Depression increases the risk of conversion from amnestic mild cognitive impairment (aMCI) to Alzheimer’s disease. Xu et al. show that the polygenic risk score for major depressive disorder-specific genetic variants predicts conversion from non-depressed aMCI to Alzheimer’s disease. The underlying mechanisms may involve modulation of hippocampal development and amyloid-beta binding.
Keywords: amnesic mild cognitive impairment conversion to Alzheimer’s disease, hippocampal volume, major depressive disorder, neurobiological mechanisms, polygenic risk scores
Abstract
Depression increases the conversion risk from amnestic mild cognitive impairment to Alzheimer’s disease with unknown mechanisms. We hypothesize that the cumulative genomic risk for major depressive disorder may be a candidate cause for the increased conversion risk. Here, we aimed to investigate the predictive effect of the polygenic risk scores of major depressive disorder-specific genetic variants (PRSsMDD) on the conversion from non-depressed amnestic mild cognitive impairment to Alzheimer’s disease, and its underlying neurobiological mechanisms. The PRSsMDD could predict the conversion from amnestic mild cognitive impairment to Alzheimer’s disease, and amnestic mild cognitive impairment patients with high risk scores showed 16.25% higher conversion rate than those with low risk. The PRSsMDD was correlated with the left hippocampal volume, which was found to mediate the predictive effect of the PRSsMDD on the conversion of amnestic mild cognitive impairment. The major depressive disorder-specific genetic variants were mapped into genes using different strategies, and then enrichment analyses and protein–protein interaction network analysis revealed that these genes were involved in developmental process and amyloid-beta binding. They showed temporal-specific expression in the hippocampus in middle and late foetal developmental periods. Cell type-specific expression analysis of these genes demonstrated significant over-representation in the pyramidal neurons and interneurons in the hippocampus. These cross-scale neurobiological analyses and functional annotations indicate that major depressive disorder-specific genetic variants may increase the conversion from amnestic mild cognitive impairment to Alzheimer’s disease by modulating the early hippocampal development and amyloid-beta binding. The PRSsMDD could be used as a complementary measure to select patients with amnestic mild cognitive impairment with high conversion risk to Alzheimer’s disease.
Introduction
The amnestic mild cognitive impairment (aMCI) is a state of cognitive deficit that is not severe enough to fulfil the criteria of dementia (Winblad et al., 2004) and shows a much higher probability of developing into Alzheimer’s disease (Palmer et al., 2008). Identifying biological measures with the potential to predict the conversion from aMCI to Alzheimer’s disease is clinically important for early interventions of Alzheimer’s disease. In the past decades, a variety of demographic (Tokuchi et al., 2014), clinical (Mazzeo et al., 2016), cognitive (Julayanont et al., 2014), neuroimaging (Yuan et al., 2009), and genetic measures (Rodriguez-Rodriguez et al., 2013; Adams et al., 2015) have been proposed as candidate measures for the prediction. Among these measures, a lifetime history of major depressive disorder (MDD) (Jorm, 2001), the presence of depressive symptom (Kida et al., 2016; Mourao et al., 2016; Sacuiu et al., 2016; Barca et al., 2017), or the coexistence of a diagnosis of MDD (Modrego and Ferrandez, 2004) has been found to increase the conversion risk from aMCI to Alzheimer’s disease, despite of non-significant findings (Palmer et al., 2010; De Roeck et al., 2016). However, few studies have investigated why depression could increase the conversion risk from aMCI to Alzheimer’s disease.
Besides social-psychological factors, such as stressful life events (Park et al., 2015) and interpersonal dysfunction (Hames et al., 2013), genome-wide association studies (GWAS) reveal a role of genetic factors in MDD (Ripke et al., 2013). In terms of genetics, one possibility for the association between MDD and the conversion of aMCI is that MDD and Alzheimer’s disease may share common genetic variants. This idea is supported by candidate gene studies (Skoog et al., 2015; Ye et al., 2016), but is not supported by the polygenic score analyses (Bulik-Sullivan et al., 2015a; Power et al., 2017). An alternative possibility is that the MDD- and Alzheimer’s disease-specific genetic variants may affect the same biological pathways, processes, cells, or structures via different mechanisms at different developmental periods. Thus, this study aimed to investigate if the cumulative MDD-specific genetic risk could predict the conversion from aMCI to Alzheimer’s disease. If so, we explored the possible neurobiological mechanisms underlying the predictive effect further.
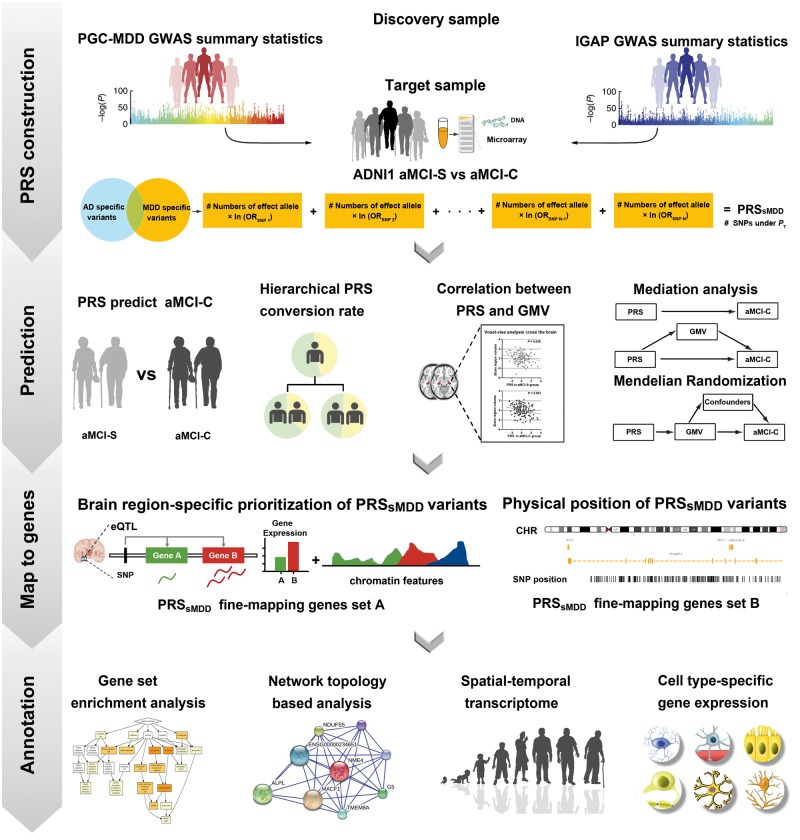
The cumulative genetic risk for MDD could be assessed by the polygenic risk scores (PRS) for this disorder (PRSMDD) (Milaneschi et al., 2016) and the cumulative genetic risk for Alzheimer’s disease could be evaluated by the PRSAD (Sabuncu et al., 2012). Although many studies have used the PRSMDD to predict MDD (Holmes et al., 2012; Milaneschi et al., 2016; Gibson et al., 2017; Qiu et al., 2017) and applied the PRSAD to predict Alzheimer’s disease (Escott-Price et al., 2015; Mormino et al., 2016) and the conversion from aMCI to Alzheimer’s disease (Rodriguez-Rodriguez et al., 2013; Adams et al., 2015), no studies have used the PRSMDD to predict the conversion from aMCI to Alzheimer’s disease. To exclude the possibility that the predictive effect of the PRSMDD on the conversion is driven by genetic variants common to MDD and Alzheimer’s disease, we only used genetic variants specific to MDD to calculate PRSsMDD by excluding common genetic variants of the two disorders. If a significant predictive effect existed, we investigated neuroanatomical substrates underlying such a prediction. The identified genetic variants were fine-mapped into genes using different strategies, and then enrichment and protein–protein interaction network analyses were performed to identify potential functions of these genes. Temporal- and cell type-specific expression analyses were finally used to explore in which developmental periods and cell types these genes are significantly expressed. A schematic summary of the study design is shown in Fig. 1.
Figure 1.
A schematic summary of the study design. ADNI = Alzheimer’s Disease Neuroimaging Initiative; CHR = chromosome; eQTL = expression quantitative trait loci; GMV = grey matter volume; GWAS = genome-wide association studies; IGAP = International Genomics of Alzheimer’s Project; OR = odd ratios; PGC-MDD = major depressive disorder working group of Psychiatric Genomics Consortium; PT = P-values threshold of genome-wide association studies.
