Abstract
Mechanical heterogeneity in biological tissues, in particular stiffness, can be used to distinguish between healthy and diseased states. However, it is often difficult to explore relationships between cellular-level properties and tissue-level outcomes when biological experiments are performed at a single scale only. To overcome this difficulty, we develop a multi-scale mathematical model which provides a clear framework to explore these connections across biological scales. Starting with an individual-based mechanical model of cell movement, we subsequently derive a novel coarse-grained system of partial differential equations governing the evolution of the cell density due to heterogeneous cellular properties. We demonstrate that solutions of the individual-based model converge to numerical solutions of the coarse-grained model, for both slowly-varying-in-space and rapidly-varying-in-space cellular properties. We discuss applications of the model, such as determining relative cellular-level properties and an interpretation of data from a breast cancer detection experiment.
Keywords: cell-based model, partial differential equation, continuum-limit, multi-scale, discrete model
1. Introduction
Biological tissues are heterogeneous and multi-scale by their very nature (figure 1a). This heterogeneity exists at all scales from sub-cellular to cellular, and from cellular to tissue levels [1–3]. We focus on cellular
Figure 1.
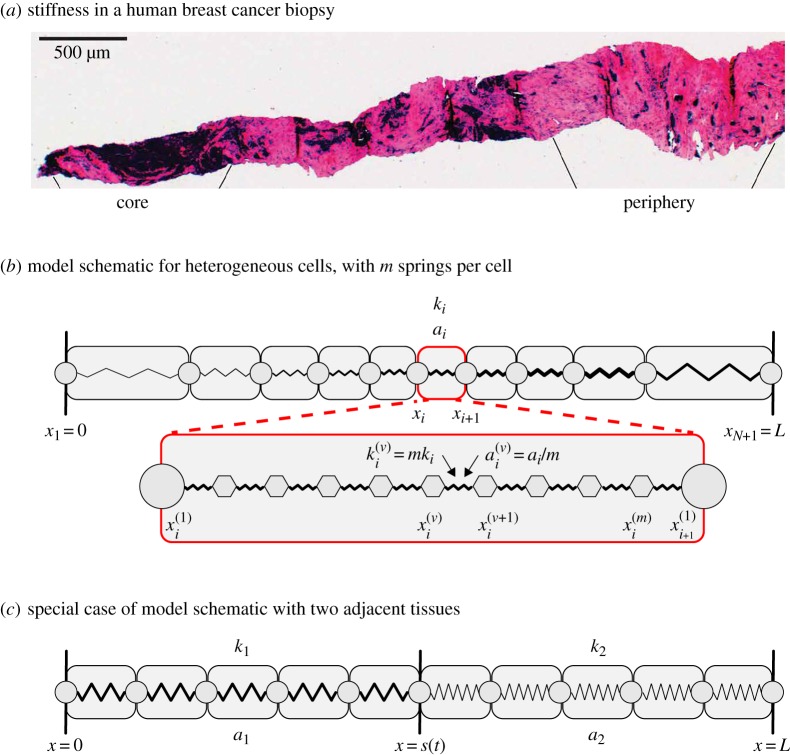
Stiffness heterogeneity in biological tissues. (a) Post atomic force microscopy histological overview of an entire breast cancer biopsy, where dark regions and pink regions are associated with low and high cell stiffness, respectively. Reproduced with permission from [6]. (b) Individual-based model schematic for arbitrarily heterogeneous tissue with N cells and m identical springs per cell. Cell i occupies the region xi(t) < x < xi+1(t) has cell stiffness ki and resting cell length ai. Spring ν in cell i, occupies the region x(ν)i(t) < x < x(ν+1)i(t), is prescribed with spring stiffness k(ν)i = mki and resting spring length a(ν)i = ai/m. The first and final spring boundaries in cell i coincide with the cell boundary positions so that x(1)i(t) = xi(t) and x(1)i+1(t) = xi+1(t) for all time. The cell and spring boundaries are shown as discs and hexagons, respectively. (c) Individual-based model schematic for a special case with two adjacent tissues, similarly this could model a heterogeneous tissue with two cell types. Cells in tissue i are prescribed with cell stiffness ki and resting cell length ai for i = 1, 2. Here each cell is represented with a single spring. The position of the interface between the two tissues is at x = s(t). (Online version in colour.)
interactions driven by mechanical stiffness which is of great importance in a variety of applications including epithelial tissue mechanics, cancer progression [4], cancer invasion and metastasis [5], stiffness as a biomarker in cancer detection [6–9], wound healing [10] and morphogenesis [11]. Tissue-level stiffness information [12] has been available for much longer than cellular-level stiffness data which requires advanced technology, such as atomic force microscopy [13–16]. However, difficulties arise in relating cellular-level data with tissue-level information when experiments are conducted and analysed at a single scale only. Mathematical modelling with in silico simulations provides a clear framework to explore these connections across biological scales.
Mathematical models of cell populations are broadly classified as either discrete or continuum. Discrete models, reviewed in [17,18], include cellular automata models, cellular Potts models, cell-centre models [17], vertex models, subcellular-element models [19] and tensegrity models [20]. Discrete models explicitly describe cellular-level interactions but often lack macroscopic intuition. Continuum models on the other hand often provide no cellular-level information [21] but can be more adept at including concepts of macroscopic stiffness [22,23] and, for large numbers of cells, as in epithelial tissues, tend to be less computationally expensive. Hybrid intermediate models also exist which consider the multi-scale nature of the problem [24–26]. A range of models specifically examine the role of mechanics [27,28]. However, in this work, we focus on models which relate cellular-level details to tissue-level outcomes. These models have been developed with a variety of coarse-graining techniques and assumptions, including the use of slowly varying and periodic assumptions on the heterogeneity in the model [29,30], correlation functions [31,32] and interaction forces from potentials [33]. Few of these models explore the role of stiffness. The work of Murray et al. [34–37] explicitly incorporates cell stiffness; they derive a nonlinear diffusion equation governing the evolution of the cell density in space and time; however, the framework focuses exclusively on homogeneous cell populations. Here, we extend this framework to heterogeneous cell populations.
The key focus of this work is to present a novel coarse-grained system of partial differential equations governing the evolution of the cell density, cell stiffness and resting cell length, from a heterogeneous cell-based model of epithelial tissue mechanics. The cell stiffness and resting cell length are constant for each cell and are simply transported in space by cell movements. The motion in this model is governed by cell–cell interaction forces modelled with Hooke's Law. In extending the work of Murray et al. [34], we provide a more general derivation of the governing equations, see §2, which is robust to the inclusion of both slowly-varying-in-space and rapidly-varying-in-space cellular properties, see §3. We show that solutions from the discrete model converge to the corresponding continuum model solution, under appropriate scalings. Additional results in §3 show the model can be applied to interpret experimental and clinical observations relating to breast cancer detection. Key algorithms used to generate results are available on GitHub.
