Summary
We report that mouse LSL-KrasG12D;Trp53fl/fl (KP)-mediated lung adenocarcinoma (LADC) tumorigenesis can proceed through both PKCι-dependent and PKCι-independent pathways. The predominant pathway involves PKCι-dependent transformation of bronchioalveolar stem cells (BASCs). However, KP mice harboring conditional knock out Prkci alleles (KPI mice) develop LADC tumors through PKCι-independent transformation of Axin2+ alveolar type 2 (AT2) stem cells. Transformed growth of KPI, but not KP, tumors is blocked by Wnt pathway inhibition in vitro and in vivo. Furthermore, a KPI-derived genomic signature predicts sensitivity of human LADC cells to Wnt inhibition, and identifies a distinct subset of primary LADC tumors exhibiting a KPI-like genotype. Thus, LADC can develop through both PKCι-dependent and PKCι-independent pathways, resulting in tumors exhibiting distinct oncogenic signaling and pharmacologic vulnerabilities.
Keywords: lung cancer, bronchioalveolar stem cells, alveolar type 2 cells, protein kinase Cι signaling, Wnt signaling, lung adenocarcinoma molecular subtypes, therapeutic vulnerability, Kras/Trp53 transformation
In Brief
Yin et al. discover that KrasG12D;Trp53−/− (KP)-driven mouse lung adenocarcinomas (LADC) are phenotypically different from KP LADC without PKCι (KPI) expression. A KPI-derived genomic signature identifies KPI-like human LADC and predicts susceptibility to Wnt inhibitors.
Graphical Abstract

Introduction
Lung cancer is the leading cause of cancer death in the world (Siegel et al., 2018). Lung adenocarcinoma (LADC) is the most common subtype of lung cancer, accounting for ~40% of diagnoses. The most prevalent oncogenic driver mutations in LADC are activating mutations in KRAS, which occur in ~30% of LADC tumors, and loss of function mutations in TP53, which occur in ~70% of LADC tumors (Zappa and Mousa, 2016). Despite recent advances in targeted therapies for distinct subtypes of lung cancer, no approved or highly effective targeted therapies exist for KRAS/TP53-driven LADC. Thus, there is a strong need to better understand the molecular mechanisms that drive KRAS/TP53 LADC and translate this knowledge into new and improved therapeutic strategies.
We previously identified PRKCI (encoding for PKCι) as an oncogene that is frequently overexpressed and associated with poor outcome in LADC (Regala et al., 2009; Regala et al., 2005b). PKCι drives an aggressive stem-like tumor-initiating cell (TIC) phenotype in both KRAS-driven LADC (Ali et al., 2016), and in lung squamous cell carcinoma (LSCC) (Justilien et al., 2014), the second most frequent form of lung cancer. In KRAS LADC, PKCι establishes and maintains a TIC phenotype by activating a PKCι-ELF3-NOTCH3 signaling axis that drives transformed growth and tumor initiation (Ali et al., 2016). Pharmacologic blockade of PKCι signaling potently inhibits KRAS LADC cell proliferation in vitro and tumorigenicity in vivo, demonstrating the critical role of PKCι-ELF3-NOTCH3 signaling in LADC tumorigenesis.
The KrasG12D;Trp53fl/fl (KP) LADC mouse model recapitulates many aspects of human KRAS/TP53 LADC, including tumor heterogeneity inherent in the human disease. Several distinct cells of origin are capable of being transformed in KP mice (Chen et al., 2014; Chung et al., 2017). Bronchioalveolar stem cells (BASCs) are multi-functional stem cells residing at the bronchioalveolar duct junction (BADJ) that can give rise, through lineage-restricted differentiation, to either bronchial club (Clara) cells or alveolar pneumocytes (Kim et al., 2005; Liu et al., 2019). In response to oncogenic Kras BASCs expand to give rise to LADC cells (Kim et al., 2005). More recent studies using adenovirus with cell type-specific promoters driving Cre expression demonstrate that oncogenic Kras can also transform AT2 cells to give rise to LADC (Mainardi et al., 2014; Sutherland et al., 2014; Xu et al., 2012). These observations have led to the hypothesis that subtypes of human LADC may also arise from distinct cells of origin, and account for the molecular, pathological and perhaps pharmacological heterogeneity exhibited by LADC tumors. However, little is known about the specific oncogenic pathways driving tumorigenesis in these alternative cells of origin, or whether specific molecular subsets of human LADC tumors resemble tumors arising from them.
We previously demonstrated that Prkci deficiency disrupts oncogenic Kras-mediated BASC expansion and subsequent lung adenoma formation in vivo (Regala et al., 2009), and that PKCι is required for the transformed phenotype of human LADC cells in vitro and tumorigenesis in vivo (Ali et al., 2016). In LADC cells, PKCι establishes and maintains a highly aggressive stem-like phenotype through activation of a PKCι-ELF3-NOTCH3 signaling axis (Ali et al., 2016). Here we investigate the role of Prkci in KP LADC tumorigenesis.
Results
PKCι-dependent and -independent pathways to KP LADC
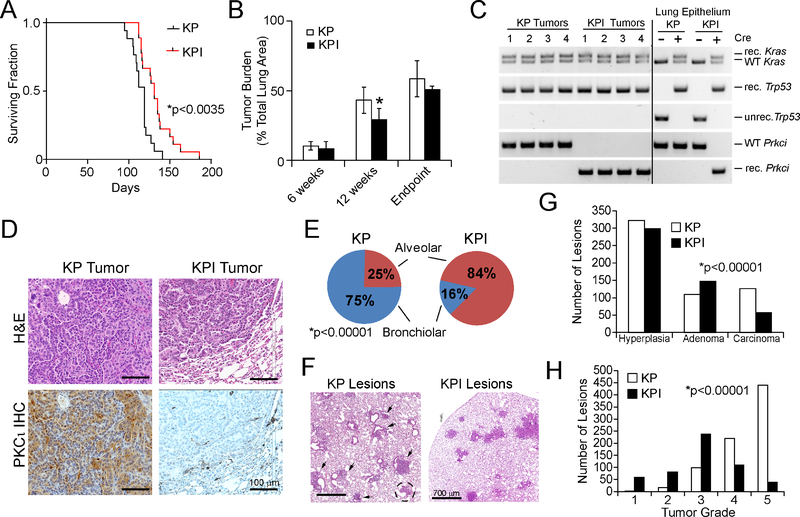
PKCι is required for transformed growth of KRAS LADC cells (Ali et al., 2016; Regala et al., 2005a), and ablation of Prkci in mouse lung inhibits mutant Kras-mediated BASC expansion and adenoma formation (Regala et al., 2009). However, the role of PKCι in KP LADC tumorigenesis has not been investigated. Therefore, we crossed Prkcifl/fl mice with KP mice to generate KPI mice. Tumors were initiated in these mice by intratracheal instillation of adenovirus expressing Cre-recombinase (Ad-Cre) as described previously (Regala et al., 2009). Both KP and KPI mice developed extensive tumor burden, but KPI mice exhibited a significantly longer survival time (p<0.0035; mean survival = 131 days for KPI; 119 days for KP mice) (Figure 1A). KPI mice had a significantly lower tumor burden than KP mice at 12 weeks post-Ad-Cre instillation, though tumor burden at six weeks and at endpoint was not significantly different between genotypes (Figure 1B). PCR of genomic DNA from KP and KPI tumors verified recombination of the LSL-KrasG12D, Trp53fl and Prkcifl alleles (Figure 1C), and immunohistochemical (IHC) staining confirmed that KP tumors expressed elevated PKCι, whereas KPI tumor cells expressed no detectable PKCι (Figure 1D). Histopathological analysis at six weeks revealed that KP mice harbor mostly bronchiolar lesions (75%) and fewer alveolar lesions (25%), whereas KPI mice harbor predominantly alveolar lesions (84%) (Figure 1E and F). Classifying tumors into three categories of increasing grade, hyperplasia, adenoma and carcinoma (Sutherland et al., 2014), revealed that KP mice exhibited more carcinomas than KPI mice (Figure 1G). Analysis at 12 weeks using the tumor grading system described by Jackson et al. (Jackson et al., 2005) revealed that both KP and KPI mice exhibited mostly adenomas and carcinomas, though KP tumors were of significantly more advanced grade than KPI lesions (Figure 1H). Thus, KP tumorigenesis can proceed in the presence or absence of Prkci, albeit with different distribution of lesions within the lung, and at different rates of progression.
Figure 1. Effect of Prkci deletion on KP tumorigenesis.
(A)Effect of genetic loss of Prkci on survival of KP and KPI mice after Ad-Cre-induced LADC formation. *p<0.0035 between KPI and KP by Student’s t-test. n=17 (KP), n=33 (KPI).
(B) Tumor burden in KP and KPI mice at 6 weeks, 12 weeks and at endpoint after tumor induction. Tumor burden (tumor area/total lung area) is expressed as % Total Lung Area. Results represent the mean ± SD; n≥5; *p<0.05 by Student’s t-test.
(C) PCR of genomic DNA from KP and KPI mice for Cre-mediated recombination of LSL-KrasG12D, Trp53fl, and Prkcifl alleles. unrec.=unrecombined; rec.=recombined; WT, wild-type.
(D) Representative images of KP and KPI tumor tissue sections stained with hematoxylin and eosin (H&E; upper panels) or for PKCι by IHC (lower panels).
(E) Distribution of preneoplastic and malignant lung lesions in KP and KPI mice. *p<0.00001 by Chi-square.
(F) Representative H&E images of lung lesions in KP and KPI mice six weeks after tumor initiation. Arrowheads indicate bronchiolar lesions; dotted circle indicates alveolar lesion in KP mouse.
(G) Classification of lesions in KP and KPI mice six weeks after tumor initiation. Lesions were counted and classified pathologically as hyperplasia, adenoma or carcinoma. Data represent number of lesions of each grade. n=5 mice/genotype. *p<0.00001 by Chi-square.
(H) Tumor grade analysis of lung lesions in KP and KPI mice 12 weeks after tumor initiation. Data represent number of lesions of each grade. n=5 mice/genotype. p<0.00001 by Chi-square.
