Abstract
α6-containing (α6*) nicotinic acetylcholine receptors (nAChRs) are expressed throughout the periphery and the central nervous system and constitute putative therapeutic targets in pain, addiction and movement disorders. The α6β2* nAChRs are relatively well studied, in part due to the availability of target specific α-conotoxins (α-Ctxs). In contrast, all native α-Ctxs identified that potently block α6β4 nAChRs exhibit higher potencies for the closely related α6β2β3 and/or α3β4 subtypes. In this study, we have identified a novel peptide from Conus ventricosus with pronounced selectivity for the α6β4 nAChR. The peptide-encoding gene was cloned from genomic DNA and the predicted mature peptide, α-Ctx VnIB, was synthesized. The functional properties of VnIB were characterized at rat and human nAChRs expressed in Xenopus oocytes by two-electrode voltage clamp electrophysiology. VnIB potently inhibited ACh-evoked currents at rα6β4 and rα6/α3β4 nAChRs, displayed ~20-fold and ~250-fold lower potencies at rα3β4 and rα6/α3β2β3 receptors, respectively, and exhibited negligible effects at eight other nAChR subtypes. Interestingly, even higher degrees of selectivity were observed for hα6/α3β4 over hα6/α3β2β3 and hα3β4 receptors. Finally, VnIB displayed fast binding kinetics at rα6/α3β4 (on-rate t½ = 0.87 min−1, off-rate t½ = 2.7 min−1). The overall preference of VnIB for β4* over β2* nAChRs is similar to the selectivity profiles of other 4/6 α-Ctxs. However, in contrast to previously identified native α-Ctxs targeting α6* nAChRs, VnIB displays pronounced selectivity for α6β4 nAChRs over both α3β4 and α6β2β3 receptors. VnIB thus represents a novel molecular probe for elucidating the physiological role and therapeutic properties of α6β4* nAChRs.
Keywords: nicotinic acetylcholine receptor (nAChR), α6-containing nAChRs, α6β4 nAChRs, α-conotoxin (α-Ctx), Conus, α-Ctx VnIB
1. Introduction
The neurotransmitter acetylcholine (ACh) mediates its fast neurotransmission through a highly heterogeneous family of ligand-gated ion channels, the nicotinic ACh receptors (nAChRs) (Albuquerque et al., 2009; Taly et al., 2009). The nAChRs are expressed throughout the periphery and the central nervous system (CNS), where they are involved in numerous distinct physiological functions, and the receptors have been targets for drug development for a wide range of neurological, psychiatric and cognitive disorders as well as for various forms of pain and addiction (Bertrand et al., 2015; Bertrand and Terry Jr, 2018; Hone et al., 2018; Picciotto et al., 2015; Quik and Wonnacott, 2011). A prominent recent example is the nAChR agonist varenicline, a smoking cessation drug with annual global sales in 2018 of $1,085 million (Pfizer Inc., 2019).
The nAChRs belong to the Cys-loop receptor superfamily, which also includes the 5-HT3 serotonin, γ-aminobutyric acidA and the glycine receptors (Chua and Chebib, 2017; Lynch et al., 2017; Walstab et al., 2010). Like other members of this receptor family, nAChRs are composed of five subunits surrounding a central ion pore. Signal transduction through the nAChR is initiated by agonist binding to orthosteric sites located in the extracellular subunit interfaces in the pentameric complex, which triggers the opening of the central ion pore, thus enabling cation influx into and depolarization of the neuron (Albuquerque et al., 2009; Taly et al., 2009).
The heterogeneity of native nAChRs arises from the existence of a total of 17 receptor subunits. In mammals, neuronal nAChRs are composed of different combinations of α (α2–7, α9 and α10) and β (β2–4) subunits, which assemble to form homomeric (composed exclusively of α7 or α9 subunits) or heteromeric (various α2–6/β2–4 combinations, α7β2 and α9α10) receptor complexes (Albuquerque et al., 2009; Taly et al., 2009). This gives rise to a wide range of receptor subtypes with distinct functional properties and expression patterns in vivo. Therefore, subtype-selective ligands, capable of discriminating between this multitude of receptor subtypes, are crucial for studies of the specific physiological roles of individual nAChRs.
The α4β2* and α7* (asterisk indicates the possible presence of additional subunits) nAChRs are the most abundant nAChR subtypes and are essentially expressed in all CNS regions (Taly et al., 2009). In contrast, α6* nAChRs display a much more restricted CNS distribution, as these receptors are confined mainly to the catecholaminergic pathways and the visual system (Azam et al., 2002; Le Novère et al., 1996). The α6 subunit forms functional receptors in combination with α4, β2, β3 and/or β4 subunits in vivo (Kuryatov et al., 2000; Letchworth and Whiteaker, 2011). α6β2* receptors (α6β2, α6β2β3, α4α6β2β3) are the predominant α6* nAChRs in the visual system (Gotti et al., 2005; Quik et al., 2011) and have been demonstrated to be involved in modulation of dopamine release from the striatum and nucleus accumbens (Azam et al., 2002; Letchworth and Whiteaker, 2011). These findings have sparked considerable interest in these receptors as potential therapeutic targets in Parkinson’s disease and nicotine addiction (Champtiaux et al., 2002; Miwa et al., 2011; Quik and Wonnacott, 2011). In contrast, albeit less studied, α6β4* nAChRs have been reported to regulate exocytosis in human and monkey adrenal chromaffin cells (Hernández-Vivanco et al., 2014; Pérez-Alvarez et al., 2012), to control the release of norepinephrine in mouse hippocampus (Azam and McIntosh, 2006), and to be expressed in rodent dorsal root ganglia neurons, where they may be involved in pain signaling (Hone et al., 2012a; Wieskopf et al., 2015).
Predatory marine snails of the Conus species feeds on mollusks, marine worms and fish and utilize their complex venoms to immobilize and capture their prey (Olivera et al., 2008). The genes encoding the toxins can be grouped into superfamilies based on similarities in the highly conserved signal sequence region of the peptide precursor. The mature conopeptides encoded by these genes can be characterized by the arrangements of their cysteine residues and by their pharmacological activity (Kaas et al., 2012; Robinson and Norton, 2014). α-Conotoxins (α-Ctx) are the predominant members of the A-superfamily of genes; these disulfide-rich peptides specifically target different nAChR subtypes and generally have a framework I cysteine pattern of CC-C-C with a Cys1-Cys3 and Cys2-Cys4 disulfide bond connectivity (Abraham and Lewis, 2018). α-Ctxs can be further subdivided based on the number of non-cysteine residues in the first and second inter-cysteine loop. The 3/5 α-Ctxs (three residues in the first cysteine-loop, five in the second) are selective blockers of the muscle nAChRs, while the 4/3, 4/4, 4/6 and 4/7 α-Ctxs mainly target neuronal nAChRs (Azam and McIntosh, 2009; Terlau and Olivera, 2004). Other members of the A-superfamily possess the same signal sequence homology, but display a framework IV cysteine pattern (CC-C-C-C-C) and target either voltage-gated K+ or Na+ channels (Craig et al., 1998; Kelley et al., 2006; Santos et al., 2004; Teichert et al., 2007).
Due to their pronounced selectivity for specific nAChR subtypes, α-Ctxs have been used in a large variety of studies to elucidate the physiological functions governed by different neuronal and muscle-type nAChRs (reviewed in (Giribaldi and Dutertre, 2018; Lebbe et al., 2014)). Relatively few of the α-Ctxs published to date target α6* nAChRs (Azam et al., 2005; Cartier et al., 1996; Dowell et al., 2003; Luo et al., 2013a; Luo et al., 2013b). α-Ctx MII was the first α-Ctx found to potently block the α6β2β3 receptor (tested using the α6/α3 chimera as a surrogate α6 subunit) and the closely related α3β2 nAChR with comparable nanomolar antagonist potencies (Cartier et al., 1996). In contrast, both α-Ctxs PIA and BuIA inhibit α6β2β3-signaling more potently than the signaling of α6β4 and α3β2, and this has made them valuable tools in studies of α6β2* nAChRs (Azam et al., 2005; Dowell et al., 2003). Interestingly, there are no reports to date of native α-Ctxs that preferentially target α6β4* nAChRs over the closely related α6β2* and α3β4* subtypes (Azam et al., 2005; Dowell et al., 2003; Luo et al., 2013b; Smith et al., 2013). In the present study we report the cloning, synthesis and pharmacological characterization of a novel α-Ctx, VnIB, from the venom of Conus ventricosus, which is found in shallow waters of the Mediterranean and hunts polychaete worms (Romeo et al., 2008). α-Ctx VnIB is the first native α-Ctx that potently and preferentially antagonizes the signaling through the neuronal α6β4 nAChR.
