Abstract
How genetic coding differentiated biology from chemistry is a long-standing challenge in Biology, for which there have been few experimental approaches, despite a wide-ranging speculative literature. We summarize five coordinated areas—experimental characterization of functional approximations to the minimal peptides (protozymes and urzymes) necessary to activate amino acids and acylate tRNA; showing that specificities of these experimental models match those expected from the synthetase Class division; population of disjoint regions of amino acid sequence space via bidirectional coding ancestry of the two synthetase Classes; showing that the phase transfer equilibria of amino acid side chains that form a two-dimensional basis set for protein folding are embedded in patterns of bases in the tRNA acceptor stem and anticodon; and identification of molecular signatures of ancestral synthetases and tRNAs necessary to define the earliest cognate synthetase:tRNA pairs—that now compose an extensive experimentally testable paradigm for progress toward understanding the coordinated emergence of the codon table and viable mRNA coding sequences. We briefly discuss recent progress toward identifying the remaining outstanding questions—the nature of the earliest amino acid alphabets and the origin of binding discrimination via distinct amino acid sequence-independent protein secondary structures—and how these, too, might be addressed experimentally.
Keywords: bidirectional genetic coding, protein folding, aminoacyl-tRNA synthetase:tRNA cognate pairs, self-organization of molecular recognition
1. Introduction
A long standing challenge to biology has been to find a plausible and experimentally testable path for the development of the genetic code from prebiotic chemistry. Developing such a path has been so difficult that little in the extensive literature on this problem (Koonin and Novozhilov, 2017) goes beyond speculation. The four conceptual pillars on which such speculation rests include: i) a “frozen accident” in which refinement of an initial code was precluded by the loss of meaning caused by re-defining codon assignments (Crick, 1968); ii) sequential specialization of the coding table to minimize translation errors (Archetti, 2004; Barbieri, 2019a); iii) incorporation of amino acids into the coding table in parallel with development of their biosynthetic pathways (Wong, 2005; Wong et al., 2016); and stereochemical complementarity between amino acids and base triplets representing codons and/or anticodons (Yarus et al., 2009). Even in combination, these ideas have been singularly ineffective at generating a sustainable experimental framework to address fundamental questions posed by the symbolic molecular coding necessary for genetic coding to emerge. The first, (i), is logically inconsistent with the others, and all are marginal in the sense that they separately fail to address the central question: coordinate evolution of aminoacyl-tRNA synthetase (aaRS) gene sequences and their cognate tRNAs with the codon assignments.
We have argued (Carter and Wills, 2018b; Wills and Carter, 2018) that much of that speculation is also misdirected at trying to understand how codon-directed protein synthesis arose from an RNA world (Koonin and Novozhilov, 2017). Our view is that the fundamental division of aminoacyl-tRNA synthetases (aaRS) into two distinct classes, their respective origin on opposite strands of one ancestral bidirectional gene, major molecular surgery to excerpt increasingly highly conserved active-site fragments of the aaRS, and paleoenzymology to test their properties, provide a far more fruitful platform from which to answer those questions.
1.1. Codon-dependent translation is the nexus of the phenotype/genotype distinction and consequently of genetics itself: why are so few curious about how it happened?
Answers to this question seem to fall into at least three categories:
It poses very challenging, multidisciplinary questions. How did molecules ‘learn’ to cooperate so that they could function like computers (Nieselt-Struwe and Wills, 1997; Wills, 1993, 2001, 2014a, b, 2016; Wills et al., 2015)? What coordinated the diverse steps necessary for molecular reflexivity to emerge (Carter and Wills, 2018b; Wills, 2016)? How did the genetic code come so close to being optimal (Freeland and Hurst, 1998; Stephenson and Freeland, 2013)?
It is easy to take translation for granted. Since the codon table was worked out in the 1960’s, not understanding how coding originated posed few barriers to further progress in biology and medicine. Biological research progressed because so much of genomics and biology can be understood without knowing how the code arose.
Perhaps the main reason is that despite the near universality of attempts to do so, the code cannot be even approximately explained by the “RNA World” (Wills and Carter, 2018). The idea that RNA blossomed on earth quite suddenly, bringing with it, ex nihilo, all the necessary tools for creating life is so guilelessly seductive, that there is intense resistance to alternative thinking.
Yet, the origin of genetic coding is of foundational importance because how it arose very likely contains keys to understanding the organization and functions of the proteome and that, in turn, has fundamental implications that enable protein engineering and medicine.
1.2. The history of codon-dependent translation
We should clarify here that we are interested in a restricted phase of the evolutionary history of codon-dependent protein synthesis. We address here only what the experimental evidence says about the earliest stages of the process, emphasizing what can now be described with reasonable confidence about the earliest events necessary for coding to emerge, and the various constraints that likely influenced its early evolution. Collins-Hed and Ardell (2018) give an excellent overview of the more recent expansion of the code and its adaptive variation as living organisms speciated, with potential implications for our subject matter. Template-directed protein synthesis now takes place on a sophisticated platform, the ribosome, equipped with a “decoding” subunit that matches codons in mRNA with cognate anticodons of aminoacyl-tRNAs. The history of genetic coding thus also encompasses that of the ribosome itself, which has been examined by others (Bokov and Steinberg, 2009; Davidovich et al., 2010; Fox, 2012; Hsiao et al., 2013; Petrov et al., 2014; Petrov et al., 2015). These studies suggest conclusions analogous to some of our own—identification of a small conserved core that may retain catalytic activity—albeit without experimental validation.
Perhaps the key observation for our purposes is that the exit tunnel may have been necessary to ensure processive assembly of polypeptides from aminoacylated tRNAs (Fox, 2012). Although the ribosome as a platform is obviously relevant to the origin of protein synthesis, its role in genetic coding is only accessory, most essentially maintaining collinearity between stepwise transpeptidation and progressive ratcheting along the mRNA, leaving open the possibility that mechanisms of coding may have preceded or emerged simultaneously with the earliest coded peptides. The covalent bond between amino acid and tRNA, however, is determined entirely by a family of proteins called aminoacyl-tRNA synthetases (aaRS), which have evolved mechanisms for conjointly recognizing both a specific amino acid and a specific tRNA. The actual decoding of genetic information therefore precedes ribosomal processing as translation results from the specific matching between aaRS and their cognate amino acids and tRNAs.
Because the assembly of an aminoacyl-tRNA ester bond is both slow and thermodynamically unfavorable, its formation in biological systems is driven by first forming a mixed anhydride, aminoacyl-5’AMP at the expense of ATP. Formation of the mixed anhydride intermediate is ~103-fold slower than the subsequent acyl transfer to tRNA. Thus, the aaRS represent not only the emergence of specific molecular recognition, but also the catalytic mechanisms for both activating the amino acid carboxyl group and transferring it to the tRNA 3’ terminal adenosine (Li et al., 2011; Martinez et al., 2015; Pham et al., 2010; Pham et al., 2007).
Considerable attention has focused on the ancestry of the ribosomal peptidyl-transferase center (PTC) (Agmon, 2009; Belousoff et al., 2010; Fox, 2012; Hsiao et al., 2013; Petrov et al., 2014; Petrov et al., 2015). One possible role of such a center in the absence of any coding mechanism is the generation of randomly or semi-randomly sequenced peptides (Belousoff et al., 2010). Whereas this is a possibility, it nonetheless has two substantial problems: (i) without genetic continuity, such peptides cannot be among the ancestors of the contemporary proteome—leaving moot how they might have been selected—and (ii) the rate-limiting step in protein synthesis is amino acid activation. The assembly of non-coded peptides would thus have been extremely slow without the benefit of ancestral aaRS. Thus, we believe that any non-coding functions of a primordial PTC lie outside our purview here, as are the origin and early evolution of the PTC.
For that reason, we focus here on the need for the simultaneous emergence of (i) at least two ancestral aaRS that differentiated not only the set of extant amino acids but also tRNAs into disjoint sets; and (ii) ancestral—and likely RNA—aaRS genes encoding the required aaRS enzymes according to the rules of a primitive, presumably binary, code. We review and analyze the experimental data that lead to the conclusion that Class I and II aaRS ancestors were encoded on the complementary strands of a single bidirectional gene. We identify how tRNAs came to be selectively differentiated and assigned by the separate Class I and II ancestral aaRS types for aminoacylation with, respectively, larger or smaller amino acid substrates.