Isolation and characterization of KP and KPI LADC cells
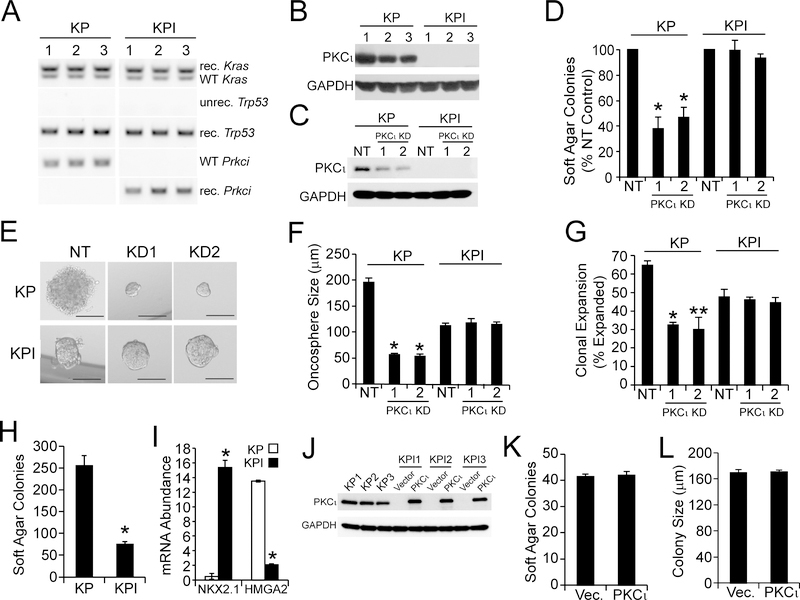
We next established KP and KPI LADC cell lines from three independent KP (KP1–3) and KPI (KPI1–3) tumors. PCR of genomic DNA confirmed complete recombination of the LSL-KrasG12D, Trp53fl and Prkcifl alleles in each cell line (Figure 2A). Immunoblot analysis confirmed that KP cells express abundant PKCι, whereas KPI cells express no PKCι (Figure 2B). Two lentiviral shRNA-mediated PKCι knock down (KD) constructs significantly decreased PKCι in KP cells (Figure 2C), and inhibited growth of KP cells in soft agar (Figure 2D) and as oncospheres in non-adherent culture (Figure 2E and F) while having no effect on KPI cells. Interestingly, KP cells formed larger oncospheres (Figure 2E and F), exhibited more efficient clonal expansion (Figure 2G), and a higher plating efficiency in soft agar than KPI cells (Figure 2H). These behaviors are consistent with the more aggressive transformed behavior of KP tumors in vivo. Consistent with their more aggressive behavior, KP cells expressed low NKX2.1 and high HMG2A2 levels compared to KPI cells (Figure 2I). NKX2.1 suppresses malignant progression of KP tumors, at least in part, through suppression of HMGA2 expression (Winslow et al., 2011). Thus, KP and KPI cells display genetics and transformed behaviors consistent with KP and KPI tumors in vivo.
Figure 2. Characterization of KP and KPI LADC cell lines.
(A) PCR of genomic DNA from three independent KP and KPI LADC cell lines (1, 2 and 3) Cre-mediated recombination of LSL-KrasG12D, Trp53fl, and Prkcifl alleles. unrec.=unrecombined; rec.=recombined; WT, wild-type.
(B) Immunoblot of KP and KPI LADC cell lines for PKCι. GAPDH is a loading control.
(C) Immunoblot analysis confirming lentiviral shRNA-mediated PKCι knock down (PKCι KD) in KP and KPI cell lines using two independent lentiviral shRNAs targeting mouse PKCι (1 and 2) or a non-target control (NT) as indicated. GAPDH is a loading control.
(D) Relative soft agar colony formation in NT control or PKCι KD KP and KPI LADC cells, results expressed as mean % NT control ±SEM. n=3. *p<0.05 compared with NT by Student’s t-test.
(E and F) Representative micrographs (E) and oncosphere size (F) in NT control and PKCι KD KP and KPI cells. (E) Scale bars represent 100 μm. (F) Results expressed as oncosphere diameter (μm) mean ±SEM. *p<0.05 compared with NT by Student’s t-test.
(G) Relative clonal expansion of NT control or PKCι KD KP and KPI cells. Results are expressed as mean % of single cells generating oncospheres ±SEM. Number of cells counted: 65 (KP/NT); 65 (KP/KD1); 70 (KP/KD2); 49 (KPI/NT); 50 (KPI/KD1); and 56 (KPI/KD2). *p<0.0002 (KP/NT vs. KP/KD1); **p=0.0001 (KP/NT vs. KP/KD2) by Chi square.
(H) Anchorage-independent growth of KP and KPI cells. Results represent mean soft agar colonies ±SEM. n=5. *p<0.05 compared to KP by Student’s t-test.
(I) Expression of NXK2.1 and HMGA2 in KP and KPI cells. Results expressed as relative mRNA abundance and represent means in three independent KP or KPI cell lines ±SEM. * p<0.05 compared to KP by Student’s t-test. n=3.
(J) Immunoblot of KPI cells transfected with adenovirus containing PKCι or with control empty adenovirus (Vector). Three untransfected KP cell lines served as a positive control for PKCι expression. GAPDH is a loading control.
(K and L) Soft agar colony number (K) and size (L) of KPI cells transfected with vector or PKCι adenovirus. Results expressed as mean ±SEM. n=3. No significant differences were observed by Student’s t-test.
We next assessed whether the less aggressive behavior of KPI cells was reversed by restoring PKCι to levels comparable to KP cells (Figure 2J). Interestingly, re-expression of PKCι had no significant effect on either the number (Figure 2K) or size (Figure 2L) of soft agar colonies. Thus, transformed growth of KPI cells is PKCι-independent, and PKCι re-expression does not affect the transformed phenotype of KPI cells.
PKCι-ELF3-NOTCH3 signaling is decreased in KPI tumor cells
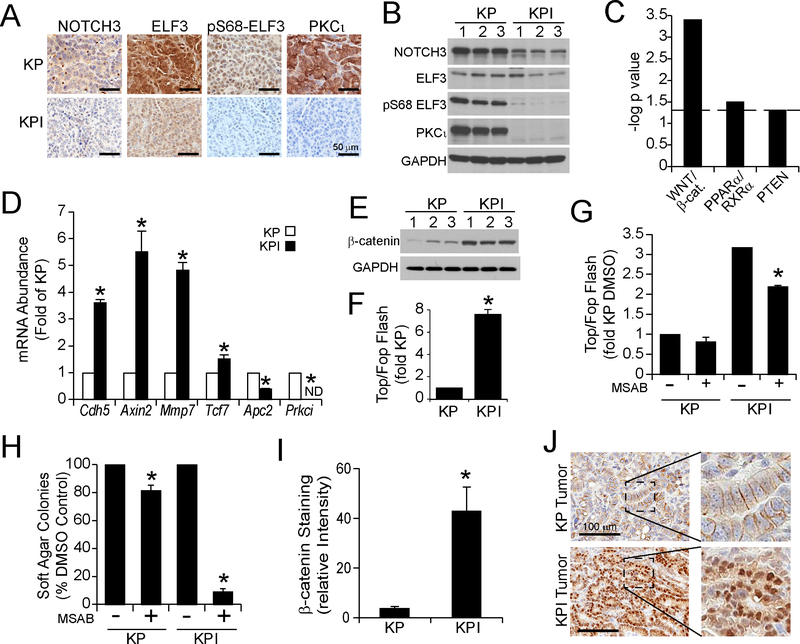
We recently described a PKCι-ELF3-NOTCH3 signaling axis that drives the growth of human KRAS-transformed LADC cells (Ali et al., 2016). In this pathway, PKCι phosphorylates the oncogenic transcription factor ELF3, which stimulates ELF3-mediated NOTCH3 transcription. NOTCH3 in turn activates cell-autonomous Notch signaling to stimulate transformed growth. IHC staining revealed that KP tumors express higher levels of NOTCH3, ELF3 and PKCι-phosphorylated ELF3 (pS68-ELF3) than KPI tumors (Figure 3A). Immunoblot analysis of KP and KPI cells verified these results (Figure 3B). We conclude that PKCι-ELF3-NOTCH3 signaling is active in KP tumor cells, but is dramatically reduced in KPI tumor cells both in vivo and in vitro.
Figure 3. Characterization of oncogenic signaling in KP and KPI tumor cells.
(A) IHC of KP and KPI tumors for NOTCH3, ELF3, p-ELF3 and PKCι. Representative images are shown.
(B) Immunoblot of KP and KPI cells for NOTCH3, ELF3, p-ELF3 and PKCι. GAPDH is a loading control.
(C) Ingenuity Pathway Analysis of RNA-seq of KP and KPI cells. Results plotted as −log p value for pathway enrichment in KPI versus KP cells. Dotted line indicates −log p value 1.3 (p value 0.05). β-cat. = β-catenin.
(D) Expression of Wnt/β-catenin pathway genes in KP and KPI cells. qPCR for Cdh5, Axin2, Mmp7, Tcf7 and Apc2 mRNA. Prkci included as a control. Results expressed as mRNA abundance (fold of KP cells) ±SEM. n=3. *p<0.005 by Student’s t-test.
(E) Immunoblot of KP and KPI LADC cells for β-catenin. GAPDH is a loading control.
(F) β-catenin transcriptional activity in KP and KPI cells. Results show the ratio of Top/Fop Flash activity expressed as fold of KP cells ±SEM. n=3. *p<0.05 by Student’s t-test.
(G) Effect of the β-catenin inhibitor MSAB on Top/Fop Flash activity in KP and KPI cells. Results represent Top/Fop Flash ratio expressed as fold DMSO-treated KP cells ±SEM. n=3. *p<0.05 compared to DMSO-treated KPI cells by Student’s t-test.
(H) Effect of MSAB on soft agar growth of KP and KPI cells. Results represent mean % DMSO control for KP and KPI cells ±SEM. n=3. *p<0.05 compared to DMSO control by Student’s t-test.
(I) Quantitative analysis of β-catenin IHC staining intensity in KP and KPI tumors. Results represent relative intensity of β-catenin in KP and KPI tumors ±SEM. n=6. *p<0.05 by Student’s t-test.
