Abstract
Despite the propensity for gastric and esophageal adenocarcinomas to select for recurrent missense mutations in TP53, the precise functional consequence of these mutations remains unclear. Here we report that endogenous mRNA and protein levels of mutant p53 were elevated in cell lines and patients with gastric and esophageal cancer. Functional studies showed that mutant p53 was sufficient, but not necessary, for enhancing primary tumor growth in vivo. Unbiased genome-wide transcriptome analysis revealed that hypoxia signaling was induced by mutant p53 in 2 gastric cancer cell lines. Using real-time in vivo imaging, we confirmed that hypoxia reporter activity was elevated during the initiation of mutant p53 gastric cancer xenografts. Further investigation revealed that, like mutant p53, the HIF1/ARNT hypoxia pathway was not required for the primary tumor functions of advanced mutant p53 gastric cancer. These findings indicate that recurrent p53 mutations in gastroesophageal adenocarcinoma are unlikely to serve as effective therapeutic targets in advanced cancer. However, in elucidating the contribution of missense mutant p53 and hypoxia signaling, the results suggest hypotheses regarding how these recurrent genomic events may contribute to gastric and esophageal adenocarcinoma formation.
Keywords: Oncology
Keywords: Gastric cancer, P53, hypoxia
Recurrent missense mutations in TP53 lead to induction of hypoxia signaling in gastric and esophageal adenocarcinoma in vivo.
Introduction
Gastric and esophageal adenocarcinoma (GEA) is one of the main causes of cancer-related mortality worldwide (1). Even with several treatment regimens available, the overall survival of patients with GEA remains dismal. Recent genomic studies revealed key molecular features in common among GEAs. Within these analyses, esophageal adenocarcinomas, which are typically found in the distal esophagus immediately adjacent to the stomach, were found to have a striking resemblance to the chromosomal instability (CIN) class of gastric adenocarcinoma, suggesting that these cancers could largely be considered a single disease entity (2). This group, CIN-GEAs, are the most common form of both gastric and esophageal adenocarcinomas and are especially prevalent in the proximal stomach/distal esophagus, where rates of disease have risen dramatically in the Western population in recent decades.
Genomic analyses of both stomach and esophageal adenocarcinomas have found TP53 to be the most frequently mutated gene, present in 46% and 77% of total tumors, respectively (3, 4). Within the CIN populations, TP53 mutations are found in 71% and greater than 90%. Furthermore, new data suggest that TP53 mutations often emerge in the early process of gastric (5) and esophageal carcinogenesis (6) — a contrast to sporadic colorectal cancer (7) or pancreatic cancer (8), where TP53 mutations have been described as developing later in the neoplastic process (i.e., following oncogene activation). Given the propensity for structural chromosomal aberrations in the development of GEA (especially with CIN), the strong propensity for loss of p53’s tumor suppressor function is an expected finding. However, as with many other cancer types, deeper review of the patterns of TP53 alteration in these cancers have found highly recurrent specific missense mutations. These recurrent missense mutations of TP53 have been suggested to possess 2 different properties: dominant-negative and gain-of-function (9). It has long been established that recurrent “hotspot” missense TP53 mutations such as those at codons 175, 248, and 273 possess a dominant-negative capacity by attenuating the transcriptional activity of WT p53 protein through aberrant tetramer complexes (9, 10). More recently, several reports have suggested that these mutants are also imbued with gain-of-function activity, which is an alternative but not mutually exclusive explanation underlying their recurrence in human cancer (11). However, the effect of these recurrent missense TP53 mutations in GEA remains uncertain. Furthermore, a critical question relates to the potential dependence of established GEAs on mutant TP53 activity and whether targeting any gain-of-function behavior may be a therapeutic vulnerability in these cancers.
We therefore investigated the function of hotspot missense mutations of TP53 in GEA using both in vivo and in vitro models. These studies demonstrated that genetic targeting of mutant TP53 does not impact cell line or tumor growth, arguing strongly that these mutations do not present an immediate therapeutic liability. However, we found that these mutations can promote tumor growth. Our studies identified a connection between mutant p53 and hypoxia signaling. Deregulation of the hypoxia pathway plays an important role in tumor progression (12, 13). In GEA, several mechanisms of hypoxia/HIF1α-related invasion and metastasis have been reported (14–16). However, the relationship between hypoxia signaling and TP53 in GEA is not well elucidated. This study establishes a functional connection between TP53 mutation and hypoxia signaling during primary tumor growth of GEA and demonstrates that while missense mutations of TP53 promote tumor progression, they are not essential for tumor formation.
Results
Frequent missense mutations in TP53 lead to elevated transcript and protein expression in GEA.
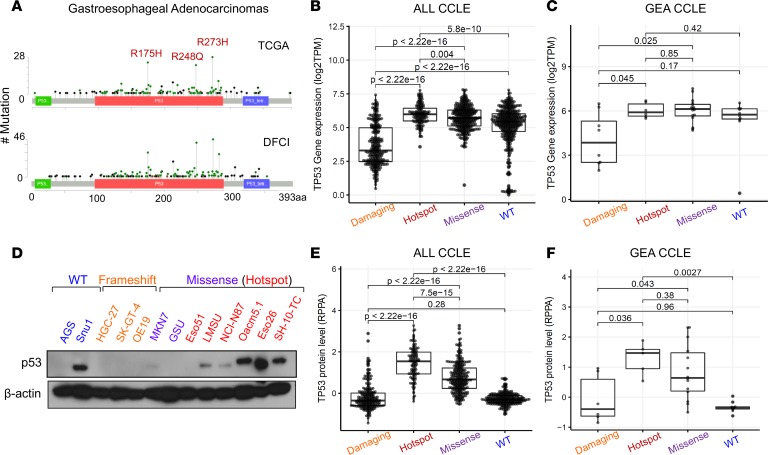
To investigate the role of TP53 mutations in GEA, we started with focused analysis of the patterns of mutations in data sets derived from patients seen at the DFCI and in the Cancer Genome Atlas (TCGA) study (4). Consistent with studies in other solid tumors (17, 18), we found that 3 specific codons within the DNA-binding domain of p53 harbored a disproportionately higher number of missense mutations, leading to the following amino acid changes: R175H, R248Q/W, and R273C/H (Figure 1A). Missense mutations, and particularly the hotspot subset, are associated with higher transcript levels of TP53 in the comprehensive catalog of cell lines included in the Cancer Cell Line Encyclopedia (CCLE) (Figure 1, B and C, and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128439DS1). Many GEA cell lines with hotspot TP53 mutations also showed higher protein expression levels of p53, most of which indicated mutant protein expression given loss of heterozygosity (LOH) in these models (Figure 1, D–F). Furthermore, analysis of reverse-phase protein array (RPPA) data from patients in the TCGA cohort allowed us to correlate p53 protein levels with genotype in primary patient samples and showed a consistent increase in protein expression with codon 175, 248, or 273 mutant p53 in patients with GEA (Supplemental Figure 1B and ref. 4). The elevated protein levels of mutant p53 are consistent with prior mechanistic models whereby these mutants are weak inducers of p53’s negative regulator, E3 ubiquitin ligase MDM2, leading to enhanced stability of mutant p53 protein (19). There is, however, evidence that mutant p53 can stabilize MDM2 expression in cancer (20), opening the possibility for other explanations for mutant p53 overexpression in cancer. It is conceivable that elevated protein expression of mutant p53 could also reflect potential pro-oncogenic activity, as previously reported in several tumors (21–28). The observation that the majority of patients with TP53 mutant Barrett’s metaplasia, a precursor to esophageal adenocarcinoma, and GEA selectively lose the remaining WT TP53 copy through LOH also raises the potential of a gain-of-function activity in addition to an established dominant-negative property (Supplemental Figure 1C). Together, these data indicate that TP53 is recurrently mutated at specific loci, highly expressed at both the transcript and proteins levels, and associated with LOH in GEA.
Figure 1. p53 is frequently mutated and overexpressed in human gastroesophageal cancer.
(A) Mutation plots of p53 derived from patients with gastroesophageal cancers collected through TCGA (top) and the DFCI (bottom). Box plot showing mRNA expression of TP53 across (B) all and (C) gastroesophageal cell lines found in the CCLE annotated by type of p53 mutation (damaging: nonsense and frameshift, n = 274 and n = 8; hotspot: R175H, R248Q/W, and R273C/H, n = 133 and n = 6; missense: all other missense excluding hotspot mutations, n = 331 and n = 16; WT: n = 394 and n = 10). P values are from pairwise Wilcoxon’s rank-sum tests. (D) Immunoblot analysis showing protein expression levels of p53 across various gastroesophageal cells lines annotated by p53 mutation status. (E and F) Box plot showing protein expression of p53 (RPPA data) across (E) all and (F) gastroesophageal cell lines found in CCLE annotated by p53 mutation status (damaging: n = 231 and n = 6; hotspot, n = 114 and 5; missense, n = 278 and n = 14; WT, n = 263 and n = 10. Lines within boxes represent median, the bounds of the boxes represent the 25th and 75th percentiles, and the whiskers extend to the lowest/largest values within 1.5 IQR from the lower and upper quartiles, respectively. P values were calculated by Wilcoxon’s rank-sum test.
