Artificial intelligence validates evolution’s oldest mathematical model by learning about butterfly mimicry.
Abstract
Traditional anatomical analyses captured only a fraction of real phenomic information. Here, we apply deep learning to quantify total phenotypic similarity across 2468 butterfly photographs, covering 38 subspecies from the polymorphic mimicry complex of Heliconius erato and Heliconius melpomene. Euclidean phenotypic distances, calculated using a deep convolutional triplet network, demonstrate significant convergence between interspecies co-mimics. This quantitatively validates a key prediction of Müllerian mimicry theory, evolutionary biology’s oldest mathematical model. Phenotypic neighbor-joining trees are significantly correlated with wing pattern gene phylogenies, demonstrating objective, phylogenetically informative phenome capture. Comparative analyses indicate frequency-dependent mutual convergence with coevolutionary exchange of wing pattern features. Therefore, phenotypic analysis supports reciprocal coevolution, predicted by classical mimicry theory but since disputed, and reveals mutual convergence as an intrinsic generator for the unexpected diversity of Müllerian mimicry. This demonstrates that deep learning can generate phenomic spatial embeddings, which enable quantitative tests of evolutionary hypotheses previously only testable subjectively.
INTRODUCTION
Analyses of biological phenotype have traditionally used only subjective (1), categorical descriptions, although variation in many traits is fundamentally quantitative (2, 3). Consequently, evolutionary questions that have been the subject of speculation for more than 150 years (4, 5) have never before been addressed definitively because of the lack of phenotypic trait quantification. More recent, quantitative approaches to phenotypic analysis have included geometric measurement, outline or landmark analysis (geometric morphometrics) (6, 7), spectrophotometry (3), and image pixel comparisons. While such standard analyses have underpinned comparative anatomy, taxonomic designation (3), investigation of phenotypic evolution, and morphological phylogenetics (2), even the largest studies capture only a small fraction of real phenomic information, require laborious manual processing (8), may be sensitive to feature conjunction (e.g., pattern element duplication) or minor misalignment (7, 9), can produce descriptive statistics that are nonreplicable (1) or internally inconsistent (10), and produce evolutionary trees with consistent topological differences to genomic phylogenies (11). For such reasons, real-world evolutionary systems, such as the classic case study of mimicry in Heliconius butterflies (4, 12, 13), have previously been beyond the reach of comprehensive phenomic analysis. In consequence, fundamental questions in evolutionary theory have remained outstanding, including (i) the statistical congruence between phenomic and genomic phylogenies (11); (ii) the statistical significance and quantitative extent, during mutualistic Müllerian mimicry, of phenotypic convergence, the principal prediction of evolutionary biology’s first mathematical model (4, 13, 14); (iii and iv) the quantitative effects of hybridization (15, 16) and biogeographic distance (17, 18) on phenotypic convergence in mimicry; (v) the existence and quantitative extent of mutual phenotypic convergence [and therefore strict coevolution (19)] in mimicry (4, 13); and (vi) the effects of mimetic convergence on the coevolution of phenotypic novelty.
New advances in machine learning using deep neural networks (deep learning) (20) enable automated quantification of total similarity across large and diverse data samples, in Euclidean spaces of moderate dimensionality (21, 22). Within biology, however, deep learning has been applied primarily to image classification tasks (20). Here, we apply deep learning using a convolutional triplet neural network (21, 22) to additionally quantify total visible phenotypic similarity among dorsal and ventral photographs of 38 subspecies of Neotropical butterfly from the species Heliconius erato and Heliconius melpomene. The analysis includes 1234 butterfly specimens, widely sampling the total geographic range (fig. S2) and extensive wing pattern polymorphism within each species (fig. S4). This enables the first comprehensive, quantitative analysis of Müllerian mimicry across this classic (12, 23), but controversial (13) coevolutionary system. This provides quantitative answers to major outstanding questions in evolutionary theory, as well as the most phenotypically inclusive investigation, to date, of evolutionary convergence (24, 25) and a fully automated, objective data-capture method for phenotypic (morphological) phylogenetics.
RESULTS
Deep learning network output and accuracy
To quantify phenotypic distances between Heliconius butterflies, a deep convolutional neural network (fig. S1) was trained to classify photographs of Heliconius butterflies by subspecies, with 1500 of 2468 total images used for network training and the remainder for testing. The training method (21, 22) used triplets of images, each replicate showing the network two images sampled from the same subspecies and one sampled from a different subspecies. The deep convolutional neural network (which we name ButterflyNet) learnt both to correctly classify images by subspecies and to calculate an internally consistent set of Euclidean distances between input images, with those of the same subspecies closer together and those of different subspecies further apart. The trained network achieved 86% accuracy in classifying test images by subspecies (compared to a chance value of 5%). Crucially, the network also calculated coordinates of all 2468 images within a phenotypic spatial embedding, with dimensionality set to 64 (table S3). In doing so, the network identifies and uses a subset of information from the input images that is sufficient to discriminate the taxonomic groups with the reported accuracy. A support vector classifier (SVC) trained on the spatial embeddings similarly achieved 87% subspecies classification accuracy, confirming that the spatial embedding preserves information sufficient for successful classification.
Phylogenetic results
Neighbor-joining trees, reconstructed from phenotypic distances between subspecies (Fig. 1 and fig. S6), group interspecies mimics rather than separating the species, consistent with evolutionary convergence, as discussed below (although species are successfully separated by specific embedding axes; Fig. 2F). However, recovered intraspecific relationships, in particular, show significant similarities to independent, published phylogenies based on gene sequences (for an overlapping subset of 25 subspecies) (Fig. 1) (26, 27). For both species (separately and combined), the phenotypic trees are significantly more similar to color pattern gene trees than are randomly generated trees (Mann-Whitney pairwise comparisons, P ≤ 2.59 × 10−256; table S10). This demonstrates that the phenotypic distances generated by deep learning contain significant phylogenetic information relative to a null hypothesis of random similarity. On average, intraspecific trees of phenotypic distance are significantly more similar to trees reconstructed from genes associated with wing color pattern (27) than they are to neutral gene loci (“housekeeping genes”) or to a control sample of random phylogenetic topologies (Mann-Whitney pairwise comparisons: H. erato, P ≤ 7.128 × 10−35; H. melpomene, P ≤ 1.645 × 10−13; and all subspecies, P ≤ 0.0066; Fig. 1 and table S10). For H. erato (Fig. 1E) phenotypic phylogenies are more similar to color gene phylogenies than color gene phylogenies are to neutral gene phylogenies. Statistics repeated after exclusion of hybrid specimens (table S2) confirm these results (table S11). We can note that the differences between gene histories are expected because of standard population genetic processes and a greater difference between genetic partitions is apparent in H. erato (Fig. 1E), which tends to have larger population sizes and carries more genetic variation (26, 28, 29). Overall, these results show that, despite a strong signal from mimicry between the species, the phenotypic spatial embedding has successfully recovered additional phylogenetic information within the image dataset [originating from the combination of lineal descent and historical hybridization (26, 28)].