2. Materials and methods
2.1. Identification and sequencing of α-Ctx VnIB
Genomic DNA was isolated from 10 mg frozen hepatopancreas tissue from C. ventricosus using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN), following the standard protocol provided by the manufacturer. 10 ng of the resulting genomic DNA was used as a template for PCR using primers targeting conserved regions located at the 3’ end of the intron and 3’ UTR sequence following the mature toxin (forward primer: 5’ TGT GTG TGT GTG GTT CTG GGT 3’, reverse primer: 5’ ACG TCG TGG TTC AGA GGG TCC TGG 3’). The PCR amplification protocol consisted of an initial denaturation step (95 °C, 60 sec) followed by 40 cycles of denaturation (95 °C, 20 sec), annealing (62 °C, 20 sec) and extension (72 °C, 30 sec) steps. The PCR product was purified using the High Pure PCR Product Purification Kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturers protocol and the eluted DNA fragment was inserted into a pAMP vector using the CloneAmp pAMP System for Rapid Cloning of Amplification Products (LifeTechnologies/Gibco BRL, Grand Island, NY) using the protocol suggested by the manufacturer. The resulting DNA products were transformed into DH5α competent E. coli. The sequence of resulting α-Ctx encoding clones were determined according to the ABI automated sequencing protocols (Core Sequencing Facility, University of Utah, UT) initiated by the M13 universal reverse primer. Sequence alignments were made using MacVector version 16.0.8 (MacVector, Inc., Apex, NC). The sequence encoding α-Ctx VnIB was deposited in GenBank (accession number MK120500).
2.2. Peptide synthesis
The peptide was synthesized applying standard solid-phase 9-fluorenylmethyloxycarbonyl (Fmoc) methods on an Apex 396 automated peptide synthesizer (AAPPTec, Louisville, KY). All amino acids with standard side chain protection were purchased from AAPPTec. The cysteine residues were orthogonally protected with a trityl group (Cys3 and Cys9) or an acetamidomethyl group (Cys4 and Cys16) to facilitate stepwise oxidative folding. Ten-fold excess of amino acid was used, and the coupling activation was achieved with 1 eq of 0.4 M benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate and 2 eq of 2 M N,N-diisopropylethyl amine in N-methyl-2-pyrrolidone. Each coupling reaction was carried out for 60 min. The Fmoc deprotection reaction was carried out for 20 min with 20% (v/v) piperidine in dimethylformamide. Amidated peptide was cleaved from the resin using Reagent K (trifluoroacetic acid/phenol/ethanedithiol/thioanisol/H2O; 9:0.75:0.25:0.5:0.5 by volume) and reverse-phase HPLC (Waters, Milford, MA) was utilized to collect the pure linear peptide. To selectively form the desired disulfide bond configuration, the peptide was folded using a two-step oxidation protocol. The first disulfide bridge was closed using 20 mM potassium ferricyanide (FeCN) and 0.1 M Tris-HCl, pH 7.5. The reaction was allowed to proceed for 45 min and was subsequently halted by diluting the peptide with buffer A (0.1% TFA, remainder H2O) to lower pH. The FeCN solution was passed through a disposable C18 cartridge and the peptide was eluted using B60 (0.092% TFA, 60% acetonitrile, remainder H2O). Linear gradients of 1% B60/min were utilized. The monocyclic peptide was dripped into an equal volume of 10 mM iodine in H2O/trifluoroacetic acid/acetonitrile (75:3:25 by volume) for simultaneous removal of the acetamidomethyl groups and closure of the second disulfide bridge. The reaction was conducted for 10 min and quenched with a freshly prepared 1 M aqueous solution of ascorbic acid. Afterwards, the bicyclic peptide product was purified by reverse-phase HPLC using a C18-column. The mass of the peptide was confirmed by MALDI-TOF/MS (monoisotopic MH+: calculated: 1734.66 Da; observed: 1734.60 Da).
2.3. Two-electrode voltage clamp (TEVC) electrophysiology
cDNA clones encoding rat or human neuronal nAChR subunits and human muscle-type nAChR subunits were used to produce cRNA for injection into Xenopus laevis oocytes as described previously (Azam et al., 2005; Hone et al., 2015; Hone et al., 2009; McIntosh et al., 2004). The human muscle-type nAChR subunit cDNAs were obtained from A. G. Engel (Mayo Clinic College of Medicine, Rochester, MN). The chimeric α6/α3 subunit, composed of the extracellular N-terminal domain of the α6 subunit (amino acids 1–237) and the transmembrane and C-terminal domain of the α3 subunit (amino acids 233–499), was used as an α6 surrogate subunit to improve functional expression of the α6* nAChRs as previously reported (Dowell et al., 2003; Kuryatov et al., 2000; Letchworth and Whiteaker, 2011; McIntosh et al., 2004). In brief, Xenopus laevis oocytes were harvested and injected with equal ratios of cRNA encoding rat or human nAChR subunits using a Drummond microdispenser (Drummond Scientific, Broomall, PA). For the majority of nAChR subunits, 0.3–10 ng cRNA was injected into oocytes. For α6/α3- and α6-containing subtypes, up to 110 ng of cRNA was injected to improve functional expression. The oocytes were subsequently incubated at 17 °C in ND96 (96.0 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5 mM HEPES, pH 7.5) containing antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml gentamycin, 2.5 μM sodium pyruvate). Oocytes recordings were made 18 h to 7 days post injection.
Oocyte recordings were obtained by gravity-perfusing a cylindrical ~30 μl recording chamber constructed of Sylgard with HEPES-buffered ND96 containing 0.1 mg/ml BSA at a rate of ~2 ml/min. BSA was added to reduce non-specific adsorption of the peptide. The oocyte membranes were clamped at a holding potential of −70 mV and ACh-gated currents were recorded using an OC-725 series TEVC amplifier (Warner Instruments, Hamden, CT) filtered through a 5-Hz low-pass filter (model FIB1; Frequency Devices, Ottawa, IL), and digitalized at 50 Hz using a digital to analog converter (model USB-6009; National Instruments, Austin, TX). The oocytes were automatically stimulated with 1 s pulses of ACh at 1 min intervals to establish a baseline control response. A concentration of 100 μM ACh was used for all receptors except for α7 (300 μM), α9α10 (10 μM) and α1β1εδ (10 μM). Upon establishment of a steady baseline response, the perfusion solution was changed to one containing ascending concentrations of α-Ctx VnIB diluted in ND-96. A concentration of 20 nM VnIB was used for the kinetic studies. Ctx was applied until a steady ACh-induced response was obtained. The percentage of response [Response (%)] was calculated by normalizing the average peak amplitude of the last three peaks at steady state in the presence of toxin to the average peak amplitude of three control responses just preceding the exposure to toxin. For toxin concentrations above 1 μM, the flow was turned off and the toxin was applied directly to the bath followed by 5 min incubation. The first subsequent ACh application was normalized to the average of a preceding control response obtained after 5 min incubation with no toxin present. All solution changes were controlled by a series of connected three-way solenoid valves connected via an OPTO 22 relay board (Newark element 14, Chicago, IL) and LabVIEW software (National Instruments).
2.4. Data analysis
All TEVC data was processed to obtain the Response (%) by normalizing the current amplitude to the baseline response as described above. The results were analyzed using GraphPad Prism version 7.0 (GraphPad Software Inc., La Jolla, CA). Concentration-response curves were fitted using a non-linear equation with a four-parameter variable slope (Eq. 1):
| Eq. 1 |
Top and bottom of the fit were constrained to 100 and 0, respectively. In this study any difference in IC50 ≤ 2-fold was not considered significant despite non-overlapping confidence intervals. Data for observed on-and off-rates (Kobs and Koff) were fitted with an exponential one-phase decay and one-phase association (Eq. 2 and 3, respectively):
| Eq. 2 |
| Eq. 3 |
Where R is the response (in percentage) at time (t) zero (R0) and when minimum or maximum plateau is reached (Rmin and Rmax, respectively). All data are given as mean ±SD values and represent data from at least three replicates.