Mutant p53 is not required for GEA tumor proliferation and primary tumorigenesis.
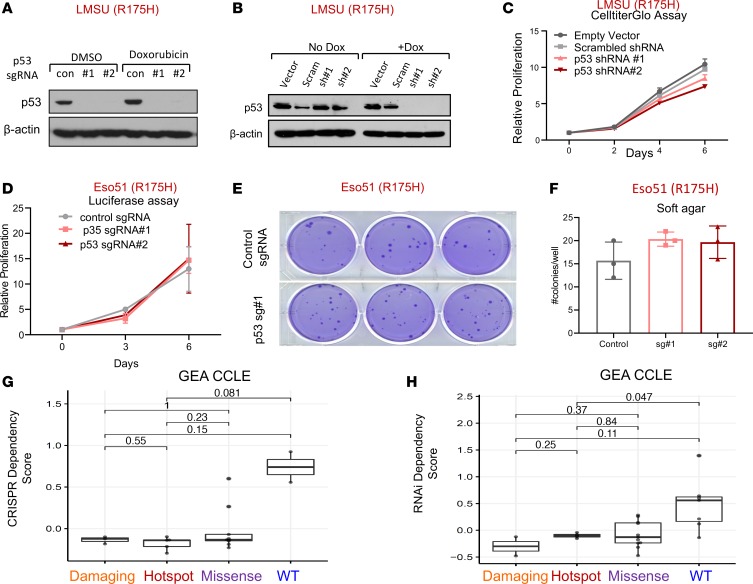
We next tested the necessity for hotspot mutant p53 in established GEA cellular models given the reports supporting its requirement in other cancer types (29, 30). Cognizant that mutations in TP53 occur early in the progression of precancerous lesions to GEA (6), we asked whether advanced GEA cells harboring endogenous mutations in TP53 still required its activity to determine whether mutant p53 may represent a therapeutic liability. To this end, we used multiple targeting short guide RNAs (sgRNAs) in conjunction with stable Cas9 expression for CRISPR-mediated gene KO and 2 distinct shRNAs to knock down (KD) mutant TP53 in several GEA cancer cell lines with TP53 hotspot mutations (Figure 2, A and B, and Supplemental Figure 2, A–C). Constitutive or inducible KD of mutant TP53 did not significantly impact adherent or ultra-low attachment proliferation of the LMSU (endogenous R175H) gastric cancer cell line by CellTiter-Glo (Figure 2C and Supplemental Figure 2, D and E). Disruption of mutant TP53 in 2 esophageal adenocarcinoma cell lines with distinct hotspot TP53 mutations confirmed these results. Mutant TP53-KO in luciferase-labeled Eso51 (endogenous R175H) and inducible KD of mutant p53 in SH10TC (endogenous R273C) cell lines did not affect nonadherent or adherent proliferation capacity (Figure 2D and Supplemental Figure 2F). We next tested the requirement for mutant p53 in soft agar colony growth assays, examining its importance in anchorage-independent growth. Consistent with the previous results, mutant p53 was expendable for soft agar growth of Eso51 and LMSU GEA cell lines (Figure 2, E and F, and Supplemental Figure 2G). As SH10TC cells did not form robust soft agar colonies, we could not evaluate the effects of p53 manipulation in these cells.
Figure 2. Mutant p53 is not required for primary tumor functions of gastroesophageal cancer cells.
(A) Immunoblot showing protein expression levels of p53 in the LMSU gastric adenocarcinoma cell line, which harbors an endogenous R175H p53 mutation, stably expressing a Cas9 control (con) vector or 2 distinct targeting sgRNAs in addition to Cas9. (B) Immunoblot showing protein expression levels of p53 in LMSU gastric cancer cells expressing an inducible vector control, scrambled (Scram) control, or 2 targeting shRNAs with or without doxycycline (Dox). (C) Proliferation of LMSU gastric adenocarcinoma cells expressing a constitutively active vector control, scrambled control, or 2 p53-targeting shRNAs using CellTiter-Glo. (D) Proliferation of Eso51 nonadherent esophageal cancer cells (R175H) stably expressing firefly luciferase in addition to a constitutively active Cas9 control vector or 2 p53-targeting sgRNAs using a luciferase assay. (E) Soft agar colony formation assay of Eso51 esophageal adenocarcinoma cells (R175H) stably expressing a constitutively active Cas9 control vector or 1 p53-targeting sgRNA. (F) Quantification of soft agar colony formation assay shown in E. (G and H) TP53 dependency scores for GEA cell lines derived from the CCLE, with damaging (nonsense/frameshift) mutations, hotspot (R175H, R248Q/W, R273C/H) mutations, or missense mutations or WTTP53 using (G) CRISPR and (H) RNAi dependency data. Lines within boxes represent median, the bounds of the boxes represent the 25th and 75th percentiles, and the whiskers extend to the lowest/largest values within 1.5 IQR from the lower and upper quartiles, respectively. P values were calculated by Wilcoxon’s rank-sum test
To more broadly evaluate the relationship between TP53 mutation and genetic dependence across solid tumor cancer cell lines, we evaluated data from 2 comprehensive data sets in which genome-scale CRISPR or RNAi libraries were introduced to hundreds of cancer cell lines to estimate gene dependencies across tumor models. As anticipated, deletion or attenuation of WT TP53 provided a modest selective growth advantage for a subset of cancer cell lines (Figure 2, G and H, and Supplemental Figure 3, A and B). We experimentally confirmed these findings in at least one gastric cancer cell line that is WT for p53. Although undetectable by immunoblot analysis (Supplemental Figure 3C), deletion of WT p53 in AGS gastric cancer cells provided a slight proliferation advantage during adherent culture and soft agar growth (Supplemental Figure 3, D and E). By contrast, loss of hotspot, missense, or damaging (defined as frameshift or nonsense mutations) mutant p53 did not impact proliferation or survival of GEA cancer cell lines (Figure 2, G and H, and ref. 31). Interestingly, there was a modest yet statistically significant detrimental effect of deleting hotspot mutant p53 by CRISPR when all CCLE cell lines were pooled (Supplemental Figure 3A). Collectively, these data provide evidence that mutant p53 is not required for GEA cells during adherent or nonadherent in vitro growth.
Enforced expression of mutant p53 promotes GEA primary tumorigenesis.
We next evaluated whether forced expression of recurrent missense mutant p53 can promote proliferation of GEA cancer cells. To test the sufficiency of mutant p53, we generated cDNA constructs overexpressing R175H, R248Q, and R273H mutant p53. Next, using site-directed mutagenesis, a synonymous mutation was introduced into each plasmid at the protospacer adjacent motif (PAM) site for p53 sgRNA#1, the precise location where sgRNA recruits Cas9 for genome editing (Supplemental Figure 4A). As a result, mutant p53 containing the silent point mutation (e.g., R175H′) would be insensitive to p53 sgRNA#1-mediated DNA editing, whereas endogenous mutant (and WT) p53 would remain susceptible to CRISPR-mediated inactivation (Supplemental Figure 4B). Using this system, we could reexpress mutant p53 in a GEA cell line that lost endogenous mutant TP53 via Cas9/p53 sgRNA#1 activity. (We attempted constitutive overexpression of WT p53, but this perturbation was not tolerated in any of the GEA cell lines tested). To demonstrate the specificity of this approach for manipulating TP53 expression, we showed that ectopically expressed versions of p53-R175H with and without the PAM site mutation were unaffected by a nontargeting control sgRNA. By contrast, only the p53-R175H′ construct containing the silent mutation was expressed in GEA cells with TP53 sgRNA#1. Both versions were effectively deleted in GEA cells expressing p53 sgRNA#2, which targets a distinct, unaltered PAM site (Supplemental Figure 4B).
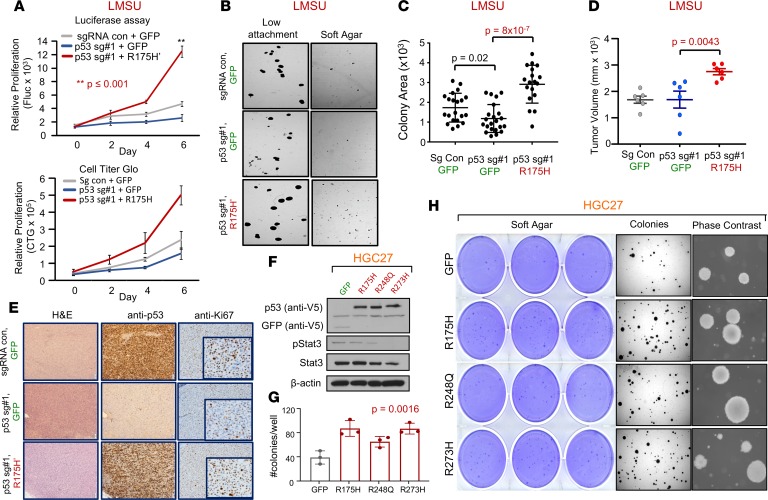
Using this robust system, we generated 3 genetically modified isogenic versions of the LMSU (endogenous R175H) cell line: a double control that expressed a nontargeting sgRNA/Cas9 and GFP (sgRNA con + GFP); a mutant p53-KO cell line that expressed p53 sgRNA#1 and GFP (p53 sg#1 + GFP); and a mutant p53 cell line that expressed p53 sgRNA#1 and a cDNA construct overexpressing p53-R175H′ (p53 sg#1 + R175H′). The results showed that, consistent with our previous data, deletion of endogenous mutant p53-R175H did not significantly alter adherent proliferation of luciferase-labeled LMSU gastric cancer cells (Figure 3A and Supplemental Figure 4C). However, enforced expression of p53-R175H′ in LMSU cells that lost endogenous mutant p53 via sgRNA#1 promoted proliferation by luciferase and CellTiter-Glo assays (Figure 3A). A similar pattern was observed when these genetically modified cells were cultured in ultra-low attachment and soft agar conditions (Figure 3, B and C).
