Abstract
Cadherins, including E-cadherin, N-cadherin, VE-cadherin, etc., are important adhesion molecules mediating intercellular junctions. The abnormal expression of cadherins is often associated with tumor development and progression. Epithelial–mesenchymal transition (EMT) is the most important step in the metastasis cascade and is accompanied by altered expression of cadherins. Recent studies reveal that as a cargo for intercellular communication, exosomes—one type of extracellular vesicles that can be secreted by tumor cells—are involved in a variety of physiological and pathological processes, especially in tumor metastasis. Tumor-derived exosomes play a crucial role in mediating the cadherin instability in recipient cells by transferring bioactive molecules (oncogenic microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), EMT-related proteins, and others), modulating their local and distant microenvironment, and facilitating cancer metastasis. In turn, aberrant expression of cadherins in carcinoma cells can also affect the biogenesis and release of exosomes. Therefore, we summarize the current research on the crosstalk between tumor-derived exosomes and aberrant cadherin signals to reveal the unique role of exosomes in cancer progression.
Keywords: cadherins, exosomes, epithelial–mesenchymal transition, microRNAs, long non-coding RNAs, tumor development
1. Introduction of Exosomes
Extracellular vesicles (EVs) are nanoscale vesicles secreted by cells, including apoptotic bodies, exosomes, and microvesicles. Exosomes are small EVs (sEVs) with a cup-shaped morphology and diameters between 40 and 150 nm [1]. The differences between exosomes and other EVs have been comprehensively revealed. Recently, researchers used high-resolution density gradient centrifugation and immune affinity capture to isolate highly purified exosomes, which showed that the DNA is absent in sEVs, and exosomes do not contain glycolytic enzymes, nor do they contain cytoskeletal proteins [2].
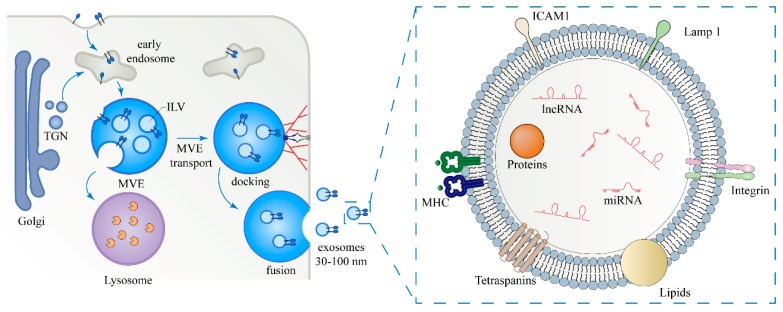
The biogenesis pathway of exosomes has been widely studied. It is well accepted that exosomes are derived from the early endosome formed by budding inwards through the plasma membrane. Then, with help from the endosomal sorting complex required for transport (ESCRT), the transition of exosomes from early endosomes to late endosomes, known as multivesicular endosomes (MVEs), is completed. ESCRT consists of ~30 proteins that form four complexes including ESCRT-0, I, II, and III, and is critical for the release of exosomes. MVEs have two destinies: one is to enter lysosome and be degraded; the other is to fuse with the plasma membrane and release intraluminal vesicles (ILVs), i.e., exosomes, into extracellular environment [3,4] (Figure 1). Although it is known that MVEs have different destinies, the molecular mechanisms by which MVEs are sorted remain unclear.
Figure 1.
Biogenesis, secretion, and cargos of exosomes. Exosomes are formed as intraluminal vesicles (ILVs) by endocytosis at the plasma membrane (PM), budding into early endosomes and multivesicular body (MVB). Cargos, which are targeted to MVEs, originate from endocytosis at the PM or from the early endosomes via the trans-Golgi network (TGN). The fate of MVBs can be either fusion with lysosomes or docking and fusion with the plasma membrane, which allows the release of exosomes to the extracellular milieu. Exosome cargo contains non-coding RNAs (including microRNAs and lncRNAs), proteins, and lipids. TGN: trans-Golgi network; ICAM-1: intercellular cell adhesion molecule-1; and MHC: major histocompatibility complex.
Exosomes are found in cell culture supernatants and in different biological fluids, including blood, urine, breast milk, amniotic fluid, malignant ascites, etc. [5,6,7]. Exosomes are widely distributed, but how to get high quality exosomes has become another thorny problem. Exosome extraction methods include ultracentrifugation, density gradient centrifugation, immunocapture, etc., and each method has its pros and cons [8]. Ultracentrifugation has been most widely used for exosome extraction by virtue of its simplicity and rapidity, but purity of exosomes obtained by this method is not ideal enough. With the deepening cognition of exosomes, it is considered that exosomes with higher purity can better reveal their functions. Compared to ultracentrifugation, density gradient centrifugation has the advantage of obtaining exosomes with higher purity, but its disadvantage is that it takes more time and energy, and few exosomes are isolated [9]. Using magnetic beads which conjugated with specific antibodies to capture exosomes is another better strategy, but this method is expensive and produces few exosomes [10]. After isolation, purified exosomes need to be further identified. Currently, exosomes are identified by three complementary methods: exosomes morphology is observed by electron microscope, the particle size is detected by nanoparticle tracer analysis (NTA), and exosomal markers including CD81, CD63, and CD9, etc., are detected by Western blot [2,11].
Like cells, exosomes have the structure of phospholipid bilayer, which can better protect the enveloped materials. Exosomes are encapsulated by proteins and lipids, and contain functional proteins and genetic material, such as mRNAs, microRNAs (miRNAs), and long non-coding RNAs (lncRNAs). An exosome’s source determines the difference in exosomal composition, which reflects the physiological status and functionality of the host cells [12,13,14] (Figure 1). Initially, exosomes were thought to be “trash cans” that transported cellular waste into the extracellular environment and were then metabolized with the circulatory system. Later, it was found that exosomes have a greater role in transferring proteins, lipids, and nucleic acids from host cells to recipient cells [15]. Exosomes are involved in many biological functions including antigen-presentation, cell-cell communication, immunomodulation, microenvironment remodeling, and tumor metastasis [16,17,18]. Usually, patients with different types of cancer release exosomes which contain different signatures of miRNAs into the circulation, which can be measured as diagnostic biomarkers [19].
Current research has shown that abnormalities in cadherin signals are strongly associated with exosomes, especially those derived from tumors [20,21]. Therefore, we summarize recent studies on the instability of cadherins and tumor progression mediated by tumor-derived exosomes.
2. Key Role of Cadherins in EMT
Cadherins, a type of cell adhesion molecule, are a class of type-1 transmembrane proteins, and their functions are highly dependent on calcium (Ca2+) ions in a homophilic manner. The cadherin superfamily shares the extracellular cadherin repeats, a single transmembrane domain, and a cytoplasmic domain [22]. Cadherins play important roles in the formation of adhesion junctions and maintenance of the cell–cell adhesion. Typically, epithelial cells express E-cadherin, whereas mesenchymal cells express various cadherins, including N-cadherin, R-cadherin, and cadherin-11 [23]. Specifically, endothelial cells express VE-cadherin [24].
