Abstract
TDP-43 proteinopathy is very common among the elderly (affecting at least 25% in individuals over 85 years of age) and is associated with substantial cognitive impairment. Risk factors implicated in age-related TDP-43 proteinopathy include commonly inherited gene variants, comorbid Alzheimer’s disease pathology, and thyroid hormone dysfunction. To test parameters that are associated with aging-related TDP-43 pathology, we performed exploratory analyses of pathologic, genetic, and biochemical data derived from research volunteers in the University of Kentucky Alzheimer’s Disease Center autopsy cohort (n=136 subjects). Digital pathologic methods were used to discriminate and quantify both neuritic and intracytoplasmic pathology in the hippocampal formation. Overall, 46.4% of the cases were positive for TDP-43 intracellular inclusions, which is consistent with results in other prior community-based cohorts. The pathologies were correlated with hippocampal sclerosis of aging (HS-Aging) linked genotypes. We also assayed brain parenchymal thyroid hormone (triiodothyronine [T3] and thyroxine [T4]) levels. In cases with SLCO1A2/IAPP or ABCC9 risk associated genotypes, the T3/T4 ratio tended to be reduced (p=0.051 using 2-tailed statistical test), and in cases with low T3/T4 ratios (bottom quintile), there was a higher likelihood of HS-Aging pathology (p=0.025 using 2-tailed statistical test). This is intriguing because the SLCO1A2/IAPP and ABCC9 risk associated genotypes have been associated with altered expression of the astrocytic thyroid hormone receptor (product of the nearby gene SLCO1C1). These data indicate that dysregulation of thyroid hormone signaling may play a role in TDP-43 proteinopathy.
Keywords: Neuropathology, ScanScope, SNP, GRN, TMEM106B, SLCO1A2, dementia
Introduction
Hippocampal sclerosis of aging (HS-Aging) is a neurodegenerative disease, distinct from Alzheimer’s disease (AD) (Montine et al., 2012; Nelson et al., 2013), yet often mimicking AD clinically (Brenowitz et al., 2014; Murray et al., 2014; Nelson et al., 2013; Pao et al., 2011; Zarow et al., 2012). HS-Aging is a very prevalent condition, affecting 10-25% of persons over age 85 (Brenowitz et al., 2014; Kero et al., 2018; Leverenz et al., 2002; Nelson et al., 2011; Zarow et al., 2012). The defining characteristics of HS-Aging are neuronal loss, astrocytosis, and transactive response DNA binding protein of 43 kDa (TDP-43) pathology in the hippocampus (Amador-Ortiz and Dickson, 2008; Nelson et al., 2011), and these pathologic features are specifically associated with cognitive impairment (Brenowitz et al., 2014; Murray et al., 2014; Nag et al., 2017; Nelson et al., 2010). “TDP-43 pathology” refers to phosphorylated TDP-43 detected via immunohistochemistry, and localized in cytoplasmic, nuclear, perivascular, and/or neurite-like structures. Currently, the upstream disease-driving mechanisms associated with TDP-43 pathology and HS-Aging are not well understood. Thyroid hormone (TH) signaling, and/or comorbid AD-type plaques and tangles, may affect persons’ vulnerability to HS-Aging pathology, and disease susceptibility factors may also overlap with frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) (Dickson et al., 1994; Josephs et al., 2008; Katsumata et al., 2018; Nelson et al., 2016a; Nelson et al., 2013; Zarow et al., 2008; Zarow et al., 2012).
The identification of genetic risk factors for HS-Aging has provided insights into disease mechanisms. Specific variants in genes (GRN and TMEM106B) that previously were associated with risk for FTLD-TDP, were later also linked with risk for HS-Aging pathology and age-related TDP-43 pathology, which probably represent a disease continuum previously referred to together as cerebral age-related TDP-43 pathology with sclerosis (Cykowski et al., 2017;Dickson et al., 2010; Dickson et al., 2015; Murray et al., 2014; Nelson et al., 2016b; Rutherford et al., 2012; Van Langenhove et al., 2012). These prior studies support the hypothesis that GRN and TMEM106B gene variants influence the brain’s vulnerability to developing TDP-43 pathology. There also is compelling evidence that TDP-43 pathology is comorbid with advanced AD-type plaques and tangles in the medial temporal lobe structures in many persons (Davidson et al., 2011; Josephs et al., 2016; Josephs et al., 2008; Nelson et al., 2018). However, the associations between GRN/TMEM106B gene variants, AD-type pathology, and TDP-43 pathology are inconsistent, so other factors must also be relevant.
The only genome-wide association study published to date that focused exclusively on autopsy-confirmed HS-Aging pathology as the endophenotype identified a single nucleotide polymorphism (SNP) in ABCC9 that was associated with risk for HS-Aging pathology (Nelson et al., 2014). In a non-overlapping group of autopsied subjects, the association between the same ABCC9 SNP and HS-Aging pathology was replicated (Nelson et al., 2015b). These same gene variants in ABCC9 are associated with brain atrophy (Nho et al., 2016), and nearby polymorphisms were also linked to dementia risk in APOE ε4(−) individuals (D'Introno et al., 2006; Mayeux et al., 2002; Pericak-Vance et al., 1997; Scott et al., 1999).
Approximately one million base-pairs away on Chr. 12p12 from ABCC9 is SLCO1C1 (Jansen et al., 2005), which encodes the brain’s main thyroid hormone (TH) importing protein. Importantly, Roostaei et al. discovered a novel Chr. 12p12 SNP based on its association with non-β neurodegeneration (Roostaei et al., 2016), and this SNP status also is associated with variability in SLCO1C1 expression in human brain (Nelson et al., 2016a). Both ABCC9 and SLCO1C1 transcripts are highly expressed in human astrocytes (Nelson et al., 2016a), and their functions appear interconnected. Astrocytes import TH from blood into brain, process blood-borne thyroxine (T4) hormone into the biologically active triiodothyronine (T3), and deliver T3 to neurons (Heuer and Visser, 2009; Morte and Bernal, 2014). The ABCC9 polypeptide (also known as SUR2) serves as a metabolic sensor that helps couple energy needs with blood flow and intracellular signaling (Nelson et al., 2015a; Nichols, 2006; Nichols et al., 2013), whereas the TH importing SLCO1C1 polypeptide is also known as OATP1C1 and Thyroxine transporter (Bernal et al., 2015; Jansen et al., 2005). TH is a potent metabolic regulating agent (McAninch and Bianco, 2014; Mullur et al., 2014; Trentin, 2006; van der Deure et al., 2008), whereas TH dysregulation has been previously associated with dementia risk (Chaker et al., 2016; Mafrica and Fodale, 2008; Sampaolo et al., 2005; Tan and Vasan, 2009) and HS-Aging specifically (Nelson et al., 2016a; Trieu, 2018). Together, these observations provide the basis for a plausible mechanistic hypothesis: gene variants that affect TH levels in the brain appear to be relevant to TDP-43/HS-Aging pathology. Much remains unknown about the biochemical landscape of the aged brain, and the impact of genetics. For example, a separate GWAS found that a SNP (rs9637454) in a gene encoding a distinct K+ channel modifying protein – KCNMB2 -- was also linked to HS-Aging risk (Beecham et al., 2014).
The main objectives of the present study were to analyze the pathologic, genetic, and biochemical parameters that can be correlated with TDP-43/HS-Aging pathology in the aged human hippocampus. We wanted to test whether gene variants that alter HS-Aging risk also are associated with altered brain TH processing. The data were gathered from research volunteers followed to autopsy in the University of Kentucky Alzheimer's Disease Center (UK-ADC) cohort. Results of these analyses provide support of the hypothesis that TH related pathways in the brain may alter vulnerability to TDP-43 pathology and HS-Aging.
Materials and methods
UK-ADC autopsy cohort:
Details of UK-ADC research volunteers’ recruitment, the overall cohort inclusion/exclusion criteria, and clinical assessments were described previously (Jicha et al., 2012; Schmitt et al., 2012). Briefly, older adult volunteers (most recruited while cognitively normal) agreed to be followed annually for cognitive, physical, and neurological examination and to donate their brain at the time of death, with a provision to allow genetic testing for research purposes. Protocols and informed consent procedures were approved by the University of Kentucky Institutional Review Board. Additional inclusion criteria for this study were the availability of neuropathology data and snap-frozen cerebellum with postmortem interval (PMI) before autopsy of <24hrs. Other inclusion/exclusion criteria for this convenience sample were established to include cognitively normal subjects as well as a relatively broad spectrum of ages and comorbid pathologies; age was >60 year at death, and some cases with α-synucleinopathies were included (see below). However, cases with rare conditions (FTLD-TDP, FTLD-Tau, brain tumor, triplet repeat disorders, or prion disease) were excluded. All the included cases were assessed systematically for pathology, genotyping, and biochemical analyses. No additional cases were analyzed for TH ELISAs or genotype after the initial samples were assayed and processed.
