Abstract
Background:
While the genetic contribution to obesity is well-established, few studies have examined how genetic variants influence standardized body mass index (BMIz) in Hispanics/Latinos, especially across childhood and adolescence.
Objectives:
We estimated the effect of established BMIz loci in Chilean children of the Santiago Longitudinal Study [SLS].
Methods:
We examined associations with BMIz at age 10 for 15 loci previously identified in European children. For significant loci, we performed association analyses at ages 5 and 16 years, for which we have smaller sample sizes. We tested associations of unweighted genetic risk scores (GRS) for previously-identified tag variants (GRS_EUR) and from the most significant variants in SLS at each locus (GRS_SLS).
Results:
We generalized five variants at age 10 (P<0.05 and directionally consistent), including rs543874 that reached Bonferroni-corrected significance. The effect on BMIz was greatest at age 10 for all significant loci, except FTO, which exhibited an increase in effect from ages 5 to 16. Both GRSs were associated with BMIz (P<0.0001), but GRS_SLS explained a much greater proportion of the variation (13.63%).
Conclusion:
Our results underscore the importance of conducting genetic investigations across life stages and selecting ancestry appropriate tag variants in future studies for disease prediction and clinical evaluation.
Keywords: BMI, pediatric cohort, obesity, genetic risk score, Latin America
INTRODUCTION
Obesity is a leading risk factor for cardiovascular disease (CVD), and its global prevalence has more than doubled since the 1980’s [1]. Latin Americans are at high risk [2, 3], with rapid expansion of obesity and CVD in the context of accelerating urbanization. This study’s participants are from Chile, where dramatic socioeconomic improvement accompanied by changes in diet, physical activity and socio-cultural environment has resulted in obesity prevalence among the highest in the world [4, 5]. Indeed, according to Chile’s Third National Health Survey, 74.2% of individuals over 15 years of age are currently overweight or obese [6]. Also, Hispanic/Latinos transition from normal weight to obesity at younger ages than age-matched members of other race-ethnic groups [7].
Genetic factors are a well-established contributor to obesity risk [8, 9]. Although, most genome-wide association studies (GWAS) of obesity-related traits have largely been in adult, European descent (EUR) populations [8]. Recent studies suggest that genetic effects on obesity-related phenotypes, like body mass index (BMI), may differ across life stages [10, 11]. To date, few genetic studies have focused on the effects of genetics across childhood [9, 11, 12], or Hispanic/Latinos [12], including Chileans, although they shoulder a high burden of obesity. These observations highlight the need to characterize the relevance of established genetic loci from infancy into adulthood. Also, genetic risk scores (GRS) are used to summarize phenotype – genotype relationships and for prediction of disease and clinical evaluation [13], and have been used to explore the genetic contribution to weight, weight gain, and BMIz in infants and young EUR children less than 5 years old [14, 15]. Yet, the applicability and appropriateness of using EUR tag variants in GRS’ constructed in ancestrally diverse populations has not been widely evaluated.
Using data available in the Santiago Longitudinal Study [SLS] of Chilean children, we aimed to 1) estimate the association between established BMIz genetic risk markers first identified in European children; 2) determine if genetic effects for significantly-associated single nucleotide polymorphisms (SNPs) differ by age in our SLS cohort; and, 3) investigate the association of BMIz with cumulative GRS derived using EUR tag SNPs and potential SLS population-specific tag SNPs.
MATERIALS AND METHODS
Santiago Longitudinal Study (SLS)
SLS participants were recruited during infancy in Santiago, Chile and have been followed through childhood and adolescence into early adulthood. The parent nutrition study, which began as an iron deficiency anemia (IDA) preventive trial, has been described elsewhere [16]. Briefly, from 1991 to 1996, 1798 infants with no major health problems, born at term and weighing ≥ 3.0 kg, participated in a randomized trial of iron supplementation to prevent IDA, a neurophysiology study of those with IDA, and nonanemic controls. Children were followed up after infancy at ages 5, 10, and 16 years of age. [17, 18] The children’s families were generally lower/middle-class, literate, and of mixed American Indian and Spanish descent. [16, 19] All parents of participants provided informed consent at all visits, and participants themselves provided assent at age 16. The current study has been approved by Institutional Review Boards (IRBs) at the University of California at San Diego, University of Michigan, University of North Carolina at Chapel Hill, Geisinger Health, and Institute of Nutrition and Food Technology, University of Chile.
Anthropometry and laboratory assessment
Childhood weight and height were measured in duplicate by a study nurse or physician using standard techniques at assessment at 5, 10, and 16 years old. Weight was measured to the nearest 0.1 kg using a SECA scale and height to the nearest 0.1 cm using a Holtain stadiometer. BMI was calculated as weight (kg)/height2 (m2), then transformed into Z-scores relative to CDC anthropometric reference data (2007-2010) [20]. These BMIz scores were compared with those estimated using the World Health Organization (BMIzWHO) reference data and the United Kingdom (BMIzUK) reference data for all three ages and for boys, girls, and combined sexes. Overall, the correlation (R2) reported was ≥ 98% (BMIzCDC vs. BMIzUK) and ≥ 94% (BMIzCDC vs. BMIzWHO), respectively, and highest among Age 10 group (99%) for both comparisons (Figure S1). As estimated BMIz was highly concordant across all panels, all results presented herein are for BMIzCDC.
