Abstract
Cardiovascular disease (CVD) remains the leading cause of death in the Western world. Despite advances in the prevention and in the management of CVD, the role of RNA epigenetics in the cardiovascular system has been until recently unexplored. The rapidly expanding research field of RNA modifications has introduced a novel layer of gene regulation in mammalian cells. RNA modifications may control all aspects of RNA metabolism, and their study reveals previously unrecognized regulatory pathways that may determine gene expression at a post-transcriptional level. Understanding the role of RNA modifications in CVD may lead towards a better understanding of disease mechanisms and the development of novel biomarkers or therapeutic strategies. In this review, we highlight the most recent and major reports in the field of RNA methylation and adenosine to inosine RNA editing related to the cardiovascular field and we discuss how this breakthrough will advance the field of precision medicine.
1. Introduction
Cardiovascular disease (CVD) is the leading cause of death in Western society, and places a significant health and economic burden on the US and the world. As instances of obesity, high cholesterol, high blood pressure, and diabetes / metabolic syndrome increase, so too does the risk of CVD [1]. CVD encompasses a wide range of disorders, including coronary heart disease and atherosclerosis, genetic cardiomyopathies, congenital cardiovascular defects, heart rhythm disorders, valvular diseases, and many others. Despite recent advancements in the early diagnosis and management of many of the above disorders, CVD mortality throughout the world remains higher than cancer. Treatment strategies for CVD have not often been tailored to the specific needs of the patient in the past, but the landscape for precision medicine in CVD is changing. In recent years, genetic editing and gene-based therapeutics have been applied in the clinic, and remarkable advances are being made in patients with Duchenne muscular dystrophy, spinal muscular atrophy, and CVD [2–8]. The notion that messenger RNA (mRNA) is not a static molecule, but is in fact dynamically regulated by a diverse array of modifications, has broadened the potential for RNA-based therapeutics (antisense oligonucleotides, aptamers, siRNAs, miRNAs, and CRISPR/Cas9) in multiple disease systems including CVD, and many clinical trials for RNA therapies are in progress [9]. Despite the attractiveness of these RNA-based therapeutics for disease management, however, problems with stability, ease-of-delivery, and off-target effects have pushed ongoing research to discover additional, novel mechanisms to regulate RNA and influence disease pathophysiology [10].
RNA contains over 140 distinct chemical modifications, many of which were first identified in tRNAs and other non-coding RNAs in a diverse range of organisms [11]. RNA methylation is the most prevalent chemical addition to RNA nucleotides, and can affect guanosines (7-methylguanosine, m7G [12]) cytosines (5-methylcytosine, m5C [13]; 5-hydroxymethylcytosine, hm5C [14]), adenosines (N1-methyladenosine, m1A [15, 16]; N6-methyladenosine, m6A [17, 18]; N6,2’-O-dimethyladenosine, m6Am [19]), and ribose (2’-O’methylation, 2’-OMe or Nm [20]). The most prevalent of these, m6A, was first identified in 1974, and in recent years has captured scientific attention as a conserved and dynamic moiety differentially regulated in many cellular- and state-dependent contexts [21–23]. m6A occurs in ribosomal RNA (rRNA) [17], transfer RNA (tRNA) [24], small nucleolar RNA (snoRNA) [25], long noncoding RNA (lncRNA) [26], circular RNA (circRNA) [27], microRNA (miRNA) [28], and messenger RNA (mRNA) [22]; it is present on over 7000 (roughly 20%) human mRNAs, distributed throughout the coding and untranslated regions, and enriched in long last exons and upstream of the stop codon [17]. m6A locations throughout mRNA, and especially in the untranslated regions, hint to the modification’s function – a putative connection to RNA processing and translation. Ongoing studies are aimed at defining the precise role of m6A in cell processes and disease states, as well as investigating the potential of manipulating m6A to alter these events.
Although m6A is the most prevalent mRNA modification, it is not the only form of methylation to influence RNA metabolism. m1A, known to regulate the stability of tRNA and rRNA, can dramatically alter mRNA-protein interactions through electrostatic effects [15, 16]. Its position near translation start sites may upregulate translation, and its deposition is, like m6A, dynamically regulated during stress [16, 29]. m6Am can also have a prominent effect upon translation efficiency by preventing decapping and miRNA-mediated mRNA degradation, thereby stabilizing mRNA [19]. m5C modifications in tRNA can control cell growth and differentiation, and although also deposited on mRNA and lncRNA by the methyltransferase NSUN2, the precise function of this modification in biological function remains unknown [13, 30]. Finally, like m1A, 2’-O-Me on ribose can affect RNA secondary structure and accessibility to RNA-binding proteins [31]. Although a great deal of work has focused on identifying the role of methyl modifications during RNA metabolism, additional studies are necessary to elucidate the full range of regulatory mechanisms afforded by RNA methylation.
Another class of modifications are the substitutional RNA modifications. In RNA editing, RNA molecules are enzymatically modified on specific nucleic acids following transcription. RNA editing was first identified more than 30 years ago as an unwinding activity of transfected RNA duplexes in Xenopus laevis embryos [32, 33]. This was the result of covalent modification of RNA and was specific to double-stranded RNAs [34]. Editing is an important mechanism regulating gene expression at the RNA level, but only recently have methodological advances enabled a thorough investigation and highlighted an essential role for editing in cell physiology [35]. RNA editing is a ubiquitous and crucial posttranscriptional modification of the genome-encoded RNA sequence that modifies primary RNA transcripts and provides an additional layer of gene regulation to expand both the protein-coding potential of the transcriptome and the range of RNA transcript functions.
The most prominent form of RNA editing is adenosine to inosine (A-to-I) deamination, catalysed by the adenosine deaminases acting on RNA-1 and −2 (ADAR-1/−2) in mammals [36]. Another less prevalent form of RNA editing is the hydrolytic deamination of cytidine to uracil (C-to-U), catalysed by the cytosine deaminases of the apolipoprotein B mRNA editing catalytic polypeptide family APOBEC1 and APOBEC3A [37]. RNA editing occurs only on double-stranded RNAs (dsRNAs), with the majority of editing sites present in repetitive sequences such as Alu and long interspersed elements (LINEs) located in introns and 5’ and 3’ untranslated regions (UTR). Different editing enzymes demonstrate specific editing preferences: for example, ADAR1 appears to preferentially edit the Alu elements in non-coding regions while ADAR2 targets more non-repetitive sequences in exons or primary/precursor microRNAs [38]. Recently, millions of adenosines (mostly within Alu repeats) in the human transcriptome have been mapped as potential sites of ADAR-mediated A-to-I editing, demonstrating the likely wide-ranging impacts of this modification [39].
