USA100 health care-associated MRSA isolates are highly antibiotic resistant and can cause invasive disease across all patient populations. Even though USA100 strains are some of the most frequently identified causes of infections, little is known about virulence regulation in these isolates. Our study demonstrates that the USA100 agr quorum-sensing system is important for the control of toxin and exoenzyme production and that the agr system has a key role in skin infection. In some USA100 isolates, the agr system is important for growth in the presence of low levels of antibiotics. Altogether, our findings demonstrate that the USA100 agr system is a critical regulator of virulence and that it may make a contribution to the optimal survival of these MRSA strains in the presence of antibiotics.
KEYWORDS: MRSA, USA100, agr, quorum sensing
ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) infections impact all patient populations both in the community and in health care settings. Despite advances in our knowledge of MRSA virulence, little is known about the regulatory mechanisms of USA100 health care-associated MRSA isolates, which are the second most frequently identified MRSA isolates found in all infections. This work focused on the contribution of the USA100 agr type II quorum-sensing system to virulence and antibiotic resistance. From a MRSA strain collection, we selected 16 representative USA100 isolates, constructed mutants with Δagr mutations, and characterized selected strain pairs for virulence factor expression, murine skin infection, and antibiotic resistance. For each strain pair, hemolysis and extracellular protease expression were significantly greater in the wild-type (WT) strains than in the Δagr mutants. Similarly, mice challenged with the WT strains had larger areas of dermonecrosis and greater weight loss than those challenged with the Δagr mutants, demonstrating that the USA100 agr system regulates virulence. Although USA100 isolates exhibit a high level of antibiotic resistance, the WT and Δagr strain pairs showed no difference in MICs by MIC testing. However, in the presence of a sub-MIC of vancomycin, most of the USA100 Δagr mutants exhibited slower growth than the WT isolates, and a couple of the Δagr mutants also grew more slowly in the presence of a sub-MIC of cefoxitin. Altogether, our findings demonstrate that the USA100 agr system is a critical regulator of virulence, and it may have a contribution to the optimal survival of these MRSA strains in the presence of antibiotics.
IMPORTANCE USA100 health care-associated MRSA isolates are highly antibiotic resistant and can cause invasive disease across all patient populations. Even though USA100 strains are some of the most frequently identified causes of infections, little is known about virulence regulation in these isolates. Our study demonstrates that the USA100 agr quorum-sensing system is important for the control of toxin and exoenzyme production and that the agr system has a key role in skin infection. In some USA100 isolates, the agr system is important for growth in the presence of low levels of antibiotics. Altogether, our findings demonstrate that the USA100 agr system is a critical regulator of virulence and that it may make a contribution to the optimal survival of these MRSA strains in the presence of antibiotics.
INTRODUCTION
Staphylococcus aureus is a Gram-positive opportunistic bacterial pathogen known to cause a diverse array of acute and chronic infections (1, 2). Exposure to antibiotics has resulted in the evolution of strains, producing strains such as methicillin-resistant Staphylococcus aureus (MRSA), which has become the cause of a global health care epidemic (3, 4). Over the past 2 decades, these MRSA strains have moved into the community and have begun infecting otherwise healthy individuals, resulting in a new wave of resistant strains called community-associated MRSA (CA-MRSA). CA-MRSA strains, the most important of which are the USA300 group of isolates, have spread globally and are responsible for severe and devastating disease (1, 3–5).
Due to the attention garnered by the emergence of CA-MRSA and the subsequent spread of common CA-MRSA strains (e.g., USA300 isolates) into health care settings, recent research has focused on understanding the virulence mechanisms enabling these isolates to be so successful. Meanwhile, strain types that more commonly cause health care-associated MRSA (HA-MRSA) infections, which have been around much longer and which have remained more confined to hospital settings, have not received as much basic research attention, despite the frequency with which they are responsible for infections. The most common HA-MRSA strains are those of the USA100 pulsed-field gel type, which have a staphylococcal cassette chromosome mec (SCCmec) type II island, are spa type 2, and are often multidrug resistant (6, 7). In 2005 and 2006, 53.2% of the MRSA samples from patients with invasive disease were of the USA100 genotype (8). In 2009 and 2010, 37% of nasal MRSA isolates and 36% of blood culture MRSA isolates from one study were USA100 (7). In a related study, Richter et al. found that USA100 isolates represented 16.8% of MRSA isolates and were the most common blood culture MRSA isolates (9). In a more recent comprehensive meta-analysis, USA100 MRSA levels were found to have remained fairly steady from 2005 to 2013, ranging from 21 to 25% of MRSA collections, second only to the USA300 isolates in each time frame (10).
In comparison to USA300 isolates, the USA100 isolates demonstrate in vitro resistance to more antibiotic classes, such as fluoroquinolones, macrolides, and lincosamides (9). Additionally, most S. aureus isolates that demonstrate vancomycin resistance or intermediate resistance are of the USA100 genotype (11). However, in terms of infection potential, HA-MRSA isolates and representative USA100 isolates are generally considered less virulent and secrete cytotoxins at lower levels than their CA-MRSA counterparts (12, 13), characteristics which have resulted in limited interest in the HA-MRSA virulence mechanisms.
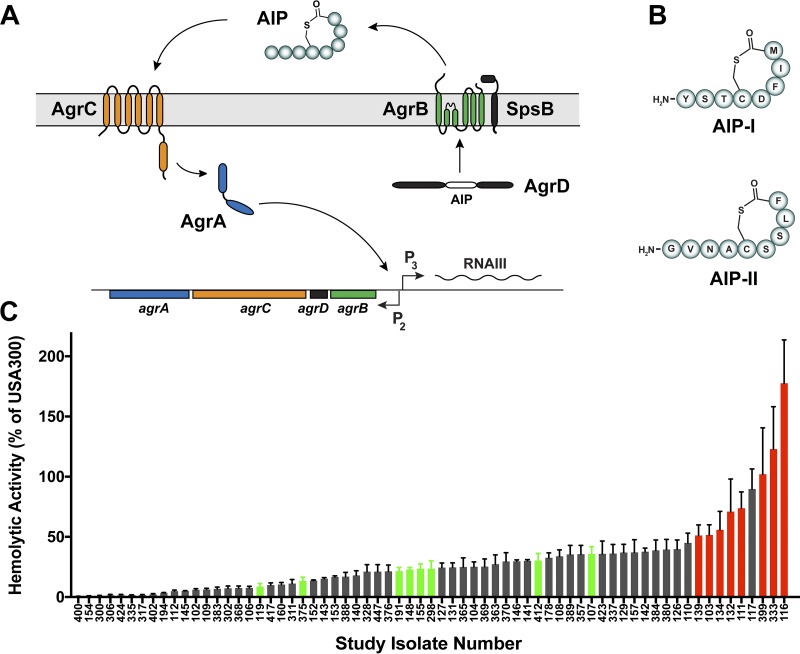
In all S. aureus strains, toxin regulation is controlled by a peptide quorum-sensing system (Fig. 1A), also called the accessory gene regulator (agr) (14–16). The system responds to a secreted autoinducing peptide (AIP) signal generated from the AgrD peptide precursor by the AgrB membrane endopeptidase. At a critical concentration, the AIP binds to a receptor on the AgrC histidine kinase, activating the kinase, which in turn phosphorylates the response regulator AgrA. Activated AgrA in turn binds the P2 and P3 promoters in the agr locus, autoinducing the system and also upregulating the RNAIII effector, which turns on toxin and exoenzyme production. Across S. aureus strains, there are hypervariable regions that exist within the agrB, agrD, and agrC genes, and this results in four different locus variants, called agr type I (agr-I), agr-II, agr-III, and agr-IV. In turn, these loci produce four different AIP types, called AIP-I through AIP-IV (17). The common CA-MRSA isolate USA300 has the agr-I allele and produces AIP-I, while USA100 isolates utilize the agr-II system and produce AIP-II (Fig. 1B). Interestingly, the type I and II systems remain the most divergent across S. aureus strains in terms of sequence (14).
