Although the high-risk human papillomavirus 16 infects anogenital and oropharyngeal sites, the cervical epithelium has a unique vulnerability to progression of cancer. Host responses during persistent infection and preneoplastic stages can shape the outcome of cancer progression in a tissue-dependent manner. Our study for the first time reports differential regulation of critical cellular functions and signaling pathways during productive HPV16 infection of cervical, foreskin, and tonsil tissues. While the virus induces hyperproliferation in infected cells, it downregulates epithelial differentiation, epidermal development, and innate immune responses, according to the tissue type. Modulation of these biological functions can determine virus fitness and pathogenesis and illuminate key cellular mechanisms that the virus employs to establish persistence and finally initiate disease progression.
KEYWORDS: HPV16, human papillomavirus, epithelial differentiation, gene expression, immune response, tissue specific
ABSTRACT
Epidemiological data confirm a much higher incidence of high-risk human papillomavirus 16 (HPV16)-mediated carcinogenesis of the cervical epithelium than for other target sites. In order to elucidate tissue-specific responses to virus infection, we compared gene expression changes induced by productive HPV16 infection of cervical, foreskin, and tonsil organotypic rafts. These rafts closely mimic persistent HPV16 infection, long before carcinogenesis sets in. The total number of gene expression changes varied considerably across the tissue types, with only 32 genes being regulated in common. Among them, we confirmed the Kelch-like family protein KLHL35 and the laminin-5 complex to be upregulated and downregulated, respectively, in all the three tissues. HPV16 infection induces upregulation of genes involved in cell cycle control, cell division, mitosis, DNA replication, and DNA damage repair in all the three tissues, indicative of a hyperproliferative environment. In the cervical and tonsil epithelium, we observe significant downregulation of genes involved in epidermis development, keratinocyte differentiation, and extracellular matrix organization. On the other hand, in HPV16-positive foreskin (HPV16 foreskin) tissue, several genes involved in interferon-mediated innate immunity, cytokine signaling, and cellular defenses were downregulated. Furthermore, pathway analysis and experimental validations identified important cellular pathways like STAT1 and transforming growth factor β (TGF-β) to be differentially regulated among the three tissue types. The differential modulation of important cellular pathways like TGF-β1 and STAT1 can explain the sensitivity of tissues to HPV cancer progression.
IMPORTANCE Although the high-risk human papillomavirus 16 infects anogenital and oropharyngeal sites, the cervical epithelium has a unique vulnerability to progression of cancer. Host responses during persistent infection and preneoplastic stages can shape the outcome of cancer progression in a tissue-dependent manner. Our study for the first time reports differential regulation of critical cellular functions and signaling pathways during productive HPV16 infection of cervical, foreskin, and tonsil tissues. While the virus induces hyperproliferation in infected cells, it downregulates epithelial differentiation, epidermal development, and innate immune responses, according to the tissue type. Modulation of these biological functions can determine virus fitness and pathogenesis and illuminate key cellular mechanisms that the virus employs to establish persistence and finally initiate disease progression.
INTRODUCTION
Human papillomaviruses (HPVs) cause more than 5% of all human cancers. HPV infections account for the most prevalent sexually transmitted infections in the United States (1). High-risk HPV16 and -18 are responsible for most HPV-associated cancers (2, 3). Besides cervical cancers, high-risk HPVs attribute to 50% of penile, 88% of anal, 43% of vulvar, 70% of vaginal, and 70% of oropharyngeal cancers (4). Epidemiological data repeatedly indicate that the cervix has a unique vulnerability and susceptibility to HPV-mediated carcinogenesis (5, 6). HPV-positive oropharyngeal cancers form a distinct clinical entity with unique epidemiology, prognostic factors, and responses to radiotherapy. Tonsils and the base of the tongue are the most affected sites in the head and neck regions, with HPV16 causing more than 85% of tumors (7). Tissue-specific differences could be attributed to epithelial cell subtypes, innate immune responses, and organ-specific microbiota, to name a few. Among patients with cervical cancer and HPV-associated oropharyngeal squamous cell carcinoma (OPSCC), there are clear differences in infiltrating lymphocyte populations (8). Furthermore, in HPV-positive OPSCC, a more effective antigen-specific T cell response was strongly related to improved survival, compared to HPV-negative OPSCCs (9).
The aim of our study is to identify tissue-specific responses in the cervix, tonsil, and foreskin that may play a critical role in the establishment of persistence during the productive stages of HPV16 infection. Previous studies have investigated high-risk HPV-mediated gene expression changes and transcriptome changes in neoplastic or invasive tumor samples from cervical cancer and head and neck cancer patients (10–20). At these later stages of disease progression, multiple cellular pathways are dysregulated, leading to a total loss in differentiation along with growth factor-independent proliferation. The part that has been missing from the puzzle is changes during productive HPV16 infection at the preneoplastic stage. Microarray analysis was carried out on organotypic raft cultures with or without HPV16 infection, obtained from cervical, foreskin, and tonsil tissues. This model system allows us to monitor the HPV life cycle in three-dimensional differentiating epithelia, which very closely mimic natural infection (21–24).
Our previous publication discusses the gene expression changes during productive HPV16 infection of the cervical tissue (25). In this paper, we perform a comparative analysis of gene expression changes induced by productive HPV16 infection of cervical, foreskin, and tonsil tissues. In all three tissues, we observe upregulation of genes involved in cell cycle regulation, cell division, DNA replication, and damage repair, indicating a hyperproliferative environment. However, we see more differences among the downregulated genes in the three tissue types. While there were several genes with function in epidermal differentiation, extracellular matrix (ECM) integrity was dampened in cervical and tonsil tissues; in the foreskin, gene categories related to immune responses, cellular defenses, and the interferon response were downregulated. Additionally, we see downregulation of important upstream regulators like transforming growth factor β (TGF-β) and STAT1 in HPV16-positive cervical and foreskin tissues, respectively. The differential regulation of key cellular pathways and regulatory networks during productive stages of infection can determine the sensitivity of the tissue to oncogenic progression.
RESULTS
Differential changes in gene expression during productive HPV16 infection of cervical, foreskin, and tonsil epithelium.
Raft tissues harvested at 10 days (day 10) in culture were used for the gene expression microarray analysis. Around this time, virus production is at its maximum, and all the late genes are expressed (26). Therefore, the gene expression changes observed at this stage reflect the effect of the replicating virus on epithelial gene regulation. Productive HPV16 infection induces differential changes in gene expression in cervical, foreskin, and tonsil tissues (Fig. 1). HPV16-positive foreskin (HPV16 foreskin) tissue had the highest number of changes, with 693 genes upregulated and 832 genes downregulated. In HPV16 cervical tissue, 594 genes were upregulated, and 651 genes were downregulated. In contrast, only 78 genes were upregulated and 88 genes were downregulated in HPV16 tonsil tissue. We observe larger numbers of downregulated genes with higher fold changes (FCs) in each tissue type, indicating an overall suppressive effect of HPV16 on the host transcriptome. The average pairwise Pearson coefficient for primary cervical sample replicates was 0.885, that for HPV16 cervical samples was 0.941, that for primary foreskin samples was 0.988, that for HPV16 foreskin samples was 0.994, that for primary tonsil samples was 0.876, and that for HPV16 tonsil samples was 0.890.
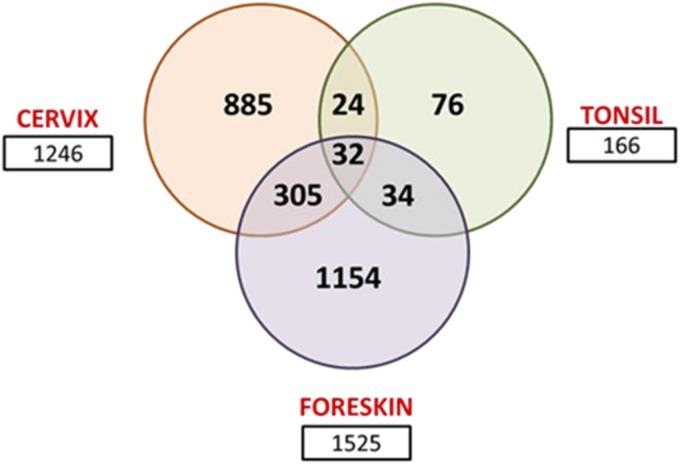
FIG 1.
Venn diagram showing total numbers of gene expression changes and overlapping changes in HPV16-positive cervix, foreskin, and tonsil. For microarray analysis of each tissue type, three primary cell lines and three HPV16-transformed cell lines were used to grow rafts. Each of these experiments was repeated at separate times, representing a total of six individually grown raft tissue for primary (n = 6) and HPV16-positive (n = 6) cell lines. Only gene expression changes of more than 1.5-fold and with a P value of <0.05 were considered significant for the analysis.
In HPV16 cervical tissue, the top upregulated genes have diverse functions, from metal binding, cell signaling, and cellular metabolism to transcriptional regulation. Genes with functional roles in epithelial differentiation, ECM assembly, cell adhesion, and immune responses were downregulated severalfold (Table 1). The top most upregulated genes in HPV16 foreskin have roles in cytoskeletal structure, ECM assembly, metabolic pathways, and cell proliferation. However, the most downregulated genes have roles in the immune response and cellular defenses against infection, indicating considerable suppression of the cellular defense mechanism (Table 1). In the infected tonsil epithelium, the top most upregulated genes have roles in cell proliferation, transcription, and metabolism. The maximally downregulated genes have functions in maintenance of ECM structure, cell-cell adhesion, and differentiation (Table 1).
TABLE 1.
Top 10 upregulated and downregulated genes in persistently HPV16-infected cervical rafts, foreskin rafts, and tonsil raftsa
| Top regulated gene | FC | Function(s) |
|---|---|---|
| Genes in HPV16-infected cervix rafts | ||
| Upregulated | ||
| MT1G | 12.03 | Heavy metal binding protein containing high no. of cysteine residues |
| MT1H | 7.7 | Heavy metal binding protein containing high no. of cysteine residues |
| TGFβR3 | 7.3 | Presents TGF-β to its receptor |
| KLHL35 | 5.81 | Kelch-like family of proteins |
| DLK2 | 5.1 | Adipogenesis |
| FBLN1 | 5.1 | Cell adhesion, migration |
| ASS1 | 4.45 | Urea cycle |
| GPER | 4.45 | Rapid estrogen signaling |
| TDRD9 | 4.4 | RNA helicase |
| TWIST1 | 4.21 | Transcriptional regulator |
| Downregulated | ||
| RPTN | −57.5 | Cornified cell envelope formation |
| SERPINB4 | −48.01 | Immune response |
| TCN1 | −34.74 | Binds vitamin B12 |
| CST6 | −27.8 | Inhibits cathepsin B |
| KLK8 | −25.8 | Peptidase |
| CEACAM6 | −24.5 | Cell adhesion |
| KLK6 | −22.1 | Peptidase |
| KLK13 | −21.5 | Peptidase |
| RNASE7 | −20.1 | Antimicrobial activity |
| PRSS3 | −20 | Defensin processing |
| Genes in HPV16-infected foreskin rafts | ||
| Upregulated | ||
| CA9 | 8.1 | pH regulation |
| NEFH | 7.7 | Neurofilament |
| DKN2A | 7.64 | Inhibits cell proliferation |
| TPM1 | 6.45 | Binds actin |
| LOX | 5.84 | ECM cross-linking |
| KLHL35 | 5.7 | Kelch-like family |
| BAIAP2L2 | 5.12 | Binds phosphoinositide |
| SPRR3 | 4.72 | Proline-rich protein |
| KCNS1 | 4.65 | Ion channel |
| ALDOC | 4.5 | Glycolysis |
| Downregulated | ||
| KRT2 | −71.05 | Keratinocyte activation |
| MX1 | −50.3 | Antiviral activity |
| HSPB8 | −50.01 | Chaperone |
| IFI27 | −49.51 | Promotes cell death |
| LCE3C | −26.16 | Formation of cornified envelope |
| DEFB4 | −22.5 | Antibacterial activity |
| OAS2 | −21.23 | Antiviral enzyme |
| IFIT1 | −14.74 | Antiviral RNA binding protein |
| IFI6 | −14.64 | Interferon inducible |
| AADAC | −12.26 | Hydrolase, lipase |
| Genes in HPV16-infected tonsil rafts | ||
| Upregulated | ||
| ARHGAP15 | 4.6 | Activates GTPase |
| PITX2 | 3.12 | Cell proliferation |
| ENO2 | 3.1 | Cell survival |
| KLHL35 | 3 | Kelch-like family |
| GSTT2 | 2.4 | Glutathione synthesis |
| TDRD9 | 2.32 | RNA helicase |
| NMU | 2.23 | Muscle contraction |
| GOLGA8B | 2.2 | Golgi structure |
| CENPQ | 2.12 | Centromere protein |
| E2F2 | 2 | Transcription activator |
| Downregulated | ||
| SAA1 | −4.8 | Serum amyloid |
| TRNP1 | −3.3 | Cell cycle |
| LPXN | −2.91 | Cell adhesion |
| CTGF | −2.8 | Differentiation |
| THBS2 | −2.61 | ECM interactions |
| LAMC2 | −2.6 | Attachment, cell organization |
| ANKRD38 | −2.5 | Cytoskeletal formation |
| SH3KBP1 | −2.4 | Lysosomal degradation |
| SNCG | −2.4 | Keratin network |
| SAA4 | −2.34 | Serum amyloid |
FC, microarray fold change.