Genotypic data and SNP Selection
We examined associations for 15 loci previously associated with BMI Z scores (BMIz) in European children aged 2 to 10 years old from Felix et al.[9] For one of the 15 loci, the previously identified tag variant was unavailable in our genotype data and no proxy SNP was available to test (R2>0.9). Therefore, we evaluated the association of BMIz at age 10 with 14 SNPs previously associated with BMIz in EUR children aged 2 to 10 years old [9] (Table 1). Illumina Infinium Multi-Ethnic Global-8 array was utilized to acquire genotypic data in unrelated participants using whole blood samples collected at the 16y follow-up visit. Quality Control (QC) included filtering on individual call rate >90%, checking for gender mismatch, relatedness, and ancestry outliers. All variants had a minor allele frequency (MAF) >0.005, Hardy-Weinberg Equilibrium (HWE; P >1x10−7), and call rate >95%.
Table 1.
Summary of association results for our Santiago Longitudinal Cohort Study (SLS) compared to previously identified effects in European descent children as reported in Felix et al (2016). SNP – single nucleotide polymorphism; CHR – chromosome; BP – base pair position; EA – effect allele; OA – other allele; EAF - effect allele frequency; β – beta coefficient; SE – standard error, P – P-value
| SLS | Felix et al. β (+/− based on EA) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nearest Gene | SNP | CHR | BP | EA/OA | EAF | Age | β | SE | P | |
| Significant Loci | ||||||||||
| SEC16B | rs543874 | 1 | 177889480 | G/A | 0.27 | 5 | 0.126 | 0.065 | 0.055 | + |
| 10 | 0.185 | 0.052 | 4.3E-04 | |||||||
| 16 | 0.164 | 0.067 | 0.015 | |||||||
| Change | 0.0014 | 0.0019 | 0.467 | |||||||
| ADCY3 | rs11676272 | 2 | 25141538 | G/A | 0.34 | 5 | 0.064 | 0.061 | 0.297 | + |
| 10 | 0.099 | 0.049 | 0.043 | |||||||
| 16 | 0.061 | 0.063 | 0.331 | |||||||
| Change | −0.0005 | 0.0018 | 0.795 | |||||||
| FAIM2 | rs7132908 | 12 | 50263148 | A/G | 0.22 | 5 | 0.033 | 0.070 | 0.639 | + |
| 10 | 0.116 | 0.058 | 0.046 | |||||||
| 16 | 0.059 | 0.072 | 0.410 | |||||||
| Change | 0.0002 | 0.0012 | 0.936 | |||||||
| OLFM4 | rs12429545 | 13 | 54102206 | A/G | 0.32 | 5 | 0.090 | 0.059 | 0.133 | + |
| 10 | 0.100 | 0.048 | 0.037 | |||||||
| 16 | 0.042 | 0.062 | 0.496 | |||||||
| Change | −0.0017 | 0.0017 | 0.324 | |||||||
| FTO | rs1421085 | 16 | 53800954 | C/T | 0.22 | 5 | 0.060 | 0.070 | 0.391 | + |
| 10 | 0.119 | 0.055 | 0.032 | |||||||
| 16 | 0.177 | 0.071 | 0.014 | |||||||
| Change | 0.0036 | 0.0020 | 0.075 | |||||||
| Non-Significant Loci | ||||||||||
| TNNI3K | rs12041852 | 1 | 75003500 | A/G | 0.35 | 10 | 0.024 | 0.049 | 0.628 | − |
| GPR61 | rs7550711 | 1 | 110082886 | C/T | 0.98 | 10 | 0.079 | 0.219 | 0.715 | − |
| TMEM18 | rs4854349/ | 2 | 647760 | G/A | 0.87 | 10 | 0.102 | 0.072 | 0.159 | + |
| rs4854348* | ||||||||||
| GNPDA2 | rs13130484 | 4 | 45175691 | C/T | 0.61 | 10 | 0.077 | 0.048 | 0.112 | − |
| TFAP2B | rs987237 | 6 | 50803050 | G/A | 0.37 | 10 | 0.088 | 0.051 | 0.087 | + |
| ELP3 | rs13253111/ | 8 | 28060690 | A/G | 0.59 | 10 | 0.043 | 0.048 | 0.374 | + |
| rs7821358* | ||||||||||
| LMX1B | rs3829849/ | 9 | 129375338 | T/C | 0.23 | 10 | 0.063 | 0.058 | 0.272 | + |
| rs62578126* | ||||||||||
| RAB27B | rs8092503/ | 18 | 52481583 | A/G | 0.29 | 10 | −0.001 | 0.052 | 0.985 | − |
| rs8085272* | ||||||||||
| MC4R | rs6567160 | 18 | 57829135 | C/T | 0.12 | 10 | 0.005 | 0.071 | 0.943 | + |
- LD (R2) between Tag and Proxy SNPs ranged from 0.94 to 1.0.
Statistical analysis
For all SNPs positioned within the 1 MB interval (+/− 500 kb from known EUR tag SNP) of 15 established BMIz loci, we conducted linear regression of BMIz at age 10 (the age with the largest sample size). Analyses were conducted in PLINK 1.07 [21] assuming an additive genetic model, adjusted for population substructure using principal components (PCs) calculated from genome-wide data in EIGENSTRAT[22]. To correct for multiple testing, we considered SNPS that displayed a Bonferroni-corrected P <0.003 as significant. For each variant that displayed a nominally significant association (P<0.05) with BMIz at age 10, we further examined the effect of that SNP at ages 5 and 16. Additionally, sensitivity analysis was performed on our data after adjusting for random clinical trial (RCT) arm of the parent infant study based on iron supplementation (“high-iron” - 12 mg/L, “low-iron” - 2.4 mg/L, or “no-iron”). The RCT variable was not a significant predictor of BMIz. Thus, all results presented herein are without adjustment for RCT.