(J) Representative images of IHC staining for β-catenin in KP and KPI tumors. Inset shows high magnification view of indicated region.
See also Table S1.
Elevated Wnt signaling drives transformed growth of KPI tumor cells in vitro
Since KPI tumors do not exhibit oncogenic PKCι-ELF3-NOTCH3 signaling, we reasoned they employ alternative, PKCι-independent oncogenic signaling pathway(s) to support transformed growth. To identify such pathways, we conducted total RNA sequencing and Ingenuity Pathway Analysis (IPA) of genes differentially-expressed in KP and KPI tumors. IPA identified many differentially-expressed key Wnt/β-catenin pathway genes (Table S1) indicating differential activation of Wnt/β-catenin signaling in KPI tumors (Figure 3C). qPCR validated increased expression of the prominent pathway components Cdh5, and direct β-catenin transcriptional targets, Axin2, Mmp7 and Tcf7, and decreased Apc2, indicative of Wnt pathway activation in KPI cells (Figure 3D). Furthermore, KPI cells expressed higher β-catenin (Figure 3E), and elevated β-catenin transcriptional activity as measured by Top/Fop Flash β-catenin transcriptional reporter assays compared to KP cells (Figure 3F).
We next assessed the effect of the potent and selective β-catenin inhibitor MSAB (Hwang et al., 2016) on KP and KPI cells. MSAB led to a significant inhibition of β-catenin transcriptional activity (Figure 3G) and soft agar growth (Figure 3H) of KPI cells while having little effect on KP cells, indicating that KPI cells are selectively dependent on Wnt/β-catenin signaling. IHC revealed that KPI tumors express higher β-catenin levels than KP tumors (Figure 3I). Furthermore, whereas β-catenin localized almost exclusively at the plasma membrane in KP tumors, a significant pool of β-catenin in KPI tumors localizes to the cytoplasm and nucleus (Figure 3J), consistent with activated β-catenin signaling in KPI tumors.
PKCι is necessary for Kras/Trp53-mediated expansion and transformed growth of BASCs
Since KPI mice develop LADC tumors with a distinct regional distribution and slower progression than KP mice, we next assessed the effect of PKCι loss on growth of lung epithelial cells. Total lung epithelial cells from KPI mice formed significantly fewer and smaller oncospheres in three dimensional Matrigel culture than cells from KP mice after Ad-Cre-mediated activation of Kras and Trp53 loss ex vivo (Figure 4A–C). Since both BASCs and AT2 cells can be transformed by oncogenic Kras and Trp53 loss (Barkauskas et al., 2013; Kim et al., 2005; Tiozzo et al., 2009; Ventura et al., 2007), we next assessed whether KPI mice exhibit inherent differences in abundance of these cells. Therefore, we isolated BASCs and AT2 cells from KP and KPI mice by fluorescence-activated cell sorting (FACS) (Lee et al., 2014) (Figure S1A). Post-sort analysis (Figure S1A) and dual immunofluorescence (Figure S1B) verified high enrichment of CCSP+/SPC+ dual positive BASC cells (93% double positive) and CCSP-/SPC+ single positive AT2 cells (96% single positive). FACS analysis also revealed no difference in abundance of BASCs and AT2 cells (expressed as % of total lung epithelial cells) between KP and KPI mice (Figure 4D). Thus, loss of either of these cell types is not likely responsible for the decreased Kras/Trp53-driven oncosphere formation in KPI mice.
Figure 4. Identification of PKCι-dependent and -independent cells of origin for LADC.
(A and B) Oncosphere number (A) and size (B) of KP and KPI cells grown in non-adherent culture. Results represent mean ±SEM. n=3; *p<0.05 by Student’s t-test.
(C) Micrographs showing the size and morphology of representative KP and KPI oncospheres. Scale bars represent 200 μm.
(D) Abundance of bronchioalveolar stem cells (BASCs) and alveolar type II (AT2) cells in KP and KPI mouse lungs. Data expressed as mean % total lung epithelial cells ±SEM. n=3.
(E) PCR of genomic DNA from KP and KPI BASC and AT2 cells after transduction with Ad-Cre ex vivo. unrec.=unrecombined; rec.=recombined; WT, wild-type. DNA from total lung epithelial cells in the absence of Ad-Cre included as a control.
(F and G) Oncosphere growth of purified BASCs and AT2 cells from KP and KPI mice transduced with Ad-Cre. Oncosphere numbers (F) and size (G) are shown as mean ±SEM. n=3. *p<0.05 compared to KP by Student’s t-test.
(H) Immunofluorescence of a bronchiolar lesion in a KP mouse stained for CCSP (green, upper left panel), SPC (red; upper right panel) or merged image CCSP, SPC and DAPI (blue, lower left panel) six weeks after induction of tumor formation. Lower right panel shows H&E image of same lung region revealing growth of bronchiolar hyperplasia from the affected terminal bronchiole. Scale bars indicate 100 μm.
(I) Immunofluorescence of an alveolar lesion in KPI mouse stained for CCSP (green, upper left panel), SPC (red; upper right panel) or merged image CCSP, SPC and DAPI (blue, lower left panel) six weeks after induction of tumor formation. Lower right panel shows H&E image of same lung region revealing an alveolar lesion without bronchiolar involvement. Scale bars indicate 100 μm.
See also Figure S1.
We next assessed the ability of oncogenic Kras and Trp53 loss to transform BASCs and AT2 cells from KP and KPI mice ex vivo. Ad-Cre transduction led to efficient recombination of the oncogenic Kras, Trp53, and Prkci alleles in both cell populations (Figure 4E). Interestingly, both BASCs and AT2 cells from KP mice exhibited robust oncosphere formation in three dimensional Matrigel culture, confirming the ability of both cell populations to undergo Kras- and Trp53-mediated transformation (Figure 4F and G). In contrast, BASCs from KPI mice exhibited a decrease in oncosphere number and size compared to BASCs from KP mice (Figure 4F and G). AT2 cells from KPI mice exhibited a smaller, but significant decrease in oncosphere formation but no change in oncosphere size compared to AT2 cells from KP mice (Figure 4F and G). We next visualized and quantified BASCs within bronchioles of KP and KPI mice in situ by dual SPC and CCSP immunofluorescence (Dovey et al., 2008; Kim et al., 2005; Regala et al., 2009). Lungs from Ad-Cre-treated KP mice at six weeks exhibited frequent clusters of SPC+/CCSP+ BASCs extending into the terminal bronchiole, consistent with the pattern of BASC expansion observed in response to oncogenic Kras (Kim et al., 2005; Yang et al., 2008) or Trp53 loss (McConnell et al., 2016) (Figure 4H). Quantitative analysis revealed a significant increase in BASCs in terminal bronchioles of KP mice compared to KPI mice (Figure S1C). Interestingly, BASC expansion in KP mice was often observed adjacent to bronchiolar hyperplasias (Figure 4H), consistent with the notion that these lesions arise from transformed BASCs. In contrast, tumors in Ad-Cre-treated KPI mice arose predominantly in the alveolar space and stained positive for SPC but negative for CCSP (Figure 4I). Thus, Kras/Trp53-induced transformation of BASCs ex vivo, and BASC expansion in vivo, is highly dependent upon PKCι. In contrast, PKCι loss leads to only a partial inhibition of AT2 cell-derived oncosphere formation ex vivo.
Identification of PKCι-dependent and PKCι-independent AT2 cell populations
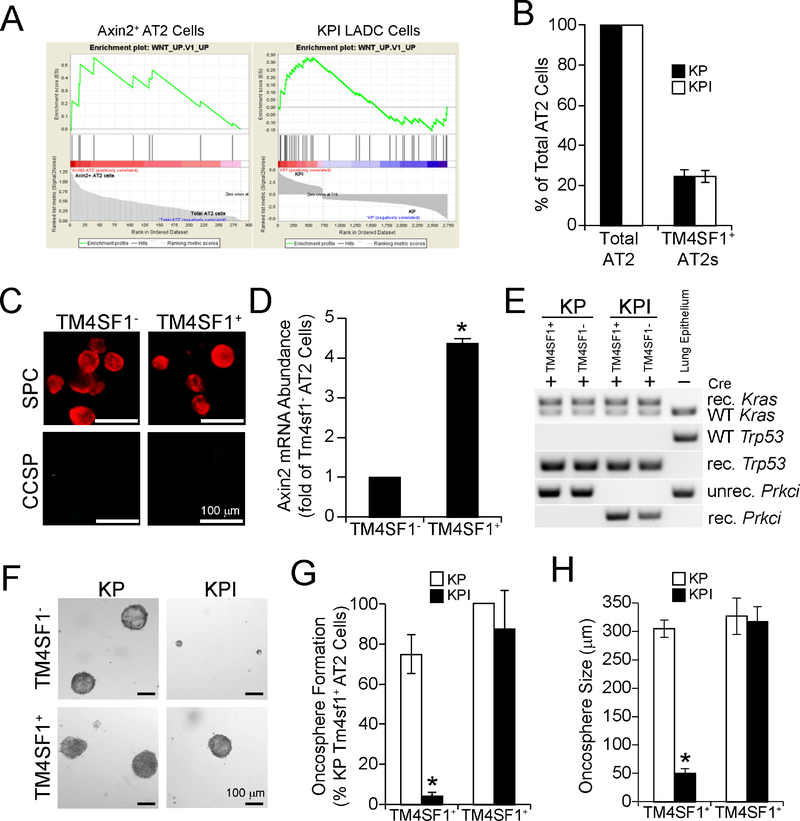
Recent studies identified a subtype of AT2 cells, Axin2+ AT2 cells, as regional lung epithelial stem/progenitor cells that proliferate during lung development to generate alveolar structures, are mobilized after lung injury to repopulate alveoli, and exhibit active Wnt signaling (Frank et al., 2016; Nabhan et al., 2018). However, the role of Axin2+ AT2 cells in LADC tumorigenesis has not been assessed. Interestingly, Gene Set Enrichment Analysis (GSEA) of RNA-seq data from Axin2+ AT2 cells and KPI LADC cells demonstrated that these cell types expressed a similar Wnt pathway signature (Figure 5A). RNA-seq analysis identified Transmembrane 4 Superfamily Member 1 (Tm4sf1) as a cell surface maker expressed in the vast majority (>90%) of Axin2+ AT2 cells but absent from Axin2− AT2 cells, making it useful for sorting Axin2+ and Axin2− AT2 cell populations (Zacharias et al., 2018). Thus, we isolated TM4SF1+ AT2 cells from total mouse lung AT2 cells by FACS (Frank et al., 2016; Zacharias et al., 2018). Consistent with published reports, ~20% of AT2 cells in both KP and KPI mice were TM4SF1+ (Figure 5B). Immunofluorescence confirmed that both TM4SF1+ and TM4SF1− AT2 cells were SPC+ but CCSP− (Figure 5C) and qPCR confirmed significantly elevated Axin2 mRNA in TM4SF1+ AT2 cells compared to TM4SF1− AT2 cells (Figure 5D). To assess transforming potential, TM4SF1+ and TM4SF1− AT2 cells were treated with Ad-Cre ex vivo to induce recombination of Kras, Trp53, and Prkci alleles (Figure 5E), followed by plating in oncosphere culture (Figure 5F). Both TM4SF1− and TM4SF1+ AT2 cells from KP mice form oncospheres with similar morphology (Figure 5F), frequency (Figure 5G) and size (Figure 5H). However, TM4SF1− AT2 cells from KPI mice exhibit a dramatic decrease in oncosphere frequency and size (Figure 5F–H). In contrast, TM4SF1+ AT2 cells from KPI mice grow as oncospheres exhibiting morphology, frequency and size indistinguishable from TM4SF1+ AT2 cells from KP mice (Figure 5 F–H). Taken together, these data indicate that LADC tumors in KPI mice arise from TM4SF1+ AT2 cells, whereas those in KP mice arise predominantly from BASCs.
