We report an alternative strategy for monitoring renal function based on administration of a pH-sensitive MRI contrast agent.
Abstract
Chronic Kidney Disease (CKD) is a cardinal feature of methylmalonic acidemia (MMA), a prototypic organic acidemia. Impaired growth, low activity, and protein restriction affect muscle mass and lower serum creatinine, which can delay diagnosis and management of renal disease. We have designed an alternative strategy for monitoring renal function based on administration of a pH sensitive MRI agent and assessed this in a mouse model. This protocol produced three metrics: kidney contrast, ~4% for severe renal disease mice compared to ~13% and ~25% for moderate renal disease and healthy controls, filtration fraction (FF), ~15% for severe renal disease mice compared to ~79% and 100% for moderate renal disease and healthy controls, and variation in pH, ~0.45 units for severe disease mice compared to 0.06 and 0.01 for moderate disease and healthy controls. Our results demonstrate that MRI can be used for early detection and monitoring of CKD.
INTRODUCTION
Organic acidemias represent a group of rare inborn errors of metabolism caused by disturbances primarily of amino acid metabolism that result in large abnormal accumulations of often toxic organic acids in tissues and body fluids including in the urine. While individually rare, collectively, this group of inborn errors of metabolism leads to notable morbidity and mortality in infancy and childhood such that many of these disorders are now included in routine newborn screening panels to enable early diagnosis and treatment. Methylmalonic acidemia (MMA) is a common and severe organic acidemia with established knockout and tissue-specific transgenic mouse models that accurately replicate key features of the human disease, including C57BL/6 Mut−/−;TgINS-Alb-Mut mice, which have been used to model the renal disease of MMA (1–3). While early identification and treatment have improved the clinical course and life expectancy in MMA, the patients remain at risk for chronic complications, particularly chronic kidney disease (CKD) and, eventually, renal failure (4, 5).
The glomerular filtration rate (GFR) is the standard index for determining renal function (6); however, direct measurements of GFR (mGFR) are invasive and cumbersome (7). Several types of clearance measurements are possible: 24-hour urine sampling, timed blood sampling, or image acquisitions. Measurements require monitoring plasma clearance of a marker, often radioisotopes [99mTc-DTPA (diethylenetriamine pentaacetic acid) and 51Cr-EDTA (ethylenediamine tetraacetic acid)] and serial blood samples (inulin, iohexol, iothalamate, and others) that require complex protocols, involving the placement of multiple intravenous lines, repeat blood sampling over up to 8 to 24 hours, timed urination, and hospital monitoring. Furthermore, the downstream measurements typically require sample shipment to highly specialized laboratories, which creates substantial administrative burden on clinics and introduces a risk for post-collection errors.
An alternative to mGFR is to estimate GFR (eGFR) using serum creatinine and/or cystatin-C (8–10), and is routinely used by clinicians (9). Computing eGFR through blood tests is much less complex because these tests can be simple to perform, are widely available, and are much less expensive than obtaining the mGFR. However, it is well recognized that routine serum biomarkers of renal disease, such as creatinine, are poor screening tests of renal function because of the dependence on muscle mass, which is substantially altered in these severely growth-impaired and protein-restricted patients (11, 12). Furthermore, eGFR is limited for patients with asymptomatic renal disease, especially because they can demonstrate normal serum creatinine levels (13). The development of alternative methods to detect the early stages of CKD, and its evolution, is therefore necessary to facilitate treatment of associated comorbidities and enable more timely planning of renoprotective or renal replacement therapies, including transplantation, in many patient populations. As a result, the development of new tools for detecting renal disease represents an unmet clinical need not only in MMA but also in the CKD population in general.
Magnetic resonance imaging (MRI) can produce high-resolution images with exquisite soft tissue contrast and has long been recognized as an outstanding tool for detecting renal tumors (14), renal cysts, and other pathologies. Additional functional information can be acquired through administration of MR contrast agents. MR contrast agents can be used to measure parameters such as kidney clearance or to count the number of glomeruli with impressive results (15, 16) and are included in abdominal imaging protocols to better characterize the kidneys. As the kidney represents the major determinant of acid-base balance in the body, pH is also a useful biomarker of renal function. MRI is also particularly well suited for pH imaging, as has been well described previously by a number of groups (17–19).
Chemical exchange saturation transfer (CEST) MRI has now emerged as the premier technology for creating pH images (20, 21). CEST detects low concentrations of contrast agent through the application of saturation pulses on labile protons to destroy their magnetization, with the resulting signal loss transferred to water through chemical exchange. The chemical shift dependence is an important feature, allowing discrimination between different agents through what has been described as multicolor (22–24) or multifrequency MRI (25, 26). For pH imaging, it is important to create pH maps that are independent of agent concentration, which can be accomplished using probes with two distinct labile protons (27, 28). We have long been interested in CEST MRI–based pH imaging using nonradioactive organic contrast agents and have developed methods to quantify chemical exchange rates as a function of pH (29), using imidazoles as CEST MRI pH sensors (30) as well as smart biomaterials, which can be used to monitor cell death through pH changes that occur following transplantation (31). Iopamidol is an excellent pH probe with renal excretion that can detect acute kidney injury (AKI), as has been shown recently (28, 32, 33).
In this study, we use CEST MRI to functionally image kidneys and investigate the progression to CKD in an MMA mouse model. We have designed a time-efficient CEST MRI protocol to study iopamidol, a nonionic contrast agent, which has yielded both perfusion and pH maps of the kidneys. Our method allows facile determination of mGFR and should be a useful method to diagnose and monitor CKD.
RESULTS
Calibration of ratiometric signal for pH mapping using iopamidol in blood serum
We first prepared phantoms to test the sensitivity of iopamidol and evaluate our pH mapping protocols over a range of pH values relevant to our studies (pH values, 5.3 to 7.3). The structure of iopamidol is shown in Fig. 1A. The Z-spectra of iopamidol in Fig. 1B display labile protons resonating at 4.2 and 5.5 parts per million (ppm), which are well resolved for a wide range of pH values.
Fig. 1. In vitro calibration plots and pH maps using iopamidol in human blood serum.
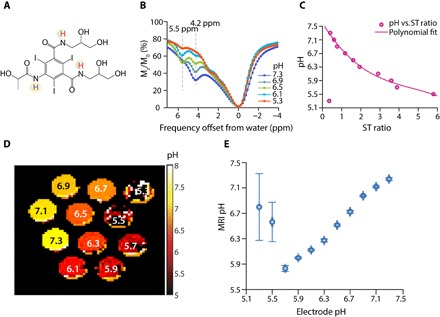
(A) Structure of iopamidol with exchangeable protons highlighted, which produce CEST contrast at 4.2 and 5.5 ppm at a relative concentration of 2:1. (B) CEST Z-spectra of iopamidol in blood serum at ω1 = 4 μT for pH = 5.3, 6.1, 6.5, 6.9, and 7.3. (C) Calibration plot used to calculate in vitro and in vivo pH; variation of experimental ST ratio at different pH values was given by pH = p1 × (ST ratio)3 + p2 × (ST ratio)2 + p3 × (ST ratio)1 + p4, with p1 = −0.01174, p2 = 0.1653, p3 = −0.927, and p4 = 7.598. The root mean square error of the fit was 0.0714 for pH values from 5.5 to 7.3. (D) pH maps of iopamidol-serum phantom. In the map, pH values below 5.5 were determined inaccurately. (E) Error bar plot representing the accuracy in MRI pH measurements compared to that of electrode pH for iopamidol-serum phantom. Error bars (blue) were obtained by calculating the mean SD in pH over a region of interest (ROI) drawn enclosing the entire tube in the phantom.