2. Model description
In this section, we describe the individual-based model, which we refer to throughout this work as the discrete model, and derive a corresponding coarse-grained approximation in the form of system of partial differential equations, which we refer to as the continuum description. The continuum limit usually assumes that the number of discrete entities that makes up the system tends to infinity [29,38], while the size of the domain also tends to infinity, as in the thermodynamic limit, or the size of a length scale tends to zero, both in such a way that the ratio of the size of length scale to the number of discrete entities is fixed. Here, to maintain a fixed total tissue length and a fixed total number of cells in the continuum limit, we instead assume that each cell is internally represented by several identical springs. We then take the continuum limit by considering that the number of springs per cell tends to infinity while the spring length tends to zero.
(a). Discrete model
In this work, the discrete model describes an epithelial tissue formed by cells in contact with each other. For simplicity, we assume that the tissue can be modelled as a one-dimensional chain of N cells with fixed total length L. Tissues in the body commonly evolve in confined spaces, for example, imposed by bone tissues, and are subjected to strong geometric constraints so we fix the left tissue boundary at x = 0 and the right tissue boundary at x = L. This also allows us to focus on internal cellular heterogeneity. Alternate free boundary conditions are possible [34,39,40] but we do not focus on such free boundary conditions in this work. Each cell can have distinct mechanical properties (figure 1). This model could be used to represent a single tissue with intrinsic heterogeneity or multiple adjacent tissues with different properties. Each cell interacts with its neighbour through an effective interaction force which could represent cell–cell adhesion [41] or compressive stresses [42]. We consider cell i, for i = 1, 2, …, N, to have its left boundary at xi(t) and its right boundary at xi+1(t), with x1(t) = 0 and xN+1(t) = L at all times. The cell has a prescribed cell stiffness, ki, and resting cell length, ai. Inside the tissue, Newton's second law of motion governs the motion of each cell boundary such that
| 2.1 |
where Mi is the mass associated with cell boundary i, Fvisci is the viscous force associated with cell boundary i, and we model interaction forces at cell boundary i using Hooke's Law
| 2.2 |
The viscous force experienced by cells, due to cell-medium and cell-matrix interactions, is modelled with Fvisci = − ηdxi(t)/dt, where η > 0 is the viscosity coefficient. Cells migrate in dissipative environments and this is commonly modelled by assuming that the motion is overdamped [34,43], hence the term on the left of equation (2.1) is zero, giving
| 2.3 |
This model, as presented thus far, considers each cell to be represented by a single spring [34,40] which is sufficient to describe the discrete model. However, when we derive the continuum model, to maintain L and N, we represent each cell internally with m identical springs and we will later consider m≫1, which corresponds to the spring length tending to zero, see §§2b. The corresponding discrete model for m springs per cell is now described. Cell i with boundaries xi and xi+1 now has m + 1 spring boundaries, x(1)i, x(2)i, …, x(m)i, x(1)i+1, with xi = x(1)i and xi+1 = x(1)i+1 (figure 1c). The cell length is related to the spring length through the scaling xi+1 − xi∼m(x(ν+1)i − x(ν)i) as m → ∞, and with equality for all m as t → ∞. Each spring ν in cell i is prescribed with a spring stiffness k(ν)i and resting spring length a(ν)i related to cell properties ki and ai through
| 2.4 |
The viscosity coefficient for a cell boundary, η, is related to the viscosity coefficient for a spring boundary, η(ν), through η(ν) = η/m. Then the corresponding spring boundary equations are
| 2.5 |
The scalings in equation (2.4) and for the viscosity coefficient are chosen such that the cell boundary velocities are maintained and are independent of m, i.e. such that dx(1)i/dt = dxi/dt. These scalings are supported by results from the discrete model with varying m, see §3.
The discrete model is governed by equation (2.3) with the fixed boundary conditions for a system with a single spring per cell, and by equation (2.5) with fixed boundary conditions for a system with m springs per cell. In each situation, the discrete model forms a deterministic coupled system of ordinary differential equations that we can solve numerically, see electronic supplementary material, §1. We can also solve each system with an eigenmode decomposition to conveniently determine the long-time steady-state solution.
(b). Derivation of continuum model
We now derive a coarse-grained system of partial differential equations describing the evolution of cell density at a larger scale. To do so, we take the continuum limit by increasing the number of springs per cell, m, while maintaining the total number of cells, N, and tissue length, L, fixed, and by performing spatial averages over length scales involving a sufficiently large number of cells to define continuous densities, but sufficiently small to retain spatial heterogeneities. We first define the microscopic cell density, , in terms of the spring boundary positions, x(ν)i(t), as
| 2.6 |
where δ is the Dirac delta function [38,44]. Integrating equation (2.6) over the tissue domain, 0 < x < L, gives the total number of cells, N, which is independent of m. We introduce a mesoscopic length scale δx such that a(ν)i≪ai≪δx≪L and define a local spatial average which, for the microscopic cell density, , is
| 2.7 |
Differentiating equation (2.7) with respect to time leads to the general conservation law
| 2.8 |
where we use properties of the Dirac delta distribution [44] and take the spatial derivative outside of the average by making use of the fact that δx is small. The averaged term on the right of equation (2.8) is the coarse-grained cell density flux, j(x, t), describing spring migration at the mesoscopic scale, expressed explicitly in terms of the spring boundary positions and velocities [38]. We now introduce three field functions, f(x, t), k(x, t), a(x, t), for the cell–cell interaction force, the cell stiffness and the resting cell length, respectively, defined such that
| 2.9 |
where the scalings for f, k and a, with respect to m, agree with the scalings from the discrete system, see equation (2.4). The field functions k(x, t) and a(x, t) capture the assumption that spring properties and respective cell properties are constant along spring boundary trajectories, x(ν)i(t). To represent the distribution of spring lengths across the domain, we introduce a continuously differentiable function, l(x, t), which we define such that
| 2.10 |
where l(ν)i≪ai≪δx≪L. Writing equation (2.5) in terms of these continuous variables, expanding each cell–cell interaction force using the small parameter l(ν)i, using the viscosity coefficient scaling, and simplifying to leading order gives
| 2.11 |
Substituting equation (2.11) into equation (2.8), relating the spring length to the cell density with l(x(ν)i(t), t) = 1/[mq(x(ν)i(t), t)], and integrating over the spatial average interval, (x − δx, x + δx) gives
| 2.12 |
where n is the number of cells in the interval (x − δx, x + δx) and i has been reset to count these cells. Then, taking the limit as m → ∞ and performing an average over the m springs per cell, gives
| 2.13 |
We apply a mean field approximation, as n≫1 in (x − δx, x + δx) due to ai≪δx, by substituting q(xi(t), t) and ∂f(xi(t), t)/∂x in the sum with the average density q(x, t) and the average interaction force gradient ∂f/∂x in the interval (x − δx, x + δx). The factor 1/q is now independent of i and cancels with the factor n/(2δx) which represents the density of cells in the spatial average interval. Then the coarse-grained cell density flux is
| 2.14 |
which provides us with an important physical interpretation and is directly related to the velocity, net force and cell–cell interaction force gradient. By inspection of equations (2.11) and (2.14), we see that the cell density flux, j, is an advective flux j = qu, where u(x, t) = 〈dxi/dt〉 is the average velocity induced by the average force gradient 〈∂f/∂x〉. We also see that the net force is given by ηj/q and the spatially averaged interaction force gradient is given by ηj.