1.3. Information transfer
The flow of molecular information in biology was first described by Crick’s “Central Dogma” (Crick, 1970). The distinction between nucleic acid and protein syntheses was articulated further by Koonin (2015) who stressed that information transmission between nucleic acids is “digital” whereas its migration into the world of functional proteins is “analog” on account of the process of protein folding. That distinction, however, overlooks a crucial aspect of the conversion: coded translation poses an especially challenging problem because it represents symbolic conversion of molecular information. That description combines two notions—“molecular” information and symbolic conversion—both of which require further comment.
Information transfer from one nucleic acid to another is governed by stereochemistry in that it relies almost exclusively on the strength of base-pairing between the strands. The nucleic acid sequences of protein-coding genes thus have no analog information to speak of, and their (potential) analog information content therefore depends entirely on the process of digital decryption given via translation.
The sequence of amino acids in a protein is not directly related to stereochemical fit, but arises from a much more complex set of symbolic relationships between amino acids and base triplets called “codons” that draw correctly acylated tRNAs by base-pairing to the mRNA on the ribosome, where the amino acid moieties are joined together. The earliest genetic coding paradigm therefore required simultaneously solving three different recognition problems—ATP, amino acid, and tRNA—and finding two related catalytic mechanisms, each in two different ways (for Class I and II aaRS) in order to implement all events necessary to accomplish the symbolic conversion.
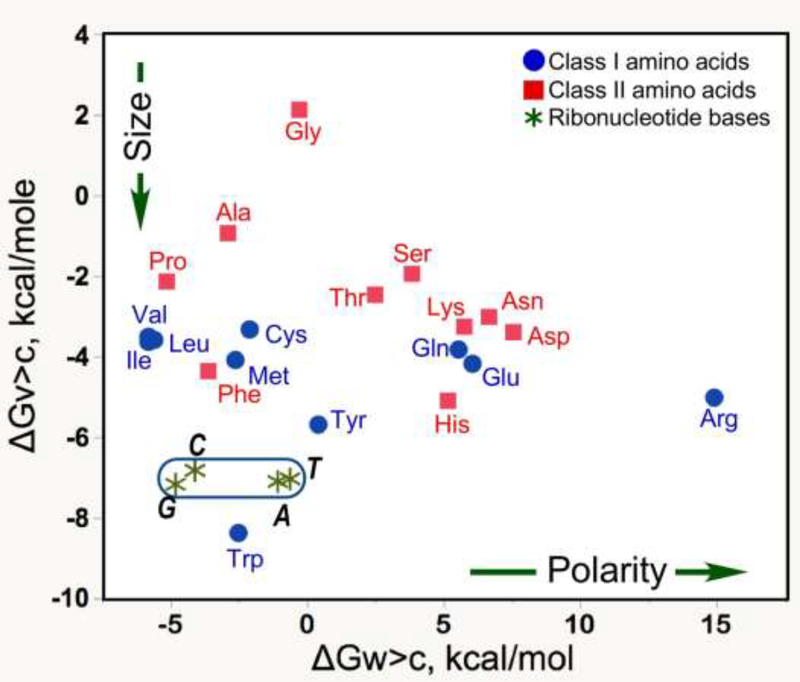
Richard Wolfenden elucidated the information content in the canonical amino acids over the course of 30 years, by measuring phase transfer equilibria of side chain models for 19 of 20 amino acids (Radzicka and Wolfenden, 1988; Wolfenden, 1983, 2007; Wolfenden et al., 2015) (Fig. 1). We eventually showed that the information in Fig. 1 is necessary and sufficient to explain both (i) the roles of amino acids in protein folding and (ii) the rules underlying aaRS specificities for their cognate tRNA substrates (Carter and Wolfenden, 2015, 2016). Those twin correlations proved a crucial link, unifying Crick’s Central Dogma with his adaptor hypothesis (Crick, 1955), which posited the existence of transfer RNA.
Figure 1.
The “information” represented in genetic coding. A two-dimensional basis set describes the canonical amino acids. Phase transfer equilibria for distribution between vapor and either water or cyclohexane turn out to describe the differences between amino acids sufficiently to explain ~90% of the variations in the surface exposure of 18 of the 20 amino acids in folded proteins. Note that because of the logarithmic dependence of free energy on equilibrium constants, the same coordinate system reveals that the four nucleic acid bases represent a tiny fraction of the overall chemical diversity of the 20 amino acids.
Nature embedded information from Fig. 1 into two separate kinds of nucleic acids. Bases in tRNA acceptor stem and anticodon sequences (Carter and Wills, 2018a; Carter and Wolfenden, 2015), together with aaRS recognition compose a programming language for writing amino acid sequences according to rules of the symbolic “look-up” table of the genetic code (Carter and Wills, 2018b; Carter and Wolfenden, 2016; Wills and Carter, 2019). Messenger RNA sequences of genes themselves represent combinations of amino acids that can fold and function—i.e. programs written in that programming language. Thus, how genetic coding began is intimately connected with how information from Fig. 1 became embedded into tRNA and mRNA sequences. We argue that the evolutionary history of the aaRS was essentially congruent to that embedding.
1.4. The central questions
This discussion positions us to frame the important questions associated with the origin of genetic coding. Launching genetic coding requires, most importantly, approximately simultaneous emergence of:
Assignment catalysts capable of biasing the synthesis of aminoacyl-tRNAs into a minimum of two disjoint sets.
Precursor tRNA molecules whose specific recognition by aaRS contain a minimum of information from Fig. 1.
Messenger RNA coding sequences for the ancestral aaRS genes that enforce the coding rules.
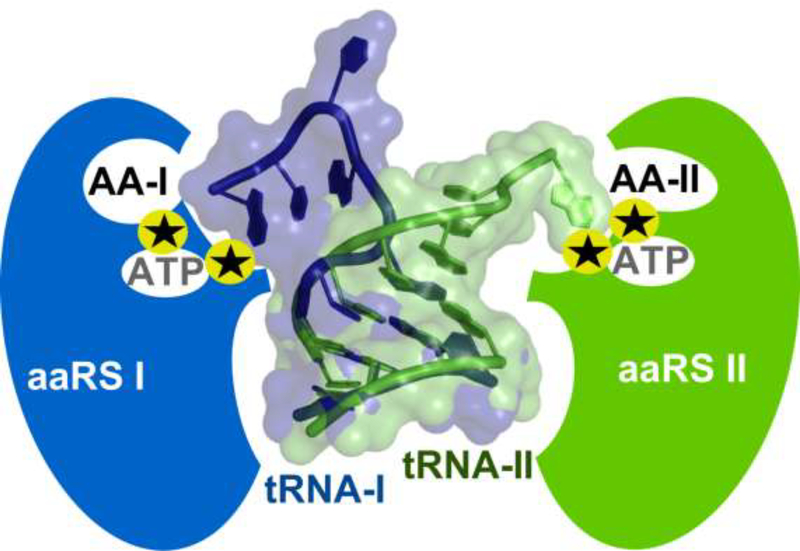
Eight stereochemical recognition events: for two different types of amino acid; for two different types of cognate tRNA; and for the transition-states of both catalytic processes required to bias the chemistry sufficiently to implement rudimentary coding. These are summarized graphically in Fig. 2.
Figure 2.
The minimum set of molecular recognition events and rate accelerations necessary to launch genetic coding. That coding necessarily requires a minimum of two letters lends a high symmetry to this scheme. For convenience, the two recognition paradigms are shown here using the aaRS Class distinction; Class I aaRS and their cognate substrates are colored blue, Class II aaRS and tRNAs are colored green. Each aaRS must be equipped with three binding sites—amino acid, ATP, cognate tRNA—requiring approximate stereochemical discrimination and two sets of catalytic machinery. The catalytic machinery must stabilize the transition states for amino acid activation and tRNA acylation (black stars). All eight binding sites have been characterized experimentally in short peptides excerpted from full length aaRS, as noted in the text.