Materials and methods
Discovery and target samples
The PRS calculation requires a discovery sample and a target sample. The discovery sample was used to identify effect size of a set of genetic variants that were nominally associated with the disease status at a predefined P-value. Then, the PRS was calculated for each subject in the target sample to estimate cumulative genetic risk of this subject for the disease. GWAS data of the Psychiatric Genomics Consortium (PGC) (Sullivan, 2010) and International Genomics of Alzheimer’s Project (IGAP) (Lambert et al., 2013) were used as the discovery samples to calculate PRSMDD and PRSAD, respectively. The target sample included 398 non-depressed (with a geriatric depression scale < 6) aMCI patients provided by the first stage of Alzheimer’s disease Neuroimaging Initiative (ADNI-1). The detailed information about the discovery and target samples is provided in the Supplementary material.
Genotyping
For the 757 subjects from ADNI-1, the genome-wide single-nucleotide polymorphisms (SNPs) were genotyped using the Illumina Human610-Quad Bead chip.
Quality control
In individual-level quality control, we excluded subjects with a missing genotyping rate >0.05, sex inconsistency, possible relative relationships, and being European population outliers identified by multidimensional scaling (MDS). In SNP-level quality control, we excluded SNPs with a missing call rate >0.05, minor allele frequency <0.01, significant deviation from Hardy-Weinberg equilibrium, and ambiguous strand. Finally, imputation was performed using MaCH (Li et al., 2010) and MiniMac (Howie et al., 2012). Detailed information of quality control and imputation is shown in the Supplementary material.
Polygenic risk score calculation
The PRS is used to assess the cumulative genetic risk for a certain disorder (Purcell et al., 2009). In the discovery sample, we calculated the association between SNPs and disease status at a predefined P threshold (PT). Under a certain PT, we then removed the effects of SNPs in linkage disequilibrium (LD) in each clumped region (excluding SNPs with r2 > 0.25 within a 250-kb window) and selected the index SNPs with the most significant P-value from each clumped association region based on the discovery sample. Thus, we obtained the information of the risk alleles and effect sizes of the index SNPs for each PT value. In the target sample, the PRS was calculated for each individual as the sum of the count of risk alleles multiplied by the corresponding effect sizes (natural log of the odds ratio) across these index SNPs.
To identify the PT value that could generate PRS with the best prediction for the conversion from aMCI to Alzheimer’s disease, the PRSice software (http://prsice.info) (Euesden et al., 2015) was used to generate 1000 PRS values for PT ranging from 0.001 to 1 with an increment of 0.001, while controlling for sex, age and educational years at baseline, the number of APOE ɛ4, and the first four MDS components for population stratification. PT = 1 indicates that all SNPs of the discovery sample are included to calculate the PRS in the target sample. By evaluating the predictive abilities, we could obtain the best PT values for calculating PRSMDD and PRSAD in the target sample. Using the best PT values, we could obtain the risk alleles and effect sizes of the index SNPs for calculating the PRSMDD and PRSAD in the target sample. The PRSMDD and PRSAD were computed for each aMCI patient and then z-transformed for visualization.
To exclude the possibility that the predictive effect of PRSMDD on the aMCI conversion is driven by genetic variants common to MDD and Alzheimer’s disease, we calculated PRS specific to MDD (PRSsMDD) only using genetic variants specific to MDD by excluding common variants of the two disorders. In the same way, we also calculated PRS specific to Alzheimer’s disease (PRSsAD). LD score regression (Bulik-Sullivan et al., 2015b) and co-localization analyses (Giambartolomei et al., 2014; Pickrell et al., 2016) were additionally performed to validate the specificity of index SNPs to MDD rather than Alzheimer’s disease (Supplementary material).
Image acquisition
Subjects were scanned with a standardized MRI protocol developed for ADNI (Jack et al., 2008). Details about the rationale and development of the standardized MRI datasets have been previously described (Wyman et al., 2013). The high resolution structural MRI data were acquired at 59 sites using 1.5 T MRI scanners with a sagittal 3D magnetization prepared rapid acquisition gradient echo sequence (http://adni.loni.ucla.edu).
Grey matter volume calculation
All structural images were visually checked by two radiologists. The preprocessing processes of structural images included segmentation, normalization, modulation, and smoothing (Supplementary material). The specific reasons for exclusion for patients with aMCI are shown in Supplementary Table 1. Finally, 322 aMCI patients with qualified genotyping and neuroimaging data were included and divided into conversion (aMCI-C, n = 187) and stable (aMCI-S, n = 135) groups. The demographic and genetic data of these patients are shown in Table 1.
Table 1.
Demographic and genetic characteristics of the target sample
| Demographic variables | aMCI-S (n = 135) | aMCI-C (n = 187) | Statistics | P |
|---|---|---|---|---|
| Age at baseline, y | 75.64 (7.16) | 74.50 (7.21) | 1.99 | 0.16 |
| APOE ɛ4 carriersa, n | 62 | 120 | 10.62 | 1.10 × 10−3 |
| Educational years | 15.50 (3.12) | 15.73 (2.89) | 0.46 | 0.50 |
| Males, n | 90 | 120 | 0.22 | 0.37 |
| GDS | 1.63 (1.38) | 1.59 (1.36) | 0.07 | 0.79 |
| PRSADb | −0.41 (0.88) | 0.30 (0.98) | 44.04 | 1.37 × 10−10 |
| PRSMDDb | −0.20 (0.97) | 0.28 (0.97) | 19.35 | 1.48 × 10−5 |
| PRSsADb | −0.40 (0.88) | 0.29 (0.98) | 42.59 | 2.65 × 10−10 |
| PRSsMDDb | −0.16 (0.97) | 0.22 (1.00) | 11.96 | 6.18 × 10−4 |
| PRSsMDD+ADb | −0.40 (0.89) | 0.31 (0.98) | 43.63 | 1.29 × 10−10 |
Data are shown as mean (SD). AD = Alzheimer’s disease; GDS = Geriatric Depression Scale.
aAPOE ɛ4 carriers include subjects with one or two copies of ɛ4 allele at the APOE locus.
bThe PRS are z-transformed for visualization.
P-values in bold indicate there are significant differences between groups.
Statistical analyses
Demographic data
The demographic data were analysed using the Statistical Package for the Social Sciences version 18.0 (SPSS Inc., Chicago, Illinois, USA). The chi-square or t-test was used to compare differences in sex, age, educational years, geriatric depression scale, and PRS measures between the aMCI-S and aMCI-C groups.
Polygenic risk analysis
The logistic regression was used to predict odds of the aMCI-C using the PRS calculated under each of the 1000 PT thresholds. The permutation test (P < 0.05) was used to correct multiple comparisons. Nagelkerke’s pseudo R2 was calculated to measure the proportion of variance explained by the PRS. A Cox proportional hazard model was used to explore the relations between the PRS and the conversion at the different time points, with the age and educational years at baseline, the sex, the number of APOE ɛ4, and the PRS as independent variables. For the best-fitting PRS, hazard ratios (HRs) for these variables were calculated using the prediction models.
The 322 aMCI patients were bisected according to their PRSsMDD or PRSsAD; we defined 161 patients with relatively low PRS as low PRS group and another 161 patients as high PRS group. The chi-square test was used to compare differences in conversion rate of aMCI among the four hierarchical risk groups (double low risk, low PRSsAD but high PRSsMDD, high PRSsAD but low PRSsMDD, and double high risk groups). The 322 aMCI patients were also trisected according to their PRSsMDD or PRSsAD, the bottom third (107 patients) was defined as the low-risk group, the middle third (107 patients) as the middle-risk group, and the upper third (108 patients) as the high-risk group. We also compared differences in conversion rate among the nine hierarchical risk groups.
Imaging data
The voxel-wise multiple regression analysis was performed to identify brain regions whose grey matter volumes were significantly correlated with PRSsMDD using Statistical Parametric Mapping software package (SPM8, http://www.fil.ion.ucl.ac.uk/spm), while controlling for the effect of neuroimaging sites. Multiple comparisons were corrected using a voxel-level family-wise error (FWE) method (Pc < 0.05, cluster size >200 voxels). The grey matter volumes of brain regions with significant correlations with the PRSsMDD were extracted for further analysis.