Fig. 1. Phylogenetic relationships between subspecies of H. erato and H. melpomene.
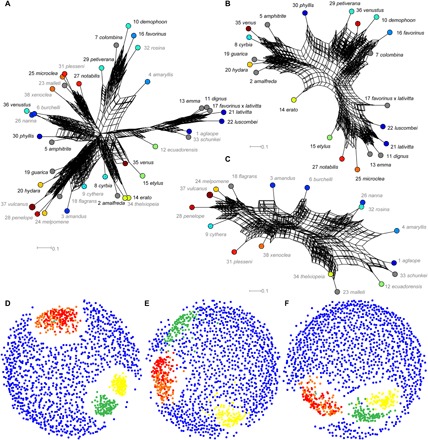
(A to C) Neighbor-joining consensus networks of phenotypic distance (eight sampled embedding axes, 100 replicates). (D to F) Tree space visualizations for phylogenies based on phenotype (orange and red, 32 embedding axes; red, hybrids excluded; cyan, 64 embedding axes), color pattern genes (green) (27), neutral genes (yellow), and randomized topologies (blue, 1000 replicates). (A and D) All subspecies. (B and E) Only H. erato. (C and F) Only H. melpomene. (A to C) Node label color indicates species (black, H. erato and gray, H. melpomene). Node colors show mimicry groups (tables S1 and S9). Node numbers show subspecies numbers (table S1).
Fig. 2. Principal component visualization of phenotypic variation among Heliconius butterflies.
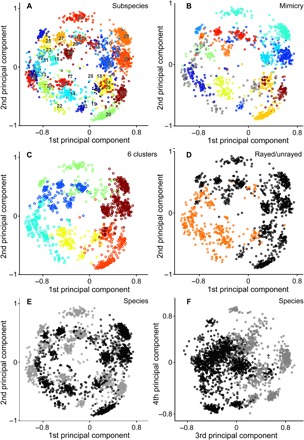
Principal component scores calculated for 2468 images of butterfly species H. erato and H. melpomene on the basis of image coordinates in a 64-dimensional phenotypic space, generated using a deep convolutional triplet network. Cumulative variance explained by displayed principal component axes: 1, 28%; 2, 50%; 3, 68%; and 4, 81%. (A) Butterfly subspecies 1 to 38 (Fig. 1 and table S7). (B) Twelve traditionally hypothesized (12) mimicry complexes (tables S1 and S9) of H. erato and H. melpomene subspecies (gray circles indicate nonmimics, not included in any of these mimicry complexes). (C) Six hierarchical clusters. (D) Two broad classes of type pattern for each subspecies (table S8), with orange rays (orange circles) or without rays (black circles). (E and F) Species, H. erato (black circles) and H. melpomene (gray circles).
Quantification of phenotypic similarity
Across the complete dataset, specimens of the same subspecies [exhaustively sampled from the Natural History Museum (NHM) collection] are clustered in a visualization of the first two principal component axes constructed from the phenotypic spatial embedding (Fig. 2A). Similarly, statistical analyses of average pairwise Euclidean distances (table S4) calculated directly from the original embedding coordinates (table S3) show that the most phenotypically similar butterfly images are those of the same subspecies (Kruskal-Wallis, P = 1.043 × 10−28; H = 128.9; Mann-Whitney pairwise, P ≤ 5.495 × 10−7; Fig. 3A, left). Alongside the high subspecies classification accuracy (≥86%), this demonstrates that named subspecies of H. erato and H. melpomene (30) are objectively distinguishable, despite phenotypic variability among individuals and incomplete reproductive isolation between subspecies within each species.
Testing phenotypic convergence
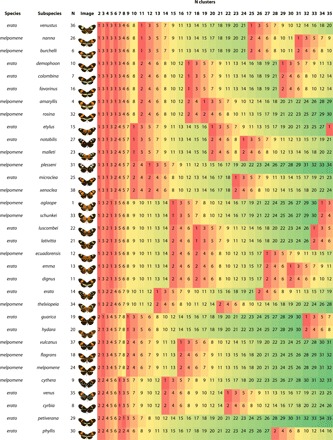
Pairwise comparisons across subspecies (Fig. 3A, middle right) show that traditionally hypothesized sympatric co-mimics (Figs. 1 and 2, fig. S3, and table S1) (12) are significantly more similar to each other than are other subspecies pairs (Mann-Whitney pairwise, P = 5.50 × 10−7). Comparisons performed separately for each species confirm these results (Kruskal-Wallis, P = 1.48 × 10−23; H = 113.2; Mann-Whitney pairwise comparisons mimicry versus identity or other, P ≤ 4.16 × 10−4; identity versus other, P ≤ 6.43 × 10−11; interspecies identity and other, nonsignificant, respectively P = 0.9532, P = 0.3285; Fig. 3B). This highlights the remarkable level of adaptive phenotypic evolution by these species, in which mimetic phenotypic similarity between the species (Figs. 2B and 3B, middle) is greater than the subspecies similarity within them (Figs. 2E and 3B, right). Across the 38 sampled subspecies, six distinct phenotypic clusters, identified by both hierarchical clustering (Fig. 2C and Table 1) and neighbor joining (fig. S6), include interspecies co-mimics.
Fig. 3. Average pairwise Euclidean phenotypic distances between subspecies of H. erato and H. melpomene.