Figure 3. Enforced expression of mutant p53 in 2 gastric adenocarcinoma cell lines confers a growth advantage.
(A) Proliferation of control (sgRNA con + GFP), p53-KO (p53 sg#1 + GFP), and p53 KO-R175H′ rescue (p53 sg#1 + R175H′) LMSU cells using a firefly luciferase assay (Fluc; top panel) and CellTiter-Glo (CTG; bottom panel). **P ≤ 0.001. (B) Low-attachment and soft agar colony formation assays of control, p53-KO, and p53 KO-R175H′ rescue LMSU cells. (C) Quantification of low-attachment colony formation assay described in B. (D) Primary tumor growth of flank xenografts of control, p53 KO, and p53 KO-R175H′ rescue LMSU cells at 6 weeks. P values were calculated by Student’s t test. (E) p53 IHC of xenografts described in D at the endpoint. (F) Immunoblot showing expression of GFP control or indicated mutant p53 in the genetically engineered HGC27 gastric cancer line (p53 null). (G) Quantification of soft agar colonies of HGC27 gastric cancer cell line expressing GFP control or the indicated p53 mutant. P value was calculated by 1-way ANOVA. (H) Crystal violet, dissection microscope, and phase-contrast images of soft agar colonies of the HGC27 gastric cancer cell line expressing GFP control or the indicated p53 mutant. All data are mean ± SD.
We next evaluated whether enforced expression of mutant p53 can promote primary tumor growth in vivo. Xenograft experiments in nude mice showed that although endogenous mutant p53-R175H was nonessential for the growth of LMSU gastric cancer cells in vivo, overexpression of mutant p53-R175H promoted primary tumor growth (Figure 3D). We ensured that transcript and protein levels of mutant p53 as determined by real-time PCR (RT-PCR) and IHC were maintained throughout the experiment as assessed at the endpoint (Figure 3E and Supplemental Figure 4D). It is noteworthy that endogenous p53-R175H was highly expressed in LMSU cells while grown in xenograft primary tumors compared with adherent culture (Figure 3E). Given these promising findings, we next wondered whether enforced expression of hotspot p53 mutants can promote primary tumor properties in HGC27, a gastric cancer cell line with an endogenous frameshift mutation in TP53 and therefore presumably defective for WT p53 activity. Unlike in LMSU cells, overexpression of hotspot p53 mutants (Figure 3F) did not promote proliferation of HGC27 cells during adherent culture. By contrast, enforced expression of p53-R175H and p53-R273H promoted ultra-low attachment (Supplemental Figure 4, E and F) and soft agar colony growth (Figure 3, G and H) to a greater extent than p53-R248Q in HGC27 cells. These data indicate that although endogenous mutant p53 is not required, overexpression can enhance primary tumor properties in GEA.
Mutant p53 expression induces a hypoxia transcriptional program in GEA.
To define genes and/or pathways that are responsible for mutant p53-mediated primary tumor growth in gastric cancer, we pursued whole genome transcriptomic analysis of TP53-manipulated GEA cell lines. We performed parallel analyses of genetic silencing of endogenous mutant TP53-R175H in the LMSU model and ectopic expression of distinct hotspot TP53 mutations in HGC27 cells. In the LMSU model, we performed RNA sequencing following TP53 KD and KO (2 shRNAs and 1 sgRNA, respectively) compared with their corresponding controls to define the transcriptional program mediated by endogenous mutant p53-R175H (Supplemental Figure 5, A and B). Gene set enrichment analysis (GSEA) of mRNA sequencing data revealed specific pathways predicted to be regulated by endogenous mutant p53-R175H (Supplemental Figure 5C). Consistent with evidence showing regulation of the mevalonate pathway by mutant p53 in breast cancer (32), genes associated with the cholesterol homeostasis pathway were upregulated in p53-R175H–expressing LMSU gastric cancer cells. Furthermore, we found enrichment of a misfolded protein response, which is consistent with the cellular presence of mutant p53 (33), and recent evidence showing its ability to regulate the proteasome pathway (28). Interestingly, the p53 pathway is also modestly upregulated in p53-R175H–expressing LMSU cells, suggesting that the mutant allele may retain partial WT p53 activity;. We also found significant enrichment of the hypoxia gene expression signature in LMSU gastric cancer cells retaining endogenous p53-R175H (normalized enrichment score [NES] = 1.84, FDR P = 0.007; Supplemental Figure 5C). Other results included upregulation of EMT genes, consistent with evidence supporting a role for mutant p53 in the mesenchymal phenotype and metastasis (21, 24, 27, 34); and upregulation of mTOR pathway signatures, a result not validated by our analysis of phosphorylated AKT in xenografted p53-R175H LMSU tumors.
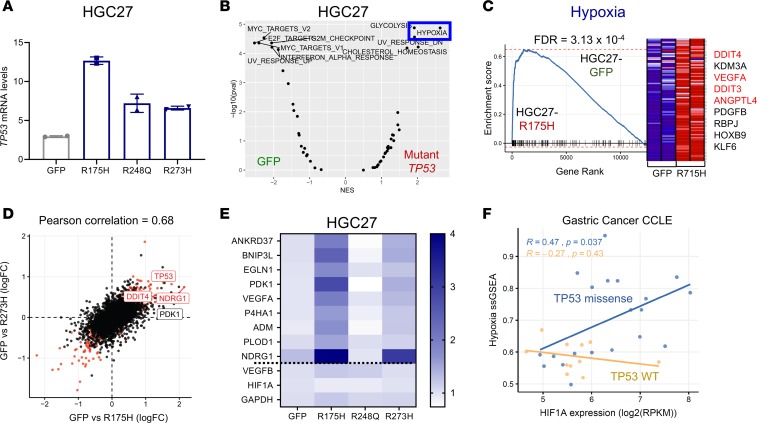
To further refine candidate genes and/or pathways regulated by mutant p53 in gastric cancer, we next performed mRNA sequencing of HGC27 gastric cells overexpressing mutant p53 (Figure 3F and Figure 4A). Comparing all 3 hotspot mutant p53–overexpressing cell lines with GFP controls, we found 3513 significantly differentially expressed genes (q < 0.2). GSEA analysis demonstrated a more robust enrichment of the hypoxia gene set in HGC27 gastric cancer cells overexpressing mutant p53 compared with GFP controls (Figure 4B; FDR = 0.0043). Indeed, multiple gene sets related to hypoxia transcriptional expression signatures and a HIF1α promoter–binding score were all enriched in HGC27 cells expressing mutant p53 (Supplemental Figure 5D). By isolating transcriptomic data from each mutant p53 allele, we found the hypoxia signature to be most strongly driven by overexpression of p53-R175H, where it was the single most significantly enriched gene set (Figure 4C; FDR = 3.13 × 10–4). p53-R273H– expressing HGC27 cells also demonstrated enrichment of the hypoxia gene expression signature, albeit to a lesser extent than those expressing p53-R175H (Supplemental Figure 5E; FDR = 1.5 × 10–2). Ectopic expression of R248Q in HGC27 did not show an enriched hypoxia signature and was an outlier, with markedly fewer differentially expressed genes relative to the ectopic GFP control. These data followed the phenotypic pattern observed during ultra-low attachment and soft agar growth of these isogenic gastric cancer cells, suggesting that these transcription associations likely contributed to the growth phenotype. There was also a strong correlation between differentially expressed genes in HGC27-R175H and HGC27-R273H compared with HGC27-GFP control cells (Pearson’s correlation, 0.68; Figure 4D and Supplemental Figure 5F). In addition, many specific hypoxia genes were co-upregulated in HGC27-R175H and HGC27-R273H cells (Figure 4D), including a 9-gene hypoxia signature (ref. 35 and Figure 4E). To show that this relationship was generalizable to gastric cancer broadly, beyond the subset represented by these 2 cell lines, we demonstrated an association among the Hallmark hypoxia gene set, HIF1α mRNA levels, and gastric cancer cell lines harboring missense mutations in TP53 (Figure 4F; P = 0.037). We also evaluated mRNA expression levels in human gastric cancer primary tumors, finding upregulation of hypoxia-associated genes in patients with TP53-altered compared with those with WT TP53 GEA (Supplemental Figure 6). These results suggest that missense mutations in p53, especially at codons 175 and 273, can regulate and induce a hypoxia transcriptional program in GEA.