Mediation analysis and Mendelian randomization
The mediation analysis was performed to test whether the grey matter volume of each significant region mediates the association between PRSsMDD and the conversion of aMCI (Preacher and Hayes, 2008; Hayes, 2013). The PRSsMDD was defined as an independent variable, the grey matter volume of each significant region as a mediator variable, and the aMCI group assignment (aMCI-S versus aMCI-C) as a binary dependent variable. In addition, both a conventional two-stage Mendelian randomization method and a Mendelian randomization-Egger sensitivity analysis (http://www.mendelianrandomization.com/index.php/software-code) were applied using PRSsMDD as instrumental variable to make causal inference between the grey matter volume of each significant region and aMCI conversion (Burgess and Thompson, 2013, 2015; Burgess, 2014) (Supplementary material).
Fine-mapping MDD-specific genetic variants into genes
The context-dependent epigenomic weighting for prioritization of variants (CEPIP) (Li et al., 2017) was used to estimate the brain region-specific regulatory probability of each PRSsMDD genetic variant by integrating the expression quantitative trait loci (eQTLs) (Brown et al., 2013) and epigenomic features in the specific human brain tissues (Kundaje et al., 2015). The PRSsMDD genetic variants were weighted by the brain region-specific regulatory probability, and were then fine-mapped into genes using gene-based association test (GATES) (within a 5-kb window) in the human reference assembly (GRch37/hg19) (Li et al., 2011). As a result, we could identify genes that increase MDD susceptibility through brain region-specific functionally regulatory mechanisms. To exclude the bias derived from introducing brain region-specific biological information, we also fine-mapped PRSsMDD genetic variants into genes only based on physical position of each variant (within a 5-kb window) and re-performed the analyses.
Gene enrichment analyses
The functions of these genes were annotated by identifying significant enrichments using the WebGestalt (http://www.webgestalt.org/option.php) (Wang et al., 2017) and Gorilla (http://cbl-gorilla.cs.technion.ac.il) (Eden et al., 2007, 2009). The Benjamini and Hochberg method for false discovery rate (FDR-BH correction) (qc < 0.05) was applied to correct for multiple comparisons. The reference gene list included 18 774 genes derived from fine-mapping of all imputed SNPs of PGC-MDD and ADNI datasets.
Network topology-based analysis
The WebGestalt software was also used to perform the network topology-based analysis based on the human protein–protein interaction (PPI) of the Biological General Repository for Interaction Datasets (BIOGRID). All fine-mapping genes were firstly mapped to the PPI network of the BIOGRID; and the tightly connected genes (a portion of the candidate genes) formed a new PPI network based on the assumption that the mechanistically important genes are likely to form tightly connected clusters whereas other genes tend to be randomly distributed in the network. The network topology-based analysis could create a score for each gene in the PPI network of the BIOGRID based on its overall proximity to the seed genes, where the proximity was measured by the random walk analysis (Kohler et al., 2008). In the PPI network of the BIOGRID, the top 10 genes with the most functional similarity with seed genes were identified as neighbouring genes and included in the new PPI network. The constituent genes of the resulted network, including the tightly connected candidate genes and the top 10 neighbouring genes, were enriched in gene ontology (GO) items using the hypergeometric test (qc < 0.05).
Temporal expression analysis
The precise regulation of the spatial and temporal gene expression plays a key role in brain development, maturation and ageing. The CSEA was used to explore in which developmental periods these PRSsMDD-related genes are specifically expressed (Xu et al., 2014) (http://genetics.wustl.edu/jdlab/csea-tool-2/). The analysis was performed using the Fisher’s exact test (qc < 0.05) across developmental stages at a specificity index threshold (pSI) threshold of 0.05. If any genes showed specific expression in any developmental stage under the most stringent threshold of pSI = 0.001, the expression pattern of these genes would be depicted using Human Brain Transcriptome (Kang et al., 2011). In our analysis, we only focused on temporal feature of these genes expressed in the significant brain regions of mediation analysis and Mendelian randomization.
Cell type-specific expression analysis
With the single-cell RNA-sequencing technique, a prior study has provided the detailed cell type-specific expression data of nine major cell types in the mouse brain regions (Zeisel et al., 2015). The Fisher’s exact test was used to identify in which cell types these PRSsMDD fine-mapping genes were specifically expressed in a certain brain region. These tests resulted in the pSI for each cell type, and the P threshold represents how likely the gene set is specifically expressed in a given cell type relative to the other cell types.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Results
PRSMDD could predict the conversion from aMCI to Alzheimer’s disease
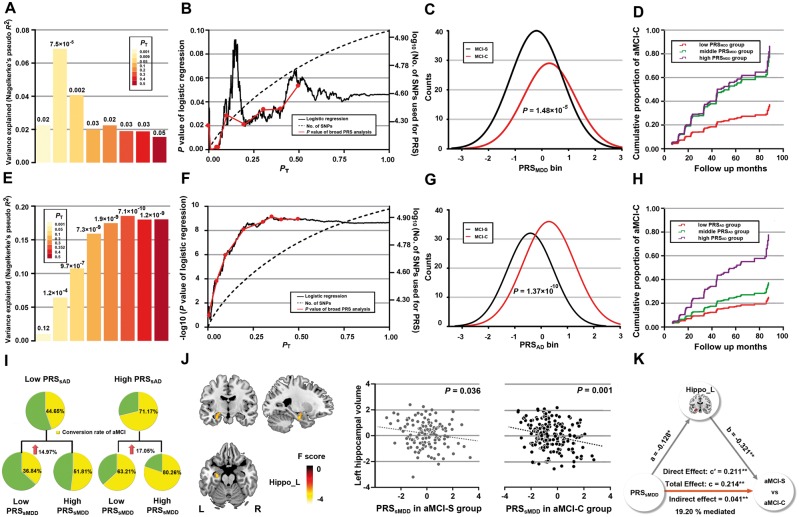
When the PRSMDD were calculated using seven broad PT values for associations between SNPs and MDD, we found the PRSMDD (PT = 0.05) could predict the status of the aMCI-C (P = 0.002) (Fig. 2A) and explained 4.08% of variance for the aMCI-C based on 10 618 index SNPs. When the PRSMDD were computed using a high resolution method, the PRSMDD calculated at PT = 0.009 showed the highest prediction (P = 7.49 × 10−5), explained 6.86% of variance based on 2559 index SNPs (Fig. 2B and Table 2). Compared with aMCI-S patients, aMCI-C patients had a significantly higher adjusted PRSMDD (P = 1.48 × 10−5) (Fig. 2C and Table 1). Compared to the low risk group, the middle [HRs = 2.45, 95% confidence interval (CI) = 1.48–4.03, P = 4.56 × 10−4] and high (HRs = 2.68, 95% CI = 1.24–5.82, P = 3.23 × 10−4) risk groups had more than two times of risk for the aMCI-C. The Cox survival analysis showed that the middle and high risk groups had a higher proportion of the aMCI-C than the low risk group (Fig. 2D). Moreover, the middle and high risk groups (mean time to conversion 35.91 months) progressed more rapidly into Alzheimer’s disease than the low risk group (mean time to conversion 42.75 months).
Figure 2.
The predictive effects of PRSMDD (A–D) and PRSAD (E–H) on the conversion from aMCI to Alzheimer’s disease, conversion rate among the bisected PRS groups (I) and mediation analysis (J–K). (A and E) The bar plots show the predictive effects of the PRS constructed by the best-fit PT and other seven broad PT values (0.001, 0.05, 0.1, 0.2, 0.3, 0.4 and 0.5) on the conversion of aMCI. The numbers above the bars are P-values for the logistic regression between the PRS and aMCI-C. The y-axis shows proportion of variance explained by the PRS. (B and F) The high resolution PRS analyses identify the best-fit PT for constructing the PRSMDD (PT = 0.009) and PRSAD (PT = 0.352). The x-axis shows the PT from 0.001 to 1 with an increment of 0.001. The black line shows the P-value of the logistic regression (left y-axis) at different PT; the dashed line shows numbers of SNPs (right y-axis) at different PT; and the red line connects points at the P values of the broad PRS analysis. (C and G) Distribution of the PRS measures for aMCI-S and aMCI-C. The y-axis shows the counts of each PRS bin (x-axis). (D and H) Cox proportional hazard model shows the associations between the PRS and the conversion from aMCI to Alzheimer’s disease at different time points (x-axis). The y-axis shows the cumulative proportion of aMCI-C for any given follow-up period on the x-axis. The red, green and purple lines show the low, middle and high PRS groups, respectively. (I) Differences in conversion rate among the bisected PRS groups. (J) Left: The brain region with significant negative correlations between the PRSsMDD and GMV. Right: The scatter plots show correlations between the PRSsMDD and GMV in the aMCI-S and aMCI-C groups. (K) The mediation analysis shows that the left hippocampal volume mediates the predictive effect of the PRSsMDD on the conversion of aMCI. AD = Alzheimer’s disease; PT = P-values threshold of genome-wide association studies; *P < 0.01; **P < 0.001.
