMCD3-Bbs1−/− cells. Bbs1 gene silencing reduces the surface levels of 5-HT2CR, but not transferrin receptor, in HEK 293 cells by immunostaining and biotin labeling assay (C), followed by Western blotting (D). E and F: Reduced [Ca2+]i response to serotonin in IMCD3-Bbs1−/− cells. Bbs1 gene silencing decreases [Ca2+]i response to serotonin (G), but not ATP (H), and reduces the membrane potential evoked by serotonin (I) in HEK 293 cells. *P < 0.01 and **P < 0.0001 vs. control group. Scale bars, 10 μm.
Next, we tested whether loss of the BBSome interferes with the membrane localization of 5-HT2CR in IMCD3 cells. Interestingly, Bbs1 gene disruption using CRISPR/Cas9 reduced 5-HT2CR–GFP in the plasma membrane (Fig. 4B), confirming the importance of the BBSome for membrane localization of the 5-HT2CR. In addition, the NPY2R and SSTR3 both failed to localize to cilia in IMCD3-Bbs1−/− cells (Supplementary Fig. 6A and B), confirming the importance of the BBSome for the trafficking of these receptors to cilia. Consistent with the observation in POMC neurons, the 5-HT2CR in the IMCD3-Bbs1−/− cells was predominantly localized in unusual large cytoplasmic vesicles (Fig. 4B).
Like CRISPR/Cas9-mediated deletion in IMCD3 cells, shRNA-induced silencing of Bbs1 gene expression reduces the membrane localization of 5-HT2CR in HEK 293 cells (Fig. 4C). Most of the GFP-tagged 5-HT2CR localizes to the plasma membrane in the control cells (transfected with scrambled shRNA), whereas the localization of this receptor is mostly in vesicles in the Bbs1-shRNA–transfected HEK 293 cells. The reduced amount of 5-HT2CR in the plasma membrane of HEK 293 cells transfected with Bbs1-shRNA was further confirmed using a cell surface biotin labeling assay, followed by Western blotting (Fig. 4D). Importantly, total 5-HT2CR expression is not changed when the Bbs1 gene is silenced. In addition, the surface level of the transferrin receptor is not affected by Bbs1 gene silencing (Fig. 4D), demonstrating the specificity of the effects evoked by BBS1 loss on receptor trafficking to the plasma membrane.
We studied the functional consequence of altered 5-HT2CR trafficking in the absence of the BBSome by assessing the response of [Ca2+]i to serotonin. Compared with control cells, the serotonin-induced increase in [Ca2+]i is substantially decreased in IMCD3-Bbs1−/− cells (Fig. 4E and F) and HEK 293 cells transfected with Bbs1-shRNA (Fig. 4G). The leptin-evoked increase in [Ca2+]i is also blunted after BBSome disruption (Supplementary Fig. 7). However, the increase in [Ca2+]i evoked by ATP was not altered by disruption of the BBSome (Fig. 4E and H). We also examined the effects of silencing the Bbs1 gene on serotonin-induced membrane potential using patch-clamp recording. Membrane potential response to serotonin is dramatically reduced in HEK 293 cells transfected with Bbs1-shRNA compared with control cells (Fig. 4I). These results demonstrate that reduction in 5-HT2CR trafficking to the plasma membrane caused by disruption of the BBSome is associated with attenuated cellular responses evoked by serotonin.
Stalled 5-HT2CR in the Late Endosome in the Absence of the BBSome
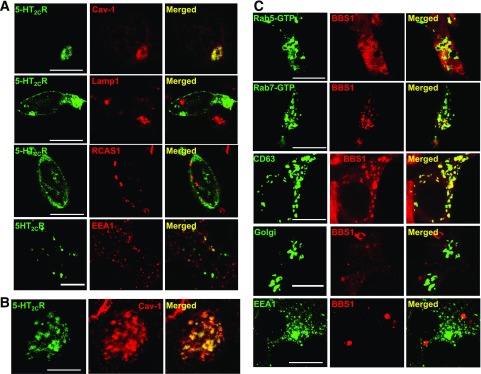
The reduced membrane localization of the 5-HT2CR without alteration in its total expression when the BBSome is disrupted indicates that the receptor is stuck in the secretory pathway. To understand the fate of 5-HT2CR in cells lacking the Bbs1 gene, we first studied the localization of the receptor in the secretory pathway using markers of key organelles, including caveolin-1 (Cav-1) (endosome), LAMP1 (lysosome), RSAC1 (Golgi), and EEA1 (early endosome) using IMCD3-Bbs1−/− cells. Interestingly, we observed that 5-HT2CR–GFP colocalizes predominantly with the endosome marker, Cav-1, when 5-HT2CR–GFP expression is low (Fig. 5A). This localization of 5-HT2CR–GFP and Cav-1 is only partial when the expression of 5-HT2CR–GFP is high (Fig. 5B).