Cadherins often undergo significant changes in the epithelial–mesenchymal transition (EMT) process. Epithelial cells undergo morphological transformation to mesenchymal phenotype and acquire stronger migration ability accompanied by cadherin dysregulation, which is called “cadherin switch” [25]. “Cadherin switch” from E-cadherin to N-cadherin is a research hotspot because it is extremely important for understanding the mechanism of tumor metastasis [26]. Loss of E-cadherin expression is the hallmark of the EMT process. E-cadherin is widely expressed in most epithelial tissues. The mature E-cadherin contains a cytoplasmic domain, transmembrane domain, and ectodomain. The cytoplasmic domain of the E-cadherin is complexed with either β-catenin or γ-catenin (plakoglobin). β-catenin and γ-catenin bind directly to α-catenin, and form the distinct cadherin–catenin complexes [27]. Plakoglobin not only binds to E-cadherin to form adherens junctions, but also interacts with desmosomal cadherins, such as desmoglein, to form desmosomal junctions which are not associated with α-catenin [28]. This adherens junction complex is linked to the actin cytoskeleton, further mediating the mechanical stability [29]. p120 catenin is a member of the catenin family and can bind to the juxta membrane domain (JBD) of E-cadherin, but not to α-catenin at the other end [30]. The expression of E-cadherin is regulated by various transcription factors, such as Slug, Snail, etc., which bind to the E-box region of the E-cadherin promoter region to inhibit the expression of E-cadherin. Recent findings have reported that lysyl oxidase-like 2 (LOXL2) could act as a modulator for Snail, and synergize with another transcription factor, bHLH transcription factor E47, to reduce E-cadherin levels [31]. The loss of E-cadherin’s homophilic binding further results in the loss of cell–cell adhesion and cell polarity [32,33]. Meanwhile, the role of E-cadherin in intracellular and intercellular signal transduction has also been widely explored [34,35,36].
Gain of mesenchymal N-cadherin expression is observed in cells undergoing EMT, and serves as another important feature of EMT. Although high expression of N-cadherin is not considered to be a promoter of tumor growth, it does correlate with tumor progression [37,38]. Other types of cadherins, such as cadherin-6 and P-cadherin, have also attracted attention in recent years. Cadherin-6 was abnormally expressed in thyroid cancer. Further studies have demonstrated that the activation of cadherin-6 could promote autophagy and enhance EMT characteristics, demonstrating a new function of cadherins [39]. In nasopharyngeal carcinoma, Epstein-Barr viral (EBV) oncoprotein LMP1 inhibited the transcription of miR-203, which specifically targeted cadherin-6 by activating NF-κB signaling. Meanwhile, cadherin-6 was a hub that linked the NF-κB and TGF-β signaling pathways to promote the development of EMT and cancer metastasis [40].
It is well known that multiple signaling pathways are involved in EMT and affect the expression of cadherins such as Wnt, TGF-β, Notch, etc. [41]. β-catenin is a messenger in EMT, connecting signals in and out of the nucleus. In the classical Wnt signaling pathway, cytoplasmic β-catenin evades degradation, accumulates in the cytoplasm, and finally translocates to the nucleus. Therefore, β-catenin is involved in the induction of EMT in physiological processes and tumorigenesis progression. Under normal conditions, E-cadherin can recruit β-catenin to adherens junctions, retard the β-catenin into the nucleus, and inhibit β-catenin activation of target gene expression. In contrast, loss of E-cadherin destructs the cadherin–catenin complexes and allows β-catenin to localize to the nucleus and exert its transcriptional activity. It is evident that both β-catenin and p120 are required for transcriptional activation of target genes—not just one in some cases [42]. N-cadherin and Wnt signaling pathways also compete for cellular β-catenin. Although it is generally believed that the increased expression of N-cadherin is positively correlated with tumorigenesis, it has also been reported that overexpression of N-cadherin transfers β-catenin from the nucleus to the cytoplasm and locates β-catenin to the plasma membrane, which may inhibit Wnt signaling and tumor development. Therefore, the role of N-cadherin in tumorigenesis needs to be further revealed [43,44], and the role of N-cadherin in tumorigenesis needs to be further revealed.
TGF-β-inducing EMT is divided into the Smad-dependent and non Smad-dependent signaling pathways. In the Smad-dependent pathway, TGF-β binding to TGF-β type I and II receptors on the cell membrane phosphorylates Smad2/3 of the Smad protein family, and then the phosphorylated Samd2/3 forms a complex with Samd4 leading to nuclear translocation from the cytoplasm. Subsequently, the Smad complex transported into the nucleus mediates the expression of relevant target transcriptional factors, which could inhibit the expression of E-cadherin [45]. MicroRNAs induced by the Smad complex suppress E-cadherin and promote the expression of characteristic proteins in tumor cells to accelerate EMT [46]. However, some miRNAs perform the opposite function. The miR-200 family is the well-known miRNA molecule that regulates tumor EMT. Studies show that expression of the miR-200 family gene was down-regulated in the breast cancer EMT model induced by TGF-β, while overexpression of the miR-200 family could reverse EMT [47].
3. Exosomal miRNAs and Cadherins
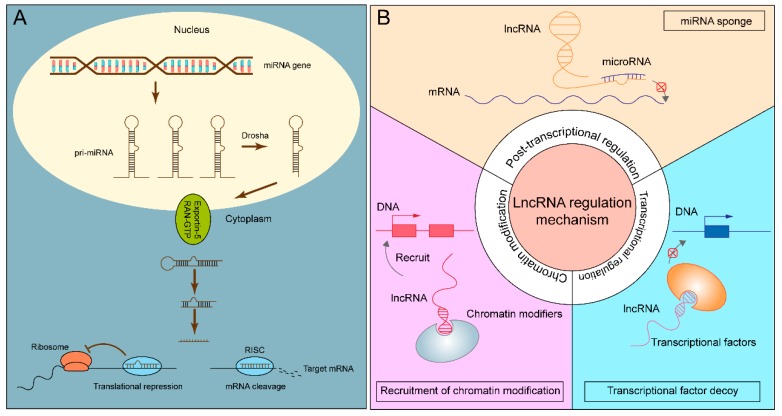
MicroRNAs are small RNAs with a length of 18–25 nucleotides. In the canonical miRNA biogenesis pathway, genes responsible for coding are transcribed into pri-miRNA, which are cut into pre-miRNA with a stem-loop structure using Drosha-RNase [48]. Subsequently, pri-miRNA is transported from the nucleus to the cytoplasm with the help of Ran-GTP and Exportin-5. Dicer, a member of the RNase III family, specifically recognizes double-stranded RNA [49]. In Dicer-dependent miRNA processing, Dicer cuts pre-miRNA into 18–25 nucleotide double-stranded miRNA. After unwinding, one of the strands of double-stranded miRNA binds to Dicer and Argonaute to form RNA-induced silencing complex (RISC) that inhibits target gene transcription [50]. However, Studies have shown that miRNAs can be produced without Drosha or Dicer, which are known as the non-classical miRNA biogenesis pathway [51,52]. Exosomes-mediated secretion of human Ago proteins, other miRNA biogenesis machinery, and RISC components remains an unsettled issue. In 2014, pre-miRNAs, Dicer, AGO2, and TRBP were found in cancer exosomes, and pre-miRNA could even be processed into mature miRNA [53]. In 2016, McKenzie et al. showed that Ago2 existed in exosomes [54]. However, the latest reference in 2019 by Jeppesen et al. demonstrated that extracellular Ago proteins resulted from the contamination of crude sEVs samples with P-body, stress granules, or GW bodies, while other miRNA-associated proteins were absent from crude sEVs. In human plasma, Ago2 was present in the gradient-purified non-vesicle (NV) pool, but absent from the sEVs pool [2]. Typically, miRNAs target the 3’UTR (untranslated region) of mRNAs, but a few can also target 5′UTR and CDS (coding sequence) [55]. There are two main mechanisms of miRNAs that cause gene silencing. miRNAs associate with their mRNA targets by base-pair complementarity. The complete complementary pairing of miRNAs and mRNAs triggers mRNA degradation, while the incomplete complementary pairing of miRNAs and mRNAs inhibits protein synthesis or causes mRNA degradation (Figure 2A). Recent studies demonstrate that miRNAs play an essential role in cell cycle, circadian rhythms, tumor development and progression, etc. [56,57,58]. Moreover, circulating miRNAs derived from exosomes, a potential biomarker for cancer diagnosis and prognosis, are widely investigated in various cancers [59,60,61].
