The histone modification reader ZCWPW1 guides male meiosis prophase I.
Abstract
Meiosis is a specialized type of cell division that creates haploid germ cells and ensures their genetic diversity through homologous recombination. We show that the H3K4me3 reader ZCWPW1 is specifically required for meiosis prophase I progression in male but not in female germ cells in mice. Loss of Zcwpw1 in male mice caused a complete failure of synapsis, resulting in meiotic arrest at the zygotene to pachytene stage, accompanied by incomplete DNA double-strand break repair and lack of crossover formation, leading to male infertility. In oocytes, deletion of Zcwpw1 only somewhat slowed down meiosis prophase I progression; Zcwpw1−/− oocytes were able to complete meiosis, and Zcwpw1−/− female mice had normal fertility until mid-adulthood. We conclude that the H3K4me3 reader ZCWPW1 is indispensable for meiosis synapsis in males but is dispensable for females. Our results suggest that ZCWPW1 may represent a previously unknown, sex-dependent epigenetic regulator of germ cell meiosis in mammals.
INTRODUCTION
Meiosis is a specialized type of cell division that creates haploid germ cells from diploid progenitors and ensures their genetic diversity through homologous recombination (1). During meiosis, recombination is initiated by programmed DNA double-strand breaks (DSBs) generated by SPO11 at sites determined by both higher-order chromosome structure and local chromatin conformation (2). Meiotic DSBs recruit a series of recombination proteins to form recombination foci and promote homologous chromosome synapsis during the zygotene stage (3). Subsequent invasion of the 3′ single-stranded DNA into the duplex of the homolog is implemented by recombinase DMC1 and RAD51 (4, 5). Recombination foci continue to mature while chromosomes are fully synapsed at the pachytene stage, and these are eventually resolved into crossover or noncrossover events (3).
During meiosis, histone H3 lysine 4 trimethylation (H3K4me3) modification is a common chromatin feature that defines recombination sites in mice and yeasts (6). The formation of meiosis-specific H3K4me3 modifications is reported to be dependent on PR domain containing 9 (PRDM9), which is the only known meiosis histone lysine methyltransferase with a PR/SET domain (7, 8). PRDM9 binds to specific DNA sequence through its zinc finger domain, and its methyltransferase domain then catalyzes H3K4me3 on the nearest nucleosome (9). Then, the DSB machinery containing SPO11 complex is recruited to hotspot sites to mediate DSB formation through a molecular mechanism that is not yet understood (3). However, the PRDM9-dependent H3K4me3 is elusive because this modification is present at many nonhotspot sites as well (10). In Prdm9−/− mice, DSBs were reported to localize improperly at PRDM9-independent H3K4me3-enriched sites in genome regulatory elements, such as promoters, and this caused a complete arrest of meiosis prophase I (11, 12).
Zinc finger CW-type and PWWP domain containing 1 (ZCWPW1) is a member of the CW domain–containing protein family. The CW domain is believed to take part in DNA binding and/or promoting protein-protein interactions in early embryonic development and in the chromatin methylation status that influences gene silencing, transcription, and other processes (13). The PWWP domain is also known to be involved in epigenetic regulation because it is present in diverse proteins involved in chromatin function, including histone-modifying enzymes, DNA-modifying enzymes, transcription factors, and DNA repair proteins (14, 15). The zinc finger CW (zf-CW) domain is a zinc-binding domain of about 60 amino acids with three conserved tryptophan and four conserved cysteine residues (13). Structural analysis showed that the zf-CW domain is a histone modification reader for the H3K4me3 (16). In the structure of the complex of the human zf-CW domain and the H3K4me3 peptide, the H3K4me3 peptide forms a three-stranded antiparallel β-sheet together with the two β-strands of the zf-CW domain (16). Chromatin pulldown analysis confirmed that the zf-CW domain recognizes H3K4me3 in vitro (17). It is noteworthy that there is structural similarity between the zf-CW domain and the plant homeo domain (PHD) finger, which plays an important role in the recognition of the histone H3 tail (18).
The zf-CW domain has been identified in several proteins that are involved in the control of the methylation state of the histone H3 tail. In Arabidopsis, the SDG8 (SET domain group 8) protein controls flowering time by recognizing methylation modifications on the histone tail and/or DNA through its zf-CW domain (19, 20). A zf-CW domain is also found in lysine demethylase 1B, which contributes to establishing DNA methylation imprints during oogenesis in mice (21). In addition, the zf-CW domain is observed in microrchidia (MORC) family members (22), and recently it was reported that MORC2B is essential for meiotic progression and fertility in mice (23). MORC2B is specifically expressed in mouse germ cells, and disruption of Morc2b causes meiotic arrest and infertility in both sexes of mice (23).
Despite our understanding of the epigenetic control mediated by zf-CW domain-containing proteins in various species, the function of ZCWPW1 in mammalian development and physiology, and especially in meiosis, is largely unknown. Our working hypothesis is that ZCWPW1 is at least partly responsible for recognizing the H3K4me3 generated by PRDM9 and thus takes part in meiosis progression in mice. By generating a Zcwpw1 knockout mouse model, we report that loss of ZCWPW1 in mice disrupted meiosis prophase I in males but had little effects on female germ cells.
RESULTS
ZCWPW1 expression and dynamic localization in meiotic germ cells
We found that Zcwpw1 mRNA was generally expressed in mouse organs including in adult testes and embryonic ovaries, where meiotic prophase I occurs (fig. S1). To further study the expression pattern of ZCWPW1 in germ cells, we isolated different types of male germ cells by bovine serum albumin (BSA) gradient sedimentation. By Western blotting, we found that ZCWPW1 expression was elevated from the leptotene to round spermatid stage during meiosis prophase I but became almost undetectable in elongated spermatids (Fig. 1A). To determine the subcellular localization of ZCWPW1, we performed cytoplasmic and nuclear fractionation and found that ZCWPW1 was only detected in the nuclear fraction of mouse testes extracts as indicated by Lamin B1 (Fig. 1B), showing that ZCWPW1 functions only in the nuclei.
Fig. 1. ZCWPW1 expression and dynamic localization in meiotic germ cells.
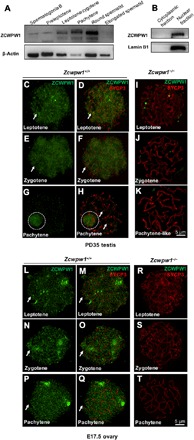
(A) Western blotting of ZCWPW1 in isolated male germ cells, demonstrating that the Zcwpw1 expression level increased from the leptotene stage to the round spermatid stage and then disappeared in the elongated spermatid. β-Actin was used as the control. (B) Western blotting of ZCWPW1 in the cytoplasmic and nuclear fractions of PD35 wild-type testes shows that ZCWPW1 was only expressed in the nuclei. Lamin B1 was used as the marker of nuclear fractions. (C to K) Chromosome spreads of spermatocytes from the testes of PD35 Zcwpw1+/+ and Zcwpw1−/− males immunostained for ZCWPW1 (green) and SYCP3 (red). ZCWPW1 was diffuse (arrows) from the leptotene to zygotene stages (C to F, arrows). In pachytene cells, ZCWPW1 was localized in the XY body (G and H, white dashed circles). (I to K) In Zcwpw1−/− spermatocytes, no ZCWPW1 signal was detected. (L to T) Chromosome spreads of oocytes from E17.5 Zcwpw1+/+ and Zcwpw1−/− ovaries immunostained for ZCWPW1 (green) and SYCP3 (red). ZCWPW1 was diffuse (arrows) from the leptotene to pachytene stages (L to Q, arrows). (R to T) In Zcwpw1−/− oocytes, no ZCWPW1 signal was detected.