Substituting equation (2.14) into equation (2.8) gives
| 2.15 |
We now return to equation (2.9) and differentiate with respect to time to derive an evolution equation for the cell stiffness function
| 2.16 |
Using equation (2.11) and similar developments, the evolution equations for the cell stiffness and resting cell length expressed in terms of mesoscopic variables become
| 2.17 |
and
| 2.18 |
Written in terms of velocity, we identify the left-hand sides of equations (2.17) and (2.18) as the convective derivatives of the cell properties.
In summary, the governing equations of the coarse-grained model are given by equations (2.15), (2.17) and (2.18) with the interaction force f given by
| 2.19 |
This results in a system of four self-consistent equations for the continuous fields q(x, t), k(x, t), a(x, t), f(x, t) in terms of spatial position rather than particle trajectories. The initial conditions for the average cell density, cell stiffness and resting cell length are
| 2.20 |
together with no flux boundary conditions for the average cell density, due to the microscopic motion constraints, and Dirichlet boundary conditions for the cell stiffness and resting cell length, as cell properties are constant along cell boundary trajectories
| 2.21 |
These governing partial differential equations (2.15), (2.17)–(2.19) are solved numerically with the initial conditions (2.20) and boundary conditions (2.21), see electronic supplementary material, §S2. With homogeneous cell populations, the governing equations reduce to the single nonlinear density diffusion equation previously derived in [34]
| 2.22 |
3. Results and discussion
In this section, we compare solutions of the continuum and discrete models with the expectation that as the number of springs per cell, m, increases solutions from the discrete model converge to the corresponding continuum solution.
(a). Homogeneous cell population
We first consider a homogeneous cell population, with one spring per cell, m = 1, to illustrate the time evolution of the cell density flux during mechanical relaxation of the tissue. To compare results from the discrete and continuum systems, we choose the initial cell configuration (figure 2a) to represent a normally distributed cell density
| 3.1 |
with mean position μ = 5 and variance σ = 3. We choose λ to satisfy so that with L = 10 the total number of cells is N = 40, see electronic supplementary material, §S1. Then, using the discrete model, we observe that the system relaxes to a uniform cell distribution (figure 2a). Figure 2b,c shows how the density and velocity, respectively, propagate along the cell boundary characteristics and demonstrate that the system undergoes temporal relaxation to a steady-state configuration. With an eigenmode decomposition of the governing equations of the discrete system, given by equation (2.3) and the fixed boundary conditions, we find all eigenvalues are negative which explains the exponential decay behaviour.
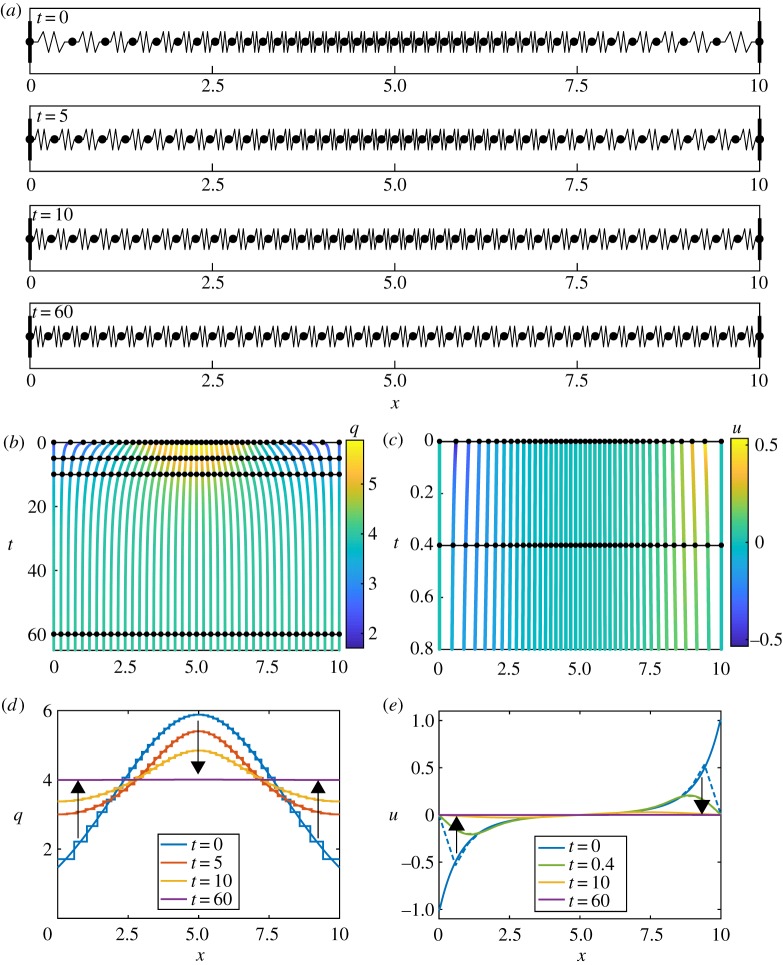
Figure 2.
Results for homogeneous k and a, with N = 40 and m = 1. (a) Snapshots of cell boundary positions and cell lengths at t = 0, 5, 15, 60. (b) Characteristic diagram for cell boundary position evolution for 0 ≤ t ≤ 65. Colour denotes the cell density. Black lines with dots represent snapshots in (a) and (d). (c) Characteristic diagram for cell position evolution for 0 ≤ t ≤ 0.8. Colour denotes velocity. Black lines and dots represent snapshots in (e). (d) Cell density snapshots at t = 0, 5, 10, 60. Results from discrete/continuum system displayed as stepped/solid lines. (e) Velocity snapshots at t = 0, 0.4, 10, 60. Results from discrete simulation and continuum system displayed as dashed/solid lines. Arrows indicate the direction of increasing time. (Online version in colour.)
We determine the discrete cell density as the inverse of the spacing between cell boundary trajectories, qi = 1/(xi+1 − xi) and we assign this value throughout the region xi < x < xi+1. We now compare this discrete information with the density from the continuum system, q, obtained by solving equations (2.15), (2.17)–(2.19). In figure 2d, we see that the initially normally distributed density tends to the uniform density , given by , which is independent of k and a. From equation (2.15), we see that this motion is driven by imbalances in the local interaction force field. We relate this to the velocity, u = (∂f/∂x)/(ηq) from §2, and we see that as the local imbalances tend to zero the cell boundary velocities tend to zero (figure 2e). Due to fast dynamics followed by slow long-term dynamics, results for t = 10 and t = 60 are mostly overlapping with the steady state (figure 2e). This agrees with the interpretation of the discrete system from equation (2.3).