Figure 2.
The role of miRNAs and lncRNAs. (A) The biogenesis and functional mechanism of miRNAs. (B) The regulatory aspects of lncRNAs.
Exosomes carrying miRNAs have a significant impact on the expression of cadherins (Table 1). Usually, tumor cells express the increased oncogenic miRNAs, and the exosomes secreted by tumor cells could cargo these oncogenic miRNAs to recipient cells and modulate cadherin levels. For example, triple-negative breast cancer derived exosomal miR-9 could be transferred to normal fibroblasts (NFs). Exosomal miR-9 affects the status of NFs and promotes the cell malignant transformation from NFs to cancer-associated fibroblasts (CAFs), which further modify the tumor microenvironment and mediate intercellular communication. In addition, the co-culture of breast cancer cells with NFs transiently transfected with miR-9 demonstrated that miR-9 enhances tumor cells motility by reducing E-cadherin [62]. In order to enter the circulatory system and form metastatic sites in the distal tissue/organs, tumor cells will break through the endothelial barrier. Among the cadherins, vascular endothelial cadherin (VE-cadherin) is a vital adhesion molecule of endothelial cells and can be directly targeted by exosomal miR-103 and miR-939. The exosomal miR-103 secreted by hepatocellular carcinoma cells (HCCs) destroyed the adhesion junctions of endothelial cells by directly inhibiting the expression of VE-cadherin. The loss of adhesion between endothelial cells enhanced vascular permeability and was more conducive to cancer metastasis [6]. Exosomes secreted by breast cancer cells may carry miR-939 to reduce the expression of VE-cadherin and decrease cell-cell adhesion, which may explain the correlation between high miR-939 expression and low survival rates [63].
On the other hand, loss of certain tumor suppressor miRNAs in exosomes may induce the occurrence of EMT. DNA methyltransferases (DNMTs), a class of important molecules in epigenetics, are abnormally expressed in cancer [64]. The high level of DNA methylation modification caused by miRNA-deficiency regulates gene expression to promote tumor development. It was found that CAFs-derived exosomal miR-148b had a low abundance which partially promoted cancer metastasis, while the increase of miR-148b could inhibit DNMT1 translation in endometrial tumor cells accompanied by up-regulated E-cadherin and down-regulated N-cadherin [65]. It provided a new sight that the deletion of tumor suppressor miRNAs in exosomes induce EMT and mediate the instability of cadherin.
Notably, environmental stimulation can alter the composition of exosomes to facilitate tumor development. The tumor microenvironment is complex and diverse, with characteristics of hypoxia, low pH, and high pressure, which provide favorable conditions for tumor migration, invasion, and angiogenesis [66,67]. For example, the increased expression of miR-21 in exosomes secreted by hypoxic oral squamous cell carcinoma (OSCC) cells downregulated the expression level of E-cadherin [68]. Wang et al. demonstrated that miR-301a-3p was enriched in exosomes from hypoxic pancreatic cancer cells. Hypoxic exosomal miR-301a-3p induced macrophage polarization by activating the PTEN/PI3Kg signaling pathway. Polarized macrophages, in turn, promoted malignant transformation of pancreatic cancer cells, characterized by increased cell migration, invasion, and other EMT phenomenon [69].
A recent study showed that the expression of exosomal miRNA would be altered following the EMT process [70]. When treated with TGF-β1, human lung cancer cells A549 obtained a mesenchymal phenotype accompanied by a decrease in E-cadherin and the elevated expression of mesenchymal markers. At the same time, the miRNA profile of M-exosomes (derived from M-phenotype A549 cells) was obviously different compared with E-exosomes (derived from E-phenotype A549 cells). The results revealed that M-exosomal miRNAs were associated with malignant transformation and metastasis [70]. Similarly, an earlier study also demonstrated TGF-β1 could induce EMT in A549 cells resulting in the discrepant expression profile of exosomal proteins and miRNAs [71]. In addition to inducible factors, drug treatment also affects the expression of exosomal miRNA secreted by tumor cells. For example, exosomal expression levels of miR-200c and miR-141 were up-regulated by decitabine treatment in human colorectal cancer (CRC) cell lines [72].
4. Exosomal lncRNAs and Cadherins
lncRNAs are a class of RNA molecules with a length greater than 200 nt. Compared to miRNAs, lncRNAs are less conserved among species and more tissue-specific [73]. LncRNAs are transcribed from intergenic regions (large intergenic ncRNAs (lincRNAs)), intragenic regions, or specific chromosomal regions [74,75,76]. lncRNAs derived from intragenic regions can be classified in antisense, overlapping, intronic, and bidirectional orientations, which are related to protein-coding genes or gene regulatory regions. Furthermore, lncRNAs are processed after transcription, such as polyadenylation, alternative splicing, RNA editing, etc. [77]. The regulatory aspects of lncRNAs are divided into chromatin modification, transcriptional regulation, and post-transcriptional regulation. In other words, lncRNAs can interact with DNA, RNA, or protein to regulate gene expression [78,79,80] (Figure 2B). In terms of epigenetic regulation, lncRNA-Xist (X-inactive specific transcript) plays a typical role in X-chromosome inactivation. Xist is transcribed by the X chromosome and then wrapped around this chromosome. By recruiting DNA methylase and histone deacetylase, Xist leads to the suppression of gene expression on the overall level of X chromosome. Tsix, the antisense transcription product of Xist, effectively prevents the accumulation of Xist, thus maintaining the X-chromosome’s activity [81]. In addition, the transcription of lncRNAs may interfere with the transcription of neighboring protein-coding genes and degrading targeted mRNAs and acting as decoy for transcriptional regulators [82].
As the emerging regulators, lncRNAs have been shown to stimulate the tumor metastasis by mediating the expression of cadherins [83,84]. For example, high levels of lncRNA-ATB inhibited the function of miR-200 family, thereby enhancing the expression of transcriptional factor ZEB1 and promoting EMT in colon cancer [85]. In contrast, lncRNA-MEG3 can adsorb miR-421, which targets E-cadherin and further inhibits cell invasion [86]. More directly, TGF-β induced EMT and enhanced the level of lncRNA-HIT which accelerated tumor development by targeting E-cadherin in NMuMG cells [87]. In addition, multiple lncRNAs can interact with the histone-lysine N- methyltransferase enzyme EZH2 (enhancer of zeste homolog 2), which modifies histone methylation and inhibits E-cadherin transcription [88,89].
RNA sequencing analysis showed that certain lncRNAs including lncUEGC1 and LINC00477, were enriched in the tumor-derived exosomes and the expression profile of lncRNAs may be used to distinguish cancer patients from healthy controls [90,91]. Furthermore, lncRNAs were found to be implicated in EMT and have an impact on tumor development [92]. Xue et al. demonstrated that lncRNA-UCA1 expression was higher in hypoxic exosomes than normoxic exosomes, and hypoxic exosomal lncRNA-UCA1 promoted tumor development through EMT [93]. In addition, the exosomal lncRNA linc-ROR from thyroid cancer stem-like cells (CSCs) was necessary to induce EMT in normal thyroid cells [94]. Li et al. found that lncRNA-Sox2ot in exosomes was associated with pancreatic ductal adenocarcinoma progression. Sox2 (SRY-related HMG-box gene 2), a transcriptional factor, has been shown to promote various tumors metastasis and drug resistance [95,96,97]. The miR-200 family not only directly targets Sox2, but also multiple transcriptional factors, such as ZEB1/2, to regulate EMT markers [98]. As competitive endogenous RNA (ceRNA), lncRNA-Sox2ot up-regulated Sox2 and promoted EMT by competitively binding to the miR-200 family [99]. Unlike miRNA sponges, exosomal lnc-MMP2-2 can bind to the upstream site of MMP-2 gene, enhance the expression of MMP-2, and promote EMT [100]. In contrast, lncRNA NONHSAT105177 inhibited EMT by up-regulating E-cadherin, down-regulating various EMT inducible transcriptional factors and mesenchymal markers [101]. Although studies on exosomal lncRNAs-mediated the expression of cadherins are relatively few (Table 1), the above findings have laid a foundation for unraveling of the mystery of the association between exosomes and cadherin.