Figure 5.
Localization of 5-HT2CR and BBS1 in the secretory pathway. A: GFP-tagged 5-HT2CR localizes in the endosome (Cav-1) but not in the lysosome (LAMP1), Golgi (RSAC1), or early endosome (EEA1) in IMCD3-Bbs1−/− cells. B: Colocalization of 5-HT2CR–GFP and Cav-1 is only partial when the expression of 5-HT2CR–GFP is high. C: RFP-tagged BBS1 colocalizes with GFP-tagged Rab5-GTP, Rab7-GTP, and CD63 (markers of the late endosome) but not with markers of Golgi or early endosome (EEA1) in HEK 293 cells. Scale bars, 10 μm.
We next analyzed the cellular localization of BBS proteins. For this, HEK 293 cells were transiently cotransfected with RFP-BBS1 along with GFP-tagged marker proteins of early endosome (Rab5 constitutive active form, Rab5–guanosine-5'-triphosphate [GTP]), late endosome (Rab7 constitutive active form Rab7-GTP or CD63), Golgi or early endosome (EEA1), and localization was visualized with confocal microscopy. We found that RFP-BBS1 colocalized with the late endosome marker proteins, Rab7-GTP and CD63, and partially colocalized with Rab5-GTP but not with Golgi or early endosome markers (Fig. 5C). Similarly, RFP-BBS7 colocalized with the late endosome proteins Rab7-GTP and CD63 (Supplementary Fig. 8A).
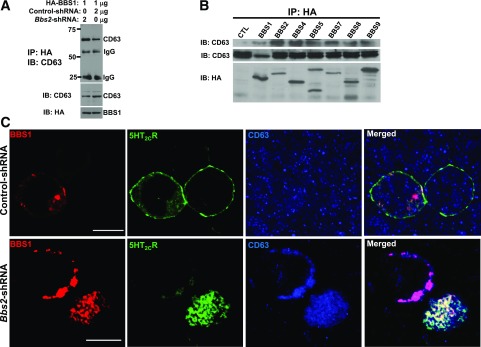
To test whether the BBSome proteins physically interact with CD63, HEK 293 cells were transiently transfected with HA-tagged BBS1 protein. BBS1 protein was then immunoprecipitated with HA-agarose beads, and the endogenous CD63 protein was detected with CD63 antibody by Western blot. As shown in Fig. 6A, CD63 protein is detected in a BBS-immunoprecipitated protein complex. Furthermore, silencing BBS2 increased BBS1 interaction with the CD63 compared with control shRNA (Fig. 6A), indicating the specificity of their interaction. We then tested whether other subunits of the BBSome also interact with the CD63 in HEK 293 cells. We found that in addition to BBS1, other BBSome proteins, including BBS2, BBS4, BBS5, BBS7, BBS8, and BBS9, interact with the endogenous CD63 (Fig. 6B). On one hand, in HEK 293 cells overexpressing BBS1 or BBS7 proteins, endogenous CD63 protein colocalizes with BBS1 or BBS7, whereas 5-HT2CR is mostly present in the plasma membrane (Fig. 6C and Supplementary Fig. 8B). On the other hand, disruption of the BBSome by silencing BBS2 in HEK 293 cells causes 5-HT2CR to colocalize with BBS1 or BBS7 with the endogenous CD63 protein. In contrast, 5-HT2CR appears in the plasma membrane when the BBSome complex is intact (Fig. 6C and Supplementary Fig. 8B). These results indicate that loss of the BBSome causes 5-HT2CR to stall in the late endosome, possibly forming a complex with CD63 and the remaining BBS proteins.
Figure 6.
BBSome proteins interact with CD63. A: Coimmunoprecipitation of the HA-tagged BBS1 with the endogenous CD63 (late endosome marker) is enhanced after Bbs2 gene silencing in HEK 293 cells. B: BBSome proteins interact with CD63 in a coimmunoprecipitation assay. C: Bbs2 gene silencing promotes localization of BBS1 with 5-HT2CR in the late endosome. Scale bars, 10 μm. IB, immunoblot; IP, immunoprecipitation.