Immunostaining of spread spermatocyte nuclei showed diffuse ZCWPW1 immunofluorescent signal from the leptotene (Fig. 1, C and D, green signal, arrows) to zygotene (Fig. 1, E and F, green signal, arrows) stages. The signal disappeared from the autosomal chromosomes (Fig. 1H, arrows) but was found only in the XY sex body of spermatocytes at the pachytene stage (Fig. 1, G and H, green signal, white dashed circles). In female germ cells at embryonic day 17.5 (E17.5), a similar diffuse ZCWPW1 signal was seen over the entire nuclear region of the oocytes from leptotene to pachytene stages (Fig. 1, L to Q, green signal, arrows). As controls, ZCWPW1 signals in male or female germ cells from mice that lack Zcwpw1 (see below) were almost undetectable (Fig. 1, I to K and R to T). All these results suggest that ZCWPW1 should work in the nuclei during the prophase I of meiosis.
ZCWPW1 is needed for maintaining fertility in a sex-dependent manner
To study in vivo functions of ZCWPW1, we generated a Zcwpw1−/−-deficient mouse model lacking exon 4 to exon 7 of Zcwpw1 (Fig. 2A). Western blotting analysis confirmed the absence of the ZCWPW1 protein in Zcwpw1−/− testes (Fig. 2B). The offspring from intercrosses of heterozygous (Zcwpw1+/−) male and female mice exhibited a normal Mendelian distribution of genotypes, and Zcwpw1−/− mice were viable and appeared to develop normally.
Fig. 2. ZCWPW1 is needed for maintaining fertility in a sex-dependent manner.
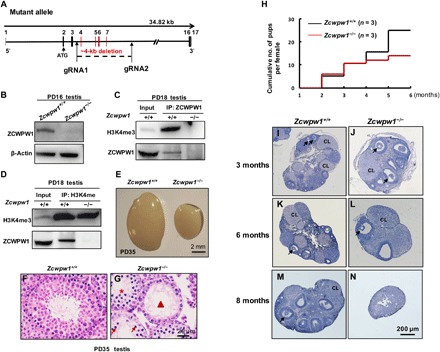
(A) Schematic representation of the CRISPR-Cas9 genome editing system to the Zcwpw1-deficient mice. (B) Western blotting showed that ZCWPW1 was deleted in PD16 Zcwpw1−/− testes compared to wild type. β-Actin was used as the loading control. (C and D) Co-immunoprecipitation analysis of ZCWPW1 and H3K4me3 from PD18 testis protein extracts. ZCWPW1 immunoprecipitated with H3K4me3. (E). Testes from PD35 Zcwpw1+/+ and Zcwpw1−/− male mice. Zcwpw1−/− male mice had reduced testis size at PD35 (n = 3, Welch’s t test analysis; P < 0.0001). (F) Hematoxylin and eosin (H&E) staining of adult Zcwpw1+/+ testis. (G) H&E staining of adult Zcwpw1−/− testis sections showed complete arrest of spermatogenesis. Arrows, apoptotic spermatocytes; arrowhead, empty seminiferous tubules; asterisk, seminiferous tubules lack of postmeiotic spermatocytes. (H) Cumulative numbers of pups per female during the defined time period. n = 3 mice for each genotype. (I to N) Histological analysis of ovaries from Zcwpw1+/+ and Zcwpw1−/− females. (I and J). Morphological studies of ovaries showed that at 3-month Zcwpw1−/− ovaries (J) exhibited similar morphologies as Zcwpw1+/+ ovaries (I). (K and L). At 6 months of age, Zcwpw1−/− ovaries (L) contained fewer but healthy follicles and corpora lutea (CL). (M and N). At around 8 months of age, the mutant ovaries (N) had no oocytes or follicles. (Photo credit: Miao Li and Tao Huang, Shandong Provincial Hospital Affiliated with Shandong University).
We performed immunoprecipitation (IP) with testicular protein extracts using the ZCWPW1 antibody and found that H3K4me3 was present in the immunoprecipitated proteins from Zcwpw1+/+ testes but was absent from Zcwpw1−/− testes (Fig. 2C). Similarly, immunoprecipitation using the H3K4me3 antibody showed pulled down ZCWPW1 protein from Zcwpw1+/+ testes protein extracts but not from Zcwpw1−/− testes (Fig. 2D). These data confirmed that ZCWPW1 might be involved in meiosis through binding to H3K4me3.
Despite their normal outward appearance, Zcwpw1−/− males were found to be completely infertile with much smaller testes (Fig. 2E). Histological analysis showed that spermatogenesis in Zcwpw1−/− males was impaired (Fig. 2G) compared to Zcwpw1+/+ males (Fig. 2F). In Zcwpw1−/− males, the seminiferous tubules lacked postmeiotic spermatocytes (Fig. 2G, asterisk) and contained apoptotic cells (Fig. 2G, arrows) or were almost empty (Fig. 2G, arrowhead). There were no spermatozoa in the Zcwpw1−/− epididymis, showing that Zcwpw1 is essential for spermatogenesis in mice.
In contrast to males, the fertility of Zcwpw1−/− females was found to be normal up to mid-adulthood (5 to 6 months of age), with comparable numbers of pups as Zcwpw1+/+ females (Fig. 2H). Morphological analysis of the ovaries showed that, at 3 and 6 months, the Zcwpw1−/− ovaries exhibited healthy ovarian morphologies (Fig. 2, J and L) as Zcwpw1+/+ ovaries (Fig. 2, I and K). However, Zcwpw1−/− ovaries were devoid of follicles at around 8 months of age (Fig. 2N), and accordingly, the female mice became infertile.
Disrupted synapsis in Zcwpw1−/− spermatocytes
To determine the reason for the disrupted meiosis in Zcwpw1−/− spermatocytes, we first observed chromosomes during meiosis prophase I by immunostaining for synaptonemal complex (SC) proteins. The SC consists of two axial elements (AEs) and a central element (CE), which are joined together by transverse filaments (24). In Zcwpw1+/+ spermatocytes, the AEs marked by synaptonemal complex protein 3 (SYCP3) began to form at the leptotene stage (Fig. 3A, red signal), and a lack of SYCP1 (green signal) indicated that CEs were not yet present (Fig. 3A). At the zygotene stage, the AEs were fully formed, and chromosomes became progressively synapsed as indicated by the appearance of CEs with the SYCP1 signals (Fig. 3B, arrow). Wild-type spermatocytes reached full synapsis on autosomes at the pachytene stage, as shown by the continuous SYCP1 signal (Fig. 3C, arrow). In Zcwpw1−/− spermatocytes, AEs formed at the leptotene stage (Fig. 3D, red signal), and at the zygotene stage, SYCP1 began to appear on the paired chromosomes, indicating that synapsis was somewhat initiated (Fig. 3E, arrow). However, in Zcwpw1−/− spermatocytes, there was always a large number of autosomes that remained unsynapsed (Fig. 3F, arrowheads, red signals) in comparison to the synapsed chromosomes (Fig. 3F, arrow), and these mutant spermatocytes remained at a stage that we called “pachytene-like” (defined by more than five synapsed chromosome pairs per cell). We quantified the synapsed chromosome pairs in each nucleus in Zcwpw1+/+ and Zcwpw1−/− testes at 8 weeks of age (Fig. 3G). In Zcwpw1+/+ testes, 153 cells (90.5%) with all chromosome pairs fully synapsed were seen, and only 16 cells (9.5%) with 4 to 18 pairs of chromosomes synapsed were present (Fig. 3G). In Zcwpw1−/− testes, however, none of the 164 spermatocytes (0%) observed in the Zcwpw1−/− testes had complete synapsis, and there was only an average of eight synapsed chromosome pairs in each cell (Fig. 3G).
Fig. 3. Disrupted chromosomal synapsis in Zcwpw1−/− spermatocytes.