We tested several radio frequency (RF) saturation powers (ω1) from 1 to 5 μΤ and found that 4 μT provided the best ST ratio calibration curve in blood serum (Fig. 1C), which has a relatively simple relationship with pH under these saturation conditions. Figure S1 shows an alternative ω1 = 3 μT, which is the next best and has a shallower dependence of ST ratio on pH for pH < 6. Figure S2 shows the ST ratio and pH measurement dependence as a function of ΔB0 for 4-μT saturation. The pH is fairly tolerant to B0 inhomogeneity at this saturation power, which is part of the reason for selecting it. For example, at pH values 5.7 and 5.9, field inhomogeneities ΔB0 = ±120 Hz generate errors of <0.1 pH units. The inhomogeneity tolerance plot for 3-μT saturation power (fig. S3) shows that ΔB0 = ±60 Hz has an error of ≥0.1 unit at pH 5.7 and 5.9, a lower tolerance, which is less desirable. While the experimental design of using electrode pH for comparing with calculated CEST MRI pH values on phantoms is a circular one, it is useful to depict the influence of experimental error using these phantoms. Figure 1D shows the resultant pH maps for our blood serum phantom, and Fig. 1E shows the SD for these pH determinations across each tube in the phantom. These data indicate that while pH values between 7.3 and 5.7 can be readily measured using our multifrequency pH mapping protocol, pH values of 5.5 and below are challenging. This can also be visualized by the spectra in Fig. 1B, which indicate very low contrast at pH 5.3. We selected these conditions for live animal studies.
Development of an in vivo CEST MRI protocol to study a mouse model of MMA
We then proceeded to testing the performance of iopamidol for CEST imaging in C57BL/6 Mut−/−;TgINS-Alb-Mut mice (referred to as Mut−/− in the remaining text and figures) and control heterozygote littermates (Mut+/−;TgINS-Alb-Mut, referred as Mut+/−). The mice were divided into four groups: regular diet (RD) control (Mut+/−), high-protein (HP) diet control (Mut+/−), mild kidney disease (RD Mut−/−), and severe kidney disease (HP Mut−/−) based on previous studies with this animal model (1). In these mice, exposure to an HP diet was previously shown to cause massive elevations of plasma (MMA), associated with decreased GFR [extensively studied at the single-nephron and whole-animal level, by plasma decay of 125I-iothalamate and fluorescein isothiocyanate (FITC) inulin, respectively], elevated creatinine, and increased expression of lipocalin-2 (Lcn2; in the kidney tissue and plasma) in the mutant animals (Mut−/−;TgINS-Alb-Mut). As shown in fig. S4, Mut+/−;TgINS-Alb-Mut mice had normal weight (20 to 30 g), low levels of plasma MMA, and low Lcn2 mRNA expression in their kidneys. For the Mut−/− mice, the plasma (MMA) and kidney Lcn2 were significantly elevated when placed on the HP diet. As expected, their weight dropped with time on this diet, replicating the findings of the previous studies. We administered iopamidol at a weight-controlled dose of 1.5 g of iodine per kilogram and acquired CEST MRI data on a single slice containing the center of both kidneys using two offsets, 4.2 and 5.5 ppm, to characterize how the contrast varies with time after injection. As shown in Fig. 2 (A and B), CEST contrast in the kidneys at 4.2 ppm increased with time over 50 min for all mice, with the strongest contrast occurring for the control mice (~23%), the weakest contrast in the mutant mice ingesting an HP diet (~4%), and intermediate changes in mutant mice fed an RD. Notably, the MMA mice at baseline did not previously manifest substantial renal pathology or decreased mGFR when studied with iohexol, rather only when tested at the single nephron level (1). In addition, the amount of contrast in the kidneys is fairly uniform in the control mice and varies more in the moderate and severe kidney disease mice (Fig. 2, C to F). This indicates that uptake of iopamidol can help distinguish between these groups early in disease progression. Furthermore, a linear correlation was observed between the weight of the RD and HP Mut−/− mice and image contrast (R = 0.93; fig. S9A).
Fig. 2. Contrast agent uptake for the four different groups of MMA mice.
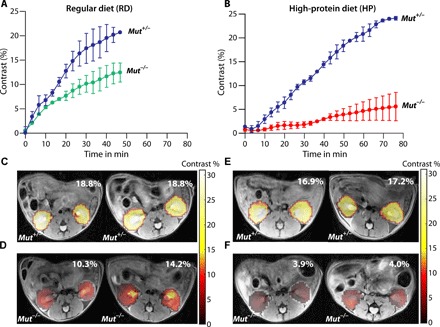
(A) Average iopamidol uptake with time for RD Mut+/− and RD Mut−/− mice. RD Mut+/− mice display higher uptake of contrast agent [>25% (n = 5)] than RD Mut−/− mice [>12% (n = 4)]. (B) Average iopamidol uptake with time for HP Mut+/− and HP Mut−/− mice. Similar to RD Mut+/− mice, >25% contrast was observed for HP Mut+/− mice. HP Mut−/− mice displayed the lowest contrast [~4% (n = 3)], indicating the lowest iopamidol uptake for these mice with more advanced kidney disease. (C and D) Corresponding time-averaged maximum contrast images of RD Mut+/− and RD Mut−/− mice. (E and F) Maximum contrast images of HP Mut+/− and HP Mut−/− mice.
Next, we evaluated a 72-offset protocol (ideal for characterizing Z-spectra) on the RD Mut+/− and RD Mut−/− mice. Representative Z-spectra from regions of interest (ROIs) drawn over both kidneys are shown in fig. S5. As can be seen, there are differences in the ST ratio for these animals, indicating that there are differences in average pH values. However, as shown in Fig. 3, using 72 offsets and our rapid imaging with refocused echoes (RARE) sequence parameters, the contrast-to-noise ratio (CNR) is too low to reliably assess filtration fraction (FF) even for the strong contrast Mut+/− control mice, with an example mouse showing FF = 84% in Fig. 3A, let alone for the lower average contrast RD Mut−/− or HP Mut−/− mice. For pH mapping, which requires calculating the ST ratio, the CNR becomes a bigger problem, as shown in Fig. 3B, as a large percentage of the pixels for the RD Mut−/− mice do not show sufficient contrast. We also characterized the B0 homogeneity for these mice, as shown in fig. S6. The B0 variation across the kidneys is very small (average variation, ~60 Hz) compared to the saturation power B1 = 4 μT (ω1 = 1073 Hz). Because of this, we chose to test a minimal saturation frequency protocol for pH mapping, two offsets, which can provide an increase in CNR by factors as large as . As seen in Fig. 3C, the FF becomes larger in the perfusion map because of this additional CNR. Furthermore, the pH maps in Fig. 3D become much sharper because of the improved contrast using this protocol. On the basis of this set of data, we decided that the two-offset protocol had excellent performance and proceeded to comparing the results of this protocol on all groups, including the lowest contrast HP Mut−/− mice. Using this two-offset protocol allows the time resolution to be very short (10 s per time point), allowing the use of a moving time average to reduce the systematic and random contributions to noise as shown. A comparison of 2-, 38-, and 72-offset protocols is shown in fig. S10.