Genetic Effects on BMIz Across Age
For variants that are nominally associated with BMIz at age 10, we performed two additional analyses to evaluate the effect of these variants on BMIz across age. First, we tested to see if there was a significant change in effect across ages 5, 10, and 16. For this test, we performed a Spearman’s rank correlation in STATA 15.1 with nominally significant BMIz-associated SNPs across age groups 5, 10, and 16 to assess a significant trend in SNP effect size across age (P<0.05). As sample size at ages 5 and 16 were lower than at age 10 (N=577, 543, and 770, respectively), we did not formally test for heterogeneity effects between ages, and instead focus our analysis on only a monotonic trend in effect across ages. Second, we evaluated the effect of each of these loci on change in BMIz as measured by the slope of BMIz score change across ages 5 to 16. Using linear mixed model regression with an unstructured variance/covariant matrix and restricted maximum likelihood (REML) estimation, BMIz was regressed with visit age as both fixed and random effects to derive the slope of adiposity change based on BLUP (best linear unbiased predictor). Only individuals with at least two measures of BMI were included in this analysis (N=574). The resulting change in BMIz was then used as the outcome variable in the genetic association analysis, assuming an additive genetic model, and adjusting for population substructure using PCs as before. These analyses were carried out using STATA v. 15.1.
Conditional Analyses
We identified the most significant SNP positioned within the 1 MB interval (+/− 500 kb from known EUR tag SNP) of 15 established EUR GWAS findings for BMIz at age 10. For each locus where the most significant SNP identified differed from the published SNP, we performed exact conditional analysis in PLINK 1.07 [21] to determine whether or not the new SNP was independent of the established association signal (e.g. whether this SNP represented a secondary signal in the known locus or was a better tag SNP for the known locus). Signals were considered attenuated if the beta decreased by more than 10% and/or did not display nominal significance following conditional analysis.
Genetic risk score (GRS)
To explore the cumulative effects of these 15 genetic regions on childhood obesity risk, we calculated GRS to estimate the variance explained in BMIz. We estimated unweighted simple-count GRS three ways: 1) GRS_EUR: by summing the number of BMIz increasing risk alleles from the 14 known GWAS SNPs; 2) GRS_SLS: by summing the number of BMIz increasing risk alleles from the top SNPs in the 15 loci in our SLS cohort. 3) GRS_ALL: by summing the number of independent BMIz increasing alleles across all 15 loci; which included the top variant at each locus and an additional 12 EUR tag SNPs that were independent of the top 15 following conditional analyses. As the true effect size and relative significance of each variant used in our GRS estimations is not well-established, we have chosen to focus on unweighted GRS (GRS ranges from 0 to 28 for GRS_EUR, 30 for GRS_SLS, and 54 for GRS_ALL). Multiple linear regressions of BMIz and GRSs were performed adjusting for PCs to control for population stratification in STATA 15.1.
RESULTS
Summary statistics
Descriptive statistics are given in Table S1 for measurements taken at ages 5, 10, and 16 (N = 577, 770, and 545, respectively; 48% girls). The mean weight and height in boys is slightly higher compared to girls at ages 5 and 16. However, BMIz was lower in girls compared to boys at ages 5 and 10, but higher by age 16. On the contrary, obesity (≥ 95th BMI percentile) prevalence was higher in girls compared to boys at ages 5 and 16 (22.7%, 14.3% vs. 19.8%, 13.8%), but lower at age 10 (14.6% vs. 18%), however, overall obesity rate decreased from age 5 (21.2%), age 10 (16.3%) to age 16 (14.1%). In general, from ages 5 to 16 BMIz and prevalence of obesity decreased for both girls and boys as age increased relative to the CDC reference panel (Table S1).
SNP Association analysis
For one of the 15 loci, the previously identified tag variant was unavailable in our genotype data and no proxy SNP was available to test (R2>0.9). Therefore, we evaluated the association of BMIz at age 10 with 14 SNPs previously associated with BMIz in EUR children aged 2 to 10 years old [9] (Table 1). Ten of the 14 SNPs were directionally consistent with previous findings, of which five (near SEC16B, FTO, OLFM4, ADCY3, and FAIM2) reached nominal significance (P<0.05), including one (rs543874, near SEC16B) that reached Bonferroni-corrected significance (P<0.0017). While there is a lack of consensus in the field on the appropriate significance threshold for interrogation of established loci for replication/generalization, our study findings support generalization of these loci to our Chilean population. However, larger sample sizes will be needed to reach convincing genome-wide significant statistical evidence.