Pathological assessments and staining methods:
See Supplemental Figure 1 for work flow. Details of sampling and assessments in the UK-ADC autopsy cohort were described previously (Abner et al., 2018; Nelson et al., 2007). Briefly, over 20 brain areas were sampled in each case using methods compatible with National Institute on Aging-Alzheimer’s Association consensus recommendation (Montine et al., 2012). The tissue was fixed in 10% formaldehyde, processed in paraffin blocks, and sections were cut at 8-micron thickness. According to consensus-based guidelines, the criterion for high level of AD pathology was Braak NFT stages V or VI, and HS was “defined by pyramidal cell loss and gliosis in CA1 and subiculum of the hippocampal formation that is out of proportion to AD neuropathologic change in the same structures” (Montine et al., 2012). Both the right and left hippocampi were evaluated for HS pathology using hematoxylin and eosin stains (H&E), and the case was deemed to be HS-Aging if either side had HS. Immunohistochemical stains for phospho-TDP-43 (1D3 clone, 1:500 dilution, from EMD Millipore, Burlington, MA) was performed as described previously (Gal et al., 2018; Smith et al., 2017). One slide of left frontal cortex (BA9) was also evaluated from each case and scored dichotomously for there either being any TDP-43 pathology detected, or none.
For digital pathologic assessments of the hippocampal formation, phospho-TDP-43 immunostained slides were loaded into an Aperio ScanScope XT™, scanned at 40X magnification via the semi-automated method, and the images stored on a dedicated server (Supplemental Figure 1). To calculate the density of TDP-43 neurites and inclusions, an Aperio GENIE (GENetic Imagery Exploitation) classifier was developed. A training set containing TDP-43 positive hippocampal slides was constructed. Annotations for neurites, inclusions, background tissue (including nuclei), and clear glass were made. These annotations were incorporated into a training montage. The montage magnification was set at 15x. The montage was subjugated to a GENIE training algorithm with 2000 iterations. Annotations were adjusted, added, or removed until the training accuracy values for each tissue class were 100. The classifier was tested on sample slides to ensure accurate identification of each tissue class. The classifier was applied to each analysis area, yielding values representing the percent of the overall analysis area covered by each tissue class. Clear glass values were removed to eliminate empty space from the final percentages, and the remaining tissue class percentages were adjusted to represent this change. This yielded the densities, represented by percentage of analysis area covered, of neurites, inclusions, and background tissue. Inclusions were also counted using an Aperio Rare Event Detection (RED) algorithm. The following modifications were made to optimize for our in-house staining protocol: hue value 0.1, hue width 0.4, color saturation threshold 0.418, intensity threshold 210, averaging radius 8, minimum pixels 100, maximum pixels 100000, and roundness threshold 0.25. The RED algorithm was then run on each analysis area. The inclusion density was calculated by dividing the total inclusion count by the overall analysis area. Nuclei were counted using the Aperio Image Analysis Toolbox™ Nuclear algorithm. The Nuclear algorithm was run on each analysis area. The number of ‘weak’ (1+), ‘moderate’ (2+), and ‘strong’ (3+) nuclei were added to calculate the total nuclei count. The nuclei density was calculated by dividing the total nuclei count by the overall analysis area.
Genetic (SNP) assays:
Genomic DNA from each case was isolated using Quick-DNA™ Miniprep Plus Kit (ZYMO RESEARCH, Irvine, CA). DNA quality and quantitation was examined using NanoDrop (ND-1000) Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and gel electrophoresis. DNA samples with A260 /A280 and A260 /A230 ratios below 1.8 were discarded or re-extracted. TaqMan® Predesigned SNP Genotyping Assays were performed; details of the SNP assays are provided in Supplemental Figure 2 and Supplemental table. Two of the SNPs previously identified as potential risk alleles did not have commercially available reagents for SNP assays. To address those SNPs, we used assays for proxy SNPs that are in very close (>.98 r2 and >.99 D') linkage disequilibrium in human (including European) populations: for rs73069071 (in SLOC1A2/IAPP), rs12301085 was assayed, and for rs9637454 (in KCNMB2), rs12496790 was assayed. For the ABCC9 gene variant, we assessed rs704180, since it has been demonstrated to be associated with HS-Aging pathology (Katsumata et al., 2017; Nelson et al., 2014; Nelson et al., 2015b). Notably, the nearby ABCC9 SNP rs4148651 was found to be an eQTL for SLCO1C1 (Nelson et al., 2016a). These two gene variants, rs4148651 and rs704180, are in linkage disequilibrium (D’=0.44 and p value of LD <0.0001 according to NIH’s “LDlink” server (Machiela and Chanock, 2015)).
TaqMan® genotyping assays were performed following manufacturer’s instructions (Life Technologies Corporation, Carlsbad, CA). Genotyping PCR and fluorescence recording were performed on a ViiA™ 7 96-well Fast Block with standard thermal cycling conditions (Hold at 95°C 10 minutes, 40 cycles of Denaturation at 95°C 15 seconds and Annealing/Extension at 60°C 1 minute) (Life Technologies Corporation, Carlsbad, CA). Data were analyzed and the corresponding alleles called using QuantStudio™ Real-Time PCR Software algorithm for autocalling and generation of allelic discrimination plots (Life Technologies Corporation, Carlsbad, CA).
TH assays:
Commercially-available total T3 and T4 enzyme-linked immunosorbent assay (ELISA) tests were used to quantify TH levels. Besides those explicitly described below, no other assays were performed on the included samples. For brain extracts, 100-300 mg of frozen cerebellum (cerebellar folia) were homogenized using a PRO200 blender (Pro Scientific, Oxford, CT) in ice-cold buffer consisting of RIPA buffer (Pierce, Rockford, IL) diluted 4X with PBS; the ratio of buffer to tissue was 4:1 v/w. In a subset of cases where both cerebellar and neocortical flash-frozen tissue was available, portions of superior and middle temporal gyri (SMTG; Brodmann Areas 21/22) were also analyzed (see below). Cells were additionally disrupted with two pulses (10 seconds each) of ultrasound using a Microsonic Cell Disruptor (Misonix, Farmingdale, NY). Cell debris was sedimented by centrifugation at 20,000 g for 30 minutes at 4°C. Supernatants containing T3 and T4 were aliquoted and stored at −80°C. Manufacturer’s ELISA protocols were followed, and T3 and T4 ELISAs were performed separately as described previously (Nelson et al., 2016a). Briefly, undiluted specimen samples and manufacturer-supplied standard solutions were loaded in duplicate into microtiter plate wells of T3 and T4 test kits (Aviva System Biology, San Diego, CA, USA) following manufacturer’s specified protocol. Samples and ELISA plates were processed blind to clinical and pathological information. Absorbance was measured at 450 nm using the SpectraMax M3 (Molecular Devices Inc., Sunnyvale, CA, USA) plate reader. Standard curves were generated with SigmaPlot software (Systat Softwre, San Jose, CA, USA), using a 4-paramter logistic fit, which was subsequently used to calculate T3 and T4 concentrations in the brain extracts (See Supplemental Figure 3). An independent standard curve was generated for each microtiter plate used. The values obtained were then standardized to tissue weight. Data were downloaded to Microsoft Excel, and only later unblinded.
Statistical analyses:
Clinical, genetic, pathologic, and biochemical covariates were tested for marginal association with each of the assayed genetic variants, HS-Aging pathology, advanced AD (defined as Braak NFT stage V or VI) pathology, and low (bottom quintile) T3/T4 ratio. Chi-square tests were used for dichotomous covariates with sufficient cell counts; Fisher’s exact test was used otherwise. T-tests were used for quantitative covariates. The distribution of digital pathologic TDP-43 inclusion densities necessitated the use of zero-inflated negative binomial regression. We modeled the probability of a non-zero inclusion and the inclusion count as two separate component processes and tested for association with inclusions using a likelihood ratio test. Nominal p-values are reported with p<0.05 deemed significant. A Bonferroni correction was used for correlation analysis, i.e., p-values are divided by the total numbers of pairwise comparison.
Results
An underlying a priori hypothesis for the current study is shown in Figure 1, and the workflow is depicted in Supplemental Figure 1. The overall goals were to analyze genetic, clinical, pathologic, and biochemical (related to TH) variables, in order to test hypotheses related to pathogenesis of TDP-43 pathology and HS-Aging. The data analyzed derived from a convenience sample based on availability of brain tissue for biochemistry, with the goal of including a relatively broad range of ages, clinical states, and neuropathologic features, to reflect the heterogeneous mix of (often comorbid) pathologies seen in advanced age. Included subject characteristics are shown in Table 1. Average age at death of included subjects was 84.0 (range 61-102) years, and 57.4% overall were female. Included were 11 cases with diffuse/neocortical Lewy body disease, whereas 56 of the included subjects (41.1%) had some subtype of Lewy body pathology. Mean and median PMI between death and autopsy were 5.6 and 3.3 hours, respectively (range 1.1-22.5 hours). APOE genotype information was available on 120/136 (88.2%) of the included subjects, and final MMSE scores were available for 126/136 (92.6%) of subjects.