Fig. 3. Comparison of 72-offset and 2-offset acquisition methods for producing FF and pH maps.
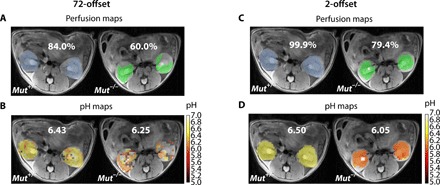
(A and B) The 72-offset and (C and D) 2-offset data of RD Mut+/− and RD Mut−/− mice, with the blue and green colors in the perfusion maps used to denote the RD Mut+/− and RD Mut−/−mice, respectively. (A) Perfusion maps to calculate FF using the 72-offset protocol. The RD Mut+/− mouse has an FF of 84%, whereas the RD Mut−/− mouse has an FF of 60%, indicating that fewer pixels were perfused with contrast agent. (B) pH maps generated using the 72-offset protocol and the calibration plot (Fig. 2). The RD Mut+/− mouse was observed to have a pH of 6.43, and the RD Mut−/− mouse has a pH of 6.25. (C) Two-offset perfusion maps for calculating FF based on taking the time average for 10 images. The RD Mut+/− mouse has an FF of 99.9%, and the RD Mut−/− mouse has an FF of 79.4%. FF calculated using the 2-offset protocol was approximately 20% higher than that calculated using the 72-offset protocol. (D) pH maps obtained using the two-offset protocol for RD Mut+/− and RD Mut−/− mice. pH values calculated were 6.50 and 6.05, respectively.
Comparison of perfusion results across all groups of MMA mice
To assess the capability of imaging iopamidol perfusion to detect kidney disease, we calculated FF using our CEST MRI data for the four groups of mice (Fig. 4). As can be seen in Fig. 4 (A, B, and D), both RD and HP Mut+/− controls (n = 5) display FF >98%. In contrast, moderate kidney disease mice (n = 4) display a moderate reduction in iopamidol perfusion (FF, ~79%), and severe kidney disease mice (n = 3) display a very substantial reduction in iopamidol perfusion (FF, <50%). Furthermore, as shown in Fig. 4A, the FF was linearly correlated with mouse weight when considering all groups (R = 0.76). The correlation improves further by considering only the RD and HP Mut−/− mice (R = 0.9; fig. S7B), which is due, in part, to the lower average weight of the control Mut+/− mice than the Mut−/− mice.
Fig. 4. In vivo FF results for MMA mice.
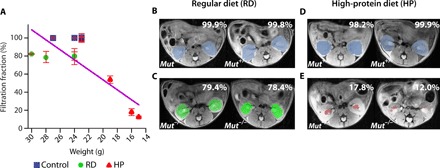
(A) FF calculated by taking the time-averaged FF for 10 images. The FF is defined as the percentage of kidney pixels with >20% of maximum contrast, and the SD in FF is calculated by comparing the 10 time points used. RD and HP Mut+/− controls display FF of ~100% (n = 5), whereas HP Mut−/− mice show the lowest FF, indicating that FF could be a CEST MRI metric for detecting renal disease. The blue, green, and red colors in the perfusion maps are used to denote the Mut+/−, RD Mut−/−, and HP Mut−/− mice, respectively. The decrease in FF was in accordance with a decrease in weight of the mice, which is a secondary indicator of disease progression for these mice. A linear correlation was observed between weights of mice and FF with R = 0.86 (for all groups of mice) and R = 0.9 (only considering the RD and HP Mut−/− mice; fig. S9B). (B and C) Time-averaged FF images of RD mice. (D and E) Time-averaged FF images of HP mice. RD Mut−/− mice (n = 4) have lower FF than RD Mut+/− controls but have higher FF than HP Mut−/− mice (n = 3).
Comparison of pH mapping results across MMA mice
To establish the suitability of our MRI protocol, we calculated pH maps using our CEST MRI data for the four groups of mice (Fig. 5). Both RD and HP Mut+/− mice displayed homogeneous pH values of 6.50 across the entire slice, as shown in Fig. 5 (A, B, D, and E) and fig. S7. In contrast, RD Mut−/− mice displayed a lower average pH (~6.1) and an order of magnitude larger range of pH values (denoted as ΔpH) in the kidney across the slice (±0.09; Fig. 5, A and C, and fig. S8, A to D). Furthermore, HP Mut−/− mice displayed a slightly lower pH (~6.0) and a significantly larger range of pH values in the kidney across the slice (±0.45; Fig. 5, D and F, and fig. S8, E to G). While the correlation between pH and mouse weight was poor (R = 0.0624; fig. S9C), the correlation between ΔpH and mouse weight was quite reasonable (R = 0.85; fig. S9D). Overall, these in vivo imaging data are consistent with the blood work and weights of these animals.
Fig. 5. In vivo pH results of two groups of mice maintained on RD and HP diets.
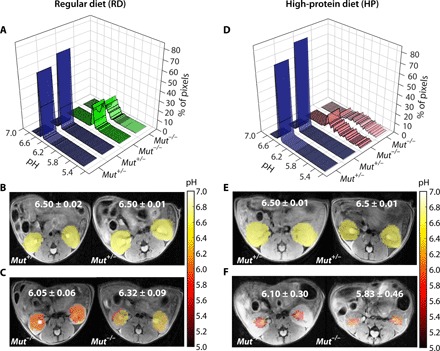
(A) pH histograms calculated for two representative RD Mut+/− and two representative RD Mut−/− mice. For Mut+/− mice (n = 5), more than 80% of the detected pixels display pH = 6.50, whereas for RD Mut−/− mice (n = 4), an acidic pH of 6.05 to 6.32 was observed for >20% of the kidney pixels. (B and C) Time-averaged pH maps of these RD Mut+/− and RD Mut−/− mice. The average pH dropped to 6.05 to 6.32 for RD Mut−/− compared to 6.50 for RD Mut+/− controls. The pH ranges of RD Mut−/− mice were ±0.06 and ±0.09 pH units compared to ±0.02 and ±0.01 pH units for RD Mut+/− mice. (D) pH histograms calculated for two representative HP Mut+/− mice and two representative HP Mut−/− mice. For HP Mut+/− mice, more than 80% of the detected pixels display a mean pH of 6.50, whereas for HP Mut−/− mice (n = 3), an acidic mean pH of 6.10 to 5.83 was observed. (E and F) Time-averaged pH images of Mut+/−and Mut−/− controls of HP mice. pH was further lowered to 5.83 for the most severely diseased mice. The pH was distributed over a narrow range of 6.50 ± 0.02 for both RD and HP Mut+/− mice, while this range significantly increased to ±0.30 and ±0.46 along with a decrease in mean pH for HP Mut−/− mice. A linear correlation was observed for RD and HP Mut−/− mice between ΔpH and mouse weight with R = 0.85 (fig. S9D).