Genetic effect of BMIz across age
To determine if the effect of nominally significant SNPs varied across ages 5, 10, and 16, we also examined the associations with BMIz for participants with measurements taken at age 5 and 16 in addition to age 10 (Table 1, Figure S2). For all five loci, the effects were directionally consistent across age, but the magnitude of the effect varied across age with a lower effect on BMIz at both ages 5 and 16 for all loci except FTO, which displayed a significant increase in effect across increasing age (Ptrend <0.0001). Of the five loci, two reached nominal significance at age 16 (rs543874 near SEC16B and rs1421085 near FTO) (Figure S3), but none reached nominal significance at age 5. The per allele effect of the SEC16B locus was greatest at age 10 (β=0.185, P=4.3E-04) compared to both age 5 (β=0.126, P=0.055) and 16 (β=0.164, P= 0.015). Although, BMIz in our Chilean cohort is higher among 5-year-olds relative to CDC reference population, there does not appear to be a strong genetic component to this.
We assessed the association of the same five SNPs on change in BMIz across ages 5 to 16 (Table 1) but did not identify any significant associations (P>0.05). Among the SNPs tested, rs1421085 near FTO displayed the largest effects on BMIz change (β=0.004, P=0.075).
Aggregated genetic risk for BMIz
We identified the most significant SNP positioned within the 1 MB interval (+/− 500 kb from known EUR tag SNP) at each of the established 15 loci for BMIz measured at age 10 (Table 2). At each of the 15 loci, we identified a more significantly associated SNP in our SLS cohort than the tag SNP or proxy SNP for EUR children. Of these 15, seven were in low linkage disequilibrium (LD) with the previous tag SNP (R2 <0.2 and D’ <1.0 in the Central European reference panel [CEU]), one tag SNP was unavailable in our data, and two were unavailable in the CEU reference data. All 15 SNPs displayed a nominally significant association with BMIz (P<0.05), including five that displayed Bonferroni-corrected statistical significance (P<0.0017, 0.05/29 variants). We performed exact conditional analysis for our most significant variants conditioning on the known EUR tag SNP. For all 14 loci where a EUR tag SNP or proxy SNP was available, our SLS tag SNP remained nominally significant following conditional analysis (Pconditional<0.05), and four of the five remained significant after multiple test correction (Pconditional<0.0017). However, for two SNPs (rs200787218 near SEC16B and rs138716876 near ADCY3) the estimated effect of the SLS SNP was somewhat attenuated (βconditional <0.9*β) after conditional analysis, indicating that these SNPs are likely the same association signal as those identified in Felix et al[9].
Table 2.
Association results of BMIz with EUR tag SNPs compared to population-specific tag SNPs selected from the 1 MB interval of these known obesity loci in SLS. CHR – chromosome; SNP – single nucleotide polymorphism; EA – effect allele; OA – other allele; EAF - effect allele frequency; β – beta coefficient; SE – standard error, P – P-value; R2 – linkage disequilibrium (LD) for SNPs; D’ – D prime; Chile – LD between SNPs within SLS cohort; CEU – LD between SNPs in European descent population
| Nearest Gene | CHR | EUR Tag SNPs | R2 | D’ | Population-Specific Tag SNPs | Conditional Analyses# | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP/Proxy | Included in GRS | Distance | SLS | CEU | CEU | SNP | Included in GRS | EA/OA | EAF | β | SE | P | β | SE | P | ||
| TNNI3K | 1 | rs12041852 | EUR, ALL | −174105 | 0.008 | 0.042 | 1 | rs61790698 | SLS, ALL | G/A | 0.047 | 0.239 | 0.109 | 0.029 | 0.237 | 0.110 | 0.032 |
| GPR61 | 1 | rs7550711 | EUR, ALL | −348505 | 0.001 | 0.001 | 1 | rs11102023 | SLS, ALL | A/C | 0.989 | 0.635 | 0.248 | 0.010 | 0.640 | 0.248 | 0.010 |