(b). Heterogeneous cell population
Here, we present results for slowly-varying-in-space and piecewise constant heterogeneous cell populations.
(i). Slowly varying cell population
For slowly-varying-in-space cellular properties, we explore how solutions of the discrete system converge to the solution of the continuum system as m increases. We consider heterogeneity in k and homogeneous a so that, on average, cells are in compression. Figure 3 depicts how the system relaxes to a non-uniform density distribution, due to cell stiffness heterogeneity, as the velocity field u tends to zero. From this simulation, we observe higher density in regions of higher k. This prediction agrees with the steady-state solution to the coarse-grained model, governed by equations (2.15), (2.17)–(2.19)
| 3.2 |
where , and , are steady-state solutions and b is a constant of integration that is related to N. We also observe that, as cell properties are constant along trajectories, the cell stiffness evolves at a fixed location in space. We see in figure 3d–h that there is close agreement between the discrete model and the continuum solutions as m increases. It is notable that even for low m we have excellent agreement between the discrete density and the continuum density at the centre of each spring. However, at spring boundaries the agreement does not hold as well for low m. We see similar discrete-continuum agreement when we consider other examples with heterogeneous k and homogeneous a, with homogeneous k and heterogeneous a, and heterogeneous k and heterogeneous a (electronic supplementary material, figures S3–S6).
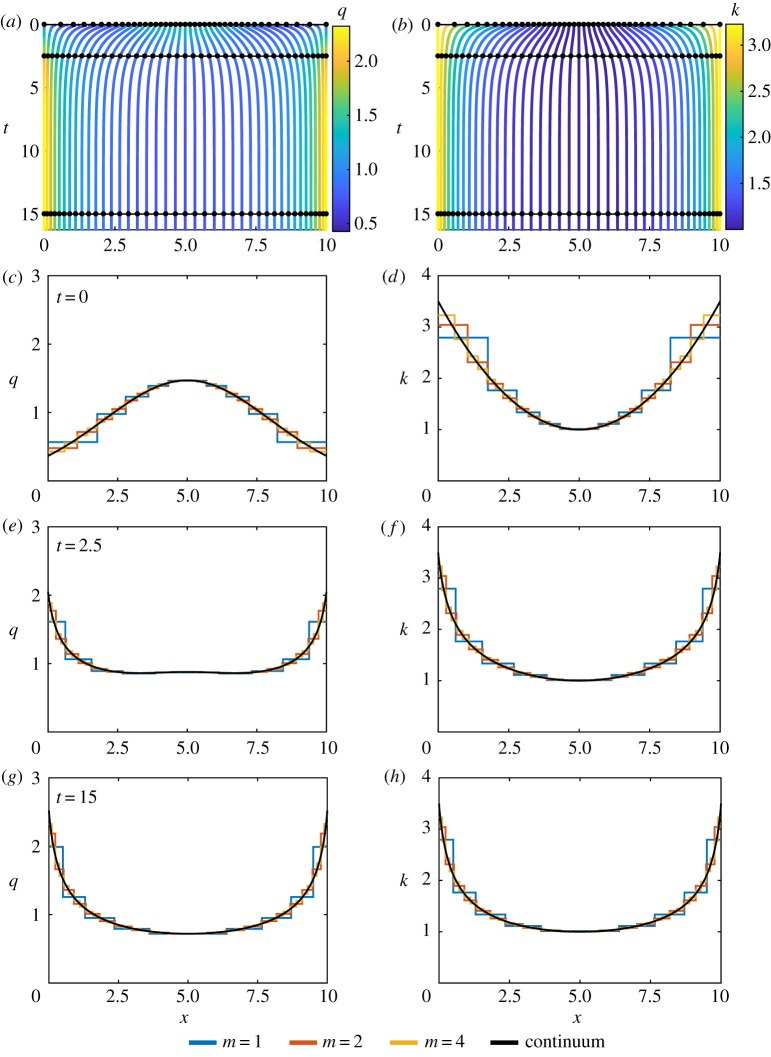
Figure 3.
Results for heterogeneous k and homogeneous a with N = 10, k0(x) = 1 + 0.1(x − 5)2 and a0(x) = 0. (a,b) Characteristic diagram for spring boundary position evolution for 0 ≤ t ≤ 16.25, with m = 4 so that every fourth trajectory represents a cell boundary. Colour denotes (a) cell density, (b) cell stiffness. In (a,b), black lines and dots represent times for snapshots in (c–h). (c,e,g) Cell density snapshots at t = 0, 2.5, 15. (d,f ,h) Cell stiffness snapshots at t = 0, 2.5, 15. In (c–h) lines display results for N = 10 with m = 1, 2, 4 and continuum system. (Online version in colour.)
(ii). Piecewise constant cell population
In this section, we consider a simple scenario with two adjacent tissues, modelled by assuming sharp inhomogeneities in cellular properties. This may represent the boundary between a malignant tissue and a normal tissue. We first explore how solutions from the discrete system converge to the corresponding continuum solution as m increases, under these rapidly-varying-in-space conditions. Each tissue has homogeneous cell properties given by cell stiffnesses k1, k2 and resting cell lengths a1, a2 in the left and right tissue, respectively, with interface position s(t) (figure 1b). For initial conditions, we choose a uniform density, q0(x) = 1, cell properties k1 = 1/2, k2 = 1, a1 = a2 = 0, L = 10 and s(0) = 5, respectively. The cell stiffness discontinuity rapidly induces a sharp change in the density at s(t) followed by slower dynamics until reaching a piecewise constant steady state as t → ∞ (figure 4). Even with these sharp inhomogeneities, we again observe close agreement between solutions of the discrete and continuum models, especially for the cell stiffness, k, where it is difficult to distinguish between the discrete model with different m and the solution of the continuum model. For the cell density, q, we again see that agreement at the spring boundaries improves as we increase m. This holds especially well given that the numerical discretization of the continuum model does not explicitly follow the location of the interface, see electronic supplementary material, §S2. It could however be determined by evaluating the velocity, ds(t)/dt = u, at the interface position.
