Although microbial activities are known to contribute to the effectiveness of biosand filtration for drinking water treatment, we have a limited understanding of what microbial groups are most effectively removed, colonize the sand, or make it through the filter. This study tracked the microbial communities in the influent, sand, and effluent of lab-scale, intermittently operated biosand filters over 8 weeks. These results represent the most detailed and time-resolved investigation of the microbial communities in biosand filters typical of those implemented at the household level in many developing countries. We show the importance of the microbial food web in biosand filtration, and we identified taxa that are preferentially removed from wastewater-impacted water sources. We found consistent patterns in filter effectiveness from source waters with differing nutrient loads and, likewise, identified specific bacterial taxa that were consistently more abundant in effluent waters, taxa that are important targets for further study and posttreatment.
KEYWORDS: biosand filtration, drinking water, microbial ecology
ABSTRACT
Biosand filtration systems are widely used for drinking water treatment, from household-level, intermittently operated filters to large-scale continuous municipal systems. While it is well-established that microbial activity within the filter is essential for the removal of potential pathogens and other contaminants, the microbial ecology of these systems and how microbial succession relates to their performance remain poorly resolved. We determined how different source waters influence the composition, temporal dynamics, and performance of microbial communities in intermittently operated biosand filters. We operated lab-scale biosand filters, adding daily inputs from two contrasting water sources with differing nutrient concentrations and found that total coliform removal increased and became less variable after 4 weeks, regardless of water source. Total effluent biomass was also lower than total influent biomass for both water sources. Bacterial community composition, assessed via cultivation-independent DNA sequencing, varied by water source, sample type (influent, effluent, or sand), and time. Despite these differences, we identified specific taxa that were consistently removed, including common aquatic and wastewater bacteria. In contrast, taxa consistently more abundant in the sand and effluent included predatory, intracellular, and symbiotic bacteria.
IMPORTANCE Although microbial activities are known to contribute to the effectiveness of biosand filtration for drinking water treatment, we have a limited understanding of what microbial groups are most effectively removed, colonize the sand, or make it through the filter. This study tracked the microbial communities in the influent, sand, and effluent of lab-scale, intermittently operated biosand filters over 8 weeks. These results represent the most detailed and time-resolved investigation of the microbial communities in biosand filters typical of those implemented at the household level in many developing countries. We show the importance of the microbial food web in biosand filtration, and we identified taxa that are preferentially removed from wastewater-impacted water sources. We found consistent patterns in filter effectiveness from source waters with differing nutrient loads and, likewise, identified specific bacterial taxa that were consistently more abundant in effluent waters, taxa that are important targets for further study and posttreatment.
INTRODUCTION
Biosand filtration is a process central to drinking water treatment at a range of scales, from household bucket systems (1, 2) to community-scale slow sand filters at municipal water works (3). Effective removal of pathogens relies on the activity of the microbial community that develops inside the sand filter (hence the term “biosand” filter) (4–6), and yet, there remain large gaps in our understanding of the microbial ecology of these systems. While a few studies have evaluated microbial removal from realistic source waters (7–10), most studies have focused on the removal of specific pathogens (11, 12) or indicator species (13–16) with pure cultures added at concentrations many orders of magnitude above what might be encountered during typical operation (5, 7, 11, 12). These studies have led to the identification of several important removal mechanisms for individual microorganisms, including physical filtration (17, 18), predation by protozoa (13, 19, 20), and lysis induced by viruses or reactive oxygen species (13). Because natural source waters contain complex microbial communities, it is necessary to understand microbial removal in a community context to inform the design, operation, and monitoring of these filtration systems.
The diversity and composition of the microorganisms in biosand filters have been characterized across a broad range of operating conditions. Most previous work on the microbial communities in biosand filters has focused on continuously operated systems representative of municipal-scale systems (5, 8, 13, 21–25), while only limited work focused on intermittently operated filters, typical of household-scale systems (26–28). This gap in our knowledge of the microbial communities in intermittently operated biosand filters is important given that many intermittently operated filters are implemented in conditions with less monitoring and often without posttreatment disinfection steps (2). Haig et al. (29) showed that similar microbial communities developed in both lab- and full-scale filters continuously operated with the same source water. Other studies have highlighted the importance of filter medium type for community diversity and filter performance (21, 23). These studies have found Planctomycetes, Bacteriodetes, Alphaproteobacteria, and Betaproteobacteria to be the most abundant bacterial taxa in drinking water filtration systems (8, 21–25, 29). While these studies have provided much-needed insight into which microorganisms are found in filtration systems, we lack information about how key operational decisions, including start-up time and source water inputs, impact filter performance and microbial community composition changes over time in intermittently operated household filters.
While filter performance has been shown to improve over time after start-up (9, 14), most operational guidelines are driven by empirical observations (30), and the factors that impact microbial succession in these filters have not been well-studied. High variability between filter replicates and overall low levels of replication have also limited the conclusions that can be drawn from previous studies (15, 28, 31). In a study of two full-scale sand filters, Haig et al. found that improvements in the performance of continuously operated filters over time correlated with increases in community evenness and the relative abundance of particular genera (including Sphingobium, Acinetobacter, and Halomonas) (9). These findings highlight that microbial colonization and successional dynamics are important to filter operation, but we lack the relevant information needed to optimize these processes. Start-up times around 30 days are common (30) but pose a challenge for adoption and implementation of these systems. Efforts to reduce this start-up time require a more detailed understanding of microbial dynamics that occur after start-up. Previous studies in natural environments have shown that nutrient levels and microbial community composition can lead to differences in successional trajectories in microbial communities (32, 33). Nutrient levels have also been shown to influence protist grazing in freshwater reservoirs (34) and microbial turnover in wastewater (35) and other ecosystems (33). Similarities in successional patterns may also arise due to the benefits of different functional traits at different stages of colonization (36) or common selection pressures within the sand filters (as has been observed in wastewater communities [37, 38]).
We hypothesized that source water nutrient levels (nitrogen and phosphorous) and microbial community composition would influence microbial succession patterns and the performance of biosand filters. We also expected that similarities observed between filters receiving different water sources would reflect common selection pressures, for example favoring biofilm formation within the sand filter. To test these hypotheses, we tracked microbial community succession and performance of intermittently operated lab-scale biosand filters over an 8-week period. We compared the influence of using two source waters with different nutrient levels on filter performance and microbial dynamics using both culture-based and culture-independent approaches. We also identified taxa that were preferentially removed during the filtration process, identifying groups with higher relative abundances in either the influent or effluent samples.
RESULTS AND DISCUSSION
To investigate the microbial community dynamics and performance of biosand filters receiving contrasting water sources, 80 lab-scale biosand filters received daily water inputs from either a pristine, low nutrient mountain reservoir or an industrially and agriculturally impacted high nutrient river water, spiked with dilute wastewater for 8 weeks. As expected, influent water, including the dilute wastewater, from the high nutrient source had higher total organic carbon nitrate, ammonia, total P, and pH than influent from low nutrient sources (see Table S1 in the supplemental material). Average values over the 8-week experiment for total organic carbon were 3.2 and 2.3 mg C liter−1 for high and low nutrient influents, respectively. Average nitrate and ammonia were 2.55 and 0.16 mg N liter−1 for high nutrient and 0.01 and 0.03 mg N liter−1 for low nutrient. The average total P was 0.27 and 0.06 mg P liter−1 for high and low nutrient sources, respectively. The average pH of the high nutrient water was 7.9, while low nutrient water was 7.3.
Biosand filter performance.