| SEC16B | 1 | rs543874 | EUR | −58402 | 0.122 | - | - | rs200787218 | SLS, ALL | A/C | 0.328 | 0.198 | 0.051 | 1.1E-04 | 0.155 | 0.054 | 0.004 |
| ADCY3 | 2 | rs11676272 | EUR | 467182 | 0.037 | - | - | rs138716876 | SLS, ALL | T/G | 0.817 | 0.178 | 0.059 | 0.003 | 0.157 | 0.061 | 0.010 |
| GNPDA2 | 4 | rs13130484 | EUR, ALL | 384441 | 0.004 | 0.029 | 1 | rs75426894 | SLS, ALL | T/C | 0.976 | 0.388 | 0.153 | 0.012 | 0.374 | 0.153 | 0.015 |
| TFAP2B | 6 | rs987237 | EUR, ALL | −398207 | 0.051 | 0.001 | 0.079 | rs2709670 | SLS, ALL | T/G | 0.223 | 0.218 | 0.055 | 9.4E-05 | 0.245 | 0.056 | 1.5E-05 |
| FAIM2 | 12 | rs7132908 | EUR, ALL | −27580 | 0.004 | 0.022 | 0.298 | rs297935 | SLS, ALL | G/A | 0.789 | 0.174 | 0.056 | 0.002 | 0.172 | 0.056 | 0.002 |
| OLFM4 | 13 | rs12429545 | EUR, ALL | 28422 | 0.160 | 0.131 | 1 | rs1072900 | SLS, ALL | C/A | 0.498 | 0.180 | 0.046 | 1.1E-04 | 0.169 | 0.051 | 9.8E-04 |
| FTO | 16 | rs1421085 | EUR, ALL | 3771 | 0.002 | 0.052 | 1 | rs62048379 | SLS, ALL | A/C | 0.060 | 0.314 | 0.095 | 9.9E-04 | 0.306 | 0.095 | 1.3E-03 |
| MC4R | 18 | rs6567160 | EUR, ALL | 339892 | 0.003 | 0.042 | 0.278 | rs62092638 | SLS, ALL | C/T | 0.173 | 0.184 | 0.062 | 0.003 | 0.186 | 0.062 | 0.003 |
| TMEM18 | 2 | rs4854349/ | EUR, ALL | −203384 | 0.007 | 0.001 | 0.072 | rs11682609 | SLS, ALL | A/C | 0.959 | 0.323 | 0.128 | 0.012 | 0.314 | 0.128 | 0.014 |
| rs4854348* | |||||||||||||||||
| ELP3 | 8 | rs13253111/ | EUR, ALL | 175550 | 0.014 | 0.049 | 0.268 | rs10097488 | SLS, ALL | C/T | 0.741 | 0.154 | 0.054 | 0.005 | 0.150 | 0.055 | 0.006 |
| rs7821358* | |||||||||||||||||
| LMX1B | 9 | rs3829849/ | EUR, ALL | −72782 | 0.009 | 0.024 | 0.329 | rs10987410 | SLS, ALL | C/T | 0.068 | 0.267 | 0.095 | 0.005 | 0.258 | 0.095 | 0.007 |
| rs62578126* | |||||||||||||||||
| RAB27B | 18 | rs8092503/ | EUR, ALL | −276712 | 0.001 | 0.005 | 0.115 | rs12961347 | SLS, ALL | T/C | 0.535 | 0.159 | 0.046 | 4.9E-04 | 0.160 | 0.046 | 4.9E-04 |
| rs8085272* | |||||||||||||||||
| ADAM23 | 2 | rs13387838/ | 217117 | - | - | - | rs77068085 | SLS, ALL | T/C | 0.957 | 0.331 | 0.115 | 0.004 | - | - | - | |
| No Proxy | |||||||||||||||||
- LD (R2) between Tag and Proxy SNPs ranged from 0.94 to 1.0.
Conditional analyses results from PLINK, the beta coefficients for the Population-Specific Tag SNPs still remained significant after entering the model with covariates pc1 to pc5 and corresponding EUR Tag SNP for each locus. These results suggest that Population-Specific Tag SNPs indeed have an effect independent of EUR Tag SNP and other covariates
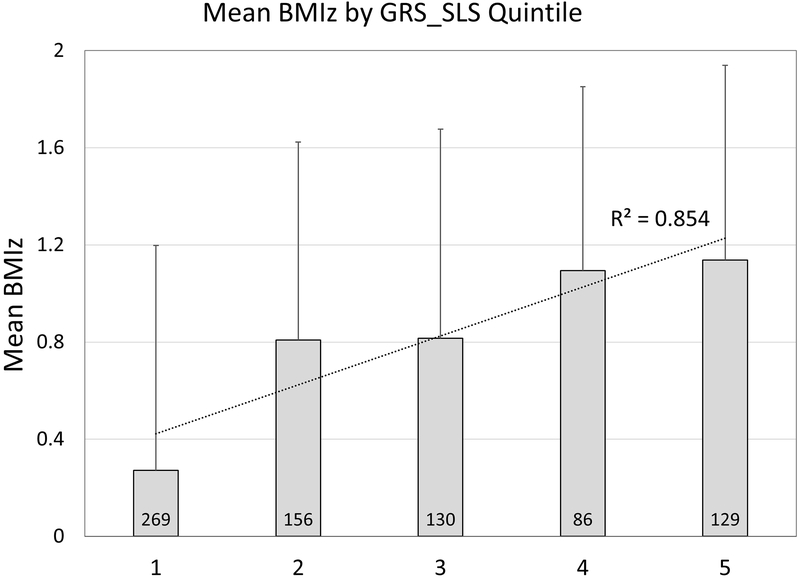
Three GRSs were constructed using the tag SNPs from Felix et al (GRS_EUR, 14 SNPs), the most significant SNP for each locus (GRS_SLS, 15 SNPs), and the GRS_SLS plus independent SNPs from GRS_EUR (GRS_ALL, 27 SNPs), as highlighted in Table 2. All three GRS were significantly associated with BMIz at age 10 (P<0.017, 0.05/3 GRS); however, the GRS_SLS was the most significant (β=0.161, P=1.4×10−26) and explained the greatest proportion of variation in BMIz (13.63%) (Table S2). Although these estimates of variance explained are likely inflated due to winner’s curse (upward bias in effect estimates of novel loci), it is still very apparent that these population-specific tag SNPs represent a marked improvement of the genetic effects of these loci on BMIz for our Chilean cohort. Additionally, we observe a difference of 0.32 BMIz units between individuals with the median number of BMIz-increasing alleles (17 alleles) and those in the upper quintile (>19 BMIz-increasing alleles) (Figure 1).
Figure 1.
Plot of mean (+SD) BMIz across quintiles of GRS_SLS, with the dashed line highlighting the predicted linear fit and correlation coefficient.