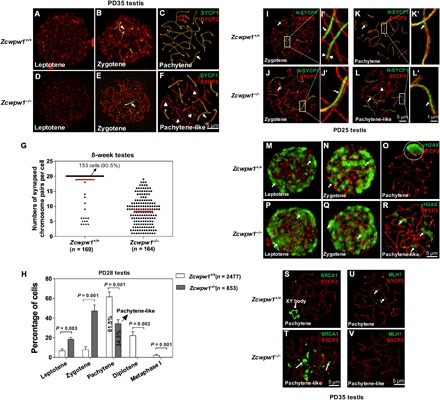
(A to F) Chromosome spreads of spermatocytes from the testes of PD35 wild-type (A to C) and Zcwpw1−/− (D to F) males were immunostained for SYCP1 (green) and SYCP3 (red). Arrows indicate synapsed chromosomes, and arrowheads indicate the single chromosome. (G). The numbers of synapsed chromosome pairs in wild-type spermatocytes and Zcwpw1−/− spermatocytes. In Zcwpw1−/− spermatocytes, the average number of synapsed chromosome pairs was 8. (H) Frequencies of meiotic stages in Zcwpw1+/+ and Zcwpw1−/− spermatocytes. The numbers marked in the bars represent the percentage of cells at indicated meiosis stage. For each genotype, three mice were analyzed. P values were calculated by Student’s t test. (I to L) SIM images of spermatocyte chromosome spreads immunostained for SYCP3 (red) and N-SYCP1 (green) from PD25 testes. Arrows indicate the synapsed region, and arrowheads indicate the AEs. (I’ to L’) Magnified views of the synapsed region show that N-SYCP1 was localized in the central region of SCs in a continuous pattern (arrows). (M to R) Chromosome spreads of spermatocytes from Zcwpw1+/+ (M to O) and Zcwpw1−/− (P and Q) males were immunostained for SYCP3 (red) and γH2AX (green). Representative images of spermatocytes at the leptotene, zygotene, and pachytene stages are shown. (S and T) Zcwpw1+/+ and Zcwpw1−/− cells immunostained for SYCP3 (red) and breast cancer 1 (BRCA1; green). Representative images of spermatocytes at pachytene (S, arrow indicating XY body) and pachytene-like (T, arrows indicating BRCA1 signal) stages are shown. (U and V) Zcwpw1+/+ and Zcwpw1−/− spermatocytes immunostained for SYCP3 (red) and MLH1 (green, arrows). Zcwpw1−/− spermatocytes lack MutL homolog 1 (MLH1) signal (V).
We further analyzed the percentage of spermatocytes at different meiotic stages using postnatal day 28 (PD28) Zcwpw1+/+ and Zcwpw1−/− testes and found that Zcwpw1−/− spermatocytes failed to proceed beyond the pachytene stage (Fig. 3H), and 34.3% of them were arrested at the pachytene-like stage (Fig. 3H, pachytene-like). This was in sharp contrast to the 61.5% of Zcwpw1+/+ spermatocytes that had reached the pachytene stage at PD28 (Fig. 3H, pachytene).
To better understand the synapsis failure in Zcwpw1−/− spermatocytes, we used high-resolution structured illumination microscopy (SIM) to observe the fine SC structures that cannot be distinguished in confocal microscopy images. In SIM imaging, the AEs appeared as two separate strands by SYCP3 immunostaining (Fig. 3I’, arrowheads, red signal). SYCP1 forms homodimers, and N termini of SYCP1 dimers were located in the CE (Fig. 3, I’ and K’, green signal, arrows) (25). The two C-terminal ends of the SCYP1 dimer localize in the AEs (26), which were observed as separate signals in two AE axes in our SIM imaging (fig. S2, A’ and C’, green signal, arrows). Another CE factor testis expressed 12 (TEX12) was also located in the central of AEs (fig. S2, E’ and G’, green signal, arrows). In Zcwpw1−/− spermatocytes, synapsis was initiated in some chromosomes, and N-SYCP1 were found in the CE on fully synapsed chromosomes (Fig. 3J’, green signal, arrow). C-SYCP1 was found along the SYCP3 axis (fig. S2B’, green signal, arrow), and TEX12 (fig. S2, F’ and H’, green signal, arrows) was found in the CE on fully synapsed chromosomes in Zcwpw1−/− spermatocytes, just as in Zcwpw1+/+ zygotene spermatocytes.
However, among the synapsed chromosomes in pachytene-like Zcwpw1−/− cells, no apparent differences in the localization pattern of N-SYCP1 (Fig. 3L’, green signal, arrow), C-SYCP1 (fig. S2D’, green signal, arrow), or TEX12 (fig. S2H’, green signal, arrow) were seen. At the same time, the unsynapsed autosomes in Zcwpw1−/− cells lacked SYCP1 signal [(Fig. 3L, arrowhead (red signals), and fig. S2D, arrowhead (red signals)] and TEX12 signal (fig. S2H, red signals, arrowhead). On the basis of these results, we conclude that some autosomes in Zcwpw1−/− spermatocytes can initiate synapsis, whereas most of the autosomes fail to synapse.
Zcwpw1−/− spermatocytes exhibit defects in meiotic recombination
We then evaluated meiotic DSB repair by analyzing the localization of γ-H2AX. Similar to the signal of γ-H2AX in Zcwpw1+/+ spermatocytes (Fig. 3, M and N, arrows), γ-H2AX in Zcwpw1−/− spermatocytes marks chromatin domains surrounding DSB sites at the leptotene (Fig. 3P, arrows) and zygotene (Fig. 3Q, arrows) stages. At the pachytene stage, in Zcwpw1+/+ cells, the γ-H2AX signal was only seen at the XY bodies (Fig. 3O, white dashed circle). In Zcwpw1−/− spermatocytes, however, the γ-H2AX signal persisted on the chromatin of unsynapsed axes (Fig. 3R, arrows), indicating a failure in the repair of DSBs. A response to asynapsed chromatin in germ cells is called meiotic silencing of unsynapsed chromatin (MSUC) (27). We further stained Zcwpw1+/+ and Zcwpw1−/− spermatocytes for the MSUC initiator factor breast cancer 1 (BRCA1), and we found that BRCA1 persisted on asynapsed Zcwpw1−/− chromosomes at the pachytene-like stage (Fig. 3T, arrows), while BRCA1 was only observed in the XY body in Zcwpw1+/+ spermatocytes at pachynema (Fig. 3S).
At the mid- to late-pachytene stage, MutL homolog 1 (MLH1) foci became apparent in Zcwpw1+/+ spermatocytes, indicating sites of crossovers (Fig. 3U, arrows). Zcwpw1−/− spermatocytes, however, failed to progress to the pachytene stage, and no crossover ever occurred, thus no MLH1 foci were found (Fig. 3V).
To more specifically assess meiotic DSB repair and recombination, we immunostained replication protein A2 (RPA2) (Fig. 4, A to F), RAD51 (Fig. 4, H to M), and DMC1 (Fig. 4, O to T). In Zcwpw1−/− spermatocytes, we defined the leptotene stage as chromatids beginning to condense and elongate but not yet synapsing (fig. S3A) and the zygotene stage as homologous chromosomes beginning to synapse but synapsis is not complete (fig. S3, B and C). We further divided the zygotene stage into “early zygotene” (fig. S3B) if there were <5 synapsed chromosome pairs and “late zygotene” (fig. S3C) if there were ≥5 synapsed chromosome pairs. We found that the initial numbers of RPA2 foci were similar between Zcwpw1+/+ and Zcwpw1−/− spermatocytes at the leptotene and zygotene stages (Fig. 4G, leptotene, early zygotene, and late zygotene). With the progression of meiotic recombination, the number of RPA2 foci decreased in Zcwpw1+/+ spermatocytes at the pachytene stage but persisted in Zcwpw1−/− spermatocytes (Fig. 4, C and F and G, pachytene/pachytene-like), showing that RPA was not efficiently removed.