Commonly regulated genes upon productive HPV16 infection of cervix, foreskin, and tonsil tissues.
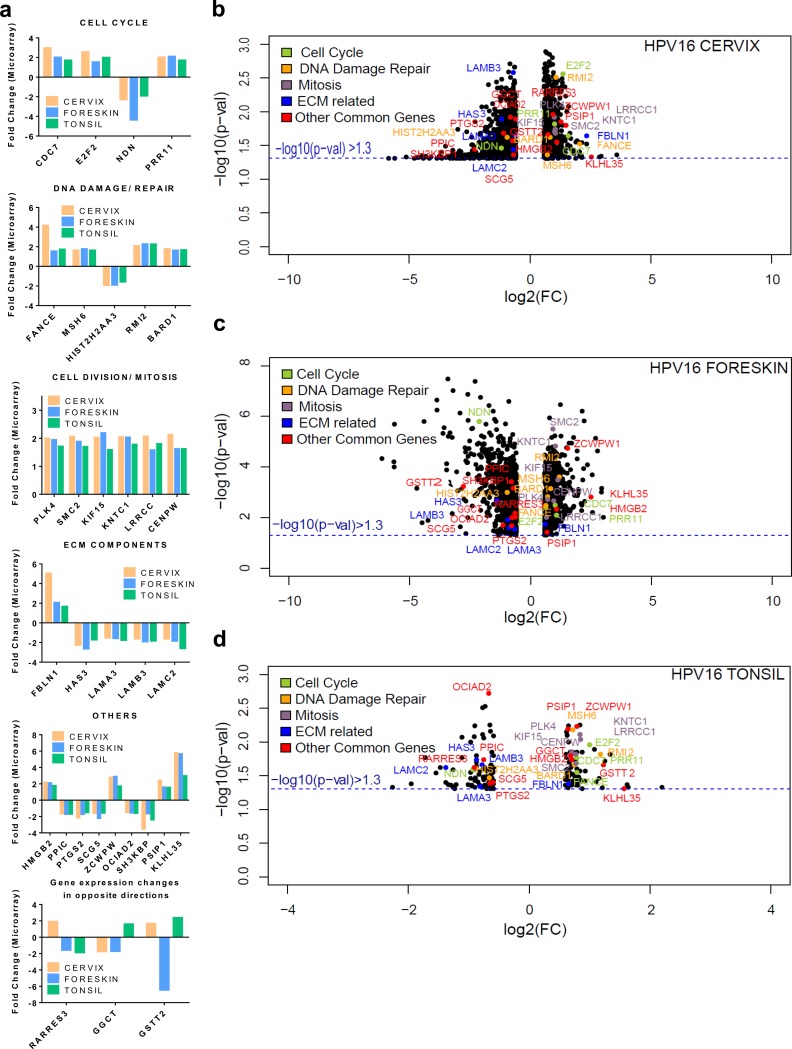
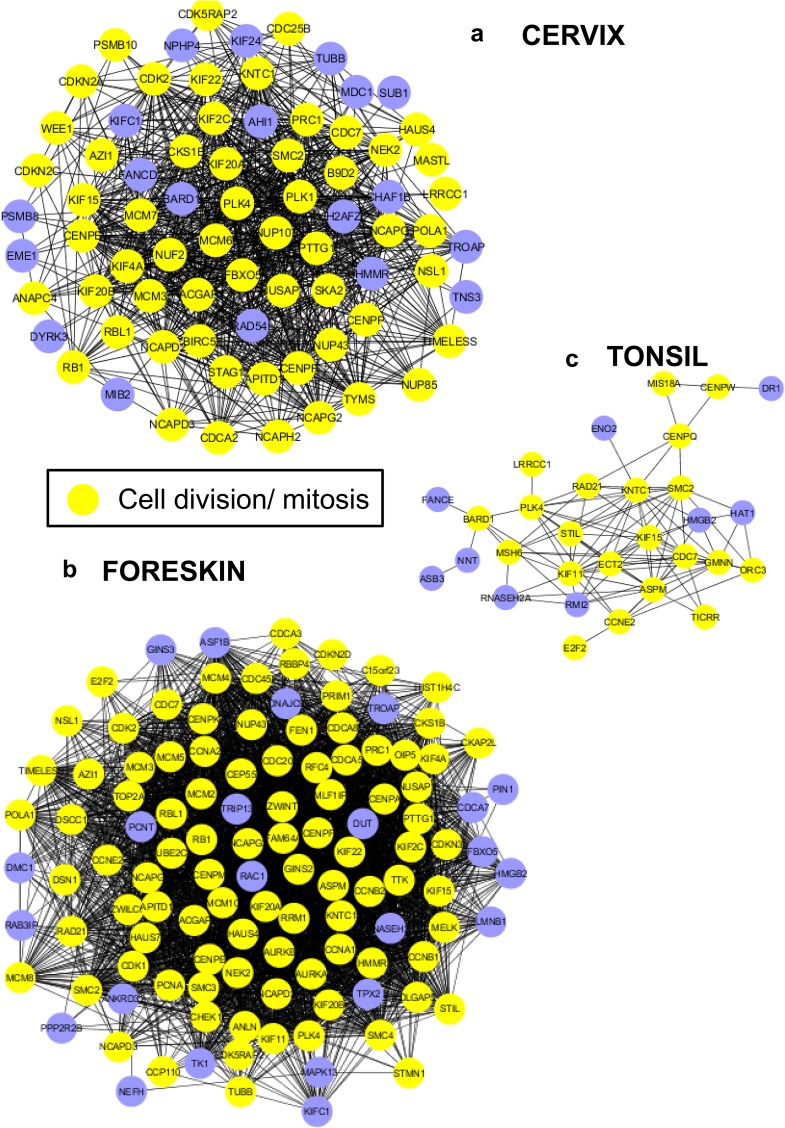
Oddly enough, only 32 genes were commonly regulated among the three tissue types. While 29 out of the 32 genes are similarly regulated in the three tissue types, 3 genes are modulated in opposing directions. Fifteen out of the 32 genes have roles in cell cycle regulation, cell division, mitosis, and DNA damage repair (Fig. 2a). Four genes have functions related to ECM maintenance and integrity. Volcano plots indicating the −log10 P value and log2 FC of significantly regulated genes are represented for each tissue type (Fig. 2b to d). The volcano plot provides a visual display of the both the FC and P value obtained from the t test, comparing uninfected and HPV16-positive tissues. Only genes with an absolute FC of ≥1.5 and a P value of <0.05 (−log10 P value of >1.3) are included in the plot, for ease of representation. The commonly regulated genes are highlighted in colors, based on their cellular functions. The majority of the genes with functions in the cell cycle, DNA damage repair, and mitosis are upregulated in all three tissue types, while 3 out of the 4 ECM-related genes are downregulated in common.
FIG 2.
Common genes regulated during productive HPV16 infection of cervical, foreskin, and tonsil epithelium. (a) Histograms representing gene expression fold changes of genes commonly regulated. We observe genes belonging to biological functions of the cell cycle, DNA damage repair, and cell division to be upregulated in all the tissues during productive infection. (b to d) Volcano plots representing log2 fold changes (x axis) plotted against the −log10 P value (y axis), obtained from CLC Genomics Workbench 4.8 package, in HPV16-positive cervical, foreskin, and tonsil rafts. The black scatter points represent all the significantly regulated genes in each tissue type. The commonly regulated genes are color-coded according to their biological functions, as indicated in the key.
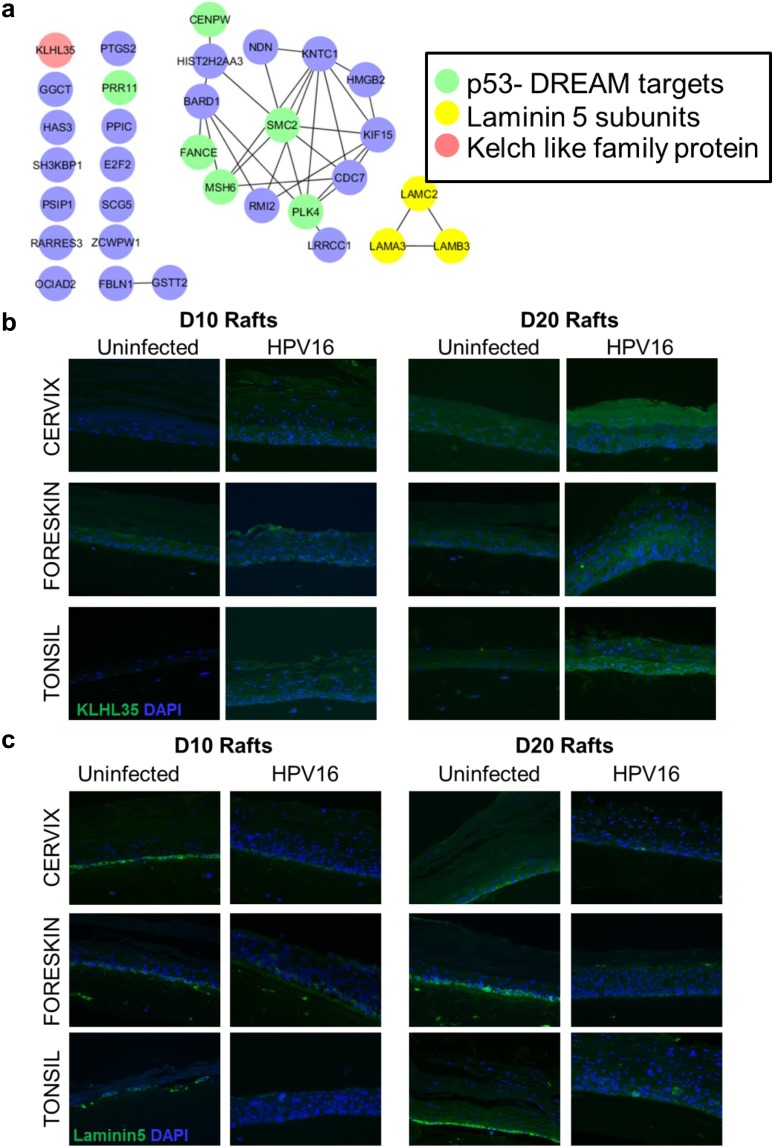
HPV16 infection results in upregulation of the p53-DREAM targets in all three tissue types (Fig. 3a). The HPV16 oncoprotein E7 disrupts the p53-DREAM repressor complex, thereby resulting in the upregulation of several cell cycle-related genes and abrogation of p53-mediated cell cycle control (27, 28). For the common genes, protein interaction networks using the STRING DB reveal a highly connected network within genes involved in cell division, mitosis, and the DNA damage response (DDR), with SMC2 acting as a central node (Fig. 3a). Another member of the family, SMC1, was shown to be critical for high-risk HPV31 genome amplification (29). We report for the first time upregulation of SMC2 in productive HPV16 infection.
FIG 3.
Productive HPV16 infection results in upregulation of KLHL35 and downregulation of the laminin-5 complex in cervical, foreskin, and tonsil rafts. (a) Protein-protein network of commonly regulated genes with productive HPV16 infection in cervix, foreskin, and tonsil tissue. Highlighted are the p53-DREAM pathway targets, KHLH35, and laminin-5 subunits. The STRING database and Cytoscape were used to derive the interacting network. (b) Immunofluorescence reveals increased expression of KLHL35 (green) in HPV16-infected cervical, foreskin, and raft tissues harvested at day 10 and day 20. DAPI (4′,6-diamidino-2-phenylindole) (blue) was used to stain the nuclei. (c) Immunofluorescence reveals decreased expression of the laminin-5 complex (green) in the basal layer cells of HPV16-infected cervical, foreskin, and tonsil rafts harvested at day 10 and day 20. DAPI (blue) was used to stain the nuclei. Images were acquired using a Nikon Eclipse 80i microscope and NIS Elements (v4.4) software at a ×200 magnification.