Figure 5. Axin2+ AT2 cells undergo PKCι-independent transformation in vitro.
(A) Gene Set Enrichment Analysis of Axin2+ AT2 cells (left panel) and KPI cells (right panel).
(B) Abundance of total and TM4SF1+ AT2 cells in KP and KPI mice. Results are expressed as mean % total AT2 ±SEM.
(C) Representative immunofluorescence staining of TM4SF1− and TM4SF1+ AT2 cells for CCSP and SPC.
(D) qPCR from TM4SF1− and TM4SF1+ AT2 cells for Axin2. Results expressed as fold of TM4SF1− AT2 cells ±SEM. n=3. *p<0.05 by Student’s t-test.
(E) PCR of genomic DNA isolated from TM4SF1− and TM4SF1+ AT2 cells from KP and KPI mice after transduction with Ad-Cre ex vivo for recombination of LSL-KrasG12D, Trp53fl, and Prkcifl alleles. unrec.=unrecombined; rec.=recombined; WT, wild-type. DNA from total lung epithelial cells in the absence of Ad-Cre included as a control.
(F) Micrographs of representative TM4SF1− and TM4SF1+ AT2 cell oncospheres from KP and KPI mice.
(G and H) Oncosphere formation (G) and growth (H) of TM4SF1− and TM4SF1+ AT2 cells from KP and KPI mice. Results represent mean ±SEM. n=3; *p<0.05 compared to KP by Student’s t-test.
KP and KPI tumor cells are differentially responsive to PKCι and Wnt inhibition
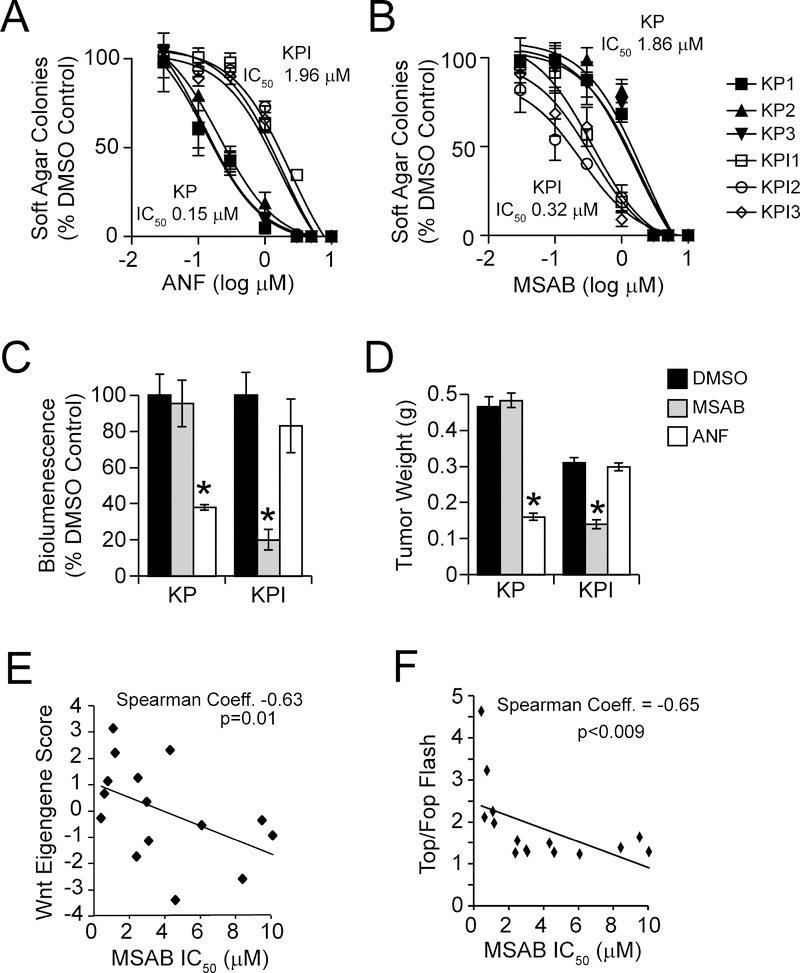
We previous demonstrated that pharmacologic inhibition of PKCι inhibits LADC growth in vitro and in vivo (Ali et al., 2016; Regala et al., 2009). Thus, we assessed the effect of the selective PKCι inhibitor auranofin (ANF) on soft agar growth of KP and KPI cells in vitro (Figure 6A and B). KP cells exhibited similar high sensitivity to the growth inhibitory effects of ANF (average IC50 ~0.15 μM), whereas KPI cells exhibited relative resistance to ANF (IC50 ~2 μM). Conversely, the Wnt inhibitor MSAB potently and selectively inhibited anchorage-independent growth of KPI cells (IC50 ~0.32 μM) but KP cells were much less responsive (IC50 ~1.9 μM). We next assessed whether orthotopic KP and KPI lung tumors implanted into the lungs of syngeneic mice exhibited selective responses to these agents in vivo. In vivo bioluminesence (IVIS) monitoring revealed that KP tumors exhibited a significant therapeutic response to ANF (10 mg/kg daily) but no response to MSAB (Figure 6C). Conversely, KPI tumors exhibited a significant response to MSAB but not to ANF (Figure 6C). Final tumor weights confirmed the differential response of KP and KPI tumors to the growth inhibitory effects of ANF and MSAB, respectively (Figure 6D).
Figure 6. KP and KPI LADC cells exhibit differential sensitivity to PKCι and β-catenin inhibition.
(A and B) Growth of KP and KPI LADC cells in soft agar treated with the PKCι inhibitor ANF (A) and the β-catenin inhibitor MSAB (B). Results represent the mean ± SD; n=3, and are plotted as % DMSO control.
(C and D) Growth of KP and KPI LADC cells as orthotopic lung tumors in syngeneic mice. Tumor growth was monitored by bioluminescence (C) and tumor weight was determined at the time of sacrifice (D). Results represent mean bioluminescence expressed as % DMSO control ±SD (C) or mean weight in grams (g) ±SEM (D), n=5/group. *p < 0.05 compared to DMSO by Student’s t-test.
(E) Plot of a Wnt Eigengene score versus MSAB IC50 in a panel of mutant KRAS human LADC cell lines. n=15.
(F) Plot of Top/Fop Flash activity versus MSAB IC50 in a panel of mutant KRAS human LADC cell lines. n=15.
See also Table S2
A subset of human KRAS/TP53 LADC cells exhibit a KPI-like genotype and Wnt pathway activation
Our data demonstrate that Kras/Trp53-driven LADC can arise from two cells of origin, resulting in LADC tumors with distinct oncogenic signaling and therapeutic vulnerabilities. To explore whether human LADC tumor cells exhibit KP- versus KPI-like genotypes, we used principal component analysis to calculate a Wnt Eigengene score based on 17 Wnt pathway genes that are differentially-expressed in KP and KPI cells, and contribute to the GSEA enrichment score (Table S2). We then calculated Wnt Eigengene scores for a cohort of established human mutant KRAS/TP53 LADC cell lines using RNA-seq data available from the Cancer Cell Line Encyclopedia (CCLE) and assessed whether the Wnt Eigengene score predicts sensitivity to MSAB. Dose response analysis revealed a significant (p=0.01) positive correlation between Wnt pathway score and sensitivity to MSAB growth inhibition (MSAB IC50; Spearman Coefficient = −0.63) (Figure 6E). Thus, LADC cell lines with high Wnt Eigengene scores exhibited higher sensitivity to MSAB (lower IC50s) than those with a low Wnt Eigengene scores (higher IC50s). Biochemical analysis revealed that MSAB sensitivity also correlated positively with β-catenin-dependent transcriptional activity (Spearman Coefficient = −0.65, p<0.009) (Figure 6F). Thus, our Wnt Eigengene score identifies KRAS/TP53 LADC cell lines exhibiting increased β-catenin transcriptional activity and enhanced sensitivity to β-catenin inhibition, consistent with a KPI-like phenotype.