3. Results
3.1. Cloning of the gene encoding α-Ctx VnIB
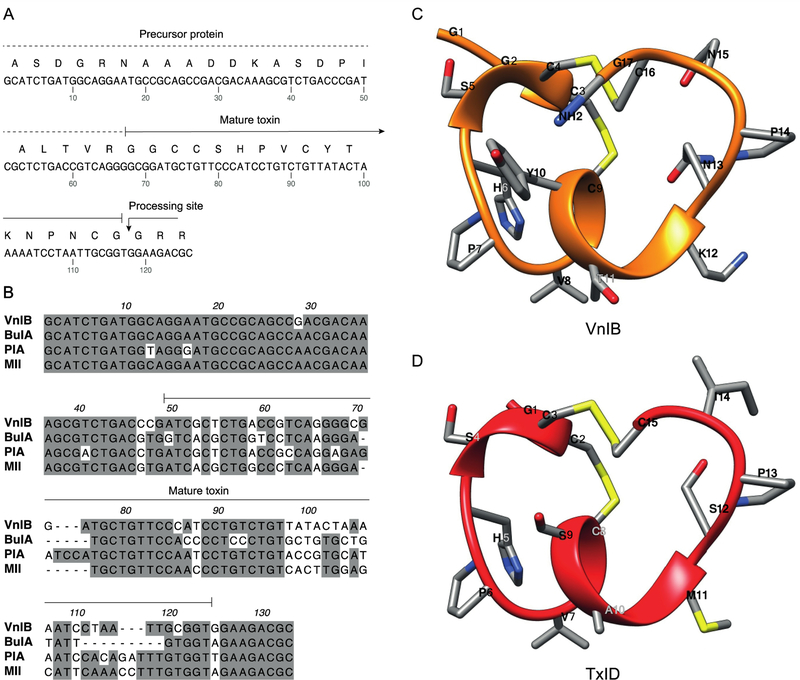
α-Ctxs are translated as prepro-peptides consisting of a N-terminal signal sequence that is highly conserved across species, followed by a conserved pro-region that separates the signal sequence from the highly divergent C-terminal mature α-Ctx region. This precursor protein undergoes proteolytic cleavage and post-translational modifications to yield the mature bioactive toxin (Abraham and Lewis, 2018; Yuan et al., 2007). In the present study, the conserved features in the intron located in the pro-region immediately preceding the toxin sequence and the 3’ UTR segment were exploited to isolate a novel peptide gene from the worm-hunting species C. ventricosus. The gene sequence was amplified by PCR directly from genomic DNA to identify a specific α-Ctx gene product composed of 17 amino acids with a characteristic CC-X4-C-X6-C framework, thus belonging to the relatively rare structural subclass of 4/6 α-Ctxs (figure 1) (Lebbe et al., 2014). As can be seen in figure 1B, where the nucleic acid sequence of α-Ctx VnIB is aligned with those of the α6β2*-targeting α-Ctxs BuIA, PIA and MII, the pro-peptide regions in the four toxins are highly conserved across the different species, whereas the mature peptide regions are substantially more divergent.
Figure 1. α-Conotoxin (α-Ctx) VnIB.
(A) The α-Ctx VnIB pro-peptide and the encoded conopeptide sequence is shown (GenBank accession number MK120500). The precursor protein is indicated by a dotted line and the mature toxin by a solid line. The presumed C-terminal amide processing site at the terminal Gly residue is indicated by an arrow. (B) Alignment of the nucleic acid sequences of α-Ctx VnIB, BuIA, PIA and MII, which all exhibit potent antagonistic activity at α6-containing (α6*) nicotinic acetylcholine receptors (nAChRs). Conserved residues are shaded in grey. Alignments were made using MacVector. (C-D) Ribbon representation of lowest energy structures of VnIB (orange) and TxID (red) with stick representations of amino acid side chains. The structural representation of VnIB was generated in USCF Chimera based on molecular-dynamics simulations of TxID in the α6β4 binding site published by J. Yu et al. (Pettersen et al., 2004; Yu et al., 2018).
3.2. Chemical synthesis of α-Ctx VnIB
The α-Ctxs are among the smallest conopeptides, and the mature toxins are generally composed of 12–20 amino acid residues, which makes them readily suitable for chemical synthesis (Abraham and Lewis, 2018; Myers et al., 1993). It is important to stress that α-Ctx VnIB has not yet been isolated from C. ventricosus venom, and the synthesis of the mature peptide was therefore performed based on some general assumptions. Firstly, the natural α-Ctx typically exhibits a globular disulfide fold with connectivity between the first and the third cysteine residue and between the second and fourth cysteine residue (Akondi et al., 2014; Dutton et al., 2002). In the case of VnIB, Cys3 would thus be expected to form a disulfide bond with Cys9 and Cys4 with Cys16. To direct the globular disulfide bond formation, the cysteine residues were orthogonally protected with acid-labile groups and acid-stable groups, respectively. The acid-labile groups were removed during peptide cleavage from the resin; the first disulfide bridge was formed using FeCN and the monocyclic peptide was isolated by solid phase extraction. Subsequently, the second disulfide bridge was formed by iodine oxidation, and the fully folded peptide was purified by HPLC. Secondly, the Gly at the C-terminus is predicted to be amidated. α-Ctxs often undergo this type of post-translational modification, and loss of the C-terminal amidation has previously been shown to alter the bioactivity of peptides (Akondi et al., 2014). The N-terminal processing site typically consists of one or more basic amino acids preceding the mature toxin sequence (figure 1A). MALDI-TOF/MS confirmed the mass of the peptide (monoisotopic MH+: calculated: 1734.66 Da; observed: 1734.60 Da) and the synthetic product was used throughout this study.
3.3. VnIB is a potent and selective α6β4 nAChR antagonist
The functional properties of VnIB were characterized by TEVC electrophysiology at a wide range of physiologically relevant nAChR subtypes heterologously expressed in Xenopus laevis oocytes. Initially, the functional properties of VnIB were characterized at 10 different rat nAChR subtypes and the human muscle-type nAChR. VnIB potently inhibited ACh-evoked signaling through rα6β4 and rα6/α3β4 receptors, displayed moderate antagonistic potency at rα3β4 and rα6/α3β2β3 receptors, and exhibited no or low activity on the human muscle-type α1β1δε nAChR and at the neuronal rα2β2, rα2β4, rα3β2, rα4β2, rα4β4, rα7 and rα9α10 subtypes (IC50 values >10 μM) (table 1, figures 2 and 3). In contrast to other native α-Ctxs, VnIB most potently blocked rα6β4 and rα6/α3β4 nAChRs signaling (rα6β4 IC50 = 12.0 nM (95% confidence interval (CI) 9.85–14.5 nM), rα6/α3β4 IC50 = 18 nM (CI 16.1–20.2 nM)), displaying ~20- and ~250-fold higher antagonist potencies at these receptors compared to rα3β4 and rα6/α3β2β2, respectively.
Table 1. Functional properties of α-Ctx VnIB at various nAChR subtypes.
The antagonist properties of VnIB were determined at nAChRs expressed in Xenopus laevis oocytes by two-electrode voltage clamp (TEVC) electrophysiology as described in Materials and methods, section 2.3. Data is given as IC50 values and Hill slopes (nH) (with 95% confidence intervals in parentheses) and were obtained from recordings on at least three individual oocytes. r, rat; h, human; ND, not determined.
| nAChR | IC50 (nM) | nH |
|---|---|---|
| rα6β4 | 12 (9.9–15) | 1.2 (0.95–1.5) |
| rα6/α3β4 | 18 (16–20) | 1.1 (0.97–1.2) |
| rα3β4 | 320 (250–390) | 1.2 (0.96–1.5) |
| rα6/α3β2β3 | 4000 (2500–6900) | 0.53 (0.42–0.66) |
| hα1β1δε | >10,000 | ND |
| rα2β2 | >10,000 | ND |
| rα2β4 | >10,000 | ND |
| rα3β2 | >10,000 | ND |
| rα4β2 | >10,000 | ND |
| rα4β4 | >10,000 | ND |
| rα7 | >10,000 | ND |
| rα9α10 | >10,000 | ND |
| hα6/α3β4 | 5.3 (4.1–6.9) | 1.1 (0.84–1.5) |
| hα3β4 | >10,000 | ND |
| hα6/α3β2β3 | >10,000 | ND |
| hα4β2 | >10,000 | ND |
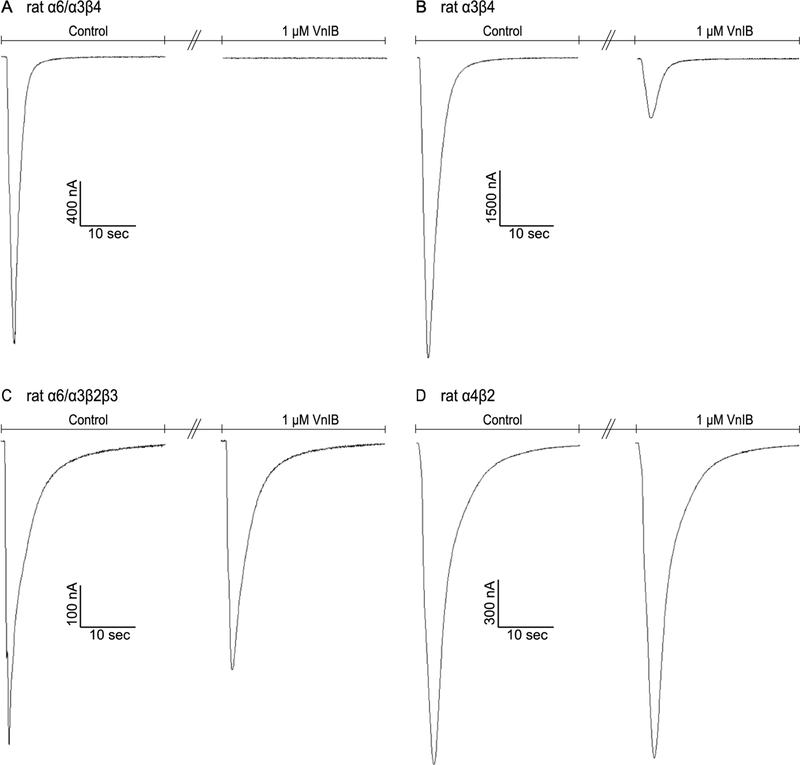
Figure 2. Comparison of α-Ctx VnIB antagonism on the signaling through various nAChR subtypes.