The Kelch-like family gene KLHL35 was highly upregulated in all three tissue types. The function of KLHL35 in epithelial tissue is unclear. However, its close paralogue KLHL24 is involved in maintenance of mechanical stability of skin and turnover of intermediate filaments, mainly keratin 14 (KRT14) (30–33). Immunofluorescence experiments confirmed the upregulation of the KLHL35 protein in HPV16 cervical, foreskin, and tonsil rafts (Fig. 3b). Interestingly, the laminin-5 complex subunit genes LAMA3, LAMB3, and LAMC2 were downregulated in all the infected tissues. Besides being an important player in epidermal-dermal stability and wound healing, laminin-5 is associated with invasive phenotypes of cancers, including cervical cancer (34–36). Most importantly, laminin-5 is required for HPV16 binding during virus attachment and entry (37). Downregulation of laminin-5 during productive HPV16 infection could prevent superinfection and affect epithelial integrity. Immunofluorescence for laminin-5 revealed decreased expression of the protein complex in the basal layer cells of HPV16 cervical, foreskin, and tonsil rafts (Fig. 3c).
Gene ontology analysis reveals enrichment of unique biological functions in HPV16 cervical, foreskin, and tonsil tissues.
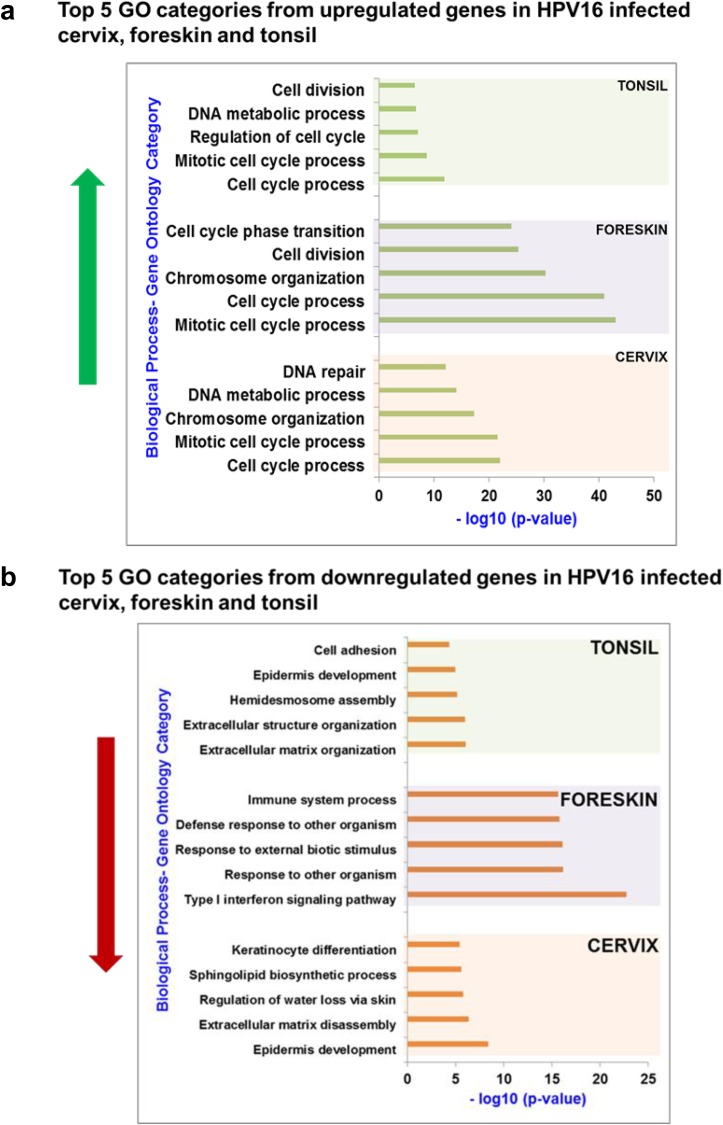
Significantly altered genes were divided into upregulated and downregulated genes, and gene ontology (GO) analysis was carried out for each list to identify enriched biological functions using GOrilla (38). In Fig. 4, histograms represent the P values for each of the top 5 GO categories among the upregulated (Fig. 4a) and downregulated (Fig. 4b) genes. The entire list of GO categories that are significantly regulated in each tissue type is summarized in Tables S1a to c in the supplemental material.
FIG 4.
(a) Gene ontology analysis reveals significant upregulation of gene categories related to increased cell proliferation in HPV16 cervical, foreskin, and tonsil epithelium. (b) Among the downregulated genes, the GO terms related to epidermis development and ECM organization were enriched in HPV16 cervical and tonsil tissues. Several GO terms related to the type I interferon response and immune responses were downregulated in HPV16 foreskin. The histograms represent the negative log10 P value obtained from GOrilla analysis for the related GO category (biological function).
Within the upregulated genes, the top 5 enriched GO terms include categories of cell division, mitosis, cell cycle processes, and DNA damage repair in HPV16-infected cervical, foreskin, and tonsil tissues (Fig. 4a). Clearly, HPV16 infection induces a hyperproliferative state in epithelial tissue types. High-risk HPV oncoproteins E6 and E7 reinitiate DNA replication in terminally growth-arrested keratinocytes, by dysregulating cell cycle checkpoints to create an S-phase-like milieu, ambient for viral DNA replication (39, 40). Unscheduled entry into S phase, along with deregulation of cell cycle checkpoints, results in DNA damage and chromosomal instability in the infected cells (41–44).
For the downregulated genes, the enriched GO terms for each tissue type varied widely among the tissue types (Fig. 4b). In HPV16 cervical tissue, genes involved in epidermal and keratinocyte development and ECM organization were significantly downregulated, whereas in HPV16 foreskin tissue, genes related to immune processes, the innate immune responses and cellular responses to viruses, and cytokine signaling were downregulated. While the total number of gene changes was minimum in HPV16 tonsil tissue, significant downregulation was observed in genes involved in ECM organization, epidermis development, cell-cell adhesion, and hemidesmosome assembly. Although overlapping GO categories are enriched among the tissue types, the genes within them are distinct. This could be due to the virus utilizing different upstream regulators to target common cellular functions in a tissue-specific manner.
Protein interaction clusters reveal important regulatory networks in HPV16 cervical, foreskin, and tonsil tissues.
Next, the highly interacting proteins or hubs from our gene list were visualized to identify regulatory molecules of interest. Using the online tool STRING (http://string-db.org/), we mapped the protein-protein associations within the upregulated and downregulated genes for each tissue type (45). Using the CLUSTER function, we identified the most stable and significant interactions in the gene list.
The protein interaction networks obtained from the upregulated gene lists reveal a dense connectivity of proteins with roles in mitosis and cell division for each tissue type (Fig. 5a to c). In line with the GO analysis, we observe an overall upregulation of cell proliferation in HPV16-infected cervical, foreskin, and tonsil tissues. It is well established that high-risk HPVs activate the cell cycle in epithelial cells to promote a replication-competent environment, which enables viral genome amplification (46, 47). Once again, we observe that although overlapping cellular functions are upregulated in each of the HPV16-infected tissues, the target genes are distinct for each tissue type, owing to tissue-specific responses to the virus.
FIG 5.
The protein interaction networks of upregulated genes depict dense interconnectivity and high representation of proteins related to mitosis and cell division in HPV16 cervical (a), foreskin (b), and tonsil (c) epithelium. The CLUSTER function (STRING database) was used to identify the most relevant protein interactions among the upregulated targets. Cytoscape software (3.6.0) was used to construct the networks, using yFiles hierarchic layout.
In addition to heightened cell proliferation, we observe significant clusters of proteins involved in metabolism (Fig. S1a and b). The increase in cellular metabolism is consistent with the increase in DNA replication and cell cycle progression induced by productive HPV16 infection. In all three tissue types, HPV16 infection results in upregulation of Fanconi anemia (FA) pathway proteins (Table S2). Especially, FANCD2 is upregulated in both HPV16 cervical and foreskin tissues in our data set. Activation of the FA pathway acts as an early host cell response to HPV16 infection to prevent oncogene-mediated chromosomal instability (48). In HPV16 tonsil tissue, only FANCE is upregulated (Fig. 5c), which is upregulated in all the tissue types (Fig. 2a).
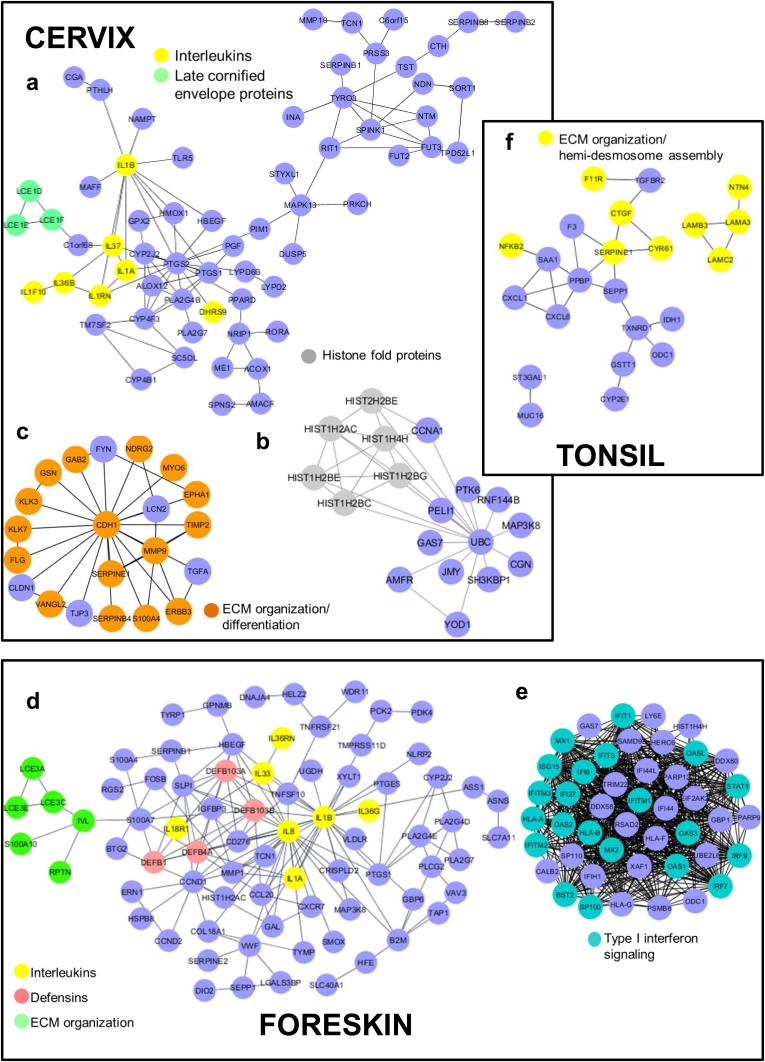
For the downregulated genes, we observe clusters with enrichment of pathways related to immune responses, cellular defenses, epidermal differentiation, and ECM organization (Fig. 6). In both HPV16 cervical and foreskin tissues, we see groups of interleukins (ILs) and interleukin receptors forming a part of significant clusters (Fig. 6a and d and Table 2). Interleukins start local inflammation and activation of adaptive immunity. IL-1β has previously been shown to be abrogated at the posttranslational level by the HPV16 E6 oncoprotein (49). IL-1β, along with several chemotactic and proinflammatory genes with a role in activation of adaptive immunity, was also downregulated in undifferentiated HPV16- and HPV18-infected keratinocytes (50). The other significant cluster of downregulated genes in HPV16-infected foreskin tissue included several proteins involved in type I interferon signaling and the 2′-5′-oligoadenylate synthetase (OAS) family of genes involved in DNA sensing (Fig. 6e). Surprisingly, we did not observe an enrichment of immune-related pathways in HPV16 tonsil tissue in spite of it being a lymphoid tissue.
FIG 6.
Protein interactome of genes downregulated upon productive HPV16 infection of epithelial tissue types. The CLUSTER function (STRING database) was used to identify the most relevant protein interactions among the upregulated targets. Cytoscape software (3.6.0) was used to construct the networks, using yFiles hierarchic layout. (a to c) In HPV16 cervical tissue, clusters representing proteins involved in epithelial differentiation and ECM organization were identified. (d and e) Several immune response-related proteins, including type I interferon signaling pathway candidates, formed the major clusters in HPV16 foreskin. Owing to a minimum number of gene expression changes, we obtain a single cluster in HPV16 tonsil that comprises proteins involved in ECM organization and hemidesmosome assembly (c).
TABLE 2.
Types of interleukins and late cornified envelope protein coding genes that are downregulated in HPV16-infected cervical and foreskin rafts
| HPV16-infected tissue | Downregulated interleukin genes | Downregulated late cornified envelope genes |
|---|---|---|
| Cervix | IL1A, IL1B, IL1F7, IL1F8, IL36B, IL37 | LCE1D, -1E, -1F, -3C, -4A, -5A |
| Foreskin | IL1A, IL1B, IL8, IL1F5, IL1F8, IL1F9, IL17C, IL33 | LCE3A, -3C, -3D, -3E |
A distinct set of late cornified envelope (LCE) protein coding genes is downregulated in HPV16 cervical and foreskin tissues (Fig. 6a and d and Table 2). The genes encoding the LCE proteins form a part of the epidermal differentiation complex (EDC) located on human chromosome 1q21 and have roles in epidermal organization and differentiation (51). Interestingly, in HPV16 foreskin, only the LCE3 family proteins are downregulated. Deletion of the LCE3B and LCE3C genes is highly associated with a risk of psoriasis development in patients (52).