DISCUSSION
We have successfully developed a CEST MRI protocol based on administration of iopamidol and using a two-offset protocol to characterize the perfusion and pH changes observed in mice and to detect differences in renal function. The metric that we use for perfusion, FF, estimates how widely iopamidol perfuses through the kidneys with units of % as opposed to GFR, which is a rate constant with units of ml/min. We chose to use FF because of the minimal modeling needed to measure this metric. To measure GFR using MRI, it is conventional to use images of the abdominal aorta to determine the arterial input function, which can lead to errors (34). Our CEST MRI protocol was designed by minimizing the number of frequencies collected to allow smoothing and signal averaging to overcome the challenges produced by the significant reduction in contrast observed for the severe kidney disease mice and kidney motion and allow for robust measurements. Measurement of GFR using iopamidol would have been more challenging for these animals. As shown in fig. S4C, the most sensitive measurement of health was mouse weight, which dropped over time when mice were placed on an HP diet. Three of the four MRI metrics that we developed correlated with this measurement (R = 0.93, 0.90, and 0.85, respectively, for contrast, FF, and ΔpH).
At this stage, a number of MRI pH imaging studies have been performed, including on the bladder, within tumors, and within kidneys (18, 30). Most of the kidney pH imaging studies have focused on testing probes on healthy control rodents, and the mean renal pH values of healthy control mice measured in this study are very similar to those reported (19, 28, 32, 35). In some cases, subtle differences were observed between calyx and cortex; we did not detect these in our maps using either the 72-offset or the 2-offset protocol. Our study was performed on a higher field strength scanner (11.7 T) than the other studies, which results in a larger frequency difference between labile protons and water. This enhances the resolution of the CEST Z-spectrum, which affects the intensity ratio measurement and, hence, the pH calculation. Additional changes include our use of a reduced set of offsets and oversampling to reduce noise. Application of a moving time average, smoothening the impact of motion on signal intensities, will result in CEST contrast smearing. On the basis of our pH mapping results of healthy control using 72- and 2-offset protocols, the smearing has a negligible effect on pH calculation. This approach is sound and necessary to observe the small contrast effects in mice with severe renal disease. Additional refinement of the CEST MRI protocol by using radial sampling or other strategies may result in further improvements in CNR or may further reduce the influence of motion on the resulting maps.
Longo and colleagues (32) also studied how AKIs created using clamp occlusion of arteries affect pH and perfusion maps. They observed increases in average pH values peaking at 7.0 ± 0.2 and for shorter occlusion times (20-min occlusion) returning to normal within 1 week; for longer occlusion times, the pH remained elevated at 1 week after AKI (6.9 ± 0.2). For our study on a mouse model of MMA-induced CKD, the average pH dropped slightly, but this was not as sensitive as the variation in pH across the kidneys in our study, which was not reported for short or long occlusion AKIs. Longo and colleagues also reported large changes in FF for AKI (lowest FF ~56% for short occlusion AKI and lowest FF ~38% for longer occlusion AKI versus FF ~84% for healthy controls); however, they used a different definition to calculate FF. Their definition of FF computed the percentage of pixels within the kidneys displaying larger than 2% contrast, whereas our definition calculated the number of pixels above 20% of the peak contrast observed in the kidneys. We prefer our definition as, in principle, our definition using this relative contrast should allow FF to be more comparable on scanners with different field strengths.
There are a number of CEST agents available (36–44); we selected iopamidol for several reasons. First, pH is an appropriate biomarker for kidney disease (21), and because iopamidol has two labile protons with distinct shifts and exchange rates, ratiometric protocols can be used to isolate pH changes from concentration. The moderately large chemical shifts (>3 ppm) enable detection on clinical 3-T MRI scanners (28, 32, 36, 45, 46). Iopamidol is a nonionic contrast agent safely administered for over 30 years to patients for angiography, including to a large number of patients known to have kidney disease (47, 48). Nonionic iodinated agents are in routine use in pediatric radiology, including pediatric computed tomography (CT) and excretory urography. In one study on more than 11,000 pediatric injections, allergic reactions were found to be rare (estimated to be 0.18% with 80% considered mild) (49, 50). The nephrotoxicity of iopamidol has been studied in the case of children with bone marrow transplantations who received nephrotoxic drugs and received between one and seven repeated injections of iopamidol and judged to be negligible (51). While we used a dose of 1.5 g of iodine per kilogram in this study, as shown in the results, the resulting contrast was well above what was needed to differentiate between the groups.
In addition to the methods we present in this work, other noninvasive imaging modalities can be used for assessment of renal function, including gadolinium-based MR renography, CT, and nuclear imaging. MR renography can derive functional parameters that estimate GFR (52); however, there are challenges for widespread adoption because of the known safety issues of gadolinium for low-GFR patients, which can lead to nephrogenic systemic fibrosis (53). The number and volume of glomeruli in the kidneys can also be assessed using MRI to detect ferritin-labeled glomerular basement membrane, which is another alternative, although it might be difficult to perform this serially (16). Dynamic CT using iodinated imaging agents can provide renal clearance data, which correlates well with GFR (54, 55); however, this method uses ionizing radiation, which can provide a notable burden in the case that this measurement should be repeated periodically and cannot measure pH, which is an additional biomarker for renal function. Nuclear imaging techniques using probes such as 99mTc-DTPA and 99mTc-MAG3 (mercaptoacetyltriglycine) are other options that are currently in clinical use; however, these probes can produce misleading results due to many factors including ROI selection, motion artifacts, and others (13, 56). MRI has regularly been used to produce high spatial resolution images of the kidneys, but the MRI metrics that we evaluated in the current study, FF and pH maps, proved more sensitive to differences between the control and moderate kidney disease mice than other renal function measurements performed previously including, importantly, mGFR in this mouse model. CEST MRI has been translated to patients for detecting endogenous biomarkers associated with tumor aggressiveness (57) and for highlighting tumor microenvironment following injection of exogenous CEST agents (36, 58). On the basis of our results, this new CEST MRI protocol is promising for assessing disease progression over time or for treatment responses in animal models of kidney disease and could be immediately translated to patients with a range of disorders that, similarly to MMA, cause CKD.
METHODS
In vitro phantom preparation
Iopamidol (molecular weight, 777.1) was obtained from Bracco Imaging (Italy). Seronorm (5 ml), a normalized human blood serum, was purchased from Sero (USA). A set of 40 mM iopamidol phantoms was prepared by carefully dissolving Seronorm in 10 ml of deionized water with 40 mM iopamidol to avoid the formation of foam. Titrations were performed to produce pH values of 5.3, 5.5, 5.7, 5.9, 6.1, 6.3, 6.5, 6.7, 6.9, 7.1, and 7.3, respectively, in aliquots of 0.3 ml. These solutions were placed in 5-mm nuclear magnetic resonance tubes and assembled along with 0.3-ml serum-only control tube to obtain the iopamidol-serum phantom for CEST MRI.