Fig. 4. Zcwpw1 is essential for male meiotic recombination.
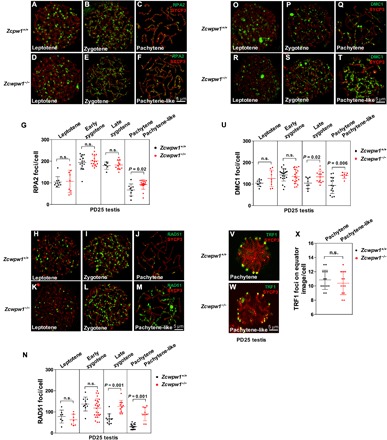
(A to U) Immunostaining for SYCP3 (red) and recombination protein (green) was performed on Zcwpw1+/+ and Zcwpw1−/− spermatocytes from PD25 mice. Representative images of spermatocytes at the leptotene, zygotene, and pachytene (pachytene-like) stages are shown. Each dot represents the number of DNA repair protein foci per cell, with black dots indicating Zcwpw1+/+ spermatocytes and red dots indicating Zcwpw1−/− spermatocytes. Solid lines show the mean and SD of foci number in each group of spermatocytes. (A to G) RPA2 foci. (H to N) RAD51 foci. (O to U) DMC1 foci. P values were calculated by Student’s t test. (V to X) Immunostaining for SYCP3 (red) and TRF1 (green) was performed on Zcwpw1+/+ and Zcwpw1−/− spermatocytes. Representative images of spermatocytes at the pachytene (pachytene-like) stages are shown. Zcwpw1−/− spermatocytes had comparable numbers of TRF1 foci on equator images as compared to Zcwpw1+/+ spermatocytes. n.s., no statistical significance.
The numbers of RAD51 and DMC1 foci were also comparable between Zcwpw1+/+ and Zcwpw1−/− spermatocytes at the leptotene [Fig. 4, H, K, O, R, N (leptotene), and U (leptotene)] and early zygotene stages [Fig. 4, I, L, P, S, N (early zygotene), and U (early zygotene)]. At late zygotene and pachytene-like stages, however, both RAD51 and DMC1 foci numbers were significantly higher in Zcwpw1−/− spermatocytes compared to Zcwpw1+/+ spermatocytes (Fig. 4, N and U), suggesting that Zcwpw1−/− spermatocytes exhibited defects in recombination and DSB repair, and their RAD51/DMC1 recombinase persisted on the chromosomes. These results indicated a defective meiotic progression in Zcwpw1−/− spermatocytes.
For homologous synapsis and recombination, telomere attachment to the nuclear envelope (NE) is a prerequisite for chromosome movement (28). As a key factor in telomere-NE attachment, signals of telomeric repeat-binding factor 1 (TRF1) that represent telomeres should be localized at the NE, which indicates a normal telomere-NE attachment. We thus stained for TRF1 to determine whether telomeres can attach normally to the NE in Zcwpw1−/− spermatocytes. Our results showed that Zcwpw1−/− spermatocytes (Fig. 4W) had comparable numbers of TRF1 foci on equator images as compared to those of the Zcwpw1+/+ spermatocytes (Fig. 4, V to X).
Successful but delayed meiosis prophase I in Zcwpw1−/− female germ cells
Compared to Zcwpw1−/− spermatocytes, Zcwpw1−/− female germ cells at E17.5 were capable of reaching the pachytene stage (Fig. 5A) and developing into oocytes, leading to normal fertility. However, they exhibited an apparent delay in meiotic progression. We found that, at E17.5, 70% of Zcwpw1+/+ oocytes had formed SC and reached pachytene stage (Fig. 5A), whereas only 19% of Zcwpw1−/− oocytes had completed synapsis at this stage, and around 75% of Zcwpw1−/− oocytes were still at zygonema (Fig. 5A). At PD1, 43% of Zcwpw1+/+ oocytes had proceeded to late diplotene stage (Fig. 5B, late diplotene), and about 16% of the Zcwpw1+/+ oocytes were in the dictyotene stage (Fig. 5B, dictyotene). However, only 20% of Zcwpw1−/− oocytes were at the late diplotene stage and only 10% were at the dictyotene stage (Fig. 5B). The slower meiosis in Zcwpw1−/− oocytes was easily seen by the 38% of Zcwpw1−/− oocytes at the late pachytene stage and the 32% of Zcwpw1−/− oocytes at the early diplotene stage at PD1 (Fig. 5B). Representative images of oocytes at the leptotene, zygotene, and pachytene stages are shown (Fig. 5, C to H, arrows indicating synapsed chromosomes).
Fig. 5. Successful but delayed meiosis prophase I in Zcwpw1−/− oocytes.
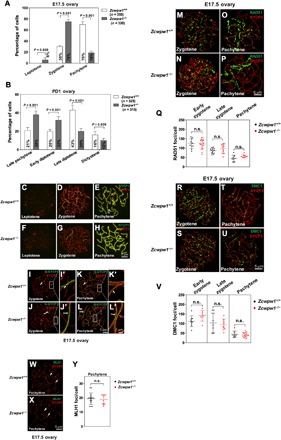
(A) Frequencies of meiotic stages in E17.5 Zcwpw1+/+ and Zcwpw1−/− oocyte chromosome spreads. The numbers marked in the bars represent the percentage of cells at the indicated meiosis stage. For each genotype, three mice were analyzed. P values were calculated by Student’s t test. (B) Frequencies of meiotic stages in PD1 Zcwpw1+/+ and Zcwpw1−/− oocyte chromosome spreads. The numbers marked in the bars represent the percentage of cells at the indicated meiosis stage. For each genotype, three mice were analyzed. P values were calculated by Student’s t test. (C to H) Chromosome spreads of E17.5 ovaries from wild-type (C to E) and Zcwpw1−/− (F to H) females were immunostained for SYCP3 (red) and SYCP1 (green). Representative images of oocytes at the leptotene, zygotene, and pachytene stages are shown. Arrows indicate the synapsed chromosomes. (I to L) Chromosome spreads of E17.5 ovaries immunostained for SYCP3 (red) and N-SYCP1 (green) using SIM at the indicated stages. Arrows indicate the synapsed region. (I’ to L’) Magnified views of the synapsed region show that N-SYCP1 localized in the central regions of SCs in a continuous pattern (arrows). (M to Y) Immunostaining for SYCP3 (red) and RAD51/DMC1/MLH1 (green) was performed on Zcwpw1+/+ and Zcwpw1−/− oocytes from E17.5 females. Representative images of oocytes in zygotene and pachytene stages are shown. Each dot represents the number of RAD51/DMC1/MLH1 foci per cell, with black dots indicating the Zcwpw1+/+ oocytes and red dots indicating Zcwpw1−/− oocytes. Solid lines show the mean and SD of foci number in each group. (M to Q) RAD51 foci. (R to V) DMC1 foci. (W to Y) MLH1 foci.
To determine whether the SC structure formed in E17.5 Zcwpw1−/− oocytes was complete and stable, we stained ovary chromosome spreads with antibodies against N-SYCP1 and TEX12. N-SYCP1 and TEX12 were localized in the CE of paired chromosomes in Zcwpw1−/− oocytes at both the zygotene and pachytene stages [N-SYCP1: Fig. 5, J, J’, L, and L’ (arrows); TEX12: fig. S4, B, B’, D, and D’ (arrows)]. This was very similar to Zcwpw1+/+ oocytes at E17.5 [N-SYCP1: Fig. 5, I, I’, K, and K′ (arrows); TEX12: fig. S4, A, A’, C, and C’ (arrows)]. These results showed that synapsis occurred normally in observed Zcwpw1−/− female germ cells.