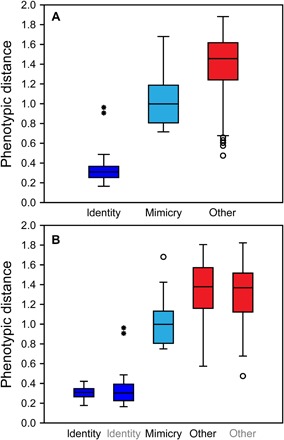
(A) Box plot of mean pairwise phenotypic distances (table S4) within subspecies (identity), between co-mimic subspecies (mimicry), and between all other subspecies (other). Sample sizes are 38, 19, and 684 subspecies pairs, respectively. (B) Separated species, H. erato (black labels, identity and other) and H. melpomene (gray) and interspecies co-mimics (mimicry). Sample sizes are 21, 17, 15, 209, and 133 pairs. Boxes show 25 to 75% quartiles; horizontal lines, medians; whiskers, inner fence within 1.5 × box height; circles and asterisks, outliers, respectively, within or beyond 3 × box height.
Table 1. Hierarchical clusters of subspecies from H. erato and H. melpomene calculated from the phenotypic spatial embedding.
Cluster membership is based on the modal specimen value for each subspecies after exclusion of hybrid specimens (table S2). Subspecies are illustrated by the dorsal photograph closest to the subspecies principal component analysis centroid (corresponding to Fig. 2A). N indicates subspecies number (table S1). Cluster numbers are colored by value to highlight divisions.
Hybridization
Statistical comparisons of phenotypic distance with hybrids excluded confirm that co-mimics (Fig. 1, fig. S3, and table S1) (12) are significantly more similar to each other than are other subspecies pairs, with a reduced average distance between the co-mimics and increased statistical significance compared to the complete dataset (hybrids excluded Mann-Whitney pairwise, P = 2.33 × 10−8; average distance mimics = 0.897, n mimics = 15; complete dataset: average distance mimics = 1.027, n mimics = 19).
Biogeography
Butterfly individuals sampled from subspecies that are traditionally hypothesized co-mimics are also confirmed to be geographically closer, on average, than those from other subspecies pairs (Kruskal-Wallis, P = 4.23 × 10−16; H = 70.8; Mann-Whitney, P ≤ 0.0066; pairwise, P = 0.0041; fig. S5). This indicates that geographic proximity has promoted interspecies convergence in mimicry (17, 18), between H. erato and H. melpomene, across their Neotropical range.
Testing mutual convergence
The extent of mutual convergence in mimicry is illustrated by a case study (Fig. 4) from three comparative analyses (covering 12 of the studied subspecies), all of which are supportive of reciprocal coevolutionary influence (fig. S7). Comparisons of the average locations of these subspecies in phenotypic space (Fig. 4) indicate statistically significant mutual convergence for this case study (with H. erato cyrbia closer to H. melpomene cythera than is H. erato venus and H. melpomene cythera closer to H. erato cyrbia than is H. melpomene vulcanus; H. erato mean distance from conspecific = 0.26; Mann-Whitney, P = 1.0195 × 10−15; H. melpomene mean distance = 0.41; P = 5.1718 × 10−31). The calculated extent of convergence by H. melpomene is 1.6 times that by H. erato (1.4 with hybrids excluded; fig. S7). Of the three comparative analyses (fig. S7), two indicate mutual convergence (Fig. 4 and fig. S7, A and B), and one indicates divergence by H. erato outside the geographic range of H. melpomene.
Fig. 4. Comparative analysis of the extent of phenotypic convergence in mimicry.
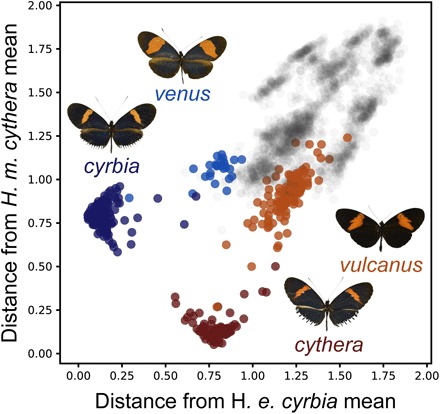
Case study from comparative analyses of 12 subspecies (fig. S7). The locations of two focal co-mimics (H. erato cyrbia, dark blue circles; H. melpomene cythera, dark red circles) in phenotypic space are compared alongside their nearest conspecifics (H. erato venus, light blue circles; H. melpomene vulcanus, light red circles). Subspecies are illustrated by dorsal photographs of the butterfly closest to the mean location for the subspecies. Gray circles indicate images of other subspecies in the dataset. Axes show the squared distance from the mean location of the focal co-mimic, summed across all 64 spatial embedding axes. Distance between subspecies means on the y axis, H. erato venus − H. erato cyrbia = 0.26. Distance between subspecies means on the x axis, H. melpomene vulcanus − H. melpomene cythera = 0.41.
Convergence on novel patterns
This case study (Fig. 4) also shows that, for both H. erato and H. melpomene, a pattern feature shared by the focal co-mimics is predominant in one of their conspecifics, but not the other, supporting mutual transfer of pattern features between these lineages. Specifically, H. erato cyrbia and H. melpomene cythera both have strongly blue, iridescent wings, a characteristic typical of H. erato venus but not of H. melpomene vulcanus [which is more black, particularly on the hindwing, e.g., individuals closest to the mean location for the subspecies (Fig. 4); see also type descriptions (31)]. Conversely, phenotypically average H. erato cyrbia and H. melpomene cythera have comparatively narrow red forewing bands, a feature more typical of H. melpomene vulcanus than of H. erato venus [average individuals (Fig. 4); type descriptions (31)]. This represents a newly demonstrated mechanism for the coevolution of novel phenotypes, as discussed below.
DISCUSSION
Quantification of phenotypic similarity using deep learning
The spatial embedding constructed by the deep convolutional neural network captures and systematizes the phenotypic variation among Heliconius butterfly subspecies, from relatively subtle differences in the size, shape, number, position and color of wing pattern features (Fig. 2A, Table 1, and fig. S4) to broad divisions between major patterns groups (Fig. 2, C and D, and Table 1). This provides a comprehensive quantification of visible phenotypic similarity and an objective test of taxonomic delimitation (3), difficult or impossible to achieve with traditional morphometric methods (6, 7) because of the scale and complexity of phenotypic variation.