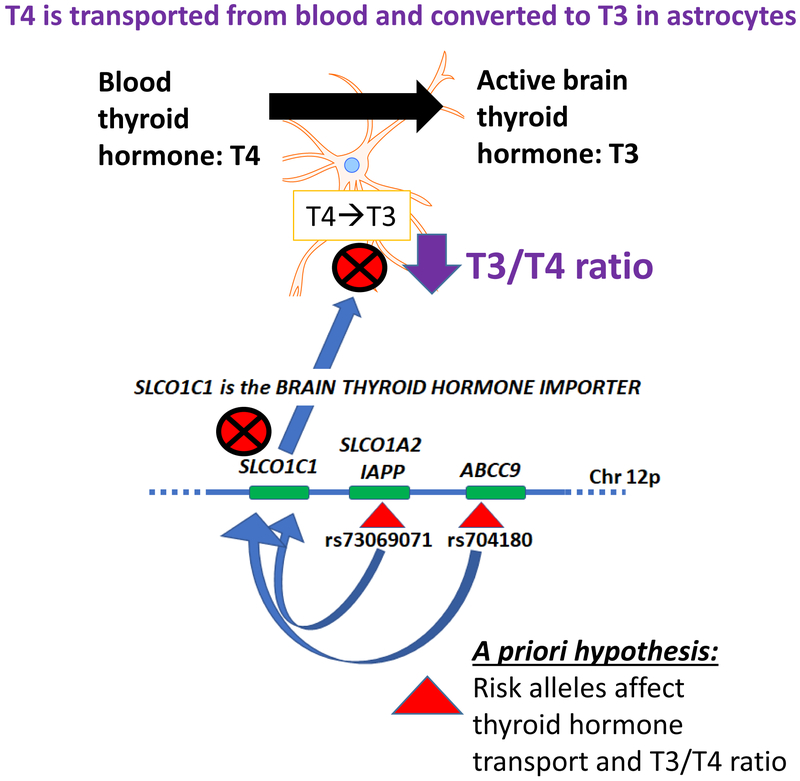
Figure 1. Schematic representation of a priori hypothesis.
An underlying hypothesis for this study is that HS-Aging linked gene variants on Chr. 12p may affect the level or activity of SLCO1C1, which is the primary brain thyroid hormone importer protein (Heuer and Visser, 2009). Rationale for this hypothesis was presented in a prior study (Nelson et al., 2016a) and the direct relevance of thyroid hormone to the brain disease was supported by data in a community-based cohort in which thyroid hormone dysregulation was found to be a risk factor for hippocampal sclerosis of aging (Trieu et al., 2018). Astrocytes import and process T4 (thyroxine) from blood, convert T4 into T3 (triiodothyronine), and then deliver T3 to neurons. A specific prediction of the study is that if SLCO1C1 functions to import T4 from blood into astrocytes for conversion into the active T3 molecules, then, a perturbation of SLCO1C1 may affect the ratio of T3 to T4 in brain parenchyma. Further, that ratio could be associated with altered risk for hippocampal sclerosis and/or TDP-43 pathology.
Table 1.
Summary information about cases included in the current study (n=136) from the University of Kentucky AD Center (UK-ADC): clinical and pathological features
| Age/Sex/post-mortem interval | |
| Age at death, Avg | 84.0 yrs (range 61-103) |
| Sex (F%) | 57.4% |
| Post-mortem interval, Avg | 5.6 hours (range 1.1-22.5) |
| Cognitive status before death | |
| Normal | 29.1% |
| MCI | 11.2% |
| Demented | 56.7% |
| Final MMSE score, Avg | 20.3 (range 0-30) |
| Pathologic features | |
| Advanced Alzheimer's Disease (Braak NFT stages V or VI) | 50.0% |
| Hippocampal sclerosis of aging | 27.2% |
| Diffuse/neocortical Lewy body disease | 8.1% |
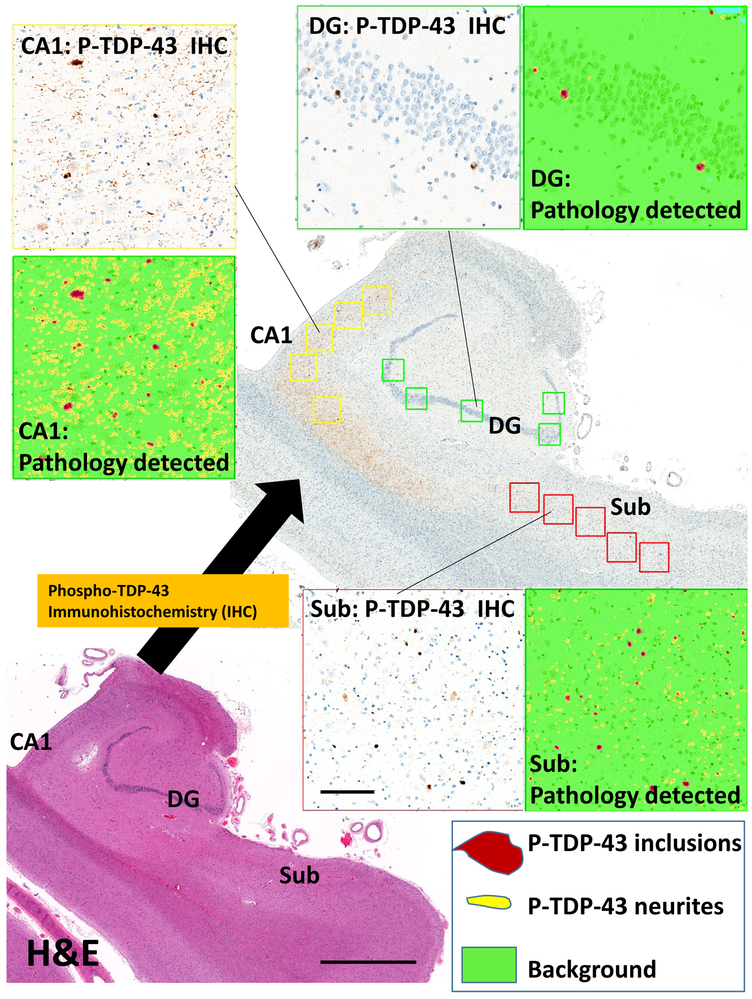
For each case, digital pathologic assessments were performed to detect and quantify TDP-43 pathology in the hippocampal formation. To help convey how these analyses were performed, Figure 2 shows a representative section of hippocampal formation with false-colored depiction of identified phospho-TDP-43 immunoreactive intracellular inclusions and neurites. These subtypes of TDP-43 pathology were detected and quantified separately.
Figure 2. Representative section of the hippocampal formation, to convey digital assessment of TDP-43 pathologies in the CA1 subfield and dentate granule (DG) cells of the hippocampus proper, and in the subiculum (Sub).
The adjacent hematoxylin and eosin (H&E)-stained section (lower-left) helps to convey the anatomic landmarks, including CA1, Sub, and DG subregions. Rightward is medial. This hippocampus had extensive Phospho-TDP-43 immunoreactivity in the CA1 (upper left), dentate granule cells (upper right), and subiculum (lower right). The immunostained pathologies were detected using digital pathologic methods that quantitatively scored the number of inclusions (false-colored maroon in the insets) and neuritic-like structures (false-colored yellow); a color key is shown in the lower right. Scale bar = 2mm for the H&E stained photomicrograph and 200 microns for the inset boxes.
SNP genotype assays were used to assess the status of gene variants that have been associated with risk for HS-Aging and/or age-related TDP-43 pathology. The SNPs evaluated, along with additional information, are depicted in Table 2. Overall results for the genotyping assays, and how those results correlated with various clinical, pathological, and biochemical parameters, are shown in Tables 3-4. Note that, as expected, the cases with HS-Aging pathology tended to have more TDP-43 pathology and a higher percentage were demented (p<0.0001), and these subjects were on average older than those without HS-Aging pathology (p=0.006) (Table 3). More detailed information about the SNP assays are presented in Supplemental material, and a representative example of the PCR genotype assay results (for SNP rs5848) is provided in Supplemental Figure 2. Detailed descriptions of how each SNP and HS-Aging linked genotype correlated with pathologies and other parameters are presented in Supplemental Tables 1-5.
Table 2.