TEVC electrophysiology traces from individual representative Xenopus laevis oocytes expressing the α6/α3β4, α3β4, α6/α3β2β3 and α4β2 nAChRs, respectively. (A) Application of 1 μM VnIB completely abolishes the current evoked by 1 s application of 100 μM ACh at the α6/α3β4 nAChR. (B-C) Application of 1 μM VnIB partially blocks the current evoked by 1 s application of 100 μM ACh at the α3β4 nAChR (80% inhibition) and the α6/α3β2β3 nAChR (23% inhibition). (D) Application of 1 μM VnIB has no significant effect on the response evoked by 1 s application of 100 μM ACh at the α4β2 nAChR.
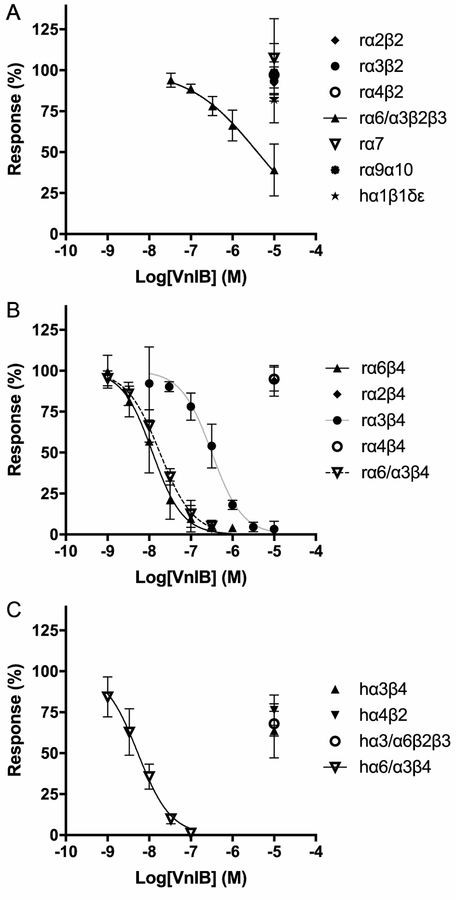
Figure 3. Functional properties of α-Ctx VnIB as a nAChR antagonist.
Xenopus laevis oocytes expressing various nAChR subtypes were subjected to TEVC electrophysiology. Oocytes were stimulated with 1 s pulses of ACh in the absence or presence of ascending concentrations of α-Ctx VnIB. (A) α-Ctx VnIB-mediated block of β2-containing rat nAChRs and the α7, α9α10 and human muscle-type α1β1δε nAChRs. VnIB blocks the ACh-evoked responses through α6/α3β2β3 with a low micromolar IC50 (3.2 μM) but has no significant effect on the other receptor subtypes (IC50 > 10 μM). (B) α-Ctx VnIB-mediated block of β4-containing rat nAChRs. VnIB blocks the ACh-evoked currents through α6β4 and α6/α3β4 nAChRs with similar antagonist potencies (IC50 12 nM and 18 nM, respectively) and the currents through the α3β4 receptor with a ~20-fold higher IC50-value (0.32 μM), but the toxin has no significant effect on α2β4 and α4β4 (IC50 > 10 μM). (C) α-Ctx VnIB-mediated block of selected human neuronal receptor subtypes. VnIB potently blocks α6/α3β4 (IC50 = 5.3 nM), but has no or low antagonistic effect at α3β4, α4β2 and α6/α3β2β3. Each data point represents mean values ± SD based on at least 3 replicates. r, rat; h, human.
The functional properties of VnIB were subsequently characterized at the human nAChR subtypes α6/α3β4, α6/α3β2β3, α3β4 and α4β2. The three former subtypes are the human orthologs of the three rat nAChR subtypes, at which VnIB displayed significant activity, and they were thus included to investigate whether the peptide displayed a similar selectivity profile at the human receptors. VnIB displayed low nanomolar potency at the hα6/α3β4 receptor (IC50 = 5.3 nM (CI 4.1–6.9 nM)), but interestingly, it was an even less potent antagonist of the ACh-evoked signaling through hα3β4 and hα6/α3β2β3 (IC50 values >10 μM) than it was at the corresponding rat nAChR subtypes (table 1, figure 3). Moreover, VnIB exhibited no activity at the hα4β2 receptor (IC 50 values >10 μM). Thus, VnIB displayed an even more pronounced selectivity towards α6/α3β4 over other nAChRs at the human receptors than at the rat receptors.
It should be noted that although it was possible to obtain functional expression of the wild-type rα6β4 nAChR, the current amplitudes recorded from these oocytes were often too small to be useable. To facilitate assessment of VnIB activity at α6*-receptors, we thus utilized an α6/α3 chimera comprising the extracellular domain of α6 and the transmembrane and intracellular domains of α3; this chimera has been widely used in previous studies of α6* nAChRs in heterologous expression systems (Dowell et al., 2003; Kuryatov et al., 2000). The IC50-value displayed by VnIB at the chimeric rα6/α3β4 nAChR did not differ substantially from that of the native rα6β4 nAChR (table 1). The α6/α3 subunit was therefore considered a good surrogate for the α6 subunit and rat and human chimeras was used throughout this study. Additionally, several previous studies have demonstrated binding of α-Ctxs to the extracellular domains of the nAChR complex (Dowell et al., 2003; Hone et al., 2012b), and based on the ~20-fold and >1,500-fold differences in inhibitory potency between α6/α3β4 and α3β4 at rat and human receptors, respectively, we conclude that this is most likely also the case for VnIB.
3.4. Kinetics of VnIB antagonism
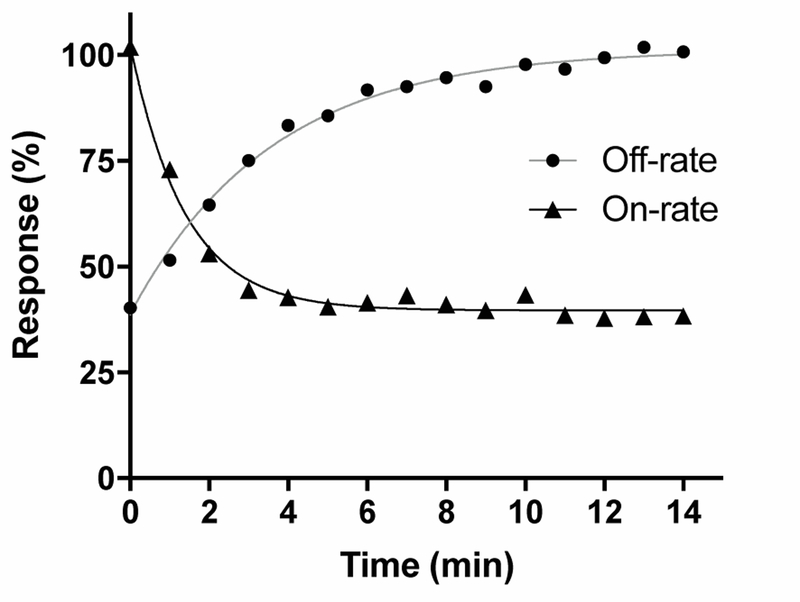
Kinetic studies were subsequently performed to evaluate the on- and off-rate of VnIB in its block of the rα6/α3β4 nAChR. Block by 20 nM VnIB (~IC50) was relatively rapid, with maximum receptor block obtained after ~5 min (t½ = 0.87 min−1) toxin application (figure 4). Washout of the toxin (20 nM) was slightly slower, with recovery of the ACh-evoked currents to baseline levels after ~8–10 min (t½ = 2.7 min−1). Based on the Kobs and Koff (0.262 ±0.042 min−1) (n=10 oocytes), the Kon (2.52∙107 ±7.72∙106 min−1∙M−1) and the inhibition constant (Ki = 11.7 ±5.01 nM) for VnIB were calculated. The Ki for the functional antagonism of VnIB at rα6/α3β4 calculated by Koff/ Kon for the individual experiments correlated well with the IC50-values determined for the toxin at rα6β4 and rα6/α3β4 based on concentration-inhibition analysis (table 1).
