Abstract
Lysosomes are acidic, membrane-bound organelles that serve as the primary catabolic compartment of the cell. They are crucial to a variety of cellular processes from nutrient storage to autophagy. Given the diversity of lysosomal functions, it is unsurprising that lysosomes are also emerging as important players in aging. Lysosomal dysfunction is implicated in several aging-related neurodegenerative diseases including Alzheimer’s, Parkinson’s, amyotrophic lateral sclerosis/frontotemporal dementia, and Huntington’s. Although the precise role of lysosomes in the aging brain is not well-elucidated, some insight into their function has been gained from our understanding of the pathophysiology of age-dependent neurodegenerative diseases. Therapeutic strategies targeting lysosomes and autophagic machinery have already been tested in several of these diseases with promising results, suggesting that improving lysosomal function could be similarly beneficial in preserving function in the aging brain.
Electronic supplementary material
The online version of this article (10.1007/s13311-019-00742-3) contains supplementary material, which is available to authorized users.
Key Words: Lysosome, aging, neurodegeneration, autophagy, therapeutic targets
Introduction
Lysosomes are dynamic organelles that function as a major catabolic center in the cell. They participate in cellular homeostasis by serving as the terminal degradation compartment in autophagy, modulating nutrient status and interacting with other organelles [1]. There is growing evidence for the role of lysosomes in aging. Some studies suggest that processes such as lysosomal acidification and autophagy decrease with age, potentially altering nutrient storage and clearance of damaged proteins and organelles [2]. The importance of lysosomes in the aging brain is also supported by the many neurodegenerative diseases presenting with lysosomal dysfunction [3]. Thus, improved knowledge of the pathophysiology of these diseases as well as identification of therapeutic targets will strengthen our understanding of and ability to preserve lysosomal function in the aging brain.
Lysosomal Functions Related to Aging
Lysosomes participate in several cellular functions including nutrient processing, autophagy, and inter-organelle contacts. There is emerging evidence that these functions may be perturbed in age-related disease processes, including those affecting the central nervous system, as well as in physiological aging.
Nutrient Storage and Sensing
Lysosomes play a critical role in nutrient homeostasis, serving as both centers for nutrient processing via a host of degradative enzymes and sites for storage of metabolites including amino acids, lipids, and ions. The transport of metabolites into the lysosome depends on transmembrane transporters and channels, whose activity is coupled to a proton gradient established by the highly conserved H+-ATPase or V-ATPase [4, 5]. V-ATPase pumps protons into the vacuolar lumen, and the subsequent extrusion of these protons drives ions and metabolites inward [6, 7].
Amino Acids
Lysosomes sense nutrient availability through the activities of V-ATPase and the conserved serine–threonine kinase, mTORC1, and both are sensitive to changes in amino acid levels [2, 8]. The activity of the V-ATPase is regulated through association–dissociation of its many subcomplexes [9]. In states of low amino acids, the V-ATPase becomes fully associated, generating a proton gradient to promote the influx of substrates into the lysosome [10]. The nutrient sensing function of the V-ATPase is additionally linked to the mTORC1 pathway via the V-ATPase, Rag-GTPases, and Ragulator complex. During states of nutrient abundance, mTORC1 associates with lysosomes to promote cellular growth and limit autophagy [11, 12]. The relationship between mTORC1 and lysosomal amino acid sensing is further evidenced by the lysosomal arginine transporter, SLC38A9, which communicates amino acid status to mTORC1 [13, 14]. This transporter mediates efflux of nonpolar amino acids from the lysosome to be utilized in processes such as protein synthesis [15]. A recent lysosomal metabolomics study identified both V-ATPase and mTORC1 as important mediators of amino acid efflux from the lysosome [16].
Evidence highlighting the importance of the V-ATPase and the mTORC1 pathways in aging is also growing. Acidification by V-ATPase or overexpression of V-ATPase components promoted long-term mitochondrial stability [17] and extended lifespan in yeast, respectively [18]. Similarly, mTORC1 inhibition was shown to increase lifespan in a range of organisms including yeast, flies, and mice [2].
Calcium
Lysosomes, along with the endoplasmic reticulum (ER), serve as main cellular reservoirs for Ca2+, an ion implicated in several signaling pathways and organelle homeostasis [19]. Although the mammalian lysosomal Ca2+ importer is unknown, various efflux channels have been characterized including transient receptor potential channel mucolipin-1 (TRPML1) and two-pore channels 1 and 2 (TPC1/2) [20]. Importantly, Ca2+ level has been correlated with lifespan as elevation of either intracellular or extracellular Ca2+ shortens replicative lifespan in yeast []. Moreover, loss of lysosomal function results in a rapid increase in cellular Ca2+ in yeast [22], and activation of the TRPML1 channel facilitates Ca2+-induced Ca2+ release from the ER, further increasing intracellular Ca2+ [23, 24]. TRPML1 has also been linked to V-ATPase function as blockage of V-ATPase or knockdown of its V0a1 subunits results in an increase in TRPML1-mediated Ca2+ efflux from lysosomes, suggesting a close interplay of the 2 proteins in Ca2+ homeostasis [25]. Regulation of cellular Ca2+ also has important implications for overall cell survival as Ca2+ overload and subsequent accumulation in the mitochondria can induce mitochondrial permeability transition pore opening, promoting the release of pro-apoptotic factors [26].
In the central nervous system, Ca2+ plays an important role in modulating electrical activity and neurotransmitter release [27]. Aged neurons show a variety of alterations in Ca2+ homeostasis including diminished efflux from the plasma membrane [28], decreased mitochondrial buffering capacity [29], and enhanced release from the ER [30, 31]. In the aging brain, this results in a net increase in cellular Ca2+, which perturbs neuronal excitability and long-term processes such as synaptic plasticity [27]. Although the function of lysosome-dependent Ca2+ homeostasis in the aging brain remains poorly understood, the ability of lysosomes to regulate cellular Ca2+ in non-neuronal cells suggests the possibility of a similar role for lysosomes in neurons.
Iron
Another key ion regulated by lysosomes is Fe2+ which enters the cell via the endolysosomal system [32, 33]. Tight regulation of cellular Fe2+ is crucial as it is an important cofactor for enzymes in the electron transport chain [34]; however, high levels may promote the formation of toxic reactive oxygen species (ROS) [35]. Of note, Fe2+ levels have been reported to be increased with age, thereby promoting mitochondrial protein aggregation and decreased lifespan in Caenorhabditis elegans [36].
Fe2+ is also important for central nervous system function via its involvement in myelin and neurotransmitter synthesis [33]. Fe2+ levels in the brain increase with age due to increased blood–brain barrier permeability, alterations in Fe2+ homeostasis, and neuroinflammation [37]. Moreover, changes to brain Fe2+ distribution in areas like the substantia nigra and basal ganglia occur with age [38]. In non-nervous tissues, free Fe2+ remains low via binding to ferritin and transferrin, whereas in catecholaminergic neurons, free Fe2+ is regulated through the formation of neuromelanin–iron complexes, which have been found to increase with age [39, 40]. A connection between lysosomes, Fe2+, and neuromelanin was recently highlighted with the characterization of neuromelanin-containing organelles as specialized autolysosomes that accumulate undegraded proteins and lipids in substantia nigra neurons [41]. The importance of iron homeostasis in the brain is also supported by a group of inherited disorders called neurodegeneration with brain iron accumulation (NBIA). Several NBIA genes are related to lysosomal function [42], and it was recently reported that iron overload in NBIA mutant cells resulted in both lysosomal and mitochondrial dysfunction [43].
Lipids and Glycoproteins
The role of lysosomes in glycoprotein and lipid homeostasis is evidenced by the plethora of lysosomal storage disorders (LSDs) that result from complete loss or decreased activity of specific lysosomal hydrolases. In LSDs, undigested lipids, glycoproteins, and mucopolysaccharides accumulate in the lysosome, leading to cellular toxicity and compromised survival [44]. A comprehensive description of the LSDs can be found in previously published reviews [45, 46]. Importantly, lysosomal dysfunction due to storage of accumulated substrates in LSDs results in decreased lysosomal degradation and various pathologies in the CNS that resemble adult-onset neurodegenerative diseases—for example, Parkinson’s disease (PD)-like Lewy body accumulation in Gaucher disease and Alzheimer’s disease (AD)-like plaques and tangles in Niemann–Pick disease type C [46]. This suggests that alterations in lysosomal degradative capacity may contribute to age-related neuropathologies.
Autophagy
Autophagy is a critical physiological process that maintains intracellular homeostasis by degrading and recycling cytoplasmic material, including damaged organelles, long-lived proteins, and protein aggregates, via delivery to the lysosome. Though the process was previously thought to occur nonselectively, studies have shown that autophagy can be stimulated under stress conditions and by cellular insults. Additionally, dysfunctional organelles like the mitochondria may also undergo degradation by the lysosome via selective autophagy.
Mechanism of Autophagy
Macroautophagy, hereafter referred to as autophagy, is 1 of 3 modes of autophagy alongside microautophagy and chaperone-mediated autophagy. It is the primary mode for degrading large parts of cytoplasm through sequestration inside double-membrane vesicles, termed autophagosomes, which then fuse with lysosomes [47]. Several highly conserved AuTophaGy-related (Atg) genes have been identified as directly involved in autophagosome formation, maturation, and fusion with the lysosome.
Autophagosome formation is initiated by the phagophore, an open double-membrane structure that forms around proteins and organelles in the cytoplasm [47]. The membrane is derived from proximal structures including the ER, endosomes, mitochondria, Golgi, and plasma membrane [48, 49]. Unc-51-like kinase 1 (ULK1) is a critical protein in phagophore induction through its interaction with autophagy proteins including mAtg13 and the scaffolding protein FIP200 [50]. Other players in autophagosome formation include a transient interaction with mAtg9 [51], which may provide additional membrane sources to the growing phagophore.
In humans, the class III phosphatidylinositol 3-kinase, vesicular protein sorting 34 (hVps34), and its complex 1 binding partners—Beclin 1, p150, and Atg14L—are also necessary for phagophore formation [52–55]. hVps34 complex 1 interactions stimulate hVps34 activity, promoting phosphatidylinositol to phosphatidylinositol 3-phosphate conversion, which facilitates recruitment of other Atgs to the forming autophagosome [52, 55]. hVps34 also has a set of complex 2 interactors, in which the conserved hVps34–Beclin 1 complex backbone interacts with ultraviolet irradiation resistant-associated gene (UVRAG) [56, 57]. UVRAG association with complex 2 modulates several functions including membrane bending, negative and positive regulation of autophagy, and promotion of autophagosome fusion with endosomes/lysosomes [58].
Elongation of the phagophore membrane involves 2 ubiquitin-dependent conjugation systems. In the first, Atg7 activates Atg12, which is transferred to Atg10 to stimulate Atg12–Atg5 covalent linkage [59–61]. The Atg12–Atg5 complex then interacts with Atg16L to form the Atg16L complex, which drives phagophore elongation [60, 61]. The second system involves the microtubule-associated protein light chain 3 (LC3) [62]. Upon autophagy induction, LC3 is cleaved to cytosolic LC3-I which is then conjugated to phosphatidylethanolamine by Atg7 and Atg3 to generate lipidated LC3-II [59, 60]. LC3-II associates with both sides of the phagophore membrane to mediate cargo selection through its recruitment of the autophagy receptor, p62, and components involved in autophagosome–lysosome fusion [63–67]. The final step involves fusion of the outer membrane of the autophagosome with the lysosome. Various SNAREs and tethering/adaptor proteins on late endosomes/lysosomes and autophagosomes have been identified and have been discussed in previous reviews [68].
Regulation of Autophagy
Studies have revealed a close interplay between lysosomes and mTORC1 in the regulation of autophagy. Under a nutrient-rich environment, mTORC1 is recruited to the lysosomal membrane via a complex consisting of the membrane-tethered Ragulator complex and Rag GTPases [12, 69], where it is activated by Ras homolog enriched in brain (Rheb) [70]. Activation of mTORC1 along with suppression of its inhibition by tuberous sclerosis tumor suppressor complex (TSC1–TSC2) in nutrient-replete conditions inhibits autophagy [71]. Autophagy initiation and lysosome biogenesis are also controlled at a transcriptional level through the transcription factor EB (TFEB). Nutrient deprivation induces nuclear translocation of TFEB to enhance expression of autophagosome-related genes [72, 73]. However, under basal conditions, activated mTORC1 phosphorylates TFEB, resulting in cytoplasmic retention of the transcription factor and inhibition of autophagy [74, 75].
mTORC1-independent mechanisms of autophagy induction have also been identified, including inhibition of inositol-1,4,5-trisphosphate (IP3) levels and attenuation of Ca2+-related pathways [76, 77]. IP3 is released from the plasma membrane upon phospholipase C processing of phosphatidylinositol 4,5-bisphosphate (PIP2) [78]. High levels of IP3 can inhibit cellular clearance [79, 80] likely by stimulating Ca2+ release through IP3 receptors and disrupting Ca2+ gradients needed for autophagosome–lysosome fusion. Elevated cytosolic Ca2+ also regulates autophagy via activation of the Ca2+-dependent cysteine protease, calpain. A negative correlation likely exists between calpain activity and autophagy initiation as calpain inhibition has been shown to promote autophagosome formation [77, 81, 82]. In addition, calpain activity could increase IP3 levels via G-protein signaling downstream of GPCRs [77]. Lastly, inhibition of TFEB through phosphorylation by the serine–threonine kinase, Akt, can block autophagy independent of mTORC1 [83]. Conversely, TFEB dephosphorylation by the Ca2+-dependent phosphatase, calcineurin, enables nuclear translocation, thereby inducing autophagy [84]. Notably, it was also shown that calcineurin is activated by local Ca2+ release from lysosomes [84].
Mitophagy
Mitophagy is a selective form of autophagy in which damaged mitochondria are eliminated, and serves as a quality control mechanism to preserve cellular homeostasis [85–87]. The best characterized pathway for mitophagy involves the Parkinson’s disease-linked proteins, PTEN-induced kinase 1 (PINK1) and Parkin [85–87]. When mitochondria are damaged, PINK1 stabilizes on the outer mitochondrial membrane to promote Parkin translocation and E3 ubiquitin ligase activity, leading to the incorporation of ubiquitinated cargo into autophagosomes [88]. In addition, PINK1 and Parkin are involved in a mechanism of mitochondrial clearance that delivers mitochondrial-derived vesicles (MDVs) enriched in oxidized proteins to lysosomes for degradation [89].
Additional pathways of mitophagy have also been identified. Transmembrane receptor-mediated mitophagy, in which LC3-interacting regions mediate recognition of damaged mitochondria, have been described for several outer mitochondrial membrane proteins including Nix [90] and FUN14 Domain containing 1 [91, 92], which were observed to be induced to drive mitophagy in response to hypoxia. Moreover, cardiolipin-mediated mitophagy, where exposure of the inner mitochondrial membrane phospholipid, cardiolipin, on the mitochondrial surface allows its interaction with LC3, has been shown to activate mitophagy independent of PINK1/Parkin [93]. Finally, other stimuli, such as iron chelators [94] and increased mitochondrial fission [95, 96], also induce mitophagy in a PINK1/Parkin-independent manner.
Although several mechanisms of mitophagy, such as the PINK1/Parkin and cardiolipin-mediated pathways, have been observed in neurons, whether additional neuronal mitophagy pathways exist and how different pathways interact and are regulated is still incompletely understood. Nevertheless, the elimination of damaged mitochondria is critical for neurons, which cannot alleviate cellular stress by division and require functional mitochondria for energy and Ca2+ homeostasis. The importance of mitophagy in neurons is also demonstrated by the various mutations in mitophagy-related genes that are causal or linked to neurodegenerative diseases such as PD and amyotrophic lateral sclerosis–frontotemporal dementia (ALS–FTD) [97] as well as by evidence suggesting that decreased lysosomal acidification impairs mitophagy in cellular models of AD [98]. Thus, understanding how mitophagy is dysregulated in neurodegeneration may further elucidate how lysosomal function in neurons is altered in age-related pathologies.