Table 1.
Summary of tumor-derived exosomal non-coding RNAs mediating the instability of cadherins.
| Exosomal non-Coding RNAs | Cancer Types | Targets | Promote (✓)/Inhibit (✕) Tumor Progression |
|---|---|---|---|
| miR-9 [62] | Breast cancer | E-cadherin | ✓ |
| miR-103 [6] | Hepatocellular carcinoma | VE-cadherin | ✓ |
| miR-939 [63] | Breast cancer | VE-cadherin | ✓ |
| miR-148b [65] | Endometrial cancer | DNMT1 | ✕ |
| miR-21 [68] | Oral squamous cell carcinoma | PTEN PDCD4 |
✓ |
| miR-200c [72] | Colorectal cancer | ZEB1 | ✕ |
| lncRNA-UCA1 [93] | Bladder cancer | ✓ | |
| linc-ROR [94] | Thyroid cancer | ✓ | |
| lncRNA-Sox2ot [99] | Pancreatic ductal adenocarcinoma | miR-200 family | ✓ |
| lnc-MMP2-2 [100] | Lung cancer | MMP-2 | ✓ |
| lncRNA NONHSAT105177 [101] | Pancreatic ductal adenocarcinoma | ✕ |
5. Exosomal Proteins and Cadherins
In addition to the aforementioned regulatory modalities of transferring RNAs, exosomes can also mediate the instability of cadherins by directly delivering certain proteins to recipient cells. Similar to nucleic acids in exosomes, proteomics analysis revealed that tumor-derived exosomes contain more oncogenic proteins, or perhaps less cancer suppressor proteins [102].
There is increasing evidence that many proteins are involved in the regulation of cadherins. It is known that the interaction of fibroblast growth factors (FGFs) with their receptors, fibroblast growth factor receptors (FGFRs), facilitates cancer progression [103]. Shi et al. found that FGF19 was highly expressed in exosomes derived from mesenchymal stem cells (MSCs). After treatment with MSC-exosomes, phosphorylated FGFR4 and ERK in nasopharyngeal carcinoma (NPC) cells increased, and the expression of EMT markers changed, presenting as down-regulation of E-cadherin, up-regulation of N-cadherin, and vimentin. It suggested that FGF19 carried by MSC-exosomes binds FGFR4 in NPC cells to activate the downstream ERK signaling pathway and thereby induce EMT [104]. Nakamura et al. demonstrated that exosomes from epithelial ovarian cancer (EOC) caused cells to more easily break through the mesothelial barrier. Further investigation revealed that high expression of CD44 in exosomes could lead to down-regulation of E-cadherin and up-regulation of MMP9, which degraded adhesive molecules in human peritoneal mesothelial cells [105]. Similarly, the epithelial growth factor receptor (EGFR)—a transmembrane glycoprotein in exosomes—was internalized by epithelial cells to induce EMT [106]. It is generally assumed that β-catenin will be degraded by ubiquitination and weaken Wnt signaling under normal conditions. Are exosomes necessary for the level of intracellular β-catenin? A novel study revealed that CD82 and CD9 contributed to the encapsulation of β-catenin in exosomes and its release into the extracellular environment, in which E-cadherin was essential for the secretion of β-catenin [107]. However, whether tumor cells suppress the release of exosomal β-catenin and thus enhance cellular motility remains to be explored.
Exosomes also contain matrix metalloproteinases (MMPs) which can degrade most protein components in the extracellular cell matrix (ECM), destroy the histological barrier of tumor cells’ invasion, and are involved in tumor metastasis [108,109]. Studies have shown that NPC-derived exosomes were able to activate AKT and ERK signaling pathways to enhance metastasis properties of tumor cells. High expression of MMP13 in exosomes secreted by NPCs cells was considered to play an important role in these processes. Exosomes released from MMP-13 silenced NPC cells reversed EMT compared with normal NPC cells, confirming the positive relationship between MMP-13 and EMT [110]. On the basis of this work, Shan et al. found that hypoxic conditions promoted the expression of MMP-13 in NPC cells, and more MMP13 was encapsulated in exosomes and delivered to recipient cells [111].
In some way, cytoskeleton proteins-rich exosomes initiated the metastasis cascade by inducing the translocation of adhesion molecules [112]. Cytoskeleton-associated proteins-enriched exosomes from metastatic colon cancer cells affected the integrity of endothelial cells by inducing a clear cytosolic delocalization of the adherens junction proteins including β-catenin, p120-catenin, and VE-cadherin, without changes of VE-cadherin expression at both mRNA and protein levels, through RhoA/Rock signaling in recipient cells. p53 deficient mouse bone marrow MSC (mBMMSC) exosomes enriched ubiquitin protein ligase E3 component N-recognin 2 (UBR2) resulted in the increased proliferation and migration of p53+/+ mBMMSC and murine foregastric carcinoma (MFC) by activating Wnt/β-catenin signaling with changed EMT marker expression including E-/N-cadherin [113]. In a separate study, the authors confirmed that many of the genes with human papilloma virus-16 (HPV-16) associated with oropharyngeal cancer (HPVOPC) had missense mutations and increased copy counts. After treatment with HPVOPC-derived exosomes, the EMT phenomenon, especially greater motility was observed in epithelial cells [114].
Notably, abnormal expression of soluble E-cadherin (sE-cadherin) in malignant ascites has been closely explored. sE-cadherin in exosomes derived from ovarian cancer cells was found to bind VE-cadherin to activate the β-catenin and NF-κB signaling pathways, and subsequently enhance angiogenesis in recipient cells [115]. It may be a novel regulatory mechanism for tumor angiogenesis.
6. Conclusions
In this review, we describe in detail how tumor-derived exosomes mediate the instability of cadherins and facilitate tumor progression. EMT occurs at the initial stage of tumor metastasis and is characterized by down-regulation of the epithelial marker E-cadherin and up-regulation of the mesenchymal marker N-cadherin. Abnormal expression of cadherins further results in the metastasis of tumor cells to distant organs or tissues. Conversely, delocalization of cadherins can also affect tumor cells to break through the endothelial barrier.