The availability of genetic distances for butterfly subspecies studied here provides a ground truth against which the phenotypic distances, generated using the deep convolutional triplet network, can be tested. This ground truth is closely related to the natural selection pressures on wing pattern phenotype that these butterflies experienced during their evolutionary history, especially for genes specifically associated with wing pattern phenotype (27). The statistically significant similarity between the phenotypic neighbor-joining trees generated using deep learning and independently reconstructed pattern gene phylogenies (26) therefore confirms the utility of this spatial embedding method (21, 22) for evolutionary analysis. This demonstrates that deep learning can achieve informative quantification of biological phenome samples at microevolutionary (intraspecies) to macroevolutionary (interspecies) scales. Our finding that deep learning can recover significant phylogenetic information from phenome samples also validates what is, to our knowledge, the first fully automated, objective method for the construction of phenotypic (or “morphological”) phylogenies. This is of particular significance since subjective morphological phylogenies and taxonomic relationships (traditionally based on morphology) have been shown to exhibit statistical nonreplicability between researchers (1) and consistent topological biases (11).
Phenotypic convergence in Müllerian mimicry
Traditional hypotheses of mimicry were based on subjective qualitative assessments of phenotypic similarity between subspecies with overlapping geographic ranges (12). The significantly lower phenotypic distances between co-mimic subspecies of H. erato and H. melpomene relative to nonmimics (Fig. 3B) now quantitatively demonstrate evolutionary convergence (24, 25) in visible phenotype during the evolution of mimicry, considering as input all phenotypic information from the 2468 dorsal and ventral butterfly photographs. Historical, qualitative discussions of convergence have also focused primarily on taxonomic type patterns (based on a small number of type specimens selected as representative examples for each subspecies) (31). Our analysis of specimens exhaustively sampled from the Natural History Museum collection demonstrates statistically significant convergence across this comprehensive sample of wild phenotypic variation. This indicates that, even with the inclusion of individual variation and natural hybrids (table S2), evolutionary convergence in visual mimicry exerts a dominant statistical signal.
Hybridization occurs naturally between some intraspecific subspecies (and with some other Heliconius species, although H. erato and H. melpomene do not interbreed) both at hybrid zones where adjacent geographic ranges meet and, sometimes, far from these range boundaries. Hybrids may deviate perceptibly from a locally common wing pattern, in which case preferential predation can provide purifying selection that stabilizes mimicry (17). Hybridization is also increasingly recognized as a source of wing pattern variation (in addition to standard mutation) that can promote reproductive isolation and ecological speciation (15). Consequently, hybridization forms an important component of this mimicry system, with potentially complex effects on mimicry evolution. Statistical comparisons of phenotypic distance with hybrids excluded demonstrate that an even stronger signal of mimicry is apparent than for the complete dataset. These findings confirm theoretical suggestions that the general effect of wild hybridization between Heliconius subspecies is to dilute the strength of phenotypic mimicry (16), although mimicry remains the dominant phenotypic signal even with hybrids included.
The two species, H. erato and H. melpomene, have independently evolved similar wing patterns multiple times, with a hierarchy of convergent pattern detail (Fig. 2, B to D, and Table 1). Across the 38 subspecies, there are six distinct and convergent phenotypic clusters (Fig. 2C, Table 1, and fig. S6), which include interspecies co-mimics. These convergent phenotypic clusters include, for example, the orange “rayed” patterns, nonrayed pattern types containing red and black “postman” patterns (Fig. 2D), with or without a yellow hindwing band (hierarchical clustering with three clusters) and with red versus yellow forewing pattern features (four clusters), as well as patterns with a yellow forewing feature of increased complexity (five clusters).
This comprehensive, quantitative demonstration of phenotypic convergence meets the principal prediction of Müller’s 1879 (4) model for the evolution of mimicry in mutually protected species (Müllerian mimics), such as unpalatable Heliconius butterflies (12). In this, the first mathematical model of Darwinian relative fitness (4, 14), Müller predicted that two equally unpalatable, co-occurring populations will come to resemble each other because both benefit by sharing the cost of predator avoidance learning, with relative fitness benefits proportional to the inverse square of abundance [with additional abundance effects suggested by later reformulations (14)] (4). Relative abundance has been estimated respectively at 1.3:1 to 2.4:1 for H. erato and H. melpomene overall (28, 29) [predicting respective benefits of 1:1.7 to 1:5.8 (4)] but is subject to marked local fluctuations, in which momentary relative abundances of the species may be equal or reversed relative to the overall trend (32).
Mutual convergence and strict coevolution
The historically controversial (13) extent of mutual convergence in mimicry is illustrated by a detailed case study (Fig. 4) from three comparative analyses (covering 12 subspecies), all of which support reciprocal influence (fig. S7), which is the definitive feature of strict coevolution (19). Focal co-mimics H. erato cyrbia and H. melpomene cythera (Fig. 4) share a white hindwing marginal fringe and strong blue iridescence of the dorsal wing surface also present in their phenotypically closest conspecifics, H. erato venus and H. melpomene vulcanus [conspecifics also recovered as sister groups in independent gene phylogenies (26)]. Both pattern features are likely to have been secondarily derived (rather than ancestral within each species) on the basis of gene phylogenies, biogeographic distribution, and phylogeographic reconstruction (12, 16, 26, 33). Geographic ranges (fig. S2) and phylogeographic reconstruction (26) also suggest that H. erato cyrbia and H. melpomene cythera are at an extreme of a west Andean subradiation (with conspecifics adjacent, north-east). This supports pattern derivation from the forms of H. erato venus and H. melpomene vulcanus to those of H. erato cyrbia and H. melpomene cythera (rather than vice versa). This shared biogeographic history is also compatible with the potential for strict interspecies coevolution, since reciprocal evolutionary influence between two taxa requires that they co-occur in both space and time (26). With regard to timing, while early phylogenetic studies cast doubt on the extent of contemporaneous diversification by H. erato and H. melpomene (34), subsequent genomic analyses (taking into account population sizes) have reconstructed their diversification over closely overlapping time ranges (28, 29).
Comparisons of the average locations of these subspecies in phenotypic space (Fig. 4) show statistically significant mutual convergence. The implied extent of convergence by H. melpomene is 1.6 times that by H. erato, in line with the general frequency-dependent fitness benefits expected in Müllerian mimicry (4) between these species (with hybrids excluded, the value is 1.4; fig. S7). Of the three comparative analyses (fig. S7), two indicate mutual convergence (Fig. 4 and fig. S7, A and B), and one indicates divergence by H. erato where this subspecies leaves the geographic range and coevolutionary influence of H. melpomene [in Central-North American H. erato petiverana, fig. S7C; see also discussion in (23)]. For comparison, reversal of evolutionary polarities (swapped conspecific foci) would imply two cases of mutual convergence (fig. S7, A and B) and one of divergence by H. melpomene (fig. S7C).