Another cluster of downregulated genes in HPV16 cervical tissue has ubiquitin C (UBC) acting as the central node interacting with a group of histones (Fig. 6b). The function of UBC within the cell is complex and ranges from proteasome-dependent proteolysis to cell signaling, cell cycle regulation, DNA repair, and apoptosis, among others (53). In both HPV16 cervical and tonsil tissues, we see clusters with proteins involved in the organization and turnover of the ECM, hemidesmosome assembly, and cell-cell adhesion (Fig. 6c and f).
Validation of gene expression changes in HPV16 cervical, foreskin, and tonsil tissues.
In order to validate gene expression data, we performed reverse transcription-quantitative PCR (RT-qPCR) followed by Western blot analysis of a few selected genes of interest. We validated expression levels of proteins involved in epithelial differentiation and organization, namely, keratin 14 (KRT14), KRT19, KRT23, Kallikrein-related peptidase 8 (KRKL8), and Repetin (RPTN) (Table 3). Validation experiments were performed with a new set of cell lines to increase the rigor. Rafts obtained from these cell lines and harvested at day 10 were used for RT-PCR to validate the microarray gene expression changes. Western blot analysis was performed on rafts harvested at day 10 and day 20. All the rafts used for validation experiments produced high titers of infectious progeny virus at day 20 (Table S3).
TABLE 3.
Fold changes and P values of genes selected for validation of microarray dataa
| Gene for validation | Microarray fold change (P value) |
||
|---|---|---|---|
| Cervix | Foreskin | Tonsil | |
| KRT14 | NS | −1.64 (0.00184) | NS |
| KRT19 | NS | 2.44 (0.000291) | NS |
| KRT23 | −11 (0.048) | NS | NS |
| GPER | 4.5 (0.048) | NS | NS |
| KLK8 | −25.8 (0.048) | NS | NS |
| RPTN | −57.48 (0.049) | −2.63 (0.000085) | NS |
P values are included in parentheses. NS, no significant change.
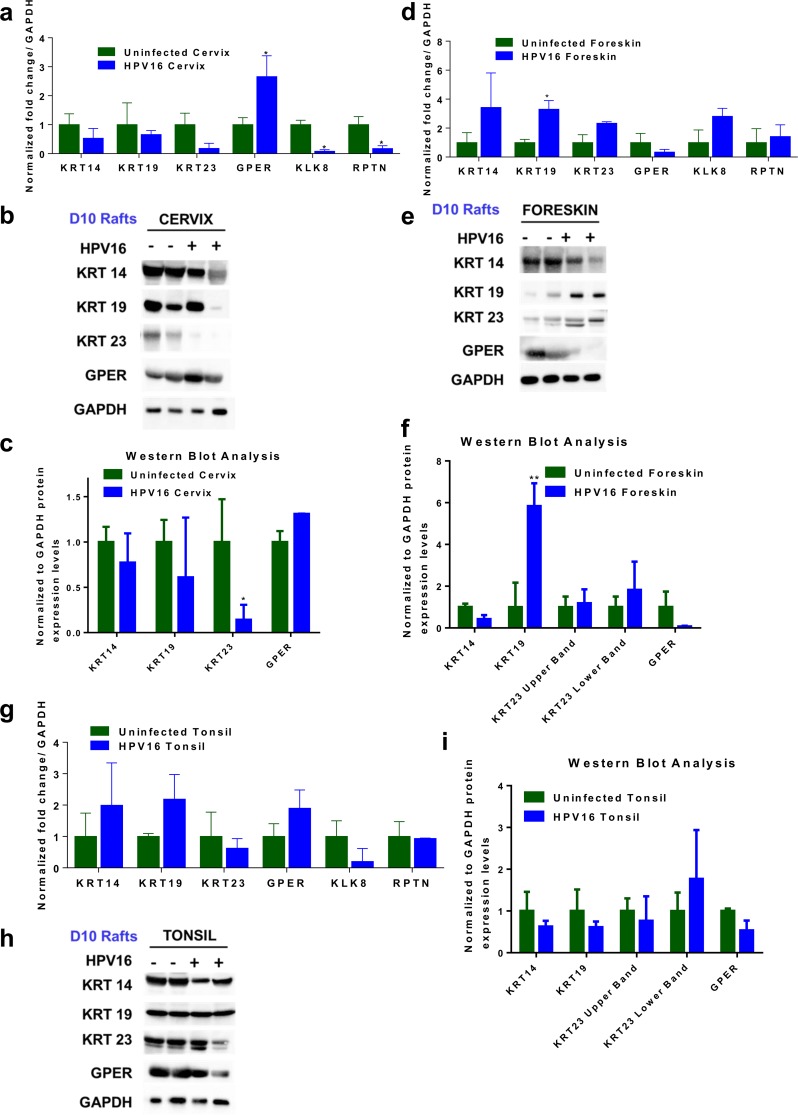
We selected keratins 14, 19, and 23 for our analysis due to their unique functions and differential expression in HPV16 cervical, foreskin, and tonsil tissues. KRT14 is expressed along with its partner KRT5 in the basal layer cells of the epithelium. KRT14 plays a role in the maintenance of cell proliferation and differentiation of basal cells, and its expression is downregulated as cells differentiate (54). Although microarray data indicated a 1.64-fold downregulation of KRT14 in HPV16 cervical tissue, we did not observe a significant change in the RT-PCR results (Fig. 7a) and protein levels (Fig. 7b and c and Fig. S3).
FIG 7.
(a, d, and g) RT-PCR validation of selected genes in HPV16-infected cervical (a), foreskin (d), and tonsil (g) rafts harvested at day 10. RNA expression levels of KRT14, KRT19, KRT23, GPER, KLK8, and RPTN were measured and normalized to GAPDH levels. The expression levels in the uninfected rafts were normalized to a value of 1. Histograms indicate average means and standard errors of the means (SEM) from three experiments. * indicates a P value of <0.05 (two-tailed t test). Western blot analysis was performed on day 10 rafts to determine protein levels of KRT14, KRT19, KRT23, and GPER. GAPDH was used as a housekeeping control. (b, e, and h) Representative blots showing protein expression of selected targets in HPV16-infected cervical (b), foreskin (e), and tonsil (h) rafts. Images were acquired with a Bio-Rad ChemiDoc MP imaging system and Image Lab (v6.0.0) software. Histograms represent the average ratios of protein levels normalized with the loading control GAPDH (means and SEM). (c, f, and i) Densitometry analysis using ImageJ software was used to calculate normalized protein levels in the cervical (c), foreskin (f), and tonsil (i) tissues. * represents a P value of <0.05, and ** represents a P value of <0.01. Rafts obtained from a new set of two uninfected and two HPV16-infected cell lines, different from those used for microarray analysis, were used for RT-PCR and Western blot analysis.
KRT19 is an important component of the oral epithelium, and its overexpression and abnormal localization are observed in high-grade cervical tumors and HPV16-positive oropharyngeal squamous cell carcinoma tumors (55, 56). In our microarray data, KRT19 is upregulated only in HPV16 foreskin tissue, during the productive stages of HPV16 infection. We confirmed a significant upregulation of KRT19 transcripts in HPV16 foreskin rafts by RT-PCR analysis (Fig. 7d). Western blot analysis also validated a significant upregulation of KRT19 protein levels only in HPV16 foreskin rafts harvested at day 10 (Fig. 7e and f) and day 20 (Fig. S3a and b).
KRT23, in addition to being a cytoskeletal protein, has recently been suggested to have roles in cell cycle regulation, apoptosis, and DNA damage repair (57, 58). Microarray data indicate an 11-fold downregulation of KRT23 only in HPV16 cervical tissue. We observed a trend toward downregulation of KRT23 mRNA levels by RT-PCR only in HPV16 cervical rafts (Fig. 7a). Furthermore, Western blot analysis confirmed a significant downregulation of KRT23 protein levels only in HPV16 cervical rafts harvested at day 10 (Fig. 7b and c) and day 20 (Fig. S3a and b).
We have previously shown that KLK8 and RPTN are downregulated during productive HPV16 infection of cervix (25). We confirmed the exclusive downregulation of KLK8 and RPTN mRNAs in HPV16 cervical tissue (Fig. 7a). Microarray data indicated a 4.5-fold upregulation of G protein-coupled estrogen receptor (GPER) exclusively in HPV16 cervical tissue (Table 3). GPER is the membrane-bound estrogen receptor that is involved in rapid signaling via the nonclassical steroid signaling pathway and is predicted to be a prognostic biomarker for cervical cancers (59). RT-PCR analysis confirmed a significant upregulation of GPER in HPV16 cervical tissue (Fig. 7a); however, no significant upregulation in protein levels was obtained from rafts harvested at day 10 (Fig. 7b and c). In rafts harvested at day 20, we observed downregulation of GPER protein levels in the HPV16-positive samples (Fig. S3a).
Overall, our validation experiments confirmed the upregulation of KRT19 (Fig. 7e and f) and GPER (Fig. 7a) mRNA levels in HPV16 foreskin and cervical rafts, respectively. KRT23, KLK8, and RPTN levels were downregulated exclusively in HPV16 cervical rafts (Fig. 7a to c). In order to increase rigor and reproducibility, we used a different set of cell lines for validation experiments than those used for microarray analysis. Owing to this, we observe some host-specific variations in the validation experiments across the samples.
Upstream regulators STAT1 and TGF-β are differentially regulated in persistently infected cervix, foreskin, and tonsil.
We next examined the common and unique upstream regulators modulated by productive HPV16 infection. Using the Ingenuity Pathway Analysis (IPA) tool, we identified the TGF-β pathway to be the most significant upstream regulator for the gene expression changes from all three tissue types (P = 4.19E−06), whereas the type I interferon pathway was significantly enriched in HPV16-infected foreskin (P = 2.18E−36). Both TGF-β and type I interferon signaling regulate important cellular functions like cell growth, differentiation, and immune responses.
