Abstract
RNA silencing or RNA interference (RNAi) is an essential mechanism in animals, fungi, and plants that functions in gene regulation and defense against foreign nucleic acids. In fungi, RNA silencing has been shown to function primarily in defense against invasive nucleic acids. We previously determined that mycoviruses are triggers and targets of RNA silencing in Sclerotinia sclerotiorum. However, recent progresses in RNAi or dsRNA-based pest control requires more detailed characterization of the RNA silencing pathways in S. sclerotiorum to investigate the utility of dsRNA-based strategy for white mold control. This study elucidates the roles of argonaute enzymes, agl-2 and agl-4, in small RNA metabolism in S. sclerotiorum. Gene disruption mutants of agl-2 and agl-4 were compared for changes in phenotype, virulence, viral susceptibility, and small RNA profiles. The Δagl-2 mutant but not the Δagl-4 mutant had significantly slower growth and virulence prior to virus infection. Similarly, the Δagl-2 mutant but not the Δagl-4 mutant, showed greater debilitation under virus infection compared to uninfected strains. The responses were confirmed in complementation studies and revealed the antiviral role of agl-2. Gene disruption mutants of agl-2, agl-4, Dicer-like (dcl)-1, and dcl-2 did not change the stability of the most abundant endogenous small RNAs, which suggests the existence of alternative enzymes/pathways for small RNA biogenesis in S. sclerotiorum. Furthermore, in vitro synthesized dsRNA targeting agl-2 showed a significantly reduced average lesion diameter (P < 0.05) on canola leaves with agl-2 down-regulated compared to controls. This is the first report describing the effectiveness of RNA pesticides targeting S. sclerotiorum RNA silencing pathway for the control of the economically important pathogen.
Keywords: RNA pesticide, argonautes, dicers, mycovirus, Sclerotinia sclerotiorum, RNA silencing, tRNA halves
Introduction
RNA silencing is a transcriptional and post-transcriptional suppression of gene expression. One of the roles that RNA silencing plays has long been identified as an adaptive defense mechanism against foreign nucleic acids, including viruses in animals, fungi, and plants (Waterhouse et al., 2001; Baulcombe, 2004, 2005). Unlike in animals and plants, the evolved RNA silencing in fungi to date has demonstrated that it is almost dispensable for endogenous gene regulation because gene disruption mutants often grow just fine. Instead, only when the mutants of RNA silencing genes are under virus infection, the antiviral role of those genes play then become evident (Segers et al., 2007; Zhang et al., 2014; Yu et al., 2018). However, studies of Neurospora crassa and other filamentous fungi have revealed diverse small RNA biogenesis pathways, suggesting that fungi adapt RNAi silencing pathways for several cellular processes with some of the RNA silencing genes playing dual roles (reviewed in Dang et al., 2011). On the other hand, fungal RNA silencing genes can also have redundant functions, such as antiviral, processing of dsRNA or transgenes (Catalanotto et al., 2004; Wang et al., 2016; Yu et al., 2018).
Sclerotinia sclerotiorum is a devastating plant fungal pathogen that causes up to 100% yield losses in crop production affecting a wide array of crops (Heffer Link and Johnson, 2007). Recent studies demonstrated that cross-kingdom RNA silencing can be blocked to control Botrytis cinerea which is closely related to S. sclerotiorum (Amselem et al., 2011). The virulence of B. cinerea can be greatly suppressed by silencing both B. cinerea Dicers at the same time (Wang et al., 2016). A similar observation was made in S. sclerotiorum following simultaneous disruption of both its Dicers to result in reduced pathogenicity (Mochama et al., 2018). Therefore, RNA silencing pathway has great potential to be manipulated to control fungal pathogens. As S. sclerotiorum has two predicted Argonautes (GenBank accession numbers Ss1G_00334 and Ss1G_11723), it is intriguing whether corresponding argonaute genes affect S. sclerotiorum virulence and whether it could add to the tool box of disease control with other novel strategies.
The Argonaute protein family constitutes four domains, N-terminal domain, Mid domain, and RNA-binding domains known as PAZ domains, and slicer domains known as PIWI domains (Poulsen et al., 2013). Argonaute proteins stabilize small dsRNA molecules produced by Dicer proteins to form RNA-induced silencing complexes (RISC) which are involved in post-transcriptional gene silencing or RNA-induced transcriptional silencing complexes involved in transcriptional gene silencing including chromatin modification in animals, plants, and insects (Irvine et al., 2006). When small dsRNA molecules produced by Dicers are incorporated into these effector complexes, one strand of the RNA molecule is removed and the remaining strand guides the complex to complementary RNA sequences which are subsequently cleaved by the Argonaute RNase H-like activity (Qihong et al., 2009).
Argonaute homologs have been identified in various fungi and they differ in function and number. The basal fungus, Mucor circinelloides, has three argonaute genes while Cryphonectria parasitica has four argonaute genes and Colletotrichum higginsianum has two (Qihong et al., 2009; Trieu et al., 2015; Campo et al., 2016). QDE-2 is a fungal argonaute homolog in N. crassa involved in quelling-the silencing of repetitive sequences such as transgenes (Fulci and Macino, 2007). In N. crassa, a separate silencing pathway called meiotic silencing of unpaired DNA (MSUD) has been characterized, and N. crassa RNA silencing components not involved in quelling have been shown to be involved in this pathway (Fulci and Macino, 2007). Similarly, in other fungi, not all components of the RNA silencing machinery are involved in RNA silencing mediated viral defense mechanisms. In Fusarium graminearum, only one of two argonaute genes, FgAgo1, is important in RNA silencing of viral nucleic acids (Yu et al., 2018) while in C. parasitica only agl-2 is required for antiviral RNA silencing, and in C. higginsianum, agl-1 but not agl-2 is essential for antiviral RNA silencing (Qihong et al., 2009; Trieu et al., 2015; Campo et al., 2016). The primary functions of the other gene homologs have not been fully characterized. As S. sclerotiorum are predicted to have two argonaute genes, agl-2 and agl-4, it presents a potential strategy to impede the proper small RNA processing after characterizing the roles of argonautes in S. sclerotiorum.