Single nucleotide polymorphisms (SNPs) that were assayed in the current study: associated chromosomes, nearby genes, and links to hippocampal sclerosis of aging and/or gene expression
| SNPs tested in current study |
Proxy for other SNP |
Chr. | Nearest gene | Linked to HS- Aging risk |
Ref. | Variants | HS-Aging risk allele(s) |
Linked to FTLD- TDP risk |
eQTL for ABCC9 and/or SLCO1C1 |
|---|---|---|---|---|---|---|---|---|---|
| rs5848 | n/a | 17q | GRN | Yes | (Dickson et al., 2010) | C/T | CT/TT | Yes | No |
| rs1990622 | n/a | 7p | TMEM106B | Yes | (Rutherford et al., 2012) | G/A | GA/AA | Yes | No |
| rs704180 | n/a | 12p | ABCC9 | Yes | (Nelson et al., 2014) | G/A | AA | No | Yes |
| rs12301085 | rs73069071 | 12p | SLCO1A2/IAPP | Yes* | (Roostaei et al., 2016) | G/A | GA/AA | No | Yes |
| rs12496790 | rs9637454 | 3q | KCNMB2 | Yes | (Beecham et al., 2014) | G/A | AA | No | No |
-Only in persons with rs704180 risk alleles, see Ref. (Nelson et al., 2016a)
Table 3.
Clinical, genetic, pathologic, and biochemical features of the present cohort (n=136), stratified by autopsy-confirmed hippocampal sclerosis of aging (HS-Aging) and advanced Alzheimer’s disease (Braak NFT stages V or VI) pathologies
| HS-Aging pathology ? → | HS-Aging no | HS-Aging yes | p-value for Braak V/VI status |
p-value for HS- Aging status |
||
|---|---|---|---|---|---|---|
| Advanced AD (Braak NFT Stages V or VI) pathology ? → |
Braak V/VI no |
Braak V/VI yes |
Braak V/VI no |
Braak V/VI yes |
||
| n | 55 | 44 | 13 | 24 | N/A | N/A |
| Age at death, Avg | 85.3 | 79.5 | 89.4 | 86.2 | 0.032 | 0.006 |
| % Female | 56.4 | 52.3 | 69.2 | 62.5 | 0.729 | 0.279 |
| % Normal cognition/clinical state | 61.8 | 7.1 | 15.4 | 0.0 | <0.001 | <0.001 |
| % Demented cognition/clinical state | 14.5 | 81.0 | 84.6 | 95.8 | <0.001 | <0.001 |
| Final MMSE Score, Avg* | 27.5 | 16.0 | 18.8 | 11.9 | <0.001 | <0.001 |
| % SLCO1A2/IAPP risk allele(s) | 29.1 | 18.2 | 16.7 | 7.2 | 0.072 | 0.992 |
| % GRN risk allele(s) | 36.4 | 43.2 | 50.0 | 29.6 | 0.296 | 0.490 |
| % TMEM106B risk allele(s) | 83.6 | 79.5 | 79.2 | 25.3 | 0.253 | 0.518 |
| % ABCC9 risk allele(s) | 25.5 | 15.9 | 30.8 | 25.0 | 0.307 | 0.472 |
| % APOE ε4 allele [+]* | 21.6 | 37.1 | 25.0 | 45.5 | 0.032 | 0.270 |
| DG: % [+] TDP-43 inclusions | 11 | 18 | 77 | 67 | 0.321 | <0.001 |
| DG: median # inclusions in [+] cases | 1.5 | 1 | 14 | 6 | ||
| CA1: % [+] TDP-43 inclusions | 24 | 18 | 85 | 75 | 0.956 | <0.001 |
| CA1: median # inclusions in [+] cases | 1 | 1 | 13 | 15.5 | ||
| CA1 TDP-43 neurites, median | 1.05E-03 | 1.06E-03 | 3.17E-02 | 1.05E-02 | 0.859 | <0.001 |
| Sub: % [+] TDP-43 inclusions | 11 | 18 | 77 | 67 | 0.526 | <0.001 |
| Sub: median # inclusions in [+] cases | 2 | 3 | 15 | 16.5 | ||
| Sub TDP-43 neurites, median | 1.04E-03 | 1.79E-03 | 1.41E-02 | 5.96E-03 | 0.807 | <0.001 |
| % with Frontal Cx TDP-43 | 1.8 | 0 | 8.3 | 4.1 | 0.441 | 0.002 |
| T4 fg/mg brain tissue, Avg | 195.8 | 183.44 | 182.7 | 211.89 | 0.984 | 0.265 |
| T3 fg/mg brain tissue, Avg | 7.58 | 7.59 | 7.83 | 7.91 | 0.885 | 0.654 |
| Average of T3/T4 ratio, Avg | 0.039 | 0.041 | 0.043 | 0.037 | 0.861 | 0.721 |
| PMI (hrs), Avg | 4.5 | 5.0 | 4.3 | 5.8 | 0.051 | 0.633 |
-APOE genotype was available on 120/136 (88.2%) of the included subjects, and final MMSE scores were available for 126/136 (92.6%) of subjects.
N/A: Not applicable.
Table 4.
Clinical, genetic, pathologic, and biochemical features of the present cohort (n=136), stratified by whether or not triiodothyronine/thyroxine (T3/T4) ratio is <0.03
| Brain parenchymal T3/T4 ratio |
p-value: T3/T4 <0.03 vs >0.03 |
||
|---|---|---|---|
| >0.03 | <0.03 (lowest quintile) |
||
| n | 109 | 27 | N/A |
| Age at death, Avg | 83.6 | 85.3 | 0.487 |
| % Female | 58.7 | 51.9 | 0.519 |
| % Normal cognition/clinical state | 29.6 | 26.9 | 0.785 |
| % Demented cognition/clinical state | 56.5 | 57.7 | 0.911 |
| MMSE, Avg* | 21.0 | 18.1 | 0.189 |
| % SLCO1A2/IAPP risk allele(s) | 20.2 | 40.7 | 0.026 |
| % GRN risk allele(s) | 41.3 | 40.7 | 0.959 |
| % TMEM106B risk allele(s) | 81.7 | 88.9 | 0.567 |
| % ABCC9 risk allele(s) | 23.9 | 18.5 | 0.554 |
| % APOE ε4 allele [+1* | 31.6 | 28 | 0.73 |
| % Braak NFT stage V or VI* | 48.6 | 55.6 | 0.519 |
| % HS-Aging | 22.9 | 44.4 | 0.025 |
| DG: % [+] TDP-43 inclusions | 27 | 41 | 0.227 |
| DG: median # inclusions in [+] cases | 2 | 6 | |
| CA1: % [+] TDP-43 inclusions | 34 | 48 | 0.499 |
| CA1: median # inclusions in [+] cases | 5 | 13 | |
| CA1 TDP-43 neurites, median | 1.44E-03 | 1.52E-03 | 0.361 |
| Sub: % [+] TDP-43 inclusions | 27 | 41 | 0.569 |
| Sub: median # inclusions in [+] cases | 6 | 17 | |
| Sub TDP-43 neurites, median | 2.42E-03 | 1.15E-03 | 0.376 |
| % with Frontal Cx TDP-43 | 4.6 | 7.4 | 0.625 |
| T4 fg/mg brain tissue, Avg | 188.51 | 213.08 | N/A |
| T3 fg/mg brain tissue, Avg | 8.46 | 4.46 | N/A |
| Average of T3/T4 ratio, Avg | 0.044 | 0.021 | N/A |
| PMI (hrs), Avg | 4.8 | 4.7 | 0.648 |
-APOE genotype was available on 120/136 (88.2%) of the included subjects, and final MMSE scores were available for 126/136 (92.6%) of subjects.
N/A: Not applicable.
Thyroid hormone (T3 and T4) ELISA assays were performed on extracts from the cerebellum of each case. To show how the T3 and T4 assay results related with the manufacturer-provided measured TH samples, standard curves from separate ELISA plates are shown in Supplemental Figure 3. The rationale for using samples of cerebellum was that the cerebellum is relatively preserved from widespread cell loss in most common neurodegenerative diseases--we sought to minimize the biochemical variation that is caused by the sometimes devastating cerebral neurodegenerative pathologies, which can also be attended by compromise of the local blood-brain barrier, possibly altering detectable TH levels. To test this hypothesis preliminarily, we performed ELISA analyses on brain parenchyma from a convenience sample of 21 cases in which snap-frozen neocortical SMTG portions were also available. In these neocortical (unlike the cerebellar) samples, there was a trend for lower T3 levels in advanced AD (Braak NFT stages V/VI); Supplemental Figure 4. These data support the hypothesis that the TH values detected in the cerebellum are affected by AD pathology than the TH values detected in the neocortex, which was the reason we used the cerebellum for our assays.
Results of T3 and T4 assays for each case are displayed in Supplemental Figure 5. Note that there was a moderate but statistically robust correlation between T3 and T4 levels in this sample (r = 0. 50, p<0.0001). Further, there was no statistically significant association between TH levels detected and PMI, nor between TH levels and the duration that the samples were stored in the −80°C freezer between autopsy and experimental assay (Supplemental Figure 6); these observations indicate that systematic technical bias from these factors was not large.