A subset of human LADC tumors exhibit a KPI-like genotype
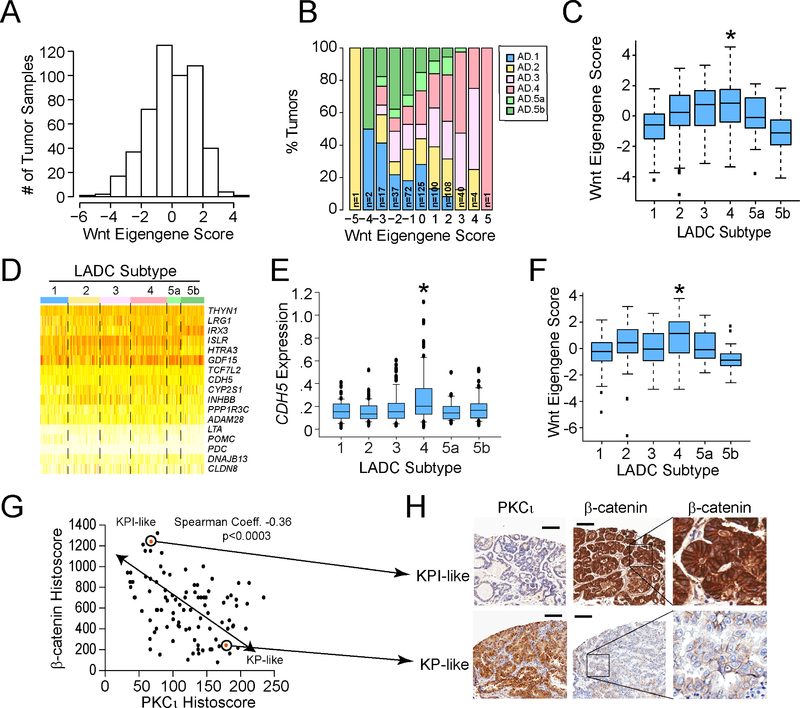
We next generated Wnt Eigengene scores for samples in the TCGA lung adenocarcinoma dataset (n=507 tumors). Wnt Eigengene scores distributed non-randomly, suggesting that a distinct subset or subset(s) of LADC tumors may exhibit high KPI-like Wnt Eigengene scores (Figure 7A). To address this possibility, we assessed whether Wnt Eigengene scores are associated with distinct molecular LADC subtypes (1, 2, 3, 4, 5a and 5b) as identified by Chen et al. using multiplatform genomic analysis (Chen et al., 2017). Analysis revealed that LADC subtype 3 and LADC subtype 4 tumors exhibited significantly higher Wnt Eigengene scores than other LADC subtypes (Figure 7B and C). Supervised clustering of the 17 genes making up the Wnt Eigengene revealed that expression of cadherin 5 (CDH5) was selectively elevated in LADC subtype 4 tumors (Figure 7D). Indeed, CDH5 expression was significantly higher in LADC subtype 4 tumors than all other LADC subtypes (Figure 7E). Interestingly, CDH5 expression also correlated with response of human KRAS/TP53 LADC cell lines to MSAB (Spearman Coefficient = −0.59, p value =0.02; data not shown). Interrogation of the TCGA data revealed that PRKCI expression was lower in LADC subtype 4 tumors than other subtypes (p value of Ad.4 vs Ad.1 = 0.04; Ad.4 vs Ad.2 = 0.07; Ad.4 vs Ad.3 = 0.09; Ad.4 vs Ad.5a = 0.01; Ad.4 vs Ad.5b = 0.04), consistent with the KPI phenotype. Finally, analysis of the subset of TCGA LADC tumor dataset harboring mutant KRAS (n=163 tumors) revealed a similar association of higher Wnt Eigengene score with mutant KRAS LADC subtype 4 tumors (Figure 7F). Taken together, these data indicate that a distinct molecular LADC subtype, LADC subtype 4, exhibits Wnt pathway activation and decreased PRKCI expression, consistent with the properties of KPI tumors.
Figure 7. Identification of a specific molecular subtype of LADC tumors with KPI-like characteristics.
(A) Distribution of Wnt Eigengene scores across the TCGA LADC tumor dataset (n=507).
(B) A stacked bar plot showing the distribution of Wnt Eigengene scores across six molecular subtypes of LADC tumors.
(C) Plot of Wnt Eigengene scores across LADC molecular subtypes. Middle lines indicate the median values, boxes show 25% and 75% confidence intervals and error bars show 95% confidence intervals. Dots indicate outliers. *indicates statistically significant differences in Eigengene score between LADC subtype 4 and other subtypes by two-sided Wilcoxon rank-sum test. p value Ad4 vs indicated subtype: Ad1 = 3.8 × 10−10; Ad2 = 0.008; Ad3 = 0.28; Ad5a = 0.01; Ad5b = 1.3 × 10−12. n=507. (D) Heat map showing expression of 17 Wnt genes comprising the Wnt Eigengene across LADC molecular subtypes.
(E) Plot of CDH5 mRNA expression across LADC molecular subtypes. Middle lines indicate the median values, boxes show 25% and 75% confidence intervals and error bars show 95% confidence intervals. Dots indicate outliers. *denotes a statistically significant difference in CHD5 expression in subtype 4 compared to other subtypes by two-sided Wilcoxon rank sum test. p value (Ad.4 vs Ad.1) = 3.5 × 10−6; (Ad.2 vs Ad.4) = 1.5 × 10−6; (Ad.3 vs Ad.4) = 0.0003; (Ad.5a vs Ad.4) = 2.2 × 10−5; (Ad.5b vs Ad.4) = 0.001.
(F) Plot of Wnt Eigengene scores in mutant KRAS LADC tumors classified by molecular subtype. A Wnt Eigengene score was calculated for LADC tumor samples in the TCGA LADC dataset harboring mutant KRAS (n=163) and plotted by molecular subtype. Middle lines indicate the median values, boxes show 25% and 75% confidence intervals and error bars show 95% confidence intervals. Dots indicate outliers. *indicates statistically significant differences in Eigengene score between LADC subtype 4 and other subtypes by two-sided Wilcoxon rank sum test. p value Ad4 vs indicated subtype: Ad1 = 0.003; Ad2 = 0.078; Ad3 = 0.072; Ad5a = 0.100; Ad5b = 7.8 × 10−5.
(G) Plot of β-catenin Histoscore versus PKCι Histoscore in human LADC tumors. TMAs of archived LADC cases were subjected to IHC for β-catenin and PKCι. Histoscores were calculated as described in STAR Methods. Spearman correlation coefficient = −0.36. p<0.0003. n=95.
(H) IHC of representative KP- and KPI-like human LADC tumors. Scale bars represent 100 μm. Inset: Higher magnification of indicated area to reveal β-catenin staining pattern.
We next assessed expression of β-catenin and PKCι by IHC in tissue microarrays from archived LADC tumors using Histoscore algorithms as described in STAR Methods. Scatter plot analysis revealed a relatively weak but significant inverse association between PKCι and β-catenin Histoscores across 95 evaluable LADC cases (Figure 7G). IHC staining of representative KP- and KPI-like LADC tumors revealed elevated cytoplasmic and nuclear localization β-catenin staining and low PKCι in KPI-like tumors, and weaker membrane-restricted β-catenin staining and high PKCι expression in KP-like tumors (Figure 7H). These data indicate that a subset of human LADC tumors exhibit KP- and KPI-like phenotypes, and reinforce the importance of PKCι in determining these phenotypes in human LADC tumors. Our data also suggest that low PKCι levels, and elevated β-catenin and CDH5 levels may be useful as biomarkers of the KPI phenotype in human LADC, and have implications for clinical management of this molecular LADC subtype.
Discussion
PRKCI is an oncogene in many human tumor types, including the most prevalent forms of lung cancer, LSCC and LADC (Ali et al., 2016; Fields et al., 2016; Justilien et al., 2014; Parker et al., 2014; Regala et al., 2005b). PKCι is a critical oncogenic driver in the vast majority of LSCC tumors (Justilien et al., 2014). 85–90% of LSCC tumors harbor chromosome 3q26 copy number gain, leading to coordinate overexpression of PKCι and a second 3q26 oncogene, SOX2. Coordinate overexpression of PKCι and SOX2 drives a highly aggressive stem-like, TIC phenotype that drives LSCC tumorigenesis by activating a PKCι-SOX2-Hedgehog signaling axis (Justilien et al., 2014).
PKCι is also a key oncogenic factor in human LADC (Ali et al., 2016; Fields et al., 2017; Justilien et al., 2017; Regala et al., 2005b). PKCι is frequently overexpressed in primary LADC tumors and human LADC cell lines, and shRNA-mediated PKCι KD leads to inhibition of transformed growth and tumor initiating activity of LADC cells (Ali et al., 2016). In LADC, PKCι activates a PKCι-ELF3-NOTCH3 signaling axis that maintains a stem-like TIC phenotype (Ali et al., 2016). However, PKCι expression is more variable in LADC tumors than in LSCC tumors. Significant subsets of LADC tumors either overexpress PKCι, or show little or no change in PKCι expression compared to normal lung epithelium. However, the significance of this variable PKCι expression pattern in LADC has been of unknown significance until now. Our data indicate that low PKCι expression is associated with a specific molecular subtype of human LADC.
We previously demonstrated that genetic ablation of Prkci blocks BASC expansion and subsequent lung adenoma formation in response to oncogenic Kras (Regala et al., 2009). Our current data extend these findings, demonstrating that Kras/Trp53 driven BASC transformation and LADC formation is highly dependent upon Prkci. Interestingly however, despite the strict dependence on Prkci for BASC-derived LADC, KPI mice develop LADCs, albeit with a distinct distribution pattern and slower rate of progression. Our molecular and cellular studies demonstrate that LADC tumors in KPI mice are PKCι-independent and arise from a distinct LADC cell-of-origin, Axin2+ AT2 cells. Importantly, restoration of PKCι expression in KPI tumor cells does not affect their transformed behavior, conclusively demonstrating that PKCι is not tumor suppressive in these cells.