In vitro MR data acquisition
MRI experiments were performed on a Bruker 11.7-T vertical bore scanner using a 25-mm transmit/receive volume coil. CEST images were acquired using continuous-wave RF saturation pulse of 3 s followed by a RARE imaging sequence. CEST data were acquired using six different saturation power (B1) from 1 to 6 μΤ for optimization. Seventy-one CEST offsets between ±7 ppm with 0.2-ppm difference were acquired to produce saturation images and Z-spectra plus 1 at +20,000 Hz as M0. Other sequence parameters include the following: centric encoding; acquisition matrix size, 64 × 48; slice thickness, 1-mm slice; and repetition time (TR)/echo time (TE), 5000/3.39 ms. For pixel-by-pixel B0 inhomogeneity calculation, water saturation shift referencing (WASSR) images were calculated using B1 = 0.5 μΤ with 42 offsets from −1.5 to +1.5 ppm.
In vitro pH calculation
We performed all postprocessing and pH calculations using MATLAB (MathWorks, USA) for our phantom. This was based on calculating the ST ratio at 4.2 and 5.5 ppm, as described previously (33). On the basis of examining the data at six B1 values, CEST images with B1 = 3 and 4 μΤ were processed to generate the pH maps. Pixel-by-pixel Z-spectra were also calculated from the CEST data. Pixel-by-pixel B0 inhomogeneity corrections were performed using the WASSR dataset and spline interpolation method, as described previously (23). The value ST = (1 − Mz/M0) was calculated pixel by pixel to generate ST maps corresponding to the two CEST peaks at 4.2 and 5.5 ppm, respectively. An ROI was drawn, and the mean ST ratio = versus pH plot was obtained and fit to a polynomial to generate a pH calibration curve. Using this calibration equation, the pH for each pixel in the phantom could then be calculated to determine how susceptible this quantity was to experimental noise.
In vitro ST ratio tolerance to B0 inhomogeneity
In vitro experiments on the iopamidol-serum phantom were analyzed to study the tolerance of our pH measurements to ΔB0 (Hz). The ST ratio of 4.2 and 5.5 ppm was calculated for each pH between 5.7 and 7.3 for ΔB0 values up to ±301.4 Hz in steps of 27.4 Hz using our experimental Z-spectra. Using the in vitro calibration equation, corresponding pH values were obtained at ΔB0 = 0, ±60, ±120, and ±180 Hz for ω1 = 3 μT and ω1 = 4 μT.
MMA mouse model for CKD
Male transgenic mice expressing methylmalonyl–coenzyme A–mutase (Mut) in the liver under the control of an albumin promoter on a knockout background Mut−/−;TgINS-Alb-Mut, designated as Mut−/− (n = 8), or littermate controls, Mut+/− (n = 5), were used. The mice were further subdivided by placing half on a regular mouse chow diet (RD) and the remainder on an HP diet, containing 70% (w/w) casein (HP), as described previously (TD.06723; Harlan Laboratories). HP mice lost weight (P < 0.0001) and had elevated plasma MMA (1330 μM; P = 0.017 compared to controls), associated with increased Lcn2 mRNA expression in their kidneys (P = 0.002), similar to previous findings in this model. Notably, no significant renal histological changes were evident on hematoxylin and eosin staining at this stage of renal disease in these mice. Exposure to an HP diet for 6 months was necessary to induce tubulointerstitial nephritis, as has been previously described (1). GFR measurements, on the other hand, were abnormal even on RD, when measured by 125I-iothalamate clearance in single-nephron microperfusion studies, and significantly decreased on an HP diet after 2 months, as assessed by the FITC inulin plasma decay method at the whole-animal level (1).
In vivo MR data acquisition and experimental design
All animal experiments were performed under a protocol approved by both the Johns Hopkins University Animal Care and Use Committee and the Animal Care and Use Committee of the National Human Genome Research Institute (NHGRI), National Institutes of Health (NIH). Each mouse was placed on a mouse tail illuminator restrainer to catheterize the tail vein. In vivo experiments were performed on an 11.7-T Bruker BioSpec horizontal scanner using an eight-channel mouse body phase array coil. The mouse was then placed on a cradle for imaging and anesthetized using 0.5 to 1.5% isoflurane, with the respiration rate continuously monitored. The body temperature was maintained at 37°C using a hot water circulation bed. A three-plane localizer sequence was used for finding the spatial location of the mouse kidneys. A multislice T2W RARE sequence was used to acquire the axial slice images. Twenty-one contiguous axial slices, each with a thickness of 1.5 mm, were collected using TE/TR of 6.6 ms/4 s, RARE factor of 8, and field of view of 128 × 128. A center slice of width 1.5 mm was chosen for CEST imaging, and high-resolution T2W image was obtained using RARE for anatomical overlay with CEST processed images. The acquisition time for high-resolution T2W images was 2 min and 8 s with a matrix size of 256 × 256. For CEST data acquisition, the RARE pulse sequence was used. Saturation pulses with B1 = 4 μΤ were applied for a total duration of 3 s, which consisted of 10 rectangular block pulses 300 ms long with a 10-μs delay between the pulses. WASSR images were collected for generating B0 maps using 42 offsets from −1.5 to +1.5 ppm and B1 = 1.5 μT. For the 72-offset CEST data acquisition, the offset frequency was incremented between ±7 ppm with an interval of 0.2 ppm for 1 to 2 hours after injection. The time taken for each set of 72 offsets was 6 min and 0 s, and the other sequence parameters were as follows: TE/TR, 3.49 ms/5 s; number of averages, 1; RARE factor, 32; matrix size, 48 × 48; field of view, 28 × 20 mm2; spatial resolution, 0.58 × 0.41 mm2; and centric encoding. Three sets of 72 offsets were collected with the same experimental parameters before administration of iopamidol. Iopamidol was injected through the tail vein at a dose of 1.5 g of iodine per kilogram. For the two-frequency offset data acquisition, the offset frequency was toggled between 4.2 and 5.5 ppm repeatedly, and CEST images at these two offsets were collected for 1 to 2 hours just after administering iopamidol. The time taken for each CEST image acquisition was 5 s. Eight to 10 pre-injection sets of 4.2 and 5.5 ppm images were collected. Field of view was 30 × 20 mm2, in-plane spatial resolution was 0.62 × 0.41 mm2, and other parameters were the same as in the 72-offset protocol.
MRI data analysis of contrast maps and FF
CEST MRI data (both before and after injection) were extracted using custom-written MATLAB functions to calculate the Z-spectra of mean pre-injection images. For the analysis of the results, the mean pre-injection Z-spectra were subtracted from all post-injection images. The “smooth” function in MATLAB was used to apply a moving average filter at each frequency. This moving average filter was applied on the time series data to remove fluctuations in signal due to motion. The smoothening factor used in the averaging process was ~40 neighboring images, with this choice based on the size of the data and the range of fluctuations. After smoothening, 10 to 20 images were averaged to generate the corresponding parameter maps (contrast, FF, and pH). Images acquired between 40 and 50 min after injection were considered for the calculation of parametric maps on Mut+/− and Mut−/− mice. For calculating the contrast and FF, CEST images at 4.2 ppm were used. Time-contrast uptake curves were obtained by calculating the average ST over pixels selected by two ROIs, one over each kidney. Maximum contrast maps were generated by calculating the pixel-by-pixel maximum ST on two ROIs, one over each kidney, overlaid on the corresponding T2W image. The time-averaged maximum contrast images were calculated by taking 10 to 20 images at corresponding post-injection time. The FF was calculated by determining the percentage of pixels in the kidneys with a contrast >20% of the maximum contrast detected at 4.2 ppm for each mouse. The time-averaged FF over 10 images was used to generate the FF bar plot.