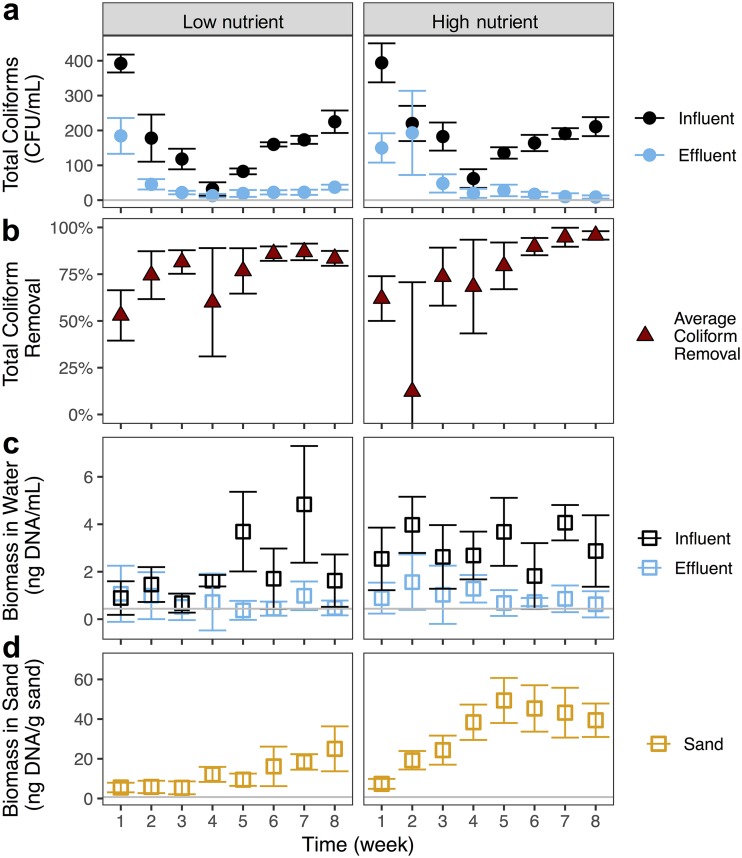
The overall performance of the biosand filters (measured by total coliform removal) improved over time and was similar between the two water sources. Effluent coliform concentrations became lower over time, reaching their lowest at week 4 for both water sources (Fig. 1a). The highest total coliform removal occurred later, at week 7 for low nutrient (87%) and week 8 for high nutrient (96%) (Fig. 1b). Over time, the variability in total coliform removal between filter replicates also decreased in both water sources (Fig. 1a). This improvement in the efficacy and consistency of coliform removal over time is in line with typical recommendations for a 30-day start-up time for “filter ripening” in intermittently operated biosand filters, commonly deployed in developing countries (30). Our results are also consistent with lab studies of larger biosand filters removing pure culture E. coli (14). Overall measures of biomass, based on DNA yield, showed that effluent DNA was, on average, 70% lower than influent DNA indicating overall removal of microorganisms as source water passed through the biosand filters (Fig. 1c). Microbial biomass in the sand filters also increased over time in both water sources (Fig. 1d). Filters receiving high nutrient river water accumulated biomass at a higher rate and reached higher total amounts of biomass (Fig. 1d), which can be explained by the higher nutrient levels.
FIG 1.
Average total coliform concentrations over time in the influent and effluent waters (a) and average percent total coliform removal (b) of filters receiving low and high nutrient source waters. Microbial biomass measured by DNA yield over time in the influent and effluent (c) and sand (d) samples. Each point represents the mean, and error bars represent the standard deviation across five replicate samples of source water (influents) or five replicate biosand filters (effluents and sand). Gray lines indicate the detection limits.
Microbial community includes common water and biofilm-forming bacteria.
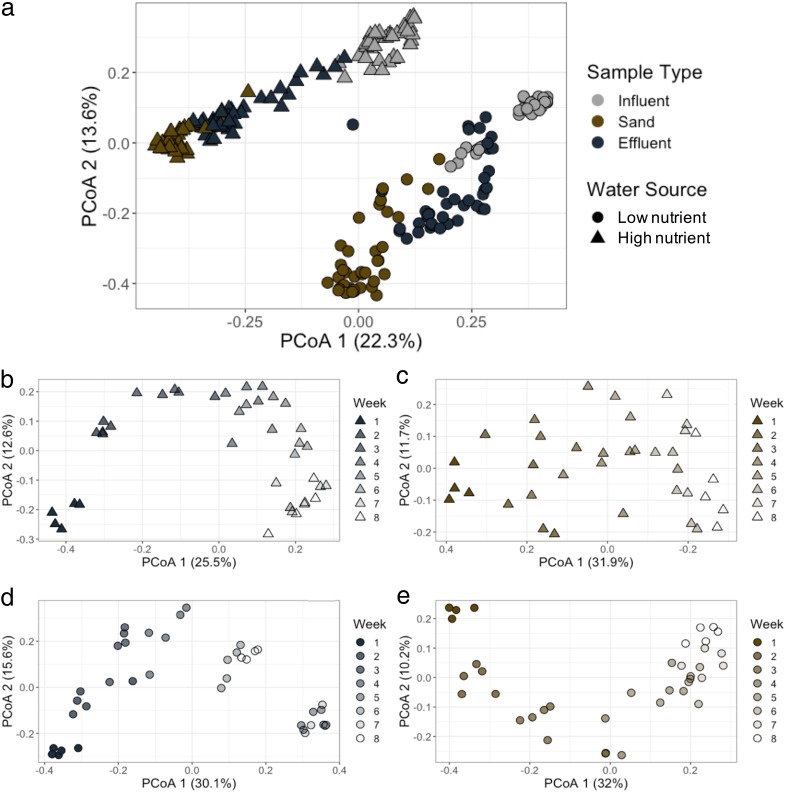
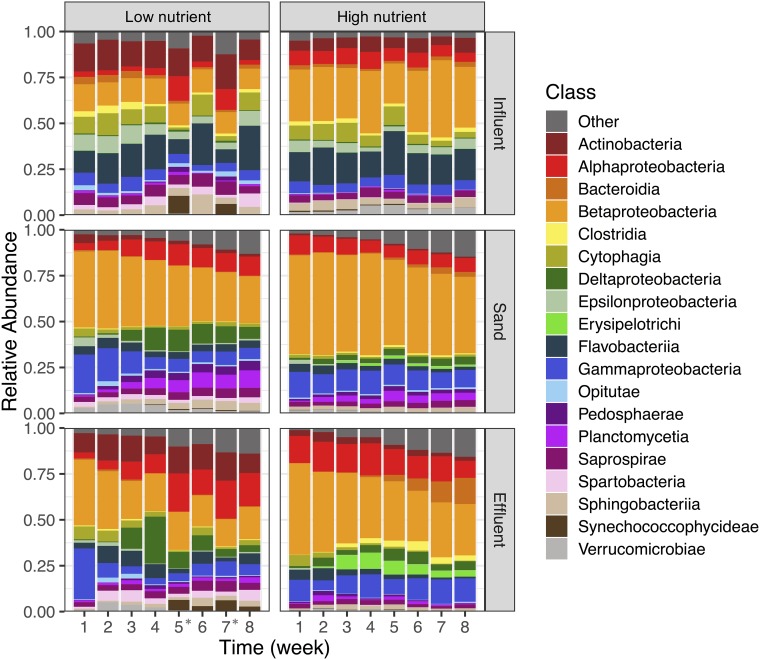
Results from 16S rRNA gene sequencing of filter influents, effluents, and sand show that microbial communities differentiate by water source, sample type, and time. As a previous comparison of full-scale and much smaller lab-scale biosand filters have shown highly reproducible microbial communities regardless of filter size (29), we anticipate that the microbial community results observed here would be broadly similar to what would be seen in full-scale systems. Water source explained a significant portion of the variation among all samples (ADONIS R2 = 0.18, P = 0.001) when comparing the microbial communities from all sample types and time points (Fig. 2a). Low nutrient reservoir samples typically had larger amounts of Actinobacteria, Sphingobacteriia, and Saprospirae than that of the high nutrient river water (Fig. 3). The most abundant Actinobacteria include unclassified taxa in the family ACK-M1, which are commonly found in lakes (39). In contrast, high nutrient water had larger amounts of Alphaproteobacteria and Betaproteobacteria (Fig. 3). One of the most abundant genera in high nutrient influent was Methylotenera which has been implicated in denitrification (40) and would be consistent with the higher levels of nitrate in the high nutrient source which averaged ∼2.5 mg N liter−1, compared with <0.01 mg N liter−1 in the low nutrient source (see Table S1 in the supplemental material).
