Abstract
In plants, the carotenoid biosynthesis pathway (CBP) is essential for the production of photosynthetic and protective pigments, plant hormones, and visual/olfactory attractants for animal pollinators and seed dispersers. The regulation of carotenoid biosynthesis at the transcriptional level is vitally important for all of these functions and has been the subject of intensive research. Many putative transcriptional regulators, both direct and indirect, have been identified through conventional mutant analysis, transcriptome profiling, yeast one-hybrid screening, and candidate gene approaches. Despite this progress, our understanding of the transcriptional regulation of carotenoid biosynthesis remains fragmented and incomplete. Frequently, a stimulus or regulator is known, but the mechanism by which it affects transcription has not been elucidated. In other cases, mechanisms have been proposed (such as direct binding of a CBP gene promoter by a transcription factor), but function was tested only in vitro or in heterologous systems, making it unclear whether these proteins actually play a role in carotenoid regulation in their endogenous environments. Even in cases where the mechanism is relatively well understood, regulators are often studied in isolation, either in a single plant species or outside the context of other known regulators. This presents a conundrum: why so many candidate regulators but so little consensus? Here we summarize current knowledge on transcriptional regulation of the CBP, lay out the challenges contributing to this conundrum, identify remaining knowledge gaps, and suggest future research directions to address these challenges and knowledge gaps.
Keywords: carotenoid biosynthesis, transcriptional regulation, photosynthetic tissue, flowers, fruits, seeds, roots
Introduction
Carotenoids are red, orange, and yellow pigments produced by photoautotrophic organisms. In the green tissues of plants, carotenoids are essential for light capture, photoprotection, and stabilization of the photosynthetic apparatus (Frank and Cogdell, 1996; Hashimoto et al., 2016). Leaf carotenoids are therefore synthesized in tight coordination with chlorophylls, and their composition is remarkably conserved across higher plants (Goodwin and Britton, 1988; Meier et al., 2011). In addition to their integral roles in photosynthesis, carotenoids accumulate as secondary metabolites in many flowers and fruits to attract pollinators and seed dispersers. Due to their dispensable nature in non-green tissues, these pigments often differ drastically in composition and concentration between species or even between varieties of the same species (Moehs et al., 2001; Bradshaw and Schemske, 2003; Nielsen et al., 2003; Giovannoni, 2007; Ha et al., 2007; Chiou et al., 2010; Yamagishi et al., 2010; Yamamizo et al., 2010; Zhang et al., 2015). Floral and fruit carotenoids can also be cleaved to produce volatile compounds (i.e., scents and flavors), which further enhance plant–animal interactions (Dudareva et al., 2006). Finally, carotenoids serve as precursors for the synthesis of the plant hormones abscisic acid (ABA) and strigolactones, as well as other apocarotenoids that are involved in many developmental processes and stress responses (Cutler et al., 2010; Hou et al., 2016; Jia et al., 2017).
Because of their critical importance in the physiology, development, ecology, and evolution of plants, carotenoid metabolism and function have been intensively studied. The highly conserved carotenoid biosynthesis pathway (CBP) has been characterized in many plants (reviewed in Ruiz-Sola and Rodríguez-Concepción, 2012). In recent years, attention has turned to the regulation of carotenoid accumulation at multiple levels: transcriptional, post-transcriptional, post-translational, storage/degradation, and feedback regulation by end products. This has led to the discovery of numerous carotenoid regulatory mechanisms such as the post-translational regulation of phytoene synthase (PSY) by Orange (Or) (Lu et al., 2006; Zhou et al., 2015), the catabolism of carotenoids by carotenoid cleavage dioxygenases (CCDs) and 9-cis-epoxycarotenoid dioxygenases (NCEDs) (e.g., Auldridge et al., 2006; Ohmiya et al., 2006; Vallabhaneni et al., 2010), and feedback regulation by apocarotenoid-derived signaling molecules (e.g., Avendaño-Vásquez et al., 2014).
In this review, we will focus on the transcriptional regulation of carotenoid biosynthesis genes. For other aspects of carotenoid regulation, we refer readers to several recent reviews (Cazzonelli and Pogson, 2010; Yuan et al., 2015; Nisar et al., 2015; Liu et al., 2015a; Hou et al., 2016; Li et al., 2016; Enfissi et al., 2017; Llorente et al., 2017; Sun et al., 2018a; Ohmiya et al., 2019). In this paper, “transcriptional regulation” of carotenoid biosynthesis genes simply refers to altered transcript abundance in response to a stimulus or as a result of the mutation, knockdown, or overexpression of another gene (e.g., transcription factor, chromatin remodeler). Additionally, we use the term “CBP genes” to refer to the core CBP, from PSY to NSY (see Figure 1 for a schematic of the CBP). Upstream non-carotenoid specific genes [mevalonate (MVA) or methylerythritol phosphate (MEP) pathways], genes of the side branches leading to the production of hormones and apocarotenoids, and genes necessary for the production of uncommon carotenoids (e.g., capsanthin, capsorubin, astaxanthin), are not discussed in detail.
Figure 1.
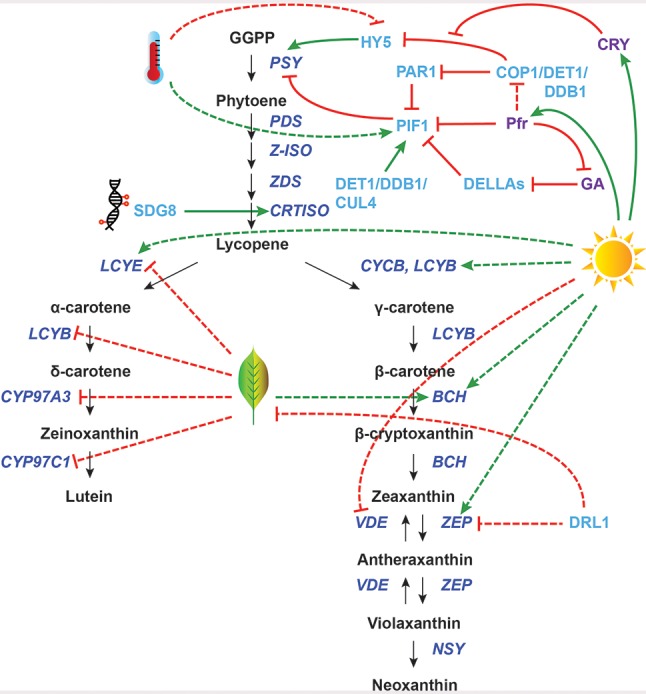
Transcriptional regulation of carotenoid biosynthesis pathway (CBP) genes in photosynthetic tissues. The regulation of CBP genes in response to light (sun), senescence (leaf), and high temperature (thermometer) and by epigenetic controls (DNA) is shown. The carotenoid biosynthesis pathway is in black, with carotenoid biosynthesis genes indicated in dark blue. Carotenoid regulators discussed in the paper are shown in light blue, with other regulators in purple. Green arrows indicate positive regulation, while blunt red arrows indicate negative regulation. Solid lines show direct interactions, while dotted lines show indirect/unknown interactions. GGPP, geranylgeranyl pyrophosphate; PSY, phytoene synthase; PDS, phytoene desaturase; Z-ISO, 15-cis-zeta-carotene isomerase; ZDS, zeta-carotene desaturase; CRTISO, carotene isomerase; LCYB, lycopene beta-cyclase; CYCB, chromoplast-specific lycopene beta-cyclase; BCH, beta-carotene hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin deepoxidase; NSY, neoxanthin synthase; LCYE, lycopene epsilon-cyclase; CYP97A3, cytochrome P450–type beta-hydroxylase; CYP97C1, cytochrome P450–type epsilon-hydroxylase.
We have organized this review by tissue type because carotenoids serve unique functions in photosynthetic tissues, fruits, flowers, seeds, and roots and because the literature is already somewhat structured in this manner. For example, tomatoes are considered the model system for carotenoid biosynthesis in fruits, and Arabidopsis for that in leaves. Even in narrowing the scope to just transcriptional regulation, this review covers about 40 putative regulators of carotenoid biosynthesis genes ( Table S1 ). However, there is little overlap of these numerous regulators between studies of different tissue types or different plant species. In other words, very few of these putative regulators seem to have a conserved function in the transcriptional control of carotenoid biosynthesis genes across tissue types or plant species. We lay out some of the challenges contributing to this conundrum, identify remaining knowledge gaps, and suggest research directions to address these challenges and knowledge gaps in the coming years.
Photosynthetic Tissues
Carotenoids are an integral part of the light harvesting apparatus, capturing light energy and protecting the photosynthetic apparatus from damaging reactive oxygen species (ROS) formed during photosynthesis (Demmig-Adams and Adams, 1996; Pogson et al., 1998; Niyogi, 1999; Baroli and Niyogi, 2000; Dall’Osto et al., 2007). These pigments may also play an important role in temperature stress by scavenging ROS produced by photosystem II (PSII) in extreme temperatures and stabilizing thylakoid membranes (Havaux, 1998; Pospíšil, 2016). Additionally, the developmental program for leaf senescence requires carotenoid precursors for the production of ABA and strigolactones (Nambara and Marion-Poll, 2005; Snowden et al., 2005; López‐Ráez et al., 2008; Alder et al., 2012; Ueda and Kusaba, 2015). Transcriptional regulation of the CBP genes in photosynthetic tissues is therefore highly influenced by light, temperature, and developmental cues.
Light
The light signaling machinery of plants has been extensively characterized in Arabidopsis (e.g., Delker et al., 2014; Dong et al., 2014; reviewed in Jiao et al., 2007; Lau and Deng, 2012), and many key regulatory genes have been identified. One such gene, Phytochrome Interacting Factor 1 (PIF1), is perhaps the best-understood transcriptional regulator of carotenoid biosynthesis. During seedling deetiolation, phytochromes are activated by red light and move from the cytoplasm into the nucleus to interact with signaling components. The bHLH transcription factor PIF1, which represses AtPSY expression in the dark, is phosphorylated by phytochromes upon photoactivation and subsequently degraded by the proteasome (Bae and Choi, 2008; Shen et al., 2008; Shin et al., 2009). This initiates the rapid de-repression of AtPSY as well as genes involved in chlorophyll biosynthesis and chloroplast development.
Toledo-Ortiz et al. (2010) showed that Arabidopsis PIF1 binds directly to a G-box element in the AtPSY promoter in both in vitro and in vivo assays and demonstrated that this binding leads to transcriptional repression. PIFs also contribute to the regulation of AtPSY in mature plants during their daily light/dark cycles. In fully deetiolated plants grown under short-day conditions, higher levels of carotenoids and AtPSY transcripts were found in pif mutants than in wild-type plants (Toledo-Ortiz et al., 2010).
Another important player in light signaling is the bZIP transcription factor Long Hypocotyl 5 (HY5), which acts antagonistically to PIF1 during photomorphogenesis. HY5 activates carotenoid and chlorophyll biosynthesis genes, as well as genes involved in chloroplast development and cotyledon expansion. Unlike PIF1, which is stabilized in the dark by the DET1/DDB1/CUL4 complex, HY5 is stabilized by light (the COP1/DDB1/CUL4 complex targets HY5 for degradation in the dark) (Shi et al., 2015; Zhu et al., 2015). Interestingly, HY5 and PIF1 bind to the same G-box element of the AtPSY promoter, which serves as a relatively simple switch to promote deetiolation upon illumination. This switch also functions in the daily light/dark cycles of mature plants (Toledo-Ortiz et al., 2014).
PIFs are also involved in shade-triggered reduction of carotenoid accumulation in Arabidopsis leaves, through an HY5-independent mechanism. When there is a low red/far red (R/FR) ratio of light in shade conditions, Phytochrome Rapidly Regulated 1 (PAR1), a bHLH co-factor, is upregulated and induces AtPSY expression by physically interacting with PIF1 and preventing it from sitting on the AtPSY promoter (Bou-Torrent et al., 2015).
Carotenoid biosynthesis is also induced when greening is de-repressed in the dark, which can be achieved through the blockage of gibberellic acid (GA) biosynthesis (Rodríguez-Villalón et al., 2009; Toledo-Ortiz et al., 2010). GA negatively regulates DELLA proteins, which in turn negatively regulate PIFs. In Arabidopsis GA biosynthesis mutants, AtPSY transcript levels in etiolated seedlings are elevated relative to the wild type. In “double” mutants lacking both GA and DELLAs, this response is repressed. Treatment of wild-type plants with a GA inhibitor reduced PIF1 binding to the G-box in the AtPSY promoter (Cheminant et al., 2011).
While the PIF1/HY5 regulatory mechanism is relatively well understood, there is still much to be learned about the transcriptional regulation of carotenoid biosynthesis during deetiolation. For instance, many other carotenoid biosynthesis genes are de-repressed during photomorphogenesis in Arabidopsis, such as AtBCH2, AtZEP, and AtLCYE (which are constitutively de-repressed in pif mutants). Of these, only AtBCH2 has a G-box in its promoter, but this G-box is not bound by PIF1 (Toledo-Ortiz et al., 2010). Additionally, truncated AtPSY genes lacking G-boxes in Arabidopsis are still light responsive (Welsch et al., 2003), indicating that there are other factors involved in light responsiveness unrelated to the PIF pathway and/or that PIFs may bind other elements.