An important question arises: can all eight requisite properties be implemented using the same, rudimentary and error-prone coding apparatus shown schematically in Fig. 2. Herein lies the nexus separating chemistry from biology. Requirements (i) and (iii) imply that the two aaRS must have been encoded reflexively, i.e., using the same coding rules that they, themselves, enforce. That is the question we address here in summarizing our work to this point and where we think it is headed.
2. Five experimental approaches to the origin of codon-dependent protein synthesis
Our work comprises five significant advances toward understanding how genetic coding created biology from chemistry:
Three lines of evidence established that the ancestors of the enzymes that translate the genetic code—aminoacyl-tRNA synthetases or aaRS—were originally encoded by opposite strands of the same ancestral bidirectional gene (Martinez et al., 2015). The unique symbolic information in a double-stranded nucleic can be interpreted in two alternative ways to give different proteins that prepare different kinds of amino acids for protein synthesis at the same rate.
The free energies governing partitioning of the 20 amino acids between water, vapor, and nonpolar solvents are tightly correlated with protein folding on the one hand, and the recognition of transfer RNA adaptors by the aaRS on the other hand (Carter and Wolfenden, 2015). The two products of the ancestral bidirectional aaRS gene likely discriminated between large and small amino acid sidechains.
Embedding patterns of bases correlated with amino acid phase transfer equilibria into tRNA bases allowed protein folding rules to rapidly separate gene products that functioned from those that did not (Carter and Wills, 2018b).
Recent identification of thermodynamic and structural signals that direct aaRS to their own tRNA substrates—how Class I aaRS find tRNAs for Class I amino acids and conversely—paved the way to understanding aaRS•tRNA recognition by the earliest cognate pairs (Carter and Wills, 2018a).
Accelerating the search for the best codon table and useful regions of amino acid sequence space, the ancestral aaRS gene resembled the “boot block” that installs a computer’s operating system. The most important property Nature discovered in this way was a set of peptide catalysts that could enforce the rules by which they were made. That minimal reflexivity, we believe, was necessary and sufficient to elaborate the entire codon table (Carter and Wills, 2018b; Wills, 2019; Wills and Carter, 2018).
3. An ancestral bidirectional gene and the molecular origins of genetic coding
The previous discussion emphasizes that the origins of codon-directed protein synthesis depended extensively on the binding and catalytic interactions diagrammed in Fig. 2.
A fundamental step toward providing an experimentally testable path to explain the origin of genetic coding was to develop three independent lines of evidence that ancestral enzymes—aminoacyl-tRNA synthetases, aaRS—that first translated the forerunner to the genetic code, emerged on opposite strands of the same original ancestral gene. Experimental evidence that such a gene gave rise to the two aaRS superfamilies is now quite strong:
Peptides from both Class I and II aaRS whose coding sequences can be aligned to form a bidirectional gene retain a full spectrum of the enzymatic activities of extant aaRS (Carter, 2014; Carter et al., 2014; Li et al., 2013).
The frequency of codon middle-base pairing is significantly elevated in bidirectional alignments of the contemporary coding sequences of extant aaRS and increases as ancestral sequences of independently reconstructed Class I and II aaRS genes approach the root node (Chandrasekaran et al., 2013).
Peptides from both strands of a designed, bidirectional gene configured to encode the ATP binding sites of Class I and II aaRS accelerate amino acid activation 106-fold as do peptides excerpted from the full length aaRS with the corresponding WT sequences. Although the WT peptides from each class have appreciable sequence identity (0.11–0.22), the corresponding designed peptides have no identity between classes, and <10% identity with the corresponding WT sequences from within their own class. The Class I and II peptides thus are drawn from essentially disjoint regions of sequence space (Martinez et al., 2015).
This remarkable, albeit bizarre result means that although the information content on opposite strands is syntactically equivalent, it can nonetheless be given two different semantic interpretations, resulting in proteins that each catalyze the single most important reaction for protein synthesis—activating amino acids—with either large (Class I) or small (Class II) side chains. The properties of the universal genetic code include its robustness to mutations on a single coding strand (Freeland and Hurst, 1998). Bidirectional coding now means that it is apparently also constrained by the need to enhance the likelihood that peptides coded on opposite strands will be functional (Opuu et al., 2017). That background introduces, in principle, the potential to construct a viable two-letter alphabet, implemented by a bidirectional gene composed of only two small but distinct sets of similar amino acids.
Properties of the ancestral bidirectional aaRS gene provide a genetic basis for the implementations depicted in Fig. 2. They also furnish solutions (Carter and Wills, 2018b) to fundamental challenges previously associated with the self-organization of specificity from a marginally biased, non-random process (Eigen, 1971a; Eigen, 1971b; Eigen et al., 1988; Eigen and Schuster, 1977). Briefly, such a bidirectional ancestral gene would have definitively differentiated amino acid sequence space into two non-intersecting regions each associated with comparable catalytic functionality, but contrasting specificity. The two coded peptides would be interdependent, each requiring the other for their own production. The severe genetic linkage would have assured that the two gene products were available at the same time and place. Finally, the small coding alphabet would also have assured high mutation rates, maximizing the genetic plasticity necessary to ensure access to a viable evolutionary path.
3.1. Amino acid phase transfer equilibria
An important reason why the origin of genetic coding remained opaque for so long is that few researchers accepted that protein folding was essential to facilitate the self-organization of coding. Thus, how the structures of the 20 canonical amino acids influenced both protein folding and the specific recognition by aaRS of cognate tRNAs were essential to that understanding. Richard Wolfenden outlined the importance of those relationships in an exceptional and prescient paper (Wolfenden et al., 1979). He and one of us solved that problem more precisely by establishing how the phase transfer equilibria of the 20 amino acids dictate both protein folding and the patterns of bases that determine how the aaRS recognize transfer RNA adaptors (Carter and Wolfenden, 2015, 2016; Wolfenden et al., 2015). Clarification of these relationships opened a new window on the origin of the code by identifying how protein folding (and function) could provide a coherent gating mechanism when combined with the emerging genetic phenomenon represented by the ancestral bidirectional—and hence bifunctional—synthetase gene.
3.2. Self-organization requires a feedback loop
Because proteins fold largely according to how amino acids separate between aqueous, vapor, and nonpolar phases (Wolfenden et al., 2015), the connection between amino acid chemistry and aaRS recognition of cognate tRNA completes a tight feedback loop (Carter and Wills, 2018b). Selecting for proteins capable of folding enabled Nature to rapidly explore alternatives for both the codon table and amino acid (and thus mRNA) sequence space, each incremental improvement, in turn, accelerating that search. Thus, because function depends on protein folding, the minimal genetic code based on discriminating between small vs large sidechains functions like a minimal set of instructions for elaborating the rest of the code by processes that became more and more like natural selection as the coding alphabet grew. Thus, the ancestral bidirectional gene was equipped to function like the “boot-block” that installs a computer’s operating system by accelerating the search for near optimal codon table and mRNA sequences, leading quickly to the logic of the central dogma.
Herein lie the most important reasons for rejecting outright RNA-World accounts of the origins of genetic coding. The feedback loop necessary to rapidly select simultaneously for a programming language—i.e. the mechanisms for evolving synthetase:tRNA cognate pairs—together with a reflexive set of “programs”—amino acid sequences capable of folding and implementing the recognition elements necessary to impose the corresponding coding rules—is, in our opinion, inconceivable if the amino acid activating enzymes are ribozymes whose function depends on RNA-, rather than protein-folding rules. Koonin (Koonin, 2011) articulated a more telling argument than we can do, why evolving such ancestral ribozymal amino acid•tRNA assignment catalysts would therefore take far, far longer, requiring multiverses to produce a single successful instance.
3.3. tRNA acceptor stem recognition
An important gap remained. To function as a boot-block the dual system of peptide catalysts would have had to have been equipped with the capacity to discriminate between potential tRNA substrates. For the original Class I aaRS to identify tRNAs for Class I amino acids, and similarly for Class II, at a time when the coding alphabet consisted of only two letters, there had to have been a code by which they could have distinguished cognate vs non-cognate tRNAs. Such a code would have been analogous to what has been called the “operational RNA code” (Schimmel et al., 1993). However, it must have been binary, in order for most or all proto-tRNAs to have been able to participate in codon-directed protein synthesis. Does evidence for such a code remain in contemporary tRNA sequences? We filled that gap by uncovering both how Class I aaRS specifically recognize a hairpin at the 3’ CCA terminus of Class I tRNA substrates and the base sequences that enable formation of that hairpin (Carter and Wills, 2018a).