This review will discuss in more detail the role of m6A and A-I editing, as these two RNA modifications have been recently implicated in crucial regulation of cardiovascular homeostasis and stress responses [40–43]. Specifically, we will discuss the impact of m6A upon RNA regulation, including the m6A ‘life cycle’ (deposition, recognition, and removal), m6A alterations during development and various diseases, including recent studies investigating m6A in cardiac disease, and finally the potential of m6A modulation in understanding CVD pathogenesis and therapeutic design. Key aspects related to m6A are also summarized in figure 1. We will also present a brief overview of A-to-I RNA editing and its impact on cardiovascular RNA epigenetics (visualized in figure 2). Recent reviews are also available for a deeper understanding of the molecular mechanisms and the pathophysiological process triggered by RNA editing [36, 44, 45].
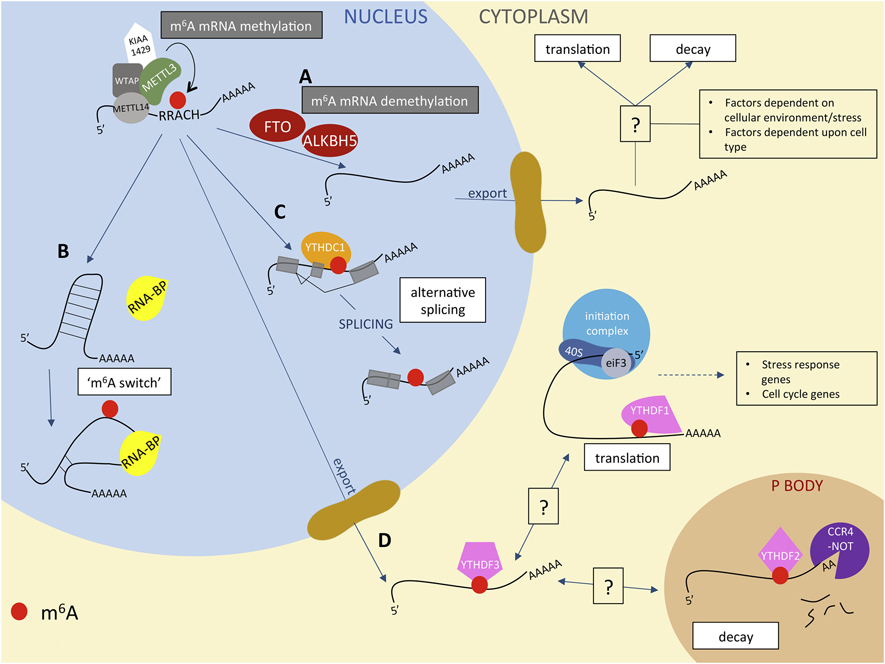
Figure 1: m6A dictates mRNA metabolism.
m6A is deposited onto mRNAs in the nucleus by the methyltransferase complex, comprised of METTL3, METTL14, WTAP, and KIAA1429. It can then be then demethylated by FTO and ALKBH5 (path A), after which it exits the nucleus and, depending on signals from the host cell, can either be translated or marked for decay (Unknown, additional signals from the cell are represented with “?”). Alternatively, m6A remains on mRNAs, allowing for additional regulation of mRNA metabolism. m6A can disrupt RNA secondary structure, exposing binding sites for RNA binding proteins, termed the ‘m6A switch’ (path B). The m6A reader YTHDC1 can bind m6A on mRNAs and dictate alternative splicing (path C). m6A-modified mRNAs can exit the nucleus and be bound by the m6A ‘readers’ YTHDF1, YTHDF2, and YTHDF3 in the cytoplasm (path D). YTHDF1 generally promotes translation of the methylated mRNA, whereas YTHDF2 promotes decay by recruiting the mRNA to P bodies and binding the CCR4-NOT deadenylase complex. YTHDF3 can promote either translation or decay, depending upon as-yet-undefined signals from the host cell.
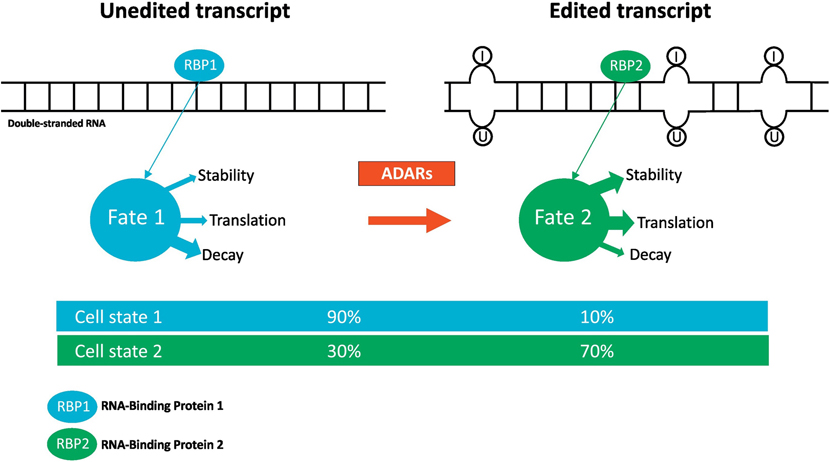
Figure2: RNA editing controls RNA-protein interaction and cellular function.
Cartoon illustrates the effect of adenosine to inosine RNA editing on double-stranded RNAs (dsRNAs) by the Adenosine Deaminases Acting on RNA (ADARs) enzymes. ADARs deaminate the adenosine residues into inosines, which no longer pair with uracil. This induces a change in RNA secondary structure, which may affect the binding of proteins to RNA molecules and thus may control several aspects of RNA metabolism and cellular function.
2. m6A and A-to-I RNA modifications control mRNA fate
2.1. m6A dictates mRNA metabolism
As the most abundant internal mRNA modification, N6-adenosine methylation can affect almost every aspect of RNA metabolism, from splicing and processing in the nucleus to translation and degradation in the cytoplasm (see Figure 1). Typically, m6A is deposited onto mRNA at a conserved consensus sequence, RRm6ACH ([G/A/U][G/A]m6AC[U/A/C]), at particular locations on mRNA that can dictate the modification’s function [23, 46]. The process of m6A deposition and removal is highly regulated, and differs significantly based on cell type, differentiation state, presence or absence of stress, and countless other factors. Furthermore, m6A recognition (‘reading’) is equally (if not more) complex, and recent studies have focused on identifying m6A reader proteins and investigating how similar reader enzymes can have separate and unique impacts on RNA fate [47–49]. In addition to the direct effect of m6A and its recognition by specific proteins, m6A also acts indirectly on RNA molecules by weakening local RNA duplexes, which opens up the sequence to additional RNA-binding proteins (termed the ‘m6A switch’) (see Figure 1, path B) [50–52].