The goals of this study were to determine the function of argonaute genes in endogenous small RNA processing and defending virus infection in S. sclerotiorum, and as a proof of concept, to demonstrate a control strategy from silencing a specific argonaute gene. To achieve them, we made gene displacement mutants of argonaute genes in this study and transfected S. sclerotiorum with a RNA virus (SsHV2-L) and compared the changes in morphology and pathogenicity. Gene displacement mutants revealed that only agl-2 is important in vegetative growth, as well as antiviral defense, whereas the biological function of agl-4 remains unknown. We further established the application of dsRNA externally targeting agl-2 as an RNA pesticide to slow the infection in a dose-dependent manner.
Materials and Methods
Fungal Culture Strains and Conditions
The wild type strain, DK3, of S. sclerotiorum was grown on potato dextrose agar (PDA) (Sigma) at 20–22Δ. The gene displaced mutants of Δagl-2 and Δagl-4 strains were grown on PDA amended with hygromycin B (Alfa Aesar) at 100 μg/mL as selection. Dicer mutants were produced in our previous study (Mochama et al., 2018).
Gene Disruption of agl-2 and agl-4
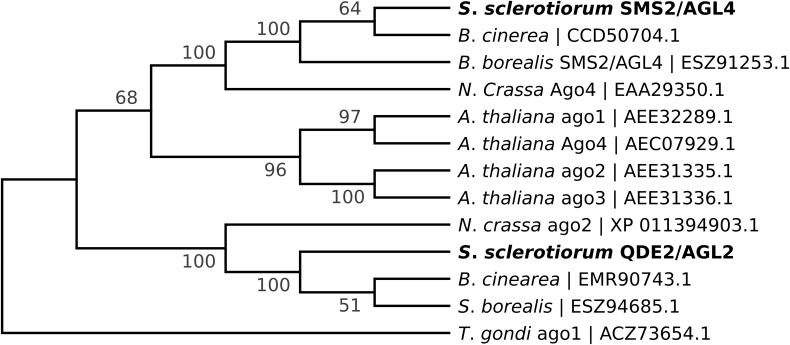
Sclerotinia sclerotiorum argonaute-like genes were predicted based on homology to those identified in N. crassa (Laurie et al., 2012). Published sequences of Ss1G_00334 and Ss1G_11723 in the GenBank (NCBI) are the putative argonaute genes coding for QDE-2/AGO-2 and SMS-2/AGO-4 in N. crassa, respectively (Figure 1). Argonaute genes were displaced by the hygromycin phosphotransferase gene (hph) using the split-marker homologous recombination cassettes as described before (Mochama et al., 2018). To generate the Δagl-2 gene displacement mutant, a 1.5 kb 5′ flanking arm of agl-2 was PCR-amplified by primers F1-AGO2 and F2-AGO2 and a 1.5 kb 3′ flanking arm of agl-2 was PCR-amplified by primers F3-AGO2 and F4-AGO2. The hph gene was amplified from pCSN43 (Fungal Genetics Stock Center) using primers PtrpC – HY and YG – TrpC to give two amplicons (1.2 and 1.3 kb) each containing part of the hph marker gene with an overlap (Table 1). The 5′- flank of the Δagl-2 gene was then connected to the partial hph amplicon containing PtrpC and the 3’ flank was connected to the hph amplicon containing TtrpC using the overlap extension PCR method. Eventually, an agl-2 gene deletion construct that included 1 kb of identical sequence to the 5′ flanking arm of the gene and 812 bp of the 3′ flanking arm sequence was derived. A similar procedure was used to generate the agl-4 deletion construct with 805 bp of sequence identical to the 5′ flanking arm and 1.1 kb of the 3′ flanking arm of agl-4.
FIGURE 1.
Phylogenetic analysis (Maximum Likelihood tree) of agl-2 and agl-4 genes depicting the relationships between the protein sequences from S. sclerotiorum (in bold), along with their orthologs in B. cinerea, S. borealis, N. crassa and Arabidopsis thaliana. The Toxoplasma gondii agl-1 amino acid sequence was used as an outgroup. Bootstrap consensus was calculated based on 100 bootstrap replicates using Mega 7.0 (Kumar et al., 2016).
TABLE 1.
Primer used in this study.
