Abstract
Inheritable cardiac disorders often associate with increased risk of sudden death in the young, as well as with structural damage to the myocardium. Early linkage analysis studies in Mendelian forms of these diseases, such as hypertrophic cardiomyopathy and long QT syndrome, uncovered large-effect genetic variants that contribute to the phenotype. In more recent years, through genotype-phenotype studies and methodological advances in genetics, it has become evident that most inheritable cardiac disorders are not monogenic but rather, have a complex genetic basis wherein multiple genetic variants contribute (oligogenic or polygenic inheritance). Conversely, studies on genes underlying these disorders uncovered pleiotropic effects, with a single gene affecting multiple and apparently unrelated phenotypes. In this review, we explore these two phenomena: on one hand, the evidence that variants in multiple genes converge to generate one clinical phenotype and on the other, the evidence that variants in one gene can lead to apparently unrelated phenotypes. While multiple conditions are addressed in order to illustrate these concepts, the experience obtained in the study of Long QT Syndrome, Brugada syndrome and of arrhythmogenic right ventricular cardiomyopathy, as well as in the study of functions related to SCN5A (the gene coding for the alpha-subunit of the most abundant sodium channel in the heart) and PKP2 (the gene coding for the desomosomal protein plakophilin-2) is discussed in more detail.
Keywords: Brugada syndrome, Long QT, ARVC, pleiotropy, complex genetics, sodium channel, plakophilin-2, gene mutation
INTRODUCTION
Our understanding of the genetic architecture of the inherited cardiac disorders associated with sudden cardiac death in the young, namely the primary electrical disorders and the cardiomyopathies, has increased markedly over the past years. Initial discoveries pertained to the identification of genes underlying disorders with a Mendelian (monogenic) inheritance. In more recent years, gene discovery efforts have evolved to consider the more complex inheritance that likely underlies the majority of cardiac phenotypes, even those previously assumed to have a simpler inheritance. Such complex genetic architecture is likely to range from oligogenic, where few genetic loci contribute to the disorder, to polygenic, with the involvement of multiple loci. Alongside the notion that inherited cardiac disorders may be the outcome of multiple discrete genetic effects, the inverse has also become clear, in that some genes have been recognized to exert remarkable pleiotropic effects, i.e., they affect multiple apparently unrelated phenotypes. This pleiotropism can lead to different disease manifestations, or contribute to different endophenotypes of the same disorder. These two topics, complex genetics and pleiotropy, which converge to advance our understanding of the relation between gene variants and inherited cardiac phenotypes, are reviewed in two separate sections. While multiple conditions are addressed, in order to illustrate these concepts, the experience obtained in the study of Long QT Syndrome, Brugada syndrome and of arrhythmogenic (right ventricular) cardiomyopathy are discussed in more detail. Although they are neither the only nor the first genes studied from this perspective, for simplicity of presentation we place particular emphasis on two genes, namely SCN5A (the gene coding for the alpha-subunit of the most abundant sodium channel in the heart) and PKP2 (the gene coding for the desomosomal protein plakophilin-2), for which studies in model systems and patients have provided strong evidence for pleiotropic effects, spanning both electrical and structural phenotypes.
COMPLEX GENETICS IN INHERITED CARDIAC DISORDERS
Genetic architecture of inherited cardiac disorders: modifier genes
Genetic variants contributing to human traits, including cardiac phenotypes, vary in their frequency and effect1. Typically, an inverse relation exists between variant frequency and effect magnitude, where large effect variants are rare, and small –but detectable- effect variants are common. This relationship was first suggested by population genetic models, and later observed in data from from association and sequencing studies in large cohorts1.
The large effect of rare genetic variants underlying Mendelian cardiac disorders, such as hypertrophic cardiomyopathy (HCM) and long QT syndrome (LQTS), facilitated the identification of genes underlying these disorders by means of linkage analysis that tracks genetic loci within large pedigrees. These discoveries, which took off in the late 1980s, ushered us into the era of cardiogenetics, which saw the identification of several genes causally related to the cardiomyopathies and the primary electrical disorders (reviewed in2). These advances have undoubtedly impacted the genetic diagnosis and clinical management of affected families. Yet, with few exceptions, knowledge of the disease-causing genes has had a relatively modest impact on the ability to predict important clinical aspects such as the age of onset of the disease, its rate of progression, or the development of major cardiac events including heart failure or sudden cardiac death. Studies in families with multiple mutation-positive individuals (i.e. positive for the familial pathogenic variant) have shown that disease penetrance (proportion of mutation carriers with the disorder) can be low, and that among those with disease manifestations, there can be broad variability in the types of symptoms and severity thereof (variable expression)3. These observations have made it clear that allocating these disorders exclusively to a mutation at a single locus might be an oversimplification of biological phenomena.
Factors such as age, gender4, environmental factors or co-morbidities may modulate the effects of the primary genetic defect. For example, a recent study has shown that hypertension may modulate arrhythmia risk in patients with the Dutch founder mutation SCN5A-1795insD, a mutation causing an overlap phenotype of LQTS and conduction disease5. Yet, besides such factors, the inheritance of other genetic variants, commonly referred to as genetic modifiers, alongside the primary genetic defect, is believed to be a potential cause of inter-individual variability in disease expression (Figure 1). Genetic modifiers may act to exacerbate the severity of the primary genetic defect or cause its presentation in childhood as opposed to adulthood, or may protect a carrier of a primary genetic defect from developing the disease. A role for genetic modifiers in modulation of the ultimate phenotypic expression of a disease-causing mutation has been firmly established in mouse genetic studies across multiple phenotypes, including those related to the heart. For example, the electrophysiological consequences of the Scn5a-1798insD variant (homologous to the human SCN5A-1795insD mutation mentioned above) was found to differ between 129P2 and FVB/NJ mouse inbred strains6.
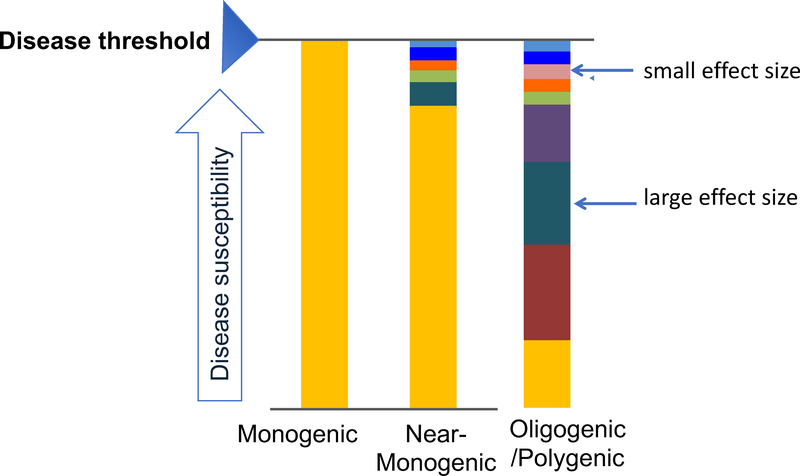
Figure 1:
Evolving model of genetic architecture of inherited arrhythmias and cardiomypathies. In the classic monogenic model, a single disease-causing variant is sufficient for phenotypic manifestation. In most families with a disease-causing variant (i.e. mutation-positive), additional common variants with small effect sizes are needed to reach the disease threshold (near-monogenic). In mutation-negative families, the genetic architecture may be more complex, comprising multiple genetic variants with different effect sizes that together determine disease risk (oligogenic/polygenic), in addition to non-genetic factors. (Reprinted with permission from2.)
The identification of genetic modifiers in the Mendelian cardiac disorders is an area of significant interest as their identification could help refine individual risk prediction for cardiac events. The nature of the genetic component that modifies the effect of the primary genetic defect in patients with inherited cardiac disorders, such as the type of variants that are operative, their number and their effect size, is unknown. It may differ across the different disorders and between populations of different ancestries (due to founder effects), but as for other inherited disorders, it is generally presumed to comprise variants of small effect that are common in the general population (commonly considered to be those with a minor allele frequency (MAF) ≥1%), and variants that are low-frequency/rare (MAF<1%) and that have an intermediate to large effect. It likely involves variants of different classes, spanning sequence variants such as single nucleotide variants to structural variants such as copy number variants and chromosomal rearrangements in coding and non-coding regions of the genome. Furthermore, such variants may occur in genes pertaining to different biological pathways that contribute to the disorder.
Genetic modifier discovery
In the inherited cardiac disorders, studies for identification of genetic modifiers have so far been primarily conducted in patients with LQTS and have largely focused on the potential modulatory effects of common single nucleotide polymorphisms (SNPs) of small effect. Initial studies tested the effect of SNPs in candidate genes (selected on the basis of known or suspected biological considerations).7 This approach (with its poor replication record across a broad range of human phenotypes8), has now been replaced by studies testing the modulatory potential of SNPs that had been robustly demonstrated to affect the QTc-interval in prior genome-wide association studies (GWAS) conducted in large samples of the general population9–11. The premise in these studies is that uncovering the genetic underpinnings of the QT-interval, a relevant endophenotype for LQTS, will inform us on genetic factors that could impact on variability in LQTS susceptibility or severity (Figure 2).
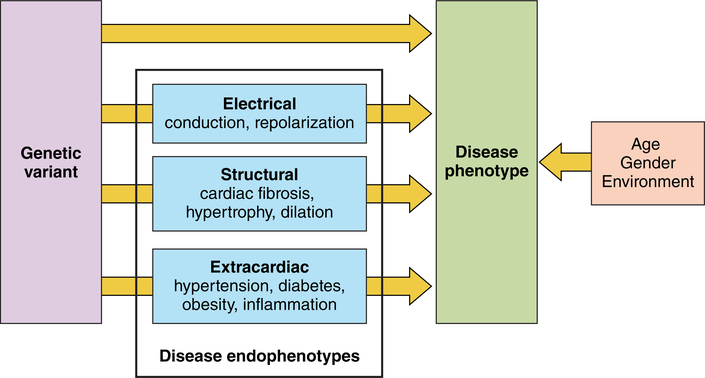
Figure 2:
Phenotypic expression is determined by genetic variation in addition to age, gender and environmental factors. The causal relation between genetic variation and disease could be either direct or through one or multiple endophenotypes.
The first QT-interval GWAS which was published in 2006 identified common genetic variants at the NOS1AP gene (encoding nitric oxide synthase adaptor protein) as modulators of the QT-interval in the general population12. Four studies subsequently demonstrated that these variants can also modulate the QT-interval and risk of arrhythmia in patients with LQTS9, 10, 13, 14. Similar data is available for SNPs at the KCNQ1 gene locus11, 13 which had been identified in later (larger) general population QT-interval GWAS15, 16. As expected for small-effect common variants (in the general population they individually modulate the QT-interval by only ~ 0.5 to 2 ms, although NOS1AP SNPs seem to be associated with a somewhat larger effect, e.g. the most significantly associated NOS1AP SNP, rs12143842, has an effect size of 3.5 ms), these ‘QT-SNPs’ individually explain only a small portion of inter-individual variability in phenotype among patients with LQTS. While clearly still far from clinical applicability, in an effort to explain a greater portion of the variability, a genetic modifier study in LQTS considered the effect of multiple SNPs in aggregate in the form of a polygenic risk score (PRS; typically calculated as the sum of trait-associated alleles, each weighted by its effect size; Figure 3) 13. In this study, a PRS based on 22 ‘QT-SNPs’ showed a strong linear relationship with QT-interval in patients with LQTS type 2 (caused by a mutation in KCNH2). This study also showed that patients with a PRS in the lowest quartile had a lower relative risk of cardiac events compared to patients in the other quartiles combined. The largest GWAS thus far for QT-interval was conducted in ~75,000 individuals of European descent and identified 68 independent SNPs at 35 loci17 that collectively explain ∼10% of QT-interval variation in the general population. The incorporation of addtional ‘QT-SNPs’ in PRS, as these become known, has the potential of explaining a greater portion of inter-individual disease variability in LQTS patients. This may in turn translate into potential clinical application of common variant genotyping in this disorder.