FIG 1.
The USA100 agr type II system. (A) Schematic of the S. aureus agr system. (B) Structures of the AIP-I signal from USA300 strains and the AIP-II signal from USA100 strains. (C) Alpha-toxin levels from a rabbit red blood cell hemolysis assay of the 67 strains that met the inclusion criteria. The data are plotted as a percentage of activity relative to that for USA300 strain LAC, which was set to 100%. The 16 USA100 strains for which results are presented in red and green were selected for further study.
Our goal in this study was to better understand the contribution of the agr-II system to USA100 MRSA antibiotic resistance and virulence. We obtained clinical USA100 isolates that represent spa type 2 and SCCmec type II and selected those that had a functional agr system, as predicted from toxin production. There is some evidence that agr contributes to antibiotic resistance (18), and since USA100 isolates are some of the most resistant isolates, we reasoned that agr-II might influence resistance levels. Since agr is a major contributor to the virulence of other MRSA isolates, we predicted that it would be important for toxin and exoenzyme production, as well as infection potential.
RESULTS
Bacterial strain selection and construction of agr mutants.
A total of 526 pulsed-field gel electrophoresis- and spa-typed S. aureus strains were provided by the University of Iowa Molecular Epidemiology Laboratory (19–21). Out of this collection, 146 were spa type 2, representing USA100 isolates. The collection was comprised of roughly equal numbers of colonizing and invasive isolates, and only the invasive ones (blood culture isolates) were considered further. Of these, only USA100 strains that contained SCCmec type II (MRSA) and that were tetracycline sensitive (for agr mutant construction) were selected for further testing, eliminating many isolates and narrowing the collection to 67 candidates.
HA-MRSA strains are known to pick up agr locus variants that result in dysfunction more commonly than CA-MRSA strains (22, 23). Since the goal was to study USA100 isolates with an intact agr system, we assessed the alpha-toxin activity of the 67 candidates as a proxy for agr function. The results of a rabbit red blood cell lysis assay serve as an indicator of alpha-toxin activity (24), and we performed this assay with the candidates and compared the results to those for USA300 strain LAC as a control (Fig. 1C). Eight USA100 isolates that had robust alpha-toxin production (marked in red in Fig. 1C; Table 1), suggesting that they had intact and functional agr type II systems, were selected. To inactivate the agr locus, the well-known Δagr::TetM (hereafter called Δagr) cassette was crossed into each strain, and the mutation was confirmed. An additional eight strains that had lower levels of alpha-toxin production (marked in green in Fig. 1C; Table 1) were selected for comparison. Table 2 summarizes the constructed USA100 wild-type (WT) and agr null strain pairs.
TABLE 1.
Characteristics of USA100 isolates selected for studya
| Study clinical isolate no. |
Source | spa type | SCCmec
type |
PVL | % of LAC RBC lysis activity |
|---|---|---|---|---|---|
| 111 | Blood | t002 | II | Negative | 73.8 |
| 103 | Blood | t002 | II | Negative | 51.5 |
| 116 | Blood | t002 | II | Negative | 177.6 |
| 132 | Blood | t002 | II | Negative | 70.9 |
| 134 | Blood | t002 | II | Negative | 55.9 |
| 139 | Blood | t002 | II | Negative | 51.1 |
| 333 | Blood | t002 | II | Positive | 123 |
| 399 | Blood | t002 | II | Positive | 102 |
| 119 | Blood | t002 | II | Negative | 8.7 |
| 375 | Blood | t002 | II | Positive | 13.5 |
| 191 | Blood | t088 | II | Negative | 21.5 |
| 148 | Blood | t002 | II | Negative | 22.9 |
| 155 | Blood | t002 | II | Negative | 23.6 |
| 298 | Blood | t002 | II | Positive | 23.6 |
| 412 | Blood | t002 | II | Positive | 30.3 |
| 107 | Blood | t002 | II | Negative | 35.7 |
PVL, Panton-Valentine leukocidin; RBC, red blood cell.
TABLE 2.
Nomenclature for the USA100 wild-type and agr mutant isolates studied
| Strain | Study clinical isolate no. |
Reference or source |
|---|---|---|
| AH1263 | LAC (Erm sensitive)a | 51 |
| AH843 | MW2 | 52 |
| AH2759 | LAC/pAmiAgrP3 | 53 |
| AH1677 | LAC/pDB59 | 27 |
| AH3185 | MW2/pAmiAgrP3 | This work |
| AH1747 | MW2/pDB59 | 27 |
| AH4489 | 103 | This work |
| AH4541 | 103 Δagr | This work |
| AH5445 | 103/pDB59 | This work |
| AH2617 | 111 | This work |
| AH2618 | 111 Δagr | This work |
| AH5510 | 111/pDB59 | This work |
| AH4490 | 116 | 31 |
| AH4542 | 116 Δagr | This work |
| AH5446 | 116/pDB59 | This work |
| AH4390 | 116/pAmiAgrP3 | This work |
| AH4491 | 132 | 31 |
| AH4543 | 132 Δagr | This work |
| AH5492 | 132/pDB59 | This work |
| AH4492 | 134 | This work |
| AH4544 | 134 Δagr | This work |
| AH5447 | 134/pDB59 | This work |
| AH5070 | 134/pAmiAgrP3 | This work |
| AH4493 | 139 | This work |
| AH4528 | 139 Δagr | This work |
| AH5448 | 139/pDB59 | This work |
| AH4494 | 333 | This work |
| AH4529 | 333 Δagr | This work |
| AH5493 | 333/pDB59 | This work |
| AH4495 | 399 | This work |
| AH4530 | 399 Δagr | This work |
| AH5299 | 412 | This work |
| AH5495 | 412/pDB59 | This work |
| AH5294 | 155 | This work |
| AH5494 | 155/pDB59 | This work |
| AH5300 | 107 | This work |
| AH5453 | 107/pDB59 | This work |
| AH5298 | 375 | This work |
| AH5452 | 375/pDB59 | This work |
| AH5296 | 298 | This work |
| AH5451 | 298/pDB59 | This work |
| AH5295 | 191 | This work |
| AH5450 | 191/pDB59 | This work |
| AH5297 | 119 | This work |
| AH5293 | 148 | This work |
Erm, erythromycin.
USA100 agr system kinetics.