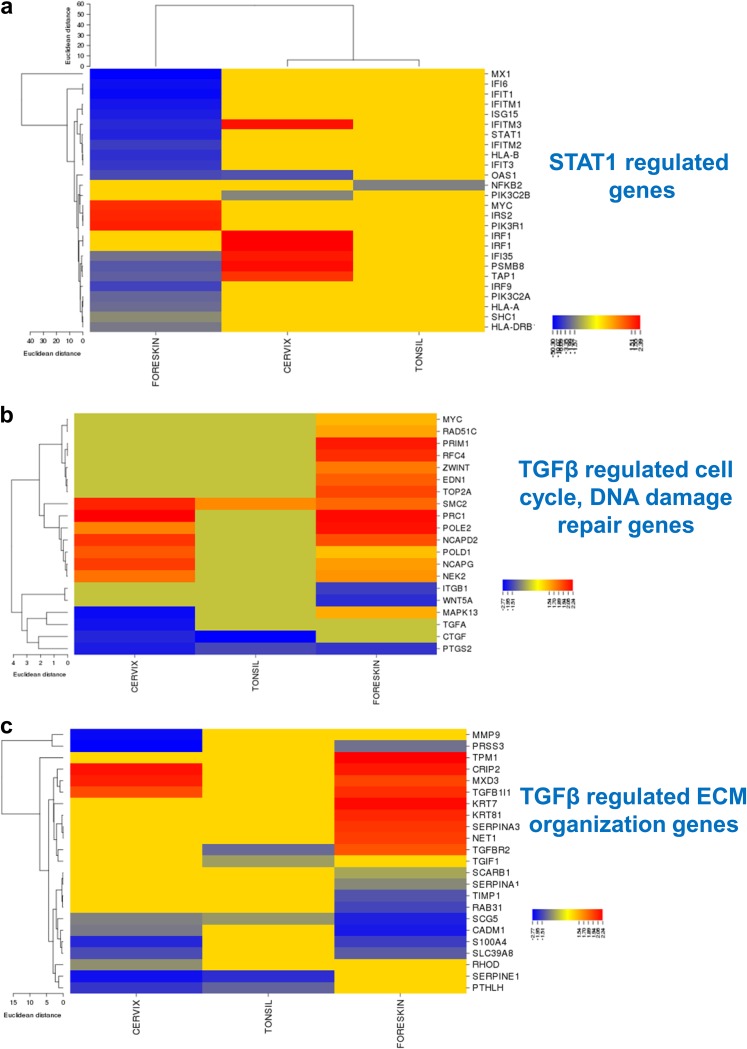
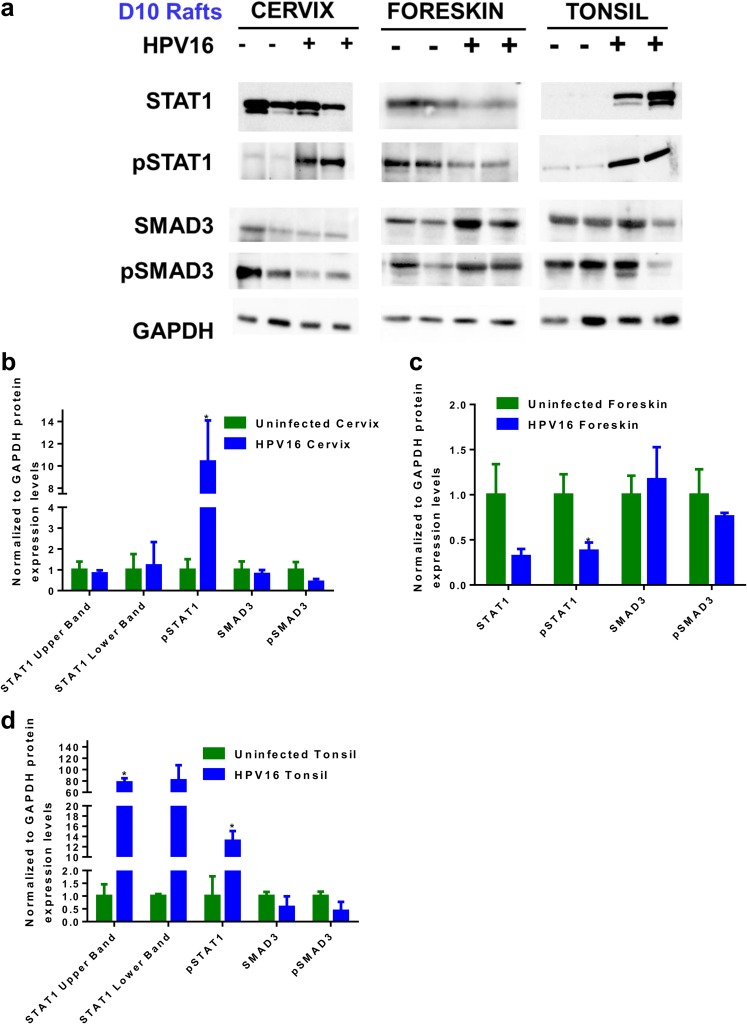
A heat map generated using CIMminer (NCI) revealed a significant downregulation of STAT1-regulated genes only in HPV16-infected foreskin (Fig. 8a). Gene expression data showed downregulation of STAT1 (−6.524-fold) in HPV16 foreskin. Western blot analysis confirmed reduced levels of total STAT1 and a significant reduction in levels of phosphorylated STAT1 (pSTAT1) in HPV16 foreskin rafts harvested at day 10 (Fig. 9a and c). Surprisingly, pSTAT1 (Y701) levels were significantly increased in the HPV16-infected cervix and tonsil rafts (Fig. 9a, b, and d). Significant suppression of antiviral responses in the foreskin during productive HPV16 infection is potentially a crucial factor that can determine virus clearance, persistence, host cell survival, and initiation of disease.
FIG 8.
Heat maps depicting expression levels of the genes regulated by the upstream regulators TGF-β and STAT1 in HPV16 foreskin, cervix, and tonsil, compared to their uninfected controls. The online tool CIMminer (NCI) was used to create the heat maps. The one-matrix method using the Euclidean distances and average linkage cluster algorithm was utilized. Heat maps represent STAT1-regulated interferon signaling genes (a), TGF-β-regulated cell cycle-related genes (b), and TGF-β regulated genes with function in ECM organization and assembly (c). IPA (Qiagen) was used to identify the target genes regulated by STAT1 and TGF-β from the data set.
FIG 9.
Expression levels of total STAT1, pSTAT1, total SMAD3, and pSMAD3 in uninfected and HPV16-infected rafts. (a) Representative blots indicating expression levels of total STAT1, pSTAT1, total SMAD3, and pSMAD3 in HPV16-infected cervical, foreskin, and tonsil rafts harvested at day 10. GAPDH was used as a loading control. (b to d) Histograms representing average ratios of protein expression levels normalized to GAPDH levels (means and SEM) in cervical (b), foreskin (c), and tonsillar (d) tissues. * indicates a P value of <0.05.
For simpler visualization, we separated the TGF-β-regulated gene list based on roles in cell cycle regulation and extracellular matrix remodeling. Among the TGF-β-regulated cell cycle-related genes, most of the targets are upregulated in HPV16 foreskin and cervical tissues (Fig. 8b), with very few gene expression changes in HPV16 tonsil. However, most of the TGF-β-regulated ECM-related genes are downregulated in infected cervix and tonsil (Fig. 8c). TGF-β is a multifunctional growth factor; HPV16 infection selectively suppresses its role in ECM maintenance and differentiation pathways. We next assessed the activation status of the TGF-β pathway by measuring the total levels of SMAD3 and phosphorylated SMAD3 (pSMAD3) (activated) (Fig. 9a to d). We observed a notable reduction of pSMAD3 levels in the HPV16-infected cervix rafts (Fig. 9a and b). This effect was more significant in the rafts harvested at day 20 (Fig. S4a and b), where we observed significant downregulation of total SMAD3 and pSMAD3 in the HPV16-positive cervical rafts.
DISCUSSION
Our study for the first time compares global gene expression changes in cervical, foreskin, and tonsil tissues during productive HPV16 infection, long before oncogenesis initiates. During this stage, cell proliferation and episomal maintenance in the basal epithelial layers are followed by genome amplification and capsid protein expression in the upper layers of the epithelium. Consequently, the virus utilizes host cellular pathways and regulatory mechanisms that ensure its efficient genome amplification, maturation, packaging, and release (40, 60, 61). Tissue-specific epithelial cell subtypes can also determine susceptibility to high-risk HPV16 infection and consequent oncogenesis. For the present study, we obtained primary cervical keratinocytes from the ectocervix, which contains squamous epithelial cells. Most cervical cancers originate in the squamocolumnar junction (SCJ) region (62, 63), which commonly expresses biomarkers like keratin 7, AGR2, MMP7, and GDA (64, 65). Keratin 19, which is an important binding partner of keratin 7, is expressed in the columnar cells of the SCJ, squamous cells at the transformation zone, and basal layer cells at the ectocervix (55). Similarly, in tonsil tissue, most HPV16-positive cancers arise in the discontinuous crypt regions (66). In nonneoplastic tonsils, KRT19 expression is preferentially observed in the reticular crypt epithelium compared to the surface epithelium (67). In our study, primary tonsil keratinocytes were obtained from both the crypt and the surface epithelium. Both primary and HPV16-positive rafts obtained from cervical and tonsillar cells used in our study express KRT19 mRNA as well as protein (Fig. 7a to c and g to i). Therefore, we strongly believe that the raft cultures used in our study closely represent natural target sites in vivo, which are susceptible to HPV16-mediated oncogenesis.
One of the most striking findings of our study was the large differences in the total numbers of gene expression changes across the HPV16-infected tissue types (Fig. 1). Only 32 genes are commonly regulated among HPV16 cervical, foreskin, and tonsil tissues, in spite of being infected by the same virus type. This list includes genes coding for cell proliferation, DNA replication, and damage repair proteins (Fig. 2), including p53-DREAM complex targets (Fig. 3a). This is not surprising considering the well-defined roles of the viral oncoproteins E6 and E7 in initiating cell cycle reentry by inhibiting the tumor suppressor genes TP53 and RB1, respectively (68–73). E7 also disrupts p53-DREAM complex-mediated repression, upregulating several cell cycle genes and p53-mediated cell cycle control and promoting cell cycle entry and mitotic proliferation (27). Accordingly, we observe significant upregulation of cell cycle-, cell division-, and mitosis-related GO categories in all three tissue types upon HPV16 infection (Fig. 4a and Fig. 5a to c). A review that reanalyzed gene expression profiles from several reported data sets found the GO categories of cell cycle, mitosis, cell division, DNA replication, and DNA repair pathways to be commonly upregulated in HPV-associated cervical cancer and head and neck cancer patient samples (74). Those authors specifically discuss how primary keratinocytes overexpressing E6/E7 fail to mimic the gene deregulation observed in situ, asserting the need to use systems that closely recapitulate proliferation and differentiation processes for in vivo systems. The cellular signaling pathways are highly dysregulated in cancerous tissue, and it becomes difficult to isolate virus-driven changes. However, in order to examine the dynamics of virus-host interactions, it is important to use a model system that would closely mimic the complete virus life cycle and enable us to focus on preneoplastic changes. HPV16-induced dysregulation of cell cycle control is an early event in pathogenesis and a common molecular signature across the tissue types.
Contrary to the upregulated genes, unique GO categories were enriched for downregulated genes for each tissue type (Fig. 4b). In the persistently infected cervical epithelium, we observe significant downregulation of genes involved in keratinocyte differentiation, epidermal development, and ECM organization (Fig. 4b). Interestingly, we observe that multiple genes located on the epidermal differentiation complex (EDC) on human chromosome 1 coding for proteins important for epidermal differentiation (75) are downregulated in infected cervical and foreskin tissues. Several members of the EDC complex are downregulated in the cervix (LCEs, S100As, and RPTN) and foreskin (LCEs and S100 proteins) (Fig. 6a and b and Table 2). In HPV16 cervical tissue, we observe downregulation of the LCE1, -2, -5, and -6 genes, which function in the maintenance of the normal skin barrier (76, 77). On the other hand, in the foreskin, only the LCE3 members are downregulated (Table 2). Members of the LCE3 group of proteins are important for skin barrier repair caused by injury and inflammation (76). Deletion of LCE3B and LCE3C genes in patients is highly associated with the development of psoriasis and other autoimmune diseases (52). Many genes that are dysregulated in psoriasis or other hyperkeratinizing skin disorders are downregulated in our microarray data, in a tissue-specific manner (78). This loss of skin barrier integrity might be important for efficient virus maturation, egress, and spread, thereby determining the establishment of persistence. Even in HPV16 tonsil epithelium, we observe downregulation of genes involved in ECM organization, cell-cell adhesion, and hemidesmosome assembly (Table 1, Fig. 4b, and Fig. 6c). We also see downregulation of TGF-β-regulated genes involved in ECM structure and maintenance in the HPV16 tissue types (Fig. 8c). Overall, HPV16 infection suppresses epidermal differentiation and organization, an effect which is more pronounced in cervical tissue.
Productive HPV16 infection differentially regulates innate immune responses in the cervical, foreskin, and tonsil epithelia. Especially in infected foreskin, we observe downregulation of several genes involved in immune responses against virus infections, such as the interferon signaling pathway and DNA and RNA virus sensing (Fig. 4b, Fig. 6e, Fig. 8a, and Fig. 9). Immune-related targets downregulated in the HPV16-infected cervical epithelium are mostly involved in the NF-κβ pathway (see Table S1a in the supplemental material for a list of GO terms). Although the innate immune response forms the first line of defense against pathogens, during virus infections, it triggers the interferon signaling cascade, resulting in an antiviral state in the cells. Several viruses have evolved strategies to suppress the interferon signaling pathway, thereby leading to virus propagation, host cell survival, and, finally, persistence (79). In a previous study, we showed resistance of HPV16-infected (acute and persistent) keratinocytes (foreskin and vaginal) to antiproliferative and necroptosis effects induced by interferon gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α) (80). Another study using HPV31-infected human foreskin lines showed reduced expression of interferon-regulated genes and total levels of STAT1 in virus-infected cell lines (81). Although these studies looked at gene expression changes in infected monolayer cells, they show a significant downregulation of the interferon responsiveness of infected foreskin cells. We also see downregulation of IL1B and related genes in HPV16 cervical and foreskin tissues (Fig. 6a and d and Table 2). Interestingly, although tonsils are lymphoid tissue, we see very few changes in immune-related genes with HPV16 infection. This could explain the enhanced adaptive immunity observed in HPV-associated oropharyngeal squamous carcinoma patients (9).
Gene expression analysis and protein expression validations confirm the preferential downregulation of TGF-β signaling in the infected cervical epithelium (Fig. 8c, Fig. 9a and b, and Fig. S4a and b). A previous study from our laboratory has shown that TGF-β treatment is able to restore the loss of differentiation induced by HPV infection in cervical rafts (82). We observed reduced levels of pSMAD3 protein levels in HPV16 cervical tissue, indicating reduced activation of the TGF-β pathway (Fig. 9a and b and Fig. S4). The TGF-β pathway is commonly dysregulated in several cancers, including cervical cancers. In cervical carcinomas, the TGF-β pathway is modulated uniquely depending on the stage of the disease (83). TGF-β overexpression is observed in cervical carcinomas, resulting in the activation of mitogen-activated protein kinase (MAPK), WNT, TNF-α, and NF-κβ pathways promoting the epithelial-to-mesenchymal transition (EMT) (84–86). Likewise, we predict that dampened TGF-β signaling during early stages of infection can play a critical role in the establishment of persistence and, consequently, progression of disease.