Figure 4. Observed on -rate and off-rate kinetics for α-Ctx VnIB on the α6/α3β4 nAChR.
Representative curves for the observed on-rate and off-rate of 20 nM α-Ctx VnIB flow-applied on Xenopus laevis oocytes expressing the α6/α3β4 nAChR. Washout data were averaged from a total of 10 oocytes. The half-time (t½) of the blocking effect is 0.87 min−1 and t½ for recovery after washout of the toxin is 2.7 min-1.
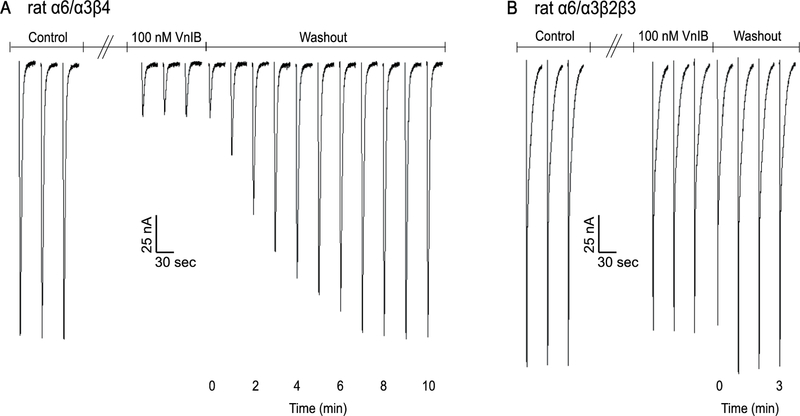
Binding kinetics were also determined for the closely related rα6/α3β2β3 nAChR subtype. Recovery from 100 nM toxin block was much more rapid compared to rα6/α3β4 upon toxin washout (figure 5). Rapid antagonist reversal is therefore another property that might be utilized to distinguish between β2* and β4* nAChRs in circumstances where both subtypes may be present.
Figure 5. Washout of VnIB-mediated block of α6/α3β4 and α6/α3β2β3 nAChRs.
Representative washout traces from recordings on Xenopus laevis oocytes expressing α6/α3β4 and α6/α3β2β3 nAChRs. Oocytes were stimulated with 1 s pulses of ACh at 1 min intervals during the washout of 100 nM α-Ctx VnIB. (A) α-Ctx VnIB reduced ACh-evoked currents through α6/α3β4 by ~85%, and the currents recovered to the original amplitude levels after 8 min washout. (B) α-Ctx VnIB reduced ACh-evoked currents through α6/α3β2β3 by ~10%, and the currents recovered to the original amplitude levels after 1 min of washout.
4. Discussion
A recurring challenge connected to explorations of the numerous nAChR subtypes present in vivo is that few subtype selective pharmacological tools are available. The pronounced nAChR subtype specificities exhibited by some α-Ctxs are unique and have made these peptides highly useful for the delineation of physiological functions mediated by different populations of nAChRs (Abraham and Lewis, 2018; Giribaldi and Dutertre, 2018). In the present study we report the discovery of the novel α-Ctx VnIB, which is the first native conopeptide that selectively targets the α6β4 nAChR subtype.
The gene encoding the DNA for α-Ctx VnIB was isolated from C. ventricosus genomic DNA and the predicted mature peptide is a 17 amino acid peptide with a CC-C-C cysteine framework, which is highly conserved in the α-Ctx family (Giribaldi and Dutertre, 2018). Note, however, that an α-Ctx-like peptide was purified from C. ventricosus venom previously, but it has not yet been pharmacologically characterized (Romeo et al., 2008). This peptide was named α-conotoxin-Vn in a prior publication and is referred to as α-Ctx Vn1A (Greek numeral “1”) in the online database ConoServer (Kaas et al., 2008). To avoid possible confusion with the Vn1A peptide, it was decided to name the peptide in the current study α-Ctx VnIB. Assignment of the Roman numeral “I” is consistent with the current naming convention and indicates that the pharmacology has been established (McIntosh et al., 1999). Vn1A and VnIB share the same cysteine framework of α-Ctxs; remarkably, however, they differ in 7 of 13 non-cysteine residues and loop-lengths, indicating large sequence variability within just this single Conus species (table 2) (Romeo et al., 2008).
Table 2. Amino acid sequences and nAChR selectivity profiles of various α-Ctxs.
Based on previously published data, up to four nAChRs subtypes, at which the given α-Ctxs has the highest potencies, are shown. α6 given in the table refers to the α6/α3 chimera. “≈” indicates a difference in IC50 < 2.5-fold. r, rat; m, mouse; ND, not determined.
| α-Ctx | Species | Sequence | nAChR preference |
|---|---|---|---|
| VnIB | C. ventricosus | GGCCSHPVCYTKNPNCG# | rα6β4 > rα3β4 >> rα6β2β3a |
| Vn1A | C. ventricosus | GGCCSYPPCAVSNPQHC | NDb |
| AuIB | C. aulicus | GCCSYPPCFATNPDC# | rα3β4 > rα6β4 ≈ rα7c |
| TxID | C. textile | GCCSHPVCSAMSPIC# | rα3β4 > rα6β4 >> rα2β4d |
| BuIA | C. bullatus | GCCSTPPCAVLYC# | rα6β2β3 > rα6β4 > rα3β2 > rα3β4e |
| MII | C. magus | GCCSNPVCHLEHSNLC# | rα6β2β3 >/≈ rα3β2f > rα6β4‡ |
| PIA | C. purpurascens | RDPCCSNPVCTVHNPQIC# | rα6β2β3 > rα6β4 >/≈ rα3β2 > rα3β4g |
| TxIB | C. textile | GCCSDPPCRNKHPDLC# | rα6β2β3h |
| OmIA | C. omaria | GCCSHPACNVNNPHICG# | rα3β2 >/≈ rα7 > rα6β2β3i |
| PeIA | C. pergrandis | GCCSHPACSVNHPELC# | rα9α10 >/≈ rα3β2 ≈ rα6β2β3 > rα6β4j |
| GI | C. geographus | ECCNPACGRHYSC# | mα1β1γδk |
C-terminal amidated
Unpublished data
Data from:
this work
α-Ctx VnIB belongs to the subfamily of 4/6 α-Ctxs. This disulfide-framework is found in less than 10% of α-Ctxs, yet it is found across all types of Conus predators (vermivorous, piscivorous and molluscivorous) (unpublished observations). The α-Ctxs AuIB and TxID share this 4/6 substructure and also potently block α6β4 nAChRs, but interestingly, the specificity is reversed compared to VnIB. AuIB and TxID display ~9.5-fold and ~7.5-fold lower antagonist potencies respectively at α6β4 compared to α3β4 nAChRs (Luo et al., 2013b; Smith et al., 2013). The 4/6 substructure itself may thus contribute to the preference for β4* over β2* nAChR subtypes exhibited by all assessed 4/6 α-Ctxs (table 2) (Dutton et al., 2002; Luo et al., 2013b; Smith et al., 2013). The influence of the 4/6 substructure for the β4-selectivity exhibited by VnIB, AuIB and TxID is further supported by the fact that the potent α3* and α6* nAChR antagonism mediated by the more prevalent 4/7 α-Ctxs, including MII, OmIA, PIA and TxIB is heavily biased towards β2-containing receptors (table 2) (Azam et al., 2005; Cartier et al., 1996; Dowell et al., 2003; Dutertre et al., 2005; Luo et al., 2013a; Luo et al., 2013b; Talley et al., 2006). Finally, the 4/4 α-Ctx BuIA, representing a third identified α-Ctx class with potent α6* nAChR activity, exerts a completely different form of β2 versus β4 discrimination. BuIA displays roughly comparable antagonist potencies at α2β2 and α2β4, at α3β2 and α3β4 and at α6/α3β2β3 and α6/α3β4 (IC50-values within 5–7-fold pairwise), but instead discriminates between β2- and β4-containing receptors through its much slower unbinding kinetics at the latter receptors (Azam et al., 2005).
In contrast to VnIB, BuIA displays reversed preference, with a 5-fold higher potency at α6β2β3 compared to α6β4. VnIB exhibited a ~250-fold higher antagonist potency at rα6/α3β4 than at rα6/α3β2β3. However, its washout time at the former receptor was ~8 fold higher than at the latter receptor subtype. The fast washout at α6/α3β2β3 did not allow an exact determination of the off-rate (figure 5), but this washout pattern is in contrast to α-Ctx BuIA, which displays a very significant (> 25 min) discrimination in washout kinetics between β2- and β4-containing nAChRs (Azam et al., 2005).