| Primer Name | Sequence | Note |
| F1-AGO2 | TGGTGAATTGTGAGTT GAATGGTG | agl-2 KO |
| F2-AGO2 | ACCCAATTCGCCCTATAGT GAGTCGTGCTGCTGGA TCAAAAGACAT | agl-2 KO |
| F3-AGO2 | AAGCCTACAGGACACACA TTCATCGTAGGTACCT GGTCATACCTTCCGCAT | agl-2 KO |
| F4-AGO2 | CAGGTCCAAGTCCT GTCCAC | agl-2 KO |
| YG-F | CGTTGCAAGACCTGC CTGAA | Both KO |
| PtrpC-F | ACGACTCACTATAGGG CGAATTGGGT | Both KO |
| TtrpC-R | TACCTACGATGAATGTGTG TCCTGTAGGCTT | Both KO |
| HY-R | GGATGCCTCCGCTCGA AGTA | Both KO |
| F1-AGO2-nested | GTTTGCAACAATC GCAGGTG | agl-2 KO |
| F4-AGO2-nested | TCTCCAACCAG CTACCGATG | agl-2 KO |
| F1-AGO4 | TTTGGTCCAGG CCTTGGTTT | agl-4 KO |
| F1-AGO4-nested | TTTTCACAACGG GTTTGGGC | agl-4 KO |
| F2-AGO4 | ACCCAATTCGCCCTATAGT GAGTCGTGAGCCATTAGCTTGG ATATTCGCA | agl-4 KO |
| F3-AGO4 | AAGCCTACAGGACACA CATTCATCGTAGGTAAGTGCCT TCATATCATAATCCTCC | agl-4 KO |
| F4-AGO4 | AAGGTTCGTCGGTT GGTAGT | agl-4 KO |
| F4-AGO4-nested | CCCTACTTGTCCC ACGTGAT | agl-4 KO |
| F1-COMP-AGO2 | ATAATAGCGGCCGCCGGA TAAACTGCCTCTTCGC | agl-2 Complementation |
| F4-COMP-AGO2 | TTTTACTGGATCCTCCT GTCCACAATCCCACAA | agl-2 Complementation |
| Ss-Ago2-T7p-1898F | TAATACGACTCACTATAG GGAGATGAATCTACA AGCCGTGCTG | dsRNA synthesis targeting agl-2 |
| Ss-Ago2-T7p-2065R | TAATACGACTCACTATAGG GAGATGAGAGGTA GCCGCTTCATT | dsRNA synthesis targeting agl-2 |
| Ss-Ago4-T7p-F | TAATACGACTCACTATAGGGAGA TTTGGTTGCAAAATCGATCA | dsRNA synthesis targeting agl-4 |
| Ss-Ago4-T7p-R | TAATACGACTCACTATAGGGA GATTGCTTGCTTTGTTGACCAG | dsRNA synthesis targeting agl-4 |
Fungal Transformation
PEG-mediated transformation method was used to transfer the gene deletion cassettes into S. Sclerotiorum DK3 protoplasts as described before (Mochama et al., 2018). Fungal DNA was extracted from mycelia and PCR-amplified by the use of primers- F1, F4, F1, HYR, YG2, and F4 to ascertain that argonaute genes were each displaced by the hph gene, confirmed by Sanger sequencing. Because S. sclerotiorum does not produce conidia, repeated hyphal tipping and nested PCR were necessary to derive a monokaryotic line of each targeted gene disruption to avoid mixed results from heterokaryotic mutants.
Complementation
To complement agl-2, protoplasts from the Δagl-2 strain was transformed with a bialaphos plasmid (pBARKS-1) cloned to express the full agl-2 gene flanked by 2.8 kb of 5′-upstream and 1.5 kb of 3′-downstream sequences to include the corresponding promoter and terminator. The agl-2 gene and flanking arms were amplified from the DNA extract of DK3 using primers F1-COMP-AGO2 and F4-COMP-AGO2 (Table 1) and inserted into the NotI and BamHI sites of pBARK-1 downstream to PTrpC and bialaphos resistance gene, bla1. Protoplasts and PEG-mediated transformation were the same as described earlier, except that the regeneration media was now supplemented with bialaphos at 10 μg/mL for selection. Multiple transformants were selected and hyphal-tipped several times to fresh PDA plates amended with bialaphos. PCR amplification using agl-2 specific primers confirmed the ectopic integration of the gene, and then four transformants were compared for the morphology on PDA between the virus-free and SsHV2-L virus-infected complemented strains.
Phenotypic Characterization of Gene Deletion Mutants
At least five replications each of DK3, Δagl-2, and Δagl-4 cultures with or without SsHV2 infections were compared for the phenotypes. Hyphal diameter was measured daily as described before (Mochama et al., 2018). A 5-mm plug was placed on a freshly cut canola leaf. More than three replicates of the leaves were inoculated on moist paper towels in covered petri dishes kept on a lab bench at room temperature. Hyphal area was measured daily at 24, 48, and 72 h post-inoculation.
In vitro dsRNA Synthesis, Inoculation, and Confirmation of Silencing by RT-qPCR
PCR amplification of the agl-2 target was performed using gene-specific primers with T7 promoter sequence added on both the forward primer and the reverse primer. Primers Ss-Ago2-T7p-1898F and Ss-Ago2-T7p-2065R were used (Table 1). dsRNAs were synthesized using MEGAscript T7 Transcription kit (Invitrogen) following the manufacturer’s procedure. To compare the suppressing effect of dsRNA on fungal pathogenicity, a 2 days actively growing plug in 3-mm diameter taken from the margin of a colony of S. sclerotiorum DK3 was placed on each canola leaf. Six replications each of different doses of abovementioned dsRNA at 200, 400, and 800 ng was pipetted to surround an agar plug in the volume of 20 μl, taking reference from the dosage of 800 ng/20 μl published by Wang et al. (2016). As controls, the same volume of water, as well as dsRNA targeting agl-4 were pipetted to surround the agar plug on canola leaves. dsRNA targeting agl-4 was produced the same way as that targeting agl-2 but with primers Ss-Ago4-T7p-F and Ss-Ago4-T7p-R. The lesion was measured length wise and at right angle across again to obtain an average for a representative diameter 2 days post-inoculation. The data was statistically analyzed using paired t-test; and using one-way ANOVA (for three or more samples), and when significant effect was determined, Tukey’s HSD test was performed to compare all pairs of means.