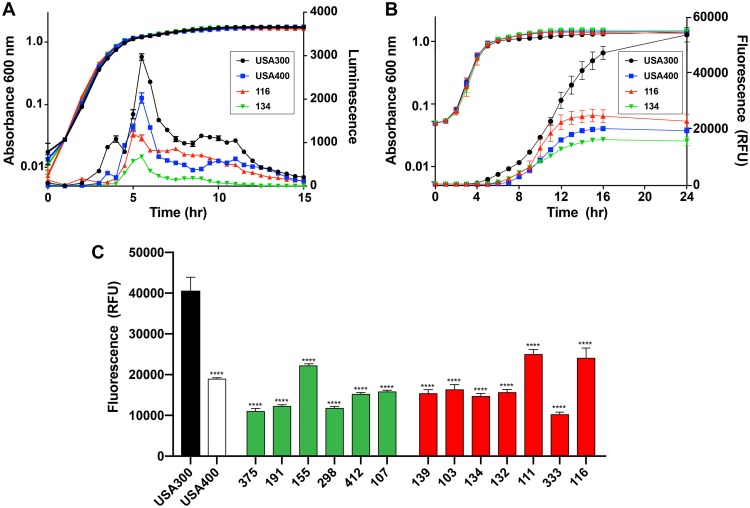
To compare the agr activation kinetics in USA100 isolates to that in other S. aureus strains, we moved agr P3-lux reporter constructs into selected USA100 strains (strains 116 and 134), USA300 LAC, and USA400 MW2. As shown in Fig. 2A, each agr P3-lux reporter strain activated the agr system in the exponential phase of growth, and maximal activity was reached at about 5 to 6 h.
FIG 2.
USA100 agr activation kinetics. (A) USA300 strain LAC, USA400 strain MW2, and USA100 strains 116 and 134 containing an agr P3-lux reporter were grown for 15 h, and the optical density at 600 nm and luminescence were measured. (B) USA300 strain LAC, USA400 strain MW2, and USA100 strains 116 and 134 containing the agr P3-YFP reporter (pDB59) were grown for 24 h, and the optical density at 600 nm and the fluorescence were measured. (C) Fluorescence measurements of all USA100 strains used in the study containing the agr P3-YFP reporter (pDB59) after 14 h of growth. Statistical significance was determined using Student's t test. ****, P < 0.0001. RFU, relative fluorescence units.
While the agr P3-lux reporter is powerful and provides useful kinetic analysis, it is somewhat challenging to use genetically, as some S. aureus strains do not take up the plasmid. To have a broader comparison, we used the pDB59 agr P3-yellow fluorescent protein (YFP) reporter, which has been our workhorse reporter for quorum-sensing assessments (25–27). We moved the pDB59 plasmid into all 16 USA100 strains (8 high-toxin producers and 8 low- to middle-level toxin producers; Tables 1 and 2). Strains 119 and 148 had no agr activity, which we confirmed with secondary methods, and strain 399 grew poorly with the pDB59 plasmid and was not tested further. Comparison of strains 116 and 134 with agr P3-lux (Fig. 2A) with strains 116 and 134 with agr P3-YFP (Fig. 2B) revealed delayed activation with the slowly folding YFP, with lux coming on at about 4 h, while YFP came on at about 10 h. The USA300 strains had the strongest agr output with both reporters, while either the USA400 or the USA100 116 strain (the highest toxin producer) had the next-strongest output (Fig. 2A and B), and the USA100 134 strain had the weakest output of this set.
Next, we compared the 13 USA100 YFP reporter strains to the CA-MRSA controls, selecting the 14-h time point to assess the maximal activity of the reporter (Fig. 2C). The activity of the USA300 reporter was consistently and significantly higher than that of all others at this time point. Some of the high-alpha-toxin-producing USA100 isolates (strains 111 and 116) maintained a high agr output, while some of the low-toxin producers (strains 191 and 375) had a low agr output or lacked agr function (strains 119 and 148). However, there was a lot of strain-dependent variability that was not always consistent (Fig. 2C). In general, the CA-MRSA strains are known to have a robust agr system function and to produce high levels of RNAIII (14), and within the CA-MRSA group, the USA300 lineage has an exceptionally strong agr function that can outperform that of isolates of other lineages, such as USA400 isolates (28). Taken together, USA100 isolates have agr kinetics of activation similar to that of CA-MSRA isolates but achieve a lower overall maximal output.
USA100 virulence factor production.
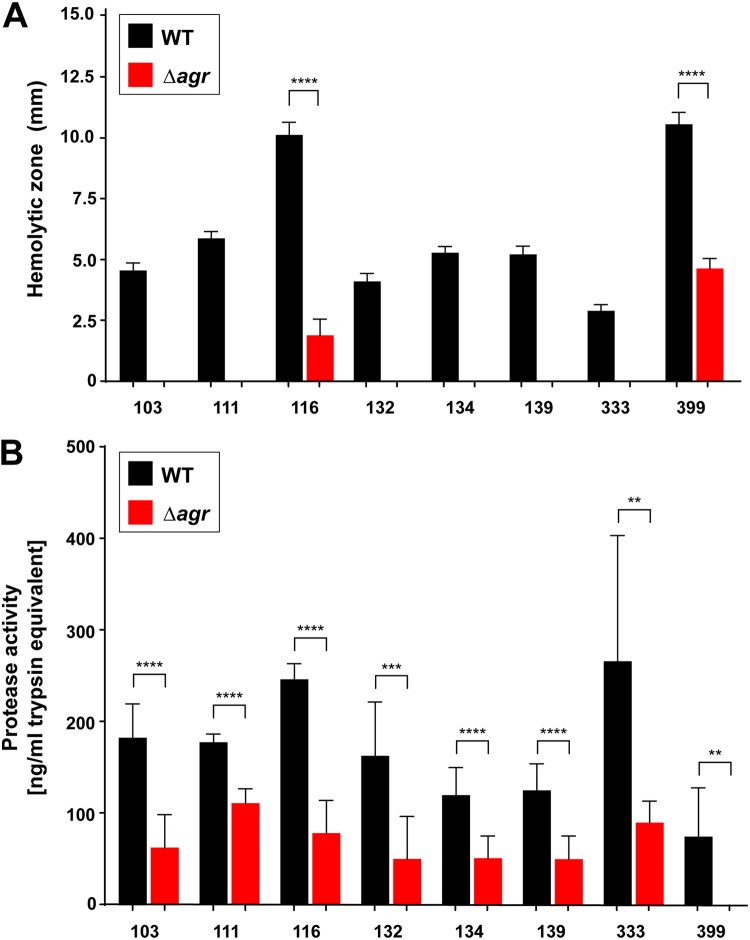
It is known that the agr system regulates hemolysins and exoenzymes that are essential for S. aureus virulence (14). The suite of agr-regulated hemolysins is diverse and includes alpha-toxin, β-hemolysin, gamma-hemolysin, phenol-soluble modulins (PSMs), and the other bicomponent leukocidins. As reported previously (29, 30), we used a qualitative sheep blood hemolysis assay to broadly assess the agr contribution to USA100 hemolysin production. For each USA100 WT and Δagr strain pair, clearing zones were measured at 24 and 48 h, and as anticipated, the WT strains demonstrated significantly more hemolysis than the agr mutant strains (Fig. 3A). Interestingly, two agr mutants, 116 Δagr and 399 Δagr, had background hemolysis at 24 and 48 h, although it was significantly less than that of the WT. None of the other Δagr mutants had background hemolysis. We reasoned that some of the hemolysis occurring in the Δagr mutants might be due to β-hemolysin, which is not as tightly agr regulated (30). The hlb gene, encoding β-hemolysin, is often disrupted in clinical isolates by an integrated bacteriophage, such as the ϕSa3-related phages (31, 32), and these phages encode additional virulence factors, such as staphylokinase (SAK) (32). To determine the presence of ϕSa3-related phages in the USA100 strains (high alpha-toxin producing), PCR was performed on all the USA100 WT strains, which confirmed the presence of the sak gene in all strains except the 399 strain. Furthermore, the hlb gene was successfully identified to be intact in the 399 WT strain.