DISCUSSION
The discovery of genetic mechanisms influencing adiposity-related traits has the potential to identify important pathways for disease prediction and treatment [23]. Yet, the bulk of similar research has focused on homogeneous middle-aged adults, with very few genetic studies on ancestrally diverse, admixed children [24]. By examining 15 established EUR childhood BMI variants in the SLS Chilean children, we found that several of the SNP-BMIz associations were directionally consistent with previous findings, and five (near SEC16B, FTO, OLFM4, ADCY3, and FAIM2) reached nominal significance with age-specific effect sizes exhibiting a lower effect on BMIz at both ages 5 and 16 for all loci except FTO, which displayed a significant increase in effect across increasing age. Further, we find that potential SLS-specific tag SNPs represent a marked improvement over EUR tag SNPs in explaining variation in BMIz for our admixed Chilean cohort.
Previous studies of the FTO locus have suggested distinct effects on BMI across the lifecourse [25, 26]. While cross-sectional studies have indicated that effects across age may be due to cohort effects [27], other studies highlight associations between FTO and longitudinal BMI measures across childhood and into adulthood [25, 26]. Both rs1558902 and rs9939609 in FTO have been associated with change in BMI in EUR-descent individuals aged 11–20 years [28] and with change in BMI for a meta-analysis, which included Hispanic/Latino children living in the U.S. [11]. In our Chilean pediatric population, we identified similar patterns of association. Nominally significant association of BMIz with FTO genetic variants was first detected at age group 10, which is consistent (at or after age 7) with other studies [29]. We observe a significant trend in the increase of the effect from age 5 to age 16 in-line with findings in both EUR descent and Asian descent populations. Studies in EUR populations have shown an increase in the effect of rs9939609 across childhood and adolescence [30] and cumulatively support a peak in effect at age 20 [25]. Similarly, the same variant exhibited higher magnitude of association with BMI and obesity in Chinese girls aged 12 to 18 years compared to 6 to 11 years [26]. Our findings provide evidence for this pattern in Hispanic/Latinos; however, additional follow-up is needed to determine if the effects of the FTO locus also reach an apex in early adulthood. More importantly, the mechanism of timing of genetic effects at FTO and other loci may be useful for informing studies of primary and secondary obesity prevention across the lifecourse.
Variants within the OLFM4, ADCY3, and FAIM2 loci are nominally associated with BMIz in SLS, but only at age 10 (our largest sample size). For these loci, effect estimates are larger at age 10 than compared to effect estimates for BMIz measured at age 5 or 16. However, one of the limitations of the current study is the smaller sample sizes at age 5 and 16, which prevented formal tests of heterogeneity between ages. Additionally, as the trend test only accounts for differences in effect estimates across age, and therefore does not consider the correlation between measurements within individuals across time. Also, while our participants were measured at early childhood (adiposity rebound period), pre-puberty and adolescence, it is possible that hormonal changes occurring between 10y and 16y visits and variation in the timing of puberty both within and across sexes have added heterogeneity to our results. Nonetheless, other studies using cross-sectional data have found similar patterns in changing allelic effect sizes across age. For example, in a study by Warrington et al.[28], the same BMI-associated significant genetic variants examined herein for ADCY3 (rs11676272) and OLFM4 (rs12429545) were associated with BMI at age 8 in a meta-analysis of two studies of EUR children, and the effect alleles illustrated a changing pattern of effect. While Warrington et al. observed an association between OLFM4 variant and change in BMI, we did not observe any significant association with change in BMIz. Also, a similar to the pattern of association across age in SLS, these two variants displayed a pattern of greater genetic effects on weight at age 10 compared to ages 5 and 16 in EUR children [28].
In this cohort, BMIz is significantly associated with the EUR tag SNP, rs543874, in SEC16B at ages 10 after multiple test correction and nominally associated with BMIz at age 16, indicating an important role for this locus in Hispanic/Latino children. Nearby variants within this locus have previously exhibited significant age interaction effects in adult cross-sectional studies [10] contrasting effects in younger adults (≤50 years) to older adults (>50 years) and exhibiting larger effects in young adults. Similarly, in a separate study the SEC16B locus displayed larger genetic effects on BMI in adolescents and young adults compared to middle-aged [31]. Also, this variant has also been significantly associated with change in BMI across adolescence and young adulthood in EUR and nominally associated in Hispanic/Latino children living in the U.S. [11]. To our knowledge, our study is the first to leverage longitudinal data to examine the differences in effects estimates of the SEC16B locus on BMIz across childhood and adolescence. Our study not only supports the relevance for this locus in Hispanic/Latinos, but underscores the importance of further investigations into the effects of this locus across the lifecourse. Further analyses are needed to determine at what age the SEC16B has the greatest effect.
At each of the 14 loci with a EUR tag SNP available, we identified a more significant BMIz-associated SNP, 12 of which were not attenuated following conditional analyses indicating independence from the EUR tag SNP. Thus, in the SLS cohort, we not only replicated [9, 28] known childhood obesity risk variants, but also identified potential population-specific alternate genetic variants in Chilean children. These variants should be followed up in subsequent analyses of obesity-related phenotypes in ethnically diverse study populations, as they may provide new insights into biology underlying childhood obesity. For the locus near TNNI3K, we identified rs61790698 as the most significant SNP, which is in a nearby gene, crystalline zeta (CRYZ). A variant near CRYZ, rs3931020 (distance = 57.7Kb, R2 = 0.016, D’=1 with rs61790698 in CEU), has been associated with circulating resistin levels and increased risk of coronary heart disease in EUR adult GWAS study [32]. Resistin levels are influenced by obesity, and along the pathway between obesity and downstream cardiometabolic consequences (e.g. insulin resistance)[33], making CRYZ an interesting biological candidate for further study. Similarly, for rs13387838 near ADAM23, we could not identify a proxy SNP, but the most significant SNP within 1 MB interval was rs77068085, which is intronic to G protein-coupled receptor1 (GPR1). GPR1 association with BMIz has not been previously reported and may represent better biological candidate as it was shown to regulate glucose homeostasis in obese mice [34].