Indeed, a chromatin immunoprecipitation–microarray (ChIP–chip) analysis in Arabidopsis seeds showed that PIF1 binds to 748 sites, only 59% of which contain G-box elements (Oh et al., 2009); additionally, only a small fraction of G-box elements in the Arabidopsis genome have been shown to be bound by PIFs (Kim et al., 2016). PIF1 has been shown to bind PIF binding E-box (PBE) elements in vitro, though this interaction is relatively weak (Kim et al., 2008; Pfeiffer et al., 2014). PIF1-interacting transcription factors may facilitate the targeting of PIF1 to specific sites, particularly those containing multiple G-boxes and/or G-box coupling elements (GCEs) (Kim et al., 2016). Thus, non-canonical PIF binding sites may play a role in PIF1 regulation of other CBP genes.
Another thing to consider is that PIF1 is certainly not a specific carotenoid regulator: it has been shown to directly regulate the chlorophyll biosynthesis gene AtPOR by binding its promoter and to indirectly regulate other genes in that pathway (Moon et al., 2008). This may account in part for the tight coordination between chlorophyll and carotenoid biosynthesis in green tissues during photomorphogenesis. However, for PIF1 to function in chromoplast-containing tissues, the regulation of carotenoid and chlorophyll biosynthesis must be decoupled (see the "Fruits" section).
The intensity of light affects both carotenoid concentration and composition (Hirschberg, 2001). High light stress produces ROS such as triplet chlorophyll and singlet oxygen, which can be deactivated by carotenoids. Additionally, excess excitation energy in the photosystems can be effectively dissipated by carotenoids, particularly zeaxanthin (Dall’Osto et al., 2012; Jahns and Holzwarth, 2012). High light causes a rapid decrease in lumen pH, which increases violaxanthin deepoxidase (VDE) enzyme activity, converting violaxanthin to zeaxanthin ( Figure 1 ). Although this interconversion between violaxanthin and zeaxanthin (i.e., the xanthophyll cycle) is regulated post-translationally by activation and inactivation of the VDE enzyme (Müller et al., 2001), high light does induce transcriptional changes of CBP genes as well. For example, the ratio of LCYB to LCYE transcripts increases fivefold in both Arabidopsis and tomato leaves in high light relative to low light (Hirschberg, 2001), which channels metabolic flux through the branch of the CBP that produces zeaxanthin ( Figure 1 ). AtBCH2 transcription is also upregulated by high light treatment in Arabidopsis (Rossel et al., 2002), likely enhancing the metabolic flow towards xanthophylls as well. However, the transcription factors responsible for these CBP gene expression changes remain unknown.
Ultraviolet B (UV-B) light also triggers the production of carotenoids, which are directly linked to photoprotection of the photosynthetic apparatus (Middleton and Teramura, 1993). Irradiation of Arabidopsis plants with UV-B causes slight decreases in lutein and β-carotene content but a substantial increase in zeaxanthin. Correspondingly, the expression of AtPSY, AtZDS, and AtBCH1/2 is enhanced. Loss-of-function AtLYCE (LUT2) mutants accumulate more β-carotene branch xanthophylls compared to the wild type and consequently show decreased DNA and oxidative damage under UV-B light (Emiliani et al., 2018). Interestingly, the UV-B response pathway and the photomorphogenesis pathway share several common components. For example, the active form of UV RESPONSE LOCUS 8 (UVR8), a UV-B–specific photoreceptor, directly interacts with COP1 and regulates HY5 expression (Brown et al., 2005; Brown and Jenkins, 2008; Cloix and Jenkins, 2008; Favory et al., 2009; Rizzini et al., 2011). Therefore, it would not be surprising if the transcriptional regulators of the CBP genes downstream of AtPSY (e.g., AtZDS, AtBCH1/2) turned out to be the same for both UV-B response and photomorphogenesis.
Temperature
The PIF1/HY5 switch can also control AtPSY expression in response to temperature cues (Toledo-Ortiz et al., 2014). In addition to being stabilized by light, HY5 is stabilized by cold temperatures (Catalá et al., 2011). In a ChIP assay, both the AtPSY and AtVDE promoters were preferentially bound by HY5 at low compared to ambient temperatures. Furthermore, the rapid increase of HY5 and decrease of PIF1 protein levels when etiolated Arabidopsis seedlings were exposed to light was more robust at lower temperature (Toledo-Ortiz et al., 2014). This would lead to higher expression of AtPSY at lower temperatures. These findings are echoed by experiments in maize (Zea mays), which showed that ZmPSY1 and ZmPSY2 expression decreases at high temperatures in both light and dark conditions (Li et al., 2008a).
The sensitivity of PSY transcript levels to temperature cues indicates that transcriptional regulation of the CBP may be partially responsible for temperature stress responses. It makes sense that high light and low temperature responses would overlap, because the consequences of these stressors are similar: they both produce ROS and prevent the repair of PSII damage (reviewed in Szymańska et al., 2017). Thus, PIF1/HY5 regulation of PSY may be an important mechanism for increasing carotenoids to scavenge ROS. However, high temperature stress, which also produces damaging ROS, reduces the expression of PSY, indicating that transcriptional regulation of this gene is not responsible for high temperature stress response.
Senescence
Leaf senescence is a developmentally controlled process leading eventually to organ death. The breakdown and recycling of macromolecules from senescing leaves allow plants to reallocate resources to reproduction or new growth (Gan and Amasino, 1997; Lim et al., 2007). One of the most prominent phenotypes during senescence is leaf yellowing due to the breakdown of pigments in chloroplasts (Ougham et al., 2005). Although all photosynthetic pigments are eventually broken down, chlorophylls are usually lost more rapidly than carotenoids. There are also changes in the composition of carotenoids during senescence: while all decline, lutein remains at relatively stable levels compared to neoxanthin, violaxanthin, and antheraxanthin (Biswal, 1995; Britton and Young, 1989). This is perhaps due to the cleavage of β-carotene branch carotenoids for the production of strigolactones and ABA, which further promote leaf senescence (Yang et al., 2003; Ueda and Kusaba, 2015).
The transcription of CBP genes changes dramatically during leaf senescence. In an Arabidopsis microarray analysis of senescing leaves, AtLCYE, AtCYP97C1, and AtCYP97A3 expression drops, reducing flux through the α-carotene branch of the pathway. This is followed by the induction of AtBCH1, which might be important for downstream hormone production (Breeze et al., 2011). Similar trends can also be seen in woody perennial plants: in aspen trees (Populus tremula), PtBCH2 is significantly induced by autumn senescence (Andersson et al., 2004).
The only known potential regulator of CBP genes during leaf senescence is DRL1 from grapevine (Vitis vinifera), encoding a NAC transcription factor. Overexpression of DRL1 in tobacco has been shown to delay leaf senescence and decrease ABA levels. The expression of NtZEP1 and carotenoid cleavage genes is reduced in these transgenic plants (Zhu et al., 2019). However, the endogenous function of DRL1 in grapevine is yet to be reported.
Other CBP Regulators in Photosynthetic Tissues
Besides ZEP, two other CBP genes downstream of PSY have potential known regulators in photosynthetic tissues. In Arabidopsis leaves, the Ethylene Response Factor (ERF) transcription factor RELATED TO AP2 2 (RAP2.2) was shown to bind the AtPSY and AtPDS promoters in vitro (Welsch et al., 2007). However, overexpression of AtRAP2.2 in Arabidopsis leaves did not result in higher AtPSY or AtPDS messenger RNA (mRNA) levels, nor a change in carotenoid concentration. A knockout mutant of AtRAP2.2 was not available, probably due to lethality. These results leave the endogenous function of AtRAP2.2 in carotenoid regulation ambiguous.
AtCRTISO is another CBP gene in photosynthetic tissues with an identified regulator: the histone methyltransferase Set Domain Group 8 (SDG8). sdg8 mutants produce low levels of CRTISO mRNA, which correlates with reduced trimethyl-H3K4 and increased dimethyl-H3K4 around the CRTISO transcription start site (Cazzonelli et al., 2009). Although this mechanism is well understood, SDG8 is certainly not a specific carotenoid regulator: mutation in this gene downregulates 85 other genes and causes broad pleiotropic effects, including increased shoot branching, reduced fertility, and early flowering. It is currently unknown whether the function of SDG8 in carotenoid biosynthesis is conserved across species.
As described above, only a few regulators of carotenoid biosynthesis in green tissues, such as PIF1 and HY5, have been well characterized and shown to directly regulate PSY in Arabidopsis. Additionally, their importance as regulators of photomorphogenesis, responses to daily light/dark cycles, and temperature has been established. However, we still know very little about what regulates most CBP genes downstream of PSY in green tissues.
Fruits
The ripening developmental program of fleshy fruits involves changes in texture (alteration of cell wall composition, reduction in turgor pressure), flavor and aroma (alteration of volatiles, sugar/starch, and acid metabolism), and color (alteration of chlorophyll, carotenoid, and flavonoid content) (Klee and Giovannoni, 2011). Many economically important fruits (e.g., tomato, orange, papaya) produce copious carotenoids during ripening, and therefore, the transcriptional control of CBP genes during fruit ripening has attracted considerable research efforts.
Tomatoes
The foremost model for carotenoid regulation during fruit ripening is tomato (Solanum lycopersicum) ( Figures 2 – 3 ), which primarily accumulates lycopene in mature fruits. During tomato fruit development, transcription of the early CBP genes SlPSY1 and SlPDS increases, while the transcription of SlLCYE and SlLCYB, which convert lycopene to other downstream products, decreases (Pecker et al., 1992; Giuliano et al., 1993; Fraser et al., 1994; Corona et al., 1996; Ronen et al., 1999; Alba et al., 2005). Tomato fruits are climacteric, and thus, ethylene biosynthesis and signaling are necessary for the onset and completion of ripening (reviewed in Liu et al., 2015b). CBP gene regulation is also tightly coupled with these processes, and many of the regulators that affect CBP gene expression also affect other aspects of ripening. These may therefore be considered general regulators of ripening, oftentimes functioning far upstream of the CBP genes.
Figure 2.
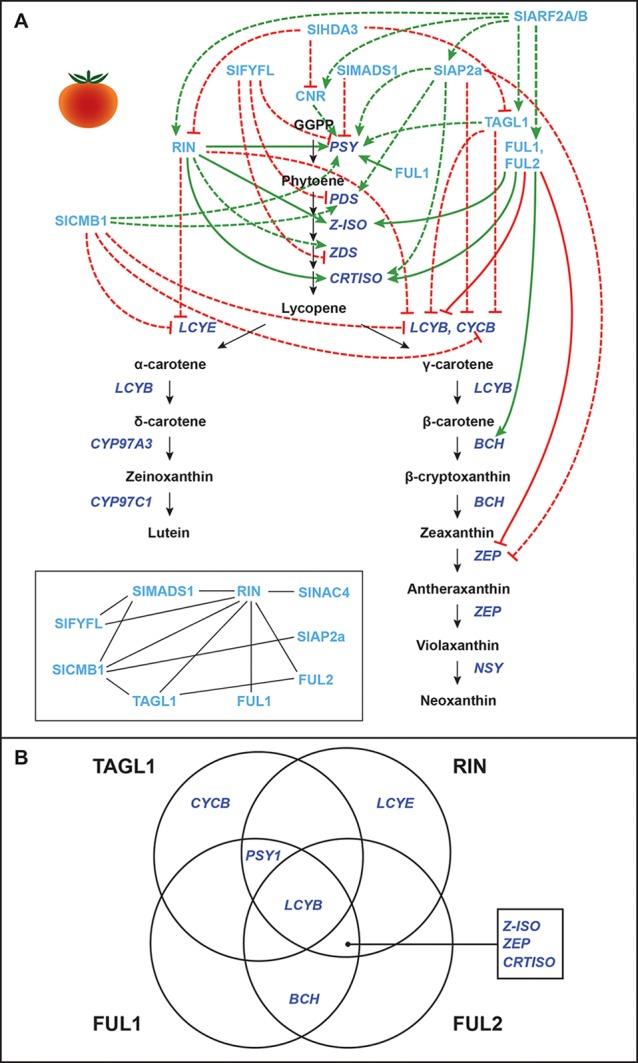
Transcriptional regulation of CBP genes in tomato fruits: “ripening quartet”–related proteins. (A) The regulation of CBP genes in tomato fruits. The carotenoid biosynthesis pathway is in black, with carotenoid biosynthesis genes indicated in dark blue. Carotenoid regulators discussed in the paper are shown in light blue. Green arrows indicate positive regulation, while blunt red arrows indicate negative regulation. Solid lines show direct interactions, while dotted lines show indirect/unknown interactions. Inset: protein–protein interactions between “ripening quartet” regulators. (B) Venn diagram showing the direct binding of TAGL1, RIN, FUL1, and FUL2 to CBP gene promoters.
Figure 3.