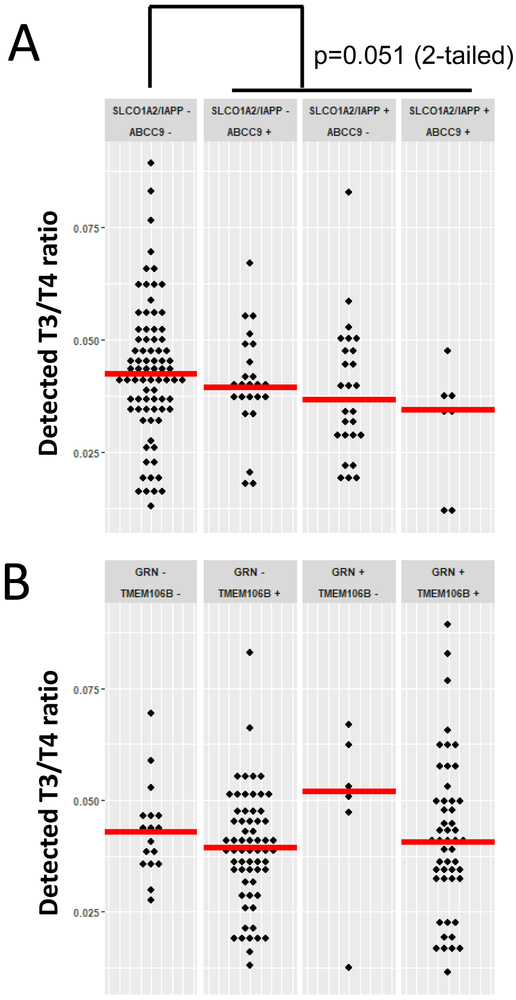
When the pathologic variables were analyzed along with the TH data, cases with either (or both) ABCC9 and SLCO1A2/IAPP risk alleles had lower-trending T3/T4 ratios than the cases that lacked either of those risk alleles (Figure 3), although this trend did not meet the statistical threshold of p<0.05 (the p-value was 0.051 using a 2-tailed statistical test). A similar trend was not observed for cases with either or both GRN or TMEM106B risk alleles. Since the T3/T4 ratio tended to be altered in association with gene variants near the brain’s TH transporter (SLCO1C1) on Chr. 12p12, we examined the association between low T3/T4 ratio and other parameters. Cases with a relatively low ratio of T3/T4 (<0.03 was chosen as a threshold to indicate low T3/T4 ratio, because this corresponded to the lowest quintile) were more likely to have HS-Aging pathology versus cases with >0.03 T3/T4 ratio (p=0.025 using a 2-tailed statistical test), as shown in Table 4. There was, however, no detected association between T3/T4 ratio and pathology across the entire spectrum of T3/T4 ratios. The association between low T3/T4 ratio and HS-Aging pathology was not attributable to clinical dementia status (independent of pathology), because the average T3/T4 ratio for subjects with all-cause dementia before death was actually higher overall than for non-demented subjects: for demented subjects (n=82) the average T3/T4 ratio was 0.043 whereas the average T3/T4 ratio for subjects cognitively normal before death (n=41) was 0.039. Further, there was no detectable correlation between T3/T4 ratios and final MMSE scores (data not shown).
Figure 3. The association between genetic risk factors and T3/T4 ratio.
A. Cases stratified by status of SLCO1A2/IAPP (rs12301085, a proxy SNP for rs73069071 which was identified as a risk allele by Roostaei and colleagues (Roostaei et al., 2016)) and ABCC9 (rs704180) genotypes. Note that there is a trend for decreased T3/T4 levels in cases in the risk allele variant of these Chr. 12p12 SNPs, with overall comparison of cases without, versus with, a risk allele (p=0.051 applying a 2-tailed test). B. Cases stratified by status of GRN (rs5848) and TMEM106B (rs1990622) genotypes. There was no detected differential variation in T3/T4 associated with these genotypes. Horizontal bars are mean values.
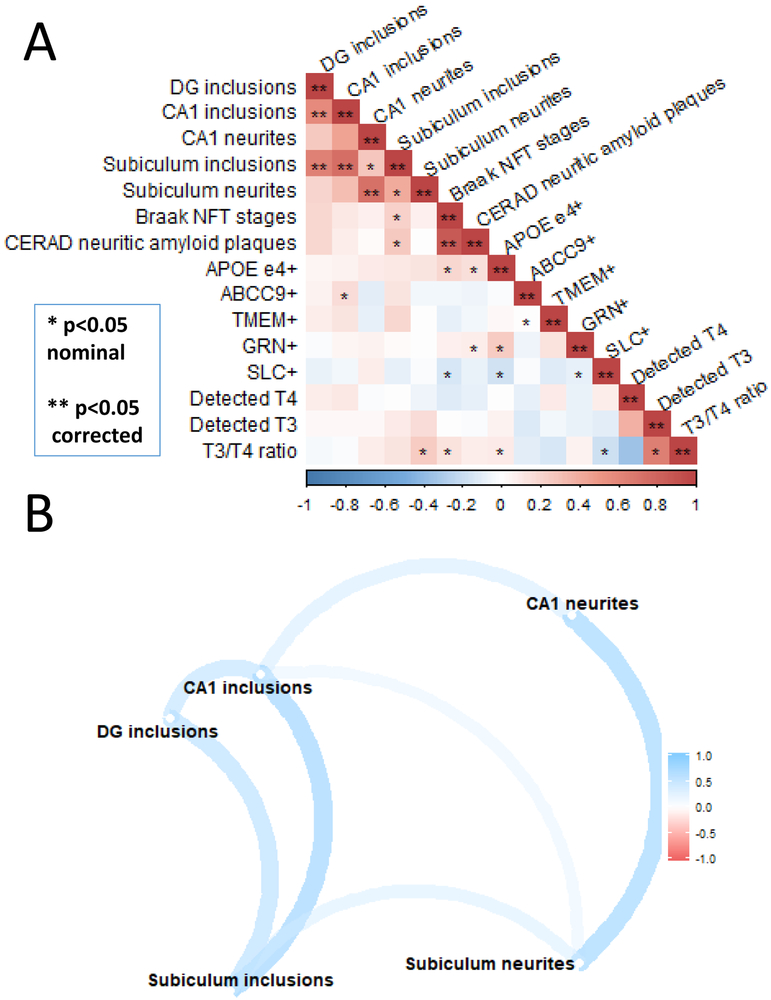
We next analyzed how pathologic and genetic parameters were correlated with each other across the entire cohort of 136 subjects (Figure 4). The strong correlations (depicted with “**” in Figure 4A) were mostly as expected – for example, cases with high densities of neuritic amyloid plaques also tended to be the same cases with high Braak NFT stages. There were numerous correlations that met criteria for nominal statistical significance, i.e., p-value below 0.05 before but not after Bonferroni-correcting for the multiple comparisons. These are depicted in Figure 4A with a “*” character. Intriguing nominally significant correlations included correlations between T3/T4 ratio and subiculum TDP-43 immunoreactive neurites, Braak NFT stages, APOE status, and SLC01A2/IAPP SNP status. The TDP-43 pathologic subtypes also tended to be correlated with each other in this cohort, as depicted via computational illustration in Figure 4B. More specifically, cases with intracytoplasmic TDP-43 inclusions in one portion of the hippocampal formation (dentate gyrus, CA1, or subiculum), also tended to have them in other anatomic areas, whereas, those with neuritic TDP-43 proteinopathy in the subiculum tended also to have the phospho-TDP-43 immunoreactive neurites – more so than the inclusions – in the hippocampal CA1 field.
Figure 4. Correlation between various genetic, pathologic, and brain thyroid hormone-related parameters among the 136 research subjects included in the current study.
A. This chart conveys the strength of correlation (blue positive, red negative) for various parameters across all the research subjects included in the current study. *−p<0.05 only before correction for multiple testing (i.e., only nominally statistically significant); **−p<0.05 after correction for multiple testing. B. A computationally-produced assessment of correlation between TDP-43 pathology subtypes in various anatomical regions of the hippocampal formation. There is a tendency for higher correlation between a particular TDP-43 pathology subtype (neuritic or intraneuronal pathology), rather than within a particular area – thus, the neuritic pathology in the subiculum is more correlated with neuritic pathology in CA1 than it is with intraneuronal pathology in the subiculum. Also note, the neuritic TDP-43 pathology correlates better with cerebrovascular pathologies than the intraneuronal TDP-43 pathology does.