Table 2.
The predictive effects of PRS on the conversion of aMCI (n = 322)
| PT | SNPs | iSNPs | P | R2, % | Specificity, % | Senstivitity, % | Accuracy, % | AUC | |
|---|---|---|---|---|---|---|---|---|---|
| PRSAD | 0.352 | 2 622 845 | 49 831 | 7.10 × 10−10 | 18.55 | 71.85 | 87.17 | 80.75 | 0.72 |
| PRSMDD | 0.009 | 13 472 | 2559 | 7.49 × 10−5 | 6.86 | 68.89 | 83.43 | 77.33 | 0.65 |
| PRSsAD | NA | 2 618 134 | 49 504 | 1.05 × 10−9 | 18.09 | 70.24 | 85.63 | 80.12 | 0.70 |
| PRSsMDD | NA | 8761 | 1806 | 1.74 × 10−3 | 4.19 | 65.11 | 81.74 | 76.01 | 0.62 |
| PRSsMDD+AD | NA | 2 631 606 | 50 527 | 2.26 × 10−10 | 19.17 | 70.37 | 88.24 | 81.06 | 0.75 |
AUC = area under curve of receiver operating characteristic curve; iSNPs = numbers of index single-nucleotide polymorphisms that constitute PRS; NA = not applicable; PT = P-values threshold of genome-wide association studies; R2 = Nagelkerke’s pseudo R2 of logistic regression; SNPs = numbers of single-nucleotide polymorphisms that constitute PRS.
We also investigated the predictive effect of the PRSAD on the aMCI-C to validate the PRS method and to provide a reference for the predictive effect of the PRSMDD. The broad analysis showed that the PRSAD calculated at PT = 0.4 could predict the aMCI-C (P = 1.20 × 10−9) (Fig. 2E), explained 18.02% of variance in the aMCI-C based on 54 106 index SNPs. The high resolution analysis revealed that the PRSAD computed at PT = 0.352 showed the highest prediction (P = 7.10 × 10−10), explained 18.55% of variance based on 49 831 index SNPs (Fig. 2F and Table 2). The aMCI-C group showed higher PRSAD than the aMCI-S group (P = 1.37 × 10−10) (Fig. 2G and Table 1). Compared to the low risk group, the middle (HRs = 1.55, 95% CI = 1.04–2.31, P = 0.032) and high (HRs = 3.35, 95% CI = 2.30–4.88, P = 3.25 × 10−10) risk groups had increased risk for the aMCI-C. Cox survival analysis showed that the high risk group had a much higher proportion of the aMCI-C than the low risk group (Fig. 2H). Sensitivity, specificity, accuracy, area under curve were also provided (Table 2) and the accuracy of the models was confirmed by 10-fold cross validation (Supplementary material and Supplementary Table 2). We also found that aMCI patients carrying APOE ɛ4 showed an increased risk for the conversion than APOE ɛ4 non-carriers (HRs = 1.78, 95% CI = 1.31–2.42, P = 2.21 × 10−4); however, the age, educational years at baseline, and sex were not associated with the conversion.
PRSsMDD could independently predict the conversion from aMCI to Alzheimer’s disease
After excluding overlapping SNPs (n = 4711) between PRSMDD and PRSAD, the PRSsMDD could significantly predict the aMCI-C (P = 0.002) and explained 4.19% of variance for the aMCI-C based on 1806 index SNPs, suggesting that MDD-specific genetic variants have an independent contribution to the aMCI-C (Table 2). Similarly, the PRSsAD could significantly predict the aMCI-C (P = 1.05 × 10−9) and explained 18.09% of variance for the aMCI-C based on 49 504 index SNPs (Table 2). Additionally, PRS for MDD-specific and Alzheimer’s disease-related genetic variants (PRSsMDD+AD) could predict the aMCI-C (P = 2.26 × 10−10) and explained 19.17% of variance for the aMCI-C based on 50 527 index SNPs (Table 2). Compared to PRSAD that explained 18.55% of variance for the aMCI-C, only 0.33% unique MDD variants were added to the PRSsMDD+AD; however, the explained variance increased by 3.34%. To exclude the potential effects of known Alzheimer’s disease genome regions, we removed SNPs located in 500 kb regions centred on the top 10 Alzheimer’s disease loci (http://www.alzgene.org/) (Supplementary Table 3) and the predictive model of each PRS was still significant (Supplementary Table 4).
To validate the specificity of these index SNPs to MDD rather than Alzheimer’s disease, LD score regression showed that there was not any significant genetic correlation [rg = 0.69, standard error (SE) = 1.32, P = 0.60] between MDD-specific genetic variants and Alzheimer’s disease-related genetic variants. Using a Bayesian posterior probability of 0.90 as a cut-off threshold, there are 1582/1806 (87.5%) index SNPs that are highly associated with MDD (Supplementary Fig. 1A), but none of them are associated with Alzheimer’s disease (Supplementary Fig. 1B), and none of these SNPs show significant colocalization between MDD and Alzheimer’s disease (Supplementary Fig. 1C).
To balance the number of index SNPs used to construct the PRS, we created (i) PRStsAD, PRSAD for the top 1806 Alzheimer’s disease-specific index SNPs with the same numbers of index SNPs as the PRSsMDD; and (ii) PRSsMDD and PRSsAD under the PT = 0.009 (the same PT as the PRSMDD). We found that the predictive effect of PRStsAD and PRSsMDD on conversion risk of aMCI were also significant (Supplementary material and Supplementary Table 5). These results indicate that the conversion from aMCI to Alzheimer’s disease is related to only a small number of MDD-specific genetic variants but to a large number of Alzheimer’s disease-specific genetic variants.
When the aMCI patients were divided into the low and high PRSsMDD groups, the high PRSsMDD group (65.38%) showed 16.25% higher conversion rate than the low PRSsMDD group (49.13%) (P = 0.002). When patients were divided into the double low risk group, low PRSsAD but high PRSsMDD group, high PRSsAD but low PRSsMDD group, and double high risk group. There were significant differences in the conversion rate of the aMCI among these hierarchical PRS groups (P = 4.26 × 10−7). In the low PRSsAD group, the aMCI patients with high PRSsMDD showed marginally higher conversion rate than those with low PRSsMDD (36.84% versus 51.81%, P = 0.05). In the high PRSsAD group, the aMCI patients with high PRSsMDD showed significantly higher conversion rate than those with low PRSsMDD (63.21% versus 80.26%, P = 0.002) (Fig. 2I and Table 3). When PRSsMDD and PRSsAD were trisected into the low, middle and high risk, there were significant differences in conversion rate among the nine hierarchical PRS groups (P = 3.23 × 10−6) (Supplementary materials andSupplementary Table 6).
Table 3.
Conversion rates of aMCI in the bisected PRS groups
| PRS groups | aMCI-S, n | aMCI-C, n | Conversion rate, % |
|---|---|---|---|
| Low PRSsAD and low PRSsMDD | 48 | 28 | 36.84 |
| Low PRSsAD and high PRSsMDD | 40 | 43 | 51.81 |
| High PRSsAD and low PRSsMDD | 32 | 55 | 63.21 |
| High PRSsAD and high PRSsMDD | 15 | 61 | 80.26 |
| All | 135 | 187 | 58.07 |
Table 4.
Top 10 neighbouring genes of the PPI network from PRSsMDD fine-mapping of 1860 genes and PRSsMDD fine-mapping of 1608 genes
| Ranking | PRSsMDD fine-mapping of 1860 genesa | PRSsMDD fine-mapping of 1608 genesb | ||
|---|---|---|---|---|
| Gene symbol | Random walk probability | Gene symbol | Random walk probability | |
| 1 | APP | 6.84 × 10−3 | APP | 6.78 × 10−3 |
| 2 | ELAVL1 | 4.49 × 10−3 | ELAVL1 | 4.49 × 10−3 |
| 3 | NTRK1 | 2.21 × 10−3 | NXF1 | 2.25 × 10−3 |
| 4 | NXF1 | 2.03 × 10−3 | NTRK1 | 2.17 × 10−3 |
| 5 | CUL3 | 1.70 × 10−3 | CUL3 | 1.71 × 10−3 |
| 6 | MOV10 | 1.50 × 10−3 | MOV10 | 1.50 × 10−3 |
| 7 | TP53 | 1.45 × 10−3 | UBC | 1.46 × 10−3 |
| 8 | EWSR1 | 1.43 × 10−3 | TR53 | 1.32 × 10−3 |
| 9 | UBC | 1.42 × 10−3 | EWSR1 | 1.19 × 10−3 |
| 10 | TMEM17 | 1.33 × 10−3 | COPS5 | 1.17 × 10−3 |
aPRSsMDD genetic variants were fine-mapped into 1860 genes based on the hippocampal-specific regulatory probability between eQTLs and epigenomic features (within a 5-kb window).
bPRSsMDD genetic variants were fine-mapped into 1608 genes based on physical position of each variant (within a 5-kb window).