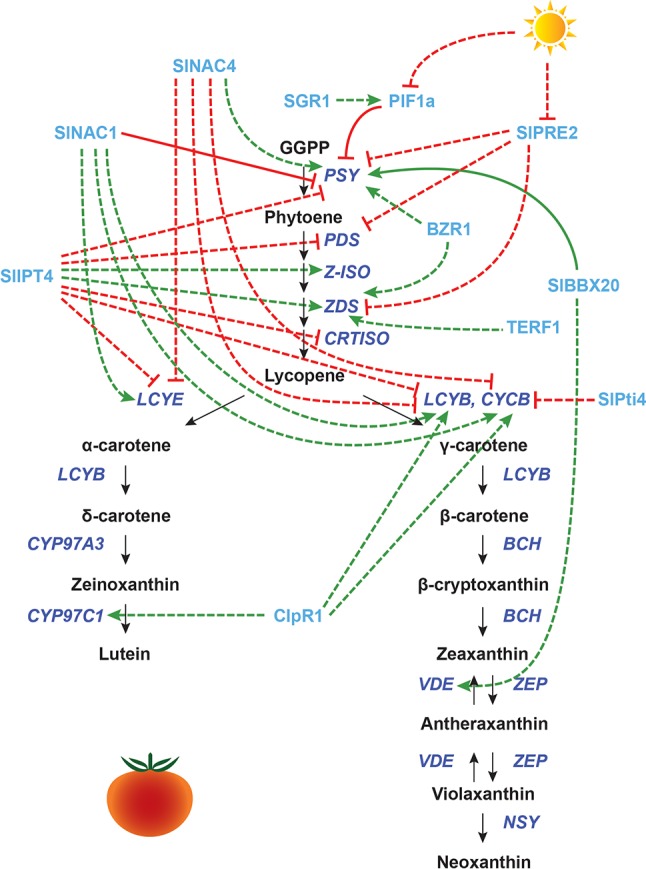
Transcriptional regulation of CBP genes in tomato fruits: other proteins. The regulation of CBP genes in tomato fruits. The carotenoid biosynthesis pathway is shown in black, with carotenoid biosynthesis genes indicated in dark blue. Carotenoid regulators discussed in the paper are shown in light blue. Green arrows indicate positive regulation, while blunt red arrows indicate negative regulation. Solid lines show direct interactions, while dotted lines show indirect interactions.
Several MADS-box ripening regulators affect the expression of CBP genes in tomatoes, and a ripening model similar to the floral quartet model has been proposed ( Figure 2 ). In this model, different combinations of MADS-box proteins bind different target genes (Bemer et al., 2012; Shima et al., 2013; Fujisawa et al., 2014). These “ripening quartet” regulators include Tomato AGAMOUS-LIKE1 (TAGL1), Ripening Inhibitor (RIN), FRUITFULL1 (FUL1), and FUL2. These transcription factors have both overlapping and individual (but never antagonistic) contributions to the expression of CBP genes, with the total effect being the positive regulation of SlPSY1, SlPSY2, SlZDS, SlZ-ISO, SlCRTISO, and SlBCH, and the negative regulation of SlLCYB, SlLCYE, and SlCYCB (the chromoplast-specific paralogue of SlLCYB) (Vrebalov et al., 2002; Ito et al., 2008; Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010; Fujisawa et al., 2011; Fujisawa et al., 2012; Fujisawa et al., 2013; Fujisawa et al., 2014; Martel et al., 2011; Bemer et al., 2012; Qin et al., 2012; Shima et al., 2013; Zhong et al., 2013). For the effects of each individual regulator, see Figure 2A .
These MADS-box proteins exert their effects over CBP gene transcription both directly by binding the promoters of some genes and indirectly by unknown mechanisms. Various studies have shown the promoter of SlLCYB to be bound by all four regulators; the promoter of SlPSY1 to be bound by TAGL1, RIN, and FUL1; the promoters of SlZ-ISO, SlCRTISO, and SlZEP by RIN, FUL1, and FUL2; the promoter of SlBCH by FUL1 and FUL2; the promoter of SlCYCB by TAGL1; and the promoter of SlLCYE by RIN (Ito et al., 2008; Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010; Fujisawa et al., 2011; Fujisawa et al., 2012; Fujisawa et al., 2013; Fujisawa et al., 2014; Martel et al., 2011; Bemer et al., 2012; Qin et al., 2012; Shima et al., 2013; Zhong et al., 2013; see Figure 2B for a graphical summary).
It should be mentioned that ChIP studies assessing RIN binding to target gene promoters have produced inconsistent results. Some studies have shown that the SlPSY1 promoter is bound by RIN (Martel et al., 2011; Zhong et al., 2013; Fujisawa et al., 2013), while others have not detected this interaction or had inconclusive results (Fujisawa et al., 2011; Fujisawa et al., 2012; Fujisawa et al., 2013). Also, though FUL1 has been shown to bind the promoter of SlPSY1 and promote its expression (Shima et al., 2013; Fujisawa et al., 2014), one study in which FUL1 and FUL2 were silenced showed that fruits did not have altered expression of SlPSY1 (Bemer et al., 2012). The loss of FUL1 function may have been compensated by RIN and TAGL1, and thus, its endogenous role in regulating carotenoid biosynthesis remains unclear.
Other CBP-regulating MADS-box genes in tomato that interact with or regulate the ripening quartet are SlMADS1, SlFYFL, and SlCMB1 ( Figure 2 ). SlMADS1 and SlFYFL are both negative regulators of carotenoid biosynthesis, suppressing SlPSY1 expression (SlFYFL also suppresses SlPDS and SlZDS) (Dong et al., 2013; Xie et al., 2014). SlCMB1 promotes the expression of SlPSY1 and SlPDS, while suppressing SlCYCB, SlLCYB, and SlLCYE transcription (Zhang et al., 2018a).
Many other regulators play a role in CBP gene regulation during fruit ripening, especially those involved in hormone synthesis and signaling. SlAP2a (an APETALA2/ERF protein) positively regulates fruit ripening, promoting the expression of SlPSY1, SlCRTISO, SlBCH, and SlPDS1, and repressing SlZEP1 and SlCYCB (Chung et al., 2010; Karlova et al., 2011). The overexpression of Tomato Ethylene Response Factor 1 (TERF1) induces SlZDS expression, perhaps via plastid-to-nucleus retrograde signaling (Wu et al., 2019). Silencing LeHB-1 (encoding an HD-Zip transcription factor) inhibits fruit ripening and lycopene production, most likely through inhibition of ethylene biosynthesis (Lin et al., 2008). However, the transcript levels of CBP genes were not assessed in the LeHB-1 silenced lines.
NAC family transcription factors involved in ethylene biosynthesis also affect the transcription of CBP genes: SlNAC4 positively regulates SlPSY1 and negatively regulates SlCYCB, SlLCYB, and SlLCYE (Zhu et al., 2014), while SlNAC1 has the opposite effect (Ma et al., 2014; Meng et al., 2016). SlNAC1 has been shown by yeast one-hybrid assay to interact with the promoters of SlPSY1 and ethylene biosynthesis genes (Ma et al., 2014).
Additionally, overexpression of SlNAC1 increases the amount of ABA, potentially by providing the carotenoid precursors for its synthesis (Ma et al., 2014). In wild-type fruits, ABA production precedes ethylene production and may be an important trigger for ripening (Zhang et al., 2009). ABA application promotes several ripening processes, including carotenoid accumulation, by regulating transcription factors for ethylene biosynthesis and signaling (Mou et al., 2016). Another regulator related to ethylene–ABA is SlPti4 (also a member of the AP2/ERF superfamily). Silencing of SlPti4 enhances ABA production while decreasing ethylene production, which induces the expression of SlCYCB and the consequent color change from red to orange (Sun et al., 2018b). Finally, the zinc finger transcription factor SlZFP2 inhibits fruit ripening by negatively regulating ABA biosynthesis (Weng et al., 2015). SlZFP2-overexpressing fruits accumulate more β-carotene and lycopene compared to the wild type, but it is unclear whether SlZFP2 actually regulates CBP gene expression, as the transcript levels of CBP genes were not reported in this study (Weng et al., 2015).
Besides ethylene and ABA, other plant hormones are also involved in tomato fruit ripening, with complex actions and interactions. Auxin acts antagonistically to ethylene, delaying ripening. Exogenous application of an auxin inhibitor to tomato fruits produces an effect similar to ethylene application, indicating that perhaps the presence vs. absence of auxin, not ethylene per se, determines ripening in tomato (Su et al., 2015). RNA interference (RNAi) mediated silencing of two paralogues of Auxin Response Factor 2, SlARF2A and SlARF2B, caused downregulation of SlPSY1, SlPDS, and SlZDS and upregulation of SlLCYB1, SlLCYB2, and SlCYCB (Hao et al., 2015). Several key ripening genes (e.g., RIN, CNR, NOR, AP2a, TAGL1, FUL1/2) were also downregulated in these RNAi lines, indicating that SlARF2A/B may regulate CBP gene expression through the ripening factors.
Brassinosteroid (BR) application to pericarp discs induces lycopene formation (Vardhini and Rao, 2002). Transgenic lines overexpressing Brassinazole Resistant 1 (BZR1) have increased transcript levels of SlPSY1 and SlZDS (Liu et al., 2014). Cytokinins (CKs) are also involved in ripening: SlIPT4, which catalyzes the rate-limiting step of CK biosynthesis, represses the expression of SlPSY1 and SlPDS, while upregulating the expression of SlZ-ISO and SlZDS (Zhang et al., 2018b). Given that both BR and CK are isoprenoids synthesized using precursors from the MEP/MVA pathways, it is perhaps not too surprising that their biosynthesis and signaling affect carotenoid biosynthesis (Sakakibara, 2006; Andrade et al., 2017). Another hormone, jasmonic acid (JA), promotes lycopene accumulation and CBP gene expression, even in ethylene-insensitive mutants (Liu et al., 2012), but the mechanism is unknown.
Light is also an important regulator of CBP genes during tomato fruit development. Interestingly, the shading response seen in Arabidopsis leaves is utilized in tomatoes for fruit ripening. In an in vivo ChIP assay, SlPIF1a binds to a PBE-box located in the promoter of the tomato SlPSY1 gene to repress its expression. This repression is only maintained when the R/FR light ratio is low. In an elegant experiment, Llorente et al. (2016) showed that the presence of chlorophyll in the immature green fruit pericarp acts as a self-shading mechanism, giving a low R/FR ratio. When developmentally triggered degradation of chlorophyll begins, the R/FR ratio increases, and SlPSY is de-repressed by phytochrome-mediated degradation of SlPIF1a.
Other light signaling components have been examined in tomato fruits. Mutations in the high pigment genes HP1 (DDB1) and HP2 (DET1) give increased amounts of chlorophyll in immature fruits and increased amounts of carotenoids in mature fruits (Mustilli et al., 1999; Levin et al., 2003; Lieberman et al., 2004). DDB1 and DET1, which are known to interact with PIF1/HY5 and regulate their protein levels in Arabidopsis leaves, do not appear to strongly affect the expression of CBP genes in tomato fruits. The high pigment levels are probably due to changes in plastid size and/or number that increase the storage capacity of carotenoids. CBP gene expression has been shown to be slightly altered in tomato det1 mutants compared to wild type: in a transcriptome study, SlPSY1, SlPDS, and SlLCYB transcript levels were all slightly elevated in immature fruits, while SlCYCB transcript levels were reduced. At the mature red stage, SlPSY1, SlZDS, and SlCYCB were upregulated, but carotenoid biosynthesis is not at peak levels in mature fruit, and thus, these differences may not be developmentally relevant (Kolotilin et al., 2007).
Another light-responsive CBP regulator in tomato is SlPRE2, an atypical bHLH transcription factor whose expression is repressed in high light. When SlPRE2 is overexpressed, it alters the growth of stems and leaves, promotes hypocotyl elongation, and downregulates chlorophyll biosynthesis genes as well as SlPSY1, SlPDS, and SlZDS. Transcript levels of HY5 are also reduced, which could explain the low level of SlPSY1 transcripts (Zhu et al., 2017a).
Other proteins appear to indirectly affect the transcription of CBP genes through plastid processes. Overexpression of the B-box protein BBX20 increases the chlorophyll and carotenoid content in tomato leaves and fruits, inducing SlPSY1 and SlVDE expression. BBX20 was found to bind to a G-box in the SlPSY1 promoter in vitro and interacts with DET1. Carotenoid content is probably enhanced because of both the increase in SlPSY1 expression and an increased number of chloroplasts. BBX20 overexpression does not affect carotenoid accumulation or CBP transcription in flowers, indicating that distinct mechanisms operate in fruits and flowers (Xiong et al., 2019).
The Clp protease ClpR1 enhances transcript levels of SlLCYB, SlCYCB, and SlCYP97C11. This gene probably affects the transcription of carotenoid biosynthesis genes through its contributions to the chloroplast-to-chromoplast transition (D’Andrea et al., 2018). The Stay Green 1 (SGR1) protein represses SlPSY1 expression (although the effect appears minor). When SGR1 is knocked down, fruits have lower PIF1 expression and altered ethylene signal transduction. SGR1 also interacts directly with SlPSY1 protein, and knockdown of this gene induces early chloroplast-to-chromoplast transition, indicating that this gene has many regulatory functions (Luo et al., 2013).
Epigenetic regulation is also crucial to fruit ripening and carotenoid biosynthesis in tomato. Zhong et al. (2013) showed that about 1% of the tomato genome shows differential methylation during fruit ripening by chemically inhibiting methyltransferases. For a review on epigenetic controls in tomato fruit ripening, see Giovannoni et al., 2017; for a review on epigenetic control of carotenoid biosynthesis, see Arango et al., 2016.