FIG 3.
Contribution of agr to USA100 virulence factor expression. (A) Hemolytic zones at 24 h, measured as the diameter (in millimeters) minus the colony size (in millimeters) for each pair. (B) Proteolytic activity, expressed as the equivalent of a trypsin standard. The WT demonstrated more hemolysis and proteolysis than the Δagr mutant in the strain pairs, but two Δagr mutants (strains 116 and 399) did show low levels of hemolysis. Statistical significance was determined using Student's t test. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01.
Secreted enzymes are another major class of S. aureus virulence factors regulated by the agr system (33, 34). We focused on the extracellular proteases, and we used a fluorescent casein cleavage assay to assess activity in the USA100 strain pairs. As shown in Fig. 3B, the USA100 WT strains had more extracellular protease activity than the Δagr mutant strains, although, surprisingly, many of the Δagr mutants had substantial background activity. It should be noted, though, that not every protease secreted by S. aureus cleaves casein with a similar efficiency. Based on the results of these different assays, overall, the USA100 agr type II system behaves similarly to that of other S. aureus strain lineages in terms of virulence factor expression.
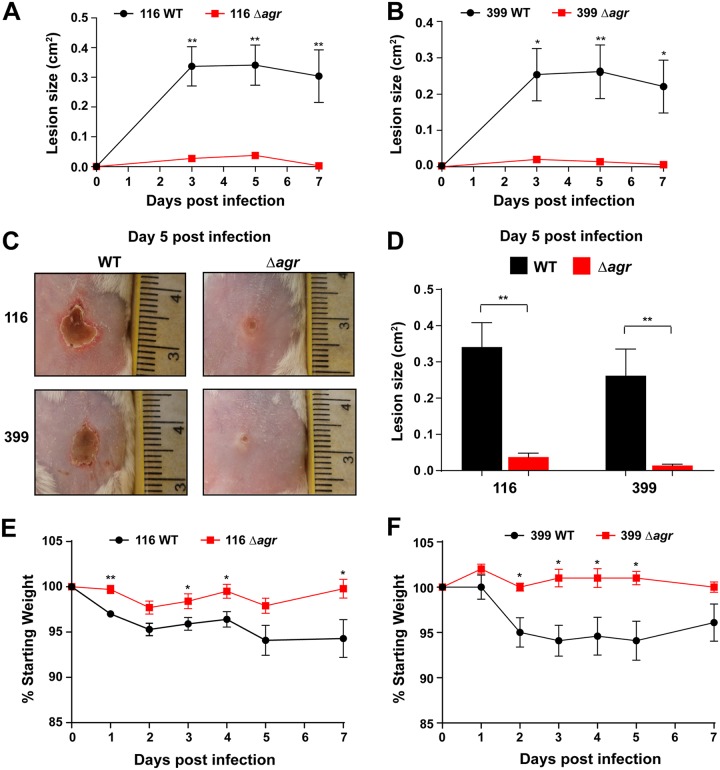
The USA100 agr system contributes to skin infection.
Numerous studies have demonstrated that the agr system of CA-MRSA USA300 strains contributes to skin infection in animal models (35–39), but its contribution is less clear in the HA-MRSA USA100 isolates. Specifically, we chose the strain pairs of the two strains 116 and 399 as USA100 strain sets that exhibited a high level of hemolysis both for the WT and at the background level for the Δagr mutant (Fig. 3A). Using an intradermal infection model that we have previously used to assess MRSA skin virulence (36–38, 40, 41), the lesion sizes were dramatically bigger in mice challenged with the 116 WT strain than in mice challenged with the Δagr mutant over time (Fig. 4A). A similar pattern was observed with the 399 strain, with the lesions in the mice challenged with the WT being consistently larger than those in the mice challenged with the Δagr mutant (Fig. 4B). Pictures of the gross lesion pathology (Fig. 4C) at 5 days and measurement of these lesion sizes (Fig. 4D) confirmed this phenotype. Consistent with this, the USA100 WT-challenged mice experienced more weight loss over 7 days than the mutant strain-challenged mice (Fig. 4E and F). Despite each of these strains having significant background levels of hemolysis on sheep blood (Fig. 3A), the contribution of this to a skin lesion was minimal. Taken together, these data suggest that when the agr system is intact, higher levels of virulence factors are expressed, resulting in a greater systemic effect of infection, in addition to a larger abscess size.
FIG 4.
Role of agr in USA100 skin infections. (A, B) Comparison of lesion size for mice inoculated with the strain 116 (A) and 399 (B) WT and Δagr mutant strain pairs over 1 week (n = 5 for each group). (C) Photographs of ventral skin lesions 5 days after inoculation with WT and Δagr mutant strains. Note the smaller areas of dermal necrosis resulting from infection with the Δagr mutants. (D) Comparison of lesion size between WT and Δagr mutant isolates 5 days after intradermal injection of mice (n = 5 for each group). (E, F) Comparison of weight loss (expressed as a percentage of the starting weight) in mice which underwent intradermal injections of the strain 116 (E) and 399 (F) WT and Δagr isolates. Statistical significance was determined using Student's t test. **, P < 0.01; *, P < 0.05.
Contribution of agr to antibiotic resistance.
Previous studies have suggested a potential difference in the survival of MRSA WT strains and Δagr strains when exposed to antibiotics (18, 23, 42). As shown in Table 3, we tested the USA100 strain pairs for resistance to 13 common antibiotics using Etest strips. Except for the tetracycline resistance due to the Δagr::TetM construct, there was no significant difference in antibiotic resistance between the USA100 WT and each paired Δagr mutant. This matches previous reports of the results of MIC testing of diverse S. aureus strains and Δagr mutants (42).
TABLE 3.