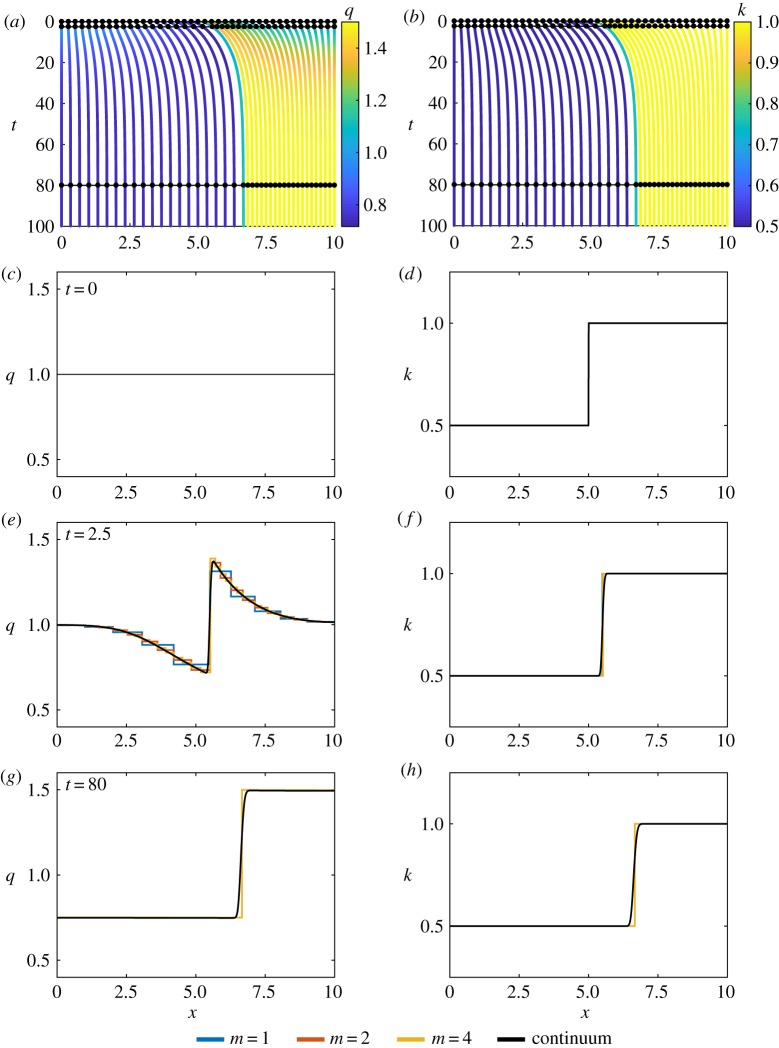
Figure 4.
Results for piecewise constant cell properties, with N = 10. (a,b) Characteristic diagram for spring boundary position evolution for 0 ≤ t ≤ 100, with m = 4 so that every fourth trajectory represents a cell boundary. Colour denotes (a) cell density, (b) cell stiffness. In (a,b), black lines and dots represent times for snapshots in (c–h). (c,e,g) Cell density snapshots at t = 0, 2.5, 80. (d,f ,h) Cell stiffness snapshots at t = 0, 2.5, 80. In (c–h) lines display results from N = 10 with m = 1, 2, 4 and continuum system. (Online version in colour.)
This simple mechanical relaxation scenario between two tissues enables us to infer some information on the cellular-level properties ki and ai by considering the evolution of the interface position, s(t). The steady-state interface position, , is given by
| 3.3 |
which depends on k1/k2, a1 and a2. Here N1 and N2 represent the total number of cells in the left and right tissues, respectively, see electronic supplementary material, §S3. We can identify and as the lengths of the left and right tissues, respectively, after their mechanical relaxation.
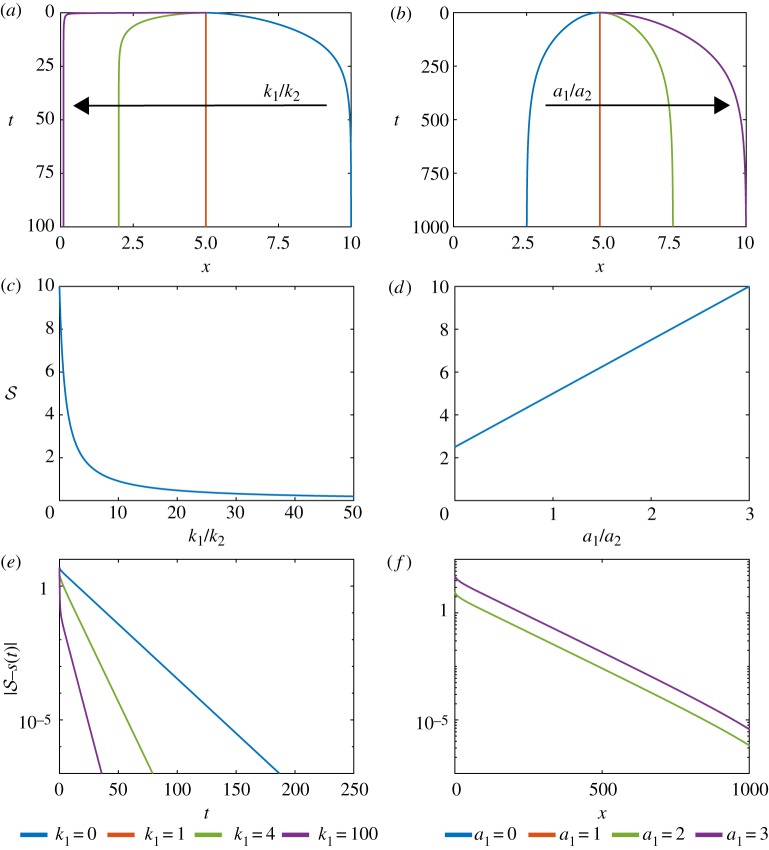
To investigate the influence of k1/k2, we vary k1 and set k2 = 1. As we have fixed boundaries at x = 0 and x = L, we set a1 = a2 = 0 to emphasize properties when we vary k1, and choose a uniform density initial condition and N1 = N2 = 5. Evaluating s(t) numerically, for efficiency with the discrete model from equation (2.3), and from equation (3.3), shows that if k1 = 0 then and the left tissue occupies the entire domain. As k1 → ∞ then the length of the left tissue decreases (figure 5a,c).
Figure 5.
Variation of relative cell stiffness, k1/k2, and relative resting cell length, a1/a2, in a model with two adjacent tissues and a constant density initial condition. (a) Characteristics of the interface position for varying k1/k2. The right tissue has fixed cell stiffness k2 = 1 while the left tissue cell stiffness is varied. (b) Characteristics of the interface particle for varying a1/a2. The right tissue has fixed resting cell length a2 = 1 while the resting cell length of the left tissue is varied. Analytical solution for the steady-state position of the interface position with given (c) relative cell stiffness and (d) relative resting cell length. (e,f ) Absolute difference between position and steady state for interface position for increasing time for varying (e) relative cell stiffness and (f ) relative resting cell length. (Online version in colour.)