Previous studies indicate that there are multiple cells of origin for Kras/Trp53 LADC. In addition to BASCs, bronchiolar club cells and AT2 cells are possible cells of origin (Kim et al., 2005; Mainardi et al., 2014; Sutherland et al., 2014), though some studies have cast doubt on the role of club cells (Xu et al., 2012). Our data are consistent with both BASCs and AT2 cells, providing compelling evidence for a specific role for Axin2+ AT2 cells, as LADC cells of origin. Axin2+ AT2 cells are regional lung epithelial stem cells that mobilize during development and after lung injury to regulate alveologenesis (Frank et al., 2016; Nabhan et al., 2018; Zacharias et al., 2018). Axin2+ AT2 cells exhibit dependency upon Wnt signaling for their growth and stem-like activity since Wnt inhibition leads to growth cessation and differentiation of Axin2+ AT2 cells into AT1 cells (Frank et al., 2016; Nabhan et al., 2018). We have found that Axin2+ AT2 cells, like KPI LADC cells, exhibit a Wnt genomic signature indicating that KPI LADC cells arise from transformed Axin2+ AT2 cells. Our data demonstrate that Axin2+ AT2 stem cells serve as cells of origin for Kras/Trp53-driven LADC, giving rise to LADC tumors that are functionally, transcriptionally and biochemically distinct from BASC-derived LADC tumors. Specifically, BASC-derived LADC tumors exhibit PKCι dependency, elevated PKCι expression and activated PKCι-ELF3-NOTCH3 signaling, whereas Axin2+ AT2 cell-derived LADC tumors exhibit PKCι-independent growth and activation of Wnt/β-catenin signaling. Consistent with their different origin and distinct oncogenic signaling mechanisms, BASC- and Axin2+ AT2-derived LADCs exhibit distinct sensitivity to pharmacologic inhibition of PKCι and Wnt signaling, respectively, both in vitro and in vivo.
Human LADC is a highly heterogeneous disease with diverse pathological, genetic and biochemical characteristics (Cancer Genome Atlas Research, 2014; Chen et al., 2014). Using a multi-platform analysis of the TCGA LADC dataset, Chen et al. classified LADC into six distinct molecular subtypes, LADC subtypes 1, 2, 3, 4, 5a and 5b (Chen et al., 2017). Interestingly, our results indicate that mouse KPI LADC tumors closely resemble the LADC subtype 4. Specifically, LADC subtype 4 tumors exhibit significantly higher Wnt Eigengene scores that define a KPI-like phenotype indicative of Wnt pathway activation. Our data indicate that this classification is of clinical significance since human KRAS/TP53 LADC cell lines with high Wnt Eigengene scores exhibit enhanced β-catenin transcriptional activity and are highly responsive to the growth inhibitory effects of the β-catenin inhibitor MSAB. Like KPI LADC cells, these human LADC cells are dependent upon Wnt/β-catenin signaling for transformed growth. In this regard, we have observed a significant subset of primary LADC tumors with elevated expression and nuclear localization of β-catenin in association with low PKCι expression, demonstrating the existence of a KPI-like phenotype in a subset of primary human LADC tumors. Our data suggest that KPI-like, LADC subtype 4 tumors may exhibit selective sensitivity to Wnt/β-catenin directed therapies, a number of which are actively being evaluated in clinical studies for the treatment of cancer. Our results indicate that expression profiling using a combination of nuclear β-catenin, high CDH5 expression and/or decreased PKCι expression may be useful in identifying patients with KPI-like, LADC subtype 4 tumors that will be more likely to respond to Wnt/β-catenin directed therapeutics.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Alan P. Fields (fields.alan@mayo.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Generation of LSL-KrasG12D;Trp53fl/fl; Prkcifl/fl Mice and Lung Adenocarcinoma Tumorigenesis
LSL-KrasG12D and Trp53fl/fl C57BL/6 mice were obtained from Jackson Laboratories and crossed to generate LSL-KrasG12D;Trp53fl/fl (KP) mice. KP mice were crossed to our previously characterized conditional knock out Prkci C57BL/6 mice (Prkcifl/fl) (Regala et al., 2009) to generate LSL-KrasG12D;Trp53fl/fl; Prkcifl/fl (KPI) mice. KP and KPI mice (6~10 weeks old) were anesthetized with ketamine and xylazine, intubated intratracheally, and instilled with two 50 μL aliquots of Ad-CMV-Cre (1.5 × 108 PFU/mL) (Vector Laboratories) as described previously (Fasbender et al., 1998). All animal experiments were approved by the Institutional Animal Care and Use Committee of the Mayo Clinic. The status of the LSL-KrasG12D, Trp53fl/fl and Prkcifl/fl alleles was determined by PCR as described (Regala et al., 2009) using the primers listed in Table S3. Animals were inspected daily for health, and moribund animals were considered to be at endpoint and were euthanized by CO2 asphyxiation for necropsy. Criteria for euthanasia were based on independent veterinary assessment using AAALAC guidelines; animals in a condition considered incompatible with continued survival were considered deaths. Animals removed at sacrifice or found dead with confirmed lung tumors were considered censored deaths.
Establishment of KP and KPI LADC tumor cell lines
Tumor tissues were dissected from Ad-Cre-instilled KP or KPI mice and processed for lung epithelial cells isolation as described in Method Details below. KP/KPI cells from 2 female mice and 1 male mouse were used in this study. Single tumor cell suspensions were plated in serum-free DMEM-F12 (1:1) medium (Life Technologies) and maintained in ultra-low attachment flasks (Corning, Corning, NY).
Human Cell Lines
Human lung adenocarcinoma cell lines (H2030 male, H2009 female, SW1573 female, H2347 female, H1373 male, H1792 male, H650 male, H358 male, H647 male, H23 male, H1573 female, H1734 female, Calu-6 female, H1355 male and H441 male) were obtained from the American Type Culture Collection, authenticated by STR profiling upon receipt, and maintained in low passage culture as recommended. Cells were grown as oncospheres in serum-free DMEM-F12 (1:1) medium (Life Technologies) and maintained in ultra-low attachment flasks (Corning, Corning, NY). Growth media was modified by adding 50 μg/ml insulin and 0.4% Albumin Bovine Fraction V from (Sigma-Aldrich St. Louis, MO), N2 Plus Media Supplement and B-27 Supplement (Life Technologies), 20 μg/ml EGF and 10 μg/ml basic FGF purchased from (PeproTech, Rocky Hill, NJ).
Orthotopic mouse LADC tumor studies
Firefly luciferase-labelled KP and KPI LADC cells were suspended in 10% Growth Factor Reduced Matrigel Matrix (BD Biosciences, San Jose, CA) in PBS and injected orthotopically into the left lungs (final volume 50 μl) of 8~10 weeks old wild type C57BL/6 mice (both male and female) using a 31-gauge needle. Tumor-bearing mice were randomly divided into 3 groups receiving PEG as a vehicle control, ANF (10 mg/Kg) or MSAB (10 mg/Kg) by IP injection daily, 6 days per week for 2 weeks. Tumor growth was monitored by bioluminescence imaging using IVIS (PerkinElmer), after intraperitoneal injection of luciferin (150 mg/kg) in live mice. Bioluminescent signals were quantified using Living Image software (PerkinElmer). All animal experiments were performed under an approved Mayo Clinic IACUC protocol.
TMA Analysis of Human LADC Tumors
Tissue microarrays (TMAs) were constructed from formalin-fixed, paraffin-embedded blocks from 124 archival, de-identified lung adenocarcinoma patient tumor samples using between 3–6 independent cores from each patient sample to provide representative coverage of each tumor. Patient samples were obtained after informed consent under a protocol approved by the Mayo Clinic Institutional Review Board (IRB) (protocol# 09–000469).
Method Details
Histology and Immunohistochemistry
Mice were sacrificed, exsanguinated, and lungs processed for histologic and immunohistochemical analysis as previously described (Regala et al., 2009). Briefly, the tissue was deparaffinized by placing slides into three changes of xylene and rehydrated in a graded ethanol series. The rehydrated tissue samples were rinsed in water and subjected to antigen retrieval in citrate buffer (pH 6.0). Slides were treated with 3% H2O2 for 5 min to reduce endogenous peroxidase activity and washed with PBS containing 0.5% (w/v) Tween 20 and incubated with the indicated antibodies: anti-PKCι, anti-ELF3, anti-p-ELF3(S68), anti-NOTCH3, anti-β-catenin (see Key Resources Table). Slide images were captured and analyzed using a ScanScope scanner and Aperio ImageScope software (Leica Biosystems). Tumor burden was calculated by determining total tumor area and dividing by total lung area using the ImageScope software on hematoxylin and Eosin (H&E) stained sections. Results were expressed as % Total Lung Area. KP and KPI lesions were analyzed at six weeks after tumor initiation using the classification system described by Sutherland et al. (Sutherland et al., 2014). Briefly, H&E stained sections were analyzed to determine lesion number, location (bronchiolar or alveolar), and classified as either hyperplasia, adenoma or carcinoma using the criteria in Sutherland et al. Differences in distribution and lesion grade were assessed by Chi-square analysis. KP and KPI tumors were also assessed at 12 weeks using the classification system described by Jackson et al. (Jackson et al., 2005). Lesions were scored as grade 1–5 based on the following criteria: Grade 1 lesions form solid tumor masses with regular nuclei; grade 2 lesions have slightly irregular nuclei and prominent nucleoli; grade 3 lesions exhibit pleomorphic nuclei and nuclear molding; grade 4 lesions have enlarged, pleomorphic nuclei, aberrant mitoses and multi-nucleated giant cells within the tumor mass; grade 5 lesions had the characteristics of grade 4 tumors plus the presence of desmoplastic stromal elements. Differences in tumor grade between KP and KPI mice were assessed by Chi-square analysis.