2.2. The m6A lifecycle: writers, readers, and erasers
The components of the m6A methyltransferase complex were discovered starting in 1994 with the work of Bokar, Rottman, and colleagues, who cloned methyltransferase-like 3 (METTL3, also known as MTA-70, MTA, or IME4) [53]. This was followed closely by the discovery of the METTL3 adaptor protein Wilms tumor 1-associated protein (WTAP) and later, METTL14 and KIAA1429 (also called VIRMA) [54–57]. METTL3 is the canonical catalytic enzyme responsible for m6A addition to mRNA, although all parts of the methyltransferase complex are required for efficient m6A deposition. Despite METTL14’s significant (approximately 43%) sequence homology with METTL3 and the presence of similar motifs required for catalytic activity, several studies have determined that METTL14 is in fact not catalytically active and instead acts as a scaffold for RNA binding to promote METTL3 activity; therefore METTL14 is dubbed a ‘pseudomethyltransferase,’ albeit an indispensable component of the methyltransferase complex [58–60]. WTAP mediates METTL3-METTL14 localization to nuclear speckles and is a similarly necessary component of the complex, as is KIAA1429/VIRMA (although the function of this component is not yet understood) [54]. Finally, recent work has identified RBM15 and RBM15B as additional components of the methyltransferase complex, which confer targeting specificity to certain adenosines [26, 61, 62].
Arguably the event that re-kindled scientific excitement towards m6A was the discovery that the fat mass and obesity-associated (FTO) protein, which regulates metabolism and energy utilization and is implicated in obesity, is an m6A demethylase or ‘eraser’, thus implicating m6A in human physiological processes [63]. FTO’s ability to erase m6A marks suggested the dynamic, regulatory, and potentially targetable nature of mRNA methylation (Fig. 1, path A). Despite initial enthusiasm for this protein as both a demethylase and a culprit in obesity, several studies have recently demonstrated that obesity-associated mutations in FTO do not affect the FTO protein, but rather the neighboring genes Irx3 and Rpgrip1l [64, 65]. In addition, several groups have hinted that m6A may not be the only FTO substrate [19]. Antibodies to m6A do not discriminate between m6A and the similar m6Am in m6A-RNA Immunoprecipitation followed by RNA sequencing (MeRIP-seq) experiments, and Mauer et al. demonstrated that FTO knockdown significantly affects m6Am, but does not appreciably increase m6A levels in vitro [19]. Despite this, there remains significant controversy about FTO substrate specificity, as Wei et al have demonstrated that FTO preferentially targets nuclear m6A in various RNAs and both m6A and m6Am in cytoplasmic mRNA [66]. They have addressed controversial FTO-m6Am specificity by demonstrating that indeed FTO demethylates m6Am with higher affinity in vitro, but in various cell lines FTO may act on both substrates, a system which depends on cell state and even RNA species [67]. A common theme surrounding methylation deposition and removal appears to be extreme complexity in regulation, which is not fully understood. In the future, this intricate system may be exploited to target particular mRNAs, or even specific m6A moieties, to regulate physiological processes.
In addition to FTO, alkB homolog 5 RNA demethylase (ALKBH5) is an m6A demethylase, but it does not act on m6Am. ALKBH5 is localized in the nucleus, hinting that demethylation can rapidly occur after methylation, before the mature mRNA is shuttled to the cytoplasm [68]. However, due to its nuclear location ALKBH5 is able to demethylate other m6A-containing noncoding RNAs, such as the lncRNA MALAT1 and some small nuclear RNAs (snRNAs) and snoRNAs [25]. Murine global ALKBH5 knockout confers a relatively mild phenotype - knockout mice have defective spermatogenesis but are otherwise normal, suggesting that ALKBH5-mediated m6A demethylation plays a subtle role in signaling that may be compensated by other pathways (Figure 1, path A) [68].
The final class of proteins directly involved in RNA metabolism are the m6A readers or YTH proteins (YT521-B homology), which fall into three major classes: the DF family (YTHDF1, 2, and 3), YTHDC1, and YTHDC2. The YTH domain of these 5 proteins is the conserved site of selective m6A binding [69]. The DF family members are highly similar to each other, reside in the cytoplasm, and have a large low-complexity domain enriched in Q, N, and P residues [69]. DC1 appears to be the major nuclear m6A reader and can mediate splicing m6A-regulated splicing events (see Figure 1, path C), whereas DC2 binds noncoding RNAs and intronic and intergenic regions, but its function is poorly understood [26, 70, 71]. As the DF readers are a major determinant of the impact of a particular m6A modification on RNA metabolism, they will be discussed in more detail below.
2.3. m6A-protein interactions specify RNA fate
The YTHDF proteins were originally thought to have separate, discrete functions on m6A-modified mRNAs. He and colleagues first studied the function of YTHDF2, demonstrating that YTHDF2 knockdown induces half-life increases in several thousand transcripts, concluding that YTHDF2-m6A binding induces mRNA instability [66]. Despite this, ribosome profiling experiments showed a negligible effect of YTHDF2-m6A binding on translation efficiency, suggesting that this reader may be influencing mRNA transcript turnover more than translation efficiency [66]. More recently, YTHDF2 has been shown to recruit the CCR4-NOT deadenylase complex to m6A-containing mRNAs, thereby directing them to cytoplasmic P bodies and promoting their decay (Figure 1, path D) [72].
Similarly, the reader YTHDF1 is also directly involved mRNA stability and translation. The He group found that, in contrast to YTHDF2, YTHDF1 acts to promote translation by directly interacting with eiF3 and other translation initiation factors, promoting cap-independent translation of m6A-containing mRNAs [49]. Although first thought to have Different mRNA targets than YTHDF2, recent work has shown that YTHDF1 in fact shares the same pool of mRNA targets as its family members YTHDF2 and YTHDF3 (Figure 1, path D) [26].
The final DF reader, YTHDF3, does not seem to have as clearly defined a function as YTHDF1 or YTHDF2. In fact, YTHDF3 interacts cooperatively with YTHDF2 to enhance mRNA decay, but can also act in concert with YTHDF1 to promote translation of targets [47, 73]. He’s group suggested a dynamic model by which m6A-methyated mRNAs are shuttled out of the cytoplasm and recognized by YTHDF3, which then acts as a “buffering agent” to mediate interactions with YTHDF1 (for translation) or to YTHDF2 (for decay) (Figure 1, path D) [73]. This system would allow for precise control of protein production during states in which cells require rapid protein expression turnover, such as cell differentiation and circadian rhythm. For more detailed reviews on m6A-binding proteins and impacts on RNA metabolism, see Refs. [61, 69, 74, 75].
2.4. A-to-I RNA editing in RNA metabolism
Similar to m6A modifications, RNA editing can affect every aspect of RNA metabolism, from transcription to RNA degradation. ADAR1 and ADAR2 exert a similar catalytic activity that modifies adenosine to inosine. RNA editing by ADARs may control RNA metabolism through Differential regulation of the binding of RNA-binding proteins to their targets [76]. This is dependent on the precise location of editing on the RNA segment. Edited transcripts have varied fates, which may be either physiological or pathological, depending on context (Figure 2). RNA editing may participate in genome recoding events at the RNA level which can in turn influence protein function [76]. Since RNA editing occurs cotranscriptionally, it can regulate alternative splicing, [77] RNA silencing, [78] trapping of the RNA in the nucleus, [79] or induce RNA degradation [80]. The many Different fates of edited RNA transcripts suggest that ADAR-mediated editing acts to regulate RNA metabolism in various ways depending on cell state (Figure 2).