The abbreviation of genes is referred to at https://www.ncbi.nlm.nih.gov/gene/.
Hippocampal volume mediates association between PRSsMDD and aMCI-C
To identify brain regions whose grey matter volumes were associated with PRSsMDD, voxel-wise correlations were performed between PRSsMDD and grey matter volume in the whole brain (Pc < 0.05, voxel-level FWE correction, cluster size > 200 voxels). The PRSsMDD was negatively correlated with the grey matter volume of the left hippocampus (Brodmann area 36; peak MNI coordinate: x = −27, y = −13.5, z = −21; peak intensity = −5.329; 227 voxels; Fig. 2J).
A total of 1806 index SNPs specific to MDD (r2 < 0.25 within 250-kb window) were included in the calculation of PRSsMDD after excluding genetic variants common to PRSMDD (PT = 0.009) and PRSAD (PT = 0.352). We set the PRSsMDD as independent variable in the mediation analysis and instrumental variable in the Mendelian randomization analysis. In the mediation analysis, we found a significant direct effect from the PRSsMDD to the aMCI group assignment (P < 0.001); from the PRSsMDD to the left hippocampal volume (P < 0.01); and from the left hippocampal volume to the aMCI group assignment (P < 0.001). A significant indirect effect was also found in the left hippocampal volume (P < 0.001), which accounted for 19.20% of variance for the aMCI-C in this mediation model (Fig. 2K). In the conventional two-stage Mendelian randomization analysis, we found a negative association between PRSsMDD and hippocampal volume (β = −0.15, SE = 0.06, P = 0.01); the predicted value of the left hippocampal volume was significantly associated with the aMCI-C (β = −2.41, OR = 0.09, SE = 0.78, P = 0.002). Mendelian randomization-Egger regression indicated no unbalanced horizontal pleiotropy (intercept = 0.011, P = 0.07) in the association between left hippocampal volume and aMCI conversion using PRSsMDD as the instrumental variable. We also found significant causal effect of the left hippocampal volume on the aMCI conversion in the Mendelian randomization-Egger regression analysis (βEgger = −2.12, OR = 0.12, SE = 0.77, P = 0.002). These results confirmed a causal chain from PRSsMDD to hippocampal volume to conversion risk of aMCI.
Enrichment analyses using genes prioritized with hippocampal biological information
The 8762 SNPs used for calculating the PRSsMDD were prioritized by integrating the hippocampus-specific eQTL and epigenomic features. The weight of the prioritization and the PGC-MDD GWAS P-value of each SNP were imported into the GATES software to fine-map these SNPs into 1860 significant genes (within a 5-kb window, 5705 SNPs located inside genes, P < 0.001).
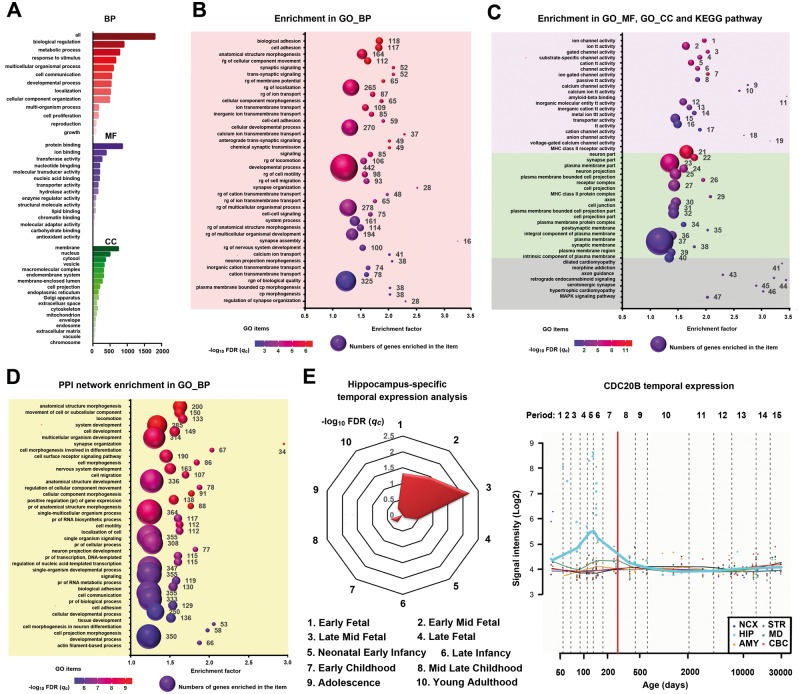
To functionally annotate the 1860 genes, WebGestalt and GOrilla were applied to identify significant enrichment in GO and pathways (Fig. 3A–C and Supplementary Tables 7–9). In the annotation of GO, 505/1860 genes were enriched in the development process, 855/1860 in the protein binding, and 738/1860 in membrane part (Fig. 3A and Supplementary Table 7). Specifically, these genes mainly over-represented in biological processes of the anatomical structure morphogenesis (qc = 4.27 × 10−5), cellular developmental process (qc = 7.08 × 10−4) and development process (qc = 1.14 × 10−3) (Fig. 3B and Supplementary Table 8), in the molecular function of the amyloid-β binding (qc = 8.64 × 10−5), and in the cellular components of the neuron part (qc = 3.10 × 10−11) and neuron projection (qc = 1.69 × 10−5) (Fig. 3C and Supplementary Table 8). In the KEGG pathway analysis, these genes were enriched in neuronal development-related axon guidance (qc = 3.95 × 10−3) (Fig. 3C and Supplementary Table 8). These functional annotations suggest that the PRSsMDD genes were mainly related to the developmental process, and involved in the molecular binding between amyloid-β protein and its precursor, which is a well-known neuropathology of Alzheimer’s disease.
Figure 3.
Gene enrichment of PRSsMDD fine-mapping 1860 genes. (A) Enrichment of the PRSsMDD genes in GO items. The x-axis shows the numbers of genes enriched in each item (y-axis). The red, purple, green bars denote the biological process, molecular function and cellular component, respectively. (B) Top 40 significant enriched GO biological process items of the PRSsMDD genes. The x-axis shows enrichment factor of each GO item (y-axis). The size of the spheres reflects the number of genes (labelled beside the balls) enriched in each item. The colour of the spheres demonstrates the significance of the enrichment analyses. (C) Top 20 significant enriched GO molecular function and cellular component items, and all significant KEGG pathway items of the PRSsMDD genes. The purple, green and grey background colours show the molecular function, cellular component and KEGG pathway, respectively. (D) Top 40 significant enriched GO biological process items of the PPI network. (E) Left: PRSsMDD genes were highly expressed in the middle-late foetal developmental period in the hippocampus. Right: CDC20B was highly expressed in the early and late mid-foetal development periods in the hippocampus. Periods 1–15 are described in Supplementary Table 11. AMY = amygdala; BP = biological process; CBC = cerebellar cortex; CC = cellular component; CDC20B = cell division cycle 20B; cp = cell projection; GO = gene ontology; HIP = hippocampus; MF = molecular function; MD = mediodorsal nucleus of the thalamus; NCX = neocortex; pr = positive regulation; rg = regulation; STR = striatum; tt = transmembrane transporter.
To combine protein-level information, we mapped 1860 genes to the PPI network (642 unmatched genes were excluded). To illustrate potential functional connectivity of the remaindering 1218 genes, we used a network-based approach to identify a tightly connected PPI network (only 846 genes were included in the network). Using the network topology-based analysis, the top 10 neighbouring genes with the most functional similarity with seed genes were also included in the construction of the final PPI network consisting of 856 genes. As the most functionally neighbouring genes (Fig. 3D), the amyloid-β precursor protein (APP) formed the protein basis of the amyloid plaques in the brain of Alzheimer’s disease patients. The 856 genes in the final PPI network were enriched mainly in development processes of the nervous system, such as nervous system development (qc = 2.83 × 10−5), neuron projection development (qc = 9.08 × 10−5), neuron projection guidance (qc = 4.20 × 10−4), neurogenesis (qc = 6.51 × 10−4) and generation of neurons (qc = 9.37 × 10−4) (Fig. 3D and Supplementary Table 10).