FIG 2.
Principal-coordinate analyses (PCoA) of microbial communities based on pairwise Bray-Curtis dissimilarity, comparing sample type and water source across all samples (a) and over time for high nutrient effluent (b), high nutrient sand (c), low nutrient effluent (d), and low nutrient sand (e). PCoA of influent microbial communities over time shown in Fig. S4. Note that the low nutrient influent was from a different source during weeks 5 and 7 (see Fig. 4).
FIG 3.
Relative abundance of the 19 most abundant classes over time for the low nutrient (left) and high nutrient (right) water sources for samples from the influent (top), sand (middle), and effluent (bottom) over time. Each bar represents combined data from four to five replicates (results from individual replicates are presented in Fig. S2). * indicates that the low nutrient influent was from a different source during weeks 5 and 7.
Sample type (influent, effluent, or sand) explained as much of the variation in microbial communities as water source (ADONIS R2 = 0.19, P = 0.001) (Fig. 2a). The influent samples had higher relative abundances of Flavobacteriia, including the genera Flavobacterium, a common aquatic organism (41), and Arcobacter, a genus often used as a marker of wastewater-impacted water sources (42). Other common aquatic bacteria that were higher in relative abundance in the influent include the groups in the classes Cytophagia (43, 44) and Sphingobacteriia (22, 45, 46). Compared with other sample types, sand samples had higher relative abundances of Proteobacteria (see Fig. S3 in the supplemental material), including the genera Zoogloea (47), Pseudomonas (46), and Dechloromonas (48), groups previously found in water filter biofilms. High nutrient sand filters harbored higher relative abundances of Aquabacterium (from the class Betaproteobacteria), another common water bacterium found in biofilms (49). Effluent samples had higher relative abundances of groups from the class Rhodospirillales. Within the class Rhodospirillales, low nutrient samples had higher relative abundance of Azospirillum, while high nutrient effluents had higher relative abundances of Novispririllum. Within the Deltaproteobacteria, low nutrient effluent had a higher relative abundance of members of the order Spirobacillales, which are commonly found in freshwater samples and are thought to be involved in the degradation of complex organics (50, 51).
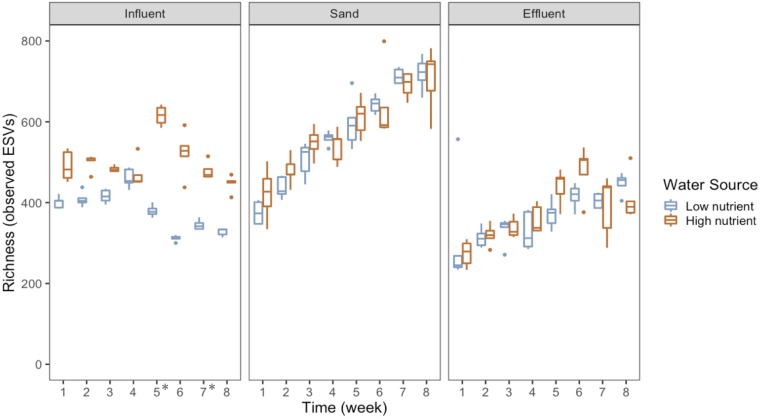
Time explained a smaller, but still significant, fraction of the overall variation in microbial communities (ADONIS R2 = 0.10, P = 0.001) (Fig. 2b to e). The changes over time in influent samples were due to natural variation in source waters between weeks, except for the unplanned switch in water sources for low nutrient filters at weeks 5 and 7 (see Fig. S4 in the supplemental material). The variation in the sand and effluent samples likely reflect the selective pressures inside the sand filter that favor the growth or colonization of some microorganisms and the removal of others. As observed during microbial succession in other environments (9, 52), the richness and evenness of the sand and effluent samples increased over time for both water sources (Fig. 4; see Fig. S5 in the supplemental material). Zoogloea spp. increased in relative abundance in the sand filters receiving either source water over time. Rhodospirillales spp. increased in relative abundance in both the effluent and sand as well. Sequences classified to the class Chlamydiia increased in relative abundance over time, especially in the effluent. All known organisms in this class are obligately intracellular bacteria and have previously been identified from drinking water sources using amoeba coculture (53, 54). The family Comamonadaceae increased in relative abundance in both the sand and effluent for both water types and are commonly found in drinking water filtration and distribution systems (22). In contrast, members of the Cytophagia and the Betaproteobacteria (mostly Burkholderiales) groups decreased in relative abundance in the effluent over time. The unplanned switch in water sources for the low nutrient biosand filters during week 5 and 7 did result in distinct influent communities for those weeks (Fig. 3). Likewise, the effluent communities during those weeks (5 and 7) and the weeks following (6 and 8) were also slightly different compared with weeks 1 to 4 (Fig. 3). The sand communities showed less variation as a result of this switch, likely because the microbial communities had already been established and the amount of biomass contributed in the new influent was insufficient to shift the sand microbial community that had developed over the preceding 4 weeks (55).
FIG 4.
Boxplot of richness, measured by the number of observed exact sequence variants (ESVs) at a rarefied sequencing depth of 2,122 reads per sample for each water source, sample type, and week. *, indicate that the low nutrient influent was from a different source during weeks 5 and 7.
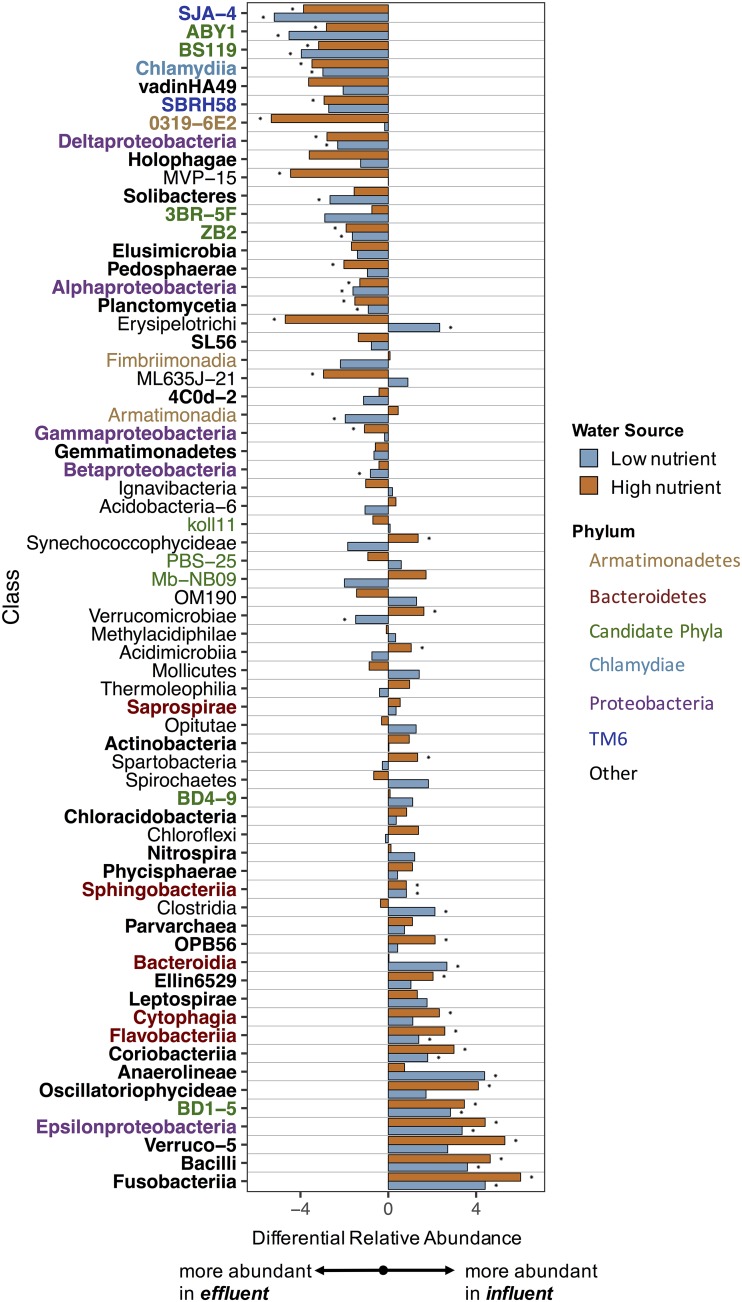
Differential relative abundances at the class level show similar trends across water sources.