Interactions with Mitochondria
The regulation of nutrient homeostasis by lysosomes is intimately associated with mitochondrial function, and the modulation of mitochondrial function by lysosomes has various connections to aging. For example, vacuolar acidity is decreased with age in yeast, resulting in reduced lifespan and impaired mitochondrial function [17]. Similarly, lifespan and mitochondrial function are compromised in cells lacking certain subunits of the V-ATPase, leading to altered mitochondrial morphology, protein import, and potential [17, 99, 100].
Recent studies have elucidated the physical interaction between lysosomes and mitochondria. A tethering complex, vCLAMP, was identified between the yeast vacuole and mitochondria and was hypothesized to regulate lipid and metabolite transport between the 2 compartments [101, 102]. Importantly, mitochondria–lysosome contact sites were recently identified in mammalian cells and were found to be regulated by GTP hydrolysis of the lysosomal GTPase, Rab7 [103]. These contact sites allow for bidirectional regulation of mitochondrial and lysosomal dynamics, whereby 1) mitochondria regulate lysosomal dynamics by modulating the GTP-binding status of Rab7 via the mitochondrial-localized Rab7-GAP (TBC1D15), and conversely 2) lysosomes regulate mitochondrial dynamics by marking the majority of mitochondrial fission sites [103]. Mitochondria–lysosome contacts may have additional functions including the transfer of metabolites such as lipids or ions [103, 104].
Lysosomes may also affect mitochondrial function indirectly through the modulation of amino acid homeostasis. High levels of cytosolic basic amino acids have been shown to induce cellular toxicity, impair mitochondrial function, and increase ROS generation [105]. Similarly, increased levels of nonpolar, unbranched amino acids in the cytosol can accumulate in the mitochondria and overload mitochondrial metabolic pathways [106, 107].
A hallmark feature of the aging brain is reduction in mitochondrial function with a concurrent increase in ROS and oxidative damage. The ability of ROS to modulate lysosomal function is mediated in part by the lysosomal TRPML1 channel, which acts as a ROS sensor. When cellular ROS is increased, activated TRPML1 releases Ca2+ into the cytosol, inducing both TFEB nuclear translocation for lysosome biogenesis and autophagy [108]. Because both TRPML1 activation and mitochondrial ROS production have been shown to increase in response to inhibition of V-ATPase, lysosomal de-acidification may serve as an upstream pathophysiological event modulating lysosomal Ca2+ release and mitochondrial function [109]. Further evidence linking ROS to lysosomal dysfunction include studies showing that loss of mitochondrial function in brain tissue following deletion of mitochondrial proteins or inhibition of oxidative phosphorylation is sufficient to impair lysosomal function in a ROS-dependent manner [110]. Although there is still debate as to whether ROS is a cause or consequence of aging, it is likely that it contributes to both; initial increases in ROS due to age further exacerbate aging by enhancing lysosomal dysfunction.
Lysosomal Function in Neurodegenerative Disease
Several studies have revealed the importance of lysosome-related functions, such as autophagy, in neuronal homeostasis. Mouse models with neural cell-specific knockout of Atg5 or Atg7, leading to impaired autophagosome-lysosome clearance, exhibited neurodegeneration with motor deficits and shorter lifespan [111, 112].
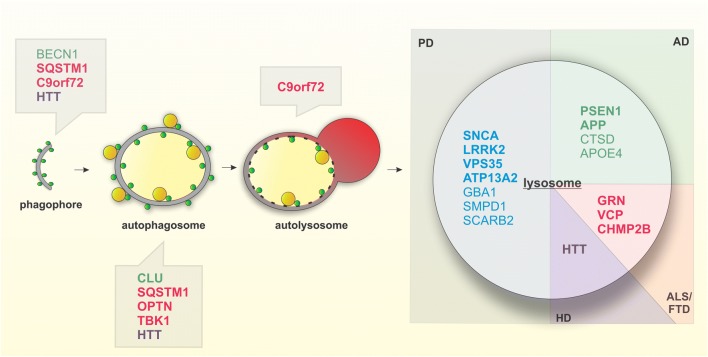
Conversely, induction of autophagy has been linked to longevity and resistance to protein aggregation and oxidative stress in neurons [71, 113]. Selective activation of lysosomal activity via TFEB was also found to clear protein aggregates and increase activation of quiescent neural stem cells during aging [114]. Although the role of lysosomes in the aging brain is still being elucidated, our understanding of age-dependent neurodegenerative diseases, many of which implicate lysosome-related genes, has provided insight into the changes in lysosomal function with age (Fig. 1).
Fig. 1.
Genes implicated in neurodegenerative disease related to autophagic and lysosomal function. Genes involved in several neurodegenerative diseases affect various steps of autophagy as well as have direct impacts on lysosomal function. Bolded correspond to causative genes, unbolded correspond to risk genes. Genes implicated in PD (blue), AD (green), ALS/FTD (red), and HD (purple)
Alzheimer’s Disease
AD is the most common neurodegenerative disease affecting more than 24 million individuals over the age of 65 [115]. The major pathological hallmarks include the accumulation of extracellular amyloid-beta (Aβ) plaques and intracellular tau-induced neurofibrillary tangles (NFTs) [115]. Normally, Aβ is trafficked to the lysosome via the autophagy pathway and degraded by cathepsin D [116]. However, recent studies suggest that release of Aβ into the extracellular space may also be in part regulated through autophagy [117]. Disruptions in autophagy were first linked to AD when autophagosome/lysosome accumulation was observed in dystrophic neurites [118]. Follow-up studies were able to show the presence of Aβ sequestered in the retained autophagosomes [119], thus highlighting the importance of the autophagy pathway in AD progression.
Clusterin, encoded by CLU, is 1 of the top candidate genes for AD development and is linked to autophagy through its involvement in autophagosome biogenesis via interaction with LC3 [120, 121]. In human studies, clusterin expression positively correlates with increasing levels of Aβ40/42 in brain areas normally afflicted in AD [122]. Furthermore, clusterin interacts directly with Aβ and reduces its aggregation, thereby protecting against toxic effects [123]. Although associations between clusterin and tau are still debated, 1 study found that tau overexpression increases clusterin expression [124], which could potentially stimulate autophagy-mediated degradation of tau.
PSEN1, another causal AD gene which encodes presenilin 1, is directly linked to autophagy through an identified role in lysosomal acidification [25, 125, 126]. Presenilin 1 is responsible for regulating gamma-secretase, which processes amyloid precursor protein (APP) into Aβ [127]. Mutations affecting presenilin 1 function may disrupt lysosomal proteolysis and promote accumulation of autophagic cargo within the cell [128, 129]. In support of this, cerebrospinal fluid (CSF) from PSEN1 patients exhibited less Aβ than normal individuals [130], suggesting that Aβ retention may lead to cellular toxicity when autophagy is impaired.
Several risk genes for AD also have links to autophagy. Atg7, which mediates the formation of the Atg12–Atg5 complex during phagophore maturation [131], is involved in incorporation of Aβ into multivesicular bodies (MVBs)/late endosomes [132, 133]. Increased transport of Aβ into MVBs could affect downstream fusion with autophagosomes. Recent studies suggest that release of Aβ into the extracellular space may be in part regulated by autophagy [117]. Inhibition of autophagy through genetic ablation of Atg7 decreases Aβ plaque formation in mouse models [117]. However, complete loss of autophagy also led to the accumulation of Aβ inside neurons [117], which can potentiate neurotoxicity. Atg7 is also linked to the tau degradation as knockout mice exhibited increased phosphorylated tau [134]. Altogether, this suggests that extreme modulation of autophagy may protect against 1 AD phenotype but exacerbate another.
Associations between Bcl-2 expression and memory have been reported in AD patients [135], highlighting an important role for Bcl-2 in AD pathogenesis. Treatment with Aβ decreases Bcl-2 expression and overexpression of Bcl-2 in APP mutant mice was found to be neuroprotective [135, 136]. Bcl-2 overexpression also decreases APP processing, extracellular Aβ deposits, and intracellular NFTs [137]. As stated previously, Beclin 1 is responsible for initiating autophagy and regulating autophagosome formation [138]. Beclin 1 heterozygous knockout mice displayed disruptions in autophagy [139], suggesting that alterations in Beclin 1 levels and function can contribute to AD progression. In agreement with this, presenilin-null cellular models and postmortem analysis of AD brains reported a decrease in Beclin 1 level [140]; however, a recent large-scale quantitative study found no significant changes in Beclin 1 gene expression or protein levels across multiple stages of AD [141].
Cathepsin D, the lysosomal protein involved in Aβ processing [116], is encoded by CTSD which confers risk in AD [142, 143]. Although the mechanism of cathepsin D in AD pathology is not well understood, 1 potential mechanism is through altered functioning of lysosomal proteolysis potentially mediated by presenilin 1 mutations which ultimately result in disrupted lysosomal acidification [25, 125, 126]. Cathepsin D levels were reportedly lower in bone marrow-derived monocytes, fibroblasts, and peripheral blood lymphocytes in AD patients [144–146], whereas elevated CTSD gene expression and protein levels were observed in AD brains and neurites [141, 147]. Interestingly, 1 additional study found that cathepsin D levels were increased in postmortem CSF albeit in an inactive form of the protein [148]. This suggests that like Beclin 1, alterations in cathepsin D function can be detrimental in AD pathogenesis when compounded with disease-associated mutations like those found in PSEN1.
Another hallmark of AD is dysfunction in the endosomal–lysosomal system, such as endosomal enlargement, which is 1 of the earliest pathological features of disease [149]. APP, in which mutations cause autosomal dominant, early-onset AD, has been implicated in AD-related endosomal aberrations. It was demonstrated that the APP β C-terminal fragment directly interacts with the adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif (APPL1), which also stabilizes active Rab5 on the endosomal membrane [150]. Overactivation of Rab5 by APP β C-terminal fragment mediates downstream events including altered endosomal motility and signaling and may also contribute to AD phenotypes such as defects in long-term potentiation [149, 151–153] and cholinergic degeneration. In addition, APP and Rab5 overactivation has been linked to defective lysosomal morphology and proteolysis as well as autophagy in various cellular and mouse models of AD [151, 152, 154].
The E4 allele of apolipoprotein E (ApoE4), a major risk factor for sporadic AD, has similarly been linked to endolysosomal dysfunction including increased endocytosis of APP [155] and impaired endosomal recycling [156, 157]. ApoE4 intermediates have also been shown to disrupt lysosomal membranes in neuronal cultures, leading to leakage of hydrolases and induction of apoptosis, which is further potentiated by Aβ(1-42) [158, 159]. A recent transcriptomics analysis in ApoE4-targeted replacement mice showed significant enrichment of genes involved in endosomal–lysosomal processing, further implicating ApoE4 in endolysosomal function [160].
Parkinson’s Disease
PD is the most common movement disorder with an estimated global prevalence of ~400 out of 100,000 by the age of 65 [161]. The cardinal motor symptoms—bradykinesia, rigidity, and tremor—are primarily attributed to dopaminergic neuron deterioration. In the majority of PD cases, Lewy bodies composed of proteins, lipids, and undegraded organelles including autolysosomes and damaged mitochondria accumulate in several brain regions [162–164].
Several autosomal dominant PD genes are directly linked to lysosomal function including SNCA, leucine-rich repeat kinase 2 (LRRK2), and vacuolar protein sorting-associated protein 35 (VPS35). The first PD gene identified, SNCA, encodes for alpha-synuclein (αSyn) [165], a major constituent of Lewy bodies [166]. Phenotypically, multiplication of the SNCA gene leads to earlier onset PD, indicating that increased expression of SNCA promotes disease pathogenesis [167, 168]. αSyn overexpression impedes autophagy by prohibiting Atg9 from initiating phagophore formation [169, 170]. Moreover, high endogenous αSyn in human iPSC-derived dopaminergic neurons reduces lysosomal proteolysis by delaying the delivery of hydrolases such as cathepsin B and glucocerebrosidase (GCase) to the lysosome [171]. SNCA missense mutations, which have an increased propensity to oligomerize, are also linked to familial PD [172, 173]. These aberrant forms of αSyn are resistant to degradation and reduce the efficiency of autophagy [174, 175]. Inhibition of autophagy and lysosomal function increase αSyn release, potentially as a compensatory mechanism [176–183].
Gain-of-function mutations in LRRK2 are the most common cause of familial late-onset PD, and polymorphisms in the locus increase PD risk [184]. LRRK2 can be localized on membranes of synaptic vesicles, late endosomes, and lysosomes [185, 186], and several Rabs involved in vesicle formation, trafficking, and docking/fusion are phosphorylated by LRRK2 [186, 187]. The consequences of pathogenic LRRK2 mutations on synaptic integrity, autophagy, and lysosomal function require further elucidation. Studies have described that patient G2019S LRRK2 fibroblasts exhibited higher lysosomal abundance and protein clearance at baseline [188], and several LRRK2 mutant fibroblasts showed decreased autophagic flux following serum/amino acid deprivation [189]. Moreover, G2019S LRRK2 iPSC-derived dopaminergic neurons were more susceptible to oxidative stress and retained higher loads of autophagic vacuoles and lipid droplets [190, 191].
VPS35, causally linked to autosomal dominant PD, is part of the retromer complex and is important for trafficking of cathepsin D, Lamp2A, and Atg9a [172, 192–194], thereby regulating autophagy and endolysosomal processing. Mitochondrial quality control also requires VPS35 for both fusion/fission dynamics and trafficking of MDVs to the lysosome for degradation [89, 195].
Loss-of-function of the cytosolic E3 ubiquitin ligase, Parkin, and the mitochondrial kinase, PINK1, are linked to early-onset autosomal recessive PD [172], and both proteins are also involved in mitochondrial quality control via mitophagy and MDVs [88, 89]. Parkin deficiency has been shown to destabilize Rab7 [196], which is crucial for both mitochondrial fission and autophagosome–lysosome fusion.
DJ-1, a chaperone with antioxidant functions, has been shown to protect dopaminergic neurons against oxidative stress in PD pathogenesis [197, 198]. iPSC-derived dopaminergic neurons from DJ-1 patients show an accelerated pathological cascade beginning with increased oxidative stress which further induces dopamine oxidation [199]. Oxidized dopamine in turn inhibits the lysosomal enzyme glucosylceramidase (GCase) and is paralleled by increased levels of soluble and insoluble αSyn [199].
The juvenile-onset atypical parkinsonism, Kufor–Rakeb syndrome, is caused by autosomal recessive mutations in PARK9, which deplete P5-type ATPase 13A2 (ATP13A2) from endolysosomes [200]. Studies using patient fibroblasts, iPSC-derived neurons, and knockout mice report mitochondrial dysfunction, lysosomal acidification and macroautophagy deficits, as well as lipid deposition preceding αSyn accumulation [201–204]. Moreover, ATP13A2 is considered to be important for maintaining intracellular ion homeostasis [205–208], a function which may be critical for autophagy/endolysosomal processing efficiency.
Various PD risk factors further implicate lysosomal dysfunction in PD pathogenesis. Autosomal recessive mutations in GBA1, which encode GCase, increase the risk for PD and cause the LSD, Gaucher disease [209, 210]. Some Gaucher patients present with parkinsonism as well as dopaminergic neurodegeneration and Lewy body pathology [209]. iPSC-derived neurons from GBA1-associated PD patients show reduced lysosomal GCase activity, macroautophagy, and Ca2+ buffering [211, 212]. Reduced GCase activity is linked to elevated glucosylceramide levels, which further promote αSyn accumulation and higher-order assembly within lysosomes [211, 213, 214]. Variants of the acid sphingomyelinase-encoding SMPD1 gene are linked to the LSDs, Niemann–Pick disease types A and B, and may follow a similar mechanism to GBA1 variants [215–217]. Furthermore, the PD risk factor SCARB2 encodes the lysosomal integral membrane protein type 2 (LIMP2), which sorts GCase to the lysosome and is important for autophagy, αSyn proteostasis, and dopaminergic viability [218–220]. Additional PD risk loci linked to lysosome-related genes (e.g., GALC, CTSB, ATP6V01A) were recently revealed by a large meta-analysis of genome-wide association studies [210]. Collectively, this points to the necessity of improving lysosomal function in PD.