Colorless non-ripening (Cnr) tomato mutants do not express SlPSY1 (Thompson et al., 1999, Eriksson et al., 2004) and thus do not produce lycopene. The CNR locus was shown to be a SQUAMOSA Promoter Binding Protein–like (SPL) gene, with the causal mutation occurring in the promoter. This mutation was an epimutation, with increased methylation in mutants (Manning et al., 2006). When wild-type tomato fruits are treated with a methylation inhibitor, they produce early-ripening red sectors (which have unmethylated CNR promoters). The sectors that remain green also remain hypermethylated, suggesting that methylation of ripening genes acts as a developmental block. SlPSY1 transcripts were isolated from early-ripening sectors, suggesting that this fruit ripening mechanism is upstream of carotenogenesis and other ripening processes (Zhong et al., 2013). The CNR SPL gene might be a conserved carotenoid regulator across species. Constitutive expression of AtmiR156b (which silences AtSPL3, a CNR SPL homologue; Wang et al., 2009) produces excess amounts of lutein and β-carotene in Brassica napus seeds, though CBP gene expression was not assessed (Gandikota et al., 2007; Wei et al., 2010; Arango et al., 2016). None of the CBP genes has a sequence complementary to AtmiR156b, so this increase in carotenoids is indirect (Wei et al., 2010), likely through the SPL gene.
Another epigenetic regulator, the tomato histone deacetylase gene SlHDA3, negatively regulates SlPSY1 expression, while positively regulating SlCYCB, SlLCYB, and SlLCYE. Ethylene biosynthesis genes and cell wall metabolism genes were also negatively regulated by SlHDA3, as were RIN, Cnr, and TAGL1 (Guo et al., 2018).
Other Climacteric Fruits
Putative transcriptional regulators have also been identified in other climacteric fruits ( Figure 4 ). In papaya (Carica papaya), in vitro and dual luciferase assays in a heterologous host (i.e., tobacco) showed that the ethylene response protein CpEIN3a binds to and activates the promoters of CpPDS4 and CpBCH. Further, its interacting partner CpNAC2 binds to and activates the promoters of CpPDS2, CpPDS4, CpZDS, CpLCYE, and CpBCH. The interaction between CpEIN3a and CpNAC2 increases activation of these genes (Fu et al., 2017). Also in papaya, the NAC family transcription factor CpNAC1 has been shown to bind to the CpPDS2 and CpPDS4 promoters in vitro and activate them in transient assays in tobacco (Fu et al., 2016). Two other papaya transcription factors, CpbHLH1 and CpbHLH2, bind to the promoters of CpCYCB and CpLCYB in vitro and in tobacco transient assays, with CpbHLH1 acting as a repressor and CpbHLH2 as an activator (Zhou et al., 2019). However, the endogenous functions of these papaya genes are unknown.
Figure 4.
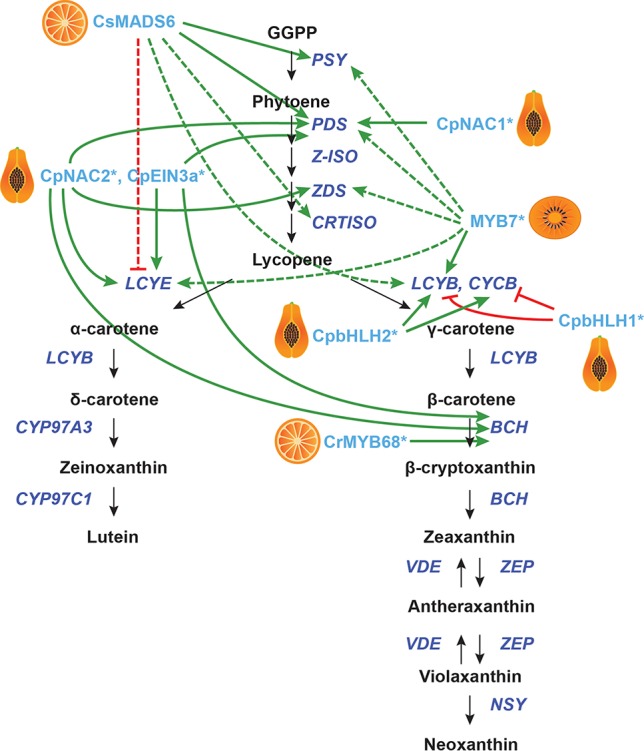
Transcriptional regulation of CBP genes in other fruits. The regulation of CBP genes in citrus, peach, papaya, and orange kiwi. The carotenoid biosynthesis pathway is shown in black, with carotenoid biosynthesis genes indicated in dark blue. Carotenoid regulators discussed in the paper are shown in light blue. Green arrows indicate positive regulation, while blunt red arrows indicate negative regulation. Solid lines show direct interactions, while dotted lines show indirect/unknown interactions.
In kiwifruit (Actinidia deliciosa), a promoter screen identified AdMYB7 (among other MYBs) as a putative regulator of AdLCYB. The authors confirmed interaction between AdMYB7 and the AdLCYB promoter in a gel mobility shift assay. When AdMYB7 was overexpressed in tobacco in a transient assay, the carotenoid content increased twofold. Stable overexpression of this gene in tobacco gave increased expression of NbPSY, NbPDS, NbZDS, NbLCYB, NbLCYE, and chlorophyll biosynthesis genes (Ampomah-Dwamena et al., 2019).
Non-Climacteric Fruits
Watermelon (Citrullus lanatus) fruits, like tomatoes, accumulate lycopene. However, they are non-climacteric, meaning that their ripening process is not concurrent with a spike of ethylene production and cellular respiration. The expression of watermelon homologues of CNR, SlAP2a, and SlERF6 was correlated with ripening and carotenoid biosynthesis; however, that of RIN, TAGL1, NAC-NOR, DET1, DDB1, and CUL4 was not (Grassi et al., 2013). This suggests that some regulators might be common to carotenoid-accumulating fruits, while others are potentially involved in other aspects of ripening, such as ethylene biosynthesis/perception and light sensing/plastid transition.
In citrus, a yeast one-hybrid screen using the promoters of CsLCYB1 and CsLYCB2 identified the MADS-box gene CsMADS6 (a homologue of TAGL1), which is expressed in flowers and fruits. Overexpression of CsMADS6 in citrus calli gave increased expression of CsPSY, CsPDS, CsCRTISO, CsLCYB2, and CsBCH, while transcription of CsLCYE was repressed. Additionally, the transcript levels of citrus HY5 and RAP2.2 homologues increased, while PIF1 levels were reduced (RIN and FUL are not expressed in citrus calli). CsMADS6 can bind the promoters of CsPSY and CsPDS in vitro to activate them (Lu et al., 2018). Another citrus gene, the R2R3-MYB CrMYB68, has been shown to directly and negatively regulate CrBCH2 by Electromobility Shift Assays and dual luciferase assays, but the endogenous function of this gene in citrus is unknown (Zhu et al., 2017b).
Flowers
The coordinated transcriptional regulation of CBP genes is largely responsible for the coloration of carotenoid-pigmented flowers (e.g., Corona et al., 1996; Moehs et al., 2001; Chiou et al., 2010; Dalal et al., 2010; Yamagishi et al., 2010; Yamamizo et al., 2010; Ohmiya, 2013; Zhang et al., 2015; Kishimoto et al., 2018; Wang and Yamagishi, 2019). However, very few genes regulating the transcription of CBP genes in flowers have been identified ( Figure 5 ). Notably, none of the regulators involved in tomato fruit ripening dramatically affect flower petal color, even though tomatoes have carotenoid-pigmented flowers. This is probably because fruit and flower carotenoid biosynthesis are differentially regulated: while tomato fruits accumulate lycopene, the main carotenoid components in tomato flowers are the xanthophylls violaxanthin and neoxanthin (Galpaz et al., 2006). Most flowers studied to date primarily store xanthophylls and/or β-carotene (Ohmiya, 2011).
Figure 5.
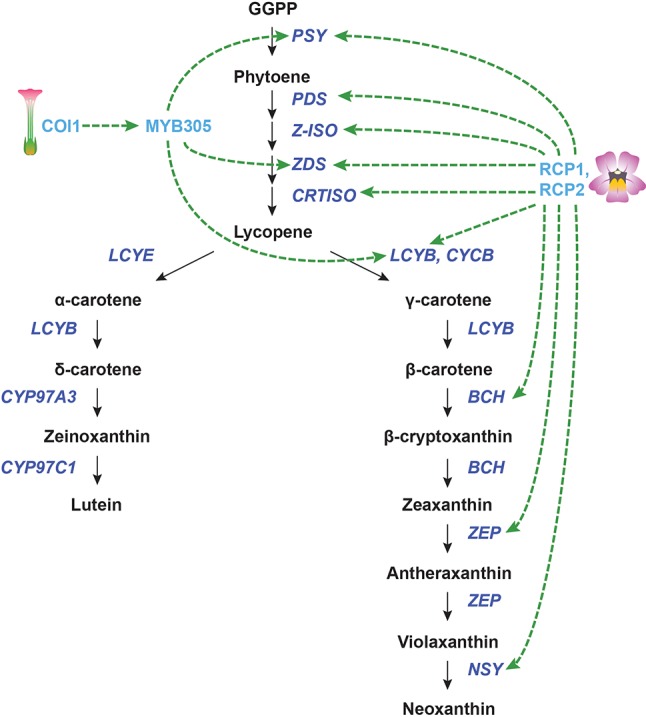
Transcriptional regulation of CBP genes in flowers. The regulation of CBP genes in Nicotiana tabacum and Mimulus species. The carotenoid biosynthesis pathway is shown in black, with carotenoid biosynthesis genes indicated in dark blue. Carotenoid regulators discussed in the paper are shown in light blue. Green arrows indicate positive regulation, while blunt red arrows indicate negative regulation. Dotted lines show indirect/unknown interactions.
The F-box protein CORONATINE INSENSITIVE 1 (COI1) is necessary for the perception of jasmonic acid JA. In addition to its many other functions, COI1-mediated JA signaling has been implicated in the production of floral and extrafloral nectar. Silencing of COI1 in Nicotiana tabacum not only suppresses nectar production in flowers but also decreases the amount of β-carotene in the floral nectary. The expression of NtPSY, NtZDS, and NtLCYB was reduced in the COI1-silenced lines during carotenoid accumulation (and persisted throughout development for NtPSY). Silencing COI1 also strongly downregulated the R2R3-MYB gene MYB305 in floral nectaries, suggesting that COI1 works upstream of this gene (Wang et al., 2014). It was previously shown that RNAi knockdown of MYB305 causes the loss of β-carotene in floral nectaries (though gene expression of the CBP genes was not analyzed) (Liu et al., 2009). MYB305, then, may mediate the transcriptional regulation of NtPSY, NtZDS, and NtLCY.
In the monkeyflower species Mimulus lewisii, an R2R3-MYB gene called Reduced Carotenoid Pigmentation 1 (RCP1) positively regulates all of the CBP genes expressed in flowers, contributing to the bright yellow coloration of the floral nectar guides (Sagawa et al., 2016). Although this seems like a promising global regulator for carotenoid biosynthesis in flowers, the DNA binding site/s of this transcription factor is/are yet to be determined. It is unknown whether RCP1 directly or indirectly activates transcription of the CBP genes. Another gene from monkeyflowers, RCP2, is also necessary for carotenoid biosynthesis in petals. RCP2 codes for a tetratricopeptide repeat protein that promotes the expression of the entire CBP, apparently through the regulation of chromoplast formation (Stanley et al., 2017). It appears that chromoplast defects in rcp2 mutants are somehow conveyed to the nucleus through retrograde signaling, which reduces transcription of all CBP genes. Again, the mechanism for this coordinated regulation of carotenoid biosynthesis genes is still unknown, and almost certainly indirect.
Seeds
Seed carotenoids are critical for ABA biosynthesis and seed dormancy, as well as protecting seeds from ROS damage. Therefore, carotenoids contribute to successful germination (Howitt and Pogson, 2006). In Arabidopsis and Nicotiana plumbaginifolia, it has been shown that ZEP transcript levels increase during seed development, peaking just before the accumulation of ABA (Audran et al., 1998; Audran et al., 2001).
Very few transcriptional regulators of seed carotenoid biosynthesis have been identified ( Figure 6 ). In maize, the endosperm P-box and AACA motif regulatory sequences are bound by P-box binding factor (PBF) and GAMYB proteins, respectively. The ZmBCH2 promoter contains both elements and is bound in vitro by PBF and GAMYB (Jin et al., 2019). A transient assay in maize showed that overexpression of each transcription factor alone increased ZmBCH2 transcript levels, but together, the effect was not additive. This regulation of ZmBCH2 is probably tied to ABA biosynthesis, and not carotenoid accumulation, as maize seeds accumulate lutein (a separate branch of the pathway).
Figure 6.