Similarly, to investigate the influence of a1, a2 we consider a1/a2, vary a1 and set a2 = 1. We set k1 = k2 = 1 which only impacts the rate at which we reach the long-time solution. In contrast to varying k1/k2, steady-state results depend on the choice of a2, not just the ratio a1/a2, see equation (3.3). For example, when a1 = 0 then which corresponds to a non-zero minimum left tissue length and a maximum length for the right tissue. We also observe that is proportional to a1 (figure 5b,d).
We find that we can use the interface boundary velocities to infer cellular-level properties. Plotting on a logarithmic scale against time shows that we can determine k1/k2 from the gradient of the linear section and we can determine a1/a2 from the y-intercept (figure 5e,f ). We find that it is easier to distinguish the ratio k1/k2 than it is to distinguish the ratio a1/a2. If the second tissue was a reference material with known k2, a2 we could then determine k1, a1.
(c). Case study: breast cancer detection
Recent experiments have proposed a new method to classify breast biopsies in situations where standard histological analysis is inconclusive [6,13,14]. The method is based on determining the stiffness histogram distribution of the tissue using atomic force microscopy. Normal tissues are associated with a single, well-defined unimodal stiffness peak, whereas malignant tissues are associated with a bimodal distribution with a prominent low-stiffness peak. Using our mathematical model, we are able to gain more insight into the differences in mechanical properties of normal and malignant tissues at the cellular level, in particular, the role of the resting cell length, which is not an easy quantity to measure experimentally. It would be impossible to interpret this experimental data with previous models that deal only with homogeneous cell populations.
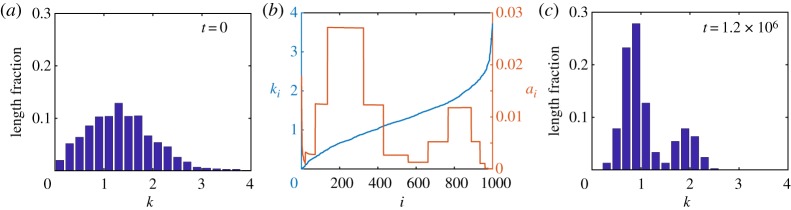
For this case study, as the experimental data are relatively discrete, we use the discrete model, which we consider to be a sufficiently simple yet insightful portrayal of the biological details. We set the initial state of the system by assuming a uniform initial density distribution and by assigning the cell stiffness of the ith cell, ki, so as to reconstruct the unimodal stiffness profile from fig. 1b (top) in [6]. To do so, we normalize the experimental stiffness histogram and interpret the normalized value as the length fraction of the tissue containing stiffness in the given histogram bin (figure 6a). This is consistent with the experimental method which implicitly assumes that the probability a cell is examined during a biopsy is proportional to its size [6]. To estimate ki, we randomly sample the unimodal stiffness distribution and arbitrarily assign them to cells i = 1, 2, …, N in ascending order. Note that the ordering of the cells does not affect our results or the interpretation of our results in any way. We assume N = 1000, m = 1 and L = 10 for illustration purposes. In order for this initial set-up to be in equilibrium despite the heterogeneity in stiffness in the tissue, the resting cell lengths ai must be chosen heterogeneously, per the steady-state system of discrete equations, see electronic supplementary material, §S4.
Figure 6.
Breast cancer detection case study. (a) Initial unimodal stiffness distribution, normalized by tissue length fraction, associated with normal tissues. (b) Initial cell stiffness ki and modified resting cell length ai for each cell i = 1, 2, …, 1000, leading to a bimodal stiffness distribution. (c) Steady-state stiffness distribution obtained with the modified resting cell lengths, exhibiting a bimodal distribution associated with malignant tissues. (Online version in colour.)
We proceed to consider how a bimodal stiffness distribution, associated with malignant tissues, could arise from such an initial state with a unimodal stiffness distribution. The simplest explanation is that a bimodal stiffness distribution may arise as a result of changes to individual cell stiffnesses, ki, [45] e.g. due to some pre-cancerous biological mechanisms. This model provides an alternative interpretation where the bimodal distribution may arise from changes not solely to the individual cell stiffnesses but to the resting cell lengths also. We now present an extreme case where the bimodal distribution may arise from changes in the resting cell lengths only. Specifically, when we simulate the discrete model with the initial conditions as above, but modify the heterogeneity in the resting cell lengths, ai, to a bimodal profile with high ai for very low ki, without changing their stiffnesses, ki, the cells redistribute themselves in the tissue in such a way that the tissue stiffness histogram develops a bimodal distribution at mechanical equilibrium (figure 6c). We note that this result is not surprising due to the coupling of cell stiffness and resting cell length in the mathematical model. However, this intuitive result may not have been clear had we relied upon experimental data and experimental observations alone. In addition, this approach assumes that cells may have very different lengths which is consistent with biological observations. Specifically, it is understood that breast cancer cells are less stiff and, in general, have a larger diameter in comparison to normal breast cells [46]. This is consistent with other areas of biology, for example, in the context of melanoma biology it is well accepted that cancer cells can be smaller than healthy cells [47,48]. We also note here that changes in the resting cell lengths have been assumed in other works [49] to model two-way feedback between mechanical tensions and signalling and here could similarly represent some unknown underlying pre-cancerous biological mechanisms.
4. Conclusion and future work
In this work, we present a one-dimensional cell-based model with heterogeneous cell properties, and its coarse-grained continuum approximation. The motion of cells is driven by cell–cell interaction forces which could represent cell–cell adhesion [41] or compressive stresses [42]. Heterogeneous cell properties, cell stiffness and resting cell length, are constant for each cell and are transported in space by cell movements. The continuum limit is taken by increasing the number of springs per cell, while maintaining the number of cells in the tissue and its fixed total length, and by considering spatial averages over length scales involving a large enough number of cells to define continuous densities but small enough to retain spatial heterogeneities.
Our results shows that solutions of the discrete model approach the solution of the continuum model as the number of springs per cell increases while the spring length tends to zero, even for rapidly varying spatial cell properties. Excellent agreement is observed even for few springs per cell at the centre of each cell. For the examples presented in this work, we find that the solution of the discrete model can be obtained much faster than the solution of the continuum model. However, the time required to simulate the discrete model increases rapidly with the number of cells. By contrast, the time required to simulate the continuum model is independent of the number of cells. Therefore, when we have large numbers of cells, as in an epithelial tissue, the continuum model is advantageous. Another advantage of the continuum model is that we can quickly develop exact closed form expressions for the long-time interface position which are more difficult to establish with the discrete model. Furthermore, the continuum model allows us to understand macroscale phenomena which are not obvious from microscopic interactions. The fact that the cell density flux in the continuum model, a macroscopic quantity, is explicitly related to the gradient of the cell–cell interaction force may have been anticipated, but it is not obvious from the microscopic interactions that this leads to an effective nonlinear diffusive transport. Finally, because the continuum model exhibits explicit relationships between macroscopic quantities, it will be more useful for inverse problems.