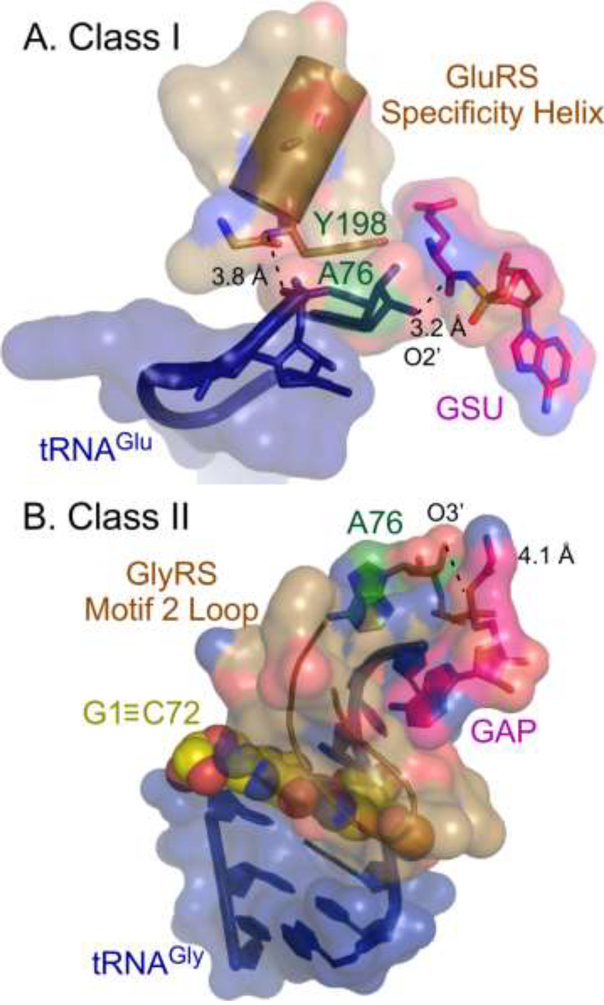
Fig. 3A illustrates the fundamentals of the synthetase•tRNA interaction that discriminate between recognition of cognate tRNAs from the minor groove of the acceptor stem by Class I aaRS. The hairpin recognized by Class I aaRS binds via the exposed 5’ phosphate group of A76 to a site at the amino terminus of what is called the specificity-determining α-helix, which exposes four unpaired amino nitrogen groups, creating a pocket of positive charge (Hol, 1985; Hol et al., 1981). That interaction is reinforced by an interaction between a conserved aromatic sidechain and both the nonpolar face of the ribose and the base of A76. The resulting complex aligns the 2’OH of A76 with the carbonyl carbon of the activated Class I amino acid.
Figure 3.
Acylation complexes of Class I and II cognate pairs. Urzymes of both Class I and II aaRS afford secondary structural features that recognize cognate tRNAs. In both complexes the terminal A76 is green, the activated amino acid analog is magenta, and the tRNA is dark blue. A. Class I GluRS complex illustrates the role played by the amino terminus of the specificity helix (sand) in recognition of the hairpin characteristic of Class I-cognate tRNAs. The recognition complex is reinforced by a conserved aromatic residue (i.e. Y198) near the N-terminus of that helix. B. Class II GlyRS complex illustrates the role of the Motif 2 loop in stabilizing the helical extension of the CCA 3’ terminus by lying atop the topmost base pair (G1≡C72; yellow) of the acceptor stem. Distances are given between the nucleophilic ribose OH group and the aminoacyl group of the activated amino acid analog and between an amino group at the N-terminus of the specificity-determining helix in GluRS and the A76 phosphate group.
Fig. 3B illustrates how a distinctly different peptide secondary structure, the Motif 2 loop, lies on top of the topmost base pair of the acceptor stem. The small dipole moment of that antiparallel peptide hairpin minimizes perturbation of the helical formation of the CCA terminus, allowing uninterrupted extension. This arrangement allows the 3’OH of the A76 ribose to interact with the carbonyl carbon of the activated Class II amino acid.
3.4. The operational code
In addition to the binary distinction between tRNAs cognate to Class I and II aaRS, the tRNA acceptor stem bases represent a much more complex network of signals to their cognate aaRS. These additional coding functions were anticipated by Giegé (Giegé, 1972) and others (Schimmel et al., 1993). Giegé’s conclusions were validated and extended by a sophisticated bioinformatic analysis (Zamudio and José, 2018), again without coherent understanding of what those signals represent. Recently, however, we have substantially clarified what kinds of information are stored in which tRNA bases (Carter and Wolfenden, 2015, 2016) and (Carter and Wills, 2018a). Those correlations contradict, to some extent, previous attempts to order the appearance of individual amino acids, consistent with our view (see discussion of impedance-matching in Carter and Wills, 2018b) that the earliest coding made rudimentary distinctions between similar types of amino acids, rather than establishing explicit connections between amino acid and codon.
4. Remaining challenges
The scenario illustrated schematically in Fig. 2 furnishes natural solutions for the most puzzling challenges—interdependence, differentiation, reflexivity, and the transition from low to high specificity—associated with the origin of genetic coding. It is utterly different from and transcends any other such proposal. It is based largely on experimental results, and so has the immense advantage of having been tested. Moreover, because it is based on elaborating the universal genetic code from a plausible and very simple ancestral code, it provides a convincing paradigm for the origin of diversity in the proteome. In the following, we assess what we feel are the most immediate and accessible of the remaining questions.
4.1. Were peptides encoded by a binary alphabet viable and functional?
Catalytic and substrate recognition by the earliest peptide catalysts likely depended not on the use of specific catalytic residues, but rather on architectural features of their secondary structures (Hol et al., 1981; Hol et al., 1978). That would imply in turn that “patterned libraries” of the sort described by Hecht (Kamtekar et al., 1993; Moffet et al., 2003; Patel et al., 2009) and Edgell (Lahr et al., 1999) based on as yet unidentified patterns within the protozyme gene should contain sufficient numbers of catalytically active peptides that some such genes might express amino acid activating peptide catalysts from both strands. Just how precisely we can validate this hypothesis in detail is uncertain. Nevertheless, recent progress suggests some answers.
The bidirectional 46-residue protozyme gene (Martinez et al., 2015) created two new protozymes to complete a 22 factorial design: two protozymes with natural sequences; two with bidirectional, designed sequences; two Class I; and two Class II. All four protozymes have nearly identical rate accelerations, kcat/KM/knon, where knon is the uncatalyzed rate constant. However, kcat and KM values for both bidirectional gene products are elevated 100 times relative to the protozymes with natural sequences. These systematic differences are statistically significant (P<0.01 on ΔGkcat and P<0.01 for ΔGKM). Enforcing bidirectional coding thus created two more active catalysts with decreased specificity, more or less what one would expect from an ancestral form.