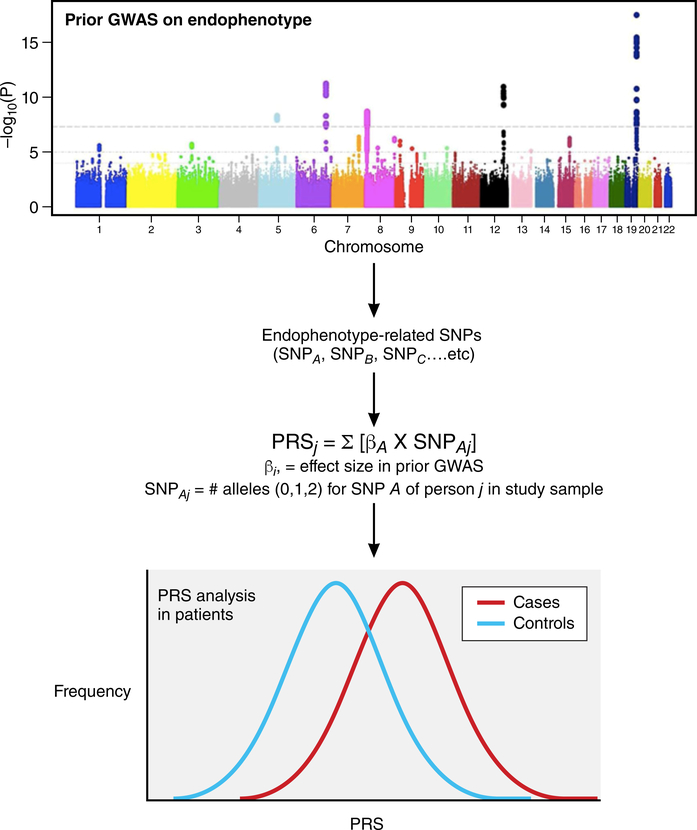
Figure 3:
Assessing the role of endophenotype-related single nucleotide polymorphisms (SNPs) in disease susceptibility and risk prediction. Large-scale genome-wide association studies (GWAS) identify endophenotype-related SNPs. Polygenic risk scores (PRS) are then calculated for cases and controls by summing the number of alleles weighted by the allele-specific effect size (β) for all endophenotype-related SNPs. The association of PRS with disease not only confirms a role for the endophenotype in question in disease susceptibility, but also contribute to the development of genetic risk predictors.
GWAS studies that have been conducted thus far for the QT-interval in the general population have generally been statistically powered to interrogate variants with a MAF >1%. The lowest-frequency variant associated with the QT-interval is the p.Asp85Asn variant in KCNE1 (MAF = 1% in populations of European ancestry; Figure 418–20). Commensurate with its lower frequency, this variant has the largest effect size of all ‘QT-SNPs’ uncovered thus far in GWAS (7.42 ms per minor allele)17. it has been shown to predispose to LQTS in at least two studies conducted respectively in European and Japanese patients7, 21 and was reported to modulate the QT-interval in Finnish males with the KCNQ1-p.Gly589Asp founder mutation22. Conversely, this variant may also be considered a low penetrance rare variant that could in turn be modulated by other genetic or non-genetic factors such as QT-prolonging drugs.
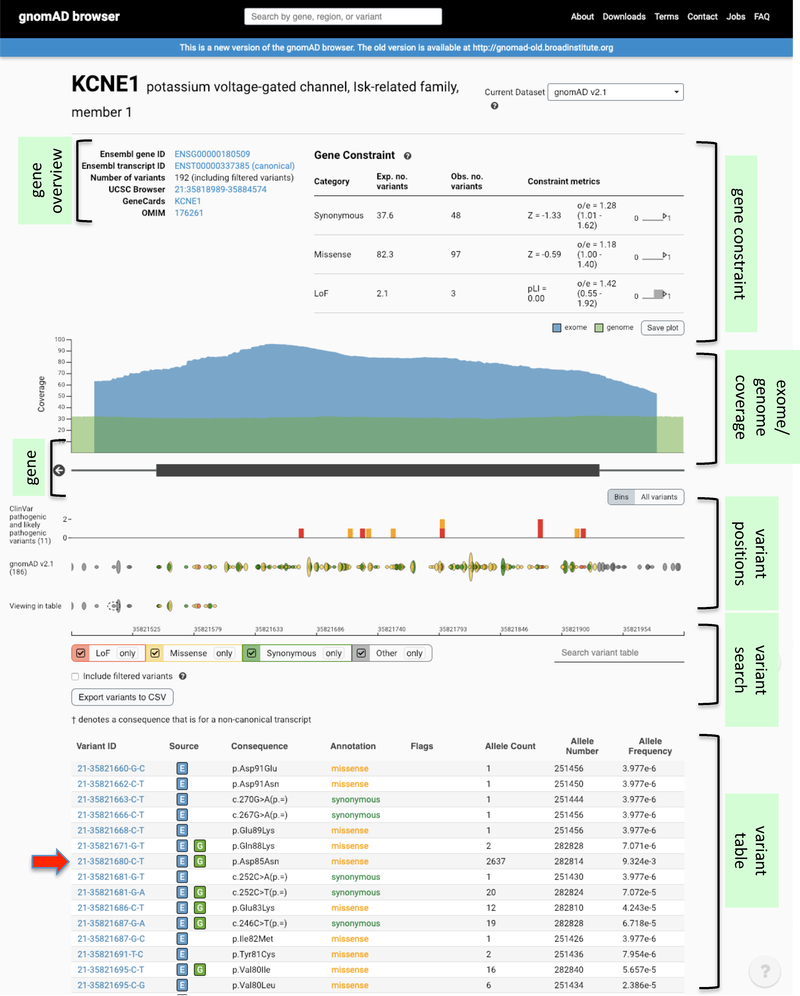
Figure 4:
Screenshot of a gnomAD database search for the KCNE1 gene. The gnomAD database aggregates and harmonizes exome and genome sequencing data from a variety of large-scale sequencing projects, and provides summary data available for the wider scientific community. As of December 2018, the gnomAD data set comprised sequence data spanning 125,748 exomes and 15,708 genomes from unrelated individuals of different ethnicities that were sequenced as part of various disease-specific and population genetic studies, totalling 141,456 individuals. The low-frequency p.Asp85Asn variant that has been associated with QT-interval and susceptibility to LQTS and drug-induced long QT syndrome, is indicated by the red arrow. Through the browser, one can appreciate aspects such as the tolerance of the gene to different types of variant categories (in the form of an observed / expected (oe) metric), as well as the genomic position, allele count and allele frequency of detected variants. More details at18–20.
Of note, p.Asp85Asn in KCNE1 and SNPs at the NOS1AP locus have also been associated with drug-induced LQTS23. Furthermore, a PRS based on 61 ‘QT-SNPs’ (of the above-mentioned 68 ‘QT-SNPs) was shown to explain a substantial proportion of QT-interval response to QT prolonging drugs in a trial of 3 QT-prolonging drugs conducted in healthy individuals, as well as risk of torsade de pointes in patients24. This work further illustrates how ‘QT-SNPs’ identified in the general population can be tested for predictive effects, individually or implemented in polygenic risk scores, in clinical cohorts. It also underscores how common genetic variation modulates both acquired and genetic risk for QT-interval prolongation.
Besides the QT-interval, GWAS has been conducted on multiple other ECG traits, including heart rate, PR-interval and QRS duration7, and although to our knowledge not yet tested, SNPs that impact on these traits are also expected to modulate inherited cardiac disorders such as sinus node dysfunction and conduction disease. However, compared to the current understanding of common genetic variants that modulate ECG traits, progress in the identification of common variants underlying endophenotypes of relevance to the cardiomyopathies, such as left ventricular dimensions, mass and function, has been slow. This is at least partly related to the fact that unlike the ECG, which is amenable to accurate measurement in large-scale cohorts, echocardiography or cardiac magnetic resonance imaging for measurement of these parameters is costly and their output is less systematically quantifiable. To circumvent these issues, one recent study sought to identify genetic determinants of myocardial mass by evaluating ECG proxy markers of this trait, identifying 52 such loci in a meta-analysis of over 73,000 individuals; yet the role of these loci in modulating disease severity in individuals with Mendelian forms of a cardiomyopathy such as HCM remains to be established. GWAS studies of cardiomyopathies have thus far focused on the identification of susceptibility loci through comparisons of patients with controls, and have been conducted in patients with HCM or DCM25–28. The role of these common susceptibility variants in modulating disease severity among patients remains to be demonstrated.
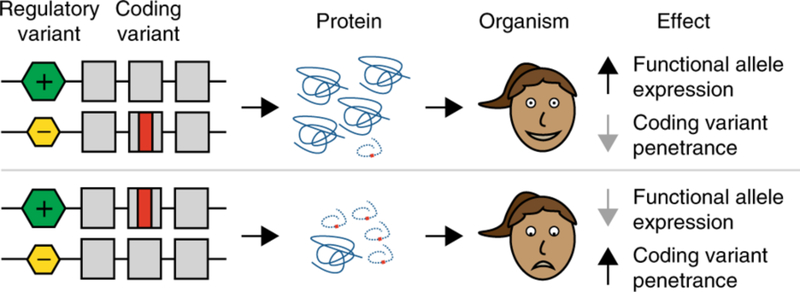
Modifier effect of cis-regulatory variation
Cis-regulatory elements are regions of non-coding DNA, such as promoter or enhancer elements, which regulate the transcription of neighboring genes. A recent study showed that genetic variants occurring in such cis-regulatory elements may act as modifiers of disease-causing variants located in the coding region of the gene, by controlling the dosage of mRNA and, in turn, the dosage of the functional gene product 29. Thus, coding region disease variants located on haplotypes that lead to increased gene product are associated with increased disease penetrance, while disease variants located on haplotypes that lead to decreased gene product are associated with decreased penetrance (Figure 5). Such a mechanism has been suggested to be operative in LQTS30. In this study, Amin and co-workers showed that genetic variation in the 3’ untranslated region of the KCNQ1 gene was associated with altered gene expression in in vitro reporter studies. They also showed that disease severity of KCNQ1 mutations depended on whether they occurred on the high-expression or low-expression haplotype30. Yet, these effects await further scrutiny as they were not replicated in another study conducted on three large families with founder mutations in KCNQ131.