MICs of USA100 strain pairs
| Strain | MICa
(μg/ml) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOX | CLI | DAP | ERY | GEN | LVX | LZD | OXA | Q-D | RIF | TET | SXT | VAN | |
| 103 WT | 96 | 0.094 | 0.25 | >256 | 0.25 | >32 | 1.5 | >256 | 0.5 | 0.012 | 0.19 | 0.064 | 1.5 |
| 103 Δagr | 48 | 0.094 | 0.125 | >256 | 0.25 | >32 | 2 | >256 | 0.5 | 0.012 | 24 | 0.064 | 1 |
| 111 WT | 12 | > 256 | 0.38 | >256 | 0.38 | >32 | 3 | >256 | 1 | 0.016 | 0.5 | 0.094 | 2 |
| 111 Δagr | 32 | >256 | 0.19 | >256 | 0.5 | >32 | 3 | 192 | 1 | 0.012 | 32 | 0.064 | 1.5 |
| 116 WT | 48 | 0.125 | 0.19 | >256 | 0.38 | >32 | 1.5 | 64 | 0.38 | 0.008 | 0.25 | 0.094 | 1 |
| 116 Δagr | 24 | 0.094 | 0.125 | >256 | 0.5 | >32 | 1.5 | 24 | 0.38 | 0.008 | 24 | 0.094 | 1 |
| 132 WT | >256 | 0.125 | 0.19 | >256 | 0.38 | >32 | 2 | >256 | 1.5 | 0.012 | 0.38 | 0.094 | 1.5 |
| 132 Δagr | >256 | 0.125 | 0.25 | >256 | 0.75 | >32 | 2 | >256 | 0.75 | 0.012 | 48 | 0.094 | 1.5 |
| 134 WT | >256 | 0.125 | 0.25 | >256 | 0.38 | >32 | 2 | >256 | 0.5 | 0.012 | 0.19 | 0.094 | 2 |
| 134 Δagr | >256 | 0.125 | 0.25 | >256 | 0.38 | >32 | 1.5 | >256 | 0.5 | 0.012 | 24 | 0.094 | 2 |
| 139 WT | >256 | >256 | 0.38 | >256 | 0.38 | >32 | 3 | >256 | 1 | 0.012 | 0.38 | 0.094 | 2 |
| 139 Δagr | >256 | >256 | 0.38 | >256 | 0.5 | >32 | 3 | >256 | 0.75 | 0.012 | 24 | 0.094 | 1.5 |
| 333 WT | 192 | 0.125 | 0.38 | 256 | 0.5 | >32 | 2 | >256 | 0.5 | 0.012 | 0.5 | 0.094 | 2 |
| 333 Δagr | 96 | 0.125 | 0.38 | >256 | 0.38 | >32 | 2 | >256 | 0.5 | 0.012 | 16 | 0.094 | 1.5 |
| 399 WT | >256 | >256 | 0.5 | >256 | 0.75 | 8 | 1.5 | >256 | 0.75 | 0.012 | 0.19 | 0.19 | 2 |
| 399 Δagr | >256 | >256 | 0.38 | >256 | 0.75 | 6 | 1.5 | >256 | 0.75 | 0.012 | 24 | 0.19 | 2 |
MICs are based on the Etest for the USA100 wild-type and agr mutant isolate pairs. Overall, there was no difference in antibiotic resistance, as determined by the use of Etest strips, with the exception of resistance to tetracycline, which was as expected, since the tetracycline resistance cassette was incorporated into the genome of each agr mutant as part of mutant construction. FOX, cefoxitin; CLI, clindamycin; DAP, daptomycin; ERY, erythromycin; GEN, gentamicin; LVX, levofloxacin; LZD, linezolid; OXA, oxacillin; Q-D, quinupristin-dalfopristin; RIF, rifampin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
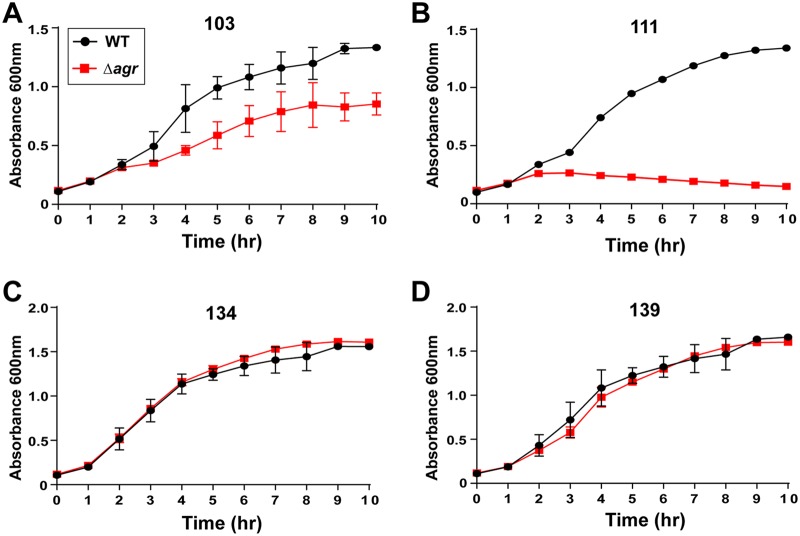
Since survival and stress assays have shown different behaviors for strains with a defective agr gene, we performed more careful growth curves for each USA100 strain pair with cefoxitin, clindamycin, daptomycin, and vancomycin. For cefoxitin at levels below the MIC (∼16 μg/ml), some differences in growth were noted. The Δagr mutant and the WT in the strain 103 strain pair demonstrated statistically significantly different inhibitory growth (Fig. 5A), and the two strains in the strain 333 strain pair grew in a fashion comparable to that for the two strains in the strain 103 strain pair (data not shown). For the strain 111 strain pair, a similar and even more pronounced pattern was apparent, with the growth of the Δagr mutant dramatically lagging behind that of the WT (Fig. 5B). The growth of the WT and the Δagr mutant in the rest of the strain pairs exhibited few differences in the presence of cefoxitin (representative plots for strains 134 and 139 are shown in Fig. 5C and D, respectively.
FIG 5.
Growth of USA100 strains in the presence of a sub-MIC of cefoxitin. (A, B) The strain 103 (A) and 111 (B) WT strains demonstrated growth superior to that of the paired Δagr mutants when exposed to cefoxitin, suggesting that a functional agr system in these strains contributes to survival under antibiotic stress. (C, D) Growth curves for representative strains 134 (C) and 139 (D) in which no differences in growth were observed between each of the strains in the strain pairs tested with the same concentration of cefoxitin.
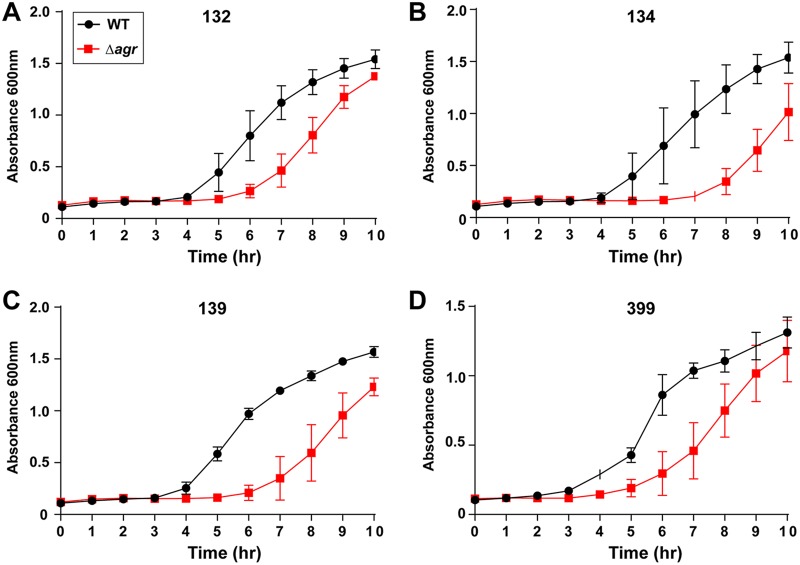
The largest differences between the pairs of USA100 strains were observed in the presence of vancomycin. At 2 μg/ml, the upper limit for the MIC (Table 3), the growth of the Δagr mutant lagged behind that of the WT for each of the strain 132, 134, 139, 333, and 399 strain pairs (the results for the strain 132, 134, 139, and 399 strain pairs are shown in Fig. 6A to D, respectively). The two strains in each of the strain 103 and 111 strain pairs did not have any obvious differences in their growth curve patterns, while the two strains in the strain 116 strain pair grew in an inconsistent manner (data not shown). Additional growth curves were performed with clindamycin (32 μg/ml) and daptomycin (20 μg/ml), and in each case, no major differences in growth between the WT and the Δagr mutant were observed.
FIG 6.