Amyotrophic Lateral Sclerosis and Frontotemporal Dementia
ALS is characterized by the progressive degeneration of motor neurons. Like other neurodegenerative diseases, ALS is a proteinopathy where pathological DNA-binding protein 43 (TDP-43) is found accumulated in the cytosol of affected neurons [221, 222]. Aggregated TDP-43 plays a pathogenic role in both ALS and FTD [221], indicating that both ALS and FTD belong to a spectrum of the same disease. To date, a large number of genes have been linked to familial ALS, but strikingly, many of these genes are directly linked to autophagic function, suggesting that improving autophagy may alleviate ALS-related pathologies.
Superoxide dismutase 1 (SOD1) was 1 of the first enzymes associated with ALS. Inhibition of autophagy led to aggregation of mutant SOD1 [223], indicating that dysfunctional SOD1 is largely degraded by autophagy. The autophagy receptor protein p62, encoded by the ALS-linked gene SQSTM1, is responsible for loading of cargo into the extending phagophore/autophagosome [224–226]. Interestingly, p62 has been shown to interact with polyubiquitinated mutant SOD1 [227, 228], which enhances its interaction with lipidated LC3-II on the phagophore membrane [228], suggesting that autophagy is activated in the presence of aggregated mutant SOD1. These interactions lead to the formation of protein inclusions within motor neurons [228]. Autophagy was also reportedly increased in mutant SOD1 mice potentially due to decreased phosphorylation of ULK1 [229], which activates autophagy [230]. In addition, proteins involved in autophagy regulation such as TFEB and Beclin 1 were found upregulated in the spinal cords of mutant SOD1 mice [231].
Several genes associated with ALS also encode RNA-binding proteins that aggregate when mutated. Both TDP-43 and fused in sarcoma (FUS) are found in intracellular aggregates in brain autopsy [232, 233]. In order to maintain homeostasis, autophagy is known to degrade aggregated TDP-43 and FUS [234–236]. Not surprisingly, increased expression of TDP-43 and FUS promote aggregation and stall autophagy in ALS models that could be rescued upon autophagy induction [234, 237, 238]. Furthermore, TDP-43 depletion increases translocation of TFEB into the nucleus, thereby stimulating autophagy [239]. However, downregulation of TDP-43 has also been shown to decrease Atg7 expression leading to reduced autophagic flux and autophagosome–lysosome fusion [240]. Thus, studies on the mechanism of action for TDP-43 in relation to autophagy have given both gain and loss of function results.
A subset of ALS-linked genes encode autophagy receptors, such as p62 and optineurin (OPTN) [241], or regulators of such receptors like Tank binding kinase 1 (TBK1) [242, 243]. As autophagy receptors, both p62 and optineurin contain a ubiquitin-associated domain (UBA) and an LC3-interacting region [241]. Generally, the proteasome is responsible for the degradation of ubiquitinated proteins. However, the discovery of autophagic receptors with UBA domains suggests that ubiquitinated cargo may also be targeted to and degraded by lysosomes. Many ALS-associated mutations affecting p62 and optineurin activity fall within the UBA domain [244, 245]. Furthermore, mutations located in the UBA domain may affect the protein’s ability to be phosphorylated by TBK1 [242, 243], suggesting that loss of TBK1 activity can enhance autophagic dysfunction in ALS.
The most commonly mutated gene in ALS, C9orf72 [246], has also been linked to autophagy via its interaction with endocytic proteins, Rab5 and Rab7 [247, 248], which regulate endosomal maturation [249]. Proper maturation and positioning of late endosomes is critical for autophagosome fusion [249]. C9orf72 also interacts with the ULK1 complex directly, and loss of C9orf72 prevents autophagy in an ULK1-mediated manner [250, 251]. As C9orf72 was previously shown to interact with Rab1a, an early autophagic regulator involved in phagophore formation [247, 251], C9orf72 may also regulate the recruitment of ULK1 to the initiation complex. Conversely, loss of C9orf72 leads to inactivation of mTORC1, increased TFEB nuclear translocation, and subsequent activation of autophagy [250, 252]. Therefore, C9orf72 is involved in several conflicting steps of autophagy but ultimately has been implicated in the maintenance of lysosomal function.
FTD is the second most common young-onset dementia and is characterized by atrophy of the frontal and temporal lobes [253, 254]. FTD is divided into subgroups based on the pathological hallmarks that develop with disease progression, including FTD with tau (FTD-tau) or TDP-43 (FTD-TDP)-positive inclusions [253]. The majority of FTD cases are attributed to mutations in microtubule-associated protein tau, progranulin (GRN), and C9orf72 [253, 254]. The remaining familial-associated FTD cases are linked to mutations in valosin-containing protein (VCP), FUS, TDP-43, and charged multivesicular body protein 2B (CHMP2B), which play distinct roles in the endolysosomal pathway [253, 254]. This suggests that disruptions in normal endosome to lysosome maturation may delay the degradation of autophagic cargo or impair the delivery of lysosomal hydrolases. For example, it was previously shown that VCP patients exhibited autophagic defects including an increase in the number of enlarged cathepsin B-positive autophagic vesicles [255]. Moreover, patients with mutations in VCP and CHMP2B were also reported to have decreased autophagic clearance and lysosomal function [255–259].
It was previously posited that FTD and LSDs were related diseases as they both shared similar common pathophysiological phenotypes. However, a concrete link was established with the discovery of a family affected by both neuronal ceroid lipofuscinosis (NCL), a young-onset LSD, and FTD [260]. Linkage analysis revealed independent heterozygous and homozygous mutations in the GRN gene [260], which encodes granulin, thus linking lysosomal dysfunction to GRN mutants and FTD. The proprotein, progranulin, is cleaved into individual granulins in the endolysosomal pathway [261, 262]. Recent work further demonstrated granulin E is responsible for modulating cathepsin D activity [263, 264], suggesting that GRN can directly alter lysosomal function through resident hydrolases. In agreement with this, GRN knockout mice displayed an increase in the levels of TMEM106B, as well as an accumulation of lipofuscin puncta in lysosomes [265], both of which are indicators of lysosomal dysfunction commonly found in NCLs. Furthermore, CTSD knockout mice exhibited increases in progranulin and TMEM106B along with saposin D, which is normally found accumulated in NCL10 patients [265]. Altogether, the genetics of ALS/FTD strongly implicate dysfunctions in both autophagy and endolysosomal systems in disease pathogenesis.
Huntington’s Disease
Huntington’s disease (HD) is an autosomal dominant, trinucleotide repeat disorder characterized by cognitive dysfunction, psychiatric disturbances, and loss of motor control [266]. The brain region most affected in HD is the striatum, an area critical to initiating and controlling movement, although other areas of the brain such as the cortex and cerebellum are also affected as the disease progresses [267]. Although defects in various pathways have been associated with HD, including alterations in Ca2+ handling, vesicle transport, and ER homeostasis [268], recent studies have implicated defective autophagy as a key feature of the disease, highlighting the potential of lysosomes and other autophagy machinery as druggable targets in HD.
The genetic basis of HD is a polyglutamine (polyQ) repeat expansion near the N-terminus of the huntingtin (htt) protein [269]. A repeat expansion of greater than 36Q is causative of disease, and like in other repeat disorders, greater repeat lengths correlate with earlier disease onset [268]. The hallmark feature of HD pathology is the accumulation and aggregation of mutant htt protein (mhtt). These aggregates form both intranuclear and intracytoplasmic neuronal inclusions, although whether these aggregates are pathogenic is still debated [270, 271]. Furthermore, htt can be proteolyzed to smaller fragments, which may also contribute to disease pathogenesis [272–275].
Although the function of wild-type htt is not fully understood, htt has been shown to interact with autophagy machinery. For instance, human and mouse HD samples exhibit an increase in autophagosomes [276, 277], and mhtt was shown to activate autophagy via sequestration and inactivation of mTORC1 [278]. Despite increasing autophagosome formation, mhtt appears to alter cargo recognition and loading through an unidentified mechanism, ultimately leading to accumulation of damaged organelles and protein aggregates [279]. Additional aberrations in autophagy are linked to effects of mhtt on vesicle trafficking including deficits in autophagosome motility [280] and autophagosome–lysosome fusion [281]. Silencing of either htt or its binding partner, huntingtin-associated protein 1, blocked retrograde transport of autophagosomes along the axonal compartment, whereas depletion of mhtt also resulted in an accumulation of autophagosomes containing undegraded cargo [280]. Alterations in mRNA of autophagy-associated proteins were also observed in the striatum of HD patients. Notably, LC3A and LAMP2 mRNA are increased, consistent with an overall increase in autophagic flux, whereas PINK1 is decreased, suggesting defects in mitophagy [282]. Finally, a polymorphism in Atg7, which is required for LC3 lipidation, is associated with earlier HD onset, thus providing strong genetic evidence for the close interplay between HD and autophagy [283].
Another important feature of htt is its ability to modulate autoclearance through posttranslational modifications. Ubiquitination of htt at K6, K9, and K15 is associated with increased degradation, whereas SUMOylation of these residues leads to decreased clearance [284]. Wild-type htt is further ubiquitinated at the K48 linkage, which is a classical marker of proteosomal degradation, whereas mhtt is ubiquitinated at K63, which is correlated with increased aggregation [275]. The ubiquitination and SUMOylation of htt is regulated through phosphorylation of S13 and S16, which are critical for mediating mhtt toxicity [285]. Acetylation at K444 is also important for regulating htt degradation and has been shown to increase mhtt clearance through enhanced binding to p62 [286].
In addition to the effects of mhtt on the autophagic pathway, recent studies have suggested a direct interaction of mhtt with lysosomes. In mouse striatum, mhtt alters lysosomal positioning via increased perinuclear localization which correlated with increased autophagic flux in response to nutrient deprivation [287]. Furthermore, it was recently shown that mhtt is able to be secreted from neurons through an unconventional endolysosomal pathway, suggesting that interactions of mhtt with the endolysosomal system may also contribute to disease pathogenesis [288].
Targeting Lysosomal Pathways in Neurodegenerative Disease
As discussed, several neurodegenerative diseases present with changes to lysosomal and autophagic function, and these alterations may in turn correlate to physiological aging processes. At present, several therapeutic approaches converge on targeting autophagy and lysosomes in neurodegenerative diseases. Therefore, combating the pathophysiological mechanisms implicated in these diseases will improve our knowledge and assist in the development of lysosome-targeted therapies to reduce disease burden and ultimately preserve lysosomal function in the aging brain.
Targeting Autophagy in Neurodegeneration
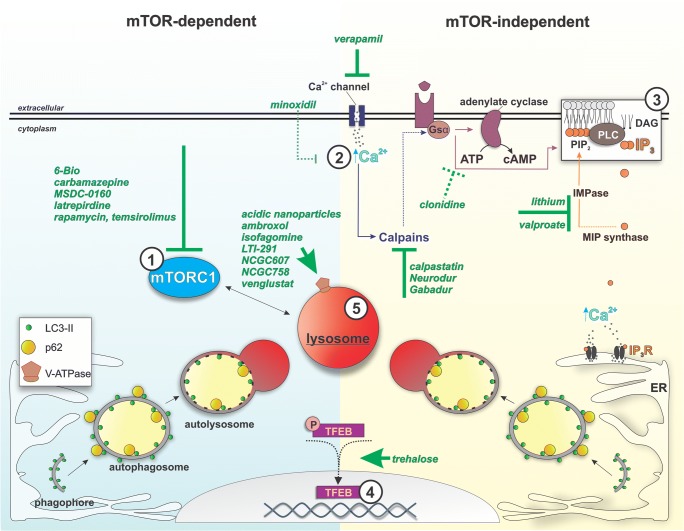
The strong association between neurodegenerative diseases and aberrations in autophagy implicates the autophagic pathway as a prime therapeutic target for the treatment of these diseases. Various therapeutic approaches targeting both mTOR-dependent and mTOR-independent pathways of autophagy have now been tested preclinically and clinically (Fig. 2, Table 1).
Fig. 2.
Pharmacological agents used in preclinical and clinical investigational therapies for neurodegeneration and their known or proposed targets related to autophagy and lysosomes. Agents whose action relates to suppression of mTORC1 activity (1), reduction of cytosolic calcium and IP3 levels as well as enhancement of TFEB activity (2-4) are expected to generally induce cellular autophagy. Targeting lysosomal acidity or glucocerebrosidase activity (5) can specifically induce lysosomal clearance efficiency
Table 1.
Therapies for neurodegenerative diseases targeting lysosomal pathways. Various therapies have been used to stimulate autophagy, both mTOR-dependent and mTOR-independent, as well as promote lysosomal function in neurodegenerative diseases
| Disease | Therapy | Pathway | Proposed mechanism | Reference(s) |
|---|---|---|---|---|
| PD | Rapamycin | mTOR-dependent | mTORC1 inhibition | [289–292] |
| Temsirolimus | ||||
| MSDC-0160 | [293] | |||
| 6-Bio | GSK3B inhibitor | [294] | ||
| Verapamil | mTOR-independent | attenuation of cytosolic Ca2+ increase | [77] | |
| Minoxidil | ||||
| Clonidine | cAMP levels reduction | |||
| Lithium, valproate | IP3 reduction | [76, 295] | ||
| Calpastatin | Calpain inhibition | [77] | ||
| Trehalose | Akt inhibitor | [296–298] | ||
| AUTEN-99 | MTMR14/Jumpy inhibitor | [299] | ||
| Acidic nanoparticles | Lysosomal | Lysosomal acidification | [300] | |
| Ambroxol** | GCase chaperone | [301–304] | ||
| Isofagomine | ||||
| NCGC758 | GCase chaperone | [214, 305] | ||
| NCGC607 | GCase chaperone | [306] | ||
| Venglustat** | Glucosylceramide synthase inhibitor | [307] | ||
| LTI-291** | GCase activator | [307] | ||
| AD | Carbamazepine | mTOR-dependent | mTORC1 inhibition | [308–311] |
| Latrepirdine | ||||
| Temsirolimus | ||||
| Lithium | mTOR-independent | IP3 reduction | [312] | |
| Resveratrol** | Unknown | [313–316] | ||
| Memantine* | NMDA receptor agonist | [317, 318] | ||
| Metformin** | Lysosomal | TFEB activation | [319, 320] | |
| ALS/FTD | Rapamycin | mTOR-dependent | mTORC1 inhibition | [237] |
| Trehalose | mTOR-independent | Akt inhibitor | [321–323] | |
| Resveratrol | Unknown | [324] | ||
| HD | Rapamycin | mTOR-dependent | mTORC1 inhibition | [278] |
| Trehalose | mTOR-independent | Akt inhibitor | [321, 325] | |
| Lithium, valproate | IP3 reduction | [326, 327] | ||
| Verapamil | attenuation of cytosolic Ca2+ increase | |||
| Clonidine, rilmenidine | cAMP levels reduction | [77, 328] | ||
| Calpastatin | Calpain inhibitor | [77, 322] | ||
| Zn finger repressors | Various | Correction of mhtt | [329] | |
| CRISPR-Cas9 | [330] | |||
| ASOs (IONIS-HTTRx)** | mhtt RNA degradation | [331, 332] |
*Memantine is FDA-approved for AD
** Currently in clinical trials
mTOR-Dependent Targets
The macrolide allosteric mTOR inhibitor, rapamycin, and its analogue, temsirolimus, have been tested in multiple neurodegenerative disease models. mTOR inhibitors were shown to induce autophagy, prevent dopaminergic loss, and revert motor, cognitive, and affective symptoms in several PD mouse models [289–292]. Moreover, rapamycin was able to improve pathology and disease phenotypes in AD, ALS, and HD models [237, 278, 333–337]. Other mTOR-dependent stimulators of autophagy, including GTM-1, carbamazepine, and latrepirdine, were shown to decrease Aβ accumulation and APP metabolites in AD cell models and also protect against memory dysfunction in AD patients [310–313].
mTOR-dependent stimulators have been most extensively tested in PD. The GSK3B inhibitor 6-Bio was neuroprotective in PD mouse model, and reduced αSyn aggregation in vitro [294]. The insulin sensitizer, MSDC-0160, prevented and restored dopaminergic loss and motor deficits in lesioned mice while additionally exerting an anti-inflammatory effect [293]. Similarly, pathways targeted by the anti-diabetic drug, exendin-4/exenatide are implicated in the preservation or restoration of dopaminergic neuron integrity in lesioned PD rodent models [338, 339], yet autophagy-related effects are unclear. Exenatide, now in phase I clinical trials for early stage PD, showed potential for sustainable motor score improvement for PD patients in a previous Phase II study [340]. Lastly, the c-Ab1 tyrosine kinase inhibitor and AMPK activator nilotinib, was shown to induce autophagy, enhance dopaminergic viability, and improve motor function in PD mouse models [341–343], yet mTOR involvement is unclear. The compound, which is currently in phase II clinical trials, showed potential to improve symptoms in both late-stage PD and Lewy body dementia patients [344].
mTOR-Independent Targets
Despite the promise of mTOR-dependent activation of autophagy, side effects associated with modulating mTOR have led to the emergence of mTOR-independent activators of autophagy as possible alternatives. Trehalose, a disaccharide, has been proposed to promote lysosomal clearance via inhibition of Akt and prevention of inhibitory TFEB phosphorylation [83]. The compound was shown to enhance clearance of mutant αSyn and Htt aggregates in cell lines, attenuate dopaminergic loss in PD mice, and protect mitochondrially challenged human iPSC-derived dopaminergic neurons [296–298, 345]. Moreover, trehalose delayed neurodegeneration while improving pathology and phenotypes in both HD and SOD1 ALS mouse models [323–325, 327].