In striking contrast to the other 4/6 α-Ctxs, VnIB displayed a substantially lower antagonist potency at α3* nAChRs than at α6* receptors, both when tested at the rat and human receptor orthologs. The loop lengths may influence the α3 and α6 > α2 and α4 subunit selectivity exhibited by α-Ctxs VnIB, AuIB and TxID, but analysis of the molecular composition of the two cysteine-loops in the peptides reveals that the α3-versus α6-selectivity determinant(s) is most likely to be found in the second cysteine-loop. The amino acid segment, SHP, found in the first cysteine-loop of VnIB, is found in a range of other conopeptides (table 3), and more strikingly, the sequence GCCSHPVC, which includes the entire first cysteine-loop in VnIB, is completely preserved in TxID, which in contrast to VnIB, shows a preference for α3* over α6* nAChRs. Thus, the pronounced α6 > α3 selectivity exhibited by VnIB is clearly rooted in the YTKNPN sequence segment constituting the second disulfide-loop. This loop sequence is not observed in any other previously characterized α-Ctxs aside from the Pro residue, which is found in a homologous position in many members of the α-Ctx family (tables 2 and 3). Therefore, the five other residues must provide the substantial structural diversity that underlies the unique α-based selectivity profile of the peptide. The residues in position 9 of AuIB and TxID, Phe and Ser respectively, has previously been shown to influence the antagonistic potency of these peptides at the α3β4 nAChR (Grishin et al., 2013; Yu et al., 2018). A recent study found that TxID[S9K] increased selectivity for α3β4 presumably by abolishing interaction between the Ser9 of TxID and Lys59 at the β4 complementary interface of α6β4 (Yu et al., 2018). α-Ctx VnIB has a Tyr in the corresponding position (Tyr10) and similar interactions in this pocket could potentially account for the selectivity of VnIB. It may be possible to augment the α6β4 selectivity of VnIB further in future analogs by modifying the residues in the second disulfide-loop segment. Altogether the above suggests that the substructure of the α-Ctxs may be a key molecular determinant for β-subunit based selectivity, while the molecular composition of the cysteine-loops is more important for the α-subunit based selectivity.
Table 3. Alignment and nAChR selectivity profiles of native and mutated α-Ctxs homologous to VnIB.
Based on previously published data, up to four nAChRs subtypes, at which the given α-Ctxs has the highest potencies, are shown. The alignment was prepared using the ConoServer (Kaas et al., 2008). Conserved residues and residues conserved in >50% of sequences are shaded dark and light gray, respectively. α6 given in the table refers to the α6/α3 chimera. “≈” indicates a difference in IC50 < 2.5-fold. r, rat; h, human; m, mouse.
| α-Ctx | Alignment | nAChR preference |
|---|---|---|
| VnIB | G G C C S H P V C Y T KN P N-C G # | rα6β4 > rα3β4 >> rα6β2β3a |
| OmIA | - G C C S H P AC N V N N P H I C G # | rα3β2 >/≈ rα7 > rα6β2β3b |
| AuIB | - G C C S Y P P C F A T N P D- C - # | rα3β4 > rα6β4 ≈ rα7c |
| PeIA[A7V,S9H,V10A,N11R] | - G C C S H P V C H A R H P E L C - # | rα6β2β3 > rα3β2‡, d |
| TxID | - G C C S H P V C S A M S P I - C - # | rα3β4 > rα6β4 >> rα2β4e |
| PeIA[A7V,S9H,V10A,N11R,E14A] | - G C C S H P V C H A R H P A L C - # | rα6β2β3 > rα6β4 >> rα3β2f |
| BnIA | - G C C S H P A C S V N N P D I C - # | hα7 > mα1β1γδ‡, g |
| RegIIA | - G C C S H P A C N V N N P H I C - # | rα3β2 > rα3β4 ≈ rα6β2* >/≈ rα7h |
C-terminal amidated
not tested at rα6β4 nAChR
Data from:
this work
As outlined above, the substructure of the 4/6 α-Ctxs may contribute to the β4 > β2 selectivity of VnIB, AuIB and TxID. However, it is interesting to note that introduction of two mutations into the 4/4 α-Ctx BuIA, BuIA[T5A;P6O], creates α6β4 > α6β2β3 preference (Azam et al., 2010). α-Ctx OmIA, in contrast, has the highest degree of similarity to VnIB, but displays a completely different selectivity profile, with preference for α3β2 and α7 and lower potency at α6β2β3. The reduced potency at α6β2β3 has been proposed to be partially mediated by the extended hydrophobic C-terminus of the peptide (table 3) (Talley et al., 2006). VnIB also has an additional glycine residue after the C-terminal cysteine, which could potentially contribute to the preference of VnIB for the α6β4 nAChR. Additionally, the N-terminus of VnIB contains an additional residue (Gly), compared to the majority of previously published α-Ctx (tables 2 and 3), which may affect potency, analogously to what has been reported for the N-terminal residues present in GID (Nicke et al., 2003). Altogether, further studies are needed to elucidate the molecular determinants in VnIB as well as in its target receptors underlying this novel receptor selectivity profile.
A substantial number of gene expression, immunoprecipitation and functional studies have demonstrated that α6, β2 and β4 subunits are co-expressed in several brain tissues, the retina and in dorsal root ganglia in addition to the presence of the α3 subunit in some of these tissues (Hone et al., 2012a; Letchworth and Whiteaker, 2011; Moretti et al., 2004). Therefore, selective ligands are needed to enable the identification of and differentiation between α6β2* and α6β4* nAChRs as well as α3* nAChRs. In tissues with potential presence of all these receptor stoichiometries, α-Ctx VnIB is the first native conopeptide antagonist that selectively targets α6β4 nAChRs, and it is considerably more potent than the previously reported α6β4-selective α-Ctx BuIA[T5A;P6O] (Azam et al., 2010). Thus, α-Ctx VnIB should serve as a useful novel research tool in the study of the in vivo distribution and pharmacology of α6* nAChRs. Additionally, VnIB could serve as scaffold for the future development of peptides with optimized antagonist potency and/or selectivity for α6β4 nAChRs.
Highlights.
α-Conotoxin VnIB is a newly identified peptide from Conus ventricosus
Unlike other native α-conotoxins, VnIB selectively inhibits α6β4 nAChRs
VnIB selectivity includes the closely related α6β2β3 and α3β4 nAChR subtypes
VnIB exhibits rapid binding and unbinding at the rat α6β4 nAChR
VnIB is a novel probe for elucidation of the structure and function of α6β4* nAChRs
Acknowledgments
This work was supported by National Institutes of Health grant GM103801 to JMM and GM48677 (BMO and JMM). MvH and AAJ were supported by a grant from the Lundbeck Foundation.
The authors would like to thank Peter N. Huynh and Arik Hone (University of Utah, Salt Lake City, UT) for their excellent technical and scientific assistance and for fruitful discussions during the preparation of the manuscript.