In vivo MRI pH calculation
In vivo pH was calculated in a similar manner to the in vitro pH calculation. The calibration equation used was pH = p1 × (ST ratio)3 + p2 × (ST ratio)2 + p3 × (ST ratio)1 + p4 with p1 = − 0.0317, p2 = 0.4463, p3 = − 2.5029, and p4 = 20.5146. The root mean square error of the fit is 0.0714. From the images corresponding to the two offsets, 4.2 and 5.5 ppm over time, ~16 images were selected, with the initial image occurring 40 to 50 min after injection for both Mut+/− and Mut−/− mice for time averaging. The pixel-by-pixel ST ratio was calculated for these images, and using the in vitro calibration equation, the pixel-by-pixel pH was calculated. This was masked using the >20% contrast criteria found for the FF pixels. Mean pH maps were generated, and the mean pH range observed was calculated by averaging the pH of the images selected. Histogram plots of pH for both RD and HP diet mice were calculated for the pH images by keeping the bin size of 200 between the pH values of 4 and 10. After producing the histograms, these data were fit to a normal distribution (Gaussian function was used) to obtain the mean or the most probable pH and range in pH for each mouse. The full width at half maximum of this Gaussian function was used to quantify the ΔpH values across the kidneys.
In vivo B0 inhomogeneity maps
In vivo B0 inhomogeneity maps were generated from the WASSR experiment performed before the injection for each mouse. WASSR Z-spectra for ROIs drawn over both the kidneys were extracted and interpolated in MATLAB using cubic spline interpolation. The water shift was measured pixel by pixel, as described previously, to obtain the ΔB0 maps over the kidney. ΔB0 maps were overlaid on the corresponding high-resolution T2W images to obtain the ΔB0 maps shown in fig. S6.
Blood and histological analysis of renal tissue
Before and after the MR imaging session, the weights of each mouse were determined. Blood samples were collected serially before and at 1 and 2 months on the RD or HP diet. The collected blood was centrifuged for 15 to 20 min at 3000 rpm to separate plasma from cells. Mice were sacrificed to extract the renal tissues, and axial cut portions of left and right kidneys were fixed separately using 4% paraformaldehyde solution and stored in a 4°C refrigerator. Other axial cut portions were frozen at −80°C for RNA extraction and real-time polymerase chain reaction to assess Lcn2 (or neutrophil gelatinase–associated lipocalin) mRNA expression (TaqMan probes used: Mm01324470_m1, for Lcn2, normalized to Gapdh expression: Mm99999915_g1), as previously described.
Supplementary Material
Acknowledgments
We thank I. Ginty and D. Romero (NHGRI, NIH) for skilled assistance with mouse maintenance and transport. Funding: This project was supported by the Maryland Stem Cell Research Foundation (grant 2829), the NIH (grant P41EB024495), and the Intramural Research Program of the NHGRI, NIH. Author contributions: K.P. and M.T.M. conceived the project, designed the experiments, and wrote the manuscript with input from all authors. K.P., A.P., and I.M. performed the experiments. Y.L. assisted with animal studies. I.M., A.P., H.J.V., and C.P.V. provided expertise on MMA, assisted with the design of animal studies, and performed disease-related measurements. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Correspondence and requests for materials should be addressed to M.T.M. The mouse models can be provided by C.P.V., pending scientific review and a completed material transfer agreement. Requests for the mouse models should be submitted to C.P.V.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaaw8357/DC1
Fig. S1. In vitro results at RF saturation field strength of 3 μΤ.
Fig. S2. In vitro pH measurement variation as a function of ΔΒ0 shift at 4-μΤ RF saturation power.
Fig. S3. In vitro pH measurement variation with ΔΒ0 shift at 3-μΤ RF saturation power.
Fig. S4. Biochemical and clinical measures in the RD and HP Mut−/−;TgINS-Alb-Mut mice as compared to heterozygote littermates.
Fig. S5. In vivo Z-spectra.
Fig. S6. Representative ΔB0 maps for RD and HP diet mice.
Fig. S7. pH-histogram plots depicting the percentage of pixels across the detectable pH range and normal distribution analysis for all control (Mut+/−) mice imaged in this study.
Fig. S8. pH-histogram plots depicting the percentage of pixels across the detectable pH range and normal distribution analysis for all Mut−/− mice imaged in this study.
Fig. S9. MRI metrics versus weight correlation plots for HP and RD Mut−/− mice.
Fig. S10. pH images calculated for a healthy control mouse using our 72, 38, and 2-offset protocols.
REFERENCES AND NOTES
- 1.Manoli I., Sysol J. R., Li L., Houillier P., Garone C., Wang C., Zerfas P. M., Cusmano-Ozog K., Young S., Trivedi N. S., Cheng J., Sloan J. L., Chandler R. J., Abu-Asab M., Tsokos M., Elkahloun A. G., Rosen S., Enns G. M., Berry G. T., Hoffmann V., DiMauro S., Schnermann J., Venditti C. P., Targeting proximal tubule mitochondrial dysfunction attenuates the renal disease of methylmalonic acidemia. Proc. Natl. Acad. Sci. U.S.A. 110, 13552–13557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler R. J., Venditti C. P., Adenovirus-mediated gene delivery rescues a neonatal lethal murine model of mut(0) methylmalonic acidemia. Hum. Gene Ther. 19, 53–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manoli I., Sysol J. R., Epping M. W., Li L., Wang C., Sloan J. L., Pass A., Gagne J., Ktena Y. P., Li L., Trivedi N. S., Ouattara B., Zerfas P. M., Hoffmann V., Abu-Asab M., Tsokos M. G., Kleiner D. E., Garone C., Cusmano-Ozog K., Enns G. M., Vernon H. J., Andersson H. C., Grunewald S., Elkahloun A. G., Girard C. L., Schnermann J., DiMauro S., Andres-Mateos E., Vandenberghe L. H., Chandler R. J., Venditti C. P., FGF21 underlies a hormetic response to metabolic stress in methylmalonic acidemia. JCI Insight 3, 124351 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.I. Manoli, J. L. Sloan, C. P. Venditti, Isolated methylmalonic acidemia, in GeneReviews, M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. Stephens, A. Amemiya, Eds. (University of Washington, 1993). [Google Scholar]