3. m6A and A-to-I RNA editing regulate cellular processes: implications for CVD
3.1. m6A in development
During development, cells respond to an array of signals dictating how they must Differentiate spatially and functionally, and this requires precise coordination of transcription and translation to guarantee that the necessary genes are expressed at appropriate times. As previously described, m6A deposition, recognition, and removal can dramatically impact the stability and translation of certain mRNAs, and therefore m6A was hypothesized to play a role in stem cell differentiation. METTL3 knockout in mouse embryonic stem cells prevented differentiation and instead maintained cells in a naïve pluripotent state [81, 82]. In addition, several transcripts essential for maintaining pluripotency in humans and mice (such as Nanog) are normally m6A methylated and thereby targeted for degradation [81–83]. The impact of m6A upon differentiation is so significant that global knockout of METTL3 in mice is lethal during early development [82]. Interestingly, as previously discussed, global knockout of the demethylase ALKBH5 produces mice with a relatively mild phenotype, namely impairments in testes development and spermatogenesis, and these defects have been hypothesized to be caused by aberrant gene expression regulation [68]. Noticeably similar phenotypes (i.e. developmental impairment) are obtained between global METTL3 knockout, global ALKBH5 knockout, and germ-cell-specific METTL3 knockout (which also causes depletion of spermatogonial stem cells and sterility) [84]. This reinforces the notion that proper m6A deposition, recognition, and removal is necessary for coordinated expression of pluripotency and lineage commitment markers and proper, timely cellular differentiation.
3.2. ADAR1 and ADAR2 in development
RNA modifications catalysed by ADAR1 or ADAR2 are an essential component of life, and therefore homeostatic and regulated levels of these enzymes are critical. Genetic ablation of either ADAR1 [85] or ADAR2 [86] in mice led to either prenatal or early postnatal lethality, highlighting the importance of RNA editing in normal physiology. ADAR1 seems to play an essential role in haematopoiesis, organ homeostasis, and suppression of innate immune system activation [87]. However, the exact mechanisms at the cellular level are poorly understood [88]. In contrast, ADAR2−/− mice are prone to seizures and die prematurely before P20, caused by neuronal death after excess influx of Ca2+ through the unedited glutamate receptor [86]. In this case, RNA editing of the glutamate receptor pre-mRNA is essential for the physiologic function of this gene. Although ADAR1 and ADAR2 possess the same deaminase domain, the site of RNA editing and the RNA molecules being targeted are subtly Different and may explain the Different phenotypes observed.
3.3. m6A in stress responses
Environmental perturbations and cellular stress have profound effects on gene expression and translation, and given the dynamic nature of m6A modifications and their ability to influence mRNA metabolism, several studies have focused on the role of m6A in cell stress. Zhou et al. discovered that following heat shock, increased m6A methylation in the 5’UTR promotes translation initiation in a cap-independent manner [89]. This response is directly linked with increased expression of YTHDF2, which the authors demonstrated directly competed with FTO in the nucleus to bind m6A on select heat-shock-response mRNAs and promote their translation [89]. Similarly, Xiang et al. showed that METTL3 activity, and therefore presence of m6A, was required for efficient repair of UV-induced DNA damage in a human osteosarcoma cell line [90]. In this scenario, m6A is necessary for timely localization of the DNA repair enzyme DNA Polymerase κ (Polκ) to the site of DNA damage [90]. Finally, a recent study has examined the roles of both m6A and m6Am (m6A/m) in stress response regulation in a mouse model of fear behavior and human Major Depressive Disorder (MDD); m6A/m changes alter transcriptome regulation following acute stress in mice, and the authors propose that m6A/m regulation in the peripheral blood of MDD patients may approximate the brain’s response to the same modifications [91]. A clear understanding of which transcripts are methylated during stress, and furthermore their larger effects on the cell, will likely be complicated by the diverse stress-response mechanisms employed by Different cell types.
m6A is crucial for stem cell fate and regulation of the stress response, and therefore seems to have a natural connection to cancer biology. Aberrant FTO expression has been described in certain subtypes of acute myeloid leukemia (AML), where forced FTO expression enhances AML cell survival and proliferation by decreasing levels of ASB2 and RARA, suggesting FTO as an ‘oncogenic m6A demethylase’ [92]. Notably, this is Different from the observation that many transcripts are stabilized by demethylation, e.g. by overexpression of FTO. Similarly, the demethylase ALKBH5 has been shown to enhance self-renewal of glioblastoma stem cells via demethylation and subsequent upregulation of FOXM1, and to promote proliferation of breast cancer stem cells via NANOG in a similar manner [93, 94]. As the major components of the methyltransferase complex, METTL3 and METTL14 has also recently been implicated in cancer pathogenesis. Lin and Choe et al first reported that METTL3 interacts with cap-dependent translation machinery, specifically eiF3, to promote the translation of oncogenes such as EGFR and TAZ, and furthermore that METTL3 is required for the proliferation of lung adenocarcinoma cells [95]. Subsequently, the methyltransferase writer complex was confirmed to be crucial for AML progression in three independent studies [96–98]. These studies, which have been reviewed extensively elsewhere, ([69, 99]) demonstrate the importance and specificity of m6A ‘writing’ and ‘erasing’ in Different cancers, especially leukemias, and the potential therapeutic benefit m6A modulation may provide. Despite this, future work is necessary to fully understand and exploit methylation-dependent transcriptome changes during stress and cancer, and these will likely vary depending on the type of cancer and the cellular environment which it creates.
3.4. A-to-I RNA editing in stress responses
Accumulating evidence suggests that cellular stress responses are critically regulated by RNA editing. The first work on this area showed that disruption of the dADAR gene in Drosophilia melanogaster leads to heat shock vulnerability and necessitates increased adaptation periods after oxygen deprivation [100]. Following this, it was suggested that A-to-I RNA editing may be one of the mechanisms cells use to regulate changes in gene expression after hypoxia, as RNA editing patterns are altered after hypoxia in mammalian cell lines. In addition to hypoxia, Differential regulation of ADAR1 has been demonstrated following inflammation [101] and increased oxidative stress [102]. Environmental factors such as energy and nutrient deprivation have been also shown to influence the expression or activity of ADARs [103, 104]. All these stress responses are firmly associated with cardiovascular disease, however there is little known at transcript level regarding the specific role of A-to-I RNA editing in these pathologies.
3.5. mRNA methylation and demethylation in cardiac disease
Given the explosion of studies investigating the role of m6A in development, stress responses, and cancer, it is not surprising that there has recently been interest in elucidating the role of m6A in cardiovascular health and disease. The importance of m6A for proper timing and regulation of the circadian rhythm hinted to its connection with cardiovascular disease, as misalignment of the circadian clock is also shown to increase CVD risk factors (hypertension, inflammatory markers, etc.) in humans [105]. In addition, METTL3 and appropriate m6A deposition is necessary for directed differentiation of mouse embryonic stem cells into cardiomyocytes and for the maintenance of MyoD in proliferative skeletal muscle myoblasts, underscoring the importance of this modification in muscle physiology [81, 106].