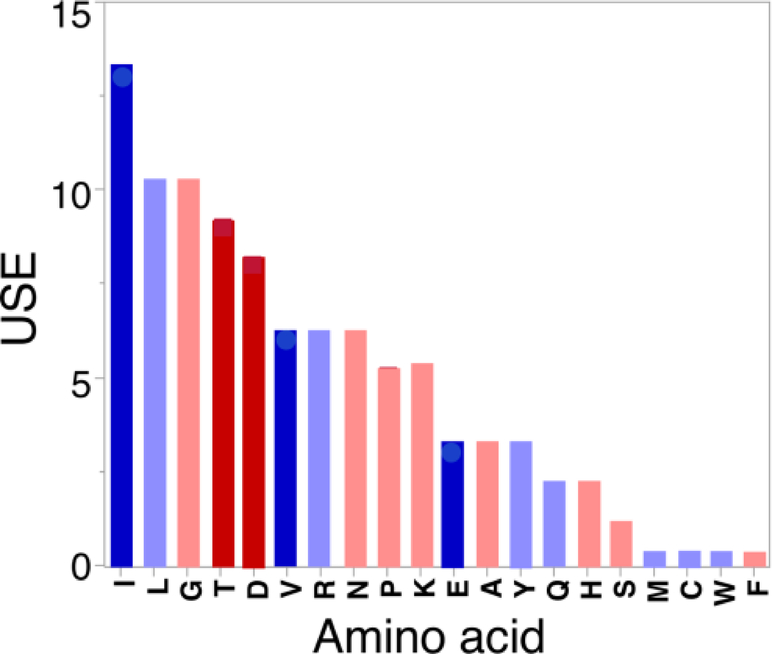
Trifonov (Trifonov, 2000) produced a consensus order for the appearance of amino acids in the genetic code. In our opinion, such efforts are susceptible to overinterpretation because the earliest coding likely produced peptides whose sequences were only marginally nonrandom (Woese, 1969; Woese, 1965); see also (Barbieri, 2019a). The bidirectional gene of Martinez et. al. (2015) is, nonetheless consistent in using codons for only 16 of the 20 canonical amino acids (Fig. 4), omitting Met, Cys, Trp, and Phe—all arguably late. Further, the frequencies of amino acids used suggest possible ways to simplify the alphabet used to construct a suitable patterned library.
Figure 4.
Amino acid usage in the bidirectional protozyme gene (Martinez et al., 2015). Blue bars are Class I amino acids; red bars are Class II amino acids. Bold colors are amino acids with specialized side chains—β-branched or carboxylate—encoded in addition to side chain size and tRNA groove recognition classifications within the operational RNA code (Carter and Wills, 2018a). They amount to more than 40% of the amino acids used to construct that gene.
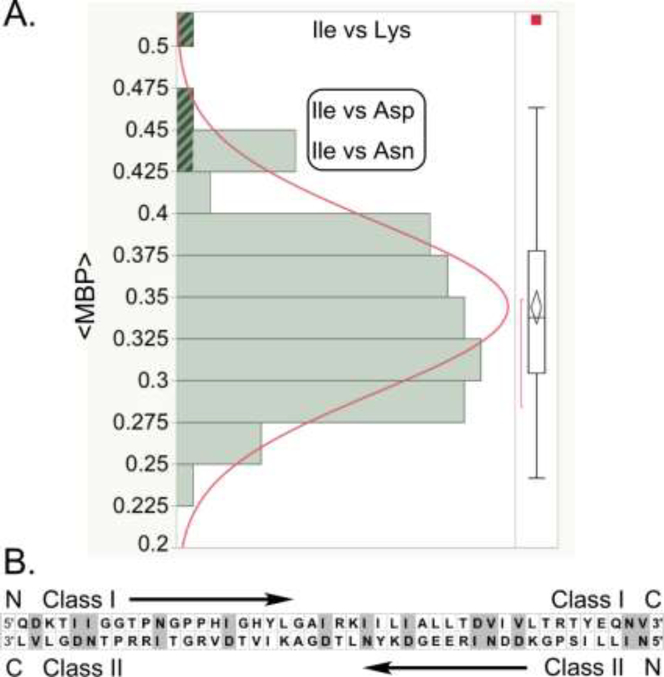
Curated structural superpositions of the Class I and Class II crystal structures (Popinga, 2019; Popinga et al., 2015) enabled us to identify with greater confidence the conserved structural motifs whose coding sequences can be aligned in opposite directions. Using only the bacterial sequences from these alignments, we repeated the analysis first described by Chandrasekaran et. al. (2013) determining middle codon-base pairing frequencies, <MBP> of antiparallel alignments of coding sequences for Class I and II aaRS. The all-by-all comparison revealed a mean <MBP> value of 0.34 ±0.005, equivalent to that observed for four aaRS by Chandrasekaran, et al. The <MBP> is arguably a measure of how far each Class I-II pair is from the ancestral bidirectional gene. The range of values (0.24 to 0.51), however, is greatly extended compared with what we previously found with a much smaller subset (Fig. 5A). At the top of the list were the Class I-II pairs (Ile-Lys; 0.51) and (Ile-Asp) (0.46). These values strongly suggest that the specificities of the first two assignment catalysts derived from the bidirectional gene were Ile-like and Asp-like. Moreover, Ile and Asp share complementary codons.
Figure 5.
Preliminary evidence concerning the nature of the earliest amino acid alphabet. A. Coding sequences were aligned for ~400 Class I and ~400 Class II aaRS with substrate specificities ranging across all twenty canonical amino acids. Middle bases were excerpted and aligned antiparallel for all-vs-all aaRS. The distribution of the codon middle-base pairing frequency shows that the IleRS sequence has uniformly high pairing frequency with Class IIb aaRS specific for Asp, Asn, and Lys. Ile codons are complementary to both Asp and Asn codons (box). The difference between mean pairing frequency of these three aaRS (0.47) and the overall mean (0.34) is 25 times the standard error of the mean of the entire population. B. The bidirectional protozyme gene (Martinez et al., 2015) makes use of the Ile-Asp pair and its close homologs Ile-Asn, Ile-Lys, Val-Asp, Val-Asn in >30% of the positions (shaded). This high percentage usage suggests that the ancestral gene could have been functional using only a highly simplified amino acid alphabet.
Curiously, 10 positions—nearly 20% of the 46-residue bidirectional gene (Fig. 5B)—represent the pair [I; D or N] whose respective coding sequences exhibit 46% codon middle-base pairing and whose codons are themselves complementary. The highest <MBP> frequency in an all-by-all comparison based on the highly curated structure-based sequence alignments is the closely related pair [I;K]; the second highest pair is [I;D]. Relaxing the criterion to include also the similar pair [V; D or N] increases the number of positions to 14, or 30%. Remarkably, testing this proposal is now experimentally within reach. Much of the designed gene can be parsed according to a simple binary pattern, so we can envision simplifying a patterned combinatorial library for the bidirectional gene, perhaps without overly corrupting the two encoded catalytic activities (Martinez et al., 2015).
Early use of codon-anticodon pairs specifying Ile (Class IA) and Asp (Class IIB) is initially an arresting observation, as it suggests that the contemporary class subdivisions do not reflect their ancestry. More than 42% of the amino acids that make up the protozyme gene are drawn from the two sets (β-branched side chains and carboxylate). However, these codon-anticodon pairs are entirely consistent with our own assessment that the relevant data on the nature of the earliest alphabets lies in the coding patterns of the tRNA acceptor-stem bases. Our analysis (Carter and Wills, 2018a, 2019; Carter and Wolfenden, 2015, 2016) implies that the distinctions were those between—in order of appearance—large and small side chains, side chains with/without a carboxylate side chain, side chains with/without a β-branched side chain, and those with/without aliphatic side chains. The definitive data relevant to this point will likely come from ancestral sequence reconstructions constrained by bidirectional coding of the two aaRS Classes and newly identified constraints on tRNA acceptor stem evolution.
4.2. How did the earliest Class I and II aaRSs discriminate between large and small amino acid side chains?
An especially challenging and still elusive question is how the earliest assignment catalysts were able to bias their selection of amino acid substrates before the amino acid alphabet of distinguishable amino acid types had grown sufficiently to permit the evolution of well-adapted amino acid binding sites with a high enough level of specificity to perform the discrimination required to keep the alphabet intact. Kaiser et. al. have described the characteristics of the ATP (Kaiser et al., 2018) and amino acid (Kaiser et al., 2019) binding sites of Class I and II aaRS. The latter analysis focused on amino acid side chains associated with high-precision binding selectivity, which arises primarily from specific side-chain interactions.
It is tempting to ask whether or not the architecture of the two aaRS Classes was responsible for their ancestral ability to differentiate between small and large amino acid side chains. Such discrimination was essential to creating the initial binary alphabet, at a time when the error frequencies of both replication and translation were severely limited, and when the amino acid alphabet could be employed only to make statistical peptides (Woese, 1969; Woese et al., 1966). A fundamental distinction between the interaction of amino acids with binding pockets built from parallel (Class I) and antiparallel (Class II) β-structures would provide a satisfying resolution to that conundrum. One therefore suspects that in the earliest stages of genetic coding, the binding discrimination might have depended not on specific recognition pockets, but on generalized differences between the Class I and II architectures that would not have depended on the precision of inserting multiple kinds of side chains in order to assure specific recognition.