Analysis of Human Lung Adenocarcinoma TMAs
Nine TMAs containing 124 diagnosed cases of LADC were sectioned and stained with antibodies to β-catenin and PKCι (as listed in the Key Resources Table). Each block was sectioned at 4.5 μm onto charged glass slides. The slides were baked, deparaffinized for 15 min in xylene, and rehydrated through graded alcohols to distilled water. Antigen retrieval was performed for 25 min in pH 9.0 retrieval solution (Dako) at 100 °C. The slides were allowed to cool in solution for 25 min, washed in tap water followed by wash buffer (Dako). Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 5 min at room temperature. The slides were also treated with Serum free protein block (Dako) for 5 min. Antibodies against β-catenin or PKCι were diluted in antibody diluent (Dako) (1:400 for β-catenin and 1:1600 for PKCι) and incubated for 60 or 35 min respectively. Following two washes with wash buffer (Dako), the slides were incubated with anti-mouse labeled polymer (Dako) for 30 min at room temperature and rinsed twice in wash buffer (Dako). A final incubation with DAB 5’ (Dako) for 5 min at room temperature was performed, followed by a rinse in distilled water. The slides were counterstained in Gills I hematoxylin for 30 s, rinsed in tap water, dehydrated and mounted. Stained TMAs were scanned at 20x magnification using an Aperio AT2 slide scanner (Leica Biosystems) and images visualized using the Aperio Slide Imaging software. PKCι staining was analyzed with the Aperio ScanScope program. Areas of interest (AOI) containing tumor tissue were marked for each case. A positive pixel algorithm was applied to each AOI, resulting in a determination of the percentage of positive staining. ScanScope reports these values, binned into categories similar to the traditional 0, 1+, 2+, and 3+ scoring system provided by a pathologist. This analysis allowed us to apply the Histo-scoring system (Hatanaka et al., 2003) which provides a more dynamic representation of the staining pattern, resulting in values ranging from 0 to 300 by applying the following formula: PKCι Histoscore = [1 × (% of tumor 1+) + 2 × (% of tumor 2+) + 3 × (% of tumor 3+)]. β-catenin staining was also visualized in the Aperio ScanScope program. AOI’s were marked on the images and plasma membrane, cytoplasmic and nuclear regions of tumor cells were manually evaluated for percentage of positive staining in 10% increments, and for staining intensity using the standard pathologist scale (0, +1, +2, +3). A Histoscore was then calculated for each case, resulting in values ranging from 0–1800 using the following formula: β-catenin Histoscore = [(1 × intensity × extent of membrane staining) + (2 × intensity × extent of cytoplasmic staining) + (3 × intensity × extent of nuclear staining)]. This β-catenin Histoscore is weighted toward cytoplasmic and nuclear β-catenin staining since such staining is indicative of Wnt pathway activation. 95 of 124 cases were evaluable for both β-catenin and PKCι IHC.
Primary Lung Epithelial Cell Isolation
Mice were anesthetized with ketamine/xylazine by intraperitoneal injection. The trachea was isolated, cannulated and the lungs perfused with 10 ml of 0.9% saline. Dispase (3 ml) was injected into the lung through the trachea followed by 0.5 ml agarose (45 °C). Lungs were immediately covered wit h ice for 2 min, removed from the mice, and incubated in 3 ml dispase for 45 min at room temperature. Lungs were subsequently transferred to tissue culture dishes with HEPES-buffered DMEM (100 U/ml DNAse I per 7 ml), and lung tissue was gently teased into small pieces. Cell suspensions were filtered through cell strainers and red blood cells were lysed by Ammonium chloride solution. Cells were further purified through CD45/CD31 negative selection as described previously (Regala et al., 2009). Total lung epithelial cells were resuspended and cultured in matrigel-coated plates.
Lentiviral RNAi Constructs and Transfections
Lentiviral vectors containing mouse PKCι or NT shRNA were obtained from Sigma, packaged into recombinant lentivirus, and used to establish stable cell transfectants as described previously (Frederick et al., 2008). An NT control vector that does not recognize any mouse or human genes was used as a negative control. Sequences of all lentiviral shRNA constructs are listed in Key Resources Table.
Anchorage-independent growth, Clonal expansion and MTT assays
Anchorage-independent growth was assessed by the ability of cells to form colonies suspended in agarose (SeaPlaque GTG Agarose from Lonza, Rockland, ME) as previously described (Ali et al., 2016; Justilien et al., 2014). To assess the effect of the PKCι inhibitor Auranofin (ANF); Santa Cruz, Cat: sc-202476) and the β-catenin inhibitor methyl 3-((4-methylphenyl)sulfonamido)benzoate (MSAB) (XcessBiosciences, Cat: M60316–2s), the drugs were added to the soft agar cultures in the concentrations specified in the Figure Legends. Plates were incubated at 37 °C in 5% CO2 and colony growth assessed after 2~3 weeks after staining with Giemsa (EMD Millipore, Darmstadt, Germany). Colony size and number were determined using ImagePro Plus version 7 software. For clonal expansion analysis, oncosphere cells were dissociated by trypsinization and cells filtered through a cell strainer. Dissociated cells were plated into 96-well ultra-low attachment tissue culture plates (Corning, Corning, NY) after dilution to achieve delivery of a single cell/100 μl/well of modified cancer stem cell media. Oncospheres were quantified and assessed for size using Image-Pro Plus version 7 software (Media Cybernetics, Bethesda, MD). Standard MTT assays were performed to measure cellular viability according to supplier’s instructions (Promega, Madison, WI) using a Spectra Max M5 (Molecular Devices, Sunnyvale, CA). Differences in soft agar colony growth, clonal expansion and MTT reduction among treatment groups were analyzed for significance by t-test.
Fluorescence-Activated Cell Sorting (FACS) Analysis
BASCs and AT2 cells were isolated from total lung epithelial cells as described previously (Lee et al., 2014; Zacharias et al., 2018). To isolate TM4SF1+ and TM4SF1− AT2 cells, we first isolated Sca-1− total lung epithelial cells using EasySep™ Mouse Sca-1 Biotin Positive Selection Kit (STEMCELL Technologies) according to the manufacturer’s instruction. These cells were then labeled with TM4SF1− Fluor647 and Epcam1-PE and subjected to FACS sorting to isolate TM4SF1+ and TM4SF1− populations essentially as described previously (Zacharias et al., 2018). SYTOX green (Thermo Fisher Scientific) was used to discriminate live and dead cells. FMO controls were used to identify and gate cells.
RNA extraction and qPCR analysis
To determine mRNA expression, cells or tissues were processed using RNAqueous phenol-free total RNA isolation protocols (Ambion, Life Technologies, Grand Island, NY). Extracted RNA was subjected to DNase treatment to eliminate DNA contamination according to provider’s instructions using TURBO DNA-freeTM (Ambion, Life Technologies). DNase-treated RNA was quantified and cDNA was synthesized using reagents (DNTPs, RT-Buffer and RT-Enzyme) from High-Capacity RNA-to-cDNA™ Kit (AB, Life Technologies) in a thermocycler (Biometra, Goettingen, Germany). Gene expression was assessed using TaqMan Fast Universal PCR Master Mix from Applied Biosystems (ABI, Life Technologies). qPCR amplification analysis was performed using Applied Biosystems ViiA7 thermal cycler (Foster City, CA). qPCR reagents were purchased from Applied Biosystems (Foster City, CA) or custom designed from Invitrogen. Relative mRNA expression values were determined using GAPDH as internal control. Changes in RNA abundance detected by QPCR were analyzed for significance by t-test. See Key Resources Table for qPCR Primers used.
Immunofluorescence microscopy
For immunofluorescence analysis, BASCs and AT2 cells were fixed in cold 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10 min on ice, then mixed with histogel (American MasterTech) and embedded in paraffin. KP/KPI tumor tissues were fixed in 10% formalin overnight and embedded in paraffin. Paraffin-embedded tissues/cells were rehydrated, washed twice with PBS and then incubated in 1X- Bovine Serum Albumin (BSA from Sigma-Aldrich) in PBS for 30 min, and then with primary antibodies diluted in PBS containing 0.5% BSA (PBS/BSA) at RT for 2 hr. After incubation, cells were washed 3 times in PBS/BSA for 5 min on ice. Washed cells were incubated with secondary Alexa Flour 488 donkey anti-rabbit and Alexa Flour 568 donkey anti-goat antibodies (Invitrogen) at RT for 1 hr, followed by four washes of 1X-PBS/BSA on ice. Coverslips were mounted on slides using Prolong Gold anti-fade from Invitrogen containing 4, 6-diamidino-2-phenylindole (DAPI). Images were captured at 100X using an oil immersion lens. Cells were counted at indicated magnification.
Ex vivo three dimensional Matrigel Culture
BASCs, total AT2 cells, TM4SF1− and TM4SF1+ AT2 cells were resuspended in Bronchial Epithelial Cell Growth Medium (BEGM; Lonza) without hydrocortisone containing 10 ng/mL keratinocyte growth factor, 5% charcoal-stripped fetal bovine serum, and 50% Matrigel (CORNING) and plated on Matrigel-coated tissue culture wells topped with BEGM. The medium was changed twice weekly. Self-renewal was assessed in oncospheres released from the Matrigel using Cell Recovery Solution (CORNING). Cells were disassociated into single cells by triturating with StemPro Accutase (Thermo Fisher Scientific) and assessed for oncosphere size/number within 2 weeks. In some experiments, total lung epithelial cells isolated from KP and KPI mice were plated as described above and infected with Ad-CMV-Cre or Ad-CMV-Null (Vector Laboratories; MOI 50). Cells were released from Matrigel culture using Cell Recovery Solution (CORNING) and total RNA and DNA were isolated for analysis. Results were analyzed for significance by t-test.
Quantification and Statistical Analysis
RNA-seq and bioinformatics analysis of mouse lung adenocarcinoma transcriptomes
Total RNA was extracted from six independent spheroid cultured KP and KPI LADC cell lines using the RNAeasy Plus mini Kit (Qiagen, Valencia, CA). Quality of RNA samples was determined using an Agilent Bioanalyzer and only samples with RINs >9.0 were used for genomic sequencing analysis. Sequencing was carried out at Mayo Clinic Advanced Genomic Technology Center at Rochester, MN. RPKM-normalized gene counts were generated and used to perform Ingenuity Pathway Analysis (IPA). After stringent filtering (p value <0.01 and log2 value >2 or <−0.6) 4,889 differentially expressed genes were subjected to pathway analysis. The core analysis was performed using default settings: direct and indirect relationships between molecules supported by experimentally observed data were considered, networks did not exceed 35 molecules, and all sources of data from human, mouse, and rat studies in the Ingenuity Knowledge Base were considered. This generated priority lists for canonical pathways. Score values were calculated from hypergeometric distribution and right-tailed Fisher’s exact test. Canonical pathways were further filtered by p value <0.05 and Z-score>1.5 or <−1.5. The results of pathway analysis were verified using another 3 independent KP (KP1~3) and KPI (KPI1~3) LADC cell lines.