Discussion
To explore evidence of disease-driving mechanisms relevant to TDP-43 proteinopathy in aged human brains, a multimodal (clinical, pathologic, genetic, and biochemical) data set was generated and analyzed from volunteers participating in a well-characterized autopsy cohort. As in another high-quality community-based cohort (Nag et al., 2018), slightly under 50% of the cases in the present study were positive for TDP-43 inclusions. A focal point of the present study was T3 and T4 levels in brain (cerebellar) parenchyma that were analyzed using ELISA assays. Patients with ABCC9 or SLCO1A2/IAPP (but not GRN or TMEM106B) gene variants tended to have lower T3/T4 ratios (compatible with the hypothesis that these gene variants alter TH processing), and, cases with low ratios of T3/T4 (lowest quintile) had a relatively high likelihood of manifesting HS-Aging pathology. Collectively, these observations are compatible with the hypothesis that genetic factors modulate brain TH hormone levels in advanced age, which may in turn affect a person’s risk for manifesting HS-Aging/TDP-43 pathology (see summary schematic, Supplemental Figure 7).
For this study, data from 136 study participants were analyzed, and there were relatively many analytic parameters. Most of the significance tests returned calculated nominal (not corrected for family-wise error rate) probabilities of p>0.05 and even the “statistically significant” results were mostly p>0.01. The lack of robust association between genotypes and TDP-43 pathology in this study – particularly for GRN and TMEM106B risk alleles that have been replicated by different labs in association with HS-Aging and TDP-43 pathology (Dickson et al., 2015; Lu et al., 2014; Murray et al., 2014; Rutherford et al., 2012; Satoh et al., 2014; Yu et al., 2015) – underscores the sample size limitation. Because of these results and the overall study design, the present work can be characterized as exploratory, and the possibilities of false-negative and false-positive results are substantial. Thus, these findings will require additional observations and replication. Nonetheless, we note that the sample size was indeed large enough to highlight some interesting and novel associations between genetics, pathology, and biochemical parameters. Other potential technical considerations apply. Whereas we found no significant correlation between ELISA-detected TH levels and either PMI or the duration the samples were stored in −80°C freezers, there are numerous other potential sources of variation, including the research subjects’ agonal conditions, drug exposures, gray/white matter ratio of the tissue used for extraction, etc. Notably, T3 levels can be decreased in serum and peripheral tissues of persons with nonthyroidal medical illness (Arem et al., 1993; Kaplan et al., 1982), and these common illnesses of aging are potential confounders. A smaller prior study (total n=24) reported that T3 (but not T4) levels were decreased in prefrontal cortical brain tissue in advanced AD (Davis et al., 2008), which is compatible with our finding in temporal neocortical tissue. Otherwise, the brain T3/T4 ratio in the present study was decreased in HS-Aging cases, but not in cases with advanced AD pathology or in clinically demented subjects overall. We emphasize that it is not a safe assumption that all brain areas have the same TH regulation; addressing this issue will require more work in the future. We also acknowledge technical sources of variation that pertain to digital pathologic methods, which we have discussed before (Attems et al., 2014; Bachstetter et al., 2015; Neltner et al., 2014; Neltner et al., 2012): for example, there are potential pitfalls related to tissue sampling, fixation duration, staining parameters, standardization, calibration, thresholding, manual region of interest selection, and various artifacts. We attempted to minimize, standardize, or control for these factors as much as possible. In the current study, the digital pathologic analyses were limited to the hippocampal formation, and the observed relationships may or may not have implications about other brain regions. Finally, the UK-ADC autopsy cohort sample was drawn from a community-based group of research volunteers who mostly enrolled while cognitively intact; however, the neuropathologic findings are not comparable to an epidemiologic cohort with regard to ethnoracial or socioeconomic characteristics (Ighodaro et al., 2017).
Despite the notes of caution, the present study also had strengths attributable to the multiple types of data that were incorporated. The different parameters enabled discovery of associations that could not be readily detected outside of a human autopsy cohort. The aged human brain is an extremely complicated milieu, so, factoring in different aspects of that complexity may be necessary to better understand how the various factors may interact. An asset in these studies was the generation and analysis of textured numerical (as opposed to dichotomous, or ordinal) parameters in both pathologic and biochemical domains. Systematic biases inherent to the study design did not relate to the researchers’ a priori hypotheses: in the genetic assessment as well as the TH ELISA detection, the workers were blinded to case identification and comorbid pathologies. The present study described correlative phenomena relevant to credible hypotheses previously linked to HS-Aging and TDP-43 (Figure 1 and Supplemental Figure 7). Specifically, those pathologies may be affected by TH dysregulation in the brain. Genetic risk factors may interact at discrete nodes along those pathways.
Thyroid derived hormones are iodinated signalling molecules that subserve evolutionarily ancient biologic functions (Crockford, 2009; Holzer et al., 2017; Taylor and Heyland, 2017), and TH plays roles in vertebrate neurodevelopment and brain disease (Bernal, 2005; Liu and Brent, 2018). Lack of TH in utero and in early life causes severe cognitive impairment, with extensive white matter pathology (Rosman, 1972). In later life, hyperthyroidism has been often associated with cognitive decline (Akintola et al., 2015; Chaker et al., 2018; Chaker et al., 2016; Moon, 2016; Moon et al., 2014; Rieben et al., 2016; Tan and Vasan, 2009; Wu et al., 2016; Yeap et al.,2012). A recent PubMed search using “(thyroid or thyroxine) and (Alzheimer’s or dementia)” returns >600 published papers. Whereas approximately 20% of persons are affected by hypothyroidism and ~5% by hyperthyroidism in advanced age (Brenowitz et al., 2018; Verburg et al., 2017), there is currently incomplete understanding as to how blood and brain T3 and T4 levels correspond in older individuals, and, ultimately, the mechanisms are still unknown as to how TH dysregulation could contribute to cognitive impairment in aging.
There is an emerging awareness that astrocytes are relevant to both TH homeostasis and TDP-43 pathology. Astrocytes import T4 from blood, convert T4 into T3, and then deliver T3 to neurons (Heuer and Visser, 2009; Morte and Bernal, 2014). TH also contributes to astrocyte development and differentiation (Bernal, 2005; Das et al., 2018). Multiple labs have reported data implicating astrocytes as a potential nexus of hippocampal TDP-43 pathology. This is demonstrated in humans with Alexander disease, which is caused by a toxic upregulation of astrocytic GFAP, often producing hippocampal TDP-43 pathology (Walker et al., 2014). In hippocampal brain extracts from HS-Aging cases, abundant GFAP peptides were found in a detergent-insoluble fraction that also contained phosphorylated TDP-43 protein (Nelson et al., 2012). Further, Lin et al. described TDP-43 pathology in astrocyte foot-processes in the hippocampus (Lin et al., 2009). The roles for astrocytes in TDP-43 pathology requires additional investigation.
Because the genomic studies pointed to TH dysregulation as a potential pathogenetic feature of HS-Aging (Nelson et al., 2016a), we previously reported results of testing TH levels in human CSF from individuals with known brain pathologies. Total T3 levels were increased in CSF samples from brains with HS-Aging (but not AD) pathology relative to controls in two separate cohorts (Nelson et al., 2016a). In the current study, parenchymal T3 and T4 levels also tended to increase in HS-Aging cases. However, there was a complex association between detected brain parenchymal TH levels, genetic risk alleles, and the observed brain pathology.
In the present study, there was a tendency for persons with ABCC9 or SLCO1A2/IAPP (but not GRN or TMEM106B) risk alleles to have lower T3/T4 ratio. This result provides support for a credible hypothesis: both ABCC9 and SLCO1A2/IAPP SNPs have been associated with variation in SLCO1C1 expression (Nelson et al., 2016a), which may lead in turn to decreased incorporation of T3 hormone in the brain for a given amount of T4 hormone in blood. Providing further support for this hypothesis, brains with lower quintile of T3/T4 ratio had elevated risk for HS-Aging pathology. Prior work from multiple laboratories has indicated that TH can serve to accelerate wound healing, and lower blood TH levels lead to worse outcomes after cerebral infarction (Genovese et al., 2013; Sadana et al., 2015; Safer, 2013; Safer et al., 2005; Suda et al., 2016; Wang et al., 2017). We interpret these data collectively to indicate that T3 in the brain may stimulate or otherwise impact compensatory mechanisms, and this influence may be modified by ABCC9 and SLCO1A2/IAPP risk alleles, yet, only a subset of HS-Aging cases are detectably affected by the mechanism(s). Adding to the complexity, one of the risk SNPs (rs73069071) exists within the IAPP gene which has also been implicated in AD and TDP-43 pathologies (Despa and Decarli, 2013; Leino et al., 2017). Further research is required to delineate mechanisms relevant to neurodegenerative diseases and, ultimately, to pursue therapeutic strategies. While some of the disease-related pathways may be studied productively in non-human experimental systems, other aspects may be specific to the aged human brain.
Supplementary Material
Highlights.
Clinical, pathological, genetic, and biochemical parameters were analyzed from 136 subjects
ABCC9 and SLCO1A2 genes are near human brain thyroid hormone receptor gene
Cases with ABCC9 and SLCO1A2 trended toward lower brain T3/T4 (thyroid hormone) ratio
Cases with lowest brain T3/T4 ratios had elevated risk for hippocampal sclerosis pathology
Thyroid hormone dysregulation is implicated in HS/TDP-43 pathology but more work is required
Acknowledgements:
We are deeply grateful to all of the study participants, clinical workers, and researchers that made this study possible. Special thanks to Sonya Anderson and Ela Patel for work in biobanking and neurohistology.