The prior analyses indicate that PRSsMDD fine-mapping genes are involved in brain developmental processes. We further explored in which developmental periods these genes were over-represented in the hippocampus. The CSEA online tool was used to explore the temporal-specific expression of these genes in the hippocampus. Under a pSI threshold of 0.05, 60 genes showed temporal-specific expression in the hippocampus in the middle-late foetal developmental period (qc = 6.02 × 10−4) (Fig. 3E). Under the most stringent threshold (pSI = 0.001), only cell division cycle 20B (CDC20B) exhibited a temporal-specific high expression in the hippocampus in the middle-late foetal stage. The Human Brain Transcriptome dataset showed that the CDC20B was highly expressed in the early and late mid-foetal development period (Fig. 3E).
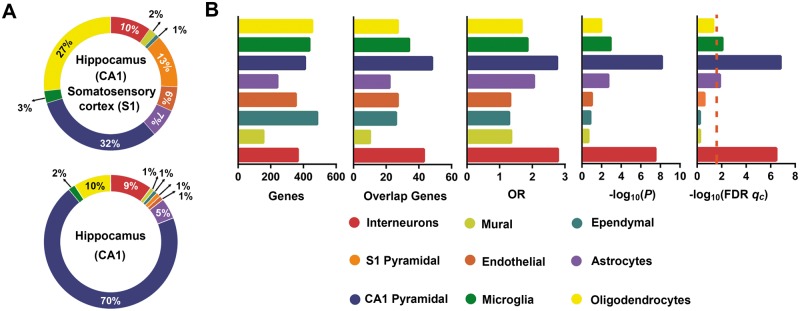
Among the nine major cell types in the mouse cortex and hippocampus, eight cell types exist in the hippocampal CA1 (Fig. 4A). The PRSsMDD genes showed substantial over-representation mainly in the hippocampal pyramidal neurons (qc = 1.60 × 10−7) and interneurons (qc = 3.61 × 10−7) (Fig. 4B). However, these genes also demonstrated specific expression in the microglia cells (qc = 0.01) and astrocytes (qc = 0.01) (Fig. 4B).
Figure 4.
Cell type-specific expression analyses. (A) The frequency of the nine major cell types in the CA1 and S1 (top) and only in the CA1 (bottom) of the mouse brains (Zeisel et al., 2015). (B) The first bar plot shows the numbers of genes in eight labelled cell types of hippocampus from the Zeisel et al. study. The second bar plot shows the numbers of overlapping genes between PRSsMDD fine-mapping 1860 genes and significantly expressed genes in each cell type of hippocampus. The last three bar plots show the cell type-specific expression results of the PRSsMDD-related genes in the eight labelled cell types of the hippocampus. The dashed red line shows the significant level under qc < 0.05 in FDR-BH correction. CA = cornu ammonis; GMV = grey matter volume; OR = odd ratios; S1 = somatosensory cortex.
Enrichment analyses using genes fine-mapped based on physical position
We also re-mapped 8762 MDD-specific SNPs into 1608 genes only based on the physical position of each variant (within 5-kb window). We found that 1468 genes were common between the two fine-mapping methods (with and without hippocampal biological information), suggesting that most genes related to these MDD-specific SNPs are associated with hippocampal gene expression. We performed enrichment analyses for the 1608 genes and found significant enrichment in developmental process and amyloid-β binding (Supplementary material, Supplementary Fig. 2 and Supplementary Tables 9 and 10).
Discussion
To our knowledge, this is the first study that integrates cross-scale neurobiological analyses and functional annotations to investigate the independent predictive effect of MDD-specific genomic variants on the conversion from aMCI to Alzheimer’s disease and the underlying neurobiological mechanisms. The PRSsMDD could predict the conversion by affecting the hippocampal volume. These genes were functionally related to the hippocampal development and amyloid-β binding. In addition, these genes showed temporal-specific expression in the hippocampus in the foetal developmental period and cell type-specific expression in the hippocampal pyramidal neurons and interneurons. These findings support a model of double attacks of hippocampus to explain for the neurobiological mechanisms of the independent contributions of MDD- and Alzheimer’s disease-specific genomic variants to the conversion from aMCI to and Alzheimer’s disease.
PRSsMDD could independently predict the conversion from aMCI to Alzheimer’s disease
That Alzheimer’s disease is a progressive polygenic disorder (Dubois et al., 2007) and no effective therapies at the late stage makes early diagnosis and treatment to be the only way to improve prognosis (Chu, 2012). As the putative premorbid state of Alzheimer’s disease (Mariani et al., 2007), patients with aMCI have a high risk of developing into Alzheimer’s disease (Palmer et al., 2008). Because not all aMCI patients would convert to Alzheimer’s disease, it is clinically important to predict the conversion from aMCI to Alzheimer’s disease for early intervention. Consistent with previous studies (Rodriguez-Rodriguez et al., 2013; Adams et al., 2015), the PRSAD was found to predict conversion. More importantly, we found that the PRSMDD could also predict the conversion of non-depressed aMCI patients. Thus, the predictive ability of the PRSMDD cannot be explained by parallel depressive symptoms (Kida et al., 2016; Mourao et al., 2016; Sacuiu et al., 2016; Barca et al., 2017) and the coexistence of MDD (Modrego and Ferrandez, 2004). To exclude the possibility that the predictive effect of the PRSMDD is driven by genetic variants common to MDD and Alzheimer’s disease, we found that the PRSsMDD could also predict the status of aMCI-C. LD score regression and co-localization analysis were performed to validate that these SNPs calculated for PRSsMDD are specific to MDD rather than Alzheimer’s disease. Moreover, aMCI patients with high PRSsMDD showed 14.97% and 17.05% higher conversion rates than those with low PRSsMDD in the low and high PRSsAD groups, respectively. These findings suggest that the PRSsMDD has an independent contribution to the conversion from aMCI to Alzheimer’s disease.
In terms of the clinical prediction for the conversion from aMCI to Alzheimer’s disease, aMCI patients with the double high risk (high PRSsMDD and high PRSsAD) showed a much higher conversion rate than those with the double low risk (low PRSsMDD and low PRSsAD) (89.65% versus 33.33%). With the increased availability and decreasing cost of the sequencing technique, the genomic data of aMCI patients could be easily obtained, which could be used to construct the PRSsMDD and PRSsAD using the method of this study. In clinical practice, one could jointly use the PRSsMDD and PRSsAD to select aMCI patients with a much greater risk for Alzheimer’s disease. Appropriate intervention procedures for these aMCI patients may prevent or delay them from progressing into Alzheimer’s disease.
Hippocampal volume mediates the prediction of PRSsMDD in aMCI-C
Correlation, mediation and Mendelian randomization analyses revealed that hippocampal volume mediated the predictive effect of the PRSsMDD on the conversion from aMCI to Alzheimer’s disease. It has been recognized that reduced hippocampal volume is the most consistent subcortical abnormality in MDD patients in a meta-analysis of 1728 patients and 7199 controls (Schmaal et al., 2016). The genetic variants have been proposed as an important cause for the reduced hippocampal volume in MDD. For example, MDD patients with risk alleles show a smaller hippocampal volume than those without risk alleles (Egan et al., 2003; Frodl et al., 2004, 2007, 2012; Kohli et al., 2011). The reduction in hippocampal volume has been attributed to reduced gene expression in the hippocampus (Egan et al., 2003; Kohli et al., 2011). In children with few life stress events, the high PRS constructed by depression-related stress system genes could predict reduced hippocampal volume (Pagliaccio et al., 2014). Moreover, in healthy individuals without depressive episodes, the depression-related risk-allele carriers also displayed a reduced hippocampal volume compared with non-carriers (Frodl et al., 2008). These findings suggest that genetic variants for MDD could result in the reduction in the hippocampal volume in different populations.
It is well known that the volumetric reduction in the hippocampus is the most prominent early pathological feature of Alzheimer’s disease (Yang et al., 2012) and one of the most reliable predictive measures for the conversion from aMCI to Alzheimer’s disease (Brueggen et al., 2015). The additional burden to the hippocampus by MDD-specific genetic variants may exacerbate hippocampal atrophy and facilitate the conversion from aMCI to Alzheimer’s disease. Thus, the model of double attacks on the hippocampus may explain the increased predictive value of the PRSsMDD on the conversion from aMCI to Alzheimer’s disease.