Two studies have been recently published investigating the role of m6A, through modulation of either FTO or METTL3, in cardiac remodeling and function following stress [40, 41]. The first of these studies, by the Sahoo group, examined the functional effect of FTO on cardiac contractile function and contractile transcripts during both cardiac homeostasis and myocardial ischemia-induced heart failure. They found that m6A is increased in failing human, pig, and mouse hearts, hinting at the modification’s clinical relevance in cardiac disease. MeRIP-seq experiments identified hypermethylation following myocardial infarction in mice in transcripts associated with cardiac hypertrophy, contraction, and sarcomere dynamics (NPPA, SERCA2a, MYH7, etc.) [40]. In addition, the authors show a corresponding decrease in FTO expression levels during heart failure, and reasoned that increased FTO expression could attenuate maladaptive cardiac remodeling following myocardial ischemia [40]. FTO knockdown induced arrhythmic events in primary isolated cardiomyocytes as well as proarrhythmic remodeling and altered ventricular repolarization in mice lacking FTO. Conversely, FTO overexpression attenuated hypoxia-induced cardiomyocyte dysfunction and restored calcium handling and sarcomere dynamics [40]. The authors of this study concluded that, due to m6A-mediated downregulation of several calcium-handling and sarcomere contractile transcripts and corresponding decreased protein expression, forced expression of FTO attenuated ischemia-induced cardiac remodeling and may therefore represent a potential therapeutic target [40]. Overall, this study presents a novel role for the demethylase FTO in the regulation of cardiac contractility and remodeling following ischemic injury.
Recently, our group has published a similar study examining the effects of m6A and its writer, METTL3, on cardiac homeostasis and hypertrophy. We have found that m6A is a dynamic modification increased in specific transcripts (including members of the Mitogen-Activated Protein Kinase (Mapk) family) in isolated primary cardiomyocytes stimulated to hypertrophy, suggesting that METTL3 and m6A act to regulate the hypertrophic response [41]. Cardiomyocyte-specific METTL3 overexpression both in vitro and in vivo stimulates physiological cardiomyocyte hypertrophy, and interestingly does not induce cardiac dysfunction in METTL3-overexpressing mice following long term pressure-overload stress [41]. In contrast, cardiomyocyte-specific METTL3 knockout prevents hypertrophy in vitro and negatively impacts cardiac homeostasis in vivo; METTL3-knockout hearts develop maladaptive eccentric remodeling and cardiac functional defects with aging and rapid, progressive dysfunction following acute pressure-overload stress [41]. Interestingly, the defects in hypertrophy seen in METTL3 knockout mice seem to affect cardiomyocyte morphology prior to the development of functional defects, suggesting that m6A content is crucial for regulating cardiomyocyte geometry and structural adaptation to stress prior to symptom onset [41]. In fact, it appears that m6A is both necessary and sufficient for cardiac hypertrophy to occur, and therefore targeting the m6A-METTL3 pathway is a potential, novel therapeutic avenue for patients in which cardiac hypertrophy is aberrant and homeostasis is perturbed.
3.6. A-to-I RNA editing in CVD
The relevance and mechanistic role of adenosine to inosine RNA editing in the cardiovascular system and disease is largely unknown, with the exception of only few reports documenting a critical role of RNA editing in cardiovascular biology. A pilot study towards this aim has previously reported that children with cyanotic congenital heart disease manifested significantly higher rates of A-to-I RNA editing in the MED13 RNA than acyanotic, suggesting that RNA modification may influence the cellular and metabolic pathways in congenital cardiac defects [42]. We have recently shown that ADAR1 plays a critical role in the regulation of proinflammatory endothelial cells. Specifically we described that ADAR-1 induced A-to-I RNA editing controls the mRNA stability of the extracellular matrix degradation enzyme cathepsin S, and that the increase of ADAR1 expression in atherosclerotic inflammatory diseases results in increased expression of cathepsin S [38]. Interesting other findings have also contributed to establishing a link between RNA editing and CVD. Fei et al unraveled a new mechanism of contractile protein repression in smooth muscle cell (SMC) dedifferentiation through ADAR1-mediated RNA editing [43]. As mentioned previously, miRNA can also be affected by RNA editing. The editing of certain miRNAs such as miR487b, which is increased in murine muscle tissue during postischemic neovascularization, results in a new proangiogenic RNA with Different target specificity [107]. More recently, Jain et al demonstrated that Filamin A pre-mRNA editing by ADAR2 triggers a Q-to-R codon exchange at the end of exon 42 which regulates the activity of key smooth muscle contraction regulators such as PLC and ROCK machinery [108]. Lack of editing in Filamin A pre-mRNA produces a Filamin A isoform that only encodes a glutamine residue (Q) leading to mislocalization of p190RhoGAP, misregulation of PLC and ROCK signaling, increased MLC phosphorylation, aortic hypercontraction, thickening of the smooth muscle layer, and increased perivascular collagen deposition [108]. Consequently, loss of Filamin A editing leads to persistently elevated diastolic blood pressure resulting in left ventricular hypertrophy in mice [108].
4. Conclusions and Future Perspectives
The above studies contribute to a field of cardiovascular epitranscriptomics that is still in its infancy, and more work will be necessary to determine how m6A writers, erasers, and readers, as well as ADAR-associated events, are able to influence cardiac function and pathophysiology. Research will no doubt require focus on unraveling the complexities associated with this field, such as the importance of m6A in cardiomyocytes versus other cell types in the heart, Different forms of cardiac injury (ischemia +/− reperfusion, pressure overload, response to neurohumoral stimulation, etc.), and the interplay between the cardiovascular system and other organ systems frequently perturbed during CVD. As it appears that a proper amount of m6A is necessary on very specific targets is necessary for proper cardiomyocyte function, both at baseline and with injury, a ‘simple solution’ of inhibiting or activating m6A writers or erasers in a particular cell type or organ would likely not provide a lasting solution for CVD in vivo. Further specificity may be achieved by manipulating the levels of particular m6A readers in a cell-type or state-dependent manner, however these types of studies are only beginning to be done. Besides m6A mRNA methylation, it is also unclear how other RNA modifications (methylations of other nucleotides or even small and noncoding RNAs, pseudouridine formation, etc.) contribute to transcript regulation in CVD, or whether these modifications will have entirely distinct functions on RNA metabolism. Also, the use of the most recent technological advances in multiplex genome-editing tools such as CRISPR/Cas [109, 110] coupled with the power of A-to-I RNA substitutional editing, may provide a new therapeutic option. Overall, current advances in sequencing and mapping technologies for RNA modifications will undoubtedly lead to the discovery of even more novel mechanisms of gene expression regulation for CVD in the future.