Comparing the ratios of relative abundance of microorganisms between the influent and the effluent at the class level allowed us to identify taxa that are preferentially found in either sample type for both water sources. Of the 65 most abundant classes (relative abundance, >0.01%), 44 classes have the same trend (i.e., more abundant in the effluent or influent across all time points) for both water sources (Fig. 5). Given the observed differences in community composition based on water source, this consistency in the taxa found preferentially in either the influent or effluent waters is noteworthy. These trends are also largely consistent across time (see Fig. S6 in the supplemental material), although the magnitude varies across time for individual taxa. Taxa that were removed consistently even at the earliest time points likely include taxa removed by physical processes, i.e., those taxa that were preferentially removed by passage through the filter or simply unable to survive within the filter. For other classes, including Epsilonproteobacteria and Flavobacteriia, the decrease in relative abundance in the effluent over time (Fig. 3 and Fig. S6) indicates that biological factors related to microbial community succession within the filters are likely important for their removal.
FIG 5.
Differential relative abundances between the influent and effluent of the most abundant microbial classes (>0.01% relative abundance) for each water source across all time points (for time resolved data see Fig. S6), ordered by the average differential relative abundance for both water sources. Bolded text labels indicate classes with the same trend in differential abundance between the two source waters. Stars indicate statistically significant differences (FDR, <1%). Text label colors indicate specific phyla of interest.
Overall reductions in relative abundances across the filters, combined with the measured reduction in total DNA (Fig. 1c), indicate that certain bacterial taxa are preferentially removed from influent source waters. Other taxa that have consistently higher relative abundances in the effluent include taxa that pass through the filter or colonize the sand. Note that some taxa with higher relative abundance in the effluent may not necessarily reflect higher total abundances and could result from the removal or decrease in relative abundance of other groups. However, the largely consistent differences in the community composition between the influent and effluent observed here (Fig. 2) indicate that the filtration process does not remove all microbial groups equally and that passing influent waters through the biosand filters shapes the effluent microbial community in a largely predictable manner.
Groups with higher relative abundances in the influent across both water sources included common aquatic and wastewater-associated bacteria. Flavobacteriia (mostly the genus Flavobacterium) and several other classes in the phylum Bacteroidetes are common aquatic bacteria (41), indicating preferential removal of these groups. A reduction in the relative abundance of Flavobacteriia and Bacteroidetes spp. has also been seen in other studies of sand filtration (8, 26). Bacteria from groups commonly associated with fecal matter that appear to be preferentially removed (higher relative abundance in the influent than the effluent) include Epsilonproteobacteria (the most abundant genus was Arcobacter) (42, 56), Fusobacteriia (57, 58), Bacilli (most abundant groups include Streptococcaceae and Carnobacteriaceae) (59, 60), and Verruco-5 (61). The class Coriobacteriia, which contains facultative anaerobes (62), was also more abundant in the influent than the effluent. Possible explanations for the removal of these groups include preferential grazing by protists, as has been shown for Flavobacterium and other Bacteroidetes spp. (34), or an inability of these taxa to survive the aerobic conditions found in the biosand filters.
Taxa that were found to have higher relative abundances in the effluent of filters from both water sources included groups containing known bacterial predators, symbionts, and obligately intracellular bacteria. While protozoan grazing has been shown to be an important removal pathway for E. coli in sand filters (13), other work has shown that amoebae and other protists can harbor potentially pathogenic microorganisms, effectively bypassing water treatment processes (63, 64). All of the bacteria in the class Chlamydiia identified here were within the order Chlamydiales, a group containing obligately intracellular bacteria that have been previously recovered by amoeba coculture from river water and other drinking water sources (53, 54). Several classes within candidate phyla (including OD1) and TM6 phyla were also found to be higher in relative abundance in the effluent. Although little is known about bacteria in these candidate phyla, several metagenome-based studies have found evidence that these organisms are likely to be symbionts (65–67). A similar pattern in the relative abundance of OD1 and other candidate phyla was also observed in previous studies of full-scale drinking water filters (22, 23). Several classes within the phylum Armatimonadetes were found to be selected for in filter effluents, a trend that has also been reported in activated carbon filters (45). Alphaproteobacteria were also higher in relative abundance in the effluent in both water sources, although different orders were more abundant in each water source. Deltaproteobacteria were preferentially found in effluent water communities, and their relative abundance increased in both the sand and effluent samples over time (Fig. 2), a trend that has been observed previously in slow sand filters (8, 29) and at a full-scale drinking water treatment plant (22). This class includes known predatory groups that were identified in these samples, including the genus Bdellovibrio and members of the order Myxococcales (order-level relative abundances of Deltaproteobacteria shown in Fig. S7 in the supplemental material) (68). While bacterial grazing may also contribute to the removal of these groups, we note that some of these groups (including OD1 and Bdellovibrio) can be quite small (<0.4 μm), and therefore, some of the increase in relative abundance of these groups could be due to physical size exclusion of larger bacteria within the filter (51) or size selection biases during protozoan grazing (69). Groups that were relatively more abundant in both the sand (see Fig. S8 in the supplemental material) and the effluent (Fig. 5) than the influent are likely colonizers of the sand and include the classes Deltaproteobacteria, Elusimicrobia, and Holophagae. These groups are likely to play a key role in the microbial community structure of filtered water (22).
These results show that biosand filters receiving distinct water sources can achieve similar levels of performance, as measured by total coliform and microbial biomass removal. Water source did significantly impact the microbial communities and the rate of accumulation of microbial biomass over time within the biosand filters. Microbial communities were also distinct depending on sample type (influent, effluent, and sand) and time. Despite these differences, there were consistent patterns in the microbial taxa that were selectively removed by filtration, mainly aquatic and wastewater-associated bacteria. The microbes found in biosand filter effluents was more likely to contain bacteria that were intracellular, symbiotic, or predatory. These findings support earlier studies with indicator organisms that found predation to be a key removal pathway during biofiltration and further highlight bacterial predation as an important area of study. While this study focused on the start-up of small-scale intermittently operated sand filters, the fundamental microbial dynamics are expected to be informative for a range of filtration applications. This work provides a foundation for understanding microbial succession, as it relates to improving filter performance, representing the most replicated and time resolved study of microbial communities in biosand filters to date. These results can be used to guide future efforts to predict and model the changes in microbial communities expected from biosand filtration during drinking water treatment. Such next steps should involve more detailed investigations of the interactions between trophic levels within complex communities. Likewise, by identifying the taxa that were preferentially found in biofilter effluents, our work guides improvements for posttreatment and monitoring strategies that target these groups of small and possibly intracellular bacteria.