Interpreted with regard to the extent of reciprocal influence in wing pattern evolution, which has been controversial and difficult to test (13, 35), mutual convergence (Figs. 3 and 4 and fig. S7, A and B) and maintenance of wing pattern similarity in sympatry (e.g., fig. S7C) suggest that H. erato and H. melpomene have influenced each other (albeit to varying extents), with each species acting as both model and mimic to some degree, meeting the essential condition of strict reciprocal (13, 19) coevolution. This is in sympathy with classical Müllerian mimicry theory (4) while in contrast to theoretical alternatives such as entirely one-sided advergence (Fig. 5) (13) or divergence of a sympatric model during parasitic, quasi-Batesian mimicry (36).
Fig. 5. Conceptual diagrams illustrating the evolutionary alternatives of mutual convergence, mutual divergence, and one-sided convergence (“advergence”).
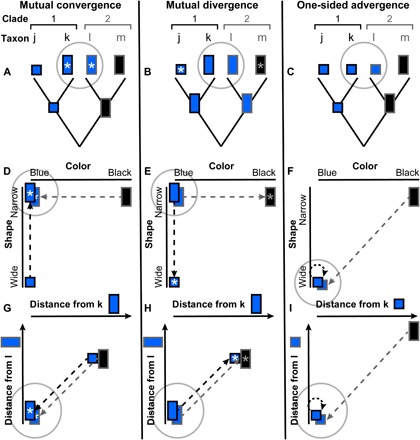
Dashed arrows indicate the direction of evolutionary change. Left (A, D, and G): Mutual convergence in focal taxa (focal taxa, gray circles) with reciprocal transfer of pattern features (e.g., forewing band shape versus wing color) between two clades (1 and 2, respectively black versus gray outlines). Middle (B, E, and H): Reversed polarity with mutual divergence from the focal taxa. Right (C, F, and I): Advergence by one clade onto another (13). Asterisks indicate new derived patterns (feature combinations). When expressed in terms of the phenotypic distance from the focal taxa (G to I), mutual convergence (G) is characterized by a decreasing distance along the arrow of evolutionary change in both clades. Mutual divergence (H) is characterized by a increasing distance in both clades. Advergence (13) is characterized by a decreasing distance (and a greater distance traveled) in one clade (I).
Coevolution of phenotypic novelty
The focal case study of H. erato cyrbia, H. melpomene cythera, and their closest conspecifics (Fig. 4) also indicates mutual coevolutionary transfer of pattern features between interspecific lineages (Fig. 4; conceptual diagram in Fig. 5). This represents a class of phenomic recombination, generating novel phenotypic combinations, but acting via predator-mediated selection on phenotype (and underlying genotype) without direct gene exchange. To our knowledge, this mechanism for the generation of evolutionary novelty represents a newly identified coevolutionary phenomenon. However, coevolutionary pattern recombination is also a logical consequence of the mutual convergence predicted by classical Müllerian mimicry theory (4, 23, 37) and exemplifies the definitive feature of coevolution: coordination of evolutionary change in genetically distinct populations (38). Mutual convergence, of the type identified here (Fig. 4), can be considered equivalent to reciprocal advergence (where, in this usage, advergence means one-sided mimicry evolution (13)). One-sided advergence involves evolution in one lineage to match a phenotypic feature already present in its mimicry model (Fig. 5). This represents a coevolutionary information transfer from one lineage to another. Consequently, mutual convergence (i.e., reciprocal advergence) in two distinct pattern features entails reciprocal information exchange, with the potential to create a novel pattern, which did not previously exist in any lineage (Figs. 4 and 5). For example, in two lineages (labeled 1 and 2) that mutually converge (Figs. 4 and 5A) across two pattern features, forewing band shape (wide or narrow) and wing color (black or blue), the ancestral patterns (here inferred as wide, blue in lineage 1 and narrow, black in lineage 2) are then supplemented by a newly derived combination (here narrow, blue in both lineages). Thus, lineage 1 has taken on the ancestral band shape of lineage 2, while lineage 2 has picked up the ancestral wing color of lineage 1, to generate a new wing pattern.
This reveals that Müllerian mimicry can generate new phenotypes by combining pattern features from different lineages (Figs. 4 and 5 and fig. S7). While it is natural to expect that divergence will generate new phenotypic traits (Fig. 5, B, E, and H), the demonstration that evolutionary convergence can also generate new phenotypes is more unexpected (Figs. 4 and 5, A, D, and G). An intuitive expectation was that mimicry would drive convergence on a single-wing pattern (13) and classical discussion of coevolution (4, 12) did not itself explain the remarkable phenotypic diversity observed among Müllerian mimics (13). Consequently, proposed diversification mechanisms have often focused on external factors such as microhabitat adaptation (13), variable predator abundance (18), and isolation in glacial refugia (12), as well as effects from additional butterfly species (12) and stochastic population genetics (13, 18) (although all have been controversial). Here, direct quantitative analysis of phenotype reveals that mutual convergence can itself increase phenotypic diversity.
MATERIALS AND METHODS
Image acquisition and preprocessing
Butterfly specimens (1269) from the species H. erato and H. melpomene were photographed at the NHM London (specimen numbers and image filenames, table S2), using consistent photographic protocols and lighting. Photographs were screened for poor image quality giving a final dataset of 1234 butterflies and 2468 photographs, including a dorsal and ventral photograph of each butterfly. For deep learning, images were then cropped and resized to a height of 64 pixels (maintaining the original image aspect ratio and padded to 140 pixels wide). The photographic dataset used for deep learning is provided in the Dryad Data Repository with filenames corresponding to joined data in table S2.
Taxonomic and locality data were recorded from NHM Heliconius butterfly specimen labels (table S2). Subspecies taxonomy follows reference (30). The complete photographic dataset covers 37 named subspecies and one-labeled cross: 21 subspecies from H. erato and 17 from H. melpomene. Specimens of these subspecies were sampled exhaustively from the NHM collection, within the limits of the data collection period. The complete photographic dataset (fig. S4, Dryad Data Repository) covers both specimens closely representative of subspecies descriptions (30, 31) (including available holotypes, syntypes, and paratypes; table S2) as well as other, naturally varying, individuals. These variants include some likely hybrid specimens showing varying levels of phenotypic admixture from other subspecies (see additional taxonomic information, table S2). Inclusion of all available specimens in machine learning covered a very broad range of the phenotypic diversity within these species, providing the deep learning network with all available information from which to learn phenotypic features correlated with subspecies identification. Two sets of statistical analyses were then conducted, one set including all 1269 photographed butterfly specimens and the second set excluding potential hybrid specimens to give a reduced dataset of 815 specimens and 1630 photographs (fig. S4 and table S2).