Figure 5:
Regulatory variants as modifiers. Coding region variants on lower-expressed haplotypes are expected to lead to decreased penetrance whereas coding region variants on higher-expressed haplotypes are expected to increase penetrance (reprinted with permission from Springer Nature: Nature Genetics, reference29).
Integrative approaches for the identification of genetic modifiers
In general, genetic studies aimed at identifying genetic modifiers in patients are challenging due to the relatively small number of patients that are available, given the rarity of these disorders. In an approach that the authors referred to as ‘physiological genomics,’ a group led by Deschênes recently combined electrophysiological studies in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) with whole exome sequencing to identify genetic modifiers operative in an LQTS family with a KCNH2 genetic defect32. They demonstrated distinct differences in cellular electrophysiology between hiPSC-CMs from severely affected patients and those of mildly affected first-degree relatives, including an increased L-type Ca2+ current (ICa,L) and more pronounced action potential (AP) prolongation. Through whole exome sequencing they identified in severely affected individuals a variant in the GTP-binding protein REM2, which was previously described to regulate voltage-gated Ca2+ channels33. Indeed, correcting this variant by genome editing reversed the enhanced ICa,L and the prolonged AP in the severely affected hiPSC-CMs, demonstrating its functional relevance in modulating disease expressivity. This work moreover shows that functional studies are not only essential to validate the role of identified modifier genes and understand the mechanism through which they act, but may also contribute to the actual identification of modifier genes.
Other investigators have resorted to alternative strategies involving system genetic studies in model organisms. These studies have exploited the defined genetic background of inbred mice or rats and the genetically determined phenotypic variability among these strains in cardiac structure and function, including cardiac mass, calcium homeostasis and electrical characteristics. Such studies have, among others, led to the identification of TNNI3K, encoding troponin 1 interacting kinase, as a modifier of cardiomyopathy (in the setting of cardiac-specific calsequestrin overexpression34) and cardiac conduction (in the setting of the Scn5a-1798insD mutation35). Of note, although the underlying mechanism remains unknown, rare genetic variants in this gene were later associated with a syndrome of conduction system disease, atrial tachyarrhythmia and dilated cardiomyopathy in several families36. In another study, comparison of cardiac gene expression between two mouse strains with the Scn5a-1798insD mutation (129P2 and FVB/N) with different disease severity, led to the identification of Scn4b, encoding the sodium channel accessory subunit β4, as a potential modifier of conduction disease severity6. Independent of the strategy used, it remains however essential to explore multiple lines of evidence before defining novel (modifier) genes as causal, ideally including both experimental and clinical (genetic) approaches.
From monogenic to polygenic
For some disorders, the initial discovery of rare genetic variants within plausible genes and the observation of familial recurrence in some cases led to the perception that Mendelian inheritance may apply. One notable disorder in this respect is Brugada Syndrome (BrS). Rare coding pathogenic variants in SCN5A are found in ~20% of cases and functional studies have provided unequivocal evidence of a loss-of-function mechanism37. Notwithstanding, genotype-phenotype studies in large families harboring rare pathogenic variants in SCN5A demonstrated incomplete penetrance, and importantly, the occurrence of phenotype-positive genotype-negative individuals, demonstrating that such variants are neither necessary nor sufficient to cause the disorder38. This observation, together with the fact that the disorder often has a sporadic presentation (i.e. absence of family history of the disorder)39, suggests that it has complex inheritance. A GWAS that was conducted in the setting of an international consortium, bringing together probands from different countries, identified 3 independent susceptibility variants, close to the SCN5A, SCN10A and HEY2 genes, providing the first evidence for the role of common genetic variation in susceptibility for the disorder40. When considered in aggregate, the identified genetic variants displayed a high cumulative effect on relative risk. Yet, the absolute risk they confer is most likely small considering the low disease prevalence, and many susceptibility variants remain to be found. These will likely include additional common variants but likely also low-frequency and rare variants, the discovery of which will require sequencing based approaches (such as exome or whole genome sequencing). Besides paving the way for clinical risk prediction in the future, the identification of novel loci in disorders with complex inheritance such as BrS provides a handle for studies aimed at understanding underlying mechanisms. For example, functional studies on the transcription factor gene HEY2 from the chromosome 6q22.31 BrS susceptibility locus identified alterations of ion channel expression across the ventricular wall as a potential disease mechanism.41
Potential polygenic basis of other inherited cardiac disorders
Genetic architecture, in terms of the number of variants needed to reach the threshold to show disease and their respective effect size, likely differs not only across disorders but also within the same disorder. For instance, around 20% of LQTS probands remain mutation-negative upon gene panel testing. Some of these cases could be due to undetected mutations in the known genes, such as mutations in deep intronic (as recently demonstrated in HCM42) or regulatory regions, that are thus far not routinely screened for. However, in cases without familial aggregation, where a highly penetrant rare variant is therefore less likely (with the exception of de novo or recessive mutations), one could argue that the genetic basis is possibly polygenic. Common variants have been shown to explain at least 21% of the QT interval variance in the general population43 and it is thus conceivable that mutation-negative LQTS cases simply carry a large burden of common QT prolonging alleles. Similarly, mutation pick-up rate of genetic testing in HCM is 40–60% and that of DCM is 15%−40% (depending on patient selection and family history44, 45). This, together with the fact that a large percentage of patients have a sporadic presentation, argues for a polygenic basis at least in some patients (alongside acquired risk factors). It is likely that a continuum of complexity of genetic architecture exists ranging from monogenic whereby the disease threshold is reached by the action of a single variant, to highly polygenic where many loci are needed (Figure 1).
Complex genetics: summary and future directions
In summary, strong evidence exists for the role of genetic modifiers in the determination of the ultimate disease manifestations in individuals with inherited cardiac disorders. Furthermore, it has become clear that some inheritable cardiac disorders may not be strongly determined by the inheritance of one large genetic defect but rather, have a more complex inheritance. Recent studies using state-of-the-art genomic approaches have started to uncover such modulatory and susceptibility genetic variants, yet much remains to be done. The rarity of the inherited cardiac disorders coupled to the large numbers of patients needed for sufficient statistical power for gene discovery in the context of complex inheritance calls for the continuation and expansion of international collaborations aimed at bringing together large numbers of highly characterized patients (as is ongoing for BrS40), and the establishment of similar networks for other disorders. Initiatives such as the European Reference Network (ERN) GUARD-Heart of the European Union (guardheart.ern-net.eu), could facilitate the establishment of such collaborations.
An extension of genetic association studies to include low-frequency variants is needed, although these studies are highly challenging as they will require a many-fold increase in sample sizes to obtain sufficient statistical power, besides the use of alternative statistical approaches such as burden tests and/or variance-component tests for rare variants. Exploring rare and low-frequency variants would shed light on new disease loci with the potential to better understand (patho)physiology, and also would increase the predictive value of polygenic risk scores since low-frequency variants typically show higher effect sizes than common ones1.
Across different fields of medicine, the application of SNP genotyping in clinical practice for risk prediction and precision prevention is promising but remains to be established46. For example, in coronary artery disease, atrial fibrillation and diabetes, genome-wide polygenic scores have been shown to predict disease with clinically relevant accuracy, and identify individuals with risk equivalent to monogenic mutations46. For these disorders, prospective outcome studies in diverse populations are needed to confirm the predictive value of such scores. Data emerging from the exploration of the PRS based on 61 ‘QT-SNPs’ in diLQTS is encouraging24 and, pending confirmation in large prospective studies in real-world collections of drug-exposed patients, such a PRS (updated as new variants are discovered) could potentially be used to individualize assessment of risks and benefits of drugs with high risk for drug-induced arrhythmias. It is hoped that as genetic loci impacting on phenotypic variability of inherited cardiac disorders are uncovered, their implementation in PRS could in the future also allow for a refined risk prediction and individualized patient management. The application of PRS could enable increased monitoring of patients that are at risk for severe cardiac events or the provision of more aggressive therapy. Also, PRS predicting rare arrhythmias and cardiomyopathies could be used to predict risk of disease occurrence within families. For instance, family members of genotype-negative cardiomyopathy cases or unaffected mutation carriers currently undergo periodic (nearly lifelong) screening using cardiac imaging47. Such an approach is associated with significant cost with an expected low yield. PRS could potentially allow for a refined risk prediction and an individualized screening program.
PLEIOTROPY: MULTIPLE TRAITS ASSOCIATED WITH PKP2 AND SCN5A
In addition to genetic complexity, pleiotropy (i.e. a single gene affecting multiple and apparently unrelated phenotypes) is an important aspect of genotype-phenotype relationships in inherited arrhythmia syndromes. This has become particularly evident in the setting of arrhythmogenic cardiomyopathy and sodium channelopathy, originally considered separate disease entities48. The conventional separation of cardiac genes as coding for “structural” or “channel” proteins suggests, at first glance, that a gene product is responsible for one function. As such, a “sodium channel protein” is commonly thought of in the context of a single task: to pass sodium through a hydrophilic pore. Similarly, “desmosomal proteins” were so named because of their participation in a cell adhesion complex (though additional functions as signaling hubs and transcriptional regulators have been defined and are mentioned later in this article). A linear extension of this one gene-one trait logic would be that mutations in one gene yield a reproducible, consistent phenotype, as the trait is manifested directly from the loss or gain of one function. This notion of one gene-one trait contrasts with advances suggesting that most genes are pleiotropic, that is, they are responsible for multiple traits 49, 50, and with research indicating that mutations in one gene can lead to multiple and sometimes seemingly unrelated phenotypes. A case in point is that of Connexin43 (Cx43) which, though best studied as an ion channel-forming protein, is associated with a highly pleiotropic inheritable disease 51. Here, we explore non-canonical functions identified for genes that, while considered “structural”, are importantly involved in arrhythmic phenotypes and also, genes coding for “channel proteins” that, when mutated, lead to structural disease.
PKP2 and Arrhythmogenic Cardiomyopathy: from the structural gene to the electrical phenotype
Plakophilin-2 (PKP2) is one of the structural components of the cardiac desmosome, part of the Armadillo family of proteins. In 2004, Gerull and colleagues52 reported for the first time the link between heterozygous mutations in the PKP2 gene, coding for plakophilin-2, and Arrhythmogenic Cardiomyopathy (ACM also known as arrhythmogenic right ventricle cardiomyopathy, ARVC). ACM is a heritable disease, characterized by high propensity for ventricular arrhythmias and sudden death and progressive fibrosis and fibrofatty infiltration of the ventricles, most often of right ventricular predominance48. The prevalence of ACM varies among regions and it is estimated at 1:2000 to 1:5000 in the general population53. Research since the first description of the disease by Marcus and Fontaine in 198254 has allowed for a more comprehensive characterization of the clinical features that can be encompassed under the single diagnosis of ACM. In general terms, the full spectrum is characterized by 4 phases: 1) an early or “concealed’ phase with high incidence of ventricular fibrillation and sudden cardiac death (11% of index patients reviewed by Groeneweg et al presented with cardiac arrest; median age at cardiac arrest was 25 years53); 2) an overt arrhythmic stage in the presence of initial structural changes predominantly in the RV; 3) global RV dysfunction and 4) biventricular dilated cardiomyopathy and heart failure55. The spectrum of manifestations has been compiled and classified under criteria that are the current standard for diagnosis55, though these criteria are likely to evolve as knowledge expands. Mutations in additional genes coding for other cardiac desmosomal or non-desmosomal proteins have been linked to ACM, but PKP2 remains the most prevalent disease gene, accounting for ~40% of genotype positive families53. Of note, in approximately 37% of index patients reported by Groeneweg et al53, no mutation could be identified. As in other diseases discussed in the previous section, ACM may present as a result of the convergence of multiple variants56.