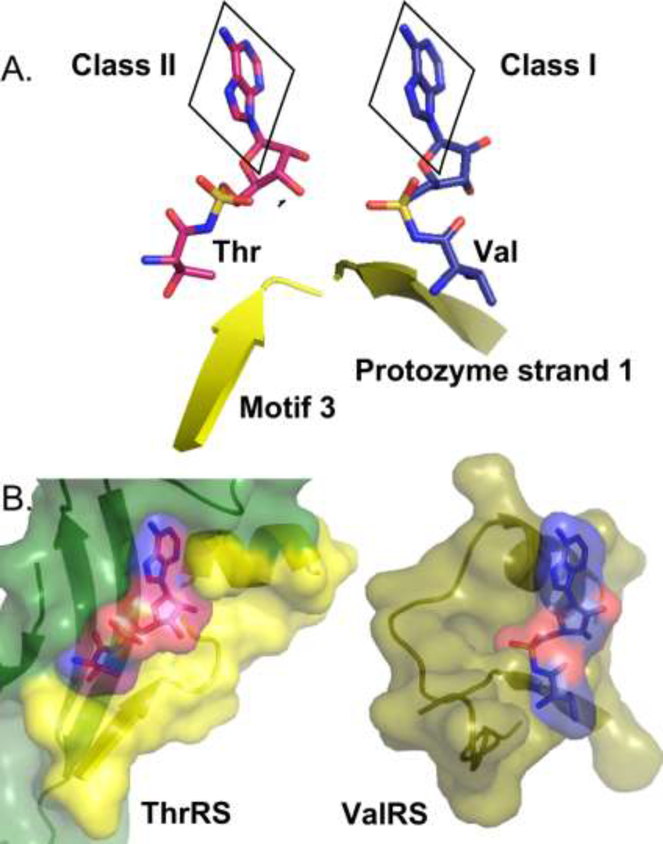
That notion was first articulated in a doctoral thesis (Belrhali, 1996; Belrhali et al., 1995). When contacted recently and asked for further details that author had lost the train of his thought, leaving the problem somewhat akin to Fermat’s marginal notation of his well-known conjecture. Another part of a possible answer appears in Fig. 4 of (Kaiser et al., 2019), in which the aminoacyl-adenylate ligands of Class I GluRS and Class II AspRS are superimposed, revealing that the two classes of side chains take off from the ribose moiety on opposite sides of the plane of the adenine ring. Inspired by that superposition, which applies generally to all Class I/II pairs, we show in Fig. 6 how this might have strongly impacted the size specificities of the Class I and II protozymes, the smallest catalytic fragments capable of catalyzing amino acid activation. We chose the valine and threonine systems for this illustration because these two β-branched side chains are both isosteric, and so have nearly identical side chain volumes.
Figure 6.
A possible origin of the side-chain size discrimination by Class I and II aaRS. A. Superposition of the activated aminoacyl-5’ adenylates (or analogs thereof) of valyl-tRNA synthetase (ValRS; 1GAX) and threonyl-tRNA synthetase (ThrRS; 1EVL). The β-strands closest to the respective amino acid β-carbon atoms run in opposite directions. Valine and Threonine have almost exactly the same volume. When the plane of the adenine ring is superimposed, however, Class I amino acids (Val) all veer to the right, whereas Class II amino acids (Thr) veer to the left. B. Complexes of the activated amino acids with ValRS and ThrRS. The direction taken by Class II side chains carries it directly into the Class II protozyme (green), thereby limiting the capacity of the binding pocket to small side chains. In Class I complexes, no polypeptide groups are present to limit the size of the side chain. Note that in Class II complexes, the β-strand arises from within Motif 3 (yellow), not the Class II protozyme.
A key difference between amino acid binding sites of Class I and II aaRS is that when the adenosine phosphate moieties of their activated aminoacyl-5’ adenylate ligands are superimposed, the amino acid side chains veer off on opposite sides of the plane defining the adenine ring (Fig. 6A). The amino acid binding sites of the protozymes, however, lie on the same side of the adenine ring (Fig. 6B). This distinction means that less room is available between the side chain and the Class II protozyme for increased side chain size, whereas the Class I protozyme offers essentially no boundary to limit side chain size.
Tantalizing as this qualitative picture may be, it raises new questions. First, it remains to be determined how Class I protozymes can reject amino acids with small side chains. Second, and equally perplexing, the fact that Class II amino acids all approach most closely to the beta strand of Motif 3 is inconsistent with the fact that Motif 3 lies outside the Class II protozyme, and in fact has no counterpart in the Class I superfamily because antiparallel orientation of Class I and II coding sequences puts Motif 3 prior to the N-terminus of many of the Class I enzymes, so it cannot have been part of any bidirectional gene. This inconsistency is highlighted by the fact that Li, et al. (2011) showed that Motif 3 enhances, but is not required for the activity of the HisRS Urzyme. One possibility, consistent with the fact that the C-terminal arginine of the arginine tweezer clamp on the adenine ring (Kaiser et al., 2018) is also in Motif 3, is that in the ancestral Class II forms, the amino acid bound more closely to the β-strand preceding the active-site arginine that forms the N-terminal half of the arginine tweezers. Under that hypothesis, an important selective advantage of the Motif 3 motif could have been to allow the amino acid binding site to migrate two β-strands away from its original binding site, enabling it to limit inclusion of larger Class II side chains into the growing amino acid alphabet, while at the same time providing the second arginine of the adenosine clamp.
4.3. Cognate pairs: how did the amino acid alphabet grow?
We have defined many of the characteristics of the earliest aaRS•tRNA combinations necessary for a “boot block” from which the genetic code can have emerged from a very simple binary alphabet capable only of slightly biased statistical peptides only some of which may have functioned (Carter and Wills, 2018b). Although many authors have alluded to processes by which the inclusion of new amino acids into the coding alphabet led to increasing functionality (reviewed by Barbieri, 2019a; Barbieri, 2019b), such commentary omits reference to a crucial aspect of how enhancing the genetic alphabet improves specificity, namely the importance of impedance matching between the errors accruing in both replication and transcription, on the one hand, and the recognition properties of the aaRS on the other hand. These arguments are summarized in (Carter and Wills, 2018b). The importance of this facet of the problem is the quantification of the error frequency as an impedance to information transfer (Wills and Carter, 2019), which implies that nature will chose couplings between the evolution of error-prone interacting processes that tend to minimize the dissipation of information and free energy.
The gold standard in answering the various questions posed under the rubric of “error reduction” is to determine with the highest possible confidence the order in which the aaRS speciated from coordinated ancestral reconstructions of aarRS•tRNA cognate pairs. To that end, we have assembled far more highly curated, structure-based multiple sequence alignments for the two aaRS superfamilies by threading sequences with unknown structure into the pdb coordinate file for structure whose sequence is closest to that of the new unknown structure. In this manner, we added ~15 new sequences per isoacceptor to those obtained from crystal structures alone, bringing the multiple sequence alignments for Class I and II aaRS to ~400 sequences (Popinga, 2019). It should also be noted that analysis of alignments derived from structural homology alone corroborates a parameterized model of amino acid substitution matrices based on the assumption that the matrix dimension matched the cardinality of the suite of aaRS types available for amino acid differentiation through every branchpoint in the phylogenies of both aaRS superfamilies (Shore et al., 2019).
Using our curated amino acid sequence alignments, we created alignments of the corresponding gene sequences, for the purpose of computing the <MBP> values for antiparallel codon middle base alignments discussed in a previous section. That <MBP> statistic will help correlate the ancestral reconstructions of the two aaRS superfamilies (Chandrasekaran et al., 2013). Finally, we have adduced substantial new constraints by clarifying not only the <MBP> values, but also the base patterns required for specific recognition of cognate tRNAS, both at the level of amino acid type (i.e., β-branched, carboxylate, aliphatic, charged, aromatic) and at the rudimentary level of amino acid Class (Carter and Wills, 2018a).