Neurobiological mechanisms underlying the predictive effect of PRSsMDD on aMCI-C
To understand why hippocampal volume could mediate the predictive effect of the PRSsMDD on conversion, the MDD-specific genetic variants were fine-mapped into the most common genes between the two different strategies (with and without hippocampal biological information), which suggested that most genes related to these MDD-specific SNPs are associated with hippocampal biological information.
The GO annotation revealed that MDD-related genes were related to amyloid-β binding, which is also supported by the PPI network analysis, showing that APP was the most important neighbouring gene in the network constructed by the MDD-related genes. The increased hippocampal amyloid-B, which is an important neuropathological change in Alzheimer's disease, is also related to depression (Rapp et al., 2006) or depressive symptoms (Donovan et al., 2018). These findings suggest that MDD-related genes may be involved in amyloid-β binding, an important neuropathological process of Alzheimer’s disease.
The enrichment analysis of the biological process revealed that MDD-related genes were mainly involved in the nervous developmental process. The temporal-specific expression analysis further indicated that these genes were mainly expressed in the middle to late foetal periods in the hippocampus, which is in line with the critical developmental period of hippocampal neurons (Tran and Kelly, 2003). For example, CDC20B showed the highest expression in the hippocampus in the middle-late foetal periods (Fig. 3E). This gene is a developmental regulator of hippocampal neurons (Kim et al., 2009; Yang et al., 2010) and plays an essential role in dendritic morphogenesis (Kim et al., 2009), axon growth (Yamada et al., 2013) and presynaptic differentiation (Yang et al., 2009), all of which are associated with hippocampal volume (Stockmeier et al., 2004; Duman et al., 2016). In addition, cell type-specific expression confirmed that these genes were mainly expressed in the hippocampal pyramidal neurons and interneurons. These findings suggest that MDD-related genes affect the hippocampal volume via modulating the gene expression of hippocampal neurons, which are the main cell types with prominent pathological impairment in Alzheimer’s disease (Blazquez-Llorca et al., 2011).
Limitations
Two limitations should be mentioned regarding this study. First, only volumetric measure was used as the intermediate phenotype. Consequently, we cannot exclude the possibility that other intermediate phenotypes may also mediate the association between PRSsMDD and aMCI conversion. Second, the cell type-specific expression analysis was based on the hippocampus of mouse rather than human, because there are no publicly available cell type-specific expression data in the human hippocampus.
Conclusion
In this study, we found that the PRSsMDD could independently predict conversion from aMCI to Alzheimer’s disease, and the combined use of the PRSsMDD and PRSsAD could select aMCI patients with a much higher risk for conversion. The predictive effect of the PRSsMDD on conversion may be mediated by the hippocampus via affecting its early developmental process and amyloid-β binding.
Supplementary Material
Acknowledgements
We are grateful to the ADNI, IGAP and PGC consortium for providing summary statistics for the discovery and target sample. Data collection and sharing was funded by ADNI (National Institutes of Health U01 AG024904).
Glossary
Abbreviations
- aMCI(‐C/S)
amnestic mild cognitive impairment (‐conversion to Alzheimer’s disease/stable diagnosis)
- GO
gene ontology
- MDD
major depressive disorder
- PPI
protein–protein interaction
- PRSAD/MDD/sAD/sMDD/stAD
polygenic risk scores for Alzheimer’s disease/major depressive disorder/Alzheimer’s disease-specific/major depressive disorder-specific/top 1806 Alzheimer’s disease-specific genetic variants
- SNP
single-nucleotide polymorphism
Funding
This work has been supported by the Natural Science Foundation of China (81425013, 81501451, 81501454, 81601476, 81701676, 81701668 and 81801687), Tianjin Key Technology R&D Program (17ZXMFSY00090), National Key Research and Development Program of China (2018YFC1314301), Postgraduate Innovation Fund of 13th Five-Year comprehensive investment of Tianjin Medical University (YJSCX201719), and Science&Technology Development Fund of Tianjin Education Commission for Higher Education (2016YD11).
Competing interests
The authors report no competing interests.
References
- Adams HH, de Bruijn RF, Hofman A, Uitterlinden AG, van Duijn CM, Vernooij MW et al. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia. Alzheimers Dement 2015; 11: 1277–85. [DOI] [PubMed] [Google Scholar]
- Barca ML, Persson K, Eldholm R, Benth JS, Kersten H, Knapskog AB et al. Trajectories of depressive symptoms and their relationship to the progression of dementia. J Affect Disord 2017; 222: 146–52. [DOI] [PubMed] [Google Scholar]
- Blazquez-Llorca L, Garcia-Marin V, Merino-Serrais P, Avila J, DeFelipe J. Abnormal tau phosphorylation in the thorny excrescences of CA3 hippocampal neurons in patients with Alzheimer’s disease. J Alzheimers Dis 2011; 26: 683–98. [DOI] [PubMed] [Google Scholar]
- Brown CD, Mangravite LM, Engelhardt BE. Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet 2013; 9: e1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggen K, Dyrba M, Barkhof F, Hausner L, Filippi M, Nestor PJ et al. Basal forebrain and hippocampus as predictors of conversion to alzheimer’s disease in patients with mild cognitive impairment - a multicenter dti and volumetry study. J Alzheimers Dis 2015; 48: 197–204. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015a; 47: 1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015b; 47: 291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 2014; 43: 922–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 2013; 42: 1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG. Mendelian randomization: methods for using genetic variants in causal estimation. New York: Chapman and Hall/CRC; 2015. [Google Scholar]
- Chu L. Alzheimer’s disease: early diagnosis and treatment. Hong Kong Med J 2012; 18: 228–37. [PubMed] [Google Scholar]
- De Roeck E, Ponjaert-Kristoffersen I, Bosmans M, De Deyn PP, Engelborghs S, Dierckx E. Are depressive symptoms in mild cognitive impairment predictive of conversion to dementia? Int Psychogeriatr 2016; 28: 921–8. [DOI] [PubMed] [Google Scholar]
- Donovan NJ, Locascio JJ, Marshall GA, Gatchel J, Hanseeuw BJ, Rentz DM et al. Longitudinal association of amyloid beta and anxious-depressive symptoms in cognitively normal older adults. Am J Psychiatry 2018; 175: 530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, DeKosky ST, Barberger-Gateau P, Cummings J et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS–ADRDA criteria. Lancet Neurol 2007; 6: 734–46. [DOI] [PubMed] [Google Scholar]
- Duman RS, Aghajanian GK, Sanacora G, Krystal JH. Synaptic plasticity and depression: new insights from stress and rapid-acting antidepressants. Nat Med 2016; 22: 238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 2007; 3: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009; 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003; 112: 257–69. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 2015; 138: 3673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics 2015; 31: 1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T, Carballedo A, Hughes MM, Saleh K, Fagan A, Skokauskas N et al. Reduced expression of glucocorticoid-inducible genes GILZ and SGK-1: high IL-6 levels are associated with reduced hippocampal volumes in major depressive disorder. Trans Psychiatry 2012; 2: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T, Koutsouleris N, Bottlender R, Born C, Jager M, Morgenthaler M et al. Reduced gray matter brain volumes are associated with variants of the serotonin transporter gene in major depression. Mol Psychiatry 2008; 13: 1093–101. [DOI] [PubMed] [Google Scholar]
- Frodl T, Meisenzahl EM, Zill P, Baghai T, Rujescu D, Leinsinger G et al. Reduced hippocampal volumes associated with the long variant of the serotonin transporter polymorphism in major depression. Arch Gen Psychiatry 2004; 61: 177–83. [DOI] [PubMed] [Google Scholar]
- Frodl T, Schule C, Schmitt G, Born C, Baghai T, Zill P et al. Association of the brain-derived neurotrophic factor Val66Met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry 2007; 64: 410–16. [DOI] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 2014; 10: e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Russ TC, Adams MJ, Clarke TK, Howard DM, Hall LS et al. Assessing the presence of shared genetic architecture between Alzheimer’s disease and major depressive disorder using genome-wide association data. Transl Psychiatry 2017; 7: e1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames JL, Hagan CR, Joiner TE. Interpersonal processes in depression. Annu Rev Clin Psychol 2013; 9: 355–77. [DOI] [PubMed] [Google Scholar]
- Hayes AF. Introduction to mediation, moderation, and conditional process analysis: a regression-based approach. New York: Guilford Press; 2013. [Google Scholar]