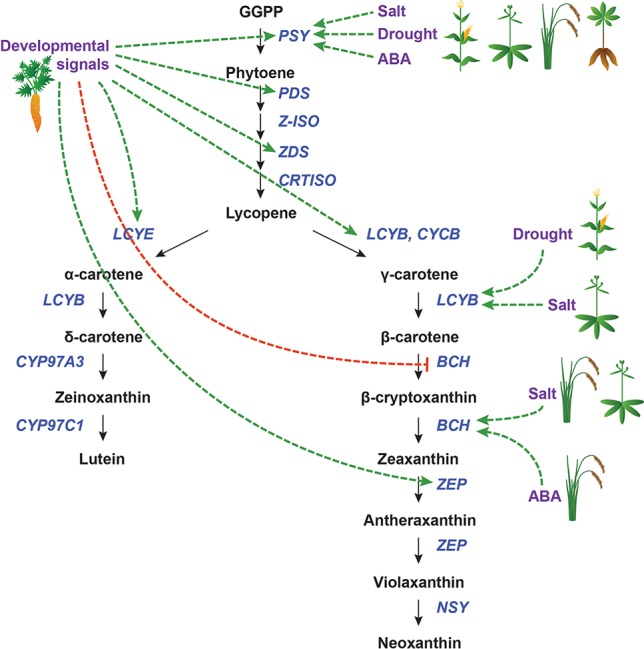
Transcriptional regulation of CBP genes in roots. The regulation of CBP genes in root tissues. Icons indicate the species (carrot, cassava, maize, rice, and Arabidopsis). Developmental and environmental cues are shown in purple. The carotenoid biosynthesis pathway is shown in black, with carotenoid biosynthesis genes indicated in dark blue. Carotenoid regulators discussed in the paper are shown in light blue. Green arrows indicate positive regulation, while blunt red arrows indicate negative regulation. Dotted lines show indirect/unknown interactions.
It is perhaps surprising that so little is known about CBP transcriptional regulation in seeds, given the developmental and economic importance of seed carotenoids. This may be because many carotenoid-containing seeds primarily accumulate lutein (e.g., wheat, maize, millet, sunflower, pumpkin, canola), and the regulation of the α-carotene branch of the pathway is little understood (Howitt and Pogson, 2006). Additionally, promoter screens for late pathway carotenoid biosynthesis genes are rarely performed and/or reported, perhaps due to a biased focus on early pathway genes like PSY.
Roots
Although most roots do not produce carotenoids in appreciable amounts, the CBP is active to provide the precursors for ABA biosynthesis (Rock and Zeevaart, 1991; Bartley and Scolnik, 1995). ABA induces expression change of stress-related genes in response to dehydration (reviewed in Shinozaki and Yamaguchi-Shinozaki, 2007). Thus, the transcriptional regulation of carotenoid biosynthesis is key to water stress responses in plants. Additionally, some crop plants (e.g., sweet potatoes and carrots) accumulate large amounts of carotenoids in storage roots, where developmental signals regulate CBP genes over the course of root maturation.
Abiotic Stress Responses
Because roots are responsible for water and nutrient acquisition, root tissues must be able to respond to environmental cues. Of particular relevance to carotenoid biosynthesis is the sensing of and response to drought and salt stress ( Figure 6 ). These mechanisms are related and overlapping (Zhu, 2002) and will thus be considered together. Rice (Oryza sativa) and maize each have three PSY paralogues, one of which (PSY3) appears to be strongly inducible by drought, salt, and exogenous ABA application. OsZEP and ZmBCH are also moderately upregulated by these stressors in rice and maize, respectively (Welsch et al., 2008; Li et al., 2008b).
In cassava (Manihot esculenta), there are also three copies of PSY. However, MePSY3 is expressed at extremely low levels, and its transcription is not affected by salt or drought stress. Instead, MePSY1 and MePSY2, which are normally expressed in photosynthetic tissues, are upregulated in roots to mediate the salt/drought response (Arango et al., 2010).
In Arabidopsis, which has only one PSY gene, salt stress upregulates AtPSY in the root but not the shoot. There is also a root-specific increase in the transcript levels of AtBCH1, AtBCH2, and AtZEP, but not other CBP genes. It was speculated that ABA signaling transcription factors might bind the AtPSY promoter preferentially in the root (Ruiz-Sola et al., 2014).
It appears that the transcriptional regulation of PSY is a conserved mechanism for drought/salt stress response in plants. When multiple paralogues of PSY are present in a genome, there may be specialization in function, perhaps mediated by differences in cis-regulatory elements (CREs). In cases where no specialization is evident, PSY regulation is tissue-specific: in both cassava and Arabidopsis, salt/drought stress upregulates PSY specifically in the root, with leaves exhibiting no change in PSY mRNA levels. Some downstream genes in the β-carotene branch of the CBP (which leads to ABA) are also altered by drought/salt stress, but the affected genes appear to be species-specific, with no later pathway genes being consistently responsive.
Storage Roots
In carrots (Daucus carota), the expression levels of most CBP genes (DcPSY1/2, DcPDS, DcZDS1/2, DcLCYE, DcLCYB, and DcZEP) increase over root development in several different carrot cultivars, including white carrots, which ultimately do not sequester carotenoids (Clotault et al., 2008). White carrots overexpressing a bacterial orthologue of PSY (crtB) in roots had significantly increased carotenoid levels (though not nearly as much β-carotene as orange carrots, suggesting that other factors also mediate this process) (Maass et al., 2009). This demonstrated that transcriptional regulation could play an important role in carotenoid accumulation in carrot roots.
In Fuentes et al. (2012), carrot roots were grown either underground or in light, and the mRNA levels of multiple CBP genes were assessed. Light-grown carrots accumulated a carotenoid profile similar to that of leaves, while dark-grown carrots accumulated mostly β-carotene. The expression patterns of most CBP genes mirrored these changes, with the exception of DcZDS1 and DcLCYB2 (which were not affected by treatment) and DcLCYE (which actually increased in both treatments). As proposed before, transcriptional regulation alone cannot account for these differences. It is important to note that light induced the formation of chloroplasts instead of chromoplasts, which indicates that plastid-to-nucleus retrograde signaling may somehow regulate CBP gene expression.
So Many Regulators, So Little Consensus
A fairly large number of putative transcriptional regulators of carotenoid biosynthesis have been identified from various species and tissue types ( Table S1 ), mostly in the past decade. A conundrum emerges from this otherwise exciting progress: why so many candidate regulators but so little consensus? Almost every tissue type in every species studied to date seems to utilize a different group of transcriptional regulators for carotenoid biosynthesis. Perhaps it is not too surprising that the CBP is differentially regulated in various tissue types, as carotenoids serve very different functions in different organs (e.g., in leaves as essential components of the photosynthetic apparatus vs. in flowers and fruits as secondary metabolites), but it is puzzling that each species seems to have evolved its own carotenoid regulators. In our opinion, this conundrum exists at least in part because of the following challenges:
The endogenous functions of some of these putative transcriptional regulators have not been verified through knockout or knockdown experiments (e.g., CpEIN3a, CpNAC1/2, CrMYB68, CsMADS6, CpbHLH1/2, AdMYB7, ZmPBF, ZmGAMYB). These regulators were usually identified through transcriptome-based co-expression analyses or yeast one-hybrid screens using promoters of CBP genes. Interactions between these regulators and their DNA binding sites in the CBP gene promoters were often tested by in vitro gel shift assays and/or dual luciferase assays. Sometimes these regulators were further characterized by transient or stable overexpression in a heterologous host (e.g., tobacco). However, one should be cautious when interpreting these results, as heterologous expression can sometimes be uninformative or even misleading (Kramer, 2015). Before knockout or knockdown data become available, we think these genes should be regarded as “candidate” instead of bona fide carotenoid regulators.
Most putative regulators were identified from ripening fruits, especially tomato (e.g., TAGL1, RIN, FUL1, FUL2, SlMADS1, SlNAC4, SlAP2a, SGR1, SlHDA3), making it very difficult to disentangle these regulators’ influence on carotenoid biosynthesis from their other ripening roles, which are largely mediated through ethylene signaling. In fact, most of the putative carotenoid regulators identified in tomato are components of the ethylene signaling network (Li et al., 2019). Therefore, it would not be unexpected if these tomato fruit ripening genes do not regulate carotenoid biosynthesis in other tissue types or in non-climacteric fruits.
Current major model systems for carotenoid regulation are somewhat unusual or at least not representative. For example, the best genetic model system, Arabidopsis, does not produce chromoplast-containing tissues. The foremost fruit model, tomato, accumulates lycopene and is climacteric, whereas fruits of many other plant species (e.g., orange, papaya) accumulate abundant downstream products (e.g., β-carotene and xanthophylls). These limitations raise the question whether the knowledge gained from these systems is widely applicable to other plant species.
Minimal effort has been put into testing whether the function of a certain regulator identified from one species is conserved in another species. So far, only the PIF1/HY5 regulatory module has been shown to play a role in carotenoid biosynthesis during both Arabidopsis photomorphogenesis and the onset of tomato fruit ripening. Even in this case, it is unclear whether PIF1/HY5 function at the onset of tomato fruit ripening is an ancestral feature of all fleshy fruits or was accidentally co-opted from the photomorphogenesis network just in tomatoes.
In addition to these challenges, there are also many gaps in our understanding of transcriptional regulation of carotenoid biosynthesis. For example, we know very little about what regulates most CBP genes downstream of PSY in photosynthetic tissues, even in Arabidopsis; we know virtually nothing about the transcriptional regulators of CBP genes in the roots of any model systems; we know only three regulators in flowers (i.e., COI1/MYB305, RCP1, and RCP2), and even for these, we know nothing about their functional mechanisms; we know a variety of phytohormones affecting CBP gene expression, but we do not know which transcriptional regulators relay their signals to CBP genes.
Future Perspectives
The challenges and knowledge gaps discussed above present wonderful opportunities for future carotenoid research. We think the following research directions will be fruitful in understanding how carotenoid biosynthesis is controlled at the transcriptional level:
Testing the function of known putative carotenoid regulators ( Table S1 ) across multiple species with well-developed genetic/genomic resources and functional tools. For example, generating knockdown transgenic lines or Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) mutants of the SDG8 orthologue in readily transformable species like tomatoes or monkeyflowers would be a straightforward way to test whether the role of this gene in CRTISO regulation is conserved across angiosperms or just an oddity in Arabidopsis. Likewise, it would be interesting to generate knockdown/knockout lines of RCP1 or RCP2 in tomatoes to see whether tomato flower color changes.
Identifying regulators of late pathway CBP genes using promoter screens. The recent study in maize (Jin et al., 2019) where two new regulators were identified using the ZmBCH2 promoter is a good example. Besides traditional yeast one-hybrid screens, recently developed methods such as CAPTURE (CRISPR Affinity Purification in siTU of Regulatory Elements) can be used to identify both transcription factors and chromatin remodelers at a particular promoter site with high specificity (Liu et al., 2017).
Discovering key CREs of the CBP genes in various species. Databases such as PLACE (Higo et al., 1999) and PlantCare (Lescot et al., 2002), in conjunction with phylogenetic shadowing methods (Blanchette and Tompa, 2002), are extremely useful in predicting CREs in silico. Additionally, for species that are transformable, a powerful new way to discover CREs in vivo is CRISPR/Cas9 genome editing with multiple guide RNAs targeted to a gene promoter (e.g., eight guide RNAs in Rodríguez-Leal et al., 2017). This method can generate a wide range of mutant alleles with deletions of various lengths within the promoter region; and because this method does not rely on a priori knowledge of sequence motifs, it allows the discovery of novel CREs.
Integrating multi-omics data (genomics, transcriptomics, proteomics, metabolomics, etc.) towards a more comprehensive understanding of CBP gene expression. With the rapid advances in generating large quantities of high-quality data as well as sophisticated bioinformatics methods and analytical tools, the systems biology approach will allow us to uncover correlations between metabolome and transcriptome profiles, to identify candidate transcriptional regulators of biosynthetic genes in co-expression modules, and to map regulatory network interactions (e.g., Amiour et al., 2012; Maruyama et al., 2014; Balazadeh et al., 2014; Larsen et al., 2016). We envision that this integrative approach will be particularly helpful in elucidating the “missing” regulators that relay various hormone cues to CBP genes. There are many “omics” resources and databases that could be used for these purposes (Mochida and Shinozaki, 2011; Rai et al., 2017), but there are also many challenges to integrating such data. Experimental design and data quality/curation must be taken into account when combining multiple omics resources (Cavill et al., 2016; Helmy et al., 2016).
Broadening the diversity of “model” systems. For example, citrus would be an excellent system to complement the existing tomato fruit model because it is non-climacteric and accumulates various carotenoids beyond lycopene. In addition, carotenoid-containing, embryogenic citrus calli can be readily produced and transformed (Lu et al., 2018), making them a powerful tool for rigorous tests of gene function. As high-quality genome assemblies and genome editing technologies become more and more accessible, it is not difficult to envision the development of even brand-new model species with interesting/economically important carotenoid phenotypes in the near future.
We believe that these research avenues will lead to many more exciting discoveries in the coming years, which will not only contribute new knowledge on the transcriptional regulation of carotenoid biosynthesis but also likely have a significant impact on carotenoid biofortification of crop plants. So far, most of the efforts towards enhancing carotenoid biosynthesis or engineering novel carotenoid products in staple crops have focused on CBP genes (e.g., aSTARice; Zhu et al., 2018). However, effective biofortification often requires transferring multiple CBP genes simultaneously to the host plant and expressing these transgenes in a coordinated fashion, the latter being a particular challenge in metabolic engineering (Nielsen and Keasling, 2016). Building both transcriptional regulators and CBP genes into a synthetic biology framework will allow us to better coordinate the expression of multiple CBP genes, to make quantitative predictions of metabolic flux, and to rationally design optimal genetic circuits with maximal phenotypic outputs.