MATERIALS AND METHODS
Biosand filter setup and water sources.
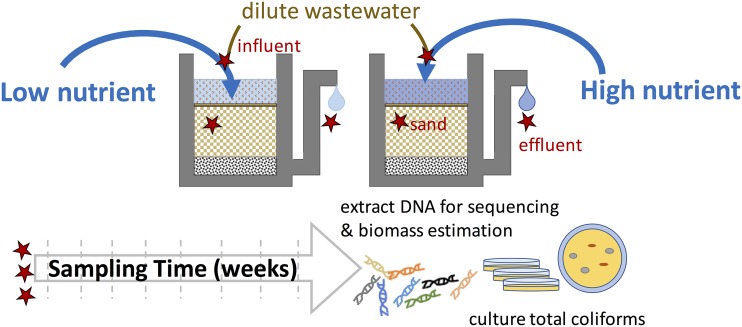
Eighty biosand filters were constructed from 2.54-cm-diameter PVC pipe with a 0.48-cm-diameter outlet tube that kept the water level 4 cm above the 10.5-cm-high sand bed at all times (Fig. 6). The bottom of each filter was filled with 18 cm3 of 3- and 6-mm-diameter glass beads. Then, 50 cm3 of commercial grade medium sand was added to each filter. The sand had an effective size of 0.17 mm and uniformity coefficient of 1.77 (Quickrete, Atlanta, GA, USA), corresponding to the smaller size range recommended for sand filters (16). The time to filter 22 ml at the start of the experiment was 36 minutes, corresponding to a filter loading rate of 366 liters h−1 m−2, which aligns with the suggested loading rate of <400 liters h−1 m−2 (30). We chose a shorter column length than typical of most full-scale filters to keep the experimental system manageable with high replication and to enable collection of biomass from the entire sand volume, as previous studies have shown differences in microbial community composition with depth (27, 29). Previous studies have also shown that the majority of microbial growth and activity takes place in the upper centimeters of the sand (12, 28, 70, 71). Glass beads and sand were washed with 10% HCl, rinsed with deionized water, and autoclaved twice before the start of the experiment.
FIG 6.
Schematic of the experimental setup. Eighty replicate biosand filters (2.54-cm diameter with a 10.5-cm filter bed depth) were constructed from PVC pipes with a standpipe to keep water 4 cm above the sand bed. Daily water inputs were added from either of two water sources (with high or low nutrients), each spiked with diluted wastewater. Samples for DNA extraction, sequencing, and biomass estimation were collected weekly from five replicate filters, including influent, sand, and effluent. Weekly coliform removal was also measured over 8 weeks.
Low nutrient source water was collected weekly from the Lakewood Reservoir (39°59′24″N, 105°30′W) and high nutrient source water from the Platte River (39°48′29″N, 104°57′32″W). Primary effluent from the Boulder Wastewater Treatment Plant was also collected weekly. Water sources and wastewater were stored at 4°C to limit growth over time. Due to construction, we could not collect Lakewood Reservoir water on weeks 5 and 7. During those weeks, Boulder Reservoir water (40°4′37″N, 105°13′12″W) was used as a low nutrient source water. Daily influent was made by adding 1 ml of wastewater (collected from the aeration basin influent) to 1 liter of each water source (low or high nutrient). The diluted wastewater was used as a source of coliforms and other wastewater-associated bacteria, targeting a final influent coliform concentration approximating values measured in the field (72, 73). The source waters spiked with wastewater were mixed thoroughly before adding 22 ml of each water (low or high nutrient) to each filter every 24 hours for the 8-week-long experiment. These filters were designed to be small, lab-scale versions of intermittently operated household biosand filters that process water in batches on a daily basis (30). The 8-week-long operation time was selected to cover the typically recommended 4-week start-up time for household biosand filters and so the filters could be operated for an additional 4 weeks without the need for cleaning during the study time frame (30).
Each day, five replicate samples of influent and effluent water from five filters for each water source were collected in 50-ml centrifuge tubes and stored frozen at −20°C for nucleic acid extractions. Coliform counts were measured using freshly collected water from five replicate samples of influent and filter effluents for each water source on the last day of each week. Total coliforms were measured using Petrifilm coliform count plates (3M, Maplewood, MN, USA) (74) according to the manufacturer’s instructions. Weekly sand filter biomass samples were collected by destructively sampling five filters from each water source. For each sand sample, 60 ml of sterile 10 mM phosphate-buffered saline (PBS) was used to wash the entire volume of sand and glass beads, using two 25-ml washes with 2 minutes of vortexing with each wash to dislodge any biofilm-associated microbes. The PBS-suspended biomass was then filtered onto a 0.2-μm filter using a Nalgene analytical test filter (Thermo Fisher Scientific, Sunnyvale, CA, USA). PBS controls were created each week by filtering the same volume of sterile PBS buffer through a clean (unused) filter. Weekly composite biomass samples were created by thawing the daily samples for each influent and effluent stored at the −20°C and combining 18 ml from each daily sample onto the same filter of a Nalgene analytical test filter for each week. This resulted in five replicate composite samples for the influent and effluent from each water source for each of the 8 weeks.
Influent water chemistry.
Influent water quality was measured each week, after the primary effluent had been added to each water source, to compare the nutrient levels. Total organic carbon, total phosphorous, nitrate, and ammonia were measured according to standard methods 5310B, 4500-P-E, 4500-NO3-D, and 4500-NH3-F, respectively (75). Water pH was measured using an accumet basic AB15 pH probe (Fisher Scientific).
DNA extraction, sequencing, quantification, and community analysis.
DNA was extracted from the biomass collected on filter paper using the DNeasy PowerSoil HTP 96 kit (Qiagen Inc., Valencia, CA, USA), as detailed previously (76). The V4-V5 region of the bacterial and archaeal 16S rRNA gene was PCR amplified in pooled duplicate reactions using GoTaq hot start colorless master mix (Promega Corporation, Madison, WI) with 515F and barcoded 806RB (77) primers that included 12-bp error-correcting barcodes and the appropriate Illumina sequencing adapters (see reference 78 for details). Amplicons were cleaned and normalized using the SequalPrep normalization plate kit (Thermo Fisher Scientific, Waltham, MA). Samples were sequenced on an Illumina MiSeq instrument running the V2 2 × 150-bp chemistry at the University of Colorado BioFrontiers Institute Next-Gen Sequencing Core Facility.
Sequences were demultiplexed, merged (minimum merged length, 200 bp), and quality filtered using USEARCH10 (79). Sequences with more than 1 error per base call were discarded. Exact 16S rRNA gene sequence variants (ESVs) were processed using UNOISE3 (80). The Ribosomal Database Project Classifier (81), trained on the Greengenes database (82) was used to determine the taxonomic affiliations of each ESV. Reads assigned to chloroplasts or mitochondria were removed. ESVs with fewer than 8 reads and samples with fewer than 2,000 reads were also removed. Out of 258 samples, 16 failed to meet this cutoff. These 16 samples included 7 of 16 PBS controls, 2 no template PCR controls, 1 extraction blank, and 6 experimental samples. The processed sequences and exact sequence variant (ESV) table are included in the supplemental material. All remaining samples were plotted in a heatmap to compare the similarity to sequenced controls (83). All of the remaining negative controls (nine PBS controls and one extraction blank) clustered separately and near only one experiment sample (an influent sample from the high nutrient source, week 5). This influent sample was subsequently removed from downstream analysis, as it was suspected to be contaminated with reagent-associated DNA based on similarity to the sequenced controls.