Gene Set Enrichment Analysis
The GSEA software is publicly available from the Broad Institute of MIT and Harvard University (http://software.broadinstitute.org/gsea/index.jsp). Axin2+ AT2 cells Wnt gene expression data were downloaded here (Frank et al., 2016). Analysis of Axin2+ AT2 cells and 4889 differentially expressed genes (KPI vs KP) was performed separately. GSEA was performed using the gene permutation option and gene sets smaller than 15 or larger than 500 were excluded.
Eigengene analysis of TCGA LADC and Quantified Cancer Cell Line Encyclopedia data
The R cqn package (Hansen et al., 2012) was applied to normalize raw read counts, which takes into consideration library size of each sample, gene length and GC content of coding region of each gene. The R edgeR package (Robinson et al., 2010) was used to perform group comparisons between KP and KPI tumors, and to identify differentially expressed (DE) genes. Up-regulated genes in KPI tumors were selected such that the false discovery rate (FDR) was less than 0.05 and log2 (fold change) was greater than 0. Down-regulated gene in KPI tumors were selected such that FDR is less than 0.05 and log2 (fold change) is less than 0. Up-regulated genes that are also in Wnt signaling pathway were used to compute the “eigengene” for KP and KPI tumor RNAseq samples. The Eigengene score represents the first principle component (PC1) of the expression matrix of those genes and positively correlates with mean expression of those genes. An Eigengene score was likewise generated for Wnt gene expression of human LADC cell lines using RNA-seq data obtained from the Cancer Cell Line Encyclopedia (CCLE), and for human LADC tumors using RNA-seq data obtained from the Cancer Genome Atlas.
Data and Software Availability
RNA-Seq data has been deposited in GEO under the accession GSE131993.
Supplementary Material
Table S1. Wnt pathway genes from Ingenuity Pathway Analysis differentially expressed in KP versus KPI LADC cells, Related to Figure 3.
Table S2. Wnt Pathway Genes used for Eigengene construction, Related to Figure 6.
Significance.
Lung cancer is the leading cause of cancer death. LADC, the major form of lung cancer, is a heterogeneous disease characterized by high relapse rates and poor prognosis. Here, we characterize two distinct tumorigenic LADC pathways in mice; PKCι-dependent transformation of BASCs and Wnt-dependent transformation of Axin2+ AT2 cells. A subset of human LADC tumors exhibit a genomic Wnt signature resembling that of Wnt-dependent mouse Axin2+ AT2 cell-derived LADCs, and LADC cells with this signature are highly sensitive to Wnt pathway inhibition. Our data provide a compelling rationale for the use of Wnt pathway-targeted therapeutics in treatment of a distinct subtype of human LADC which can be identified using Wnt pathway gene profiling.
Highlights.
Distinct PKCι- and Wnt-dependent Kras/Trp53 LADC tumorigenesis pathways exist
Axin2+ alveolar type II cells serve as the cell of origin for Wnt-dependent LADC
PKCι- and Wnt-dependent Kras/Trp53 LADC exhibit distinct therapeutic vulnerabilities
A distinct molecular sub-type of human LADC exhibits properties of Wnt-dependent LADC
Acknowledgments
We thank Ms. Kayla Lewis and Dr. Lee Jamieson for technical assistance, Ms. Brandy Edenfield and the Mayo Clinic Cancer Biology Histology Facility for processing tumor tissues for analysis, The Mayo Clinic Sequencing Facility for RNA-seq runs, and Dr. Laura J. Tuffin-Lewis for assistance with flow cytometric analysis. We also acknowledge members of the Fields laboratory for critical feedback on the manuscript. This work was supported by grants from the National Institutes of Health/National Cancer Institute (R01 CA081436–21 and R01 CA206267–03 to APF; R01 CA140290–05 to NRM). APF is the Monica Flynn Jacoby Professor of Cancer Research, an endowment fund that provides partial support for the investigator’s research program. VJ is a recipient of a Mayo Clinic Center for Biomedical Discovery Career Development Award. NY and YL are recipients of the Edward C. Kendall Fellowship in Biochemistry from the Mayo Clinic Graduate School.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali SA, Justilien V, Jamieson L, Murray NR, and Fields AP (2016). Protein Kinase Ciota Drives a NOTCH3-dependent Stem-like Phenotype in Mutant KRAS Lung Adenocarcinoma. Cancer Cell 29, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BL (2013). Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2014). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, Lu Y, Kurie JM, Gibbons DL, Mills GB, et al. (2017). Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene 36, 1384–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Fillmore CM, Hammerman PS, Kim CF, and Wong KK (2014). Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Daemen A, Cheng JH, Long JE, Cooper JE, Wang BE, Tran C, Singh M, Gnad F, Modrusan Z, et al. (2017). Kras mutant genetically engineered mouse models of human cancers are genomically heterogeneous. Proc Natl Acad Sci U S A 114, E10947–E10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey JS, Zacharek SJ, Kim CF, and Lees JA (2008). Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc Natl Acad Sci U S A 105, 11857–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasbender A, Lee JH, Walters RW, Moninger TO, Zabner J, and Welsh MJ (1998). Incorporation of adenovirus in calcium phosphate precipitates enhances gene transfer to airway epithelia in vitro and in vivo. J Clin Invest 102, 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Ali SA, Justilien V, and Murray NR (2017). Targeting oncogenic protein kinase Ciota for treatment of mutant KRAS LADC. Small GTPases 8, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Justilien V, and Murray NR (2016). The chromosome 3q26 OncCassette: A multigenic driver of human cancer. Adv Biol Regul 60, 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DB, Peng T, Zepp JA, Snitow M, Vincent TL, Penkala IJ, Cui Z, Herriges MJ, Morley MP, Zhou S, et al. (2016). Emergence of a Wave of Wnt Signaling that Regulates Lung Alveologenesis by Controlling Epithelial Self-Renewal and Differentiation. Cell Rep 17, 2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick LA, Matthews JA, Jamieson L, Justilien V, Thompson EA, Radisky DC, and Fields AP (2008). Matrix metalloproteinase-10 is a critical effector of protein kinase Ciota-Par6alpha-mediated lung cancer. Oncogene 27, 4841–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, and Wu Z (2012). Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka Y, Hashizume K, Nitta K, Kato T, Itoh I, and Tani Y (2003). Cytometrical image analysis for immunohistochemical hormone receptor status in breast carcinomas. Pathol Int 53, 693–699. [DOI] [PubMed] [Google Scholar]
- Hwang SY, Deng X, Byun S, Lee C, Lee SJ, Suh H, Zhang J, Kang Q, Zhang T, Westover KD, et al. (2016). Direct Targeting of beta-Catenin by a Small Molecule Stimulates Proteasomal Degradation and Suppresses Oncogenic Wnt/beta-Catenin Signaling. Cell Rep 16, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, and Jacks T (2005). The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 65, 10280–10288. [DOI] [PubMed] [Google Scholar]
- Justilien V, Ali SA, Jamieson L, Yin N, Cox AD, Der CJ, Murray NR, and Fields AP (2017). Ect2-Dependent rRNA Synthesis Is Required for KRAS-TRP53-Driven Lung Adenocarcinoma. Cancer Cell 31, 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, and Fields AP (2014). The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 25, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, and Jacks T (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835. [DOI] [PubMed] [Google Scholar]
- Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, Wagers AJ, Tseng YH, Ryeom S, and Kim CF (2014). Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu K, Cui G, Huang X, Yao S, Guo W, Qin Z, Li Y, Yang R, Pu W, et al. (2019). Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet 51, 728–738. [DOI] [PubMed] [Google Scholar]
- Mainardi S, Mijimolle N, Francoz S, Vicente-Duenas C, Sanchez-Garcia I, and Barbacid M (2014). Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci U S A 111, 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell AM, Yao C, Yeckes AR, Wang Y, Selvaggio AS, Tang J, Kirsch DG, and Stripp BR (2016). p53 Regulates Progenitor Cell Quiescence and Differentiation in the Airway. Cell Rep 17, 2173–2182. [DOI] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, and Desai TJ (2018). Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker PJ, Justilien V, Riou P, Linch M, and Fields AP (2014). Atypical Protein Kinase Ciota as a human oncogene and therapeutic target. Biochem Pharmacol 88, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regala RP, Davis RK, Kunz A, Khoor A, Leitges M, and Fields AP (2009). Atypical protein kinase C{iota} is required for bronchioalveolar stem cell expansion and lung tumorigenesis. Cancer Res 69, 7603–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Copland JA, Thompson EA, and Fields AP (2005a). Atypical protein kinase Ciota plays a critical role in human lung cancer cell growth and tumorigenicity. J Biol Chem 280, 31109–31115. [DOI] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, and Fields AP (2005b). Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res 65, 8905–8911. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, and Jemal A (2018). Cancer statistics, 2018. CA Cancer J Clin 68, 7–30. [DOI] [PubMed] [Google Scholar]
- Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, and Berns A (2014). Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A 111, 4952–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiozzo C, De Langhe S, Yu M, Londhe VA, Carraro G, Li M, Li C, Xing Y, Anderson S, Borok Z, et al. (2009). Deletion of Pten expands lung epithelial progenitor pools and confers resistance to airway injury. Am J Respir Crit Care Med 180, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura JJ, Tenbaum S, Perdiguero E, Huth M, Guerra C, Barbacid M, Pasparakis M, and Nebreda AR (2007). p38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet 39, 750–758. [DOI] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. (2011). Suppression of lung adenocarcinoma progression by Nkx2–1. Nature 473, 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Rock JR, Lu Y, Futtner C, Schwab B, Guinney J, Hogan BL, and Onaitis MW (2012). Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci U S A 109, 4910–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Iwanaga K, Raso MG, Wislez M, Hanna AE, Wieder ED, Molldrem JJ, Wistuba II, Powis G, Demayo FJ, et al. (2008). Phosphatidylinositol 3-kinase mediates bronchioalveolar stem cell expansion in mouse models of oncogenic K-ras-induced lung cancer. PLoS One 3, e2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappa C, and Mousa SA (2016). Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 5, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Wnt pathway genes from Ingenuity Pathway Analysis differentially expressed in KP versus KPI LADC cells, Related to Figure 3.
Table S2. Wnt Pathway Genes used for Eigengene construction, Related to Figure 6.
Data Availability Statement
RNA-Seq data has been deposited in GEO under the accession GSE131993.