When desmosomal mutations were associated with familial ACM the inference was made that a defective desmosome, due to a loss of function mutation, could facilitate fibrosis development by weakening intercellular adhesion between cardiomyocytes. Yet, other key features of the disease, such as the abundant adiposis, or the high likelihood of life-threatening arrhythmias in the early stages of the disease, remained not completely explained by thinking of PKP2 as a uni-functional molecule. The question remained as to how a molecule that is supposed to be “making Velcro” between cells could, when mutated, drastically alter the electrical stability of the heart, even prior to the onset of overt cardiomyopathic changes57. The first approach at understanding the relation between PKP2 or other desmosomal proteins and arrhythmogenesis came from the observation that hearts from patients with ACM showed a loss of gap junction plaques58. Experimental observations followed, confirming that PKP2 expression was necessary for the maintenance of normal intercellular communication through gap junctions 59. Given the physical proximity between desmosomes and gap junctions in the cardiac intercalated disc (see, e.g.,60), and the importance of cell adhesion to gap junction formation61, it was still possible to argue that the loss of gap junction plaques was a direct consequence of the loss of intercellular adhesion.
PKP2 impacts on sodium current: relevance for ACM and Brugada syndrome (BrS)
To assess whether or not the electrically-relevant functions of PKP2 in cardiomyocytes were direct consequences of intercellular adhesion properties, Sato et al measured sodium current in single, isolated cells (hence devoid of intercellular structures) in the presence or absence of PKP262. The data showed that loss of expression of PKP2 in adult and in neonatal rat ventricular myocytes (using shRNA technology) led to a decrease in sodium current (INa) density and a shift in voltage-dependent inactivation properties of the voltage-gated sodium channel NaV1.5. The relation between “desmosomal” protein abundance or sequence integrity and the properties of NaV1.5 was later confirmed for other experimental systems such as zebrafish63 and human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) from ACM patients64, 65, and for murine myocytes genetically deficient in PKP266, or expressing mutations in desmoglein-257. Immunohistochemical characterization of human ACM-affected hearts was consistent with the experimental observations67. Altogether, these studies supported the notion that one gene, though initially classified by its involvement in a structure (the desmosome) can affect parallel and seemingly independent traits, including one directly related to the electrical heart. The demonstration of the relation between loss of PKP2 and NaV1.5 function led to the hypothesis that a PKP2-dependent impairment of sodium current could lead to a phenotype classically associated with sodium current loss-of-function, namely, BrS. This hypothesis was supported by the data of Cerrone et al,65 as well as by additional reports,68, 69 and gave grounds to the possibility that ACM and BrS could be seen as clinical manifestations at the opposite end of a common disease spectrum, as it was initially suggested by the group of Corrado and colleagues70. Yet, it is also important to note that other studies have not demonstrated a burden of rare PKP2 variants in BrS37.
Transcriptional regulation is affected by loss of PKP2 function
Contact inhibition of cell growth is a well-known phenomenon in various cell systems. As a basic concept, it indicates that the cell can switch into a transcriptional program leading to cell division when contact is lost at the surface. This mechanism is fundamental to the normal continuity of cell barriers (e.g., epithelia) and its dysfunction can lead to cancerous growth71. A desmosome-to-transcription axis in the adult myocyte has been supported by observations indicating that loss of desmosomal proteins can activate the Hippo and the Wnt pathways,72, 73 both regulators of the transcription of pro-fibrosis/adiposis genes74. The Wnt/beta-catenin pathway signaling is a well-known regulator of myogenesis versus adipogenesis. In the view of Garcia Gras et al73, translocation of plakoglobin to the nucleus following loss of desmosomal integrity competitively impairs the ability of beta-catenin to transcriptionally suppress pro-fibroadipotic genes, leading to cardiac fibroadiposis. Regarding the Hippo pathway, the work of Chen et al72 shows that intercalated disc (ID) disassembly can lead to Hippo pathway activation and downstream phosphorylation (and thus inactivation) of YAP, the Hippo pathway effector, contributing to suppression of Wnt/beta-catenin signaling and enhanced adipogenesis (see also Figure 3 in74). Separate studies of Dubash et al75 show that lack of PKP2 also increases expression of TGF-beta1 and activates the p38-MAPK-dependent profibrotic pathway. These studies have shown that there is a connection between PKP2 expression, and the cardiac transcriptional program. To fully explore the relation between the expression of PKP2 in adult ventricular myocytes, and the transcriptional program of the heart, Cerrone et al developed a cardiac-specific, tamoxifen-activated murine model of PKP2 deficiency76 and performed RNAseq analysis at the time in which the mice presented a phenotype of an arrhythmogenic cardiomyopathy of right ventricular predominance76. A total of 1200 transcripts were differentially up- or down-regulated using stringent inclusion criteria. Importantly, the study revealed decreased transcripts of genes involved in intracellular calcium homeostasis, such as RyR2, Trdn, and Ank2. Functionally, these changes translated into a highly arrhythmogenic, catecholamine-dependent phenotype. These data indicate that another trait under the umbrella of the Pkp2 gene is the transcriptional control of intracellular calcium cycling, a function with major implications in arrhythmogenesis. To extend these observations in mice to the human heart, Montnach et al77 took advantage of the GTEx 7.0 database, which includes a compendium of the cardiac transcriptome obtained from 272 deceased individuals78. These authors correlated the transcript abundance of PKP2 in the left ventricle, with the abundance of every other transcript in the transcriptome for all subjects in the database. This process was followed by weighted correlation network analysis (WGCNA79), a method to identify, from the correlative data, gene groups likely to belong to a common network. The authors concluded that PKP2 is part of a gene network that includes other transcripts involved in cell adhesion (such as desmoglein-2), as well as in intracellular calcium homeostasis76, 77.
The evidence that increased sarcoplasmic calcium release can create the substrate for PKP2-dependent ventricular arrhythmias was reminiscent of arrhythmias seen in cases of CPVT, a separate heritable disease most commonly caused by gain-of-function mutations in the RyR2 channel80. Interestingly, flecainide has proven effective to control arrhythmias in CPVT patients, likely because of direct blockade of the RyR2 channel81, 82. When flecainide was tested in PKP2-deficient animals, catecholaminergic-induced ventricular arrhythmias were abolished76. Furthermore, anecdotal evidence suggests that flecainide may be effective in combination therapy for the treatment of ACM-related arrhythmias in humans83, an exciting observation that requires further consideration. These observations indicate that in addition to structural disease, desmosomal mutations can lead to arrhythmic death in the absence of altered cardiac structure. This possibility is supported by a recent report of exertion-related autopsy-negative cases of sudden unexplained death in the young (SUDY) later examined by whole-exome sequencing, where the authors identified mutations not only in genes associated with electrical dysfunction such as RyR2 or SCN5A, but also in PKP2 84. A separate study by Tester et al revealed PKP2 truncation variants in patients with clinical diagnosis of CPVT, and in decedents with exercise-associated autopsy-negative SUDY85. These and other reports do not firmly establish PKP2 variants as contributors to the CPVT phenotype (within the framework of complex genetics), but emphasize the importance of considering all multiple traits associated with one gene when considering the molecular substrate of a given phenotype, even if, as shown by Lahrouchi et al86, the ratio of rare variants of unknown significance to pathogenic or likely pathogenic cardiomyopathic variants in post-mortem genetic testing in cases of sudden arrhythmic death syndrome may be extremely unfavorable.
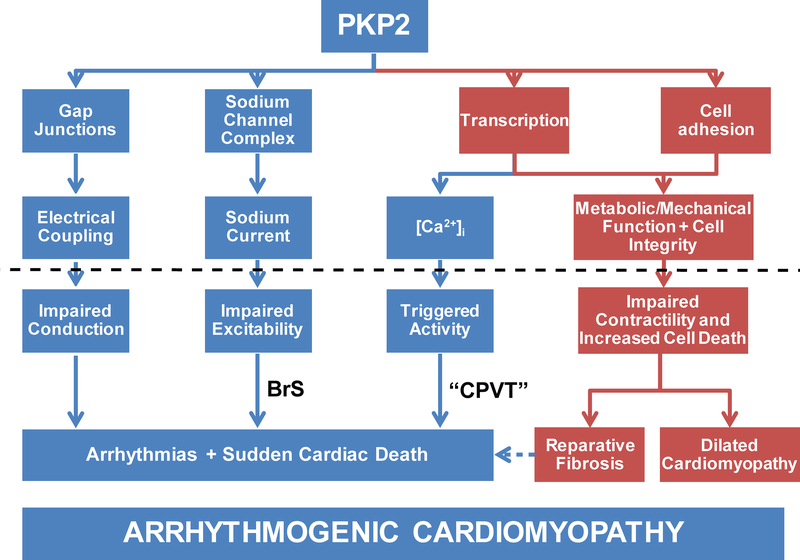
In Figure 6 we present the concepts outlined above in a diagrammatic form to illustrate the different phenotypes that could result from dysfunction in the PKP2 gene. Importantly, descending down one path or another can yield clinical manifestations that can vary from the purely arrhythmogenic (BrS or CPVT) to the purely structural (dilated cardiomyopathy), with ACM being a wider clinical profile in which both electrical and structural phenotypes get integrated under one umbrella. The hypothesis that ARVC, sodium channel disease and CPVT are all part of the same pathological spectrum is intriguing, and deserving of further consideration.