Growth of USA100 strains in the presence of a sub-MIC of vancomycin. WT strains 132 (A), 134 (B), 139 (C), and 399 (D) demonstrated growth superior to that of the paired Δagr mutants when exposed to vancomycin at 2 μg/ml. For at least these strains tested, a functional agr system contributes to improved survival in the presence of vancomycin.
DISCUSSION
The agr peptide quorum-sensing system is a well-described virulence regulator of CA-MRSA (14). In this study, our goal was to understand the contribution of agr to the virulence and antibiotic resistance of clinical HA-MRSA isolates, specifically, those of the USA100 group, which have a more divergent agr type II system and which make an AIP-II signal (Fig. 1). Overall, we found that hemolysin and exoenzyme production among the USA100 isolates was dependent on the agr system, with some strain-specific exceptions, and the USA100 isolates triggered dermonecrosis in a murine skin infection model in an agr-dependent manner. The contribution of agr to USA100 antibiotic resistance was limited in terms of MIC levels, but surprisingly, many of the agr mutants exhibited less tolerance to vancomycin than their WT counterparts.
The agr type II system is the most divergent from the common type I system in terms of sequence and signal structure (Fig. 1B). Despite the variance, the kinetics and behavior of the regulatory system in USA100 isolates shared many similarities to those published for laboratory strains and CA-MRSA. The agr activation profile between USA100 and CA-MRSA isolates was similar in timing, although it was weaker in overall output in USA100 isolates (Fig. 2). However, the size of the lesions in a skin infection model correlated well with those that we have observed with USA300 strains (36–38, 40), which could be due to our selection of USA100 isolates with higher alpha-toxin activity (Fig. 1C). Some USA100 agr mutants showed extensive background hemolysis, and in the case of strain 399, this could have been due to an intact sequence for hlb, encoding β-hemolysin (31). However, these results were not reproducible in the strain 116 strain pair, suggesting that background hemolysis is potentially due to another factor. Similarly, there was a surprising level of background protease activity in many of the USA100 Δagr mutants. The reason for this is not clear, but it could be due to variance in aureolysin (Aur) metalloprotease activity. Aur is one of the primary casein-cleaving enzymes (43) and is under only limited agr regulatory control (44). Therefore, the background casein cleavage in Δagr mutants (Fig. 3B) could be due to low levels of Aur production.
Our study also aimed to determine whether or not the agr system is involved in USA100 antibiotic resistance. There are previous data to suggest that the agr system may have a role in altering antibiotic resistance patterns, as evidenced by the increased survival of agr mutants when exposed to antibiotics (18, 23) and also the development of vancomycin-intermediate resistance (vancomycin-intermediate S. aureus) when the agr system is dysfunctional (45). Standard MIC testing of the WT strains and the Δagr mutants with 13 different antibiotics found no significant differences in resistance (Table 3), except for resistance to tetracycline, which was used to construct the Δagr::TetM mutation. In a more careful fitness assessment in the presence of low antibiotic levels, the WT and Δagr mutants displayed no growth differences in the presence of clindamycin or daptomycin, which in the case of daptomycin is contrary to the information previously published in the literature (18, 42). When the USA100 isolates were grown in the presence of low levels of vancomycin, some differences in the response were observed. For 5 of the 8 USA100 isolate pairs tested (Fig. 6), the WT grew substantially faster than the Δagr mutants, which seems in contrast to the agr dysfunction correlating with vancomycin heteroresistance (45). However, the growth assays with a sub-MIC of vancomycin performed in this study are somewhat different from a heteroresistance assessment. Of the remaining 3 USA100 isolate pairs that showed no significant differences in growth with vancomycin, the WT isolates grew faster than the Δagr mutants in the presence of a sub-MIC of cefoxitin in two of these pairs (Fig. 5). Overall, in the USA100 isolates, our findings suggest that a functional agr system is important for optimal survival in the presence of antibiotic stress. We speculate that some of the differences from findings previously reported in the literature are due to strain selection (18, 23, 45). The activity of agr across USA100 isolates varies widely (Fig. 1C), and in our study, we focused only on isolates with a robust and functional agr system.
As MRSA continues to cause infections worldwide, there is a need to understand both the virulence and resistance mechanisms of the common CA-MRSA strains and the underappreciated impact of HA-MRSA. Among the HA-MSRA strains, strains in the USA100 group are the most common and are known to be highly antibiotic resistant, and they continue to represent a significant burden to the health care system. Our work shows that the virulence properties of the agr type II system largely parallel those of the type I system, but the contribution to antibiotic resistance is less clear and in need of further investigation. Previous work has shown that blocking the agr system could be a potential target for reducing the severity of infection (36, 38, 40), and this could be applied to the treatment of USA100 infections. With the high level of antibiotic resistance in these strains and the ongoing interest in antibiotic stewardship (46), the development of new approaches to treat multidrug-resistant infections is important.
MATERIALS AND METHODS
USA100 strains.
The MRSA strains used in this study had previously been isolated from patients participating in surveillance studies coordinated by the University of Iowa Molecular Epidemiology Laboratory. These strains included both colonizing strains isolated from the nasopharynx and invasive strains, mostly from blood samples.
agr mutant construction.
The Δagr::TetM construct (47) was created by crossing Δagr::TetM by bacteriophage transduction as previously described (48). Successful transduction of the tetracycline resistance cassette was confirmed by DNA PCR. Chromosomal DNA was purified using a Qiagen PureGene kit along with a modified manufacturer protocol. DNA PCR was then performed using Crimson Taq DNA polymerase along with RNAIII downstream primer 5′-GTATAAATAAGAAGCGCCCGAAATA-3′ and agrA downstream primer 5′-CCAGCTATACAGTGCATTTGCTAGT-3′, with the expected product size for the wild type being 3,823 bp. Deletion of the agr operon and insertion of the tetracycline resistance cassette were confirmed after gel electrophoresis, which demonstrated a shorter product in the agr knockout strains, as expected.
Luminescence reporters and assay.
The agr P3-lux reporter plasmid pAmiAgrP3 (49) was purified from restriction-negative S. aureus strain RN4220 and moved by electroporation into strains LAC (USA300) and MW2 (USA400) and the USA100 strains 116 and 134, using a previously described transformation method (50). Overnight cultures were diluted 1:100 in triplicate into tryptic soy broth (TSB) supplemented with chloramphenicol (10 μg/ml) in a black 96-well plate and grown at 37°C and 1,000 rpm in a Stuart humidified microtiter plate shaker. The optical density at 600 nm (OD600) and luminescence were monitored using a Tecan Infinite M Plex plate reader.
Fluorescence reporters and assay.
The agr reporter plasmid pDB59 was transduced from restriction-negative S. aureus strain RN4220 and into strains LAC (USA300) and MW2 (USA400) and the USA100 hospital strains using phage Φ11. Overnight cultures were diluted 1:500 in triplicate into TSB supplemented with chloramphenicol (10 μg/ml) in a black 96-well plate and grown at 37°C and 1,000 rpm in a Stuart humidified microtiter plate shaker. The OD600 and the fluorescence at 535 nm following excitation at 492 nm were monitored using a Tecan Infinite M Plex plate reader.
Alpha-toxin lysis assay.
S. aureus lysis of rabbit red blood cells was performed as previously described (24).
Hemolysis assay.