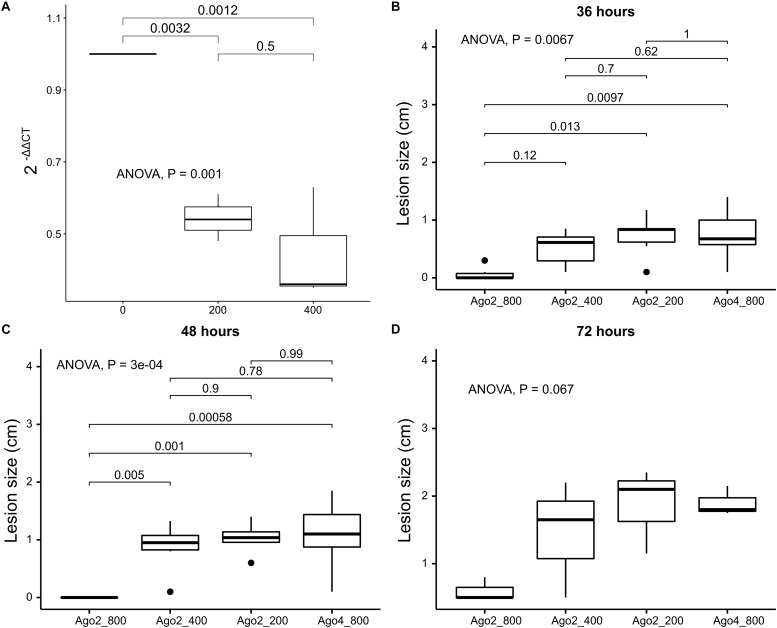
Only the lesions from 200 and 400 ng/20 μl were cut out to extract for total RNA using RNeasy Plant Mini Kit (Qiagen) since 800 ng/20 μl treatment does not produce lesions. RT-qPCR was performed to confirm the silencing of agl-2 gene in a dose-response manner using Luna Universal One-Step RT-qPCR Kit (NEB) following the manufacturer’s protocol. The comparative CT method (ΔΔCT method) was used to analyze the data. The expression levels of agl-2 and an endogenous control (actin) were evaluated with three biological replicates and four technical replicates each. The statistical significance of observed fold-difference was analyzed by ANOVA and Tukey’s test for pair-wise means separation.
Small RNA Libraries Preparation and Analysis of the Sequencing Results
MirVana miRNA Isolation kit (Thermo Fisher Scientific) was used to extract small RNAs from 4-day-old mycelia. NEBNext small RNA Library Kit (NEB, Ipswich, MA, United States) was used to construct the libraries for sequencing. The libraries were barcoded, pooled in a single lane for 50-nt single-end reads sequencing on an HiSeq4000 at the Roy J. Carver Biotechnology Center, UIUC. Three replicates of samples from SsHV2-L virus-infected DK3 as well as four replicates (two virus-infected and two virus-free mutants) each of Δdcl-1, Δdcl-2, Δagl-2, and Δagl-4 samples were sequenced. Adaptors were trimmed by BBMap tools (Bushnell, 2014). ShortStack (Axtell, 2013) was used to identify loci producing sRNAs by clustering. The number of reads aligned to S. sclerotiorum and SsHV2-L genomes were computed using bowtie (Langmead et al., 2009), and further downstream analysis were performed using in-house Perl and R scripts. tRNA encoding genes were predicted by tRNAscan-SE (Lowe and Chan, 2016).
Results
Disruption Mutants of Argonaute-Like Genes Were Generated
Argonaute-like genes were disrupted directly from wild-type strain DK3 using the same approach as described before (Mochama et al., 2018). Disruption was screened by PCR amplification using F1 and F4 primers and DNA extracts from multiple transformants as the templates to rule out ectopic integration of the hph gene. Sanger sequencing of the PCR amplicons confirmed the integration. Once a monokaryotic mutation was obtained by hyphal-tipping and confirmed by PCR that the target genes were completely deleted, further characterizations of the mutants were carried out.
Effect of Argonaute-Like Genes Disruption on Phenotype
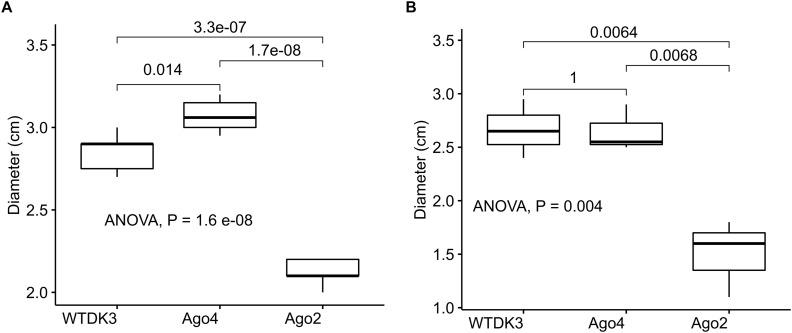
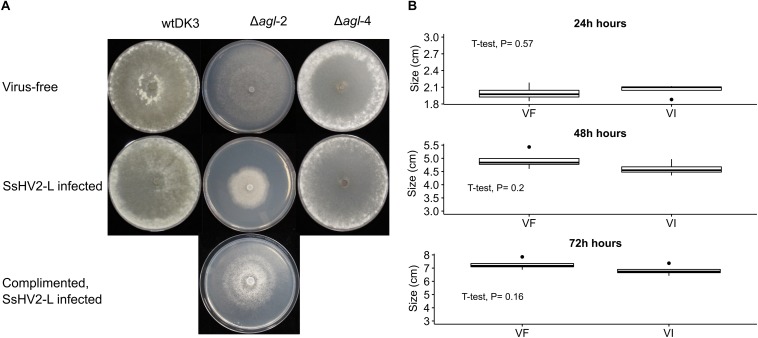
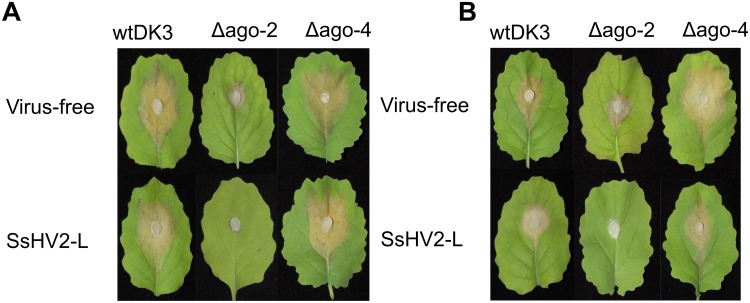
The colony morphology including the growth rate and the size of the sclerotia of the argonaute mutants and the wild-type strain DK3 on PDA were compared. Single mutant Δagl-4 and DK3 exhibited similar growth rates, whereas the Δagl-2 gene displacement mutant exhibited significantly slower growth at 24 h measured by the diameters of hyphal growth (P < 0.05) (Figure 2A). As shown in Figure 3, at 4 days post-inoculation (dpi) (Figure 3A), no change in growth rate was observed in Δagl-4, whereas Δagl-2 mutant shows a slower growth and a reduction in the size of sclerotia produced (Figure 4). After multiple times of hyphal-tipping, four agl-2 complemented transformants were assayed. The complemented strains exhibited a reversal of phenotype in antiviral defense (Figure 3B).