Mood-stabilizing agents, such as lithium and valproate, also function as mTOR-independent autophagy activators through inositol depletion via inhibition of inositol monophosphatase and inositol 3-phosphate synthase, respectively [78]. These agents have been shown to enhance clearance of aggregate-prone htt and protect dopaminergic integrity in PD mouse models [76, 295], alleviate pathology and motor phenotypes in HD models [328, 329], and improve cognitive decline in AD patients with few adverse effects [314]. Resveratrol, another autophagy stimulator, was found to delay symptoms and pathology in SOD1 ALS mice [326], as well as decrease Aβ pathology in AD models in an AMPK-dependent manner [313, 314]. Although 1 study found that AD patients treated with resveratrol show an increase in plasma Aβ40 levels [317], another study demonstrated decreased levels of Aβ42 in the CSF with long-term treatment in patients [318]. Therefore, whether resveratrol is beneficial for AD patients in the long term remains to be determined. Calpain inhibitors such as calpastatin, Gabadur, and Neurodur have also been shown to be beneficial in PD and HD mouse models in an mTOR-independent manner [346, 347].
Other putative mTOR-independent autophagy inducers have been tested in specific disease models. AUTEN-99, which inhibits the myotubularin-related phosphatase MTMR14/Jumpy, a negative regulator of autophagic membrane formation, phenotypically rescues PD-associated outcomes in Drosophila models [299]. Memantine, an NMDA receptor agonist, displays efficacy in patients with moderate to severe AD [319, 320], though the compound’s effects on autophagy remain to be explored. Rilmenidine, an antihypertensive, attenuates HD pathology and accumulation of mhtt in a mouse model via an mTOR-independent mechanism [330]. Lastly, L-type Ca2+ channel antagonist verapamil, ATP-sensitive potassium channel opener minoxidil, and the G-signaling modulator clonidine, have been proposed to promote autophagy via modulation of IP3 levels, intracellular Ca2+, and calpain activity, and have been suggested as potential therapeutic agents in HD and other neurodegenerative diseases [77].
It is not currently known whether induction of autophagy can confer long-term improvements in neuropathology, delay disease progression, or ideally prevent neurodegeneration. Apart from its central role in cellular degradation and recycling, autophagy has emerging roles in DNA repair, inflammation, and protein secretion [348–350], suggesting that upstream targeting of this mechanism can have multiple effects which cannot be precisely predicted based on current knowledge. Though autophagy is an attractive target in neurodegeneration, the prolonged and systemic administration of upstream autophagy stimulators comes with the risks. For instance, although rapamycin shows positive outcomes in preclinical studies, higher doses of rapamycin inhibit mTORC2 thereby potentially compromising survival, whereas immunosuppression and other side effects have also been reported [351]. For several other compounds, the mode of action is not thoroughly characterized, and the effect on autophagy may not be specific or direct. In addition, a very recent study in C. elegans demonstrated that increasing autophagy can also be detrimental and compromise lifespan, depending on the status of the mitochondria [352]. This becomes a concern for the systemic induction of autophagy because different cell types or tissues exhibit baseline metabolic differences and could be differentially affected by aging and neurodegeneration. An additional consideration is the timing of intervention as well as the genetic background of patients. Indeed, enhancing autophagy at early disease stages may be effective in delaying the accumulation of disease-related proteins. On the contrary, increasing autophagosome formation at later disease stages or in patients whose lysosomal function is likely already compromised may exacerbate neurodegenerative pathology due to exceeding the cellular capacity for degradation. For these reasons, more targeted approaches could have more efficacious, powerful, and specific outcomes for patients.
Targeting Lysosomes in Neurodegeneration
More direct lysosome-targeted therapies have been proposed and tested in PD, AD, and HD (Table 1). Given the selective vulnerability of substantia nigra neurons in PD, direct gene targeting is a potentially relevant therapeutic strategy. Preclinical models suggest that overexpression of Beclin 1, Lamp2A, and TFEB could induce lysosomal function and confer neuroprotection [290, 353, 354]. Another approach to target lysosomes in PD involves the direct delivery of nanoparticles to enhance lysosomal acidification, which restored lysosomal activity in ATP13A2 and GBA1 PD models [300].
Decreased lysosomal hydrolase activity has been attributed to many neurodegenerative diseases. Thus, patients may also benefit from direct targeting and enhancement of these hydrolases. For example, AAV delivery of GRN to progranulin knockout mice significantly reversed lysosomal dysfunction associated with both FTD and NCL [355]. In addition, viral expression of GBA1 or reduction of GCase substrate levels via pharmacological glucosylceramide synthase inhibition reduced αSyn pathology and rescued dopaminergic integrity and cognitive function in rodents [308, 309, 356]. Venglustat, which inhibits glucosylceramide synthase, is currently in phase II clinical trials for early-stage GBA1-PD patients [307]. Reduction of glucosylceramide levels can also be achieved via small molecule non-inhibitory chaperones. NCGC758, which activates GCase specifically in the lysosomal compartment, preserved physiological αSyn conformation and increased the efficiency of trafficking and maturation of GCase in the endolysosomal compartment in PD iPSC-derived midbrain neurons [214, 305]. Similarly, NCGC607, also a small molecule non-inhibitory chaperone was found to reduce glucosphingolipids and αSyn levels in iPSC-derived dopaminergic neurons from Gaucher patients with parkinsonism [306]. Other compounds, such as the pharmacological chaperones ambroxol and isofagomine, have also been shown to increase GCase levels and activity and preserve αSyn homeostasis in PD/GBA1-PD and also wild-type cell and animal models [301–304]. Currently, ambroxol is in phase II clinical trials for GBA1-PD and PD with dementia patients [307]. Furthermore, the compound LTI-291, which stimulates GCase activity in the brain, is being assessed in phase II trials in GBA1-PD patients [307]. Unlike enzyme replacement therapies which would require invasive CNS delivery, pharmacological chaperones target specific lysosomal hydrolases, such as GCase, and could be orally administered and cross the blood–brain barrier. Examining the distribution of these compounds in disease-relevant CNS regions is important, to which end both wild-type and lysosomal enzyme-deficient animal models are necessary. Furthermore, GCase activation via pharmacological chaperones such as isofagomine binding to the active site of the enzyme, requires washout in order to achieve maximal lysosomal activity induction [357, 358]. The administration to patients should therefore be optimized so that the chaperone is rapidly removed from the lysosome, allowing the enzyme sufficient time to interact with its endogenous substrate. Future studies focusing on the efficacy of non-inhibitory or allosteric chaperones [305, 306, 359] could provide significant advancements, because the enzyme could interact with the endogenous substrate without the necessity of compound removal.
In AD, a class of interventions is focused on directly improving lysosomal function. Current clinical trials suggest that protein phosphatase 2A agonists, such as metformin, have beneficial effects by inhibiting hyperphosphorylation of tau [321, 322]. As phosphorylated tau constitutes a major component of NFTs in neurons [321, 322], metformin in conjunction with other treatments that reduce Aβ pathology may be useful for improving both pathological hallmarks of AD. Furthermore, TFEB transduction in the hippocampi of AD mouse models reduced AD-associated pathologies by upregulating lysosome biogenesis [360].
Because of growing evidence for htt regulation of autophagy and lysosomal function in HD, direct targeting of htt was proposed as a therapeutic approach in disease. The monogenetic etiology of HD makes it particularly attractive for gene therapies, specifically those targeting mhtt DNA and mRNA. DNA-targeting therapies, such as zinc finger proteins and CRIPSR-Cas9, to transduce cells with functional htt, have been tested in animal models. Zinc finger repressors reduced mhtt expression in the brains of R6/2 mice [331], and CRISPR-Cas9 gene editing ameliorated neurotoxicity in an HD mouse model [332]. Moreover, mRNA-targeting therapies against mhtt, such as antisense oligonucleotides (ASOs), nucleotide-based therapies that bind mRNA to trigger degradation, have already entered clinical trials [361]. The first of these, IONIS-HTTRx, was well-tolerated and resulted in dose-dependent reductions in mhtt levels in the CSF of early HD patients [362] and has now entered phase III clinical trials. The development of therapies directly targeting mhtt is advantageous in that it may not only mitigate lysosomal defects in disease but improve other aspects of disease pathophysiology and symptomology as well.
Overall, lysosomal function is a promising and specific target in neurodegeneration, with the possibility to benefit patients of various genetic backgrounds at early or even later stages of disease. Further elucidation of lysosomal functions and regulation of lysosome-related pathways will be instrumental to shaping the design and specificity of future therapeutic interventions. In this endeavor, a significant step forward has been made by the development of iPSC and CRISPR/Cas9 technologies, which in combination have improved our ability to assess the efficacy of therapies targeting lysosomal function in patient-derived neurons in both sporadic and familial disease. Nevertheless, a major challenge of the future will be the identification of sensitive and specific biomarkers for both diagnosing neurodegenerative diseases early and evaluating patient responses to therapies, which may include assessment of lysosomal efficiency in disease-relevant regions of the CNS.
Conclusion
Lysosomal function in the aging brain is an area that is just beginning to be explored. The lysosome is cardinal to various cellular functions including autophagy, nutrient and ion homeostasis, and inter-organelle interactions. As our understanding of the role of lysosomes in aging continues to take shape, it will be necessary to identify how various lysosomal nutrient storage, sensing, and processing pathways are engaged in brain-specific cell types in order to facilitate the discovery of therapeutic targets. For now, the best insight into lysosomal alterations with age will come from the detailed investigation of pathophysiological mechanisms in age-related neurodegenerative diseases that often present with hallmarks of lysosomal and autophagic dysfunction. Several therapeutic strategies to combat these diseases have already emerged and entered into clinical trials, including those promoting the upregulation of autophagy and those directly aimed at rescuing lysosomal function. The promise of these treatments in neurodegeneration beckons the question of whether these therapies will be similarly beneficial in preserving lysosomal function in the aging brain.
Electronic Supplementary Material
(DOCX 460 kb)
Acknowledgments
We thank Dr. Yvette C. Wong and Dr. Joseph Mazzulli for their helpful advice. The authors were supported by National Institutes of Health grants as follows: D.K. by R01 NS076054 and R37 NS096241.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
The original version of this article was revised to include a revised version of figure 2 overlooked by the publisher during the production process.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/24/2019
This article was corrected to include a revised version of figure 2 overlooked by the Publisher during the production process.