Discussion
There are several new findings in the current study. First, we show that targeted disruption of the BBSome by deleting the Bbs1 gene in POMC neurons leads to obesity. We demonstrate that obesity in these mice is associated with an increase in food intake. Second, we provide evidence that mice lacking the BBSome in AgRP neurons also display an increase in body weight and fat mass. Third, we identify an important role for the BBSome in mediating the trafficking of the 5-HT2CR in POMC neurons. We demonstrate that loss of the BBSome reduces surface expression of the 5-HT2CR and interferes with the ability of serotonin to modulate cellular processes. In absence of the BBSome, 5-HT2CR becomes stalled in the late endosome of the secretory pathway. Together, these findings provide mechanistic insights into the contribution of the hypothalamic BBSome to the control of energy balance and handling of metabolic receptors.
Substantial evidence points to the importance of POMC and AgRP neurons as first-order neurons in the neurocircuit that controls metabolic functions (3,6). These neurons are equipped with receptors for various metabolic signals, including serotonin, and regulate the downstream neurocircuitry to enact appropriate and consequential metabolic and behavioral responses to maintain energy balance. Several cellular processes within POMC and/or AgRP neurons have previously been shown to have important implications for metabolic control. This includes gene expression (22), protein translation and processing (27,28), endoplasmic reticulum stress (29,30), and mitochondria function (31–34). The changes in the trafficking of metabolic receptors evoked by disruption of the BBSome in POMC and AgRP neurons extend the cellular processes within these neurons that influence energy homeostasis.
We demonstrate that absence of the BBSome substantially reduces surface expression of the 5-HT2CR in cultured cells and POMC neurons. In BBSome-deficient cells the 5-HT2CR is held up in large vesicles in the cytoplasm. We previously demonstrated that the LepRb is also stalled in large cellular vesicles when the BBSome is disrupted (23). To determine the identity of these vesicles, we analyzed the localization of the BBSome proteins and 5-HT2CR in key organelles of the secretary pathway. We found that BBS proteins colocalize with the late endosome markers and partially colocalize with the early endosome marker. Importantly, we have identified the late endosome as the vesicle where the 5-HT2CR is held up when the BBSome is disrupted. Future studies will determine what makes the receptors stall in this stage of the secretory pathway after loss of the BBSome. Additional studies are also needed to assess the possibility of overcoming the mistrafficking of metabolic receptors when the BBSome is disrupted.
Obesity is a major phenotype criterion in patients with BBS (15,35). Mice carrying deletion or mutations in BBS genes also display an obesity phenotype that is driven by hyperphagia and a decrease in energy expenditure (23,36). These findings point to a fundamental role of BBS proteins in the regulation of appetite and energy balance. This has triggered tremendous interest in understanding the mechanisms underlying the control of energy homeostasis by the BBSome. We previously demonstrated that loss of the BBSome in the nervous system or LepRb-containing cells, but not in adipocytes, largely recapitulated the increase in body weight and food intake associated with BBS (20). Our findings that BBSome deficiency in POMC neurons or AgRP neurons is sufficient to increase body weight and adiposity demonstrate the importance of the BBSome in these neuronal populations. The decreased hypothalamic POMC mRNA levels in mice lacking the BBS gene globally or in defined neuronal populations, such as POMC neurons, is consistent with the defective signaling of receptors that regulate Pomc gene expression, including LepRb and NPY2R (37,38). This reduction in Pomc gene expression may contribute to the hyperphagia and obesity associated with BBS. Thus, the obesity associated with BBS may emanate from loss of the BBSome in hypothalamic neurons.
We also speculate that our findings may have implications for understanding the abnormalities associated with common obesity. This is supported by the demonstration that variants of BBS genes increase susceptibility to common human obesity (12–14). Of note, a reduction in surface expression of hypothalamic receptors was reported in animals with dietary obesity (39). A possible contribution of the BBSome to the abnormalities associated with common obesity warrants further investigation.
One limitation of our study relates to the use of mice expressing constitutive POMCCre and AgRPCre to delete the Bbs1 gene in these neuronal populations since embryonic expression of Cre may have deleted the Bbs1 gene in peripheral tissues and cells that do not necessarily differentiate to POMC or AgRP neurons. Indeed, a subset of POMC-expressing progenitor cells are known to differentiate to other cell types, including AgRP neurons (40), which can result in misleading feeding responses when constitutive POMCCre mice are used (41). It should be noted, however, that constitutive POMCCre and AgRPCre were used successfully to dissociate the contribution of POMC versus AgRP neurons to leptin regulation of regional autonomic nerve activity (21). Nonetheless, the recent development of inducible POMCCre (26) and AgRPCre mice (42) that allow circumventing the issues related to constitutive Cre activation will be useful to test the effects of Bbs1 gene deletion in POMC and AgRP neurons of adult mice.