We next explored whether the slower meiosis progression in Zcwpw1−/− female germ cells was caused by defective DSB repair, chromosome recombination, and crossover formation. Immunostaining of RAD51 and DMC1, however, showed that at zygotene and pachytene stage, Zcwpw1+/+ [Fig. 5, M and O (RAD51 foci) and R and T (DMC1 foci)] and Zcwpw1−/− oocytes [Fig. 5, N and P (RAD51 foci) and S and U (DMC1 foci)] had comparable numbers of RAD51 and DMC1 foci (Fig. 5, Q and V). MLH1 foci were similar in Zcwpw1+/+ (Fig. 5W, arrows) and Zcwpw1−/− oocytes (Fig. 5X, arrows) at the pachytene stage, and no significant differences were observed (Fig. 5Y). These results indicated that the slower progression of meiosis prophase I in Zcwpw1−/− oocytes that still existed at E17.5 ovary was not accompanied by defective recombination and crossover formation.
The Zcwpw1−/− female follicle pool is reduced and leads to premature ovarian insufficiency (POI)
To determine whether the loss of oocytes and ovarian follicles at late adulthood in Zcwpw1−/− female mice was caused by the delayed meiosis progression, we quantified oocyte numbers at different ovarian developmental stages. Ovaries dissected from E13.5, PD1, and PD8 mice were fixed, sectioned, and immunostained with mouse vasa homolog (MVH) antibody. Quantification of relative numbers of oocytes at these stages showed that there were comparable numbers of female germ cells in E13.5 Zcwpw1+/+ and Zcwpw1−/− ovaries in which meiosis is just about to start or just starts (Fig. 6, A to C). At PD1, compared to Zcwpw1+/+ ovaries, Zcwpw1−/− ovaries contained only approximately 55% of oocytes, showing that the oocyte number was largely reduced when meiosis prophase I was about to finished (Fig. 6, D to F). At PD8, however, the Zcwpw1−/− ovaries only had about 20% as many oocytes as Zcwpw1+/+ ovaries (Fig. 6, G to I). Thus, the delay in meiosis I observed in Zcwpw1−/− female germ cells was likely the reason for the reduced oocyte numbers in mutant mice.
Fig. 6. The knockout of Zcwpw1 leads to POI.
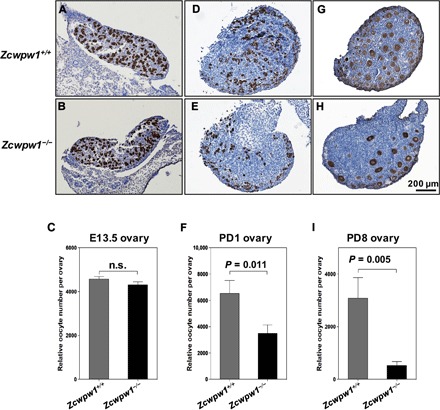
(A and B) Representative Zcwpw1+/+ (A) and Zcwpw1−/− (B) ovary sections from E13.5 females immunostained for mouse vasa homolog (MVH) with hematoxylin counterstaining. (C) Oocyte counts (relative numbers) showed that there were similar numbers of oocytes in E13.5 Zcwpw1+/+ and Zcwpw1−/− females. MVH-positive cells were counted. (D and E) Representative Zcwpw1+/+ (D) and Zcwpw1−/− (E) ovary sections from PD1 females immunostained for MVH with hematoxylin counterstaining. (F) Relative oocyte counts showed that Zcwpw1−/− ovaries contained significantly fewer oocytes than Zcwpw1+/+ ovaries. (G and H) Representative Zcwpw1+/+ (G) and Zcwpw1−/− (H) ovary sections from PD8 females immunostained for MVH with hematoxylin counterstaining. (I) Relative oocyte counts showed that Zcwpw1−/− ovaries contained significantly fewer follicles than Zcwpw1+/+ ovaries. MVH-positive oocyte nuclei with characteristic surrounding granulosa cell layers were counted. In all cases, counts were made for every section (8 μm per section) and summed to calculate the total number of oocytes per ovary. For each genotype, six ovaries from three mice were analyzed. P values were calculated by Student’s t test.
Misregulated expression of chromatin remodeling genes in Zcwpw1−/− testes
Chromatin remodeling involves the dynamic modification of the chromatin architecture and the recruitment of transcription machinery proteins and thus controls gene expression (29). We performed a proteomic analysis in PD14 Zcwpw1−/− testes by high-performance liquid chromatography–mass spectrometry (HPLC-MS) and identified 94 differentially expressed proteins (table. S1). Gene ontology analysis identified nuclear chromosome as the most up-regulated pathway (fig. S5A). For example, ubiquitin carboxyl-terminal hydrolase isozyme L5 (UCHL5) is a deubiquitylating enzyme that regulates DSB resection and repair by homologous recombination and promotes the maintenance of genome integrity (30). Among the down-regulated genes, the SWI/SNF superfamily complex was identified, including actin-related protein 5 (Actr5), chromatin complexes subunit (Bap18), chromodomain helicase DNA binding protein 5 (Chd5), and actin-related protein 8 (Actr8), and these proteins play key roles in chromatin remodeling at both promoters and enhancers and function in transcriptional regulation (31). These results suggest that ZCWPW1 may exert its effects on synapsis by affecting chromatin remodeling.
DISCUSSION
Here, we show that a histone modification reader, ZCWPW1, is specifically required for the progression of meiosis prophase I in a sex-dependent manner in mice. Zcwpw1−/− male mice are completely infertile because Zcwpw1−/− spermatocytes display meiotic arrest at around the zygotene to pachytene stage with incomplete homologous synapsis, which is accompanied by defective DSB repair. In contrast, part of the Zcwpw1−/− oocytes achieve full synapsis and complete the entire meiosis process, albeit their progression through prophase I is slower. This slower meiotic prophase I is believed to be the reason behind the death of some oocytes and the result of a smaller initial follicle pool in Zcwpw1−/− females. Thus, the Zcwpw1−/− female mice have normal fertility until mid-adulthood, after which they prematurely exhaust their ovarian follicles and exhibit a POI-like phenotype. We conclude that ZCWPW1 is indispensable for meiosis prophase I in male mice but is only partially required for meiosis in female mice.
In this study, Zcwpw1−/− spermatocytes failed to fully synapse and did not complete their DSB repair, and they completely lacked crossover. RPA2 and RAD51/DMC1 recombinase persisted on unsynapsed region and failed to be released, indicating a defect in recombination. Synapsis failure in Zcwpw1−/− spermatocytes might trigger MSUC, which is a response to asynapsis in meiotic germ cells that leads to transcriptional silencing of unsynapsed regions (32). MSUC is initiated by accumulation of the DNA damage response protein BRCA1 on unsynapsed chromosome axes (33). BRCA1 then recruits ataxia telangiectasia and Rad3 related protein (ATR) to the unsynapsed axis (27), and ATR phosphorylates the variant nucleosomal histone H2AX, thus triggering the chromatin changes that lead to transcriptional silencing (34). We found that the BRCA1 and γ-H2AX signal persists on the axes of unsynapsed chromosomes in Zcwpw1−/− spermatocytes, implying that MSUC has been initiated on unsynapsed autosomes in Zcwpw1−/− spermatocytes. Therefore, this transcriptional silencing triggered by MSUC most likely silences genes that are crucial to the progression of meiosis, as previously reported (35), leading to meiotic arrest and male infertility. Sex chromosomes undergo a special type of MSUC called meiotic sex chromosome inactivation (MSCI) (36).
ZCWPW1 protein is localized on the XY body in pachytene spermatocytes, implying that ZCWPW1 might be involved in the formation or function of the XY body, including MSCI. It is known that failed MSCI causes “pathological” expression of some sex-linked “toxic” genes that should remain inactive during meiosis, and this leads to meiotic failure in, for example, Msh5−/− and Dmc1−/− spermatocytes (37, 38). For example, two Y-encoded transcription factors, ZFY1/2 (zinc finger protein 1/2, Y-linked), have been identified as toxic proteins, and their expression during the failure of Y chromosome inactivation triggers spermatocyte apoptosis in XYY male mice (39). ZCWPW1 might regulate these sex-linked genes, and the possible functions of ZCWPW1 in the formation and regulation of sex bodies represent a major question that deserves further investigation.