Funding support
Funding included grants P30 AG028383, R01 AG057187, and R01 AG042475 from the National Institute on Aging (NIA)/National Institutes of Health (NIH).
Footnotes
Competing interests:
The authors report no competing interests.
References
- Abner EL, et al. , 2018. Diffuse Amyloid-beta Plaques, Neurofibrillary Tangles, and the Impact of APOE in Elderly Persons' Brains Lacking Neuritic Amyloid Plaques. J Alzheimers Dis. 64, 1307–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akintola AA, et al. , 2015. Subclinical hypothyroidism and cognitive function in people over 60 years: a systematic review and meta-analysis. Front Aging Neurosci. 7, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Ortiz C, Dickson DW, 2008. Neuropathology of hippocampal sclerosis. Handb Clin Neurol. 89, 569–72. [DOI] [PubMed] [Google Scholar]
- Arem R, et al. , 1993. Reduced tissue thyroid hormone levels in fatal illness. Metabolism. 42, 1102–8. [DOI] [PubMed] [Google Scholar]
- Attems J, et al. , 2014. Quantitative neuropathological assessment to investigate cerebral multi-morbidity. Alzheimers Res Ther. 6, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachstetter AD, et al. , 2015. Disease-related microglia heterogeneity in the hippocampus of Alzheimer's disease, dementia with Lewy bodies, and hippocampal sclerosis of aging. Acta Neuropathol Commun. 3, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham GW, et al. , 2014. Genome-wide association meta-analysis of neuropathologic features of Alzheimer's disease and related dementias. PLoS Genet. 10, e1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal J, 2005. Thyroid hormones and brain development. Vitam Horm. 71, 95–122. [DOI] [PubMed] [Google Scholar]
- Bernal J, et al. , 2015. Thyroid hormone transporters--functions and clinical implications. Nat Rev Endocrinol. 11, 406–17. [DOI] [PubMed] [Google Scholar]
- Brenowitz WD, et al. , 2018. Treated hypothyroidism is associated with cerebrovascular disease but not Alzheimer's disease pathology in older adults. Neurobiol Aging. 62, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz WD, et al. , 2014. Hippocampal sclerosis of aging is a key Alzheimer's disease mimic: clinical-pathologic correlations and comparisons with both alzheimer's disease and non-tauopathic frontotemporal lobar degeneration. J Alzheimers Dis. 39, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker L, et al. , 2018. Age-dependent association of thyroid function with brain morphology and microstructural organization: evidence from brain imaging. Neurobiol Aging. 61, 44–51. [DOI] [PubMed] [Google Scholar]
- Chaker L, et al. , 2016. Thyroid function and the risk of dementia: The Rotterdam Study. Neurology. 87, 1688–1695. [DOI] [PubMed] [Google Scholar]
- Crockford SJ, 2009. Evolutionary roots of iodine and thyroid hormones in cell-cell signaling. Integr Comp Biol. 49, 155–66. [DOI] [PubMed] [Google Scholar]
- Cykowski MD, et al. , 2017. Hippocampal Sclerosis in Older Patients: Practical Examples and Guidance with a FOcus on Cerebral Age-Related TDP-43 with Sclerosis. Arch Pathol Lab Med. 141,1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Introno A, et al. , 2006. Current knowledge of chromosome 12 susceptibility genes for late-onset Alzheimer's disease. Neurobiol Aging. 27, 1537–53. [DOI] [PubMed] [Google Scholar]
- Das M, et al. , 2018. Thyroid Hormone and Astrocyte Differentiation. Vitam Horm. 106, 283–312. [DOI] [PubMed] [Google Scholar]
- Davidson YS, et al. , 2011. TDP-43 pathological changes in early onset familial and sporadic Alzheimer's disease, late onset Alzheimer's disease and Down's syndrome: association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol. 122, 703–13. [DOI] [PubMed] [Google Scholar]
- Davis JD, et al. , 2008. Thyroid hormone levels in the prefrontal cortex of post-mortem brains of Alzheimer's disease patients. Curr Aging Sci. 1, 175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despa F, Decarli C, 2013. Amylin: what might be its role in Alzheimer's disease and how could this affect therapy? Expert Rev Proteomics. 10, 403–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, et al. , 2010. Common variant in GRN is a genetic risk factor for hippocampal sclerosis in the elderly. Neurodegener Dis. 7, 170–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, et al. , 1994. Hippocampal sclerosis: a common pathological feature of dementia in very old (> or = 80 years of age) humans. Acta Neuropathol. 88, 212–21. [DOI] [PubMed] [Google Scholar]
- Dickson DW, et al. , 2015. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 85, 1354–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, et al. , 2018. Detergent Insoluble Proteins and Inclusion Body-Like Structures Immunoreactive for PRKDC/DNA-PK/DNA-PKcs, FTL, NNT, and AIFM1 in the Amygdala of Cognitively Impaired Elderly Persons. J Neuropathol Exp Neurol. 77, 21–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese T, et al. , 2013. Post-ischaemic thyroid hormone treatment in a rat model of acute stroke. Brain Res. 1513, 92–102. [DOI] [PubMed] [Google Scholar]
- Heuer H, Visser TJ, 2009. Minireview: Pathophysiological importance of thyroid hormone transporters. Endocrinology. 150, 1078–83. [DOI] [PubMed] [Google Scholar]
- Holzer G, et al. , 2017. Evolution of ligands, receptors and metabolizing enzymes of thyroid signaling. Mol Cell Endocrinol. 459, 5–13. [DOI] [PubMed] [Google Scholar]
- Ighodaro ET, et al. , 2017. Challenges and Considerations Related to Studying Dementia in Blacks/African Americans. J Alzheimers Dis. 60, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J, et al. , 2005. Thyroid hormone transporters in health and disease. Thyroid. 15, 757–68. [DOI] [PubMed] [Google Scholar]
- Jicha GA, et al. , 2012. Preclinical AD Workgroup staging: pathological correlates and potential challenges. Neurobiol Aging. 33, 622 e1–622 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, et al. , 2016. Updated TDP-43 in Alzheimer's disease staging scheme. Acta Neuropathol. 131, 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, et al. , 2008. Abnormal TDP-43 immunoreactivity in AD modifies clinicopathologic and radiologic phenotype. Neurology. 70, 1850–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MM, et al. , 1982. Prevalence of abnormal thyroid function test results in patients with acute medical illnesses. Am J Med. 72, 9–16. [DOI] [PubMed] [Google Scholar]
- Katsumata Y, et al. , 2018. Dichotomous scoring of TDP-43 proteinopathy from specific brain regions in 27 academic research centers: associations with Alzheimer's disease and cerebrovascular disease pathologies. Acta Neuropathol Commun. 6, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata Y, et al. , 2017. Gene-based association study of genes linked to hippocampal sclerosis of aging neuropathology: GRN, TMEM106B, ABCC9, and KCNMB2. Neurobiol Aging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kero M, et al. , 2018. Hippocampal Sclerosis in the Oldest Old: A Finnish Population-Based Study. J Alzheimers Dis. 63, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leino M, et al. , 2017. Transactive DNA Binding Protein 43 Rather Than Other Misfolded Proteins in the Brain is Associated with Islet Amyloid Polypeptide in Pancreas in Aged Subjects with Diabetes Mellitus. J Alzheimers Dis. 59, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverenz JB, et al. , 2002. Clinical and neuropathological characteristics of hippocampal sclerosis: a community-based study. Arch Neurol. 59, 1099–106. [DOI] [PubMed] [Google Scholar]
- Lin WL, et al. , 2009. Transactivation response DNA-binding protein 43 microvasculopathy in frontotemporal degeneration and familial Lewy body disease. J Neuropathol Exp Neurol. 68, 1167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Brent GA, 2018. Thyroid hormone and the brain: Mechanisms of action in development and role in protection and promotion of recovery after brain injury. Pharmacol Ther. 186, 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu RC, et al. , 2014. TMEM106B and APOE polymorphisms interact to confer risk for late-onset Alzheimer's disease in Han Chinese. J Neural Transm (Vienna). 121, 283–7. [DOI] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ, 2015. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 31, 3555–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafrica F, Fodale V, 2008. Thyroid function, Alzheimer's disease and postoperative cognitive dysfunction: a tale of dangerous liaisons? J Alzheimers Dis. 14, 95–105. [DOI] [PubMed] [Google Scholar]
- Mayeux R, et al. , 2002. Chromosome-12 mapping of late-onset Alzheimer disease among Caribbean Hispanics. Am J Hum Genet. 70, 237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAninch EA, Bianco AC, 2014. Thyroid hormone signaling in energy homeostasis and energy metabolism. Ann N Y Acad Sci. 1311, 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montine TJ, et al. , 2012. National Institute on Aging-Alzheimer's Association guidelines for the neuropathologic assessment of Alzheimer's disease: a practical approach. Acta Neuropathol. 123, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JH, 2016. Endocrine Risk Factors for Cognitive Impairment. Endocrinol Metab (Seoul). 31, 185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JH, et al. , 2014. Lower-but-normal serum TSH level is associated with the development or progression of cognitive impairment in elderly: Korean Longitudinal Study on Health and Aging (KLoSHA). J Clin Endocrinol Metab. 99, 424–32. [DOI] [PubMed] [Google Scholar]