For Hispanic/Latinos to fully benefit from precision medicine, we need to better characterize the genetic diversity of Hispanic/Latinos and determine how this diversity may influence unique underpinnings to disease [35]. We addressed this issue by constructing an ancestry specific GRS in a Chilean cohort with extensive admixture. The estimation of GRS–BMIz associations using published tag SNPs explained 1.94% of the variation in BMIz in our SLS cohort, close to the percent variance explained reported in Felix et al (2.0%). The small difference between these two estimates may be the result of differences in genetic and environmental influences over BMIz, the absence of rs13387838 in our SLS cohort, and our focus on unweighted GRS. However, we also note that the GRS_EUR performed poorly compared to using ancestry appropriate tag SNPs identified in our Chilean population (13.63% variation compared to only 1.94%). While this result is promising, these results should be interpreted with caution, as winner’s curse may play a role in the large estimate of variance explained. Regardless, our study adds to the recent body of literature that demonstrates the importance of GRS in assessing obesity risk[14, 15], and the importance of selecting ancestry appropriate tag SNPs in future studies of disease prediction and clinical evaluation as it is shown that magnitude of association of GRS with BMI varies across different ethnic birth cohorts [13].
In summary, our study findings demonstrate an important role of genetic variants previously identified in EUR children in our Latin American pediatric population. Yet, we also demonstrate the importance of considering ancestry-specific variants for the most complete understanding of the role of genetic variation on risk of obesity during childhood. We observe distinct genetic effects across childhood and adolescence for FTO. Such patterning across the lifecourse may be informative for understanding disease pathogenesis and in particular in children. From a public health standpoint, such findings are critical as once obesity is established in childhood, it is very difficult to reverse [36]. Findings from this study yielded information on ancestry-specific alleles, biological mechanisms, and candidates for prevention efforts with population-specific genetic risk estimates that are applicable in a variety of contexts, especially in ancestrally diverse populations.
Supplementary Material
-
(1)What is already known about this subject
- Genetic contribution to BMI is well established.
- Genetic risk alleles for childhood BMI have been established in European-descent populations.
- Evidence that some adult BMI-associated loci identified in primarily European samples generalize to Hispanic/Latino children.
-
(2)What this study adds
- Estimating genetic effects on BMI in an ancestrally diverse and admixed Chilean pediatric cohort.
- Estimating genetic effects on BMI across childhood and adolescence.
- Importance of genetic risk scores in obesity prediction and clinical evaluation in an ancestrally diverse pediatric population.
ACKNOWLEDGEMENTS
This work was funded in part by University of North Carolina Nutrition Research Institute internal pilot grant, AHA grant 15GRNT25880008, and NIH award K99/R00HL130580-02. We would like to thank the participants and their family members from the Santiago Longitudinal Cohort Study (SLS).
Footnotes
Publisher's Disclaimer: This is the peer reviewed version of the following article: Justice AE, Chittoor G, Blanco E, Graff M, Wang Y, Albala C, Santos JL, Angel B, Lozoff B, Voruganti VS, North KE, Gahagan S. Genetic determinants of BMI from early childhood to adolescence: The Santiago Longitudinal Study (SLS). Pediatr Obes. 2019 Mar;14(3):e12479, which has been published in final form at https://doi.org/10.1111/ijpo.12479. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions.
CONFLICTS OF INTEREST
The authors have no conflicts to disclose.
REFERENCES
- 1.Finucane MM, Stevens GA, Cowan MJ, et al. , National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet, 2011. 377(9765): p. 557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holub CK, Elder JP, Arredondo EM, et al. , Obesity control in Latin American and U.S. Latinos: a systematic review. Am J Prev Med, 2013. 44(5): p. 529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popkin BM and Slining MM, New dynamics in global obesity facing low- and middle-income countries. Obes Rev, 2013. 14 Suppl 2: p. 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garmendia ML, Corvalan C and Uauy R, Addressing malnutrition while avoiding obesity: minding the balance. Eur J Clin Nutr, 2013. 67(5): p. 513–7. [DOI] [PubMed] [Google Scholar]
- 5.Albala C, Vio F, Kain J and Uauy R, Nutrition transition in Chile: determinants and consequences. Public Health Nutr, 2002. 5(1A): p. 123–8. [DOI] [PubMed] [Google Scholar]
- 6.Ministry of Health G.o.C. ENCUESTA NACIONAL DE SALUD 2016-2017: Primeros resultados. 2017. [cited 2018 March 23]; Available from: http://www.minsal.cl/wp-content/uploads/2017/11/ENS-2016-17_PRIMEROS-RESULTADOS.pdf.