The use of in vitro systems, including a kidney cell line (HEK 293), to understand the neuronal mechanisms of obesity in our conditional mouse models can also be viewed as a limitation to our studies. However, in vitro studies are very useful to examine the biology of the BBSome and gain mechanistic insights into its function. Importantly, we provide evidence showing that the in vitro data are recapitulated in vivo, as indicated by the similar defects in the trafficking of NPY2R and 5-HT2CR evoked by loss of the BBSome in cultured cells and hypothalamic neurons.
In conclusion, our findings point to the BBSome in POMC and AgRP neurons as a critical regulator of energy homeostasis. Our results enhance our understanding of the cellular and molecular processes that underlie obesity in BBS with potential implications for common obesity.
Supplementary Material
Article Information
Funding. This work was supported by funds from the National Institutes of Health (grant NS-096246 to Y.M.U., HL-134850 to J.L.G., EY-011298 to V.C.S., and HL-084207 to K.R.), the American Heart Association (18EIA33890055 to J.L.G. and 14EIA18860041 to K.R.), and the University of Iowa Fraternal Order of Eagles Diabetes Research Center to K.R. The Zeiss LSM710 in the University of Iowa’s Central Microscopy Research Facilities used for confocal imaging is funded by the National Institutes of Health (grant 1S10-RR-025439-01).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. D.-F.G. designed and performed the experiments and wrote the manuscript. Z.L. performed the calcium experiments. Y.W. performed the electrophysiology experiments. C.S. created the knockout cell line. D.R.T. measured body composition by MRI. G.B.R., Y.M.U., J.L.G., and V.C.S. helped with experimental design and data interpretation. J.L.G. measured energy expenditure. K.R. conceptualized, analyzed, and interpreted studies and wrote the manuscript. K.R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1088/-/DC1.
References
- 1.Willer CJ, Speliotes EK, Loos RJF, et al.; Wellcome Trust Case Control Consortium; Genetic Investigation of ANthropometric Traits Consortium . Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009;41:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locke AE, Kahali B, Berndt SI, et al.; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature 2006;443:289–295 [DOI] [PubMed] [Google Scholar]
- 4.Elmquist JK, Elias CF, Saper CB. From lesions to leptin: hypothalamic control of food intake and body weight. Neuron 1999;22:221–232 [DOI] [PubMed] [Google Scholar]
- 5.Cui H, López M, Rahmouni K. The cellular and molecular bases of leptin and ghrelin resistance in obesity. Nat Rev Endocrinol 2017;13:338–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andermann ML, Lowell BB. Toward a wiring diagram understanding of appetite control. Neuron 2017;95:757–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wyler SC, Lord CC, Lee S, Elmquist JK, Liu C. Serotonergic control of metabolic homeostasis. Front Cell Neurosci 2017;11:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nachury MV, Loktev AV, Zhang Q, et al. . A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 2007;129:1201–1213 [DOI] [PubMed] [Google Scholar]
- 9.Loktev AV, Zhang Q, Beck JS, et al. . A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell 2008;15:854–865 [DOI] [PubMed] [Google Scholar]
- 10.Seo S, Baye LM, Schulz NP, et al. . BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A 2010;107:1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Yu D, Seo S, Stone EM, Sheffield VC. Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet-Biedl syndrome protein complex, the BBSome. J Biol Chem 2012;287:20625–20635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benzinou M, Walley A, Lobbens S, et al. . Bardet-Biedl syndrome gene variants are associated with both childhood and adult common obesity in French Caucasians. Diabetes 2006;55:2876–2882 [DOI] [PubMed] [Google Scholar]
- 13.Lim ET, Liu YP, Chan Y, et al.; Go-T2D Consortium . A novel test for recessive contributions to complex diseases implicates Bardet-Biedl syndrome gene BBS10 in idiopathic type 2 diabetes and obesity. Am J Hum Genet 2014;95:509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendricks AE, Bochukova EG, Marenne G, et al.; Understanding Society Scientific Group; EPIC-CVD Consortium; UK10K Consortium . Rare variant analysis of human and rodent obesity genes in individuals with severe childhood obesity. Sci Rep 2017;7:4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo DF, Rahmouni K. Molecular basis of the obesity associated with Bardet-Biedl syndrome. Trends Endocrinol Metab 2011;22:286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H, White SR, Shida T, et al. . The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 2010;141:1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loktev AV, Jackson PK. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep 2013;5:1316–1329 [DOI] [PubMed] [Google Scholar]