Figure 6:
PKP2 as a pleiotropic gene. Deficiencies in PKP2 have been associated with at least four downstream effects: integrity of gap junctions and hence electrical coupling; function of the sodium channel complex and consequently, sodium current properties; cell adhesion to maintain mechanical integrity, and regulation of transcription which affects both, metabolic/mechanical function, as well as the regulation of intracellular calcium concentration -[Ca2+]i. If disturbed (dotted line) each of the four arms can lead to either an electrical or a structural phenotype. A dysfunction along all four arms would yield the complete arrhythmogenic cardiomyopathy phenotype. But not all arms have to be affected in all cases and as such, a walk downstream of the “sodium channel complex” arm would lead to a BrS-like phenotype; downstream of transcription-[Ca2+]i would yield a phenotype resembling CPVT, whereas a mechanical phenotype of dilated cardiomyopathy would also be possible. As such, a pleiotropic gene can result in one or more of multiple, seemingly unrelated phenotypes. (Reproduced with permission from reference76. )
SCN5A as a pleiotropic gene: from “purely electrical” to a structural phenotype
The observation that variants in one gene can result in different phenotypes extends to genes commonly associated with cellular electrophysiology, such as GJA1 (coding for Cx43 and associated with oculdentodigital dysplasia51) and SCN5A. The latter codes for NaV1.5, the α-subunit of the cardiac sodium channel, the key mediator of depolarization in the adult atrial and ventricular tissue. Mutations in SCN5A have been associated with electrical phenotypes but more recently, with structural phenotypes as well.49, 87 Among the first phenotypes linked to mutations in the SCN5A gene was a congenital form of Long QT Syndrome, annotated as LQT3.88 SCN5A mutations found in patients with LQT3 lead to augmented late sodium current (“gain-of-function mutations”) that by virtue of increasing depolarizing current during the repolarizing phase, prolong action potential duration. Peculiar features of LQT3 at variance with other forms are marked bradycardia and increased arrhythmic risk during rest conditions, i.e. when heart rate is usually slower4, 49. This is a common feature of inherited disorders linked to SCN5A mutations, probably related to the effect of the channel on cardiac conduction and impulse initiation. As discussed above, mutations that cause decreased INa amplitude were initially linked to BrS 49 though recent evidence shows features and genetic inheritance patterns more complex than originally expected. Loss of function mutations in SCN5A have also been associated to other phenotypes, mostly affecting cardiac conduction89–91. Lev-Lenegre disease or Progressive Cardiac Conduction Disease is a condition characterized by progressive P wave, PR interval and QRS interval duration lengthening. Ultimately, patients develop atrio-ventricular block and/or bundle branch block occurring at a relatively young age in adults and tends to have autosomal dominant familiar transmission90, 91. Sick sinus syndrome and some forms of familiar atrial fibrillation/atrial standstill have also been linked to the presence of SCN5A mutations, although these are rare occurrences87. Interestingly, increased fibrosis in different areas of the conduction system are often part of the clinical spectrum of these conditions, suggesting a potential role of defective NaV1.5 not only in modulating cardiac action potential, but also the structural phenotype. Pleiotropy associated with SCN5A mutations is further evidenced by the observation that a single mutation (e.g., SCN5A-1795insD5, 35, 92 and SCN5A-E1784K93) can cause an overlap syndrome comprising clinical and biophysical features of both loss and gain of sodium channel function.
While the phenotypes described above are somehow “expected” given the well-known function of NaV1.5, a number of cases of dilated cardiomyopathy (DCM) have also been reported49, 94. DCM is characterized by progressive dilation and impaired contractility of the left ventricle, leading to heart failure and ventricular arrhythmias. A small fraction of DCM cases may be familial, with an underlying genetic cause49. The majority of genes implicated in inherited DCM are coding for structural proteins of the cardiac sarcomere, cytoskeleton or nuclear lamina envelop. However, several isolated reports and familial cases of DCM have been attributed to SCN5A variants. Interestingly, > 20 different mutations have been associated to a DCM phenotype and their biophysical characteristics include loss of function, gain of function or overlap effects. One particular mutation, R222Q, was reported in several unrelated families showing elevated burden of premature ventricular complexes (PVCs)95, 96. The PVCs originated from the Purkinje fibers as multifocal ectopic complexes and it has been postulated that the high arrhythmic burden was the substrate causing the progressive ventricular dilation. Additional mutations in this or adjacent nucleotides have since then been described (R219H, R222P, R225W, R225P). The group of Chahine97 functionally characterized several of these mutations and showed that they can result in an abnormal flow of cations (“gating pore currents”) through the usually non-conductive voltage sensor domains. They also postulated that gating pore currents could contribute not only to increased arrhythmic burden but also to the observed morphological changes. Interestingly, pharmacological control of PVCs was able to revert the cardiomyopathic phenotype, adding support to the vision that some of these forms could be tachycardia-related cardiomyopathies.
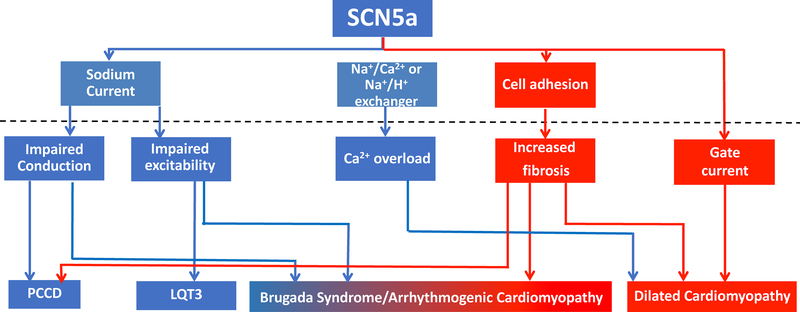
Additional SCN5A mutations, either with loss of function or overlapping effects have been linked to familial DCM. Usually these DCM forms have the peculiar feature of showing conduction defects49. The mechanistic link between dysfunctional NaV1.5 channels and increased fibrosis and dilation is complex and has not been well clarified. One hypothesis that has been proposed is that leakage of positive charges may increase the protonic pumps (Na+/H+ and Na+/Ca2+ exchangers), ultimately leading to Ca2+ overload which could impair contraction 98. An alternative (or complementary) possibility, however, is that just as adhesion complexes can modulate sodium current, NaV1.5 modulates cell adhesion. Studies in support of this hypothesis were reported by Leo-Macias et al99. These authors demonstrated that NaV1.5 at the ID is located in close physical association with N-Cadherin, the major protein that maintains cell-cell adhesion between heart cells. Using an assay to study intercellular adhesion strength, the authors determined that loss of NaV1.5 can actually weaken cell-cell adhesion. From the perspective outlined above, just as mutations in PKP2 can lead to a “channelopathy,” mutations in SCN5A could lead to a clinical manifestation most often associated with a “cell adhesion protein” such as ACM. In favor of the latter, Te Riele et al100 combined exome and gene-candidate screening in a multicenter ACM cohort of 281 unrelated probands, and identified 6 SCN5A variants. Further analysis of one of the cases, using hiPSC-CMs from the patient and CRISPR-CAS9 to reverse the mutation, showed a direct association between the mutation, and the structural integrity of the N-Cadherin clusters at sites of cell-cell junction. In summary, mutations in SCN5A are associated with diverse phenotypes, some expected for a protein that regulates sodium current but also others not expected for a “channel protein” such as NaV1.5. As illustrated in the diagram in Figure 7, we conclude that one gene can be involved in multiple traits, thus supporting the notion that a model of one gene-one disease does not apply to the structural-electrical spectrum described in this review; in fact, cases of monogenic diseases are more likely to be the exception rather than the rule.
Figure 7:
SCN5A as a pleiotropic gene. As in the case of PKP2 (Figure 6), seemingly unrelated functions related to the expression of the SCN5A gene can, if impaired, cause a clinical phenotype consequent to the balance of endophenotypes that were affected by the mutation, ranging from purely electrical (blue) to structural (red) or a combination of both “Brugada syndrome/arrhythmogenic cardiomyopathy.”
Pleiotropy: summary and future directions
A number of studies have shown that a single molecular entity can have more than one function. The notion of molecules as multi-functional units carries as an extension a departure from (or rather, an expansion of) the model of one gene-one disease, and supports an alternative model where one gene can lead to multiple traits. This pleiotropic model helps us understand disease manifestations otherwise not consistent with the “canonical” function of a gene or its product. But it also has, embedded in it, an ever-expanding range of complexities, as these multiple traits are likely not parallel and independent but rather, inter-connected through non-linear networks. Creating a broader window into the function of molecules that govern cardiac function, together with advances in our ability to acquire and analyze “big data” at all levels of biological organization (cells, organs, organisms, populations) are promising -though by no means the only- approaches to shorten the knowledge gap impeding us to understand the relation between the sequence of a genome, and the function of the heart.
Supplementary Material
Acknowledgments
FUNDING SOURCES
Supported by grants from the Dutch Heart Foundation (CVON-PREDICT2 and CVON-eDETECT; CRB; CAR), the Netherlands Organization for Scientific Research (VICI fellowship, 016.150.610, to CRB; VIDI fellowship, 91714371, to CAR), the American Heart Association (18TPA34230006 to MC; 18TPA34230042 to MD), the National Institutes of Health (RO1 HL134328 and RO1 HL136179 to MD), the Philippa and Marvin Carsley Chair in cardiology (RT) and a Fondation Leducq Transatlantic Network of Excellence (CRB; MD; CAR). Dr. Tadros is a clinical research scholar of Fonds de recherche du Québec – Santé.
Footnotes
Conflict of Interest disclosures : none.