References
- 1.Lim CY, Zoncu R. The lysosome as a command-and-control center for cellular metabolism. J Cell Biol. 2016;214(6):653–664. doi: 10.1083/jcb.201607005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmona-Gutierrez D, Hughes AL, Madeo F, Ruckenstuhl C. The crucial impact of lysosomes in aging and longevity. Ageing Res Rev. 2016;32:2–12. doi: 10.1016/j.arr.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Telpoukhovskaia MA, Bahr BA, Chen X, Gan L. Endo-lysosomal dysfunction: a converging mechanism in neurodegenerative diseases. Curr Opin Neurobiol. 2018;48:52–58. doi: 10.1016/j.conb.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Sekito T, Fujiki Y, Ohsumi Y, Kakinuma Y. Novel families of vacuolar amino acid transporters. IUBMB Life. 2008;60(8):519–525. doi: 10.1002/iub.92. [DOI] [PubMed] [Google Scholar]
- 5.Russnak R, Konczal D, McIntire SL. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J Biol Chem. 2001;276(26):23849–23857. doi: 10.1074/jbc.M008028200. [DOI] [PubMed] [Google Scholar]
- 6.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8(11):917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 7.Chapel A, Kieffer-Jaquinod S, Sagne C, Verdon Q, Ivaldi C, Mellal M, et al. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol Cell Proteomics. 2013;12(6):1572–1588. doi: 10.1074/mcp.M112.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colacurcio DJ, Nixon RA. Disorders of lysosomal acidification—the emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res Rev. 2016;32:75–88. doi: 10.1016/j.arr.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stransky LA, Forgac M. Amino acid availability modulates vacuolar H+-ATPase assembly. J Biol Chem. 2015;290(45):27360–27369. doi: 10.1074/jbc.M115.659128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18(9):524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung J, Genau HM, Behrends C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol. 2015;35(14):2479–2494. doi: 10.1128/MCB.00125-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519(7544):477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171(3):642–54.e12. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358(6364):807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492(7428):261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruckenstuhl C, Netzberger C, Entfellner I, Carmona-Gutierrez D, Kickenweiz T, Stekovic S, et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 2014;10(5):e1004347. doi: 10.1371/journal.pgen.1004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raffaello A, Mammucari C, Gherardi G, Rizzuto R. Calcium at the center of cell signaling: interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem Sci. 2016;41(12):1035–1049. doi: 10.1016/j.tibs.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsubakiyama R, Mizunuma M, Gengyo A, Yamamoto J, Kume K, Miyakawa T, et al. Implication of Ca2+ in the regulation of replicative life span of budding yeast. J Biol Chem. 2011;286(33):28681–28687. doi: 10.1074/jbc.M111.231415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohya Y, Umemoto N, Tanida I, Ohta A, Iida H, Anraku Y. Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a Pet- phenotype are ascribable to defects of vacuolar membrane H(+)-ATPase activity. J Biol Chem. 1991;266(21):13971–13977. [PubMed] [Google Scholar]
- 23.Kilpatrick BS, Yates E, Grimm C, Schapira AH, Patel S. Endo-lysosomal TRP mucolipin-1 channels trigger global ER Ca2+ release and Ca2+ influx. J Cell Sci. 2016;129(20):3859–3867. doi: 10.1242/jcs.190322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrity AG, Wang W, Collier CM, Levey SA, Gao Q, Xu H. The endoplasmic reticulum, not the pH gradient, drives calcium refilling of lysosomes. eLife. 2016;5. [DOI] [PMC free article] [PubMed]
- 25.Lee JH, McBrayer MK, Wolfe DM, Haslett LJ, Kumar A, Sato Y, et al. Presenilin 1 maintains lysosomal Ca(2+) homeostasis via TRPML1 by regulating vATPase-mediated lysosome acidification. Cell Rep. 2015;12(9):1430–1444. doi: 10.1016/j.celrep.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim Biophys Acta. 2016;1863(12):2977–2992. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Nikoletopoulou V, Tavernarakis N. Calcium homeostasis in aging neurons. Front Genet. 2012;3:200. doi: 10.3389/fgene.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaelis ML, Bigelow DJ, Schoneich C, Williams TD, Ramonda L, Yin D, et al. Decreased plasma membrane calcium transport activity in aging brain. Life Sci. 1996;59(5–6):405–412. doi: 10.1016/0024-3205(96)00319-0. [DOI] [PubMed] [Google Scholar]
- 29.Xiong J, Verkhratsky A, Toescu EC. Changes in mitochondrial status associated with altered Ca2+ homeostasis in aged cerebellar granule neurons in brain slices. J Neurosci. 2002;22(24):10761–10771. doi: 10.1523/JNEUROSCI.22-24-10761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A, Foster TC. Enhanced long-term potentiation during aging is masked by processes involving intracellular calcium stores. J Neurophysiol. 2004;91(6):2437–2444. doi: 10.1152/jn.01148.2003. [DOI] [PubMed] [Google Scholar]
- 31.Gant JC, Sama MM, Landfield PW, Thibault O. Early and simultaneous emergence of multiple hippocampal biomarkers of aging is mediated by Ca2+-induced Ca2+ release. J Neurosci. 2006;26(13):3482–3490. doi: 10.1523/JNEUROSCI.4171-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheftel AD, Zhang AS, Brown C, Shirihai OS, Ponka P. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood. 2007;110(1):125–132. doi: 10.1182/blood-2007-01-068148. [DOI] [PubMed] [Google Scholar]
- 33.Ashraf A, Clark M, So PW. The Aging of Iron Man. Front Aging Neurosci. 2018;10:65. doi: 10.3389/fnagi.2018.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Todkar K, Ilamathi HS, Germain M. Mitochondria and lysosomes: discovering bonds. Front Cell Dev Biol. 2017;5:106. doi: 10.3389/fcell.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gammella E, Recalcati S, Cairo G. Dual role of ROS as signal and stress agents: iron tips the balance in favor of toxic effects. Oxidative Med Cell Longev. 2016;2016:8629024. doi: 10.1155/2016/8629024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klang IM, Schilling B, Sorensen DJ, Sahu AK, Kapahi P, Andersen JK, et al. Iron promotes protein insolubility and aging in C. elegans. Aging (Albany NY) 2014;6(11):975–991. doi: 10.18632/aging.100689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014;13(10):1045–1060. doi: 10.1016/S1474-4422(14)70117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connor JR, Snyder BS, Arosio P, Loeffler DA, LeWitt P. A quantitative analysis of isoferritins in select regions of aged, parkinsonian, and Alzheimer’s diseased brains. J Neurochem. 1995;65(2):717–724. doi: 10.1046/j.1471-4159.1995.65020717.x. [DOI] [PubMed] [Google Scholar]
- 39.Zecca L, Bellei C, Costi P, Albertini A, Monzani E, Casella L, et al. New melanic pigments in the human brain that accumulate in aging and block environmental toxic metals. Proc Natl Acad Sci U S A. 2008;105(45):17567–17572. doi: 10.1073/pnas.0808768105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zecca L, Casella L, Albertini A, Bellei C, Zucca FA, Engelen M, et al. Neuromelanin can protect against iron-mediated oxidative damage in system modeling iron overload of brain aging and Parkinson’s disease. J Neurochem. 2008;106(4):1866–1875. doi: 10.1111/j.1471-4159.2008.05541.x. [DOI] [PubMed] [Google Scholar]
- 41.Zucca FA, Vanna R, Cupaioli FA, Bellei C, De Palma A, Di Silvestre D, et al. Neuromelanin organelles are specialized autolysosomes that accumulate undegraded proteins and lipids in aging human brain and are likely involved in Parkinson’s disease. NPJ Parkinsons Dis. 2018;4:17. doi: 10.1038/s41531-018-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider SA, Dusek P, Hardy J, Westenberger A, Jankovic J, Bhatia KP. Genetics and pathophysiology of neurodegeneration with brain iron accumulation (NBIA) Curr Neuropharmacol. 2013;11(1):59–79. doi: 10.2174/157015913804999469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seibler P, Burbulla LF, Dulovic M, Zittel S, Heine J, Schmidt T, et al. Iron overload is accompanied by mitochondrial and lysosomal dysfunction in WDR45 mutant cells. Brain. 2018;141(10):3052–3064. doi: 10.1093/brain/awy230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mony VK, Benjamin S, O'Rourke EJ. A lysosome-centered view of nutrient homeostasis. Autophagy. 2016;12(4):619–631. doi: 10.1080/15548627.2016.1147671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parenti G, Andria G, Ballabio A. Lysosomal storage diseases: from pathophysiology to therapy. Annu Rev Med. 2015;66:471–486. doi: 10.1146/annurev-med-122313-085916. [DOI] [PubMed] [Google Scholar]
- 46.Mc Donald JM, Krainc D. Lysosomal proteins as a therapeutic target in neurodegeneration. Annu Rev Med. 2017;68:445–458. doi: 10.1146/annurev-med-050715-104432. [DOI] [PubMed] [Google Scholar]
- 47.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 48.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12(9):831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 49.Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep. 2011;3:25. doi: 10.3410/B3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganley Ian G., Lam Du H., Wang Junru, Ding Xiaojun, Chen She, Jiang Xuejun. ULK1·ATG13·FIP200 Complex Mediates mTOR Signaling and Is Essential for Autophagy. Journal of Biological Chemistry. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23(10):1860–1873. doi: 10.1091/mbc.e11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119(Pt 2):259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 53.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20(6):355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2012;109(22):8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.e08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5(4):534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- 58.Munson MJ, Allen GF, Toth R, Campbell DG, Lucocq JM, Ganley IG. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 2015;34(17):2272–2290. doi: 10.15252/embj.201590992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 60.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092–2100. doi: 10.1091/mbc.e07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20(1):59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19(8):3290–3298. doi: 10.1091/mbc.e07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117(Pt 13):2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 65.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem. 2006;281(6):3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 66.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13(12):1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 67.Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53(2):167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 68.Kriegenburg F, Ungermann C, Reggiori F. Coordination of autophagosome-lysosome fusion by Atg8 family members. Curr Biol. 2018;28(8):R512–R518. doi: 10.1016/j.cub.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 69.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawrence RE, Cho KF, Rappold R, Thrun A, Tofaute M, Kim DJ, et al. A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat Cell Biol. 2018;20(9):1052–1063. doi: 10.1038/s41556-018-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 2018;20(12):1338–1348. doi: 10.1038/s41556-018-0235-8. [DOI] [PubMed] [Google Scholar]
- 72.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 73.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129(13):2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170(7):1101. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4(5):295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ju S, Greenberg ML. 1D-myo-inositol 3-phosphate synthase: conservation, regulation, and putative target of mood stabilizers. Clin Neurosci Res. 2004;4(3):181–187. doi: 10.1016/j.cnr.2004.09.010. [DOI] [Google Scholar]
- 79.Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14:1029. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 80.Vicencio JM, Ortiz C, Criollo A, Jones AWE, Kepp O, Galluzzi L, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 81.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 82.Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C, et al. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2(4):e144–e. doi: 10.1038/cddis.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, et al. Corrigendum: mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:15793. doi: 10.1038/ncomms15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17(3):288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8(1):3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 86.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524(7565):309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pickles S, Vigie P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28(4):R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33(19):2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24(7):787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14(2):177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 92.Lv M, Wang C, Li F, Peng J, Wen B, Gong Q, et al. Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy. Protein & Cell. 2017;8(1):25–38. doi: 10.1007/s13238-016-0328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14(12):1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284(20):13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Burman JL, Pickles S, Wang C, Sekine S, Vargas JNS, Zhang Z, et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J Cell Biol. 2017;216(10):3231–3247. doi: 10.1083/jcb.201612106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chu CT. Mechanisms of selective autophagy and mitophagy: implications for neurodegenerative diseases. Neurobiol Dis. 2019;122:23–34. doi: 10.1016/j.nbd.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin-Maestro P, Gargini R, Sproul AA, Garcia E, Anton LC, Noggle S, et al. Mitophagy failure in fibroblasts and iPSC-derived neurons of Alzheimer’s disease-associated presenilin 1 mutation. Front Mol Neurosci. 2017;10:291. doi: 10.3389/fnmol.2017.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13(3):847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Merz S, Westermann B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009;10(9):R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30(1):95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 102.Honscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, et al. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30(1):86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 103.Wong YC, Ysselstein D, Krainc D. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 2018;554(7692):382–386. doi: 10.1038/nature25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong YC, Kim S, Peng W, Krainc D. Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol 2019. [DOI] [PMC free article] [PubMed]
- 105.Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54(3):266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40(2):323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang X, Cheng X, Yu L, Yang J, Calvo R, Patnaik S, et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat Commun. 2016;7:12109. doi: 10.1038/ncomms12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marino ML, Fais S, Djavaheri-Mergny M, Villa A, Meschini S, Lozupone F, et al. Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis. 2010;1:e87. doi: 10.1038/cddis.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Demers-Lamarche J, Guillebaud G, Tlili M, Todkar K, Belanger N, Grondin M, et al. Loss of mitochondrial function impairs lysosomes. J Biol Chem. 2016;291(19):10263–10276. doi: 10.1074/jbc.M115.695825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 112.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J-i, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 113.Rubinsztein David C, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 114.Leeman DS, Hebestreit K, Ruetz T, Webb AE, McKay A, Pollina EA, et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science. 2018;359(6381):1277–1283. doi: 10.1126/science.aag3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mayeux R., Stern Y. Epidemiology of Alzheimer Disease. Cold Spring Harbor Perspectives in Medicine. 2012;2(8):a006239–a006239. doi: 10.1101/cshperspect.a006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McDermott JR, Gibson AM. Degradation of Alzheimer’s beta-amyloid protein by human cathepsin D. Neuroreport. 1996;7(13):2163–2166. doi: 10.1097/00001756-199609020-00021. [DOI] [PubMed] [Google Scholar]
- 117.Nilsson P, Loganathan K, Sekiguchi M, Matsuba Y, Hui K, Tsubuki S, et al. Abeta secretion and plaque formation depend on autophagy. Cell Rep. 2013;5(1):61–69. doi: 10.1016/j.celrep.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 118.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64(2):113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 119.Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, et al. Macroautophagy—a novel beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171(1):87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu JT, Ma XY, Wang YL, Sun L, Tan L, Hu N, et al. Genetic variation in clusterin gene and Alzheimer’s disease risk in Han Chinese. Neurobiol Aging. 2013;34(7):1921 e17–23. doi: 10.1016/j.neurobiolaging.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 121.Zhang F, Kumano M, Beraldi E, Fazli L, Du C, Moore S, et al. Clusterin facilitates stressinduced lipidation of LC3 and autophagosome biogenesis to enhance cancer cell survival. Nat Commun. 2014;5:5775. doi: 10.1038/ncomms6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Miners JS, Clarke P, Love S. Clusterin levels are increased in Alzheimer’s disease and influence the regional distribution of Abeta. Brain Pathol. 2017;27(3):305–313. doi: 10.1111/bpa.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beeg M, Stravalaci M, Romeo M, Carra AD, Cagnotto A, Rossi A, et al. Clusterin binds to Abeta1-42 oligomers with high affinity and interferes with peptide aggregation by inhibiting primary and secondary nucleation. J Biol Chem. 2016;291(13):6958–6966. doi: 10.1074/jbc.M115.689539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou Y, Hayashi I, Wong J, Tugusheva K, Renger JJ, Zerbinatti C. Intracellular clusterin interacts with brain isoforms of the bridging integrator 1 and with the microtubule-associated protein Tau in Alzheimer’s disease. PLoS One. 2014;9(7):e103187. doi: 10.1371/journal.pone.0103187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141(7):1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013;37(12):1949–1961. doi: 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murphy MP, Uljon SN, Fraser PE, Fauq A, Lookingbill HA, Findlay KA, et al. Presenilin 1 regulates pharmacologically distinct gamma-secretase activities. Implications for the role of presenilin in gamma-secretase cleavage. J Biol Chem. 2000;275(34):26277–26284. doi: 10.1074/jbc.M002812200. [DOI] [PubMed] [Google Scholar]