Figure 4. Mutant p53 induces a hypoxia transcriptional program in gastric cancer cells.
(A) mRNA expression of TP53 in the HGC27 cell line expressing the indicated mutant p53. Data are presented as mean ± SD. (B) Volcano plot showing gene set enrichment analyses (GSEA) for pathways enriched in GFP control or combined mutant p53–overexpressing HGC27 cells (using the Hallmark gene set collection). (C) GSEA plot of Hallmark hypoxia gene set enrichment in R175H-expressing HGC27 cells versus GFP controls. Red labels indicate select hypoxia genes. (D) Scatter plot comparing differentially expressed genes in HGC27-R175H and HGC27-R273H compared with HGC27-GFP control (Pearson’s correlation = 0.68). FC, fold change. (E) Heatmap showing 9-gene hypoxia signature across HGC27 cells expressing either GFP control or the indicated mutant p53. (F) Scatter plot showing correlation among Hallmark hypoxia single-sample gene set enrichment (ssGSEA), HIF1α mRNA expression levels, and TP53 mutation status in gastric cancer cell lines. P value was calculated by Pearson’s correlation.
Mutant p53 activates the hypoxia pathway in GEA cells.
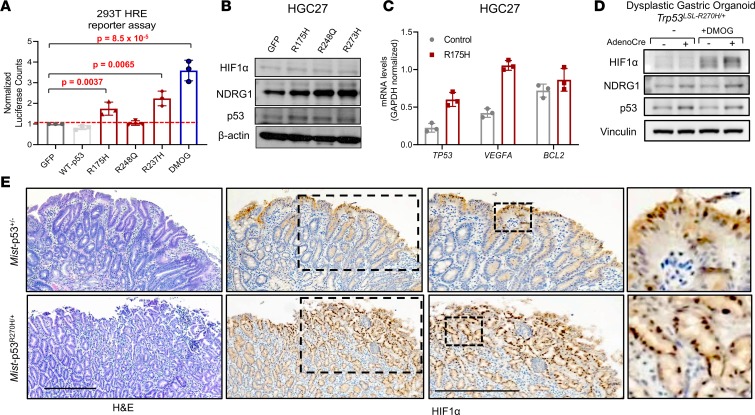
We next performed additional validation of the ability of mutant p53 to induce hypoxia signaling in GEA cells. Utilizing a luciferase reporter driven by hypoxia response elements (HREs), we first confirmed that hypoxia-inducing agents can reliably activate the reporter in vitro. Of the agents, dimethyloxalyl glycine (DMOG) was a potent inducer of HRE luciferase reporter activity in HGC27 cells, consistent with its ability to stabilize HIF1α through competitive inhibition of HIF1α prolyl hydroxylase (Supplemental Figure 7A), whereas desferrioxamine (DFO) had a more modest impact. We next cotransfected the HRE luciferase reporter with WT or mutant p53–expressing plasmids into HEK293T cells. The HRE luciferase reporter was activated in HEK293T cells overexpressing p53-R175H and p53-R273H, but not WT p53, albeit to a lesser degree than positive control DMOG-treated cells (Figure 5A). We confirmed that expression of WT p53 indeed activated the p53 pathway using a luciferase reporter driven by p53-binding elements (Supplemental Figure 7B). Consistent with previous results (Supplemental Figure 5C), p53-R175H induced a 2-fold induction in p53 reporter activity, but this was a far smaller increase than with WT p53, again suggesting hypomorphic WT p53 activity (Supplemental Figure 7B). We also confirmed that expression of mutant p53 could activate the hypoxia reporter in HGC27 cells (Supplemental Figure 7C). Moreover, expression of mutant p53 in gastric cancer cells induced or regulated downstream targets of hypoxia signaling, including protein levels of NDRG1 by immunoblot (Figure 5B and Supplemental Figure 7D) and mRNA expression levels of VEGFA by RT-PCR (Figure 5C and Supplemental Figure 7E); protein HIF1α was undetectable under endogenous conditions (Supplemental Figure 7D).
Figure 5. Mutant p53 activates downstream mediators of hypoxia in gastric dysplasia and cancer.
(A) HIF-responsive promoter (HRE) firefly luciferase reporter activity in HEK293T cells transiently transfected with the indicated mutant p53 constructs; 1 mM DMOG treatment was used as a positive control. Data were normalized to cotransfected constitutively active Renilla luciferase activity. (B) Immunoblot showing protein expression levels of NDRG1, HIF1α, and p53 in HGC27 cells expressing the indicated mutant p53. (C) Gene expression levels of TP53, VEGFA, and BCL2 in control or p53-R175H–overexpression HGC27 cells. Results are shown as mean ± SD. P values were calculated by Student’s t test (D) Protein expression of HIF1α and p53 in conditional Trp53LSL-R270H/+ dysplastic gastric organoids derived from MNU-exposed mice with and without AdenoCre virus in the presence or absence of 1 mM DMOG by immunoblot analysis. (E) Histopathology of dysplastic gastric lesions in Mist-p53+/– and Mist-p53R270H/+ mice after 1 year of DCA/MNU treatment. H&E staining (left panel); and HIF1α IHC (right panels). Scale bars: 250 μm.
To ensure that induction of hypoxia signaling by mutant p53 is relevant to other model systems, we utilized isogenic gastric organoids derived from genetically engineered mice with conditional expression of the Trp53R270H mutant allele upon Cre recombination. Endogenous expression of Trp53R270H in nondysplastic gastric organoids by ex vivo AdenoCre virus treatment increased protein levels of HIF1α in the presence and absence of DFO and DMOG (Supplemental Figure 7F). Moreover, conditional activation of endogenous Trp53R270H in dysplastic gastric organoids derived from a carcinogen-induced mouse model by ex vivo AdenoCre increased protein levels of HIF1α in the presence of DMOG (Figure 5D). Furthermore, to investigate whether this regulation occurs in vivo during the pathogenesis of gastric premalignancy, we analyzed HIF1α expression in dysplastic premalignant lesions from an integrative genetically engineered mouse model. To conditionally express Trp53R270H in gastric tissue in vivo, we utilized the Mist1-Cre driver, which is expressed in chief cells and select progenitor cells. Tamoxifen-induced Mist1-Trp53R270H and uninduced, control Mist1-Trp53+/– mice were exposed to drinking water that contained a disease-relevant dietary carcinogen (N-methyl-N-nitrosourea [MNU]) and inflammatory agent (the bile acid deoxycholic acid [DCA]) to develop premalignant gastric lesions. Dysplastic lesions from Mist1-Trp53R270H mice demonstrated greater nuclear HIF1α staining compared with Mist1-Trp53+/– mice (Figure 5E). Of importance, these results indicate that mutant p53 can activate hypoxia signaling as early as during gastric premalignancy.
To corroborate these findings, we examined the primary tumors at the endpoint of our LMSU xenograft experiment for expression levels of HIF1α by IHC. Primary tumors from LMSU control (R175H-expressing) and LMSU-mutant p53-KO mice showed no difference in HIF1α protein levels by IHC. However, p53-R175H–overexpressing LMSU primary tumors showed higher levels of HIF1α by IHC (Supplemental Figure 8; ANOVA P = 0.1). These results were, of course, confounded by the larger tumor size of p53-R175H–overexpressing LMSU primary tumors (Figure 3D). Angiogenesis as assessed by CD34 staining was not significantly different among the 3 groups (Supplemental Figure 8).
Mutant p53 activates hypoxia in vivo using real-time bioluminescent imaging in a xenograft model.