REFERENCES
- 1.Gibson G Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bezzina CR, Lahrouchi N and Priori SG. Genetics of sudden cardiac death. Circ Res. 2015;116:1919–1936. [DOI] [PubMed] [Google Scholar]
- 3.Priori SG, Napolitano C and Schwartz PJ. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99:529–533. [DOI] [PubMed] [Google Scholar]
- 4.Priori SG, Schwartz PJ, Napolitano C, Bloise R, Ronchetti E, Grillo M, Vicentini A, Spazzolini C, Nastoli J, Bottelli G, Folli R and Cappelletti D. Risk stratification in the long-QT syndrome. N Engl J Med. 2003;348:1866–1874. [DOI] [PubMed] [Google Scholar]
- 5.Rivaud MR, Jansen JA, Postema PG, Nannenberg EA, Mizusawa Y, van der Nagel R, Wolswinkel R, van der Made I, Marchal GA, Rajamani S, Belardinelli L, van Tintelen JP, Tanck MWT, van der Wal AC, de Bakker JMT, van Rijen HV, Creemers EE, Wilde AAM, van den Berg MP, van Veen TAB, Bezzina CR and Remme CA. A common co-morbidity modulates disease expression and treatment efficacy in inherited cardiac sodium channelopathy. Eur Heart J. 2018;39:2898–2907. [DOI] [PubMed] [Google Scholar]
- 6.Remme CA, Scicluna BP, Verkerk AO, Amin AS, van Brunschot S, Beekman L, Deneer VH, Chevalier C, Oyama F, Miyazaki H, Nukina N, Wilders R, Escande D, Houlgatte R, Wilde AA, Tan HL, Veldkamp MW, de Bakker JM and Bezzina CR. Genetically determined differences in sodium current characteristics modulate conduction disease severity in mice with cardiac sodium channelopathy. Circ Res. 2009;104:1283–1292. [DOI] [PubMed] [Google Scholar]
- 7.Kolder IC, Tanck MW and Bezzina CR. Common genetic variation modulating cardiac ECG parameters and susceptibility to sudden cardiac death. J Mol Cell Cardiol. 2012;52:620–9. [DOI] [PubMed] [Google Scholar]
- 8.Siontis KC, Patsopoulos NA and Ioannidis JP. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur J Hum Genet. 2010;18:832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomas M, Napolitano C, De Giuli L, Bloise R, Subirana I, Malovini A, Bellazzi R, Arking DE, Marban E, Chakravarti A, Spooner PM and Priori SG. Polymorphisms in the NOS1AP gene modulate QT interval duration and risk of arrhythmias in the long QT syndrome. J Am Coll Cardiol. 2010;55:2745–2752. [DOI] [PubMed] [Google Scholar]
- 10.Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, Greenberg DA, Schwartz PJ and George AL Jr. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation. 2009;120:1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duchatelet S, Crotti L, Peat RA, Denjoy I, Itoh H, Berthet M, Ohno S, Fressart V, Monti MC, Crocamo C, Pedrazzini M, Dagradi F, Vicentini A, Klug D, Brink PA, Goosen A, Swan H, Toivonen L, Lahtinen AM, Kontula K, Shimizu W, Horie M, George AL Jr., Tregouet DA, Guicheney P and Schwartz PJ. Identification of a KCNQ1 polymorphism acting as a protective modifier against arrhythmic risk in long-QT syndrome. Circulation Cardiovascular genetics. 2013;6:354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O’Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T and Chakravarti A. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. [DOI] [PubMed] [Google Scholar]
- 13.Kolder I, Tanck MWT, Postema PG, Barc J, Sinner MF, Zumhagen S, Husemann A, Stallmeyer B, Koopmann TT, Hofman N, Pfeufer A, Lichtner P, Meitinger T, Beckmann BM, Myerburg RJ, Bishopric NH, Roden DM, Kaab S, Wilde AAM, Schott JJ, Schulze-Bahr E and Bezzina CR. Analysis for Genetic Modifiers of Disease Severity in Patients With Long-QT Syndrome Type 2. Circ Cardiovas Genet. 2015;8:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earle N, Yeo Han D, Pilbrow A, Crawford J, Smith W, Shelling AN, Cameron V, Love DR and Skinner JR. Single nucleotide polymorphisms in arrhythmia genes modify the risk of cardiac events and sudden death in long QT syndrome. Heart Rhythm. 2014;11:76–82. [DOI] [PubMed] [Google Scholar]
- 15.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JC, Hofman A, Heckbert SR, O’Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG and Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, Ehret GB, Orru M, Pattaro C, Kottgen A, Perz S, Usala G, Barbalic M, Li M, Putz B, Scuteri A, Prineas RJ, Sinner MF, Gieger C, Najjar SS, Kao WH, Muhleisen TW, Dei M, Happle C, Mohlenkamp S, Crisponi L, Erbel R, Jockel KH, Naitza S, Steinbeck G, Marroni F, Hicks AA, Lakatta E, Muller-Myhsok B, Pramstaller PP, Wichmann HE, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kaab S, Abecasis GR and Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, Sotoodehnia N, Rossin EJ, Morley M, Wang X, Johnson AD, Lundby A, Gudbjartsson DF, Noseworthy PA, Eijgelsheim M, Bradford Y, Tarasov KV, Dorr M, Muller-Nurasyid M, Lahtinen AM, Nolte IM, Smith AV, Bis JC, Isaacs A, Newhouse SJ, Evans DS, Post WS, Waggott D, Lyytikainen LP, Hicks AA, Eisele L, Ellinghaus D, Hayward C, Navarro P, Ulivi S, Tanaka T, Tester DJ, Chatel S, Gustafsson S, Kumari M, Morris RW, Naluai AT, Padmanabhan S, Kluttig A, Strohmer B, Panayiotou AG, Torres M, Knoflach M, Hubacek JA, Slowikowski K, Raychaudhuri S, Kumar RD, Harris TB, Launer LJ, Shuldiner AR, Alonso A, Bader JS, Ehret G, Huang H, Kao WH, Strait JB, Macfarlane PW, Brown M, Caulfield MJ, Samani NJ, Kronenberg F, Willeit J, Consortium CA, Consortium C, Smith JG, Greiser KH, Meyer Zu Schwabedissen H, Werdan K, Carella M, Zelante L, Heckbert SR, Psaty BM, Rotter JI, Kolcic I, Polasek O, Wright AF, Griffin M, Daly MJ, Dcct/Edic, Arnar DO, Holm H, Thorsteinsdottir U, e MC, Denny JC, Roden DM, Zuvich RL, Emilsson V, Plump AS, Larson MG, O’Donnell CJ, Yin X, Bobbo M, D’Adamo AP, Iorio A, Sinagra G, Carracedo A, Cummings SR, Nalls MA, Jula A, Kontula KK, Marjamaa A, Oikarinen L, Perola M, Porthan K, Erbel R, Hoffmann P, Jockel KH, Kalsch H, Nothen MM, Consortium H, den Hoed M, Loos RJ, Thelle DS, Gieger C, Meitinger T, Perz S, Peters A, Prucha H, Sinner MF, Waldenberger M, de Boer RA, Franke L, van der Vleuten PA, Beckmann BM, Martens E, Bardai A, Hofman N, Wilde AA, Behr ER, Dalageorgou C, Giudicessi JR, Medeiros-Domingo A, Barc J, Kyndt F, Probst V, Ghidoni A, Insolia R, Hamilton RM, Scherer SW, Brandimarto J, Margulies K, Moravec CE, del Greco MF, Fuchsberger C, O’Connell JR, Lee WK, Watt GC, Campbell H, Wild SH, El Mokhtari NE, Frey N, Asselbergs FW, Mateo Leach I, Navis G, van den Berg MP, van Veldhuisen DJ, Kellis M, Krijthe BP, Franco OH, Hofman A, Kors JA, Uitterlinden AG, Witteman JC, Kedenko L, Lamina C, Oostra BA, Abecasis GR, Lakatta EG, Mulas A, Orru M, Schlessinger D, Uda M, Markus MR, Volker U, Snieder H, Spector TD, Arnlov J, Lind L, Sundstrom J, Syvanen AC, Kivimaki M, Kahonen M, Mononen N, Raitakari OT, Viikari JS, Adamkova V, Kiechl S, Brion M, Nicolaides AN, Paulweber B, Haerting J, Dominiczak AF, Nyberg F, Whincup PH, Hingorani AD, Schott JJ, Bezzina CR, Ingelsson E, Ferrucci L, Gasparini P, Wilson JF, Rudan I, Franke A, Muhleisen TW, Pramstaller PP, Lehtimaki TJ, Paterson AD, Parsa A, Liu Y, van Duijn CM, Siscovick DS, Gudnason V, Jamshidi Y, Salomaa V, Felix SB, Sanna S, Ritchie MD, Stricker BH, Stefansson K, Boyer LA, Cappola TP, Olsen JV, Lage K, Schwartz PJ, Kaab S, Chakravarti A, Ackerman MJ, Pfeufer A, de Bakker PI and Newton-Cheh C. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG and Exome Aggregation C. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genome Aggregation Database (gnomAD). http://gnomad.broadinstitute.org. Accessed July 9, 2019.
- 20.Francioli L, Tiao G, Karczewski K, Solomonson M, Watts N. gnomAD v2.1. MacArthur Lab. October 17, 2018. https://macarthurlab.org/2018/10/17/gnomad-v2-1. Accessed July 9, 2019.
- 21.Nishio Y, Makiyama T, Itoh H, Sakaguchi T, Ohno S, Gong YZ, Yamamoto S, Ozawa T, Ding WG, Toyoda F, Kawamura M, Akao M, Matsuura H, Kimura T, Kita T and Horie M. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. J Am Coll Cardiol. 2009;54:812–819. [DOI] [PubMed] [Google Scholar]
- 22.Lahtinen AM, Marjamaa A, Swan H and Kontula K. KCNE1 D85N polymorphism--a sex-specific modifier in type 1 long QT syndrome? BMC Med Genet. 2011;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaab S, Crawford DC, Sinner MF, Behr ER, Kannankeril PJ, Wilde AA, Bezzina CR, Schulze-Bahr E, Guicheney P, Bishopric NH, Myerburg RJ, Schott JJ, Pfeufer A, Beckmann BM, Martens E, Zhang T, Stallmeyer B, Zumhagen S, Denjoy I, Bardai A, Van Gelder IC, Jamshidi Y, Dalageorgou C, Marshall V, Jeffery S, Shakir S, Camm AJ, Steinbeck G, Perz S, Lichtner P, Meitinger T, Peters A, Wichmann HE, Ingram C, Bradford Y, Carter S, Norris K, Ritchie MD, George AL Jr. and Roden DM. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circulation Cardiovascular genetics. 2012;5:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strauss DG, Vicente J, Johannesen L, Blinova K, Mason JW, Weeke P, Behr ER, Roden DM, Woosley R, Kosova G, Rosenberg MA and Newton-Cheh C. Common Genetic Variant Risk Score Is Associated With Drug-Induced QT Prolongation and Torsade de Pointes Risk: A Pilot Study. Circulation. 2017;135:1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wooten EC, Hebl VB, Wolf MJ, Greytak SR, Orr NM, Draper I, Calvino JE, Kapur NK, Maron MS, Kullo IJ, Ommen SR, Bos JM, Ackerman MJ and Huggins GS. Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circulation Cardiovascular genetics. 2013;6:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, Ruppert V, Arbustini E, Wichter T, Germain M, Dubourg O, Tavazzi L, Aumont MC, DeGroote P, Fauchier L, Trochu JN, Gibelin P, Aupetit JF, Stark K, Erdmann J, Hetzer R, Roberts AM, Barton PJ, Regitz-Zagrosek V, Cardiogenics C, Aslam U, Duboscq-Bidot L, Meyborg M, Maisch B, Madeira H, Waldenstrom A, Galve E, Cleland JG, Dorent R, Roizes G, Zeller T, Blankenberg S, Goodall AH, Cook S, Tregouet DA, Tiret L, Isnard R, Komajda M, Charron P and Cambien F. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meder B, Ruhle F, Weis T, Homuth G, Keller A, Franke J, Peil B, Lorenzo Bermejo J, Frese K, Huge A, Witten A, Vogel B, Haas J, Volker U, Ernst F, Teumer A, Ehlermann P, Zugck C, Friedrichs F, Kroemer H, Dorr M, Hoffmann W, Maisch B, Pankuweit S, Ruppert V, Scheffold T, Kuhl U, Schultheiss HP, Kreutz R, Ertl G, Angermann C, Charron P, Villard E, Gary F, Isnard R, Komajda M, Lutz M, Meitinger T, Sinner MF, Wichmann HE, Krawczak M, Ivandic B, Weichenhan D, Gelbrich G, El-Mokhtari NE, Schreiber S, Felix SB, Hasenfuss G, Pfeufer A, Hubner N, Kaab S, Arbustini E, Rottbauer W, Frey N, Stoll M and Katus HA. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014;35:1069–1077. [DOI] [PubMed] [Google Scholar]