Deep learning
To quantify phenotypic distances between Heliconius butterflies, a deep convolutional neural network was trained to classify photographs of Heliconius butterflies by subspecies, with 1500 of the 2468 total images used for network training and the remainder for testing. The training method (21, 22) used triplets of images, each replicate showing the network two images sampled from the same subspecies and one sampled from a different subspecies. Image classification and spatial embedding were performed using a 15-layer deep learning network (Supplementary Computer Code), which we name ButterflyNet (fig. S1). This makes use of a triplet embedding loss function (21, 22) to train a network to organize its inputs (images) in a space such that proximity in that space (Euclidean distance) is highly correlated with identity (in this case subspecies). The learned embedding was then passed through an additional small network to perform direct categorical subspecies classification. Overall, the total network optimizes the sum of the triplet loss and the categorical cross entropy (eqs. S1 and S2). The computer code used for machine learning is provided as a Python script (Supplementary Computer Code), which makes use of the PyTorch, Scikit-l arn, and Adam packages (for further details, see Supplementary Methods).
After the network was trained on 1500 images randomly sampled from the 2468 images in the dataset, network testing was performed on the remainder (968 images). Testing presents the trained network with new images, which it has not encountered before. The network then classifies the new images by subspecies, image classifications are compared to the known subspecies identities, and the overall accuracy of test classifications is reported. Additional testing was performed using an SVC trained on the embeddings from the main network (ButterflyNet) to determine the accuracy of classification of specimens to subspecies based on their locations in the phenotypic spatial embedding.
Quantification of phenotypic similarity
Pairwise Euclidean phenotypic distances between all images were calculated from the coordinates of all 2468 images within a spatial embedding with 64 dimensions, generated using the network (table S3). These distances were then used to calculate the average pairwise Euclidean phenotypic distances between all subspecies (table S4). These were then used in statistical comparisons between sets of unique unordered subspecies pairs (Fig. 3; for further details, see Supplementary Methods). Average phenotypic distances for subspecies sets and principal component scores were calculated using MATLAB scripts. Nonparametric statistical analyses were conducted using the program PAST 3, after Shapiro-Wilk’s tests indicated that distances for some subspecies sets were non-normally distributed (using an α value of 0.05). These analyses included Kruskal-Wallis tests for equal medians and, where this overall test was significant, subsequent Mann-Whitney pairwise comparisons of statistical distributions between groups.
Phylogenetic analyses
Neighbor-joining trees for subspecies were constructed on the basis of phenotypic distances using MATLAB scripts (sampling either all 64 embedding axes with 1 replicate or subsamples of 8 or 32 axes with 100 replicates). Neighbor joining is a simple and fast algorithm for phylogenetic reconstruction, which reconstructs relationships based on phenetic (overall) similarity (39) e.g., Euclidean phenotypic distance, as applied here. Neighbor joining does not require an a priori mechanistic model for the evolutionary process in question (e.g., evolution of butterfly wing pattern phenotype), in contrast to maximum likelihood models of DNA substitution, for example. This method is therefore suitable for this first phylogenetic study of phenomic distances calculated using deep learning on butterfly photographs. The correlation was then statistically compared between the resultant phenotypic neighbor-joining trees and genetic phylogenies reconstructed with Bayesian methods (which incorporate DNA substitution models) (27). To test the phylogenetic informativeness of the phenotypic distances against such independent data sources, sets of neighbor-joining phenotypic trees (of either all subspecies, H. erato only, or H. melpomene only) were compared against random tree topologies and phylogenies (26) reconstructed from published gene sequences (27) from gene loci (sampled from a different, smaller set of 127 butterfly individuals), which were either associated with Heliconius wing color pattern (optix, bves, kinesin, GPCR, and VanGogh) (27) or were neutral markers (mt COI-COII, SUMO, Suz12, 2654, and CAT). Pairwise distances between trees from the different sets were calculated using the Robinson-Foulds (symmetric distance) metric in the program PAUP and statistically compared using nonparametric Mann-Whitney tests in the program PAST (after Shapiro-Wilk’s tests indicated non-normal distributions). Tree space visualizations of tree similarity were produced, based on the Robinson-Foulds distance, using the tree set visualization package in the program Mesquite. Consensus networks were constructed to visualize all splits (taxon partitions) implied among sets of trees using the program SplitsTree 4.
Testing evolutionary convergence
Quantitative tests of evolutionary convergence were applied using an operational definition of convergence essentially the same as that for discrete traits (independent derivation of the same morphological state, e.g., within two species), with quantitative analysis used to test relative phenotypic similarity across the complete dataset (24, 25) and the number of quantitatively distinct phenotypic states, e.g., clusters (for further details, see Supplementary Methods).
To further explore the extent of reciprocal convergence in mimicry between H. erato and H. melpomene, detailed comparative analyses (40) were then conducted using 12 selected subspecies (fig. S7). First, sets of four subspecies were identified, with each set consisting of two pairs of interspecies co-mimics in which conspecifics were nearest neighbors in the phenotypic spatial embedding, permitting phenotypic sister-group comparisons (fig. S7, A and B). Focal co-mimics, with pattern features that are potentially derived, rather than ancestral, were identified for each comparative analysis based on all available independent information from gene phylogenies, biogeographic distribution (fig. S2), and phylogeographic reconstruction (12, 16, 26, 33). For comparison, the analyses were then repeated with reversed polarity. In each comparative analysis, the position in phenotypic space of each of the focal subspecies was compared to that of their nearest conspecific [a type of sister-group comparison (25)]. Mann-Whitney tests for equal medians tested whether conspecifics differed significantly in their distance from the focal co-mimic of the other species (after Shapiro-Wilk’s tests indicated that some subspecies values were non-normally distributed).