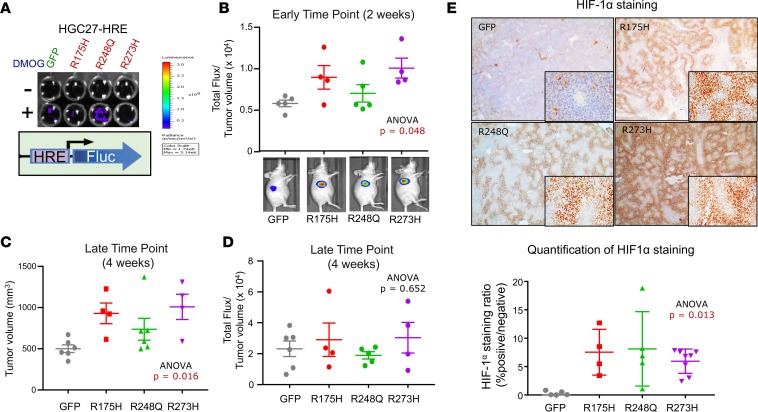
To address whether mutant p53 can induce hypoxia in vivo without interference of tumor size, we engineered HGC27 gastric cancer cells to stably express the HRE–firefly luciferase reporter (HGC27-HRE), followed by stable expression of GFP control, p53-R175H, p53-R248Q, or p53-R273H (Figure 6A). These cells were also designed to constitutively express Renilla luciferase as a surrogate measure of tumor size by bioluminescence; however, expression levels were too low for in vivo detection. We therefore relied on manual caliper measurements to control for tumor size. We observed that the ratio of hypoxia reporter activity to tumor size was higher in mutant p53–expressing HGC27-HRE cells at early time points (~2 weeks) when tumor volumes were not significantly different (Figure 6B; ANOVA P = 0.048). Enforced expression of mutant p53-R175H and p53-R273H compared with p53-R248Q in HGC27-HRE cells led to a greater increase in hypoxia reporter activity relative to GFP (Figure 6B), consistent with our previous results. Furthermore, overexpression of mutant p53 in HGC27-HRE cells promoted primary tumor growth over time (Figure 6C; ANOVA P = 0.016), corroborating our findings in LMSU cells (Figure 3, D and E). Once again, the stronger effects were observed in p53-R175H– and p53-R273H–expressing HGC27-HRE cells. As the experiment progressed, the increased hypoxia reporter activity of mutant p53–expressing HGC27-HRE primary tumors was diluted, as indicated by either saturated bioluminescent reporter measurements or larger tumor sizes (Figure 6, C and D). Protein levels of HIF1α, however, remained elevated in mutant p53–expressing HGC27-HRE cells by IHC at the experiment endpoint (Figure 6E; ANOVA P = 0.013), suggesting that perhaps the reporter system was less reliable at larger tumor volumes. Together, these data provide an in vivo validation of the impact of hotspot missense TP53 mutations upon induction of hypoxia signaling.
Figure 6. Mutant p53 induces hypoxia during the initiation of gastric cancer primary tumor xenografts using real-time in vivo imaging.
(A) Luciferase assay of adherent cultured HGC27 cells stably expressing HRE–firefly reporter and either GFP or the indicated mutant p53. (B) Ratio of total flux (HRE-reporter activity) to tumor volume in HGC27-HRE xenografts at the 2-week time point. Representative images of mice are shown. (C) Tumor volume of HGC27-HRE xenografts at the 4-week time point. (D) Ratio of total flux (HRE-reporter activity) to tumor volume in HGC27-HRE xenografts at the 2-week time point. Representative images of mice are shown. (E) Representative images and quantification of HIF1α IHC of HGC27-HRE xenograft tumors. All data are presented as mean± SEM.
Disruption of the HIF1/ARNT hypoxia program is expendable in mutant p53 gastric cancer primary tumors.
To investigate whether the HIF-mediated hypoxia signaling axis was necessary for mutant p53-mediated augmentation of tumor growth in gastric cancer, we first genetically deleted the essential cofactor aryl hydrocarbon receptor nuclear translocator (ARNT, also known as HIFβ) in the HGC27 model. ARNT heterodimerizes with HIF1α, HIF2α, and HIF3α in order to execute transcriptional control of hypoxia-responsive genes. Using 3 different sgRNAs against ARNT, we showed that disruption of HIF-dependent signaling attenuates HRE luciferase reporter activity in HGC27 cells treated with DMOG (Supplemental Figure 9A). Consistent with these results, NDRG1 expression levels were reduced in ARNT-KO HGC27 cell lines in the presence and absence of DMOG by immunoblot analysis (Supplemental Figure 9B). As expected, HIF1α levels were unaffected by elimination of ARNT (Supplemental Figure 9B). We next tested whether deleting ARNT impacted hypoxia reporter activity in HGC27-HRE cells overexpressing mutant p53-R175H (HGC27-HRE-R175H cells). ARNT KO led to reduced reporter activity in HGC27-HRE-R175H cells in the presence of DMOG, although the decrease was smaller than in parental HGC27-HRE cells (Supplemental Figure 9C). Interestingly, NDRG1 levels were not affected by ARNT KO in HGC27-HRE-R175H cells (Supplemental Figure 9D), indicating a potential HIF-independent mechanism of hypoxia transcriptional activation. We next tested whether ARNT deletion negatively impacted primary tumor growth properties observed in HGC27-HRE-R175H cells. ARNT KO did not affect ultra-low attachment proliferation of HGC27-HRE-R175H cells in vitro (Supplemental Figure 10A). Disruption of HIF/ARNT signaling led to a modest reduction in hypoxia reporter activity in HGC27-HRE-R175H cells in vivo, which corresponded to no significant impact on primary tumor growth (Supplemental Figure 9, E–G). These findings were corroborated by the lack of difference in protein levels of phospho-NDRG1 and VEGF in the xenograft tumors by IHC at the experimental endpoint (data not shown), suggesting that ARNT inhibition may not effectively disrupt hypoxia signaling in vivo. To generalize this finding, we asked whether ARNT was required for proliferation of gastric adenocarcinoma cell lines in CCLE. Although hypoxia signaling activity as measured by single-sample GSEA (ssGSEA) was elevated in gastric cancer cell lines harboring hotspot mutations in TP53, consistent with our previous results (Figure 4C), they were inversely dependent on ARNT (Supplemental Figure 9H).
Direct inhibition of HIF1α in mutant p53 gastric cancer diminishes primary tumor growth in vivo.
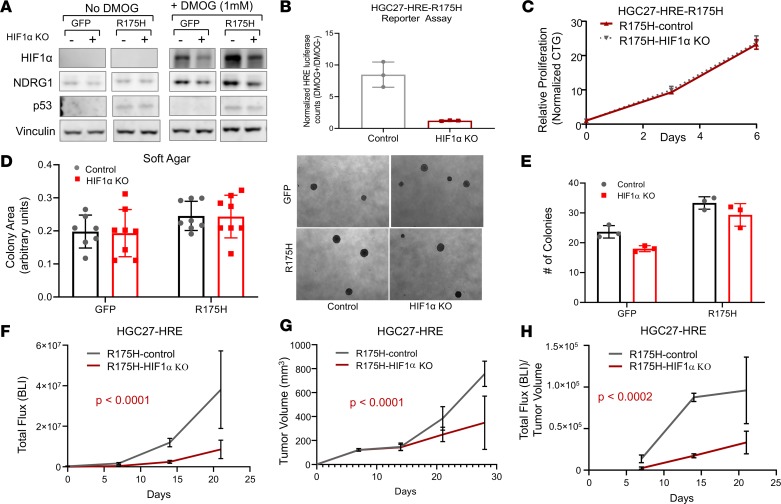
Given the modest attenuation of HIF signaling by ARNT inhibition, and the ability of ARNT to impair transcriptional activity of HIF1α, HIF2α, and HIF3α — each of which can affect distinct cancer functions (36–38) — we next directly tested whether disruption of HIF1α impacts primary tumor function of mutant p53 gastric cancer cells in the HGC27 model. CRISPR/Cas9-mediated deletion of HIF1α in HGC27-HRE GFP control cells led to a potent reduction in protein expression of HIF1α and downstream target NDRG1 in the presence of DMOG by immunoblot analysis (Figure 7A). Despite robust induction of HIF1α in HGC27-HRE-R175H cells (Figure 7A), HIF1α KO led to decreased protein expression of HIF1α and NDRG1 by immunoblot analysis and HRE-reporter activity by luciferase assay (Figure 7B). Disruption of HIF1α did not significantly impact adherent proliferation of HGC27-HRE-R175H cells by CellTiter-Glo assay (Figure 7C), consistent with dependency data in other gastric cancer cell lines (Supplemental Figure 10, B and C) or anchorage-independent colony growth in soft agar (Figure 7, D and E). These data indicate that inhibition of HIF1α-mediated hypoxia signaling does not significantly impact proliferative properties in vitro.
Figure 7. Disruption of HIF1α impairs primary tumor growth of mutant p53 gastric cancer.
(A) Immunoblot of HIF1α and NDRG1 expression levels in HGC27-HRE cells stably expressing either control or HIF1α sgRNA in the presence and absence of 1 mM DMOG. (B) Luciferase assay of HGC27-HRE-R175H cells stably expressing either control or HIF1α sgRNA displayed as a ratio of 1 mM DMOG/no DMOG. (C) Proliferation of HGC27-HRE-R175H cells stably expressing either control or HIF1α sgRNA under adherent culture conditions as measured by CTG; normalized to baseline counts on day 0. (D and E) Soft agar growth of HGC27-HRE-GFP and HGC27-HRE-R175H cells stably expressing either control or HIF1α sgRNA. Right panel in D shows representative images of colonies. (F) Total HRE flux (bioluminescence) of HGC27-HRE-R175H–expressing control (n = 5) or HIF1α sgRNA (n = 5) xenografts; comparison of fits, P < 0.0001. (G) Tumor volume of HGC27-HRE-R175H–expressing control (n = 5) or HIF1α sgRNA (n = 5) xenografts; comparison of fits, P < 0.0032. (H) Total HRE flux/tumor volume of HGC27-HRE-R175H–expressing control (n = 5) or HIF1α sgRNA (n = 5) xenografts; comparison of fits, P < 0.0002. All data are presented as mean ± SD.