Analysis of H3K4me3 in Spo11−/− spermatocytes showed that H3K4me3 modifications were present at the potential hotspot loci even in the absence of SPO11, indicating that the hotspot-associated H3K4me3 is indeed a marker before the initiation of recombination but not as a consequence of DSB formation (40). The number of H3K4me3 modifications is greater than the actual number of DSB hotspots in whole genome (12, 41), because H3K4me3 is a general marker of active transcription and is primarily associated with gene promoters and enhancers and possibly with other functional genomic elements (42). In meiosis, DSB locations are determined by PRDM9-dependent H3K4me3 modification in mice, and the latter are usually directed away from functional genomic elements such as gene promoter regions (12). In this study, our data suggest that ZCWPW1 is another important player related to PRDM9-mediated meiosis progression, and ZCWPW1 might play a role in mediating DSB hotspot distribution in mouse spermatocytes through its recognition of H3K4me3 modifications. In Saccharomyces cerevisiae, DSBs also occur in H3K4me3-enriched sites, and suppressor of PRP protein 1 (SPP1) linked the H3K4me3 modification and DSB machinery. The SPP1 protein recognized the modification by PHD finger and interacted with other proteins required for SPO11 activity (43, 44). Similar to SPP1, ZCWPW1 may read the H3K4me3 deposited by PRDM9 and recruit DSB machinery to hotspot sites by some unknown mechanisms. In the absence of ZCWPW1, the PRDM9-dependent H3K4me3 may not be properly recognized, and SPO11 complex may not be recruited to proper sites, leading to meiosis arrest. It is also possible that ZCWPW1 directly activates the transcription of a group of genes that are crucial for the progression of meiotic prophase I because the H3K4me3 modification is a common transcription activation marker in the genome (45).
The different phenotypes between male and female are another potential topic to us. Recently, it was reported that the different hotspot usage led to extensive differences in the initiation of recombination in male and female mice (46). Hotspot usage is influenced by PRDM9 affinity for its binding sites and local chromatin/DNA modifications (47). There are differences indeed in DNA modification between male and female—the genome is globally demethylated at the time of DSB formation in females but not in males (46). ZCWPW1 likely takes part in chromosome remodeling by its H3K4me3 reading function, and this might partly explain our results that ZCWPW1 is not absolutely required for meiosis progression in female germ cells. Furthermore, male germ cells have increased stringency of checkpoint mechanisms in relation to their female counterparts (48).
In summary, we have pinpointed ZCWPW1 as a histone modification reader that is indispensable in guiding meiosis prophase I in male mice. Further studies identifying the ZCWPW1-H3K4me3–mediated DSB landscape, recombination events, and ZCWPW1-downstream genes will be of great importance in understanding the epigenetic regulation of mammalian meiosis.
MATERIALS AND METHODS
Generation of Zcwpw1 knockout mice
The Zcwpw1-deficient mouse model in a C57BL/6 genetic background was generated by deleting the genomic DNA fragment covering exon 4 to exon 7 using the CRISPR-Cas9–mediated genome editing system (Cyagen Biosciences, USA). The founders were genotyped by polymerase chain reaction (PCR), followed by DNA sequencing analysis. The mice were housed under controlled environmental conditions with free access to water and food, and illumination was on between 6:00 am to 6:00 pm. All experimental protocols were approved by the Regional Ethics Committee of Shandong University.
Genotyping was performed by PCR amplification of genomic DNA extracted from mouse tails. PCR primers for the Zcwpw1 mutant allele were 5′- AGC TGCTGGGATTAAATGTCTGTTCC-3′ (forward) and 5′-CTACACTGTGCCTTCTACCTTCTTTGAGA-3′ (reverse), yielding a 690–base pair (bp) fragment. PCR primers for the Zcwpw1 wild-type allele were 5′-CAAGATGGAGGAGATATGCAGTACATG-3′ (forward) and 5′-CTACACTGTGCCTTCTACCTTCTTTGAGA-3′ (reverse), yielding a 617-bp fragment.
Production of the ZCWPW1 antibody
Antibodies to mouse ZCWPW1 were produced by Dia-an Biological Technology Incorporation (Wuhan, China). Briefly, a complementary DNA (cDNA) fragment encoding amino acids 448 to 622 of mouse Zcwpw1 was inserted into the p-ET-32a + vector (EMD Millipore) and transfected into BL21-CodonPlus(DE3) Escherichia coli cells. The cells were cultured at 37°C overnight and induced by addition of 0.2 mM isopropyl-1-thio-β-d-galactoside (Sigma-Aldrich) for 4 hours at 28°C. Cells were harvested by centrifugation and disrupted by sonication, and the soluble homogenates were purified by Ni-nitrilotriacetic acid (NI-NTA) Agarose (Qiagen) according to the manufacturer’s instructions. The protein was dialyzed in phosphate-buffered saline (PBS) and used to immunize rabbits, and the antiserum was affinity-purified on antigen-coupled CNBr-activated agarose (GE Healthcare).
Purification of male germ cells
The purification of male germ cells was performed as described previously (49). The cell type and purity in each fraction were assessed on the basis of their diameters and morphological characteristics under a light microscope.
Tissue collection and histological analysis
Testes and ovaries from more than three mice for each genotype were dissected immediately after euthanasia, fixed in 4% paraformaldehyde (P1110, Solarbio, Beijing, China) for up to 24 hours, stored in 70% ethanol, and embedded in paraffin after dehydration. Sections (5 μm) were prepared and mounted on glass slides. After deparaffinization, slides were stained with hematoxylin and eosin for histological analysis.
Immunocytology and antibodies
Spermatocyte and oocyte chromosome spreading was prepared as previously described (50, 51). Structurally preserved spermatocytes were prepared as described previously (52).
Primary antibodies used for immunofluorescence were as follows: rabbit anti-SYCP3 (1:500 dilution; Abcam #ab15093), mouse anti-SYCP3 (1:500 dilution; Abcam #ab97672), rabbit anti-SYCP1 (C-terminal) (1:2000 dilution; Abcam #ab15090), rabbit anti-SYCP1 (N-terminal) (1:2000 dilution; Abclonal #A12139), rabbit anti-RPA2 (1:200 dilution; Abcam #ab76420), rabbit anti-RAD51 (1:200 dilution; Thermo Fisher Scientific #PA5-27195), rabbit anti-DMC1 (1:100 dilution; Santa Cruz Biotechnology #sc-22768), mouse anti–phospho-histone H2AX (pSer139) (1:300 dilution; Millipore #05-636), mouse anti-MLH1 (1:50 dilution; BD Biosciences #550838), rabbit anti-TEX12 (1:1000 dilution; Proteintech #17068-1-AP), mouse anti-TRF1 (1:1000 dilution; homemade), and rabbit anti-BRCA1 (1:500 dilution; a gift from L.-Y. Lu, Zhejiang University). Primary antibodies were detected with Alexa Fluor 488– or 594–conjugated secondary antibodies (1:500 dilution; Abcam #ab150084, #ab150077, #ab150113, and #ab150120) for 1 hour at room temperature. The slides were washed several times with PBS and mounted using VECTASHIELD antifade mounting medium with DAPI (Vector Laboratories, #H-1200).