Using productive organotypic raft cultures, we were able to identify virus-induced changes in a differentiating epithelium, which closely mimics in vivo infection. As epithelial differentiation is integral to the virus life cycle, this system helps us understand the effect of replicating and maturing virus on the host tissue. A recent publication reports differential changes in gene expression from HPV16-infected undifferentiated and differentiated keratinocytes of cervical and foreskin origins in cell culture studies (87). Using W12 cells lines and near-diploid immortalized keratinocytes (NIKs), those authors investigated the effect of high-calcium-induced differentiation and HPV16 infection on the cellular transcriptome. W12 cells are HPV16-positive cervical epithelial cells obtained from a patient with a low-grade cervical lesion (88). On the other hand, NIKs are a spontaneously immortalized human keratinocyte cell line isolated from the BC-1-Ep strain of neonatal foreskin keratinocytes (89). Similar to our results, gene categories related to keratinocyte differentiation, extracellular matrix integrity, and cell-cell adhesion were downregulated upon HPV16 infection. Although our study clearly indicates significant upregulation of GO categories related to the cell cycle, mitosis, and DNA damage repair, in this study, the type I interferon response, cell matrix adhesion, and endopeptidase inhibitor activity were significantly upregulated upon HPV16 infection of differentiated NIKs. Similar to our results, proinflammatory genes like IL1A and IL1B were downregulated in HPV16 NIKs. Interestingly, viral restriction factors like IFIT1, IFIT2, IFIT3, IFIT5, OASL, and interferon regulatory factor 1 (IRF1) were upregulated severalfold in the differentiated HPV16 NIK cell lines. This is in contrast to our results, as we observed genes with roles as restriction factors and in viral DNA sensing to be downregulated in HPV16 foreskin rafts (Fig. 6e). Our data show that tissue-specific responses to virus infection representing the preneoplastic state could determine virus clearance or persistence, immune evasion, and susceptibility to oncogenesis. Differential regulation of important cellular mechanisms like cell proliferation, differentiation, innate immunity, and upstream regulators like STAT1 and TGF-β can determine viral fitness and pathogenesis. In the future, it would be interesting to analyze how these pathways and upstream regulators are modulated during the progression to oncogenesis. These tissue-specific responses could provide important clues for identifying early biomarkers, understanding virus pathogenesis, and devising therapeutic interventions to prevent HPV-associated cancers.
MATERIALS AND METHODS
Cell culture.
Isolation and culture of primary keratinocytes were performed as previously described (90). Keratinocyte lines from multiple hosts were used for all the studies to remove any individual host bias. The Human Subjects Protection Office of the Institutional Review Board (IRB) at the Penn State University College of Medicine screened our study design for exempt status, as no human participants were involved, as defined by federal regulations.
Primary cervical and tonsil keratinocytes derived from deidentified tissues obtained from routine surgeries were used for the study. Primary foreskin was obtained from deidentified newborn circumcisions. For cervical keratinocyte isolation, we used tissue from the ectocervix. For isolation of keratinocytes from tonsillar specimens, we use the entire epithelial tissue obtained from the donor. Therefore, keratinocytes obtained from the tonsil samples are a mix from the surface and the crypt region. Primary keratinocyte lines obtained from multiple hosts were used for all studies to remove any individual host bias, using previously described protocols (91, 92). HPV16 epithelial lines were obtained via electroporation of the HPV16 genome and maintained according to established protocols in the laboratory (93, 94). In short, pBSHPV16 plasmid DNA was digested with BamHI to linearize the viral DNA and separate it from the vector sequence. A total of 10 μg viral DNA was electroporated into primary cells using a Gene Pulser (Bio-Rad Laboratories, Hercules, CA). Immortalized keratinocytes stably maintaining HPV16 genomes following electroporation were cultured with mitomycin C-treated J2 3T3 feeder cells and maintained in E medium.
Organotypic raft cultures were obtained from primary keratinocytes at passages 1 to 2 and from HPV16 keratinocytes at passages 6 to 9, as previously described (94, 95). Primary cells and immortalized epithelial cells were seeded onto collagen matrices consisting of rat tail type I collagen containing J2 3T3 feeder cells. Following cell attachment and growth to confluence, the matrices were lifted onto stainless steel grids and fed with E medium supplemented with 10 μM 1,2-dioctanoyl-sn-glyserol (C8:0; Sigma Chemical Company). Rafts were harvested after 10 days (day 10) for microarray analysis and qPCR validations and after 20 days (day 20) for Western blotting and immunofluorescence staining. Although viral late genes are expressed by day 10 (26), it is only by day 20 that mature virions are produced (92). For microarray analysis, a total of six individually grown raft tissue samples (n = 6) obtained from three independently derived cell lines were used, each for primary and HPV16-positive sets. For validation experiments, a new set of primary cell lines (n = 2) and HPV16 cell lines (n = 2) were used to grow raft tissues to increase rigor and for wide sample representation. Virus titers were determined using established protocols as described previously (95).
Microarray analysis.
For each tissue type, three individually derived primary cell lines and HPV16-transformed cell lines were used to grow raft tissue. Each of these experiments was repeated at separate times, representing a total of six individually grown raft tissues for primary (n = 6) and HPV16-positive (n = 6) cell lines. RNA was extracted from rafts harvested at day 10 using the RNeasy fibrous tissue midi kit (Qiagen). Microanalyses were performed using the Illumina human HT-12 v4 expression bead chip (Illumina, San Diego, CA), which targets over 31,000 annotated genes with more than 47,000 probes derived from National Center for Biotechnology Information (NCBI) RefSeq release 38 (7 November 2009) and other sources. An Agilent 2100 bioanalyzer with an RNA Nano LabChip (Agilent, Santa Clara, CA) was used to asses RNA concentration and quality. cRNA was synthesized from 500 ng of RNA according to the manufacturer’s instructions by TotalPrer amplification (Ambion, Austin, TX). T7 oligo(dT) primed reverse transcription was used to produce first-strand cDNA, followed by second-strand synthesis and RNA degradation by RNase H and filtration cleanup. Multiple copies of biotinylated cRNA were generated by in vitro transcription (IVT), purified using filtration, and quantified using a NanoDrop instrument, and the volume was adjusted for a total of 750 ng/sample. Furthermore, samples were fragmented and denatured before hybridization for 18 h at 58°C. After hybridization, the bead chips were washed and fluorescently labeled. A BeadArray reader (Illumina, San Diego, CA) was used to scan the bead chips.
The CLC Genomics Workbench 4.8 package (Qiagen) was used to determine significantly differentially expressed genes of the HPV16-positive versus primary tissue microarray samples. Significantly differentially expressed genes were considered to be those with a P value of <0.05 and an absolute fold change of ≥1.5.
Gene ontology analysis and protein association network.
For gene ontology (GO) analysis, the GOrilla database was used (38). Significantly altered genes were divided into upregulated and downregulated genes, and GO analysis was carried out for each list. Two unranked lists of genes, target (significant genes) and background (all the Illumina genes in the array), were used to identify significantly enriched GO terms. Gene lists were categorized based on the GO subontologies of biological process (BP). Furthermore, REVIGO (http://revigo.irb.hr/) was used to summarize long lists of GO terms by removing redundant GO terms using a similarity coefficient of 0.7 (96).
STRING (http://string-db.org/) was used to identify protein interactions within the upregulated and downregulated gene lists for each tissue type. (45). Further CLUSTER function was used to rearrange the network and identify the most stable and significant interactions in the list. Pathway analysis was performed using Ingenuity Pathway Analysis (Qiagen). Heat maps were made using the online tool CIMminer (Genomics and Pharmacology Facility, NCI). We used the one-matrix method using the Euclidean distances and average linkage cluster algorithm to create the heat map.
RT-qPCR.
RT-qPCR was performed to validate the microarray gene expression changes for selected genes from rafts harvested at day 10 from uninfected and HPV16-infected cervical, foreskin, and tonsil tissues. RNA was extracted using the ZR-Duet DNA/RNA Miniprep plus kit (Zymo Research). First-strand cDNA was synthesized using the Accuscript high-fidelity first-strand cDNA synthesis kit (Agilent), according to the manufacturer’s instructions, from 500 ng of RNA. cDNA synthesis was carried out using the following protocol: 42°C for 60 min, followed by 70°C for 15 min and 4°C for 2 min. PCR was performed using the QuantiTect SYBR green PCR kit (Qiagen). All primers were synthesized by Integrated DNA Technologies (IDT). The PCRs were performed using a C1000 thermal cycler (Bio-Rad) programmed for 30 min at 50°C and then for 15 min at 95°C and 42 cycles of 15 s at 94°C and 1 min at 54.5°C. Table 4 summarizes the sequences of the primers used for the study.
TABLE 4.
Primers used
| Gene | Primer | Reference or source |
|---|---|---|
| KRT14 | Forward primer 5′-GGCCTGCTGAGATCAAAGACTAC-3′ | 97 |
| Reverse primer 5′-CACTGTGGCTGTGAGAATCTTGTT-3′ | ||
| KRT19 | Forward primer 5′-AGCATGAAAGCTGCCTTGGA-3′ | |
| Reverse primer 5′-GCTCACTATCAGCTCGCACA-3′ | ||
| KRT23 | Forward primer 5′-CCACCTGGAGAAGGAAATCA-3′ | |
| Reverse primer 5′-TTGATGGTCTCCTGGGTTATG-3′ | ||
| GPER | Forward primer 5′-GGCCACGTCATGTCTCTAAA-3′ | |
| Reverse primer 5′-CCAGTCGTGAGGTTTCCTAAG-3′ | ||
| KLK8 | Forward primer 5′-TGGGTCCGAATCAGTAGGT-3′ | 25 |
| Reverse primer 5′-GCAGGAACATCCACGTCTT-3′ | ||
| RPTN | Forward primer 5′-CACAAATATGCCAAAGGGAATG-3′ | 25 |
| Reverse primer 5′-GTCATTTGGTCTCTGGAGGATG-3′ | ||
| GAPDH | Forward primer 5′-ACCACAGTCCATGCCATCAC-3′ | Readymade primer, IDT |
| Reverse primer 5′-TCCACCACCCTGTTGCTGTA-3′ | ||
Western blotting.
Raft tissues harvested at day 20 were used to perform Western blotting using a protocol described previously (98). Total protein concentrations were estimated using the Peterson protein assay method (99), and extracts were electrophoresed using a sodium dodecyl sulfate-polyacrylamide gel (8 to 10%) and transferred to a nitrocellulose membrane. After blocking the membrane in 5% bovine serum albumin (BSA) in Tris-buffered saline–Tween (TBST), blots were incubated overnight at 4°C with the primary antibodies listed below. The membranes were then washed with TBST and incubated with a horseradish peroxidase-linked secondary antibody (NA931VS/NA934VS; GE Healthcare) at room temperature for 1 h. The membranes were washed with TBST (3 times for 15 min each) and developed with the Amersham ECL Prime Western blot detection reagent (GE Healthcare). Images were acquired with a Bio-Rad ChemiDoc MP imaging system and Image Lab (v6.0.0) software. The ImageJ tool Analyze Gel was used to perform densitometry. GraphPad Prism was used to plot and analyze quantification data. Table 5 summarizes the antibodies used for the study.
TABLE 5.
Antibodies used
| Proteina | Dilution | Catalogue number; manufacturer |
|---|---|---|
| KRT14 | 1:3,000 | C-8791; Sigma |
| KRT19 | 1:2,000 | sc-6278; Santa Cruz |
| KRT23 | 1:2,000 | H00025984-M01; Abnova |
| GPER | 1:1,000 | MAB5534; R&D Systems |
| STAT1 | 1:2,000 | 9172; Cell Signaling Technology |
| pSTAT1 | 1:2,000 | sc-8394; Santa Cruz |
| SMAD3 | 1:2,000 | 9513; Cell Signaling Technology |
| pSMAD3 | 1:2,000 | 9520; Cell Signaling Technology |
| GAPDH | 1:2,000 | sc-47724; Santa Cruz |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Immunofluorescence.