- 28.Esslinger U, Garnier S, Korniat A, Proust C, Kararigas G, Muller-Nurasyid M, Empana JP, Morley MP, Perret C, Stark K, Bick AG, Prasad SK, Kriebel J, Li J, Tiret L, Strauch K, O’Regan DP, Marguiles KB, Seidman JG, Boutouyrie P, Lacolley P, Jouven X, Hengstenberg C, Komajda M, Hakonarson H, Isnard R, Arbustini E, Grallert H, Cook SA, Seidman CE, Regitz-Zagrosek V, Cappola TP, Charron P, Cambien F and Villard E. Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One. 2017;12:e0172995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castel SE, Cervera A, Mohammadi P, Aguet F, Reverter F, Wolman A, Guigo R, Iossifov I, Vasileva A and Lappalainen T. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat Genet. 2018;50:1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amin AS, Giudicessi JR, Tijsen AJ, Spanjaart AM, Reckman YJ, Klemens CA, Tanck MW, Kapplinger JD, Hofman N, Sinner MF, Muller M, Wijnen WJ, Tan HL, Bezzina CR, Creemers EE, Wilde AA, Ackerman MJ and Pinto YM. Variants in the 3’ untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur Heart J. 2012;33:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crotti L, Lahtinen AM, Spazzolini C, Mastantuono E, Monti MC, Morassutto C, Parati G, Heradien M, Goosen A, Lichtner P, Meitinger T, Brink PA, Kontula K, Swan H and Schwartz PJ. Genetic Modifiers for the Long-QT Syndrome: How Important Is the Role of Variants in the 3’ Untranslated Region of KCNQ1? Circulation Cardiovascular genetics. 2016;9:330–339. [DOI] [PubMed] [Google Scholar]
- 32.Chai S, Wan X, Ramirez-Navarro A, Tesar PJ, Kaufman ES, Ficker E, George AL Jr. and Deschenes I. Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. J Clin Invest. 2018;128:1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang T and Colecraft HM. Regulation of voltage-dependent calcium channels by RGK proteins. Biochem Biophys Acta. 2013;1828:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wheeler FC, Tang H, Marks OA, Hadnott TN, Chu PL, Mao L, Rockman HA and Marchuk DA. Tnni3k modifies disease progression in murine models of cardiomyopathy. PLoS genetics. 2009;5:e1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lodder EM, Scicluna BP, Milano A, Sun AY, Tang H, Remme CA, Moerland PD, Tanck MW, Pitt GS, Marchuk DA and Bezzina CR. Dissection of a quantitative trait locus for PR interval duration identifies Tnni3k as a novel modulator of cardiac conduction. PLoS genetics. 2012;8:e1003113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theis JL, Zimmermann MT, Larsen BT, Rybakova IN, Long PA, Evans JM, Middha S, de Andrade M, Moss RL, Wieben ED, Michels VV and Olson TM. TNNI3K mutation in familial syndrome of conduction system disease, atrial tachyarrhythmia and dilated cardiomyopathy. Hum Mol Genet. 2014;23:5793–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Scouarnec S, Karakachoff M, Gourraud JB, Lindenbaum P, Bonnaud S, Portero V, Duboscq-Bidot L, Daumy X, Simonet F, Teusan R, Baron E, Violleau J, Persyn E, Bellanger L, Barc J, Chatel S, Martins R, Mabo P, Sacher F, Haissaguerre M, Kyndt F, Schmitt S, Bezieau S, Le Marec H, Dina C, Schott JJ, Probst V and Redon R. Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Hum Mol Genet. 2015;24:2757–2763. [DOI] [PubMed] [Google Scholar]
- 38.Probst V, Wilde AA, Barc J, Sacher F, Babuty D, Mabo P, Mansourati J, Le Scouarnec S, Kyndt F, Le Caignec C, Guicheney P, Gouas L, Albuisson J, Meregalli PG, Le Marec H, Tan HL and Schott JJ. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circulation Cardiovascular genetics. 2009;2:552–557. [DOI] [PubMed] [Google Scholar]
- 39.Schulze-Bahr E, Eckardt L, Breithardt G, Seidl K, Wichter T, Wolpert C, Borggrefe M and Haverkamp W. Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum Mutat. 2003;21:651–652. [DOI] [PubMed] [Google Scholar]
- 40.Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud JB, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F, Guicheney P, Fressart V, Leenhardt A, Antzelevitch C, Bartkowiak S, Schulze-Bahr E, Zumhagen S, Behr ER, Bastiaenen R, Tfelt-Hansen J, Olesen MS, Kaab S, Beckmann BM, Weeke P, Watanabe H, Endo N, Minamino T, Horie M, Ohno S, Hasegawa K, Makita N, Nogami A, Shimizu W, Aiba T, Froguel P, Balkau B, Lantieri O, Torchio M, Wiese C, Weber D, Wolswinkel R, Coronel R, Boukens BJ, Bezieau S, Charpentier E, Chatel S, Despres A, Gros F, Kyndt F, Lecointe S, Lindenbaum P, Portero V, Violleau J, Gessler M, Tan HL, Roden DM, Christoffels VM, Le Marec H, Wilde AA, Probst V, Schott JJ, Dina C and Redon R. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45:1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veerman CC, Podliesna S, Tadros R, Lodder EM, Mengarelli I, de Jonge B, Beekman L, Barc J, Wilders R, Wilde AAM, Boukens BJ, Coronel R, Verkerk AO, Remme CA and Bezzina CR. The Brugada Syndrome Susceptibility Gene HEY2 Modulates Cardiac Transmural Ion Channel Patterning and Electrical Heterogeneity. Circ Res. 2017;121:537–548. [DOI] [PubMed] [Google Scholar]
- 42.Bagnall RD, Ingles J, Dinger ME, Cowley MJ, Ross SB, Minoche AE, Lal S, Turner C, Colley A, Rajagopalan S, Berman Y, Ronan A, Fatkin D and Semsarian C. Whole Genome Sequencing Improves Outcomes of Genetic Testing in Patients With Hypertrophic Cardiomyopathy. J Am Coll Cardiol. 2018;72:419–429. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME and Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Authors/Task Force m, Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, McKenna WJ, Mogensen J, Nihoyannopoulos P, Nistri S, Pieper PG, Pieske B, Rapezzi C, Rutten FH, Tillmanns C and Watkins H. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J. 2014;35:2733–2779. [DOI] [PubMed] [Google Scholar]
- 45.Weintraub RG, Semsarian C and Macdonald P. Dilated cardiomyopathy. Lancet. 2017;390:400–414. [DOI] [PubMed] [Google Scholar]
- 46.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT and Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, Morales A, Taylor MRG, Vatta M and Ware SM. Genetic Evaluation of Cardiomyopathy-A Heart Failure Society of America Practice Guideline. J Card Fail. 2018;24:281–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corrado D, Link MS and Calkins H. Arrhythmogenic Right Ventricular Cardiomyopathy. N Engl J Med. 2017;376:61–72. [DOI] [PubMed] [Google Scholar]
- 49.Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C, Blom N, Brugada J, Chiang CE, Huikuri H, Kannankeril P, Krahn A, Leenhardt A, Moss A, Schwartz PJ, Shimizu W, Tomaselli G and Tracy C. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013;10:1932–1963. [DOI] [PubMed] [Google Scholar]
- 50.Verbanck M, Chen CY, Neale B and Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laird DW. Syndromic and non-syndromic disease-linked Cx43 mutations. FEBS letters. 2014;588:1339–1348. [DOI] [PubMed] [Google Scholar]
- 52.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E and Thierfelder L. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36:1162–1164. [DOI] [PubMed] [Google Scholar]
- 53.Groeneweg JA, Bhonsale A, James CA, te Riele AS, Dooijes D, Tichnell C, Murray B, Wiesfeld AC, Sawant AC, Kassamali B, Atsma DE, Volders PG, de Groot NM, de Boer K, Zimmerman SL, Kamel IR, van der Heijden JF, Russell SD, Jan Cramer M, Tedford RJ, Doevendans PA, van Veen TA, Tandri H, Wilde AA, Judge DP, van Tintelen JP, Hauer RN and Calkins H. Clinical Presentation, Long-Term Follow-Up, and Outcomes of 1001 Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Patients and Family Members. Circulation Cardiovascular genetics. 2015;8:437–446. [DOI] [PubMed] [Google Scholar]
- 54.Marcus FI, Fontaine GH, Guiraudon G, Frank R, Laurenceau JL, Malergue C and Grosgogeat Y. Right ventricular dysplasia: a report of 24 adult cases. Circulation. 1982;65:384–398. [DOI] [PubMed] [Google Scholar]
- 55.Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, Calkins H, Corrado D, Cox MG, Daubert JP, Fontaine G, Gear K, Hauer R, Nava A, Picard MH, Protonotarios N, Saffitz JE, Sanborn DM, Steinberg JS, Tandri H, Thiene G, Towbin JA, Tsatsopoulou A, Wichter T and Zareba W. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation. 2010;121:1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu T, Yang Z, Vatta M, Rampazzo A, Beffagna G, Pilichou K, Scherer SE, Saffitz J, Kravitz J, Zareba W, Danieli GA, Lorenzon A, Nava A, Bauce B, Thiene G, Basso C, Calkins H, Gear K, Marcus F, Towbin JA and Multidisciplinary Study of Right Ventricular Dysplasia I. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55:587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzo S, Lodder EM, Verkerk AO, Wolswinkel R, Beekman L, Pilichou K, Basso C, Remme CA, Thiene G and Bezzina CR. Intercalated disc abnormalities, reduced Na(+) current density, and conduction slowing in desmoglein-2 mutant mice prior to cardiomyopathic changes. Cardiovasc Res. 2012;95:409–418. [DOI] [PubMed] [Google Scholar]
- 58.Kaplan SR, Gard JJ, Protonotarios N, Tsatsopoulou A, Spiliopoulou C, Anastasakis A, Squarcioni CP, McKenna WJ, Thiene G, Basso C, Brousse N, Fontaine G and Saffitz JE. Remodeling of myocyte gap junctions in arrhythmogenic right ventricular cardiomyopathy due to a deletion in plakoglobin (Naxos disease). Heart Rhythm. 2004;1:3–11. [DOI] [PubMed] [Google Scholar]
- 59.Oxford EM, Musa H, Maass K, Coombs W, Taffet SM and Delmar M. Connexin43 remodeling caused by inhibition of plakophilin-2 expression in cardiac cells. Circ Res. 2007;101:703–711. [DOI] [PubMed] [Google Scholar]