- 128.Xia D, Watanabe H, Wu B, Lee SH, Li Y, Tsvetkov E, et al. Presenilin-1 knockin mice reveal loss-of-function mechanism for familial Alzheimer’s disease. Neuron. 2015;85(5):967–981. doi: 10.1016/j.neuron.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Coffey EE, Beckel JM, Laties AM, Mitchell CH. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 2014;263:111–124. doi: 10.1016/j.neuroscience.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Portelius E, Andreasson U, Ringman JM, Buerger K, Daborg J, Buchhave P, et al. Distinct cerebrospinal fluid amyloid beta peptide signatures in sporadic and PSEN1 A431E-associated familial Alzheimer’s disease. Mol Neurodegener. 2010;5:2. doi: 10.1186/1750-1326-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakatogawa H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013;55:39–50. doi: 10.1042/bse0550039. [DOI] [PubMed] [Google Scholar]
- 132.Nilsson P, Sekiguchi M, Akagi T, Izumi S, Komori T, Hui K, et al. Autophagy-related protein 7 deficiency in amyloid beta (Abeta) precursor protein transgenic mice decreases Abeta in the multivesicular bodies and induces Abeta accumulation in the Golgi. Am J Pathol. 2015;185(2):305–313. doi: 10.1016/j.ajpath.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 133.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, et al. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103(30):11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Inoue K, Rispoli J, Kaphzan H, Klann E, Chen EI, Kim J, et al. Macroautophagy deficiency mediates age-dependent neurodegeneration through a phospho-tau pathway. Mol Neurodegener. 2012;7:48. doi: 10.1186/1750-1326-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Paradis E, Douillard H, Koutroumanis M, Goodyer C, LeBlanc A. Amyloid beta peptide of Alzheimer’s disease downregulates Bcl-2 and upregulates bax expression in human neurons. J Neurosci. 1996;16(23):7533–7539. doi: 10.1523/JNEUROSCI.16-23-07533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Karlnoski R, Wilcock D, Dickey C, Ronan V, Gordon MN, Zhang W, et al. Up-regulation of Bcl-2 in APP transgenic mice is associated with neuroprotection. Neurobiol Dis. 2007;25(1):179–188. doi: 10.1016/j.nbd.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rohn TT, Vyas V, Hernandez-Estrada T, Nichol KE, Christie LA, Head E. Lack of pathology in a triple transgenic mouse model of Alzheimer’s disease after overexpression of the anti-apoptotic protein Bcl-2. J Neurosci. 2008;28(12):3051–3059. doi: 10.1523/JNEUROSCI.5620-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118(6):2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Neely KM, Green KN, LaFerla FM. Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a gamma-secretase-independent manner. J Neurosci. 2011;31(8):2781–2791. doi: 10.1523/JNEUROSCI.5156-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bordi M, Berg MJ, Mohan PS, Peterhoff CM, Alldred MJ, Che S, et al. Autophagy flux in CA1 neurons of Alzheimer hippocampus: increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy. 2016;12(12):2467–2483. doi: 10.1080/15548627.2016.1239003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Albayrak O, Tirniceriu A, Riemenschneider M, Kurz A, Scherag A, Egensperger R. The cathepsin D (224C/T) polymorphism confers an increased risk to develop Alzheimer’s disease in men. J Gerontol A Biol Sci Med Sci. 2010;65(3):219–224. doi: 10.1093/gerona/glp209. [DOI] [PubMed] [Google Scholar]
- 143.Mariani E, Seripa D, Ingegni T, Nocentini G, Mangialasche F, Ercolani S, et al. Interaction of CTSD and A2M polymorphisms in the risk for Alzheimer’s disease. J Neurol Sci. 2006;247(2):187–191. doi: 10.1016/j.jns.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 144.Urbanelli L, Emiliani C, Massini C, Persichetti E, Orlacchio A, Pelicci G, et al. Cathepsin D expression is decreased in Alzheimer’s disease fibroblasts. Neurobiol Aging. 2008;29(1):12–22. doi: 10.1016/j.neurobiolaging.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 145.Tian L, Zhang K, Tian ZY, Wang T, Shang DS, Li B, et al. Decreased expression of cathepsin D in monocytes is related to the defective degradation of amyloid-beta in Alzheimer’s disease. J Alzheimers Dis. 2014;42(2):511–520. doi: 10.3233/JAD-132192. [DOI] [PubMed] [Google Scholar]
- 146.Straface E, Matarrese P, Gambardella L, Vona R, Sgadari A, Silveri MC, et al. Oxidative imbalance and cathepsin D changes as peripheral blood biomarkers of Alzheimer disease: a pilot study. FEBS Lett. 2005;579(13):2759–2766. doi: 10.1016/j.febslet.2005.03.094. [DOI] [PubMed] [Google Scholar]
- 147.Adamec E, Mohan PS, Cataldo AM, Vonsattel JP, Nixon RA. Up-regulation of the lysosomal system in experimental models of neuronal injury: implications for Alzheimer’s disease. Neuroscience. 2000;100(3):663–675. doi: 10.1016/S0306-4522(00)00281-5. [DOI] [PubMed] [Google Scholar]
- 148.Schwagerl AL, Mohan PS, Cataldo AM, Vonsattel JP, Kowall NW, Nixon RA. Elevated levels of the endosomal-lysosomal proteinase cathepsin D in cerebrospinal fluid in Alzheimer disease. J Neurochem. 1995;64(1):443–446. doi: 10.1046/j.1471-4159.1995.64010443.x. [DOI] [PubMed] [Google Scholar]
- 149.Nixon RA. Amyloid precursor protein and endosomal-lysosomal dysfunction in Alzheimer’s disease: inseparable partners in a multifactorial disease. FASEB Journal: official publication of the Federation of American Societies for Experimental Biology. 2017;31(7):2729–2743. doi: 10.1096/fj.201700359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim S, Sato Y, Mohan PS, Peterhoff C, Pensalfini A, Rigoglioso A, et al. Evidence that the rab5 effector APPL1 mediates APP-betaCTF-induced dysfunction of endosomes in Down syndrome and Alzheimer’s disease. Mol Psychiatry. 2016;21(5):707–716. doi: 10.1038/mp.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lauritzen I, Pardossi-Piquard R, Bauer C, Brigham E, Abraham JD, Ranaldi S, et al. The beta-secretase-derived C-terminal fragment of betaAPP, C99, but not Abeta, is a key contributor to early intraneuronal lesions in triple-transgenic mouse hippocampus. J Neurosci. 2012;32(46):16243–1655a. doi: 10.1523/JNEUROSCI.2775-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lauritzen I, Pardossi-Piquard R, Bourgeois A, Pagnotta S, Biferi MG, Barkats M, et al. Intraneuronal aggregation of the beta-CTF fragment of APP (C99) induces Abeta-independent lysosomal-autophagic pathology. Acta Neuropathol. 2016;132(2):257–276. doi: 10.1007/s00401-016-1577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tamayev R, Matsuda S, Arancio O, D'Adamio L. beta- but not gamma-secretase proteolysis of APP causes synaptic and memory deficits in a mouse model of dementia. EMBO Mol Med. 2012;4(3):171–179. doi: 10.1002/emmm.201100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hung COY, Livesey FJ. Altered gamma-secretase processing of APP disrupts lysosome and autophagosome function in monogenic Alzheimer’s disease. Cell Rep. 2018;25(13):3647–60.e2. doi: 10.1016/j.celrep.2018.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.He X, Cooley K, Chung CH, Dashti N, Tang J. Apolipoprotein receptor 2 and X11 alpha/beta mediate apolipoprotein E-induced endocytosis of amyloid-beta precursor protein and beta-secretase, leading to amyloid-beta production. J Neurosci. 2007;27(15):4052–4060. doi: 10.1523/JNEUROSCI.3993-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Heeren J, Grewal T, Laatsch A, Becker N, Rinninger F, Rye KA, et al. Impaired recycling of apolipoprotein E4 is associated with intracellular cholesterol accumulation. J Biol Chem. 2004;279(53):55483–55492. doi: 10.1074/jbc.M409324200. [DOI] [PubMed] [Google Scholar]
- 157.Rellin L, Heeren J, Beisiegel U. Recycling of apolipoprotein E is not associated with cholesterol efflux in neuronal cells. Biochim Biophys Acta. 2008;1781(5):232–238. doi: 10.1016/j.bbalip.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 158.Ji ZS, Miranda RD, Newhouse YM, Weisgraber KH, Huang Y, Mahley RW. Apolipoprotein E4 potentiates amyloid beta peptide-induced lysosomal leakage and apoptosis in neuronal cells. J Biol Chem. 2002;277(24):21821–21828. doi: 10.1074/jbc.M112109200. [DOI] [PubMed] [Google Scholar]
- 159.Ji ZS, Mullendorff K, Cheng IH, Miranda RD, Huang Y, Mahley RW. Reactivity of apolipoprotein E4 and amyloid beta peptide: lysosomal stability and neurodegeneration. J Biol Chem. 2006;281(5):2683–2692. doi: 10.1074/jbc.M506646200. [DOI] [PubMed] [Google Scholar]
- 160.Nuriel T, Peng KY, Ashok A, Dillman AA, Figueroa HY, Apuzzo J, et al. The endosomal-lysosomal pathway is dysregulated by APOE4 expression in vivo. Front Neurosci. 2017;11:702. doi: 10.3389/fnins.2017.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dorsey ER, Elbaz A, Nichols E, Abd-Allah F, Abdelalim A, Adsuar JC, et al. Global, regional, and national burden of Parkinson’s disease, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018;17(11):939–953. doi: 10.1016/S1474-4422(18)30295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Anglade P, Vyas S, Hirsch EC, Agid Y. Apoptosis in dopaminergic neurons of the human substantia nigra during normal aging. Histol Histopathol. 1997;12(3):603–610. [PubMed] [Google Scholar]
- 163.Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper J, et al. Chaperone-mediated autophagy markers in parkinson disease brains. Arch Neurol. 2010;67(12):1464–1472. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- 164.Shahmoradian SH, Lewis AJ, Genoud C, Graff-Meyer A, Hench J, Moors T, et al. Lewy pathology in Parkinson’s disease consists of a crowded organellar, membranous medley. bioRxiv. 2018:137976. [DOI] [PubMed]
- 165.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 166.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alphasynuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 167.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, et al. alphaSynuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 168.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, et al. Alphasynuclein locus duplication as a cause of familial Parkinson’s disease. Lancet (London, England) 2004;364(9440):1167–9. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 169.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313(5785):324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190(6):1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mazzulli JR, Zunke F, Isacson O, Studer L, Krainc D. alpha-Synuclein-induced lysosomal dysfunction occurs through disruptions in protein trafficking in human midbrain synucleinopathy models. Proc Natl Acad Sci U S A. 2016;113(7):1931–1936. doi: 10.1073/pnas.1520335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hernandez DG, Reed X, Singleton AB. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J Neurochem. 2016;139(Suppl 1):59–74. doi: 10.1111/jnc.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lazaro DF, Rodrigues EF, Langohr R, Shahpasandzadeh H, Ribeiro T, Guerreiro P, et al. Systematic comparison of the effects of alpha-synuclein mutations on its oligomerization and aggregation. PLoS Genet. 2014;10(11):e1004741. doi: 10.1371/journal.pgen.1004741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tanik SA, Schultheiss CE, Volpicelli-Daley LA, Brunden KR, Lee VM. Lewy body-like alpha-synuclein aggregates resist degradation and impair macroautophagy. J Biol Chem. 2013;288(21):15194–15210. doi: 10.1074/jbc.M113.457408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305(5688):1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 176.Alvarez-Erviti L, Seow Y, Schapira AH, Gardiner C, Sargent IL, Wood MJ, et al. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol Dis. 2011;42(3):360–367. doi: 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, et al. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30(20):6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Danzer KM, Kranich LR, Ruf WP, Cagsal-Getkin O, Winslow AR, Zhu L, et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ejlerskov P, Rasmussen I, Nielsen TT, Bergstrom AL, Tohyama Y, Jensen PH, et al. Tubulin polymerization-promoting protein (TPPP/p25alpha) promotes unconventional secretion of alphasynuclein through exophagy by impairing autophagosome-lysosome fusion. J Biol Chem. 2013;288(24):17313–17335. doi: 10.1074/jbc.M112.401174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lee H-J, Cho E-D, Lee KW, Kim J-H, Cho S-G, Lee S-J. Autophagic failure promotes the exocytosis and intercellular transfer of α-synuclein. Exp Mol Med. 2013;45:e22. doi: 10.1038/emm.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Poehler AM, Xiang W, Spitzer P, May VE, Meixner H, Rockenstein E, et al. Autophagy modulates SNCA/alpha-synuclein release, thereby generating a hostile microenvironment. Autophagy. 2014;10(12):2171–2192. doi: 10.4161/auto.36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fussi N, Höllerhage M, Chakroun T, Nykänen N-P, Rösler TW, Koeglsperger T, et al. Exosomal secretion of α-synuclein as protective mechanism after upstream blockage of macroautophagy. Cell Death Dis. 2018;9(7):757. doi: 10.1038/s41419-018-0816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Minakaki G, Menges S, Kittel A, Emmanouilidou E, Schaeffner I, Barkovits K, et al. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy. 2018;14(1):98–119. doi: 10.1080/15548627.2017.1395992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Alessi DR, Sammler E. LRRK2 kinase in Parkinson’s disease. Science. 2018;360(6384):36. doi: 10.1126/science.aar5683. [DOI] [PubMed] [Google Scholar]
- 185.Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18(21):4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Eguchi T, Kuwahara T, Sakurai M, Komori T, Fujimoto T, Ito G, et al. LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc Natl Acad Sci. 2018;115(39):E9115. doi: 10.1073/pnas.1812196115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Steger M, Diez F, Dhekne HS, Lis P, Nirujogi RS, Karayel O, et al. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 2017;6. [DOI] [PMC free article] [PubMed]
- 188.Bravo-San Pedro JM, Niso-Santano M, Gomez-Sanchez R, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A, et al. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell Mol Life Sci. 2013;70(1):121–136. doi: 10.1007/s00018-012-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Manzoni C, Mamais A, Dihanich S, McGoldrick P, Devine MJ, Zerle J, et al. Pathogenic Parkinson’s disease mutations across the functional domains of LRRK2 alter the autophagic/lysosomal response to starvation. Biochem Biophys Res Commun. 2013;441(4):862–866. doi: 10.1016/j.bbrc.2013.10.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8(3):267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol Med. 2012;4(5):380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Follett J, Norwood SJ, Hamilton NA, Mohan M, Kovtun O, Tay S, et al. The Vps35 D620N mutation linked to Parkinson’s disease disrupts the cargo sorting function of retromer. Traffic (Copenhagen, Denmark) 2014;15(2):230–244. doi: 10.1111/tra.12136. [DOI] [PubMed] [Google Scholar]
- 193.Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, et al. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat Commun. 2014;5:3828. doi: 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tang FL, Erion JR, Tian Y, Liu W, Yin DM, Ye J, et al. VPS35 in dopamine neurons is required for endosome-to-golgi retrieval of Lamp2a, a receptor of chaperone-mediated autophagy that is critical for alpha-synuclein degradation and prevention of pathogenesis of Parkinson’s disease. J Neurosci. 2015;35(29):10613–10628. doi: 10.1523/JNEUROSCI.0042-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tang FL, Liu W, Hu JX, Erion JR, Ye J, Mei L, et al. VPS35 deficiency or mutation causes dopaminergic neuronal loss by impairing mitochondrial fusion and function. Cell Rep. 2015;12(10):1631–1643. doi: 10.1016/j.celrep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Song P, Trajkovic K, Tsunemi T, Krainc D. Parkin modulates endosomal organization and function of the endo-lysosomal pathway. J Neurosci. 2016;36(8):2425–2437. doi: 10.1523/JNEUROSCI.2569-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 198.Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, et al. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468(7324):696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Burbulla LF, Song P, Mazzulli JR, Zampese E, Wong YC, Jeon S, et al. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson’s disease. Science. 2017;357(6357):1255–1261. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lees AJ, Singleton AB. Clinical heterogeneity of ATP13A2 linked disease (Kufor-Rakeb) justifies a PARK designation. Neurology. 2007;68(19):1553–1554. doi: 10.1212/01.wnl.0000265228.66664.f4. [DOI] [PubMed] [Google Scholar]