Glossary
- m6A
N6-methyladenosine
- mRNA
messenger RNA
- METTL3
Methyltransferase-like 3
- METTL14
Methyltransferase-like 14
- WTAP
Wilms tumor-1 associated protein
- KIAA1429
vir like m6A methyltransferase associated
- FTO
fat mass and obesity-associated
- ALKBH5
AlkB homolog 5, RNA demethylase
- RNA-BP
RNA binding protein
- YTHDC1
YTH domain-containing protein 1
- YTHDF
YTH domain-containing family protein
- P body
processing body
- CCR4-NOT
CCR4-NOT deadenylase complex
- eiF3
elongation initiation factor 3
- 40S
40S ribosomal subunit
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, de Ferranti SD, Ferguson JF, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Lutsey PL, Mackey JS, Matchar DB, Matsushita K, Mussolino ME, Nasir K, O’Flaherty M, Palaniappan LP, Pandey A, Pandey DK, Reeves MJ, Ritchey MD, Rodriguez CJ, Roth GA, Rosamond WD, Sampson UKA, Satou GM, Shah SH, Spartano NL, Tirschwell DL, Tsao CW, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P, A.H.A.C.o.E.a.P.S.C.a.S.S. Subcommittee, Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association, Circulation 137(12) (2018) e67–e492. [DOI] [PubMed] [Google Scholar]
- [2].Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN, Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy, Science 351(6271) (2016) 400–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].El Refaey M, Xu L, Gao Y, Canan BD, Adesanya TMA, Warner SC, Akagi K, Symer DE, Mohler PJ, Ma J, Janssen PML, Han R, In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice, Circ Res 121(8) (2017) 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, den Dunnen JT, Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations, Hum Mutat 30(3) (2009) 293–9. [DOI] [PubMed] [Google Scholar]
- [5].Corey DR, Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy, Nat Neurosci 20(4) (2017) 497–499. [DOI] [PubMed] [Google Scholar]
- [6].Hulot JS, Salem JE, Redheuil A, Collet JP, Varnous S, Jourdain P, Logeart D, Gandjbakhch E, Bernard C, Hatem SN, Isnard R, Cluzel P, Le Feuvre C, Leprince P, Hammoudi N, Lemoine FM, Klatzmann D, Vicaut E, Komajda M, Montalescot G, Lompre AM, Hajjar RJ, Effect of intracoronary administration of AAV1/SERCA2a on ventricular remodelling in patients with advanced systolic heart failure: results from the AGENT-HF randomized phase 2 trial, Eur J Heart Fail 19(11) (2017) 1534–1541. [DOI] [PubMed] [Google Scholar]
- [7].Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, Yaroshinsky A, Zsebo KM, Dittrich H, Hajjar RJ, Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure, Circulation 124(3) (2011) 304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kaneko M, Hashikami K, Yamamoto S, Matsumoto H, Nishimoto T, Phospholamban Ablation Using CRISPR/Cas9 System Improves Mortality in a Murine Heart Failure Model, PLoS One 11(12) (2016) e0168486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Laina A, Gatsiou A, Georgiopoulos G, Stamatelopoulos K, Stellos K, RNA Therapeutics in Cardiovascular Precision Medicine, Front Physiol 9 (2018) 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dowdy SF, Overcoming cellular barriers for RNA therapeutics, Nat Biotechnol 35(3) (2017) 222–229. [DOI] [PubMed] [Google Scholar]
- [11].Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM, MODOMICS: a database of RNA modification pathways. 2017 update, Nucleic Acids Res 46(D1) (2018) D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Furuichi Y, LaFiandra A, Shatkin AJ, 5’-Terminal structure and mRNA stability, Nature 266(5599) (1977) 235–9. [DOI] [PubMed] [Google Scholar]
- [13].Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T, Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA, Nucleic Acids Res 40(11) (2012) 5023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, Wetzel C, Kreher J, Soin R, Creppe C, Limbach PA, Gueydan C, Kruys V, Brehm A, Minakhina S, Defrance M, Steward R, Fuks F, RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine, Science 351(6270) (2016) 282–5. [DOI] [PubMed] [Google Scholar]
- [15].Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Dore LC, Amariglio N, Rechavi G, He C, The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA, Nature 530(7591) (2016) 441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C, Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome, Nat Chem Biol 12(5) (2016) 311–6. [DOI] [PubMed] [Google Scholar]
- [17].Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G, Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq, Nature 485(7397) (2012) 201–6. [DOI] [PubMed] [Google Scholar]
- [18].Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jarey SR, Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons, Cell 149(7) (2012) 1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, Gross SS, Elemento O, Debart F, Kiledjian M, Jarey SR, Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability, Nature 541(7637) (2017) 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, Dominissini D, He C, Nm-seq maps 2’-O-methylation sites in human mRNA with base precision, Nat Methods 14(7) (2017) 695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perry R, Kelley D, Existence of methylated messenger RNA in mouse L cells, Cell, 1974, pp. 37–42.4370211 [Google Scholar]
- [22].Rottman F, Shatkin AJ, Perry RP, Sequences containing methylated nucleotides at the 5’ termini of messenger RNAs: possible implications for processing, Cell 3(3) (1974) 197–9. [DOI] [PubMed] [Google Scholar]
- [23].Narayan P, Rottman FM, An in vitro system for accurate methylation of internal adenosine residues in messenger RNA, Science 242(4882) (1988) 1159–62. [DOI] [PubMed] [Google Scholar]
- [24].Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, Kellner S, Holter SM, Garrett L, Wurst W, Becker L, Klopstock T, Fuchs H, Gailus-Durner V, Hrabe de Angelis M, Karadottir RT, Helm M, Ule J, Gleeson JG, Odom DT, Frye M, Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders, Embo j 33(18) (2014) 2020–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jarey SR, Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome, Nat Methods 12(8) (2015) 767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jarey SR, m(6)A RNA methylation promotes XIST-mediated transcriptional repression, Nature 537(7620) (2016) 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z, Extensive translation of circular RNAs driven by N(6)-methyladenosine, Cell Res 27(5) (2017) 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B, N6-adenosine methylation in MiRNAs, PLoS One 10(2) (2015) e0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Doré LC, Amariglio N, Rechavi G, He C, The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA, Nature 530(7591) (2016) 441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M, Characterizing 5-methylcytosine in the mammalian epitranscriptome, Genome Biol 14(11) (2013) 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y, Illumina-based RiboMethSeq approach for mapping of 2’-O-Me residues in RNA, Nucleic Acids Res 44(16) (2016) e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bass BL, Weintraub H, A developmentally regulated activity that unwinds RNA duplexes, Cell 48(4) (1987) 607–13. [DOI] [PubMed] [Google Scholar]
- [33].Rebagliati MR, Melton DA, Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity, Cell 48(4) (1987) 599–605. [DOI] [PubMed] [Google Scholar]
- [34].Bass BL, Weintraub H, Cattaneo R, Billeter MA, Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA, Cell 56(3) (1989) 331. [DOI] [PubMed] [Google Scholar]
- [35].Stellos K, The rise of epitranscriptomic era: implications for cardiovascular disease, Cardiovasc Res 113(5) (2017) e2–e3. [DOI] [PubMed] [Google Scholar]