Author Contributions
LS and Y-WY wrote the manuscript.
Funding
Our work on carotenoids is supported by the National Science Foundation (IOS-1558083, IOS-1827645).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of our laboratory and Drs. Foen Peng, Pam Diggle, Jeff Seemann, and Qinlong Zhu for discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01017/full#supplementary-material
References
- Alba R., Payton P., Fei Z., McQuinn R., Debbie P., Martin G. B., et al. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 (11), 2954–2965. 10.1105/tpc.105.036053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335 (6074), 1348. 10.1126/science.1218094 [DOI] [PubMed] [Google Scholar]
- Amiour N., Imbaud S., Clément G., Agier N., Zivy M., Valot B., et al. (2012). The use of metabolomics integrated with transcriptomic and proteomic studies for identifying key steps involved in the control of nitrogen metabolism in crops such as maize. J. Exp. Bot. 63 (14), 5017–5033. 10.1093/jxb/ers186 [DOI] [PubMed] [Google Scholar]
- Ampomah-Dwamena C., Thrimawithana A. H., Dejnoprat S., Lewis D., Espley R. V., Allan A. C. (2019). A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 221 (1), 309–325. 10.1111/nph.15362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A., Keskitalo J., Sjödin A., Bhalerao R., Sterky F., Wissel K., et al. (2004). A transcriptional timetable of autumn senescence. Genome Biol. 5 (4), R24. 10.1186/gb-2004-5-4-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade P., Caudepón D., Altabella T., Arró M., Ferrer A., Manzano D. (2017). Complex interplays between phytosterols and plastid development. Plant Signal Behav. 12 (11), e1387708. 10.1080/15592324.2017.1387708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango J., Wüst F., Beyer P., Welsch R. (2010). Characterization of phytoene synthases from cassava and their involvement in abiotic stress-mediated responses. Planta 232 (5), 1251–1262. 10.1007/s00425-010-1250-6 [DOI] [PubMed] [Google Scholar]
- Arango J., Beltrán J., Nuñez J., Chavarriaga P. (2016). “Evidence of epigenetic mechanisms affecting carotenoids,” in Carotenoids in Nature (Cham, Switzerland: Springer; ) 295–307. [DOI] [PubMed] [Google Scholar]
- Audran C., Borel C., Frey A., Sotta B., Meyer C., Simonneau T., et al. (1998). Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia . J. Plant Physiol. 118 (3), 1021–1028. 10.1104/pp.118.3.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran C., Liotenberg S., Gonneau M., North H., Frey A., Tap-Waksman K., et al. (2001). Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drought stress and during seed development. Funct. Plant Biol. 28 (12), 1161–1173. [Google Scholar]
- Auldridge M. E., McCarty D. R., Klee H. J. (2006). Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9 (3), 315–321. 10.1016/j.pbi.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Avendaño-Vázquez A., Cordoba E., Llamas E., San Román C., Nisar N., Torre D. L., et al. (2014). An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis . Plant Cell 26 (6), 2524–2537. 10.1105/tpc.114.123349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59 (1), 281–311. 10.1146/annurev.arplant.59.032607.092859 [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Schildhauer J., Araújo W. L., Munné-Bosch S., Fernie A. R., Proost S., et al. (2014). Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: transcriptomic and metabolomic consequences. J. Exp. Bot. 65 (14), 3975–3992. 10.1093/jxb/eru119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli I., Niyogi K. K. (2000). Molecular genetics of xanthophyll dependent photoprotection in green algae and plants. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 355 (1402), 1385–1394. 10.1098/rstb.2000.0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley G. E., Scolnik P. A. (1995). Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell 7 (7), 1027–1038. 10.1105/tpc.7.7.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M., Karlova R., Ballester A. R., Tikunov Y. M., Bovy A. G., Wolters-Arts M., et al. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24 (11), 4437–4451. 10.1105/tpc.112.103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B. (1995). Carotenoid catabolism during leaf senescence and its control by light. J. Photochem. Photobiol. B, Biol. 30 (1), 3–13. 10.1016/1011-1344(95)07197-A [DOI] [Google Scholar]
- Blanchette M., Tompa M. (2002). Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 12 (5), 739–748. 10.1101/gr.6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J., Toledo-Ortiz G., Ortiz-Alcaide M., Cifuentes-Esquivel N., Halliday K. J., Martinez-García J., et al. (2015). Regulation of carotenoid biosynthesis by shade relies on specific subsets of antagonistic transcription factors and cofactors. J. Plant Physiol. 169 (3), 1584. 10.1104/pp.15.00552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw H., Jr., Schemske D. W. (2003). Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426 (6963), 176. 10.1038/nature02106 [DOI] [PubMed] [Google Scholar]
- Breeze E., Harrison E., Mchattie S., Hughes L., Hickman R., Hill C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23 (3), 873–894. 10.1105/tpc.111.083345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton G., Young A. J. (1989). “Chloroplast carotenoids: function, biosynthesis and effects of stress and senescence,” in Trends in Photosynthesis Research. Eds. Biswal U. C., Britton G. (India: Agro Botanical Publishers; ), 303–319. [Google Scholar]
- Brown B. A., Cloix C., Jiang G. H., Kaiserli E., Herzyk P., Kliebenstein D. J., et al. (2005). A UV-B–specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci 102 (50), 18225–18230. 10.1073/pnas.0507187102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. A., Jenkins G. I. (2008). UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. J. Plant Physiol. 146 (2), 576–588. 10.1104/pp.107.108456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R., Medina J., Salinas J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis . Proc. Natl. Acad. Sci. U. S. A. 108 (39), 16475. 10.1073/pnas.1107161108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavill R., Jennen D., Kleinjans J., Briedé J. J. (2016). Transcriptomic and metabolomic data integration. Brief. Bioinformatics 17 (5), 891–901. 10.1093/bib/bbv090 [DOI] [PubMed] [Google Scholar]
- Cazzonelli C. I., Cuttriss A. J., Cossetto S. B., Pye W., Crisp P., Whelan J., et al. (2009). Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21 (1), 39–53. 10.1105/tpc.108.063131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C. I., Pogson B. J. (2010). Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15 (5), 266–274. 10.1016/j.tplants.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J., Erhardt M., et al. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis . Plant Cell 23 (5), 1849. 10.1105/tpc.111.085233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou C., Pan H., Chuang Y., Yeh K. (2010). Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 232 (4), 937–948. 10.1007/s00425-010-1222-x [DOI] [PubMed] [Google Scholar]
- Chung M., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J., et al. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64 (6), 936–947. 10.1111/j.1365-313X.2010.04384.x [DOI] [PubMed] [Google Scholar]
- Cloix C., Jenkins G. I. (2008). Interaction of the Arabidopsis UV-B–specific signaling component UVR8 with chromatin. Mol. Plant 1 (1), 118–128. 10.1093/mp/ssm012 [DOI] [PubMed] [Google Scholar]
- Clotault J., Peltier D., Berruyer R., Thomas M., Briard M., Geoffriau E. (2008). Expression of carotenoid biosynthesis genes during carrot root development. J. Exp. Bot. 59 (13), 3563–3573. 10.1093/jxb/ern210 [DOI] [PubMed] [Google Scholar]
- Corona V., Aracri B., Kosturkova G., Bartley G. E., Pitto L., Giorgetti L., et al. (1996). Regulation of a carotenoid biosynthesis gene promoter during plant development. Plant J. 9 (4), 505–512. [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- D’Andrea L., Simon-Moya M., Llorente B., Llamas E., Marro M., Loza-Alvarez P., et al. (2018). Interference with clp protease impairs carotenoid accumulation during tomato fruit ripening. J. Exp. Bot. 69 (7), 1557–1568. 10.1093/jxb/erx491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal M., Chinnusamy V., Bansal K. C. (2010). Isolation and functional characterization of lycopene β-cyclase (CYC-B) promoter from Solanum habrochaites . BMC Plant Biol. 10 (1), 61. 10.1186/1471-2229-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., North H., Marion-Poll A., Bassi R. (2007). The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell 19 (3), 1048. 10.1105/tpc.106.049114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Holt N. E., Kaligotla S., Fuciman M., Cazzaniga S., Carbonera D., et al. (2012). Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. J. Biol. Chem. 287 (50), 41820–41834. 10.1074/jbc.M112.405498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C., Sonntag L., James G., Janitza P., Ibañez C., Ziermann H., et al. (2014). The DET1–COP1–HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 9 (6), 1983–1989. 10.1016/j.celrep.2014.11.043 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B., Adams W. W., III (1996). The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1 (1), 21–26. 10.1016/S1360-1385(96)80019-7 [DOI] [Google Scholar]
- Dong J., Tang D., Gao Z., Yu R., Li K., He H., et al. (2014). Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26 (9), 3630. 10.1105/tpc.114.130666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T., Hu Z., Deng L., Wang Y., Zhu M., Zhang J., et al. (2013). A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. J. Plant Physiol. 163 (2), 1026–1036. 10.1104/pp.113.224436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N., Negre F., Nagegowda D. A., Orlova I. (2006). Plant volatiles: recent advances and future perspectives. Crit. Rev. Plant Sci. 25 (5), 417–440. 10.1080/07352680600899973 [DOI] [Google Scholar]
- Emiliani J., D’Andrea L., Ferreyra M. L. F., Maulión E., Rodriguez E., Rodriguez-Concepción M., et al. (2018). A role for β, β-xanthophylls in Arabidopsis UV-B photoprotection. J. Exp. Bot. 69 (20), 4921–4933. 10.1093/jxb/ery242 [DOI] [PubMed] [Google Scholar]
- Enfissi E. M. A., Nogueira M., Bramley P. M., Fraser P. D. (2017). The regulation of carotenoid formation in tomato fruit. Plant J. 89 (4), 774–788. 10.1111/tpj.13428 [DOI] [PubMed] [Google Scholar]
- Eriksson E. M., Bovy A., Manning K., Harrison L., Andrews J., De Silva J., et al. (2004). Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. J. Plant Physiol. 136 (4), 4184–4197. 10.1104/pp.104.045765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory J. J., Stec A., Gruber H., Rizzini L., Oravecz A., Funk M., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B–induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28 (5), 591–601. 10.1038/emboj.2009.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank H. A., Cogdell R. J. (1996). Carotenoids in photosynthesis. Photochem. Photobiol. 63 (3), 257–264. 10.1111/j.1751-1097.1996.tb03022.x [DOI] [PubMed] [Google Scholar]
- Fraser P. D., Truesdale M. R., Bird C. R., Schuch W., Bramley P. M. (1994). Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). J. Plant Physiol. 105 (1), 405–413. 10.1104/pp.105.1.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Han Y., Fan Z., Chen J., Chen W., Lu W., et al. (2016). The papaya transcription factor CpNAC1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening. J. Agric. Food Chem. 64 (27), 5454–5463. 10.1021/acs.jafc.6b01020 [DOI] [PubMed] [Google Scholar]
- Fu C., Han Y., Kuang J., Chen J., Lu W. (2017). Papaya CpEIN3a and CpNAC2 co-operatively regulate carotenoid biosynthesis-related genes CpPDS2/4, CpLCY-e and CpCHY-b during fruit ripening. Plant Cell Physiol. 58 (12), 2155–2165. 10.1093/pcp/pcx149 [DOI] [PubMed] [Google Scholar]
- Fuentes P., Pizarro L., Moreno J. C., Handford M., Rodriguez-Concepcion M., Stange C. (2012). Light-dependent changes in plastid differentiation influence carotenoid gene expression and accumulation in carrot roots. Plant Mol. Biol. 79 (1–2), 47–59. 10.1007/s11103-012-9893-2 [DOI] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Ito Y. (2011). Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 11 (1), 26. 10.1186/1471-2229-11-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Shima Y., Higuchi N., Nakano T., Koyama Y., Kasumi T., et al. (2012). Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235 (6), 1107–1122. 10.1007/s00425-011-1561-2 [DOI] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Shima Y., Ito Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25 (2), 371–386. 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Shima Y., Nakagawa H., Kitagawa M., Kimbara J., Nakano T., et al. (2014). Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26 (1), 89–101. 10.1105/tpc.113.119453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N., Ronen G., Khalfa Z., Zamir D., Hirschberg J. (2006). A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18 (8), 1947–1960. 10.1105/tpc.105.039966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S., Amasino R. M. (1997). Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). J. Plant Physiol. 113 (2), 313. 10.1104/pp.113.2.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R. P., Höhmann S., Cardon G. H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49 (4), 683–693. 10.1111/j.1365-313X.2006.02983.x [DOI] [PubMed] [Google Scholar]
- Giménez E., Pineda B., Capel J., Antón M. T., Atarés A., Pérez-Martín F., et al. (2010). Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS One 5 (12), e14427. 10.1371/journal.pone.0014427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10 (3), 283–289. 10.1016/j.pbi.2007.04.008 [DOI] [PubMed] [Google Scholar]
- Giovannoni J., Nguyen C., Ampofo B., Zhong S., Fei Z. (2017). The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 68, 61–84. [DOI] [PubMed] [Google Scholar]