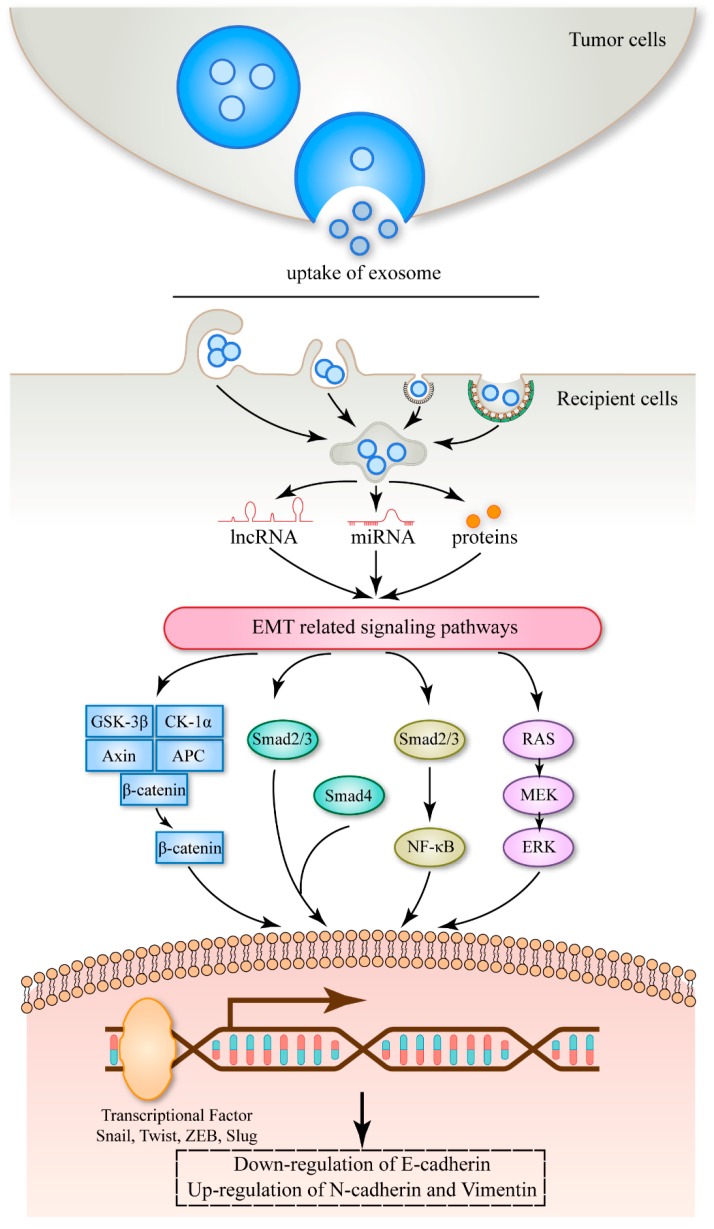
Exosomes, a cargo of intercellular communication, are encapsulated with many bioactive substances. Usually, tumor-derived exosomes may carry more carcinogens or less tumor suppressors. Tumor-derived exosomes tend to transfer EMT-related RNAs and proteins to recipient cells, mediate the instability of cadherins, and promote cancer progression (Figure 3).
Figure 3.
A schematic model of tumor-derived exosomes dysregulating cadherin expression. Tumor-derived exosomes deliver miRNA, lncRNA, and proteins to recipient cells, stimulate EMT-related signaling pathways, down-regulate the expression of E-cadherin, and up-regulate the expression of N-cadherin and vimentin, and further promote tumor progression.
Acknowledgments
The authors are grateful to Dosher James Logan for performing English editing of the manuscript.
Funding
This research was funded by the National Science Foundation of China (No. 81672537, 81802654), National Science and Technology Major Project of China (No. 2018ZX10302205), the Natural Science Foundation of Shaanxi Province, China (2019JZ-22), and the Youth Innovation Team of Shaanxi Universities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. Reassessment of exosome composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thery C., Zitvogel L., Amigorena S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 4.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Im H., Shao H., Park Y.I., Peterson V.M., Castro C.M., Weissleder R., Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang J.H., Zhang Z.J., Shang L.R., Luo Y.W., Lin Y.F., Yuan Y., Zhuang S.M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459–1475. doi: 10.1002/hep.29920. [DOI] [PubMed] [Google Scholar]
- 7.Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;30:3–22. doi: 10.1002/0471143030.cb0322s30. Chapter 3. [DOI] [PubMed] [Google Scholar]
- 8.Tauro B.J., Greening D.W., Mathias R.A., Ji H., Mathivanan S., Scott A.M., Simpson R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line lim1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Bobrie A., Colombo M., Krumeich S., Raposo G., Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greening D.W., Xu R., Ji H., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 11.Baranyai T., Herczeg K., Onodi Z., Voszka I., Modos K., Marton N., Nagy G., Mager I., Wood M.J., El Andaloussi S., et al. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE. 2015;10:e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathivanan S., Fahner C.J., Reid G.E., Simpson R.J. Exocarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tkac J., Bertok T., Hires M., Jane E., Lorencova L., Kasak P. Glycomics of prostate cancer: Updates. Expert Rev. Proteomics. 2018;16:1–12. doi: 10.1080/14789450.2019.1549993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skotland T., Sandvig K., Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Wan Z., Gao X., Dong Y., Zhao Y., Chen X., Yang G., Liu L. Exosome-mediated cell-cell communication in tumor progression. Am. J. Cancer Res. 2018;8:1661–1673. [PMC free article] [PubMed] [Google Scholar]
- 16.Arslan F., Lai R.C., Smeets M.B., Akeroyd L., Choo A., Aguor E.N., Timmers L., van Rijen H.V., Doevendans P.A., Pasterkamp G., et al. Mesenchymal stem cell-derived exosomes increase atp levels, decrease oxidative stress and activate pi3k/akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013;10:301–312. doi: 10.1016/j.scr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto K., Ochi H., Sunamura S., Kosaka N., Mabuchi Y., Fukuda T., Yao K., Kanda H., Ae K., Okawa A., et al. Cancer-secreted hsa-mir-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting arhgap1 and fam134a. Proc. Natl. Acad. Sci. USA. 2018;115:2204–2209. doi: 10.1073/pnas.1717363115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y., Xue C., Li X., Ba L., Gu J., Sun Z., Han Q., Zhao R.C. Effects of gastric cancer cell-derived exosomes on the immune regulation of mesenchymal stem cells by the nf-κb signaling pathway. Stem Cells Dev. 2019;28:464–476. doi: 10.1089/scd.2018.0125. [DOI] [PubMed] [Google Scholar]
- 19.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 20.Steinbichler T.B., Dudas J., Riechelmann H., Skvortsova I.I. The role of exosomes in cancer metastasis. Semin. Cancer Biol. 2017;44:170–181. doi: 10.1016/j.semcancer.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Dioufa N., Clark A.M., Ma B., Beckwitt C.H., Wells A. Bi-directional exosome-driven intercommunication between the hepatic niche and cancer cells. Mol. Cancer. 2017;16:172. doi: 10.1186/s12943-017-0740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angst B.D., Marcozzi C., Magee A.I. The cadherin superfamily: Diversity in form and function. J. Cell Sci. 2001;114:629–641. doi: 10.1242/jcs.114.4.629. [DOI] [PubMed] [Google Scholar]
- 23.Wheelock M.J., Shintani Y., Maeda M., Fukumoto Y., Johnson K.R. Cadherin switching. J. Cell Sci. 2008;121:727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 24.Lagendijk A.K., Hogan B.M. Ve-cadherin in vascular development: A coordinator of cell signaling and tissue morphogenesis. Curr. Top. Dev. Biol. 2015;112:325–352. doi: 10.1016/bs.ctdb.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Scarpa E., Szabo A., Bibonne A., Theveneau E., Parsons M., Mayor R. Cadherin switch during emt in seural crest cells leads to contact inhibition of locomotion via repolarization of forces. Dev. Cell. 2015;34:421–434. doi: 10.1016/j.devcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gravdal K., Halvorsen O.J., Haukaas S.A., Akslen L.A. A switch from e-cadherin to n-cadherin expression indicates epithelial to mesenchymal transition and is of strong and independent importance for the progress of prostate cancer. Clin. Cancer Res. 2007;13:7003–7011. doi: 10.1158/1078-0432.CCR-07-1263. [DOI] [PubMed] [Google Scholar]
- 27.Schmalhofer O., Brabletz S., Brabletz T. E-cadherin, beta-catenin, and zeb1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 28.Aktary Z., Pasdar M. Plakoglobin: Role in tumorigenesis and metastasis. Int. J. Cell Biol. 2012;2012:189521. doi: 10.1155/2012/189521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankar J., Nabi I.R. Actin cytoskeleton regulation of epithelial mesenchymal transition in metastatic cancer cells. PLoS ONE. 2015;10:e0119954. doi: 10.1371/journal.pone.0119954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kourtidis A., Ngok S.P., Anastasiadis P.Z. P120 catenin: An essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog. Mol. Biol. Transl. Sci. 2013;116:409–432. doi: 10.1016/B978-0-12-394311-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canesin G., Cuevas E.P., Santos V., Lopez-Menendez C., Moreno-Bueno G., Huang Y., Csiszar K., Portillo F., Peinado H., Lyden D., et al. Lysyl oxidase-like 2 (loxl2) and e47 emt factor: Novel partners in e-cadherin repression and early metastasis colonization. Oncogene. 2015;34:951–964. doi: 10.1038/onc.2014.23. [DOI] [PubMed] [Google Scholar]