The changes in the microbial communities were analyzed using R (v3.4.1) (84) and the phyloseq package (85) with plots generated using the ggplot2 package (86). Community composition was compared using principal-coordinate analysis based on the Bray-Curtis dissimilarity metric and the permutational analysis of variance tests were conducted using ADONIS (87) in the R package vegan (88). To compare differential relative abundances of the most abundant classes (>0.01% relative abundance) between influent and effluent samples for each water source, the ALDEx2 R package was used to determine statistically significant differences based on the Welch’s t test with a Benjamini-Hochberg false discovery rate (FDR) correction of centered log-transformed (base 2) data using 128 Monte-Carlo instances drawn from the Dirichlet distribution (89, 90).
DNA yield on all samples was measured using the Quant-iT PicoGreen double-stranded DNA (dsDNA) assay (Invitrogen, Carlsbad, CA) according the manufacturer’s instructions. The detection limit for this method was determined to be 0.28 ng/μl. The concentration of the 16S rRNA gene was compared with DNA yield for a subset of samples, using quantitative PCR. The primers were the unbarcoded 515F and 806RB, the same as used for sequencing. The fast plus EvaGreen quantitative PCR (qPCR) master mix (Biotium, Fremont, CA, USA) was used with PCR conditions as follows: 2 minutes at 95°C; then 40 cycles of 5 s at 95°C, 5 s at 50°C, and 30 s at 72°C; followed by 60 s at 72°C. Genomic DNA extracted from Escherichia coli K-12 was serially diluted and used as a standard. Results from DNA yield and qPCR were well-correlated (see Fig. S1 in the supplemental material); therefore, DNA yield was used as a proxy for bacterial biomass in these samples. The mass of DNA was normalized by the volume of water or mass of sand the sample was extracted from. This approach provides a conservative metric of cell concentrations as some of the DNA detected is likely extracellular or derived from dead cells (91). Additionally, reads from chloroplasts and mitochondria made up less than 7% of the 16S rRNA gene reads, indicating low contributions of eukaryotic organisms to the DNA pools.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jessica Hanley and Lady Grant for their help with sample preparation and the University of Colorado’s Center for Limnology for assistance in water collection. We are grateful to the City of Boulder Public Works for water samples and especially the help of Zach Lelwica, Cole Sigmon, and Conor Tyler.
T.M.W. was supported by funding provided by the Cooperative Institute for Research in Environmental Sciences Postdoctoral Fellowship program.
The authors declare no competing financial interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01142-19.
REFERENCES
- 1.Sobsey MD, Stauber CE, Casanova LM, Brown JM, Elliott MA. 2008. Point of use household drinking water filtration: a practical, effective solution for providing sustained access to safe drinking water in the developing world. Environ Sci Technol 42:4261–4267. doi: 10.1021/es702746n. [DOI] [PubMed] [Google Scholar]
- 2.Ngai T, Coff B, Baker D, Lentz R. 2014. Global review of the adoption, use and performance of the biosand filter, p 309–317. In Progress in slow sand and alternative biofiltration processes. International Water Association, London, UK. [Google Scholar]
- 3.Haig SJ, Collins G, Davies RL, Dorea CC, Quince C. 2011. Biological aspects of slow sand filtration: past, present and future. Water Supply 11:468–472. doi: 10.2166/ws.2011.076. [DOI] [Google Scholar]
- 4.Elliott MA, Digiano FA, Sobsey MD. 2011. Virus attenuation by microbial mechanisms during the idle time of a household slow sand filter. Water Res 45:4092–4102. doi: 10.1016/j.watres.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Weber-Shirk ML, Dick RI. 1997. Biological mechanisms in slow sand filters. J Am Water Works Assoc 89:72. doi: 10.1002/j.1551-8833.1997.tb08180.x. [DOI] [Google Scholar]
- 6.Chan S, Pullerits K, Riechelmann J, Persson KM, Rådström P, Paul CJ. 2018. Monitoring biofilm function in new and matured full-scale slow sand filters using flow cytometric histogram image comparison (chic). Water Res 138:27–36. doi: 10.1016/j.watres.2018.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Stauber CE, Elliott MA, Koksal F, Ortiz GM, Digiano FA, Sobsey MD. 2006. Characterisation of the biosand filter for E. coli reductions from household drinking water under controlled laboratory and field use conditions. Water Sci Technol 54:1–7. doi: 10.2166/wst.2006.440. [DOI] [PubMed] [Google Scholar]
- 8.Pfannes KR, Langenbach KMW, Pilloni G, Stührmann T, Euringer K, Lueders T, Neu TR, Müller JA, Kästner M, Meckenstock RU. 2015. Selective elimination of bacterial faecal indicators in the schmutzdecke of slow sand filtration columns. Appl Microbiol Biotechnol 99:10323–10332. doi: 10.1007/s00253-015-6882-9. [DOI] [PubMed] [Google Scholar]
- 9.Haig S-J, Quince C, Davies RL, Dorea CC, Collins G. 2015. The relationship between microbial community evenness and function in slow sand filters. mBio 6:e00729-15. doi: 10.1128/mBio.00729-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeger EM, Braeckevelt M, Reiche N, Müller JA, Kästner M. 2016. Removal of pathogen indicators from secondary effluent using slow sand filtration: optimization approaches. Ecol Eng 95:635–644. doi: 10.1016/j.ecoleng.2016.06.068. [DOI] [Google Scholar]
- 11.Bomo AM, Ekeberg D, Stevik TK, Hanssen JF, Frostegard A. 2004. Retention and removal of the fish pathogenic bacterium Yersinia ruckeri in biological sand filters. J Appl Microbiol 97:598–608. doi: 10.1111/j.1365-2672.2004.02342.x. [DOI] [PubMed] [Google Scholar]
- 12.Hijnen WAM, Dullemont YJ, Schijven JF, Hanzens-Brouwer AJ, Rosielle M, Medema G. 2007. Removal and fate of Cryptosporidium parvum, Clostridium perfringens and small-sized centric diatoms (Stephanodiscus hantzschii) in slow sand filters. Water Res 41:2151–2162. doi: 10.1016/j.watres.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 13.Haig S-J, Schirmer M, D'amore R, Gibbs J, Davies RL, Collins G, Quince C. 2015. Stable-isotope probing and metagenomics reveal predation by protozoa drives E. coli removal in slow sand filters. ISME J 9:797–808. doi: 10.1038/ismej.2014.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott MA, Stauber CE, Koksal F, Digiano FA, Sobsey MD. 2008. Reductions of E. coli, echovirus type 12 and bacteriophages in an intermittently operated household-scale slow sand filter. Water Res 42:2662–2670. doi: 10.1016/j.watres.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Napotnik JA, Baker D, Jellison KL. 2017. Effect of sand bed depth and medium age on Escherichia coli and turbidity removal in biosand filters. Environ Sci Technol 51:3402–3409. doi: 10.1021/acs.est.6b05113. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins MW, Tiwari SK, Darby J. 2011. Bacterial, viral and turbidity removal by intermittent slow sand filtration for household use in developing countries: experimental investigation and modeling. Water Res 45:6227–6239. doi: 10.1016/j.watres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 17.Weber-Shirk ML, Dick RI. 1997. Physical-chemical mechanisms in slow sand filters. J Am Water Works Assoc 89:87. doi: 10.1002/j.1551-8833.1997.tb08164.x. [DOI] [Google Scholar]
- 18.Aronino R, Dlugy C, Arkhangelsky E, Shandalov S, Oron G, Brenner A, Gitis V. 2009. Removal of viruses from surface water and secondary effluents by sand filtration. Water Res 43:87–96. doi: 10.1016/j.watres.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Weber-Shirk ML, Dick RI. 1999. Bacterivory by a chrysophyte in slow sand filters. Water Res 33:631–638. doi: 10.1016/S0043-1354(98)00272-3. [DOI] [Google Scholar]