Supplementary Material
Acknowledgments
Data compilation received additional assistance from volunteer research assistant S. Sjosten. We thank J. Mallet and S. Conway Morris for discussion of the manuscript as well as J. Turner and two anonymous referees for highly constructive reviews. Funding: Funding was received from an ELSI Origins Network (EON) Research Fellowship (J.F.H.C.) supported by a grant from the John Templeton Foundation. Author contributions: This study was originally designed by J.F.H.C. and N.G. S.L. and R.C. collected data, as supervised by B.H. and J.F.H.C. N.G. and J.F.H.C. wrote the computer code. J.F.H.C. performed statistical analyses. J.F.H.C. wrote the manuscript with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional photographic data related to this paper are available on the Dryad data repository, DOI: 10.5061/dryad.2hp1978.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaaw4967/DC1
Supplementary Methods
Fig. S1. Diagram of the architecture of the deep learning network ButterflyNet used in this study.
Fig. S2. Geographic localities for sampled butterfly specimens from the polymorphic mimicry complex of H. erato and H. melpomene.
Fig. S3. Heatmap showing mean pairwise phenotypic and geographic distances between 38 subspecies of H. erato (black labels) and H. melpomene (gray labels).
Fig. S4. Collections of specimen photographs used in this study, grouped by subspecies.
Fig. S5. Average pairwise Euclidean geographic distances between subspecies of H. erato and H. melpomene.
Fig. S6. Neighbor-joining trees of phenotypic distance between subspecies of H. erato and H. melpomene.
Fig. S7. Comparative analyses of the extent of phenotypic convergence in mimicry.
Fig. S8. Principal component visualization of Heliconius butterflies.
Table S1. Traditionally hypothesized co-mimic subspecies of H. erato and H. melpomene.
Table S2. Taxonomic and locality data recorded for historical specimens of H. erato and H. melpomene held in the collections of the NHM London.
Table S3. Coordinates of butterfly images on 64 axes of a Euclidean phenotypic space constructed using a deep convolutional network with triplet training.
Table S4. Mean pairwise Euclidean phenotypic distances between subspecies from H. erato and H. melpomene.
Table S5. Mean pairwise squared Euclidean phenotypic distances between subspecies from H. erato and H. melpomene.
Table S6. Mean pairwise Euclidean geographic distances between subspecies from H. erato and H. melpomene.
Table S7. Number of sampled butterfly individuals for each subspecies.
Table S8. Broad pattern class of the type specimen of each subspecies.
Table S9. Traditionally hypothesized mimicry complexes of H. erato and H. melpomene subspecies.
Table S10. Statistical comparisons of pairwise Robinson-Foulds distances between sets of phylogenetic trees.
Table S11. Statistical comparisons with hybrids excluded of pairwise Robinson-Foulds distances between sets of phylogenetic trees.
Supplementary Computer Code in ipynb Format
Supplementary Computer Code in PDF Format
Reference (41)
REFERENCES AND NOTES
- 1.J. A. Hawkins, A survey of primary homology assessment: Different botanists perceive and define characters in different ways, in Homology & Systematics: Coding characters for phylogenetic analysis (Taylor and Francis, 2000), pp. 22–53. [Google Scholar]
- 2.Wiens J. J., Character analysis in morphological phylogenetics: Problems and solutions. Syst. Biol. 50, 689–699 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Tobias J. A., Seddon N., Spottiswoode C. N., Pilgrim J. D., Fishpool L. D. C., Collar N. J., Quantitative criteria for species delimitation. Ibis 152, 724–746 (2010). [Google Scholar]
- 4.Müller F., Ituna and Thyridia; a remarkable case of mimicry in butterflies. Trans. Entomol. Soc. Lond. 12–29 (1879). [Google Scholar]
- 5.Merrill R. M., Dasmahapatra K. K., Davey J. W., Dell'Aglio D. D., Hanly J. J., Huber B., Jiggins C. D., Joron M., Kozak K. M., Llaurens V., Martin S. H., Montgomery S. H., Morris J., Nadeau N. J., Pinharanda A. L., Rosser N., Thompson M. J., Vanjari S., Wallbank R. W. R., Yu Q., The diversification of Heliconius butterflies: What have we learned in 150 years? J. Evol. Biol. 28, 1417–1438 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Adams D. C., Rohlf F. J., Slice D. E., Geometric morphometrics: Ten years of progress following the 'revolution'. Ital. J. Zool. 71, 5–16 (2004). [Google Scholar]
- 7.Le Poul Y., Whibley A., Chouteau M., Prunier F., Llaurens V., Joron M., Evolution of dominance mechanisms at a butterfly mimicry supergene. Nat. Commun. 5, 5644 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooney C. R., Bright J. A., Capp E. J. R., Chira A. M., Hughes E. C., Moody C. J. A., Nouri L. O., Varley Z. K., Thomas G. H., Mega-evolutionary dynamics of the adaptive radiation of birds. Nature 542, 344–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Cramon-Taubadel N., Frazier B. C., Lahr M. M., The problem of assessing landmark error in geometric morphometrics: Theory, methods, and modifications. Am. J. Phys. Anthropol. 134, 24–35 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Gerber S., The geometry of morphospaces: Lessons from the classic Raup shell coiling model. Biol. Rev. 92, 1142–1155 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Goldenfeld N., Looking in the right direction: Carl Woese and evolutionary biology. RNA Biol. 11, 248–253 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Sheppard P. M., Turner J. R. G., Brown K. S., Benson W. W., Singer M. C., Genetics and the evolution of muellerian mimicry in heliconius butterflies. Phil. Trans. R. Soc. Lond. (Biol.) 308, 433–610 (1985). [Google Scholar]
- 13.Mallet J., Causes and consequences of a lack of coevolution in Müllerian mimicry. Evol. Ecol. 13, 777–806 (1999). [Google Scholar]
- 14.Mallet J., Mimicry: An interface between psychology and evolution. Proc. Natl. Acad. Sci. U.S.A. 98, 8928–8930 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Heliconius Genome Consortium , Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. Mallet. Speciation, raciation, and color pattern evolution in Heliconius butterflies: Evidence from hybrid zones, in Hybrid Zones and the Evolutionary Process, R. G. Harrison, Ed. (Oxford Univ. Press, 1993), pp 226–260. [Google Scholar]