Acknowledgments
This work was supported by NIGMS R01-78227. We thank S. N. Chandrasekaran for providing programs and scripts for computing the <MBP> metric and F. Kaiser for providing a manuscript ahead of submission and for discussions of the analytical properties of Class I and II aaRS amino acid binding sites.
1. Abbreviations
- aaRS
aminoacyl-tRNA synthetases
- Y
pyrimidine
- R
purine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agmon I, 2009. The dimeric proto-ribosome: structural details and possible implications on the origin of life. Int. J. Mol. Sci 30, 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archetti M, 2004. Selection on codon usage for error minimization at the protein level. J. Mol. Evol 59, 400–415. [DOI] [PubMed] [Google Scholar]
- Barbieri M, 2019a. Evolution of the Genetic Code: the Ambiguity-reduction Theory. Biological Theory In review. [DOI] [PubMed] [Google Scholar]
- Barbieri M, 2019b. A General Model on the Origin of Biological Codes. BioSystems In Press. [DOI] [PubMed] [Google Scholar]
- Belousoff MJ, Davidovich C, Bashan A, Yonath A, 2010. On the development towards the modern world: a plausible role of uncoded peptides in the RNA world, In: Ruiz-Mirazo K, Luisi PL (Ed.), Origins of Life and Evolution of Biospheres. Springer, pp. 415–419. [Google Scholar]
- Belrhali H, 1996. Détermination par Cristallographie aux Rayons X des Mécanismes de Formation du Seryl-Adenylate et du bis(5’-Adenosyl) Tetraphosphate par la Seryl-aRNT Synthetase de Thermus Thermophilus. [Determination by X-ray Crystallographie of the Mechanisms by which Seryl-Adenylate and bis(5’-Adenosyl) Tetraphosphate are formed by the Seryl-tRNA Synthetase from Thermus Thermophilus] Sciences Biologiques Fondamentales et Appliquées. European Synchrotron Radiation Facility, Grenoble. [Google Scholar]
- Belrhali H, Yaremchuk A, Tukalo M, Berthet-Colominas C, Rasmussen B, Bosecke P, Diat O, Cusack S, 1995. The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure 3, 341–352. [DOI] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV, 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457, 977–980. [DOI] [PubMed] [Google Scholar]
- Carter CW Jr, Wills PR, 2018a. Hierarchical groove discrimination by Class I and II aminoacyl-tRNA synthetases reveals a palimpsest of the operational RNA code in the tRNA acceptor-stem bases. Nucleic Acids Research 46, 9667–9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr, Wills PR, 2018b. Interdependence, Reflexivity, Fidelity, and Impedance Matching, and the Evolution of Genetic Coding. Molecular Biology and Evolution 35, 269–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr, Wills PR, 2019. Class I and II aminoacyl-tRNA synthetase tRNA groove discrimination created the first synthetase•tRNA cognate pairs and was therefore essential to the origin of genetic coding. IUBMB Life In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr., 2014. Urzymology: Experimental Access to a Key Transition in the Appearance of Enzymes. J. Biol. Chem 289, 30213–30220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr., Li L, Weinreb V, Collier M, Gonzales-Rivera K, Jimenez-Rodriguez M, Erdogan O, Chandrasekharan SN, 2014. The Rodin-Ohno Hypothesis That Two Enzyme Superfamilies Descended from One Ancestral Gene: An Unlikely Scenario for the Origins of Translation That Will Not Be Dismissed. Biology Direct 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr., Wolfenden R, 2015. tRNA Acceptor-Stem and Anticodon Bases Form Independent Codes Related to Protein Folding. Proc. Nat. Acad. Sci. USA 112 7489–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr., Wolfenden R, 2016. Acceptor-stem and anticodon bases embed amino acid chemistry into tRNA. RNA Biology 13, 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran SN, Yardimci G, Erdogan O, Roach JM, Carter CW Jr, 2013. Statistical Evaluation of the Rodin-Ohno Hypothesis: Sense/Antisense Coding of Ancestral Class I and II Aminoacyl-tRNA Synthetases. Molecular Biology and Evolution 30, 1588–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Hed A, Ardell DH, 2018. Adaptive Partitioning of the tRNA Interaction Interface by Aminoacyl-tRNA-Synthetases. Theoretical Population Biology In Press. [DOI] [PubMed] [Google Scholar]
- Crick FHC, 1955. On Degenerate Templates and the Adaptor Hypothesis. Unpublished; https://profiles.nlm.nih.gov/ps/retrieve/Narrative/SC/p-nid/153.
- Crick FHC, 1968. The Origin of the Genetic Code. J. Mol. Biol 38, 367–379. [DOI] [PubMed] [Google Scholar]
- Crick FHC, 1970. Central Dogma of Molecular Biology. Nature 227, 561–563. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Belousoff M, Wekselman I, Shapira T, Krupkin M, Zimmerman E, Bashan A, Yonath A, 2010. The Proto-Ribosome: an ancient nano-machine for peptide bond formation. Isr J. Chem 50, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M, 1971a. Molecular self-organisation and the early stages of evolution. Quart Rev Biophys 4, 149–212. [PubMed] [Google Scholar]
- Eigen M, 1971b. Selforganization of Matter and the Evolution of Biological Macromolecules. Naturwissenschaften 58, 465–523. [DOI] [PubMed] [Google Scholar]
- Eigen M, McCaskill JS, Schuster P, 1988. Molecular Quasi-Species. J. Phys. Chem 92, 6881–6891. [Google Scholar]
- Eigen M, Schuster P, 1977. The Hypercycle: A Principle of Natural Self-Organization Part A: Emergence of the Hypercycle. Naturwissenschaften 64, 541–565. [DOI] [PubMed] [Google Scholar]
- Fox GE, Tran Q and Yonath A, 2012. An exit cavity was crucial to the polymerase activity of the early ribosome. Astrobiology 12, 57–60;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeland SJ, Hurst LD, 1998. The Genetic Code is One in a Million. J. Mol. Evol 47, 238–248. [DOI] [PubMed] [Google Scholar]
- Giegé R, 1972. Recherches sur la spécificité de reconnaissance des acides ribonucléiques de transfert par les aminoacyl-tRNA synthétases [Study on the specificity of recognition of transfer ribonucleic acids by aminoacyl-tRNA synthetases], Biological Chemistry. Université Louis Pasteur, Strasbourg, France. [Google Scholar]
- Hol WGJ, 1985. The Role of the α-Helix Dipole in Protein Function and Structure. Prog. Biophys. molec. Biol 45, 149–195. [DOI] [PubMed] [Google Scholar]
- Hol WJG, Halie LM, Sander C, 1981. Dipoles of the α-helix and β-sheet: their role in protein folding. Nature 294, 532–536. [DOI] [PubMed] [Google Scholar]
- Hol WJG, van Duijnen PT, Berensen HJC, 1978. The α-helix dipole and the properties of proteins. Nature 273, 443–446. [DOI] [PubMed] [Google Scholar]
- Hsiao C, Lenz K, Peters T, Fang JK, Schneider P-Y, Anderson DM, Preeprem EJ, Bowman T, O’Neill JC, Lie EB, S. Athavale L, Gossett S, Trippe JJ, Murray C, Petrov J, Wartell AS, Harvey RM, Hud SC, Williams NV, L.D., 2013. Molecular paleontology: a biochemical model of the ancestral ribosome. Nucleic Acids Res. 41, 3373–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser F, Bittrich S, Salentin S, Leberecht C, Haupt VJ, Krautwurst S, Schroeder M, Labudde D, 2018. Backbone Brackets and Arginine Tweezers delineate Class I and Class II aminoacyl tRNA synthetases. PLoS Comput Biol 14, e1006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser F, Krautwurst S, Salentin S, Haupt VJ, Leberecht C, Bittrich S, Labudde D, Schroeder M, 2019. Characterization of Amino Acid Recognition in Aminoacyl-tRNA Synthetases. BioRxiv 606459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH, 1993. Protein Design by Binary Patterning of Polar and Non-polar Amino Acids. Science 262, 1680–1685. [DOI] [PubMed] [Google Scholar]