Rafts harvested at day 20 were fixed in 10% buffered formalin and embedded in paraffin, and 4-μm cross sections were prepared. For immunofluorescence staining, the slides were deparaffinized in xylene, followed by washes in ethanol and rehydration. Slides were submerged in Tris-EDTA buffer (pH 9) in a 90°C water bath for 10 min for optimal antigen retrieval. The slides were then rinsed with TBST and blocked with the Background Sniper blocking reagent (Biocare Medical). Further slides were incubated overnight at 4°C with antibodies against KLHL35 (1:250 to 1:500) (catalogue number NBP1-89837; Novus) and laminin-5 (1:1,000) (catalogue number ab14509; Abcam). The slides were rinsed in TBST and incubated with secondary antibodies (1:2,000) (Alexa Fluor 488; Life Technologies) at room temperature for 1 h, followed by a TBST wash and incubation with Hoechst nuclear stain (1:5,000) for 15 min. This was followed by two washes with TBST, and coverslips were loaded with Prolong gold antifade (Life Technologies). A Nikon Eclipse 80i microscope and NIS Elements (v4.4) software were used to acquire images at a ×200 magnification.
Statistical analysis.
For the microarray gene expression data analysis, quantile-normalized means were determined using the CLC Genomics Workbench (v4.8) software package. Statistical significance was determined with the homogeneous t test. For the qPCR and Western blot analyses, a nonparametric unpaired t test with Welch’s correction was performed, and a P value cutoff of <0.05 was considered significant. The plotting tool GraphPad Prism was used to plot and analyze data.
Data availability.
The GEO accession number for our microarray data is GSE129159.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lynn Budgeon and Debra Shearer for processing, embedding the raft tissues, sectioning, and hematoxylin and eosin staining.
Work in C.M.’s group was supported by National Institutes of Health grants R01CA225268 and R01DE018305-03S1 (NIDCR-ARRA supplement).
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00915-19.
REFERENCES
- 1.Satterwhite CL, Torrone E, Meites E, Dunne EF, Mahajan R, Ocfemia MC, Su J, Xu F, Weinstock H. 2013. Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sex Transm Dis 40:187–193. doi: 10.1097/OLQ.0b013e318286bb53. [DOI] [PubMed] [Google Scholar]
- 2.Lowy DR, Schiller JT. 2012. Reducing HPV-associated cancer globally. Cancer Prev Res (Phila) 5:18–23. doi: 10.1158/1940-6207.CAPR-11-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2012. Human papillomavirus-associated cancers—United States, 2004–2008. MMWR Morb Mortal Wkly Rep 61:258–261. [PubMed] [Google Scholar]
- 4.Giuliano AR, Tortolero-Luna G, Ferrer E, Burchell AN, de Sanjose S, Kjaer SK, Munoz N, Schiffman M, Bosch FX. 2008. Epidemiology of human papillomavirus infection in men, cancers other than cervical and benign conditions. Vaccine 26(Suppl 10):K17–K28. doi: 10.1016/j.vaccine.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiller JT, Lowy DR. 2012. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat Rev Microbiol 10:681–692. doi: 10.1038/nrmicro2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham SV. 2017. The human papillomavirus replication cycle, and its links to cancer progression: a comprehensive review. Clin Sci (Lond) 131:2201–2221. doi: 10.1042/CS20160786. [DOI] [PubMed] [Google Scholar]
- 7.Ndiaye C, Mena M, Alemany L, Arbyn M, Castellsagué X, Laporte L, Bosch FX, de Sanjosé S, Trottier H. 2014. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol 15:1319–1331. doi: 10.1016/S1470-2045(14)70471-1. [DOI] [PubMed] [Google Scholar]
- 8.Santegoets SJ, van Ham VJ, Ehsan I, Charoentong P, Duurland CL, van Unen V, Höllt T, van der Velden LA, van Egmond SL, Kortekaas KE, de Vos van Steenwijk PJ, van Poelgeest MIE, Welters MJP, van der Burg SH. 2019. The anatomical location shapes the immune infiltrate in tumors of same etiology and affects survival. Clin Cancer Res 25:240–252. doi: 10.1158/1078-0432.CCR-18-1749. [DOI] [PubMed] [Google Scholar]
- 9.Welters MJP, Ma W, Santegoets SJAM, Goedemans R, Ehsan I, Jordanova ES, van Ham VJ, van Unen V, Koning F, van Egmond SI, Charoentong P, Trajanoski Z, van der Velden LA, van der Burg SH. 2018. Intratumoral HPV16-specific T cells constitute a type I-oriented tumor microenvironment to improve survival in HPV16-driven oropharyngeal cancer. Clin Cancer Res 24:634–647. doi: 10.1158/1078-0432.CCR-17-2140. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Plasencia C, Vázquez-Ortiz G, López-Romero R, Piña-Sanchez P, Moreno J, Salcedo M. 2007. Genome wide expression analysis in HPV16 cervical cancer: identification of altered metabolic pathways. Infect Agents Cancer 2:16. doi: 10.1186/1750-9378-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Miller C, Mosher R, Zhao X, Deeds J, Morrissey M, Bryant B, Yang D, Meyer R, Cronin F, Gostout BS, Smith-McCune K, Schlegel R. 2003. Identification of cervical cancer markers by cDNA and tissue microarrays. Cancer Res 63:1927–1935. [PubMed] [Google Scholar]
- 12.Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA. 2007. Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer 43:415–432. doi: 10.1016/j.ejca.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, Ahlquist P. 2007. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res 67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosty C, Sheffer M, Tsafrir D, Stransky N, Tsafrir I, Peter M, de Crémoux P, de La Rochefordière A, Salmon R, Dorval T, Thiery JP, Couturier J, Radvanyi F, Domany E, Sastre-Garau X. 2005. Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene 24:7094–7104. doi: 10.1038/sj.onc.1208854. [DOI] [PubMed] [Google Scholar]
- 15.Schlecht NF, Burk RD, Adrien L, Dunne A, Kawachi N, Sarta C, Chen Q, Brandwein-Gensler M, Prystowsky MB, Childs G, Smith RV, Belbin TJ. 2007. Gene expression profiles in HPV-infected head and neck cancer. J Pathol 213:283–293. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- 16.Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, Netterville JL, Levy S, Yarbrough WG, Chung CH. 2006. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res 12:701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 17.Wong YF, Cheung TH, Tsao GS, Lo KW, Yim SF, Wang VW, Heung MM, Chan SC, Chan LK, Ho TW, Wong KW, Li C, Guo Y, Chung TK, Smith DI. 2006. Genome-wide gene expression profiling of cervical cancer in Hong Kong women by oligonucleotide microarray. Int J Cancer 118:2461–2469. doi: 10.1002/ijc.21660. [DOI] [PubMed] [Google Scholar]
- 18.Wong YF, Selvanayagam ZE, Wei N, Porter J, Vittal R, Hu R, Lin Y, Liao J, Shih JW, Cheung TH, Lo KW, Yim SF, Yip SK, Ngong DT, Siu N, Chan LK, Chan CS, Kong T, Kutlina E, McKinnon RD, Denhardt DT, Chin KV, Chung TK. 2003. Expression genomics of cervical cancer: molecular classification and prediction of radiotherapy response by DNA microarray. Clin Cancer Res 9:5486–5492. [PubMed] [Google Scholar]
- 19.Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL, Fearon ER, Cho KR. 2007. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res 67:10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 20.Gius D, Funk MC, Chuang EY, Feng S, Huettner PC, Nguyen L, Bradbury CM, Mishra M, Gao S, Buttin BM, Cohn DE, Powell MA, Horowitz NS, Whitcomb BP, Rader JS. 2007. Profiling microdissected epithelium and stroma to model genomic signatures for cervical carcinogenesis accommodating for covariates. Cancer Res 67:7113–7123. doi: 10.1158/0008-5472.CAN-07-0260. [DOI] [PubMed] [Google Scholar]
- 21.Ryndock EJ, Biryukov J, Meyers C. 2015. Replication of human papillomavirus in culture. Methods Mol Biol 1249:39–52. doi: 10.1007/978-1-4939-2013-6_3. [DOI] [PubMed] [Google Scholar]
- 22.Meyers C, Frattini MG, Hudson JB, Laimins LA. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 23.Anacker D, Moody C. 2012. Generation of organotypic raft cultures from primary human keratinocytes. J Vis Exp 2012:3668. doi: 10.3791/3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson R, Laimins LA. 2005. Differentiation of HPV-containing cells using organotypic “raft” culture or methylcellulose. Methods Mol Med 119:157–169. doi: 10.1385/1-59259-982-6:157. [DOI] [PubMed] [Google Scholar]
- 25.Kang SD, Chatterjee S, Alam S, Salzberg AC, Milici J, van der Burg SH, Meyers C. 2018. Effect of productive human papillomavirus 16 infection on global gene expression in cervical epithelium. J Virol 92:e01261-18. doi: 10.1128/JVI.01261-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozbun MA, Meyers C. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol 71:5161–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer M, Uxa S, Stanko C, Magin TM, Engeland K. 2017. Human papilloma virus E7 oncoprotein abrogates the p53-p21-DREAM pathway. Sci Rep 7:2603. doi: 10.1038/s41598-017-02831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engeland K. 2018. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ 25:114–132. doi: 10.1038/cdd.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta K, Gunasekharan V, Satsuka A, Laimins LA. 2015. Human papillomaviruses activate and recruit SMC1 cohesin proteins for the differentiation-dependent life cycle through association with CTCF insulators. PLoS Pathog 11:e1004763. doi: 10.1371/journal.ppat.1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Has C. 2017. The “Kelch” surprise: KLHL24, a new player in the pathogenesis of skin fragility. J Invest Dermatol 137:1211–1212. doi: 10.1016/j.jid.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Maier K, Leppert J, Hausser I, Schwieger-Briel A, Weibel L, Theiler M, Kiritsi D, Busch H, Boerries M, Hannula-Jouppi K, Heikkilä H, Tasanen K, Castiglia D, Zambruno G, Has C. 2016. Monoallelic mutations in the translation initiation codon of KLHL24 cause skin fragility. Am J Hum Genet 99:1395–1404. doi: 10.1016/j.ajhg.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JYW, Liu L, Hsu CK, Aristodemou S, Ozoemena L, Ogboli M, Moss C, Martinez AE, Mellerio JE, McGrath JA. 2017. Mutations in KLHL24 add to the molecular heterogeneity of epidermolysis bullosa simplex. J Invest Dermatol 137:1378–1380. doi: 10.1016/j.jid.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Lin Z, Li S, Feng C, Yang S, Wang H, Ma D, Zhang J, Gou M, Bu D, Zhang T, Kong X, Wang X, Sarig O, Ren Y, Dai L, Liu H, Zhang J, Li F, Hu Y, Padalon-Brauch G, Vodo D, Zhou F, Chen T, Deng H, Sprecher E, Yang Y, Tan X. 2016. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat Genet 48:1508–1516. doi: 10.1038/ng.3701. [DOI] [PubMed] [Google Scholar]
- 34.Nishiyama T, Amano S, Tsunenaga M, Kadoya K, Takeda A, Adachi E, Burgeson RE. 2000. The importance of laminin 5 in the dermal-epidermal basement membrane. J Dermatol Sci 24(Suppl 1):S51–S59. doi: 10.1016/S0923-1811(00)00142-0. [DOI] [PubMed] [Google Scholar]
- 35.Skyldberg B, Salo S, Eriksson E, Aspenblad U, Moberger B, Tryggvason K, Auer G. 1999. Laminin-5 as a marker of invasiveness in cervical lesions. J Natl Cancer Inst 91:1882–1887. doi: 10.1093/jnci/91.21.1882. [DOI] [PubMed] [Google Scholar]