FIGURE 2.
Average hyphal diameter of S. sclerotiorum wild type and argonaute gene disruption mutants (Δagl-2 and Δagl-4) grown (A) on PDA for 1 dpi, and (B) on detached canola leaves 2 dpi.
FIGURE 3.
Colony morphology of virus-free and virus-infected gene deletion mutants. (Top row) Virus-free DK3, Δagl-2 and Δagl-4. (Bottom row) wild-type and mutant strains infected with hypovirus SsHV2-L. Cultures were grown for (A) 4 days on PDA. The virus-infected agl-2 mutant displays significantly slower growth and altered colony morphology. (B) Comparison of the complemented strains with and without virus infection (P > 0.05).
FIGURE 4.
Comparison of sclerotial morphology in wild-type DK3 and mutant strains, Δagl-2 and Δagl-4. The Δagl-2 mutant produces smaller sclerotia on average.
Effects of Argonaute-Like Gene Disruptions on S. sclerotiorum Pathogenicity
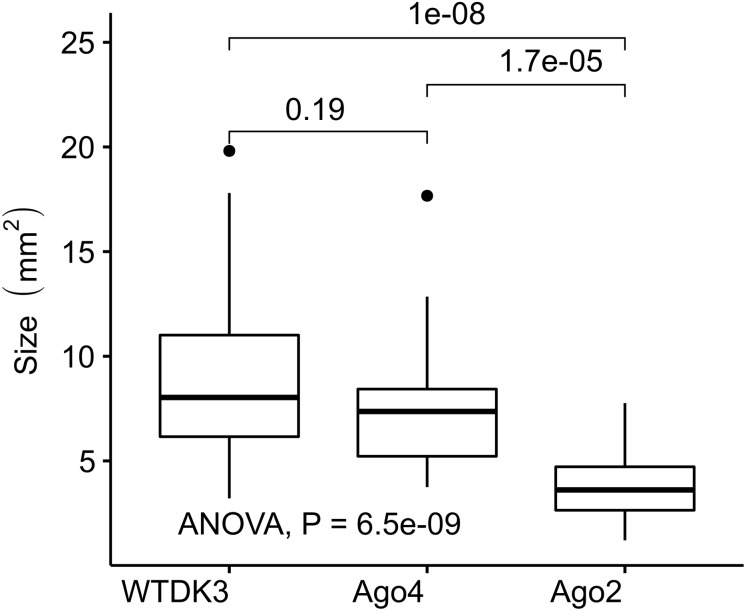
The virulence of S. sclerotiorum argonaute mutants was evaluated by inoculating detached leaves with agar plugs of mycelia. Lesion size data was collected at 1, 2, and 3 dpi showed that no difference in the lesion size produced by the mutant Δagl-4, but a significantly smaller lesion produced by Δagl-2 mutant compared to those produced by DK3 (Figures 2B, 5) (P < 0.05).
FIGURE 5.
Virulence assays on detached canola. Plugs were taken from the edge of actively growing DK3, Δagl-2, and Δagl-4 cultures and inoculated onto detached leaves. Lesion size was measured (A) 2 dpi, (B) 3 dpi.
Effects of Argonaute Gene on Antiviral Defense
To examine the effect of viral infection on strains containing null-mutations of agl-2 and agl-4, mutants were transfected through hyphal fusion with SsHV2-L infected mycelia. As shown in Figure 3, no significant differences were observed in growth and morphology in the agl-4 mutant infected with the mycovirus compared to virus-infected DK3, whereas the agl-2 mutant showed a significantly debilitated growth (Figures 3, 5) (P < 0.05).
In vitro Synthesized dsRNA Targeting agl-2 Shows Reduced Virulence of S. sclerotiorum
Once we determined that agl-2 plays an important role in endogenous small RNA processing, exemplified by a debilitated growth even without virus infection, the agl-2 was then targeted using in vitro synthesized dsRNA constructs in order to disrupt the fungal small RNA processing. RT-qPCR confirmed that agl-2 was silenced at the level of 800 ng in 20 μl volume but not at the lower doses of 200 or 400 ng (Figure 6A). As shown in Figures 6B–D, strains in which in vitro 800 ng of dsRNA was applied externally to target agl-2 exhibited a slower spread on canola leaves up to 3 days post-infection compared to lower doses at 200 and 400 ng applied or the targeting of agl-4 by the corresponding dsRNA (Supplementary Material).
FIGURE 6.
Effects of external RNA pesticide on inhibiting S. sclerotiorum from causing lesions on canola leaves comparing dsRNA targeting agl-2 at 800 ng/20 μl, 400 ng/20 μl, 200 ng/20 μl, dsRNA targeting agl-4 at 800 ng/20 μl as control (from left to right), confirmed by (A) RT-qPCR with reduced expression levels of agl-2 at 200 and 400 ng, and lesion comparison at (B) 36, (C) 48, and (D) 72 h post-inoculation.