To test the hypothesis of agr system control of virulence factor expression, hemolysis assays were performed. Each WT strain and agr knockout strain pair was plated on sheep blood agar and incubated overnight at 37°C; single colonies were subcultured in TSB and again incubated overnight at 37°C. Each WT and agr mutant culture was then diluted 1:100 in TSB and grown to reach an OD600 of 3 (approximately 3 to 4 h). Triplicate samples were grown for each strain pair. Subsequently, three samples were plated for each WT and mutant strain on sheep blood agar; each sample contained 2 μl of the culture described above. The samples were allowed to dry and were then incubated for 24 h at 37°C. At 24 h, the diameter of each bacterial colony and the diameter of hemolysis were measured with calipers. These measurements were taken again at 48 h, after the plates were allowed to incubate at 37°C for an additional 24 h. A total of 9 replicates were performed for each WT strain and its paired agr mutant strain.
Protease assay.
Protease assays were completed to further test the hypothesis of agr control of virulence factor expression. These experiments were performed with a Pierce fluorescent protease assay kit (Thermo Scientific), which assesses protease activity using fluorescence resonance energy transfer (FRET). For these assays, each WT and mutant strain was cultured overnight in TSB. Each of these cultures was then diluted 1:100 in TSB and grown to an OD600 of 1.5. One milliliter of culture at an OD600 of 1.5 for each WT and mutant sample was centrifuged at 13,000 × g for 1 min. One hundred microliters of the supernatant was transferred to a 96-well plate in triplicate for each wild type and mutant pair; 100 μl of a working reagent containing fluorescent-labeled casein was then added to each well, and the plates were allowed to incubate for 1 h, at which time the plates were analyzed in a fluorescent plate reader (Tecan Infinite 200) at wavelengths of between 485 and 538 nm. Tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin (0.5 μg/ml) was used to generate a standard curve, which was then used to calculate protease activity for casein as a trypsin equivalent for each WT and mutant strain. Biological and technical replicates were performed, for a total of 9 data points for each WT and mutant isolate.
Murine infection model.
Murine models were used to test the virulence of both the WT and agr mutant strains as we previously reported (36–38, 40). Cultures were grown overnight at 37°C in a shaking incubator set at 200 rpm from frozen glycerol stocks for two selected wild-type and agr mutant strain pairs (strains 116 and 399) in 5 ml of TSB. Subcultures were prepared to a total volume of 350 ml, consisting of 34.65 ml of TSB and 350 μl of an overnight culture, and then grown to an OD600 of 0.5 at 37°C in a shaking incubator at 200 rpm. Ten milliliters of this culture was then added to 40 ml of Dulbecco’s phosphate-buffered saline (DPBS) and centrifuged at 3,500 rpm for 10 min. The pelleted cells were then resuspended in sterile saline to reach a concentration of 2 × 107 CFU/50 μl and placed on ice. Five BALB/c mice were used to test each strain, resulting in the use of 20 mice in total. Mice were anesthetized with 1 to 5% isoflurane. The abdominal skin of each mouse was shaven with a microtome blade; the mice were then labeled, weighed, and prepped for sterile injection. Insulin syringes were used to intradermally inoculate each mouse with 50 μl of a specified S. aureus suspension. The mice were monitored daily, with the lesion size (in square centimeters) as well as weight loss being recorded as markers of disease for up to 2 weeks. All mouse experiments were approved by the University of Iowa Institutional Animal Care and Use Committee.
Determination of MIC and antibiotic resistance patterns.
Comparison of antibiotic resistance patterns was performed using Etest strips (bioMérieux USA) to determine the MICs of cefoxitin, clindamycin, daptomycin, erythromycin, gentamicin, levofloxacin, linezolid, oxacillin, quinupristin-dalfopristin, rifampin, tetracycline, trimethoprim-sulfamethoxazole, and vancomycin. Triplicate testing for each isolate (the wild-type and null mutant isolates) was performed per the manufacturer’s protocol, and a methicillin-sensitive Staphylococcus aureus subsp. aureus Rosenbach (ATTC 29213) strain served as a quality control for these experiments.
To examine more subtle differences between these isolates in the presence of selected antibiotics, growth curves were performed for all strain pairs. The antibiotics selected for these studies were cefoxitin, clindamycin, daptomycin, and vancomycin. The WT and agr mutant strains were cultured overnight in TSB at 37°C. Ninety-six-well plates were prepared with serially diluted concentrations of antibiotic added to TSB. For each antibiotic, seven different concentrations were tested; in addition, one control sample without antibiotic was used for each strain. The concentrations tested for cefoxitin were 1 mg/ml, 500 μg/ml, 250 μg/ml, 125 μg/ml, 62.5 μg/ml, 31.25 μg/ml, and 15.625 μg/ml; those tested for clindamycin were 16 μg/ml, 8 μg/ml, 4 μg/ml, 2 μg/ml, 1 μg/ml, 0.5 μg/ml, and 0.25 μg/ml; those tested for daptomycin were 100 μg/ml, 50 μg/ml, 20 μg/ml, 16 μg/ml, 8 μg/ml, 4 μg/ml, and 2 μg/ml; and those tested for vancomycin were 2 μg/ml, 1 μg/ml, 0.5 μg/ml, 0.25 μg/ml, 0.125 μg/ml, and 0.625 μg/ml. Bacteria were grown from a 1:50 dilution in each well containing the respective concentration of antibiotic. These plates were then incubated at 37°C and shaken at 1,000 rpm. The absorbance (OD600) was measured by use of a microtiter plate reader (Tecan Infinite 200) starting at hour 0 and then hourly thereafter until the bacteria had reached stationary phase (8 to 10 h); measurements were then used to generate growth curves for each strain at the various antibiotic concentrations.
Statistical analysis.
GraphPad Prism (version 7) software was used to perform statistical analysis of the hemolysis, protease, and animal data. Student's t test was chosen to calculate the difference in hemolysis and proteolysis between the wild-type strain and paired agr mutant as well as the murine data comparing lesion size and weight loss between the groups challenged with the WT and the agr mutant. A P value of <0.05 was chosen to designate a statistically significant difference between values. Throughout the work, error bars represent standard deviations.
ACKNOWLEDGMENTS
Funding for this project was provided by University of Iowa Stead Family Children’s Hospital discretionary funds. A.R.H. was supported by a merit award (award I01 BX002711) from the U.S. Department of Veteran Affairs.