- 5.Hörster F., Baumgartner M. R., Viardot C., Suormala T., Burgard P., Fowler B., Hoffmann G. F., Garbade S. F., Kölker S., Baumgartner E. R., Long-term outcome in methylmalonic acidurias is influenced by the underlying defect (mut0, mut-, cblA, cblB). Pediatr. Res. 62, 225–230 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Stevens L. A., Levey A. S., Measured GFR as a confirmatory test for estimated GFR. J. Am. Soc. Nephrol. 20, 2305–2313 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Schwartz G. J., Furth S. L., Glomerular filtration rate measurement and estimation in chronic kidney disease. Pediatr. Nephrol. 22, 1839–1848 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Miller W. G., Reporting estimated GFR: A laboratory perspective. Am. J. Kidney Dis. 52, 645–648 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Filler G., Lepage N., Should the Schwartz formula for estimation of GFR be replaced by cystatin C formula? Pediatr. Nephrol. 18, 981–985 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Bouvet Y., Bouissou F., Coulais Y., Seronie-Vivien S., Tafani M., Decramer S., Chatelut E., GFR is better estimated by considering both serum cystatin C and creatinine levels. Pediatr. Nephrol. 21, 1299–1306 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Walter J. H., Michalski A., Wilson W. M., Leonard J. V., Barratt T. M., Dillon M. J., Chronic renal failure in methylmalonic acidaemia. Eur. J. Pediatr. 148, 344–348 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Kruszka P. S., Manoli I., Sloan J. L., Kopp J. B., Venditti C. P., Renal growth in isolated methylmalonic acidemia. Genet. Med. 15, 990–996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan X., Zhang J., Tang K., Quan C., Tian Y., Li H., Ao G., Qiu L., Determination of glomerular filtration rate with CT measurement of renal clearance of iodinated contrast material versus 99mTc-DTPA dynamic imaging “gates” method: A validation study in asymmetrical renal disease. Radiology 282, 552–560 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Zhang J. L., Morrell G., Rusinek H., Sigmund E. E., Chandarana H., Lerman L. O., Prasad P. V., Niles D., Artz N., Fain S., Vivier P. H., Cheung A. K., Lee V. S., New magnetic resonance imaging methods in nephrology. Kidney Int. 85, 768–778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusinek H., Boykov Y., Kaur M., Wong S., Bokacheva L., Sajous J. B., Huang A. J., Heller S., Lee V. S., Performance of an automated segmentation algorithm for 3D MR renography. Magn. Reson. Med. 57, 1159–1167 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Beeman S. C., Zhang M., Gubhaju L., Wu T., Bertram J. F., Frakes D. H., Cherry B. R., Bennett K. M., Measuring glomerular number and size in perfused kidneys using MRI. Am. J. Physiol. Renal Physiol. 300, F1454–F1457 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Gillies R. J., Raghunand N., Garcia-Martin M. L., Gatenby R. A., pH imaging. A review of pH measurement methods and applications in cancers. IEEE Eng. Med. Biol. Mag. 23, 57–64 (2004). [DOI] [PubMed] [Google Scholar]
- 18.van Sluis R., Bhujwalla Z. M., Raghunand N., Ballesteros P., Alvarez J., Cerdán S., Galons J. P., Gillies R. J., In vivo imaging of extracellular pH using 1H MRSI. Magn. Reson. Med. 41, 743–750 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Raghunand N., Howison C., Sherry A. D., Zhang S., Gillies R. J., Renal and systemic pH imaging by contrast-enhanced MRI. Magn. Reson. Med. 49, 249–257 (2003). [DOI] [PubMed] [Google Scholar]
- 20.M. T. McMahon, A. A. Gilad, J. W. M. Bulte, P. C. M. van Zijl, Chemical Exchange Saturation Transfer Imaging: Advances and Applications (Pan Stanford Publishing, ed. 1, 2017), 479 pp. [Google Scholar]
- 21.Pavuluri K., McMahon M. T., pH imaging using chemical exchange saturation transfer (CEST) MRI. Isr. J. Chem. 57, 862–879 (2017). [Google Scholar]
- 22.Liu G., Moake M., Har-el Y.-e., Long C. M., Chan K. W. Y., Cardona A., Jamil M., Walczak P., Gilad A. A., Sgouros G., van Zijl P. C. M., Bulte J. W. M., McMahon M. T., In vivo multicolor molecular MR imaging using diamagnetic chemical exchange saturation transfer liposomes. Magn. Reson. Med. 67, 1106–1113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu G., Gilad A. A., Bulte J. W. M., van Zijl P. C. M., McMahon M. T., High-throughput screening of chemical exchange saturation transfer MR contrast agents. Contrast Media Mol. Imaging 5, 162–170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon M. T., Gilad A. A., DeLiso M. A., Berman S. M., Bulte J. W., van Zijl P. C., New “multicolor” polypeptide diamagnetic chemical exchange saturation transfer (DIACEST) contrast agents for MRI. Magn. Reson. Med. 60, 803–812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terreno E., Castelli D. D., Aime S., Encoding the frequency dependence in MRI contrast media: The emerging class of CEST agents. Contrast Media Mol. Imaging 5, 78–98 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Viswanathan S., Ratnakar S. J., Green K. N., Kovacs Z., De León-Rodriguez L. M., Sherry A. D., Multi-frequency PARACEST agents based on europium(III)-DOTA-tetraamide ligands. Angew. Chem. Int. Ed. Engl. 48, 9330–9333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward K., Balaban R., Determination of pH using water protons and chemical exchange dependent saturation transfer (CEST). Magn. Reson. Med. 44, 799–802 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Wu Y., Zhou I. Y., Igarashi T., Longo D. L., Aime S., Sun P. Z., A generalized ratiometric chemical exchange saturation transfer (CEST) MRI approach for mapping renal pH using iopamidol. Magn. Reson. Med. 79, 1553–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon M. T., Gilad A. A., Zhou J. Y., Sun P. Z., Bulte J. W. M., van Zijl P. C. M., Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): pH calibration for poly-L-lysine and a starburst dendrimer. Magn. Reson. Med. 55, 836–847 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X., Song X., Ray Banerjee S., Li Y., Byun Y., Liu G., Bhujwalla Z. M., Pomper M. G., McMahon M. T., Developing imidazoles as CEST MRI pH sensors. Contrast Media Mol. Imaging 11, 304–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan K. W., Liu G., Song X., Kim H., Yu T., Arifin D. R., Gilad A. A., Hanes J., Walczak P., van Zijl P. C., Bulte J. W., McMahon M. T., MRI-detectable pH nanosensors incorporated into hydrogels for in vivo sensing of transplanted-cell viability. Nat. Mater. 12, 268–275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longo D. L., Cutrin J. C., Michelotti F., Irrera P., Aime S., Noninvasive evaluation of renal pH homeostasis after ischemia reperfusion injury by CEST-MRI. NMR Biomed. 30, (2017). [DOI] [PubMed] [Google Scholar]
- 33.Longo D. L., Busato A., Lanzardo S., Antico F., Aime S., Imaging the pH evolution of an acute kidney injury model by means of iopamidol, a MRI-CEST pH-responsive contrast agent. Magn. Reson. Med. 70, 859–864 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Cutajar M., Mendichovszky I. A., Tofts P. S., Gordon I., The importance of AIF ROI selection in DCE-MRI renography: Reproducibility and variability of renal perfusion and filtration. Eur. J. Radiol. 74, E154–E160 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Wu Y. K., Zhang S. R., Soesbe T. C., Yu J., Vinogradov E., Lenkinski R. E., Sherry A. D., pH imaging of mouse kidneys in vivo using a frequency-dependent paraCEST agent. Magn. Reson. Med. 75, 2432–2441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones K. M., Randtke E. A., Yoshimaru E. S., Howison C. M., Chalasani P., Klein R. R., Chambers S. K., Kuo P. H., Pagel M. D., Clinical translation of tumor acidosis measurements with AcidoCEST MRI. Mol. Imaging Biol. 19, 617–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anemone A., Consolino L., Longo D. L., MRI-CEST assessment of tumour perfusion using x-ray iodinated agents: Comparison with a conventional Gd-based agent. Eur. Radiol. 