- 20.Haig S-J, Gauchotte-Lindsay C, Collins G, Quince C. 2016. Bioaugmentation mitigates the impact of estrogen on coliform-grazing protozoa in slow sand filters. Environ Sci Technol 50:3101–3110. doi: 10.1021/acs.est.5b05027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vignola M, Werner D, Wade MJ, Meynet P, Davenport RJ. 2018. Medium shapes the microbial community of water filters with implications for effluent quality. Water Res 129:499–508. doi: 10.1016/j.watres.2017.09.042. [DOI] [PubMed] [Google Scholar]
- 22.Pinto AJ, Xi C, Raskin L. 2012. Bacterial community structure in the drinking water microbiome is governed by filtration processes. Environ Sci Technol 46:8851–8859. doi: 10.1021/es302042t. [DOI] [PubMed] [Google Scholar]
- 23.Lautenschlager K, Hwang C, Ling F, Liu W-T, Boon N, Köster O, Egli T, Hammes F. 2014. Abundance and composition of indigenous bacterial communities in a multi-step biofiltration-based drinking water treatment plant. Water Res 62:40–52. doi: 10.1016/j.watres.2014.05.035. [DOI] [PubMed] [Google Scholar]
- 24.Lin W, Yu Z, Zhang H, Thompson IP. 2014. Diversity and dynamics of microbial communities at each step of treatment plant for potable water generation. Water Res 52:218–230. doi: 10.1016/j.watres.2013.10.071. [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Vikram A, Casson L, Bibby K. 2017. Centralized drinking water treatment operations shape bacterial and fungal community structure. Environ Sci Technol 51:7648–7657. doi: 10.1021/acs.est.7b00768. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee N, Bartelli D, Patra C, Chauhan BV, Dowd SE, Banerjee P. 2016. Microbial diversity of source and point-of-use water in rural Haiti—a pyrosequencing-based metagenomic survey. PLoS One 11:e0167353. doi: 10.1371/journal.pone.0167353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nitzsche KS, Weigold P, Lösekann-Behrens T, Kappler A, Behrens S. 2015. Microbial community composition of a household sand filter used for arsenic, iron, and manganese removal from groundwater in Vietnam. Chemosphere 138:47–59. doi: 10.1016/j.chemosphere.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Narihiro T, Straub AP, Pugh CR, Tamaki H, Moor JF, Bradley IM, Kamagata Y, Liu W-T, Nguyen TH. 2014. Ms2 bacteriophage reduction and microbial communities in biosand filters. Environ Sci Technol 48:6702–6709. doi: 10.1021/es500494s. [DOI] [PubMed] [Google Scholar]
- 29.Haig S-J, Quince C, Davies RL, Dorea CC, Collins G. 2014. Replicating the microbial community and water quality performance of full-scale slow sand filters in laboratory-scale filters. Water Res 61:141–151. doi: 10.1016/j.watres.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Centre for Affordable Water and Sanitation Technology. 2012. Biosand filter construction manual. Centre for Affordable Water and Sanitation Technology, Calgary, Alberta. [Google Scholar]
- 31.Young-Rojanschi C, Madramootoo C. 2014. Intermittent versus continuous operation of biosand filters. Water Res 49:1–10. doi: 10.1016/j.watres.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Dini-Andreote F, Stegen JC, Van Elsas JD, Salles JF. 2015. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci U S A 112:E1326. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fierer N, Nemergut D, Knight R, Craine JM. 2010. Changes through time: integrating microorganisms into the study of succession. Res Microbiol 161:635–642. doi: 10.1016/j.resmic.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Jezbera J, Hornak K, Simek K. 2006. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ Microbiol 8:1330–1339. doi: 10.1111/j.1462-2920.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 35.Van Der Gast CJ, Ager D, Lilley AK. 2008. Temporal scaling of bacterial taxa is influenced by both stochastic and deterministic ecological factors. Environ Microbiol 10:1411–1418. doi: 10.1111/j.1462-2920.2007.01550.x. [DOI] [PubMed] [Google Scholar]
- 36.Ortiz-Álvarez R, Fierer N, De Los Ríos A, Casamayor EO, Barberán A. 2018. Consistent changes in the taxonomic structure and functional attributes of bacterial communities during primary succession. ISME J 12:1658–1667. doi: 10.1038/s41396-018-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin JS, Wells GF. 2017. Regional synchrony in full-scale activated sludge bioreactors due to deterministic microbial community assembly. ISME J 11:500. doi: 10.1038/ismej.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ju F, Zhang T. 2015. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISME J 9:683–695. doi: 10.1038/ismej.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindström ES, Kamst-Van Agterveld MP, Zwart G. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl Environ Microbiol 71:8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mustakhimov I, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. 2013. Insights into denitrification in Methylotenera mobilis from denitrification pathway and methanol metabolism mutants. J Bacteriology 195:2207–2211. doi: 10.1128/JB.00069-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirchman DL. 2002. The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 42.Engberg J, On SLW, Harrington CS, Gerner-Smidt P. 2000. Prevalence of Campylobacter Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for Campylobacters. J Clin Microbiol 38:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sack ELW, Van Der Wielen PWJJ, Van Der Kooij D. 2014. Polysaccharides and proteins added to flowing drinking water at microgram-per-liter levels promote the formation of biofilms predominated by Bacteroidetes and Proteobacteria. Appl Environ Microbiol 80:2360. doi: 10.1128/AEM.04105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashidan KK, Bird DF. 2001. Role of predatory bacteria in the termination of a cyanobacterial bloom. Microb Ecol 41:97–105. [DOI] [PubMed] [Google Scholar]
- 45.Liao X, Chen C, Wang Z, Wan R, Chang C-H, Zhang X, Xie S. 2013. Changes of biomass and bacterial communities in biological activated carbon filters for drinking water treatment. Process Biochem 48:312–316. doi: 10.1016/j.procbio.2012.12.016. [DOI] [Google Scholar]
- 46.Norton CD, Lechevallier MW. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl Environ Microbiol 66:268–276. doi: 10.1128/AEM.66.1.268-276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Upadhyaya G, Yuen W, Brown J, Morgenroth E, Raskin L. 2010. Changes in the structure and function of microbial communities in drinking water treatment bioreactors upon addition of phosphorus. Appl Environ Microbiol 76:7473–7481. doi: 10.1128/AEM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emtiazi F, Schwartz T, Marten SM, Krolla-Sidenstein P, Obst U. 2004. Investigation of natural biofilms formed during the production of drinking water from surface water embankment filtration. Water Res 38:1197–1206. doi: 10.1016/j.watres.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 49.Kalmbach S, Manz W, Bendinger B, Szewzyk U. 2000. In situ probing reveals aquabacterium commune as a widespread and highly abundant bacterial species in drinking water biofilms. Water Res 34:575–581. doi: 10.1016/S0043-1354(99)00179-7. [DOI] [Google Scholar]
- 50.Lezcano MÁ, Velázquez D, Quesada A, El-Shehawy R. 2017. Diversity and temporal shifts of the bacterial community associated with a toxic cyanobacterial bloom: an interplay between microcystin producers and degraders. Water Res 125:52–61. doi: 10.1016/j.watres.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 51.Proctor CR, Besmer MD, Langenegger T, Beck K, Walser J-C, Ackermann M, Bürgmann H, Hammes F. 2018. Phylogenetic clustering of small low nucleic acid-content bacteria across diverse freshwater ecosystems. ISME J 12:1344–1359. doi: 10.1038/s41396-018-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santegoeds CM, Ferdelman TG, Muyzer G, de Beer D. 1998. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl Environ Microbiol 64:3731–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corsaro D, Feroldi V, Saucedo G, Ribas F, Loret J-F, Greub G. 2009. Novel Chlamydiales strains isolated from a water treatment plant. Environ Microbiol 11:188–200. doi: 10.1111/j.1462-2920.2008.01752.x. [DOI] [PubMed] [Google Scholar]