- 201.Dehay B, Ramirez A, Martinez-Vicente M, Perier C, Canron MH, Doudnikoff E, et al. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc Natl Acad Sci U S A. 2012;109(24):9611–9616. doi: 10.1073/pnas.1112368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Grunewald A, Arns B, Seibler P, Rakovic A, Munchau A, Ramirez A, et al. ATP13A2 mutations impair mitochondrial function in fibroblasts from patients with Kufor-Rakeb syndrome. Neurobiol Aging. 2012;33(8):1843.e1–7. doi: 10.1016/j.neurobiolaging.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 203.Usenovic M, Tresse E, Mazzulli JR, Taylor JP, Krainc D. Deficiency of ATP13A2 leads to lysosomal dysfunction, alpha-synuclein accumulation, and neurotoxicity. J Neurosci. 2012;32(12):4240–4246. doi: 10.1523/JNEUROSCI.5575-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schultheis PJ, Fleming SM, Clippinger AK, Lewis J, Tsunemi T, Giasson B, et al. Atp13a2-deficient mice exhibit neuronal ceroid lipofuscinosis, limited alpha-synuclein accumulation and agedependent sensorimotor deficits. Hum Mol Genet. 2013;22(10):2067–2082. doi: 10.1093/hmg/ddt057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gitler AD, Chesi A, Geddie ML, Strathearn KE, Hamamichi S, Hill KJ, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet. 2009;41(3):308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kong SM, Chan BK, Park JS, Hill KJ, Aitken JB, Cottle L, et al. Parkinson’s disease-linked human PARK9/ATP13A2 maintains zinc homeostasis and promotes alpha-Synuclein externalization via exosomes. Hum Mol Genet. 2014;23(11):2816–2833. doi: 10.1093/hmg/ddu099. [DOI] [PubMed] [Google Scholar]
- 207.Tsunemi T, Krainc D. Zn(2)(+) dyshomeostasis caused by loss of ATP13A2/PARK9 leads to lysosomal dysfunction and alpha-synuclein accumulation. Hum Mol Genet. 2014;23(11):2791–2801. doi: 10.1093/hmg/ddt572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Narayanaswamy N, Chakraborty K, Saminathan A, Zeichner E, Leung K, Devany J, et al. A pH-correctable, DNA-based fluorescent reporter for organellar calcium. Nat Methods. 2019;16(1):95–102. doi: 10.1038/s41592-018-0232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bultron G, Kacena K, Pearson D, Boxer M, Yang R, Sathe S, et al. The risk of Parkinson’s disease in type 1 Gaucher disease. J Inherit Metab Dis. 2010;33(2):167–173. doi: 10.1007/s10545-010-9055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chang D, Nalls MA, Hallgrimsdottir IB, Hunkapiller J, van der Brug M, Cai F, et al. A metaanalysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49(10):1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schondorf DC, Aureli M, McAllister FE, Hindley CJ, Mayer F, Schmid B, et al. iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun. 2014;5:4028. doi: 10.1038/ncomms5028. [DOI] [PubMed] [Google Scholar]
- 212.Fernandes HJ, Hartfield EM, Christian HC, Emmanoulidou E, Zheng Y, Booth H, et al. ER stress and autophagic perturbations lead to elevated extracellular alpha-synuclein in GBA-N370S Parkinson’s iPSC-derived dopamine neurons. Stem Cell Reports. 2016;6(3):342–356. doi: 10.1016/j.stemcr.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, et al. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146(1):37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zunke Friederike, Moise Alexandra C., Belur Nandkishore R., Gelyana Eilrayna, Stojkovska Iva, Dzaferbegovic Haris, Toker Nicholas J., Jeon Sohee, Fredriksen Kristina, Mazzulli Joseph R. Reversible Conformational Conversion of α-Synuclein into Toxic Assemblies by Glucosylceramide. Neuron. 2018;97(1):92-107.e10. doi: 10.1016/j.neuron.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gan-Or Z, Ozelius LJ, Bar-Shira A, Saunders-Pullman R, Mirelman A, Kornreich R, et al. The p.L302P mutation in the lysosomal enzyme gene SMPD1 is a risk factor for Parkinson disease. Neurology. 2013;80(17):1606–1610. doi: 10.1212/WNL.0b013e31828f180e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dagan E, Schlesinger I, Ayoub M, Mory A, Nassar M, Kurolap A, et al. The contribution of Niemann-Pick SMPD1 mutations to Parkinson disease in Ashkenazi Jews. Parkinsonism Relat Disord. 2015;21(9):1067–1071. doi: 10.1016/j.parkreldis.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 217.Deng S, Deng X, Song Z, Xiu X, Guo Y, Xiao J, et al. Systematic genetic analysis of the SMPD1 gene in Chinese patients with Parkinson’s disease. Mol Neurobiol. 2016;53(7):5025–5029. doi: 10.1007/s12035-015-9426-5. [DOI] [PubMed] [Google Scholar]
- 218.Hopfner F, Schulte EC, Mollenhauer B, Bereznai B, Knauf F, Lichtner P, et al. The role of SCARB2 as susceptibility factor in Parkinson’s disease. Mov Disord. 2013;28(4):538–540. doi: 10.1002/mds.25349. [DOI] [PubMed] [Google Scholar]
- 219.Blanz J, Groth J, Zachos C, Wehling C, Saftig P, Schwake M. Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand β-glucocerebrosidase. Hum Mol Genet. 2010;19(4):563–572. doi: 10.1093/hmg/ddp523. [DOI] [PubMed] [Google Scholar]
- 220.Rothaug M, Zunke F, Mazzulli JR, Schweizer M, Altmeppen H, Lullmann-Rauch R, et al. LIMP-2 expression is critical for beta-glucocerebrosidase activity and alpha-synuclein clearance. Proc Natl Acad Sci U S A. 2014;111(43):15573–15578. doi: 10.1073/pnas.1405700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30(2):639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.van Eersel J, Ke YD, Gladbach A, Bi M, Gotz J, Kril JJ, et al. Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLoS One. 2011;6(7):e22850. doi: 10.1371/journal.pone.0022850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281(41):30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 224.Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68(11):1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 225.Teyssou E, Takeda T, Lebon V, Boillee S, Doukoure B, Bataillon G, et al. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathol. 2013;125(4):511–522. doi: 10.1007/s00401-013-1090-0. [DOI] [PubMed] [Google Scholar]
- 226.Peng H, Yang J, Li G, You Q, Han W, Li T, et al. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27(5):657–674. doi: 10.1038/cr.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gal J, Strom AL, Kilty R, Zhang F, Zhu H. p62 accumulates and enhances aggregate formation in model systems of familial amyotrophic lateral sclerosis. J Biol Chem. 2007;282(15):11068–11077. doi: 10.1074/jbc.M608787200. [DOI] [PubMed] [Google Scholar]
- 228.Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111(4):1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bandyopadhyay U, Nagy M, Fenton WA, Horwich AL. Absence of lipofuscin in motor neurons of SOD1-linked ALS mice. Proc Natl Acad Sci U S A. 2014;111(30):11055–11060. doi: 10.1073/pnas.1409314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chen Y, Liu H, Guan Y, Wang Q, Zhou F, Jie L, et al. The altered autophagy mediated by TFEB in animal and cell models of amyotrophic lateral sclerosis. Am J Transl Res. 2015;7(9):1574–1587. [PMC free article] [PubMed] [Google Scholar]
- 232.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ito D, Suzuki N. Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNAbinding proteins TDP-43 and FUS. Neurology. 2011;77(17):1636–1643. doi: 10.1212/WNL.0b013e3182343365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang X, Fan H, Ying Z, Li B, Wang H, Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci Lett. 2010;469(1):112–116. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 235.Ryu HH, Jun MH, Min KJ, Jang DJ, Lee YS, Kim HK, et al. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging. 2014;35(12):2822–2831. doi: 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 236.Brady OA, Meng P, Zheng Y, Mao Y, Hu F. Regulation of TDP-43 aggregation by phosphorylation and p62/SQSTM1. J Neurochem. 2011;116(2):248–259. doi: 10.1111/j.1471-4159.2010.07098.x. [DOI] [PubMed] [Google Scholar]
- 237.Wang IF, Guo BS, Liu YC, Wu CC, Yang CH, Tsai KJ, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A. 2012;109(37):15024–15029. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10(8):677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Xia Q, Wang H, Hao Z, Fu C, Hu Q, Gao F, et al. TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion. EMBO J. 2016;35(2):121–142. doi: 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bose JK, Huang CC, Shen CK. Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem. 2011;286(52):44441–44448. doi: 10.1074/jbc.M111.237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Deng Z, Purtell K, Lachance V, Wold MS, Chen S, Yue Z. Autophagy receptors and neurodegenerative diseases. Trends Cell Biol. 2017;27(7):491–504. doi: 10.1016/j.tcb.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 242.Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A. 2016;113(15):4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24(15):4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 244.Rubino E, Rainero I, Chio A, Rogaeva E, Galimberti D, Fenoglio P, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79(15):1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Liu Z, Li H, Hong C, Chen M, Yue T, Chen C, et al. ALS-associated E478G mutation in human OPTN (optineurin) promotes inflammation and induces neuronal cell death. Front Immunol. 2018;9:2647. doi: 10.3389/fimmu.2018.02647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23(13):3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Aoki Y, Manzano R, Lee Y, Dafinca R, Aoki M, Douglas AGL, et al. C9orf72 and RAB7L1 regulate vesicle trafficking in amyotrophic lateral sclerosis and frontotemporal dementia. Brain. 2017;140(4):887–897. doi: 10.1093/brain/awx024. [DOI] [PubMed] [Google Scholar]
- 249.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2016;2(9):e1601167. doi: 10.1126/sciadv.1601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Webster CP, Smith EF, Bauer CS, Moller A, Hautbergue GM, Ferraiuolo L, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016;35(15):1656–1676. doi: 10.15252/embj.201694401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ugolino J, Ji YJ, Conchina K, Chu J, Nirujogi RS, Pandey A, et al. Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 2016;12(11):e1006443. doi: 10.1371/journal.pgen.1006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Irwin DJ, Cairns NJ, Grossman M, McMillan CT, Lee EB, Van Deerlin VM, et al. Frontotemporal lobar degeneration: defining phenotypic diversity through personalized medicine. Acta Neuropathol. 2015;129(4):469–491. doi: 10.1007/s00401-014-1380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Olszewska DA, Lonergan R, Fallon EM, Lynch T. Genetics of frontotemporal dementia. Curr Neurol Neurosci Rep. 2016;16(12):107. doi: 10.1007/s11910-016-0707-9. [DOI] [PubMed] [Google Scholar]
- 255.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6(2):217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Urwin H, Authier A, Nielsen JE, Metcalf D, Powell C, Froud K, et al. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19(11):2228–2238. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Bayraktar O, Oral O, Kocaturk NM, Akkoc Y, Eberhart K, Kosar A, et al. IBMPFD disease-causing mutant VCP/p97 proteins are targets of autophagic-lysosomal degradation. PLoS One. 2016;11(10):e0164864. doi: 10.1371/journal.pone.0164864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Clayton EL, Milioto C, Muralidharan B, Norona FE, Edgar JR, Soriano A, et al. Frontotemporal dementia causative CHMP2B impairs neuronal endolysosomal traffic-rescue by TMEM106B knockdown. Brain. 2018;141(12):3428–3442. doi: 10.1093/brain/awy284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Clayton EL, Mizielinska S, Edgar JR, Nielsen TT, Marshall S, Norona FE, et al. Frontotemporal dementia caused by CHMP2B mutation is characterised by neuronal lysosomal storage pathology. Acta Neuropathol. 2015;130(4):511–523. doi: 10.1007/s00401-015-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90(6):1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zhou X, Paushter DH, Feng T, Sun L, Reinheckel T, Hu F. Lysosomal processing of progranulin. Mol Neurodegener. 2017;12(1):62. doi: 10.1186/s13024-017-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lee CW, Stankowski JN, Chew J, Cook CN, Lam YW, Almeida S, et al. The lysosomal protein cathepsin L is a progranulin protease. Mol Neurodegener. 2017;12(1):55. doi: 10.1186/s13024-017-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Valdez C, Wong YC, Schwake M, Bu G, Wszolek ZK, Krainc D. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet. 2017;26(24):4861–4872. doi: 10.1093/hmg/ddx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhou X, Paushter DH, Feng T, Pardon CM, Mendoza CS, Hu F. Regulation of cathepsin D activity by the FTLD protein progranulin. Acta Neuropathol. 2017;134(1):151–153. doi: 10.1007/s00401-017-1719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, et al. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 2014;127(6):845–860. doi: 10.1007/s00401-014-1262-6. [DOI] [PubMed] [Google Scholar]
- 266.Novak MJ, Tabrizi SJ. Huntington’s disease. BMJ. 2010;340:c3109. doi: 10.1136/bmj.c3109. [DOI] [PubMed] [Google Scholar]
- 267.Croce Katherine R., Yamamoto Ai. A role for autophagy in Huntington's disease. Neurobiology of Disease. 2019;122:16–22. doi: 10.1016/j.nbd.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Martin DD, Ladha S, Ehrnhoefer DE, Hayden MR. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 2015;38(1):26–35. doi: 10.1016/j.tins.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 269.MACDONALD M. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 270.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–558. doi: 10.1016/S0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 271.Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martinez-Sanchez A, Klein R, et al. In situ architecture and cellular interactions of polyQ inclusions. Cell. 2017;171(1):179–187. doi: 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 272.Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, et al. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci U S A. 2001;98(22):12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Mende-Mueller LM, Toneff T, Hwang SR, Chesselet MF, Hook VY. Tissue-specific proteolysis of Huntingtin (htt) in human brain: evidence of enhanced levels of N- and C-terminal htt fragments in Huntington’s disease striatum. J Neurosci. 2001;21(6):1830–1837. doi: 10.1523/JNEUROSCI.21-06-01830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lunkes A, Lindenberg KS, Ben-Haiem L, Weber C, Devys D, Landwehrmeyer GB, et al. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell. 2002;10(2):259–269. doi: 10.1016/S1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- 275.Bhat KP, Yan S, Wang CE, Li S, Li XJ. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc Natl Acad Sci U S A. 2014;111(15):5706–5711. doi: 10.1073/pnas.1402215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20(19):7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Petersen A, Larsen KE, Behr GG, Romero N, Przedborski S, Brundin P, et al. Expanded CAG repeats in exon 1 of the Huntington’s disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum Mol Genet. 2001;10(12):1243–1254. doi: 10.1093/hmg/10.12.1243. [DOI] [PubMed] [Google Scholar]
- 278.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 279.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13(5):567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wong YC, Holzbaur EL. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci. 2014;34(4):1293–1305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.del Toro D, Alberch J, Lazaro-Dieguez F, Martin-Ibanez R, Xifro X, Egea G, et al. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell. 2009;20(5):1478–1492. doi: 10.1091/mbc.e08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15(6):965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 283.Metzger S, Walter C, Riess O, Roos RA, Nielsen JE, Craufurd D, et al. The V471A polymorphism in autophagy-related gene ATG7 modifies age at onset specifically in Italian Huntington disease patients. PLoS One. 2013;8(7):e68951. doi: 10.1371/journal.pone.0068951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304(5667):100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 285.Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64(6):828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137(1):60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Erie C, Sacino M, Houle L, Lu ML, Wei J. Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur J Neurosci. 2015;42(3):1941–1951. doi: 10.1111/ejn.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Trajkovic K, Jeong H, Krainc D. Mutant Huntingtin is secreted via a late endosomal/lysosomal unconventional secretory pathway. J Neurosci. 2017;37(37):9000–9012. doi: 10.1523/JNEUROSCI.0118-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson’s disease. J Neurosci. 2010;30(3):1166–1175. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110(19):E1817–E1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Masini D, Bonito-Oliva A, Bertho M, Fisone G. Inhibition of mTORC1 signaling reverts cognitive and affective deficits in a mouse model of Parkinson’s disease. Front Neurol. 2018;9:208. doi: 10.3389/fneur.2018.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Siracusa R, Paterniti I, Cordaro M, Crupi R, Bruschetta G, Campolo M, et al. Neuroprotective effects of temsirolimus in animal models of Parkinson’s disease. Mol Neurobiol. 2018;55(3):2403–2419. doi: 10.1007/s12035-017-0496-4. [DOI] [PubMed] [Google Scholar]
- 293.Ghosh A, Tyson T, George S, Hildebrandt EN, Steiner JA, Madaj Z, et al. Mitochondrial pyruvate carrier regulates autophagy, inflammation, and neurodegeneration in experimental models of Parkinson’s disease. Sci Transl Med. 2016;8(368):368ra174. doi: 10.1126/scitranslmed.aag2210. [DOI] [PubMed] [Google Scholar]