- 17.Kapan D. D., Three-butterfly system provides a field test of müllerian mimicry. Nature 409, 338–340 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Sherratt T. N., Spatial mosaic formation through frequency-dependent selection in Müllerian mimicry complexes. J. Theor. Biol. 240, 165–174 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Janzen D. H., When is it coevolution? Evolution 34, 611–612 (1980). [DOI] [PubMed] [Google Scholar]
- 20.Webb S., Deep learning for biology. Nature 554, 555–557 (2018). [DOI] [PubMed] [Google Scholar]
- 21.E. Hoffer, N. Ailon, Deep metric learning using triplet network, in Similarity-based pattern recognition (Third International Workshop, SIMBAD 2015, Copenhagen, Denmark; Lecture Notes in Computer Science, vol. 9370), A. Feragen, M. Pelillo, M. Loog, Eds. (Springer, Cham, 2015), pp 84–92. [Google Scholar]
- 22.F. Schroff, D. Kalenichenko, J. Philbin, Facenet: A unified embedding for face recognition and clustering, in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR, 2015), pp. 815–823. [Google Scholar]
- 23.L. E. Gilbert, Coevolution and mimicry, in Coevolution, D. M. Futuyma, M. Slatkin, Eds. (Sinauer Associates Inc., 1983). [Google Scholar]
- 24.Arbuckle K., Bennett C. M., Speed M. P., A simple measure of the strength of convergent evolution. Methods Ecol. Evol. 5, 685–693 (2014). [Google Scholar]
- 25.Stayton C. T., Testing hypotheses of convergence with multivariate data: Morphological and functional convergence among herbivorous lizards. Evolution 60, 824–841 (2006). [PubMed] [Google Scholar]
- 26.Hoyal Cuthill J. F., Charleston M., Wing patterning genes and coevolution of Müllerian mimicry in Heliconius butterflies: Support from phylogeography, cophylogeny, and divergence times. Evolution 69, 3082–3096 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Hines H. M., Counterman B. A., Papa R., de Moura P. A., Cardoso M. Z., Linares M., Mallet J., Reed R. D., Jiggins C. D., Kronforst M. R., Owen McMillan W., Wing patterning gene redefines the mimetic history of Heliconius butterflies. Proc. Natl. Acad. Sci. U.S.A. 108, 19666–19671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak K. M., Wahlberg N., Neild A. F. E., Dasmahapatra K. K., Mallet J., Jiggins C. D., Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst. Biol. 64, 505–524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoyal Cuthill J., Charleston M., Phylogenetic codivergence supports coevolution of mimetic Heliconius butterflies. PLOS ONE 7, e36464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.G. Lamas, Atlas of Neotropical Lepidoptera: Checklist Pt. 4a Hesperioidea-Papilionoidea (Association for Tropical Lepidoptera and Scientific Publishers, 2004). [Google Scholar]
- 31.H. Holzinger, R. Holzinger, Heliconius and Related Genera (Sciences Nat, 1994). [Google Scholar]
- 32.L. E. Gilbert, The Biology of Butterfly Communities (Academic Press, 1984). [Google Scholar]
- 33.Parnell A. J., Bradford J. E., Curran E. V., Washington A. L., Adams G., Brien M. N., Burg S. L., Morochz C., Fairclough J. P. A., Vukusic P., Martin S. J., Doak S., Nadeau N. J., Wing scale ultrastructure underlying convergent and divergent iridescent colours in mimetic Heliconius butterflies. J. R. Soc. Interface 15, 20170948 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brower A. V. Z., Parallel race formation and the evolution of mimicry in Heliconius butterflies: A phylogenetic hypothesis from mitochondrial DNA sequences. Evolution 50, 195–221 (1996). [DOI] [PubMed] [Google Scholar]
- 35.J. N. Thompson, From the entangled bank to the evolutionary synthesis, in The Coevolutionary Process (University of Chicago Press, 1994), pp. 23–35. [Google Scholar]
- 36.Speed M. P., Batesian, quasi-Batesian or Müllerian mimicry? Theory and data in mimicry research. Evol. Ecol. 13, 755–776 (1999). [Google Scholar]
- 37.Turner J. R. G., The evolutionary dynamics of batesian and muellerian mimicry: Similarities and differences. Ecol. Entomol. 12, 81–95 (1987). [Google Scholar]
- 38.Thompson J. N., Concepts of coevolution. Trends Ecol. Evol. 4, 179–183 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 40.P. H. Harvey, M. D. Pagel, The comparative method for studying adaptation, in The Comparative Method in Evolutionary Biology (Oxford Univ. Press, 1991), pp. 1–34. [Google Scholar]
- 41.J. F. Hoyal Cuthill, S. J. Braddy, P. C. J. Donoghue, A formula for maximum possible steps in multistate characters: Isolating matrix parameter effects on measures of evolutionary convergence. 26, 98–102 (2010). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaaw4967/DC1
Supplementary Methods
Fig. S1. Diagram of the architecture of the deep learning network ButterflyNet used in this study.
Fig. S2. Geographic localities for sampled butterfly specimens from the polymorphic mimicry complex of H. erato and H. melpomene.
Fig. S3. Heatmap showing mean pairwise phenotypic and geographic distances between 38 subspecies of H. erato (black labels) and H. melpomene (gray labels).
Fig. S4. Collections of specimen photographs used in this study, grouped by subspecies.
Fig. S5. Average pairwise Euclidean geographic distances between subspecies of H. erato and H. melpomene.
Fig. S6. Neighbor-joining trees of phenotypic distance between subspecies of H. erato and H. melpomene.
Fig. S7. Comparative analyses of the extent of phenotypic convergence in mimicry.
Fig. S8. Principal component visualization of Heliconius butterflies.
Table S1. Traditionally hypothesized co-mimic subspecies of H. erato and H. melpomene.
Table S2. Taxonomic and locality data recorded for historical specimens of H. erato and H. melpomene held in the collections of the NHM London.
Table S3. Coordinates of butterfly images on 64 axes of a Euclidean phenotypic space constructed using a deep convolutional network with triplet training.
Table S4. Mean pairwise Euclidean phenotypic distances between subspecies from H. erato and H. melpomene.
Table S5. Mean pairwise squared Euclidean phenotypic distances between subspecies from H. erato and H. melpomene.
Table S6. Mean pairwise Euclidean geographic distances between subspecies from H. erato and H. melpomene.
Table S7. Number of sampled butterfly individuals for each subspecies.
Table S8. Broad pattern class of the type specimen of each subspecies.
Table S9. Traditionally hypothesized mimicry complexes of H. erato and H. melpomene subspecies.
Table S10. Statistical comparisons of pairwise Robinson-Foulds distances between sets of phylogenetic trees.
Table S11. Statistical comparisons with hybrids excluded of pairwise Robinson-Foulds distances between sets of phylogenetic trees.
Supplementary Computer Code in ipynb Format
Supplementary Computer Code in PDF Format
Reference (41)