- 32.Petrova Y.I., Schecterson L., Gumbiner B.M. Roles for e-cadherin cell surface regulation in cancer. Mol. Biol. Cell. 2016;27:3233–3244. doi: 10.1091/mbc.E16-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diepenbruck M., Christofori G. Epithelial-mesenchymal transition (emt) and metastasis: Yes, no, maybe? Curr. Opin. Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Chen H.N., Yuan K., Xie N., Wang K., Huang Z., Chen Y., Dou Q., Wu M., Nice E.C., Zhou Z.G. Pdlim1 stabilizes the e-cadherin/β-catenin complex to prevent epithelial–mesenchymal transition and metastatic potential of colorectal cancer cells. Cancer Res. 2016;76:1122. doi: 10.1158/0008-5472.CAN-15-1962. [DOI] [PubMed] [Google Scholar]
- 35.Brouxhon S.M., Kyrkanides S., Teng X., Athar M., Ghazizadeh S., Simon M., O’Banion M.K., Ma L. Soluble e-cadherin: A critical oncogene modulating receptor tyrosine kinases, mapk and pi3k/akt/mtor signaling. Oncogene. 2013;33:225–235. doi: 10.1038/onc.2012.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Q.P., Kuang J.Y., Yang Q.K., Bian X.W., Yu S.C. Beyond a tumor suppressor: Soluble e-cadherin promotes the progression of cancer. Int. J. Cancer. 2016;138:2804–2812. doi: 10.1002/ijc.29982. [DOI] [PubMed] [Google Scholar]
- 37.Su Y., Li J., Shi C., Hruban R.H., Radice G.L. N-cadherin functions as a growth suppressor in a model of k-ras-induced panin. Oncogene. 2016;35:3335–3341. doi: 10.1038/onc.2015.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hulit J., Suyama K., Chung S., Keren R., Agiostratidou G., Shan W., Dong X., Williams T.M., Lisanti M.P., Knudsen K., et al. N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 2007;67:3106–3116. doi: 10.1158/0008-5472.CAN-06-3401. [DOI] [PubMed] [Google Scholar]
- 39.Gugnoni M., Sancisi V., Gandolfi G., Manzotti G., Ragazzi M., Giordano D., Tamagnini I., Tigano M., Frasoldati A., Piana S., et al. Cadherin-6 promotes emt and cancer metastasis by restraining autophagy. Oncogene. 2017;36:667–677. doi: 10.1038/onc.2016.237. [DOI] [PubMed] [Google Scholar]
- 40.Zuo L.L., Zhang J., Liu L.Z., Zhou Q., Du S.J., Xin S.Y., Ning Z.P., Yang J., Yu H.B., Yue W.X., et al. Cadherin 6 is activated by epstein-barr virus lmp1 to mediate emt and metastasis as an interplay node of multiple pathways in nasopharyngeal carcinoma. Oncogenesis. 2017;6:402. doi: 10.1038/s41389-017-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Zoni E., van der Pluijm G., Gray P.C., Kruithof-de Julio M. Epithelial plasticity in cancer: Unmasking a microRNA network for tgf-beta-, notch-, and wnt-mediated emt. J. Oncol. 2015;2015:198967. doi: 10.1155/2015/198967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCrea P.D., Maher M.T., Gottardi C.J. Nuclear signaling from cadherin adhesion complexes. Curr. Top. Dev. Biol. 2015;112:129–196. doi: 10.1016/bs.ctdb.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M., Ren D., Guo W., Huang S., Wang Z., Li Q., Du H., Song L., Peng X. N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via erbb signaling in prostate cancer cells. Int. J. Oncol. 2016;48:595–606. doi: 10.3892/ijo.2015.3270. [DOI] [PubMed] [Google Scholar]
- 44.Sadot E., Simcha I., Shtutman M., Ben-Ze’ev A., Geiger B. Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proc. Natl. Acad. Sci. USA. 1998;95:15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurioka K., Wato M., Iseki T., Tanaka A., Morita S. Differential expression of the epithelial mesenchymal transition factors snail, slug, twist, tgf-beta, and e-cadherin in ameloblastoma. Med. Mol. Morphol. 2017;50:68–75. doi: 10.1007/s00795-016-0149-0. [DOI] [PubMed] [Google Scholar]
- 46.Chen W., Zhou S., Mao L., Zhang H., Sun D., Zhang J., Li J., Tang J.H. Crosstalk between tgf-beta signaling and miRNAs in breast cancer metastasis. Tumor Biol. 2016;37:10011–10019. doi: 10.1007/s13277-016-5060-8. [DOI] [PubMed] [Google Scholar]
- 47.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The mir-200 family and mir-205 regulate epithelial to mesenchymal transition by targeting zeb1 and sip1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 48.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNAse iii drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 49.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 50.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamura K., Hagen J.W., Duan H., Tyler D.M., Lai E.C. The mirtron pathway generates microRNA-class regulatory RNAs in drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKenzie A.J., Hoshino D., Hong N.H., Cha D.J., Franklin J.L., Coffey R.J., Patton J.G., Weaver A.M. Kras-mek signaling controls ago2 sorting into exosomes. Cell Rep. 2016;15:978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou X., Duan X., Qian J., Li F. Abundant conserved microRNA target sites in the 5’-untranslated region and coding sequence. Genetica. 2009;137:159–164. doi: 10.1007/s10709-009-9378-7. [DOI] [PubMed] [Google Scholar]
- 56.Tajima K., Yae T., Javaid S., Tam O., Comaills V., Morris R., Wittner B.S., Liu M., Engstrom A., Takahashi F., et al. Setd1a modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat. Commun. 2015;6:8257. doi: 10.1038/ncomms9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S., Fesler A., Ju J. Implications of circadian rhythm regulation by microRNAs in colorectal cancer. Cancer Transl. Med. 2016;2:1–6. doi: 10.4103/2395-3977.177555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui R., Meng W., Sun H.L., Kim T., Ye Z., Fassan M., Jeon Y.J., Li B., Vicentini C., Peng Y., et al. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc. Natl. Acad. Sci. USA. 2015;112:E4288–E4297. doi: 10.1073/pnas.1502068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madhavan D., Peng C., Wallwiener M., Zucknick M., Nees J., Schott S., Rudolph A., Riethdorf S., Trumpp A., Pantel K., et al. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016;37:461–470. doi: 10.1093/carcin/bgw008. [DOI] [PubMed] [Google Scholar]