- Giuliano G., Bartley G. E., Scolnik P. A. (1993). Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5 (4), 379–387. 10.1105/tpc.5.4.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin T., Britton G. W. (1988). “Distribution and analysis of carotenoids,” in Plant Pigments Ed. Goodwin T. W. (London: Academic Press; ), 62–132. [Google Scholar]
- Grassi S., Piro G., Lee J. M., Zheng Y., Fei Z., Dalessandro G., et al. (2013). Comparative genomics reveals candidate carotenoid pathway regulators of ripening watermelon fruit. BMC Genomics 14 (1), 781. 10.1186/1471-2164-14-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hu Z., Yu X., Li A., Li F., Wang Y., et al. (2018). A histone deacetylase gene, SlHDA3, acts as a negative regulator of fruit ripening and carotenoid accumulation. Plant Cell Rep. 37 (1), 125–135. 10.1007/s00299-017-2211-3 [DOI] [PubMed] [Google Scholar]
- Ha S., Kim J., Park J., Lee S., Cho K. (2007). A comparison of the carotenoid accumulation in capsicum varieties that show different ripening colours: deletion of the capsanthin–capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. J. Exp. Bot. 58 (12), 3135–3144. 10.1093/jxb/erm132 [DOI] [PubMed] [Google Scholar]
- Hao Y., Hu G., Breitel D., Liu M., Mila I., Frasse P., et al. (2015). Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS Genetics 11 (12), e1005649. 10.1371/journal.pgen.1005649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Uragami C., Cogdell R. J. (2016). “Carotenoids and photosynthesis,” in Carotenoids in Nature (Cham, Switzerland: Springer; ), 111–139. [DOI] [PubMed] [Google Scholar]
- Havaux M. (1998). Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci. 3 (4), 147–151. 10.1016/S1360-1385(98)01200-X [DOI] [Google Scholar]
- Helmy M., Crits-Christoph A., Bader G. D. (2016). Ten simple rules for developing public biological databases. PLoS Comput. Biol. 12 (11), e1005128. 10.1371/journal.pcbi.1005128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 (1), 297–300. 10.1093/nar/27.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4 (3), 210–218. 10.1016/S1369-5266(00)00163-1 [DOI] [PubMed] [Google Scholar]
- Hou X., Rivers J., León P., McQuinn R. P., Pogson B. J. (2016). Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 21 (9), 792–803. 10.1016/j.tplants.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Howitt C. A., Pogson B. J. (2006). Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ. 29 (3), 435–445. 10.1111/j.1365-3040.2005.01492.x [DOI] [PubMed] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60 (6), 1081–1095. 10.1111/j.1365-313X.2009.04064.x [DOI] [PubMed] [Google Scholar]
- Ito Y., Kitagawa M., Ihashi N., Yabe K., Kimbara J., Yasuda J., et al. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55 (2), 212–223. 10.1111/j.1365-313X.2008.03491.x [DOI] [PubMed] [Google Scholar]
- Jahns P., Holzwarth A. R. (2012). The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. BBA-Bioenergetics 1817 (1), 182–193. 10.1016/j.bbabio.2011.04.012 [DOI] [PubMed] [Google Scholar]
- Jia K., Baz L., Al-Babili S. (2017). From carotenoids to strigolactones. J. Exp. Bot. 69 (9), 2189–2204. 10.1093/jxb/erx476 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Lau O. S., Deng X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 (3), 217. 10.1038/nrg2049 [DOI] [PubMed] [Google Scholar]
- Jin X., Bai C., Bassie L., Nogareda C., Romagosa I., Twyman R. M., et al. (2019). ZmPBF and ZmGAMYB transcription factors independently transactivate the promoter of the maize (Zea mays) β-carotene hydroxylase 2 gene. New Phytol. 222 (2), 793–804. 10.1111/nph.15614 [DOI] [PubMed] [Google Scholar]
- Karlova R., Rosin F. M., Busscher-Lange J., Parapunova V., Do P. T., Fernie A. R., et al. (2011). Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23 (3), 923–941. 10.1105/tpc.110.081273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., et al. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20 (5), 1260–1277. 10.1105/tpc.108.058859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kang H., Park J., Kim W., Yoo J., Lee N., et al. (2016). PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 28 (6), 1388–1405. 10.1105/tpc.16.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto S., Oda-Yamamizo C., Ohmiya A. (2018). Regulation of carotenoid pigmentation in corollas of petunia. Plant Mol. Biol. Rep. 36 (4), 632–642. 10.1007/s11105-018-1107-x [DOI] [Google Scholar]
- Klee H. J., Giovannoni J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45 (1), 41–59. 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- Kolotilin I., Koltai H., Tadmor Y., Bar-Or C., Reuveni M., Meir A., et al. (2007). Transcriptional profiling of high pigment-2dg tomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. J. Plant Physiol. 145 (2), 389–401. 10.1104/pp.107.102962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E. M. (2015). A stranger in a strange land: the utility and interpretation of heterologous expression. Front. Plant Sci. 6, 734. 10.3389/fpls.2015.00734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S., Deng X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17 (10), 584–593. 10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Larsen P. E., Sreedasyam A., Trivedi G., Desai S., Dai Y., Cseke L. J., et al. (2016). Multi-omics approach identifies molecular mechanisms of plant–fungus mycorrhizal interaction. Front. Plant Sci. 6, 1061. 10.3389/fpls.2015.01061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin I., Frankel P., Gilboa N., Tanny S., Lalazar A. (2003). The tomato dark green mutation is a novel allele of the tomato homolog of the DEETIOLATED1 gene. Theor. Appl. Genet. 106 (3), 454–460. 10.1007/s00122-002-1080-4 [DOI] [PubMed] [Google Scholar]
- Li F., Vallabhaneni R., Yu J., Rocheford T., Wurtzel E. T. (2008. a). The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. J. Plant Physiol. 147 (3), 1334–1346. 10.1104/pp.108.122119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Vallabhaneni R., Wurtzel E. T. (2008. b). PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress–induced root carotenogenesis. J. Plant Physiol. 146 (3), 1333–1345. 10.1104/pp.107.111120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yuan H., Zeng Y., Xu Q. (2016). “Plastids and carotenoid accumulation,” in Carotenoids in Nature (Cham, Switzerland: Springer; ), 273–293. [DOI] [PubMed] [Google Scholar]
- Li S., Chen K., Grierson D. (2019). A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 221 (4), 1724–1741. 10.1111/nph.15545 [DOI] [PubMed] [Google Scholar]
- Lieberman M., Segev O., Gilboa N., Lalazar A., Levin I. (2004). The tomato homolog of the gene encoding UV-damaged DNA binding protein 1 (DDB1) underlined as the gene that causes the high pigment-1 mutant phenotype. Theor. Appl. Genet. 108 (8), 1574–1581. 10.1007/s00122-004-1584-1 [DOI] [PubMed] [Google Scholar]
- Lim P. O., Kim H. J., Gil Nam H. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55 (2), 301–310. 10.1111/j.1365-313X.2008.03505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Ren G., Guirgis A., Thornburg R. W. (2009). The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell 21 (9), 2672–2687. 10.1105/tpc.108.060079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Jia C., Zhang M., Chen D., Chen S., Guo R., et al. (2014). Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol. J. 12 (1), 105–115. 10.1111/pbi.12121 [DOI] [PubMed] [Google Scholar]
- Liu L., Wei J., Zhang M., Zhang L., Li C., Wang Q. (2012). Ethylene independent induction of lycopene biosynthesis in tomato fruits by jasmonates. J. Exp. Bot. 63 (16), 5751–5761. 10.1093/jxb/ers224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Shao Z., Zhang M., Wang Q. (2015. a). Regulation of carotenoid metabolism in tomato. Mol. Plant 8 (1), 28–39. 10.1016/j.molp.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Liu M., Pirrello J., Chervin C., Roustan J., Bouzayen M. (2015. b). Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. J. Plant Physiol. 169 (4), 2380. 10.1104/pp.15.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang Y., Chen Y., Li M., Zhou F., Li K., et al. (2017). In situ capture of chromatin interactions by biotinylated dCas9. Cell 170 (5), 1028–1043. 10.1016/j.cell.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B., D’andrea L., Ruiz-Sola M. A., Botterweg E., Pulido P., Andilla J., et al. (2016). Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J. 85 (1), 107–119. 10.1111/tpj.13094 [DOI] [PubMed] [Google Scholar]
- Llorente B., Martinez-Garcia J., Stange C., Rodriguez-Concepcion M. (2017). Illuminating colors: regulation of carotenoid biosynthesis and accumulation by light. Curr. Opin. Plant Biol. 37, 49–55. 10.1016/j.pbi.2017.03.011 [DOI] [PubMed] [Google Scholar]
- López-Ráez J. A., Charnikhova T., Gómez-Roldán V., Matusova R., Kohlen W., De Vos R., et al. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178 (4), 863–874. 10.1111/j.1469-8137.2008.02406.x [DOI] [PubMed] [Google Scholar]
- Lu S., Van Eck J., Zhou X., Lopez A. B., O’Halloran D. M., Cosman K. M., et al. (2006). The cauliflower or gene encodes a DnaJ cysteine-rich domain–containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18 (12), 3594–3605. 10.1105/tpc.106.046417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Zhang Y., Zhu K., Yang W., Ye J., Chai L., et al. (2018). The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. J. Plant Physiol. 176 (4), 2657–2676. 10.1104/pp.17.01830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Zhang J., Li J., Yang C., Wang T., Ouyang B., et al. (2013). A STAY-GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 198 (2), 442–452. 10.1111/nph.12175 [DOI] [PubMed] [Google Scholar]
- Ma N., Feng H., Meng X., Li D., Yang D., Wu C., et al. (2014). Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 14 (1), 351. 10.1186/s12870-014-0351-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass D., Arango J., Wüst F., Beyer P., Welsch R. (2009). Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PloS One 4 (7), e6373. 10.1371/journal.pone.0006373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A. J., King G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38 (8), 948. 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- Martel C., Vrebalov J., Tafelmeyer P., Giovannoni J. J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING–dependent manner. J. Plant Physiol. 157 (3), 1568. 10.1104/pp.111.181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Urano K., Yoshiwara K., Morishita Y., Sakurai N., Suzuki H., et al. (2014). Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. J. Plant Physiol. 164 (4), 1759–1771. 10.1104/pp.113.231720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S., Tzfadia O., Vallabhaneni R., Gehring C., Wurtzel E. T. (2011). A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana . BMC Syst. Biol. 5 (1), 77. 10.1186/1752-0509-5-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C., Yang D., Ma X., Zhao W., Liang X., Ma N., et al. (2016). Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J. Plant Physiol. 193, 88–96. 10.1016/j.jplph.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Middleton E. M., Teramura A. H. (1993). The role of flavonol glycosides and carotenoids in protecting soybean from ultraviolet-B damage. J. Plant Physiol. 103 (3), 741–752. 10.1104/pp.103.3.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K., Shinozaki K. (2011). Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol. 52 (12), 2017–2038. 10.1093/pcp/pcr153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehs C. P., Tian L., Osteryoung K. W., DellaPenna D. (2001). Analysis of carotenoid biosynthetic gene expression during marigold petal development. Plant Mol. Biol. 45 (3), 281–293. 10.1023/A:1006417009203 [DOI] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis . Proc. Natl. Acad. Sci. U. S. A. 105 (27), 9433. 10.1073/pnas.0803611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou W., Li D., Bu J., Jiang Y., Khan Z. U., Luo Z., et al. (2016). Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening. PLoS One 11 (4), e0154072. 10.1371/journal.pone.0154072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Li X. P., Niyogi K. K. (2001). Non-photochemical quenching. A response to excess light energy. J. Plant Physiol. 125 (4), 1558–1566. 10.1104/pp.125.4.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A. C., Fenzi F., Ciliento R., Alfano F., Bowler C. (1999). Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11 (2), 145–157. 10.1105/tpc.11.2.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. 10.1146/annurev.arplant.56.032604.144046 [DOI] [PubMed] [Google Scholar]
- Nielsen J., Keasling J. D. (2016). Engineering cellular metabolism. Cell 164 (6), 1185–1197. 10.1016/j.cell.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Nielsen K. M., Lewis D. H., Morgan E. R. (2003). Characterization of carotenoid pigments and their biosynthesis in two yellow flowered lines of Sandersonia aurantiaca (Hook). Euphytica 130 (1), 25–34. 10.1023/A:1022328828688 [DOI] [Google Scholar]
- Nisar N., Li L., Lu S., Khin N., Pogson B. (2015). Carotenoid metabolism in plants. Mol. Plant 8 (1), 68–82. 10.1016/j.molp.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Niyogi K. K. (1999). Photoprotection Revisited: Genetic and molecular approaches. Annu. Rev. Plant Biol. 50 (1), 333–359. 10.1146/annurev.arplant.50.1.333 [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., et al. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis . Plant Cell 21 (2), 403–419. 10.1105/tpc.108.064691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A., Kishimoto S., Aida R., Yoshioka S., Sumitomo K. (2006). Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. J. Plant Physiol. 142 (3), 1193–1201. 10.1104/pp.106.087130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya A. (2011). Diversity of carotenoid composition in flower petals. Jpn. Agric. Res. Q. 45 (2), 163–171. 10.6090/jarq.45.163 [DOI] [Google Scholar]