- Koonin EV, 2011. The Logic of Chance: The Nature and Origin of Biological Evolution Pearson Education; FT Press Science, Upper Saddle River, NJ. [Google Scholar]
- Koonin EV, 2015. Why the Central Dogma: on the nature of the great biological exclusion principl. Biology Direct 10, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Novozhilov AS, 2017. Origin and Evolution of the Universal Genetic Code. Annu. Rev. Genet 51, 45–62. [DOI] [PubMed] [Google Scholar]
- Lahr SJ, Broadwater A, Carter CW Jr., Collier ML, Hensley L, Waldner JC, Pielak GJ, Edgell MH, 1999. Patterned library analysis: A method for the quantitative assessment of hypotheses concerning the determinants of protein structure. Proceedings of the National Academy of Sciences, USA 96, 14860–14865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Francklyn C, Carter CW Jr, 2013. Aminoacylating Urzymes Challenge the RNA World Hypothesis. J. Biol. Chem 288, 26856–26863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Weinreb V, Francklyn C, Carter CW, Jr, 2011. Histidyl-tRNA Synthetase Urzymes: Class I and II Aminoacyl-tRNA Synthetase Urzymes have Comparable Catalytic Activities for Cognate Amino Acid Activation. J. Biol. Chem 286, 10387–10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez L, Jimenez-Rodriguez M, Gonzalez-Rivera K, Williams T, Li L, Weinreb V, Chandrasekaran SN, Collier M, Ambroggio X, Kuhlman B, Erdogan O, Carter CWJ, 2015. Functional Class I and II Amino Acid Activating Enzymes Can Be Coded by Opposite Strands of the Same Gene. J. Biol. Chem 290, 19710–19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffet DA, Foley J, Hecht MH, 2003. Midpoint reduction potentials and heme binding stoichiometries of de novo proteins from designed combinatorial libraries. Biophys. Chem 105, 231–239. [DOI] [PubMed] [Google Scholar]
- Nieselt-Struwe K, Wills PR, 1997. The Emergence of Genetic Coding in Physical Systems. J. theor. Biol 187, 1–14. [DOI] [PubMed] [Google Scholar]
- Opuu V, Silvert M, Simonson T, 2017. Computational design of fully overlapping coding schemes for protein pairs and triplets. Scientific REPORTS 7, 15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SC, Bradley LH, Jinadasa SP, Hecht MH, 2009. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Prot. Sci 18, 1388–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, Hud NV, Williams LD, 2014. Evolution of the Ribosome at Atomic Resolution. Proc. Nat. Acad. Sci. USA 111 10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD, 2015. History of the ribosome and the origin of translation. PNAS 112 15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Y, Kuhlman B, Butterfoss GL, Hu H, Weinreb V, Carter CW Jr, 2010. Tryptophanyl-tRNA synthetase Urzyme: a model to recapitulate molecular evolution and investigate intramolecular complementation. J. Biol. Chem 285, 38590–38601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Y, Li L, Kim A, Erdogan O, Weinreb V, Butterfoss G, Kuhlman B, Carter CW Jr, 2007. A Minimal TrpRS Catalytic Domain Supports Sense/Antisense Ancestry of Class I and II Aminoacyl-tRNA Synthetases. Mol Cell 25, 851–862. [DOI] [PubMed] [Google Scholar]
- Popinga A, 2019. From the Origins of Life to Epidemics: Bayesian Inference, Stochastic Simulation, and Dynamics of Bioinformatic Systems., Computer Science. University of Auckland: Supplementary Data http://github.com/alexpopinga/aaRS-Pipeline, accessed 11 April 2019, Auckland, NZ. [Google Scholar]
- Popinga A, Bouckaert R, Wills PR, 2015. Complex phylogeny of aminoacyl-tRNA synthetases. XVth ESEB Meeting, Lausanne, Switzerland, p. Abstract 52521. [Google Scholar]
- Radzicka A, Wolfenden R, 1988. Comparing the Polarities of the Amino Acids: Side-Chain Distribution Coefficients between the Vapor Phase, Cyclohexane, 1–0ctanol, and Neutral Aqueous Solution. Biochem. 27, 1664–1670. [Google Scholar]
- Schimmel P, Giegé R, Moras D, Yokoyama S, 1993. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Nat. Acad. Sci. USA 90, 8763–8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore J, Holland BR, Sumner JG, Nieselt K, Wills PR, 2019. Substitution Matrices Recapitulate Amino Acid Specificity of AARS Phylogenies. Mol. Biol. Evol Submitted. [DOI] [PubMed] [Google Scholar]
- Stephenson JD, Freeland SJ, 2013. Unearthing the Root of Amino Acid Similarity. J Mol Evol 77, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov EN, 2000. Consensus temporal order of amino acids and evolution of the triplet code. Gene 261, 139–151. [DOI] [PubMed] [Google Scholar]
- Wills PR, 1993. Self-organization of genetic coding. J. Theor. Biol. 162, 267–287. [DOI] [PubMed] [Google Scholar]
- Wills PR, 2001. Autocatalysis, information, and coding. BioSystems 50, 49–57. [DOI] [PubMed] [Google Scholar]
- Wills PR, 2014a. Genetic Information, Physical Interpreters and Thermodynamics; The Material-Informatic Basis of Biosemiosis. Biosemiotics 7, 141–165. [Google Scholar]
- Wills PR, 2014b. Spontaneous Mutual Ordering of Nucleic Acids and Proteins. Orig Life Evol Biosph 44, 293–298. [DOI] [PubMed] [Google Scholar]
- Wills PR, 2016. The generation of meaningful information in molecular systems. Phil. Trans. R. Soc. A A374, 20150016. [DOI] [PubMed] [Google Scholar]
- Wills PR, 2019. Reflexivity, Coding, and Quantum Biology. BioSystems In preparation. [DOI] [PubMed] [Google Scholar]
- Wills PR, Carter CW Jr, 2018. Insuperable problems of an initial genetic code emerging from an RNA World. BioSystems 164, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills PR, Carter CW Jr., 2019. Impedance matching in an electrical circuit furnishes a detailed model for the coupling of error rates and avoidance of dissipative losses in biological information transfer. J.Theor. Biol. In Preparation. [Google Scholar]
- Wills PR, Nieselt K, McCaskill JS, 2015. Emergence of Coding and its Specificity as a Physico-Informatic Problem. Orig. Life Evol. Biosph 45, 249–255. [DOI] [PubMed] [Google Scholar]
- Woese C, 1969. Models for the Evolution of Codon Assignments. J. Mol. Biol 43, 235–240. [DOI] [PubMed] [Google Scholar]
- Woese CR, 1965. Order in the Origin of the Genetic Code. Proc. Nat. Acad. Sci. USA 54, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Dugre DH, Saxinger WC, Dugre SA, 1966. The molecular basis for the genetic code. Proc. Natl. Acad. Sci. USA 55, 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R, 1983. Waterlogged Molecules. Science 222, 1087–1093. [DOI] [PubMed] [Google Scholar]
- Wolfenden R, 2007. Experimental Measures of Amino Acid Hydrophobicity and the Topology of Transmembrane and Globular Proteins. Journal of General Physiology 129, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R, Cullis PM, Southgate CCF, 1979. Water, Protein Folding, and the Genetic Code. Science 206, 575–577. [DOI] [PubMed] [Google Scholar]
- Wolfenden R, Lewis CA, Yuan Y, Carter CW Jr., 2015. Temperature dependence of amino acid hydrophobicities. Proc. Nat. Acad. Sci. USA 112 7484–7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JT-F, 2005. Coevolution theory of the genetic code at age thirty. BioEssays 27, 416–425. [DOI] [PubMed] [Google Scholar]
- Wong JT-F, Ng S-K, Mat W-K, Hu T, Xue H, 2016. Coevolution Theory of the Genetic Code at Age Forty: Pathway to Translation and Synthetic Life. Life 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M, Widmann J, Knight R, 2009. RNA-amino acid binding: A stereochemical era for the genetic code. J. Mol. Evol 69, 406–429. [DOI] [PubMed] [Google Scholar]
- Zamudio GS, José MVI, 2018. Identity elements of tRNA as derived from information analysis. Orig. Life Evol. Biosph 48, 73–81 [DOI] [PubMed] [Google Scholar]