- 60.Aherne S.T., Madden S.F., Hughes D.J., Pardini B., Naccarati A., Levy M., Vodicka P., Neary P., Dowling P., Clynes M. Circulating miRNAs mir-34a and mir-150 associated with colorectal cancer progression. BMC Cancer. 2015;15:329. doi: 10.1186/s12885-015-1327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larrea E., Sole C., Manterola L., Goicoechea I., Armesto M., Arestin M., Caffarel M.M., Araujo A.M., Araiz M., Fernandez-Mercado M., et al. New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int. J. Mol. Sci. 2016;17:627. doi: 10.3390/ijms17050627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baroni S., Romero-Cordoba S., Plantamura I., Dugo M., D’Ippolito E., Cataldo A., Cosentino G., Angeloni V., Rossini A., Daidone M.G., et al. Exosome-mediated delivery of mir-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016;7:e2312. doi: 10.1038/cddis.2016.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Modica M., Regondi V., Sandri M., Iorio M.V., Zanetti A., Tagliabue E., Casalini P., Triulzi T. Breast cancer-secreted mir-939 downregulates ve-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017;384:94–100. doi: 10.1016/j.canlet.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 64.Robertson K.J.O. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 65.Li B.L., Lu W., Qu J.J., Ye L., Du G.Q., Wan X.P. Loss of exosomal mir-148b from cancer-associated fibroblasts promotes endometrial cancer cell invasion and cancer metastasis. J. Cell Physiol. 2019;234:2943–2953. doi: 10.1002/jcp.27111. [DOI] [PubMed] [Google Scholar]
- 66.Oudin M.J., Weaver V.M. Physical and chemical gradients in the tumor microenvironment regulate tumor cell invasion, migration, and metastasis. Cold Spring Harb. Symp. Quant. Biol. 2016;81:189–205. doi: 10.1101/sqb.2016.81.030817. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y., Davis C., Shah S., Hughes D., Ryan J.C., Altomare D., Pena M.M. Il-33 promotes growth and liver metastasis of colorectal cancer in mice by remodeling the tumor microenvironment and inducing angiogenesis. Mol. Carcinog. 2017;56:272–287. doi: 10.1002/mc.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li L., Li C., Wang S., Wang Z., Jiang J., Wang W., Li X., Chen J., Liu K., Li C., et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver mir-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.CAN-15-1625. [DOI] [PubMed] [Google Scholar]
- 69.Wang X., Luo G., Zhang K., Cao J., Huang C., Jiang T., Liu B., Su L., Qiu Z. Hypoxic tumor-derived exosomal mir-301a mediates m2 macrophage polarization via pten/pi3kgamma to promote pancreatic cancer metastasis. Cancer Res. 2018;78:4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 70.Tang Y.T., Huang Y.Y., Li J.H., Qin S.H., Xu Y., An T.X., Liu C.C., Wang Q., Zheng L. Alterations in exosomal miRNA profile upon epithelial-mesenchymal transition in human lung cancer cell lines. BMC Genomics. 2018;19:802. doi: 10.1186/s12864-018-5143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J., Kim T.Y., Lee M.S., Mun J.Y., Ihm C., Kim S.A. Exosome cargo reflects tgf-beta1-mediated epithelial-to-mesenchymal transition (emt) status in a549 human lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2016;478:643–648. doi: 10.1016/j.bbrc.2016.07.124. [DOI] [PubMed] [Google Scholar]
- 72.Tanaka S., Hosokawa M., Ueda K., Iwakawa S. Effects of decitabine on invasion and exosomal expression of mir-200c and mir-141 in oxaliplatin-resistant colorectal cancer cells. Biol. Pharm. Bull. 2015;38:1272–1279. doi: 10.1248/bpb.b15-00129. [DOI] [PubMed] [Google Scholar]
- 73.Liz J., Esteller M. LncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 74.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakaya H.I., Amaral P.P., Louro R., Lopes A., Fachel A.A., Moreira Y.B., El-Jundi T.A., da Silva A.M., Reis E.M., Verjovski-Almeida S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 77.Qureshi I.A., Mehler M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cajigas I., Leib D.E., Cochrane J., Luo H., Swyter K.R., Chen S., Clark B.S., Thompson J., Yates J.R., 3rd, Kingston R.E., et al. Evf2 lncRNA/brg1/dlx1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development. 2015;142:2641–2652. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang A., Zhao J.C., Kim J., Fong K.W., Yang Y.A., Chakravarti D., Mo Y.Y., Yu J. LncRNA hotair enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13:209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang N., Wang X., Xie X., Liao Y., Liu N., Liu J., Miao N., Shen J., Peng T. LncRNA dancr promotes tumor progression and cancer stemness features in osteosarcoma by upregulating axl via mir-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 81.Sado T., Hoki Y., Sasaki H. Tsix silences xist through modification of chromatin structure. Dev. Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 82.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 83.Xiao C., Wu C.H., Hu H.Z. LncRNA uca1 promotes epithelial-mesenchymal transition (emt) of breast cancer cells via enhancing wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2016;20:2819–2824. [PubMed] [Google Scholar]
- 84.Wang Y.J., Liu J.Z., Lv P., Dang Y., Gao J.Y., Wang Y. Long non-coding RNA ccat2 promotes gastric cancer proliferation and invasion by regulating the e-cadherin and lats2. Am. J. Cancer Res. 2016;6:2651–2660. [PMC free article] [PubMed] [Google Scholar]
- 85.Yue B., Qiu S., Zhao S., Liu C., Zhang D., Yu F., Peng Z., Yan D. LncRNA-atb mediated e-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J. Gastroenterol. Hepatol. 2016;31:595–603. doi: 10.1111/jgh.13206. [DOI] [PubMed] [Google Scholar]
- 86.Zhang W., Shi S., Jiang J., Li X., Lu H., Ren F. LncRNA meg3 inhibits cell epithelial-mesenchymal transition by sponging mir-421 targeting e-cadherin in breast cancer. Biomed. Pharmacother. 2017;91:312–319. doi: 10.1016/j.biopha.2017.04.085. [DOI] [PubMed] [Google Scholar]
- 87.Richards E.J., Zhang G., Li Z.P., Permuth-Wey J., Challa S., Li Y., Kong W., Dan S., Bui M.M., Coppola D., et al. Long non-coding RNAs (lncRNA) regulated by transforming growth factor (tgf) beta: LncRNA-hit-mediated tgfbeta-induced epithelial to mesenchymal transition in mammary epithelia. J. Biol. Chem. 2015;290:6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo M., Li Z., Wang W., Zeng Y., Liu Z., Qiu J. Long non-coding RNA h19 increases bladder cancer metastasis by associating with ezh2 and inhibiting e-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 89.Sun N.X., Ye C., Zhao Q., Zhang Q., Xu C., Wang S.B., Jin Z.J., Sun S.H., Wang F., Li W. Long noncoding RNA-ebic promotes tumor cell invasion by binding to ezh2 and repressing e-cadherin in cervical cancer. PLoS ONE. 2014;9:e100340. doi: 10.1371/journal.pone.0100340. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Lin L.Y., Yang L., Zeng Q., Wang L., Chen M.L., Zhao Z.H., Ye G.D., Luo Q.C., Lv P.Y., Guo Q.W., et al. Tumor-originated exosomal lncuegc1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer. 2018;17:84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berrondo C., Flax J., Kucherov V., Siebert A., Osinski T., Rosenberg A., Fucile C., Richheimer S., Beckham C.J. Expression of the long non-coding RNA hotair correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS ONE. 2016;11:e0147236. doi: 10.1371/journal.pone.0147236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun Z., Yang S., Zhou Q., Wang G., Song J., Li Z., Zhang Z., Xu J., Xia K., Chang Y., et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer. 2018;17:82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xue M., Chen W., Xiang A., Wang R., Chen H., Pan J., Pang H., An H., Wang X., Hou H., et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-uca1. Mol. Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hardin H., Helein H., Meyer K., Robertson S., Zhang R., Zhong W., Lloyd R.V. Thyroid cancer stem-like cell exosomes: Regulation of emt via transfer of lncRNAs. Lab. Invest. 2018;98:1133–1142. doi: 10.1038/s41374-018-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li X., Xu Y., Chen Y., Chen S., Jia X., Sun T., Liu Y., Li X., Xiang R., Li N. Sox2 promotes tumor metastasis by stimulating epithelial-to-mesenchymal transition via regulation of wnt/beta-catenin signal network. Cancer Lett. 2013;336:379–389. doi: 10.1016/j.canlet.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 96.Mu P., Zhang Z., Benelli M., Karthaus W.R., Hoover E., Chen C.-C., Wongvipat J., Ku S.-Y., Gao D., Cao Z., et al. Sox2 promotes lineage plasticity and antiandrogen resistance in tp53- and rb1-deficient prostate cancer. Science. 2017;355:84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang K., Ji W., Yu Y., Li Z., Niu X., Xia W., Lu S. Fgfr1-erk1/2-sox2 axis promotes cell proliferation, epithelial-mesenchymal transition, and metastasis in fgfr1-amplified lung cancer. Oncogene. 2018;37:5340–5354. doi: 10.1038/s41388-018-0311-3. [DOI] [PubMed] [Google Scholar]
- 98.Park S.M., Gaur A.B., Lengyel E., Peter M.E. The mir-200 family determines the epithelial phenotype of cancer cells by targeting the e-cadherin repressors zeb1 and zeb2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Z., Jiang P., Li J., Peng M., Zhao X., Zhang X., Chen K., Zhang Y., Liu H., Gan L., et al. Tumor-derived exosomal lnc-sox2ot promotes emt and stemness by acting as a cerna in pancreatic ductal adenocarcinoma. Oncogene. 2018;37:3822–3838. doi: 10.1038/s41388-018-0237-9. [DOI] [PubMed] [Google Scholar]
- 100.Wu D.M., Deng S.H., Liu T., Han R., Zhang T., Xu Y. Tgf-beta-mediated exosomal lnc-mmp2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting mmp2 expression. Cancer Med. 2018;7:5118–5129. doi: 10.1002/cam4.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang X., Li H., Lu X., Wen C., Huo Z., Shi M., Tang X., Chen H., Peng C., Fang Y., et al. Melittin-induced long non-coding RNA nonhsat105177 inhibits proliferation and migration of pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9:940. doi: 10.1038/s41419-018-0965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dutta S., Reamtong O., Panvongsa W., Kitdumrongthum S., Janpipatkul K., Sangvanich P., Piyachaturawat P., Chairoungdua A. Proteomics profiling of cholangiocarcinoma exosomes: A potential role of oncogenic protein transferring in cancer progression. Biochim. Biophys. Acta. 2015;1852:1989–1999. doi: 10.1016/j.bbadis.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 103.Katoh M., Nakagama H. Fgf receptors: Cancer biology and therapeutics. Med. Res. Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 104.Shi S., Zhang Q., Xia Y., You B., Shan Y., Bao L., Li L., You Y., Gu Z. Mesenchymal stem cell-derived exosomes facilitate nasopharyngeal carcinoma progression. Am. J. Cancer Res. 2016;6:459–472. [PMC free article] [PubMed] [Google Scholar]
- 105.Nakamura K., Sawada K., Kinose Y., Yoshimura A., Toda A., Nakatsuka E., Hashimoto K., Mabuchi S., Morishige K.I., Kurachi H., et al. Exosomes promote ovarian cancer cell invasion through transfer of cd44 to peritoneal mesothelial cells. Mol. Cancer Res. 2017;15:78–92. doi: 10.1158/1541-7786.MCR-16-0191. [DOI] [PubMed] [Google Scholar]
- 106.Fujiwara T., Eguchi T., Sogawa C., Ono K., Murakami J., Ibaragi S., Asaumi J., Calderwood S.K., Okamoto K., Kozaki K. Carcinogenic epithelial-mesenchymal transition initiated by oral cancer exosomes is inhibited by anti-egfr antibody cetuximab. Oral Oncol. 2018;86:251–257. doi: 10.1016/j.oraloncology.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 107.Chairoungdua A., Smith D.L., Pochard P., Hull M., Caplan M.J. Exosome release of beta-catenin: A novel mechanism that antagonizes wnt signaling. J. Cell Biol. 2010;190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bekes E.M., Schweighofer B., Kupriyanova T.A., Zajac E., Ardi V.C., Quigley J.P., Deryugina E.I. Tumor-recruited neutrophils and neutrophil timp-free mmp-9 regulate coordinately the levels of tumor angiogenesis and efficiency of malignant cell intravasation. Am. J. Pathol. 2011;179:1455–1470. doi: 10.1016/j.ajpath.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sandri S., Faiao-Flores F., Tiago M., Pennacchi P.C., Massaro R.R., Alves-Fernandes D.K., Berardinelli G.N., Evangelista A.F., de Lima Vazquez V., Reis R.M., et al. Vemurafenib resistance increases melanoma invasiveness and modulates the tumor microenvironment by mmp-2 upregulation. Pharmacol. Res. 2016;111:523–533. doi: 10.1016/j.phrs.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 110.You Y., Shan Y., Chen J., Yue H., You B., Shi S., Li X., Cao X. Matrix metalloproteinase 13-containing exosomes promote nasopharyngeal carcinoma metastasis. Cancer Sci. 2015;106:1669–1677. doi: 10.1111/cas.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shan Y., You B., Shi S., Shi W., Zhang Z., Zhang Q., Gu M., Chen J., Bao L., Liu D., et al. Hypoxia-induced matrix metalloproteinase-13 expression in exosomes from nasopharyngeal carcinoma enhances metastases. Cell Death Dis. 2018;9:382. doi: 10.1038/s41419-018-0425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schillaci O., Fontana S., Monteleone F., Taverna S., Di Bella M.A., Di Vizio D., Alessandro R. Exosomes from metastatic cancer cells transfer amoeboid phenotype to non-metastatic cells and increase endothelial permeability: Their emerging role in tumor heterogeneity. Sci. Rep. 2017;7:4711. doi: 10.1038/s41598-017-05002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mao J., Liang Z., Zhang B., Yang H., Li X., Fu H., Zhang X., Yan Y., Xu W., Qian H. Ubr2 enriched in p53 deficient mouse bone marrow mesenchymal stem cell-exosome promoted gastric cancer progression via wnt/beta-catenin pathway. Stem Cells. 2017;35:2267–2279. doi: 10.1002/stem.2702. [DOI] [PubMed] [Google Scholar]
- 114.Kannan A., Hertweck K.L., Philley J.V., Wells R.B., Dasgupta S. Genetic mutation and exosome signature of human papilloma virus associated oropharyngeal cancer. Sci. Rep. 2017;7:46102. doi: 10.1038/srep46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tang M.K.S., Yue P.Y.K., Ip P.P., Huang R.L., Lai H.C., Cheung A.N.Y., Tse K.Y., Ngan H.Y.S., Wong A.S.T. Soluble e-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat. Commun. 2018;9:2270. doi: 10.1038/s41467-018-04695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]