- 18.Siljee JE, Wang Y, Bernard AA, et al. . Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet 2018;50:180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starks RD, Beyer AM, Guo DF, et al. . Regulation of insulin receptor trafficking by Bardet Biedl syndrome proteins. PLoS Genet 2015;11:e1005311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo DF, Cui H, Zhang Q, et al. . The BBSome controls energy homeostasis by mediating the transport of the leptin receptor to the plasma membrane. PLoS Genet 2016;12:e1005890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell BB, Harlan SM, Morgan DA, Guo DF, Cui H, Rahmouni K. Differential contribution of POMC and AgRP neurons to the regulation of regional autonomic nerve activity by leptin [published correction appears in Mol Metab 2018;14:158]. Mol Metab 2018;8:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry FE, Sugino K, Tozer A, Branco T, Sternson SM. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. eLife 2015;4:e09800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo S, Guo DF, Bugge K, Morgan DA, Rahmouni K, Sheffield VC. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum Mol Genet 2009;18:1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fath MA, Mullins RF, Searby C, et al. . Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum Mol Genet 2005;14:1109–1118 [DOI] [PubMed] [Google Scholar]
- 25.Davis RE, Swiderski RE, Rahmouni K, et al. . A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci U S A 2007;104:19422–19427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berglund ED, Liu C, Sohn JW, et al. . Serotonin 2C receptors in pro-opiomelanocortin neurons regulate energy and glucose homeostasis [published correction appears in J Clin Invest 2014;124:1868]. J Clin Invest 2013;123:5061–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Creemers JW, Pritchard LE, Gyte A, et al. . Agouti-related protein is posttranslationally cleaved by proprotein convertase 1 to generate agouti-related protein (AGRP)83-132: interaction between AGRP83-132 and melanocortin receptors cannot be influenced by syndecan-3. Endocrinology 2006;147:1621–1631 [DOI] [PubMed] [Google Scholar]
- 28.Yasuda A, Jones LS, Shigeri Y. The multiplicity of post-translational modifications in pro-opiomelanocortin-derived peptides. Front Endocrinol (Lausanne) 2013;4:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Zhang G, Zhang H, Karin M, Bai H, Cai D. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell 2008;135:61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams KW, Liu T, Kong X, et al. . Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab 2014;20:471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietrich MO, Liu ZW, Horvath TL. Mitochondrial dynamics controlled by mitofusins regulate Agrp neuronal activity and diet-induced obesity. Cell 2013;155:188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneeberger M, Dietrich MO, Sebastián D, et al. . Mitofusin 2 in POMC neurons connects ER stress with leptin resistance and energy imbalance. Cell 2013;155:172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santoro A, Campolo M, Liu C, et al. . DRP1 suppresses leptin and glucose sensing of POMC neurons. Cell Metab 2017;25:647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez S, Gómez-Valadés AG, Schneeberger M, et al. . Mitochondrial dynamics mediated by mitofusin 1 is required for POMC neuron glucose-sensing and insulin release control. Cell Metab 2017;25:1390–1399.e6 [DOI] [PubMed] [Google Scholar]
- 35.Mujahid S, Hunt KF, Cheah YS, et al. . The endocrine and metabolic characteristics of a large Bardet-Biedl syndrome clinic population. J Clin Endocrinol Metab 2018;103:1834–1841 [DOI] [PubMed] [Google Scholar]
- 36.Rahmouni K, Fath MA, Seo S, et al. . Leptin resistance contributes to obesity and hypertension in mouse models of Bardet-Biedl syndrome. J Clin Invest 2008;118:1458–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz MW, Seeley RJ, Woods SC, et al. . Leptin increases hypothalamic pro-opiomelanocortin mRNA expression in the rostral arcuate nucleus. Diabetes 1997;46:2119–2123 [DOI] [PubMed] [Google Scholar]
- 38.Sainsbury A, Schwarzer C, Couzens M, et al. . Important role of hypothalamic Y2 receptors in body weight regulation revealed in conditional knockout mice. Proc Natl Acad Sci U S A 2002;99:8938–8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irani BG, Dunn-Meynell AA, Levin BE. Altered hypothalamic leptin, insulin, and melanocortin binding associated with moderate-fat diet and predisposition to obesity. Endocrinology 2007;148:310–316 [DOI] [PubMed] [Google Scholar]
- 40.Padilla SL, Carmody JS, Zeltser LM. Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med 2010;16:403–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Q, Krolewski DM, Moore S, et al. . Uneven balance of power between hypothalamic peptidergic neurons in the control of feeding. Proc Natl Acad Sci U S A 2018;115:E9489–E9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Liu C, Uchida A, et al. . Arcuate AgRP neurons mediate orexigenic and glucoregulatory actions of ghrelin. Mol Metab 2013;3:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.