To examine whether deletion of HIF1α impacted primary tumor growth in vivo, isogenic HGC27-HRE-R175H cells were injected into the flanks of nude mice. Unlike ARNT inhibition, disruption of HIF1α led to a greater than 5-fold reduction in hypoxia signaling in vivo (Figure 7F). Direct inhibition of HIF1α corresponded to a significant attenuation in primary growth of HGC-HRE-R175H cells in vivo (Figure 7G). Importantly, the ratio of hypoxia reporter activity to tumor volume was preserved in each arm throughout the experiment, suggesting that these results could not be explained by tumor volume alone (Figure 7H). These data indicate that disruption of HIF1α-mediated hypoxia signaling may be required for mutant p53 gastric cancer in vivo.
Discussion
GEAs are lethal cancers owing to their aggressive and chemoresistant behavior, the molecular basis of which requires further elucidation. TP53, the most frequently altered gene in GEAs, usually sustains missense mutations in the DNA-binding domain. Given the extensive literature on potential gain-of-function properties of select, disproportionately frequent TP53 missense mutations in many cancers (21–24, 27, 30, 39, 40), we designed an experimental system to investigate the functional importance of hotspot missense TP53 mutations in GEAs, ultimately to test the hypothesis that targeting of these gain-of-function properties may be of therapeutic value. Despite being overexpressed in GEAs, we found that endogenous mutant p53 was not necessary for proliferation, anchorage-independent growth, and/or in vivo primary tumor growth in several GEA cell lines. These results suggest that although missense mutations in TP53 are an early, enabling event in the pathogenesis of GEAs, gain-of-function properties may not be required during advanced stages of tumor progression. Select mouse models, such as those involving T cell lymphomas (30) and non–small cell lung cancer (41), have shown a requirement for mutant p53, which is thought to be secondary to an addiction to its gain-of-function activity. A comprehensive analysis of the requirement for mutant p53 across many cancer types suggests that this is less frequently the case (Figure 2, G and H, Supplemental Figure 3, A and B, and ref. 31). These findings suggest that the therapeutic value of directly inhibiting mutant p53 may be limited in advanced GEAs.
Instead, we found that overexpression of mutant p53 is sufficient to drive primary tumor cancer properties in GEAs, consistent with findings in other tumor types (21–28). Unbiased transcriptomic analysis evaluating genes regulated by endogenous mutant p53 in one gastric cancer cell line and overexpression of mutant p53 in another revealed enrichment for a hypoxia-associated transcriptional program. A hypoxic microenvironment is a hallmark of cancer pathogenesis that enables cancer cells to utilize HIF-dependent transcriptional programs to their advantage (42). Angiogenesis, metabolism, stemness, and metastasis are some of the advanced tumor properties implicated in hypoxia/HIF regulation. Hypoxia/HIF signaling is a poor prognostic marker in human gastric cancer (43), and activation of the pathway has been shown to promote gastric cancer progression in experimental models (43–46). Despite the evidence that hypoxia plays a pro-cancer function in gastric cancer, the precise mechanisms have yet to be elucidated.
There is expanding literature on the relationship between p53 and HIF/hypoxia in physiological and pathological settings (47). Early studies pointed to the ability of HIF/hypoxia to induce WT p53 (48, 49) and serve as a selective pressure for neoplastic cells with defects in apoptosis (50). In a negative feedback role, WT p53 was shown to attenuate angiogenesis by promoting the degradation of HIF1α (51, 52). There is now evidence to suggest that genes negatively regulated by WT p53 are often activated by mutant p53 (23). Consistent with this notion, we found that 2 mutant p53 alleles that strongly correlate in transcriptional output can promote activation of hypoxia signaling in GEA cell lines. Similar findings were most recently reported in non–small cell lung cancer, in which mutant p53 physically engaged HIF1α to regulate extracellular matrix components to promote cancer progression (41). Mutant p53 has also been shown to increase tumor vascularization via ROS-mediated activation of the HIF1α /VEGF-A pathway in colon tumor cells (53).
We show for the first time to our knowledge the ability of mutant p53 to induce hypoxia during primary tumor growth using real-time in vivo luciferase reporter–based imaging. Importantly, we found that the greatest impact of mutant p53 on hypoxia occurred during the early stages of primary tumor growth, a phenomenon that attenuated as the tumors grew larger over time. These results could represent a technical limitation of the in vivo reporter system or the strong propensity to activate hypoxia signaling during tumor growth, especially with the rapid growth kinetics of mouse xenografts. However, these results may also indicate the potential pathological importance of mutant p53–mediated hypoxia induction during the initiation/early formation of primary tumor growth or even in the preneoplastic phase, providing a growth advantage to the p53 mutant cell prior to oncogene activation. Once the tumor reaches a sufficient size, the effect of mutant p53 on hypoxia induction is difficult to discern. There is evidence to support the importance of hypoxia-induced angiogenesis during these early stages of tumorigenesis: progression from hyperplasia to neoplasia requires neovascularization, enabling the transition from dysplastic lesions to overt cancer (54). It will be important for future studies to investigate the importance of hypoxia induction by mutant p53 using models of gastroesophageal premalignancy or early neoplasia.
Ultimately, we found that inhibiting HIF1α-dependent hypoxia signaling impaired primary tumor growth in mutant p53 gastric cancer. Importantly, the strongest effect of HIF1α deletion on mutant p53 gastric cancer appeared to occur under hypoxic conditions, such as during primary tumor growth; these detrimental effects were not observed while cells were cultured in vitro. Collectively, these data suggest that activation of a hypoxia transcriptional program by mutant p53 under endogenous conditions may not be a significant liability in gastric cancers. In addition, these results may motivate the pursuit of new areas of investigation to elucidate the context under which mutant p53 and hypoxia activation impact the development of upper gastrointestinal cancers. For example, based on the data from the premalignant gastric lesions using the carcinogenesis genetically engineered mouse model, it is possible that the importance of mutant p53–induced hypoxia activity may be at early stages of gastric tumorigenesis. In contrast to the HIF1α studies, inhibition of ARNT did not significantly impact primary tumor growth of mutant p53 gastric cancer. One possible explanation is that disruption of multiple HIF proteins, which can exert distinct cancer functions, may have confounded the results. Another explanation is that hypoxia signaling was not attenuated below the critical amount required to observe an impact on primary tumor growth; deletion of HIF1α was a stronger detriment to hypoxia signaling than ARNT inhibition. A deeper investigation into mutant p53 biology and its intersection with hypoxia signaling will improve our understanding of upper gastrointestinal cancers, which it is hoped will inspire new ideas for therapeutic intervention.
Methods
Mice.
To activate endogenous expression of p53R270H in gastric tissue, we crossed Trp53LSL-R270H mice (provided by Kwok Wong, NYU Langone Hospital, New York, New York, USA) to Mist1-CreERT2 mice (provided by Timothy Wang, Columbia University Medical Center, New York, New York, USA). Experimental mice aged 6–8 weeks received 5 consecutive daily 200-mL i.p. injections of tamoxifen in sunflower oil at 10 mg/mL for activation. Control mice did not receive tamoxifen. All treated mice received 240 ppm MNU in drinking water every other week and 0.3% DCA continuously for the indicated time period. Mice were euthanized at the endpoint of the experiment, and stomachs were harvested for histopathological and immunohistochemical analyses.
Tumor xenografts and bioluminescence analysis.
1 × 106 tumor cells were injected into flanks of athymic Ncr-nu/nu mice. Tumor measurements were made by caliper, and tumor volumes were calculated using the following formula: volume = length × width2 × 0.5. Hypoxia reporter activity was monitored by total flux of luciferase activity as measured by bioluminescence imaging (BLI). Anesthetized mice were injected i.p. with d-luciferin. Bioluminescence images were acquired with a Xenogen IVIS 200 Imaging System (PerkinElmer). Analysis was performed with Living Image software 4.2 (PerkinElmer) by measuring photon flux in tumors.
Cell lines and culture conditions.
The human cancer cell lines LMSU, HGC27, AGS, ESO51, and SH10TC were obtained from the CCLE core facility, which obtained them directly from commercial sources and authenticated the lines using standard short tandem repeat analysis. LMSU is a gastric adenocarcinoma cell line harboring an endogenous R175H TP53 mutation cultured in F10 medium supplemented with 10% FBS. HGC27 is a gastric adenocarcinoma cell line harboring an endogenous P152fs TP53 mutation cultured in αMEM supplemented with 10% FBS and nonessential amino acids. AGS is a gastric adenocarcinoma cell line that is TP53 WT cultured in Ham’s F12 medium supplemented with 10% FBS. ESO51 is a suspension esophageal adenocarcinoma cell line harboring an endogenous R175H TP53 mutation cultured in DMEM supplemented with 10% FBS. SH10TC is an esophageal adenocarcinoma cell line harboring an endogenous R273C TP53 mutation cultured in RPMI medium supplemented with 10% FBS.
Cell proliferation assays.