Immunohistochemistry and quantification of ovarian follicles
Immunohistochemistry analysis was performed on ovary sections to identify the oocytes. Briefly, the ovaries from E13.5, PD1, and PD8 were fixed with 4% paraformaldehyde for 24 hours, then washed with PBS, and stored in 70% ethanol. The samples were embedded in paraffin, and 8-μm sections were prepared. After deparaffinization and rehydration through a graded ethanol series, the slides were incubated with 5% BSA for 1 hour at room temperature and incubated with anti-MVH (1:5000 dilution; Abcam #ab27591) antibody overnight at 4°C. Quantification of ovarian follicles was performed as previously described (53). Briefly, MVH-positive oocyte nuclei with characteristic surrounding granulosa cell layers were counted for each section. Counts were added to calculate the total number of oocytes per ovary. For each genotype, three to six ovaries were analyzed.
Reverse transcription PCR
Total RNA was isolated from various tissues of wild-type and knockout adult mice. To analyze the expression level of Zcwpw1 mRNAs in various tissues, equal amounts of cDNA were synthesized using the PrimeScript RT reagent kit with genomic DNA Eraser (Takara). Qualitative PCR was performed using TB Green Premix Ex Taq (Takara) and specific forward and reverse primers as follows: Zcwpw1 primer pair, 5′-GGGGACATCTAAGCTAGGCC-3′ (forward) and 5′-CTGCACTCACGGCCATCTT-3′ (reverse). β-Actin was amplified as a housekeeping gene with the primers 5′-CATCCGTAAAGACCTCTATGCCAAC-3′ (forward) and 5′-ATGGAGCCACCGATCCACA-3′ (reverse). All reverse transcription PCR reactions were performed with an initial denaturation at 95°C for 10 min, followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 5 min using a T100 Thermal Cycler (Bio-Rad).
Western blotting
To prepare extracts, tissues were collected from male C57BL/6 mice and suspended in lysis buffer (51) [50 mM HEPES-KOH (pH 7.5), 100 mM KCl, 2 mM EDTA, 10% glycerol, 0.1% NP-40, 10 mM NaF, 0.25 mM Na3VO4, and 50 mM β-glycerolphosphate] supplemented with cOmplete Protease Inhibitor (Roche #04693116001). After homogenization, the cell extracts were centrifuged at 20,000g for 20 min at 4°C. The supernatant extracts were used for immunoprecipitation and Western blots. Equal amounts of protein were electrophoresed on 10% SDS–polyacrylamide gels, and the bands were transferred to polyvinylidene fluoride membranes (Millipore, USA). Immunoreactive bands were detected and analyzed with a Bio-Rad ChemiDoc MP imaging System and Image Lab Software (Bio-Rad, USA). Relative protein levels in each sample were normalized to β-Actin to standardize the loading variations. The primary antibodies for immunoblotting included anti–β-Actin (1:10,000 dilution; Cell Signaling Technology, #4970) and anti-ZCWPW1 (1:1000 dilution; produced as described above).
Immunoprecipitation
Immunoprecipitation was performed with PD18 testes protein extracts using anti-ZCWPW1 (1:100 dilution) and anti-H3K4me3 (1:100 dilution; Abcam #ab8580) antibodies. Antibodies and Zcwpw1+/+ or Zcwpw1−/− testes extracts were incubated overnight at 4°C, followed by incubation with protein A agarose (Roche, #11719408001) and protein G agarose (Roche, #11719416001) for 1 hour at 4°C. The Western blots were repeated at least three times.
Imaging
Immunolabeled chromosome spreads were imaged by confocal microscopy using either a Leica TCS SP5 resonant scanning confocal microscope driven by the Leica Application Suite Software or an Andor Dragonfly spinning disc confocal microscope driven by Fusion Software. Projection images were then prepared using ImageJ Software (National Institutes of Health, version 1.6.0-65) or Bitplane Imaris (version 8.1) software. Histology samples were analyzed with an epifluorescence microscope (BX52, Olympus) and processed using Photoshop (Adobe) software packages. Super-resolution SIM analysis was performed with the Acquire SR software on a DeltaVision OMX SR SIM system (GE Healthcare) equipped with a 60×/1.42 oil objective, and the images were further computationally reconstructed and processed with SoftWoRx software (GE Healthcare) to generate super-resolution optical series sections with twofold extended resolution in both the xy and z axes.
Proteomic analysis
HPLC-MS was carried out using a protocol as described previously by Su et al. (54). Briefly, testes from wild-type and Zcwpw1−/− mice at PD14 were collected (three replicates). Testes were lysed in protein extraction buffer consisting of 75 mM NaCl, 50 mM tris (pH 8.2), 8 M urea, 1 mM NaF, 1% (v/v) EDTA-free protease inhibitor cocktail, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, and 1 mM phenylmethylsulfonyl fluoride. The samples were then subjected to tandem mass tags labeling. Aliquots of the same samples were combined, lyophilized, resuspended, and then loaded onto an XBridge BEH130 C18 column (2.1 mm by 150 mm, 3.5 μm; Waters) with an UltiMate 3000 HPLC system at a flow rate of 200 μl/min. For MS evaluation, 30 fractions were sequentially resuspended in 0.1% formic acid, and an LTQ Orbitrap Velos mass spectrometer (Thermo Finnigan, San Jose, CA) coupled on-line to a Proxeon EASY-nLC 1000 was used for analysis.
Statistical analysis
All data are presented as means ± SEM. The statistical significance of the differences between the mean values for the different genotypes was measured by Welch’s t test with a paired, two-tailed distribution. The data were considered significant when the P value was less than 0.05.
Supplementary Material
Acknowledgments
We are grateful for interesting discussions with colleagues from Zhejiang University, China, in the very initial phase of the project. Funding: This work was supported by the National Key Research and Development Programs of China (2018YFC1003700 and 2016YFC1000600), the National Natural Science Foundation of China (81771538), and the Young Scholars Program of Shandong University (2016WLJH50). Author contributions: Conceptualization: K.L., H.-B.L., and M.L. Methodology and investigation: M.L., T.H., M.-J.L., and C.-X.Z. Writing (original draft): M.L. and T.H. Writing (review and editing): M.L. and K.L. with assistance from all other authors. Funding acquisition: K.L., H.-B.L., and Z.-J.C. Supervision: H.-B.L. and K.L. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaax1101/DC1
Fig. S1. Zcwpw1 is generally expressed in different tissues.
Fig. S2. The SC structure appeared normal in Zcwpw1−/− spermatocytes.
Fig. S3. Representative images of leptotene and zygotene stages in Zcwpw1−/− spermatocytes.
Fig. S4. The SC structure appeared normal in Zcwpw1−/− oocytes.
Fig. S5. Protein profiling analysis of PD14 wild-type and Zcwpw1−/− testes.
Table S1. List of 94 differentially expressed proteins in PD14 Zcwpw1+/+ and Zcwpw1−/− testes by HPLC-MS.