Abbreviations
- ACh
acetylcholine
- nAChR
nicotinic acetylcholine receptor
- α-Ctx
α-conotoxin
- CNS
central nervous system
- TEVC
two-electrode voltage clamp
- IC50
inhibitory concentration at 50% response
- CI
confidence interval
- SD
standard deviation
- FeCN
ferricyanide
- r
rat
- h
human
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Abraham N, Lewis RJ, 2018. Neuronal Nicotinic Acetylcholine Receptor Modulators from Cone Snails. Mar Drugs 16, 208 10.3390/md16060208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondi KB, Muttenthaler M, Dutertre S. b., Kaas Q, Craik DJ, Lewis RJ, Alewood PF, 2014. Discovery, synthesis, and structure-activity relationships of conotoxins. Chem Rev 114, 5815–5847. 10.1021/cr400401e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque EX, Pereira EF, Alkondon M, Rogers SW, 2009. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev 89, 73–120. 10.1152/physrev.00015.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, Dowell C, Watkins M, Stitzel JA, Olivera BM, McIntosh JM, 2005. α-Conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J Biol Chem 280, 80–87. 10.1074/jbc.M406281200 [DOI] [PubMed] [Google Scholar]
- Azam L, Maskos U, Changeux JP, Dowell CD, Christensen S, De Biasi M, McIntosh JM, 2010. α-Conotoxin BuIA[T5A;P6O]: a novel ligand that discriminates between α3β4 and α3β2 nicotinic acetylcholine receptors and blocks nicotine-stimulated norepinephrine release. Faseb j 24, 5113–5123. 10.1096/fj.10-166272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, McIntosh JM, 2006. Characterization of nicotinic acetylcholine receptors that modulate nicotine-evoked [3H]norepinephrine release from mouse hippocampal synaptosomes. Mol Pharmacol 70, 967–976. 10.1124/mol.106.024513 [DOI] [PubMed] [Google Scholar]
- Azam L, McIntosh JM, 2009. α-conotoxins as pharmacological probes of nicotinic acetylcholine receptors. Acta Pharmacol Sin 30, 771–783. 10.1038/aps.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, Winzer-Serhan UH, Chen Y, Leslie FM, 2002. Expression of neuronal nicotinic acetylcholine receptor subunit mRNAs within midbrain dopamine neurons. J Comp Neurol 444, 260–274. 10.1002/cne.10138 [DOI] [PubMed] [Google Scholar]
- Bertrand D, Lee CH, Flood D, Marger F, Donnelly-Roberts D, 2015. Therapeutic Potential of α7 Nicotinic Acetylcholine Receptors. Pharmacol Rev 67, 1025–1073. 10.1124/pr.113.008581 [DOI] [PubMed] [Google Scholar]
- Bertrand D, Terry AV Jr, 2018. The wonderland of neuronal nicotinic acetylcholine receptors. Biochem Pharmacol 151, 214–225. 10.1016/j.bcp.2017.12.008 [DOI] [PubMed] [Google Scholar]
- Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM, 1996. A new α-conotoxin which targets α3β2 nicotinic acetylcholine receptors. J Biol Chem 271, 7522–7528. 10.1074/jbc.271.13.7522 [DOI] [PubMed] [Google Scholar]
- Champtiaux N, Han ZY, Bessis A, Rossi FM, Zoli M, Marubio L, McIntosh JM, Changeux JP, 2002. Distribution and pharmacology of α6-containing nicotinic acetylcholine receptors analyzed with mutant mice. J Neurosci 22, 1208–1217. 10.1523/jneurosci.22-04-01208.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HC, Chebib M, 2017. GABA(A) Receptors and the Diversity in their Structure and Pharmacology. Adv Pharmacol 79, 1–34. 10.1016/bs.apha.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Craig AG, Zafaralla G, Cruz LJ, Santos AD, Hillyard DR, Dykert J, Rivier JE, Gray WR, Imperial J, DelaCruz RG, Sporning A, Terlau H, West PJ, Yoshikami D, Olivera BM, 1998. An O-glycosylated neuroexcitatory Conus peptide. Biochemistry 37, 16019–16025. 10.1021/bi981690a [DOI] [PubMed] [Google Scholar]
- Dowell C, Olivera BM, Garrett JE, Staheli ST, Watkins M, Kuryatov A, Yoshikami D, Lindstrom JM, McIntosh JM, 2003. α-Conotoxin PIA is selective for α6 subunit-containing nicotinic acetylcholine receptors. J Neurosci 23, 8445–8452. 10.1523/jneurosci.23-24-08445.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Nicke A, Lewis RJ, 2005. β2 subunit contribution to 4/7 α-conotoxin binding to the nicotinic acetylcholine receptor. J Biol Chem 280, 30460–30468. 10.1074/jbc.M504229200 [DOI] [PubMed] [Google Scholar]
- Dutton JL, Bansal PS, Hogg RC, Adams DJ, Alewood PF, Craik DJ, 2002. A new level of conotoxin diversity, a non-native disulfide bond connectivity in α-conotoxin AuIB reduces structural definition but increases biological activity. J Biol Chem 277, 48849–48857. 10.1074/jbc.M208842200 [DOI] [PubMed] [Google Scholar]
- Franco A, Kompella SN, Akondi KB, Melaun C, Daly NL, Luetje CW, Alewood PF, Craik DJ, Adams DJ, Marí F, 2012. RegIIA: an α4/7-conotoxin from the venom of Conus regius that potently blocks α3β4 nAChRs. Biochem Pharmacol 83, 419–426. 10.1016/j.bcp.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Giribaldi J, Dutertre S, 2018. α-Conotoxins to explore the molecular, physiological and pathophysiological functions of neuronal nicotinic acetylcholine receptors. Neurosci Lett 679, 24–34. 10.1016/j.neulet.2017.11.063 [DOI] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Zanardi A, Gaimarri A, Champtiaux N, Changeux JP, Whiteaker P, Marks MJ, Clementi F, Zoli M, 2005. Heterogeneity and selective targeting of neuronal nicotinic acetylcholine receptor (nAChR) subtypes expressed on retinal afferents of the superior colliculus and lateral geniculate nucleus: identification of a new native nAChR subtype α3β2(α5 or beta3) enriched in retinocollicular afferents. Mol Pharmacol 68, 1162–1171. 10.1124/mol.105.015925 [DOI] [PubMed] [Google Scholar]
- Grishin AA, Cuny H, Hung A, Clark RJ, Brust A, Akondi K, Alewood PF, Craik DJ, Adams DJ, 2013. Identifying key amino acid residues that affect α-conotoxin AuIB inhibition of α3β4 nicotinic acetylcholine receptors. J Biol Chem 288, 34428–34442. 10.1074/jbc.M113.512582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Vivanco A, Hone AJ, Carmona-Hidalgo B, McIntosh JM, Albillos A, 2014. Monkey adrenal chromaffin cells express α6β4* nicotinic acetylcholine receptors. PloS one 9, e94142 10.1371/journal.pone.0094142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, McIntosh JM, Azam L, Lindstrom J, Lucero L, Whiteaker P, Passas J, Blázquez J, Albillos A, 2015. α-Conotoxins Identify the α3β4* Subtype as the Predominant Nicotinic Acetylcholine Receptor Expressed in Human Adrenal Chromaffin Cells. Mol Pharmacol 88, 881–893. 10.1124/mol.115.100982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Meyer EL, McIntyre M, McIntosh JM, 2012a. Nicotinic acetylcholine receptors in dorsal root ganglion neurons include the α6β4* subtype. Faseb j 26, 917–926. 10.1096/fj.11-195883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Ruiz M, Scadden M, Christensen S, Gajewiak J, Azam L, McIntosh JM, 2013. Positional scanning mutagenesis of α-conotoxin PeIA identifies critical residues that confer potency and selectivity for α6/α3β2β3 and α3β2 nicotinic acetylcholine receptors. J Biol Chem 288, 25428–25439. 10.1074/jbc.M113.482059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Scadden M, Gajewiak J, Christensen S, Lindstrom J, McIntosh JM, 2012b. α-Conotoxin PeIA[S9H,V10A,E14N] potently and selectively blocks α6β2β3 versus α6β4 nicotinic acetylcholine receptors. Mol Pharmacol 82, 972–982. 10.1124/mol.112.080853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Servent D, McIntosh JM, 2018. α9-containing nicotinic acetylcholine receptors and the modulation of pain. Br J Pharmacol 175, 1915–1927. 10.1111/bph.13931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Whiteaker P, Christensen S, Xiao Y, Meyer EL, McIntosh JM, 2009. A novel fluorescent α-conotoxin for the study of α7 nicotinic acetylcholine receptors. J Neurochem 111, 80–89. 10.1111/j.1471-4159.2009.06299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfizer inc., 2019. Pfizer Reports Fourth-Quarter and Full-Year 2018 Results https://www.pfizer.com/news/press-release/press-release-detail/pfizer_reports_fourth_quarter_and_full_year_2018_results (accessed 13 March 2019).