- Ohmiya A. (2013). Qualitative and quantitative control of carotenoid accumulation in flower petals. Sci. Hortic. 163, 10–19. 10.1016/j.scienta.2013.06.018 [DOI] [Google Scholar]
- Ohmiya A., Kato M., Shimada T., Nashima K., Kishimoto S., Nagata M. (2019). Molecular basis of carotenoid accumulation in horticultural crops. Hortic. J. 88 (2), 135–149. 10.2503/hortj.UTD-R003 [DOI] [Google Scholar]
- Ougham H. J., Morris P., Thomas H. (2005). The colors of autumn leaves as symptoms of cellular recycling and defenses against environmental stresses. Curr. Top. Dev. Biol. 66, 136–161. [DOI] [PubMed] [Google Scholar]
- Pecker I., Chamovitz D., Linden H., Sandmann G., Hirschberg J. (1992). A single polypeptide catalyzing the conversion of phytoene to zeta-carotene is transcriptionally regulated during tomato fruit ripening. Proc. Natl. Acad. Sci. U. S. A. 89 (11), 4962–4966. 10.1073/pnas.89.11.4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J. M., Zhang Y., Quail P. H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis . Mol. Plant 7 (11), 1598–1618. 10.1093/mp/ssu087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B. J., Niyogi K. K., Bjorkman O., Dellapenna D. (1998). Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. 95 (22), 13324–13329. 10.1073/pnas.95.22.13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospíšil P. (2016). Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 7, 1950. 10.3389/fpls.2016.01950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Wang Y., Cao B., Wang W., Tian S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70 (2), 243–255. 10.1111/j.1365-313X.2011.04861.x [DOI] [PubMed] [Google Scholar]
- Rai A., Saito K., Yamazaki M. (2017). Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. 90 (4), 764–787. 10.1111/tpj.13485 [DOI] [PubMed] [Google Scholar]
- Rizzini L., Favory J. J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332 (6025), 103–106. 10.1126/science.1200660 [DOI] [PubMed] [Google Scholar]
- Rock C. D., Zeevaart J. A. (1991). The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 88 (17), 7496–7499. 10.1073/pnas.88.17.7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Leal D., Lemmon Z. H., Man J., Bartlett M. E., Lippman Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171 (2), 470–480. 10.1016/j.cell.2017.08.030 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Villalón A., Gas E., Rodríguez-Concepción M. (2009). Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 60 (3), 424–435. 10.1111/j.1365-313X.2009.03966.x [DOI] [PubMed] [Google Scholar]
- Ronen G., Cohen M., Zamir D., Hirschberg J. (1999). Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant delta. Plant J. 17 (4), 341–351. 10.1046/j.1365-313X.1999.00381.x [DOI] [PubMed] [Google Scholar]
- Rossel J. B., Wilson I. W., Pogson B. J. (2002). Global changes in gene expression in response to high light in Arabidopsis . J. Plant Physiol. 130 (3), 1109–1120. 10.1104/pp.005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola M., Arbona V., Gómez-Cadenas A., Rodríguez-Concepción M., Rodríguez-Villalón A. (2014). A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in Arabidopsis . PLoS One 9 (3), e90765. 10.1371/journal.pone.0090765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola M. A., Rodríguez-Concepción M. (2012). Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10, e0158. 10.1199/tab.0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagawa J. M., Stanley L. E., LaFountain A. M., Frank H. A., Liu C., Yuan Y. (2016). An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 209 (3), 1049–1057. 10.1111/nph.13647 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. 10.1146/annurev.arplant.57.032905.105231 [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20 (6), 1586. 10.1105/tpc.108.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang X., Mo X., Tang C., Zhong S., Deng X. W. (2015). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc. Natl. Acad. Sci. U. S. A. 112 (12), 3817. 10.1073/pnas.1502405112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y., Kitagawa M., Fujisawa M., Nakano T., Kato H., Kimbara J., et al. (2013). Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 82 (4-5), 427–438. 10.1007/s11103-013-0071-y [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I. S., Bae G., Lee C., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U. S. A. 106 (18), 7660. 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 (2), 221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Snowden K. C., Simkin A. J., Janssen B. J., Templeton K. R., Loucas H. M., Simons J. L., et al. (2005). The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17 (3), 746. 10.1105/tpc.104.027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley L. E., Ding B., Sun W., Mou F., Hill C., Chen S., et al. (2017). A tetratricopeptide repeat protein regulates carotenoid biosynthesis and chromoplast development in monkeyflowers (Mimulus). Biorxiv 171249. 10.1101/171249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Diretto G., Purgatto E., Danoun S., Zouine M., Li Z., et al. (2015). Carotenoid accumulation during tomato fruit ripening is modulated by the auxin–ethylene balance. BMC Plant Biol. 15 (1), 114. 10.1186/s12870-015-0495-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Yuan H., Cao H., Yazdani M., Tadmor Y., Li L. (2018. a). Carotenoid metabolism in plants: the role of plastids. Mol. Plant 11 (1), 58–74. 10.1016/j.molp.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Sun Y., Liang B., Wang J., Kai W., Chen P., Jiang L., et al. (2018. b). SlPti4 affects regulation of fruit ripening, seed germination and stress responses by modulating ABA signaling in tomato. Plant Cell Physiol. 59 (10), 1956–1965. 10.1093/pcp/pcy111 [DOI] [PubMed] [Google Scholar]
- Szymańska R., Ślesak I., Orzechowska A., Kruk J. (2017). Physiological and biochemical responses to high light and temperature stress in plants. Environ. Exp. Bot. 139, 165–177. 10.1016/j.envexpbot.2017.05.002 [DOI] [Google Scholar]
- Thompson A. J., Tor M., Barry C. S., Vrebalov J., Orfila C., Jarvis M. C., et al. (1999). Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. J. Plant Physiol. 120 (2), 383–390. 10.1104/pp.120.2.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Rodríguez-Concepción M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. U. S. A. 107 (25), 11626. 10.1073/pnas.0914428107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Johansson H., Lee K. P., Bou-Torrent J., Stewart K., Steel G., et al. (2014). The HY5–PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription (light and temperature photosynthetic gene transcription regulation). PLoS Genetics 10 (6), e1004416. 10.1371/journal.pgen.1004416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Kusaba M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis . J. Plant Physiol. 169 (1), 138. 10.1104/pp.15.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni R., Bradbury L. M. T., Wurtzel E. T. (2010). The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 504 (1), 104–111. 10.1016/j.abb.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhini B. V., Rao S. S. R. (2002). Acceleration of ripening of tomato pericarp discs by brassinosteroids. Phytochemistry 61 (7), 843–847. 10.1016/S0031-9422(02)00223-6 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296 (5566), 343–346. 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Pan I. L., Arroyo A. J. M., Mcquinn R., Chung M., Poole M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21 (10), 3041–3062. 10.1105/tpc.109.066936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana . Cell 138 (4), 738–749. 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- Wang W., Liu G., Niu H., Timko M. P., Zhang H. (2014). The F-box protein COI1 functions upstream of MYB305 to regulate primary carbohydrate metabolism in tobacco (Nicotiana tabacum L. cv. TN90). J. Exp. Bot. 65 (8), 2147–2160. 10.1093/jxb/eru084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yamagishi M. (2019). Mechanisms suppressing carotenoid accumulation in flowers differ depending on the hybrid groups of lilies (Lilium spp.). Sci. Hortic. 243, 159–168. 10.1016/j.scienta.2018.08.025 [DOI] [Google Scholar]
- Wei S., Yu B., Gruber M. Y., Khachatourians G. G., Hegedus D. D., Hannoufa A. (2010). Enhanced seed carotenoid levels and branching in transgenic Brassica napus expressing the Arabidopsis miR156b gene. J. Agric. Food Chem. 58 (17), 9572–9578. 10.1021/jf102635f [DOI] [PubMed] [Google Scholar]
- Welsch R., Medina J., Giuliano G., Beyer P., von Lintig J. (2003). Structural and functional characterization of the phytoene synthase promoter from Arabidopsis thaliana. Planta 216 (3), 523–534. 10.1007/s00425-002-0885-3 [DOI] [PubMed] [Google Scholar]
- Welsch R., Maass D., Voegel T., Dellapenna D., Beyer P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. J. Plant Physiol. 145 (3), 1073–1085. 10.1104/pp.107.104828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R., Wust F., Bar C., Al-Babili S., Beyer P. (2008). A third phytoene synthase is devoted to abiotic stress–induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. J. Plant Physiol. 147 (1), 367–380. 10.1104/pp.108.117028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L., Zhao F., Li R., Xu C., Chen K., Xiao H. (2015). The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. J. Plant Physiol. 167 (3), 931–949. 10.1104/pp.114.255174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Liu L. L., Yan Y. C. (2019). TERF1 regulates nuclear gene expression through chloroplast retrograde signals. Russ. J. Plant Physiol., 66 (1), 22–28. 10.1134/S1021443719010205 [DOI] [Google Scholar]
- Xie Q., Hu Z., Zhu Z., Dong T., Zhao Z., Cui B., et al. (2014). Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Sci. Rep. 4, 4367. 10.1038/srep04367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C., Luo D., Lin A., Zhang C., Shan L., He P., et al. (2019). A tomato B-box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome–mediated degradation. New Phytol. 221 (1), 279–294. 10.1111/nph.15373 [DOI] [PubMed] [Google Scholar]
- Yamagishi M., Kishimoto S., Nakayama M. (2010). Carotenoid composition and changes in expression of carotenoid biosynthetic genes in tepals of Asiatic hybrid lily. Plant Breed. 129 (1), 100–107. 10.1111/j.1439-0523.2009.01656.x [DOI] [Google Scholar]
- Yamamizo C., Kishimoto S., Ohmiya A. (2010). Carotenoid composition and carotenogenic gene expression during Ipomoea petal development. J. Exp. Bot. 61 (3), 709–719. 10.1093/jxb/erp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. C., Zhang J. H., Wang Z. Q., Zhu Q. S., Liu L. J. (2003). Involvement of abscisic acid and cytokinins in the senescence and remobilization of carbon reserves in wheat subjected to water stress during grain filling. Plant Cell Environ. 26 (10), 1621–1631. 10.1046/j.1365-3040.2003.01081.x [DOI] [Google Scholar]
- Yuan H., Zhang J., Nageswaran D., Li L. (2015). Carotenoid metabolism and regulation in horticultural crops. Hortic Res. 2, 15036. 10.1038/hortres.2015.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Liu C., Wang Y., Yao X., Wang F., Wu J., et al. (2015). Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol. 206 (4), 1513–1526. 10.1111/nph.13335 [DOI] [PubMed] [Google Scholar]
- Zhang J., Hu Z., Yao Q., Guo X., Nguyen V., Li F., et al. (2018. a). A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 8 (1), 3413. 10.1038/s41598-018-21672-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Yuan B., Leng P. (2009). The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 60 (6), 1579–1588. 10.1093/jxb/erp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li Z., Tu Y., Cheng W., Yang Y. (2018. b). Tomato (Solanum lycopersicum) SlIPT4, encoding an isopentenyltransferase, is involved in leaf senescence and lycopene biosynthesis during fruit ripening. BMC Plant Biol. 18 (1), 107. 10.1186/s12870-018-1327-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Yun-Ru C., Zheng Y., Huang M., Vrebalov J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31 (2), 154. 10.1038/nbt.2462 [DOI] [PubMed] [Google Scholar]
- Zhou X., Welsch R., Yang Y., Alvarez D., Riediger M., Yuan H., et al. (2015). Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 112 (11), 3558–3563. 10.1073/pnas.1420831112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Shen Y., Zhou P., Fatima M., Lin J., Yue J., et al. (2019). Papaya CpbHLH1/2 regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic Res. 6 (1), 80. 10.1038/s41438-019-0162-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Luo T., Liu C., Wang Y., Yang H., Yang W., et al. (2017. a). An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of citrus reticulate. New Phytol. 216 (1), 178–192. 10.1111/nph.14684 [DOI] [PubMed] [Google Scholar]
- Zhu J. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 (1), 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Bu Q., Xu X., Paik I., Huang X., Hoecker U., et al. (2015). CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat. Commun. 6, 7245. 10.1038/ncomms8245 [DOI] [PubMed] [Google Scholar]
- Zhu M., Chen G., Zhou S., Tu Y., Wang Y., Dong T., et al. (2014). A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 55 (1), 119–135. 10.1093/pcp/pct162 [DOI] [PubMed] [Google Scholar]
- Zhu Q., Zeng D., Yu S., Cui C., Li J., Li H., et al. (2018). From Golden Rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm. Mol. Plant 11 (12), 1440–1448. 10.1016/j.molp.2018.09.007 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Chen G., Guo X., Yin W., Yu X., Hu J., et al. (2017. b). Overexpression of SlPRE2, an atypical bHLH transcription factor, affects plant morphology and fruit pigment accumulation in tomato. Sci. Rep. 7 (1), 5786. 10.1038/s41598-017-04092-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Li G., Yan C., Liu L., Zhang Q., Han Z., et al. (2019). DRL1, encoding a NAC transcription factor, is involved in leaf senescence in grapevine. Int. J. Mole. Sci. 20 (11), 2678. 10.3390/ijms20112678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