By dealing explicitly with heterogeneous cell populations, this model has many potential applications. The first application we consider is a simple tissue relaxation simulation, where we track the position of the interface between two distinct adjacent tissues as the system mechanically relaxes, to infer cellular-level properties. Results suggest it is easier to determine the relative cell stiffnesses than it is to determine the relative resting cell lengths. Results also show that when cells are, on average, in tension a tissue with lower stiffness extends and compresses a tissue with higher stiffness. In the second application, we use the model to interpret recent experiments in breast cancer detection which reveal distinct stiffness profiles associated with normal, benign and malignant tissues [6]. We show that a bimodal stiffness distribution, associated with a malignant tissue, could arise from a unimodal stiffness distribution, associated with a normal tissue, from changes not just in cell stiffnesses but from changes in the resting cell length's only. The resting cell length is not an easily measured experimental quantity and these results suggest that this could be an important variable to consider.
Many extensions of this work are possible, both mathematically and biologically. Important extensions will be to introduce cell proliferation, apoptosis and free boundaries where the continuum limit is less obvious [34,39,40]. Another interesting extension will be to generalize the cell–cell interaction force law to include nonlinear effects for large separations [35,40]. These extensions will be the subject of future works. Finally, the model's ability to relate cellular-level stiffness data and tissue-level information has many potential extensions biologically including applying the model to particular scenarios such as epithelial tissue mechanics, cancer progression [4,5], cancer detection [6–8], wound healing [10] and morphogenesis [11].
Supplementary Material
Acknowledgements
We thank the three anonymous referees for their comments.
Data accessibility
This article does not contain any additional data. Key algorithms used to generate results are available on https://github.com/ryanmurphy42/Murphy2019.git.
Authors' contributions
All authors conceived and designed the study; R.J.M. performed numerical simulations and drafted the article; all authors provided comments and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work is supported by the Australian Research Council (DP170100474). R.E.B. is a Royal Society Wolfson Research Merit Award holder, would like to thank the Leverhulme Trust for a Research Fellowship and also acknowledges the BBSRC for funding via grant no. BB/R000816/1.
References
- 1.Trepat X, Sahai E. 2018. Mesoscale physical principles of collective cell organisation. Nat. Phys. 14, 671–682. ( 10.1038/s41567-018-0194-9) [DOI] [Google Scholar]
- 2.Altschuler SJ, Wu LF. 2010. Cellular heterogeneity: when do differences make a difference? Cell 141, 559–563. ( 10.1016/j.cell.2010.04.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallaher JA, Enriquez-Navas PM, Luddy KA, Gatenby RA, Anderson ARA. 2018. Spatial heterogeneity and evolutionary dynamics modulate time to recurrence in continuous and adaptive cancer therapies. Cancer Res. 78, 2127–2139. ( 10.1158/0008-5472.CAN-17-2649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuel MS, Koch S. 2011. Actomyosin-mediated cellular tension drives increased tissue stiffness and β-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell 19, 776–791. ( 10.1016/j.ccr.2011.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen AV. et al. 2016. Stiffness of pancreatic cancer cells is associated with increased invasive potential. Integr. Biol. 8, 1232–1245. ( 10.1039/C6IB00135A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plodinec M. et al. 2012. The nanomechanical signature of breast cancer. Nat. Nanotechnol. 7, 757–764. ( 10.1038/nnano.2012.167) [DOI] [PubMed] [Google Scholar]
- 7.Huo CW, Chew GL, Britt KL, Ingman WV, Henderson MA, Hopper JL, Thompson EW. 2014. Mammographic density—a review on the current understanding of its association with breast cancer. Breast Cancer Res. Treat. 144, 479–502. ( 10.1007/s10549-014-2901-2) [DOI] [PubMed] [Google Scholar]
- 8.Suresh S. 2007. Elastic clues in cancer detection. Nat. Nanotechnol. 2, 748–749. ( 10.1038/nnano.2007.397) [DOI] [PubMed] [Google Scholar]
- 9.Alibert C, Goud B, Manneville J-B. 2017. Are cancer cells really softer than normal cells? Biol. Cell 109, 167–189. ( 10.1111/boc.201600078) [DOI] [PubMed] [Google Scholar]
- 10.Evans ND, Oreffo ROC, Healy E, Thurner PJ, Man YH. 2013. Epithelial mechanobiology, skin wound healing, and the stem cell niche. J. Mech. Behav. Biomed. 28, 397–409. ( 10.1016/j.jmbbm.2013.04.023) [DOI] [PubMed] [Google Scholar]
- 11.Fletcher AG, Cooper F, Baker RE. 2017. Mechanocellular models of epithelial morphogenesis. Phil. Trans. R. Soc. B 372, 20150519 ( 10.1098/rstb.2015.0519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levental I, Georges PC, Janmey PA. 2006. Soft biological materials and their impact on cell function. Soft Matter 3, 299–306. ( 10.1039/B610522J) [DOI] [PubMed] [Google Scholar]
- 13.Lekka M. et al. 2012. Cancer cell detection in tissue sections using AFM. Arch. Biochem. Biophys. 518, 151–156. ( 10.1016/j.abb.2011.12.013) [DOI] [PubMed] [Google Scholar]
- 14.Lekka M. 2012. Atomic force microscopy: a tip for diagnosing cancer. Nat. Nanotechnol. 7, 691–692. ( 10.1038/nnano.2012.196) [DOI] [PubMed] [Google Scholar]
- 15.Lekka M. 2016. Discrimination between normal and cancerous cells using AFM. BioNanoScience 6, 65–80. ( 10.1007/s12668-016-0191-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross SE, Jin Y, Rao J, Gimzewski JK. 2007. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2, 780–783. ( 10.1038/nnano.2007.388) [DOI] [PubMed] [Google Scholar]
- 17.Pathmanathan P, Cooper J, Fletcher A, Mirams G, Murray P, Osborne J, Pitt-Francis J, Walter A, Chapman SJ. 2009. A computational study of discrete mechanical tissue models. Phys. Biol. 6, 036001 ( 10.1088/1478-3975/6/3/036001) [DOI] [PubMed] [Google Scholar]
- 18.Osborne JM, Fletcher AG, Pitt-Francis JM, Maini PK, Gavaghan DJ. 2017. Comparing individual-based approaches to modelling the self-organization of multicellular tissues. PLoS Comput. Biol. 13, e1005387 ( 10.1371/journal.pcbi.1005387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandersius SA, Weijer CJ, Newman TJ. 2011. Emergent cell and tissue dynamics from subcellular modeling of active biomechanical processes. Phys. Biol. 8, 045007 ( 10.1088/1478-3975/8/4/045007) [DOI] [PubMed] [Google Scholar]