- Holmes AJ, Lee PH, Hollinshead MO, Bakst L, Roffman JL, Smoller JW et al. Individual differences in amygdala-medial prefrontal anatomy link negative affect, impaired social functioning, and polygenic depression risk. J Neurosci 2012; 32: 18087–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012; 44: 955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008; 27: 685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorm AF. History of depression as a risk factor for dementia: an updated review. Aust N Z J Psychiatry 2001; 35: 776–81. [DOI] [PubMed] [Google Scholar]
- Julayanont P, Brousseau M, Chertkow H, Phillips N, Nasreddine ZS. Montreal Cognitive Assessment Memory Index Score (MoCA-MIS) as a predictor of conversion from mild cognitive impairment to Alzheimer’s disease. J Am Geriatr Soc 2014; 62: 679–84. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M et al. Spatio-temporal transcriptome of the human brain. Nature 2011; 478: 483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida J, Nemoto K, Ikejima C, Bun S, Kakuma T, Mizukami K et al. Impact of depressive symptoms on conversion from mild cognitive impairment subtypes to alzheimer’s disease: a community-based longitudinal study. J Alzheimers Dis 2016; 51: 405–415. [DOI] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell 2009; 136: 322–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet 2008; 82: 949–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron 2011; 70: 252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518: 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Li M, Liu Z, Yan B, Pan Z, Huang D et al. cepip: context-dependent epigenomic weighting for prioritization of regulatory variants and disease-associated genes. Genome Biol 2017; 18: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet 2011; 88: 283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani E, Monastero R, Mecocci P. Mild cognitive impairment: a systematic review. J Alzheimers Dis 2007; 12: 23–35. [DOI] [PubMed] [Google Scholar]
- Mazzeo S, Santangelo R, Bernasconi MP, Cecchetti G, Fiorino A, Pinto P et al. Combining cerebrospinal fluid biomarkers and neuropsychological assessment: a simple and cost-effective algorithm to predict the progression from mild cognitive impairment to alzheimer’s disease dementia. J Alzheimers Dis 2016; 54: 1495–508. [DOI] [PubMed] [Google Scholar]
- Milaneschi Y, Lamers F, Peyrot WJ, Abdellaoui A, Willemsen G, Hottenga JJ et al. Polygenic dissection of major depression clinical heterogeneity. Mol Psychiatry 2016; 21: 516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrego PJ, Ferrandez J. Depression in patients with mild cognitive impairment increases the risk of developing dementia of Alzheimer type: a prospective cohort study. Arch Neurol 2004; 61: 1290–3. [DOI] [PubMed] [Google Scholar]
- Mormino EC, Sperling RA, Holmes AJ, Buckner RL, De Jager PL, Smoller JW et al. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology 2016; 87: 481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourao RJ, Mansur G, Malloy-Diniz LF, Castro Costa E, Diniz BS. Depressive symptoms increase the risk of progression to dementia in subjects with mild cognitive impairment: systematic review and meta-analysis. Int J Geriatr Psychiatry 2016; 31: 905–11. [DOI] [PubMed] [Google Scholar]
- Pagliaccio D, Luby JL, Bogdan R, Agrawal A, Gaffrey MS, Belden AC et al. Stress-system genes and life stress predict cortisol levels and amygdala and hippocampal volumes in children. Neuropsychopharmacology 2014; 39: 1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K, Backman L, Winblad B, Fratiglioni L. Mild cognitive impairment in the general population: occurrence and progression to Alzheimer disease. Am J Geriatr Psychiatry 2008; 16: 603–11. [DOI] [PubMed] [Google Scholar]
- Palmer K, Di Iulio F, Varsi AE, Gianni W, Sancesario G, Caltagirone C et al. Neuropsychiatric predictors of progression from amnestic-mild cognitive impairment to Alzheimer’s disease: the role of depression and apathy. J Alzheimers Dis 2010; 20: 175–83. [DOI] [PubMed] [Google Scholar]
- Park S, Hatim A, Si TM, Jeon HJ, Srisurapanont M, Bautista D et al. Stressful life events preceding the onset of depression in Asian patients with major depressive disorder. Int J Soc Psychiatry 2015; 61: 735–42. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 2016; 48: 709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power RA, Tansey KE, Buttenschon HN, Cohen-Woods S, Bigdeli T, Hall LS et al. Genome-wide association for major depression through age at onset stratification: major depressive disorder working group of the psychiatric genomics consortium. Bio Psychiatry 2017; 81: 325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preacher KJ, Hayes AF. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behav Res Methods 2008; 40: 879–91. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Shen M, Buss C, Chong YS, Kwek K, Saw SM et al. Effects of antenatal maternal depressive symptoms and socio-economic status on neonatal brain development are modulated by genetic risk. Cereb Cortex 2017; 27: 3080–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp MA, Schnaider-Beeri M, Grossman HT, Sano M, Perl DP, Purohit DP et al. Increased hippocampal plaques and tangles in patients with Alzheimer disease with a lifetime history of major depression. Arch Gen Psychiatry 2006; 63: 161–7. [DOI] [PubMed] [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013; 18: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez E, Sanchez-Juan P, Vazquez-Higuera JL, Mateo I, Pozueta A, Berciano J et al. Genetic risk score predicting accelerated progression from mild cognitive impairment to Alzheimer’s disease. J Neural Transm 2013; 120: 807–12. [DOI] [PubMed] [Google Scholar]
- Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex 2012; 22: 2653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacuiu S, Insel PS, Mueller S, Tosun D, Mattsson N, Jack CR Jr et al. chronic depressive symptomatology in mild cognitive impairment is associated with frontal atrophy rate which hastens conversion to alzheimer dementia. Am J Geriatr Psychiatry 2016; 24: 126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L, Veltman DJ, van Erp TG, Samann PG, Frodl T, Jahanshad N et al. Subcortical brain alterations in major depressive disorder: findings from the ENIGMA major depressive disorder working group. Mol Psychiatry 2016; 21: 806–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog I, Waern M, Duberstein P, Blennow K, Zetterberg H, Borjesson-Hanson A et al. A 9-year prospective population-based study on the association between the APOE*E4 allele and late-life depression in Sweden. Bio Psychiatry 2015; 78: 730–6. [DOI] [PubMed] [Google Scholar]
- Stockmeier CA, Mahajan GJ, Konick LC, Overholser JC, Jurjus GJ, Meltzer HY et al. Cellular changes in the postmortem hippocampus in major depression. Bio Psychiatry 2004; 56: 640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron 2010; 68: 182–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuchi R, Hishikawa N, Kurata T, Sato K, Kono S, Yamashita T et al. Clinical and demographic predictors of mild cognitive impairment for converting to Alzheimer’s disease and reverting to normal cognition. J Neurol Sci 2014; 346: 288–92. [DOI] [PubMed] [Google Scholar]
- Tran TD, Kelly SJ. Critical periods for ethanol-induced cell loss in the hippocampal formation. Neurotoxicol Teratol 2003; 25: 519–28. [DOI] [PubMed] [Google Scholar]
- Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 2017; 45: W130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund LO et al. Mild cognitive impairment–beyond controversies, towards a consensus: report of the international working group on mild cognitive impairment. J Intern Med 2004; 256: 240–6. [DOI] [PubMed] [Google Scholar]
- Wyman BT, Harvey DJ, Crawford K, Bernstein MA, Carmichael O, Cole PE et al. Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimers Dement 2013; 9: 332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci 2014; 34: 1420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Bonni A. Spatial organization of ubiquitin ligase pathways orchestrates neuronal connectivity. Trends Neurosci 2013; 36: 218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Pan P, Song W, Huang R, Li J, Chen K et al. Voxelwise meta-analysis of gray matter anomalies in Alzheimer’s disease and mild cognitive impairment using anatomic likelihood estimation. J Neurol Sci 2012; 316: 21–9. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Bonni A. The dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr Opin Neurobiol 2010; 20: 92–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y et al. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science 2009; 326: 575–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Bai F, Zhang Z. shared genetic risk factors for late-life depression and Alzheimer’s disease. J Alzheimers Dis 2016; 52: 1–15. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Gu ZX, Wei WS. Fluorodeoxyglucose-positron-emission tomography, single-photon emission tomography, and structural MR imaging for prediction of rapid conversion to Alzheimer disease in patients with mild cognitive impairment: a meta-analysis. AJNR Am J Neuroradiol 2009; 30: 404–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015; 347: 1138–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.