- 60.Leo-Macias A, Liang FX and Delmar M. Ultrastructure of the intercellular space in adult murine ventricle revealed by quantitative tomographic electron microscopy. Cardiovasc Res. 2015;107:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer RA, Laird DW, Revel JP and Johnson RG. Inhibition of gap junction and adherens junction assembly by connexin and A-CAM antibodies. J Cell Biol. 1992;119:179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato PY, Musa H, Coombs W, Guerrero-Serna G, Patino GA, Taffet SM, Isom LL and Delmar M. Loss of plakophilin-2 expression leads to decreased sodium current and slower conduction velocity in cultured cardiac myocytes. Circ Res. 2009;105:523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asimaki A, Kapoor S, Plovie E, Karin Arndt A, Adams E, Liu Z, James CA, Judge DP, Calkins H, Churko J, Wu JC, MacRae CA, Kleber AG and Saffitz JE. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Science translational medicine. 2014;6:240ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP and Chen HS. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerrone M, Lin X, Zhang M, Agullo-Pascual E, Pfenniger A, Chkourko Gusky H, Novelli V, Kim C, Tirasawadichai T, Judge DP, Rothenberg E, Chen HS, Napolitano C, Priori SG and Delmar M. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a brugada syndrome phenotype. Circulation. 2014;129:1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerrone M, Noorman M, Lin X, Chkourko H, Liang FX, van der Nagel R, Hund T, Birchmeier W, Mohler P, van Veen TA, van Rijen HV and Delmar M. Sodium current deficit and arrhythmogenesis in a murine model of plakophilin-2 haploinsufficiency. Cardiovasc Res. 2012;95:460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noorman M, Hakim S, Kessler E, Groeneweg JA, Cox MG, Asimaki A, van Rijen HV, van Stuijvenberg L, Chkourko H, van der Heyden MA, Vos MA, de Jonge N, van der Smagt JJ, Dooijes D, Vink A, de Weger RA, Varro A, de Bakker JM, Saffitz JE, Hund TJ, Mohler PJ, Delmar M, Hauer RN and van Veen TA. Remodeling of the cardiac sodium channel, connexin43, and plakoglobin at the intercalated disk in patients with arrhythmogenic cardiomyopathy. Heart Rhythm. 2013;10:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forkmann M, Tomala J, Huo Y, Mayer J, Christoph M, Wunderlich C, Salmas J, Gaspar T and Piorkowski C. Epicardial Ventricular Tachycardia Ablation in a Patient With Brugada ECG Pattern and Mutation of PKP2 and DSP Genes. Circ Arrhythm Electrophysiol. 2015;8:505–507. [DOI] [PubMed] [Google Scholar]
- 69.Peters S Arrhythmogenic right ventricular dysplasia-cardiomyopathy and provocable coved-type ST-segment elevation in right precordial leads: clues from long-term follow-up. Europace. 2008;10:816–820. [DOI] [PubMed] [Google Scholar]
- 70.Corrado D, Buja G, Basso C, Nava A and Thiene G. What is the Brugada syndrome? Cardiol Rev. 1999;7:191–195. [DOI] [PubMed] [Google Scholar]
- 71.Moh MC and Shen S. The roles of cell adhesion molecules in tumor suppression and cell migration: a new paradox. Cell Adh Migr. 2009;3:334–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT and Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res. 2014;114:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS and Marian AJ. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116:2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoorntje ET, Te Rijdt WP, James CA, Pilichou K, Basso C, Judge DP, Bezzina CR and van Tintelen JP. Arrhythmogenic cardiomyopathy: pathology, genetics, and concepts in pathogenesis. Cardiovasc Res. 2017;113:1521–1531. [DOI] [PubMed] [Google Scholar]
- 75.Dubash AD, Kam CY, Aguado BA, Patel DM, Delmar M, Shea LD and Green KJ. Plakophilin-2 loss promotes TGF-beta1/p38 MAPK-dependent fibrotic gene expression in cardiomyocytes. J Cell Biol. 2016;212:425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cerrone M, Montnach J, Lin X, Zhao YT, Zhang M, Agullo-Pascual E, Leo-Macias A, Alvarado FJ, Dolgalev I, Karathanos TV, Malkani K, Van Opbergen CJM, van Bavel JJA, Yang HQ, Vasquez C, Tester D, Fowler S, Liang F, Rothenberg E, Heguy A, Morley GE, Coetzee WA, Trayanova NA, Ackerman MJ, van Veen TAB, Valdivia HH and Delmar M. Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nature communications. 2017;8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Montnach J, Agullo-Pascual E, Tadros R, Bezzina CR and Delmar M. Bioinformatic analysis of a plakophilin-2-dependent transcription network: implications for the mechanisms of arrhythmogenic right ventricular cardiomyopathy in humans and in boxer dogs. Europace. 2018;20:iii125–iii132. [DOI] [PubMed] [Google Scholar]
- 78.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langfelder P and Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cerrone M, Napolitano C and Priori SG. Catecholaminergic polymorphic ventricular tachycardia: A paradigm to understand mechanisms of arrhythmias associated to impaired Ca(2+) regulation. Heart Rhythm. 2009;6:1652–1659. [DOI] [PubMed] [Google Scholar]
- 81.Watanabe H, Chopra N, Laver D, Hwang HS, Davies SS, Roach DE, Duff HJ, Roden DM, Wilde AA and Knollmann BC. Flecainide prevents catecholaminergic polymorphic ventricular tachycardia in mice and humans. Nat Med. 2009;15:380–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kannankeril PJ, Moore JP, Cerrone M, Priori SG, Kertesz NJ, Ro PS, Batra AS, Kaufman ES, Fairbrother DL, Saarel EV, Etheridge SP, Kanter RJ, Carboni MP, Dzurik MV, Fountain D, Chen H, Ely EW, Roden DM and Knollmann BC. Efficacy of Flecainide in the Treatment of Catecholaminergic Polymorphic Ventricular Tachycardia: A Randomized Clinical Trial. JAMA Cardiol. 2017;2:759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ermakov S, Gerstenfeld EP, Svetlichnaya Y and Scheinman MM. Use of flecainide in combination antiarrhythmic therapy in patients with arrhythmogenic right ventricular cardiomyopathy. Heart Rhythm. 2017;14:564–569. [DOI] [PubMed] [Google Scholar]
- 84.Anderson JH, Tester DJ, Will ML and Ackerman MJ. Whole-Exome Molecular Autopsy After Exertion-Related Sudden Unexplained Death in the Young. Circulation Cardiovascular genetics. 2016;9:259–265. [DOI] [PubMed] [Google Scholar]
- 85.Tester DJ, Ackerman JP, Giudicessi JR, Ackerman NC, Cerrone M, Delmar M and Ackerman MJ. Plakophilin-2 Truncation Variants in Patients Clinically Diagnosed With Catecholaminergic Polymorphic Ventricular Tachycardia and Decedents With Exercise-Associated Autopsy Negative Sudden Unexplained Death in the Young. JACC Clin Electrophysiol. 2019;5:120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lahrouchi N, Raju H, Lodder EM, Papatheodorou E, Ware JS, Papadakis M, Tadros R, Cole D, Skinner JR, Crawford J, Love DR, Pua CJ, Soh BY, Bhalshankar JD, Govind R, Tfelt-Hansen J, Winkel BG, van der Werf C, Wijeyeratne YD, Mellor G, Till J, Cohen MC, Tome-Esteban M, Sharma S, Wilde AAM, Cook SA, Bezzina CR, Sheppard MN and Behr ER. Utility of Post-Mortem Genetic Testing in Cases of Sudden Arrhythmic Death Syndrome. J Am Coll Cardiol. 2017;69:2134–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilde AAM and Amin AS. Clinical Spectrum of SCN5A Mutations: Long QT Syndrome, Brugada Syndrome, and Cardiomyopathy. JACC Clin Electrophysiol. 2018;4:569–579. [DOI] [PubMed] [Google Scholar]
- 88.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA and Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. [DOI] [PubMed] [Google Scholar]
- 89.Zhang ZS, Tranquillo J, Neplioueva V, Bursac N and Grant AO. Sodium channel kinetic changes that produce Brugada syndrome or progressive cardiac conduction system disease. Am J Physiol Heart Circ Physiol. 2007;292:H399–407. [DOI] [PubMed] [Google Scholar]
- 90.Probst V, Kyndt F, Potet F, Trochu JN, Mialet G, Demolombe S, Schott JJ, Baro I, Escande D and Le Marec H. Haploinsufficiency in combination with aging causes SCN5A-linked hereditary Lenegre disease. J Am Coll Cardiol. 2003;41:643–652. [DOI] [PubMed] [Google Scholar]
- 91.Kyndt F, Probst V, Potet F, Demolombe S, Chevallier JC, Baro I, Moisan JP, Boisseau P, Schott JJ, Escande D and Le Marec H. Novel SCN5A mutation leading either to isolated cardiac conduction defect or Brugada syndrome in a large French family. Circulation. 2001;104:3081–3086. [DOI] [PubMed] [Google Scholar]
- 92.Bezzina C, Veldkamp MW, van Den Berg MP, Postma AV, Rook MB, Viersma JW, van Langen IM, Tan-Sindhunata G, Bink-Boelkens MT, van Der Hout AH, Mannens MM and Wilde AA. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85:1206–1213. [DOI] [PubMed] [Google Scholar]
- 93.Makita N, Behr E, Shimizu W, Horie M, Sunami A, Crotti L, Schulze-Bahr E, Fukuhara S, Mochizuki N, Makiyama T, Itoh H, Christiansen M, McKeown P, Miyamoto K, Kamakura S, Tsutsui H, Schwartz PJ, George AL Jr. and Roden DM. The E1784K mutation in SCN5A is associated with mixed clinical phenotype of type 3 long QT syndrome. J Clin Invest. 2008;118:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McNair WP, Sinagra G, Taylor MR, Di Lenarda A, Ferguson DA, Salcedo EE, Slavov D, Zhu X, Caldwell JH and Mestroni L. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol. 2011;57:2160–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laurent G, Saal S, Amarouch MY, Beziau DM, Marsman RF, Faivre L, Barc J, Dina C, Bertaux G, Barthez O, Thauvin-Robinet C, Charron P, Fressart V, Maltret A, Villain E, Baron E, Merot J, Turpault R, Coudiere Y, Charpentier F, Schott JJ, Loussouarn G, Wilde AA, Wolf JE, Baro I, Kyndt F and Probst V. Multifocal ectopic Purkinje-related premature contractions: a new SCN5A-related cardiac channelopathy. J Am Coll Cardiol. 2012;60:144–156. [DOI] [PubMed] [Google Scholar]
- 96.Mann SA, Castro ML, Ohanian M, Guo G, Zodgekar P, Sheu A, Stockhammer K, Thompson T, Playford D, Subbiah R, Kuchar D, Aggarwal A, Vandenberg JI and Fatkin D. R222Q SCN5A mutation is associated with reversible ventricular ectopy and dilated cardiomyopathy. J Am Coll Cardiol. 2012;60:1566–1573. [DOI] [PubMed] [Google Scholar]
- 97.Moreau A, Gosselin-Badaroudine P, Mercier A, Burger B, Keller DI and Chahine M. A leaky voltage sensor domain of cardiac sodium channels causes arrhythmias associated with dilated cardiomyopathy. Scientific reports. 2018;8:13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beckermann TM, McLeod K, Murday V, Potet F and George AL Jr. Novel SCN5A mutation in amiodarone-responsive multifocal ventricular ectopy-associated cardiomyopathy. Heart Rhythm. 2014;11:1446–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leo-Macias A, Agullo-Pascual E, Sanchez-Alonso JL, Keegan S, Lin X, Arcos T, Feng Xia L, Korchev YE, Gorelik J, Fenyo D, Rothenberg E and Delmar M. Nanoscale visualization of functional adhesion/excitability nodes at the intercalated disc. Nature communications. 2016;7:10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Te Riele AS, Agullo-Pascual E, James CA, Leo-Macias A, Cerrone M, Zhang M, Lin X, Lin B, Sobreira NL, Amat-Alarcon N, Marsman RF, Murray B, Tichnell C, van der Heijden JF, Dooijes D, van Veen TA, Tandri H, Fowler SJ, Hauer RN, Tomaselli G, van den Berg MP, Taylor MR, Brun F, Sinagra G, Wilde AA, Mestroni L, Bezzina CR, Calkins H, Peter van Tintelen J, Bu L, Delmar M and Judge DP. Multilevel analyses of SCN5A mutations in arrhythmogenic right ventricular dysplasia/cardiomyopathy suggest non-canonical mechanisms for disease pathogenesis. Cardiovasc Res. 2017;113:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.