- 294.Suresh SN, Chavalmane AK, Dj V, Yarreiphang H, Rai S, Paul A, et al. A novel autophagy modulator 6-Bio ameliorates SNCA/alpha-synuclein toxicity. Autophagy. 2017;13(7):1221–1234. doi: 10.1080/15548627.2017.1302045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Monti B, Gatta V, Piretti F, Raffaelli SS, Virgili M, Contestabile A. Valproic acid is neuroprotective in the rotenone rat model of Parkinson’s disease: involvement of alpha-synuclein. Neurotox Res. 2010;17(2):130–141. doi: 10.1007/s12640-009-9090-5. [DOI] [PubMed] [Google Scholar]
- 296.Rodriguez-Navarro JA, Rodriguez L, Casarejos MJ, Solano RM, Gomez A, Perucho J, et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010;39(3):423–438. doi: 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 297.Sarkar S, Chigurupati S, Raymick J, Mann D, Bowyer JF, Schmitt T, et al. Neuroprotective effect of the chemical chaperone, trehalose in a chronic MPTP-induced Parkinson’s disease mouse model. Neurotoxicology. 2014;44:250–262. doi: 10.1016/j.neuro.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 298.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTORindependent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282(8):5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 299.Kovács T, Billes V, Komlós M, Hotzi B, Manzéger A, Tarnóci A, et al. The small molecule AUTEN-99 (autophagy enhancer-99) prevents the progression of neurodegenerative symptoms. Sci Rep. 2017;7:42014. doi: 10.1038/srep42014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Bourdenx M, Daniel J, Genin E, Soria FN, Blanchard-Desce M, Bezard E, et al. Nanoparticles restore lysosomal acidification defects: implications for Parkinson and other lysosomal-related diseases. Autophagy. 2016;12(3):472–483. doi: 10.1080/15548627.2015.1136769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Migdalska-Richards A, Daly L, Bezard E, Schapira AH. Ambroxol effects in glucocerebrosidase and alpha-synuclein transgenic mice. Ann Neurol. 2016;80(5):766–775. doi: 10.1002/ana.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Migdalska-Richards A, Ko WKD, Li Q, Bezard E, Schapira AHV. Oral ambroxol increases brain glucocerebrosidase activity in a nonhuman primate. Synapse (New York, NY) 2017;71(7):e21967. doi: 10.1002/syn.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Richter F, Fleming SM, Watson M, Lemesre V, Pellegrino L, Ranes B, et al. A GCase chaperone improves motor function in a mouse model of synucleinopathy. Neurotherapeutics. 2014;11(4):840–856. doi: 10.1007/s13311-014-0294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Sanchez-Martinez A, Beavan M, Gegg ME, Chau KY, Whitworth AJ, Schapira AH. Parkinson disease-linked GBA mutation effects reversed by molecular chaperones in human cell and fly models. Sci Rep. 2016;6:31380. doi: 10.1038/srep31380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Mazzulli JR, Zunke F, Tsunemi T, Toker NJ, Jeon S, Burbulla LF, et al. Activation of beta-glucocerebrosidase reduces pathological alpha-synuclein and restores lysosomal function in Parkinson’s patient midbrain neurons. J Neurosci. 2016;36(29):7693–7706. doi: 10.1523/JNEUROSCI.0628-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Aflaki E, Borger DK, Moaven N, Stubblefield BK, Rogers SA, Patnaik S, et al. A new glucocerebrosidase chaperone reduces alpha-synuclein and glycolipid levels in iPSC-derived dopaminergic neurons from patients with Gaucher disease and Parkinsonism. J Neurosci. 2016;36(28):7441–7452. doi: 10.1523/JNEUROSCI.0636-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Charvin Delphine, Medori Rossella, Hauser Robert A., Rascol Olivier. Therapeutic strategies for Parkinson disease: beyond dopaminergic drugs. Nature Reviews Drug Discovery. 2018;17(11):804–822. doi: 10.1038/nrd.2018.136. [DOI] [PubMed] [Google Scholar]
- 308.Sardi SP, Clarke J, Kinnecom C, Tamsett TJ, Li L, Stanek LM, et al. CNS expression of glucocerebrosidase corrects alpha-synuclein pathology and memory in a mouse model of Gaucher-related synucleinopathy. Proc Natl Acad Sci U S A. 2011;108(29):12101–12106. doi: 10.1073/pnas.1108197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Sardi SP, Viel C, Clarke J, Treleaven CM, Richards AM, Park H, et al. Glucosylceramide synthase inhibition alleviates aberrations in synucleinopathy models. Proc Natl Acad Sci. 2017;114(10):2699. doi: 10.1073/pnas.1616152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Li L, Zhang S, Zhang X, Li T, Tang Y, Liu H, et al. Autophagy enhancer carbamazepine alleviates memory deficits and cerebral amyloid-beta pathology in a mouse model of Alzheimer’s disease. Curr Alzheimer Res. 2013;10(4):433–441. doi: 10.2174/1567205011310040008. [DOI] [PubMed] [Google Scholar]
- 311.Steele JW, Kim SH, Cirrito JR, Verges DK, Restivo JL, Westaway D, et al. Acute dosing of latrepirdine (Dimebon), a possible Alzheimer therapeutic, elevates extracellular amyloid-beta levels in vitro and in vivo. Mol Neurodegener. 2009;4:51. doi: 10.1186/1750-1326-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Steele JW, Lachenmayer ML, Ju S, Stock A, Liken J, Kim SH, et al. Latrepirdine improves cognition and arrests progression of neuropathology in an Alzheimer’s mouse model. Mol Psychiatry. 2013;18(8):889–897. doi: 10.1038/mp.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhang L, Wang L, Wang R, Gao Y, Che H, Pan Y, et al. Evaluating the effectiveness of GTM-1, rapamycin, and carbamazepine on autophagy and alzheimer disease. Med Sci Monit. 2017;23:801–808. doi: 10.12659/MSM.898679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Matsunaga S, Kishi T, Annas P, Basun H, Hampel H, Iwata N. Lithium as a treatment for Alzheimer’s disease: a systematic review and meta-analysis. J Alzheimers Dis. 2015;48(2):403–410. doi: 10.3233/JAD-150437. [DOI] [PubMed] [Google Scholar]
- 315.Vingtdeux V, Giliberto L, Zhao H, Chandakkar P, Wu Q, Simon JE, et al. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-beta peptide metabolism. J Biol Chem. 2010;285(12):9100–9113. doi: 10.1074/jbc.M109.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Chiang MC, Nicol CJ, Cheng YC. Resveratrol activation of AMPK-dependent pathways is neuroprotective in human neural stem cells against amyloid-beta-induced inflammation and oxidative stress. Neurochem Int. 2018;115:1–10. doi: 10.1016/j.neuint.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 317.Turner RS, Thomas RG, Craft S, van Dyck CH, Mintzer J, Reynolds BA, et al. A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology. 2015;85(16):1383–1391. doi: 10.1212/WNL.0000000000002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Moussa C, Hebron M, Huang X, Ahn J, Rissman RA, Aisen PS, et al. Resveratrol regulates neuro-inflammation and induces adaptive immunity in Alzheimer’s disease. J Neuroinflammation. 2017;14(1):1. doi: 10.1186/s12974-016-0779-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Robinson DM, Keating GM. Memantine: a review of its use in Alzheimer’s disease. Drugs. 2006;66(11):1515–1534. doi: 10.2165/00003495-200666110-00015. [DOI] [PubMed] [Google Scholar]
- 320.van Marum RJ. Update on the use of memantine in Alzheimer’s disease. Neuropsychiatr Dis Treat. 2009;5:237–247. doi: 10.2147/NDT.S4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Kickstein E, Krauss S, Thornhill P, Rutschow D, Zeller R, Sharkey J, et al. Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proc Natl Acad Sci U S A. 2010;107(50):21830–21835. doi: 10.1073/pnas.0912793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Barini E, Antico O, Zhao Y, Asta F, Tucci V, Catelani T, et al. Metformin promotes tau aggregation and exacerbates abnormal behavior in a mouse model of tauopathy. Mol Neurodegener. 2016;11:16. doi: 10.1186/s13024-016-0082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Castillo K, Nassif M, Valenzuela V, Rojas F, Matus S, Mercado G, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9(9):1308–1320. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 324.Wang Y, Liu FT, Wang YX, Guan RY, Chen C, Li DK, et al. Autophagic modulation by trehalose reduces accumulation of TDP-43 in a cell model of amyotrophic lateral sclerosis via TFEB activation. Neurotox Res. 2018;34(1):109–120. doi: 10.1007/s12640-018-9865-7. [DOI] [PubMed] [Google Scholar]
- 325.Zhang X, Chen S, Song L, Tang Y, Shen Y, Jia L, et al. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy. 2014;10(4):588–602. doi: 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Mancuso R, del Valle J, Modol L, Martinez A, Granado-Serrano AB, Ramirez-Nunez O, et al. Resveratrol improves motoneuron function and extends survival in SOD1(G93A) ALS mice. Neurotherapeutics. 2014;11(2):419–432. doi: 10.1007/s13311-013-0253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, et al. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10(2):148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 328.Chiu CT, Liu G, Leeds P, Chuang DM. Combined treatment with the mood stabilizers lithium and valproate produces multiple beneficial effects in transgenic mouse models of Huntington’s disease. Neuropsychopharmacology. 2011;36(12):2406–2421. doi: 10.1038/npp.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Pouladi MA, Brillaud E, Xie Y, Conforti P, Graham RK, Ehrnhoefer DE, et al. NP03, a novel low-dose lithium formulation, is neuroprotective in the YAC128 mouse model of Huntington disease. Neurobiol Dis. 2012;48(3):282–289. doi: 10.1016/j.nbd.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 330.Rose C, Menzies FM, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O, et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington's disease. Hum Mol Genet. 2010;19(11):2144–2153. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Garriga-Canut M, Agustin-Pavon C, Herrmann F, Sanchez A, Dierssen M, Fillat C, et al. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc Natl Acad Sci U S A. 2012;109(45):E3136–E3145. doi: 10.1073/pnas.1206506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 editing of the mutant Huntingtin allele in vitro and in vivo. Mol Ther. 2017;25(1):12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Tramutola A, Lanzillotta C, Barone E, Arena A, Zuliani I, Mosca L, et al. Intranasal rapamycin ameliorates Alzheimer-like cognitive decline in a mouse model of Down syndrome. Transl Neurodegener. 2018;7:28. doi: 10.1186/s40035-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Van Skike CE, Jahrling JB, Olson AB, Sayre NL, Hussong SA, Ungvari Z, et al. Inhibition of mTOR protects the blood-brain barrier in models of Alzheimer’s disease and vascular cognitive impairment. Am J Physiol Heart Circ Physiol. 2018;314(4):H693–H703. doi: 10.1152/ajpheart.00570.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Lin AL, Zheng W, Halloran JJ, Burbank RR, Hussong SA, Hart MJ, et al. Chronic rapamycin restores brain vascular integrity and function through NO synthase activation and improves memory in symptomatic mice modeling Alzheimer’s disease. J Cereb Blood Flow Metab. 2013;33(9):1412–1421. doi: 10.1038/jcbfm.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Lin AL, Jahrling JB, Zhang W, DeRosa N, Bakshi V, Romero P, et al. Rapamycin rescues vascular, metabolic and learning deficits in apolipoprotein E4 transgenic mice with pre-symptomatic Alzheimer’s disease. J Cereb Blood Flow Metab. 2017;37(1):217–226. doi: 10.1177/0271678X15621575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Harkavyi A, Abuirmeileh A, Lever R, Kingsbury AE, Biggs CS, Whitton PS. Glucagon-like peptide 1 receptor stimulation reverses key deficits in distinct rodent models of Parkinson’s disease. J Neuroinflammation. 2008;5(1):19. doi: 10.1186/1742-2094-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Bertilsson G, Patrone C, Zachrisson O, Andersson A, Dannaeus K, Heidrich J, et al. Peptide hormone exendin-4 stimulates subventricular zone neurogenesis in the adult rodent brain and induces recovery in an animal model of Parkinson’s disease. J Neurosci Res. 2008;86(2):326–338. doi: 10.1002/jnr.21483. [DOI] [PubMed] [Google Scholar]
- 340.Athauda D, Maclagan K, Skene SS, Bajwa-Joseph M, Letchford D, Chowdhury K, et al. Exenatide once weekly versus placebo in Parkinson's disease: a randomised, double-blind, placebocontrolled trial. Lancet (London, England) 2017;390(10103):1664–1675. doi: 10.1016/S0140-6736(17)31585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Karuppagounder SS, Brahmachari S, Lee Y, Dawson VL, Dawson TM, Ko HS. The c-Abl inhibitor, nilotinib, protects dopaminergic neurons in a preclinical animal model of Parkinson’s disease. Sci Rep. 2014;4:4874. doi: 10.1038/srep04874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Hebron ML, Lonskaya I, Moussa CE. Tyrosine kinase inhibition facilitates autophagic SNCA/alpha-synuclein clearance. Autophagy. 2013;9(8):1249–1250. doi: 10.4161/auto.25368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Hebron M. L., Lonskaya I., Moussa C. E.- H. Nilotinib reverses loss of dopamine neurons and improves motor behavior via autophagic degradation of -synuclein in Parkinson's disease models. Human Molecular Genetics. 2013;22(16):3315–3328. doi: 10.1093/hmg/ddt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Pagan F, Hebron M, Valadez EH, Torres-Yaghi Y, Huang X, Mills RR, et al. Nilotinib effects in Parkinson’s disease and dementia with Lewy bodies. J Park Dis. 2016;6(3):503–517. doi: 10.3233/JPD-160867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Siddiqui A., Bhaumik D., Chinta S. J., Rane A., Rajagopalan S., Lieu C. A., Lithgow G. J., Andersen J. K. Mitochondrial Quality Control via the PGC1 -TFEB Signaling Pathway Is Compromised by Parkin Q311X Mutation But Independently Restored by Rapamycin. Journal of Neuroscience. 2015;35(37):12833–12844. doi: 10.1523/JNEUROSCI.0109-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Menzies FM, Garcia-Arencibia M, Imarisio S, O'Sullivan NC, Ricketts T, Kent BA, et al. Calpain inhibition mediates autophagy-dependent protection against polyglutamine toxicity. Cell Death Differ. 2015;22(3):433–444. doi: 10.1038/cdd.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Getaw Worku Hassen, Leo Kesner, Alfred Stracher, Abraham Shulman, Edward Rockenstein, Michael Mante, Anthony Adame, Cassia Overk, Robert A. Rissman, Eliezer Masliah. Effects of Novel Calpain Inhibitors in Transgenic Animal Model of Parkinson’s disease/dementia with Lewy bodies. Sci Rep 2018;8(1). [DOI] [PMC free article] [PubMed]
- 348.Hewitt G, Korolchuk VI. Repair, reuse, recycle: the expanding role of autophagy in genome maintenance. Trends Cell Biol. 2017;27(5):340–351. doi: 10.1016/j.tcb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 349.Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–116. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19(3):170–183. doi: 10.1038/s41577-018-0095-2. [DOI] [PubMed] [Google Scholar]
- 351.Bove J, Martinez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12(8):437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 352.Zhou B, Kreuzer J, Kumsta C, Wu L, Kamer KJ, Cedillo L, et al. Mitochondrial permeability uncouples elevated autophagy and lifespan extension. Cell. 2019;177(2):299–314. doi: 10.1016/j.cell.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Xilouri M, Brekk OR, Landeck N, Pitychoutis PM, Papasilekas T, Papadopoulou-Daifoti Z, et al. Boosting chaperone-mediated autophagy in vivo mitigates α-synuclein-induced neurodegeneration. Brain. 2013;136(7):2130–2146. doi: 10.1093/brain/awt131. [DOI] [PubMed] [Google Scholar]
- 354.Xilouri M, Brekk OR, Polissidis A, Chrysanthou-Piterou M, Kloukina I, Stefanis L. Impairment of chaperone-mediated autophagy induces dopaminergic neurodegeneration in rats. Autophagy. 2016;12(11):2230–2247. doi: 10.1080/15548627.2016.1214777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Arrant AE, Onyilo VC, Unger DE, Roberson ED. Progranulin gene therapy improves lysosomal dysfunction and microglial pathology associated with frontotemporal dementia and neuronal ceroid lipofuscinosis. J Neurosci. 2018;38(9):2341–2358. doi: 10.1523/JNEUROSCI.3081-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Rocha EM, Smith GA, Park E, Cao H, Brown E, Hayes MA, et al. Glucocerebrosidase gene therapy prevents α-synucleinopathy of midbrain dopamine neurons. Neurobiol Dis. 2015;82:495–503. doi: 10.1016/j.nbd.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 357.Steet RA, Chung S, Wustman B, Powe A, Do H, Kornfeld SA. The iminosugar isofagomine increases the activity of N370S mutant acid beta-glucosidase in Gaucher fibroblasts by several mechanisms. Proc Natl Acad Sci U S A. 2006;103(37):13813–13818. doi: 10.1073/pnas.0605928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Valenzano KJ, Khanna R, Powe AC, Boyd R, Lee G, Flanagan JJ, et al. Identification and characterization of pharmacological chaperones to correct enzyme deficiencies in lysosomal storage disorders. Assay and Drug Development Technologies. 2011;9(3):213–235. doi: 10.1089/adt.2011.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Patnaik S, Zheng W, Choi JH, Motabar O, Southall N, Westbroek W, et al. Discovery, structure-activity relationship, and biological evaluation of noninhibitory small molecule chaperones of glucocerebrosidase. J Med Chem. 2012;55(12):5734–5748. doi: 10.1021/jm300063b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, et al. Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Abeta generation and amyloid plaque pathogenesis. J Neurosci. 2015;35(35):12137–12151. doi: 10.1523/JNEUROSCI.0705-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Wild EJ, Tabrizi SJ. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017;16(10):837–847. doi: 10.1016/S1474-4422(17)30280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Tabrizi S, Leavitt B, Kordasiewicz H, Czech C, Swayze E, Norris DA, et al. Effects of IONIS-HTTRx in patients with early Huntington’s disease, results of the first HTT-lowering drug trial (CT.002) Neurology. 2018;90(15 Supplement):CT.002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 460 kb)