- 54.Thomas V, Casson N, Greub G. 2006. Criblamydia sequanensis, a new intracellular Chlamydiales isolated from seine river water using amoebal co-culture. Environ Microbiol 8:2125–2135. doi: 10.1111/j.1462-2920.2006.01094.x. [DOI] [PubMed] [Google Scholar]
- 55.Mallon CA, Elsas JDV, Salles JF. 2015. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol 23:719–729. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Lee C, Agidi S, Marion JW, Lee J. 2012. Arcobacter in Lake Erie beach waters: an emerging gastrointestinal pathogen linked with human-associated fecal contamination. Appl Environ Microbiol 78:5511. doi: 10.1128/AEM.08009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett KW, Eley A. 1993. Fusobacteria: new taxonomy and related diseases. J Med Microbiol 39:246–254. doi: 10.1099/00222615-39-4-246. [DOI] [PubMed] [Google Scholar]
- 58.Ilhan ZE, Dibaise JK, Isern NG, Hoyt DW, Marcus AK, Kang D-W, Crowell MD, Rittmann BE, Krajmalnik-Brown R. 2017. Distinctive microbiomes and metabolites linked with weight loss after gastric bypass, but not gastric banding. ISME J 11:2047. doi: 10.1038/ismej.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawson PA, Caldwell ME. 2014. The family Carnobacteriaceae, p 19–65. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: Firmicutes and Tenericutes. Springer Berlin Heidelberg, Berlin, Germany. [Google Scholar]
- 60.Lory S. 2014. The family Streptococcaceae, p 367–370. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: Firmicutes and Tenericutes. Springer Berlin Heidelberg, Berlin, Germany. [Google Scholar]
- 61.Steelman SM, Chowdhary BP, Dowd S, Suchodolski J, Janečka JE. 2012. Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet Res 8:231. doi: 10.1186/1746-6148-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta RS, Chen WJ, Adeolu M, Chai Y. 2013. Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int J Syst Evol Microbiol 63:3379–3397. doi: 10.1099/ijs.0.048371-0. [DOI] [PubMed] [Google Scholar]
- 63.Thomas V, McDonnell G, Denyer SP, Maillard J-Y. 2010. Free-living amoebae and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev 34:231–259. doi: 10.1111/j.1574-6976.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 64.Berry D, Xi C, Raskin L. 2006. Microbial ecology of drinking water distribution systems. Curr Opin Biotechnol 17:297–302. doi: 10.1016/j.copbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Castelle CJ, Brown CT, Thomas BC, Williams KH, Banfield JF. 2017. Unusual respiratory capacity and nitrogen metabolism in a parcubacterium (OD1) of the candidate phyla radiation. Sci Rep 7:40101. doi: 10.1038/srep40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson WC, Stegen JC. 2015. The reduced genomes of parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol 6:713. doi: 10.3389/fmicb.2015.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McLean JS, Lombardo M-J, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, Vyahhi N, Hall AP, Yang Y, Dupont CL, Ziegler MG, Chitsaz H, Allen AE, Yooseph S, Tesler G, Pevzner PA, Friedman RM, Nealson KH, Venter JC, Lasken RS. 2013. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci U S A 110:E2390–E2399. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davidov Y, Jurkevitch E. 2004. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax–Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol 54:1439–1452. doi: 10.1099/ijs.0.02978-0. [DOI] [PubMed] [Google Scholar]
- 69.Šimek K, Chrzanowski TH. 1992. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl Environ Microbiol 58:3715–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jellison KL, Dick RI, Weber-Shirk ML. 2000. Enhanced ripening of slow sand filters. J Environ Eng 126:1153–1157. doi: 10.1061/(ASCE)0733-9372(2000)126:12(1153). [DOI] [Google Scholar]
- 71.Calvo-Bado LA, Pettitt TR, Parsons N, Petch GM, Morgan JAW, Whipps JM. 2003. Spatial and temporal analysis of the microbial community in slow sand filters used for treating horticultural irrigation water. Appl Environ Microbiol 69:2116–2125. doi: 10.1128/AEM.69.4.2116-2125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hong H, Qiu J, Liang Y. 2010. Environmental factors influencing the distribution of total and fecal coliform bacteria in six water storage reservoirs in the Pearl River Delta Region, China. J Environ Sci 22:663–668. doi: 10.1016/S1001-0742(09)60160-1. [DOI] [PubMed] [Google Scholar]
- 73.Karn SK, Harada H. 2001. Surface water pollution in three urban territories of Nepal, India, and Bangladesh. Environ Manage 28:483–496. doi: 10.1007/s002670010238. [DOI] [PubMed] [Google Scholar]
- 74.Schraft H, Watterworth LA. 2005. Enumeration of heterotrophs, fecal coliforms and Escherichia coli in water: comparison of 3M Petrifilm plates with standard plating procedures. J Microbiol Methods 60:335–342. doi: 10.1016/j.mimet.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 75.American Public Health Association, American Water Works Association Water Environment Federation. 2012. Standard methods for the examination of water and wastewater, 22nd ed American Public Health Association, Washington, DC. [Google Scholar]
- 76.Bowers RM, Clements N, Emerson JB, Wiedinmyer C, Hannigan MP, Fierer N. 2013. Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ Sci Technol 47:12097–12106. doi: 10.1021/es402970s. [DOI] [PubMed] [Google Scholar]
- 77.Apprill A, McNally S, Parsons R, Weber L. 2015. Minor revision to v4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 78.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2010. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC. 2010. Search and clustering orders of magnitude faster than blast. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 80.Edgar RC. 2018. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34:2371–2375. doi: 10.1093/bioinformatics/bty113. [DOI] [PubMed] [Google Scholar]
- 81.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McDonald D, Price MN, Goodrich J, Nawrocki EP, Desantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, Peacock SJ, Smith GCS, Parkhill J. 2018. Recognizing the reagent microbiome. Nat Microbiol 3:851–853. doi: 10.1038/s41564-018-0202-y. [DOI] [PubMed] [Google Scholar]
- 84.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 85.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wickham H. 2016. Ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 87.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecology 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 88.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2018. Vegan: community ecology package. https://CRAN.R-project.org/package=vegan.
- 89.Fernandes AD, Reid JN, Macklaim JM, Mcmurrough TA, Edgell DR, Gloor GB. 2014. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2:15. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macklaim JM, Clemente JC, Knight R, Gloor GB, Reid G. 2015. Changes in vaginal microbiota following antimicrobial and probiotic therapy. Microb Ecol Health Dis 26:27799. doi: 10.3402/mehd.v26.27799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chiao T-H, Clancy TM, Pinto A, Xi C, Raskin L. 2014. Differential resistance of drinking water bacterial populations to monochloramine disinfection. Environ Sci Technol 48:4038–4047. doi: 10.1021/es4055725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.