- 36.Imura J, Uchida Y, Nomoto K, Ichikawa K, Tomita S, Iijima T, Fujimori T. 2012. Laminin-5 is a biomarker of invasiveness in cervical adenocarcinoma. Diagn Pathol 7:105. doi: 10.1186/1746-1596-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards KF, Mukherjee S, Bienkowska-Haba M, Pang J, Sapp M. 2014. Human papillomavirus species-specific interaction with the basement membrane-resident non-heparan sulfate receptor. Viruses 6:4856–4879. doi: 10.3390/v6124856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Münger K, Howley PM. 2002. Human papillomavirus immortalization and transformation functions. Virus Res 89:213–228. doi: 10.1016/S0168-1702(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 40.Longworth MS, Laimins LA. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev 68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JJ. 2010. Genomic instability induced by human papillomavirus oncogenes. N Am J Med Sci (Boston) 3:43–47. doi: 10.7156/v3i2p043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bristol ML, Das D, Morgan IM. 2017. Why human papillomaviruses activate the DNA damage response (DDR) and how cellular and viral replication persists in the presence of DDR signaling. Viruses 9:E268. doi: 10.3390/v9100268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong SY. 2017. DNA damage response is hijacked by human papillomaviruses to complete their life cycle. J Zhejiang Univ Sci B 18:215–232. doi: 10.1631/jzus.B1600306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prati B, Marangoni B, Boccardo E. 2018. Human papillomavirus and genome instability: from productive infection to cancer. Clinics (Sao Paulo) 73:e539s. doi: 10.6061/clinics/2018/e539s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moody C. 2017. Mechanisms by which HPV induces a replication competent environment in differentiating keratinocytes. Viruses 9:E261. doi: 10.3390/v9090261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. 2015. Human papillomavirus molecular biology and disease association. Rev Med Virol 25(Suppl 1):2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spardy N, Duensing A, Charles D, Haines N, Nakahara T, Lambert PF, Duensing S. 2007. The human papillomavirus type 16 E7 oncoprotein activates the Fanconi anemia (FA) pathway and causes accelerated chromosomal instability in FA cells. J Virol 81:13265–13270. doi: 10.1128/JVI.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niebler M, Qian X, Höfler D, Kogosov V, Kaewprag J, Kaufmann AM, Ly R, Böhmer G, Zawatzky R, Rösl F, Rincon-Orozco B. 2013. Post-translational control of IL-1β via the human papillomavirus type 16 E6 oncoprotein: a novel mechanism of innate immune escape mediated by the E3-ubiquitin ligase E6-AP and p53. PLoS Pathog 9:e1003536. doi: 10.1371/journal.ppat.1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karim R, Meyers C, Backendorf C, Ludigs K, Offringa R, van Ommen GJ, Melief CJ, van der Burg SH, Boer JM. 2011. Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PLoS One 6:e17848. doi: 10.1371/journal.pone.0017848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. 1996. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol 106:989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- 52.Shen C, Gao J, Yin X, Sheng Y, Sun L, Cui Y, Zhang X. 2015. Association of the late cornified envelope-3 genes with psoriasis and psoriatic arthritis: a systematic review. J Genet Genomics 42:49–56. doi: 10.1016/j.jgg.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Lucia R, Marcia B, Rita C, Mauro M. 2013. Ubiquitin C gene: structure, function and transcriptional regulation. Adv Biosci Biotechnol 4:1057–1062. [Google Scholar]
- 54.Alam H, Sehgal L, Kundu ST, Dalal SN, Vaidya MM. 2011. Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol Biol Cell 22:4068–4078. doi: 10.1091/mbc.E10-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee H, Lee H, Cho YK. 2017. Cytokeratin7 and cytokeratin19 expression in high grade cervical intraepithelial neoplasm and squamous cell carcinoma and their possible association in cervical carcinogenesis. Diagn Pathol 12:18. doi: 10.1186/s13000-017-0609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santoro A, Pannone G, Ninivaggi R, Petruzzi M, Santarelli A, Russo GM, Lepore S, Pietrafesa M, Laurenzana I, Leonardi R, Bucci P, Natalicchio MI, Lucchese A, Papagerakis S, Bufo P. 2015. Relationship between CK19 expression, deregulation of normal keratinocyte differentiation pattern and high risk-human papilloma virus infection in oral and oropharyngeal squamous cell carcinoma. Infect Agents Cancer 10:46. doi: 10.1186/s13027-015-0041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Birkenkamp-Demtröder K, Hahn SA, Mansilla F, Thorsen K, Maghnouj A, Christensen R, Øster B, Ørntoft TF. 2013. Keratin23 (KRT23) knockdown decreases proliferation and affects the DNA damage response of colon cancer cells. PLoS One 8:e73593. doi: 10.1371/journal.pone.0073593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang N, Zhang R, Zou K, Yu W, Guo W, Gao Y, Li J, Li M, Tai Y, Huang W, Song C, Deng W, Cui X. 2017. Keratin 23 promotes telomerase reverse transcriptase expression and human colorectal cancer growth. Cell Death Dis 8:e2961. doi: 10.1038/cddis.2017.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friese K, Kost B, Vattai A, Marmé F, Kuhn C, Mahner S, Dannecker C, Jeschke U, Heublein S. 2018. The G protein-coupled estrogen receptor (GPER/GPR30) may serve as a prognostic marker in early-stage cervical cancer. J Cancer Res Clin Oncol 144:13–19. doi: 10.1007/s00432-017-2510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanley MA. 2012. Epithelial cell responses to infection with human papillomavirus. Clin Microbiol Rev 25:215–222. doi: 10.1128/CMR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinidis P, Tsikouras P, Iatrakis G, Zervoudis S, Koukouli Z, Bothou A, Galazios G, Vladareanu S. 2016. Human papilloma virus’ life cycle and carcinogenesis. Maedica (Buchar) 11:48–54. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Marsh M. 1956. Original site of cervical carcinoma; topographical relationship of carcinoma of the cervix to the external os and to the squamocolumnar junction. Obstet Gynecol 7:444–452. [PubMed] [Google Scholar]
- 63.Richart RM. 1973. Cervical intraepithelial neoplasia. Pathol Annu 8:301–328. [PubMed] [Google Scholar]
- 64.Herfs M, Yamamoto Y, Laury A, Wang X, Nucci MR, McLaughlin-Drubin ME, Münger K, Feldman S, McKeon FD, Xian W, Crum CP. 2012. A discrete population of squamocolumnar junction cells implicated in the pathogenesis of cervical cancer. Proc Natl Acad Sci U S A 109:10516–10521. doi: 10.1073/pnas.1202684109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang EJ, Quick MC, Hanamornroongruang S, Lai K, Doyle LA, McKeon FD, Xian W, Crum CP, Herfs M. 2015. Microanatomy of the cervical and anorectal squamocolumnar junctions: a proposed model for anatomical differences in HPV-related cancer risk. Mod Pathol 28:994–1000. doi: 10.1038/modpathol.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang SYC, Kannan N, Zhang L, Martinez V, Rosin MP, Eaves CJ. 2015. Characterization of epithelial progenitors in normal human palatine tonsils and their HPV16 E6/E7-induced perturbation. Stem Cell Rep 5:1210–1225. doi: 10.1016/j.stemcr.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark MA, Wilson C, Sama A, Wilson JA, Hirst BH. 2000. Differential cytokeratin and glycoconjugate expression by the surface and crypt epithelia of human palatine tonsils. Histochem Cell Biol 114:311–321. [DOI] [PubMed] [Google Scholar]
- 68.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 69.Werness BA, Levine AJ, Howley PM. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 70.Dyson N, Howley PM, Münger K, Harlow E. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol 75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu W, Putral L, Hengst K, Minto K, Saunders NA, Leggatt G, McMillan NA. 2006. Inhibition of cervical cancer cell growth in vitro and in vivo with lentiviral-vector delivered short hairpin RNA targeting human papillomavirus E6 and E7 oncogenes. Cancer Gene Ther 13:1023–1032. doi: 10.1038/sj.cgt.7700971. [DOI] [PubMed] [Google Scholar]
- 73.Hall AH, Alexander KA. 2003. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol 77:6066–6069. doi: 10.1128/jvi.77.10.6066-6069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buitrago-Pérez A, Garaulet G, Vázquez-Carballo A, Paramio JM, García-Escudero R. 2009. Molecular signature of HPV-induced carcinogenesis: pRb, p53 and gene expression profiling. Curr Genomics 10:26–34. doi: 10.2174/138920209787581235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kypriotou M, Huber M, Hohl D. 2012. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the “fused genes” family. Exp Dermatol 21:643–649. doi: 10.1111/j.1600-0625.2012.01472.x. [DOI] [PubMed] [Google Scholar]
- 76.Bergboer JG, Tjabringa GS, Kamsteeg M, van Vlijmen-Willems IM, Rodijk-Olthuis D, Jansen PA, Thuret JY, Narita M, Ishida-Yamamoto A, Zeeuwen PL, Schalkwijk J. 2011. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am J Pathol 178:1470–1477. doi: 10.1016/j.ajpath.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ishitsuka Y, Huebner AJ, Rice RH, Koch PJ, Speransky VV, Steven AC, Roop DR. 2016. Lce1 family members are Nrf2-target genes that are induced to compensate for the loss of loricrin. J Invest Dermatol 136:1656–1663. doi: 10.1016/j.jid.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stawczyk-Macieja M, Szczerkowska-Dobosz A, Rębała K, Purzycka-Bohdan D. 2015. Genetic background of skin barrier dysfunction in the pathogenesis of psoriasis vulgaris. Postepy Dermatol Alergol 32:123–126. doi: 10.5114/pdia.2014.44003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schulz KS, Mossman KL. 2016. Viral evasion strategies in type I IFN signaling—a summary of recent developments. Front Immunol 7:498. doi: 10.3389/fimmu.2016.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma W, Tummers B, van Esch EM, Goedemans R, Melief CJ, Meyers C, Boer JM, van der Burg SH. 2016. Human papillomavirus downregulates the expression of IFITM1 and RIPK3 to escape from IFNgamma- and TNFalpha-mediated antiproliferative effects and necroptosis. Front Immunol 7:496. doi: 10.3389/fimmu.2016.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang YE, Laimins LA. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol 74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ozbun MA, Meyers C. 1996. Transforming growth factor beta1 induces differentiation in human papillomavirus-positive keratinocytes. J Virol 70:5437–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glick AB. 2012. The role of TGFbeta signaling in squamous cell cancer: lessons from mouse models. J Skin Cancer 2012:249063. doi: 10.1155/2012/249063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kloth JN, Fleuren GJ, Oosting J, de Menezes RX, Eilers PH, Kenter GG, Gorter A. 2005. Substantial changes in gene expression of Wnt, MAPK and TNFalpha pathways induced by TGF-beta1 in cervical cancer cell lines. Carcinogenesis 26:1493–1502. doi: 10.1093/carcin/bgi110. [DOI] [PubMed] [Google Scholar]
- 85.Lee MY, Chou CY, Tang MJ, Shen MR. 2008. Epithelial-mesenchymal transition in cervical cancer: correlation with tumor progression, epidermal growth factor receptor overexpression, and snail up-regulation. Clin Cancer Res 14:4743–4750. doi: 10.1158/1078-0432.CCR-08-0234. [DOI] [PubMed] [Google Scholar]
- 86.Qureshi R, Arora H, Rizvi MA. 2015. EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett 356:321–331. doi: 10.1016/j.canlet.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 87.Klymenko T, Gu Q, Herbert I, Stevenson A, Iliev V, Watkins G, Pollock C, Bhatia R, Cuschieri K, Herzyk P, Gatherer D, Graham SV. 2017. RNA-Seq analysis of differentiated keratinocytes reveals a massive response to late events during human papillomavirus 16 infection, including loss of epithelial barrier function. J Virol 91:e01001-17. doi: 10.1128/JVI.01001-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stanley MA, Browne HM, Appleby M, Minson AC. 1989. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int J Cancer 43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- 89.Allen-Hoffmann BL, Schlosser SJ, Ivarie CAR, Sattler CA, Meisner LF, O’Connor SL. 2000. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J Invest Dermatol 114:444–455. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 90.McLaughlin-Drubin ME, Wilson S, Mullikin B, Suzich J, Meyers C. 2003. Human papillomavirus type 45 propagation, infection, and neutralization. Virology 312:1–7. doi: 10.1016/S0042-6822(03)00312-X. [DOI] [PubMed] [Google Scholar]
- 91.Conway MJ, Alam S, Christensen ND, Meyers C. 2009. Overlapping and independent structural roles for human papillomavirus type 16 L2 conserved cysteines. Virology 393:295–303. doi: 10.1016/j.virol.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Conway MJ, Alam S, Ryndock EJ, Cruz L, Christensen ND, Roden RB, Meyers C. 2009. Tissue-spanning redox gradient-dependent assembly of native human papillomavirus type 16 virions. J Virol 83:10515–10526. doi: 10.1128/JVI.00731-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meyers C, Mayer TJ, Ozbun MA. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol 71:7381–7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McLaughlin-Drubin ME, Meyers C. 2005. Propagation of infectious, high-risk HPV in organotypic “raft” culture. Methods Mol Med 119:171–186. doi: 10.1385/1-59259-982-6:171. [DOI] [PubMed] [Google Scholar]
- 95.Biryukov J, Cruz L, Ryndock EJ, Meyers C. 2015. Native human papillomavirus production, quantification, and infectivity analysis. Methods Mol Biol 1249:317–331. doi: 10.1007/978-1-4939-2013-6_24. [DOI] [PubMed] [Google Scholar]
- 96.Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smits JPH, Niehues H, Rikken G, van Vlijmen-Willems IMJJ, van de Zande GWHJ, Zeeuwen PLJM, Schalkwijk J, van den Bogaard EH. 2017. Immortalized N/TERT keratinocytes as an alternative cell source in 3D human epidermal models. Sci Rep 7:11838. doi: 10.1038/s41598-017-12041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meyers C, Alam S, Mane M, Hermonat PL. 2001. Altered biology of adeno-associated virus type 2 and human papillomavirus during dual infection of natural host tissue. Virology 287:30–39. doi: 10.1006/viro.2001.0968. [DOI] [PubMed] [Google Scholar]
- 99.Peterson GL. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem 83:346–356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession number for our microarray data is GSE129159.