Cell viability was quantified by measuring cellular ATP content using the CellTiter-Glo Cell Viability Assay (Promega) according to the manufacturer’s instructions. All experiments were performed in triplicate in 96-well plates. Adherent culture refers to cancer cells grown on traditional cell culture plates upon which cells attach to surface and require trypsinization for dissociation and passage. Ultra-low attachment is a method for culturing cells under nonadherent conditions. This method can be used to assess proliferation, colony-forming ability, and anchorage-independent growth. Typically, between 1 × 103 and 1 × 104 cells were plated in each well of a 24-well or 6-well plate (Corning, 3471). Area measurements and quantification of low-attachment colonies was measured using ImageJ software (NIH).
Generation of CRISPR-mediated KO cell lines.
All genetically engineered gastric cancer cell lines were generated using the protocol described by Shalem et al. (55). In brief, sgRNAs targeting TP53 and ARNT were designed, amplified, and cloned into lentiCRISPR v2 (56). shRNAs targeting TP53 were cloned into PLKO.1 and TET-PLKO vectors. Lentivirus was generated using standard protocols. In brief, HEK293T cells were plated in 10-cm2 plates with fresh medium without antibiotics. Subsequently, 5 μg of the lentiviral vector, 100 ng of the envelope plasmid, and 900 ng of the packaging plasmid were diluted in Opti-MEM (Thermo Fisher Scientific, 31985070); 20 μl X-tremeGENE 9 DNA Transfection Reagent (MilliporeSigma, 036335779001) was added dropwise; and this mixture was incubated for 20 minutes. The DNA complexes were added dropwise to the cells and incubated for 12 hours before aspiration and addition of 6 mL fresh medium. After 24 hours, the virus-containing medium was collected and filtered through a 0.45-μm syringe, and the lentivirus was stored at −80°C. The oligonucleotide sequences used for cloning TP53 sgRNAs were as follows: sgRNA control (distal U6 promoter): GAGGCTAAGCGTCGCAA; TP53sg#F2F-F: GGGCAGCTACGGTTTCCGTC, TP53sg#F2F-R: GACGGAAACCGTAGCTGCCC (57); TP53sg#1-F: CCATTGTTCAATATCGTCCG, TP53sg#1-R: CGGACGATATTGAACAATGG; TP53sg#2-F: CCCCGGACGATATTGAACAA, TP53sg#2-R: TTGTTCAATATCGTCCGGGG. shRNA sequences for TP53 are listed in Supplemental Table 1. The oligonucleotide sequences used for cloning ARNT and HIF1α sgRNAs were as follows: ARNTsg#1-F: 5′-CACCGGGCTATTAAGCGACG GTCA-3′, ARNTsg#1-R: 5′-AAACTGACCGTCGCTTAATAGCCC-3; ARNTsg#3-F: 5′-CACCGAGAAACGGCCATGCGTAAGA-3′, ARNTsg#3R: 5′-AAACTCTTACGCATGGCCGTT TCTC-3′; sgHIF1α forward: 5′-CACCGTGTGAGTTCGCATCTTGATA-3′, sgHIF1α reverse: 5′-AAACTATCAAGATGCGAACTCACAC-3′.
RNA isolation and RT-PCR.
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN), and cDNA was synthesized using the TaqMan Reverse Transcription Reagents Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Gene-specific primers for SYBR Green Real-Time PCR were designed by use of Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized by Integrated DNA Technologies. RT-PCR was performed and data were analyzed using the CFX96 Real-Time PCR Detection System (Bio-Rad) and Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. Relative mRNA expression was determined by normalizing to GAPDH expression, which served as an internal control. See Supplemental Table 2 for primers used for RT-PCR.
Immunoblots and antibodies.
Western blot analysis was performed as previously described (58). The following antibodies were used for Western blotting (from Cell Signaling Technology, except where indicated): anti-p53 (2524, 1:1,000), anti-V5, anti–phospho-Stat3, anti-Stat3, anti-NDRG1, and anti–β-actin (A5441, 1:1,000, MilliporeSigma). See complete unedited blots in the supplemental material.
Immunohistochemistry.
Xenograft tumors were excised and fixed with 10% formalin overnight and embedded in paraffin (FFPE). Unstained sections were stained using the following protocol. All IHC was performed on the Leica Bond automated staining platform. HIF1α antibody from Cell Signaling Technology (catalog 22204s, clone E1Q6W) was run at 1:30,000 dilution using the Leica Biosystems Refine Detection Kit with EDTA antigen retrieval and TSA amplification (PerkinElmer). Antibody from Beckman Coulter (catalog IM0787, clone QBEnd10) was run at 1:70 dilution using the Leica Biosystems Refine Detection Kit with citrate antigen retrieval. p53 antibody from Cell Signaling Technology (catalog 48818, clone DO-7) was run at 1:100 dilution using the Leica Biosystems Refine Detection Kit with citrate antigen retrieval. Representative images were taken using a Leica DM1000 LED light microscope camera.
Quantification of HIF1α.
Three to 5 representative ×20 images were taken from each xenograft tumor FFPE slide stained with HIF1α. Images were then uploaded into ImageJ, and color threshold was adjusted from color space to HSB, threshold color to red, and thresholding method to default. Positively stained cells were gated with a filter of approximate hue 0 to 45, whereas unstained cells were gated with a filter from approximate hue 60 to 215. To save images, the threshold color was changed from red to black and white. The ImageJ-processed image was opened in Adobe Photoshop to use the histogram function. The colors were normalized by using the caution symbol in the panel. The leftmost spike, representing black, is the percent total cell cover of positive/negative results.
Differential expression analysis.
We first excluded genes that had fewer than 1 count per million in at least 6 samples in each experiment. The weighted trimmed mean of M-values method (59) was used to normalize the library size of each sample, using the calcNormFactors function in the R package: edgeR (60). To estimate log fold change differences, and associated P values, between sample groups, we used the R package limma (61). Read counts data were transformed using the limma function “voom” prior to model fitting, in order to model the mean-variance relationship of the log counts data (62). For LMSU, we compared expression in the samples with sgRNA- or shRNA-mediated TP53-KO/KD versus the controls (NCBI Gene Expression Omnibus [GEO], GSE132331). For HGC27, we compared expression in the samples with mutant p53 overexpression allele R175H, R248Q, or R273H with GFP control (GEO, GSE132333). Correlations between replicates were modeled using the duplicateCorrelation function of limma. P values were estimated from the empirical Bayes moderated t statistics, and q values were estimated using the Benjamini-Hochberg method (63). GSEA (64) was run to test for gene sets that were up- or downregulated in each experimental condition in a subset of experiments. For the majority of experiments, we used the R package fgsea (65) to estimate normalized enrichment statistics, and associated P values, for each gene set in the Hallmark collection of the Molecular Signatures Database (66). The GSEA algorithm was run using log fold change values as the gene-level statistics, 100,000 random permutations, and a GSEA parameter of 1.
Cancer cell line analysis.
RNAi gene dependency data were derived from the combined Achilles/DRIVE/Marcotte data set processed using DEMETER2 (67). CRISPR/Cas9 dependency data, processed using CERES (68), were taken from the Dependency Map 18Q4 data release (DEMETER2 data; doi.org/10.6084/m9.figshare.6025238.v4). Cancer cell line omics data were taken from the CCLE mutation calls, and mRNA expression data were taken from the Dependency Map 18Q4 data release (69). All cancer cell line gene dependency and omics data sets can be accessed at https://depmap.org/portal/ CCLE cell line TP53 status was obtained from the DepMap 18Q4 mutation calls file. Cell lines that did not have any (nonsilent) mutation in TP53 were classified as WT; cell lines with mutations resulting in the protein changes R248Q/W, R273H/C, and R175H were classified as hotspot; cell lines with other missense mutations were classified as missense; and cell lines with a TP53 mutation classified as deleterious in the CCLE mutation calls file were classified as damaging. Cell lines with damaging and hotspot mutations were classified as damaging. Two-group comparisons of cancer cell line omics and gene dependency data were performed using Wilcoxon’s rank-sum tests. Single-sample GSEA enrichment scores were calculated for the Hallmark hypoxia gene set (66) using the R package GSVA (70). All of the cell line omics and gene dependency data used in the analysis are provided in Supplemental Table 3.
Statistics.
Experiments were performed in triplicate. Data are presented as mean ± SD unless indicated otherwise. For each experiment, independent biological experiments or technical replicates are as noted in the figure legends and were repeated with similar results. Statistical analysis was performed using Microsoft Office statistical tools or in Prism 7.0 (GraphPad). Pairwise comparisons between groups (experimental versus control) were performed using an unpaired 2-tailed Student’s t test or Kruskal-Wallis test as appropriate. P < 0.05 was considered to be statistically significant. For all experiments, the variance between comparison groups was found to be equivalent. For xenograft experiments, data are presented as mean ± SD, and statistical comparisons were performed using unpaired, 2-tailed Student’s t tests with Welch’s correction or comparison of fits. Sample sizes and animal numbers were determined from pilot laboratory experiments and previously published literature. Animals were excluded from analysis if they were euthanized due to health reasons unrelated to tumor volume end point. For in vivo experiments, all mice were randomized before studies.
Study approval.
All procedures involving mice and experimental protocols were approved by the IACUC of the DFCI (no. 11-009).