- [36].Nishikura K, A-to-I editing of coding and non-coding RNAs by ADARs, Nat Rev Mol Cell Biol 17(2) (2016) 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gagnidze K, Rayon-Estrada V, Harroch S, Bulloch K, Papavasiliou FN, A New Chapter in Genetic Medicine: RNA Editing and its Role in Disease Pathogenesis, Trends Mol Med 24(3) (2018) 294–303. [DOI] [PubMed] [Google Scholar]
- [38].Stellos K, Gatsiou A, Stamatelopoulos K, Perisic Matic L, John D, Lunella FF, Jae N, Rossbach O, Amrhein C, Sigala F, Boon RA, Furtig B, Manavski Y, You X, Uchida S, Keller T, Boeckel JN, Franco-Cereceda A, Maegdefessel L, Chen W, Schwalbe H, Bindereif A, Eriksson P, Hedin U, Zeiher AM, Dimmeler S, Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation, Nat Med 22(10) (2016) 1140–1150. [DOI] [PubMed] [Google Scholar]
- [39].Bazak L, Levanon EY, Eisenberg E, Genome-wide analysis of Alu editability, Nucleic Acids Res 42(11) (2014) 6876–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N, Jha D, Zhang S, Kohlbrenner E, Chepurko E, Chen J, Trivieri MG, Singh R, Bouchareb R, Fish K, Ishikawa K, Lebeche D, Hajjar RJ, Sahoo S, FTO-Dependent m6A Regulates Cardiac Function During Remodeling and Repair, Circulation (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dorn LE, Lasman L, Chen J, Xu X, Hund TJ, Medvedovic M, Hanna JH, van Berlo JH, Accornero F, The N(6)-Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy, Circulation 139(4) (2019) 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Borik S, Simon AJ, Nevo-Caspi Y, Mishali D, Amariglio N, Rechavi G, Paret G, Increased RNA editing in children with cyanotic congenital heart disease, Intensive Care Med 37(10) (2011) 1664–71. [DOI] [PubMed] [Google Scholar]
- [43].Fei J, Cui XB, Wang JN, Dong K, Chen SY, ADAR1-Mediated RNA Editing, A Novel Mechanism Controlling Phenotypic Modulation of Vascular Smooth Muscle Cells, Circ Res 119(3) (2016) 463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gatsiou A, Vlachogiannis N, Lunella FF, Sachse M, Stellos K, Adenosine-to-Inosine RNA Editing in Health and Disease, Antioxid Redox Signal 29(9) (2018) 846–863. [DOI] [PubMed] [Google Scholar]
- [45].Gatsiou A, Stellos K, Dawn of Epitranscriptomic Medicine, Circ Genom Precis Med 11(9) (2018) e001927. [DOI] [PubMed] [Google Scholar]
- [46].Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F, Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex, J Biol Chem 269(26) (1994) 17697–704. [PubMed] [Google Scholar]
- [47].Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, Bhattarai DP, Zhao YL, Sun BF, Yang YG, Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation, Cell Res 27(3) (2017) 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian SB, Ignatova Z, Dynamic m(6)A methylation facilitates mRNA triaging to stress granules, Life Sci Alliance 1(4) (2018) e201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C, N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency, Cell 161(6) (2015) 1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T, N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions, Nature 518(7540) (2015) 560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kierzek E, Kierzek R, The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines, Nucleic Acids Res 31(15) (2003) 4472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T, N(6)-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding, J Mol Biol 428(5 Pt A) (2016) 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM, Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase, Rna 3(11) (1997) 1233–47. [PMC free article] [PubMed] [Google Scholar]
- [54].Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Rendtlew Danielsen JM, Liu F, Yang YG, Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase, Cell Res 24(2) (2014) 177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C, A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation, Nat Chem Biol 10(2) (2014) 93–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, Sanjana NE, Freinkman E, Pacold ME, Satija R, Mikkelsen TS, Hacohen N, Zhang F, Carr SA, Lander ES, Regev A, Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites, Cell Rep 8(1) (2014) 284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bujnicki JM, Feder M, Radlinska M, Blumenthal RM, Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase, J Mol Evol 55(4) (2002) 431–44. [DOI] [PubMed] [Google Scholar]
- [58].Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P, Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex, Nature 534(7608) (2016) 575–8. [DOI] [PubMed] [Google Scholar]
- [59].Wang P, Doxtader KA, Nam Y, Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases, Mol Cell 63(2) (2016) 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wang X, Huang J, Zou T, Yin P, Human m(6)A writers: Two subunits, 2 roles, RNA Biol 14(3) (2017) 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Meyer KD, Jarey SR, Rethinking m(6)A Readers, Writers, and Erasers, Annu Rev Cell Dev Biol 33 (2017) 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villasenor R, Hess D, Andrade-Navarro MA, Biggiogera M, Helm M, Soller M, Buhler M, Roignant JY, Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d, Genes Dev 32(5–6) (2018) 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C, N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO, Nat Chem Biol 7(12) (2011) 885–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez-Skarmeta JL, Nóbrega MA, Obesity-associated variants within FTO form long-range functional connections with IRX3, Nature 507(7492) (2014) 371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Stratigopoulos G, Burnett LC, Rausch R, Gill R, Penn DB, Skowronski AA, LeDuc CA, Lanzano AJ, Zhang P, Storm DR, Egli D, Leibel RL, Hypomorphism of Fto and Rpgrip1l causes obesity in mice, J Clin Invest 126(5) (2016) 1897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C, N6-methyladenosine-dependent regulation of messenger RNA stability, Nature 505(7481) (2014) 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, Jia G, Chen J, He C, Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm, Mol Cell 71(6) (2018) 973–985.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C, ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility, Mol Cell 49(1) (2013) 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nachtergaele S, He C, Chemical Modifications in the Life of an mRNA Transcript, Annu Rev Genet (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, Yang Y, Huang N, Jiang GB, Wang HL, Zhou Q, Wang XJ, Zhao YL, Yang YG, Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing, Mol Cell 61(4) (2016) 507–519. [DOI] [PubMed] [Google Scholar]
- [71].Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ, Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development, PLoS Genet 14(5) (2018) e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L, YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex, Nat Commun 7 (2016) 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C, YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA, Cell Res 27(3) (2017) 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Patil DP, Pickering BF, Jarey SR, Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins, Trends Cell Biol 28(2) (2018) 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Roignant JY, Soller M, m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression, Trends Genet 33(6) (2017) 380–390. [DOI] [PubMed] [Google Scholar]
- [76].Bhalla T, Rosenthal JJ, Holmgren M, Reenan R, Control of human potassium channel inactivation by editing of a small mRNA hairpin, Nat Struct Mol Biol 11(10) (2004) 950–6. [DOI] [PubMed] [Google Scholar]