- 20.Ingber DE. 2003. Tensegrity I. Cell structure and hierarchical systems biology. J. Cell Sci. 116, 1157–1173. ( 10.1242/jcs.00359) [DOI] [PubMed] [Google Scholar]
- 21.Armstrong NJ, Painter KJ, Sherratt JA. 2006. A continuum approach to modelling cell-cell adhesion. J. Theor. Biol. 243, 98–113. ( 10.1016/j.jtbi.2006.05.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collis J, Brown DL, Hubbard ME, O'Dea RD. 2017. Effective equations governing an active poroelastic medium. Proc. R. Soc. A 473, 20160755 ( 10.1098/rspa.2016.0755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penta R, Ambrosi D, Shipley RJ. 2013. Effective governing equations for poroelastic growing media. Q. J. Mech. Appl. Math. 67, 69–91. ( 10.1093/qjmam/hbt024) [DOI] [Google Scholar]
- 24.Osborne JM. et al. 2010. A hybrid approach to multi-scale modelling of cancer. Phil. Trans. R. Soc. A 368, 5013–5028. ( 10.1098/rsta.2010.0173) [DOI] [PubMed] [Google Scholar]
- 25.Anderson ARA. 2005. A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion. Math. Med. Biol. 22, 163–186. ( 10.1093/imammb/dqi005) [DOI] [PubMed] [Google Scholar]
- 26.Vavourakis V, Wijeratne PA, Shipley R, Loizidou M, Stylianopoulos T, Hawkes DJ. 2017. A validated multiscale in-silico model for mechano-sensitive tumour angiogenesis and growth. PLoS Comput. Biol. 13, e1005259 ( 10.1371/journal.pcbi.1005259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajagopal V, Holmes WR, Lee PVS. 2018. Computational modeling of single-cell mechanics an cytoskeletal mechanobiology. Wires. Syst. Biol. Med. 10, 5–7. ( 10.1002/wsbm.1407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tambe DT. et al. 2011. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10, 469–475. ( 10.1038/nmat3025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fozard JA, Byrne HM, Jensen OE, King JR. 2010. Continuum approximations of individual-based models for epithelial monolayers. Math. Med. Biol. 27, 39–74. ( 10.1093/imammb/dqp015) [DOI] [PubMed] [Google Scholar]
- 30.O'Dea RD, King JR. 2012. Continuum limits of pattern formation in hexagonal-cell monolayers. J. Math. Biol. 64, 579–610. ( 10.1007/s00285-011-0427-3) [DOI] [PubMed] [Google Scholar]
- 31.Middleton AM, Fleck C, Grima R. 2014. A continuum approximation to an off-lattice individual-cell based model of cell migration and adhesion. J. Theor. Biol. 359, 220–232. ( 10.1016/j.jtbi.2014.06.011) [DOI] [PubMed] [Google Scholar]
- 32.Matsiaka OM, Penington CJ, Baker RE, Simpson MJ. 2018. Discrete and continuum approximations for collective cell migration in a scratch assay with cell size dynamics. Bull. Math. Biol. 80, 738–757. ( 10.1007/s11538-018-0398-2) [DOI] [PubMed] [Google Scholar]
- 33.Bodnar M, Velazquez JJL. 2005. Derivation of macroscopic equations for individual cell-based models: a formal approach. Math. Method Appl. Sci. 28, 1757–1779. ( 10.1002/mma.638) [DOI] [Google Scholar]
- 34.Murray PJ, Edwards CM, Tindall MJ, Maini PK. 2009. From a discrete to a continuum model of cell dynamics in one dimension. Phys. Rev. E 80, 031912 ( 10.1103/PhysRevE.80.031912) [DOI] [PubMed] [Google Scholar]
- 35.Murray PJ, Edwards CM, Tindall MJ, Maini PK. 2012. Classifying general nonlinear force laws in cell-based models via the continuum limit. Phys. Rev. E 85, 021921 ( 10.1103/PhysRevE.85.021921) [DOI] [PubMed] [Google Scholar]
- 36.Murray PJ, Kang J, Mirams GR, Shin S-Y, Byrne HM, Maini PK, Cho KH. 2010. Modelling spatially regulated β-catenin dynamics and invasion in intestinal crypts. Biophys. J. 99, 716–725. ( 10.1016/j.bpj.2010.05.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray PJ, Walter A, Fletcher AG, Edwards CM, Tindall MJ, Maini PK. 2011. Comparing a discrete and continuum model of the intestinal crypt. Phys. Biol. 8, 026011 ( 10.1088/1478-3975/8/2/026011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans DJ, Morriss G. 2008. Statistical mechanics of nonequilibrium liquids. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 39.Lorenzi T, Murray PJ, Ptashnyk M. 2019. From individual-based mechanical models of multicellular systems to free-boundary problems. arxiv Preprint.
- 40.Baker RE, Parker A, Simpson MJ. 2018. A free boundary model of epithelial dynamics. J. Theor. Biol. ( 10.1016/j.jtbi.2018.12.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston ST, Simpson MJ, Plank MJ. 2013. Lattice-free descriptions of collective motion with crowding and adhesion. Phys. Rev. E 88, 062720 ( 10.1103/PhysRevE.88.062720) [DOI] [PubMed] [Google Scholar]
- 42.Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Minn LL. 2011. Mechanical compression drives cancer cells toward invasive phenotype. Proc. Natl Acad. Sci. USA 109, 911–916. ( 10.1073/pnas.1118910109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meineke FA, Potten CS, Loeffler M. 2001. Cell migration and organisation in the intestinal crypt using a lattice-free model. Cell Prolif. 34, 253–266. ( 10.1046/j.0960-7722.2001.00216.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lighthill MJ. 1958. An introduction to Fourier analysis and generalised functions. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 45.Guck J. et al. 2005. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 88, 689–3698. ( 10.1529/biophysj.104.045476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.TruongVo TN. et al. 2017. Microfluidic channel for characterizing normal and breast cancer cells. J. Micromech. Microeng. 27, 035017 ( 10.1088/1361-6439/aa5bbb) [DOI] [Google Scholar]
- 47.Haridas P, Penington CJ, McGovern JA, McElwain DLS. 2017. Quantifying rates of cell migration and cell proliferation in co-culture barrier assays reveals how skin and melanoma cells interact during melanoma spreading and invasion. J. Theor. Biol. 423, 13–25. ( 10.1016/j.jtbi.2017.04.017) [DOI] [PubMed] [Google Scholar]
- 48.Haridas P, McGovern JA, McElwain DLS. 2017. Quantitative comparison of the spreading and invasion of radial growth phase and metastatic melanoma cells in an three-dimensional human skin equivalent model. Peer J 5, e3754 ( 10.7717/peerj.3754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zmurchok C, Bhaskar D, Edelstein-Keshet L. 2018. Coupling mechanical tension and GTPase signaling to generate cell and tissue dynamics. Phys. Biol. 15, 046004 ( 10.1088/1478-3975/aab1c0) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article does not contain any additional data. Key algorithms used to generate results are available on https://github.com/ryanmurphy42/Murphy2019.git.