27, 2170–2179 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Longo D. L., Michelotti F., Consolino L., Bardini P., Digilio G., Xiao G., Sun P. Z., Aime S., In vitro and in vivo assessment of nonionic iodinated radiographic molecules as chemical exchange saturation transfer magnetic resonance imaging tumor perfusion agents. Invest. Radiol. 51, 155–162 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Yang X., Yadav N. N., Song X., Ray Banerjee S., Edelman H., Minn I., van Zijl P. C., Pomper M. G., McMahon M. T., Tuning phenols with intra-molecular bond shifted hydrogens (IM-SHY) as diaCEST MRI contrast agents. Chemistry 20, 15824–15832 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song X., Yang X., Ray Banerjee S., Pomper M. G., McMahon M. T., Anthranilic acid analogs as diamagnetic CEST MRI contrast agents that feature an intramolecular-bond shifted hydrogen. Contrast Media Mol. Imaging 10, 74–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bar-Shir A., Liu G., Liang Y., Yadav N. N., McMahon M. T., Walczak P., Nimmagadda S., Pomper M. G., Tallman K. A., Greenberg M. M., van Zijl P. C. M., Bulte J. W. M., Gilad A. A., Transforming thymidine into a magnetic resonance imaging probe for monitoring gene expression. J. Am. Chem. Soc. 135, 1617–1624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castelli D. D., Terreno E., Longo D., Aime S., Nanoparticle-based chemical exchange saturation transfer (CEST) agents. NMR Biomed. 26, 839–849 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Hingorani D. V., Montano L. A., Randtke E. A., Lee Y. S., Cardenas-Rodriguez J., Pagel M. D., A single diamagnetic catalyCEST MRI contrast agent that detects cathepsin B enzyme activity by using a ratio of two CEST signals. Contrast Media Mol. Imaging 11, 130–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Evbuomwan O. M., Tieu M., Zhao P., Martins A. F., Sherry A. D., Protonation of carboxyl groups in EuDOTA-tetraamide complexes results in catalytic prototropic exchange and quenching of the CEST signal. Philos. Trans. A Math. Phys. Eng. Sci. 375, 20170113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller-Lutz A., Khalil N., Schmitt B., Jellus V., Pentang G., Oeltzschner G., Antoch G., Lanzman R. S., Wittsack H. J., Pilot study of iopamidol-based quantitative pH imaging on a clinical 3T MR scanner. MAGMA 27, 477–485 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Sun P. Z., Longo D. L., Hu W., Xiao G., Wu R., Quantification of iopamidol multi-site chemical exchange properties for ratiometric chemical exchange saturation transfer (CEST) imaging of pH. Phys. Med. Biol. 59, 4493–4504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomsen H. S., Morcos S. K., Risk of contrast-medium-induced nephropathy in high-risk patients undergoing MDCT—A pooled analysis of two randomized trials. Eur. Radiol. 19, 891–897 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Solomon R. J., Natarajan M. K., Doucet S., Sharma S. K., Staniloae C. S., Katholi R. E., Gelormini J. L., Labinaz M., Moreyra A. E., Cardiac angiography in renally impaired patients (CARE) study: A randomized double-blind trial of contrast-induced nephropathy in patients with chronic kidney disease. Circulation 115, 3189–3196 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Beckett K. R., Moriarity A. K., Langer J. M., Safe use of contrast media: What the radiologist needs to know. Radiographics 35, 1738–1750 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Dillman J. R., Strouse P. J., Ellis J. H., Cohan R. H., Jan S. C., Incidence and severity of acute allergic-like reactions to i.v. nonionic iodinated contrast material in children. AJR Am. J. Roentgenol. 188, 1643–1647 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Haight A. E., Kaste S. C., Goloubeva O. G., Xiong X. P., Bowman L. C., Nephrotoxicity of iopamidol in pediatric, adolescent, and young adult patients who have undergone allogeneic bone marrow transplantation. Radiology 226, 399–404 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Zhang J. L., Rusinek H., Bokacheva L., Lerman L. O., Chen Q., Prince C., Oesingmann N., Song T., Lee V. S., Functional assessment of the kidney from magnetic resonance and computed tomography renography: Impulse retention approach to a multicompartment model. Magn. Reson. Med. 59, 278–288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanal E., Tweedle M. F., Residual or retained gadolinium: Practical implications for radiologists and our patients. Radiology 275, 630–634 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Gaspari F., Perico N., Ruggenenti P., Mosconi L., Amuchastegui C. S., Guerini E., Daina E., Remuzzi G., Plasma clearance of nonradioactive iohexol as a measure of glomerular filtration rate. J. Am. Soc. Nephrol. 6, 257–263 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Tsushima Y., Blomley M. J. K., Kusano S., Endo K., Use of contrast-enhanced computed tomography to measure clearance per unit renal volume: A novel measurement of renal function and fractional vascular volume. Am. J. Kidney Dis. 33, 754–760 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Qi Y., Hu P., Xie Y., Wei K., Jin M., Ma G., Li Q., Xu B., Chen X., Glomerular filtration rate measured by (99m)Tc-DTPA renal dynamic imaging is significantly lower than that estimated by the CKD-EPI equation in horseshoe kidney patients. Nephrology 21, 499–505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou J. Y., Zhu H., Lim M., Blair L., Quinones-Hinojosa A., Messina S. A., Eberhart C. G., Pomper M. G., Laterra J., Barker P. B., van Zijl P. C. M., Blakeley J. O., Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J. Magn. Reson. Imaging 38, 1119–1128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X., Yadav N. N., Knutsson L., Hua J., Kalyani R., Hall E., Laterra J., Blakeley J., Strowd R., Pomper M., Barker P., Chan K. W. Y., Liu G., McMahon M. T., Stevens R. D., van Zijl P. C. M., Dynamic glucose-enhanced (DGE) MRI: Translation to human scanning and first results in glioma patients. Tomography 1, 105–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaaw8357/DC1
Fig. S1. In vitro results at RF saturation field strength of 3 μΤ.
Fig. S2. In vitro pH measurement variation as a function of ΔΒ0 shift at 4-μΤ RF saturation power.
Fig. S3. In vitro pH measurement variation with ΔΒ0 shift at 3-μΤ RF saturation power.
Fig. S4. Biochemical and clinical measures in the RD and HP Mut−/−;TgINS-Alb-Mut mice as compared to heterozygote littermates.
Fig. S5. In vivo Z-spectra.
Fig. S6. Representative ΔB0 maps for RD and HP diet mice.
Fig. S7. pH-histogram plots depicting the percentage of pixels across the detectable pH range and normal distribution analysis for all control (Mut+/−) mice imaged in this study.
Fig. S8. pH-histogram plots depicting the percentage of pixels across the detectable pH range and normal distribution analysis for all Mut−/− mice imaged in this study.
Fig. S9. MRI metrics versus weight correlation plots for HP and RD Mut−/− mice.
Fig. S10. pH images calculated for a healthy control mouse using our 72, 38, and 2-offset protocols.