- Morte B, Bernal J, 2014. Thyroid hormone action: astrocyte-neuron communication. Front Endocrinol (Lausanne). 5, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullur R, et al. , 2014. Thyroid hormone regulation of metabolism. Physiol Rev. 94, 355–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray ME, et al. , 2014. Differential clinicopathologic and genetic features of late-onset amnestic dementias. Acta Neuropathol. 128, 411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, et al. , 2018. TDP-43 pathology in anterior temporal pole cortex in aging and Alzheimer's disease. Acta Neuropathol Commun. 6, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, et al. , 2017. TDP-43 pathology and memory impairment in elders without pathologic diagnoses of AD or FTLD. Neurology. 88, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2018. The Amygdala as a Locus of Pathologic Misfolding in Neurodegenerative Diseases. J Neuropathol Exp Neurol. 77, 2–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2010. Modeling the association between 43 different clinical and pathological variables and the severity of cognitive impairment in a large autopsy cohort of elderly persons. Brain Pathol. 20, 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2014. ABCC9 gene polymorphism is associated with hippocampal sclerosis of aging pathology. Acta Neuropathol. 127, 825–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2007. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles "do count" when staging disease severity. J Neuropathol Exp Neurol. 66, 1136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2015a. ABCC9/SUR2 in the brain: Implications for hippocampal sclerosis of aging and a potential therapeutic target. Ageing Res Rev. 24, 111–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2016a. Genomics and CSF analyses implicate thyroid hormone in hippocampal sclerosis of aging. Acta Neuropathol. 132, 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2011. Hippocampal sclerosis in advanced age: clinical and pathological features. Brain. 134, 1506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2013. Hippocampal sclerosis of aging, a prevalent and high-morbidity brain disease. Acta Neuropathol. 126, 161–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2016b. "New Old Pathologies": AD, PART, and Cerebral Age-Related TDP-43 With Sclerosis (CARTS). J Neuropathol Exp Neurol. 75, 482–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, et al. , 2015b. Reassessment of risk genotypes (GRN, TMEM106B, and ABCC9 variants) associated with hippocampal sclerosis of aging pathology. J Neuropathol Exp Neurol. 74, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neltner JH, et al. , 2014. Arteriolosclerosis that affects multiple brain regions is linked to hippocampal sclerosis of ageing. Brain. 137, 255–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neltner JH, et al. , 2012. Digital pathology and image analysis for robust high-throughput quantitative assessment of Alzheimer disease neuropathologic changes. J Neuropathol Exp Neurol. 71, 1075–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho K, et al. , 2016. Hippocampal Sclerosis of Aging, a Common Alzheimer's Disease 'Mimic': Risk Genotypes are Associated with Brain Atrophy Outside the Temporal Lobe. J Alzheimers Dis. 52, 373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CG, 2006. KATP channels as molecular sensors of cellular metabolism. Nature. 440, 470–6. [DOI] [PubMed] [Google Scholar]
- Nichols CG, et al. , 2013. KATP channels and cardiovascular disease: suddenly a syndrome. Circ Res. 112, 1059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao WC, et al. , 2011. Hippocampal Sclerosis in the Elderly: Genetic and Pathologic Findings, Some Mimicking AlzheimerDisease Clinically. Alzheimer Dis Assoc Disord. 25, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericak-Vance MA, et al. , 1997. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 278, 1237–41. [PubMed] [Google Scholar]
- Rieben C, et al. , 2016. Subclinical Thyroid Dysfunction and the Risk of Cognitive Decline: a Meta-Analysis of Prospective Cohort Studies. J Clin Endocrinol Metab. 101, 4945–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostaei T, et al. , 2016. Genome-wide interaction study of brain beta-amyloid burden and cognitive impairment in Alzheimer's disease. Mol Psychiatry. 22, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosman NP, 1972. The neuropathology of congenital hypothyroidism. Adv Exp Med Biol. 30, 337–66. [PubMed] [Google Scholar]
- Rutherford NJ, et al. , 2012. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology. 79, 717–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadana P, et al. , 2015. Anti-edema action of thyroid hormone in MCAO model of ischemic brain stroke: Possible association with AQP4 modulation. J Neurol Sci. 354, 37–45. [DOI] [PubMed] [Google Scholar]
- Safer JD, 2013. Thyroid hormone and wound healing. J Thyroid Res. 2013, 124538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safer JD, et al. , 2005. Topical thyroid hormone accelerates wound healing in mice. Endocrinology. 146, 4425–30. [DOI] [PubMed] [Google Scholar]
- Sampaolo S, et al. , 2005. Increased cerebrospinal fluid levels of 3,3',5'-triiodothyronine in patients with Alzheimer's disease. J Clin Endocrinol Metab. 90, 198–202. [DOI] [PubMed] [Google Scholar]
- Satoh J, et al. , 2014. TMEM106B expression is reduced in Alzheimer's disease brains. Alzheimers Res Ther. 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt FA, et al. , 2012. University of Kentucky Sanders-Brown Healthy Brain Aging Volunteers: Donor Characteristics, Procedures, and Neuropathology. Curr Alzheimer Res. 9, 724–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, et al. , 1999. Further evidence linking late-onset Alzheimer disease with chromosome 12. JAMA. 281, 513–4. [DOI] [PubMed] [Google Scholar]
- Smith VD, et al. , 2017. Overlapping but distinct TDP-43 and tau pathologic patterns in aged hippocampi. Brain Pathol. 28, 264–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda S, et al. , 2016. Low free triiodothyronine predicts poor functional outcome after acute ischemic stroke. J Neurol Sci. 368, 89–93. [DOI] [PubMed] [Google Scholar]
- Tan ZS, Vasan RS, 2009. Thyroid function and Alzheimer's disease. J Alzheimers Dis. 16, 503–7. [DOI] [PubMed] [Google Scholar]
- Taylor E, Heyland A, 2017. Evolution of thyroid hormone signaling in animals: Non-genomic and genomic modes of action. Mol Cell Endocrinol. 459, 14–20. [DOI] [PubMed] [Google Scholar]
- Trentin AG, 2006. Thyroid hormone and astrocyte morphogenesis. J Endocrinol. 189, 189–97. [DOI] [PubMed] [Google Scholar]
- Trieu T, et al. , 2018. Risk factors of hippocampal sclerosis in the oldest old: The 90+ Study. Neurology. 91, e1788–e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu T. e. a., 2018. Risk Factors of Hippocampal Sclerosis in the Oldest Old: The 90+ Study. Neurology. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Deure WM, et al. , 2008. Thyroid hormone transport and metabolism by organic anion transporter 1C1 and consequences of genetic variation. Endocrinology. 149, 5307–14. [DOI] [PubMed] [Google Scholar]
- Van Langenhove T, et al. , 2012. The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann Med. 44, 817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verburg FA, et al. , 2017. Prevalence of thyroid disorders in elderly people in Germany. A screening study in a country with endemic goitre. Nuklearmedizin. 56, 9–13. [DOI] [PubMed] [Google Scholar]
- Walker AK, et al. , 2014. Astrocytic TDP-43 pathology in Alexander disease. J Neurosci. 34, 6448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. , 2017. Low T3 levels as a predictor marker predict the prognosis of patients with acute ischemic stroke. Int J Neurosci. 127, 559–566. [DOI] [PubMed] [Google Scholar]
- Wu Y, et al. , 2016. Higher FT4 or TSH below the normal range are associated with increased risk of dementia: a meta-analysis of 11 studies. Sci Rep. 6, 31975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap BB, et al. , 2012. Higher free thyroxine levels predict increased incidence of dementia in older men: the Health in Men Study. J Clin Endocrinol Metab. 97, E2230–7. [DOI] [PubMed] [Google Scholar]
- Yu L, et al. , 2015. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 84, 927–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarow C, et al. , 2008. Understanding hippocampal sclerosis in the elderly: epidemiology, characterization, and diagnostic issues. Curr Neurol Neurosci Rep. 8, 363–70. [DOI] [PubMed] [Google Scholar]
- Zarow C, et al. , 2012. Prevalence, laterality, and comorbidity of hippocampal sclerosis in an autopsy sample. Brain Behav. 2, 435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.