- Johnson DS, Martinez J, Elgoyhen AB, Heinemann SF, McIntosh JM, 1995. α-Conotoxin ImI exhibits subtype-specific nicotinic acetylcholine receptor blockade: preferential inhibition of homomeric α7 and α9 receptors. Mol Pharmacol 48, 194–199 [PubMed] [Google Scholar]
- Kaas Q, Westermann JC, Halai R, Wang CK, Craik DJ, 2008. ConoServer, a database for conopeptide sequences and structures. Bioinformatics 24, 445–446. 10.1093/bioinformatics/btm596 [DOI] [PubMed] [Google Scholar]
- Kaas Q, Yu R, Jin AH, Dutertre S, Craik DJ, 2012. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res 40, D325–330. 10.1093/nar/gkr886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WP, Schulz JR, Jakubowski JA, Gilly WF, Sweedler JV, 2006. Two toxins from Conus striatus that individually induce tetanic paralysis. Biochemistry 45, 14212–14222. 10.1021/bi061485s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kompella SN, Hung A, Clark RJ, Marí F, Adams DJ, 2015. Alanine scan of α-conotoxin RegIIA reveals a selective α3β4 nicotinic acetylcholine receptor antagonist. J Biol Chem 290, 1039–1048. 10.1074/jbc.M114.605592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov A, Olale F, Cooper J, Choi C, Lindstrom J, 2000. Human α6 AChR subtypes: subunit composition, assembly, and pharmacological responses. Neuropharmacology 39, 2570–2590. 10.1016/S0028-3908(00)00144-1 [DOI] [PubMed] [Google Scholar]
- Le Novère N, Zoli M, Changeux JP, 1996. Neuronal nicotinic receptor a6 subunit mRNA is selectively concentrated in catecholaminergic nuclei of the rat brain. Eur J Neurosci 8, 2428–2439. 10.1111/j.1460-9568.1996.tb01206.x [DOI] [PubMed] [Google Scholar]
- Lebbe EK, Peigneur S, Wijesekara I, Tytgat J, 2014. Conotoxins targeting nicotinic acetylcholine receptors: an overview. Mar Drugs 12, 2970–3004. 10.3390/md12052970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchworth SR, Whiteaker P, 2011. Progress and challenges in the study of α6-containing nicotinic acetylcholine receptors. Biochem Pharmacol 82, 862–872. 10.1016/j.bcp.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Kulak JM, Cartier GE, Jacobsen RB, Yoshikami D, Olivera BM, McIntosh JM, 1998. α-conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J Neurosci 18, 8571–8579. 10.1523/jneurosci.18-21-08571.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhangsun D, Wu Y, Zhu X, Hu Y, McIntyre M, Christensen S, Akcan M, Craik DJ, McIntosh JM, 2013a. Characterization of a novel α-conotoxin from Conus textile that selectively targets α6/α3β2β3 nicotinic acetylcholine receptors. J Biol Chem 288, 894–902. 10.1074/jbc.M112.427898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhangsun D, Zhu X, Wu Y, Hu Y, Christensen S, Harvey PJ, Akcan M, Craik DJ, McIntosh JM, 2013b. Characterization of a novel α-conotoxin TxID from Conus textile that potently blocks rat α3β4 nicotinic acetylcholine receptors. J Med Chem 56, 9655–9663. 10.1021/jm401254c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JW, Zhang Y, Talwar S, Estrada-Mondragon A, 2017. Glycine Receptor Drug Discovery. Adv Pharmacol 79, 225–253. 10.1016/bs.apha.2017.01.003 [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Azam L, Staheli S, Dowell C, Lindstrom JM, Kuryatov A, Garrett JE, Marks MJ, Whiteaker P, 2004. Analogs of α-conotoxin MII are selective for α6-containing nicotinic acetylcholine receptors. Mol Pharmacol 65, 944–952. 10.1124/mol.65.4.944 [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Plazas PV, Watkins M, Gomez-Casati ME, Olivera BM, Elgoyhen AB, 2005. A novel α-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat α9α10 and α7 nicotinic cholinergic receptors. J Biol Chem 280, 30107–30112. 10.1074/jbc.M504102200 [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Santos AD, Olivera BM, 1999. Conus peptides targeted to specific nicotinic acetylcholine receptor subtypes. Annu Rev Biochem 68, 59–88. 10.1146/annurev.biochem.68.1.59 [DOI] [PubMed] [Google Scholar]
- Miwa JM, Freedman R, Lester HA, 2011. Neural systems governed by nicotinic acetylcholine receptors: emerging hypotheses. Neuron 70, 20–33. 10.1016/j.neuron.2011.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti M, Vailati S, Zoli M, Lippi G, Riganti L, Longhi R, Viegi A, Clementi F, Gotti C, 2004. Nicotinic acetylcholine receptor subtypes expression during rat retina development and their regulation by visual experience. Mol Pharmacol 66, 85–96. 10.1124/mol.66.1.85 [DOI] [PubMed] [Google Scholar]
- Myers RA, Cruz LJ, Rivier JE, Olivera BM, 1993. Conus peptides as chemical probes for receptors and ion channels. Chem Rev 93, 1923–1936. 10.1021/cr00021a013 [DOI] [Google Scholar]
- Nguyen B, Le Caer JP, Aráoz R, Thai R, Lamthanh H, Benoit E, Molgó J, 2014. Isolation, purification and functional characterization of α-BnIA from Conus bandanus venom. Toxicon 91, 155–163. 10.1016/j.toxicon.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Nicke A, Loughnan ML, Millard EL, Alewood PF, Adams DJ, Daly NL, Craik DJ, Lewis RJ, 2003. Isolation, structure, and activity of GID, a novel α4/7-conotoxin with an extended N-terminal sequence. J Biol Chem 278, 3137–3144. 10.1074/jbc.M210280200 [DOI] [PubMed] [Google Scholar]
- Olivera BM, Quik M, Vincler M, McIntosh JM, 2008. Subtype-selective conopeptides targeted to nicotinic receptors: Concerted discovery and biomedical applications. Channels 2, 143–152. 10.4161/chan.2.2.6276 [DOI] [PubMed] [Google Scholar]
- Pérez-Alvarez A, Hernández-Vivanco A, McIntosh JM, Albillos A, 2012. Native α6β4* nicotinic receptors control exocytosis in human chromaffin cells of the adrenal gland. Faseb j 26, 346–354. 10.1096/fj.11-190223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Lewis AS, van Schalkwyk GI, Mineur YS, 2015. Mood and anxiety regulation by nicotinic acetylcholine receptors: a potential pathway to modulate aggression and related behavioral states. Neuropharmacology 96, 235–243. 10.1016/j.neuropharm.2014.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quik M, Perez XA, Grady SR, 2011. Role of α6 nicotinic receptors in CNS dopaminergic function: relevance to addiction and neurological disorders. Biochem Pharmacol 82, 873–882. 10.1016/j.bcp.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quik M, Wonnacott S, 2011. α6β2* and α4β2* nicotinic acetylcholine receptors as drug targets for Parkinson’s disease. Pharmacol Rev 63, 938–966. 10.1124/pr.110.003269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SD, Norton RS, 2014. Conotoxin gene superfamilies. Mar Drugs 12, 6058–6101. 10.3390/md12126058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo C, Di Francesco L, Oliverio M, Palazzo P, Massilia GR, Ascenzi P, Polticelli F, Schininà ME, 2008. Conus ventricosus venom peptides profiling by HPLC-MS: a new insight in the intraspecific variation. J Sep Sci 31, 488–498. 10.1002/jssc.200700448 [DOI] [PubMed] [Google Scholar]
- Santos AD, McIntosh JM, Hillyard DR, Cruz LJ, Olivera BM, 2004. The A-superfamily of conotoxins: structural and functional divergence. J Biol Chem 279, 17596–17606. 10.1074/jbc.M309654200 [DOI] [PubMed] [Google Scholar]
- Smith NJ, Hone AJ, Memon T, Bossi S, Smith TE, McIntosh JM, Olivera BM, Teichert RW, 2013. Comparative functional expression of nAChR subtypes in rodent DRG neurons. Front Cell Neurosci 7, 225 10.3389/fncel.2013.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley TT, Olivera BM, Han KH, Christensen SB, Dowell C, Tsigelny I, Ho KY, Taylor P, McIntosh JM, 2006. α-Conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as α3β2 and α7 nicotinic acetylcholine receptors. J Biol Chem 281, 24678–24686. 10.1074/jbc.M602969200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly A, Corringer PJ, Guedin D, Lestage P, Changeux JP, 2009. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat Rev Drug Discov 8, 733–750. 10.1038/nrd2927 [DOI] [PubMed] [Google Scholar]
- Teichert RW, Jacobsen R, Terlau H, Yoshikami D, Olivera BM, 2007. Discovery and characterization of the short κA-conotoxins: a novel subfamily of excitatory conotoxins. Toxicon 49, 318–328. 10.1016/j.toxicon.2006.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H, Olivera BM, 2004. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev 84, 41–68. 10.1152/physrev.00020.2003 [DOI] [PubMed] [Google Scholar]
- Walstab J, Rappold G, Niesler B, 2010. 5-HT(3) receptors: role in disease and target of drugs. Pharmacol Ther 128, 146–169. 10.1016/j.pharmthera.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Wieskopf JS, Mathur J, Limapichat W, Post MR, Al-Qazzaz M, Sorge RE, Martin LJ, Zaykin DV, Smith SB, Freitas K, Austin JS, Dai F, Zhang J, Marcovitz J, Tuttle AH, Slepian PM, Clarke S, Drenan RM, Janes J, Al Sharari S, Segall SK, Aasvang EK, Lai W, Bittner R, Richards CI, Slade GD, Kehlet H, Walker J, Maskos U, Changeux JP, Devor M, Maixner W, Diatchenko L, Belfer I, Dougherty DA, Su AI, Lummis SC, Imad Damaj M, Lester HA, Patapoutian A, Mogil JS, 2015. The nicotinic α6 subunit gene determines variability in chronic pain sensitivity via cross-inhibition of P2X2/3 receptors. Sci Transl Med 7, 287ra272 10.1126/scitranslmed.3009986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhu X, Harvey PJ, Kaas Q, Zhangsun D, Craik DJ, Luo S, 2018. Single Amino Acid Substitution in α-Conotoxin TxID Reveals a Specific α3β4 Nicotinic Acetylcholine Receptor Antagonist. J Med Chem 61, 9256–9265. 10.1021/acs.jmedchem.8b00967 [DOI] [PubMed] [Google Scholar]
- Yuan DD, Han YH, Wang CG, Chi CW, 2007. From the identification of gene organization of α-conotoxins to the cloning of novel toxins. Toxicon 49, 1135–1149. 10.1016/j.toxicon.2007.02.011 [DOI] [PubMed] [Google Scholar]