- [77].Rueter SM, Dawson TR, Emeson RB, Regulation of alternative splicing by RNA editing, Nature 399(6731) (1999) 75–80. [DOI] [PubMed] [Google Scholar]
- [78].Bass BL, How does RNA editing affect dsRNA-mediated gene silencing?, Cold Spring Harb Symp Quant Biol 71 (2006) 285–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kumar M, Carmichael GG, Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts, Proc Natl Acad Sci U S A 94(8) (1997) 3542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Scadden AD, The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage, Nat Struct Mol Biol 12(6) (2005) 489–96. [DOI] [PubMed] [Google Scholar]
- [81].Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, Carter AC, Flynn RA, Zhou C, Lim KS, Dedon P, Wernig M, Mullen AC, Xing Y, Giallourakis CC, Chang HY, m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells, Cell Stem Cell 15(6) (2014) 707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-Haim MS, Eyal E, Yunger S, Pinto Y, Jaitin DA, Viukov S, Rais Y, Krupalnik V, Chomsky E, Zerbib M, Maza I, Rechavi Y, Massarwa R, Hanna S, Amit I, Levanon EY, Amariglio N, Stern-Ginossar N, Novershtern N, Rechavi G, Hanna JH, Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation, Science 347(6225) (2015) 1002–6. [DOI] [PubMed] [Google Scholar]
- [83].Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC, N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells, Nat Cell Biol 16(2) (2014) 191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, Zhang Y, Song W, Jia G, Yang X, He C, Tong MH, Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis, Cell Res 27(10) (2017) 1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wang Q, Khillan J, Gadue P, Nishikura K, Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis, Science 290(5497) (2000) 1765–8. [DOI] [PubMed] [Google Scholar]
- [86].Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH, Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2, Nature 406(6791) (2000) 78–81. [DOI] [PubMed] [Google Scholar]
- [87].Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR, RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself, Science 349(6252) (2015) 1115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, Jantsch MF, Dorin J, Adams IR, Scadden AD, Ohman M, Keegan LP, O’Connell MA, The RNA-editing enzyme ADAR1 controls innate immune responses to RNA, Cell Rep 9(4) (2014) 1482–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhou J, Wan J, Gao X, Zhang X, Jarey SR, Qian SB, Dynamic m(6)A mRNA methylation directs translational control of heat shock response, Nature 526(7574) (2015) 591–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, Ling D, Hsu PH, Zou L, Jambhekar A, He C, Shi Y, RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response, Nature 543(7646) (2017) 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Engel M, Eggert C, Kaplick PM, Eder M, Roh S, Tietze L, Namendorf C, Arloth J, Weber P, Rex-Haner M, Geula S, Jakovcevski M, Hanna JH, Leshkowitz D, Uhr M, Wotjak CT, Schmidt MV, Deussing JM, Binder EB, Chen A, The Role of m(6)A/m-RNA Methylation in Stress Response Regulation, Neuron 99(2) (2018) 389–403.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, Hong GM, Wang X, Chen P, Gurbuxani S, Arnovitz S, Li Y, Li S, Strong J, Neilly MB, Larson RA, Jiang X, Zhang P, Jin J, He C, Chen J, FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase, Cancer Cell 31(1) (2017) 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL, Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA, Proc Natl Acad Sci U S A 113(14) (2016) E2047–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, Majumder S, He C, Huang S, m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program, Cancer Cell 31(4) (2017) 591–606.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lin S, Choe J, Du P, Triboulet R, Gregory RI, The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells, Mol Cell 62(3) (2016) 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, Schulman J, Famulare C, Patel M, Klimek VM, Garrett-Bakelman FE, Melnick A, Carroll M, Mason CE, Jarey SR, Kharas MG, The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells, Nat Med 23(11) (2017) 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, De Braekeleer E, Ponstingl H, Hendrick A, Vakoc CR, Vassiliou GS, Kouzarides T, Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control, Nature 552(7683) (2017) 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, Sheng Y, Wang Y, Wunderlich M, Zhang B, Dore LC, Su R, Deng X, Ferchen K, Li C, Sun M, Lu Z, Jiang X, Marcucci G, Mulloy JC, Yang J, Qian Z, Wei M, He C, Chen J, METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification, Cell Stem Cell 22(2) (2018) 191–205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Pinello N, Sun S, Wong JJ, Aberrant expression of enzymes regulating m(6)A mRNA methylation: implication in cancer, Cancer Biol Med 15(4) (2018) 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ma E, Gu XQ, Wu X, Xu T, Haddad GG, Mutation in pre-mRNA adenosine deaminase markedly attenuates neuronal tolerance to O2 deprivation in Drosophila melanogaster, J Clin Invest 107(6) (2001) 685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Patterson JB, Samuel CE, Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase, Mol Cell Biol 15(10) (1995) 5376–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wang Y, Men M, Xie B, Shan J, Wang C, Liu J, Zheng H, Yang W, Xue S, Guo C, Inhibition of PKR protects against H2O2-induced injury on neonatal cardiac myocytes by attenuating apoptosis and inflammation, Sci Rep 6 (2016) 38753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Bhansali P, Dunning J, Singer SE, David L, Schmauss C, Early life stress alters adult serotonin 2C receptor pre-mRNA editing and expression of the alpha subunit of the heterotrimeric G-protein G q, J Neurosci 27(6) (2007) 1467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Gan Z, Zhao L, Yang L, Huang P, Zhao F, Li W, Liu Y, RNA editing by ADAR2 is metabolically regulated in pancreatic islets and beta-cells, J Biol Chem 281(44) (2006) 33386–94. [DOI] [PubMed] [Google Scholar]
- [105].Morris CJ, Purvis TE, Hu K, Scheer FA, Circadian misalignment increases cardiovascular disease risk factors in humans, Proc Natl Acad Sci U S A 113(10) (2016) E1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kudou K, Komatsu T, Nogami J, Maehara K, Harada A, Saeki H, Oki E, Maehara Y, Ohkawa Y, The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation, Open Biol 7(9) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].van der Kwast R, van Ingen E, Parma L, Peters HAB, Quax PHA, Nossent AY, Adenosine-to-Inosine Editing of MicroRNA-487b Alters Target Gene Selection After Ischemia and Promotes Neovascularization, Circ Res 122(3) (2018) 444–456. [DOI] [PubMed] [Google Scholar]
- [108].Jain M, Mann TD, Stulic M, Rao SP, Kirsch A, Pullirsch D, Strobl X, Rath C, Reissig L, Moreth K, Klein-Rodewald T, Bekeredjian R, Gailus-Durner V, Fuchs H, Hrabe de Angelis M, Pablik E, Cimatti L, Martin D, Zinnanti J, Graier WF, Sibilia M, Frank S, Levanon EY, Jantsch MF, RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure, Embo j 37(19) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, Fellows MD, Porritt MJ, Firth MA, Carreras A, Baccega T, Seeliger F, Bjursell M, Tsai SQ, Nguyen NT, Nitsch R, Mayr LM, Pinello L, Bohlooly YM, Aryee MJ, Maresca M, Joung JK, In vivo CRISPR editing with no detectable genome-wide off-target mutations, Nature 561(7723) (2018) 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F, RNA editing with CRISPR-Cas13, Science 358(6366) (2017) 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]