REFERENCES
- 1.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Chambers HF, DeLeo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLeo FR, Chambers HF. 2009. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest 119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46(Suppl 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH, MRSA Consortium. 2012. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States hospitals. Antimicrob Agents Chemother 56:1324–1330. doi: 10.1128/AAC.05804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limbago B, Fosheim GE, Schoonover V. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol 47:1344–1351. doi: 10.1128/JCM.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter SS, Heilmann KP, Dohrn CL, Riahi F, Costello AJ, Kroeger JS, Biek D, Critchley IA, Diekema DJ, Doern GV. 2011. Activity of ceftaroline and epidemiologic trends in Staphylococcus aureus isolates collected from 43 medical centers in the United States in 2009. Antimicrob Agents Chemother 55:4154–4160. doi: 10.1128/AAC.00315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrel M, Perencevich EN, David MZ. 2015. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000-2013. Emerg Infect Dis 21:1973–1980. doi: 10.3201/eid2111.150452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts JC. 2013. Community-associated methicillin-resistant Staphylococcus aureus epidemic clone USA100; more than a nosocomial pathogen. Springerplus 2:133. doi: 10.1186/2193-1801-2-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Cheung GY, Hu J, Wang D, Joo HS, Deleo FR, Otto M. 2010. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis 202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol 175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 14.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem Rev 111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavanaugh JS, Horswill AR. 2016. Impact of environmental cues on Staphylococcal quorum sensing and biofilm development. J Biol Chem 291:12556–12564. doi: 10.1074/jbc.R116.722710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 17.Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, Etienne J, Lina G. 2002. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol 184:1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pader V, Hakim S, Painter KL, Wigneshweraraj S, Clarke TB, Edwards AM. 2016. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat Microbiol 2:16194. doi: 10.1038/nmicrobiol.2016.194. [DOI] [PubMed] [Google Scholar]
- 19.Andrews JI, Fleener DK, Messer SA, Kroeger JS, Diekema DJ. 2009. Screening for Staphylococcus aureus carriage in pregnancy: usefulness of novel sampling and culture strategies. Am J Obstet Gynecol 201:396.e1–396.e5. doi: 10.1016/j.ajog.2009.06.062. [DOI] [PubMed] [Google Scholar]
- 20.Van De Griend P, Herwaldt LA, Alvis B, DeMartino M, Heilmann K, Doern G, Winokur P, Vonstein DD, Diekema D. 2009. Community-associated methicillin-resistant Staphylococcus aureus, Iowa, USA. Emerg Infect Dis 15:1582–1589. doi: 10.3201/eid1510.080877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diekema DJ, Richter SS, Heilmann KP, Dohrn CL, Riahi F, Tendolkar S, McDanel JS, Doern GV. 2014. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol 35:285–292. doi: 10.1086/675283. [DOI] [PubMed] [Google Scholar]
- 22.Chong YP, Kim ES, Park SJ, Park KH, Kim T, Kim MN, Kim SH, Lee SO, Choi SH, Woo JH, Jeong JY, Kim YS. 2013. Accessory gene regulator (agr) dysfunction in Staphylococcus aureus bloodstream isolates from South Korean patients. Antimicrob Agents Chemother 57:1509–1512. doi: 10.1128/AAC.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuji BT, MacLean RD, Dresser LD, McGavin MJ, Simor AE. 2011. Impact of accessory gene regulator (agr) dysfunction on vancomycin pharmacodynamics among Canadian community and health-care associated methicillin-resistant Staphylococcus aureus. Ann Clin Microbiol Antimicrob 10:20. doi: 10.1186/1476-0711-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. 2010. agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun 2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. 2009. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods 77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchdoerfer RN, Garner AL, Flack CE, Mee JM, Horswill AR, Janda KD, Kaufmann GF, Wilson IA. 2011. Structural basis for ligand recognition and discrimination of a quorum-quenching antibody. J Biol Chem 286:17351–17358. doi: 10.1074/jbc.M111.231258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis 198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 29.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnside K, Lembo A, de Los Reyes M, Iliuk A, Binhtran NT, Connelly JE, Lin WJ, Schmidt BZ, Richardson AR, Fang FC, Tao WA, Rajagopal L. 2010. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One 5:e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salgado-Pabon W, Herrera A, Vu BG, Stach CS, Merriman JA, Spaulding AR, Schlievert PM. 2014. Staphylococcus aureus beta-toxin production is common in strains with the beta-toxin gene inactivated by bacteriophage. J Infect Dis 210:784–792. doi: 10.1093/infdis/jiu146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deghorain M, Van Melderen L. 2012. The staphylococci phages family: an overview. Viruses 4:3316–3335. doi: 10.3390/v4123316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, Stevens SM, Horswill AR, Shaw LN. 2013. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, Smeltzer MS. 2012. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol 86:1183–1196. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhs A, Lyles JT, Parlet CP, Nelson K, Kavanaugh JS, Horswill AR, Quave CL. 2017. Virulence inhibitors from Brazilian peppertree block quorum sensing and abate dermonecrosis in skin infection models. Sci Rep 7:42275. doi: 10.1038/srep42275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todd DA, Parlet CP, Crosby HA, Malone CL, Heilmann KP, Horswill AR, Cech NB. 2017. Signal biosynthesis inhibition with ambuic acid as a strategy to target antibiotic-resistant infections. Antimicrob Agents Chemother 61:e00263-17. doi: 10.1128/AAC.00263-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paharik AE, Parlet CP, Chung N, Todd DA, Rodriguez EI, Van Dyke MJ, Cech NB, Horswill AR. 2017. Coagulase-negative staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22:746–756. doi: 10.1016/j.chom.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quave CL, Lyles JT, Kavanaugh JS, Nelson K, Parlet CP, Crosby HA, Heilmann KP, Horswill AR. 2015. Castanea sativa (European chestnut) leaf extracts rich in ursene and oleanene derivatives block Staphylococcus aureus virulence and pathogenesis without detectable resistance. PLoS One 10:e0136486. doi: 10.1371/journal.pone.0136486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parlet CP, Kavanaugh JS, Crosby HA, Raja HA, El-Elimat T, Todd DA, Pearce CJ, Cech NB, Oberlies NH, Horswill AR. 2019. Apicidin attenuates MRSA virulence through quorum-sensing inhibition and enhanced host defense. Cell Rep 27:187–198.e6. doi: 10.1016/j.celrep.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar K, Chen J, Drlica K, Shopsin B. 2017. Tuning of the lethal response to multiple stressors with a single-site mutation during clinical infection by Staphylococcus aureus. mBio 8:e01476-17. doi: 10.1128/mBio.01476-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlsson A, Saravia-Otten P, Tegmark K, Morfeldt E, Arvidson S. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect Immun 69:4742–4748. doi: 10.1128/IAI.69.8.4742-4748.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mootz JM, Malone CL, Shaw LN, Horswill AR. 2013. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect Immun 81:3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakoulas G, Eliopoulos GM, Moellering JRC, Novick RP, Venkataraman L, Wennersten C, DeGirolami PC, Schwaber MJ, Gold HS. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J Infect Dis 187:929–938. doi: 10.1086/368128. [DOI] [PubMed] [Google Scholar]
- 46.Spellberg B, Bartlett JG, Gilbert DN. 2013. The future of antibiotics and resistance. N Engl J Med 368:299–302. doi: 10.1056/NEJMp1215093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olson ME. 2016. Bacteriophage transduction in Staphylococcus aureus, p 69–74. In Bose JL. (ed), The genetic manipulation of staphylococci: methods and protocols. Springer New York, New York, NY. doi: 10.1007/7651_2014_186. [DOI] [Google Scholar]
- 49.Subrt N, Mesak LR, Davies J. 2011. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J Antimicrob Chemother 66:979–984. doi: 10.1093/jac/dkr043. [DOI] [PubMed] [Google Scholar]
- 50.Lofblom J, Kronqvist N, Uhlen M, Stahl S, Wernerus H. 2007. Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J Appl Microbiol 102:736–747. doi: 10.1111/j.1365-2672.2006.03127.x. [DOI] [PubMed] [Google Scholar]
- 51.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 53.Figueroa M, Jarmusch AK, Raja HA, El-Elimat T, Kavanaugh JS, Horswill AR, Cooks RG, Cech NB, Oberlies NH. 2014. Polyhydroxyanthraquinones as quorum sensing inhibitors from the guttates of Penicillium restrictum and their analysis by desorption electrospray ionization mass spectrometry. J Nat Prod 77:1351–1358. doi: 10.1021/np5000704. [DOI] [PMC free article] [PubMed] [Google Scholar]