Profiles of sRNAs in Distinct S. sclerotiorum Strains
The raw sequence reads were uploaded to NCBI SRA database under accessions SRR8844548 (WT-1), SRR8844549 (WT-2), SRR8785208 (WT-3), SRR8785205 (Δdcl1-1), SRR8785204 (Δdcl1-2), SAMN12129781 (Δdcl1-VF1), SAMN12129782 (Δdcl1-VF2), SRR8785203 (Δdcl2-1), SRR8785202 (Δdcl2-2), SAMN12129783 (Δdcl2-VF1), SAMN12129784 (Δdcl2-VF2), SRR8785201 (Δagl2-1), SRR8785200 (Δagl2-2), SAMN12129777 (Δagl2-VF1), SAMN12129778 (Δagl2-VF2), SRR8785199 (Δagl4-1), SRR8785198 (Δagl4-2), SAMN12129779 (Δagl4-VF1), and SAMN12129780 (Δagl4-VF2). Table 2 summarized the numbers of aligned small RNA sequence reads from the mutants and the WT sample that passed the ShortStack filtering parameters to allow the clustered reads for downstream analysis.
TABLE 2.
Numbers of aligning small RNA sequence reads from hypovirus-transfected wild type and mutants of Sclerotinia sclerotiorum.
| Read counts | Percent of aligned reads | ||||||
| Samples | Filtered read | IGR (% of tRNA derived small RNA) | CDS | Retrotransposons | Mitochondria | rRNA | Other |
| Ago2_SsHV2L_1 | 6510462 | 46.4 (42.7%) | 10.6 | 3.3 | 5.7 | 5.6 | 28.5 |
| Ago2_SsHV2L_2 | 6599083 | 46.7 (46.73%) | 9.3 | 2.9 | 5.6 | 5.8 | 29.7 |
| Ago2_VF_1 | 5895665 | 48.4 (31.46%) | 12.7 | 4.4 | 5.6 | 4.3 | 24.6 |
| Ago2_VF_2 | 5646987 | 49.1 (32.7%) | 13.5 | 4.8 | 5.2 | 3.9 | 23.5 |
| Ago4_SsHV2L_1 | 15140898 | 58.1 (68.9%) | 5.9 | 2.2 | 2.8 | 1.9 | 29.1 |
| Ago4_SsHV2L_2 | 8083962 | 49.4 (56.8%) | 8.2 | 3.0 | 4.5 | 8.0 | 26.9 |
| Ago4_VF_1 | 18251848 | 80.1 (78.6%) | 3.9 | 0.5 | 1.6 | 4.2 | 9.7 |
| Ago4_VF_2 | 13039613 | 83.0 (80.6%) | 3.5 | 0.5 | 1.8 | 2.8 | 8.4 |
| Dcl1_SsHV2L_1 | 6932872 | 32.9 (77.6%) | 2.2 | 0.2 | 1.7 | 24.9 | 38.1 |
| Dcl1_SsHV2L_2 | 7040569 | 52.5 (62.7%) | 5.3 | 2.3 | 2.6 | 9.2 | 28.1 |
| Dcl1_VF1 | 9213301 | 50.4 (57.8%) | 6.3 | 1.5 | 4.5 | 8.0 | 29.4 |
| Dcl1_VF2 | 9087471 | 32.7 (28.6%) | 5.6 | 0.9 | 4.2 | 19.8 | 36.8 |
| Dcl2_SsHV2L_1 | 14427517 | 43.6 (62.5%) | 5.6 | 2.3 | 4.6 | 4.9 | 38.9 |
| Dcl2_SsHV2L_2 | 11097593 | 43.2 (59.2%) | 6.7 | 3.0 | 4.2 | 5.5 | 37.3 |
| Dcl2_VF1 | 17041328 | 40.6 (35.4%) | 9.8 | 3.4 | 4.7 | 6.7 | 34.7 |
| Dcl2_VF2 | 13619703 | 50.0 (59.2%) | 5.6 | 1.8 | 4.7 | 7.2 | 30.8 |
| WTDK3_SsHV2L_1 | 15565013 | 63.7 (76.0%) | 5.1 | 1.7 | 3.1 | 3.4 | 23 |
| WTDK3_SsHV2L_2 | 14313982 | 39.2 (41.4%) | 9.4 | 4.4 | 2.8 | 1.2 | 43 |
| WTDK3_SsHV2L_3 | 14553278 | 40.8 (25.4%) | 11.6 | 4.5 | 8.5 | 2.8 | 31.8 |
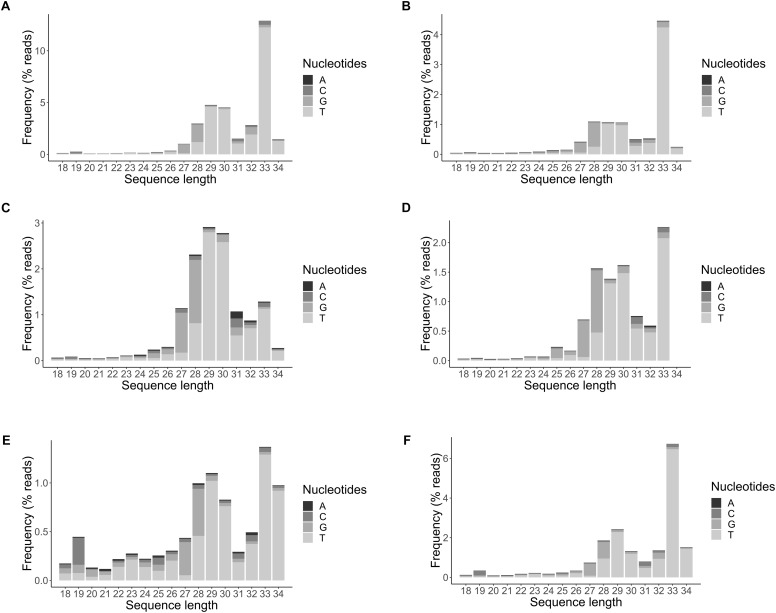
A major portion of endogenous small RNAs were found to represent the same small RNAs identified from our previous study (Lee Marzano et al., 2018), predominantly tRNA halves (tRFs): tRF5-Glu(GAA), 5′-TCCGAATTAGTGTAGGGGTTAACATAACTC-3′, and tRF5-Asp(GAC), 5′-TCTTTGATGGTCTAACGGTCATGATTTCC-3′, derived from tRNAs delivering glutamic acid and aspartic acid at 1.1 and 1.3% of total filtered reads in average, respectively. Homologs of the abovementioned two tRFs range in size from 29 to 35 bases with a major peak at 33 nt as TCTTTGATGGTCTAACGGTCATGATTTCCGTCC (Figures 7A–D) (underlined are the bases when expanding the size of small RNAs up to 34 nt long). tRF5-Glu(GAA) is predicted to be produced from SS1G_14562 and SS1G_14600 on chromosomes 7 and 4, respectively, whereas tRF5-Asp(GAC) is produced from SS1G_14527 on chromosome 14. The clustering result showed that these two small RNAs were mapped solely to intergenic regions. The BLASTn search (Altschul et al., 1990) indicates their homology to specific loci on chromosomes 4, 7, 14, and 16 for tRF5-Glu(GAA), and on chromosomes 1, 5, 11, 12, and 14 for tRF5-Asp(GAC). These small RNAs and their homologs are derived from mature tRNAs and contribute to the major peak at 29 or 33 nt. Compared to the wild type (Figure 7A), the productions of the two species were not affected by either single dicer mutations (Figures 7B,C), nor double dicer mutations (Figure 7D) (Mochama et al., 2018), and the stability of these small RNAs were not drastically affected by argonaute mutations (Figures 7E,F). Therefore, the reanalyzed data of double dicer mutant (previously published in Mochama et al., 2018) revealed that the production of these tRFs were not produced by either dicer.