- 7.Gordon-Larsen P, Adair LS, Nelson MC and Popkin BM, Five-year obesity incidence in the transition period between adolescence and adulthood: the National Longitudinal Study of Adolescent Health. Am J Clin Nutr, 2004. 80(3): p. 569–75. [DOI] [PubMed] [Google Scholar]
- 8.Locke AE, Kahali B, Berndt SI, et al. , Genetic studies of body mass index yield new insights for obesity biology. Nature, 2015. 518(7538): p. 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felix JF, Bradfield JP, Monnereau C, et al. , Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet, 2016. 25(2): p. 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler TW, Justice AE, Graff M, et al. , The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet, 2015. 11(10): p. e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graff M, North KE, Richardson AS, et al. , BMI loci and longitudinal BMI from adolescence to young adulthood in an ethnically diverse cohort. Int J Obes (Lond), 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Wilkinson A, Shen J, Wu X and Chow WH, Genetic polymorphisms in genes related to risk-taking behaviours predicting body mass index trajectory among Mexican American adolescents. Pediatr Obes, 2017. 12(5): p. 356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belsky DW, Moffitt TE, Sugden K, et al. , Development and evaluation of a genetic risk score for obesity. Biodemography Soc Biol, 2013. 59(1): p. 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makela J, Lagstrom H, Pitkanen N, et al. , Genetic risk clustering increases children’s body weight at 2 years of age - the STEPS Study. Pediatr Obes, 2016. 11(6): p. 459–467. [DOI] [PubMed] [Google Scholar]
- 15.Li A, Robiou-du-Pont S, Anand SS, et al. , Parental and child genetic contributions to obesity traits in early life based on 83 loci validated in adults: the FAMILY study. Pediatr Obes, 2018. 13(3): p. 133–140. [DOI] [PubMed] [Google Scholar]
- 16.Lozoff B, De Andraca I, Castillo M, Smith JB, Walter T and Pino P, Behavioral and developmental effects of preventing iron-deficiency anemia in healthy full-term infants. Pediatrics, 2003. 112(4): p. 846–54. [PubMed] [Google Scholar]
- 17.Pacheco LS, Blanco E, Burrows R, Reyes M, Lozoff B and Gahagan S, Early Onset Obesity and Risk of Metabolic Syndrome Among Chilean Adolescents. Prev Chronic Dis, 2017. 14: p. E93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.East P, Lozoff B, Blanco E, et al. , Infant iron deficiency, child affect, and maternal unresponsiveness: Testing the long-term effects of functional isolation. Dev Psychol, 2017. 53(12): p. 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose O, Blanco E, Martinez SM, et al. , Developmental scores at 1 year with increasing gestational age, 37–41 weeks. Pediatrics, 2013. 131(5): p. e1475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fryar CD, Gu Q and Ogden CL, Anthropometric reference data for children and adults: United States, 2007-2010. Vital Health Stat 11, 2012(252): p. 1–48. [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. , PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet, 2007. 81(3): p. 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA and Reich D, Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet, 2006. 38(8): p. 904–9. [DOI] [PubMed] [Google Scholar]
- 23.Green ED, Guyer MS and National I Human Genome Research, Charting a course for genomic medicine from base pairs to bedside. Nature, 2011. 470(7333): p. 204–13. [DOI] [PubMed] [Google Scholar]
- 24.Popejoy AB and Fullerton SM, Genomics is failing on diversity. Nature, 2016. 538(7624): p. 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy R, Wills AK, Wong A, et al. , Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet, 2010. 19(3): p. 545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Zhao X, Cheng H, et al. , Age- and sex-dependent association between FTO rs9939609 and obesity-related traits in Chinese children and adolescents. PLoS One, 2014. 9(5): p. e97545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenquist JN, Lehrer SF, O’Malley AJ, Zaslavsky AM, Smoller JW and Christakis NA, Cohort of birth modifies the association between FTO genotype and BMI. Proc Natl Acad Sci U S A, 2015. 112(2): p. 354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warrington NM, Howe LD, Paternoster L, et al. , A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol, 2015. 44(2): p. 700–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hakanen M, Raitakari OT, Lehtimaki T, et al. , FTO genotype is associated with body mass index after the age of seven years but not with energy intake or leisure-time physical activity. J Clin Endocrinol Metab, 2009. 94(4): p. 1281–7. [DOI] [PubMed] [Google Scholar]
- 30.Hallman DM, Friedel VC, Eissa MA, et al. , The association of variants in the FTO gene with longitudinal body mass index profiles in non-Hispanic white children and adolescents. Int J Obes (Lond), 2012. 36(1): p. 61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graff M, Ngwa JS, Workalemahu T, et al. , Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet, 2013. 22(17): p. 3597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi Q, Menzaghi C, Smith S, et al. , Genome-wide association analysis identifies TYW3/CRYZ and NDST4 loci associated with circulating resistin levels. Hum Mol Genet, 2012. 21(21): p. 4774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steppan CM and Lazar MA, The current biology of resistin. J Intern Med, 2004. 255(4): p. 439–47. [DOI] [PubMed] [Google Scholar]
- 34.Rourke JL, Muruganandan S, Dranse HJ, McMullen NM and Sinal CJ, Gpr1 is an active chemerin receptor influencing glucose homeostasis in obese mice. J Endocrinol, 2014. 222(2): p. 201–15. [DOI] [PubMed] [Google Scholar]
- 35.Carlson CS, Matise TC, North KE, et al. , Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol, 2013. 11(9): p. e1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The NS, Suchindran C, North KE, Popkin BM and Gordon-Larsen P, Association of adolescent obesity with risk of severe obesity in adulthood. JAMA, 2010. 304(18): p. 2042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.