REFERENCES AND NOTES
- 1.Handel M. A., Schimenti J. C., Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11, 124–136 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Baudat F., Imai Y., de Massy B., Meiotic recombination in mammals: Localization and regulation. Nat. Rev. Genet. 14, 794–806 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., Morita T., The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell 1, 707–718 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Cloud V., Chan Y.-L., Grubb J., Budke B., Bishop D. K., Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borde V., Robine N., Lin W., Bonfils S., Géli V., Nicolas A., Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28, 99–111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powers N. R., Parvanov E. D., Baker C. L., Walker M., Petkov P. M., Paigen K., The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLOS Genet. 12, e1006146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohenauer T., Moore A. W., The Prdm family: Expanding roles in stem cells and development. Development 139, 2267–2282 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Berg I. L., Neumann R., Lam K.-W. G., Sarbajna S., Odenthal-Hesse L., May C. A., Jeffreys A. J., PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 42, 859–863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K., High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., Przeworski M., Coop G., de Massy B., PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry J., Zhao Y., The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem. Sci. 28, 576–580 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., Reddy B., Thompson J., Wang H., Noma K.-i., Yates J. R. III, Jia S., Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol. Cell 33, 428–437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stec I., Nagl S. B., van Ommen G.-J. B., den Dunnen J. T., The PWWP domain: A potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473, 1–5 (2000). [DOI] [PubMed] [Google Scholar]
- 16.He F., Umehara T., Saito K., Harada T., Watanabe S., Yabuki T., Kigawa T., Takahashi M., Kuwasako K., Tsuda K., Matsuda T., Aoki M., Seki E., Kobayashi N., Guntert P., Yokoyama S., Muto Y., Structural insight into the zinc finger CW domain as a histone modification reader. Structure 18, 1127–1139 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Hoppmann V., Thorstensen T., Kristiansen P. E., Veiseth S. V., Rahman M. A., Finne K., Aalen R. B., Aasland R., The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 30, 1939–1952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams-Cioaba M. A., Min J., Structure and function of histone methylation binding proteins. Biochem. Cell Biol. 87, 93–105 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z., Yu Y., Meyer D., Wu C., Shen W.-H., Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7, 1256–1260 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Huang Y., Uncovering the mechanistic basis for specific recognition of monomethylated H3K4 by the CW domain of Arabidopsis histone methyltransferase SDG8. J. Biol. Chem. 293, 6470–6481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciccone D. N., Su H., Hevi S., Gay F., Lei H., Bajko J., Xu G., Li E., Chen T., KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Li D.-Q., Nair S. S., Kumar R., The MORC family: New epigenetic regulators of transcription and DNA damage response. Epigenetics 8, 685–693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi B., Xue J., Zhou J., Kasowitz S. D., Zhang Y., Liang G., Guan Y., Shi Q., Liu M., Sha J., Huang X., Wang P. J., MORC2B is essential for meiotic progression and fertility. PLOS Genet. 14, e1007175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schücker K., Holm T., Franke C., Sauer M., Benavente R., Elucidation of synaptonemal complex organization by super-resolution imaging with isotropic resolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2029–2033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkel K., Alsheimer M., Ollinger R., Benavente R., Protein SYCP2 provides a link between transverse filaments and lateral elements of mammalian synaptonemal complexes. Chromosoma 118, 259–267 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Cahoon C. K., Yu Z., Wang Y., Guo F., Unruh J. R., Slaughter B. D., Hawley R. S., Superresolution expansion microscopy reveals the three-dimensional organization of the Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. U.S.A. 114, E6857–E6866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner J. M. A., Aprelikova O., Xu X., Wang R., Kim S., Chandramouli G. V. R., Barrett J. C., Burgoyne P. S., Deng C.-X., BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol. 14, 2135–2142 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Tu Z., Bayazit M. B., Liu H., Zhang J., Busayavalasa K., Risal S., Shao J., Satyanarayana A., Coppola V., Tessarollo L., Singh M., Zheng C., Han C., Chen Z., Kaldis P., Gustafsson J.-Å., Liu K., Speedy A-Cdk2 binding mediates initial telomere-nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. Proc. Natl. Acad. Sci. U.S.A. 114, 592–597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teif V. B., Rippe K., Predicting nucleosome positions on the DNA: Combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Res. 37, 5641–5655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishi R., Wijnhoven P., le Sage C., Tjeertes J., Galanty Y., Forment J. V., Clague M. J., Urbe S., Jackson S. P., Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 16, 1016–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alver B. H., Kim K. H., Lu P., Wang X., Manchester H. E., Wang W., Haswell J. R., Park P. J., Roberts C. W. M., The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 8, 14648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schimenti J., Synapsis or silence. Nat. Genet. 37, 11–13 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Turner J. M. A., Mahadevaiah S. K., Fernandez-Capetillo O., Nussenzweig A., Xu X., Deng C.-X., Burgoyne P. S., Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37, 41–47 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Bellani M. A., Romanienko P. J., Cairatti D. A., Camerini-Otero R. D., SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell Sci. 118, 3233–3245 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Shiu P. K. T., Raju N. B., Zickler D., Metzenberg R. L., Meiotic silencing by unpaired DNA. Cell 107, 905–916 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Ichijima Y., Ichijima M., Lou Z., Nussenzweig A., Camerini-Otero R. D., Chen J., Andreassen P. R., Namekawa S. H., MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 25, 959–971 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner J. M. A., Meiotic sex chromosome inactivation. Development 134, 1823–1831 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Burgoyne P. S., Mahadevaiah S. K., Turner J. M. A., The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 10, 207–216 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Royo H., Polikiewicz G., Mahadevaiah S. K., Prosser H., Mitchell M., Bradley A., de Rooij D. G., Burgoyne P. S., Turner J. M. A., Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 20, 2117–2123 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Buard J., Barthès P., Grey C., de Massy B., Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 28, 2616–2624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grey C., Barthes P., Chauveau-Le Friec G., Langa F., Baudat F., de Massy B., Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLOS Biol. 9, e1001176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A., A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Acquaviva L., Székvölgyi L., Dichtl B., Dichtl B. S., de La Roche Saint Andre C., Nicolas A., Geli V., The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339, 215–218 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Sommermeyer V., Béneut C., Chaplais E., Serrentino M. E., Borde V., Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell 49, 43–54 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J. C., Ferrier P., Spicuglia S., H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 30, 4198–4210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brick K., Thibault-Sennett S., Smagulova F., Lam K.-W. G., Pu Y., Pratto F., Camerini-Otero R. D., Petukhova G. V., Extensive sex differences at the initiation of genetic recombination. Nature 561, 338–342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker M., Billings T., Baker C. L., Powers N., Tian H., Saxl R. L., Choi K., Hibbs M. A., Carter G. W., Handel M. A., Paigen K., Petkov P. M., Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin 8, 31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roeder G. S., Bailis J. M., The pachytene checkpoint. Trends Genet. 16, 395–403 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Gan H., Wen L., Liao S., Lin X., Ma T., Liu J., Song C.-x., Wang M., He C., Han C., Tang F., Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 4, 1995 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Peters A. H., Plug A. W., van Vugt M. J., de Boer P., A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Liu H., Huang T., Li M., Li M., Zhang C., Jiang J., Yu X., Yin Y., Zhang F., Lu G., Luo M.-C., Zhang L.-R., Li J., Liu K., Chen Z.-J., SCRE serves as a unique synaptonemal complex fastener and is essential for progression of meiosis prophase I in mice. Nucleic Acids Res. 47, 5670–5683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page J., Suja J. A., Santos J. L., Rufas J. S., Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res. 6, 639–642 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Liu L., Rajareddy S., Reddy P., Du C., Jagarlamudi K., Shen Y., Gunnarsson D., Selstam G., Boman K., Liu K., Infertility caused by retardation of follicular development in mice with oocyte-specific expression of Foxo3a. Development 134, 199–209 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Guo J., Zhang T., Guo Y., Sun T., Li H., Zhang X., Yin H., Cao G., Yin Y., Wang H., Shi L., Guo X., Sha J., Eppig J. J., Su Y.-Q., Oocyte stage-specific effects of MTOR determine granulosa cell fate and oocyte quality in mice. Proc. Natl. Acad. Sci. U.S.A. 115, E5326–E5333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaax1101/DC1
Fig. S1. Zcwpw1 is generally expressed in different tissues.
Fig. S2. The SC structure appeared normal in Zcwpw1−/− spermatocytes.
Fig. S3. Representative images of leptotene and zygotene stages in Zcwpw1−/− spermatocytes.
Fig. S4. The SC structure appeared normal in Zcwpw1−/− oocytes.
Fig. S5. Protein profiling analysis of PD14 wild-type and Zcwpw1−/− testes.
Table S1. List of 94 differentially expressed proteins in PD14 Zcwpw1+/+ and Zcwpw1−/− testes by HPLC-MS.