FIGURE 7.
tRNA-derived small RNAs profiled for their size distribution in panel (A) wild type strain DK3, (B) Δdcl-1, (C) Δdcl-2, (D) Δdcl-1/dcl-2, (E) Δagl-2, (F) Δagl-4.
Discussion
Studies conducted on a number of fungal species have uncovered robust RNA silencing mechanisms with important roles in fungal antiviral defense. Similarly, this study elucidates the RNA silencing mechanisms in S. sclerotiorum and establishes the significant roles played by Argonaute-like genes in this devastating plant pathogenic fungus. Primarily, these findings clearly demonstrate that while the wild-type strain displayed reduced virulence following SsHV2-L virus infection (Marzano et al., 2015), RNA-silencing-deficient mutants [specifically Δagl-2 mutant in this study, and previously reported Δdcl-1/dcl-2 double mutant (Mochama et al., 2018)] displayed an even more significantly debilitated growth and reduced virulence under virus infection.
The slower growth of Δagl-2 without virus infection also suggested that agl-2 contributes to cellular gene regulation through the prevention of RISC formation with endogenous small RNAs. Specifically, we found that the deletion of agl-2 gene but not agl-4 resulted in compromised growth and virulence prior to virus infection, suggesting the contributions made by agl-2 to physiological and developmental processes. The agl-2 mutant exhibited slower growth, smaller sclerotia, and reduced virulence. Therefore, the changes observed in the agl-2 mutant may be attributed to a significant reduction in small RNA loading and stabilization of endogenous small RNAs. As expected, size distribution of small RNAs is not greatly affected upon the deletions of agl-2 or agl-4 genes when the Dicers are functional.
The great debilitation observed in the Δagl-2 mutant caused by virus infection was not detected in the virus-infected Δagl-4. This suggests that the AGL-2 protein is solely responsible for incorporating vsiRNAs into the RISC complex as part of the viral RNA silencing mechanism leading to the silencing of viral RNA. Argonaute proteins have been shown to associate with vsiRNAs in plants to target complementary viral mRNAs and in some cases host genes as well (Mengji et al., 2014; Carbonell and Carrington, 2015). miRNA-like molecules with possible gene regulation functions have been found to associate with fungal Argonaute proteins like the QDE-2 protein in N. crassa (Lee et al., 2010). Our study suggests that AGL-2 protein in S. sclerotiorum may also contribute to endogenous gene regulation. While AGL-4 protein’s function remains unknown, it is likely to play important roles including miRNA degradation (Sheu-Gruttadauria et al., 2019). Moreover, as single argonaute mutants do not have drastic changes in small RNA stability, this suggests possible functional redundancy in the two Argonautes.
Single gene disruption mutants of dcl-1, dcl-2, agl-2, agl-4, and double dicer mutants of dcl-1/dcl-2 did not alter the accumulation of tRFs, which suggests the existence of alternative enzymes or pathways for the biogenesis of this class of small RNA in S. sclerotiorum. Other endonuclease exist in S. sclerotiorum, such as RNaseL-like endonuclease that share similarities with yeast Ire1p proteins which are said to be involved in fungal mRNA splicing (Dong et al., 2001). Another endonuclease, Rny1 in yeast, is a ribonuclease T2-like precursor, and disruption of Rny1 lead to usually large cells that are temperature-sensitive for growth in yeast (MacIntosh et al., 2001). Also, the sizes of small RNA for tRFs were much larger than the dicer-processed ∼22 nt ones, supporting the speculation of different endonuclease(s) in action. Therefore, generating Rny1 and RNase L mutants to assess any disruption in the fungal growth and development and most importantly quantify the changes in the levels of tRFs will answer the pending questions brought up by this study. Moreover, the biological function of this under-characterized class of small RNA in this major pathogen of all dicots demands further study. Questions such as whether tRFs are induced by virus infection or simply a stress response, whether they target the IGRs as Blastn results suggested, or whether tRFs can be manipulated to debilitate S. sclerotiorum remain to be answered in the future.
The results derived from this study pave the way for the development of new control strategies that exploit RNA silencing mechanisms. The external RNA pesticide developed suggests the occurrence of external uptake of RNA in S. sclerotiorum. Furthermore, host-induced gene silencing (HIGS), virus-induced gene silencing (VIGS) approaches or heterologous expression of dsRNA sprays (spray-induced gene silencing) targeting agl-2 in S. sclerotiorum are expected to be effective in reducing the virulence, adding to the tool box of disease control.