Author contributions
NS and AJB conceived the study and designed experiments. NS and OK performed in vitro experiments. NS, OK, and NK performed RT-PCR experiments. NS, OK, and YZ performed mouse experiments. NS and MC performed IHC analysis. NS, JM, and MI performed computational analysis of transcriptome data. BL, AC, and WK provided reagents, helped with experimental design, and interpreted data from hypoxia experiments. NS, OK, and AJB wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Adam Sperling, Doug Micalizzi, David Liu, Harshabad Singh, Ankur Nagaraja, Matthew Oser, Sagar Koduri, and Sam McBrayer for insightful discussions. We thank the Dana-Farber/Harvard Cancer Center, especially Benjamin Ferland, for the use of the Specialized Histopathology Core, which provided histology and IHC service. We thank Blanca Pineda, Daisy Moreno, Catherine Sypher, and the rest of the DFCI Animal Resource staff members for technical assistance and animal care. The Dana-Farber/Harvard Cancer Center is supported in part by National Cancer Institute (NCI) Cancer Center Support Grant NIH 5 P30 CA06516. This work was funded by NCI grant P01 CA098101 to AJB and American Cancer Society Postdoctoral Fellowship and KL2/CMERIT Harvard Catalyst Award to NS.
Version 1. 08/08/2019
Electronic publication
Version 2. 08/16/2019
The abstract was corrected.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(15):e128439. https://doi.org/10.1172/jci.insight.128439.
Contributor Information
Nilay Sethi, Email: nilay_sethi@dfci.harvard.edu.
Osamu Kikuchi, Email: osamu_kikuchi@dfci.harvard.edu.
Yanxi Zhang, Email: Yanxi_Zhang@DFCI.HARVARD.EDU.
Max Chung, Email: max_chung@brown.edu.
Nicholas Kafker, Email: nick.kafker@gmail.com.
Mirazul Islam, Email: mislam@broadinstitute.org.
Adam J. Bass, Email: adam_bass@dfci.harvard.edu.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45(5):478–86. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu T, et al. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology. 2014;147(2):407–417. doi: 10.1053/j.gastro.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 6.Stachler MD, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47(9):1047–1055. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135(4):1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2017;14(5):296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hainaut P, Pfeifer GP. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb Perspect Med. 2016 Nov 1;6(11):a026179. doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bargonetti J, Reynisdóttir I, Friedman PN, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6(10):1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 11.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science. 2016;352(6282):175–180. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22(2):239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato Y, et al. Establishment and characterization of a new hypoxia-resistant cancer cell line, OCUM-12/Hypo, derived from a scirrhous gastric carcinoma. Br J Cancer. 2010;102(5):898–907. doi: 10.1038/sj.bjc.6605543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohwer N, et al. HIF-1alpha determines the metastatic potential of gastric cancer cells. Br J Cancer. 2009;100(5):772–781. doi: 10.1038/sj.bjc.6604919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, et al. Subcellular distribution of S100A4 and its transcriptional regulation under hypoxic conditions in gastric cancer cell line BGC823. Cancer Sci. 2010;101(5):1141–1146. doi: 10.1111/j.1349-7006.2010.01533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sande CM, Chang B, Monga V, Bossler AD, Ma D. Biallelic TP53 gain of function mutations in rapidly progressing solid tumors. Cancer Genet. 2018;222–223:20–24. doi: 10.1016/j.cancergen.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Glazko GV, Koonin EV, Rogozin IB. Mutation hotspots in the p53 gene in tumors of different origin: correlation with evolutionary conservation and signs of positive selection. Biochim Biophys Acta. 2004;1679(2):95–106. doi: 10.1016/j.bbaexp.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Villuendas R, et al. p21WAF1/CIP1 and MDM2 expression in non-Hodgkin’s lymphoma and their relationship to p53 status: a p53+, MDM2–, p21-immunophenotype associated with missense p53 mutations. J Pathol. 1997;181(1):51–61. doi: 10.1002/(SICI)1096-9896(199701)181:1<51::AID-PATH689>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 20.Peng Y, Chen L, Li C, Lu W, Agrawal S, Chen J. Stabilization of the MDM2 oncoprotein by mutant p53. J Biol Chem. 2001;276(9):6874–6878. doi: 10.1074/jbc.C000781200. [DOI] [PubMed] [Google Scholar]
- 21.Dong P, et al. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2013;32(27):3286–3295. doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148(1–2):244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogan-Sakin I, et al. Mutant p53(R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells. Cell Death Differ. 2011;18(2):271–281. doi: 10.1038/cdd.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 26.Nigro JM, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342(6250):705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 27.Schofield HK, et al. Mutant p53R270H drives altered metabolism and increased invasion in pancreatic ductal adenocarcinoma. JCI Insight. 2018;3(2):e97422. doi: 10.1172/jci.insight.97422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walerych D, et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol. 2016;18(8):897–909. doi: 10.1038/ncb3380. [DOI] [PubMed] [Google Scholar]
- 29.Zhu J, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525(7568):206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandrova EM, et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature. 2015;523(7560):352–356. doi: 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giacomelli AO, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat Genet. 2018;50(10):1381–1387. doi: 10.1038/s41588-018-0204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freed-Pastor W, Prives C. Targeting mutant p53 through the mevalonate pathway. Nat Cell Biol. 2016;18(11):1122–1124. doi: 10.1038/ncb3435. [DOI] [PubMed] [Google Scholar]
- 33.Parrales A, et al. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol. 2016;18(11):1233–1243. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohashi S, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70(10):4174–4184. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briggs KJ, et al. Paracrine induction of HIF by glutamate in breast cancer: EglN1 senses cysteine. Cell. 2016;166(1):126–139. doi: 10.1016/j.cell.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakraborty AA, et al. HIF activation causes synthetic lethality between the VHL tumor suppressor and the EZH1 histone methyltransferase. Sci Transl Med. 2017;9(398):eaal5272. doi: 10.1126/scitranslmed.aal5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho H, et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature. 2016;539(7627):107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim WY, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119(8):2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kollareddy M, et al. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat Commun. 2015;6:7389. doi: 10.1038/ncomms8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfister NT, et al. Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells. Genes Dev. 2015;29(12):1298–1315. doi: 10.1101/gad.263202.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amelio I, et al. p53 mutants cooperate with HIF-1 in transcriptional regulation of extracellular matrix components to promote tumor progression. Proc Natl Acad Sci U S A. 2018;115(46):E10869–E10878. doi: 10.1073/pnas.1808314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaelin WG., Jr How oxygen makes its presence felt. Genes Dev. 2002;16(12):1441–1445. doi: 10.1101/gad.1003602. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths EA, et al. Hypoxia-associated markers in gastric carcinogenesis and HIF-2alpha in gastric and gastro-oesophageal cancer prognosis. Br J Cancer. 2008;98(5):965–973. doi: 10.1038/sj.bjc.6604210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffiths EA, Pritchard SA, Welch IM, Price PM, West CM. Is the hypoxia-inducible factor pathway important in gastric cancer? Eur J Cancer. 2005;41(18):2792–2805. doi: 10.1016/j.ejca.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Kitajima Y, Miyazaki K. The critical impact of HIF-1a on gastric cancer biology. Cancers (Basel) 2013;5(1):15–26. doi: 10.3390/cancers5010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Ni Z, Duan Z, Wang G, Li F. Altered expression of hypoxia-inducible factor-1α (HIF-1α) and its regulatory genes in gastric cancer tissues. PLoS One. 2014;9(6):e99835. doi: 10.1371/journal.pone.0099835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem Sci. 2015;40(8):425–434. doi: 10.1016/j.tibs.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 48.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392(6674):405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 49.Graeber TG, Peterson JF, Tsai M, Monica K, Fornace AJ, Jr, Giaccia AJ. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14(9):6264–6277. doi: 10.1128/MCB.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graeber TG, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379(6560):88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 51.Ravi R, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14(1):34–44. [PMC free article] [PubMed] [Google Scholar]
- 52.Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273(20):11995–11998. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- 53.Khromova NV, Kopnin PB, Stepanova EV, Agapova LS, Kopnin BP. p53 hot-spot mutants increase tumor vascularization via ROS-mediated activation of the HIF1/VEGF-A pathway. Cancer Lett. 2009;276(2):143–151. doi: 10.1016/j.canlet.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 54.Folkman J, Watson K, Ingber D, Hanahan D. Induction of angiogenesis during the transition from hyperplasia to neoplasia. Nature. 1989;339(6219):58–61. doi: 10.1038/339058a0. [DOI] [PubMed] [Google Scholar]
- 55.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520(7549):697–701. doi: 10.1038/nature14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong GS, et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med. 2018;24(7):968–977. doi: 10.1038/s41591-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 64.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sergushichev AA, et al. GAM: a web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res. 2016;44(W1):W194–W200. doi: 10.1093/nar/gkw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McFarland JM, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9(1):4610. doi: 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meyers RM, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49(12):1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cancer Cell Line Encyclopedia Consortium. Genomics of Drug Sensitivity in Cancer Consortium Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528(7580):84–87. doi: 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.