Author Contributions
AN, PM, and S-YLM conceived and designed the experiments and wrote the manuscript. PM, CF, and HS performed the experiments. AN analyzed the small RNA data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported in part by National Sclerotinia Initiative Grant SA1800330 (to S-YLM) and SDSU startup from USDA Hatch fundSD00H606-16 project number with accession number 1009451.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00976/full#supplementary-material
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amselem J., Cuomo C. A., van Kan J. A., Viaud M., Benito E. P., Couloux A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M. J. (2013). ShortStack: comprehensive annotation and quantification of small RNA genes. Rna 19 740–751. 10.1261/rna.035279.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2005). RNA silencing. Trends Biochem. Sci. 30 290–293. [DOI] [PubMed] [Google Scholar]
- Bushnell B. (2014). Bbmap: a Fast, Accurate, Splice-Aware Aligner. Berkeley, CA: Lawrence Berkeley National Laboratory (LBNL). [Google Scholar]
- Campo S., Gilbert K. B., Carrington J. C. (2016). Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum. PLoS Pathog. 12:e1005640. 10.1371/journal.ppat.1005640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A., Carrington J. C. (2015). Antiviral roles of plant Argonautes. Curr. Opin. Plant Biol. 27 111–117. 10.1016/j.pbi.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C., Pallotta M., ReFalo P., Sachs M. S., Vayssie L., Macino G., et al. (2004). Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24 2536–2545. 10.1128/mcb.24.6.2536-2545.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Yang Q., Xue Z., Liu Y. (2011). RNA interference in fungi: pathways, functions, and applications. Eukary. Cell 10 1148–1155. 10.1128/EC.05109-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Niwa M., Walter P., Silverman R. H. (2001). Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. Rna 7 361–373. 10.1017/s1355838201002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulci V., Macino G. (2007). Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr. Opin. Microbiol. 10 199–203. 10.1016/j.mib.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Heffer Link V., Johnson K. (2007). White mold. Plant Health Instruct. 10.1094/PHI-I-2007-0809-01 [DOI] [Google Scholar]
- Irvine D. V., Zaratiegui M., Tolia N. H., Goto D. B., Chitwood D. H., Vaughn M. W., et al. (2006). Argonaute slicing is required for heterochromatic silencing and spreading. Science 313 1134–1137. 10.1126/science.1128813 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie J. D., Ali S., Linning R., Mannhaupt G., Wong P., Güldener U., et al. (2012). Genome comparison of barley and maize smut fungi reveals targeted loss of RNA silencing components and species-specific presence of transposable elements. Plant Cell 24 1733–1745. 10.1105/tpc.112.097261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-C., Li L., Gu W., Xue Z., Crosthwaite S. K., Pertsemlidis A., et al. (2010). Diverse pathways generate microRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol. Cell 38 803–814. 10.1016/j.molcel.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Marzano S. Y., Neupane A., Domier L. (2018). Transcriptional and small RNA responses of the white mold fungus Sclerotinia sclerotiorum to infection by a virulence-attenuating hypovirus. Viruses 10:E713. 10.3390/v10120713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Chan P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44 W54–W57. 10.1093/nar/gkw413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh G. C., Bariola P. A., Newbigin E., Green P. J. (2001). Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: unexpected functions for ancient enzymes? Proc. Natl. Acad. Sci. 98 1018–1023. 10.1073/pnas.98.3.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzano S.-Y., Hobbs H. A., Nelson B. D., Hartman G. L., Eastburn D. M., McCoppin N. K., et al. (2015). Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus. J. Virol. 89 5060–5071. 10.1128/jvi.03199-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengji C., Du P., Wang X., Yu Y.-Q., Qiu Y.-H., Li W., et al. (2014). Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111:14613. 10.1073/pnas.1407131111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochama P., Jadhav P., Neupane A., Marzano S.-Y. L. (2018). Mycoviruses as triggers and targets of RNA silencing in white mold fungus Sclerotinia sclerotiorum. Viruses 10:E214. 10.3390/v10040214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C., Vaucheret H., Brodersen P. (2013). Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell 25 22–37. 10.1105/tpc.112.105643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qihong S., Gil H. C., Donald L. N. (2009). A single argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. U.S.A. 106:17927. 10.1073/pnas.0907552106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers G. C., Zhang X., Deng F., Sun Q., Nuss D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104 12902–12906. 10.1073/pnas.0702500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J., Pawlica P., Klum S. M., Wang S., Yario T. A., Oakdale N. T. S., et al. (2019). Structural Basis for Target-Directed MicroRNA Degradation. Available at SSRN: https://ssrn.com/abstract=3318944. (accesed January 24, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu T. A., Calo S., Nicolás F. E., Vila A., Moxon S., Dalmay T., et al. (2015). A non-canonical RNA silencing pathway promotes mRNA degradation in basal fungi (An RdRP-dependent degradation mechanism of mRNA). PLoS Genet. 11:e1005168. 10.1371/journal.pgen.1005168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Lin F.-M., Thomma B. P. H. J., Huang H.-D., Jin H., et al. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2:16151. 10.1038/nplants.2016.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse P. M., Wang M. B., Lough T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411 834–842. 10.1038/35081168 [DOI] [PubMed] [Google Scholar]
- Yu J., Lee K.-M., Cho W. K., Park J. Y., Kim K.-H. (2018). Differential contribution of RNA interference components in response to distinct fusarium graminearum virus infections. J. Virol. 92 e1756–e1717. 10.1128/JVI.01756-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.-X., Spiering M. J., Nuss D. L. (2014). Characterizing the roles of Cryphonectria parasitica RNA-dependent RNA polymerase-like genes in antiviral defense, viral recombination and transposon transcript accumulation. PLoS One 9:e108653. 10.1371/journal.pone.0108653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.